Abstract
Arabinogalactan proteins (AGPs) comprise a family of hydroxyproline-rich glycoproteins that are implicated in plant growth and development. In this study, 69 AGPs are identified from the rice genome, including 13 classical AGPs, 15 arabinogalactan (AG) peptides, three non-classical AGPs, three early nodulin-like AGPs (eNod-like AGPs), eight non-specific lipid transfer protein-like AGPs (nsLTP-like AGPs), and 27 fasciclin-like AGPs (FLAs). The results from expressed sequence tags, microarrays, and massively parallel signature sequencing tags are used to analyse the expression of AGP-encoding genes, which is confirmed by real-time PCR. The results reveal that several rice AGP-encoding genes are predominantly expressed in anthers and display differential expression patterns in response to abscisic acid, gibberellic acid, and abiotic stresses. Based on the results obtained from this analysis, an attempt has been made to link the protein structures and expression patterns of rice AGP-encoding genes to their functions. Taken together, the genome-wide identification and expression analysis of the rice AGP gene family might facilitate further functional studies of rice AGPs.
Keywords: Arabinogalactan protein, expression analysis, genome, rice
Introduction
Proteoglycans or glycoproteins are basal components of the cell wall implicated in various processes of plant growth and development throughout the plant kingdom. A large number of these proteins are rich in proline (Pro) or hydroxyproline (Hyp) and are named Pro-rich/Hyp-rich glycoproteins (P/HRGPs). The P/HRGPs share common features that consist of regions containing Hyp residues, which are called ‘glycomodules’ and in which most Hyp residues are usually glycosylated by a large branched arabinogalactan (AG) polysaccharide or by small non-branched arabinooligosaccharides (arabinosides). According to the ‘Hyp contiguity hypothesis’, contiguous and non-contiguous clustered Hyp residues are the sites attached by arabinoside and AG polysaccharide, respectively. Evidence for the ‘Hyp contiguity hypothesis’ was obtained using synthetic genes. In synthetic HRGPs composed entirely of (Ser-Hyp-Ser-Hyp)n or (Ala-Hyp-Ala-Hyp)n repeats, 100% of the Hyp residues have AG polysaccharides (Shpak et al., 1999, 2001), but Hyp that occurs singly is rarely glycosylated by AG polysaccharide. Short arabinosides are added to Hyp residues in (Ser/Thr)-Hyp-Hyp-Hyp extensin-like motifs and large arabinogalactan chains are added to the non-contiguous Hyp residues in Ser-Hyp-Thr-Hyp-Thr AGP-like motifs (Goodrum et al., 2000). The P/HRGP superfamily is classified into three subfamilies distinguished by characteristic repetitive structural motifs yielding different degrees of O-glycosylation: the minimally glycosylated Pro-rich proteins (PRPs), the moderately glycosylated extensins, and the highly glycosylated arabinogalactan proteins (AGPs) (Schultz et al., 2002).
Using monoclonal antibodies (JIM4, JIM13, LM2, etc.) and a synthetic chemical reagent called β-glucosyl Yariv reagent (β-GlcY) that specifically binds to AGPs and blocks their normal functions, many studies have indicated that AGPs are implicated in various aspects of plant growth and development, including cell proliferation (Serpe and Nothnagel, 1994; Langan and Nothnagel, 1997; Yang et al., 2007), cell expansion (Willats and Knox, 1996), programmed cell death (Gao and Showalter, 1999), pollen tube growth (Wu et al., 1995; Qin et al., 2007), xylem differentiation (Gao and Showalter, 2000; Motose et al., 2001a, b; Zhang et al., 2003), somatic embryogenesis (van Hengel et al., 2002), zygotic division, and embryo development (Hu et al., 2006; Qin and Zhao, 2006, 2007).
Reverse genetics approaches such as knockout mutants and RNA interference (RNAi) represent powerful means to investigate functions of individual AGPs. Molecular functions of AGPs have mainly been studied in Arabidopsis thaliana. AtAGP6 and AtAGP11 are two homologous Arabidopsis genes encoding classical AGPs specifically expressed in stamens, pollen grains, and pollen tubes. Loss of function of AtAGP6 and AtAGP11 leads to reduced fertility, indicating that AtAGP6 and AtAGP11 play an important role in pollen tube growth and stamen function (Levitin et al., 2008). There are three Lys-rich AGPs in Arabidopsis (Sun et al., 2005). The mutant of AtAGP17 with one insertion in its promoter, named rat1 (resistance to Agrobacterium), is defective in binding of Agrobacterium to its roots (Nam et al., 1999; Gaspar et al., 2004). In AtAGP18 RNAi plants, functional megaspores fail to enlarge and divide, resulting in the abortion of ovules and reduction of seeds, which proves that AtAGP18 is essential for female gametogenesis (Acosta-Garcia and Vielle-Calzada, 2004). In the atagp19 mutant, cell division and expansion are defective, indicating that AtAGP19 has functions in various aspects of plant growth and development (Yang et al., 2007). The mutant of AtFLA4, also called sos5 (salt overly sensitive5), results in abnormal cell expansion, thinner cell walls, and increased sensitivity to salt (Shi et al., 2003). The double mutant of two xylogen homologues AtXYP1 and AtXYP2 shows defective vascular development. Moreover, xylogen was recognized as ‘chimeric’ AGP, as indicated by its large molecular weight range that exceeds its predicted peptide mass, and its reactivity to β-GlcY reagent and JIM13 antibody (Motose et al., 2004). AtAGP30, a non-classical AGP containing six cysteines in the C-terminus, is required for root regeneration and seed germination (van Hengel and Roberts, 2003). RNAi plants of PpAGP1 in Physcomitrella patens resulted in cell length reduction (Lee et al., 2005). CsAGP1 in cucumber (Cucumis sativus) is responsive to gibberellin (GA), and overexpression of CsAGP1 results in a taller stature and earlier flowering than the wild-type plants (Park et al., 2003). LeAGP1 from tomato (Solanum lycopersicum, formerly Lycopersicon esculentum) is up-regulated by cytokinin, and its overexpressing transgenic lines have cytokinin-overexpressing phenotypes (Sun et al., 2004). By using the RNAi technique and attAGP-targeted virus-induced gene silencing, it was proved that the expression of tomato attAGP is induced by infection with Cuscuta reflexa, which promotes the parasite's adherence (Albert et al., 2006).
Based on the structure of AGP protein backbones, they have also been divided into several subclasses: classical AGPs, Lys-rich AGPs, AG peptides, fasciclin-like AGPs (FLAs), non-classical AGPs, and ‘chimeric’ AGPs (Gaspar et al., 2001; Schultz et al., 2002). Previously, 14 classical AGPs, three Lys-rich AGPs, 13 AG peptides, 21 FLAs, two non-classical AGPs, and several ‘chimeric’ AGPs had been identified in Arabidopsis (Schultz et al., 2002, 2004; Borner et al., 2003; van Hengel and Roberts, 2003; Liu and Mehdy, 2007). Recently, 22 early nodulin-like proteins (ENODLs) have been identified in Arabidopsis; 18 of them are regarded as early nodulin-like (eNod-like) AGPs (Mashiguchi et al. 2009).
Most AGPs were identified in Arabidopsis based on biased amino acid composition analysis and a hidden Markov model built from the alignment of 88 fasciclin domains (Schultz et al., 2002; Johnson et al., 2003). Completion of the rice genome (International Rice Genome Sequencing Project, 2004) makes it possible to identify AGP-encoding genes from rice, while most genes are annotated as hypothetical. Although extensive studies had revealed the roles of some AGPs and the underlying mechanisms in Arabidopsis, no AGP-encoding gene was functionally analysed in rice. Therefore, there is an urgent need for a thorough bioinformatic analysis and characterization of AGPs in rice genomes. Previously, 33 and 21 FLAs had been identified in wheat (Triticum aestivum) and rice, respectively (Faik et al., 2006). In this study, the predicted AGP-encoding genes were searched in the rice genome and a phylogenetic analysis was conducted. Furthermore, publicly available resources such as microarrays were evaluated to help in the selection of specific experiments for target genes. The expression patterns of selected target genes in rice organs and tissues at different developmental stages, and the regulation of their expression under abiotic stresses and phytohormone treatments were also examined. The studies indicate important physiological functions of AGPs and can be a base and guide for research into the AGP gene family in rice.
Materials and methods
Plant materials and treatment methods
The rice (Oryza sativa L. japonica cv. Nipponbare) plants were grown in a greenhouse at Wuhan University. Rice seeds were soaked in sterile water for 2 d, and then transferred to containers with sponges as supporting materials in sterile water; this is the first day of the seedling age. Plant materials for expression pattern analysis were: (i) 14-day-old root (YR, young root) and leaf (YL, young leaf); (ii) panicle <3 cm (P1), 5 cm (P2), 10 cm (P3), 15 cm (P4), 20 cm (P5), and 28 cm (P6); (iii) anther (An), and stigma and ovary (SO) from a 28 cm panicle; (iv) ovaries from 1 DAP (day after pollination) (O1) and 3 DAP (O2) seeds; and (v) embryos from 5 DAP (E1), 10 DAP (E2), and 30 DAP (E3) seeds. The temperature for plant growth was 30/25 °C under a photoperiod of 16 h light and 8 h dark. For hormone treatments, 14-day-old seedlings were transferred into containers and treated with deionized water, 25 μM abscisic acid (ABA), and 10 μM GA for 3–24 h. For stress treatments, the 14-day-old seedlings were carefully transferred onto filter papers in a 28 °C illumination incubator as drought stress, placed in 400 mM NaCl solution in a 28 °C illumination incubator as salt stress, and kept at 4 °C as low temperature stress, for 3–24 h, as described previously (Yuan et al., 2008). All materials were taken and quickly frozen in liquid nitrogen, and stored at –80 °C until RNA extraction.
Finding AGPs based on biased amino acid composition, length, and conversed domain
A Perl script (amino acid bias) was downloaded to calculate the PAST (Pro, Ala, Ser, Thr) percentage for all of the proteins in rice (http://www.agwine.adelaide.edu.au/plant/cgb/pcf/bio/). All of the rice proteins (.pep files) were downloaded from RGAP (Rice Genome Annotation Project) annotation release 5 (http://rice.plantbiology.msu.edu/data_download.shtml) and RAP-DB (Rice Annotation Project Database) annotation release 2 (http://rapdb.dna.affrc.go.jp/download/index.html). The Perl program is available as free software for UNIX, Windows, and Mac operating systems (http://www.activeperl.com/). For Windows XP, ActivePerl-5.6.1.638 was used. The protein bias program generated two different lists of proteins. The output files of the Perl script are text files that include ‘long’ proteins >75 amino acid residues in length and above a certain PAST threshold (50% or 55%), and ‘short’ proteins between 55 and 75 amino acid residues in length and below a certain PAST percentage (35% or 40%). Additional output files that contained the first 50 amino acid residues were generated for both the ‘long’ and the ‘short’ proteins. To be AG modified, the protein has to enter the endoplasmic reticulum in eukaryotes. Typically, the existence of a signal peptide is necessary. Therefore, the additional output files were subjected to SignalP 3.0 (http://www.cbs.dtu.dk/services/SignalP/) to check for the presence of N-terminal signal sequences. To check for the presence of a C-terminal GPI (glycosylphosphatidylinositol) additional signal, both the ‘long’ and ‘short’ files were subjected to the big-PI Plant Predictor website: GPI Modification Site Prediction in Plants (http://mendel.imp.ac.at/gpi/plant_server.html) (Eisenhaber et al. 2003).
The search criteria for rice AGPs were as follows. (i) The proteins were classified as AGPs if they contained predominantly Ala-Pro, Ser-Pro, or Thr-Pro throughout the protein with no more than 11 amino acid residues between consecutive Pro residues, but did not contain repeats associated with extensins or PRPs (e.g. Ser-Pro4 or Pro-Pro-Xaa-Yaa-Lys) (Schultz et al., 2002). (ii) The exceptions were the Lys-rich AGPs that were a subclass of classical AGPs. These AGPs have a Lys-rich domain of ∼16 amino acid residues that separate AGP glycomodules. (iii) AGPs were defined as non-classical AGPs if they contained an AGP-like region and other atypical regions except N- and C-terminal signals. (iv) The proteins were classified as AG peptides if their encoded protein backbone was between 55 and 75 residues in length and had XP (X=Ala, Ser, Thr) motifs. (v) The proteins were classified as nsLTP-like AGPs and eNod-like AGPs if they contained a conserved domain such as a non-specific lipid transfer (nsLTP)-like domain and eNod-like domain which was detected by ‘Search Conserved Domains on a protein’ at NCBI (http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi). (vi) The proteins were classified as extensins if they contain Ser-Pro3 and/or Ser-Pro4 repeats which were mostly separated by Tyr, Lys, and Val residues. (vii) The proteins were classified as PRPs if they were rich in proline and possessed none of the characteristics mentioned above. (viii) Several prediction algorithms (http://www.ebi.ac.uk/interpro/, IPR000782, http://pfam.sanger.ac.uk/, PF02469, and http://smart.embl-heidelberg.de/, SM00554) were used for finding FLAs.
Chromosomal localization and gene duplications
The chromosomal distributions of AGP-encoding genes were determined by searching the physical positions of their corresponding locus numbers in the TIGR database. Physical maps of each japonica rice chromosome were downloaded from the IRGSP (International Rice Genome Sequencing Project) (http://rgp.dna.affrc.go.jp/IRGSP/download). BACs (bacterial artificial chromosomes) or PACs (bacteriophage P1 artificial chromosomes) containing AGP-encoding genes were searched. All the sequenced contigs of japonica cv Nipponbare were physically constructed as pseudomolecules by IRGSP (http://rgp.dna.affrc.go.jp/IRGSP), representing 12 rice chromosomes, and are available in GenBank (accession nos AP008207–AP008218). Each of the AGP-encoding genes was positioned on these rice chromosome pseudomolecules by BLASTN search.
Genes separated by ≤5 genes were considered to be tandem duplicates. Genes belonging to segmental duplicates were detected by searching the ‘Segmental genome duplication of rice’ in the TIGR database (http://rice.plantbiology.msu.edu/segmental_dup/index.shtml).
Alignment and phylogenetic analysis
Multiple sequence alignments of amino acid sequences were generated using ClustalX (1.83) and were manually corrected. The obtained sequence alignments were used as input to construct phylogenetic trees with the Neighbor–Joining algorithm within MEGA 4.0 (Tamura et al., 2007). Bootstrapping was performed 10 000 times to obtain support values for each branch. Branches corresponding to partitions reproduced in <50% bootstrap replicates were collapsed.
Digital expression analysis: EST expression profile, microarrays, and MPSS tags
EST (expressed sequence tag) expression profiles were obtained from UniGene at NCBI websites. The total numbers of ESTs were: callus, 164 803; flower, 136 501; leaf, 171 750; panicle, 138 119; root, 68 198; seed, 32 358; stem, 126 877; and vegetative meristem, 4594 (Supplementary Table S3 available at JXB online). Genes were defined as specifically expressed in one tissue if the ESTs of any tissue contributed more than half of the EST frequency.
The results of rice microarrays are available in the Gene Expression Omnibus database at the NCBI website (http://www.ncbi.nlm.nih.gov/geo/) and the Rice Functional Genomic Express Database (http://signal.salk.edu/cgi-bin/RiceGE). For temporal and spatial expression analysis (GSE6893), different stages of panicle and seed development were categorized according to panicle length and days after pollination, respectively, based on landmark developmental events as follows. (i) Panicle: up to 0.5 mm, shoot apical meristem (SAM); 0–3 cm, floral transition and floral organ development (P1); 3–5 cm and 5–10 cm, meiotic stage (P2 and P3); 10–15 cm, young microspore stage (P4); 15–22 cm, vacuolated pollen stage (P5); 22–30 cm, mature pollen stage (P6). (ii) Seeds: 0–2 DAP, early globular embryo (S1); 3–4 DAP, middle and late globular embryo (S2); 5–10 DAP, embryo morphogenesis (S3); 11–20 DAP, embryo maturation (S4); 21–29 DAP, dormancy and desiccation tolerance (S5) (Itoh et al., 2005, Jain et al., 2007). For abiotic stress analysis (GSE6901), rice seedlings were transferred to a beaker containing 200 mM NaCl solution for salt stress, dried between folds of tissue paper at 28±1 °C in a culture room for drought stress, and kept at 4±1 °C for cold stress, for 3 h (Jain et al., 2007). For hormone response (ABA and GA; GSE661), the callus which had been cultured for 30 d was transferred to a medium containing either 50 mM ABA or 50 mM GA. The absolute signal values of Arabidopsis AGP-encoding genes were downloaded using ‘Bulk Gene Download’ at Nottingham Arabidopsis Stock Centre's microarray database, and the results of developmental stages (GSE5629–5633) and stress treatments (GSE5620–5621 and 5623–5624) were used to analyse the expression of AGP-encoding genes in Arabidopsis (http://affymetrix.arabidopsis.info/narrays/help/psp-wbubn.html). In order to make these absolute signal values suitable for cluster display, the absolute values were divided by the average of all absolute values, and the logarithmic values of the ratios in previous step were used as input for cluster display (Supplementary Table S5 at JXB online). Hierarchical cluster displays were generated from the logarithmic values of all AGP-encoding genes, using Cluster and Treeview (Eisen et al., 1998).
MPSS (massively parallel signature sequencing) tags of rice AGPs were obtained from the MPSS project (http://mpss.udel.edu/rice/) mapped to TIGR gene models (Nobuta et al., 2007). The signature was considered to be significant if it uniquely identifies an individual gene and shows a perfect match (100% identity over the tag length). The normalized abundance (tpm, tags per million) of these signatures for a given gene in a given library represents a quantitative estimate of expression of that gene. MPSS expression data for 17-base and 20-base signatures representing 12 different organs and tissues or treatments of rice were used for the analysis. The description of these organs and tissues is: NCA, 35 d callus; NGD, 10 d germinating seedlings grown in the dark; NGS, 3 d germinating seed; NIP, 90 d immature panicle; NME, 60 d meristem tissue; NOS, ovary and mature stigma; NPO, mature pollen; NST, 60 d stem; NYL, 14 d young leaves; and NYR, 14 d young roots (Supplementary Table S4 at JXB online).
Real-time PCR analysis
To confirm the expression of representative AGP-encoding genes in rice organs and tissues at different developmental stages, and stress and hormone reactions from microarray data, real-time PCR analysis was performed using gene-specific primers (Supplementary Table S1 at JXB online). At least two independent biological replicates and three technical replicates of each biological replicate were made for real-time PCR analysis. The first-strand cDNA was synthesized from 1 μg of total RNA using reverse transcriptase (ReverTra Ace, TOYOBO). Real-time PCR was carried out by SYBR-green fluorescence using a Rotor-Gene 6000 real-time PCR machine (Corbett Research). The real-time quantitative PCR was performed on equal amounts of cDNA which were prepared from the various materials. The expression of each gene in different RNA samples was normalized to the expression of an internal gene, UBQ5 (Jain et al., 2006). The relative expression levels were analysed as described previously (Yuan et al., 2008).
Results
Identification of 69 AGP-encoding genes in the rice genome
The AGP gene family is generally classified into classical AGPs, Lys-rich AGPs, AG peptides, FLAs, non-classical AGPs, and ‘chimeric’ AGPs (Gaspar et al., 2001; Schultz et al., 2002). In order to identify classical AGPs, AG peptides, and ‘chimeric’ AGPs, the method derived from Arabidopsis was mainly adopted by calculating the proportion of PAST (Schultz et al., 2002). First, text files that contained all protein sequences were downloaded from RGAP and RAP-DB, and an amino acid bias Perl script (http://agwine.adelaide.edu.au/plant/cgb/pcf/bio/) was used to calculate the PAST percentage of proteins. All the proteins >55% PAST were selected for further analysis, and 77 and 103 PAST-rich proteins were identified in RGAP and RAP-DB, respectively (Table 1 and Fig. 1). AGP protein backbones are expected to have an N-terminal secretion signal for targeting to the endoplasmic reticulum where glycosylation occurs, and the addition of a GPI anchor may also take place. Therefore, all PAST-rich proteins were scanned for the presence of N-terminal secretion signals using SignalP 3.0. Only 44 and 36 PAST-rich sequences were predicted to be secreted in RGAP and RAP-DB, respectively (Table 1). Not all known AGPs were identified using 55% PAST for Arabidopsis (Schultz et al 2002), so, as previously done for Arabidopsis, the threshold was reduced to 50% PAST, and 246 and 271 proteins were identified, of which 128 and 99 were predicted to be secreted (Table 1 and Fig. 1). The procedure used to identify AG peptides was similar to the one used by Schultz et al. (2002); proteins between 50 and 75 amino acid residues in length were searched with a 35% PAST. A total of 127 and 42 proteins that are predicted to be secreted were found in RGAP and RAP-DB, respectively (Table 1 and Fig. 1).
Table 1.
AGPs predicted from the rice genome based on the biased amino acid composition and length
| Databasea | PASTb | Total | Signal peptidec | AGPs | AG peptides | Extensins | PRPs | Othersd |
| RGAP | >55% | 77 | 44 | 11 | 3 | 4 | 9 | 17 |
| >50% | 246 | 128 | 19 | 4 | 7 | 12 | 86 | |
| >40% (50–75) | 161 | 76 | ND | 10 | 2 | ND | 64 | |
| >35% (50–75) | 346 | 127 | ND | 11 | 2 | ND | 114 | |
| RAP-DB | >55% | 103 | 36 | 9 | 4 | 3 | 9 | 11 |
| >50% | 271 | 99 | 17 | 5 | 7 | 13 | 57 | |
| >40% (50–75) | 85 | 27 | ND | 9 | 2 | ND | 16 | |
| >35% (50–75) | 153 | 42 | ND | 10 | 2 | ND | 30 |
RGAP, Rice Genome Annotation Project; RAP-DB, Rice Annotation Project Database.
PAST, the proportion of Pro, Ala, Ser, and Thr.
N-terminal signal peptides were determined by SignalP (Nielsen et al., 1997).
Others, proteins rich in Ala, Ser, and Thr. ND, not determined.
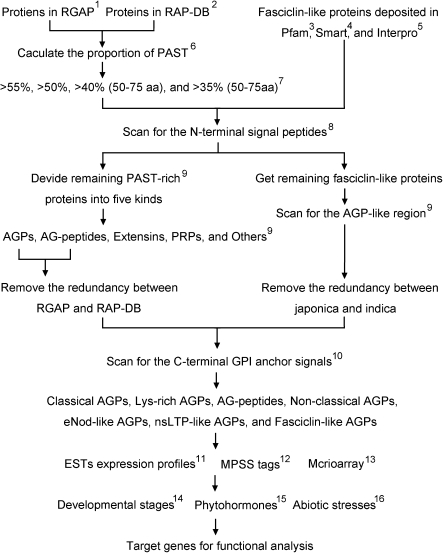
Fig. 1.
The workflow and parameters of AGP identification and data mining. 1, RGAP, Rice Genome Annotation Project; 2, RAP-DB, Rice Annotation Project Database; 3, PF02469 at http://pfam.sanger.ac.uk/; 4, SM00554 at http://smart.embl-heidelberg.de/; 5, IPR000782 at http://www.ebi.ac.uk/interpro/; 6, run a Perl script that divides proteins into several parts according to their amino acid composition; 7, aa, the length of candidate proteins in amino acids; 8, signal peptide prediction, http://www.cbs.dtu.dk/services/SignalP/; 9, the criteria to separate different kinds of HRGPs are detailed in Materials and methods; 10, GPI anchor signal prediction, http://mendel.imp.ac.at/gpi/plant_server.html; 11, EST expression profiles from UniGene at http://www.ncbi.nlm.nih.gov/unigene/; 12, MPSS tags, http://mpss.udel.edu/rice/; 13, the absolute signal values were downloaded at http://signal.salk.edu/cgi-bin/RiceGE; 14, GSE6893, expression at various developmental stages; 15, GSE661, expression under ABA and GA treatments; 16, GSE6901, expression under abiotic stresses treatments.
All PAST-rich proteins used for final analysis were divided into AGPs, AG peptides, extensins, PRPs, and others according to the features of their backbones. Based on the criteria listed in the Materials and methods, there are 19 AGPs (classical AGPs, Lys-rich AGPs, non-classical AGPs, and ‘chimeric’ AGPs) and 11 AG peptides identified in TIGR, and 17 AGPs and 10 AG peptides in RAP-DB. The nsLTP-like AGPs and eNod-like AGPs were regarded as ‘chimeric’ AGPs before, but organized as two subfamilies of AGPs in the present study. After removing the redundancy, 13 classical AGPs, 15 AG peptides, three non-classical AGPs, three eNod-like AGPs, and eight nsLTP-like AGPs were identified in rice (Table 2). Whereas the length of OsAGP5 is 298 amino acids, much longer than other classical AGPs, OsAGP10 has two Ser-rich domains that block AGP glycomodules. Usually, the length of AG peptides is between 55 and 75 residues. One exception is AGP26, which contains 77 amino acids. OsAGP28 is an AG peptide that is not detected in RGAP and RAP-DB, but was identified by Mashiguchi et al. (2004).
Table 2.
AGP-encoding genes identified in the rice genome
| Genea | Subfamilyb | RGAP locusc | RAP-DB locusd | Chromosome locatione | Size (aa)f | GPIg | Evidenceh |
| OsAGP1 | C | NA | Os08g0482300 | Chr8:23845140, 23845908 | 146 | Y | A B C D E |
| OsAGP2 | C | LOC_Os01g71170 | Os01g0937900 | Chr1:41178390, 41179324 | 158 | Y | B D E |
| OsAGP3 | C | NA | Os03g0188500 | Chr3: 4633965, 4634593 | 124 | Y | A B C D E |
| OsAGP4 | C | LOC_Os04g37770 | NA | Chr4:22266907, 22267580 | 131 | Y | C D |
| OsAGP5 | C | NA | Os04g0545250 | Chr4:27141445, 27142670 | 298 | Y | A B C |
| OsAGP6 | C | LOC_Os05g37110 | Os05g0443200 | Chr5:21629021, 21629428 | 135 | Y | C |
| OsAGP7 | C | LOC_Os06g17450 | Os06g0284900 | Chr6:10125749, 10126413 | 128 | Y | B C E |
| OsAGP8 | C | LOC_Os07g28660 | Os07g0469800 | Chr7:16774201, 16774828 | 179 | Y | C |
| OsAGP9 | C | NA | Os10g0377150 | Chr10:12281284, 12281703 | 139 | Y | NA |
| OsAGP10 | C | LOC_Os08g38250 | Os08g0490400 | Chr8:24242332, 24242931 | 199 | Y | C E |
| OsAGP11 | C | LOC_Os04g41480 | Os04g0492200 | Chr4:24404213, 24404920 | 235 | Y | C E |
| OsAGP12 | KC | LOC_Os01g02010 | Os01g0110200 | Chr1:541987, 543510 | 240 | Y | A B C E |
| OsAGP13 | KC | LOC_Os07g40130 | Os07g0590800 | Chr7:24077193, 24079058 | 230 | Y | A B C D E |
| OsAGP14 | Pep | LOC_Os01g37950 | Os01g0559900 | Chr1:21260852, 21261347 | 70 | N | A B C E |
| OsAGP15 | Pep | LOC_Os01g40950 | Os01g0592500 | Chr1:23166562, 23167281 | 72 | N | A B C D E |
| OsAGP16 | Pep | Os01g46850 | Os01g0657000 | Chr1:26721684, 26723875 | 69 | Y | A B C E |
| OsAGP17 | Pep | Os01g55220 | Os01g0756900 | Chr1:31765586, 31765798 | 70 | Y | A B C E |
| OsAGP18 | Pep | LOC_Os01g57030 | Os01g0778300 | Chr1:32943973, 32944508 | 68 | Y | B C D |
| OsAGP19 | Pep | LOC_Os01g57040 | Os01g0778400 | Chr1:32945415, 32945896 | 68 | Y | C |
| OsAGP20 | Pep | LOC_Os02g16500 | Os02g0264800 | Chr2:9417103, 9418631 | 73 | N | A B C E |
| OsAGP21 | Pep | LOC_Os02g48710 | Os02g0718600 | Chr2:29807343, 29807886 | 69 | Y | A B C D E |
| OsAGP22 | Pep | NA | Os02g0718100 | Chr2:29799080, 29799676 | 60 | Y | A B C D E |
| OsAGP23 | Pep | LOC_Os05g12580 | Os05g0217000 | Chr5:7219499, 7219837 | 67 | Y | A B C E |
| OsAGP24 | Pep | LOC_Os06g21400 | Os06g0318800 | Chr6:12358337, 12359021 | 65 | Y | A B C D E |
| OsAGP25 | Pep | LOC_Os06g21410 | Os06g0318900 | Chr6:12365595, 12367161 | 74 | N | C D E |
| OsAGP26 | Pep | LOC_Os06g30920 | Os06g0505700 | Chr6:17983456, 17983849 | 77 | Y | A B C E |
| OsAGP27 | Pep | LOC_Os07g38630 | Os07g0573900 | Chr7:23201833, 23202319 | 64 | Y | B C D E |
| OsAGP28i | Pep | NA | NA | Chr5:26814680, 26815094 | 63 | Y | A |
| OsAGP29 | NC | LOC_Os01g42210 | Os01g0607100 | Chr1:23918314, 23920085 | 221 | Y | C E |
| OsAGP30 | NC | LOC_Os02g10240 | Os02g0196100 | Chr2:5364379, 5366226 | 229 | N | A B C E |
| OsAGP31 | NC | LOC_Os03g63540 | Os03g0852400 | Chr3:35888283, 35889672 | 219 | N | A B C D E |
| OsELA1 | eNod | LOC_Os01g54430 | Os01g0748150 | Chr1:31316996, 31318114 | 237 | Y | A B D E |
| OsELA2 | eNod | LOC_Os02g06670 | Os02g0162200 | Chr2:3353562, 3354820 | 261 | Y | C E |
| OsELA3 | eNod | LOC_Os06g46740 | Os06g0681200 | Chr6:28387298, 28388576 | 279 | Y | A B C E |
| OsLLA1 | nsLTP I | LOC_Os03g26820 | Os03g0385400 | Chr3:15306262, 15308462 | 178 | Y | A B C D E |
| OsLLA2 | nsLTP I | LOC_Os03g46180 | Os03g0664600 | Chr3:26112985, 26114326 | 228 | Y | C D E |
| OsLLA3 | nsLTP II | LOC_Os04g52250 | Os04g0612300 | Chr4:30866056, 30866925 | 202 | N | B C |
| OsLLA4 | nsLTP II | LOC_Os06g46780 | Os06g0681500 | Chr6:28405288, 28406220 | 268 | N | C |
| OsLLA5 | nsLTP II | LOC_Os06g46870 | Os06g0682750 | Chr6:28447781, 28448967 | 270 | N | C E |
| OsLLA6 | nsLTP I | LOC_Os07g07790 | Os07g0174400 | Chr7:3909569, 3910639 | 188 | Y | C D E |
| OsLLA7 | nsLTP I | LOC_Os07g43290 | Os07g0625800 | Chr7:25919683, 25921877 | 177 | Y | A B C D E |
| OsLLA8 | nsLTP II | LOC_Os10g11370 | Os10g0191100 | Chr10:6319745, 6322898 | 357 | N | C E |
| OsFLA1 | Fas III | LOC_Os04g48490 | Os04g0574200 | Chr4:28725108, 28726790 | 431 | Y | A B C E |
| OsFLA2 | Fas III | LOC_Os03g03600 | Os03g0128000 | Chr3:1578312, 1579906 | 401 | Y | A B C D E |
| OsFLA3 | Fas III | LOC_Os08g23180 | Os08g0321000 | Chr8:13982326, 13985506 | 415 | Y | A B C E |
| OsFLA4 | Fas I | LOC_Os08g38270 | Os08g0490600 | Chr8:24249970, 24251221 | 271 | Y | A B C E |
| OsFLA5 | Fas I | LOC_Os08g39270 | Os08g0502400 | Chr8:24815594, 24817738 | 274 | Y | A B C E |
| OsFLA6 | Fas I | LOC_Os05g48900 | Os05g0563600 | Chr5:27989048, 27990300 | 272 | Y | A B C E |
| OsFLA7 | Fas I | LOC_Os01g47780 | Os01g0668100 | Chr1:27339855, 27340906 | 247 | Y | A B C D E |
| OsFLA8 | Fas I | LOC_Os01g06580 | Os01g0159200 | Chr1:3096346, 3097527 | 269 | Y | A B C D E |
| OsFLA9 | Fas I | LOC_Os05g07060 | Os05g0163300 | Chr5:3715739, 3716868 | 275 | Y | A B C D E |
| OsFLA10 | Fas I | LOC_Os09g30010 | Os09g0476800 | Chr9:18256643, 18257801 | 273 | Y | A B C E |
| OsFLA11 | Fas III | LOC_Os09g07350 | Os09g0248100 | Chr9:3653972, 3657804 | 401 | Y | A B C E |
| OsFLA12 | Fas III | LOC_Os01g62380 | Os01g0841100 | Chr1:36098359, 36103250 | 427 | Y | A B C D E |
| OsFLA13 | Fas I | LOC_Os04g39600 | Os04g0472200 | Chr4:23414397, 23415562 | 263 | Y | A B C E |
| OsFLA14 | Fas I | LOC_Os04g39590 | Os04g0472100 | Chr4:23412950, 23414118 | 277 | Y | A B C E |
| OsFLA15 | Fas I | LOC_Os02g20560 | Os02g0308800 | Chr2:12127908, 12129258 | 268 | Y | A B C E |
| OsFLA16 | Fas IV | LOC_Os07g06680 | Os07g0160600 | Chr7:3257238, 3261152 | 479 | N | A B C D E |
| OsFLA17 | Fas IV | LOC_Os03g57490 | Os03g0788700 | Chr3:32771257, 32775727 | 479 | N | A B C E |
| OsFLA18 | Fas I | LOC_Os05g48890 | Os05g0563550 | Chr5:27984690, 27985698 | 251 | Y | A B C E |
| OsFLA19 | Fas I | LOC_Os02g20540 | Os02g0308400 | Chr2:12113830, 12114971 | 267 | Y | A B C E |
| OsFLA20 | Fas II | LOC_Os02g26320 | Os02g0461500 | Chr2:15453153, 15454310 | 304 | Y | A B C E |
| OsFLA21 | Fas II | LOC_Os02g49420 | Os02g0726000 | Chr2:30192724, 30193979 | 266 | Y | A B C E |
| OsFLA22 | Fas II | LOC_Os02g26290 | Os02g0461000 | Chr2:15430699, 15431764 | 213 | N | A B C E |
| OsFLA23 | Fas I | LOC_Os06g17460 | Os06g0285100 | Chr6:10130034, 10130805 | 211 | Y | E |
| OsFLA24 | Fas IV | LOC_Os03g57460 | Os03g0788600 | Chr3:32749877, 32753506 | 474 | N | A B C E |
| OsFLA25 | Fas II | LOC_Os06g44660 | Os06g0656800 | Chr6:26956539, 26957614 | 289 | Y | A B C E |
| OsFLA26 | Fas III | LOC_Os05g38500 | Os05g0459700 | Chr5:22524615, 22526317 | 472 | N | A B C E |
| OsFLA27 | Fas I | LOC_Os09g30486 | Os09g0482780 | Chr9:18610276, 18612149 | 267 | N | A B C E |
Systematic designation given to rice AGPs. FLAs that have previously been identified by bioinformatic analysis are indicated in bold (Faik et al, 2006); β-GlcY-reactive AGPs are indicated in bold and underlined (Mashiguchi et al., 2004).
C, classical AGP; KC, Lys-rich classical AGP; Pep, AG peptide; NC, non-classical AGP; eNod, early nodulin-like AGP; nsLTP, non-specific lipid transfer protein-like AGP; Fas, fasciclin-like AGP.
c,dLocus identity no. assigned by cRGAP and dRAP-DB, which could be converted by ID converter (http://rapdb.dna.affrc.go.jp/tools/converter/).
Chromosomal localization of rice AGP-encoding genes.
Length of the open reading frame in amino acids.
GPI anchor signal predicted by big-PI (http://mendel.imp.ac.at/gpi/plant_server.html); Y, yes, N, no.
Evidence for gene expression from (A) full-length cDNA, (B) ESTs, (C) MPSS tags, (D) serial analysis of gene expression (SAGE), and (E) microarray data.
OsAGP28 does not exist in RGAP and RAP-DB.
NA, not available.
It is difficult to find FLAs using the amino acid bias (Perl script) because of their low proportion of PAST (from 31.74 to 45.35) (Supplementary Table S2 at JXB online). In previous research, 21 rice FLA genes were identified together with 33 wheat FLA genes (Faik et al., 2006). All rice fasciclin-like proteins from Pfam (PF02469, 34 proteins from japonica), Smart (SM00554, 58 proteins from indica and japonica), and Interpro (IPR000782, 89 proteins from indica and japonica) were downloaded as a source, the proteins from indica and the redundant proteins were deleted, then they were scanned for the presence of N-terminal secretion signals using SignalP 3.0. After excluding proteins that have no signal peptides, the remaining fasciclin-like proteins were scanned for the presence of the putative AG polysaccharide additional domain, where Pro/Hyp residues are organized in non-contiguous clusters. To predict the occurrence of such glycomodules in these putative rice FLAs, Pro/Hyp-containing sequence motifs in each of them were manually counted. Finally, 27 OsFLAs genes were identified by integrating research results and annotation advances. At least 77.78% of rice FLAs are predicted to be GPI anchored to the plasma membrane (Table 2).
Protein structure and phylogenetic analysis
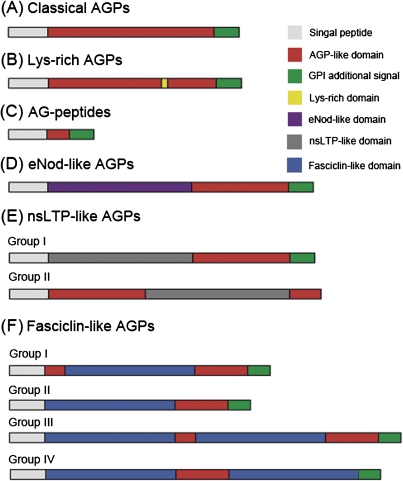
According to the structural characteristics of rice AGP backbones, they can be classified into seven subfamilies, including classical AGPs, Lys-rich AGPs, AG peptides, non-classical AGPs, eNod-like AGPs, nsLTP-like AGPs, and FLAs. The protein backbones of OsAGPs have been organized into domains leading to their classification as either ‘classical’ or ‘non-classical’ (Showalter, 1993, 2001; Gaspar et al., 2001). Classical AGPs consist of a core protein of highly varying length and domain complexity, and often contain a GPI lipid anchor (Fig. 2A). Non-classical AGPs are defined as having regions that are atypical of an AGP-like region, for example regions rich in Asn or Cys residues in addition to regions containing Pro/Hyp (Du et al., 1996). Three non-classical AGPs were found in rice; they have an atypical region in addition to the AGP-like region. OsAGP29 contains a GPI lipid anchor signal, but OsAGP30 and OsAGP31 do not. Some OsAGPs, designated as lysine-rich AGPs, contain a short lysine-rich domain between the Pro-rich domain and the hydrophobic C-terminus (Figs 2B, 3B). Several rice AGP backbones are only 50–75 residues long and are termed AG peptides (Fig. 2C). There are many kinds of ‘chimeric’ AGPs, such as OsELAs, OsLLAs, and OsFLAs. An early nodulin (eNod)-like domain related to copper binding is present between the signal peptide and the AGP-like region in ELAs (Figs 2D, 3C). Similarly, an nsLTP-like domain exists between the signal peptide and the AGP-like region in LLAs (or xylogen) (Figs 2E, 3D). The eight OsLLAs were divided into two groups, the group I OsLLAs have an nsLTP-like domain between the signal peptide and the AGP-like region; and the AGP-like regions of group II OsLLAs are separated into two parts by nsLTP-like domains, and no GPI anchor signals are present (Fig. 2E). Moreover, 27 rice FLAs were divided into four groups due to the numbers of AGP-like regions and fasciclin domains (Fig. 2F). There are two AGP-like regions and one fasciclin domain in group I OsFLAs, one AGP-like region and one fasciclin domain in group II OsFLAs, two AGP-like regions and two fasciclin domains in group III OsFLAs, and one AGP-like region and two fasciclin domains in group IV OsFLAs.
Fig. 2.
Schematic representations of the different subclasses of rice AGP protein backbones. Not drawn to scale
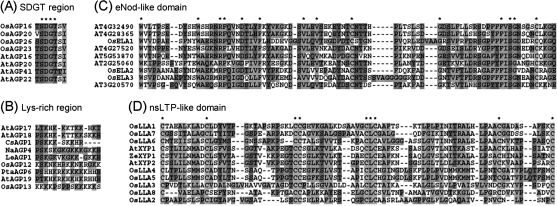
Fig. 3.
Multiple sequence alignments of the SDGT region (A), Lys-rich region (B), eNod-like domain (C), and nsLTP-like domain (D).
To investigate the phylogenetic relationship of AGPs, six unrooted trees of classical AGPs, Lys-rich AGPs, AG peptides, ELAs, LLAs, and FLAs were constructed from alignments of their full-length amino acid sequences from rice, Arabidopsis, and other plants (Fig. 4). Classical AGPs expand in an overall species-specific manner; 11 proteins in the first clade are from Arabidopsis, 10 proteins in the second clade are from rice, and the proteins in the third clade are three from Arabidopsis and one from rice (Fig. 4A). The phylogenetic relationship of two family members of Lys-rich AGPs (OsAGP12 and OsAGP13) is close to AtAGP19 (Fig. 4B). There is one clade of AG peptides termed SDGT region-containing AG peptides because they all have four conversed residues (Ser, Asp, Gly, and Thr) in their protein backbones (Figs 3A, 4C). Although OsFLA21 was phylogeneticly close to the members of group III FLAs, it was recognized as a member of group II FLAs, because there is one fasciclin-like domain and two AGP-like regions in its protein backbone. Group II of LLAs is absent in Arabidopsis, so they may specifically exist in monocots (Fig. 4E). However, the family members in clades of AG peptides, group I of OsLLAs, OsELAs, and OsFLAs are present in both Arabidopsis and rice, indicating that they may play similar roles in the two plants (Fig. 4C–F).
Fig. 4.
Phylogenetic relationship of AGPs between rice and other species. (A) Classical AGPs, (B) Lys-rich AGPs, (C) AG peptides, (D) eNod-like AGPs, (E) nsLTP-like AGPs, and (F) fasciclin-like AGPs. The phylogenetic tree is based on the multiple sequence alignments of AGPs and was generated using MEGA 4.0 by the Neighbor–Joining method. Bootstrap support from 10 000 reiterations is indicated above the branches.
Chromosomal localization and gene duplication
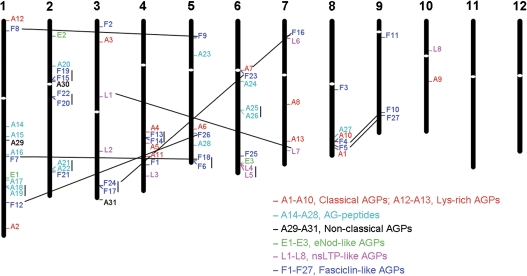
The approximate positions of AGP-encoding genes were marked on the physical map of the 12 rice chromosomes. According to the genomic localization of AGP-encoding genes, the 69 AGP-encoding genes of rice distribute in 10 of the 12 chromosomes: 13 genes on chromosome 1, 10 genes on chromosome 2, nine genes on chromosome 6, seven genes each on chromosomes 3 and 4, six genes each on chromosomes 5 and 8, five genes on chromosome 7, three genes on chromosome 9, and two genes on chromosome 10 (Fig. 5).
Fig. 5.
Genomic localization of AGP-encoding genes on rice chromosomes. AGP-encoding genes classified into different subfamilies are shown in different colours. White ovals on the chromosomes indicate the position of centromeres. Chromosome numbers are indicated at the top of each chromosome. The AGP-encoding genes presented on duplicated chromosomal segments between two chromosomes are connected by black lines, and tandem duplicated genes on the same chromosome are connected by a vertical bar.
Nine pairs of rice AGP-encoding genes are arranged in tandem repeats (Fig. 5), representing localized gene duplications, which are separated by a maximum of five intervening genes. The tandem duplicated genes belong to three subfamilies, including five pairs of OsFLAs, three pairs of AG peptides, and one pair of OsLLAs. The duplication of the AGP-encoding genes is also associated with chromosomal segment duplications. Six pairs of OsFLAs and one pair of OsLLAs are located on the duplicated segmental regions of rice chromosomes mapped by TIGR when the maximal length between collinear gene pairs is 500 kb. All of these duplicated genes exhibit high sequence similarity in both the AGP-like regions and the additional domains.
The gene duplication analysis shows that 22 of 27 OsFLA genes expand through gene duplication (Fig. 5). Interestingly, OsFLA6 and OsFLA18, and OsFLA17 and OsFLA24 are gene pairs of tandem duplication, and OsFLA7 and OsFLA18, and OsFLA16 and OsFLA24 are gene pairs of chromosomal segmental duplication, revealing that these gene clusters expand through both tandem and segmental duplications. In addition, one pair of tandem duplicated genes (OsLLA4 and OsLLA5) and one pair of chromosomal duplicated genes (OsLLA1 and OsLLA7) exist in the nsLTP-like AGPs. It is noteworthy that tandem duplication plays important roles in the expansion of the AG peptide subfamily; three pairs of tandem duplicated genes are found in AG peptides (OsAGP18 and OsAGP19, OsAGP21 and OsAGP22, and OsAGP25 and OsAGP26) (Fig. 6).
Fig. 6.
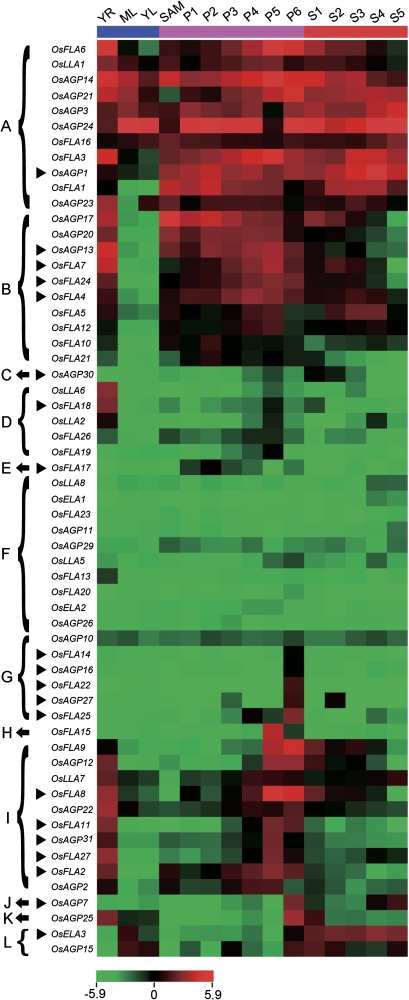
Expression profiles of AGP-encoding genes in various rice organs and tissues at different development stages. The microarray data sets (GSE6893) of gene expression at various developmental stages were used for cluster display. A heat map representing hierarchical clustering of average log signal values of all the AGP-encoding genes in various organs and tissues at different developmental stages (indicated at the top of each lane). The genes which share similar expression patterns are divided into 12 clusters: (A) all examined tissues; (B) all examined tissues except ML and YL; (C) S1 and S2; (D) YR and P5; (E) P3; (F) low expression in all examined tissues; (G) P6; (H) P5; (I) YR, P5, and P6; (J) P6, S4, and S5; (K) YR, P6, and S1; (L) YL, ML, P6, and S1–S6. The representative AGP-encoding genes differentially expressed during various stages of development for which real-time PCR analysis was performed are indicated by a black triangle on the left. The colour scale (representing average log signal values) is shown at the bottom.
Expression analysis of rice AGP-encoding genes in various organs and tissues at different developmental stages
The gene expression pattern provides important clues for investigating gene function. The expression of rice AGP-encoding genes was analysed using three publicly available resources: EST expression profiles, microarrays, and MPSS tags.
EST expression profiles of AGP-encoding genes were obtained by searching the RAP-DB locus across the UniGene database at NCBI. It was found that 52 of the 69 AGP-encoding genes have at least one corresponding full-length cDNA and/or EST, 47 AGP-encoding genes have both full-length cDNA and EST evidence, whereas five genes have only EST evidence (Table 2). According to EST abundance data of rice organs and tissues, 48 of 52 AGP-encoding genes can be transcriptionally analysed using EST expression profiles. A large number of AGP-encoding genes show high expression abundance in stem, vegetative meristem, panicle, and seed. Some of them show tissue-specific expression patterns: OsFLA15 and OsFLA19 in stem; OsAGP13, OsAGP17, OsAGP20, and OsFLA2 in vegetative meristem; OsAGP7 and OsFLA22 in panicle; and OsAGP1 in seed (Supplementary Table S3 at JXB online).
Microarrays provide a high-throughput means to analyse the expression of genes of interest at the transcriptional level. Sixty of 69 AGP-encoding genes have at least one probe on Affymetrix rice whole-genome arrays (GPL2025). The results of microarray analysis revealed that most of the AGP-encoding genes were expressed in at least one of the reproductive and vegetative developmental stages (SAM, P1–P6, S1–S5, young root and leaf, and mature leaf) (Fig. 6). Eleven genes are abundantly expressed in all examined tissues (Fig. 6A). Ten genes are abundantly expressed in root, SAM, panicles, and seed (Fig. 6B). OsAGP30 is abundantly expressed in early stages of seed development (Fig. 6C). Five genes are abundantly expressed in roots and panicles at the stage of vacuolated pollen (P5) (Fig. 6D); OsFLA17 was specifically expressed in early stages of panicle development (Fig. 6E). The expression levels of 10 genes were relatively low in all examined tissues (Fig. 6F). Interestingly, five genes (OsAGP16, OsAGP27, OsFLA14, OsFLA22, and OsFLA25) and one gene (OsFLA15) were specifically expressed in panicles at the stage of vacuolated pollen (P5) and at the stage of mature pollen (P6), respectively, indicating that these genes may be correlated to pollen development (Fig. 6G, H). Ten genes were abundantly expressed in roots, late stages of panicle development, and early stages of seed development (Fig. 6I). OsAGP7 was dominantly expressed in panicles at the stage of mature pollen (P6) and seeds at stages of embryo maturation (S4) and desiccation (S5) (Fig. 6J), OsAGP25 in roots, panicles at the stage of mature pollen (P6), and seeds at the stage of early globular embryo (S1) (Fig. 6K), and OsELA3 and OsAGP15 in leaves, panicles at the stage of mature pollen (P6), and seeds (Fig. 6L).
Moreover, differential expression analysis was performed to identify AGP-encoding genes which have more abundant expression in the processes of seedling, panicle, and seed development by comparing the expression between vegetative and reproductive tissues (Table 3). A gene is considered as differentially expressed if its expression level at a certain stage is significantly lower or higher (<0.5 or >2) than in the compared tissues (Table 3). A large number (30) of rice AGP-encoding genes are differentially expressed in reproductive tissues compared with vegetative tissues, indicating that these genes may play important roles in panicle and seed development (Table 3). Six rice AGP-encoding genes are differentially expressed in vegetative tissues, and OsLLA2 and OsLLA6 were dominantly expressed in root (Fig. 6D and Table 3).
Table 3.
Differential expression analysis of rice AGP-encoding genes in vegetative and reproductive tissues
| Gene | Probea | MOVb | MORc | MOR/MOVd | DEVe | DERf |
| OsAGP1 | Os-21950-1-S1_at | 1264.8 | 21 933.7 | 17.34 | Y | |
| OsAGP10 | OsAffx-6065-1-S1_x_at | 252.9 | 311.5 | 1.23 | ||
| OsAGP11 | OsAffx-26415-1-S1_at | 58.3 | 165.4 | 2.84 | Y | |
| OsAGP12 | Os-56224-1-S1-at | 3766.0 | 6611.0 | 1.76 | ||
| OsAGP13 | Os-5095-1-S1-at | 14 091.4 | 7888.9 | 0.56 | ||
| OsAGP14 | Os-6768-1-S1_at | 16 850.3 | 21 308.1 | 1.26 | ||
| OsAGP15 | Os-57295-1-S1_at | 1961.8 | 2595.2 | 1.32 | ||
| OsAGP16 | Os-30521-1-S1_at | 53.9 | 763.9 | 14.17 | Y | |
| OsAGP17 | Os-5831-1-S1_at | 8865.8 | 19 596.6 | 2.21 | Y | |
| OsAGP2 | OsAffx-11813-1-S1_at | 1180.7 | 2311.9 | 1.96 | ||
| OsAGP20 | Os-12412-1-S1_at | 4099.2 | 5674.4 | 1.38 | ||
| OsAGP21 | Os-5592-1-S1_at | 10 774.2 | 16 628.3 | 1.54 | ||
| OsAGP22 | Os-5823-1-S1_at | 7667.5 | 2714.3 | 0.35 | Y | |
| OsAGP23 | Os-11520-1-S1_at | 6995.2 | 3350.9 | 0.48 | Y | |
| OsAGP24 | Os-5384-1-S1_at | 26 966.3 | 48 847.0 | 1.81 | ||
| OsAGP25 | OsAffx-27752-1-S1_s_at | 4398.8 | 7348.5 | 1.67 | ||
| OsAGP26 | Os-50508-1-S1_at | 63.2 | 94.2 | 1.49 | ||
| OsAGP27 | OsAffx-5568-1-S1_at | 52.1 | 1817.6 | 34.89 | Y | |
| OsAGP29 | OsAffx-23631-1-S1_at | 86.2 | 219.3 | 2.54 | Y | |
| OsAGP30 | Os-55753-1-S1_at | 44.0 | 876.7 | 19.93 | Y | |
| OsAGP3 | Os-55382-1-S1_at | 5047.5 | 14 647.8 | 2.90 | Y | |
| OsAGP31 | Os-5927-1-S1_at | 2157.1 | 4118.3 | 1.91 | ||
| OsAGP7 | OsAffx-15474-1-S1_s_at | 96.6 | 4359.5 | 45.13 | Y | |
| OsELA1 | Os-34474-2-S1_at | 44.5 | 140.4 | 3.16 | Y | |
| OsELA2 | OsAffx-24136-1-S1_at | 60.5 | 98.2 | 1.62 | ||
| OsELA3 | Os-55380-1-S1_at | 1639.5 | 5631.9 | 3.44 | Y | |
| OsFLA1 | OsAffx-14326-1-S1_s_at | 878.5 | 12 342.8 | 14.05 | Y | |
| OsFLA10 | Os-56233-1-S1_s_at | 477.3 | 1726.2 | 3.62 | Y | |
| OsFLA11 | Os-49544-1-S1_at | 4495.4 | 4285.7 | 0.95 | ||
| OsFLA12 | Os-42475-1-S1_at | 597.8 | 2387.9 | 3.99 | Y | |
| OsFLA13 | Os-5963-1-S1_at | 353.6 | 81.4 | 0.23 | Y | |
| OsFLA14 | Os-53497-1-S1_at | 62.3 | 731.3 | 11.74 | Y | |
| OsFLA15 | Os-17961-1-S1_a_at | 66.3 | 10 701.1 | 161.40 | Y | |
| OsFLA16 | Os-17949-1-S1_a_at | 2138.4 | 4617.1 | 2.16 | Y | |
| OsFLA17 | Os-23180-1-S1_at | 36.2 | 850.9 | 23.51 | Y | |
| OsFLA18 | OsAffx-15160–1-S1_at | 4545.8 | 582.5 | 0.13 | Y | |
| OsFLA19 | Os-14430-1-S1_at | 77.2 | 728.6 | 9.44 | Y | |
| OsFLA2 | Os-5826-2-S1_a_at | 3439.2 | 5314.1 | 1.55 | ||
| OsFLA20 | Os-55769-1-S1_at | 47.8 | 100.4 | 2.10 | Y | |
| OsFLA21 | Os-53492-1-S1_at | 243.2 | 1278.5 | 5.26 | Y | |
| OsFLA22 | Os-6347-1-S1_at | 33.9 | 1932.2 | 57.00 | Y | |
| OsFLA23 | OsAffx-15475–1-S1_at | 50.5 | 76.0 | 1.50 | ||
| OsFLA24 | Os-22627-1-S1_at | 3010.4 | 5033.8 | 1.67 | ||
| OsFLA25 | Os-20322-1-S1_at | 53.8 | 4595.4 | 85.42 | Y | |
| OsFLA26 | OsAffx-27225-1-S1_at | 224.5 | 474.7 | 2.11 | Y | |
| OsFLA27 | Os-52641-1-S1_at | 6064.7 | 4457.6 | 0.74 | ||
| OsFLA3 | Os-5500-1-S1_s_at | 13 961.0 | 17 519.0 | 1.25 | ||
| OsFLA4 | Os-55554-1-S1_at | 1780.7 | 6680.5 | 3.75 | Y | |
| OsFLA5 | Os-54871-1-S1_at | 1362.3 | 3671.7 | 2.70 | Y | |
| OsFLA6 | Os-53704-1-S1_x_at | 19 535.7 | 20 590.0 | 1.05 | ||
| OsFLA7 | Os-5908-1-S1_at | 11 698.0 | 8593.8 | 0.73 | ||
| OsFLA8 | Os-1437-1-S1_at | 6607.4 | 17 431.6 | 2.64 | Y | |
| OsFLA9 | Os-54555-1-S1_at | 897.7 | 15 115.8 | 16.84 | Y | |
| OsLLA1 | Os-9710-1-S1_at | 4940.3 | 4113.9 | 0.83 | ||
| OsLLA2 | OsAffx-13377-1-S1_at | 1117.9 | 539.3 | 0.48 | Y | |
| OsLLA5 | OsAffx-15958-1-S1_at | 95.7 | 300.0 | 3.13 | Y | |
| OsLLA6 | OsAffx-28335-1-S1_at | 5696.2 | 233.3 | 0.04 | Y | |
| OsLLA7 | Os-9342-1-S1_at | 3120.1 | 1746.4 | 0.56 | ||
| OsLLA8 | OsAffx-18220-1-S1_x_at | 87.0 | 212.5 | 2.44 | Y |
The probe ID is used on microarray plate GPL2025. The experiment GSE6893 was used for differential expression analysis.
MOV, the maximum absolute values of vegetative tissues.
MOR, the maximum absolute values of reproductive tissues.
The values of MOR divided by MOV. Values considered significant (i.e. <0.5 or >2) are in bold.
DEV, differential expression in vegetative tissues.
DER, differential expression in reproductive tissues.
MPSS generates thousands of molecules per reaction and provides a sensitive quantitative measure of gene expression for nearly all genes in the genome (Brenner et al., 2000). It was found that MPSS signatures are available for 58 rice AGP-encoding genes in at least one of the libraries (Supplementary Table S4 at JXB online). This further strengthens the idea that most rice AGP-encoding genes are expressed. Differences are displayed by number of tags (tpm, transcripts per million), being low in <50, moderate in between 50 tpm and 500 tpm, and strong in >500 tpm. A large percentage of AGP-encoding genes (37) are expressed at a high level, while 14 and seven AGP-encoding genes exhibit moderate and low expression, respectively (Supplementary Table S4). Further, both 17-base and 20-base signature data sets are used to compare the differential expression of AGP-encoding genes in different MPSS libraries. Thirteen AGP-encoding genes were identified with tissue-specific expression patterns, suggesting that these genes may play important roles in pollen, stigmas and ovaries, immature panicles, and germinating seeds. Moreover, the highest number of genes (eight) show preponderant expression in mature pollen (Supplementary Table S4).
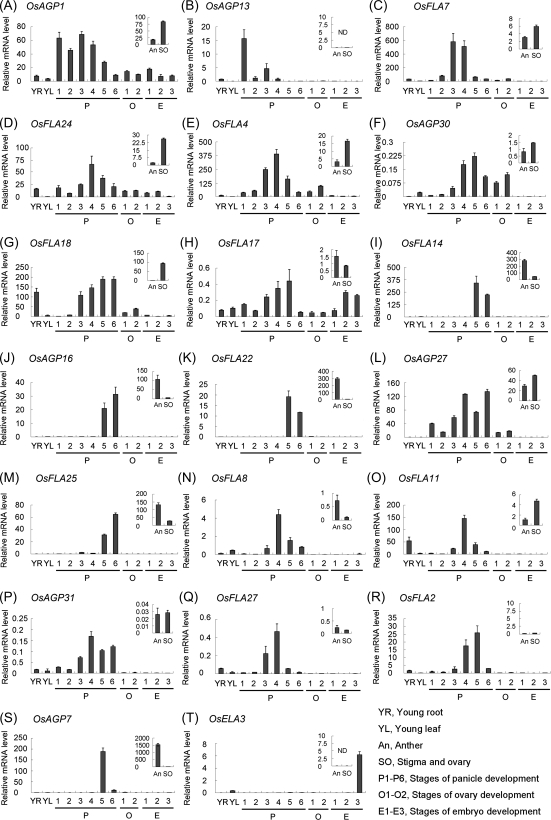
In order to validate the results of digital expression analysis, real-time PCR analysis was performed for some representative genes. The results of real-time PCR showed that the expression patterns of these genes were in general agreement with the data of microarrays and MPSS tags. For example, OsAGP1, OsFLA7, OsAGP30, and OsAGP27 are expressed during the processes of panicle and ovary development, and also predominantly in stigmas and ovaries as compared with anther (Fig. 7A, C, F, L). OsAGP13 is mainly expressed in panicles at the stage of floral transition (P1) (Fig. 7B), but was not detected in either anther or stigmas and ovaries. OsFLA24, OsFLA4, OsFLA18, OsFLA17, OsFLA11, OsAGP31, and OsFLA2 were mainly expressed in panicles at the stages of megaspore and microspore development (P2–P5), and OsFLA24, OsFLA4, OsFLA18, and OsFLA11 were also predominantly expressed in stigmas and ovaries (Fig. 7D, E, G, H, O, P, R). OsFLA14, OsAGP16, OsFLA22, and OsFLA25 were specifically expressed in panicles at the stages of flower maturation (P5 and P6), and predominantly in anther (Fig. 7I, J, K, M). The expression levels of OsFLA8 and OsFLA27 were highest in panicles at the stage of the young microspore (P4) and were mainly expressed in anther (Fig. 7N, Q), while OsAGP7 was specifically expressed in P5 and anther, and OsELA3 in 30 DAP embryos (Fig. 7T).
Fig. 7.
Real-time PCR analysis for confirmation of the differential expression of representative AGP-encoding genes in various organs and tissues at different development stages. The small pictures inserted in the figures represent their relative expression levels in anther (An) and stigma and ovary (SO) from 28 cm panicles.
Expression analysis of rice AGP-encoding genes under abiotic stress, ABA, and GA treatments
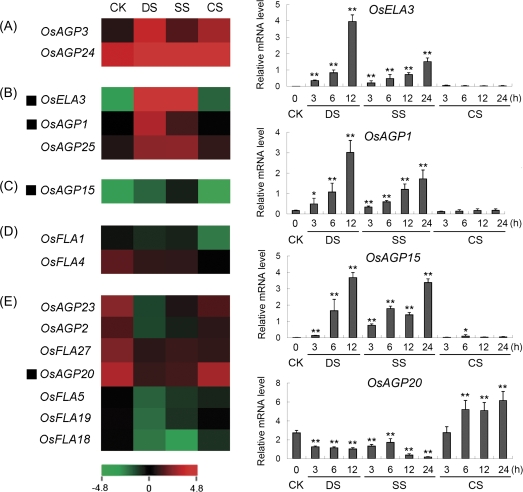
To investigate the abiotic stress response of rice AGP-encoding genes, the results of microarray (GSE6901) from 7-day-old seedlings subjected to drought, salt, and cold stresses were analysed. The data revealed that a total of 15 genes were significantly down- or up-regulated (<0.5 or >2) as compared with the control in at least one of the stress conditions examined (Fig. 8). The transcript levels of two genes (OsAGP3 and OsAGP24) were up-regulated by all three stresses (Fig. 8A), three genes (OsELA3, OsAGP1, and OsAGP25) were up-regulated significantly by drought and salt stresses (Fig. 8B), one gene (OsAGP15) was up-regulated by salt stress (Fig. 8C), two genes (OsFLA1 and OsFLA4) were down-regulated by cold stress (Fig. 8D), and seven genes (OsAGP23, OsAGP2, OsFLA27, OsAGP20, OsFLA5, OsFLA19, and OsFLA18) were down-regulated by drought and salt stresses (Fig. 8E). By using the real-time PCR technique, the expression levels of four representative AGP-encoding genes were investigated under various stress conditions. The expression of OsAGP15, OsELA3, and OsAGP1 was induced by drought and salt stresses, and showed a gradual increased expression pattern after the stresses were applied (Fig. 8). On the other hand, the expression level of OsAGP20 was up-regulated by cold at 6, 12, and 24 h, and down-regulated by drought and salt stresses (Fig. 8).
Fig. 8.
Expression profiles of rice AGP-encoding genes differentially expressed under abiotic stresses. The microarray data sets (GSE6901) of gene expression under various abiotic stresses were used for cluster display. The average log signal values of AGP-encoding genes under control and various stress conditions (indicated at the top of each lane) are presented by a heat map. Only those genes that exhibited ≥2-fold differential expression, under any of the given abiotic stress conditions, are shown. (A) Up-regulated by drought, salt, and cold stresses; (B) up-regulated by drought and salt stresses; (C) up-regulated by salt stress; (D) down-regulated by cold stress; (E) down-regulated by drought and salt stresses. The representative AGP-encoding genes differentially expressed under different abiotic stress conditions for which real-time PCR analysis was performed are indicated as a black triangle to the left. The results of real-time PCR analysis for confirmation of the differential expression of rice AGP-encoding genes under abiotic stresses are shown on the right. Two asterisks (**, P <0.01) and one asterisk (*, 0.01<P < 0.05) represent significant differences between the controls and treatments as determined by Origin 7.5. The mRNA levels for each candidate gene in different organ and tissue samples were calculated relative to its expression in control seedlings. The colour scale (representing average log signal values) is shown at the bottom. CK, control; DS, drought stress; SS, salt stress; CS, cold stress.
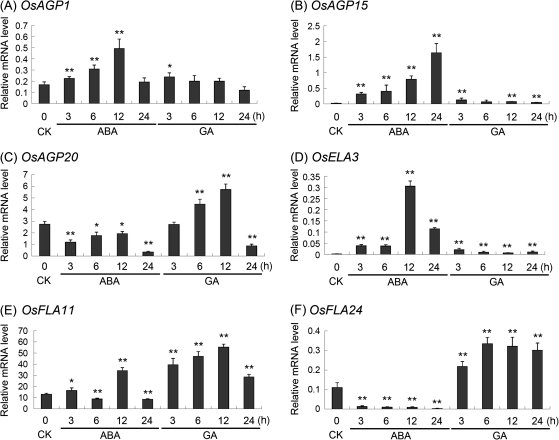
The results of ABA- and GA-treated callus were used to analyse the regulation of AGP-encoding genes (GSE661). It was found that many AGP-encoding genes were regulated by ABA and GA (Supplementary Table S5 at JXB online). To verify the results of microarray under ABA and GA treatments, the transcriptional level of six representative AGP-encoding genes in young seedlings treated with ABA and GA were investigated by using real-time PCR (Fig. 9). The expression levels of OsAGP1, OsAGP15, and OsELA3 were markedly increased in seedlings under ABA treatment (Fig. 9A, B, D). The expression level of OsAGP20 was down-regulated by ABA, but up-regulated by GA (Fig. 9C). The expression of OsFLA24 was almost totally inhibited by ABA, but induced by GA (Fig. 9F). However, the expression of OsFLA11 was more complex, being up-regulated by GA at 3–12 h and transiently up-regulated by ABA at 12 h (Fig. 9E).
Fig. 9.
Real-time PCR analysis for confirmation of the differential expression of rice AGP-encoding genes under ABA and GA treatments. Two asterisks (**, P <0.01) and one asterisk (*, 0.01<P < 0.05) represent significant differences between the controls and treatments as determined by Origin 7.5. CK, control.
Expression analysis of AGP-encoding genes in rice and Arabidopsis
Gene expression patterns can provide important clues for the study of gene function. Therefore, a comparative analysis of the expression patterns of the rice and Arabidopsis AGP gene family has been performed. Using microarray and MPSS data, the expression of rice and Arabidopsis AGP-encoding genes was examined in inflorescence, leaf, root, and seed/silique, and under ABA, GA, and abiotic stress treatments (Fig. 10).
Fig. 10.
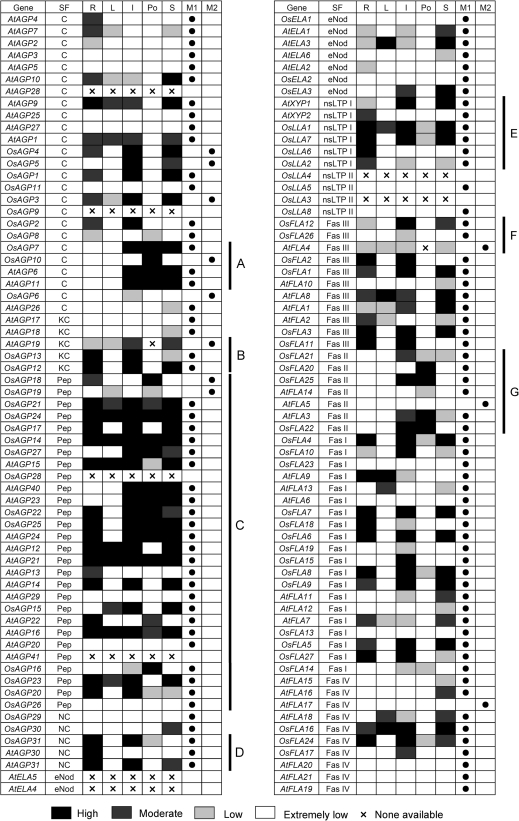
The expression patterns in different tissues for Arabidopsis and rice AGP-encoding genes. AGP-encoding genes are presented in the same order as in the corresponding phylogenetic trees. The expression data of AGP-encoding genes in different tissues were combined from microarrays and MPSS tags. The expression data in pollen of rice AGP-encoding genes were extracted from MPSS tags. Letters on the top indicate different tissues and databases. M1 and M2 represent microarrays and MPSS tags, respectively. The ratios of the absolute values divided by the average of all microarray values were used for analysis (Supplementary Table S5 at JXB online). Light grey, dark grey, and black boxes indicate low (between 0.5 and 1 or the signature numbers between 0 tpm and 50 tpm), moderate (between 1 and 2 or between 50 tpm and 500 tpm), and high (>2 or >500 tpm) expression levels, respectively. The white boxes indicate that no expression could be detected (<0.5). ‘×’ represents no probe or signature on microarray and MPSS. SF, subfamily; R, root; L, leaf; I, inflorescence; Po, pollen; S, silique or seed.
OsAGP7 and OsAGP10 have similar expression patterns in flower, seed, and pollen to AtAGP6 and AtAGP11 which are essential for pollen function (Levitin et al., 2008) (Fig. 10A). The two rice Lys-rich AGPs (OsAGP12 and OsAGP13) have similar expression patterns in root, flower, and seed and a close phylogenetic relationship with AtAGP19 which functions in various aspects of plant growth, including cell division and expansion (Yang et al., 2007) (Fig. 10B).
There are 15 and 13 AG peptide-encoding genes in rice and Arabidopsis, respectively. The expression results of OsAGP28 and AtAGP41 are not available in both the microarray and MPSS, AtAGP20 and OsAGP26 are expressed at an extremely low level in all examined tissues, and most of the remaining AG peptide-encoding genes (24) have high expression levels in root, flower, pollen, and seed (Fig. 10C). It is interesting that 15 of 24 AG peptide-encoding genes are expressed in pollen, OsAGP16 and OsAGP18 are specifically expressed in pollen, and AtAGP23 and AtAGP40 are highly expressed in flower, pollen, and seed (Fig. 10C).
Although the amino acid sequence similarity of non-classical AGPs is relatively low, OsAGP31, AtAGP30, and AtAGP31 are highly expressed in roots (Fig. 10D), and AtAGP30 plays a role in root regeneration (Hengel and Roberts, 2003).
There are two xylogen genes in Arabidopsis, AtXYP1 and AtXYP2 (Motose et al., 2004), and four xylogen-like genes (nsLTP-AGPs) in rice, OsLLA1, OsLLA2, OsLLA6, and OsLLA7. OsLLA1 and OsLLA7 have similar expression patterns in root, flower, and seed to AtXYP1, and OsLLA2 and OsLLA6 have similar expression patterns in root to AtXYP2 (Fig. 10E).
OsFLA12 and OsFLA26 are the rice homologues of AtFLA4 which is also characterized as SOS5 (Shi et al., 2003), OsFLA12 is similarly expressed to AtFLA4, and OsFLA26 is specifically expressed in flower (Fig. 10F).
Interestingly, all of the group II FLAs, except AtFLA5, were expressed in the reproductive processes; OsFLA20, OsFLA22, OsFLA25, AtFLA3, and AtFLA14 were predominantly expressed in flower and pollen (Fig. 10G).
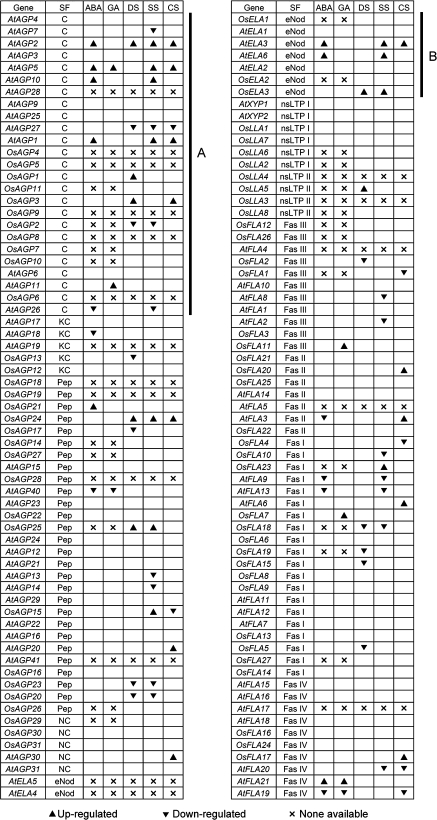
Four Arabidopsis classical AGPs (AtAGP1, AtAGP2, AtAGP5, and AtAGP10) were up-regulated by ABA and various abiotic stresses, and two rice classical AGPs (OsAGP1 and OsAGP3) were up-regulated by drought stress (Fig. 11A). AtELA3 and AtELA6 were up-regulated by ABA and salt stress, and OsELA3 was up-regulated by drought and salt stresses (Fig. 11B). These results indicated that classical AGPs and eNod-like AGPs may play important roles in ABA signalling and abiotic stresses response.
Fig. 11.
The expression patterns under ABA, GA, and abiotic stress treatments for Arabidopsis and rice AGP-encoding genes. AGP-encoding genes are presented in the same order as in the corresponding phylogenetic trees. The expression data of AGP-encoding genes under ABA, GA, drought stress, salt stress, and cold stress treatments were from microarrays. Upright triangles and inverted triangle represent values that are significantly lower (<0.5) and higher (>2) than the control, and ‘×’ represents no probe on microarray. SF, subfamily; DS, drought stress; SS, salt stress; CS, cold stress.
To sum up, the gene expression patterns provide a solid foundation for future functional studies of AGPs in both rice and Arabidopsis.
Discussion
AGP gene family in rice
It is widely accepted that AGPs are a type of P/HRGP with an N-terminal signal sequence and a C-terminal GPI anchor additional signal, can react with β-GlcY, and are recognized by a series of monoclonal antibodies (Yariv et al., 1962; Eisenhaber et al., 2003; Schultz et al., 2002, 2004). A similar number of AGP-encoding genes exist in rice (69 genes) and Arabidopsis (>61 genes, including six putative Arabidopsis eNod-like AGPs), although the rice genome size is ∼3.7 times, and the gene number is ∼1.5 times, more than those of Arabidopsis. Most AGPs identified in the rice genome are classical AGPs >50% PAST, AG peptides >35% PAST, and FLAs that contain one or two fasciclin-like domains. However, only three non-classical AGPs, three ELAs, and eight LLAs are identified together with classical AGPs; other proteins with a lower PAST proportion cannot be identified using the amino acid biased method. Therefore, efforts to identify AGPs with a <50% PAST proportion will be the focus of AGP identification. Moreover, OsELA1 has two putative AG glycomodules (TPPP and APPP) and one probable AG polysaccharide attachment site (APEPA), but not typical AG glycomodules such as XP (X=A, S, T) repeats. The idea that (A/T) PPP could work as AG glycomodules was also validated in other AGPs of Arabidopsis (Schultz et al., 2000). Proteins with putative AG glycomodules could be recognized as AGPs, which broadens the definition of AGPs.
The eNod-like and nsLTP-like AGPs were first grouped as subfamilies of the AGP gene family. It had also been reported that there are at least 248 GPI-anchored proteins in Arabidopsis, 40% of them having putative AG glycomodules, including 10 eNod-like proteins and 11 nsLTP-like proteins (Borner et al., 2003). There is sufficient evidence to show that the eNod-like and nsLTP-like AGPs really existed in both Arabidopsis and rice. In Arabidopsis, a hybrid protein termed a xylogen is homologous to nsLTPs and contains AG glycomodules and a GPI anchor additional signal. Xylogen is considered as an AGP because its molecular weight ranges from 50 kDa to 100 kDa which exceeds its predicted molecular mass of 16 kDa, and it can react with β-Yariv reagent and JIM13 antibody (Motose et al., 2004).
Six AGPs, namely a classical AGP (OsAGP1), an nsLTP-like AGP (OsLTPL1, designated as OsLLA1), an eNod-like AGP (OsENODL1, designated as OsELA1), and three AG peptides (OsAGPEP1, OsAGPEP2, and OsAGPEP3) were identified as β-Yariv reagent-reactive glycopeptides from rice aleurone cells (Mashiguchi et al., 2004). One of them, OsAGPEP3 (designated as OsAGP28), was not identified in the present whole-genome analysis, because it does not exist in either RGAP or RAP-DB, but five of them were obtained in the present study (designated OsAGP1, OsLLA1, OsELA1, OsAGP14, and OsAGP19).
The results of EST abundance, microarray, and MPSS signature analysis revealed that most AGP-encoding genes from the present prediction are expressed, except OsAGP9 which is suggested to be a pseudogene (Table 2). The expression levels of 10 AGP-encoding genes were relatively low in all organs and tissues at various developmental stages examined by microarray (Fig. 6F). The reasons for this are complicated; some of them might be pseudogenes, some might be expressed at relatively lower levels than other family members, and some might be expressed in other organs and tissues or developmental stages that were not selected for microarray analysis, such as stem, etc. The present expression analysis provides evidences that rice AGP-encoding genes with different expression patterns may play different functional roles in distinct developmental stages.
Putative functions of rice AGPs in various developmental processes
The expression of the majority of the AGP-encoding genes was spatially regulated, and many rice AGP-encoding genes were mainly expressed during sexual reproduction, especially in the processes of pollen development. This observation is consistent with the role of AGP-encoding genes in controlling pollen function (Coimbra et al., 2007, 2009; Levitin et al., 2008; Anand and Tyagi, 2009).
The two classical AGP-encoding genes, OsAGP7 and OsAGP10, are highly expressed in pollen, similar to AtAGP6 and AtAGP11 (Fig. 10A). The phylogenetically closest rice gene is OsAGP6, but it has a different expression pattern from that of AtAGP6 and AtAGP11. In this case, it is possible that the genes sharing the same function are not those with higher identity. Therefore, OsAGP7 and OsAGP10 may play a conserved role in pollen development, like AtAGP6 and AtAGP11 which redundantly control pollen development and fertility (Table 4) (Levitin et al., 2008; Coimbra et al., 2009, 2010). The group III FLA genes are mainly expressed in the sexual reproductive processes. In anther, 15 of 24 AG peptide-encoding genes are expressed (Fig. 10C, G), implying their important functions in pollen development. Six AG peptide-encoding genes (AtAGP16, AtAGP21, AtAGP22, AtAGP23, AtAGP24, and AtAGP40) are also found to be expressed in mature pollen by transcriptomic analysis of GPI-anchored protein genes (Lalanne et al., 2004). To sum up, these results suggest that classical AGPs, AG peptides, and group III FLAs may play an important role in rice anther and pollen development.
Table 4.
Comparison between rice AGPs and their Arabidopsis homologues
| Subfamily | Groups | Rice | Arabidopsis | Known function |
| C | Fig. 9A | OsAGP7 OsAGP10 | AtAGP6 AtAGP11 | Pollen development |
| KC | Fig. 9B | AtAGP17 | Agrobacterium infection | |
| AtAGP18 | Macrosporogenesis | |||
| OsAGP12 OsAGP13 | AtAGP19 | Cell division | ||
| NC | Fig. 9D | AtAGP30 | Root regeneration | |
| AtAGP31 | Defence response | |||
| nsLTP I | Fig. 9E | OsLLA1 OsLLA7 | AtXYP1 | Vascular differentiation |
| OsLLA2 OsLLA6 | AtXYP2 | Vascular differentiation | ||
| Fas III | Fig. 9F | OsFLA12 OsFLA26 | AtFLA4 | Cell expansion |
However, the physiological and biochemical processes and molecular mechanism of how AGPs regulated anther and pollen development are still unclear. The complex structure of the sugar chains of AGPs makes them a potential source of small signalling molecules, such as biologically active oligosaccharides. A novel tetrasaccharide that accumulated specifically in rice anthers during microsporogenesis, with a structure similar to a tetrasaccharide unit found in the glycan chain of AGP, played an important role in the development of rice anther (Kawaguchi et al., 1996). It is notable that AGPs might be partially deglycosylated by glycosidases after being glycosylated in endoplasmic reticulum, and the released oligosaccharides might execute the function of AGPs, such as the tetrasaccharide mentioned above. Furthermore, the expression and activity of AGP-specific glycosylhydrolases might therefore provide mechanisms to control the biological activity of AGPs.
The Lys-rich AGP-encoding genes are essential for Arabidopsis: AtAGP17, AtAGP18, and AtAGP19 participate in Agrobacterium infection, macrosporogenesis, and cell division, respectively (Acosta-Garcia and Vielle-Calzada, 2004; Gaspar et al., 2004; Yang et al., 2007). There are two Lys-rich AGP-encoding genes in rice, OsAGP12 and OsAGP13, which are similarly expressed and phylogenetically close to AtAGP19 (Figs 4C, 10B), indicating that OsAGP12 and OsAGP13 may function like AtAGP19 which controls cell division (Table 4). Moreover, the EST expression profile of OsAGP13 shows a specific expression in the SAM, indicating that it may control the cell division of the SAM.
The situation for group I nsLTP-like AGPs is more complicated. Double knockouts of two xylogen-encoding genes (AtXYP1 and AtXYP2) in Arabidopsis show defects of vascular development (Motose et al., 2004). There are two homologues of AtXYP1 in rice, OsLLA1 and OsLLA7, and two homologues of AtXYP2 in rice, OsLLA2 and OsLLA6, respectively (Fig. 5E). OsLLA1, OsLLA7, and AtXYP1 are similarly expressed in root, flower, and seed; OsLLA2, OsLLA6, and AtXYP2 are highly expressed in root. OsLLA1 and OsLLA7 are gene pairs from chromosomal duplication (Fig. 6). The redundancy of group I nsLTP-like AGPs will increase the difficulties in identifying their functions.
OsFLA12 and OsFLA26 are gene pairs from chromosomal duplication; OsFLA12 show similar expression patterns to its Arabidopsis homologue, AtFLA4 (SOS5), but OsFLA26 is expressed in flower. OsFLA12 may be the rice homologue of SOS5, and OsFLA26 may change its function in the course of evolution.
Phytohormone- and abiotic stress-responsive AGP-encoding genes
ABA is an important hormone that regulates many downstream genes via ABREs (ABA-responsive elements), and is involved in response to abiotic stresses throughout the plant kingdom (Busk and Pages, 1998). Several classical AGPs (OsAGP1), AG peptides (OsAGP15), and eNod-like AGPs (OsELA3) are responsive to ABA and drought and salt stresses. The relative expression levels of OsELA3 in young seedlings under drought and salt treatments are >400 and 200 times those of non-treated seedlings, respectively, suggesting that it responds markedly to drought and salt stresses, and OsAGP1 and OsAGP15 were significantly up-regulated by drought and salt stresses (Fig. 8). Moreover, OsAGP1, OsAGP15, and OsELA3 are also up-regulated by ABA, indicating that they may be stress-inducible genes and participate in the ABA signalling pathway (Fig. 9A, B, D). The findings are consistent with those in Arabidopsis, such as the atagp30 mutant which shows suppression of the ABA-induced delay in germination and altered expression of some ABA-regulated genes (van Hengel and Roberts, 2003), and a non-classical AGP gene, AtAGP31, which showed a decreased mRNA level in response to ABA treatment (Liu and Mehdy, 2004). The molecular mechanism by which AGPs are involved in drought and salt responses remains to be elucidated. It is probable that the polysaccharide chains of AGPs which could be deglycosylated by glycosidases might be a source of oligosaccharides, and these oligosaccharides might increase the intracellular osmotic pressure, and reduce the speed of dehydration. The expression level of an AG peptide gene, OsAGP20, was down-regulated by drought and salt stresses, but up-regulated by cold stress. It had been reported that a tetrasaccharide with similar structural characters to the sugar chains of AGP might play an important role in both the development of anther and its response to chilling (Kawaguchi et al., 1996), and AGPs that were induced by cold stresses, such as OsAGP20, might be a potential resource which could be degraded into tetrasaccharides.
It had been previously reported that GA signalling was specifically inhibited by β-GlcY treatment in barley aleurone protoplasts (Mashiguchi et al., 2008). Microarray analysis of β-GlcY-treated aleurone cells revealed that β-GlcY was largely effective in repressing GA-induced gene expression. It is also probable that AGPs are involved in the perception of stimuli causing defence responses. Five genes (AtAGP5, AtAGP11, OsFLA7, OsFLA11, and AtFLA21) were significantly up-regulated by GA (Fig. 11), indicating that GA plays important roles in regulation of the functions of AGPs. CsAGP1, a GA-responsive classical AGP, was isolated from cucumber hypocotyls (Park et al., 2003). However, direct proof of how AGPs work as regulators that control the expression of GA-induced genes is still lacking.
Conclusion
The detailed structural, phylogenetic, and expression analyses in this study provide insights into the functions of AGPs in rice. For example, several AGP-encoding genes displayed specific expression during various stages of panicle and seed development and were induced by ABA, GA, and abiotic stresses, implying their important functions in the processes of reproductive development, ABA and GA signalling pathways, and abiotic stress responses. Moreover, genes of orthologous groups are similarly expressed in rice and Arabidopsis, suggesting conserved roles for AGP-encoding genes between monocots and dicots. In the future, reverse genetics methods such as RNAi and T-DNA insertion mutants could be used for functional analysis of rice AGP-encoding genes. However, gene redundancies should be considered, as several gene pairs with similar expression patterns and a high degree of homology might have overlapping functions. Therefore, specific RNAi constructs that silence two genes at a time could be used and sometimes double and triple mutants should be screened for a notable phenotype. Extensive studies will improve our understanding of the functions of AGPs in relation to various aspects of reproductive development and abiotic stresses in monocots.
Supplementary data
Supplementary data are available at JXB online.
Table S1. Primers used in real-time PCR.
Table S2. Proline, alanine, serine, and threonine (PAST) proportion of rice AGPs.
Table S3. EST expression profile analysis of rice AGP-encoding genes.
Table S4. MPSS analysis of rice AGP-encoding genes in various organs and tissues.
Table S5. Expression analysis of rice and Arabidopsis AGP-encoding genes in various developmental stages and under phytohormone and abiotic stress treatments.
Supplementary Material
Acknowledgments
This work was supported by the National Natural Science Foundation of China (30821064, 30770132), the Chinese 111 Project # B06018, and the Special Doctorial Program Funds of the Ministry of Education of China (20090141110035).
Glossary
Abbreviations
- ABA
abscisic acid
- AGP
arabinogalactan protein
- ELA
early nodulin-like arabinogalactan protein
- EST
expressed sequence tag
- MPSS
massively parallel signature sequencing
- FLA
fasciclin-like AGP
- GA
gibberellic acid
- β-GlcY
β-glucosyl Yariv reagent
- LLA
non-specific lipid transfer protein-like AGP
References
- Albert M, Belastegui-Macadam X, Kaldenhoff R. An attack of the plant parasite Cuscuta reflexa induces the expression of attAGP, an attachment protein of the host tomato. The Plant Journal. 2006;48:548–556. doi: 10.1111/j.1365-313X.2006.02897.x. [DOI] [PubMed] [Google Scholar]
- Acosta-Garcia G, Vielle-Calzada JP. A classical arabinogalactan protein is essential for the initiation of female gametogenesis in. Arabidopsis. The Plant Cell. 2004;16:2614–2628. doi: 10.1105/tpc.104.024588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anand S, Tyagi AK. Characterization of a pollen-preferential gene OSIAGP from rice (Oryza sativa L. subspecies indica) coding for an arabinogalactan protein homologue, and analysis of its promoter activity during pollen development and pollen tube growth. Transgenic Research. 2009 doi: 10.1007/s11248-009-9319-3. (in press) [DOI] [PubMed] [Google Scholar]
- Borner GH, Lilley KS, Stevens TJ, Dupree P. Identification of glycosylphosphatidylinositol-anchored proteins in arabidopsis. A proteomic and genomic analysis. Plant Physiology. 2003;132:568–577. doi: 10.1104/pp.103.021170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brenner S, Johnson M, Bridgham J, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nature Biotechnology. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- Busk PK, Pagès M. Regulation of abscisic acid-induced transcription. Plant Molecular Biology. 1998;37:425–435. doi: 10.1023/a:1006058700720. [DOI] [PubMed] [Google Scholar]
- Coimbra S, Almeida J, Junqueira V, Costa ML, Pereira LG. Arabinogalactan proteins as molecular markers in Arabidopsis thaliana sexual reproduction. Journal of Experimental Botany. 2007;58:4027–4035. doi: 10.1093/jxb/erm259. [DOI] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Jones B, Mendes MA, Pereira LG. Pollen grain development is compromised in Arabidopsis agp6 agp11 null mutants. Journal of Experimental Botany. 2009;60:3133–3142. doi: 10.1093/jxb/erp148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coimbra S, Costa M, Mendes MA, Pereira AM, Pinto J, Pereira LG. Early germination of Arabidopsis pollen in a double null mutant for the arabinogalactan protein genes AGP6 and AGP11. Sexual Plant Reproduction. 2010 doi: 10.1007/s00497-010-0136-x. (in press) [DOI] [PubMed] [Google Scholar]
- Du H, Clarke AE, Bacic A. Arabinogalactan-proteins: a class of extracellular matrix proteoglycans involved in plant growth and development. Trends in Cell Biology. 1996;6:411–414. doi: 10.1016/s0962-8924(96)20036-4. [DOI] [PubMed] [Google Scholar]
- Eisenhaber B, Wildrpaner M, Schultz CJ, Borner GH, Dupree P, Eisenhaber F. Glycosylphosphatidylinositol lipid anchoring of plant proteins. Sensitive prediction from sequence- and genome-wide studies for Arabidopsis and rice. Plant Physiology. 2003;133:1691–1701. doi: 10.1104/pp.103.023580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proceedings of the National Academy of Sciences, USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faik A, Abouzouhair J, Sarhan F. Putative fasciclin-like arabinogalactan-proteins (FLA) in wheat (Triticum aestivum) and rice (Oryza sativa): identification and bioinformatic analysis. Molecular Genetics and Genomics. 2006;276:478–494. doi: 10.1007/s00438-006-0159-z. [DOI] [PubMed] [Google Scholar]
- Gao M, Showalter AM. Yariv reagent treatment induces programmed cell death in Arabidopsis cell cultures and implicates arabinogalactan protein involvement. The Plant Journal. 1999;19:321–331. doi: 10.1046/j.1365-313x.1999.00544.x. [DOI] [PubMed] [Google Scholar]
- Gao M, Showalter AM. Immunolocalization of LeAGP-1, a modular arabinogalactan-protein, reveals its developmentally regulated expression in tomato. Planta. 2000;210:865–874. doi: 10.1007/s004250050691. [DOI] [PubMed] [Google Scholar]
- Gaspar Y, Johnson KL, McKenna JA, Bacic A, Schultz CJ. The complex structures of arabinogalactan-proteins and the journey towards understanding function. Plant Molecular Biology. 2001;47:161–176. [PubMed] [Google Scholar]
- Gaspar Y, Nam J, Schultz CJ, Lee LY, Gilson PR. Characterization of the Arabidopsis lysine-rich arabinogalactan-protein AtAGP17 mutant (rat1) that results in a decreased efficiency of agrobacterium transformation. Plant Physiology. 2004;135:2162–2171. doi: 10.1104/pp.104.045542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodrum LJ, Patel A, Leykam JF, Kieliszewski MJ. Gum Arabic glycoprotein contains glycomodules of both extensin and arabinogalactan glycoproteins. Phytochemistry. 2000;54:99–106. doi: 10.1016/s0031-9422(00)00043-1. [DOI] [PubMed] [Google Scholar]
- Hu Y, Qin Y, Zhao J. Localization of an arabinogalactan protein epitope and the effects of Yariv phenylglycoside during zygotic embryo development of Arabidopsis thaliana. Protoplasma. 2006;229:21–31. doi: 10.1007/s00709-006-0185-z. [DOI] [PubMed] [Google Scholar]
- International Rice Genome Sequencing Project. The map-based sequence of the rice genome. Nature. 2005;436:793–800. doi: 10.1038/nature03895. [DOI] [PubMed] [Google Scholar]
- Itoh J, Nonomura K, Ikeda K, Yamaki S, Inukai Y, Yamagishi H, Kitano H, Nagato Y. Rice plant development: from zygote to spikelet. Plant and Cell Physiology. 2005;46:23–47. doi: 10.1093/pcp/pci501. [DOI] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Arora R, Agarwal P, Ray S, Sharma P, Kapoor S, Tyagi AK, Khurana JP. F-box proteins in rice. Genome-wide analysis, classification, temporal and spatial gene expression during panicle and seed development, and regulation by light and abiotic stress. Plant Physiology. 2007;143:1467–1483. doi: 10.1104/pp.106.091900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain M, Nijhawan A, Tyagi AK, Khurana JP. Validation of housekeeping genes as internal control for studying gene expression in rice by quantitative real-time PCR. Biochemical and Biophysical Research Communications. 2006;345:646–651. doi: 10.1016/j.bbrc.2006.04.140. [DOI] [PubMed] [Google Scholar]
- Johnson KL, Jones BJ, Bacic A, Schultz CJ. The fasciclin-like arabinogalactan proteins of Arabidopsis. A multigene family of putative cell adhesion molecules. Plant Physiology. 2003;133:1911–1925. doi: 10.1104/pp.103.031237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawaguchi K, Shibuya N, Ishii T. A novel tetrasaccharide, with a structure similar to the terminal sequence of an arabinogalactan-protein, accumulates in rice anthers in a stage-specific manner. The Plant Journal. 1996;9:777–785. doi: 10.1046/j.1365-313x.1996.9060777.x. [DOI] [PubMed] [Google Scholar]
- Lalanne E, Honys D, Johnson A, Borner GH, Lilley KS, Dupree P, Grossniklaus U, Twell D. SETH1 and SETH2, two components of the glycosylphosphatidylinositol anchor biosynthetic pathway, are required for pollen germination and tube growth in Arabidopsis. The Plant Cell. 2004;16:229–240. doi: 10.1105/tpc.014407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langan KJ, Nothnagel EA. Cell surface arabinogalactan-proteins and their relation to cell proliferation and viability. Protoplasma. 1997;196:87–98. [Google Scholar]
- Lee KJD, Sakata Y, MauSL Pettolino F, Bacic A. Arabinogalactan proteins are required for apical cell extension in the moss Physcomitrella patens. The Plant Cell. 2005;17:3051–3065. doi: 10.1105/tpc.105.034413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitin B, Richter D, Markovich I, Zik M. Arabinogalactan proteins 6 and 11 are required for stamen and pollen function in Arabidopsis. The Plant Journal. 2008;56:351–363. doi: 10.1111/j.1365-313X.2008.03607.x. [DOI] [PubMed] [Google Scholar]
- Liu C, Mehdy MC. A nonclassical arabinogalactan protein gene highly expressed in vascular tissues, AGP31, is transcriptionally repressed by methyl jasmonic acid in Arabidopsis. Plant Physiology. 2007;145:863–874. doi: 10.1104/pp.107.102657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mashiguchi K, Asami T, Suzuki Y. Genome-wide identification, structure and expression studies, and mutant collection of 22 early nodulin-like protein genes in Arabidopsis. Bioscience, Biotechnology, and Biochemistry. 2009;73:2452–2459. doi: 10.1271/bbb.90407. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Urakami E, Hasegawa M, Sanmiya K, Matsumoto I, Yamaguchi I, Asami T, Suzuki Y. Defense-related signaling by interaction of arabinogalactan proteins and β-glucosyl Yariv reagent inhibits gibberellin signaling in barley aleurone cells. Plant and Cell Physiology. 2008;49:178–190. doi: 10.1093/pcp/pcm175. [DOI] [PubMed] [Google Scholar]
- Mashiguchi K, Yamaguchi I, Suzuki Y. Isolation and identification of glycosylphosphatidylinositol-anchored arabinogalactan proteins and novel β-glucosyl Yariv-reactive proteins from seeds of rice (Oryza sativa) Plant and Cell Physiology. 2004;45:1817–1829. doi: 10.1093/pcp/pch208. [DOI] [PubMed] [Google Scholar]
- Motose H, Fukuda H, Sugiyama M. Involvement of local intercellular communication in the differentiation of zinnia mesophyll cells into tracheary elements. Planta. 2001a;213:121–131. doi: 10.1007/s004250000482. [DOI] [PubMed] [Google Scholar]
- Motose H, Sugiyama M, Fukuda H. An arabinogalactan protein(s) is a key component of a fraction that mediates local intercellular communication involved in tracheary element differentiation of zinnia mesophyll cells. Plant and Cell Physiology. 2001b;42:129–137. doi: 10.1093/pcp/pce014. [DOI] [PubMed] [Google Scholar]
- Motose H, Sugiyama M, Fukuda H. A proteoglycan mediates inductive interaction during plant vascular development. Nature. 2004;429:873–878. doi: 10.1038/nature02613. [DOI] [PubMed] [Google Scholar]
- Nam J, Mysore KS, Zheng C, Knue MK, Matthysse AG, Gelvin SB. Identification of T-DNA tagged Arabidopsis mutants that are resistant to transformation by agrobacterium. Molecular and General Genetics. 1999;261:429–438. doi: 10.1007/s004380050985. [DOI] [PubMed] [Google Scholar]
- Nielsen H, Engelbrecht J, Brunak S, von Heijne G. Identification of prokaryotic and eukaryotic signal peptides and prediction of their cleavage sites. Protein Engineering. 1997;10:1–6. doi: 10.1093/protein/10.1.1. [DOI] [PubMed] [Google Scholar]
- Nobuta K, Venu RC, Lu C, et al. An expression atlas of rice mRNAs and small RNAs. Nature Biotechnology. 2007;25:473–477. doi: 10.1038/nbt1291. [DOI] [PubMed] [Google Scholar]
- Park MH, Suzuki Y, Chono M, Knox JP, Yamaguchi I. CsAGP1, a gibberellin responsive gene from cucumber hypocotyls, encodes a classical arabinogalactan protein and is involved in stem elongation. Plant Physiology. 2003;131:1450–1459. doi: 10.1104/pp.015628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qin Y, Chen D, Zhao J. Localization of arabinogalactan proteins in anther, pollen, and pollen tube of Nicotiana tabacum L. Protoplasma. 2007;231:43–53. doi: 10.1007/s00709-007-0245-z. [DOI] [PubMed] [Google Scholar]
- Qin Y, Zhao J. Localization of arabinogalactan proteins in egg cells, zygotes, and two-celled proembryos and effects of β-d-glucosyl Yariv reagent on egg cell fertilization and zygote division in Nicotiana tabacum L. Journal of Experimental Botany. 2006;57:2061–2074. doi: 10.1093/jxb/erj159. [DOI] [PubMed] [Google Scholar]
- Qin Y, Zhao J. Localization of arabinogalactan-proteins in different stages of embryos and their role in cotyledon formation of Nicotiana tabacum L. Sexual Plant Reproduction. 2007;20:213–224. [Google Scholar]
- Schultz CJ, Johnson KL, Currie G, Bacic A. The classical arabinogalactan protein gene family of Arabidopsis. The Plant Cell. 2000;12:1751–1768. doi: 10.1105/tpc.12.9.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Rumsewicz MP, Johnson KL, Jones BJ, Gaspar YM, Bacic A. Using genomic resources to guide research directions. The arabinogalactan protein gene family as a test case. Plant Physiology. 2002;129:1448–1463. doi: 10.1104/pp.003459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz CJ, Ferguson KL, Lahnstein J, Bacic A. Post-translational modifications of arabinogalactan-peptides of Arabidopsis thaliana. Endoplasmic reticulum and glycosylphosphatidylinositol-anchor signal cleavage sites and hydroxylation of proline. Journal of Biological Chemistry. 2004;279:45503–45511. doi: 10.1074/jbc.M407594200. [DOI] [PubMed] [Google Scholar]
- Serpe MD, Nothnagel EA. Effects of Yariv phenylglycosides on rose cell-suspensions—evidence for the involvement of arabinogalactan-proteins in cell proliferation. Planta. 1994;193:542–550. [Google Scholar]
- Shi H, Kim Y, Guo Y, Stevenson B, Zhu JK. The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. The Plant Cell. 2003;15:19–32. doi: 10.1105/tpc.007872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM. Structure and function of plant-cell wall proteins. The Plant Cell. 1993;5:9–23. doi: 10.1105/tpc.5.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Showalter AM. Arabinogalactan-proteins: structure, expression and function. Cellular and Molecular Life Sciences. 2001;58:1399–1417. doi: 10.1007/PL00000784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shpak E, Barbar E, Leykam JF, Kieliszewski MJ. Contiguous hydroxyproline residues direct hydroxyproline arabinosylation in Nicotiana tabacum. Journal of Biological Chemistry. 2001;276:11272–11278. doi: 10.1074/jbc.M011323200. [DOI] [PubMed] [Google Scholar]
- Shpak E, Leykam JF, Kieliszewski MJ. Synthetic genes for glycoprotein design and the elucidation of hydroxyproline-O-glycosylation codes. Proceedings of the National Academy of Sciences, USA. 1999;96:14736–14741. doi: 10.1073/pnas.96.26.14736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun W, Kieliszewski MJ, Showalter AM. Overexpression of tomato LeAGP-1 arabinogalactan-protein promotes lateral branching and hampers reproductive development. The Plant Journal. 2004;40:870–881. doi: 10.1111/j.1365-313X.2004.02274.x. [DOI] [PubMed] [Google Scholar]
- Sun W, Xu J, Yang J, Kieliszewski MJ, Showalter AM. The lysine-rich arabinogalactan-protein subfamily in Arabidopsis: gene expression, glycoprotein purification and biochemical characterization. Plant and Cell Physiology. 2005;46:975–984. doi: 10.1093/pcp/pci106. [DOI] [PubMed] [Google Scholar]
- Tamura K, Dudley J, Nei M, Kumar S. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Molecular Biology and Evolution. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. Fucosylated arabinogalactan-proteins are required for full root cell elongation in Arabidopsis. The Plant Journal. 2002;32:105–113. doi: 10.1046/j.1365-313x.2002.01406.x. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, Roberts K. AtAGP30, an arabinogalactan-protein in the cell walls of the primary root, plays a role in root regeneration and seed germination. The Plant Journal. 2003;36:256–270. doi: 10.1046/j.1365-313x.2003.01874.x. [DOI] [PubMed] [Google Scholar]
- van Hengel AJ, van Kammen A, de Vries SC. A relationship between seed development, arabinogalactan-proteins (AGPs) and the AGP mediated promotion of somatic embryogenesis. Physiologia Plantarum. 2002;114:637–644. doi: 10.1034/j.1399-3054.2002.1140418.x. [DOI] [PubMed] [Google Scholar]
- Willats WG, Knox JP. A role for arabinogalactan-proteins in plant cell expansion: evidence from studies on the interaction of β-glucosyl Yariv reagent with seedlings of Arabidopsis thaliana. The Plant Journal. 1996;9:919–925. doi: 10.1046/j.1365-313x.1996.9060919.x. [DOI] [PubMed] [Google Scholar]
- Wu HM, Wang H, Cheung AY. A pollen tube growth stimulatory glycoprotein is deglycosylated by pollen tubes and displays a glycosylation gradient in the flower. Cell. 1995;82:395–403. doi: 10.1016/0092-8674(95)90428-x. [DOI] [PubMed] [Google Scholar]
- Yang J, Sardar HS, McGovern KR, Zhang Y, Showalter AM. A lysine-rich arabinogalactan protein in Arabidopsis is essential for plant growth and development, including cell division and expansion. The Plant Journal. 2007;49:629–640. doi: 10.1111/j.1365-313X.2006.02985.x. [DOI] [PubMed] [Google Scholar]
- Yariv J, Rapport MM, Graf L. The interaction of glycosides and saccharides with antibody to the corresponding phenylazo glycosides. Biochemistry Journal. 1962;85:383–388. doi: 10.1042/bj0850383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yuan J, Chen D, Ren Y, Zhang X, Zhao J. Characteristic and expression analysis of a metallothionein gene, OsMT2b, down-regulated by cytokinin suggests functions in root development and seed embryo germination of rice. Plant Physiology. 2008;146:1637–1650. doi: 10.1104/pp.107.110304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Brown G, Whetten R, Loopstra CA, Neale D. An arabinogalactan protein associated with secondary cell wall formation in differentiating xylem of loblolly pine. Plant Molecular Biology. 2003;52:91–102. doi: 10.1023/a:1023978210001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.