Abstract
Male-sterile plants are used in hybrid breeding as well as for gene confinement for genetically modified plants in field trials and agricultural production. Apart from naturally occurring mutations leading to male sterility, biotechnology has added new possibilities for obtaining male-sterile plants, although so far only one system is used in practical breeding due to limitations in propagating male-sterile plants without segregations in the next generation or insufficient restoration of fertility when fruits or seeds are to be harvested from the hybrid varieties. Here a novel mechanism of restoration for male sterility is presented that has been achieved by interference with extracellular invertase activity, which is normally specifically expressed in the anthers to supply the developing microspores with carbohydrates. Microspores are symplastically isolated in the locular space of the anthers, and thus an unloading pathway of assimilates via the apoplasmic space is mandatory for proper development of pollen. Antisense repression of the anther-specific cell wall invertase or interference with invertase activity by expressing a proteinacious inhibitor under the control of the anther-specific invertase promoter results in a block during early stages of pollen development, thus causing male sterility without having any pleiotropic effects. Restoration of fertility was successfully achieved by substituting the down-regulated endogenous plant invertase activity by a yeast invertase fused to the N-terminal portion of potato-derived vacuolar protein proteinase II (PiII–ScSuc2), under control of the orthologous anther-specific invertase promoter Nin88 from tobacco. The chimeric fusion PiII–ScSuc2 is known to be N-glycosylated and efficiently secreted from plant cells, leading to its apoplastic location. Furthermore, the Nin88::PiII-ScSuc2 fusion does not show effects on pollen development in the wild-type background. Thus, such plants can be used as paternal parents of a hybrid variety, thereby the introgression of Nin88::PiII-ScSuc2 to the hybrid is obtained and fertility is restored. In order to broaden the applicability of this male sterility/restoration system to other plant species, a phylogenic analysis of plant invertases(β-fructofuranosidases) and related genes of different species was carried out. This reveals a specific clustering of the cell wall invertases with anther-specific expression for dicotyl species and another cluster for monocotyl plants. Thus, in both groups of plants, there seems to be a kind of co-evolution, but no recent common ancestor of these members of the gene family. These findings provide a helpful orientation to classify corresponding candidate genes in further plant species, in addition to the species analysed so far (Arabidopsis, tobacco, tomato, potato, carrots, rice, and wheat).
Keywords: Genetic engineering, hybrid breeding, invertase, male sterility, restoration
Introduction
Among breeding strategies, F1 hybrids have several advantages over open-pollinated varieties. F1 hybrids are obtained by crossing parental inbred lines, and their special importance is due to the uniformity that is based on the resulting genetic homogeneity as well as on higher yields through heterosis or ‘hybrid vigour’, due to their heterozygous nature (Brewbaker, 1964; Feistritzer and Kelly, 1987). The heterozygous nature of the hybrids requires seeds to be obtained from breeding companies.
A further important application for male sterility systems, apart from hybrid breeding, is their use for gene confinement for the increasing number of genetically modified crop plants used in field trials and agricultural production (Daniell, 2002). Although the risk of outcrossing to wild species depends on the mode of pollination (self-pollination or outcrossing by wind or insects) as well as the presence of related species in the surrounding ecosystem, the availability and use of such biological safety precautions will help to cope with public awareness and fears regarding the potential spread of transgenes, and concerns about the environmental impact of genetically modified plants (GMPs). Nowadays, the most widespread are ‘first-generation’ GMPs such as herbicide-tolerant soybean and rapeseed, and insect-tolerant corn and cotton. Perhaps the public awareness and fears of potential risks of GMPs arise from these modifications, since they are associated with toxins and the commercial interests of a few companies. The so-called second generation of GMPs is that of plants in which the quality of the product has been changed and improved; examples are rice with increased provitamin A levels, rapeseed varieties with modified fatty acid composition, or lysine-rich soybean. The ‘third generation’ of GMPs producing pharmaceutical or industrial proteins or vaccines is under development.
Naturally occurring genetic systems such as sex inheritance, self-incompatibility, and, in the first instance, cytoplasmic male sterility (CMS), have been used for a long time in practical breeding strategies. In the case of CMS, the maternal transmission of sterility-inducing cytoplasm, specified by mitochondrial mutations, in combination with Mendelian nuclear genes, permits the efficient control of pollination. However, CMS is not available in all crops and, if available, the requirement to maintain three lines (male-sterile, maintainer, and restorer) as well as the transfer of these traits to locally adapted varieties is a time-consuming process. Problems may also arise due to unstable male-sterile lines resulting in the pollution of the hybrid with sibs, or instability of the restorer, with the consequence of decreased pollination and thus decreased fruit or seed setting (reviewed in Engelke et al., 2004a, b). Biotechnology has added new possibilities of obtaining male-sterile plants. Various successful approaches to engineer male sterility have been described, although only a single system is ready to be used in agriculture and indeed is already in practical use (reviewed in Roitsch and Engelke 2006).
Developing microspores are symplastically isolated in the locular space of the anthers, and thus an unloading pathway of assimilates via the apoplasmic space is mandatory for proper development of pollen. Therefore, sucrose, the most ubiquitous transport sugar, is released from the sieve elements of the phloem into the apoplast via a sucrose transporter, where irreversible hydrolysis occurs by an extracellular invertase, which is ionically bound to the cell wall (Roitsch and González, 2004). Uptake of the hexose monomers (glucose and, with a lower preference, fructose) into the sink cell is realized by high-affinity hexose transporters. The importance of cell wall-bound invertases (cwINVs) during this process was demonstrated in previous studies involving model plant species (Goetz et al., 2001; Proels et al., 2006; Hirsche et al., 2009). In the plants investigated so far, Nicotiana tabacum, Solanum lycopersicum, and Arabidopsis thaliana, repression of cwINV activity (Nin88, Lin7, and AtcwINV2, respectively) by anther-specific RNA interference turned out to be an efficient method to circumvent carbohydrate supply of the symplastically isolated pollen with a subsequent strong decrease of pollen germination ability and seed setting. Comparable results were also obtained by expressing a proteinaceous invertase inhibitor. The specific involvement of invertases during anther development was also suggested for other plant species, for example Lilium longiflorum (Clément et al., 1996; Ranwala and Miller, 1998; Hsu et al., 2007), potato (Maddison et al., 1999), and carrot (Lorenz et al., 1995), indicating the crucial function of cwINVs in providing carbohydrates for male gametophyte development. In wheat, an arrest in pollen development because of water deficiency correlated with alterations in carbohydrate metabolism, a drastic decrease in invertase activity, and subsequently the failure to accumulate starch (Saini et al., 1984; Dorion et al., 1996; Lalonde et al., 1997). Comparable results were found for rice under cold stress (Oliver et al., 2005). These findings demonstrate the highly specific effects leading to male sterility in different dicotyledenous and monocotyledenous plants, and support the contention that generating male-sterile plants by anther-specific interference with cwINV activity should be generally applicable to different plant species.
The question arises of whether the cwINVs with anther-specific expression are closely related to each other, or if there is some kind of co-evolution in different plant species. To answer this question, the number of known sequences coding for acid invertases in N. tabacum was broadened and an alignment was carried out of the anther-specific invertases of different species with other members of the gene family (β-fructofuranosidases EC 3.2.1.26) found in the cell wall (cwINV), the vacuole (vacINV), or the cytoplasm (cytINV), as well as with related genes coding for fructan-building enzymes (FBEs) and fructan exohydrolases (FEHs). The consequences of the phylogenic relationships for future applications of the described transgenic approach to produce male sterility in other species are discussed.
As a pre-condition for practical application of the described male sterility in hybrid breeding or as a biological safety precaution, the system has to be extended to include a mechanism that allows the multiplication of male-sterile plants without segregation in the offspring (maintenance) as well as for a restorer mechanism when seeds or fruits are to be harvested from the crops. To achieve these goals, a strategy is devised in the present study: replacement of the down-regulated natural plant invertase activity by expressing the distantly related isoenzyme Suc2 of yeast (Saccharomyces cerevisiae), which was not expected to interfere with the plant antisense invertase or with the plant invertase inhibitor due to low sequence homology.
Materials and methods
Male-sterile tobacco plants, generated by expressing an antisense construct of anther-specific cell wall invertase (Nin88::Nin88-antisense) or a proteinacious invertase inhibitor (Nin88::NtCIF)
Two male-sterile plants expressing the antisense construct Nin88::Nin88-antisense (102K and 102KK) as well as one plant expressing the proteinacious inhibitor Nin88::NtCIF (105L) were selected in the ‘SNN’ background. As has been previously described, these plants are characterized by strongly reduced total pollen amount, pollen vitality, and germination efficiency, as well as their inability to produce seeds by self-pollination (Hirsche et al., 2009).
Construction of anther-specific promoter–yeast invertase fusions (Nin88::ScSuc2 and Nin88::PiII-ScSuc2) and transformation into tobacco
The constructs were derived from the previously described reporter construct originally used for identification of tobacco Nin88 promoter activity (Nin88 fused to the uidA gene in the pBI101 binary vector; Goetz et al., 2001) by replacing the uidA gene with the cDNA of the yeast (S. cerevisiae) invertase Suc2.
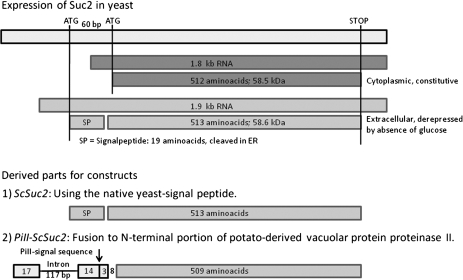
In yeast, the invertase gene Suc2 encodes two differently regulated mRNAs (1.8 kb and 1.9 kb) that differ at their 5′ ends. The smaller RNA encodes an intracellular form that is constitutively expressed at low levels in yeast. The larger RNA contains a signal peptide-coding sequence leading to a secreted form of invertase (Fig. 1) (Carlson et al., 1983). Two different constructs have been made under control of the anther-specific invertase promoter of tobacco, Nin88, using either Suc2 with its native yeast signal peptide (Nin88::ScSuc2) or a shorter part of Suc2 fused to the N-terminal portion of potato-derived vacuolar protein proteinase II (Nin88::PiII-ScSuc2; Fig. 1). This latter chimeric fusion is known to be N-glycosylated and efficiently secreted from plant cells, leading to its apoplastic location (von Schaewen et al., 1990). These constructs were used for Agrobacterium tumefaciens strain LBA4404-mediated transformation of tobacco leave discs (N. tabacum cv. Samsun) by using standard procedures (Horsch et al., 1985). Transformed plants were selected on Murashige–Skoog (MS) medium containing 2% sucrose, 0.3% gelrite, and 100 mg l−1 kanamycin. In a first approach both constructs have been tested in the wild-type tobacco background, in order to see the effects on pollen germination efficiencies.
Fig. 1.
Scheme showing the expression of yeast (Saccharomyces cerevisiae) invertase Suc2 (Carlson et al., 1983, with modifications) and the derived parts of the constructs used for supertransformation of male-sterile plants (ScSuc2 with the native signal sequence leading to extracellular location in yeast; PiII-ScSuc2, that is secreted from the plant cells leading to its apoplastic location, according to PI-3-INV in von Schaewen et al., 1990).
Further investigations focused on the PiII–ScSuc2 sequence, fused to the Nin88 promoter to obtain stable restoration of fertility in the formerly male-sterile plants. Supertransformation of the kanamycin-resistant male-sterile plants expressing Nin88::Nin88-antisense (102K and 102KK) or Nin88::NtCIF (105L) was achieved by transferring the cassette Nin88::PiII-Suc2-nosTerm to pCambia1300, a vector that allows selection on hygromycin (50 mg l−1 in MS as described above).
Plants were grown in the greenhouse at 22 °C in the light and 18 °C in the dark with 12 h light under additional illumination. Transformation was confirmed by PCR, amplifying parts of the specific constructs.
In vitro germination tests of tobacco pollen
The germination of pollen was tested on sitting drops of medium according to Shivanna and Sawhney (1995) containing 290 mM sucrose, 1.5 mM calcium nitrate, 1.6 mM boric acid, 1 mM potassium nitrate, 0.8 mM magnesium sulphate, and 15% (w/v) PEG4000, solidified to sitting drops with 1.5% (w/v) low melt agarose. Pollen germination rates reflect the mean of three independent flowers with at least 100 pollen each.
In situ staining of invertase activity in tobacco pollen
Anthers were collected, frozen in liquid nitrogen, and stored at –80 °C. Before staining, 500 μl of buffer (200 mM HEPES, 3 mM MgCl2, 1 mM EDTA, and 2% glycerin) were added to the anthers, giving a pollen suspension from which anthers were removed by using a pipette tip. Pollen were centrifuged for 5 min at 8000 g, and the pellet resuspended in 500 μl of buffer. This washing step was repeated to remove sugars which might give false-positive results. Washed pollen was resuspended in 40 μl of buffer and each sample was separated into two incubation tubes. Invertase activity was determined according to a modified protocol of Doehlert and Felker (1987). The incubation medium contained 70 mM K2HPO4+40 mM citrate, resulting in pH 4.5, 20 U ml−1 glucose oxidase (GOD; Boehringer), 0.014% phenazine methosulphate (PMS), 0.024% nitroblue tetrazolium salt (NBT), and 0.5% sucrose. A 1 ml aliquot of this reaction mixture was added to pollen and incubated for 20 h at 26 °C in the dark with shaking (600 rpm). In the control reactions sucrose was omitted. After centrifugation for 5 min at 8000 g, the incubation medium was replaced with 500 μl of 70% EtOH. For light microscopic investigation of the staining, pollen was centrifuged again and resuspended in 100 μl of water. Photographs were taken with 100-fold magnification.
Partial cloning of additional tobacco invertases
In order to broaden knowledge of the invertase gene family of tobacco, parts of exon 3 of invertases from N. tabacum were amplified from genomic DNA or cDNA from anthers using degenerate primers that have been designed by alignment of different known invertase sequences. Primers oin3 (CCTTCACYTNTTYTAYCARTAYAAYCC) and oin4 (CCTTTCRWARAARGTYTTDGWWGCGTA) amplified PCR products of ∼750 bp in size (Roitsch et al., 1995). The PCR products were ligated in pGEM-T Easy vector and transformed in Escherichia coli DH5a, according to the instructions of the supplier (Promega, Madison, WI, USA). Due to the degenerate nature of the primers used for amplification, a mixture of different invertases was expected among the transformed cells. For that reason, single colonies were picked, and plasmids were isolated and characterized by test restriction with enzymes having 4 bp recognition sites (BsuRI, HpaII, and TaqI; MBI Fermentas, St Leon-Rot, Germany). Plasmids with different restriction patterns were sequenced. Putative invertase sequences of N. tabacum were identified using the Blast N program (NCBI; Altschul et al., 1997) and aligned with the reported coding sequences (CDS) from other species, using the Clustal W program (Higgens et al., 1994).
Phylogenic tree construction of anther-specific invertases and related enzymes of different monocotyledenous and dicotyledenous plants
The phylogram is a branching diagram (tree) assumed to be an estimate of a phylogeny; branch lengths are proportional to the amount of inferred evolutionary change (EMBL; Larkin et al., 2007). After the abbreviation for the coding sequences, the pI value of the corresponding protein is given, calculated with Protein Calculator v3.3 (www.scripps.edu/∼cdputnam/protcalc.html). The complete invertase gene families (acid and cytoplasmic) from A. thaliana and Oryza sativa are the backbone of the phylogenic analyses, extended with already described and newly isolated sequences of the acid invertases from tobacco, as well as related sequences from the well-studied fructan-accumulating species Cichorium intybus and Triticum aestivum (with FBE and FEH genes). CwINVs with known anther-specific expression from S. lycopersicum, Solanum tuberosum, and Daucus carota are added. The accession numbers of the sequences used to construct the phlogenetic tree are given below
Non-fructan plants:
Cell wall invertases. A. thaliana: AtcwINV1–AtcwINV6 (Sherson et al., 2003), At3g13790, At3g52600, At1g55120, At2g36190, At3g13784, At5g11920. O. sativa: OsCIN1–OsCIN8 (Cho et al., 2005), AY578158–65 [corresponding to OsCIN1–OsCIN3 and OsCIN5–OsCIN9 in Ji et al. (2005) since an additional pseudogene is integrated as OsCIN4 in Ji et al. (2005)]; AY575551 (Os03g0735800) referred to as OsCIN4(Ji)_pseudo. N. tabacum: Ntβfruc1 (NtcwINV1) (Greiner et al., 1995), X81834; NtNin88 (NtcwINV2) (Goetz et al., 2001), AF376773; and newly isolated NtcwINV3, HM022265; NtcwINV4, HM022266; NtcwINV5-FEH?, HM022267; NtcwINV6-FEH?, HM022268. S. lycopersicum: SlLin7 (Godt and Roitsch, 1997; Proels et al., 2003, 2006), AY173050. S. tuberosum: StInvGF (Maddison et al., 1999), AJ133765. D. carota: DcInvDc2 (Lorenz et al., 1995), X78424.
Vacuolar invertases. A. thaliana: Atβfruct3–Atβfruct4 (Tymowska-Lalanne and Kreis, 1998), At1g62660, At1g12240. O. sativa: OsVIN1–OsVIN2 (Ji et al., 2005), AF276703, AF276704). N. tabacum: NtVI (NtvacINV1) (Lauer, 2006), AJ305044, and newly isolated NtvacINV2, HM022269.
Cytoplasmic (alkaline/neutral) invertases. A. thaliana: At-A/N-InvA–At-A/N-InvI (Vargas et al., 2003), At1g56560, At4g34860, At3g06500, At1g22650, At5g22510, At1g72000, At1g35580, At3g05820, At4g09510. O. sativa: OsNIN1–OsNIN8 (Ji et al., 2005), NM_001056471, NM_001049466, NM_001053547, NM_001059261, AP005311, NM_001072410, NM_001059365, NM_001053630.
Fructan-accumulating plants:
Cell wall invertases. T. aestivum: TaIVR1 and TaIVR3 (Minhas and Saini, 1998; Koonjul et al., 2004), AF030420, AF030421.
Vacuolar invertases. C. intybus: CivacINV (van den Ende et al., 2002), AJ419971. T. aestivum, TaVIN2 (L Schroeven, personal communication), AJ635225, TaVIN3 (=TaIVR5, Koonjul et al., 2004), AF069309.
Cytoplasmic (alkaline/neutral) invertases. T. aestivum: TaAInv (Vargas et al., 2007), AM295169.
Fructan-building enzymes (FBEs). C. intybus: Ci1-FFT (JP Goblet, L Canon, and Van PJ Cutsem, personal communication), U84398; Ci1-SST (de Halleux and van Cutsem, 1997), U81520. T. aestivum: Ta1-SST and Ta6-FST (Kawakami and Yoshida, 2002), AB029888, AB029887; Ta1-FFT (Kawakami and Yoshida, 2005), B088409.
Fructan exohydrolases (FEHs). C. intybus: Ci1-FEH-1 (van den Ende et al., 2000), AJ242538; Ci1-FEH-IIa and Ci1-FEH-IIb (van den Ende et al., 2001), AJ295033, AJ295034. T. aestivum: Ta1-FEHw1–Ta1-FEHw3 (Zhang et al., 2008), FJ184989, FJ184991, FJ184990; Ta6-FEH (van Riet et al., 2006), AM075205; Ta1&6-FEH (Kawakami et al., 2005), AB089269.
Results
Transformation of wild-type tobacco SNN with two different anther-specific promoter–yeast invertase fusions (Nin88::ScSuc2 or Nin88::PiII-ScSuc2) does not result in drastic changes in pollen germination
Repression of cwINV activity by anther-specific RNA interference turned out to be an efficient method to circumvent carbohydrate supply of the symplastically isolated pollen with a subsequent strong decrease of pollen germination ability and seed setting. Comparable results were also obtained by expressing a proteinaceous invertase inhibitor (see Introduction). The present study devises a strategy for restoration of fertility by replacing the down-regulated natural plant invertase activity through the distantly related isoenzyme Suc2 of yeast. Therefore, two constructs have been made and tested in the wild-type background in a first approach.
In yeast, the invertase gene Suc2 encodes two differently regulated mRNAs (1.8 kb and 1.9 kb) that differ at their 5′ ends. The smaller RNA encodes an intracellular form that is constitutively expressed at low levels in yeast. The larger RNA contains a signal peptide-coding sequence leading to a secreted form of invertase that is regulated by derepression of glucose (Fig. 1) (Carlson et al., 1983). The secreted form is glycosylated, with ∼50% of the molecular mass attributed to N-linked oligosaccharide residues (in the form of nine high-mannose oligosaccharide chains, each linked to an asparagine residue on the 60 kDa polypeptide backbone). In yeast, the secreted form has been found in the periplasmic space as oligomers which result from an aggregation of the dimer, the smallest enzymically active unit. Dimers, tetramers, and hexamers have a molecular mass of ∼260, 360, and 560 kDa, respectively (Chu et al., 1983).
Using this information, two different constructs with the yeast invertase gene Suc2 under the control of the anther-specific invertase promoter of tobacco, Nin88, have been engineered (Fig. 1): (i) the yeast invertase Suc2 with its native signal peptide resulting in the construct Nin88::ScSuc2; and (ii) an N-terminally shortened version of Suc2 fused to the N-terminal portion of potato-derived vacuolar protein proteinase II (von Schaewen et al. 1990), giving rise to Nin88::PiII-ScSuc2. The chimeric fusion of Suc2 to the N-terminal portion of potato-derived vacuolar protein proteinase II was known to be N-glycosylated and efficiently secreted from plant cells, leading to its apoplastic location (von Schaewen et al., 1990).
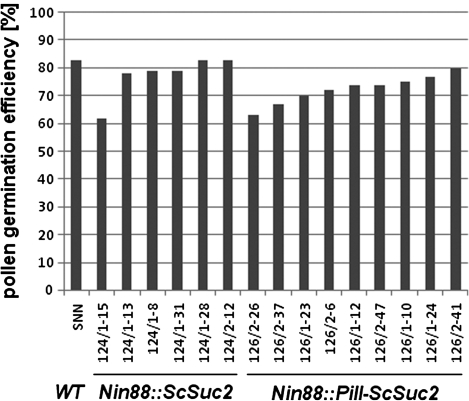
Both these constructs were transformed into wild-type tobacco SNN, and six and nine independently regenerated plants were investigated, respectively. Phenotypic analyses of vegetative growth and determination of pollen germination efficiencies, calculated as the percentage of germinating pollen in the visible pollen population, did not reveal any apparent changes in relation to the wild-type controls. The pollen germination efficiency of the wild type was determined to be ∼80%, while the transformants, independently from the construct used, showed slight variations of 60–80% (Fig. 2).
Fig. 2.
Influence of the constructs Nin88::ScSuc2 and Nin88::PiII-ScSuc2 on pollen germination efficiency in the wild-type SNN background.
Male sterility due to antisense or inhibitor interference with anther-specific cwINV is restored by supertransformation with the chimeric yeast invertase construct Nin88::PiII-ScSuc2
Since neither construct, either that with the native yeast signal peptide (Nin88::ScSuc2) or that with the potato-derived leader sequence (Nin88::PiII-ScSuc2), had an apparent effect when transformed into wild-type tobacco (see above), the construct Nin88::PiII-ScSuc2 was investigated in order to test the strategy for restoration of fertility, for the following reason: the fusion of the N-terminal portion of potato-derived vacuolar protein proteinase II (leader sequence PiII) with Suc2 was already known to be efficiently secreted from plant cells, as previous studies with protoplasts had revealed (von Schaewen et al., 1990). Thus, this construct was the more promising one for replacing the down-regulated natural plant cwINV activity through the distantly related isoenzyme Suc2 of yeast.
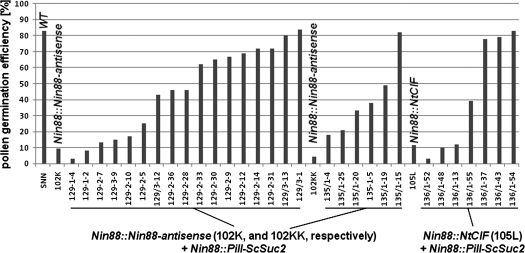
This construct was supertransformed into three originally male-sterile tobacco plants which were selected from a previous study (Hirsche et al., 2009). These three male-sterile plants had originated by transformation with the antisense construct against the anther-specific cwINV Nin88 (Nin88::Nin88-antisense), plant 102K and 102KK, or by expressing the proteinacious invertase inhibitor NtCIF (Nin88::NtCIF), plant 105L. The strong phenotypes of these three plants have been characterized by reduced total numbers of pollen per anther (from ∼50 000 in the wild type to <10 000) and, furthermore, by drastically reduced pollen germination efficiencies in the visible pollen population [from ∼80% in the wild type to <10% in the male-sterile plants; Hirsche et al. (2009), see also Figs 3a, b, 4]. These three male-sterile plants were supertransformed with the yeast invertase construct Nin88::PiII-ScSuc2 and 17 plants from 102K, six from 102KK, and seven from 105L regenerated, and the pollen germination efficiencies were determined. Among the supertransformed plants, fertility is restored to different extents (Figs 3c–f, 4). In about two-thirds of the plants, pollen germination efficiencies of >30% were determined. Independently from the construct originally used for creating the male-sterile plants (antisense or inhibitor), the best phenotypes of the supertransformants showed pollen germination efficiencies of ∼80%, thus reaching the level of the wild type (Fig. 4).
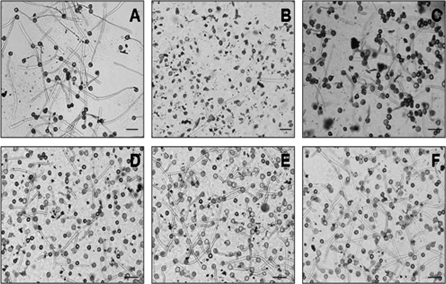
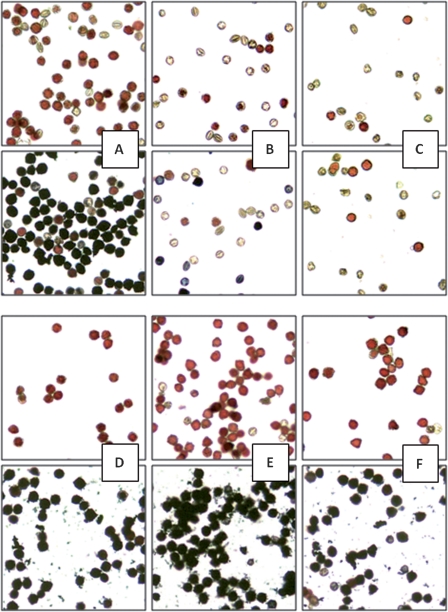
Fig. 3.
In vitro pollen germination of wild-type tobacco SNN (A), Nin88::Nin88-antisense (B, plant Nt102K), and of plants supertransformed with Nin88::PiII-ScSuc2 (C–F, plants Nt129-2-5, Nt129-2-28, Nt129-2-30, and Nt129-2-31). Bars=100 μm.
Fig. 4.
Pollen germination efficiencies (calculated as the relative part of germinating pollen in the visible pollen population) of wild-type tobacco SNN, male-sterile plants 102K, 102KK (Nin88::Nin88-antisense), and 105L (Nin88::NtCIF), as well as their supertransformed (Nin88::PiII-ScSuc2) descendants.
In order to visualize the reduced endogenous invertase activity in the male-sterile plants and its substitution in the supertransformants by the yeast invertase, histochemical in situ invertase activity stains were performed. The staining method uses sucrose as substrate. The glucose liberated by invertase activity is oxidized by GOD, thereby reducing PMS and finally NBT. The reaction yields an intensely blue insoluble formazan which is easily visible microscopically, while NBT itself is soluble and yields a practically colourless (slightly yellow) solution (Dahlqvist and Brun, 1962). While pollen of the wild type SNN (Fig. 5A, lower row) shows a dark blue-black staining in the incubation medium, pollen of male-sterile plants expressing Nin88::Nin88-antisense (Fig. 5B) or Nin88::NtCIF (Fig. 5C) largely failed to become stained. Even in the control, where sucrose as the substrate is omitted, pollen of male-sterile plants do not show the reddish background staining normally observed in fertile pollen. This verifies the degradation of the pollen in the male-sterile plants previously detected by acetocarmine staining (Hirsche et al., 2009). However, when supertransformed with the yeast invertase (Nin88::PiII-ScSuc2), the staining in the control (Fig. 5D–F, upper row) as well as in the incubation medium (Fig. 5D–F, lower row) resembles that of the wild type. Thus, invertase activity is restored to the level of the wild type through tissue-specific expression of the heterologous invertase, reflecting the normal development of pollen and the ability for normal pollen germination efficiencies observed in these genotypes (see above).
Fig. 5.
In situ stains of invertase activity of wild-type tobacco, male-sterile plants, and supertransformed plants. Upper rows, controls (– sucrose); lower rows, dark staining indicates sucrose cleavage. (A) Wild type SNN; (B) male-sterile plant 102K, expressing Nin88::Nin88-antisense; (C) male-sterile plant 105L, expressing Nin88::Nin88-NtCIF; (D) Nt129-2-14 (antisense plant 102K, supertransformed with Nin88::PiIIScSuc2); (E) Nt135-1-15 (antisense plant 102KK, supertransformed with Nin88::PiIIScSuc2); (F) Nt136-1-54 (inhibitor plant 105L, supertransformed with Nin88::PiIIScSuc2).
Nicotiana tabacum has a similar number of genes in the acid invertase gene family to Arabidopsis
So far the investigations of anther-specific cwINVs have focused on the model plants N. tabacum, S. lycopersicum, and A. thaliana (Goetz et al., 2001; Proels et al., 2006; Hirsche et al., 2009). However, candidates for invertases with an impact on pollen development are known from other dicotyledenous as well as from monocotyledenous plants (see Introduction). A phylogenic analysis of these genes within the invertase gene family was done with the aim to provide insights into whether the cwINVs with anther-specific expression are closely related to each other, or if there is some type of co-evolution in the different plant species. This knowledge will provide an impact on future applications of the described transgenic approach to produce male sterility in other species.
Therefore, it was decided to extend knowledge about the acid invertase family from tobacco in order to broaden the basis for the phylogenic analysis. This basis is provided by the known invertase gene families from Arabidopsis and rice.
In the genome of A. thaliana six putative cwINV genes have been identified. Two of them, originally referred to as Atβfruct1 and Atβfruct2 (Tymowska-Lalanne and Kreis, 1998), were renamed AtcwINV1 and AtcwINV2, respectively by Sherson et al. (2003). Later, AtcwINV3 and AtcwINV6 with lower pI values of 5.5 and 4.8 compared with 8.1–9.7 for the remaining cwINVs, turned out to be FEHs that split one terminal fructose unit from a longer fructan chain, instead of sucrose cleavage (de Coninck et al., 2005). It was decided to adapt the nomenclature of the Arabidopsis invertases suggested by Sherson et al. (2003) for the tobacco invertases, but, in order to accommodate the findings of de Coninck et al. (2005), FEH? was added after the name of such tobacco sequences with lower pI values to indicate that these are most probably FEHs. A set of four cwINVs and two cwINVs(FEH?) comparable with that in Arabidopsis was found in tobacco. Two cwINVs have previously been described: following the numbering in Arabidopsis, Ntβfruc1 (Greiner et al., 1995, accession no. X81834) would correspond to NtcwINV1, and NtNin88 (Goetz et al., 2001, accession no. AF376773) would correspond to NtcwINV2. Four additional sequences have been identified during this study: NtcwINV3 (accession no. HM022265), NtcwINV4 (accession no. HM022266), NtcwINV5-FEH? (accession no. HM022267), and NtcwINV6-FEH? (accession no. HM022268). The latter two most probably have to be considered as FEHs, according to their low pI values (Table 1).
Table 1.
The acid invertase gene family from Nicotiana tabacum
| Suggested nomenclature | GenBank accession no. | pI (calculated from the region amplified) | Highest homology | |
| % homology | Gene | |||
| NtcwINV1 | Identical to Ntβfruc1a | – | – | Ntβfruc1a (X81834) pI 9.1 |
| NtcwINV2 | Identical to Nin88b | 9.1 | 100 % | Nin88b (AF376773) pI 8.4 |
| NtcwINV3 | HM022265 | 9.3 | 100 % | Nin77c pI 9.3 |
| NtcwINV4 | HM022266 | 8.9 | 98 % | CIN1 (X81792) pI 9.1 |
| NtcwINV5 FEH? | HM022267 | 6.2 | 60 % | GmCWINV (CAD91338) pI 8.7 |
| NtcwINV6 FEH? | HM022268 | 4.6 | – | – |
| NtvacINV1 | Identical toNt-VId | 5.0 | 100 % | Nt-VId (AJ305044) pI 5.8 |
| NtvacINV2 | HM022269 | 5.4 | 86 % | Lin9 (AM050394) pI 6.7 |
Previously identified by Greiner et al. (1995) (accession no. X81834).
Previously identified by Goetz et al. (2001) (accession no. AF376773).
Previously identified by D Godt; sequence not available in databases.
Previously identified by Lauer (2006) (accession no. AJ305044).
vacINVs have lower pI values of ∼5–6, compared with cwINVs with pI values of 8–9 and they can be distinguished from each other by a single amino acid difference in their cysteine catalytic sites (WEC-P/V-DF): cwINVs have a proline residue in the sequence motif and vacINVs possess a valine residue (Goetz and Roitsch, 1999). In Arabidopsis, two vacINV genes are known (Haouazine et al., 1997; Tymowska-Lalanne and Kreis, 1998) and have been referred to as Atβfruct3 and Atβfruct4 (also by Sherson et al., 2003). Since both, cwINVs and vacINVs are β-fructofuranosidases, and the numbers 3 and 4 in Arabidopsis are somewhat misleading because only two vacINVs are present, it is suggested to designate the tobacco invertases genes as NtvacINV. A comparable set of two vacINV genes has also been identified by the same approach in tobacco: NtvacINV1 (previously described as NtVI; Lauer, 2006, accession no. AJ305044) and NtvacINV2 (accession no. HM022269) (Table 1).
Phylogenic relationship of the anther-specific invertases in comparison with related enzymes from different species
The complete invertase gene families (acid and cytoplasmic) from A. thaliana and O. sativa are the backbone of the phylogenic analyses, extended with the sequences of the acid invertases from tobacco, as well as related sequences from the well-studied fructan-accumulating species C. intybus and T. aestivum with FBE and FEH genes. CwINVs with known anther-specific expression from S. lycopersicum, S. tuberosum, and D. carota are also added.
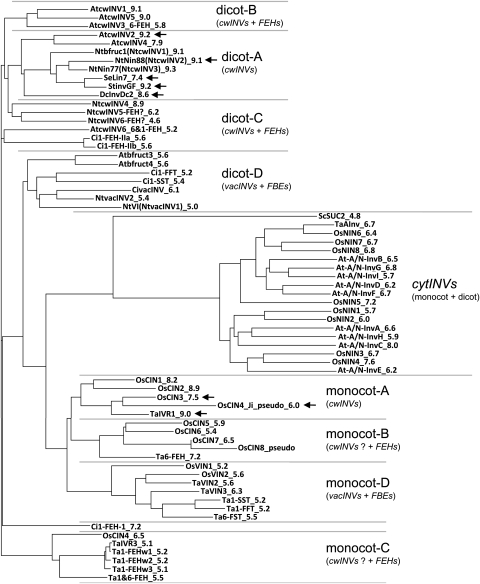
All anther-specific invertases of dicotyledenous plants cluster within one phylogenic group, referred to as dicot-A. The anther-specific invertases of monocotyledenous plants cluster within another phylogenic group, referred to as monocot-A (Fig. 6, marked with arrowheads). Although these clusters are only distantly related to each other, both consist of cwINVs only, some of them with other expression profiles; however, no other enzymes investigated are located within these clusters.
Fig. 6.
Phylogenic tree showing the evolutionary relationships among invertases, fructan exohydrolases, and fructosyltransferases. Each node with descendants represents the most recent common ancestor of the invertases, and the branch lengths are proportional to the amount of inferred evolutionary change and thus correspond to time estimates. Arrowheads indicate anther-specifically expressed cell wall-bound invertases. CwINVs, cell wall-bound invertases; vacINVs, vacuolar invertases; cytINVs, cytoplasmic invertases; FEHs, fructan exohydrolases; FBEs, fructan-building enzymes. For abbreviations and references for the single coding sequences of the different species see Materials and methods; the number at the end of the abbreviation indicates the pI value of the corresponding protein.
The remaining cwINVs of dicotyledenous and moncotyledenous plants group in two different clusters each, together with FEHs (Fig. 6, dicot-B and -C, moncot-B and -C). In the case of the monocot-B and -C clusters, all enzymes have low pI values, thus it is debatable whether the enzymes described as cwINVs are in fact FEHs (this problem is indicated by cwINVs? in Fig. 6).
The vacINVs cluster together with FBEs in the two specific clusters dicot-D and monocot-D.
Finally, the cytoplasmic INVs (cytINVs) of Arabidopsis and rice form a common cluster that shows only a distant relationship to the sequences mentioned above and thus this cluster was designated cytINVs (monocot+dicot) (Fig. 6).
Discussion
The phylogenic classification of invertases to related genes in different species
In plant cells, invertases (β-fructofuranosidases; EC 3.2.1.26) are found in the cell wall (cwINV), vacuole (vacINV), and cytoplasm (cytINV). cwINVs and vacINVs are both acid invertases with optimum pH ∼5.0 and their amino acid sequences are more closely related to each other than to that of cytINV (alkaline/neutral, optimum pH between 6.5 and 8.0), whose origin is believed to be in cyanobacteria (Sturm, 1999; Sturm and Tang, 1999; Vargas et al., 2003). This distinct evolutionary origin is clearly visible in Fig. 6, where cytINVs from monocotyledons and dicotyledons build a common cluster apart from acid invertases. While cytINVs were thought for a long time to be exclusively localized in the cytosol, recent reports point to a subcellular location with targets to mitochondria and chloroplasts (Ji et al., 2005; Murayama and Handa, 2007; Vargas et al., 2008).
Six putative cwINVs have been identified in the A. thaliana genome (AtcwINV1–6; Sherson et al. 2003), two of them, AtcwINV3 and AtcwINV6, later turned out to be FEHs that split one terminal fructose unit from a longer fructan chain, instead of carrying out sucrose cleavage (de Coninck et al., 2005). They possess lower pI values of 5.5 and 4.8 compared with 8.1–9.7 for the remaining cwINVs. Comparable results were found in the present study by analysing the tobacco sequences: among the group of six cwINVs in tobacco, two possess lower pI values and most probably have to be considered as FEHs: NtcwINV5-FEH? and NtcwINV6-FEH? As in A. thaliana and as predicted for N. tabacum, other species such as sugar beet (Beta vulgaris) also possess FEHs, but apparently lack endogenous fructan substrates. The most plausible function for a specific 6-FEH in these non-fructan plants would be to degrade (and/or prevent the formation of) exogenous levan-type fructans of bacterial origin (van den Ende et al., 2003).
In contrast to non-fructan plants, ∼15% of flowering plants use fructans as reserve carbohydrates. Fructan-accumulating species mainly belong to the dicot families Asteraceae, Campanulaceae, and Boraginaceae [with (2,1)-type fructans, referred to as inulin] and the monocot families Poaceae and Liliaceae [with predominantly (2,6)-type fructans, referred to as levans, as in bacteria] (reviewed in van den Ende et al., 2004). The close relationship between the functions of the genes encoding FEHs and cwINVs was shown by Le Roy et al. (2007): a single amino acid exchange in AtcwINV1 is sufficient to switch the invertase function to an FEH function. Both types of enzymes, cwINVs and FEHs, partly do not cluster in different branches of the pyhlogenetic tree, indicating that they evolved from each other rather than from different ancestors.
The cwINVs and FEHs from monocotyl plants form clusters, which can be distinguished from the clusters from dicotyledenous plants, a further hint that FEHs evolved independently from each other in both groups of plants. The rice cwINVs form one main cluster that is separated into two subclusters monocot-A and monocot-B (Fig. 6). Both groups have been previously distinguished (Ji et al.. 2005, designated as α and β). Interestingly, only the sequences in monocot-A possess pI values of 7.5–9.0, while the remaining rice sequences of the neighbouring monocot-B cluster have pI values between 5.9 and 6.5. The low pI values might be a hint that these latter genes encode FEHs rather than cwINVs. Indeed, these sequences show high similarities to Ta6-FEH from wheat. OsCIN4 was located in the β-group by Ji et al. (2005; designated as OsCIN5, cf. Materials and methods); however, in the present broader phylogenic analysis, this gene groups in the cluster monocot-C together with TaIVR3 from wheat. TaIVR3 (originally described as cell wall invertase in Koonjul et al., 2004) corresponds to Ta1-FEHw1 (Zhang et al., 2008) and thus turned out to be an FEH gene, as all the remaining genes of this cluster in Fig. 6 are FEH genes from wheat (Zhang et al., 2008). The calculated pI for the related OsCIN4 is 6.5, a hint that this might also be a FEH rather than an invertase. Though the well characterized FEHs clearly evolved from cwINVs in both monocotyledenous and dicotyledenous plants, recently high FEH activities were also described for both rice vacINVs, especially for OsVIN1 (Ji et al., 2007).
The vacINVs from both Arabidosis and tobacco are characterized by lower pI values of ∼5–6 and a single amino acid difference in their cysteine catalytic sites (WEC-V-DF, see above), and together comprose the cluster dicot-D (Fig. 6). Interestingly, the same number of vac-INVs was described in the monocotyledenous species rice (OsVIN1 and OsVIN2; Ji et al., 2005). Thus the set of vacINVs seems to be somewhat conserved in monocots and dicots, though the sequences do not group in the same cluster. Closely related to vacINVs are FBEs (fructosyltransferases) which are involved in the biosynthesis of fructan in fructan-accumulating plants, whereas these genes are lacking in non-fructan plants such as Arabidopsis and rice. The corresponding enzymes from C. intybus and T. aestivum were integrated in the phylogenic analyses, and, as expected, they group together with vacINVs in the clusters dicot-D and monocot-D, respectively (Fig. 6). The evolutionary steps from vacINVs to fructosyltransferases occurred independently in different species. The initial step started with water in a vacINV ancestor as the fructosyl group acceptor was replaced by a second sucrose molecule, resulting in sucrose:sucrose-fructosyltransferases 1-SST (Vijn et al., 1998; Ritsema et al., 2006; Ji et al., 2007). In further evolution, there were changes in the donor substrate and/or the acceptor substrate to give the remaining forms of fructosyltransferases.
Relationship of anther-specific cwINV genes and consequences for identifying corresponding genes in other species in order to engineer male sterility
The anther-specific expressed isoenzymes indicate the crucial function of extracellular invertases in providing carbohydrates to the male gametophyte in different species, namely in Arabidopsis (AtcwINV2; Hirsche et al., 2009), carrots (DcInvDc2; Lorenz et al., 1995), potato (StInvGF; Maddison et al., 1999), tobacco (Nin88; Goetz et al., 2001; Hirsche et al., 2009), and tomato (ScLin7; Godt and Roitsch, 1997; Fridman and Zamir, 2003; Proels et al. 2006). Interestingly, all these anther-specific cwINVs from dicotyledenous plants cluster in the distinct group dicot-A in the phylogenic tree (Fig. 6). Within this cluster, additional cwINVs with other expression profiles are found, but no FEHs, which group together with the remaining cwINVs in the additional clusters dicot-B and -C.
In monocotyledenous plants most investigations of the involvement of invertases in pollen development have been done in wheat and rice, considering abiotic stresses (drought and cold). Although vacINV and cwINV activities are involved in pollen abortion due to drought stress in wheat, the most pollen- and tapetum-specific expression profile was evident for the cell wall-bound TaIVR1 (Koonjul et al., 2004). Comparable results were found in rice under drought stress (Saini,1997) as well as under cold stress (Oliver et al., 2005). Cho et al. (2005) suggested that OsCIN3 is the most important cwINV gene for pollen development in rice, comparable with Nin88 in tobacco, since all the other cwINV genes are not specifically expressed in the flower. These findings were supported by further investigations showing the specific expression profile of OsCIN3 in tapetum and pollen and a distinct cold stress reaction (Oliver et al., 2005; designated as OsINV4). Furthermore OsCIN3 and TaIVR1 show a close relationship to each other (Fig. 6, cluster monocot-A). Within this cluster of the monocotyledenous species two further cwINV genes (OsCIN1 and OsCIN2) are located, with their transcripts all having the characteristic high pI values. In contrast to the findings of Oliver et al. (2005) and Cho et al. (2005), Ji et al. (2005) also detected transcripts of OsCIN3 in other tissues by RT-PCR; nevertheless the highest transcript level was also found in the panicle. Apart from OsCIN3, Ji et al. (2005) found a flower-specific expression for the truncated cwINV gene OsCIN4(Ji)_pseudo (designated as OsCIN4 in Ji et al., 2005). The coding sequence of this truncated invertase shows a close relationship to OsCIN3 and TaIVR1, all grouping in cluster monocot-A, which can be clearly distinguished from the cluster with the anther-specific cwINVs of dicotyl plants (Fig. 6).
Summing up the above-mentioned findings on the phylogenic relationships of anther-specific cwINVs, the most important is the fact that sequences of monocotyl and dicotyl plants fall in one specific cluster each, monocot-A and dicot-A, respectively, but these clusters can be clearly distinguished from each other and both contain further cwINV genes with different expression profiles. Thus, in both groups of plants, there is some kind of co-evolution, but no recent common ancestor of anther-specific cwINVs. This knowledge of the relationship might be helpful for identifying corresponding genes from other species by sequence alignment and thus for the application of the male sterility system by anther-specific interference with cwINV activity. It should be mentioned that in spite of the close relationship among the anther-specific cwINVs within both clusters, a promoter compatibility of these invertases cannot necessarily be assumed for distantly related species, as previous studies revealed for Arabidopsis and tobacco (Hirsche et al., 2009).
Development of a restorer system for metabolically engineered male sterility
For the practical application of engineered male sterility in a hybrid breeding programme, the F1 hybrid varieties in the farmer's field usually have to be fertile to produce the required grain or seed. In naturally occurring CMS systems, this restoration of fertility is achieved by introducing a restorer gene from the paternal parent to the hybrid. By far most restorer genes suppress male-sterile phenotypes by alterations of transcript patterns of the mitochondrial mutations causing CMS, thereby decreasing the expression of the related proteins, and they belong to the pentatricopeptide repeat-containing gene family (PPR), as in petunia [Rf of petunia (Bentolila et al., 2002); Rfo of Ogura radish (Brown et al., 2003); Rfk1 of Kosena radish (Koizuka et al., 2003); Rf-1 in BT rice (Kazama and Toriyama, 2003; Komori et al., 2004]. Although various successful approaches to engineer male sterility by transformation of the nucleus or the plastids have been described, most of them fail to provide an effective means of restoration, comparable with the natural restorer genes of CMS (reviewed in Roitsch and Engelke, 2006). Thus, the only system that is already in practical use is the barstar–barnase system. Mariani et al. (1990) have interfered with tapetal development using the 5' region of a tobacco tapetum-specific gene (TA29) to drive expression of recombinant RNase genes (RNase T1 and barnase) within the tapetal cells of transgenic tobacco and oilseed rape plants, thus preventing pollen formation. To produce a fertile hybrid for sale to farmers, the male-sterile line is grown alongside a second line containing the barstar gene which codes for a protein that blocks the action of barnase. This system was used by PlantGeneticSystems (Ghent, Belgium) for oilseed rape that is grown commercially mainly in Canada.
Here a restorer system is presented for the previously described engineered male sterility system, using anther-specific interference with extracellular invertase activity. The applicability of this system has been shown for tobacco, tomato, and Arabidosis (Goetz et al., 2001; Proels et al., 2006; Hirsche et al., 2009), and the crucial function of cwINVs in providing carbohydrates for male gametophyte development and the specific involvement of invertases during anther development was also suggested for other dicotyledenous and monocotyledenous plant species (see above). Restoration of fertility is now successfully achieved by substituting the down-regulated natural plant invertase activity for a yeast invertase under the control of the same promoter. The yeast invertase is fused to the N-terminal portion of potato-derived vacuolar protein proteinase II (PiII–ScSuc2), leading to its apoplastic location. The by-passing of the anther-specific reduced carbohydrate supply results in a drastic increase of pollen germination rates; the best phenotypes in the present investigation showed pollen germination efficiencies comparable with those of untransformed wild types. Moreover, the transgene does not affect pollen germination rates when transformed to wild types, thus such plants can be used as paternal parents in a hybrid breeding programme in order to introgress the yeast invertase leading to restoration of fertility in the hybrid variety.
Apart from difficulties concerning the restoration of the hybrid, further difficulties in propagating the male-sterile lines seriously limit the practical application of many transgenic male sterilities. The challenge of propagating male-sterile plants was elegantly solved by Ruiz and Daniell (2005) by imitating the naturally occurring CMS systems. In many (but not all) plants, during formation of pollen, plastids are excluded or degraded so pollen does not contain plastid DNA which is inherited maternally through the ovum. Since transformation of plastids was successful at least for some species, engineering CMS via the chloroplast genome was suggested: a β-ketothiolase gene was introduced and hyperexpressed in chloroplasts, leading to male sterility by depleting a substrate needed for fatty acid synthesis essential for pollen production (Ruiz and Daniell, 2005). This male sterility was reversible by prolonged light exposure, a mechanism which can theoretically be used for restoration of fertility in the hybrid; however, this approach seems not to be feasible under field conditions and limits the practical use of this system.
The male-sterile line can also be reproduced and maintained by selection of male-sterile plants from a segregating offspring, as is done with the above-mentioned barstar–barnase system: linkage of the barnase gene with the bar (Streptomyces hygroscopicus) marker gene, which encodes a phosphinothricin acetyltransferase enzyme that inactivates glufosinate, permits identification of the male-sterile line before flowering. Another possibility would be a reversible suppression of the sterility. Since induction, especially under field conditions, is always leaky to some extent, an induction of the sterility-inducing gene, in this case the plant invertase antisense or invertase inhibitor, would lead to pollen production to some extent and the hybrids would be wasted with sibs. On the other hand, this leakiness is a minor problem when the gene bringing back the fertility, in the present case the yeast invertase, is connected to an inducible promoter. For propagating the male-sterile line it does not matter whether pollen production is complete or not, as long as pollination is ensured. This offers a way for propagation of the male-sterile line, as an alternative to the linkage to herbicide tolerance.
Acknowledgments
We are grateful to Professor Dr Uwe Sonnewald for providing us with a clone containing the chimeric PiII–ScSuc2 construct. The skilful technical assistance of Christine Hampp, and financial support by the Bundesministerium für Bildung und Forschung, is gratefully acknowledged.
References
- Altschul SF, Madden TA, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila S, Alfonso AA, Hanson MR. A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proceedings of the National Academy of Sciences, USA. 2002;99:10887–10892. doi: 10.1073/pnas.102301599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brewbaker JL. Agricultural genetics. Englewood Cliffs, NJ: Prentice-Hall; 1964. [Google Scholar]
- Brown GG, Formanova H, Jin H, Wargachuk R, Dendy C, Patil P, Laforest M, Zhang J, Cheung WY, Landry BS. The radish Rfo restorer gene of Ogura cytoplasmic male sterility encodes a protein with multiple pentatricopeptide repeats. The Plant Journal. 2003;35:262–272. doi: 10.1046/j.1365-313x.2003.01799.x. [DOI] [PubMed] [Google Scholar]
- Carlson M, Taussig R, Kustu S, Botstein D. The secreted form of invertase in Saccharomyces cerevisiae is synthesized from RNA encoding a signal sequence. Molecular and Cellular Biology. 1983;3:439–447. doi: 10.1128/mcb.3.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cho JI, Lee SK, Ko S, et al. Molecular cloning and expression analysis of the cell-wall invertase gene family in rice (Oryza sativa L.) Plant Cell Reports. 2005;24:225–236. doi: 10.1007/s00299-004-0910-z. [DOI] [PubMed] [Google Scholar]
- Chu FK, Watorek W, Maley F. Factors affecting the oligomeric structure of yeast external invertase. Archives of Biochemistry and Biophysics. 1983;223:543–555. doi: 10.1016/0003-9861(83)90619-7. [DOI] [PubMed] [Google Scholar]
- Clément C, Burrus M, Audran JC. Floral organ growth and carbohydrate content during pollen development in Lilium. American Journal of Botany. 1996;83:459–469. [Google Scholar]
- Dahlqvist A, Brun A. A method for the histochemical demonstration of disaccharidase activities: application to invertase and trehalase in some animal tissues. Journal of Histochemistry and Cytochemistry. 1962;10:294–302. [Google Scholar]
- Daniell H. Molecular strategies for gene containment in transgenic crops. Nature Biotechnology. 2002;20:581–586. doi: 10.1038/nbt0602-581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Coninck B, Le Roy K, Francis I, Clerens S, Vergauwen R, Halliday AM, Smith SM, Van Laere A, Van den Ende W. Arabidopsis AtcwINV3 and 6 are not invertases but are fructan exohydrolases (FEHs) with different substrate specificities. Plant, Cell and Environment. 2005;28:432–443. [Google Scholar]
- De Halleux S, van Cutsem P. Cloning and sequencing of the 1-SST cDNA from chicory root (Accession No. U81520) (PGR97-036) Plant Physiology. 1997;113:1003. [Google Scholar]
- Doehlert DC, Felker FC. Characterization and distribution of invertase activity in developing maize (Zea mays) kernels. Physiologia Plantarum. 1987;70:51–57. [Google Scholar]
- Dorion S, Lalonde S, Saini HS. Induction of male sterility in wheat by meiotic-stage water deficit is preceded by a decline in invertase activity and changes in carbohydrate metabolism in anthers. Plant Physiology. 1996;111:137–145. doi: 10.1104/pp.111.1.137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engelke T, Agbicodo E, Tatlioglu T. Mitochondrial genome variation in Allium ampeloprasum and its wild relatives. Euphytica. 2004b;137:181–191. [Google Scholar]
- Engelke T, Gera G, Tatlioglu T. Determination of the frequencies of restorer- and maintainer-alleles involved in CMS1 and CMS2 in German chive varieties. Plant Breeding. 2004b;123:51–59. [Google Scholar]
- Feistritzer WR, Kelly AF. Hybrid seed production of selected cereal oil and vegetable crops. Rome: Food and Agriculture Organization of the United Nations; 1987. [Google Scholar]
- Fridman E, Zamir D. Functional divergence of a syntenic invertase gene family in tomato, potato, and Arabidopsis. Plant Physiology. 2003;131:603–609. doi: 10.1104/pp.014431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godt DE, Roitsch T. Regulation and tissue-specific distribution of mRNAs for three extracellular invertase isoenzymes of tomato suggests an important function in establishing and maintaining sink metabolism. Plant Physiology. 1997;115:273–282. doi: 10.1104/pp.115.1.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Godt DE, Guivarc′h A, Kahmann U, Chriqui D, Roitsch T. Induction of male sterility in plants by metabolic engineering of the carbohydrate supply. Proceedings of the National Academy of Sciences, USA. 2001;98:6522–6527. doi: 10.1073/pnas.091097998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goetz M, Roitsch T. The different pH-optima and substrate specificities of extracellular and vacuolar invertases are determined by a single amino acid substitution. The Plant Journal. 1999;20:707–711. doi: 10.1046/j.1365-313x.1999.00628.x. [DOI] [PubMed] [Google Scholar]
- Greiner S, Weil M, Krausgrill S, Rausch T. A tobacco cDNA coding for cell-wall invertase. Plant Physiology. 1995;108:825–826. doi: 10.1104/pp.108.2.825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haouazine-Takvorian N, Tymowska-Lalanne Z, Takvorian A, Tregear J, Lejeune A, Kreis M. Characterization of two members of the Arabidopsis thaliana gene family, Atβfruct3 and Atβfruct4, coding for vacuolar invertases. Gene. 1997;197:239–251. doi: 10.1016/s0378-1119(97)00268-0. [DOI] [PubMed] [Google Scholar]
- Higgens D, Thompson J, Gibson T, Thompson JD, Higgens DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hirsche J, Engelke T, Völler D, Götz M, Roitsch T. Interspecies compatibility of the anther specific cell wall invertase promoters from Arabidopsis and tobacco for generating male sterile plants. Theoretical and Applied Genetics. 2009;118:235–245. doi: 10.1007/s00122-008-0892-2. [DOI] [PubMed] [Google Scholar]
- Horsch RB, Fry JB, Hoffmann NL, Wallroth M, Eichholtz D, Rogers SG, Fraley RT. A simple and general method for transferring genes into plants. Science. 1985;227:1229–1231. doi: 10.1126/science.227.4691.1229. [DOI] [PubMed] [Google Scholar]
- Hsu YF, Wang CS, Raja R. Gene expression pattern at desiccation in the anther of Lilium longiflorum. Planta. 2007;226:311–322. doi: 10.1007/s00425-007-0483-5. [DOI] [PubMed] [Google Scholar]
- Ji X, Van den Ende W, Schroeven L, Clerens S, Geuten K, Cheng S, Bennett J. The rice genome encodes two vacuolar invertases with fructan exohydrolase activity but lacks the related fructan biosynthesis genes from Pooideae. New Phytologist. 2007;173:50–62. doi: 10.1111/j.1469-8137.2006.01896.x. [DOI] [PubMed] [Google Scholar]
- Ji X, Van den Ende W, Van Laere A, Cheng S, Bennett J. Structure, evolution, and expression of the two invertase gene families of rice. Journal of Molecular Evolution. 2005;60:615–634. doi: 10.1007/s00239-004-0242-1. [DOI] [PubMed] [Google Scholar]
- Kawakami A, Yoshida M. Molecular characterization of sucrose:sucrose1-fructosyltransferase and sucrose:fructan 6-fructosyltransferase associated with fructan accumulation in winter wheat during cold hardening. Bioscience, Biotechnology, and Biochemistry. 2002;66:2297–2305. doi: 10.1271/bbb.66.2297. [DOI] [PubMed] [Google Scholar]
- Kawakami A, Yoshida M. Fructan:fructan 1-fructosyltransferase, a key enzyme for biosynthesis of graminan oligomers in hardened wheat. Planta. 2005;223:90–104. doi: 10.1007/s00425-005-0054-6. [DOI] [PubMed] [Google Scholar]
- Kawakami A, Yoshida M, van den Ende W. Molecular cloning and functional analysis of a novel 6&1-FEH from wheat (Triticum aestivum L.) preferentially degrading small graminans like bifurcose. Gene. 2005;358:93–101. doi: 10.1016/j.gene.2005.05.029. [DOI] [PubMed] [Google Scholar]
- Kazama T, Toriyama K. A pentatricopeptide repeat-containing gene that promotes the processing of aberrant atp6 RNA of male-sterile rice. FEBS Letters. 2003;544:99–102. doi: 10.1016/s0014-5793(03)00480-0. [DOI] [PubMed] [Google Scholar]
- Koizuka N, Imai R, Fujimoto H, Hayakawa T, Kimura Y, Kohno-Murase J, Sakai T, Kawasaki S, Imamura J. Genetic characterization of a pentatricopeptide repeat protein gene, orf687, that resores fertility in the cytoplasmic male-sterile Kosena radish. The Plant Journal. 2003;34:407–415. doi: 10.1046/j.1365-313x.2003.01735.x. [DOI] [PubMed] [Google Scholar]
- Komori T, Ohta S, Murai N, Takakura Y, Kuraya Y, Suzuki S, Hiei Y, Imaseki H, Nitta N. Map-based cloning of a fertility restorer gene, Rf-1, in rice (Oryza sativa L.) The Plant Journal. 2004;37:315–325. doi: 10.1046/j.1365-313x.2003.01961.x. [DOI] [PubMed] [Google Scholar]
- Koonjul PK, Minhas JS, Nunes C, Sheoran IS, Saini HS. Selective transcriptional down-regulation of anther invertases precedes the failure of pollen development in water-stressed wheat. Journal of Experimental Botany. 2004;56:179–190. doi: 10.1093/jxb/eri018. [DOI] [PubMed] [Google Scholar]
- Lalonde S, Beebe D, Saini HS. Early signs of disruption of wheat anther development associated with the induction of male sterility by meiotic-stage water deficit. Sexual Plant Reproduction. 1997;10:40–48. [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, et al. ClustalW and ClustalX version 2. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Lauer K. Universität Heidelberg. Heidelberg, Germany: 2006. Nt-VIF—ein proteinogener Inhibitor der vakuolären Invertasen in Tabak. Thesis, Fakultät für Biologie. [Google Scholar]
- Le Roy K, Lammens W, Verhaest M, De Coninck B, Rabijns A, Van Laere A, Van den Ende W. Unraveling the difference between invertases and fructan exohydrolases: a single amino acid (Asp-239) substitution transforms Arabidopsis cell wall invertase1 into a fructan 1-exohydrolase. Plant Physiology. 2007;145:616–625. doi: 10.1104/pp.107.105049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lorenz K, Lienhard S, Sturm A. Structural organization and differential expression of carrot beta-fructofuranosidase genes: identification of a gene coding for a flower bud-specific isozyme. Plant Molecular Biology. 1995;28:189–194. doi: 10.1007/BF00042049. [DOI] [PubMed] [Google Scholar]
- Maddison AL, Hedley PE, Meyer RC, Aziz N, Davidson D, Machray GC. Expression of tandem invertase genes associated with sexual and vegetative growth cycles in potato. Plant Molecular Biology. 1999;41:741–751. doi: 10.1023/a:1006389013179. [DOI] [PubMed] [Google Scholar]
- Mariani C, DeBeuckeleer M, Truettner J, Leemans J, Goldberg RB. Induction of male sterility in plants by a chimaeric ribonuclease gene. Nature. 1990;347:737–741. [Google Scholar]
- Minhas JS, Saini HS. Cloning and characterization of a cDNA (Accession No. AF030420) from wheat anthers that encodes the cell wall form of invertase (PGR 98-206) Plant Physiology. 1998;118:1535. [Google Scholar]
- Murayama H, Handa S. Genes for alkaline/neutral invertases are located in plant mitochondria and also in plastids. Planta. 2007;225:1193–1203. doi: 10.1007/s00425-006-0430-x. [DOI] [PubMed] [Google Scholar]
- Oliver SN, van Dongen JT, Alfred SC, et al. Cold-induced repression of the rice anther-specific cell wall invertase gene OSINV4 is correlated with sucrose accumulation and pollen sterility. Plant, Cell and Environment. 2005;28:1534–1551. [Google Scholar]
- Proels RK, González MC, Roitsch T. Gibberellin-dependent induction of tomato extracellular invertase Lin7 is required for pollen development. Functional Plant Biology. 2006;33:547–554. doi: 10.1071/FP04146. [DOI] [PubMed] [Google Scholar]
- Proels RK, Hause B, Berger S, Roitsch T. Novel mode of hormone induction of tandem tomato invertase genes in floral tissues. Plant Molecular Biology. 2003;52:191–201. doi: 10.1023/a:1023973705403. [DOI] [PubMed] [Google Scholar]
- Ranwala AP, Miller WB. Sucrose cleaving enzymes and carbohydrate pools in Lilium longiflorum floral organs. Physiologia Plantarum. 1998;103:541–550. [Google Scholar]
- Ritsema T, Hernández L, Verhaar A, Altenbach D, Boller T, Wiemken A, Smeekens S. Developing fructan-synthesizing capability in a plant invertase via mutations in the sucrose-binding box. The Plant Journal. 2006;48:228–237. doi: 10.1111/j.1365-313X.2006.02862.x. [DOI] [PubMed] [Google Scholar]
- Roitsch T, Bittner M, Godt DE. Induction of apoplastic invertase of Chenopodium rubrum by D-glucose and a glucose analog and tissue-specific expression suggest a role in sink–source regulation. Plant Physiology. 1995;108:285–294. doi: 10.1104/pp.108.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roitsch T, Engelke T. Cytoplasmic, genic and transgene induced male sterility. In: da Silva JAT, editor. Floriculture, ornamental and plant biotechnology. London, UK: Global Science Books, Ltd; 2006. pp. 512–522. [Google Scholar]
- Roitsch T, González M. Function and regulation of invertases in higher plants: sweet sensations. Trends in Plant Science. 2004;9:607–613. doi: 10.1016/j.tplants.2004.10.009. [DOI] [PubMed] [Google Scholar]
- Ruiz O, Daniell H. Engineering cytoplasmic male sterility via the chloroplast genome. Plant Physiology. 2005;138:1232–1246. doi: 10.1104/pp.104.057729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini HS. Effects of water stress on male gametophyte development in plants. Sexual Plant Reproduction. 1997;10:67–73. [Google Scholar]
- Saini HS, Sedgley M, Aspinall D. Developmental anatomy in wheat of male sterility induced by heat stress, water deficit or abscisic acid. Australian Journal of Plant Physiology. 1984;11:243–253. [Google Scholar]
- Schaewen von A, Stitt M, Schmidt R, Sonnewald U, Willmitzer L. Expression of a yeast-derived invertase in the cell wall of tobacco and Arabidopsis plants leads to accumulation of carbohydrate and inhibition of photosynthesis and strongly influences growth and phenotype of transgenic tobacco plants. EMBO Journal. 1990;9:3033–3044. doi: 10.1002/j.1460-2075.1990.tb07499.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherson SM, Alford HL, Forbes SM, Wallace G, Smith SM. Roles of cell-wall invertases and monosaccharide transporters in the growth and development of Arabidopsis. Journal of Experimental Botany. 2003;54:552–531. doi: 10.1093/jxb/erg055. [DOI] [PubMed] [Google Scholar]
- Shivanna KR, Sawhney VK. Polyethylene glycol improves the in vitro growth of Brassica pollen tubes without loss in germination. Journal of Experimental Botany. 1995;46:1771–1774. [Google Scholar]
- Sturm A. Invertases. Primary structures, functions, and roles in plant development and sucrose partitioning. Plant Physiology. 1999;121:1–7. doi: 10.1104/pp.121.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sturm A, Tang GQ. The sucrose-cleaving enzymes of plants are crucial for development, growth and carbon partitioning. Trends in Plant Science. 1999;4:401–407. doi: 10.1016/s1360-1385(99)01470-3. [DOI] [PubMed] [Google Scholar]
- Tymowska-Lalanne Z, Kreis M. Expression of the Arabidopsis thaliana invertase gene family. Planta. 1998;207:259–265. doi: 10.1007/s004250050481. [DOI] [PubMed] [Google Scholar]
- Van den Ende W, De Coninck B, Clerens S, Vergauwen R, Van Laere A. Unexpected presence of fructan 6-exohydrolases (6-FEHs) in non-fructan plants. Characterization, cloning, mass mapping and functional analysis of a novel ‘cell-wall invertase-like’ specific 6-FEH from sugar beet (Beta vulgaris L.) The Plant Journal. 2003;36:697–710. doi: 10.1046/j.1365-313x.2003.01912.x. [DOI] [PubMed] [Google Scholar]
- Van den Ende W, De Coninck B, Van Laere A. Plant fructan exohydrolases: a role in signaling and defense? Trends in Plant Science. 2004;9:523–528. doi: 10.1016/j.tplants.2004.09.008. [DOI] [PubMed] [Google Scholar]
- Van den Ende W, Michiels A, De Roover J, Verhaert P, Van Laere A. Cloning and functional analysis of chicory root fructan1-exohydrolase I (1-FEH I): a vacuolar enzyme derivedfrom a cell-wall invertase ancestor? Mass fingerprint of the 1-FEH I enzyme. The Plant Journal. 2000;24:447–456. doi: 10.1046/j.1365-313x.2000.00890.x. [DOI] [PubMed] [Google Scholar]
- Van den Ende W, Michiels A, Le Roy K, Van Laere A. Cloning of a vacuolar invertase from Belgian endive leaves (Cichorium intybus) Physiologia Plantarum. 2002;115:504–512. doi: 10.1034/j.1399-3054.2002.1150404.x. [DOI] [PubMed] [Google Scholar]
- Van den Ende W, Michiels A, Van Wonterghem D, Clerens SP, De Roover J, Van Laere AJ. Defoliation induces fructan 1-exohydrolase II in Witloof chicory roots. Cloning and purification of two isoforms, fructan 1-exohydrolase IIa and fructan 1-exohydrolase IIb. Mass fingerprint of the fructan 1-exohydrolase II enzymes. Plant Physiology. 2001;126:1186–1195. doi: 10.1104/pp.126.3.1186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Riet L, Nagaraj V, Van den Ende W, Clerens S, Wiemken A, Van Laere A. Purification, cloning and functional characterization of a fructan 6-exohydrolase from wheat (Triticum aestivum L.) Journal of Experimental Botany. 2006;57:213–223. doi: 10.1093/jxb/erj031. [DOI] [PubMed] [Google Scholar]
- Vargas W, Cumino A, Salerno GL. Cyanobacterial alkaline/neutral invertases. Origin of sucrose hydrolysis in the plant cytosol? Planta. 2003;216:951–960. doi: 10.1007/s00425-002-0943-x. [DOI] [PubMed] [Google Scholar]
- Vargas WA, Pontis HG, Salerno GL. Differential expression of alkaline and neutral invertases in response to environmental stresses: characterization of an alkaline isoform as a stress-response enzyme in wheat leaves. Planta. 2007;226:1535–1545. doi: 10.1007/s00425-007-0590-3. [DOI] [PubMed] [Google Scholar]
- Vargas WA, Pontis HG, Salerno GL. New insights on sucrose metabolism: evidence for an active A/N-Inv in chloroplasts uncovers a novel component of the intracellular carbon trafficking. Planta. 2008;277:795–807. doi: 10.1007/s00425-007-0657-1. [DOI] [PubMed] [Google Scholar]
- Vijn I, van Dijken A, Luscher M, Bos A, Smeets E, Weisbeek P, Wiemken A, Smeekens S. Cloning of sucrose:sucrose 1-fructosyltransferase from onion and synthesis of structurally defined fructan molecules from sucrose. Plant Pysiology. 1998;117:1507–1513. doi: 10.1104/pp.117.4.1507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J, Huang S, Fosu-Nyarko J, Dell B, McNeil M, Waters I, Moolhuijzen P, Conocono E, Appels R. The genome structure of the 1-FEH genes in wheat (Triticum aestivum L.): new markers to track stem carbohydrates and grain filling QTLs in breeding. Molecular Breeding. 2008;22:339–351. [Google Scholar]