Abstract
Cinnamyl alcohol dehydrogenase (CAD) catalyses the final step in the biosynthesis of monolignols. In the present study, a cDNA encoding a CAD was isolated from wheat, designated as TaCAD1. A genome-wide data mining in the wheat EST database revealed another 10 CAD-like homologues, namely TaCAD2 to TaCAD11. A phylogenetic analysis showed that TaCAD1 belonged to the bona fide CAD group involved in lignin synthesis. Two other putative CADs from the wheat genome (TaCAD2 and TaCAD4) also belonged to this group and were very close to TaCAD1, but lacked C-terminal domain, suggesting that they are pseudogenes. DNA gel blot analysis for the wheat genome showed two to three copies of CAD related to TaCAD1, but RNA gel blot analysis revealed only single band for TaCAD1, which was highly expressed in stem, with quite low expression in leaf and undetectable expression in root. The predicted three-dimension structure of TaCAD1 resembled that of AtCAD5, but two amino acid substitutions were identified in the substrate binding region. Recombinant TaCAD1 protein used coniferyl aldehyde as the most favoured substrate, also showed high efficiencies toward sinapyl and p-coumaryl aldehydes. TaCAD1 was an enzyme being pH-dependent and temperature-sensitive, and showing a typical random catalysing mechanism. At the milky stage of wheat, TaCAD1 mRNA abundance, protein level and enzyme activity in stem tissues were higher in a lodging-resistant cultivar (H4546) than in lodging-sensitive cultivar (C6001). These properties were correlated to the lignin contents and lodging indices of the two cultivars. These data suggest that TaCAD1 is the predominant CAD in wheat stem for lignin biosynthesis and is critical for lodging resistance.
Keywords: Triticum aestivum L., cinnamyl alcohol dehydrogenase (CAD), lodging resistance, lignin biosynthesis
Introduction
Lignin is frequently a major structural component of secondary cell walls in vascular plants, conferring mechanical strength as well as hydrophobicity to the plant body. Lignin is derived from dehydrogenative polymerization of lignin subunits (monolignols); the three main monolignols being p-coumaryl, coniferyl, and sinapyl alcohols. The biochemical pathway leading to the formation of monolignols consists of successive hydroxylation and O-methylation of the aromatic ring before conversion of the side-chain carboxyl to an alcohol function. Cinnamyl alcohol dehydrogenase (CAD, EC 1.1.1.195) catalyses the conversion of the corresponding cinnamyl aldehydes to cinnamyl alcohols; this is the last step in the synthesis of monolignols before their polymerization in cell walls. Hence CAD is a key enzyme in monolignol biosynthesis (Baucher et al., 1996), which has been characterized in many plant species, including Aralia cordata (Hibino et al., 1993), tobacco (Halpin et al., 1992), Eucalyptus gunnii and E. globulus (Hawkins and Boudet, 1994; Melis et al., 1999), loblolly pine (O'Malley et al., 1992), lucerne (Brill et al., 1999), poplar (Van Doorsselaere et al., 1995), and strawberry (Blanco-Portales et al., 2002). According to these data, CAD displays distinct features between gymnosperms and angiosperms. Gymnosperm CAD is encoded by a single gene, and is believed to be highly specific for the reduction of coniferyl aldehyde, and with lower catalytic activity toward sinapyl aldehyde (O'Malley et al., 1992; Galliano et al., 1993). On the contrary, angiosperm CAD has multiple isoforms that have significant affinity for both coniferyl and sinapyl aldehydes (Brill et al., 1999). An in silico analysis of the Arabidopsis genome has identified nine CAD-homologues, but only three (AtCAD5, AtCAD4, and AtCAD1) were demonstrated to be the main enzymes involved in monolignol biosynthesis (Kim et al., 2004; Sibout et al., 2005; Eudes et al., 2006). The functions of other isoforms remain elusive.
There are only a few reports on CAD from monocot plants. CAD cDNAs have been isolated from ryegrass (Lynch et al., 2002), sugarcane (Selman-Housein et al., 1999), and sorghum (Tsuruta et al., 2007). Some brown-midrib mutants from maize and sorghum have been characterized; these mutations were shown directly to affect CAD expression (Halpin et al., 1998; Saballos et al., 2009; Sattler et al., 2009). By a map-based cloning approach to the gh2 (Gold Hull and Internode 2) rice mutant, this gene was shown to encode a CAD (Zhang et al., 2006). Recently, 12 and 14 CAD homologues were identified in the genomes of rice (Tobias and Chowk, 2005) and sorghum (Saballos et al., 2009), respectively. OsCAD2 and SbCAD2 have been shown to be responsible for lignin biosynthesis, being related to hull and internode phenotypes in rice (Zhang et al., 2006) and sorghum (Sattler et al., 2009), respectively. There is much to learn about CADs in monocot plants.
We are particularly interested in lignin biosynthesis in wheat (Triticum aestivum L.). Wheat crops provide not only the number one food grain for humans, but also an important lignocellulosic biomass, the straw. In China alone, annual production of wheat straw exceeds 147 mt (Liu et al., 2008). Lignin determines the quality of wheat straw as a valuable biomass. More importantly, lignin is a factor of crop lodging (Ma, 2009). The biochemical characterization and physiological analyses of cinnamoyl CoA reductase (CCR) and caffeic acid O-methyltransferase (COMT) from wheat (Ma, 2007; Ma and Xu, 2008) have previously been reported. Here, the characterization of another important lignin biosynthetic gene, CAD, and its functions in wheat stem development and lodging-resistance is reported.
Materials and methods
Plant materials
Two wheat cultivars, C6001 and H4564, were used; C6001 is lodging-sensitive while H4564 is lodging-resistant (Ma, 2009). Plants were grown in a naturally lit greenhouse with standard irrigation and fertilization. Our first collection of stem, leaf, and root tissues was done from young plants with 2 or 3 visible internodes, and then more tissues were collected at 3-week intervals until one week before anthesis. For each collection, tissues were immediately frozen and stored in liquid nitrogen until being used.
RNA and DNA isolation
Total RNA was isolated from wheat tissues using TRI reagent (Molecular Research Center, Cincinnati, USA). Poly(A)+ RNA was isolated using PolyAT tractR mRNA Isolation Kit (Promega). Genomic DNA was purified from wheat leaf tissues according to Dellaporta et al. (1983).
Isolation of a wheat CAD gene
A wheat stem cDNA library in Lambda Uni-Zap XR Vector was constructed with wheat cultivar H4564 using Stratagene kits (Ma, 2007). A wheat CAD probe was isolated by the rapid amplification of cDNA ends method (Ma, 2007). RT-PCR was carried out using the following primers:
forward primers:
A1: 5′-TA(T/C)CC(T/C/A/G)ATGGT(T/C/A/G)CC(T/C/A/G)GG(T/C/A/G)C-3′,
A2: 5′-AA(T/C)GA(T/C)GT(T/C/A/G)(T/A)A(T/C)AC(T/C/A/G)GA(T/C)G-3′,
and reverse primers:
A3: 5′-GACTCGAGTCGACATCG-3′,
A4: 5′-AC(A/G)TC(T/C/A/G)AC(T/C/A/G)AC(A/G)AA(T/C)C(T/G)(A/G)TA-3′. Two rounds of PCR reactions were conducted to amplify the specific CAD sequence. The first round PCR used the primers A1 and A3, and second round PCR (nested PCR) used primers A2 and A3. About 2×105 recombinant phages were transferred onto Hybond-N+ membrane (Amersham Bioscience, Piscataway, NJ, USA) and hybridized with a 32P-labelled CAD probe using standard procedure at 42 °C (Sambrook et al., 1989). Positive isolates were purified by three rounds of plating and hybridization. The insert cDNA was cloned into pBluescript SK+ vector and sequenced using an ABI 377 DNA Sequencer.
Bioinformatics analysis
Sequence similarities were analysed using the SIM-Alignment Tool (Altschul et al., 1997). Evolutionary relationships were studied using the Clustal W method with PAM 250 residue weight table (Thompson et al., 1994). Secondary and three-dimensional structures were predicted using SWISS-MODEL Workspace (http://swissmodel.expasy.org/workspace) and a three-dimensional figure was prepared with the program MOLMOL (Koradi et al., 1996) (http://www.mol.biol.ethz.ch/wuthrich/software/molmol). The wheat EST database (http://compbio.dfci.harvard.edu/cgi-bin/tgi/gimain.pl?gudb=wheat) was blasted to search for CAD-like sequences.
DNA and RNA gel blot analysis
Wheat genomic DNA was digested with EcoRI, BamHI, HindIII, or XbaI and resolved on a 1.0% agarose gel. Ten μg of total RNA was run on 1.4% (w/v) formaldehyde agarose gels. DNA and RNA were blotted and hybridized according to the procedure described by Ma (2007). RNA hybridization signals were normalized by a soybean 18S ribosomal RNA (Eckenrode et al., 1985).
Functional expression of the wheat CAD in E. coli
PCR was used to introduce an EcoRI site at the 5′ end and a NotI site at the 3′ end of the TaCAD1 coding region. The primers used were: 5′-CGGAATTCATGGGCAGCGTCGACGCCTC-3′ (forward primer) and 5′-ATAAGAATGCGGCCGCTCAGGCGGCGTCCTCGATG-3′ (reverse primer). The accuracy of the PCR amplification was confirmed by sequencing. The amplified product was double-digested with EcoRI/NotI and the resultant fragment was cloned into the same sites of the pET-28a vector (Novagen). The engineered plasmid was transformed into E. coli BL21 (DE3) cells (Novagen). As a control, BL21 (DE3) cells were transformed with the empty pET-28a vector. The growth and induction of bacterial cells were optimized according to the manufacturer's instructions. Proteins were extracted and then subjected to denaturing SDS-PAGE TRIS-glycine gels (Novex, San Diego, CA). A Kaleidoscope pre-stained standard (Bio-Rad) was used as a molecular weight marker. The gels were stained with Coomassie blue R250. Purification of recombinant protein was performed using Ni-NTA His-Bind Resin (Novagen) according to the procedure of Ma and Tian (2005).
CAD enzyme kinetic assay
CAD enzyme activity was measured according to the method of Goffner et al. (1992). For each reaction, 10 min of declination at OD340 was monitored automatically with 1 min intervals. Km and Vmax values were determined by extrapolation from Lineweaver-Burke plots. The effects of pH on enzyme activity were determined after the reaction mixture was incubated in phosphate potassium buffer with the required pH for 15 min. The stability of enzyme was analysed after incubating the reaction mixture at different temperatures.
Protein extraction from wheat tissues and gel blot analysis
Wheat stem tissues were ground in liquid nitrogen. The powdered tissues were extracted for 1 h at 4 °C in extraction buffer (100 mM TRIS-HCl pH 7.5, 0.2 mM MgCl2, 2 mM DTT, and 10% glycerol). The samples were spun at 12 000 g for 10 min at 4 °C and the supernatant was desalted on PD-10 columns (Pharmacia, Piscataway, NJ, USA). Protein samples were resolved by 12% SDS-PAGE, and then electro-transferred onto a nitrocellulose membrane. The protein gel blot analysis was carried out according to the procedure described by Ma (2009). The blot was probed with polyclonal anti-maize CAD serum (Halpin et al., 1998) followed by a peroxidase-conjugated goat anti-rabbit polyclonal antibody (Bio-Rad Laboratories Beijing Ltd., Beijing 100086, China) and detected using the ECL kit (Amersham).
CAD enzyme activity in wheat was determined in the soluble protein fraction from stem tissues. Protein concentrations were determined by the Bradford assay (Bradford, 1979) with BSA as standard.
Results
Characterization of the wheat CAD
RT-PCR with degenerate primers for CAD produced a 660 bp cDNA fragment. Using this fragment as a probe, coupled with high-stringency hybridization, five positive plaques were detected in the wheat stem cDNA library. Restriction analysis showed that all these isolates had the same restriction fragment pattern. DNA sequence analysis showed that they belonged to the same cDNA, which was designated as TaCAD1 (GenBank accession no GU563724). This cDNA was 1311 nucleotides in length with a 1083-nucleotide open reading frame (ORF), flanked by 5′ and 3′ untranslated regions of 87 and 141 nucleotides, respectively. The GC content of the ORF was 66.1%, which is similar to that of other monocot CAD genes (Campbell and Gowri, 1990). The deduced translation product of TaCAD1 consists of 360 amino acids with a relative molecular mass of 38 624 Da and a theoretical isolectric point of 5.926.
The deduced amino acid sequence contained a highly conserved motif in CAD, 188GLGGVG193, which is known to participate in binding the pyrophosphate group of NADP+ through a helical dipole (Youn et al., 2006). In AtCAD5 (from Arabidopsis), there is a putative, substrate-binding pocket of 12 residues: T49, Q53, L58, M60, C95, W119, V276, P286, M289, L290, F299, and I300 (Youn et al., 2006). In TaCAD1, only two residues were different; A60 and V95.
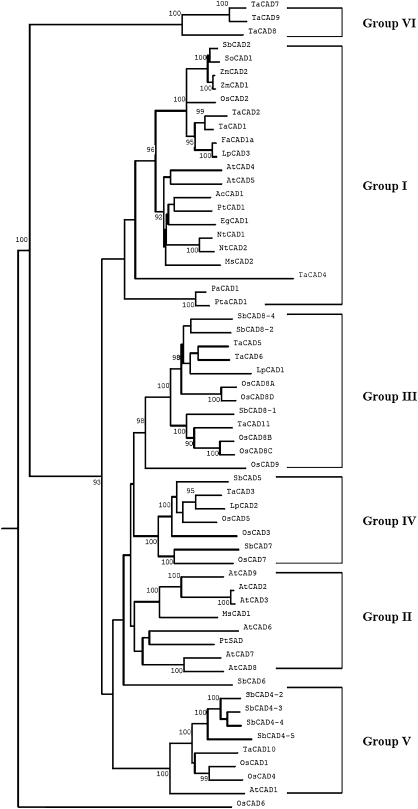
Since the wheat genome is not yet fully sequenced, the wheat EST database was used to identify CAD-like sequences with the TaCAD1 amino acid sequence as a query. Ten putative CAD of full-length isoforms were obtained, they were named TaCAD2, TaCAD3,… TaCAD11. These 11 wheat CAD sequences were used to construct a phylogenetic tree, along with CAD sequences from a few other plants. The tree revealed six groups of CAD (nomenclature of Saballos et al., 2009). Group I contained sequences identified as the bona fide CAD enzymes from gymnosperms, monocot and dicot angiosperms. A majority of these CADs have been demonstrated to be involved in lignin biosynthesis. This group is further divided into three subgroups, which included CADs from dicots, monocots, and gymnosperms, respectively. Group II contained CAD sequences from Arabidopsis, alfalfa, and poplar. It is worth noting that a sinapyl alcohol dehydrogenase (SAD) from poplar (Li et al., 2001) is a member of this group, although this type of enzyme has not been identified from other angiosperms. Groups III and IV represent CAD sequences from monocot plants. OsCAD7 in group IV has been shown to affect the culm strength of rice in a recent report (Li et al., 2009). Group V contained CAD sequences from rice, sorghum, and wheat, and also included AtCAD1. The latter has been demonstrated partly to complement AtCAD4 and AtCAD5 mutants (Eudes et al., 2006). Group VI, which was a new group identified in the present analysis, included OsCAD6, TaCAD7, TaCAD8, and TaCAD9. It is still questionable for this group as the sequence resources are probably too limited for definitive conclusions.
There were three wheat CAD sequences in Group I. TaCAD2 and TaCAD1 were closely related. Although sequence comparison revealed a major difference between the two, with TaCAD2 lacking about 40 amino acids in its C-terminus, the identity in the rest of the amino acid sequences was over 94%. TaCAD2 is possibly a pseudogene of TaCAD1. A third wheat CAD in this group is TaCAD4. It has only 217 amino acids and has diverged from other bona fide CAD enzymes from monocot plants. The functions of this CAD warrant investigation.
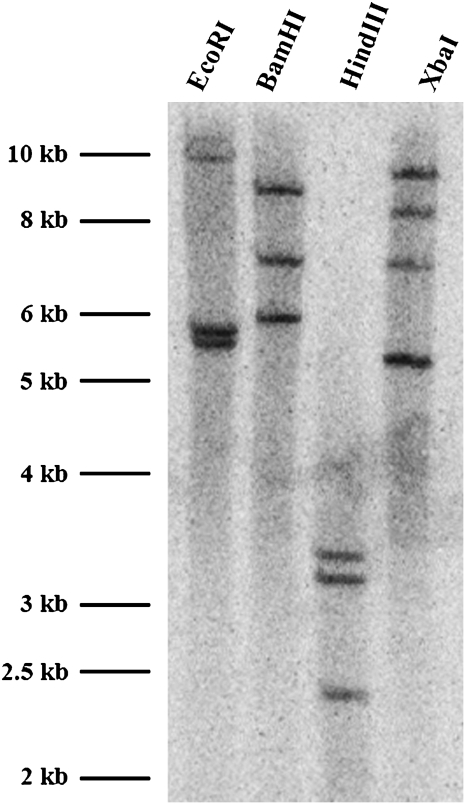
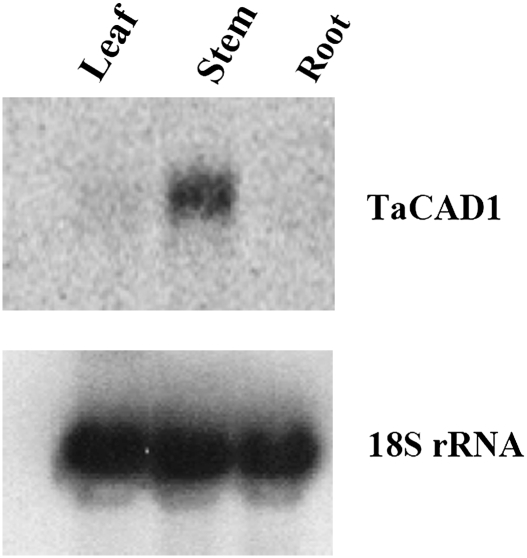
Genomic DNA of wheat was digested with restriction enzymes EcoRI, BamHI, HindIII, or XbaI, to determine the copy number of TaCAD1. In TaCAD1 cDNA, there were no restriction sites for these enzymes. The blot was probed with the coding region of TaCAD1, and two strong bands were detected in EcoRI digestion, three bands each in BamH1 and HindIII digestions, and four bands in the XbaI digestion (Fig. 2). These digestion patterns suggested that two or three copies of CAD genes were present in the wheat genome. This result is consistent with data from the wheat EST database, where two possible CAD isoforms (TaCAD2 and TaCAD4) are close to TaCAD1 (Fig. 1). Indeed, Northern blot analyses revealed a single band of the size of TaCAD1 cDNA (Fig. 3). This indicated that TaCAD2 and TaCAD4 were either not expressed or could not be recognized by the TaCAD1 probe. Results also showed that the TaCAD1 transcript was most abundant in stem tissues and with a much lower abundance in leaf tissues. No signal was detected in root tissues (Fig. 3).
Fig. 2.
DNA gel blot analysis of genomic DNA isolated from wheat leaf tissues. Each lane was loaded with 10 μg DNA, and the gel was probed with TaCAD1 cDNA. The restriction enzymes used are shown.
Fig. 1.
Dendrogram for phylogenetic relationships of CAD proteins from various plants. The tree was constructed using Clustal W method with PAM 250 residue weight table and boot-strap values were shown in each branch. GenBank accession numbers are as follows: Arabidopsis thaliana: AtCAD1 (AY288079), AtCAD2 (AY302077), AtCAD3 (AY302078), AtCAD4 (AY302081), AtCAD5 (AY302082), AtCAD6 (AY302075), AtCAD7 (AY302079), AtCAD8 (AY302080), and AtCAD9 (AY302076); Oryza sativa: OsCAD1 (AAN09864), OsCAD2 (DQ234272), OsCAD3 (AAP53892), OsCAD4 (BK003970), OsCAD5 (BK003971), OsCAD6 (CAD39907), OsCAD7 (CAE05206), OsCAD8A to D (BK003972), and OsCAD9 (AAN05338); Sorghum bicolor: SbCAD2 (Sb04g005950), SbCAD4-2 (Sb10g006300), SbCAD4-3 (Sb10g006290), SbCAD4-4 (Sb10g006280), SbCAD4-5 (Sb10g006270), SbCAD5 (Sb07g006090), SbCAD6 (Sb06g001430), SbCAD7 (Sb06g028240), SbCAD8-1 (Sb02g024220), SbCAD8-2 (Sb02g024210), and SbCAD8-4 (Sb02g024190); Triticum aestivum: TaCAD1 (GU563724), TaCAD2 (TC143210), TaCAD3 (TC143265), TaCAD4 (TC144004), TaCAD5 (TC149391), TaCAD6 (TC149393), TaCAD7 (TC170425), TaCAD8 (TC170426), TaCAD9 (TC170429), TaCAD10 (TC172690), and TaCAD11 (TC179401); Aralia cordata: AcCAD1 (D13991); Eucalyptus globulus: EgCAD1 (AF038561); Festuca arundinacea: FaCAD1a (AF188292); Lolium perenne: LpCAD1 (AF472591), LpCAD2 (AF472592), and LpCAD3 (AF010290); Medicago sativa: MsCAD1 (AF083333) and MsCAD2 (AF083332); Nicotiana tabacum: NtCAD1 (X62343) and NtCAD2 (X62344); Picea abies: PaCAD1 (X72675); Populus tremuloides: PtCAD1 (AF217957) and PtSAD (AF273256); Pinus taeda: PtaCAD1 (Z37992); Saccharum officinarum: SoCAD1 (AJ231135); Zea mays: ZmCAD1 (AJ005702) and ZmCAD2 (Y13733).
Fig. 3.
RNA gel blot analysis of TaCAD1 gene expression in wheat leaf, stem, and root tissues. Each lane was loaded with 10 μg total RNA. Hybridization with an 18S rDNA probe was included for the intactness of the RNA preparations and to serve as an internal control for gel loading and blotting.
Predicted secondary and three-dimensional structures of TaCAD1
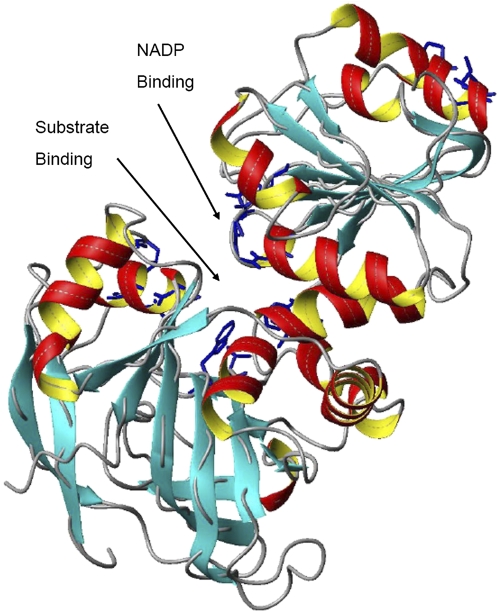
The predicted secondary and three-dimensional structures of TaCAD1 were very similar to AtCAD5 that had been analysed by crystallography (Youn et al., 2006, http://www.rcsb.org/pdb/cgi/ explore.cgi; pdbId: 2CF6). α helices and β sheets were the basic elements of the TaCAD1 structure, which were interlaced with random coils, bends, and turns (Fig. 4). The whole protein of TaCAD1 was composed of the catalytic and nucleotide-binding domains, each with six helices plus 12 sheets and five helices plus five sheets of secondary elements. One major difference was that, in TaCAD1, a new helix element was predicted in residues 286SPMVLG292, which was located between βE and βF. This interface was mainly due to several amino acid substitutions, which included L285 to S, T286 to S, L288 to M, and L289 to V. However, this new helix had merged with βE and appeared to have no influence on the whole TaCAD1 structure. Another difference between the two CADs involved two amino acids in the substrate binding site, where M60 and C95 in AtCAD5 were replaced by A60 and V95 in TaCAD1.
Fig. 4.
The predicted three-dimensional structure of TaCAD1. Data were analysed by the SWISS-MODEL software, and the figure was prepared with the program MOLMOL. α-helices are indicated in red and yellow, β-sheets by arrows, and turns and coils by curled lines. The NADPH binding and substrate binding sites are also indicated. (This figure is available in colour at JXB online.)
Properties of the recombinant TaCAD1 enzyme
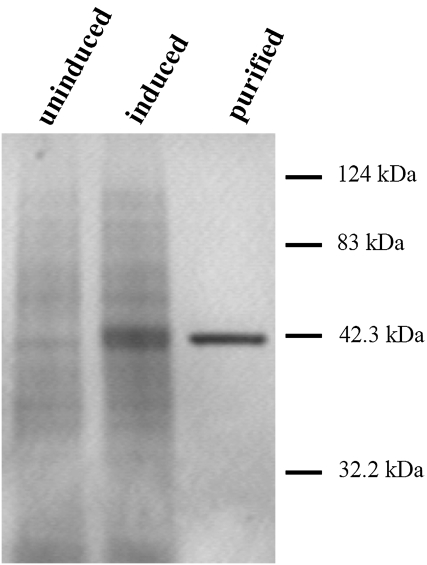
TaCAD1 was expressed in E. coli as a His-tagged protein with a relative molecular mass of about 42.5 kDa. This protein was purified to homogeneity (Fig. 5), and its kinetics toward the five possible substrates was analysed. The overall catalytic properties were revealed by Km, Vmax, and Kenz (Kcat/Km) values (Table 1). According to the calculated catalytic efficiency (Kenz) of TaCAD1, it showed the lowest catalytic efficiencies towards 5-OH-coniferyl and caffeoyl aldehydes. Higher catalytic efficiencies were observed for sinapyl and p-coumaryl aldehydes, and an even higher efficiency was documented for coniferyl aldehyde. These results were consistent with the proposed pathway of these three substrates in the synthesis of syringyl, coumaryl, and guaiacyl subunits of lignin.
Fig. 5.
Purification of the recombinant TaCAD1 protein from E. coli. Total protein fractions were extracted from uninduced and IPTG-induced E. coli cultures harbouring the TaCAD1 expression vector. The TaCAD1 protein was purified using His-tag resin and was resolved by SDS-PAGE. The gel was stained with Coomassie blue R-250.
Table 1.
Kinetic parameters of the recombinant TaCAD1 protein
| Substrates | Vmax (nkat mg−1 protein) | Km (μM) | Kenz (S−1 M−1) |
| p-Coumaraldehyde | 159.92±12.20 | 27.16±4.23 | 250,210 |
| 5-OH-coniferaldehyde | 97.24±11.42 | 24.39±3.72 | 169,364 |
| Coniferaldehyde | 458.83±52.91 | 21.94±3.21 | 896,194 |
| Caffeoylaldehyde | 172.33±20.43 | 89.06±9.27 | 82,151 |
| Sinapaldehyde | 273.74±31.32 | 40.63±5.56 | 286,185 |
Values represent means of three independent replicates±standard deviation.
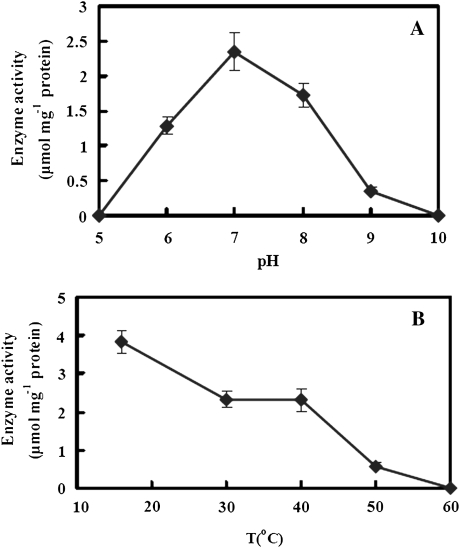
The enzymatic activity of TaCAD1 was pH-dependent, as evaluated in phosphate potassium buffer using coniferyl aldehyde and NADPH as substrates (Fig. 6A). TaCAD1 activity increased abruptly from pH 6.0 up to the optimal pH of 7.0. The enzyme still had >70% of its maximum activity at pH 8.0, but at pH 9.0 its activity was almost completely stopped. This suggests that TaCAD1 is strictly pH-dependent and this optimal pH fit well with the cytosolic localization of CAD. A similar result was reported for native CAD enzyme from Japanese black pine (Kutsuki et al., 1982).
Fig. 6.
Effects of pH (A) and temperature (B) on the recombinant TaCAD1 activity. Substrates were coniferyl aldehyde and NADPH, and a 200 mM phosphate potassium buffer was used. In (A), the temperature was 30 °C, and in (B), the pH was 7.0. All reaction mixtures were incubated in water bath for 15 min and then activity was determined. Each value is the mean of three independent samples and vertical bars represent standard errors.
TaCAD1 is also temperature-sensitive. An increase in temperature from 16 °C to 30 °C resulted in a decay in activity, but no further decay till 40 °C. However, a further increase in temperature to 50 °C resulted in about 85% of enzyme activity loss, and at 60 °C the enzyme lost its activity completely (Fig. 6B).
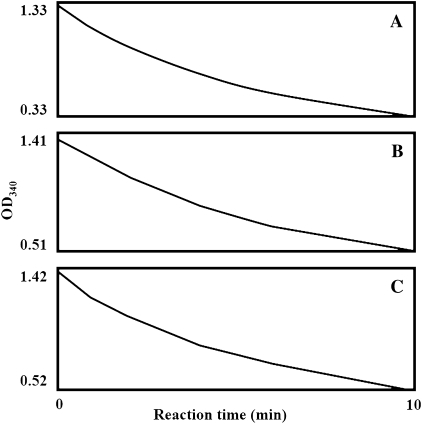
The initial reaction velocity was monitored immediately after the addition of all reaction components (coniferyl aldehyde, NADPH, and TaCAD1 protein). Reactions had taken place immediately, regardless of the order of the components being added (Fig. 7A, B, C). This was suggestive of the typical random mechanism of the TaCAD1 reaction.
Fig. 7.
Analysis of the reaction mechanism catalysed by TaCAD1. Each final reaction mixture contained 100 μM NADPH, 40 μM coniferyl aldehyde, and 1 μg purified recombinant TaCAD1 protein. For each reaction, a two-component mixture was prepared first and a third component was then added. The third (and last) component added was (A) TaCAD1 protein, (B) NADPH, and (C) coniferyl aldehyde. The X axis shows the time lapse after addition of the third component.
TaCAD1 gene expression and enzyme activity in relation to stem lodging resistance
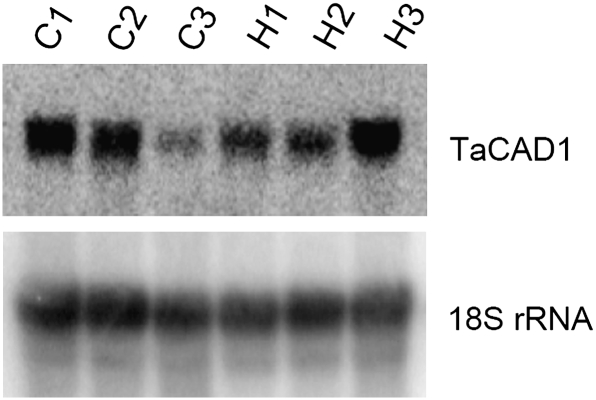
It was worth investigating the actions of TaCAD1 that relate to stem-specific biological functions such as mechanical support, since it is highly expressed in the stem (Fig. 3). Two wheat cultivars were examined; H4564 was lodging-resistant and C6001 was lodging-sensitive but the two cultivars had a similar developmental process (Ma, 2009). It is already known that H4564 had stronger stems and a lower lodging index than C6001, and these were attributable to the higher lignin synthesis in H4564 (Ma, 2009). In the present study, TaCAD1 expression was analysed in stems of different developmental stages (Fig. 8). TaCAD1 mRNA was highly expressed in stems at the elongation and heading stages in both cultivars. However, TaCAD1 mRNA levels declined at the milky stage in C6001, but remained high in H4564.
Fig. 8.
RNA gel blot analysis of TaCAD1 gene expression at different developmental stages of wheat stem. Ten μg total RNA was loaded in each lane. C1, C2, and C3: elongation, heading and milky stages of C600; H1, H2, and H3: elongation, heading and milky stages of H4564. Hybridization with an 18S rDNA probe was included for the intactness of the RNA preparations and to serve as an internal control for gel loading and blotting.
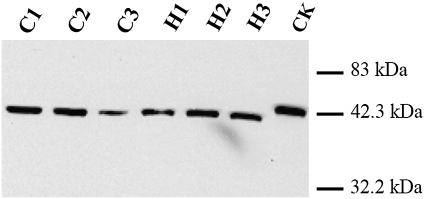
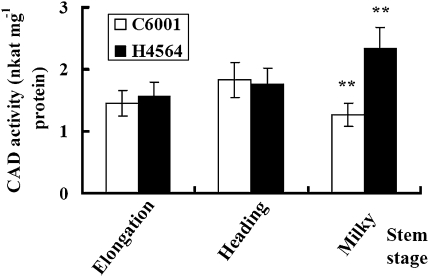
TaCAD1 protein levels in wheat stem tissues were determined by gel blot using a polyclonal antibody against maize CAD (Fig. 9). A single band was detected from wheat stem samples as for the recombinant TaCAD1 protein. Thus, TaCAD1 appeared to be the main isozyme in wheat stems. H4564 and C6001 were similar in the TaCAD1 protein level at the elongation and heading stages. At the milky stage, H4564 had a higher TaCAD1 protein level than C6001 (Fig. 9). In accordance with this observation, H4564 and C6001 displayed little difference in CAD enzymatic activity at the elongation and heading stages, but H4564 had 84.3% more CAD activity than C6001 at the milky stage (Fig. 10).
Fig. 9.
Protein gel blot analysis of TaCAD1 protein from wheat stem tissues. Each lane was loaded with 3.0 μg protein, and the recombinant TaCAD1 protein (CK, 10 ng) was used a positive control. The blot was probed with a polyclonal antiserum against maize CAD protein. C1, C2, and C3: elongation, heading and milky stages of C6001; H1, H2, and H3: elongation, heading and milky stages of H4564.
Fig. 10.
Comparison of wheat cultivars H4564 and C6001 in CAD activity (nkat mg−1 protein). Protein was from of stem tissues at three stages of development. Values are the means of four experiments, with standard errors. Probability values between H4564 and C6001 were estimated by the Student's t test and significant differences at P0.01 level was indicated by **.
Discussion
TaCAD1 is the predominant CAD isoform responsible for lignin synthesis in the wheat stem
CAD-like genes may exist in multiple isoforms, as revealed by genome-wide analyses from Arabidopsis, rice, and sorghum, but the numbers of the bona fide CAD enzymes known to be involved in lignin biosynthesis are rather limited. It is believed that there is one CAD enzyme in gymnosperms (O'Malley et al., 1992), and two or three CAD isoforms in each dicot species. For example, AtCAD5, AtCAD4, and AtCAD1 are the three main CADs that are responsible for lignin biosynthesis in Arabidopsis (Sibout et al., 2005; Eudes et al., 2006).
With Southern blot hybridization for wheat, two or three bands were detected (Fig. 2), suggesting the existence of two or three closely related TaCAD1 in the wheat genome. According to the wheat EST database, there are two possible CAD isoforms (TaCAD2 and TaCAD4) that may not have enzyme activity due to an incomplete C-terminus sequence (TaCAD2) or too short a peptide (TaCAD4). In addition, Northern blot (Fig. 3) and Western blot (Fig. 9) hybridizations showed one band corresponding to TaCAD1 in wheat. Similar situations are reported in other monocot plants. For example, three CAD cDNAs were identified from Lolium perenne, but only one (LpCAD3; Lynch et al., 2002) belongs to the bona fide CAD family (Fig. 1). From rice (Tobias and Chowk, 2005) and sorghum (Saballos et al., 2009), 12 and 14 CAD-like homologues were identified, respectively, but only one was showed to be responsible for lignin biosynthesis in either species (Zhang et al., 2006; Sattler et al., 2009). Taken together, it seems to be generally true that only one CAD is involved in lignin biosynthesis in monocot plants, as in gymnosperms, but unlike that in dicot plants.
A recent report, however, showed the complex situation for CAD (Li et al., 2009). A flexible culm1 (fc1) mutant was obtained from rice and this mutation was located on the gene that was previously identified as OsCAD7 (Fig. 1). The fc1 mutant caused a reduction in culm strength, as well as a decrease in CAD activity and lignin content. However, the amount of cellulose in fc1 culms was almost equally reduced to that of lignin. The culm strength in the second internode was reduced more than that in the first internode, but CAD activity exhibited the inverse proportion to culm strength. Since OsCAD7 does not belong the bona fide CAD family (Fig. 1), its biochemical nature needs further identification. In Arabidopsis, the AtCAD1 gene was shown partially to restore the AtCAD4/AtCAD5 double mutation (Eudes et al., 2006), but AtCAD1 protein lacked detectable CAD catalytic activities in vitro (Kim et al., 2007). In other experiments, a single mutation in AtCAD5 showed only a slight reduction of S lignin content (Sibout et al., 2003), only the AtCAD4/AtCAD5 double mutation was necessary to reduce the S lignin unit drastically (Sibout et al., 2005). In vitro analysis, however, showed that AtCAD4 could not use sinapyl aldehyde, the precursor for the S lignin unit (Kim et al., 2004). Clearly, more in-depth investigations on CAD functions are required, particularly in relation to in planta experiments (such as transgenic plants and mutation) and in vitro biochemical analysis.
TaCAD1 is very similar to other bona fide CADs, in both primary amino acid sequence (Fig. 1) and three-dimensional structure (Fig. 4). However, a major difference was found between TaCAD1 and AtCAD5: M60 and C95 in the substrate binding site of AtCAD5 were replaced by A and V, respectively, in TaCAD1. Interestingly, these two substitutions are conserved in all known monocot CADs. Saballos et al. (2009) suggested that these substitutions were likely to reduce substrate binding efficiency, and alter substrate specificity. According to our enzyme kinetic analyses, TaCAD1 (Kenz=896 000 S−1 M−1; Table 1) was comparable to both AtCAD5 (1 091 000 S−1 M−1) and AtCAD4 (74 000 S−1 M−1). Km values of TaCAD1 were also close to those of AtCAD5 and AtCAD4 (Kim et al., 2004). These data suggest that two amino acid substitutions in TaCAD1’s substrate binding site are not likely to reduce substrate binding efficiency. But the scenario for substrate specificity was different. Both AtCAD5 and AtCAD4 used p-coumaryl aldehyde as the most preferred substrate. AtCAD5 also used sinapyl and coniferyl aldehydes, with 64% and 32% activity, respectively, of the activity of p-coumaryl aldehyde. AtCAD4 readily used coniferyl aldehyde with 43% activity of p-coumaryl aldehyde but could not use sinapyl aldehyde. It was found that TaCAD1 used coniferyl aldehyde as the most preferred substrate, and its catalytic efficiencies for sinapyl and p-coumaryl aldehydes were only 32% and 28%, respectively, of the activity of coniferyl aldehyde. Apparently, the two amino acid substitutions in the TaCAD1 may be responsible for the observed differences in substrate specificity.
CAD's substrate specificity may link to the composition of lignin. The p-coumaryl, coniferyl, and sinapyl alcohols are monolignols that will give rise to the p-hydroxyphenyl (H), guaiacyl (G), and syringyl (S) units, respectively, of the lignin polymer. Lignin in dicot plants is mainly composed of both G and S units. In Arabidopsis, for example, the G, S, and H monomers in stem tissues accounted for 69.8%, 28.3%, and 1.9%, respectively, of the total lignin (Sibout et al., 2003). In wheat stem tissues, these monomer units were approximately 52.3%, 35.1%, and 12.6%, respectively, of the total lignin (Zhang et al., 2010). Therefore, the catalytic efficiencies of TaCAD1 towards the three substrates are nearly proportional to the final monomer compositions.
TaCAD1 was an enzyme whose activity is pH-dependent and temperature-sensitive (Fig. 6). Its temperature-sensitivity is particularly intriguing when compared with TaCCR1 (wheat cinnamoyl-CoA reductase 1) (Ma, 2007) and TaCM (wheat caffeic acid 3-O-methyltransferase) (Ma and Xu, 2008) in the lignin synthetic pathway of the same species. The latter enzymes did not exhibit such temperature sensitivity. It is unclear whether this is due to the binary complex required for CAD actions.
TaCAD1 showed a typical random reaction mechanism; in other words, TaCAD1 protein would randomly bind its aldehyde substrates and the co-enzyme NADPH to initiate the reaction (Fig. 7). This is in contrast with TaCCR2 (wheat cinnamoyl-CoA reductase 2), another NADP-dependent dehydrogenase in lignin biosynthesis, which needs to bind the co-enzyme NADPH ahead of its substrates (Ma and Tian, 2005). It is worth noting that a functional CAD exists as a dimmer and its catalytic and nucleotide-binding domains are located inside a cleft formed between the two subunits of the protein (Youn et al., 2006). This structural configuration could explain the random reaction mechanism of TaCAD1.
TaCAD1 is related to lodging resistance of wheat
TaCAD1 exhibited a unique organ-specific expression pattern. Its expression was high in stem, very low in the leaf, and undetectable in the root (Fig. 3). This is in contrast to situations in most other monocot plants examined. For example, CAD genes from rice, sugarcane (Selman-Housein et al., 1999), and ryegrass (Lynch et al., 2002) were expressed in both stem and root tissues, and ryegrass LpCAD3 was particularly abundant in root tissues. Both sorghum SbCAD2 (Saballos et al., 2009) and maize ZmCAD1 (Halpin et al., 1998) were expressed in most tissues, including leaf, stem, and root. The physiological implications of the different spatial distributions of the enzyme activities are not known. Nevertheless, it was noticed that the expression pattern of TaCAD1 was similar to TaCCR1 (Ma, 2007), another important wheat lignin gene. It would be interesting to know if these two enzymes are functional in the same physiological process, such as lodging-resistance.
Results for mRNA transcript abundance (Fig. 8), protein levels (Fig. 9), and enzyme activity (Fig. 10) of TaCAD1 all pointed to a connection of this enzyme with stem lodging-resistance. H4564 (lodging-resistant) and C6001 (lodging-sensitive) are remarkably different in all these aspects at the milky stage which is the time when lodging could be devastating in the field. A high TaCAD1 gene expression would lead to enhanced lignin synthesis and, consequently, increase the strength and lodging resistance of stem tissues. This physiological role of CAD has been confirmed in maize ((Halpin et al., 1998) and sorghum (Sattler et al., 2009) by using CAD mutants.
Earlier, it was reported that gene expression and enzyme activity of TaCCR1 (Ma, 2007) and TaCM (Ma, 2009), two other wheat lignin genes that are related to lodging-resistance, are higher in H4564 than in C6001 at the heading and milky stages. Now we know that expression of TaCAD1 is critical at the later stages. This temporal difference in gene expression coincides with the enzymes’ positions in the monolignol synthesis pathway: CCR and COMT in the early stage and CAD in the late stage of the pathway. Accumulation of lignin is the end result of well-orchestrated gene expression events. Clearly, the issue of lignin gene actions with stem lodging resistance is complex and warrants further intensive investigations.
Acknowledgments
This work was supported by the National High Technology and Research Development Program of PR China (‘863’ Program, No. 2007AA10Z101), the National Natural Science Foundation of China (No. 30970261), the Chinese National Special Foundations for Transgenic Plant Research and Commercialization (No. 2008ZX08002-003), and the Innovation Project of the Chinese Academy of Sciences. The author thanks Dr Claire Halpin, University of Dundee, Dundee, UK, for providing the anti-maize CAD serum, and Dr Fengshan Ma, Wilfrid Laurier University, Waterloo, Canada, for critical reading of the manuscript.
References
- Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman JD. Gapped Blast and PSI-Blast: a new generation of protein database search programs. Nucleic Acids Research. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baucher M, Chabbert B, Pilate G, et al. Red xylem and higher lignin extractability by down-regulating a cinnamyl alcohol dehydrogenase in poplar. Plant Physiology. 1996;112:1479–1490. doi: 10.1104/pp.112.4.1479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blanco-Portales R, Medina-Escobar N, López-Ráez JA, González-Reyes JA, Villalba JM, Moyano E, Caballero JL, Muñoz-Blanco J. Cloning, expression and immunolocalization pattern of a cinnamyl alcohol dehydrogenase gene from strawberry (Fragaria×ananassa cv. Chandler) Journal of Experimental Botany. 2002;53:1723–1734. doi: 10.1093/jxb/erf029. [DOI] [PubMed] [Google Scholar]
- Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein–dye binding. Analytical Biochemistry. 1976;72:248–254. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- Brill EM, Abrahams S, Hayes CM, Jenkins CLD, Watson JM. Molecular characterization and expression of a wound-inducible cDNA encoding a novel cinnamyl-alcohol dehydrogenase enzyme in lucerne. Plant Molecular Biology. 1999;41:279–291. doi: 10.1023/a:1006381630494. [DOI] [PubMed] [Google Scholar]
- Campbell WH, Gowri G. Codon usage in higher plants, green algae, and cyanobacteria. Plant Physiology. 1990;92:1–11. doi: 10.1104/pp.92.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dellaporta SL, Wood J, Hicks JB. A plant DNA minipreparation: version II. Plant Molecular Biology Reporter. 1983;1:19–21. [Google Scholar]
- Eckenrode VK, Arnold J, Meagher RB. Comparison of the nucleotide sequence of soybean 18S rRNA with the sequences of other small subunit rRNAs. Journal of Molecular Evolution. 1985;21:259–269. doi: 10.1007/BF02102358. [DOI] [PubMed] [Google Scholar]
- Eudes A, Pollet B, Sibout R, Do CT, Séguin A, Lapierre C, Jouanin L. Evidence for a role of AtCAD1 in lignification of elongating stems of Arabidopsis thaliana. Planta. 2006;225:23–39. doi: 10.1007/s00425-006-0326-9. [DOI] [PubMed] [Google Scholar]
- Galliano H, Cabane M, Eckerskorn C, Lottspeich F, Sandermann H, Ernst D. Molecular cloning, sequence analysis and elicitor-/ozone-induced accumulation of cinnamyl alcohol dehydrogenase from Norway spruce (Picea abies) Plant Molecular Biology. 1993;23:145–156. doi: 10.1007/BF00021427. [DOI] [PubMed] [Google Scholar]
- Goffner D, Joffroy I, Grima-Pettenati J, Halpin C, Knight ME, Schuch W, Boudet AM. Purification and characterization of isoforms of cinnamyl alcohol dehydrogenase (CAD) from eucalyptus xylem. Planta. 1992;188:48–53. doi: 10.1007/BF00198938. [DOI] [PubMed] [Google Scholar]
- Halpin C, Holt K, Chojecki J, Oliver D, Chabbert B, Monties B, Edwards K, Barakats A, Foxon GA. Brown-midrib maize (bm1): a mutation affecting the cinnamyl alcohol dehydrogenase gene. The Plant Journal. 1998;14:545–553. doi: 10.1046/j.1365-313x.1998.00153.x. [DOI] [PubMed] [Google Scholar]
- Halpin C, Knight ME, Grima-Pettenati J, Goffner D, Boudet A, Schuch W. Purification and characterization of cinnamyl alcohol dehydrogenase from tobacco stems. Plant Physiology. 1992;98:12–16. doi: 10.1104/pp.98.1.12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawkins SW, Boudet AM. Purification and characterization of cinnamyl alcohol dehydrogenase isoforms from the periderm of Eucalyptus gunnii Hook. Plant Physiology. 1994;104:75–84. doi: 10.1104/pp.104.1.75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibino T, Shibata D, Chen JQ, Higuchi T. Cinnamyl alcohol dehydrogenase from Aralia cordata: cloning of the cDNA and expression of the gene in lignified tissues. Plant and Cell Physiology. 1993;34:659–665. [Google Scholar]
- Kim SJ, Kim KW, Cho MH, Franceschi VR, Davin LB, Lewis NG. Expression of cinnamyl alcohol dehydrogenases and their putative homologues during Arabidopsis thaliana growth and development: lessons for database annotations? Phytochemistry. 2007;68:1957–1974. doi: 10.1016/j.phytochem.2007.02.032. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Kim MR, Bedgar D, Moinuddin SGA, Cardenas CL, Davin LB, Kang CCK, Lewis NG. Functional reclassification of the putative cinnamyl alcohol dehydrogenase multigene family in Arabidopsis. Proceedings of the National Academy of Sciences, USA. 2004;101:1455–1460. doi: 10.1073/pnas.0307987100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koradi R, Billeter M, Wuthrich K. MOLMOL: a program for display and analysis of macromolecular structures. Journal of Molecular Graphics. 1996;14:51–55. doi: 10.1016/0263-7855(96)00009-4. [DOI] [PubMed] [Google Scholar]
- Kutsuki H, Shimada M, Higuchi T. Regulatory role of cinnamyl alcohol dehydrogenease in the formation of guaiacyl and syringyl lignins. Phytochemistry. 1982;21:19–23. [Google Scholar]
- Li L, Cheng XF, Leshkevich J, Umezawa T, Harding SA, Chiang VL. The last step of syringyl monolignol biosynthesis in angiosperms is regulated by a novel gene encoding sinapyl alcohol dehydrogenase. The Plant Cell. 2001;13:1567–1585. doi: 10.1105/TPC.010111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Yang Y, Yao JL, Chen GX, Li XH, Zhang QF, Wu CY. FLEXIBLE CULM 1 encoding a cinnamyl-alcohol dehydrogenase controls culm mechanical strength in rice. Plant Molecular Biology. 2009;69:685–697. doi: 10.1007/s11103-008-9448-8. [DOI] [PubMed] [Google Scholar]
- Liu H, Jiang GM, Zhuang HY, Wang KJ. Distribution, utilization structure and potential of biomass resources in rural China: with special references of crop residues. Renewable and Sustainable Energy Reviews. 2008;12:1402–1418. [Google Scholar]
- Lynch D, Lidgett A, McInnes R, Huxley H, Jones E, Mahoney N, Spangenberg G. Isolation and characterisation of three cinnamyl alcohol dehydrogenase homologue cDNAs from perennial ryegrass (Lolium perenne L.) Journal of Plant Physiology. 2002;159:653–660. [Google Scholar]
- Ma QH. Characterization of a cinnamoyl-CoA reductase that is associated with stem development in wheat. Journal of Experimental Botany. 2007;58:2011–2021. doi: 10.1093/jxb/erm064. [DOI] [PubMed] [Google Scholar]
- Ma QH. The expression of caffeic acid 3- O-methyltransferase in two wheat genotypes differing in lodging resistance. Journal of Experimental Botany. 2009;60:2763–2771. doi: 10.1093/jxb/erp132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma QH, Tian B. Biochemical characterization of a cinnamoyl-CoA reductase from wheat. Biological Chemistry. 2005;386:553–560. doi: 10.1515/BC.2005.065. [DOI] [PubMed] [Google Scholar]
- Ma QH, Xu Y. Characterization of a caffeic acid 3- O-methyltransferase from wheat and its function in lignin biosynthesis. Biochimie. 2008;90:515–524. doi: 10.1016/j.biochi.2007.09.016. [DOI] [PubMed] [Google Scholar]
- Melis DLE, Whiteman PH, Stevenson TW. Isolation and characterisation of a cDNA clone encoding cinnamyl alcohol dehydrogenase in Eucalyptus globulus Labill. Plant Science. 1999;143:173–182. [Google Scholar]
- O'Malley DM, Porter S, Sederoff RR. Purification, characterization, and cloning of cinnamyl alcohol dehydrogenase in loblolly pine (Pinus taeda L. Plant Physiology. 1992;98:1364–1371. doi: 10.1104/pp.98.4.1364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saballos A, Ejeta G, Sanchez E, Kang C, Vermerris W. A genomewide analysis of the cinnamyl alcohol dehydrogenase family in sorghum (Sorghum bicolor (L.) Moench) identifies SbCAD2 as the brown midrib6 gene. Genetics. 2009;181:783–795. doi: 10.1534/genetics.108.098996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. Cold Spring Harbor: New York Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- Sattler SE, Saathoff AJ, Haas EJ, Palmer NA, Funnell-Harris DL, Sarath G, Pedersen JF. A nonsense mutation in a cinnamyl alcohol dehydrogenase gene is responsible for the sorghum brown midrib6 phenotype. Plant Physiology. 2009;150:584–595. doi: 10.1104/pp.109.136408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Selman-Housein G, Lopez MA, Hernandez D, Civardi L, Miranda F, Rigau J, Puigdomenech P. Molecular cloning of cDNAs coding for three sugarcane enzymes involved in lignification. Plant Science. 1999;143:163–171. [Google Scholar]
- Sibout R, Eudes A, Mouille G, Pollet B, Lapierre C, Jouanin L, Seguin A. CINNAMYL ALCOHOL DEHYDROGENASE-C and - D are the primary genes involved in lignin biosynthesis in the floral stem of Arabidopsis. The Plant Cell. 2005;17:2059–2076. doi: 10.1105/tpc.105.030767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sibout R, Eudes A, Pollet B, Goujon T, Mila I, Granier F, Seguin A, Lapierre C, Jouanin L. Expression pattern of two paralogs encoding cinnamyl alcohol dehydrogenases in Arabidopsis. Isolation and characterization of the corresponding mutants. Plant Physiology. 2003;132:848–860. doi: 10.1104/pp.103.021048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thompson JD, Higgins DG, Gibson TJ, Clustal W. Improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position specific gap penalties and weight matrix choices. Nucleic Acids Research. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tobias CM, Chowk EK. Structure of the cinnamyl alcohol dehydrogenase gene family in rice and promoter activity of a member associated with lignification. Planta. 2005;220:678–688. doi: 10.1007/s00425-004-1385-4. [DOI] [PubMed] [Google Scholar]
- Tsuruta S, Ebina M, Nakagawa H, Kawamura O, Akashi R. Isolation and characterization of cDNA encoding cinnamyl alcohol dehydrogenase (CAD) in sorghum (Sorghum bicolor (L.) Moench) Japanese Society of Grassland Science. 2007;53:103–109. [Google Scholar]
- Van Doorsselaere J, Baucher M, Feuillet C, Boudet AM, Van Montagu M, Inzé D. Isolation of cinnamyl alcohol dehydrogenase cDNAs from two important economic species: alfalfa and poplar. Demonstration of a high homology of the gene within angiosperms. Plant Physiology and Biochemistry. 1995;33:105–109. [Google Scholar]
- Youn B, Camacho R, Moinuddin SGA, Lee C, Davin LB, Lewis NG, Kang C. Crystal structures and catalytic mechanism of the Arabidopsis cinnamyl alcohol dehydrogenases AtCAD5 and AtCAD4. Organic and Biomolecular Chemistry. 2006;4:1687–1697. doi: 10.1039/b601672c. [DOI] [PubMed] [Google Scholar]
- Zhang JH, Deng HB, Lin L, Sun Y, Pan CS, Liu SJ. Isolation and characterization of wheat straw lignin with a formic acid process. Bioresource Technology. 2010;101:2311–1216. doi: 10.1016/j.biortech.2009.11.037. [DOI] [PubMed] [Google Scholar]
- Zhang KW, Qian Q, Huang ZJ, Wang YQ, Li M, Hong LL, Zeng DL, Gu MH, Chu CC, Cheng ZK. GOLD HULL AND INTERNODE2 encodes a primarily multifunctional cinnamyl-alcohol dehydrogenase (CAD) in Oryza sativa. Plant Physiology. 2006;140:972–983. doi: 10.1104/pp.105.073007. [DOI] [PMC free article] [PubMed] [Google Scholar]