Abstract
The function of monomeric GTPases of the RAS superfamily in fruit development and ripening has been partially characterized. Here the identification of peach (Prunus persica) small GTPases of the RAS superfamily expressed in fruit and the characterization of their expression profiles during fruit development are described. Extensive searches on expressed sequence tag (EST) databases led to the selection of a total of 24 genes from peach encoding proteins with significant similarity to Arabidopsis small GTPases. Sequence similarity analyses and identification of conserved motifs, diagnostic of specific RAS families and subfamilies, enabled bona fide assignment of fourteen PpRAB, seven PpARF/ARL/SAR, two PpROP and one PpRAN GTPases. Transcriptional expression profiles of peach monomeric GTPases, analysed by real-time quantitative reverse transcription-PCR, were obtained for mesocarp samples, collected in two consecutive years. Reproducible patterns of expression could be identified for five peach RAB-encoding genes (PpRABA1-1, PpRABA2, PpRABD2-1, PpRABD2-2, and PpRABC2), two ARFs (PpARFA1-1 and PpARLB1), and two ROPs (PpROP3 and PpROP4). Interestingly, the transient transcriptional up-regulation of PpARF genes and of PpRAB genes of the A and D clades, putatively controlling the exocytic delivery of cell wall components and modifying enzymes, appeared to coincide with peaks of growth speed and sugar accumulation and with the final phases of ripening. To our knowledge, this is the first description of the co-ordinated differential expression of a set of genes encoding small GTPases of the ARF and RAB families which takes place during key moments of fruit development and maturation.
Keywords: Fruit, GTPase, peach, ripening, trafficking, vesicle
Introduction
Small GTPases, proteins with molecular masses of between 21 kDa and 30 kDa, are monomeric guanine nucleotide-binding proteins related to the α subunit of heterotrimeric G proteins (Yang, 2002). They are universal molecular switches, regulating several cellular processes such as vescicle trafficking, cytoskeletal dynamics, cell polarity, and gene expression, through their cycling between an ‘activated’ state, when bound to GTP, and a GDP-bound inactive state. Upon stimulation by an upstream signal, the GTP-bound active form interacts with specific downstream effector proteins, which leads to the regulation of cellular responses and developmental processes that are highly conserved throughout eukaryotes. Monomeric GTPases are grouped into the RAS superfamily, named after the founding members of human Ras genes, which are homologous to the viral Ras oncogene (Yang, 2002). Members of this superfamily are structurally and functionally classified into the RAS, RHO, RAB, ARF, and RAN families. These families perform specific essential functions in the cell and are found in all eukaryotes. One notable exception is that plant genomes do not encode RAS proteins (Vernoud et al., 2003; Molendijk et al., 2004; Ma, 2007).
RHO proteins, organized into RHO, RAC, and CDC42 subfamilies in yeast and animals, are involved in the regulation of a variety of cellular processes ranging from the organization of the actin cytoskeleton, the establishment of cell polarity, and vescicle trafficking, to the control of gene expression. In plants, all small GTP-binding proteins segregating within the RHO family appear to be members of a unique subfamily called the ROPs (for RHO-related proteins of plants). ROPs are distinct from other RHO proteins in several aspects. Their effector domain contains several amino acid residues unique to ROPs and their insert region consists of 8–10 amino acid residues that share little homology with those (12 residues) found in other RHO proteins (Ma, 2007; Berken and Wittinghofer, 2008). These unique features suggest that plant ROPs may have unique effectors, distinct from those of animals (Zheng and Yang, 2000; Nibau et al., 2006; Yang and Fu, 2007; Berken and Wittinghofer, 2008). Studies in different plant species have established that ROPs regulate cellular oxidative environments and behave as important signalling modules in the integration of plant responses to external stimuli (Berken, 2006; Nibau et al., 2006). In Arabidopsis, oxygen deprivation rapidly and transiently activates ROP, resulting in NADPH oxidase-dependent H2O2 accumulation (Baxter-Burrell et al., 2002). ROP-mediated control of H2O2 production is required to trigger defence responses to various biotic and abiotic stress factors (Ono et al., 2001; Agrawal et al., 2003; Berken, 2006). Finally, ROP GTPases are also involved in signal transduction pathways mediated by plant hormones such as abscisic acid (ABA), auxin, and brassinolide (reviewed by Molendijk et al., 2004; Berken, 2006; Nibau et al., 2006).
RAB and ARF GTPases are major components of the vescicle trafficking machinery. The RAB family is the largest family of small GTP-binding proteins, and RABs have been shown to play important roles in the specification of membrane identity and vesicle trafficking via both the exocytotic and endocytotic routes (Vernoud et al., 2003; Woollard and Moore, 2008). Fifty-seven and 47 distinct RAB proteins are present in the Arabidopsis and rice genomes, respectively (Rutherford and Moore, 2002; Vernoud et al., 2003; Jiang and Ramachandran, 2006). Examination of the phylogenetic trees generated with protein sequences from human, yeast, and Arabidopsis has shown that the plant RAB family can be further subdivided into eight subfamilies, designated as RABA–RABH, each with counterparts in yeast and animals (Rutherford and Moore, 2002; Vernoud et al., 2003; Woollard and Moore, 2008). The correlation between sequence similarity among eukaryotes and regulation of membrane trafficking, through related compartments, appears to be a conserved feature in the RAB GTPase family. Experimental evidence is accumulating which links bioinformatic predictions with the involvement of diverse RAB GTPases in different steps of the endocytic and biosynthetic endoplasmic reticulum (ER)–Golgi and post-Golgi membrane trafficking pathways in the regulation of developmental processes (reviewed by Lycett, 2008; Nielsen et al., 2008; Woollard and Moore, 2008).
The ARF family of small GTPases were first identified as ADP ribosylation factors, and 21 ARF GTPase family members have been identified in Arabidopsis based on amino acid sequence identity (Vernoud et al., 2003). Members of the ARF family belong to the SAR, ARF, and ARF-like (ARL) subfamilies and are regulators of vesicle budding in different steps of membrane trafficking. The SAR subgroup is necessary for coat protein complex II (COPII)-dependent transport from the ER to the Golgi, whereas the ARF subgroup regulates both COPI-dependent retrograde transport to the ER and clathrin-dependent budding from the trans-Golgi and the plasma membrane (PM). ARLs are not as well characterized as SARs and ARFs, although an ARL-knockout phenotype has been described in Arabidopsis (Molendijk et al., 2004).
The RAN (Ras-related nuclear) protein was originally isolated as a homologue to Ras proteins (Drivsa et al., 1990), and proteins of this group play important roles in nucleocytoplasmic transport and microtubule organization (Jiang and Ramachandran, 2006).
Accumulating evidence shows that plant small GTP-binding proteins have common and diverse functional roles, when compared with their animal counterparts. In fact, reports on Arabidopsis and rice have indicated that the basic structures and functions of small GTP-binding proteins are conserved in plants. However, plants have the capacity to use small GTP-binding proteins as unique key molecular switches for the modulation of many plant-specific signalling pathways (Ma, 2007; Lycett, 2008). This is particularly true if the sessile nature of plants is taken into account, thus justifying a growing interest in the characterization of plant small GTPases.
Even though significant efforts are being made which are progressively shedding light on the function of small GTPases in plants, very little is known about their role in fruit development and ripening, a classical plant-specific process. Interestingly, an increasing body of evidence, recently reviewed by Lycett (2008), is accumulating in support of the hypothesis that proteins of the RABA/RAB11 subclade may be involved in the regulation of trafficking vescicles carrying cell wall-modifying enzymes to the apoplast. In fact, a member of the RABA subfamily was identified in mango as a protein expressed during fruit ripening and thought to be probably involved in enzyme trafficking to the cell wall (Zainal et al., 1996). Antisense inhibition of an orthologue clone (LeRab11a) isolated from tomato fruit resulted in delayed fruit ripening (Lu et al., 2001), probably due to a block in the vesicle-mediated delivery of wall-modifying enzymes to the apoplast, a process required for the softening process of ripening fruit (Lu et al., 2001). This hypothesis has been supported by evidence showing that the protein encoded by LeRab11a is involved in trafficking between the Golgi apparatus and the PM (Rehman et al., 2008).
Two recent studies have undertaken the first characterization of the grape ROP and RAB GTPase gene families during fruit development and ripening (Abbal et al., 2007, 2008). The authors have shown that the amount of mRNAs encoding Vitis vinifera ROPs (VvRops) were high at the early ‘green’ stages of fruit development and decreased progressively towards fruit ripening (Abbal et al., 2007). Among the VvRop genes, VvRop9 was reported to exhibit strong berry specificity, and a particular response to ABA (Abbal et al., 2007). In contrast, no single expression pattern was observed for genes encoding V. vinifera RAB proteins (VvRabs) during berry development and ripening (Abbal et al., 2008), even if the transcription of RABA- and RABD-encoding genes was reported to be up-regulated in tomato fruit during growth (Lycett, 2008, and references therein).
To our knowledge, no reports have dealt systematically with the expression analysis of members of small GTPase families during fruit development. Here the identification of expressed sequence tag (EST) clones encoding small GTPases of the RAS superfamily and the characterization of their expression profile during development and ripening of peach (Prunus persica, Batsch), a climacteric fruit showing significant accumulation of sucrose in its mesocarp cells (Vizzotto et al., 1996; Tonutti et al., 1997; Ruperti et al., 2001), are reported. It is shown that at least some members of the peach RAS superfamily display significant fluctuations of their transcripts during key moments of the last stages of peach fruit development and ripening.
Materials and methods
Plant material
Peach fruits were harvested from adult Redhaven (P. persica L. Batsch) trees at the Experimental Farm of Udine University, north-eastern Italy (46.01'N, 13.13E'). Trees received routine horticultural care. Experiments were carried out on fruits collected in 2004, and were repeated in 2005. In order to randomize biological variations, samples were pooled from 10 different plants. Fruits were collected to represent a developmental series of seven stages, from 72 after anthesis (DAA) in 2004 and 73 DAA in 2005, until harvest, frozen in liquid nitrogen, and stored at –80 °C for the subsequent analyses. Fruit growth was monitored on 20 fruits every week by measuring the transverse diameter, and the first derivative of the growth curve was calculated, as described by Connors (1919) and Chalmers and van den Ende (1975), to represent growth dynamics.
Determination of mesocarp sucrose content
Sugars were extracted from 1 g of fruit mesocarp slices with several washing steps with ethanol–water solution as described by Nonis et al. (2008). Sucrose content in mesocarp tissue was measured at weekly intervals starting from 72 DAA and 73 DAA (in 2004 and 2005, respectively). Determinations were carried out through enzymatic assay (Vizzotto et al., 1996), based on the increase in NADPH absorbance at 340 nm, stoichiometric to the amount of glucose and fructose. The efficiency of the methods was tested by using known amounts (10, 5, 2.5, and 1 mM) of carbohydrates.
Sucrose content was calculated from the difference of the glucose concentrations before and after an enzymatic hydrolysis (catalysed by invertase) to glucose and fructose.
In silico sequence analysis
Small GTPase-coding sequences of the Arabidopsis thaliana genome, as determined by Vernoud et al. (2003), were used to perform a homology search on NCBI (http://www.ncbi.nlm.nih.gov/blast/Blast.cgi), ESTree (http://www.itb.cnr.it/estree/), and TIGR (http://plantta.tigr.org/) databases.
ESTs were studied individually, by comparison with Arabidopsis sequences, to check whether they represented full-length cDNAs and to eliminate transcript redundancy. The resulting sequences were assembled into contigs using the CAP-assembler of Bioedit (http://www.mbio.ncsu.edu/BioEdit/bioedit.html), and start and stop codons were identified. A search in the P. persica NCBI Trace Archive server of the shotgun peach genome sequencing project (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi) was performed to obtain full-length cDNAs from incomplete contigs.
The predicted peach GTPase proteins were assigned to one of four families on the basis of their similarity to the sequences of Arabidopsis and were named according to their closest similarity to Arabidopsis proteins. Where more than one peach GTPase was present in the same subclade, a nomenclature based on numbers was adopted.
The amino acid sequences of small GTPases from peach and Arabidopsis were compared using the ClustalW tool (http://www.ebi.ac.uk/Tools/clustalw2/index.html). The multiple alignment resulted in an unrooted distance tree produced by using UPGMA algorithms of MEGA version 4 (Kumar et al., 2008). The reliability of the tree was examined using bootstrap analyses (1000 replicates).
RNA extraction, DNase treatment, and cDNA synthesis
Total RNA was obtained from peach fruit mesocarp according to the method described by Nonis et al. (2007). The final RNA pellet was resuspended in RNAse-free water and checked for integrity on a 1% agarose gel. RNA samples were stored at –80 °C.
A 10 μg aliquot of total RNA was treated with DNase enzyme (Promega) to remove contamination by genomic DNA. The reaction mix was incubated at 37 °C for 30 min, and the RNA was finally purified and concentrated with an RNeasy MinElute cleanup kit (Qiagen), according to the manufacturer's instructions. An aliquot of RNA was quantified spectrophotometrically, and electophoretically separated on a 1% agarose gel to check integrity. RNA (1 μg) was retrotranscribed in a total volume of 20 μl as described by Quaggiotti et al. (2004).
Quantitative real-time PCR
Real-time quantitative PCR experiments were performed on an MJ Opticon 2 using a Real Master Mix (Eppendorf) SYBR Green kit and gene-specific primers. In order to discriminate between the genes diplaying significant sequence similarity, primers were designed on the most divergent regions, mostly on the 3' untranslated region (UTR), on the basis of multiple sequence alignments performed with ClustalW (http://www.ebi.ac.uk/Tools/clustalw/index.html). Primer specificity was further confirmed by melting curve analysis. Furthermore, in order to assess the efficiency of the primers, serial dilutions of cDNA were tested using a concentration series of 6.4, 3.2, 1.6, 0.8, 0.4, and 0.2 ng μl−1. The full list of primer sequences employed in this study for peach small GTPases is given in Table 1. The reaction mix (20 μl) contained 5 μl of cDNA, 0.4 μl of each primer (10 μM), 9 μl of Master Mix, and 5.2 μl of RNase-free water. All the experiments were performed in triplicate with the same thermal cycling conditions: a first denaturation step at 95 °C for 3 min, followed by 41 cycles (94 °C 15 s, 56 °C 20 s, 68 °C 30 s). Gene expression analyses were carried out in triplicate with 6.4 ng μl−1 and 3.2 ng μl−1 dilutions of cDNA, producing comparable results. All quantifications were normalized to ubiquitin-conjugating enzyme (accession number BF717254), amplified with the primers 5′-CCCACCTGATTACCCTTTCA-3′ and 5′-GGATCTGTCAGCAGTGAGCA-3′ (Nonis et al., 2007). Ct numbers were extracted for both the reference gene and the target gene, with baselines subtracted by using the average-over-cycle range method (3–7 cycles) and a threshold set at 0.016. The mean Ct values were normalized against the reference gene. Since primer efficiencies were approximately equal, the expression was calculated by the 2–ΔΔCt method (Livak and Schmittgen, 2001).
Table 1.
3′ End primers used for real-time quantitative RT-PCR analysis
| Forward primer sequence | Reverse primer sequence | |
| PpARFA1-1 | TTGAGGGCTTAGACTGGTTGA | TCCAAAATCGGAACAAAAGAA |
| PpARFA1-2 | TCTTCATTCTCTCCGCCAAC | CCCCAGAAACTCCAAGTCAA |
| PpARFC1 | TAGGCAGCAACGTGGAAGAG | GCCAGCAACCTGAAGAGTTC |
| PpARLA1 | ATGTTGTGGATGCTGCTGAT | GCAAACTTCCCCTATCGGTAA |
| PpARLB1 | TTTTCGTTGTTTTATGGACTCTG | CAAAGTCGTTTTCCCAGCTT |
| PpARLC1 | CCCGACTTTCATCGAATCAT | GAATTGTCAAGCCCAACCT |
| PpSARA1 | GGAATTGAGCATTGGGAAGA | ACCACTGCATCCACCTTAGC |
| PpRABA1-2 | ACCACACGGATTCCAACATT | TTCTTTGCTTCCAGTGCAGA |
| PpRABA1-1 | CAAAGGACAGACGATCAACG | GCAATTTCGTCGAGTCAGGT |
| PpRABA2 | CAGGGTTATGCCGAAAAAGA | CCTCCCGTGACTGCAATAGT |
| PpRABA4 | CTAATGGCAACCCTGCATCT | AACGGTCCAATGAAATCACG |
| PpRABA5 | CTGCCCTGGACTCAACAAAT | AAACCATATCACCCGGAACA |
| PpRABA6 | GATTGGGGACTCTGGTGTTG | TGATGTTCCGGTAAGCGAAT |
| PpRABB1 | TGCTCATAGAAGGGCTGTCA | TCCAACCTTGATGCCAGAT |
| PpRABC1 | TTCCAAAGAGCTTCGGTCAG | AGAATCCCCAATCAGCAACA |
| PpRABC2 | CACCATCACCATCACCACAT | GGGCAAGATCGTCAACAGAG |
| PpRABD2-2 | CAGATCCGAGGACAACCTGT | TATGGTTTCACAAGGCACGA |
| PpRABD2-1 | CAACAAACCATCCACTGTGC | TAAGGGTCCAATGCAAAACC |
| PpRABE1 | TAGGAACTGGATTCGCAACA | CACAGCCCTTTTGCTTTCAT |
| PpRABF2 | TCCCTCAATTCTGCTCGATT | CAAAGCGCAACACCAGACTA |
| PpRABG3 | TGGTGTGCATCAAAAGGAAA | ATCTTGGCTGATTGCTGCTT |
| PpRAN3 | GGTCAAGGCAAAGCAGGTTA | CTGTTGTTGTGCAGCCAGAT |
| PpROP3 | TTCAGTGCAAATGTGGTCGT | CACCGCGATAACTCAAAGGT |
| PpROP4 | CGGAAGGAGGCTAGTGTGTC | CGCAAGTTAGGTTCGGAGAG |
Results
Identification of genes encoding small GTPases from peach and analysis of their similarity to Arabidopsis small GTPases
A search of the currently available ESTs expressed during peach fruit development was performed to identify sequences encoding proteins with a significant similarity to small monomeric GTPases of the RAS superfamily. All sequences encoding small GTPases from Arabidopsis (Vernoud et al., 2003) were used to carry out BLASTn and tBLASTx searches against the available databases of peach ESTs (ESTree, http://www.itb.cnr.it/estree/; NCBI, http://www.ncbi.nlm.nih.gov; http://plantta.jcvi.org/). A cut-off value of E−20 was chosen for sequence selection, and 200 peach EST cDNAs were identified that displayed significant similarity in their deduced amino acid sequence to known small GTPses from Arabidopsis. After filtering for redundancy, 24 genes were selected. The most abundant classes of genes were those encoding the RAB small GTPases (14 genes) and ARF/ARL/SAR GTPases (seven genes), correlating with their relative abundance in Arabidopsis. Two genes encoding ROP and one encoding a RAN GTPase were also found. Alignment with the closest related Arabidopsis sequences enabled the identification of bona fide start and stop codons. Twenty peach sequences covered the complete coding region, while four were partial and the corresponding full-length sequences were obtained by searching (Megablast), the P. persica NCBI Trace Archive server of the shotgun peach genome sequencing project (http://www.ncbi.nlm.nih.gov/Traces/trace.cgi). A full list of the peach small GTPases identified in this work is given in Table 2, together with the accession numbers of sequences assembled to obtain their complete cDNA and the closest corresponding Arabidopsis genes.
Table 2.
Putative small GTPases from peach (Prunus persica), accession numbers, and their closest Arabidopsis homologues
| Prunus persica GTPases | Prunus persica accession no.a | Arabidopsis closest homologue | Arabidopsis AGI gene |
| PpRABA1-1 | TA5861_3760b | AtRABA1f | At5g60860 |
| PpRABA1-2 | TA7012_3760b | AtRABA1c | At5g45750 |
| PpRABA2 | TA4712_3760b | AtRABA2a | At1g09630 |
| PpRABA4 | TA4817_3760b | AtRABA4a | At5g65270 |
| PpRABA5 | TA5151_3760b | AtRABA5e | At1g05810 |
| PpRABA6 | AJ823264c | AtRABA6a | At1g73640 |
| 2200746346d | |||
| 2167819167d | |||
| PpRABB1 | TA4959_3760b | AtRABB1b | At4g35860 |
| PpRABC1 | TA6278_3760b | AtRABC1 | At1g43890 |
| 2187916258d | |||
| PpRABC2 | TA5305_3760b | AtRABC2a | At5g03530 |
| PpRABD2-2 | TA3537_3760b | AtRABD2c | At4g17530 |
| PpRABD2-1 | TA4413_3760b | AtRABD2c | At4g17530 |
| PpRABE1 | TA5649_3760b | AtRABE1a | At3g53610 |
| PpRABF2 | BUO42728c | AtRABF2a | At5g45130 |
| PpRABG3 | TA6078_3760b | AtRABG3f | At3g18820 |
| PpARFA1-1 | TA3041_3760b | AtARFA1f | At1g10630 |
| PpARFA1-2 | TA4389_3760b | AtARFA1b | At5g14670 |
| PpARLA1 | BU044827c | AtARLA1c | At3g49870 |
| PpARFC1 | TA5387_3760b | AtARFC1 | At3g22950 |
| PpARLB1 | BU043462c | AtARLB1 | At5g52210 |
| 2167362775d | |||
| 2167505212d | |||
| PpSARA1 | TA5313_3760b | AtSAR1 | At4g02080 |
| PpARLC1 | DY653778c | AtARLC1 | At2g18390 |
| 2126540771d | |||
| AJ825266c | |||
| PpROP3 | TA3784_3760b | AtROP3 | At2g17800 |
| PpROP4 | TA6502_3760b | AtROP4 | At1g75840 |
| AJ870547c | |||
| PpRAN3 | TA3256_3760b | AtRAN3 | At5g55190 |
The names of peach GTPases, belonging to the RAB, ARF/ARL/SAR, ROP, and RAN families, are listed in the first column.
Details of the accession numbers of sequences employed to obtain the corresponding GTPase coding sequences. Accession numbers identified by different superscript letters refer to different databases:
sequences from the TIGR database, including plant transcript assemblies (TAs) from expressed transcripts collected from dbEST (ESTs) and from the NCBI GenBank nucleotide database;
singletons that are not assembled into TAs and retain their GenBank accession numbers as identifiers;
sequences from the WGS-Prunus persica Trace Archive.
In the third and fourth column the closest, Arabidopsis homologues and their corresponding AGI numbers are given.
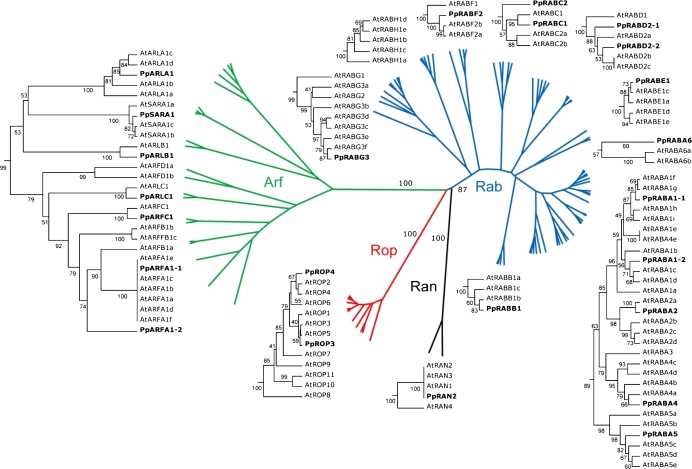
An UPGMA-based distance tree was produced to visualize the overall similarity between protein sequences of peach and Arabidopsis small GTPases. In detail, the 24 deduced amino acid sequences from peach, obtained from the translation of assembled ESTs, were aligned with A. thaliana known monomeric GTPase proteins (Vernoud et al., 2003). The resulting tree (Fig. 1) showed four main clades (fully supported by bootstrap analysis), corresponding to the four small GTPases families. It also confirmed subgroups previously identified in the Arabidopsis families (Vernoud et al., 2003). Compared with Arabidopsis and grapevine (Abbal et al., 2007, 2008), peach presented fewer homologous members of small GTPases; nevertheless, the number of sequences in the four families appeared comparable for the three species. PpRABs, as with AtRAB proteins, could be grouped into the eight subfamilies A–H. As in V. vinifera (Abbal et al., 2008), about half of the PpRABs (six) belonged to the RABA group, also the most abundant group in Arabidopsis. The remaining eight PpRAB members were distributed among seven other groups, with one member in RABB, RABE, RABF, and RABG subfamilies, two members in RABC and RABD branches, and no representatives in the RABH group.
Fig. 1.
UPGMA-based distance tree showing the similarity between P. persica and Arabidopsis small GTPase proteins and their distribution within the RAB, ARF/ARL/SAR, ROP, and RAN families. Two different graphic representations of the same tree are combined to show both the general partition in families and the detail of each group. Families and subfamilies of Arabidopsis small GTPases, previously identified by Vernoud et al. (2003), and the 24 sequences from peach (in bold type) are shown. The numbers above and below branches indicate bootstrap support percentages, based on 1000 replicates. (This figure is available in colour at JXB online.)
The PpARF GTPases segregated into ARF, ARL, and SAR subfamilies, in agreement with Vernoud et al. (2003). Comparison between P. persica and Arabidopis ARF sequences showed that peach amino acid sequences grouped into all Arabidopsis clusters (one for each), with the exception of the AtARFB and AtARFD groups.
Where more than one peach GTPase appeared in the same subclade, a nomenclature based on numbers was adopted (PpRABA1-1, PpRABA1-2, PpRABD1-1 PpRABD1-2, PpARFA1-1, and PpARFA1-2 in Fig. 1) to avoid the misleading identification of putative peach genes orthologous to Arabidopsis GTPases, in the absence of supporting functional data.
As expected, the two PpROP amino acid sequences grouped in the same clade with the 11 encoded AtROP proteins, and they are distributed exclusively in one of the four groups, as defined by Zheng and Yang (2000), clustering in group IV.
The only PpRAN protein identified showed its highest identity with AtRAN3.
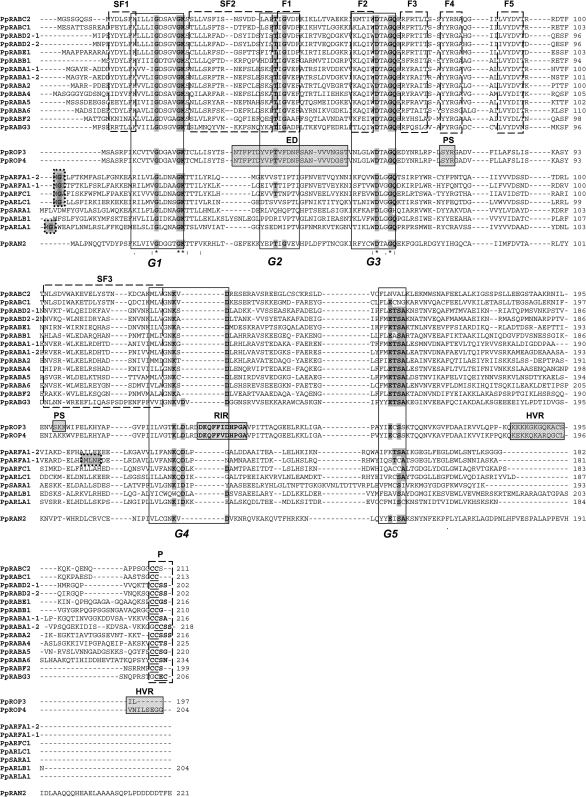
Identification of conserved family- and subfamily-specific protein sequence motifs of peach monomeric GTPases
Multiple sequence alignments, carried out with the deduced amino acid sequences of peach putative GTPases, allowed the identification of family- and subfamily-specific domains and motifs, and further supported their assignement to specific (sub) families. Overall, five typical GTPase domains were identified (G1–G5 in Fig. 2). The presence of the amino acid stretches which are diagnostic for RAB family members, termed F1–F5 and described by Pereira-Leal and Seabra (2000), together with that of the C-terminal prenylation motifs, including typical two cysteine residues, could be seen in PpRAB GTPases. The G residue of the IGVDF domain (F1 mammalian RAB family-specific domain), almost absolutely conserved in mammalian RAB, ARF, and RAN GTPases (Pereira-Leal and Seabra, 2000), also appeared to be conserved in PpRABs and PpRAN3, but not in PpARFs (Fig. 2). In peach proteins putatively assigned as ROPs, sequence analyses identified RHO-specific domains previously described by Zheng and Yang (2000). These included the effector domain of ROP proteins (ED), the Rho insert region (RIR), the hypervariable region (HVR) at the C-terminal region of the proteins, in addition to putative serine/theronine-dependent phosphorylation sites (motifs SYR and SKK) characteristic of different ROP groups (PS) (Fig. 2) (Zheng and Yang, 2000; Berken, 2006; Berken and Wittinghofer, 2008). The conserved arginine present in ROP proteins (R76), found in the putative recognition site SYR, present exclusively in plant RHO GTPases (Zheng and Yang, 2000; Berken, 2006; Berken and Wittinghofer, 2008), could be identified in both P. persica sequences. The conserved glycine residue (G in position 2), acceptor for myristate in ARF GTPases, was found in five of the seven deduced PpARF proteins, while the MXXE motif at position 110–113, responsible for the post-Golgi and PM targeting of ARF1 (Matheson et al., 2008), was found only in PpARFA1-1.
Fig. 2.
Multiple sequence alignment of deduced amino acid sequences of P. persica small GTPases. The five conserved motifs named ‘G box’ sequences are identified and boxed with rectangles (G1, G2, G3, G4, and G5) (according to Jiang and Ramachandran, 2006). Residues highlighted in grey show general conservation within most species. Asterisks (*) indicate the amino acid residues with 100% similarity in all sequences. RHO-specific domains were identified as described by Zheng and Yang (2000) and are highlighetd in grey rectangles: ED, effector domain of Rac/ROP proteins; RIR, Rho insert region; PS, putative serine/theronine-dependent phosphorylation sites (motifs SYR and SKK) distinctive for different ROP groups (Zheng and Yang, 2000) (ProSite program prediction); HVR, hypervariable region. RAB family- (F) and subfamily- (SF) specific domains and motifs (defined according to Pereira-Leal and Seabra, 2000) are indicated at the top of the alignment and are included in boxes with dashed lines: F1–F5, RAB family-specific domains; SF1–SF3, RAB subfamily-specific domains; P, RAB-specific prenylation motif; the double cysteine residues located in the C-termini of RAB GTPases representing the geranylgeranylation regions are highlighted in bold and underlined. ARF-specific motifs are in boxes with dotted lines highlighted in grey: G in position 2 is the conserved glycine residue acceptor for myristate; MLNE, conserved MxxE motif at position 110–113 involved in ARF1 Golgi targeting (Matheson et al., 2008).
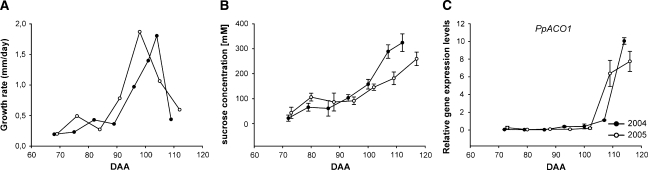
Peach fruit developmental dynamics, mesocarp sugar content, and PpACO1 expression
In order to characterize the developmental stages of fruits upon which expression analyses were conducted, data about growth kinetics and sugar content were collected during two different years (2004 and 2005). To facilitate further the comparison among fruit developmental stages in both years, a prediction of the ethylene climacteric was carried out. This determined the onset and progression of the ripening syndrome on the basis of the expression of the PpACO1 gene (accession number AF319166). Previous work demonstrated that accumulation of mRNA encoding 1-aminocyclopropane-1-carboxylic acid (ACC) oxidase (ACO1) from peach correlates with the ethylene levels synthesized by peach fruit during the ripening progress (Callahan et al., 1992; Ruperti et al., 2001).
Fruit size, expressed as a transverse diameter, was monitored throughout development, on the mid-season cultivar Redhaven, starting from 30 DAA in 2004 and 37 DAA in 2005, until 114 DAA and 116 DAA, respectively. Fruits showed in both years the typical double-sigmoid growth pattern (Connors, 1919; Chalmers and van den Ende, 1975) (data not shown). In Fig. 3A, the daily increase in fruit diameter is reported starting from ∼68 DAA, corresponding to the second exponential phase of peach fruit growth (Connors, 1919; Chalmers and van den Ende, 1975). During this period fruit growth is exclusively attributable to cell expansion, as mesocarp cell divisions had already ceased. Concomitantly, at this stage of development, the endocarp tissue had already reached its final size and become fully lignified. In addition, sucrose starts to accumulate, marking the onset of fruit maturation prior to ripening (DeJong and Goudriaan, 1989; Masia et al., 1992; Ognjanov et al., 1995).
Fig. 3.
Peach fruit growth dynamics (mm d−1) (A), sucrose accumulation (mM) (B) and PpACO1 relative gene expression levels, evaluated in 2004 (filled circles) and 2005 (open circles) (C). DAA, days after anthesis.
A first increase of growth rate was measured between 74 and 82 DAA, and 69 and 76 DAA in 2004 and 2005, respectively. This was followed by a short lag phase (between 82 and 90 DAA in 2004, and 76 and 84 DAA in 2005) before a final period of increasing growth rate could be seen, peaking at 98 DAA and 104 DAA in 2004 and 2005, respectively (Fig. 3A). Based on these data, the dynamics of fruit growth rate in 2005 appeared to occur ∼6 d earlier than they did in 2004.
Sucrose content of mesocarp tissue, measured throughout fruit growth in both years, was present at negligible levels during the early phases of fruit growth (data not shown). It then underwent a gradual increase starting from 72 DAA and 73 DAA in 2004 and 2005, respectively, up to the end of fruit growth and until ripening (Fig. 3B). The sucrose accumulation process also appeared to be biphasic and interrupted by a lag phase taking place between 79 and 86 DAA, and 80 and 95 DAA in 2004 and in 2005, respectively, before a significant increase in sucrose levels could be detected (Fig. 3B). Overall, the absolute sucrose concentration in the mesocarp appeared to be significantly higher in fruits collected in 2004 after 100 DAA.
The expression profile of PpACO1 in peach fruit mesocarp, as determined by real-time PCR, showed that the climacteric increase in the accumulation of its transcript and, therefore, the onset of the ethylene peak, took place at 114 DAA in 2004 and 109 DAA in 2005 (Fig. 3C). In addition, ACO1 transcript accumulation showed a tendency to decline in samples collected after 110 DAA in 2005, suggesting that in these samples the ethylene climacteric was recorded fully. These data, together with those on fruit growth dynamics, confirmed that fruit development and ripening in 2005 took place earlier in the year than in 2004.
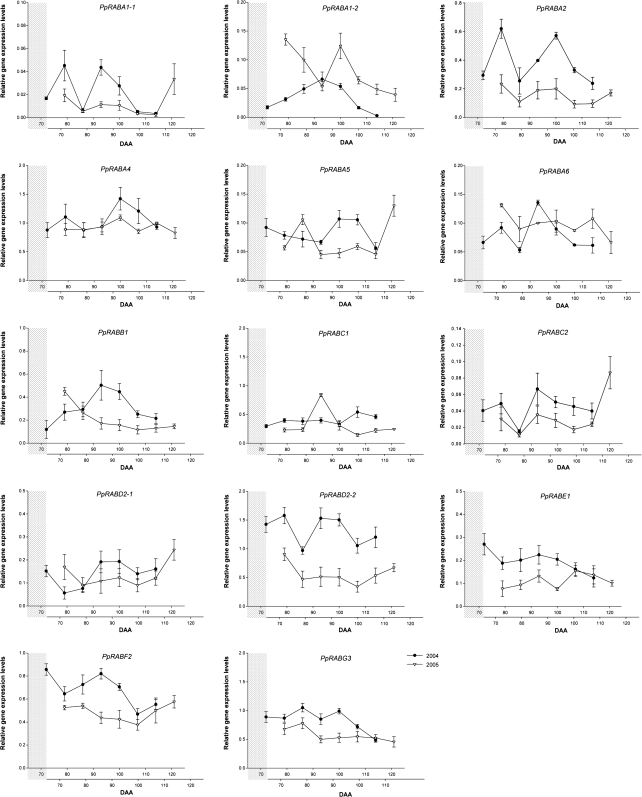
Expression of small GTPase-encoding genes during peach fruit development and ripening
To gain information about the possible physiological functions of different members of the peach small GTPase family, the transcriptional expression pattern of their encoding genes has been evaluated by real-time PCR during the last stages of peach fruit development. Only the phase of fruit expansion, coincident with the onset of sucrose accumulation, was considered for expression pattern analysis. The gene encoding ubiquitin-conjugating enzyme was chosen as a standard for constitutive expression (Nonis et al., 2007). The obtained expression profiles were the result of three different experiments carried out on samples collected weekly, in two different years (2004 and 2005), beginning from 72 and 73 DAA until 114 and 116 DAA, respectively.
Interestingly, a delay of a week was measured for the expression time course of a significant number of genes in samples collected in 2004. This was in agreement with data showing a slightly delayed fruit development and onset of ripening in 2004. For this reason, expression data of 2005 are represented in Figs. 4–6 with their x-axis (DAA) shifted 1 week ahead, as indicated by the grey shaded area, to facilitate comparisons between years.
Fig. 4.
Relative gene expression levels of PpRAB GTPase-encoding genes in peach fruit mesocarp (PpRABA–PpRABG), evaluated by real-time PCR in 2004 (filled circles) and 2005 (open triangles). DAA, days after anthesis. The lower x-axis shows DAA from 2005 shifted ahead, as shown by the grey shaded area, to allow the best alignment of gene expression profiles from the two years, as explained in the text.
Fig. 5.
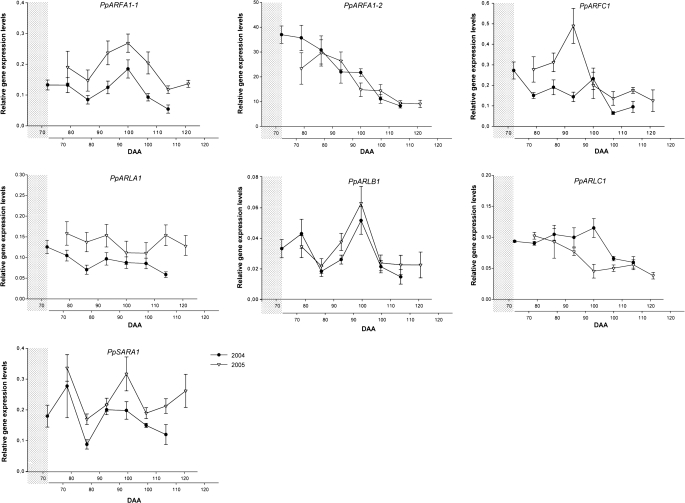
Relative expression levels of PpARF/ARL/SAR GTPase-encoding genes in peach fruit mesocarp, evaluated by real-time PCR in 2004 (filled circles) and 2005 (open triangles). DAA, days after anthesis. The lower x-axis shows DAA from 2005 shifted ahead, as shown by the grey shaded area, to allow best alignment of gene expression profiles from the two years, as explained in the text.
Fig. 6.
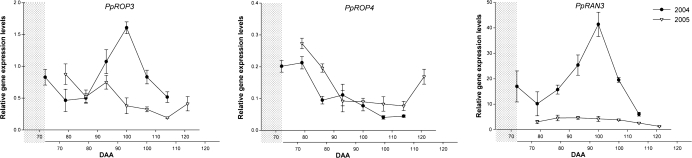
Relative gene expression levels of PpROP and PpRAN GTPase-encoding genes in peach fruit mesocarp, evaluated in 2004 (filled circles) and 2005 (open circles). DAA, days after anthesis. The lower x-axis shows DAA from 2005 shifted ahead to allow best alignment of gene expression profiles from the two years, as explained in the text and shown by the grey shaded area.
As far as peach RAB GTPase genes are concerned, the pattern of expression was complex and overlapped only partially between the two years analysed. Nevertheless, the expression dynamics of some PpRAB genes appeared remarkably consistent (shown in Fig. 4). PpRABA1-2, PpRABA4, PpRABA5, PpRABA6, PpRABB1, PpRABC1, PpRABE1, PpRABF2, and PpRABG3 displayed significantly different rates of transcript accumulation in the two years. On the other hand, PpRABD2-1, PpRABD2-2, and PpRABC2 showed reproducible expression patterns in the two years, with a minimum at 86 DAA and 80 DAA, and a steady increase, ranging from between a 2.5 and 3 factor increase for RABD2-1, and between a 4 and 7 factor increase for PpRABC2 towards ripening (at 107 DAA and 102 DAA in 2004 and 2005, respectively). Relative expression of PpRABD2-2 was higher throughout all stages of fruit development from samples collected in 2004. A transcriptional up-regulation towards the late phases of ripening, seen only for samples collected in 2005, could also be shown for PpRABA1-1 (13 times), PpRABA5 (∼3 times) and, to a lesser extent, for PpRABA2. In addition, the patterns of expression of PpRABA1-1, PpRABA2, PpRABD2-1, PpRABD2-2, and PpRABC2 exhibited two transient up-regulations, at 72–86 DAA and 86–107 DAA (2004), and at 73–80 and 80–102 DAA (2005), that appeared more pronounced in 2004, with significantly higher expression levels.
The relative level of PpARF transcript accumulation was consistent over both years (Fig. 5), with PpARFA1-2 and PpARLC1 expression decreasing progressively throughout fruit development and ripening. This decrease in transcript accumulation appeared dramatic (a 4-fold decrease) for PpARFA1-2. PpARFA1-2 expression resulted in amounts significantly higher than all other genes, during earlier stages of fruit development. Concomitantly, the transcription of PpARFA1-1, PpARLB1, and, to some extent, PpSARA1 genes was very similar, with nearly overlapping accumulation profiles and amounts in both years. Their pattern of expression appeared similar to those of PpRABA1-1, PpRABA2, PpRABD2-1, PpRABD2-2, and PpRABC2, with a minimun at 86 (2004) and 80 (2005) DAA and a transient peak at 100 DAA in 2004 and at 95 DAA in 2005, with an average 2.8-fold up-regulation for PpARFA1-1 and 3-fold for PpARLB1 in the two years. PpARFC1 also showed a transient minor transcriptional enhancement at 100 DAA in 2004 and at 88 DAA in 2005. PpARLA1 gene expression did not show significant differences in either year (Fig. 5).
PpROP3 and PpRAN3 gene expression was influenced significantly by year, showing a significantly higher peak of expression at 100 DAA in 2004 that was not detected in 2005. A minor increase in PpROP3 transcription was measured at 88 DAA in 2005. The expression profile of PpROP4 was similar in both years, declining throughout fruit development until, in 2005, a final increase that was also seen for PpROP3 (Fig. 6).
Discussion
Small monomeric GTPases of the RAS superfamily are important universal signalling switches in eukaryotes, controlling a set of diverse cellular processes including vescicle trafficking, cytoskeletal organization, signal transduction, and gene expression. Research in recent years has shed light on many aspects of the conserved and specific functions of small GTPases in plants; however, little is known about their role in fruit development and ripening, a plant-specific process.
Here the identification of 24 genes encoding small GTPases belonging to the RAS superfamily, expressed during peach (P. persica, Batsch) fruit development and ripening, has been described. BLASTn and BLASTx searches conducted on databases containing ESTs of genes expressed at different stages of peach fruit development have enabled the identification of 24 unigenes encoding bona fide peach small GTPases. Analyses of sequence similarities and of the presence of specific family and subfamily conserved motifs in their primary sequence (shown in Fig. 2) allowed the identification of the closest homologues from Arabidopsis and the assignment of 14 peach sequences to the RAB family, seven to the ARF/ARL/SAR family, two to the ROP family, and one to the RAN family. This result was in agreement with and proportional to the number of small GTPases found in Arabidopsis (Vernoud et al., 2003) and grape (Abbal et al., 2007, 2008). Within the peach RAB family, the most abundant subclass was the RABA subclade which included six PpRABA genes. This is consistent with the significant expansion of the RABA group in plants compared with their RAB11 counterparts in animals (Vernoud et al., 2003; Woollard and Moore, 2008). No PpRAB genes belonging to the H subclass could be identified.
To gain deeper insights into the putative role played by peach monomeric GTPases during peach fruit development and ripening, their mode of expression was studied in mesocarp samples collected during two seasons (2004 and 2005) from the mid-season cultivar Redhaven. Mesocarp sampling was restricted to the period of fruit development after endocarp lignification and was coincident with the onset of sucrose accumulation, at 72 DAA and 73 DAA in 2004 and 2005, respectively. The experimental design enabled the focus to be exclusively on correlations between changes in expression of small GTPases and growth dynamics during the last phases of fruit development: maturation and ripening. The peach fruit growth curve reflects complex interactions between mesocarp and endocarp, displaying two phases of exponential growth separated by a lag phase, associated with endocarp lignification (Connors, 1919; Chalmers and Van den Ende, 1975). By the time endocarp lignification is completed, mesocarp cells have already stopped dividing and have started to undergo expansion and maturation, marked by the onset of sucrose accumulation, finally leading to fruit ripening (Vizzotto et al., 1989, 1996; Tonutti et al., 1991; Ognjanov et al., 1995). In this phase, the exponential growth dynamics of peach fruits reflect solely the effects of sustained expansion of mesocarp cells (Ognjanov et al., 1995). During this period (ranging from ∼72 DAA until 120 DAA), fruits from both years displayed nearly overlapping growth dynamics and ethylene climacteric. Both the growth rate and sucrose accumulation appeared to be biphasic, with a first peak interrupted by a lag period (taking place between ∼80 DAA and 90 DAA), before exponential growth and sugar accumulation again started after 90–95 DAA. These data confirmed those reported by others in different environments and years for the mid-season cv Redhaven (Vizzotto et al., 1989; Liverani and Cangini, 1991; Tonutti et al., 1991; Ognjanov et al., 1995), and showed that this growth pattern is highly conserved and under strict genetic control. Indeed, in 2005, fruits developed earlier (∼6 d) and, as a consequence, also ripened earlier. Their ethylene climacteric, evaluated on the basis of the expression of the PpACO1 gene and previously shown to parallel ethylene biosynthesis (Callahan et al., 1992; Ruperti et al., 2001), also started earlier and peaked at 109 DAA. Accordingly, in order to enable comparisons between gene expression patterns in 2004 and 2005, an adjustment in the timing of the expression profiles of small GTPase-encoding genes from the two years was made. Figures 4–6 show that, at least for some genes, this shift resulted in nearly overlapping expression profiles, suggesting that their transcriptional regulation was strictly dependent on developmental cues and independent of the influence of environmental factors. On the other hand, the expression of some genes appeared significantly influenced by the year considered and therefore by environmental clues: this was particularly evident for PpROP3 and PpRAN3. Both genes only displayed a significant up-regulation in their relative expression level in 2004. The different regulation of PpROP3 and PpRAN3 may be explained as a response to abiotic (climatic) factors, as 2004 was significantly warmer than 2005 (data not shown). Though we are unable to draw conclusions on the specific factors controlling their gene expression, an involvement of PpROP3 and PpRAN3 proteins in regulating the plant's response to environmental stresses may be hypothesized on the basis of similar findings in their closest homologues in Arabidopsis (Molendijk et al., 2004; Berken, 2006). A similar effect could be claimed to account for the significantly higher expression levels of some PpRAB-encoding genes (such as PpRABA1-1, PpRABA2, and PpRABD2-2) or the significantly different expression profile (such as that of PpRABA1-2, PpRABA5, PpRABA6, and PpRABF2) in 2004, considering the increasing body of evidence showing the involvement of RAB GTPases in mediating responses to environmental factors and stresses (reviewed by Molendijk et al., 2004).
Interestingly, the expression of a group of genes appeared remarkably reproducible in the two seasons analysed, irrespective of the differences in climatic factors, with overlapping dynamics and, in the case of most PpARF genes, nearly identical relative transcriptional levels. Among these, five peach RAB- (PpRABA1-1, PpRABA2, PpRABD2-1, PpRABD2-2, and PpRABC2), three ARF- (PpARFA1-1, PpARFA1-2, and PpARLB1), and two ROP- (PpROP3 and PpROP4) encoding genes showed a modulated pattern of expression.
Transcripts of both PpROP genes were up-regulated during the last stages of fruit ripening in 2005, after the ethylene climacteric had peaked, suggesting their possible involvement downstream of ethylene, in regulating late events of ripening, and also confirming previous works reporting an effect of ethylene on ROP expression (Molendijk et al., 2004).
PpRAB transcripts showed a pattern of expression with two transient up-regulations during fruit growth and a final one during ripening. Interestingly, the first two changes appeared at almost the same time as peaks in growth rate and sugar accumulation of peach fruits reported in Fig. 3A and B. This up-regulation was conserved in terms of patterns of expression during both years, though it was more evident in 2004. A similar trend was also evident for the PpARF/ARL/SAR genes, PpARFA1-1, PpARLB1, and, to some extent, PpSARA1. These data would support the conclusion that a concerted co-expression of genes encoding RAB and ARF GTPases, involved in the regulation of diverse steps of vescicle trafficking, may be required for the phases of sustained cell expansion, sugar accumulation, and fruit growth. Even if the precise role played individually by these GTPases cannot be clarified at this stage, their involvement in driving the cell wall rearrangements necessary for cell expansion may nevertheless be hypothesized, at least for PpRAB genes. In fact, on the basis of homology, RAB GTPases of the D and A subclades exert functions in ER to Golgi, and Golgi to PM trafficking steps, respectively, in both mammalian and plant cells (Vernoud et al., 2003; Nielsen et al., 2008; Woollard and Moore, 2008), therefore fulfilling complementary and subsequent steps of the exocytic synthetic route of vescicle trafficking. In addition, several reports have shown that the trafficking of secretory and vacuolar markers in Arabidopsis relies on RABD2a regulation (reviewed by Woollard and Moore, 2008). Evidence from diverse plant systems is accumulating which supports the involvement of RABD and RABA GTPases in this synthetic route, by delivering cell wall material, or enzymes involved in cell wall remodelling, during cell expansion and wall loosening (reviewed by Lycett, 2008). Our data further support this hypothesis, showing a dynamic co-regulation of RABD and RABA transcripts with peach fruit growth and mesocarp cell expansion. Proteins of the RABC class are similar to mammalian RAB18, and evidence in support of their precise role in plant endomembrane oganization is still very scarse (Lycett, 2008; Woollard and Moore, 2008). Nevertheless, the present data, which suggest a co-regulation of PpRABC2 and PpRAB genes of the A and D groups, may suggest a possible involvement of PpRABC2 in a common process. The increased expression of PpRABA1-1, PpRABA2, PpRABA5, and PpRABC2 genes during the last stages of ripening may reflect their involvement in mediating delivery to the apoplast of cell wall-depolymerizing enzymes required for fruit softening (Lycett, 2008). This poses the question of whether the same RAB proteins may play a role in driving the cell wall rearrangements which are required for cell expansion and cell wall loosening during ripening.
As far as the differential expression of PpARFA1-1, PpARFA1-2, and PpARLB1 GTPases is concerned, their profiles were the most reproducible in the two years among all genes analysed, also in terms of relative levels of transcript accumulation. These data suggest that these genes may play central roles in fruit development since their expression appeared independent from environmental perturbations and, as a consequence, is strictly dependent on developmental cues. It is possible to hypothesize that PpARFA1-1 may play a similar role to that of AtARFA1a in Arabidopsis, in regulating trafficking between the trans-Golgi network and the PM (Xu and Scheres, 2005; Matheson et al., 2008), since they share a conserved localization motif (the MXXE motif at position 110–113, Fig. 2), responsible for post-Golgi and PM targeting (Matheson et al., 2008). The remarkable co-regulation of PpARFA1-1 and PpARLB1 genes in both years may point to their involvement in a so far uncharacterized common pathway. Even though the precise role of the PpARFA1-2 protein cannot be inferred, its involvement in regulating earlier stages of fruit development can be hypothesized on the basis of the significantly higher relative transcription of its encoding gene in comparison with all other genes during early phases of fruit development, and of the significant decrease in its transcription towards fruit ripening.
An additional intriguing possibility comes from recent reports hypothesizing that, concomitantly with carrier-mediated uptake, heterotrophic cells, such as those of fruit mesocarp, may also take up sugars by fluid endocytosis (Etxeberria et al., 2005a, b). The correlations shown herein between growth speed, sucrose accumulation, and expression of A and D class PpRAB genes, PpARF1-1, and PpARLB1 may prompt speculation as to whether the same proteins may also be involved in mediating sucrose accumulation in the vacuole. Indeed, RABA and RABD proteins have been shown to be involved in regulating trafficking between the Golgi and the pre-vacuolar compartment (Woollard and Moore, 2008) and ARF1 in mediating endocytosis in Arabidopsis (Xu and Scheres, 2005). These hypotheses will need direct experimental evidence in their support if they are to be used to dissect the exact roles played by the peach small GTPases described herein. Nevertheless, the data show for the first time that during the last phases of fruit mesocarp cell expansion and maturation and coincident with the onset of sugar accumulation, mesocarp cells actively reorganize their endomembrane system and vesicle trafficking machinery, by coordinating expression of GTPases of the RAB and ARF families.
Acknowledgments
The authors are greatly indebted to Dr William Teale for critical reading of the manuscript. RF was supported by a grant of the Italian Ministry of Education, Research and University (MIUR). AN was supported by a grant of the Regional Government of Friuli Venezia Giulia. TM was supported by a grant of the MIUR, Cofin (PRIN) project number 2003079333_003. This work was funded by the MIUR, Cofin (PRIN) projects numbers 2004079422_004 and 2003079333_003.
Glossary
Abbreviations
- DAA
days after anthesis
- EST
expressed sequence tag
- PM
plasma membrane
References
- Abbal P, Pradal M, Muniz L, Sauvage F-X, Chatelet P, Ueda T, Tesniere C. Molecular characterisation and expression analysis of the Rab GTPase family in Vitis vinifera reveal the specific expression of a VvRabA protein. Journal of Experimental Botany. 2008;59:2403–2416. doi: 10.1093/jxb/ern132. [DOI] [PubMed] [Google Scholar]
- Abbal P, Pradal M, Sauvage FX, Chatelet P, Paillard S, Canaguier A, Adam-Blondon A-F, Tesniere C. Molecular characterization and expression analysis of the Rop GTPase family in Vitis vinifera. Journal of Experimental Botany. 2007;58:2641–2652. doi: 10.1093/jxb/erm113. [DOI] [PubMed] [Google Scholar]
- Agrawal GK, Iwahashi H, Rakwal R. Small GTPase ‘Rop’: molecular switch for plant defense responses. FEBS Letters. 2003;546:173–180. doi: 10.1016/s0014-5793(03)00646-x. [DOI] [PubMed] [Google Scholar]
- Baxter-Burrell A, Yang Z, Springer PS, Bailey-Serres J. Rop GAP4-dependent Rop GTPase rheostat control of Arabidopsis oxygen deprivation tolerance. Science. 2002;296:2026–2028. doi: 10.1126/science.1071505. [DOI] [PubMed] [Google Scholar]
- Berken A. ROPs in the spotlight of plant signal transduction. Cellular and Molecular Life Sciences. 2006;63:2446–2459. doi: 10.1007/s00018-006-6197-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berken A, Wittinghofer A. Structure and function of Rho-type molecular switches in plants. Plant Physiology and Biochemistry. 2008;46:380–393. doi: 10.1016/j.plaphy.2007.12.008. [DOI] [PubMed] [Google Scholar]
- Callahan AM, Morgens PH, Wright P, Nichols KE., Jr Comparison of Pch313 (pTOM13 homolog) RNA accumulation during fruit softening and wounding of two phenotypically different peach cultivars. Plant Physiology. 1992;100:482–488. doi: 10.1104/pp.100.1.482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalmers DJ, van den Ende B. A reappraisal of the growth and development of peach fruit. Australian Journal of Plant Physiology. 1975;2:623–634. [Google Scholar]
- Connors CH. Growth of fruits of peach. New Jersey Agriculture Experiment Station Annual Report. 1919;40:82–88. [Google Scholar]
- DeJong TM, Goudriaan J. Modelling peach fruit growth and carbohydrate requirements: reevaluation of the double-sigmoid growth pattern. Journal of the American Society for Horticultural Science. 1989;114:800–804. [Google Scholar]
- Drivsa GT, Shih A, Coutavas E, Rush MG, D'Eustachio P. Characterization of four novel ras-like genes expressed in a human teratocarcinoma cell line. Molecular and Cellular Biology. 1990;10:1793–1798. doi: 10.1128/mcb.10.4.1793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Etxeberria E, Baroja-Fernandez E, Muñoz FJ, Pozueta-Romero J. Sucrose-inducible endocytosis as a mechanism for nutrient uptake in heterotrophic plant cells. Plant and Cell Physiology. 2005a;46:474–481. doi: 10.1093/pcp/pci044. [DOI] [PubMed] [Google Scholar]
- Etxeberria E, González P, Tomlinson P, Pozueta-Romero J. Existence of two parallel mechanisms for glucose uptake in heterotrophic plant cells. Journal of Experimental Botany. 2005b;56:1905–1912. doi: 10.1093/jxb/eri185. [DOI] [PubMed] [Google Scholar]
- Jiang SY, Ramachandran S. Comparative and evolutionary analysis of genes encoding small GTPases and their activating proteins in eukaryotic genomes. Physiological Genomics. 2006;24:235–251. doi: 10.1152/physiolgenomics.00210.2005. [DOI] [PubMed] [Google Scholar]
- Kumar S, Nei M, Dudley J, Tamura K. MEGA: a biologist-centric software for evolutionary analysis of DNA and protein sequences. Briefings in Bioinformatics. 2008;9:299–306. doi: 10.1093/bib/bbn017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Livak J, Schmittgen T. Analysis of relative gene expression data using realtime quantitative PCR and the 2–ΔΔCtmethod. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Liverani A, Cangini A. Ethylene evolution and changes in carbohydrates and organic acid during maturation of two white and two yellow fleshed peach cultivars. Advances in Horticultural Science. 1991;5:59–63. [Google Scholar]
- Lu C, Zainal Z, Tucker GA, Lycett GW. Developmental abnormalities and reduced fruit softening in tomato plants expressing an antisense Rab11 GTPase gene. The Plant Cell. 2001;13:1819–1833. doi: 10.1105/TPC.010069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lycett G. The role of Rab GTPases in cell wall metabolism. Journal of Experimental Botany. 2008;59:4061–4074. doi: 10.1093/jxb/ern255. [DOI] [PubMed] [Google Scholar]
- Ma QU. Small GTP-binding proteins and their functions in plants. Journal of Plant Growth Regulation. 2007;26:369–388. [Google Scholar]
- Masia A, Zanchin A, Rascio N, Ramina A. Some biochemical and ultrastructural aspects of peach fruit development. Journal of the American Society for Horticultural Science. 1992;117:808–815. [Google Scholar]
- Matheson LA, Suri SS, Hanton SL, Chatre L, Brandizzi F. Correct targeting of plant ARF GTPases relies on distinct protein domains. Traffic. 2008;9:103–120. doi: 10.1111/j.1600-0854.2007.00671.x. [DOI] [PubMed] [Google Scholar]
- Molendijk AJ, Ruperti B, Palme K. Small GTPases in vesicle trafficking. Current Opinion in Plant Biology. 2004;7:694–700. doi: 10.1016/j.pbi.2004.09.014. [DOI] [PubMed] [Google Scholar]
- Nibau C, Wu HM, Cheung AY. RAC/ROPGTPases: ‘hubs’ for signal integration and diversification in plants. Trends in Plant Science. 2006;11:309–315. doi: 10.1016/j.tplants.2006.04.003. [DOI] [PubMed] [Google Scholar]
- Nielsen E, Cheung AY, Ueda T. The regulatory RAB and ARF GTPases for vesicular trafficking. Plant Physiology. 2008;147:1516–1526. doi: 10.1104/pp.108.121798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonis A, Ruperti B, Falchi R, Casatta E, Thamashebi SE, Vizzotto G. Differential expression and regulation of a neutral invertase encoding gene from peach (Prunus persica): evidence for a role in fruit development. Physiologia Plantarum. 2007;129:436–446. [Google Scholar]
- Nonis A, Ruperti B, Pierasco A, Canaguier A, Adam-Blondon A-F, Di Gaspero G, Vizzotto G. Neutral invertases in grapevine and comparative analysis with Arabidopsis, poplar and rice. Planta. 2008;229:129–142. doi: 10.1007/s00425-008-0815-0. [DOI] [PubMed] [Google Scholar]
- Ognjanov V, Vujanic-Varga D, Misic PD, Veresbaranji I, Macet K, Tesovic Z, Krstic M, Petrovic N. Anatomical and biochemical studies of fruit development in peach. Scientia Horticulturae. 1995;64:33–48. [Google Scholar]
- Ono E, Wong HL, Kawasaki T, Hasegawa M, Kodama O, Shimamoto K. Essential role of the small GTPase Rac in disease resistance of rice. Proceedings of the National Academy of Sciences, USA. 2001;98:759–764. doi: 10.1073/pnas.021273498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pereira-Leal JB, Seabra MC. The mammalian Rab family of small GTPases: definition of family and subfamily sequence motifs suggests a mechanism for functional specificity in the Ras superfamily. Journal of Molecular Biology. 2000;301:1077–1087. doi: 10.1006/jmbi.2000.4010. [DOI] [PubMed] [Google Scholar]
- Quaggiotti S, Ruperti B, Pizzeghello D, Francioso O, Tugnoli V, Nardi S. Effect of low molecular size humic substances on nitrate uptake and expression of genes involved in nitrate transport in maize (Zea mays L.) Journal of Experimental Botany. 2004;55:803–813. doi: 10.1093/jxb/erh085. [DOI] [PubMed] [Google Scholar]
- Rehman RU, Stigliano E, Lycett GW, Sticher L, Sbano F, Faraco M, Dalessandro G, Di Sansebastiano GP. Tomato Rab11a characterization evidenced a difference between SYP121-dependent and SYP122-dependent exocytosis. Plant and Cell Physiology. 2008;49:751–766. doi: 10.1093/pcp/pcn051. [DOI] [PubMed] [Google Scholar]
- Ruperti B, Bonghi C, Rasori A, Ramina A, Tonutti P. Characterization and expression of two members of the peach 1-aminocyclopropane-1-carboxylate oxidase gene family. Physiologia Plantarum. 2001;111:336–344. doi: 10.1034/j.1399-3054.2001.1110311.x. [DOI] [PubMed] [Google Scholar]
- Rutherford S, Moore I. The Arabidopsis Rab GTPase family: another enigma variation. Current Opinion in Plant Biology. 2002;5:518–528. doi: 10.1016/s1369-5266(02)00307-2. [DOI] [PubMed] [Google Scholar]
- Tonutti P, Bonghi C, Ruperti B, Tornielli GB, Ramina A. Ethylene evolution and 1-aminocyclopropane-1-carboxylate oxidase gene expression during early development and ripening of peach fruit. Journal of the American Society for Horticultural Science. 1997;122:642–647. [Google Scholar]
- Tonutti P, Casson P, Ramina A. Ethylene biosynthesis during peach fruit development. Journal of the American Society for Horticultural Science. 1991;116:274–279. [Google Scholar]
- Vernoud V, Horton AC, Yang ZB, Nielsen E. Analysis of the small GTPase gene superfamily of Arabidopsis. Plant Physiology. 2003;131:1191–1208. doi: 10.1104/pp.013052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vizzotto G, Masia A, Bonghi C, Tonutti P, Ramina A. IAA levels in Prunus persica (L.) Batsch in relation to fruit growth and development. Acta Horticulturae. 1989;239:387–390. [Google Scholar]
- Vizzotto G, Pinton R, Varanini Z, Costa G. Sucrose accumulation in developing peach fruit. Physiologia Plantarum. 1996;96:225–230. [Google Scholar]
- Woollard AD, Moore I. The functions of Rab GTPases in plant membrane traffic. Current Opinion in Plant Biology. 2008;11:1–10. doi: 10.1016/j.pbi.2008.09.010. [DOI] [PubMed] [Google Scholar]
- Xu J, Scheres B. Dissection of Arabidopsis ADP-RIBOSYLATION FACTOR 1 function in epidermal cell polarity. The Plant Cell. 2005;17:525–536. doi: 10.1105/tpc.104.028449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z. Small GTPases: versatile signaling switches in plants. The Plant Cell. 2002;14:S375–S388. doi: 10.1105/tpc.001065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang Z, Fu Y. ROP/RAC GTPase signalling. Current Opinion in Plant Biology. 2007;10:490–495. doi: 10.1016/j.pbi.2007.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zainal Z, Tucker GA, Lycett GW. A rab11-like gene is developmentally regulated in ripening mango (Mangifera indica L.) fruit. Biochimica et Biophysica Acta. 1996;1314:187–190. doi: 10.1016/s0167-4889(96)00133-4. [DOI] [PubMed] [Google Scholar]
- Zheng ZL, Yang Z. The Rop GTPase: an emerging signaling switch in plants. Plant Molecular Biology. 2000;44:1–9. doi: 10.1023/a:1006402628948. [DOI] [PubMed] [Google Scholar]