Abstract
In an effort to understand the epigenetic regulation of ribosomal RNA gene (rDNA) expression we have previously demonstrated the role of DNA methyltransferases and methyl CpG binding proteins in rRNA synthesis. Here, we studied the role of protein arginine methyltransferase PRMT5 and the two methylated histones H3R8Me2 and H4R3Me2, in rDNA expression in Epstein Barr virus- transformed primary B-cells (LCLs) and in HeLa cells responding to serum-regulated growth. Chromatin immunoprecipitation assay showed that histones H3 and H4 associated with rRNA promoters were differentially methylated at arginine residues 8 and 3, respectively, depending on its transcriptional activity. Association of PRMT5 and methylated H3 with the unmethylated promoters in resting B-cells was significantly reduced in rapidly growing LCLs. Unlike PRMT5 and H3R8Me2, histone H4 associated with both methylated and unmethylated rRNA promoters in resting B-cells was methylated at the R3 residue. However, a dramatic decrease in R3 methylation of H4 recruited to the unmethylated rRNA promoters was observed in LCLs while it remained unaltered in the fraction bound to the methylated promoters. Differential interaction of PRMT5 and methylation of H3 and H4 associated with the rRNA promoters was also observed when serum starved HeLa cells were allowed to grow in serum replenished media. Ectopic expression of PRMT5 suppressed activity of both unmethylated and methylated rRNA promoter in transient transfection assay whereas siRNA mediated knockdown of PRMT5 increased rRNA synthesis in HeLa cells. These data suggest a key role of PRMT5 and the two methylated histones in regulating rRNA promoter activity.
Keywords: RNA POLYMERASE 1, PRMT5, H3R8ME2, H4R3ME2, EBV TRANSFORMATION
While most studies on epigenetic regulation focused on genes transcribed by RNA polymerase II (Pol II), recent studies from a few laboratories including our own, have demonstrated that RNA Polymerase I-directed ribosomal RNA gene (rDNA) transcription is also modulated by epigenetic mechanisms [Grummt and Pikaard, 2003; Ghoshal et al., 2004; Majumder et al., 2006; McStay and Grummt, 2008]. DNA methylation, histone modifications and chromatin remodeling constitute the epigenetic mechanisms underlying alterations in gene expression. Although diploid somatic cells harbor 300–400 copies of the rRNA genes, nearly half of these genes are transcriptionally silenced largely due to epigenetic mechanisms. Most studies on epigenetic regulation of ribosomal gene expression have centered on DNA methylation [Chen and Pikaard, 1997; Santoro and Grummt, 2001; Ghoshal et al., 2004; Majumder et al., 2006]. We showed that human rDNA is differentially regulated in normal and cancer cells by significant hypomethylation of rRNA promoters in primary human hepato-cellular carcinomas relative to matching liver tissues [Ghoshal et al., 2004]. Consistent with this observation, all three DNA methyltransferases (DNMT1, 3A, 3B) are associated with inactive rDNA in the nucleolus, and significant elevation in rDNA primary transcript is observed in DNMT 1 and DNMT3B null human colon carcinoma cells [Majumder et al., 2006]. Unlike human ribosomal genes that contain CpG island within its promoter regions, mouse rDNA contains only a single CpG at −113 position within the upstream control element (UCE) of the promoter. The cytosine in this dinucleotide when methylated, prevents access of the key transcription factor UBF to the promoter, resulting in transcriptional suppression [Santoro and Grummt, 2001].
Methyl CpG binding proteins (MBDs) with highly homologous methyl CpG binding domains can modulate rDNA suppression [Ghoshal et al., 2004; Brown and Szyf, 2007; McStay and Grummt, 2008]. Methylation of DNA usually results in binding of MBDs, which, in turn, recruits repressor complexes containing histone methyltransferases and histone deacetylases [Fuks et al., 2003; Sarraf and Stancheva, 2004]. One of these proteins MBD2 specifically repressed methylated rRNA promoters, and chromatin immunoprecipitation assay showed its preferential association with the methylated promoters [Ghoshal et al., 2004]. Further, all MBDs were found in the nucleolus as well as nucleoplasm [Ghoshal et al., 2004], consistent with their potential roles in rDNA transcription.
Some efforts have been made to understand the role of posttranslational modifications of histones in rDNA expression [for reviews see Grummt and Pikaard, 2003; McStay and Grummt, 2008]. As observed for Pol II-transcribed genes, acetylated histones H3 and H4 and histone H3 methylated at lysine 4 (H3K4Me2) are associated with active rDNA whereas inactive ribosomal RNA genes are associated with heterochromatin [McStay and Grummt, 2008]. While DNA methylation generally results in recruitment of post-translationally modified histones to the methylated promoter regions, it has been suggested that trimethylation of H3 K9 and K27 as well as H4K20 is required for subsequent DNA methylation in fungi, plants and mammals [Tamaru et al., 2003; Schotta et al., 2004; Fuks, 2005]. A recent investigation has indeed found a link between arginine methylation of histones and DNA methylation that leads to gene silencing [Zhao et al., 2009]. This study has shown that symmetric methylation of histone H4 arginine (H4R3Me2) by the protein arginine methyltransferase PRMT5 serves as a direct target for DNMT3A binding, which then methylates CpG rich regions causing gene silencing.
PRMTs are emerging as important histone methyltransferases. The two types of evolutionarily conserved PRMTs differ in the nature of methylation of arginine on one of the terminal guanidino nitrogen atoms. PRMT5, one of the type II arginine methyltransferases, catalyzes monomethylation and symmetric dimethylation of arginine [Bedford and Richard, 2005; Pal et al., 2007] and is involved in a variety of cellular processes, including transcriptional regulation and germ cell development [Pal et al., 2003, 2004; Ancelin et al., 2006]. Recent reports indicate that PRMT5 can regulate gene expression by modifying histones or indirectly by modulating the activity of specific transcription factors [Hosohata et al., 2003; Pal et al., 2004; Dacwag et al., 2007]. Since all studies on the transcriptional regulation by PRMT5 were performed on the genes transcribed by Pol II, it was of considerable interest to study its role in the Pol I transcription of rRNA genes. This was particularly relevant in light of the reports that rRNA genes are also modulated by epigenetic mechanisms. In the present study, we explored the role of PRMT5 in the control of rDNA expression in response to Epstein-Barr virus (EBV) infection and in response to serum deprivation/enrichment. Here, we report that differential association of PRMT5 with the methylated and unmethylated rRNA promoters, and methylation status of histones H3 and H4 (methylated at arginine 8 and 3, respectively) at the rRNA promoters, play a key role in modulating rDNA expression under different growth conditions.
MATERIALS AND METHODS
EBV TRANSFORMATION OF B-LYMPHOCYTES
Normal B lymphocytes were isolated from freshly drawn blood using ficoll density gradient centrifugation and enriched using the Rosette-Sep kit from Stem Cell Technologies (Vancouver, British Columbia, Canada) according to the manufacturer’s instructions. Isolated cells were incubated in RPMI 1640 media containing 10% fetal bovine serum, 2 mM l-glutamine and 100 U/ml penicillin-100 mg/ml streptomycin (Invitrogen, Rockville, MD) at 37°C in a 5% CO2 incubator. The B-cell enriched fraction consisted of >95% B-cells, that was determined by routine CD19 and CD3 expression analysis.
EBV-immortalized lymphoblastoid cell lines were obtained by in vitro infection of normal B-lymphocytes with EBV-containing supernatant from B95.8 cell line following standard protocols [Rickinson et al., 1984; Young and Murray, 2003]. Briefly, 5 × 105 B cells/well were treated with 2 µg/ml Cyclosporine A in 96 wells plates to inhibit the T cell compartment. Virus containing supernatant obtained from TPA stimulated (20 ng/ml) B95.8 marmoset cells after 1 week in culture, was used to infect the B cells. The medium was replaced by fresh medium every week and after 3 weeks the microcultures were transferred into flasks. B-cells prior to transfection and after 35 days of infection (immortalized) were collected for day 0 and day 35 time points.
The fully transformed B-cell lines (LCLs) were derived from hu-PBL-SCID mouse tumors as previously described [Dierksheide et al., 2005]. Briefly, peripheral blood lymphocytes (PBL) from EBV-seropositive donors were injected into severe combined immunodeficiency (SCID) mice. Recipient mice were monitored for up to 6 months for engraftment of human cells and development of widely metastatic EBV-driven lymphoproliferative tumors. These tumors were confirmed to be of human B-cell origin using flow cytometry (human CD45+, CD19/CD20+). To isolate neoplastic B-cells, infiltrated tissues were minced extensively in RPMI 1640 supplemented with 10% FBS and strained through 70 µm pore size cell strainers (BD Biosciences, San Jose, CA). Tumor B cells were then isolated using ficoll density gradient centrifugation and cultured in RPMI 1640 supplemented with 20% FBS.
CELL CULTURE AND TRANSFECTION ASSAYS
HeLa cells were grown in DMEM with 10% FBS.
The transfection studies were performed as described [Ghoshal et al., 2004].
IN VITRO DNA METHYLATION
M.Hha I methylation of pHrD-IRES-Luc was performed as described [Ghoshal et al., 2004].
RNA ISOLATION, REVERSE TRANSCRIPTION AND REAL-TIME RT-PCR ANALYSIS
Total RNA was isolated using Trizol (Invitrogen). Reverse transcription was carried out with random hexamers and M-MuLV reverse transcriptase from 3 µg of total RNA in 20 µl of total volume following the manufacturer’s protocol. An aliquot of the cDNA was used for real-time PCR analysis. All real-time PCR reactions were carried out using the Mx3000 Multiplex Quantitative PCR System (Stratagene). The optimum primer concentration was 150 nM. All PCR amplifications were performed using Brilliant® SYBR® Green QPCR Master Mix (Applied Biosystems) with ROX as a reference dye in a 10 ml reaction volume. A standard curve for each cDNA was first generated using 10-fold serial dilutions (108 to 102 copies) of the respective cDNAs as template. The copy number of each cDNA expressed was calculated from the standard curve and normalized to that of β-actin. Dissociation profile of the amplified products indicated that none of the primer pairs generated dimers. The primer sequences are the following:
47S (product size 92 bp).
47S-F: 5′-CCTGTCGTCGGAGAGGTTGG-3′, 47S-R: 5′-ACCCCACGCCTTCCCACAC-3′.
Annealing temperature 60°C.
β-Actin (product size: 276 bp).
β-Actin-F: 5′-TTTGAGACCTTCAACACCCC AGCC-3′, β-actin-R: 5′-AATGTCACGCACG ATTTCCCGC-3′.
Annealing temperature 60°C.
WESTERN BLOT ANALYSIS
Whole cell extract prepared from HeLa cells overexpressing PRMT5-Flag [Pal et al., 2003] was separated on a 10% SDS–polyacrylamide gel, and transferred to nitrocellulose membrane, blocked with 3% milk in Tris buffered (pH 7.5) saline (TBS). For the detection of the overexpressed proteins the membrane was subjected to immunoblot analysis with anti-Flag M2 antibody (Sigma) [Ghoshal et al., 2002; Majumder et al., 2002] and HRP-conjugated anti-mouse IgG as the secondary antibody. The antigen-antibody complex was detected using ECL™ kit (Amersham).
CHROMATIN IMMUNOPRECIPITATION
Formaldehyde cross-linked chromatin was prepared as described [Pal and Sif, 2007]. For ChIP analysis, antibody against UBF (Santa Cruz), antisera against PRMT5, H3R8Me2 and H4R3Me2 raised in our laboratory, were used [Pal et al., 2004, 2007]. The chromatin was first pre-cleared with preimmune sera coupled to protein A/G beads followed by overnight incubation with preimmune or immune sera. The immune complex was then captured by protein A beads, washed successively with buffers as described [Ghoshal et al., 2002]. The immunoprecipitated DNA-protein complex was eluted, de-cross-linked, treated with RNase A and proteinase K and purified as described [Ghoshal et al., 2002]. The immunoprecipitated DNA and the input DNA were digested with Hpa II or Msp I and the digests along with equal amount of the undigested immunoprecipitated DNA were subjected to QPCR with human rRNA promoter specific primers [Ghoshal et al., 2004]. The results are expressed as the ratio of methylated or unmethylated DNA precipitated with the antibodies to the respective input. All ChIP experiments were repeated at least twice (see figure legends to Fig. 2F for additional details of this procedure).
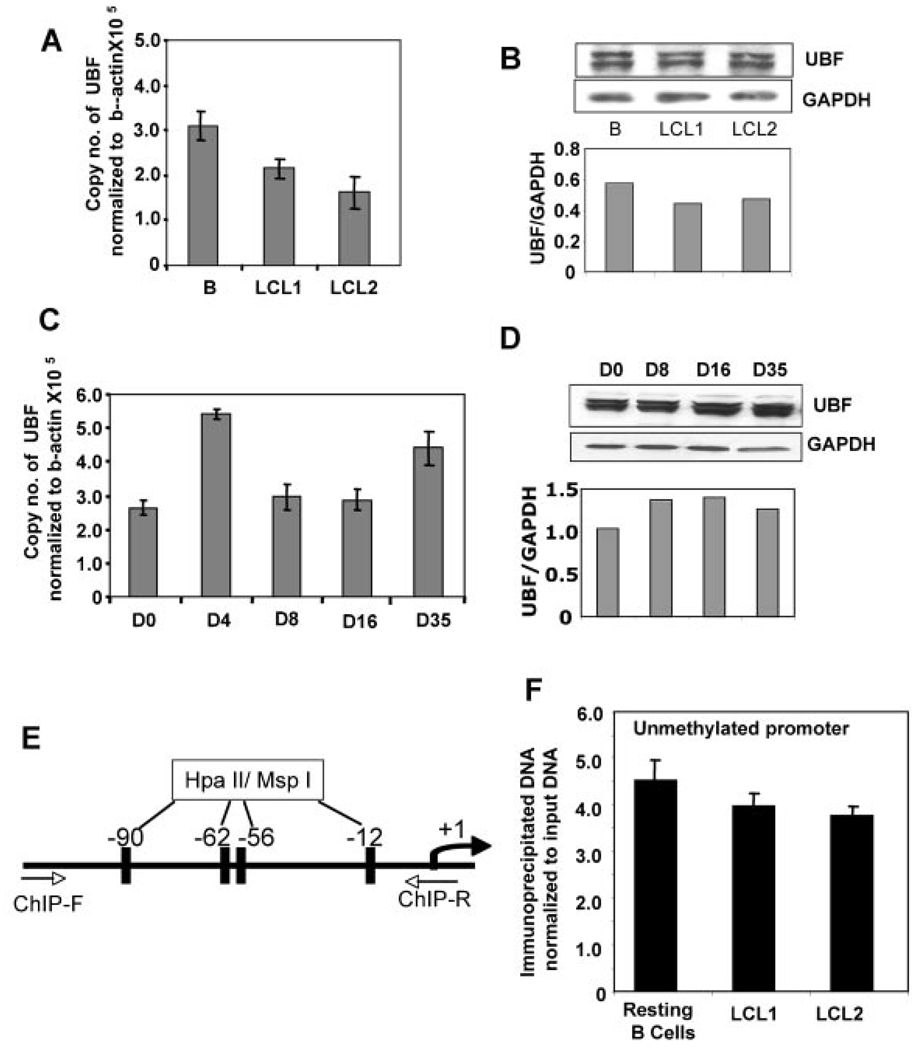
Fig. 2.
UBF expression and association with unmethylated (active) rRNA promoter are not altered during increased rRNA synthesis in EBV infected B-cells. UBF mRNA levels in resting B cells and LCLs (Panel A), and EBV-infected cells (Panel C) were monitored by real-time PCR and protein levels were analyzed by Western blot (Panels B,D). Copy number of UBF was normalized to b-actin and UBF protein level was normalized to GAPDH. E: Schematic diagram of PCR amplified rRNA promoter. F: Resting B-cells and exponentially growing EBV-transformed cells (LCL1, LCL2) were cross-linked with formaldehyde and chromatin prepared from these cells was immunoprecipitated with anti-UBF antibody. DNA pulled down by the antibody as well as input DNA were divided into three identical fractions that were either mock-digested or digested with Hpa II (H) or Msp I (M). An aliquot of the product from each digestion was subjected to QPCR. The reaction products were separated on polyacrylamide and Kodak software was used to quantify the PCR products. Association with methylated promoter = Hpa II signal in ChIP DNA/Hpa II signal in input (1:200 dilution). Association with unmethylated promoter = signal in undigested minus signal in Hpa II digested ChIP DNA/ input signal in undigested minus signal in Hpa II digested DNA (1:200 dilution).
INDIRECT IMMUNOFLUORESCENCE ANALYSIS
HeLa cells were grown overnight on cover slips and fixed with paraformaldehyde for 15 min at room temperature, washed with PBS, and permeabilized with 0.3% Triton X-100 in PBS for 10 min at room temperature. Next the cells were incubated with 1% BSA for 1 h to block nonspecific binding. The blocked chambers were subsequently washed and incubated overnight at 4°C with mixture of anti-nucleolin monoclonal antibody (anti-C23, Santa Cruz) and anti-PRMT5 antibody. FITC-conjugated anti-rabbit (for PRMT5) antibodies were used for green channel detection while monoclonal anti-C23 antibody was detected by TRITC-conjugated monoclonal anti-mouse antibody (Sigma) for detection in red channel. Nuclei were stained using DAPI in the mounting fluid.
PRMT5 siRNA AND ITS TRANSFECTION
HeLa cells were transfected at 70% confluency with 100 nM PRMTS siRNA (Santa Cruz, SC37022) or the control scrambled siRNA (Santa Cruz, SC37007) using Lipofectamine 2000. The cells were harvested at 24, 48, and 72 h from the time of transfection. Whole cell extracts from these cells were subjected to Western blot analysis and total RNA isolated was used for real-time PCR.
RESULTS
RIBOSOMAL GENE EXPRESSION IS SIGNIFICANTLY ELEVATED IN EPSTEIN BARR VIRUS (EBV) TRANSFORMED B-CELLS COMPARED TO RESTING B-CELLS
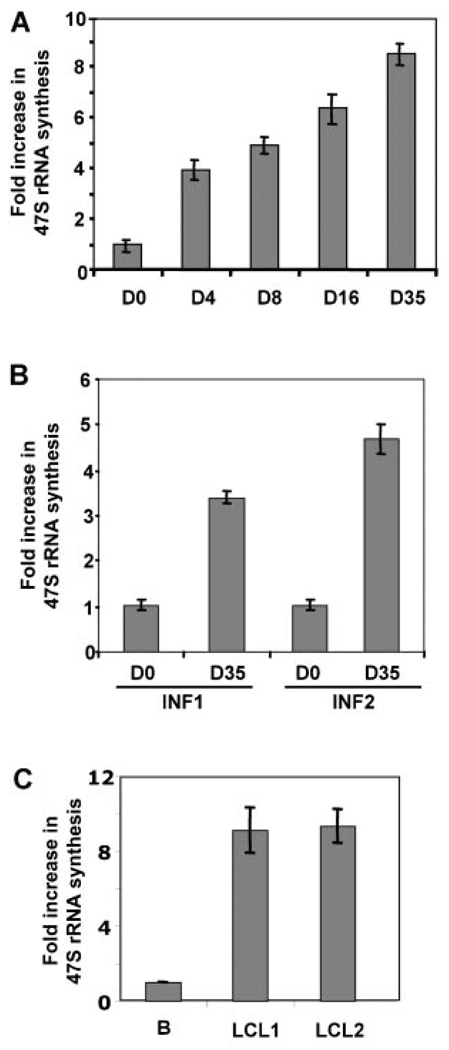
EBV, an oncogenic herpes virus capable of immortalizing primary B-lymphocytes into indefinitely proliferating lymphoblastoid cell lines in vitro, is associated with the development of endemic Burkitt’s lymphoma, some forms of Hodgkin’s lymphoma, and immune deficiency-associated lymphoproliferative disorders [Kieff and Rickinson, 2006]. As early as 5 days following primary infection, surviving B-cells show constitutive activation of multiple signal transduction pathways leading to cellular proliferation, resistance to apoptosis and upregulation of cell adhesion molecules [Rickinson and Kieff, 2001]. The events associated with EBV-driven progression of the B cell from resting lymphocyte to an immortalized, and eventually, a transformed state (LCLs) provide a unique system to study the regulation of ribosomal RNA synthesis. Analysis of ribosomal RNA (rRNA) synthesis by real-time PCR revealed a gradual increase in rRNA synthesis from day 0 to day 35 when the temporal change was followed from the day of infection (Fig. 1A). Similarly, a three- to fourfold increase in rRNA synthesis was detected in EBV-immortalized B-cells 35 days post-infection when compared to cells prior to infection (Fig. 1B). The difference in the level of rRNA synthesis 35 days post-infection in different samples can be attributed to the variability between different B-cell donors. As expected, exponentially growing established EBV-transformed B-cell lines (LCLs) demonstrate similar increase in rRNA synthesis compared to resting B-cells (Fig. 1C).
Fig. 1.
EBV transformation of B-cells results in increased ribosomal RNA synthesis compared to resting B-cells. A: Total RNA was isolated from EBV infected B-cells harvested on Days 0, 4, 8, 16, and 35 and ribosomal gene transcription was measured by real-time PCR. Fold increase in 47S rRNA synthesis was calculated after normalization to β-actin. B: Freshly isolated B-cells from two individual donors were infected with EBV and total RNA was isolated on day 0 and Day 35. 47S rRNA synthesis was analyzed by real-time PCR in B-cells from two individual donors (INF1 and INF2) normalized to β-actin and compared between day 0 and EBV infected B-cell populations on Day 35. C: Total RNA isolated from EBV transformed B-cell lines was subjected to Real-time PCR. 47S rRNA synthesis normalized to b-actin in resting B-cells (B) was compared with that in two different EBV transformed lymphoblastoid cell lines (LCL1 and LCL2). Error bars represents standard deviation of triplicate measurements.
UPSTREAM BINDING FACTOR (UBF) IS EXCLUSIVELY ASSOCIATED WITH THE UNMETHYLATED rRNA PROMOTERS IN BOTH RESTING AND EBV TRANSFORMED B-CELLS
The recruitment of UBF, a key regulatory factor required for rDNA transcription, to the rRNA promoters is one of the first steps in rRNA synthesis. Since rDNA expression dramatically increased upon B-cell transformation, we analyzed the expression of UBF transcript and protein during the time course of EBV immortalization of resting B-cells and in established EBV transformed LCLs. Although variations in both UBF mRNA and protein level were observed in LCLs and during the course of EBV-infection, UBF protein was abundant in the resting B-cells where rRNA synthesis was negligible (Fig. 2A–D). This observation reiterates the importance of UBF for basal rDNA transcription, and the requirement of other chromatin remodeling proteins and transcription factors in regulating rDNA transcription in response to growth stimuli.
The majority of approximately 400 copies of rDNA present in the mammalian cells is methylated and silenced in somatic cells [Leary and Huang, 2001; McStay et al., 2002]. Because of our interest in regulation of rDNA transcription in EBV-transformed lymphoma cells, we compared the nature of UBF association with the rRNA promoters in EBV transformed cells (LCLs) with that in resting B-cells, using the ChIP-CHOP assay as described [Majumder et al., 2006]. In brief, DNA pulled down with anti-UBF antibody as well as the input DNA from each cell line, was digested with methylation sensitive enzyme Hpa II, its methylation insensitive isoschizomer Msp I (digestion control), or mock digested prior to amplification with rDNA specific primers (fragment shown in Fig. 2E). Since this amplified promoter region harbors four Hpa II/Msp I sites, the Hpa II-resistant PCR product represents the proportion of the rRNA promoter methylated at all four sites (represented as methylated DNA in the bar diagram). The PCR product from the undigested DNA minus that from the Hpa II-resistant product represents the level of unmethylated promoter (represented as unmethylated DNA in the bar diagram). Because Msp I is a methylation insensitive enzyme, PCR product was not generated from Msp I-digested DNA. Although ChIP-CHOP assay revealed 90% and ~75% of the rRNA promoters to be methylated in resting B-cells and the LCLs, respectively (data not shown), UBF was exclusively associated with the unmethylated promoters in both resting B-cells and the two cell lines tested (Fig. 2F, see legends for details of this procedure). Anti-UBF antibody failed to pull down any detectable methylated rRNA promoter. The lack of rDNA transcription in resting B-cells despite considerable UBF association with the unmethylated promoters reinforces the notion that post-translational modification of UBF [O’Mahony et al., 1992; Voit and Grummt, 2001] is required for transcription initiation.
PRMT5 REGULATES rRNA PROMOTER ACTIVITY IN EBV TRANSFORMED B-CELLS
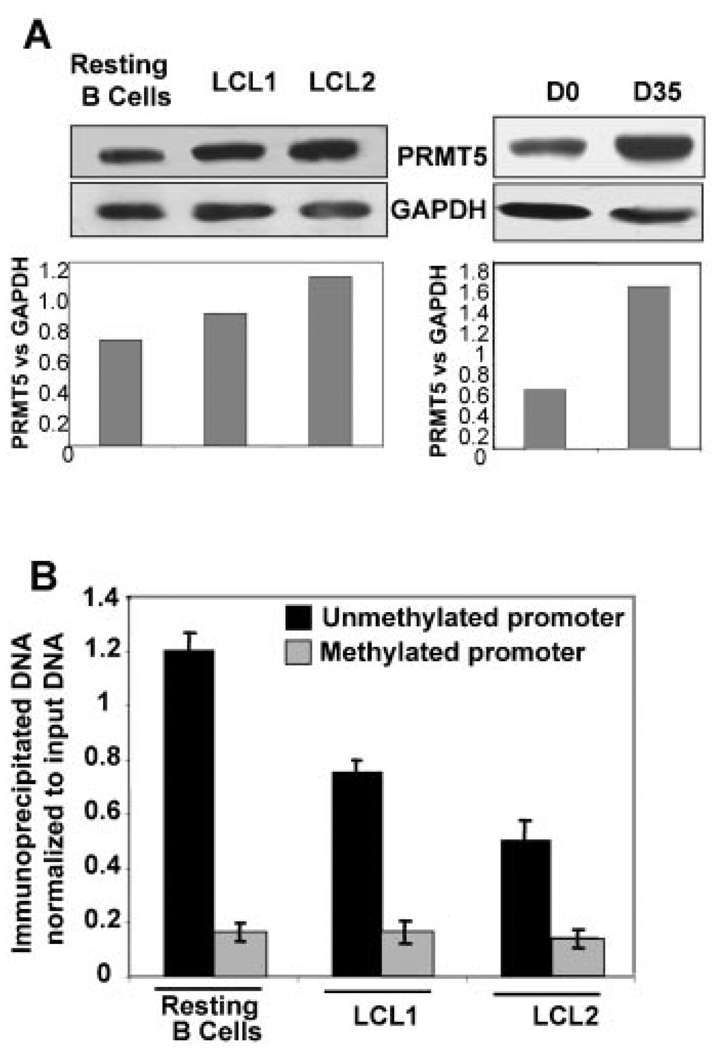
N-methylation of protein arginine residues is a post-translational modification that has been a subject of intense investigation, particularly with regard to its role in cell function. Protein arginine methyltransferases (PRMT) catalyze the methylation of arginine residues of a diverse group of proteins that can be monomethylated, symmetrically dimethylated or asymmetrically dimethylated at the terminal nitrogen atoms of the guanidinium side chain of the arginine residue. PRMT5 is one of the several newly cloned members of the PRMT family that is known to function as a transcriptional repressor of genes transcribed by RNA Polymerase II. The role of this protein as a transcriptional repressor is largely due to its ability to methylate the arginine 8 residue of histone H3 (H3R8Me2) and the arginine 3 residue of histone H4 (H4R3Me2), the two histone post-translational modifications relevant to transcriptional regulation mediated by PRMT5 [Pal and Sif, 2007]. Here, we explored the potential role of PRMT5 in regulating RNA polymerase I –mediated transcription of rDNA. Analysis of PRMT5 expression revealed abundant expression of the protein in resting B-cells and increased expression of the protein in LCLs and EBV-immortalized B-cells (Fig. 3A), as reported earlier [Dacwag et al., 2007].
Fig. 3.
Association of PRMT5 with rRNA promoter is reduced in EBV transformed B-cells. A: PRMT5 level was monitored by Western blot analysis in EBV transformed cells (LCLs), EBV-infected and resting B cells. B: Resting B-cells and exponentially growing EBV transformed cells (LCL1, LCL2) were cross-linked with formaldehyde and chromatin from these cells was immunoprecipitated with anti-PRMT5 antibody. The pulled down DNA was either left uncut (U) or digested with Hpa II (H) or Msp I (M). rRNA promoter was amplified from all three sets of precipitated and digested DNA and analyzed by semi-quantitative PCR. The bar diagram represents association of PRMT5 with unmethylated and methylated rRNA promoter (as described in Fig. 2F).
We next analyzed association of PRMT5 with the methylated and unmethylated promoters in resting B-cells and LCLs by ChIP-CHOP assay. The data revealed almost exclusive association of PRMT5 with unmethylated rRNA promoters and a small but reproducible association with the methylated promoters in resting B-cells (Fig. 3B). A similar pattern of PRMT5 association with the methylated and unmethylated rRNA promoters was also observed in both LCLs. PRMT5 association with the unmethylated promoters in LCLs was, however, significantly lower (42–59%) than that in resting B-cells (Fig. 3B), which was consistent with higher rRNA promoter activity in the LCLs compared to the resting B-cells.
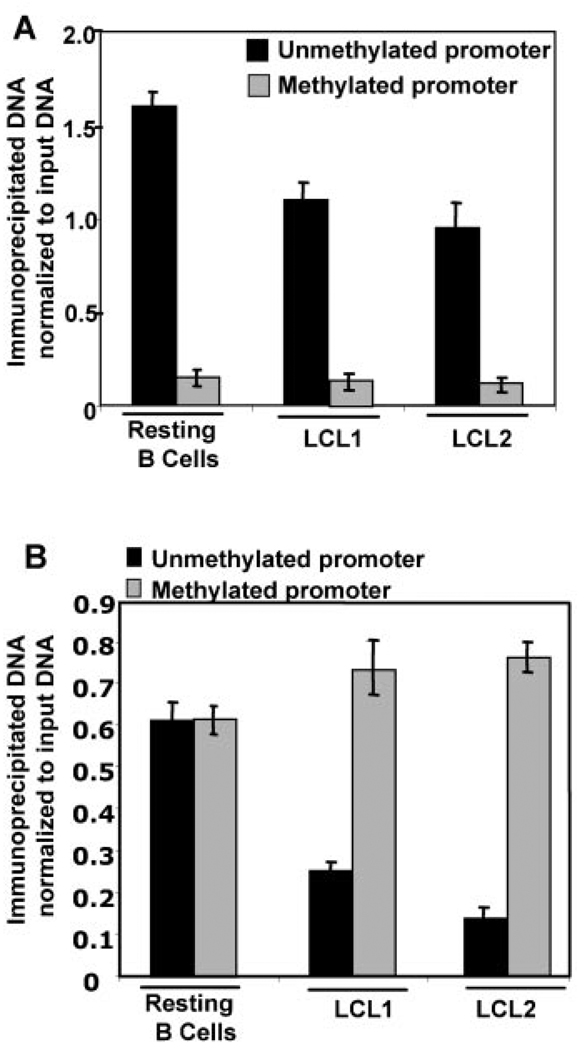
Transcriptional repression of the Rb and ST7 tumor suppressor genes is regulated by PRMT5-induced methylation of R8 residue of H3 and R3 residue of H4 associated with the respective promoters [Pal et al., 2004]. First, we investigated the possibility that the reduced level of PRMT5 mediated rDNA transcription is caused by specific R8 methylation of histone H3 recruited to rRNA promoters. To test this possibility, DNA pulled down with antisera specific for the methylated histone was subjected to digestion with methylation sensitive restriction enzyme Hpa II, followed by amplification with gene-specific primers. Indeed, the methylation of histone H3 at R8 associated with the unmethylated rRNA promoters in resting B-cells was significantly higher (1.5- to 2-fold) than that in rapidly proliferating LCLs (Fig. 4A). The extent of H3R8 methylation associated with the methylated promoter in both cell types was fiveto eightfold less than that observed with the unmethylated promoter. This data suggests that H3R8Me2 is largely involved in suppression of the unmethylated rRNA promoter in B-lymphocytes and LCLs with minimal effect on the methylated promoter, and that H3R8Me2 is a key repressor histone involved in PRMT5-mediated repression of transcription- ready (unmethylated) rRNA promoter.
Fig. 4.
A: H3R8Me2 associates preferentially with unmethylated rRNA promoter in resting B-cells, which is reduced in EBV transformed B-cells. Formaldehyde cross-linked chromatin prepared from resting B-cells and exponentially growing EBV transformed cells (LCL1, LCL2) was immunoprecipitated with anti-H3R8Me2 antibody. The pulled down DNA was either left uncut (U) or digested with Hpa II (H) or Msp I (M). rRNA promoter was amplified from all three sets of precipitated and digested DNA and analyzed by semi-quantitative PCR. Association of H3R8Me2 with unmethylated and methylated rRNA promoter is represented in the bar diagram (as described in Fig. 2F). B: H4R3Me2 associates preferentially with methylated rRNA promoter in the EBV transformed B-lymphocytes. Chromatin from resting B-cells and exponentially growing EBV transformed cells (LCL1, LCL2) was immunoprecipitated with anti-H4R3Me2 antibody, and digested with Hpa II (H) or Msp I (M). rRNA promoter was amplified from uncut (U) and restriction enzyme digested DNA by semi-quantitative PCR. H4R3Me2 binding to unmethylated and methylated rRNA promoter is represented in the bar diagram (as described in Fig. 2F).
We then used ChIP-CHOP assay to determine the methylation pattern of another key substrate of PRMT5, histone H4 at R3 residue, recruited to the rRNA promoter. In contrast to H3R8Me2, histone H4 associated with both unmethylated and methylated promoters in the resting B-cells was methylated at R3 residue (Fig. 4B). A marginal increase in the methylation of H4R3 associated with the methylated promoters in LCLs was observed compared to the resting B-cells. However, methylation of H4 at R3 associated with the unmethylated promoters was reduced significantly (>50%) in the highly proliferative LCLs, implying that this repressive histone modification is one of the key regulators of rDNA transcription. Similarly, consistent methylation of the H4 fraction recruited to the methylated rRNA promoters in both resting B-cells and LCLs also indicates that this histone modification probably facilitates suppression of the methylated rRNA promoter irrespective of proliferating status of the cells.
PRMT5 PREFERENTIALLY ASSOCIATES WITH UNMETHYLATED rRNA PROMOTERS IN SERUM STARVED HeLa CELLS
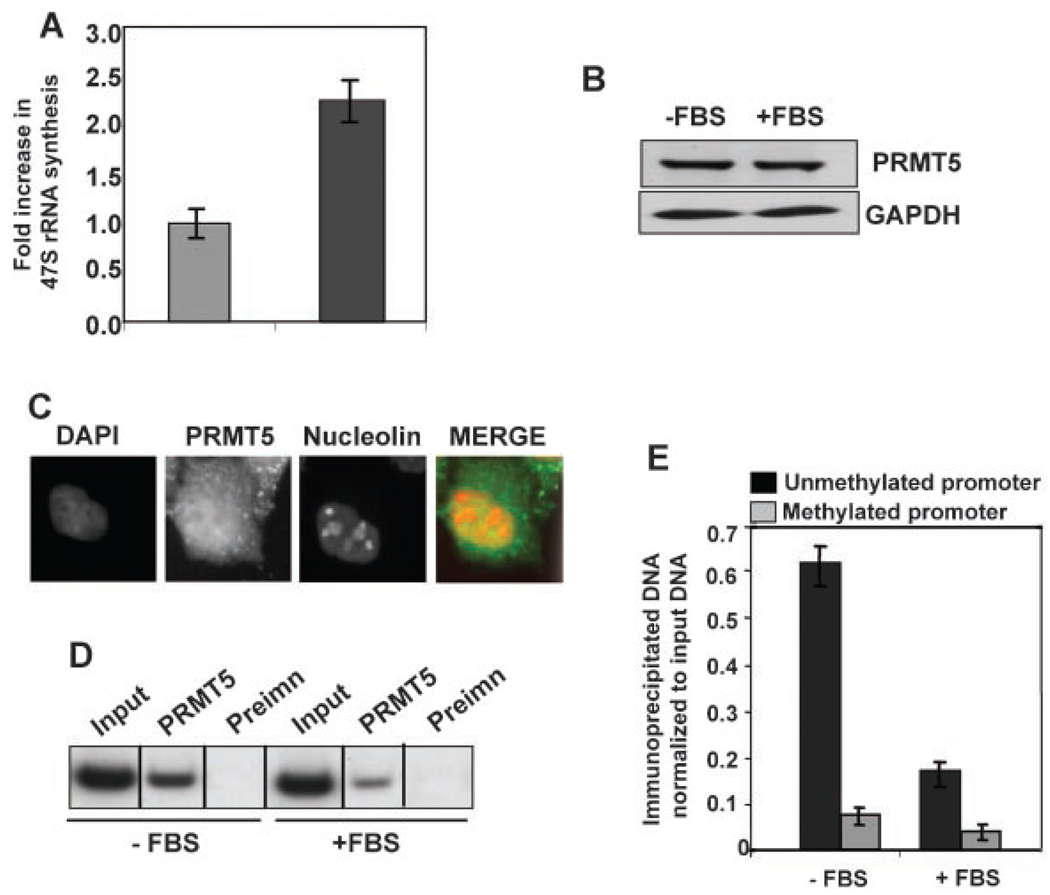
To determine if regulation of rDNA transcription by PRMT5 is a general phenomenon, we used a classical cell culture model for studying rapid alterations in rRNA synthesis. Ribosomal RNA synthesis is usually at its minimum in serum starved non-proliferating cells and a dramatic increase occurs immediately upon supplementation with serum [O’Mahony et al., 1992]. We took advantage of this “OFF” and “ON” state of rDNA transcription in HeLa cells to elucidate the role of PRMT5 and the modified histones in ribosomal gene expression. A twofold increase in ribosomal RNA synthesis was observed in cells grown in serum-enriched medium, as measured by increase in the 47S rRNA synthesis (Fig. 5A). The level of PRMT5 remained essentially unaltered in serum starved and serum supplemented HeLa cells (Fig. 5B). In addition, immunofluorescence studies showed that PRMT5 is present in both cytoplasm and nuclear compartment of HeLa cells. Co-localization of PRMT5 with nucleolin, a nucleolus specific protein, confirmed that PRMT5 is also present in the HeLa cell nucleolus, the site of rRNA synthesis (Fig. 5C).
Fig. 5.
PRMT5 expression remains unaltered in serum starved and serum replenished HeLa cells. A: Exponentially growing HeLa cells were serum starved (0%) for 72 h (−FBS) and then allowed to grow in presence of 10% serum for 3 h. 47S rRNA synthesis in these cells was compared by real time PCR and normalized to β-actin. B: Whole cell extracts from HeLa cells (−FBS and +FBS) were subjected to Western blot analysis with anti-PRMT5 antibody. The blot was reprobed with anti-GAPDH antibody for protein normalization. C: PRMT5 co-localizes with nucleolin in HeLa cell nucleolus. HeLa cells were stained with TRITC tagged mouse monoclonal antibody against nucleolin and with FITC-tagged rabbit polyclonal antibody against PRMT5. The cells were also stained with DAPI and visualized under fluorescence microscope. D: PRMT5 preferentially associates with unmethylated rRNA promoter in serum starved HeLa cells. Formaldehyde crosslinked chromatin prepared from serum starved (−FBS) and serum replenished (+FBS) HeLa cells was immunoprecipitated with anti-PRMT5 antibody (PRMT5) or preimmune sera (Preimn), and rRNA promoter was amplified from immunoprecipitated DNA and input DNA (Input). E: The immunoprecipitated DNA was either left undigested, or digested with Hpa II or Msp I. rRNA promoter was amplified from uncut, Hpa II and Msp I digested DNA by semi-quantitative PCR. The bar diagram represents PRMT5 binding to unmethylated and methylated rRNA promoter (as described in Fig. 2F).
To establish the role of PRMT5 in silencing rDNA transcription, rRNA promoter was amplified from the DNA of serum starved and serum supplemented HeLa cells immunoprecipitated with anti-PRMT5 antibody. PCR amplification of the immunoprecipitated DNA (undigested) showed that association of PRMT5 with rRNA promoters in serum-starved cells was 3.5-fold higher than that in the serum-replenished cells (Fig. 5D). Quantification of PRMT5 association with unmethylated and methylated rRNA promoter demonstrated predominant association of PRMT5 with the unmethylated promoters in serum-starved cells that was reduced by 70% in serum-replenished cells (Fig. 5F). Although the association of PRMT5 with methylated rRNA promoters under both conditions was minimal, a 50% reduction in its association was observed in serum-supplemented cells compared to the serum-starved cells. Based on this data we conclude that PRMT5 plays a pivotal role in suppressing rDNA transcription from the unmethylated promoters in HeLa cells when the cells are in the stationary phase or non-proliferating state.
HISTONES H3 AND H4 ASSOCIATED WITH THE METHYLATED AND UNMETHYLATED rRNA PROMOTERS IN HELA CELLS ARE DIFFERENTIALLY METHYLATED
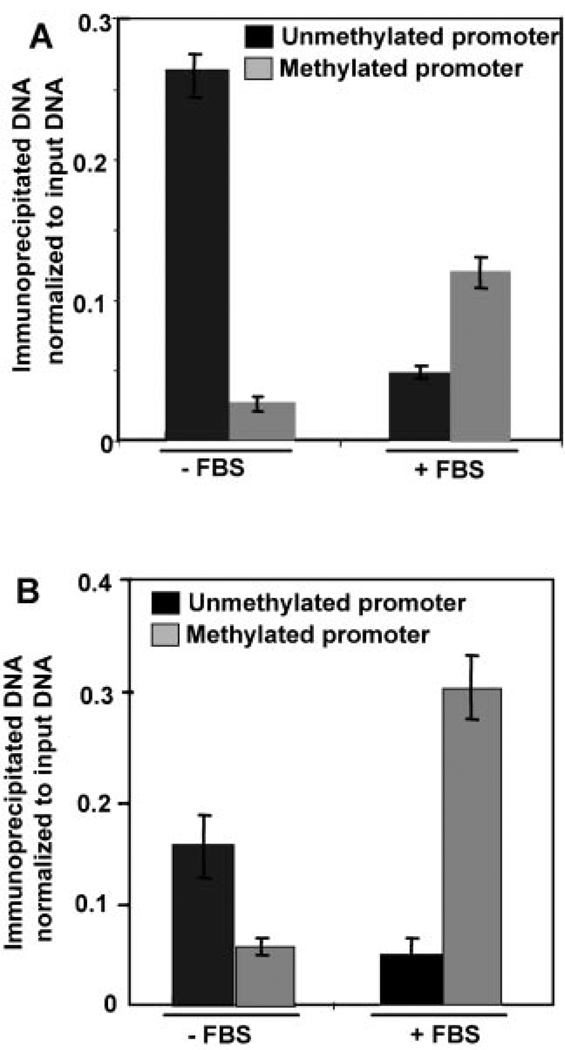
As observed with PRMT5, ChIP-CHOP assay with anti-H3R8Me2 antibody revealed minimal R8 methylation of histone H3 associated with actively transcribing rRNA promoters in the serum replenished HeLa cells. Arginine 8 methylation of H3 bound with rRNA promoters in serum starved HeLa cells was at least twice that observed in serum-enriched cells (Fig. 6A). Interestingly, major fraction of H3R8Me2 associated rRNA promoter in the serum-starved cells were unmethylated, and only 10% of the H3R8Me2 associated rRNA promoters in the serum starved cells were methylated. The extent of methylation of H3 at R8 within the methylated rRNA promoter region in the serum-enriched cells was relatively small and was twice that associated with the unmethylated promoter (Fig. 6A). These data suggest that H3R8Me2 suppresses rDNA expression from the transcriptionally competent unmethylated promoter, but it is demethylated in cells actively transcribing rDNA.
Fig. 6.
H3R8Me2 preferentially associates with the unmethylated rRNA promoter in serum starved HeLa cells and its association is reduced upon serum supplementation. A: Serum starved and serum supplemented HeLa cells were cross-linked with formaldehyde and chromatin prepared from these cells was immunoprecipitated with anti-H3R8Me2 antibody. The pulled down DNA was either left uncut (U) or digested with Hpa II (H) or Msp I (M). rRNA promoter was amplified from all three sets of precipitated and digested DNA and analyzed by semi-quantitative PCR. H3R8Me2 binding to unmethylated and methylated rRNA promoter is represented in the bar diagram (as described in Fig. 2F). B: H4R3Me2 preferentially associates with unmethylated promoter in serum starved and methylated promoter in serum-supplemented cells. Chromatin prepared from serum starved and serum supplemented HeLa cells was immunoprecipitated with anti-H4R3Me2 antibody. The pulled down DNA was either left uncut (U) or digested with Hpa II (H) or Msp I (M). rRNA promoter was amplified from all three sets of precipitated and digested DNA and analyzed by semi-quantitative PCR. H4R3Me2 binding to unmethylated and methylated rRNA promoter is represented after normalization to input DNA (as described in Fig. 2F).
As observed with H3R8Me2, H4 associated with the unmethylated promoter in the serum-starved cells in which rRNA promoter is in inactive state was significantly methylated at R3 residue (Fig. 6B). Unlike H3R8Me2, a larger proportion of H4R3Me2 was associated with transcriptionally inactive methylated rRNA promoter in the serum supplemented HeLa cells suggesting that the suppression of the methylated promoter by PRMT5 is manifested essentially by methylating its substrate H4 at R3 recruited to this promoter (Fig. 6B). It is noteworthy that, there is a remarkable difference in methylation of H4R3 associated with the rRNA promoters in the resting B-cells, and in the serum starved HeLa cells. A significantly high level of methylation of H4R3 recruited to the rRNA promoter in resting B-cells implicates that this modified histone is a repressor mark on long term inactive promoter, but may not play a significant role in exponentially growing cells in culture. It is also interesting to note that despite reduced association of PRMT5 with the rRNA promoter in actively dividing cells, H4R3Me2 is enriched on the methylated promoters in these cells. It is conceivable that arginine 3 residue of H4 histone is also a substrate of other arginine methyltransferases that may be involved in regulating rRNA promoter function.
ALTERED EXPRESSION OF PRMT5 IN VIVO AFFECTS THE PROMOTER ACTIVITY OF BOTH ENDOGENOUS AND TRANSFECTED rDNA
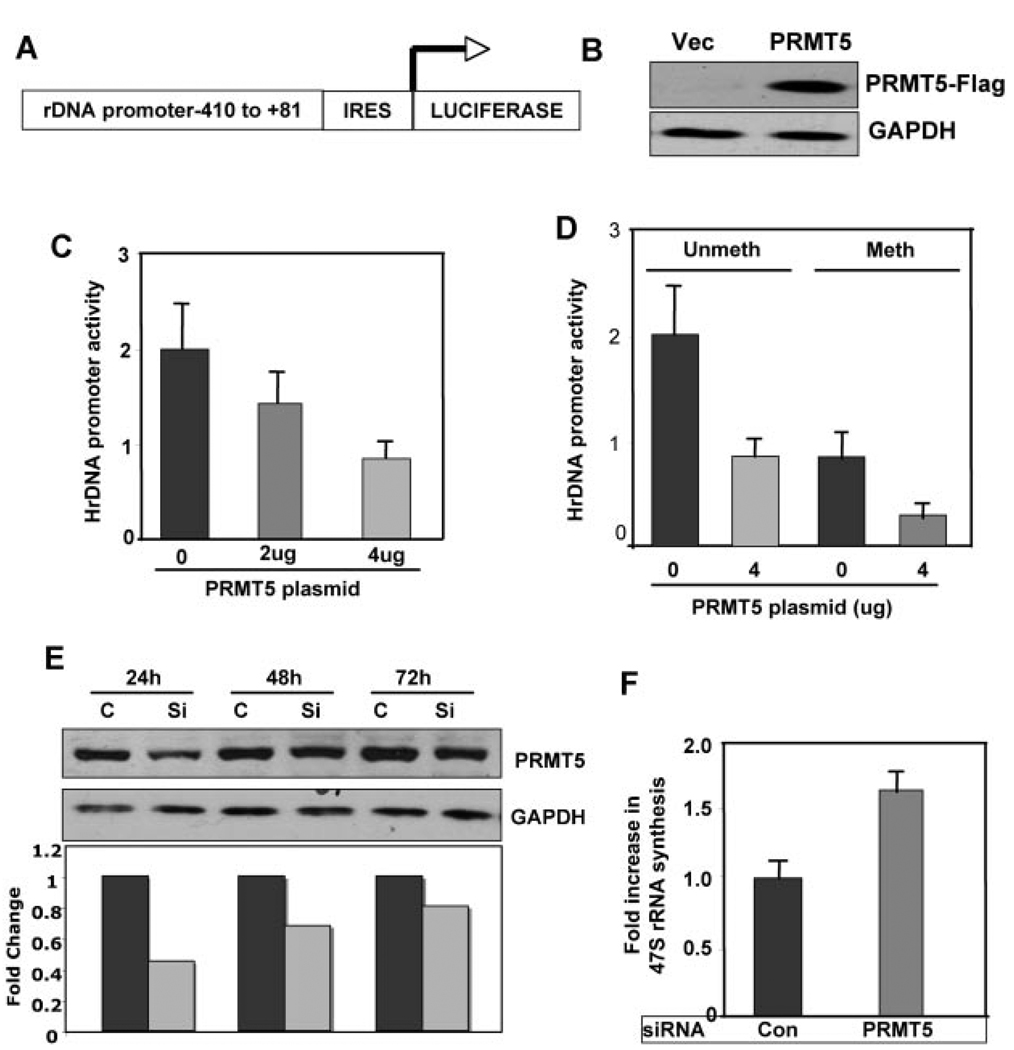
To confirm that PRMT5 directly affects ribosomal gene transcription, transient transfection assays were performed with the rRNA promoter driven Luciferase expression vector pHrD-IRES-Luc (Fig. 7A) [Ghoshal et al., 2004]. HeLa cells were transfected with PRMT5 expression vector or the corresponding empty vector along with pHrD-IRES-Luc and the internal control pRLTK and the expression of Flag-tagged PRMT5 was detected with anti-Flag antibody (Fig. 7B). A dose dependent decrease in rRNA promoter driven Luciferase activity was observed upon ectopic expression of PRMT5 compared to the vector transfected HeLa cells (Fig. 7C). Since we observed a significant association of PRMT5 with the methylated fraction of the rRNA promoter in HeLa cells, normal B-cells and LCLs, we next explored the effect of PRMT5 on the methylated rRNA promoter. A 50% inhibition of the methylated rRNA promoter was observed upon PRMT5 overexpression (Fig. 7D).
Fig. 7.
Ectopic expression of PRMT5 in HeLa cells inhibits the activity of both unmethylated and methylated rRNA promoter. A: Schematic representation of the rRNA promoter/luciferase reporter plasmid (pHrD-IRES-Luc) used in the transfection study. B: HeLa cells were transiently transfected with Flag tagged PRMT5 expression vector. The expression of PRMT5 protein was monitored by Western blot analysis using anti-Flag M2-monoclonal antibody. C: HeLa cells were transiently transfected with empty vector (0) or 2 and 4 µg of PRMT5 expression vector along with pHrD-IRES-Luc and the internal control pRL-TK. Cells harvested 48 h after transfection were analyzed for luciferase activity and normalized to pRL-TK. D: HeLa cells were transiently transfected with empty vector (0) and 4 µg of PRMT5 expression vector along with unmethylated or methylated pHrD-IRES-Luc and pRL-TK. Cells harvested 48 h after transfection were analyzed for luciferase activity and normalized to pRL-TK. E: HeLa cells were transfected with PRMT5 siRNA or control siRNA and expression of PRMT5 was monitored by western blot analysis at 24, 48, and 72 h after transfection. F: HeLa cells transfected with PRMT5 siRNA or control siRNA were harvested 24 h posttransfection and rRNA synthesis was monitored by real time PCR. Error bars represents standard deviation of triplicate measurements.
We next sought to determine if depletion of PRMT5 has any effect on endogenous ribosomal RNA synthesis. For this purpose, PRMT5 was depleted from HeLa cells by siRNA that resulted in 50% reduction in PRMT5 protein within 24 h posttransfection compared to the scrambled siRNA transfected cells (Fig. 7E). The extent of PRMT5 depletion was followed for 72 h, when only a 25% reduction in PRMT5 level was observed compared to the scrambled siRNA transfected cells. Since the effect of siRNA was maximal at 24 h we analyzed rDNA transcription at this time point by measuring ribosomal RNA synthesis in a 70% confluent cell population. A consistent increase in rRNA promoter activity was observed in PRMT5 depleted cells compared to the scrambled siRNA transfected cells (Fig. 7F). These data suggest that PRMT5 plays a direct role in regulating rDNA transcription by virtue of its ability to methylate histones H3 and H4.
DISCUSSION
The primary objective of this study was to establish the role of arginine methyltransferase PRMT5 in ribosomal gene expression. Although recent studies have revealed a relationship between histone H4 methylation by arginine methyltransferase PRMT5 and DNA methylation, and its role in gene silencing, these studies were confined to genes transcribed by RNA polymerase II [Pal et al., 2003, 2004; Zhao et al., 2009]. The present study has demonstrated that PRMT5 can also regulate Pol I-directed transcription in response to EBV transformation or to altered serum levels influencing cell growth. While previous studies have established the role of DNA methylation in the regulation of rDNA expression [Santoro and Grummt, 2001, 2005; Ghoshal et al., 2004; Majumder et al., 2006], the present study has shown that methylation of histones H3 and H4 to H3R8Me2 and H4R3Me2 by PRMT5 plays a role in the suppression of methylated rDNA and in modulating the promoter activity of unmethylated rDNA. This observation implicates a relationship between specific histone methylation and DNA methylation in rDNA expression. The localization of PRMT5 in the nucleolus, the site of rRNA synthesis, in addition to extranucleolar fraction, further supports the potential role of this arginine methyltransferase in rDNA expression. It would be of interest to explore the possibility that some or most of this enzyme may be transported from the nucleolus to other cellular compartments in cancer cells.
An interesting finding was that while adequate levels of the essential Pol I transcription factor UBF was associated with the functional unmethylated rDNA, rDNA expression was minimal at best in resting B cells. This result further supports the notion that UBF association with functional rDNA is not sufficient for active rRNA synthesis. This conclusion is not inconsistent with the recent report that depletion of UBF leads to stable and reversible methylation-independent silencing of rRNA genes, and that UBF levels regulate active rRNA gene chromatin during growth and differentiation [Sanij et al., 2008]. This study does not rule out requirement of UBF activation and the need for unmethylated promoters for active transcription of ribosomal genes. Therefore, in addition to UBF loading on the promoter, it is highly likely that activation of rRNA promoter requires dissociation of repressors such as PRMT5 and its target methylated histones from the unmethylated rDNA.
A key observation was the differential association of PRMT5 with methylated and unmethylated promoters depending upon the growth rate of the cells. This arginine methyltransferase bound almost exclusively to the unmethylated functionally competent rRNA promoter in resting B cells, resulting in methylation of histones and subsequent repressor complex formation which is likely to prevent recruitment of positive transcription factors to the promoter. In response to EBV infection or growth stimulation, the association of PRMT5 with the unmethylated promoter was dramatically reduced, which exposes the transcriptionally competent promoter to factors essential for active transcription. The dissociation of histone methyltransferase from the unmethylated promoter under these growth-promoting conditions was consistent with the increased rRNA synthesis in vivo. In this context, it is noteworthy that PRMT5 can form complexes with the hSW1/SNF chromatin remodeling factors BRG and BRM, and this association increases the methyltransferase activity [Pal et al., 2004]. It should be noted that there are exceptions to the notion that PRMT5 along with dimethylated histone marks functions only as a transcriptional repressor. Thus, RNAi-induced reduction of PRMT levels leads to decreased IL-2 expression [Richard et al., 2005]. Another example is the requirement of PRMT5 for myogenin transcription associated with muscle differentiation [Dacwag et al., 2007]. It is conceivable that methylated H3R8 and H4R3 may recruit specific transactivators depending upon the cell type and physiological state. It is evident from the present study that PRMT5 is involved in the suppression of rRNA synthesis. While it would be of interest to follow the kinetics of association/dissociation of these repressor molecules with the rRNA promoter at different stages of EBV infection it is technically challenging due to the lack of sufficient chromatin from cells at early time points.
Finally, PRMT5 has recently been implicated in multiple events that contribute to cellular transformation including transcriptional repression of tumor suppressor genes and modulation of p53 [Jansson et al., 2008] and NFkB activity [Tanaka et al., 2009]. PRMT5 over expression has been documented in multiple non-Hodgkin’s lymphoma cell lines and primary mantle cell lymphoma tumor samples [Dacwag et al., 2007]. The results reported here provide a comprehensive example of how in spite of PRMT5 over expression differential association of this enzyme and its substrates with different population of rRNA promoter may contribute towards cell growth and transformation.
ACKNOWLEDGMENTS
We thank Dr. Kalpana Ghoshal for helpful discussion. This work was supported in part, by the grants CA101956 and CA086978 from the National Institutes of Health.
Grant sponsor: National Institutes of Health; Grant numbers: CA101956, CA086978.
Abbreviations used
- pol II
RNA polymerase II
- pol I
RNA polymerase I
- rDNA
ribosomal DNA
- rRNA
ribosomal RNA
- MBD
methyl CpG domain binding protein
- PRMT
protein arginine methyl transferase
REFERENCES
- Ancelin K, Lange UC, Hajkova P, Schneider R, Bannister AJ, Kouzarides T, Surani MA. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat Cell Biol. 2006;8:623–630. doi: 10.1038/ncb1413. [DOI] [PubMed] [Google Scholar]
- Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- Brown SE, Szyf M. Epigenetic programming of the rRNA promoter by MBD3. Mol Cell Biol. 2007;27:4938–4952. doi: 10.1128/MCB.01880-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZJ, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription: A role for DNA methylation and histone modification in nucleolar dominance. Genes Dev. 1997;11:2124–2136. doi: 10.1101/gad.11.16.2124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dacwag CS, Ohkawa Y, Pal S, Sif S, Imbalzano AN. The protein arginine methyltransferase Prmt5 is required for myogenesis because it facilitates ATP-dependent chromatin remodeling. Mol Cell Biol. 2007;27:384–394. doi: 10.1128/MCB.01528-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dierksheide JE, Baiocchi RA, Ferketich AK, Roychowdhury S, Pelletier RP, Eisenbeis CF, Caligiuri MA, VanBuskirk AM. IFN-gamma gene polymorphisms associate with development of EBV + lymphoproliferative disease in hu PBL-SCID mice. Blood. 2005;105:1558–1565. doi: 10.1182/blood-2003-07-2476. [DOI] [PubMed] [Google Scholar]
- Fuks F. DNA methylation and histone modifications: Teaming up to silence genes. Curr Opin Genet Dev. 2005;15:490–495. doi: 10.1016/j.gde.2005.08.002. [DOI] [PubMed] [Google Scholar]
- Fuks F, Hurd PJ, Wolf D, Nan X, Bird AP, Kouzarides T. The methyl-CpG-binding protein MeCP2 links DNA methylation to histone methylation. J Biol Chem. 2003;278:4035–4040. doi: 10.1074/jbc.M210256200. [DOI] [PubMed] [Google Scholar]
- Ghoshal K, Datta J, Majumder S, Bai S, Dong X, Parthun M, Jacob ST. Inhibitors of histone deacetylase and DNA methyltransferase synergistically activate the methylated metallothionein I promoter by activating the transcription factor MTF-1 and forming an open chromatin structure. Mol Cell Biol. 2002;22:8302–8319. doi: 10.1128/MCB.22.23.8302-8319.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghoshal K, Majumder S, Datta J, Motiwala T, Bai S, Sharma SM, Frankel W, Jacob ST. Role of human ribosomal RNA (rRNA) promoter methylation and of methyl-CpG-binding protein MBD2 in the suppression of rRNA gene expression. J Biol Chem. 2004;279:6783–6793. doi: 10.1074/jbc.M309393200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grummt I, Pikaard CS. Epigenetic silencing of RNA polymerase I transcription. Nat Rev Mol Cell Biol. 2003;4:641–649. doi: 10.1038/nrm1171. [DOI] [PubMed] [Google Scholar]
- Hosohata K, Li P, Hosohata Y, Qin J, Roeder RG, Wang Z. Purification and identification of a novel complex which is involved in androgen receptor-dependent transcription. Mol Cell Biol. 2003;23:7019–7029. doi: 10.1128/MCB.23.19.7019-7029.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansson M, Durant ST, Cho EC, Sheahan S, Edelmann M, Kessler B, La Thangue NB. Arginine methylation regulates the p53 response. Nat Cell Biol. 2008;10:1431–1439. doi: 10.1038/ncb1802. [DOI] [PubMed] [Google Scholar]
- Kieff E, Rickinson A. Epstein-Barr virus and its replication. In: Howley DMKaPM., editor. Fields virology. Philadelphia, PA: Lippincott, Williams, and Wilkins; 2006. pp. 2603–2654. [Google Scholar]
- Leary DJ, Huang S. Regulation of ribosome biogenesis within the nucleolus. FEBS Lett. 2001;509:145–150. doi: 10.1016/s0014-5793(01)03143-x. [DOI] [PubMed] [Google Scholar]
- Majumder S, Ghoshal K, Datta J, Bai S, Dong X, Quan N, Plass C, Jacob ST. Role of de novo DNA methyltransferases and methyl CpG-binding proteins in gene silencing in a rat hepatoma. J Biol Chem. 2002;277:16048–16058. doi: 10.1074/jbc.M111662200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Majumder S, Ghoshal K, Datta J, Smith DS, Bai S, Jacob ST. Role of DNA methyltransferases in regulation of human ribosomal RNA gene transcription. J Biol Chem. 2006;281:22062–22072. doi: 10.1074/jbc.M601155200. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- McStay B, Grummt I. The epigenetics of rRNA genes: From molecular to chromosome biology. Annu Rev Cell Dev Biol. 2008;24:131–157. doi: 10.1146/annurev.cellbio.24.110707.175259. [DOI] [PubMed] [Google Scholar]
- McStay B, Paule M, Schultz MC, Willis I, Pikaard CS. Oddpols united: New insights into transcription by RNA polymerases I and III. Gene Expr. 2002;10:263–269. doi: 10.3727/000000002783992415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Mahony DJ, Xie WQ, Smith SD, Singer HA, Rothblum LI. Differential phosphorylation and localization of the transcription factor UBF in vivo in response to serum deprivation. In vitro dephosphorylation of UBF reduces its transactivation properties. J Biol Chem. 1992;267:35–38. [PubMed] [Google Scholar]
- Pal S, Sif S. Interplay between chromatin remodelers and protein arginine methyltransferases. J Cell Physiol. 2007;213:306–315. doi: 10.1002/jcp.21180. [DOI] [PubMed] [Google Scholar]
- Pal S, Yun R, Datta A, Lacomis L, Erdjument-Bromage H, Kumar J, Tempst P, Sif S. mSin3A/histone deacetylase 2- and PRMT5-containing Brg1 complex is involved in transcriptional repression of the Myc target gene cad. Mol Cell Biol. 2003;23:7475–7487. doi: 10.1128/MCB.23.21.7475-7487.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Vishwanath SN, Erdjument-Bromage H, Tempst P, Sif S. Human SWI/SNF-associated PRMT5 methylates histone H3 arginine 8 and negatively regulates expression of ST7 and NM23 tumor suppressor genes. Mol Cell Biol. 2004;24:9630–9645. doi: 10.1128/MCB.24.21.9630-9645.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pal S, Baiocchi RA, Byrd JC, Grever MR, Jacob ST, Sif S. Low levels of miR-92b/96 induce PRMT5 translation and H3R8/H4R3 methylation in mantle cell lymphoma. EMBO J. 2007;26:3558–3569. doi: 10.1038/sj.emboj.7601794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard S, Morel M, Cleroux P. Arginine methylation regulates IL-2 gene expression: A role for protein arginine methyltransferase 5 (PRMT5) Biochem J. 2005;388:379–386. doi: 10.1042/BJ20040373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rickinson AB, Kieff E. Epstein-Barr virus. In: Howley DMKaPM., editor. Fields virology. Philadelphia: Lippincott Williams & Wilkins; 2001. [Google Scholar]
- Rickinson AB, Rowe M, Hart IJ, Yao QY, Henderson LE, Rabin H, Epstein MA. T-cell-mediated regression of “spontaneous” and of Epstein-Barr virus-induced B-cell transformation in vitro: Studies with cyclosporin A. Cell Immunol. 1984;87:646–658. doi: 10.1016/0008-8749(84)90032-7. [DOI] [PubMed] [Google Scholar]
- Sanij E, Poortinga G, Sharkey K, Hung S, Holloway TP, Quin J, Robb E, Wong LH, Thomas WG, Stefanovsky V, Moss T, Rothblum L, Hannan KM, McArthur GA, Pearson RB, Hannan RD. UBF levels determine the number of active ribosomal RNA genes in mammals. J Cell Biol. 2008;183:1259–1274. doi: 10.1083/jcb.200805146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santoro R, Grummt I. Molecular mechanisms mediating methylation-dependent silencing of ribosomal gene transcription. Mol Cell. 2001;8:719–725. doi: 10.1016/s1097-2765(01)00317-3. [DOI] [PubMed] [Google Scholar]
- Santoro R, Grummt I. Epigenetic mechanism of rRNA gene silencing: Temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol Cell Biol. 2005;25:2539–2546. doi: 10.1128/MCB.25.7.2539-2546.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarraf SA, Stancheva I. Methyl-CpG binding protein MBD1 couples histone H3 methylation at lysine 9 by SETDB1 to DNA replication and chromatin assembly. Mol Cell. 2004;15:595–605. doi: 10.1016/j.molcel.2004.06.043. [DOI] [PubMed] [Google Scholar]
- Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G, Reinberg D, Jenuwein T. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev. 2004;18:1251–1262. doi: 10.1101/gad.300704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamaru H, Zhang X, McMillen D, Singh PB, Nakayama J, Grewal SI, Allis CD, Cheng X, Selker EU. Trimethylated lysine 9 of histone H3 is a mark for DNA methylation in Neurospora crassa. Nat Genet. 2003;34:75–79. doi: 10.1038/ng1143. [DOI] [PubMed] [Google Scholar]
- Tanaka H, Hoshikawa Y, Oh-hara T, Koike S, Naito M, Noda T, Arai H, Tsuruo T, Fujita N. PRMT5, a novel TRAIL receptor-binding protein, inhibits TRAIL-induced apoptosis via nuclear factor-kappaB activation. Mol Cancer Res. 2009;7:557–569. doi: 10.1158/1541-7786.MCR-08-0197. [DOI] [PubMed] [Google Scholar]
- Voit R, Grummt I. Phosphorylation of UBF at serine 388 is required for interaction with RNA polymerase I and activation of rDNA transcription. Proc Natl Acad Sci USA. 2001;98:13631–13636. doi: 10.1073/pnas.231071698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young LS, Murray PG. Epstein-Barr virus and oncogenesis: From latent genes to tumours. Oncogene. 2003;22:5108–5121. doi: 10.1038/sj.onc.1206556. [DOI] [PubMed] [Google Scholar]
- Zhao Q, Rank G, Tan YT, Li H, Moritz RL, Simpson RJ, Cerruti L, Curtis DJ, Patel DJ, Allis CD, Cunningham JM, Jane SM. PRMT5-mediated methylation of histone H4R3 recruits DNMT3A, coupling histone and DNA methylation in gene silencing. Nat Struct Mol Biol. 2009;16:304–311. doi: 10.1038/nsmb.1568. [DOI] [PMC free article] [PubMed] [Google Scholar]