Abstract
Biomedical researchers often work with massive, detailed and heterogeneous datasets. These datasets raise new challenges of information organization and management for scientific interpretation, as they demand much of the researchers’ time and attention. The current study investigated the nature of the problems that researchers face when dealing with such data. Four major problems identified with existing biomedical scientific information management methods were related to data organization, data sharing, collaboration, and publications. Therefore, there is a compelling need to develop an efficient and user-friendly information management system to handle the biomedical research data. This study evaluated the implementation of an information management system, which was introduced as part of the collaborative research to increase scientific productivity in a research laboratory. Laboratory members seemed to exhibit frustration during the implementation process. However, empirical findings revealed that they gained new knowledge and completed specified tasks while working together with the new system. Hence, researchers are urged to persist and persevere when dealing with any new technology, including an information management system in a research laboratory environment.
Keywords: biomedical data, bioscience, information management, implementation, collaboration
1. Introduction
Biomedical informatics, an inherently interdisciplinary and integrative field, has great amounts of data ranging from the public health clinical research to the genomic research. Biomedical research when coupled with the high speed processing technologies results in highly detailed data sets (Roos, 2001). With increased emphasis on translational and collaborative research, vast amounts of heterogeneous data are generated. Managing and sharing these data is vital for subsequent analysis in the biomedical domain (Lyons-Weiler, 2005). The following topics on biomedical research information management are the most studied in literature: Database architectures, development of ontologies, and data integration techniques (Topaloglou, 2004). While previous researchers have conducted needs assessments of biomedical researchers from a system design perspective (Anderson et al., 2007), few studies have examined the impact of existing laboratory data management practices on bioscience research.
The primary purpose of this current study was to identify and categorize the shortcomings of the existing laboratory data management practices from the perspective of the principal investigators and laboratory members. In addition, the authors emphasized the importance of new technology in response to discovered limitations and analyzed the implementation challenges when such new system was introduced into the laboratory environment.
2. Biomedical Research Data
Biomedical data can be of various forms drawn from a wide range of sources such as images from CAT and MRI scans; signals from EEG; laboratory data from blood, specimen analysis; and clinical data from patients. Growing barriers between clinical and basic research are making it more difficult to translate newly generated scientific knowledge at the bench into clinical practice at the bedside. With recent National Institute of Health (NIH) priority for translational research, organization of the basic laboratory data and clinical data has become significant (NIH, 2008). In this paper, we primarily focus on basic laboratory data and its management. However, section 2.2 gives some insight into the nature of clinical data organization.
2.1 Bioscience Laboratory Data and its Organization
Genomic research laboratories are one of the primary data sources in biomedical research. They are data intensive as evidenced by the immense databases generated by the Human Genome project (ConsortiumIHGS, 2001). The data processed in a genomic laboratory range across the DNA sequence, mutation, expression arrays, assays, antibodies, and oligonucleotides to name a few. The challenge of genomic medicine lies in analyzing and integrating these diverse and voluminous data sources to elucidate normal and abnormal physiology (Louie, Mork, Sanchez, Halevy, & Hornoch, 2007). The manner these data are organized in a research laboratory plays a key role in aiding and driving the research coherently. Current laboratory data management methods primarily include handwritten laboratory notebooks, paper files, home-grown small databases and spreadsheet files (Anderson et al., 2007). The impact of these techniques on the bioscience laboratory research is discussed in section 3.2.
2.2 Clinical Data and its Organization
Like scientific laboratory data, clinical data need to be well-organized to generate adequately balanced results in the realm of translational research. Most of the clinical data often appear in free-text form with little or no structure (Schweiger et al., 2003). In their raw form, clinical records consist of hundreds of test results, medication and appointment notes. Illegibility of handwritten documents and inability to access data from various clinical sources greatly limit the effectiveness and efficiency of traditional paper based clinical records. Such drawbacks of paper based clinical records triggered the advent of computer-based medical records (Shortliffe & Barnett, 2006). Contrary to traditional paper records, data recorded in electronic medical record (EMR) systems is legible, remotely accessible and better organized because of the structure imposed on the data input (Tang & McDonald, 2006). Electronic medical record systems, however, are not flawless. Studies show that the use of computer-based patient record technology may cause unintended problems such as loss of relevant and critical information(Patel, Arocha, & Kushniruk, 2002).
In summary, handwritten paper files and homegrown databases are usually used for managing the basic research data, while electronic medical records are increasingly used for handling clinical data. Next, the authors will examine the problems that bioscience researchers often face with the current methods of basic laboratory data organization.
3. Current Trends and Issues of Biomedical Data Management
Like other basic sciences, recent advances in genetics and general laboratory methods have led to a tremendous increase in the amount of research data captured and analyzed by research teams. Unfortunately, existing commercial software and LIMS (Laboratory Information Management Systems) are unable to organize such data collected from modern bioscience research laboratories, and meet individual researcher’s needs (Anderson et al., 2007). The authors investigated two scientific laboratories (referred to as labs now on) to understand the influence of these data management methods on bioscience research. While six candidate labs were considered, two test labs were selected based on their responsiveness, motivation of the lab’s Principal Investigator (referred to as PI now on), and the richness of lab environment in terms of its ability to represent the manifold changes of use of information technology to improve scientific productivity and laboratory researcher’s satisfaction in the realm of bioscience research.
3.1. Data Collection Methods
Ethnographic observations were conducted. A trained researcher unobtrusively observed the activities at different times in the test labs and took observational notes(Van Maanen, 1996). The purpose of ethnographic observations was to understand workflow of the bioscience labs, and gain insight into interaction strategies among lab members in order to guide and improve efficiency of data collection in the next phase. The important concepts identified during the ethnographic phase were used to design web-based questionnaires. Two questionnaires (Q 1, Q 2) were used in this study with the lab PIs to understand the information management practices followed in the test labs. Q 1 was administered to all six candidate lab PIs during the test lab selection process, while Q 2 was given only to the PIs of the two selected test labs. Both the questionnaires included open-ended and closed specific questions. The questionnaires served as a means -
to gain knowledge about the overall state of labs in terms of 1) magnitude and nature of data handled, and 2) data management techniques
to create an account of current data handling and communication practices in the two test labs
The participants responded to all the questions and based on their questionnaire responses the themes for the semi-structured interviews were framed. Unlike the questionnaire framework, where detailed questions were formulated ahead of time, semi structured interviews began with more general unstructured questions (Bernard, 2002; Crabtree & Miller, 1992). Semi-structured interviews provided an opportunity to learn more about the laboratory goals and practices. There interviews allowed us to collect detailed descriptions to understand the reasons behind the problems faced by current day bioscience researchers. A number of new questions were generated during these interviews, allowing both the interviewer and interviewee to probe further on a particular issue(s). The four interview areas of interest were laboratory data storage, laboratory data management, queries on stored data, and collaboration. Nine test lab members in different professional roles such as lab manager, computer support specialist, and bench molecular biology investigators were interviewed. These interviews contained rich descriptive accounts of specific team members’ roles and activities. All interview data were audio recorded and transcribed for analysis. Beyond in-person interviews, we plan to conduct online semi-structured interviews. This “mixed mode” interviewing strategy will be included in the next phase of the study (Meho, 2006). Finally, Google documents were utilized to record and track the progress of the study. Detailed analysis and discussion of Google documents is presented in sections 5 and 6.
3.2. Data Analysis
According to the data collected during the initial recruitment survey (Q1), both the test labs have been dealing with human subjects, paper medical records, locally made DNA and RNA proteins, tissue blocks, microscopic slides, radiogram films and a wide range of biomaterials such as oligonucleotides and antibodies. Table 1 presents an overview of the data collected during Q1. As indicated in Table 1, test lab II has been established for 15 years, while test lab I only has been in operation for seven years. Therefore the magnitude of data handled by test lab II is greater than that of test lab I. For this reason, the evaluation of information management practices and implementation process presented in the later sections of this paper focuses primarily on test lab II.
Table 1.
Quantitative measures of various data and computing power in the test laboratories
| Test Laboratory I |
Test Laboratory II |
|
|---|---|---|
| Years in operation | 7 | 15 |
| Number of human subjects involved in research | 900 | 1500 |
| Number of workstations | 8 | 24 |
| Servers | 5 | 3 |
| Tubes of locally made DNA, RNA, protein | 5000 | 5000 |
| Tubes of purchased reagents | 250 | 1000 |
| Frozen tissue blocks | 1500 | 8000 |
| Microscopic slides | 500 | 50000 |
| Radiogram films | 100 | 1000 |
| Paper medical records | 25 | 25 |
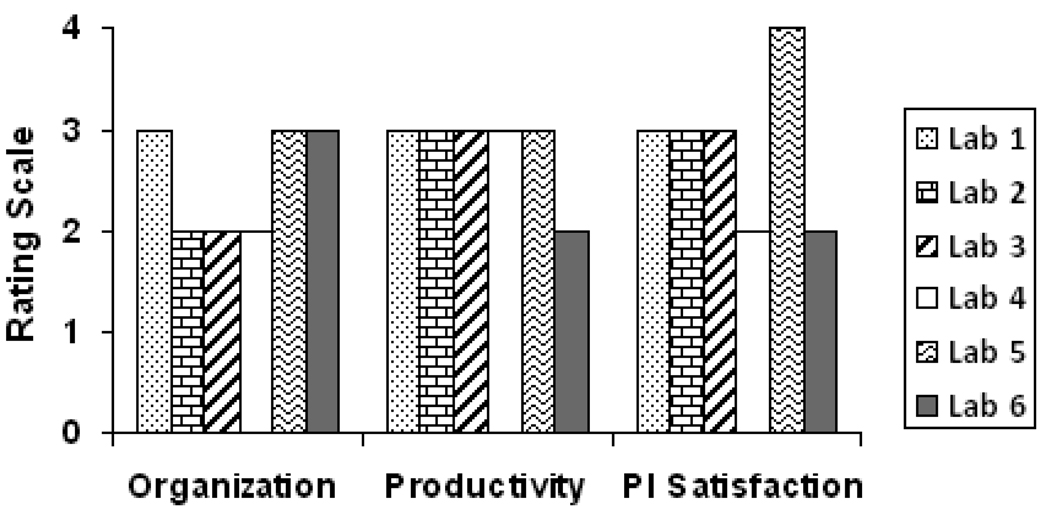
In Q1, the PIs of the six candidate labs were asked to summarize their own lab’s productivity, satisfaction and organization on a scale ranging from 1=poor to 4=excellent. Of the 18 responses shown in Figure 1, only one lab was self-identified as excellent in terms of satisfaction. Organization of lab data was rated as the most problematic compared to productivity and satisfaction by five of the six PIs. Based upon the findings, data management in the test labs could be improved.
Figure 1.
Ratings given by the six candidate laboratory principal investigators for their own laboratory’s data organization, productivity, and satisfaction on a scale ranging from 1= poor to 4= excellent.
This derivation was further bolstered by the findings from the analysis of the semi-structured interviews conducted with the test lab PIs and lab members. Analysis of interview data also uncovered numerous problems emerging from the existing information management methods used in the two test labs. These issues are elaborated below and pertinent quotes from the interviews are included.
3.2.1. Problems of data maintenance by an individual
Basic maintenance of records is typically performed by individual researchers. In the test labs, researchers often kept scientific data in spreadsheets as well as handwritten notebooks and logs. The following quote illustrates the problems that may surface when data are organized in such idiosyncratic ways.
“ I mean she’s very smart and she keeps good notes, but first she will go to the computer here and then she’ll go to her written notes, I mean without her, it would be very hard to back trace”-- PI, Test lab-II.
Although the context was clear to the creator of the notes and organized to facilitate personal efficiency, the structure was not transparent to other researchers. Similar concern was expressed by one of the lab members as shown below.
“Yeah, I’d have to train somebody, and that’s a big concern for me. I have started writing down protocols for different actions taken by the database, but I haven’t certainly completed it.”-- Lab member, Test lab-II.
When asked about transferability of work, the lab member questioned the feasibility of a smooth transition of duties unless the newcomer was considerably trained. Such personally customized data organization coupled with no established convention left the research data cryptic to co-researchers.
3.2.2. Problems of data sharing within the laboratory
Findings revealed that portions of data were created and maintained by some members (e.g. lab managers) that were required to be shared with the other members of the test labs. These scientific data and experiment results were shared formally in weekly lab meetings (e.g., verbal presentations) or informally (e.g., peer discussions). However, there were concerns about database access permissions, security and protection of individual contributions with this approach.
“And just the normal human aspect of how, who does what, who’s an author where, has not really fully been worked out and it’s complicated. So they are hoping that every group will put the data into that system, there’s been a lot of resistance. Well, we’re not sure we want to put it in because who is going to have access to it…”-- PI, Test lab-II
Certain procedures were followed for diagnosis assessments in the test labs as mentioned below in the quote. Such inconsistent methods had every possibility of something going wrong and thus increased the probability of error.
“Well, we have then XX and I meet once a week and we review what we’ve done. Then we meet a second time and we go over the grading. What we do there is, once the grading gets completed, the forms are filled out, then I send the forms to her, she enters the data and then we have a database meeting and we will pull up these patients and I have the form in front of me and she reads off from the database, what’s in the database and that’s how we confirm the grading. And at that point, we assign a final diagnosis to the patient and she…you know, I say out loud what the final diagnosis is and she confirms it and we put it in the database.”-- Lab member, Test lab-II.
Similarly, the other data management and data sharing practices followed by the test labs were decentralized and lacked coordination. For instance, ordering of materials was handled by each lab member individually by updating a common database. Although data access and data search in the test labs was often controlled by the managers, discrepancies of record maintenance were obvious with the existing methods.
3.2.3. Problems of data sharing with other laboratories, collaborators and experts in diverse domains
Generally in the two test labs, databases were kept at each research site. Copies were transformed (e.g., format conversion) and forwarded in the form of spreadsheets or delimited files for interpreting and integrating with destination databases. However, there were frequent problems in representing and communicating context which is crucial when working with collaborators in the same domain as well as with experts in other domains (e.g., biostatisticians). The problems that the researchers faced when collaborating with other members of their research team are illustrated using the following quotes.
“I can give data that I think are appropriate to answer a question to a biostatistician, but when they look at it, they see it from a different point of view. And that spread sheet does not really encapsulate where it came from very well, how was it generated, was it random, how was this data collected. You would run a series of queries that you think are pertinent to what this biostatistician would want to know. They become a part of the exploration and not just a receiver of whatever I decided to put in my spreadsheet on the day. What I get back is almost never fully documented in any way that I can really understand and add more to the process.”-- PI, Test lab-I
“But right now, I ‘m actually trying to figure out with these people in Europe that we’re beginning to collaborate with, their person wants the identified phenotype information, just for an example, I’m in the process of contacting him because just like you said, he’s not going to know what our variables mean. So, what do I do? I send him an email and I said, “These are our forms so you can see how it’s attached to the tables, but what exactly do you want? Basically, I have to keep kind of playing with it until I give them what they need”-- Lab member, Test lab- II.
From the two quotes above, it can be inferred that data sharing with the experts from other domains was greatly affected due to limited communication options of information management practices followed in the test labs. Indeed, issues arose when data were shared with the collaborators in the same domain because of inadequate universal terminology. This point can be exemplified with the following quote stated by one of the lab PIs.
“The only common context which we have is just basic language, that is, in terms of disease terminologies, which of course are slippery. But I think on the data level, the common things that people agree on are the names of genes but there’re synonyms, pseudo genes… There’s no common framework. There are still many gene names that are being changed.”--PI, Test lab-I
Representational heterogeneity across the databases resulting from the decentralized scientific community frustrates efforts to integrate them (Sujansky, 2001). Given the challenge with data sharing and integration across multiple sites, the collaborative research may not be appealing.
3.2.4. Limitations to publication success
Research findings and scientific knowledge are largely disseminated to the scientific community through publications. Because of data organization issues, the data collection in the test labs was confined. The researchers had to narrow down their research questions and in turn their projects resulted in less comprehensive solutions. Publishing the thus derived findings and results would not be an easy task. This issue was discussed by one of the lab PI’s as shown below.
“Not really recording exactly how they did do it, but they’ll get as close as they can in the publication because they don’t have good records…usually the level of detail…there are many things that labs cannot even attempt… because of their lack of organization. They narrow the focus of their question so that it involves one molecular entity and one small set of clinical data. They assume that the types of assays that they are running are the pertinent assays. And they don’t have time to do any other kinds of data collection, so they take the data that they do have and interpret it as best they can.”-- PI, Test lab-I
Data loss occasionally occurred in the test labs due to disorganization and unstructured records of research activities. Such loss of information may result in unsubstantiated findings- a primary concern of translational research (Unger, 2007). According to the PI of test lab II, retrieving the lost data by re-conducting the same study was time-consuming and frustrating.
“But we’ve re-made a lot of things just because either we don’t know where something is, or even if we find it, it’s about papers, but a little more trivial detail, we don’t know exactly what sequence is in there, we don’t know exactly what restricted enzymes. So that is frustrating and it’s a big waste of time. Or somebody writes to us, can I have such and such a (cannot understand) and I have to write back, “you know I really apologize, I’m embarrassed, but we can’t find it” and that’s ridiculous that shouldn’t happen, but it does happen.”--PI, Test lab-II
Thus data loss, in conjunction with incomplete methods and unstructured information management limited the publication rate, as well as, restricted the interpretation of novel designs. Analysis of the semi-structured interviews with the two test lab PIs and lab members revealed several issues that are imperative to resolve in order to enhance laboratory productivity. The interviewees expressed their views on the data management trends prevailing in their respective lab, depending upon their professional role and ongoing projects. The major problems identified with scientific research data management were related to data organization, data sharing, collaboration, and publications. Limitations to cross study comparisons and data integration were the other problems identified with the current information management methods. In summary, poor data organization can potentially lead to substantial data loss as well as degraded security and privacy levels.
4. Need for a Biomedical Research Information Management System
Modern genomics research demonstrates the power of computing and communication tools to facilitate rapid progress for information exchange and collaboration. Previous researchers have advocated for the development of interoperating systems that support secure gathering, interchange, and analysis of high-quality information (Rindfleish & Brutlag, 1998). Such systems should also provide a spectrum of options to accommodate a range of unique individual researcher needs (Anderson et al., 2007). Section 3 of this paper outlines the well-documented need for an information management system that allows researchers to adhere to standardized data management practices. Collaboration among researchers should also be facilitated by the proposed system. It is important to keep in mind, though, that collaborative projects produce massive heterogeneous databases. Future information management systems should be able to effectively handle such data by resolving representational heterogeneities using uniform conceptual schemas (Sujansky, 2001). Several approaches such as warehouse integration, mediator-based integration, and navigational integration have been adopted to deal with heterogeneous biological data (Hernandez & Kambhampati, 2004). Data integration challenges posed by bioscience research require deeper analysis, which is beyond the scope of this paper. Baralis and Fiori (2008) discussed bioscience data integration and provided several insights; readers are encouraged to review their work for more information.
Many systems have been developed for efficient research data management, but these systems are constrained by lab size and/or research concentration (Birkland & Yona, 2006; Droit et al., 2007; Li, Gennari, & Brinkely, 2006; Maurer et al., 2005; Navarange et al., 2005; Viksna et al., 2007). However, the development rate of such systems reflects the mandatory need for efficient information management in bioscience research. According to their responses to Q2, PIs of the two test labs also insisted upon an efficient tool for managing their research data. A portion of the questions and responses from Q2 are provided below to illustrate their position. Both the PIs commented that the current methods used to track research projects were inefficient and lacked organization. One of the PIs noted that data created by a lab member were not easily searchable or interpretable in his or her absence, while the other PI mentioned of one data management system (called Labmatrix), which facilitated completion of similar tasks with fewer hassles.
Question1: How do you identify and evaluate potential new laboratory projects? Do you keep a list of ideas that you would like to pursue? If so, how do you manage this list?
Response: "After starting many projects and not completing them, I do not take action on new projects (although I constantly think of new ones that I would like to pursue) until I get some of my existing ones on a more sustainable track. I do maintain a list of projects as part of my list of things to do, which is in Outlook, but is not well integrated with my overall lab work"-PI, Test Lab I
Response: "New ideas are always coming up either from myself or from people in my lab. Desire of people to pursue particular new ideas, within reason, is a major determinant of what we work on. I do not have an efficient way to keep track of ideas, although I do sometimes email myself notes. Could definitely use a more disciplined approach. I get a little more organized around grant time."-PI, Test Lab II
Question2: Are the data, techniques, and protocols that have been generated by an individual searchable in their absence? Please comment
Response: "Yes, thanks to ‘Labmatrix’v1 database (version running in my lab), and to maintenance of file directories"-PI, Test Lab I
Response: "It varies, but this is often a MAJOR problem."-PI, Test Lab II
Question3: How do you keep track of the history (source, how it was made, what data has been generated etc) for each experimental biomaterial used in your laboratory?
Response: "This is all managed through our ‘Labmatrix’v1 database, prior to database, could not be managed"-PI, Test Lab I
Response: "Done differently be each individual"-PI, Test Lab II
It is clear from the above statements that utilizing a data management system would likely organize data and improve the lab practices and productivity. Studies demonstrated that Labmatrix, a web-based research information management system, facilitates effective data management (Gopalan et al., 2008; Suh et al., 2008). Rubin and colleagues (Rubin et al., 2008) summarized the advantages of Labmatrix. Some of the benefits are outlined below:
Consistent laboratory practices
Enhanced data integrity and data standardization
Collaborative data access
90% time savings for specimen and data retrieval
These findings suggest that an information management system would reduce the time taken to perform certain tasks. The direct and indirect savings associated with reduced time over the long term could offset the initial time invested to set up the system. In conclusion, the authors urge for an information management system to alleviate the problems that the researchers face with existing research data management practices. In section 5, the authors introduce the Bioscience Research Integration Software Platform (BRISP) research group. A closer look at the collaboration between the BRISP team members reveals some non-intuitive findings related to the implementation of a new system for efficient research data organization.
5. Bioscience Research Integration Software Platform (BRISP) Team Collaboration
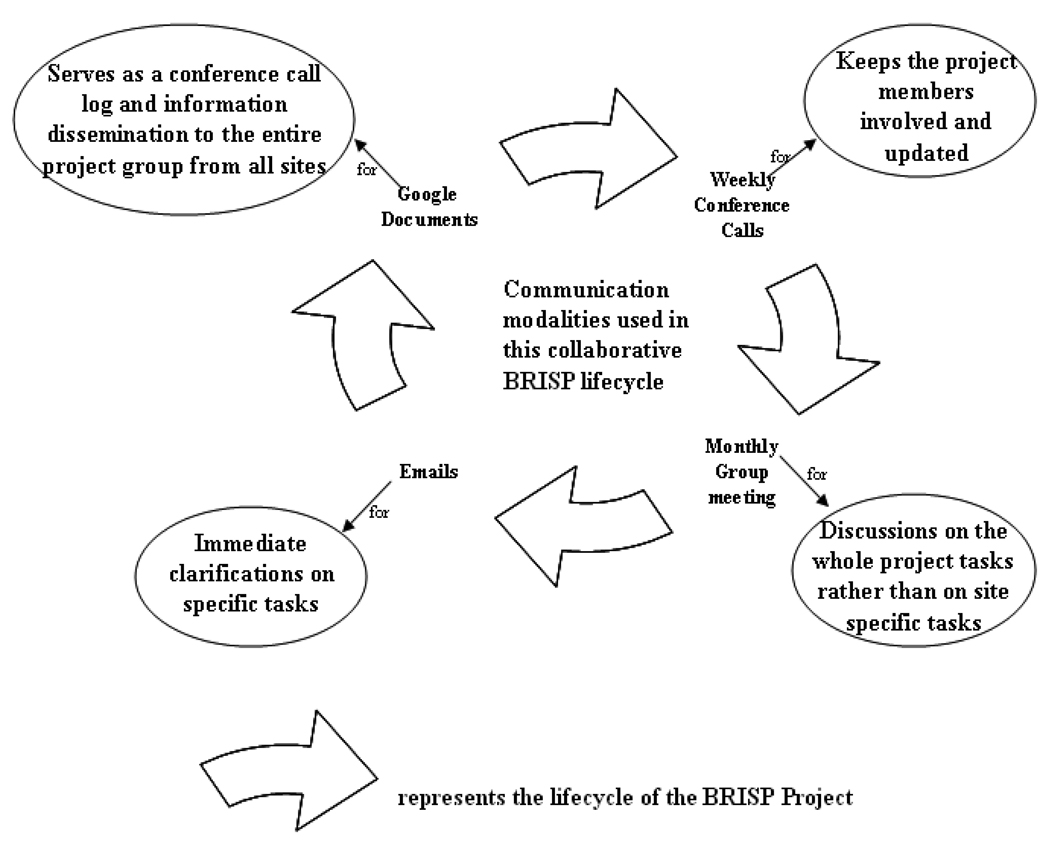
One of the objectives of BRISP team is to research, develop and implement a system for efficient research data management in test lab II. A distributed group of experts from different fields collaborated to achieve this goal. The group used four communication modalities to progress with the project (see Figure 2). Figure 2 characterizes the context of each of the four modalities: Emails, Weekly teleconference calls, Google Documents and Monthly group meetings. The arrows in Figure 2 depict the life cycle of BRISP project. During the BRISP project life cycle, a new research topic was usually introduced by the team leader. A typical execution plan was drafted and forwarded to the involved research team members, often via email. As portrayed in Figure 2, emails were also used for immediate clarifications on certain tasks. Weekly conference calls took place to discuss the progress of the BRISP project. Contents of these conference calls were archived using Google Documents (also referred to as “Google Docs”). Google Docs is a free, web-based word processor, spreadsheet and presentation, application offered by Google. This application allows users to create and edit documents online while collaborating in real time with other users (Google, 2009).Lab members who were unavailable during a particular call used Google Docs to quickly review the contents of that call. Monthly group meetings were held to discuss the status of the study at distributed BRISP research sites as well as to brainstorm future steps.
Figure 2.
Typical communication modalities used in Bioscience Research Integration Software Platform (BRISP) project life cycle.
As explained earlier, Google documents served as a teleconference call log and as a means to track progress at other research sites. Figure 3 presents a typical conference call log recorded in the Google Docs. It can be understood from Figure 3 that the calls were not archived verbatim. Rather, a gist of the discussions was documented. The contents in Google Docs were organized into several subgroups. Items for completion were underscored in each of the sub-groups. These items informed lab members of their pending and upcoming tasks. Because Google Docs can be shared, opened, and edited simultaneously by multiple users at the same time, members were able to update the application when their respective tasks were completed. These timely updates allowed the BRISP group to evaluate the status of the project objectives in a timely manner. The details of conference call schedules, attendees, and to-do tasks recorded in Google Docs (see Figure 3) altogether served as a great resource to establish timeline detail of the BRISP study.
Figure 3.
An illustration of the Bioscience Research Integration Software Platform (BRISP) conference call recording in Google Documents.
The weekly conference call recordings archived in the Google documents were helpful in generating conclusions about the later phase of the project- the implementation of the data management system in test lab II. Section 6 covers the implementation aspects of a research information management system based on the information contained in Google documents.
6. Introduction of a new information management system into a laboratory environment
Whenever a new system is introduced into a lab environment, the researchers should be able to use the system with ease, and it should not negatively impact existing workflow. In biological labs, integration of new technology has been shown to positively and negatively affect workflow and research culture (Dennis, 2002, May 2; Kaminski & Friedman, 2002). Special emphasis on selection, implementation, training, and support for lab information management systems (Klien, 2003) has remained critical for bioscience research. Analysis of the teleconference call content archived in Google Docs revealed that investment of time, additional technical resources, and willingness of the lab members to utilize the new technology were the key factors affecting the implementation of the new system in the test lab. These findings are consistent with other studies in the literature (Ash, Anderson, & Hornoch, 2008).
During the implementation process the test lab used this new system for managing various data such as- 1) experiment-related data (e.g. information related to antibodies, plasmids, oligonucleotides, and 2) administrative information (e.g. lab protocols). Given the scope of the paper, however, the authors chose to describe the test lab's attempts to manage oligonucleotides (referred to as “oligos” now on) using the new system during the implementation process. The key elements identified during the implementation of the new system in the test lab are summarized below:
Implementing the new system in the test lab required personnel involvement. This process demanded more time than one can usually afford in a busy lab. To ensure involvement, the PI had to promote direct advantages from working with the system to his/her lab members. Without such leadership, the lab members probably would have exerted little effort to learn and use the new tool to complete their work. Without the motivation and skills to use new systems, researchers may be unable to manage the reservoir of rapidly growing scientific data, and they may unintentionally hamper scientific discovery (Anderson, Ash, & Hornoch, 2007).
-
With the implementation of the new data management system, the test lab was on the verge of transforming its decentralized information management practices to a centralized system. Table 2 outlines the laboratory practices before and during the implementation of the new system. As indicated in Table 2, prior to the introduction of the new system, the test lab did not follow any naming convention. Handwritten labels were used which could be easily misinterpreted. The storage locations of the oligos were not clearly recorded, and in turn the probability of lost/misplaced inventory seemed to be high. During the implementation of the new system the test lab made a shift towards established naming norms. The lab used printed barcode labels instead of handwritten ones. With the new system the lab were also able to record the storage locations, thus minimizing the possibility of inventory loss, misplacement, and/or misinterpretation.
The transformation illustrated in Table 2 indicates that the test lab used additional resources like barcode labels, labeling software and separate storage locations. Lab members were trained to use the new system with these integrated additional resources to manage the oligos. As evidenced, implementation of the new system required advanced accessories and utilization training.
Analysis of the teleconference call content in Google Docs revealed that the lab members became frustrated with the new system. Despite constant dissatisfaction displayed by the lab members with the new technology, the test lab progressed with the implementation of the new system. After all, the overall objective was to organize the information of oligos using the new system. To achieve this objective, completion of several intermediate tasks was necessary. For example, barcode scanners needed to be installed, details (e.g., name and location) of the oligos needed to be uploaded, and storage space needed to be allotted. The lab members finished three intermediate tasks and had the fourth one in progress, and thus, the research group completed 90% of their overall objective. However, analysis of Google Docs showed that there were signs of frustration from the lab members during this time period. Although time consuming and frustrating for lab members, the test lab made substantial progress with the new system.
The implementation of the new system in the test lab was a learning experience for the lab members. Inspection of Google Docs revealed that lab members learned various new methods such as barcode scanners, labeling software, and more broadly manipulating the new system. This learning process was implicit in Table 2, which depicts the transformation of the test lab before as well as during the implementation of the new system.
Regular contact with the system creators and customization of the system to meet specific needs of the lab will greatly condense the period of implementation and improve the implementation experience. Based on content from the teleconference calls, industry personnel did in fact offer guidance at various points during the implementation. They also tailored the system to meet the needs of the lab. Such assistance from the industry enabled lab members to accomplish their tasks in a timely fashion. Without such interaction and accommodations, the implementation procedure would have been more time consuming. Regular feedback on the system’s performance also helped the industry personnel to augment the system to better respond to the needs of the test lab.
Table 2.
Management of oligonucleotides in the test laboratory before and during implementation of the new information management system
| Before implementation of the new system |
During implementation of the new system |
|---|---|
| No naming convention | Naming convention followed |
| Handwritten labels | Accompanied with labeling software |
| Storage locations not recorded | Storage locations recorded |
| Decentralized laboratory inventory ordering | Centralized laboratory inventory ordering |
New technology adoption depends on its relative advantage, compatibility with the existing systems, results demonstratability and ease of use. In addition to these technological factors, the organization’s voluntariness, ongoing commitment, offered trainings and support to its employees influence the success of implementation (Karsh & Holden, 2007). New technology is generally well accepted when its possible short-term and long term gains are emphasized and clear.
6. Conclusions and Future Implications
Prior research has found that the inability of bioscience research laboratories to adequately store, retrieve, share, manage, query, analyze and interpret collected data greatly impedes the research productivity. This paper describes the nature of the heterogeneous biomedical research data and probes into the problems often encountered by researchers handling such voluminous data. Our study identified four major problems with current scientific data management applications in the two test laboratories. They were related to data maintenance, data sharing, collaboration, and publications. It was learnt that poor data organization can potentially lead to substantial data loss as well as degraded security and privacy levels, which may have negative influences on research. We advocate that a data management system would likely assist in organizing data efficiently and in improving the laboratory practices and productivity.
This study raises the next question of intervention to incorporate certain features into information management systems for improved collaboration among distributed research sites as well as within the laboratories. Such interventions should at least enable us to-
Achieve common terminologies, distributed workflows: This can solve some problems of collaboration between geographically distributed researchers from same domain;
Utilize visualization functionalities: Such visualization aids can address the needs of researchers collaborating with specialists from other domains.
Develop newer tools to support publication lifecycle, literature review, authoring, and simultaneous easy access to research data, results, and interpretations;
Deploy metadata like entities to cover a range of heterogeneous biomedical data derived from several distributed sources;
Personalize research information while imposing proper structure on data handled by a laboratory researcher, such an option would reduce problems arising from idiosyncratic ways of information management Our study shows that implementation of an information management system in a bioscience laboratory is a challenging task. Although time consuming during implementation, an information management system would in the long run reduce the time taken to perform certain tasks in operation phase. The direct and indirect savings associated with reduced time over the long term could offset the initial time invested to set up the system. Findings from the analysis of the implementation process of a new information management system in one of the test laboratories show that a laboratory progresses with its tasks despite constant dissatisfaction displayed by the laboratory members with the new technology. This suggests that there is a strong need for perseverance on the part of the researchers when dealing with any new technology in the laboratory environment, including a research data management system. Implementation of such a system requires time, involvement and patience of the laboratory personnel. During implementation, which itself can be viewed as a learning experience, the research laboratory members gain new knowledge. The future information management systems should be designed such that the researchers can retain some of the skills they acquire while using existing systems. We define this ability of a system’s design to allow human beings to use other similar systems with minimal training, showing some generalizable skills as “Human Interoperability”.
System usability and system interoperability may be the other aspects worthy of designers’ consideration when developing an information management system. Usability is a well- understood concept in the scientific literature and refers to a system’s capacity to allow users to carry out their tasks safely, effectively, efficiently, and enjoyably (Nielsen, 1993 ; Preece, Rogers, & Sharp, 2002). The larger issue of system acceptability can be viewed as being dependent on the system’s usability. System interoperability is defined as “the ability of two or more systems or components to exchange information and to use the information that has been exchanged” (IEEE, 1990). Analysis of existing systems’ usability and interoperability enables researchers to establish standards for the next generation of research information management systems.
Acknowledgements
We graciously acknowledge the support from Drs. Steven G. Bova, Donald Zack, Jian Wang and other members of our collaborative study. We thank Jan Horsky for his help with the early stages of data collection and analysis. The study was supported in part by a grant (1R41CA105217-01A1-STTR) from National Institute of Health/National Cancer Institute (NIH/NCI).
Biographies
Sahiti Myneni is a Faculty Research Associate in Department of Biomedical Informatics at Arizona State University. She conducts her research activities at the Center for Decision Making and Cognition. Her research interests include usability assessment and interoperability evaluation of information management systems and web-portals. She is particularly interested in the evaluation of healthcare information systems used in critical care.
Vimla L. Patel, PhD, DSc, FRSC: Patel is Professor and Vice-Chair of the Arizona State University’s Department of Biomedical Informatics and the Director of the Center for Decision Making and Cognition in the Ira A. Fulton School of Engineering. She is also a professor of Basic Medical Sciences in the University of Arizona, College of Medicine in Phoenix. She received her graduate training at McGill University in Montreal, Canada, in Educational Psychology and Medical Cognition. Her research interests include cognitive mechanisms underlying human performance, decision-making, complexity and medical errors, and human-computer interaction in health-care domains. http://www.fulton.asu.edu/~patel/
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures: The authors have nothing to disclose and have no Conflict of Interest.
References
- Anderson NR, Ash JS, Hornoch PT. A qualitative study of the implementation of a bioinformatics tool in a biological research laboratory. International Journal of Medical Informatics. 2007;76:821–828. doi: 10.1016/j.ijmedinf.2006.09.022. [DOI] [PubMed] [Google Scholar]
- Anderson NR, Lee ES, Brockenbrough JS, Minie ME, Fuller SJB, et al. Issues in biomedical research data management and analysis: Needs and Barriers. Journal of American Medical Informatics Association. 2007;14:478–488. doi: 10.1197/jamia.M2114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ash JS, Anderson NR, Hornoch PT. People and Organizational Issues in Research Systems Implementation. Journal of American Medical Informatics Association. 2008;15:283–289. doi: 10.1197/jamia.M2582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baralis E, Fiori A. Exploring Heterogeneous Biological Data Sources; Paper presented at the IEEE 19th International Conference on Database and Expert Systems Application, 2008. DEXA '08; Turin. 2008. Sep 1–5, [Google Scholar]
- Bernard R. Research Methods in Anthropology: Qualitative and Quantitative Methods. Walnut Creek: AltaMira Press; 2002. [Google Scholar]
- Birkland A, Yona G. BIOZON: a system for unification, management and analysis of heterogeneous biological data. BMC Bioinformatics. 2006:7–70. doi: 10.1186/1471-2105-7-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ConsortiumIHGS. The human genome. Nature. 2001:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- Crabtree B, Miller W. Doing Qualitative Research. Newbury Park, CA: Sage; 1992. [Google Scholar]
- Dennis C. Biology Databases: Information overload [Electronic Version] Nature. 2002 May 2;14:417. doi: 10.1038/417014a. [DOI] [PubMed] [Google Scholar]
- Droit A, Hunter JM, Rouleau M, Ethier C, Cloutier AP, Bourgais D, et al. PARPs database: A LIMS system for protein-protein interaction data mining or laboratory information management system. BMC Bioinformatics. 2007:8–483. doi: 10.1186/1471-2105-8-483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Google. 2009 www.docs.google.com. Retrieved February 1, 2009.
- Gopalan B, Orloff M, Stanuch K, Waite K, Platzer P, Eng C. Integration of clinical, molecular and cellular phenotypes to unravel the mechanisms of complex diseases; Paper presented at the American Medical Informatics Association Translational Bioinformatics Summit; San Francisco, CA. 2008. Mar 10–12, [Google Scholar]
- Hernandez T, Kambhampati S. Integration of biological sources: current systems and challenges ahead. ACM SIGMOD Records. 2004;33(3):51–60. [Google Scholar]
- IEEE. IEEE (Institute of Electrical and Electronics Engineers): Standard Computer Dictionary- A Compilation of IEEE Standard Computer Glossaries. 1990 [Google Scholar]
- Kaminski N, Friedman N. Practical approaches to analyzing results of micro array experiments. American Journal of Respiratory Cell and Molecular Biology. 2002;27(2):125–132. doi: 10.1165/ajrcmb.27.2.f247. [DOI] [PubMed] [Google Scholar]
- Karsh BT, Holden R. New Technology Implementation in Health Care. In: Carayon P, editor. Handbook of Human Factors and Ergonomics in Health Care and Patient Safety. Mahwah, New Jersey: Lawrence Erlbaum Associates, Publishers; 2007. pp. 393–410. [Google Scholar]
- Klien CS. LIMS user acceptance testing. Quality Assurance: Good Practice, Regulation, and Law. 2003;10(2):91–106. doi: 10.1080/10529410390262736. [DOI] [PubMed] [Google Scholar]
- Li H, Gennari JH, Brinkely JF. Model Driven Laboratory Information Management Systems; Paper presented at the American Medical Informatics Association Symposium Proceedings; Washington, DC. 2006. Nov 11–15, [PMC free article] [PubMed] [Google Scholar]
- Louie B, Mork P, Sanchez FM, Halevy A, Hornoch PT. Data integration and genomic medicine. Journal of Biomedical Informatics. 2007;40:5–16. doi: 10.1016/j.jbi.2006.02.007. [DOI] [PubMed] [Google Scholar]
- Lyons-Weiler J. Standards of Excellence and Open Questions in Cancer Biomarker Research. Cancer Informatics. 2005;1(1):1–7. [PMC free article] [PubMed] [Google Scholar]
- Maurer M, Molidor R, Sturn A, Hartler J, Hackl H, Stocker G, et al. MARS: Microarray analysis, retrieval and storage system. BMC Bioinformatics. 2005:6–101. doi: 10.1186/1471-2105-6-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meho LI. E-Mail Interviewing in Qualitative Research: AMethodological Discussion. Journal of the American Society for Information Science and Technology. 2006;57(10):1285–1295. [Google Scholar]
- Navarange M, Game L, Fowler D, Wadekar V, Banks H, Cooley N, et al. MiMiR:a comprehensive solution for storage, annotation and exchange of microarray data. BMC Bioinformatics. 2005:6–268. doi: 10.1186/1471-2105-6-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nielsen J. Usability Engineering. New York: Academic Press; 1993. [Google Scholar]
- NIH. 2008 http://nihroadmap.nih.gov/clinicalresearch/overview-translational.asp Retrieved March 25, 2009.
- Patel VL, Arocha JF, Kushniruk A. Patients' and Physicians' Understanding of Health and Biomedical Concepts: Relationship to the Design of EMR Systems. Journal of Biomedical Informatics. 2002;35:8–16. doi: 10.1016/s1532-0464(02)00002-3. [DOI] [PubMed] [Google Scholar]
- Preece J, Rogers Y, Sharp H. Interaction design: beyond human–computer interaction. New York: Wiley; 2002. [Google Scholar]
- Rindfleish TC, Brutlag DL. Directions for Clinical Research and Genomic Research into the Next Decade: Implications for Informatics. Journal of American Medical Informatics Association. 1998;5:404–411. doi: 10.1136/jamia.1998.0050404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos DS. Computational Biology: Bioinformatics--Trying to swim in a sea of data. Science. 2001;291:1260–1261. doi: 10.1126/science.291.5507.1260. [DOI] [PubMed] [Google Scholar]
- Rubin E, Zenklusen JC, Worrell R, Chen SH, Wang J, Peterson J, et al. Translational Bridge for Personalized Medicine: Biospecimen Information Management; Paper presented at the American Medical Informatics Association Translational Bioinformatics Summit; San Francisco, CA. 2008. Mar 10–12, [Google Scholar]
- Schweiger R, Hoelzer S, Rudolf D, Rieger J, Dudeck J. Linking clinical data using XML Topic Maps. Artificial Intelligence in Medicine. 2003;28:105–115. doi: 10.1016/s0933-3657(03)00038-1. [DOI] [PubMed] [Google Scholar]
- Shortliffe EH, Barnett GO. Biomedical Data: Their Acquisition, Storage and Use. In: Shortliffe E, Cimino J, editors. Biomedical Informatics. New York: Springer Science; 2006. pp. 46–79. [Google Scholar]
- Suh KS, Remache YK, Patel JS, Chen SH, Shaikh AM, Wang J, et al. Translational Bioinformatics-Guided Workflow for Procurement of Patient Samples; Paper presented at the American Medical Informatics Association Translational Bioinformatics Summit; San Francisco, CA. 2008. [Google Scholar]
- Sujansky W. Heterogeneous Database Integration in Biomedicine. Journal of Biomedical Informatics. 2001;34:285–298. doi: 10.1006/jbin.2001.1024. [DOI] [PubMed] [Google Scholar]
- Tang PC, McDonald CJ. Electronic Health Record Systems. In: Shortliffe E, Cimino J, editors. Biomedical Informatics. New York: Springer Science; 2006. pp. 447–475. [Google Scholar]
- Topaloglou T. Biological Data Management: Research, Practice and Opportunities; Paper presented at the Proceeding of 30th International Conference on Very Large Data Bases; Toronto, Canada. 2004. [Google Scholar]
- Unger E. All is not well in the world of teanslational research. Journal of American College of Cardiology. 2007;50:738–740. doi: 10.1016/j.jacc.2007.04.067. [DOI] [PubMed] [Google Scholar]
- Van Maanen J. Ethnography. In: Kuper A, Kuper J, editors. The Social Science Encyclopedia. 2nd ed. London: Routledge; 1996. pp. 263–265. [Google Scholar]
- Viksna J, Celms E, Opmanis M, Podnieks K, Rucevskis P, Zarins A, et al. PASSIM- an open source software system for managing information in biomedical studies. BMC Bioinformatics. 2007;8:52. doi: 10.1186/1471-2105-8-52. [DOI] [PMC free article] [PubMed] [Google Scholar]