Abstract
Type II topoisomerases are required for the management of DNA tangles and supercoils1, and are targets of clinical antibiotics and anti-cancer agents2. These enzymes catalyze the ATP-dependent passage of one DNA duplex (the transport or T-segment) through a transient, double-stranded break in another (the gate or G-segment), navigating DNA through the protein using a set of dissociable internal interfaces, or “gates”3,4. For more than 20 years, it has been established that a pair of dimer-related tyrosines, together with divalent cations, catalyze G-segment cleavage5–7. Recent efforts have proposed that strand scission relies on a “two-metal mechanism”8–10, a ubiquitous biochemical strategy that supports vital cellular processes ranging from DNA synthesis to RNA self-splicing11,12. Here we present the structure of the DNA-binding and cleavage core of Saccharomyces cerevisiae topo II covalently linked to DNA through its active-site tyrosine at 2.5 Å resolution, revealing for the first time the organization of a cleavage-competent type II topoisomerase configuration. Unexpectedly, metal-soaking experiments indicate that cleavage is catalyzed by a novel variation of the classic two-metal approach. Comparative analyses extend this scheme to explain how distantly-related type IA topoisomerases cleave single-stranded DNA, unifying the cleavage mechanisms for these two essential enzyme families. The structure also highlights a hitherto undiscovered allosteric relay that actuates a molecular “trapdoor” to prevent subunit dissociation during cleavage. This connection illustrates how an indispensable chromosome-disentangling machine auto-regulates DNA-breakage to prevent the aberrant formation of mutagenic and cytotoxic genomic lesions.
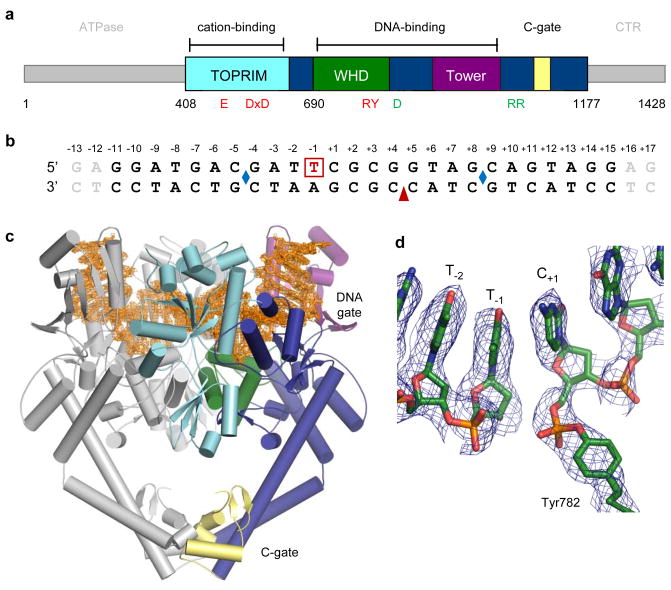
A wealth of structural and biochemical efforts have illuminated key aspects of type II topoisomerase mechanism; however, a biophysical understanding of their control of DNA cleavage chemistry has remained persistently out of reach. A prior DNA-bound, type II topoisomerase structure showed how these enzymes engage and bend substrate duplexes13, but captured a pre-cleavage intermediate that could not comment on strand scission. Because type II topoisomerases disfavour cleavage when bound to short oligonucleotides, we used bridging phosphorothiolate chemistry to create a suicide substrate that leaves a free thiol unable to support religation14. This approach has been used at the 5′-ribose position to trap and image a 3′-end cleaving type I topoisomerase (topo IB) linked to DNA15. To form an irreversible cleavage complex with a 5′-end cleaving type II topoisomerase, we incubated the principal DNA binding and cleavage domain of S. cerevisiae topo II (residues 408–1177) with a DNA substrate containing a nick at one cleavage site and a 3′-bridging phosphorothiolate at the other (Figs. 1a–b). Following co-crystallization, we determined the structure of this complex to 2.5 Å resolution using molecular replacement. Electron density permitted building and refinement of a model to an Rwork/Rfree of 23.9%/25.9% (Figs. 1c, S1; Table S1), and showed clear evidence for a covalent 5′ link between DNA and the active-site tyrosine (Y782) resident in topo II’s Winged Helix Domain (WHD) (Figs. 1a, 1d).
Fig. 1. Structure of a topo II/DNA cleavage complex.
a. S. cerevisiae topo II domain arrangement. Functional regions are coloured and labelled. Active-site residues are red; “trapdoor” residues green.
b. DNA substrate. One strand (top) contains a 3′-bridging phosphorothiolate between the −1/+1 positions (red, boxed). Two complementary strands (bottom) adjoin at a nick (red arrowhead). Blue diamonds indicate where DNA bend points. The terminal two base-pairs (grey) are disordered.
c. The cleavage complex. One topo II monomer is coloured as in (a). 2Fo-Fc density (1σ contour, orange) is shown around the DNA.
d. The 5′-phosphotyrosine link modelled into a composite simulated-annealing omit map (1 σ contour).
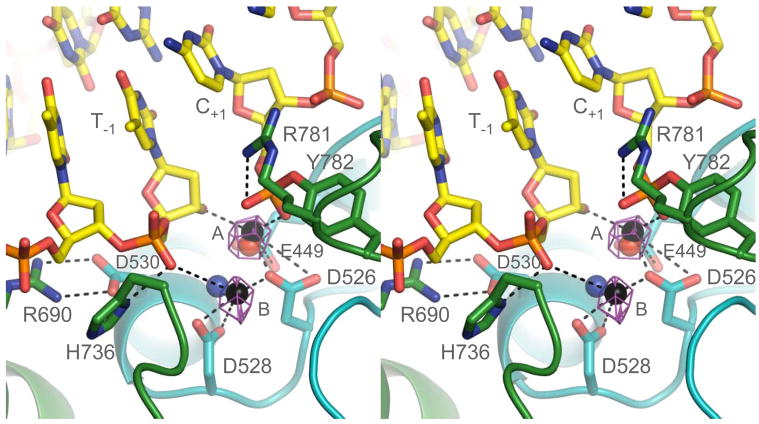
The structure reveals the constellation of amino acids required to form a cleavage-competent active site. However, given the known metal dependence for DNA cleavage by type II topoisomerases6, we were at first puzzled by a lack of electron density for these cofactors, despite the presence of 2.5 mM MgCl2 in the crystallization milieu. Suspecting this absence might be due to the low pH (4.5) of crystallization, we solved a second structure of an isomorphous crystal soaked at a higher pH (6.5) with zinc chloride (Mg2+, Mn2+, and Ca2+ did not produce suitable diffraction, and glutaraldehyde cross-linking was required to stabilize the crystals for pH exchange). Soaking strategies have been used successfully to gain insights into other divalent cation-dependent polymerase and phosphodiesterase systems such as DNA Pol I and RNaseH16,17. Anomalous difference maps for this structure, which was determined by molecular replacement to 3.0 Å resolution, show clear evidence for two zinc ions (7 σ peaks) in the active site. As anticipated from prior studies of topo II and its related bacterial counterpart, DNA gyrase, the ions are coordinated by highly-conserved acidic residues within the enzyme’s Topoisomerase/Primase (TOPRIM) domain18, with E449 and D526 contacting metal A, and D526 and D528 engaging metal B (Figs. 1a, 2).
Fig. 2. A cleavage-competent active site.
Close-up showing DNA (yellow), catalytic amino acids (cyan – TOPRIM, green – WHD) and metal coordination (black spheres). A Zn-anomalous difference map is shown as purple mesh (5σ contour). The two modelled zinc ions are spaced 3.5 Å apart. Hydrogen bonds and metal interactions are displayed as dashed lines. Both nonbridging phosphotyrosyl oxygens are liganded, one by metal A and the other by Arg781. Mg2+ ions seen in a non-covalent topo II/DNA complex13 and in the DNA-free topo VI A-subunit21 are shown as blue and red spheres, respectively.
In classic two-metal polymerase/phosphodiesterase reactions, one ion activates a catalytic water or ribose hydroxyl for nucleophilic attack, while the other coordinates a leaving group12. Both cofactors stabilize a pentavalent transition state. Because of the frequent occurrence of this mechanism across many unrelated biomolecular systems, we were surprised to observe that the metal organization in topo II does not recapitulate such a configuration (Fig. S2). Metal A interacts with both a non-bridging oxygen of the phosphotyrosine and the 3′-ribose position (Fig. 2), and is thus well-positioned to assist with transition state chemistry. However, metal B sits farther afield, contacting only a non-bridging oxygen of the (−1/−2) phosphate upstream of the cleavage site.
Although we cannot rule out the possibility that some positional shift could take place to reorient the active site and metal ions into a more canonical two-metal arrangement, several lines of evidence indicate that the structure observed here recapitulates a catalytically-relevant configuration. Replacement of an invariant arginine (R781), which contacts the phosphotyrosine moiety, dramatically reduces DNA cleavage and relaxation activity19, except when substituted with lysine20. Metal binding at either the A or B locus has been seen previously in two different type II topoisomerase structures (Fig. 2)13,21, and mutation of associated liganding groups severely compromises strand scission in both eukaryotic and bacterial enzymes8–10. The relative position of the metal-binding cluster with respect to DNA and the active-site tyrosine is further supported by: 1) patterns of thiol-dependent DNA cleavage seen in cysteine-mutagenesis and metal-ion rescue experiments consistent with the bonding arrangement seen here9,10, 2) the formation of an ion pair between two highly-conserved amino acids (D530 and R690) required for wild-type function19, and 3) a new role for an invariant histidine (H736), which engages the (−1) phosphate bound by metal B. Finally, in the metal-free structure, the thio-ribose 3′-SH adopts a C2′ endo (B-form DNA) sugar pucker non-optimally oriented for promoting religation, whereas in the zinc-bound structure, the thiol flips to a C3′ endo state (A form) similar to its neighboring nucleotides, moving within 2.0 Å of metal A and taking an in-line position with respect to the phosphotyrosyl linkage.
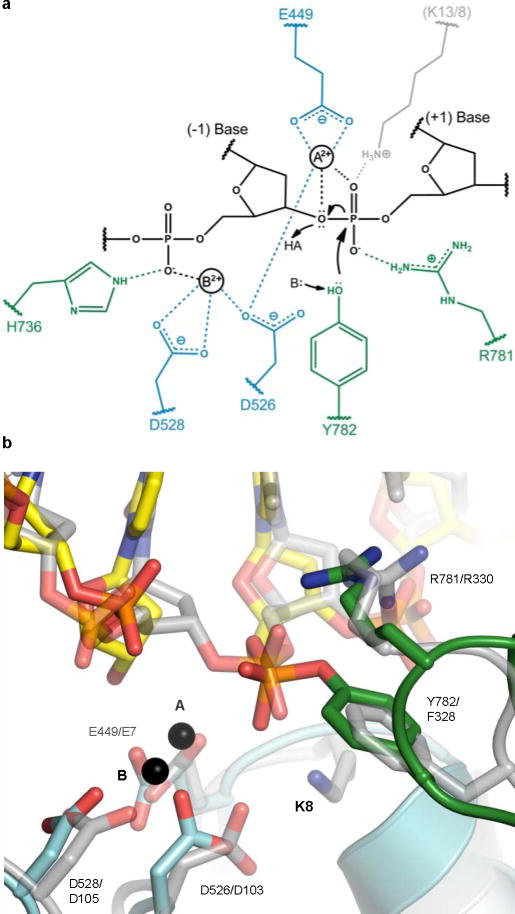
Collectively, these interactions indicate that type II topoisomerases employ a distinct variation of canonical two-metal mechanisms (Fig. 3a). In this scheme, one ion (metal A) partakes directly in transition state stabilization and in promoting the leaving and attacking of the ribose 3′-OH during DNA cleavage and religation. The other (metal B) plays a role in anchoring the DNA. The reason why type II topoisomerases may have lost the need for a second metal in performing chemistry likely is due to: 1) the lower pKa (~10–11) of the attacking tyrosine nucleophile during cleavage, which makes it easier to deprotonate compared to water or an incoming ribose hydroxyl (pKa ~13–15), and 2) the acquisition of a positively-charged arginine, which takes the place of the second metal to assist with transition state stabilization and to depress the pKa on the tyrosine. Consistent with this reasoning, basic side-chain groups are not typically found to directly coordinate the reactive phosphate in classic two-metal ion systems.
Fig. 3. DNA cleavage by type IA and II topoisomerases.
a. Proposed cleavage mechanism. The general base (B:) and acid (HA) are unknown, but may be metal-associated waters. Metal A and Arg781 stabilize the transition state; metal B and His736 anchor the (−1) phosphate. Amino acids (blue – TOPRIM, green – WHD) are labelled based on yeast topo II, except for a lysine residue present in type IA (but not type II) topoisomerases (grey, E. coli topo I/III numbering).
b. Type IA/II topoisomerase active site superposition. A DNA-bound (and uncleaved) topo III structure24 (grey) is overlaid on topo II complex (cyan/green). Catalytic residues are labelled (yeast topo II/E. coli topo III numbering).
Our structure further reconciles the type II topoisomerase cleavage mechanism with that used by type IA topoisomerases, enzymes that cleave single, rather than double, DNA strands. Prior studies have noted that both groups of topoisomerases employ the same collection of active site residues and conserved domains to cut DNA22. Moreover, both type II and IA topoisomerases form 5′-phosphotyrosyl bonds and require metal ions for full activity7,23. However, one perplexing difference between the two families is that DNA cleavage by type IA topoisomerases is not strictly metal-dependent: although religation requires metal ions, their presence is needed only to stimulate, but not execute, strand scission7. Comparison of the type II and IA24 topoisomerase active sites reveals the existence of an extra lysine in type IA topoisomerases that contacts the scissile phosphate (Fig. 3b). This lysine is found in all IA enzymes, although it switches between two adjacent locations in topo I and topo III paralogs (Fig. S3). Significantly, mutation of this lysine in topo I abrogates metal-independent cleavage25, suggesting that this basic amino acid is able to promote strand scission by fulfilling the role of metal A. Moreover, because this amino acid is incapable of polarizing the resultant 3′-OH, it would not be expected to support religation. Consistent with this reasoning, substitution of the amino acid equivalent to D526 in our structure with asparagine in topo I (D117N) creates a dominant lethal enzyme that does not efficiently religate DNA, even in the presence of metals26, likely because a key metal-A binding residue is disrupted and hence favours lysine-mediated cleavage. Similarly, type II topoisomerases, which lack this lysine, cannot cleave DNA without metal ions present. Thus, the structure unifies previously conflicting lines of evidence to establish a common cleavage mechanism between two distantly-related and functionally distinct topoisomerase families.
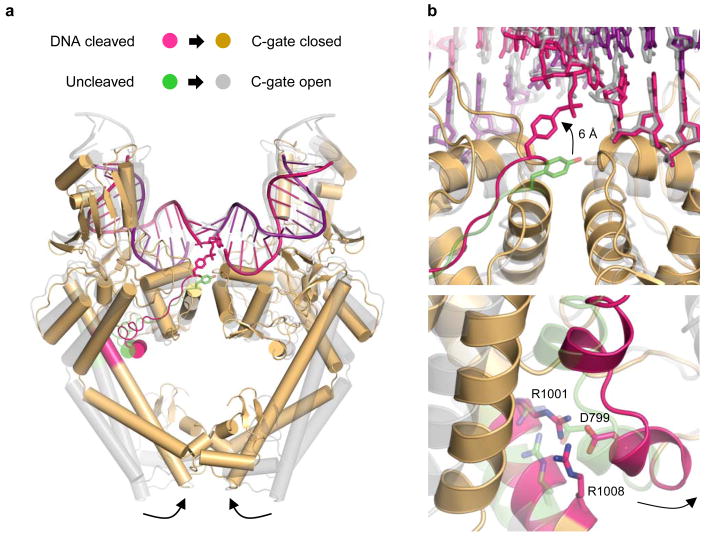
Finally, our structure establishes a new molecular connection by which DNA breakage is coordinated with the association and dissociation of inter-subunit interfaces during strand scission to prevent the accidental formation of double-strand DNA breaks. One major difference between a previous, noncovalent DNA-topo II structure and the present study is that the primary dimer interface of the enzyme (the “C-gate”) switches from an open to a closed state (Fig. 4a). Comparison of the structures reveals why: formation of a covalent attachment between Y782 and DNA pulls the tyrosine upward from its uncleaved position by ~6 Å (Fig. 4b). This movement is made possible by a rocking of the WHD domains (Fig. S4a), which in turn tugs on a long linker and small α-helix (residues 797–802) that engages a set of distal coiled-coil “lever arms” through a conserved salt-bridge network (D799, R1001, and R1008) (Figs. S4b–c). Motions at this elbow joint have been shown previously to open and close the DNA-binding regions of topo II (Fig. S4d)27– 29. Here, however, formation of the phosphotyrosyl bond acts like the drawstring on a latch to pull the coiled coils inward, driving C-gate closure. This transition highlights how the status of the active site is communicated to DNA gating element over 50 Å away, and reveals that type IIA topoisomerases retain a molecular “trapdoor” as a failsafe to ensure that protein/protein interactions are maximized at times when DNA is cleaved (Fig. S5). Such tight control over DNA cleavage and inter-subunit associations helps explain how the fidelity of the type II topoisomerase breakage/rejoining reaction is maximized to protect the genome from the aberrant formation of potentially damaging and cytotoxic DNA lesions during supercoiling management and chromosome disentanglement.
Fig. 4. Cleavage-dependent control of C-gate dynamics.
a. Superposition of noncovalent (grey)13 and cleavage (orange) complexes between topo II and DNA reveal how C-gate opening and closure is linked to active site status. The connection from the active-site tyrosines to the coiled-coil arms is coloured green/magenta. For clarity, the TOPRIM domains are hidden.
b. Close-up of positional shifts. (Upper) Upward movement of the active-site tyrosine upon becoming attached to the DNA. (Lower) Concomitant inward movement of the coiled-coil joint through a conserved salt bridge network.
Methods Summary
The covalent cleavage complex was trapped as described previously30 and purified by tandem cation-exchange and size-exclusion chromatography. Crystals were grown by hanging-drop vapour diffusion, and diffraction data collected on BL8.3.1 (Advanced Light Source, Lawrence Berkeley National Laboratory). Initial phases were calculated by molecular replacement. For soaks, crystals were glutaraldehyde cross-linked in situ and exchanged into higher pH by vapour diffusion, prior to the addition of ZnCl2.
Full methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
This work was supported the NIH (GM033944 and GM053960 for N.O.; T32CA09592 for J.E.D.; GM08295 for B.H.S.; and CA077373 for J.M.B.).
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions B.H.S. purified the complex, grew the crystals, and solved the structures. J.M.B. assisted with refinement and inspection of the molecular models. A.B.B. synthesized the phosphorothiolate reagent. J.E.D. assisted the design of the DNA substrate and optimizing cleavage conditions. N.O. and J.M.B. designed the experiments. All authors contributed to the manuscript.
Author Information Coordinates for the apo and Zn-bound complexes have been deposited in the RSCB PDB under the accession numbers 3L4J and 3L4K, respectively. Reprints and permissions information is available at npg.nature.com/reprintsandpermissions. The authors declare no competing financial interests.
References
- 1.Schoeffler AJ, Berger JM. DNA topoisomerases: harnessing and constraining energy to govern chromosome topology. Q Rev Biophys. 2008;41:41–101. doi: 10.1017/S003358350800468X. [DOI] [PubMed] [Google Scholar]
- 2.Deweese JE, Osheroff N. The DNA cleavage reaction of topoisomerase II: wolf in sheep’s clothing. Nucleic Acids Res. 2009;37:738–48. doi: 10.1093/nar/gkn937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Roca J, Wang JC. The capture of a DNA double helix by an ATP-dependent protein clamp: a key step in DNA transport by type II DNA topoisomerases. Cell. 1992;71:833–40. doi: 10.1016/0092-8674(92)90558-t. [DOI] [PubMed] [Google Scholar]
- 4.Roca J, Berger JM, Harrison SC, Wang JC. DNA transport by a type II topoisomerase: direct evidence for a two-gate mechanism. Proc Natl Acad Sci U S A. 1996;93:4057–62. doi: 10.1073/pnas.93.9.4057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Worland ST, Wang JC. Inducible overexpression, purification, and active site mapping of DNA topoisomerase II from the yeast Saccharomyces cerevisiae. J Biol Chem. 1989;264:4412–6. [PubMed] [Google Scholar]
- 6.Goto T, Wang JC. Yeast DNA topoisomerase II. An ATP-dependent type II topoisomerase that catalyzes the catenation, decatenation, unknotting, and relaxation of double-stranded DNA rings. J Biol Chem. 1982;257:5866–72. [PubMed] [Google Scholar]
- 7.Goto T, Laipis P, Wang JC. The purification and characterization of DNA topoisomerases I and II of the yeast Saccharomyces cerevisiae. J Biol Chem. 1984;259:10422–9. [PubMed] [Google Scholar]
- 8.West KL, et al. Mutagenesis of E477 or K505 in the B′ domain of human topoisomerase II beta increases the requirement for magnesium ions during strand passage. Biochemistry. 2000;39:1223–33. doi: 10.1021/bi991328b. [DOI] [PubMed] [Google Scholar]
- 9.Noble CG, Maxwell A. The role of GyrB in the DNA cleavage-religation reaction of DNA gyrase: a proposed two metal-ion mechanism. J Mol Biol. 2002;318:361–71. doi: 10.1016/S0022-2836(02)00049-9. [DOI] [PubMed] [Google Scholar]
- 10.Deweese JE, Guengerich FP, Burgin AB, Osheroff N. Metal ion interactions in the DNA cleavage/ligation active site of human topoisomerase IIalpha. Biochemistry. 2009;48:8940–7. doi: 10.1021/bi900875c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Steitz TA, Steitz JA. A general two-metal-ion mechanism for catalytic RNA. Proc Natl Acad Sci U S A. 1993;90:6498–502. doi: 10.1073/pnas.90.14.6498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yang W, Lee JY, Nowotny M. Making and breaking nucleic acids: two-Mg2+-ion catalysis and substrate specificity. Mol Cell. 2006;22:5–13. doi: 10.1016/j.molcel.2006.03.013. [DOI] [PubMed] [Google Scholar]
- 13.Dong KC, Berger JM. Structural basis for gate-DNA recognition and bending by type IIA topoisomerases. Nature. 2007;450:1201–5. doi: 10.1038/nature06396. [DOI] [PubMed] [Google Scholar]
- 14.Burgin AB, Jr, Huizenga BN, Nash HA. A novel suicide substrate for DNA topoisomerases and site-specific recombinases. Nucleic Acids Res. 1995;23:2973–9. doi: 10.1093/nar/23.15.2973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Redinbo MR, Stewart L, Kuhn P, Champoux JJ, Hol WG. Crystal structures of human topoisomerase I in covalent and noncovalent complexes with DNA. Science. 1998;279:1504–13. doi: 10.1126/science.279.5356.1504. [DOI] [PubMed] [Google Scholar]
- 16.Wang J, Yu P, Lin TC, Konigsberg WH, Steitz TA. Crystal structures of an NH2-terminal fragment of T4 DNA polymerase and its complexes with single-stranded DNA and with divalent metal ions. Biochemistry. 1996;35:8110–9. doi: 10.1021/bi960178r. [DOI] [PubMed] [Google Scholar]
- 17.Nowotny M, Yang W. Stepwise analyses of metal ions in RNase H catalysis from substrate destabilization to product release. EMBO J. 2006;25:1924–33. doi: 10.1038/sj.emboj.7601076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Aravind L, Leipe DD, Koonin EV. Toprim--a conserved catalytic domain in type IA and II topoisomerases, DnaG-type primases, OLD family nucleases and RecR proteins. Nucleic Acids Res. 1998;26:4205–13. doi: 10.1093/nar/26.18.4205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu Q, Wang JC. Identification of active site residues in the “GyrA” half of yeast DNA topoisomerase II. J Biol Chem. 1998;273:20252–60. doi: 10.1074/jbc.273.32.20252. [DOI] [PubMed] [Google Scholar]
- 20.Okada Y, et al. Assignment of functional amino acids around the active site of human DNA topoisomerase IIalpha. J Biol Chem. 2000;275:24630–8. doi: 10.1074/jbc.M003243200. [DOI] [PubMed] [Google Scholar]
- 21.Nichols MD, DeAngelis K, Keck JL, Berger JM. Structure and function of an archaeal topoisomerase VI subunit with homology to the meiotic recombination factor Spo11. EMBO J. 1999;18:6177–88. doi: 10.1093/emboj/18.21.6177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Berger JM, Fass D, Wang JC, Harrison SC. Structural similarities between topoisomerases that cleave one or both DNA strands. Proc Natl Acad Sci U S A. 1998;95:7876–81. doi: 10.1073/pnas.95.14.7876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Domanico PL, Tse-Dinh YC. Mechanistic studies on E. coli DNA topoisomerase I: divalent ion effects. J Inorg Biochem. 1991;42:87–96. doi: 10.1016/0162-0134(91)80035-g. [DOI] [PubMed] [Google Scholar]
- 24.Changela A, DiGate RJ, Mondragon A. Crystal structure of a complex of a type IA DNA topoisomerase with a single-stranded DNA molecule. Nature. 2001;411:1077–81. doi: 10.1038/35082615. [DOI] [PubMed] [Google Scholar]
- 25.Strahs D, Zhu CX, Cheng B, Chen J, Tse-Dinh YC. Experimental and computational investigations of Ser10 and Lys13 in the binding and cleavage of DNA substrates by Escherichia coli DNA topoisomerase I. Nucleic Acids Res. 2006;34:1785–97. doi: 10.1093/nar/gkl109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cheng B, et al. Asp-to-Asn substitution at the first position of the DxD TOPRIM motif of recombinant bacterial topoisomerase I is extremely lethal to E. coli. J Mol Biol. 2009;385:558–67. doi: 10.1016/j.jmb.2008.10.073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Berger JM, Gamblin SJ, Harrison SC, Wang JC. Structure and mechanism of DNA topoisomerase II. Nature. 1996;379:225–32. doi: 10.1038/379225a0. [DOI] [PubMed] [Google Scholar]
- 28.Morais Cabral JH, et al. Crystal structure of the breakage-reunion domain of DNA gyrase. Nature. 1997;388:903–6. doi: 10.1038/42294. [DOI] [PubMed] [Google Scholar]
- 29.Fass D, Bogden CE, Berger JM. Quaternary changes in topoisomerase II may direct orthogonal movement of two DNA strands. Nat Struct Biol. 1999;6:322–6. doi: 10.1038/7556. [DOI] [PubMed] [Google Scholar]
- 30.Deweese JE, Burgin AB, Osheroff N. Using 3′-bridging phosphorothiolates to isolate the forward DNA cleavage reaction of human topoisomerase IIalpha. Biochemistry. 2008;47:4129–40. doi: 10.1021/bi702194x. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.