Abstract
Exposure to dieldrin induces neurotoxic effects in the vertebrate CNS and disrupts reproductive processes in teleost fish. Reproductive impairment observed in fish by dieldrin is likely the result of multiple effects along the hypothalamic-pituitary-gonadal axis but the molecular signaling cascades are not well characterized. To better elucidate the mode of action of dieldrin in the hypothalamus, this study measured neurotransmitter levels and examined the transcriptomic response in female largemouth bass (LMB) to an acute treatment of dieldrin. Male and female LMB were injected with either vehicle or 10 mg dieldrin/kg and sacrificed after seven days. There were no significant changes in dopamine or DOPAC concentrations in the neuroendocrine brain of males and females after treatment but GABA levels in females were moderately increased 20–30% in the hypothalamus and cerebellum. In the female hypothalamus, there were 227 transcripts (p<0.001) identified as being differentially regulated by dieldrin. Functional enrichment analysis revealed transcription, DNA repair, ubiquitin-proteasome pathway, and cell communication, as biological processes over-represented in the microarray analysis. Pathway analysis identified DNA damage, inflammation, regeneration, and Alzheimer’s disease as major cell processes and diseases affected by dieldrin. Using multiple bioinformatics approaches, this study demonstrates that the teleostean hypothalamus is a target for dieldrin-induced neurotoxicity and provides mechanistic evidence that dieldrin activates similar cell pathways and biological processes that are also associated with the etiology of human neurological disorders.
1. Introduction
Organochlorine pesticides (OCPs) are a broad class of environmental contaminants that are found in polluted water systems, in sediments, and in tissues of aquatic organisms worldwide. The OCP dieldrin (1,2,3,4,10,10-hexachloro-6,7-epoxy-1,4,4a,5,6,7,8,8a-octahydro-1,4,5,8-dimethanonaphthalene) remains persistent in sediment in agricultural regions where it was previously used despite being banned over 30 years ago in North America. Jorgenson (2001) reported that dieldrin, and the related pesticide aldrin, were ranked second behind DDT in agricultural use in the United States in the 1960s and dieldrin was used on approximately 90 crop species that included corn, hay, rye, and oat. Many OCPs are transported into aquatic ecosystems via runoff from these agricultural sites, and in some cases, dieldrin continues to be detected in close proximity to these sites at levels high enough to pose a risk to aquatic organisms.
Dieldrin is a potent γ-aminobutyric acid (GABA)-A receptor modulator that suppresses GABAergic signaling by preventing Cl− influx into the neuron (Gant et al., 1987; Bloomquist et al., 1992; Ikeda et al., 1998). Dieldrin-mediated receptor antagonism causes the loss of GABAergic inputs within the central nervous system (CNS). Neurotoxic effects of dieldrin have been reported in both in vitro and in vivo mammalian models. Experimental data investigating neurophysiological responses to dieldrin in mammalian models suggests that dieldrin induces apoptosis and cell death in the human dopaminergic PC12 cell line (Kitazawa et al., 2001; Kitazawa et al., 2004), adversely affects the ubiquitin-proteasome pathway in rat mesencephalic dopaminergic neuronal (Sun et al., 2005) and SK-N-MC (human, caucasian, neuroblastoma) neuroblastoma cells (Wang et al., 2006), and increases oxidative stress in mouse nigrostriatal dopamine neurons in vivo (Hatcher et al., 2007). Similar to mammals, dieldrin-mediated effects also occur in the teleostean CNS. In catfish, dieldrin has been shown to bind GABA-A receptors in P2 membrane preparations from brain with relatively high affinity (IC50 = 592.5 ± 105 nM; Carr et al., 1999). In addition, dieldrin is highly lipophilic and studies in teleost fishes have demonstrated that dieldrin bioaccumulates in fatty tissues such as the brain and liver (Lamai et al., 1999; Satyanarayan et al., 2005). There is also evidence that dieldrin-mediated neurotoxicity occurs in the teleostean CNS. During zebrafish development at 48 and 96 h post-fertilization, dieldrin caused apoptosis, disorganized motor neuron axon formation, and significant loss of dopaminergic neurons in the brain (Ton et al., 2006). These studies support the hypotheses that dieldrin directly binds GABA-A receptors, bioaccumulates in the brain, and induces neurotoxicity in teleosts.
To better understand the molecular cascades underlying dieldrin exposure in the CNS, we measured neurotransmitter levels and the genomic response in the hypothalamus of largemouth bass (Micropterus salmoides) (LMB) to an acute intraperitoneal (i.p.) injection of 10 mg dieldrin/kg. LMB was chosen as a model because wild LMB located in Lake Apopka, Florida are exposed to high groundwater and sediment concentrations of dieldrin, as well as to other OCPs due to the region being a previously heavily used agricultural area. LMB are top predators in the ecosystem and accumulate high levels of OCPs in their natural environment (Denslow, unpub. data). A previously described LMB oligonucleotide microarray (Garcia-Reyero and Griffitt et al., 2008) was used to profile gene expression in the LMB hypothalamus. Similar to mammals, the hypothalamus of teleost fish has a high abundance of both GABAergic (Martyniuk et al., 2007) and dopaminergic (Vetillard et al., 2002) cells and is a potential target for dieldrin neurotoxicity. Complementary bioinformatics approaches (functional enrichment and pathway analysis) were also utilized to identify major pathways that underlie dieldrin neurotoxicity in the teleostean hypothalamus.
2. Materials and Methods
2.1. Fish and experimental design
Largemouth bass were purchased from the American Sportfish Hatchery (Montgomery, Alabama) and maintained at the Aquatic Toxicology Laboratory at the Center for Environmental and Human Toxicology (CEHT) at the University of Florida (Gainesville, Florida). Fish were acclimated in aerated 104–250 gallon fiberglass tanks under constant conditions of 21±2 °C for a minimum of 1 week prior to exposure. Fish were fed Silvercup floating pellets (Ziegler Bros., Murray, UT) once a day ad libitum. In January, female and male LMB at comparable stages of gonadal development were given a single intraperitoneal (i.p.) injection of either 10 mg dieldrin/kg with sesame oil as the carrier or a single injection of sesame oil alone (control). A dose of dieldrin (10 mg/kg) was chosen for this study to prevent mortality and induce a significant transcriptional response in the hypothalamus. Other studies in mammals (Thiffault et al., 200; Hatcher et al., 2007; Sava et al., 2007) and fish (Pedrajas et al., 1995) have used comparable doses (ranging 1–80 mg/kg) and observed biological responses such as modulation of DA levels in the CNS and biochemical stress responses in the liver. Based on our observation throughout the experiment, there were no marked changes in the behavior of the bass that were attributable to the injection of dieldrin. After 7 days, female and male LMB were sacrificed. Hypothalamus, telencephalon, and c erebellum were rapidly dissected, frozen in liquid nitrogen, and stored at −80 °C until processed for GABA measurements and microarray hybridizations. A subset of fresh brain tissues were dissected and used immediately to determine dopamine (DA) levels by HPLC. All animals were treated as per the protocol approved by University of Florida Institutional Animal Care and Use Committee.
2.2. High performance liquid chromatography (HPLC)
Measurement of GABA was conducted using electrochemical detection coupled with HPLC as per Peinado et al. (1986). Briefly, pre-column o-phthalaldehyde (OPA) derivatization (27 mg of OPA in 0.5 ml of methanol, 10 µl of 2-methyl-2-propanethiol and 0.1 M sodium borate to 5 mL total volume) was used before chemical separation at a flow-rate of 1.0 ml/min with a detector potential of +700 mV. A 150 × 4.6 mm, 4.0-µm particle size, Synergi 4µ Max RP analytical column provided a retention time of under 5 min., using a mobile phase which consisted of 0.1 M sodium acetate, 1 mM EDTA, in 50% acetonitrile + 2% (v/v) tetrahydrofuran (pH adjusted to 6.0 with glacial acetic acid). Peaks were quantified based on a GABA standard curve with a linear range of 0.5–10 µM.
Dopamine and 3,4-dihydroxyphenylacetic acid (DOPAC, a major DA metabolite) levels were determined by HPLC using electrochemical detection according to the method described by Gesto et al. (2006) with modifications. Hypothalamus, telencephalon, and cerebellum were obtained from both control and treated fish, and were weighed and immediately placed into a cold solution containing 0.1 mM EDTA in 0.1M perchloric acid (with L-DOPA added as an internal standard). Samples were sonicated on ice and centrifuged at 12,000 × g for 15 min. at 4°C. A 50-µl aliquot of the supernatant was injected onto a 150 × 4.6 mm, 4 µm particle size, Synergi 4u Max RP analytical column (Phenomenex, Torrance, CA) with approximate retention times of 2.5, 4.4 and 6.6 min for L-DOPA, DOPAC, and DA, respectively. The mobile phase contained 1 mM octane sulfonic acid, sodium salt, 0.1 mM EDTA, and 0.25 mM triethylamine in 90% 0.1 M anhydrous monobasic sodium phosphate (NaH2PO4) and 10% methanol (pH adjusted to 3.85 with 5M phosphoric acid). Separations were performed at a flow-rate of 0.30 ml/min at a detector potential of +450 mV (range 20 nA/V). Peaks were quantified based on DA and DOPAC standard curves with a linear range of 10–100 ng/ml. All statistical tests were performed using the SAS program JMP Genomics v3.2 (SAS, Cary, NC). Statistical tests were performed in each sex separately. An ANOVA followed by Tukey’s post hoc test was used to determine if there were significant differences in the levels of GABA, DA, and DOPAC across brain region in the control animals only. An ANOVA followed by a Bonferroni correction was performed to determine if there were differences between control and dieldrin treated animals in each brain region examined.
2.3. Gene expression analysis using the LMB oligonucleotide 4 x 44K microarray platform
The LMB oligonucleotide microarray was generated by 454-Life Sciences GS 20 pyrosequencing and has been previously described and validated (Garcia-Reyero and Griffitt et al., 2008). Microarrays were printed in the format of 4 × 44K by Agilent (Palo Alto, CA). Gene expression analysis was per formed with four biological replicates for the control and four biological replicates for the treatment. Total RNA for microarray analysis was extracted from LMB hypothalamus using RNA STAT-60 reagent (TEL-TEST Inc., Friendswood, TX, USA). RNA quantity for microarray analysis was measured using the NanoDrop ND-1000 (Nanodrop Technologies, Wilmington, DE) and RNA quality was evaluated using the Agilent 2100 BioAnalyzer with the RNA 6000 Nanochip. RNA integrity values (RIN) were > 8.8 for all samples used in the analysis with an average RIN of 9.4.
Microarray hybridizations were performed according to the Agilent One-Color Microarray-Based Gene Expression Analysis protocol using Cyanine 3 (Cy3) (Agilent, Palo Alto, CA). Briefly, 1 µg total RNA per sample was used for the production of cDNA and labeled/amplified cRNA as per the Agilent Low RNA Input Fluorescent Amplification Kit. The one-color spike mix was also prepared and added according to the protocol for 1 µg total RNA. Amplified cRNA containing the incorporated Cy3 label was purified using the Qiagen® RNeasy mini-spin columns (Qiagen, Valencia, CA, USA), following the instructions provided by the manufacturer. After purification, samples were evaluated for yield and incorporation of Cy3 dye using the NanoDrop ND-1000. Each sample contained a specific activity > 9.0 pmol Cy3/µL, and amounts were adjusted to a final mass of 1.65 µg for 4 × 44K microarray hybridizations. Fragmentation of the cRNA was performed in the recommended blocking agent and volume of 2x GE Hybridization Buffer (Agilent, Gene Expression Hybridization Kit) and the reaction proceeded for 30 min. at 60°C. A final volume of 100 µl containing fragmented cRNA was added to the 4 × 44K microarrays, and hybridization proceeded for 17 h at 65°C.
Microarrays were washed the following day according to the supplemental Agilent protocol “Preventing Ozone-Related Problems”. Slides were washed in GE Wash Buffer 1 at room temperature for 1 min, followed by GE Wash Buffer 2 at 37°C for 1 min, acetonitrile wash at room temperature for 1 min, and Stabilizing and Drying Solution wash at room temperature for 30 s. Microarrays were kept in the dark until scanned at 5 µm at both 10 and 100 PMT (Agilent G2505 B Microarray Scanner). Agilent Feature Extraction Software (v9.5) formed a composite of the two scans and calculated parameters for Extended Dynamic Range. The quality of microarray data were evaluated by manual inspection of the quality control reports provided from the Agilent software and hybridizations were deemed to be of high quality.
2.4. Microarray analysis, pathway analysis, and gene categorization
Raw expression data were imported into JMP® Genomics v3.2. Raw intensity data for each microarray were normalized using Loess normalization with a smoothing factor of 0.2. After microarray normalization, differentially regulated transcripts were identified using a one-way analysis of variance (ANOVA) with a False Discovery Rate (FDR=5%) to control for false positives. Raw microarray data for this experiment have been deposited into the NCBI Gene Expression Omnibus (GEO) database (series GSE12698; platform GPL6527).
Gene expression data were subjected to hierarchal clustering. Distance calculations were performed using the program Cluster (Eisen et al., 1998) and visualized using the Java TreeView program (Saldanha, 2004). Clustering was based on complete linkage and absolute correlation. For functional enrichment analysis, FatiGO in Babelomics (Al-Shahrour et al., 2005, 2006) was used to identify biological processes and molecular functions that were over-represented in the list containing regulated transcripts (p<0.05) compared to a second list of all gene probes on the LMB microarray. GO level analysis was inclusive, annotation was propagated to upper levels, and the GO levels included 3–9. The file contained specific annotations and gene ontology for LMB probes.
For highly significant altered transcripts (p<0.001; fold change > ±1.5), pathway analysis was performed using Pathway Studio® v5.0 (Ariadne Genomics, Rockville, MD, USA) to identify cell processes and disease relationships that are associated with transcripts that are mediated by dieldrin (Nikitin et al. 2003). The closest human homologs (NCBI) for LMB genes were manually obtained (RefSeq in NCBI) and Entrez Gene identifiers were retrieved using the ID mapping service in Pathway Studio® (MD Anderson GeneLink, University of Texas, Houston, TX, USA). Pathways were built by query against human diseases. All gene relationships were supported by at least 2 references from the literature and connectivity among entities was >1600.
Pathway analysis was further performed on transcripts involved in the biological process of DNA repair (p<0.05) because this process was identified as an over-represented gene ontology. The database was queried to construct a pathway based on all shortest biological interactions that included protein-protein binding and regulation of expression. Connectivity between entities (genes) was restricted to two or more literature references from NCBI PubMed that supported the interaction.
2.5. Cloning of largemouth bass genes for real-time SYBR green assay
Cloning strategies for LMB genes have been described in detail elsewhere (Martyniuk et al., 2009). Briefly, RNA STAT-60™ (Friendswood, TX) was used to extract total RNA from homogenized LMB whole brain as per the manufacturer’s protocol. Primers with optimal annealing temperature ~58–60°C were designed to amplify sequences of 66–173 base pairs (bp) (Table 1). PCR cycling conditions follow that of Martyniuk et al. (2009). Amplicons were cloned into a pGEM-® T easy vector and transformed in TOP-10 cells (Invitrogen). Clones were sequenced at the Interdisciplinary Center for Biotechnology Research (ICBR; University of Florida) to verify target specificity. Standard curves relating initial template copy number to fluorescence and amplification cycle were generated using pGEM-® T easy vector containing the gene target as a template. Linearity of standard curves showed R2 > 0.98 and efficiencies ranged between 95–105%.
Table 1.
Primers used for real-time PCR. Note that the forward primer is provided as 5’ to 3’ orientation in relation to the 5’ to 3’ coding strand (sense) deposited in GenBank and the reverse primer is 5’ to 3’ in relation to the complementary strand (anti-sense).
| Gene | Forward primer (5' - 3') | Reverse primer (5' - 3') | Amplicon size (bp) |
|---|---|---|---|
| androgen receptor | CAC CAC AGA GAA TGT GCC TGA | CAG GTG AGT GCG CCG TAA | 66 |
| estrogen receptor beta b | CCG ACA CCG CCG TGG TGG ACT C | AGC GGG GCA AGG GGA GCC TCA A | 96 |
| glutathione S-transferase | AAC TTT TCG CTG GCT GAT GT | TCT TGT CCC TGT GGG TTC TC | 173 |
| heat shock cognate 70 | CAG TGA TGA AGA CAA GCA GAA GA | GCC ACC AGC ACT CTG ATA CA | 163 |
| Niemann-Pick disease C2 | GAT GGC TGC AAG TCT GGA AT | ACT GGG AAC CTG ATG CAG AA | 164 |
| ribosomal 18s | CGG CTA CCA CAT CCA AGG AA | CCT GTA TTG TTA TTT TTC GTC ACT ACC T | 86 |
Transcript levels of nuclear androgen receptor (ar), estrogen receptor beta b (esr-beta b; AY211021), glutathione S-transferase (gst; AY335905), glutathione peroxidase (gpx; FJ030930), heat shock protein 70 (hsp70; FJ751227), and Niemann-Pick disease type C2 (npc2; FJ751228) were measured in the hypothalamus (n=6) using real-time PCR (iCycler Thermal Cycler; Bio-Rad). LMB genes follow nomenclature rules set out for zebrafish (www.ZFIN.org). LMB ar and esr-beta b were investigated because of previous evidence that dieldrin regulates the abundance of these transcripts in LMB gonad and liver (Garcia-Reyero et al., 2006). LMB gst, gpx, hsp70 were chosen because each is involved in the general and oxidative stress response and dieldrin induces these responses in tissues of both fish (Pedrajas et al., 1995) and mammals (Hatcher et al., 2007). The LMB hsp70 is more closely homologous to the HSP cognate 70 found in other teleost fish. LMB npc2 was chosen because deficiencies in this protein are associated with improper cholesterol metabolism and apoptosis in the CNS (Wu et al., 2005). Microarray analysis identified all six transcripts as being altered by dieldrin in the hypothalamus of LMB (p<0.05).
For real-time PCR, each sample of total RNA was DNase treated using the TURBO DNA-free kit (Ambion, Austin, TX, USA) as per manufacturer’s protocol and cDNA was synthesized using SuperScript™ II Reverse Transcriptase (Invitrogen). Real-time PCR analysis was carried out using 1X iQ SYBR Green Supermix (Bio-Rad, Hercules, CA), 1 µl of each gene specific primer (10 mM), and 100 ng cDNA. The two-step thermal cycling parameters were as follows: initial 1-cycle Taq activation at 95°C for 3 min, followed by 40 cycles at 95°C for 15 s, and 60°C for 1 min. After 40 cycles, a dissociation curve was produced starting at 55°C (+1°C/30 seconds) to 95°C. Transcripts were assayed on an iCycler Thermal Cycler (Bio-Rad). 18S ribosomal RNA (IQ Supermix, Bio-Rad) was used as the reference gene to normalize expression data. Each gene analysis included 1) two samples that did not receive reverse transcriptase (no RT control) and 2) two samples that did not receive the cDNA template to ensure there was no genomic contamination. Melting curves for each gene indicated a single product being formed. Gene expression data were not normally distributed (Shapiro-Wilk W test) and a non-parametric Mann-Whitney U-test was used to determine if there were significant differences (p<0.05) in mRNA levels between control and treatment.
3. Results
3.1 Neurotransmitter levels in female and male LMB
GABA concentrations in each sex and tissue were normally distributed as determined by a Goodness of Fit test (Shapiro–Wilk test). When considering each brain region within sex, there was significant variation in GABA levels across control female LMB brain region (d.f.=2; F=4.27; p=0.034). The telencephalon of control female LMB had significantly more GABA compared to the cerebellum but neither tissue differed in GABA levels compared to the hypothalamus. There were no detectable differences in GABA levels in control male LMB (d.f.=2; F=2.41, p=0.13). However, similar to females, there was a trend for more GABA in the telencephalon compared to the cerebellum.
Acute exposure to dieldrin affected brain GABA content differently depending on sex and brain region. When comparing GABA levels between control and dieldrin treated LMB within tissue, GABA levels in the hypothalamus (d.f.=1; F=6.46; p=0.028) and cerebellum (d.f.=1; F=6.70; p=0.033) of females were 20–30 % higher after dieldrin treatment (Table 2). However, these differences were no longer significant after correction for multiple hypotheses testing (Bonferroni adjusted p-value). There were no significant changes in GABA levels in female telencephalon. There were no changes in GABA levels in any male brain tissue exposed to dieldrin compared to controls.
Table 2.
Concentrations of GABA, DOPAC, and dopamine (nmol/g ) in both female and male cerebellum, telencephalon, and hypothalamus in control and dieldrin treated LMB. In females, there was a significant elevation of approximately 20–30% of GABA in both the hypothalamus and cerebellum.
| Sex | Brain Region | Group | n | GABA | n | DOPAC | n | Dopamine |
|---|---|---|---|---|---|---|---|---|
| Female | cerebellum | control | 6 | 22.98 ± 2.53 | 7 | 1.06 ± 0.07 | 7 | 0.63 ± 0.04 |
| treated | 4 | 31.13 ± 0.49* | 4 | 1.14 ± 0.12 | 4 | 0.72 ± 0.10 | ||
| telencephalon | control | 5 | 42.63 ± 8.17 | 6 | 1.30 ± 0.09 | 6 | 1.80 ± 0.10 | |
| treated | 6 | 44.98 ± 4.74 | 6 | 1.28 ± 0.10 | 6 | 2.10 ± 0.18 | ||
| hypothalamus | control | 7 | 29.55 ± 2.96 | 7 | 1.37 ± 0.07 | 7 | 3.15 ± 0.19 | |
| treated | 6 | 39.30 ±2.22* | 6 | 1.39 ± 0.14 | 6 | 3.15 ± 0.29 | ||
| Male | cerebellum | control | 5 | 28.99 ± 4.92 | 5 | 1.41 ± 0.14 | 5 | 0.78 ± 0.07 |
| treated | 5 | 29.10 ± 3.00 | 7 | 1.30 ± 0.10 | 7 | 0.64 ± 0.04 | ||
| telencephalon | control | 6 | 56.13 ± 4.16 | 6 | 1.31 ± 0.06 | 6 | 1.82 ± 0.05 | |
| treated | 4 | 47.65 ± 4.10 | 4 | 1.16 ± 0.06 | 4 | 1.59 ± 0.13 | ||
| hypothalamus | control | 5 | 35.92 ± 6.87 | 5 | 1.48 ± 0.11 | 5 | 2.29 ± 0.57 | |
| treated | 7 | 40.58 ± 4.55 | 7 | 1.49 ± 0.07 | 7 | 2.92 ± 0.54 |
p<0.05 before correction for multiple hypotheses testing
DOPAC and DA in each sex and tissue were normally distributed as determined by a Goodness of Fit test (Shapiro–Wilk test). In control female LMB, there were significant differences in DOPAC levels (d.f.=2; F= 4.85; p=0.022), with significantly more DOPAC in the hypothalamus compared to the cerebellum. In control male LMB, brain regions did not significantly differ in DOPAC levels. In control female LMB, DA also varied significantly across brain region (d.f.=2; F=97.53; p<0.001) with significantly higher levels of DA being found in hypothalamus compared to the telencephalon and cerebellum. Furthermore, there were significantly higher levels of DA in the telencephalon compared to the cerebellum. In control male LMB, DA also significantly varied across brain regions (d.f.=2, F=5.78, p0.016). There were higher levels of DA in the hypothalamus compared to the cerebellum. After dieldrin treatment, there were no significant changes in DA or DOPAC levels of any female or male brain region examined (Table 2).
3.2. Microarray analysis
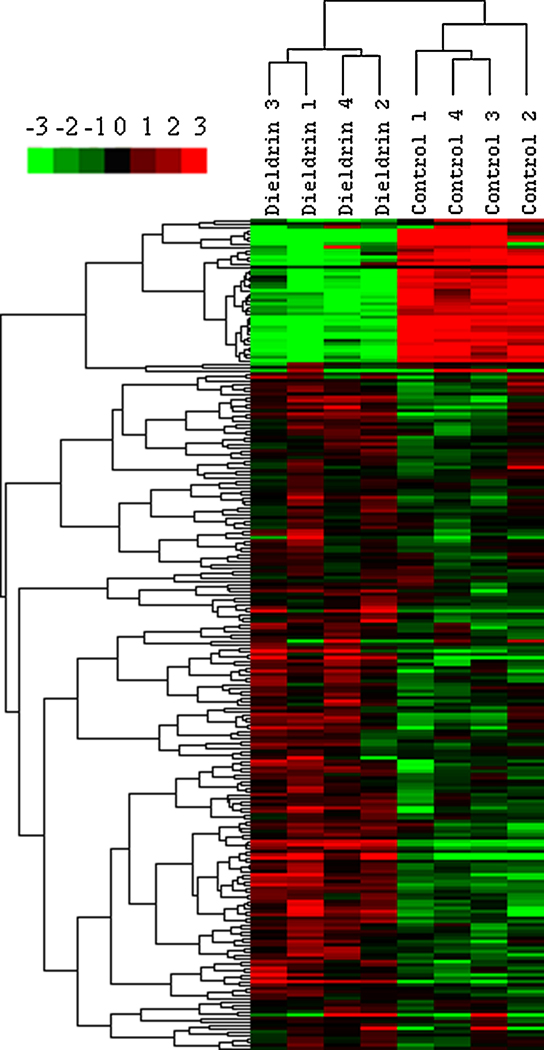
Cluster analysis of regulated transcripts (p<0.001; fold change > ±1.5) revealed that there was a clear distinction between control and dieldrin treated females, with the vast majority of genes being induced by dieldrin (Figure 1). There were 227 genes (199 induced and 28 reduced) identified as differentially expressed in the hypothalamus of LMB exposed to dieldrin (Appendix 1; p<0.001). There were significantly more genes induced by dieldrin compared to genes reduced by dieldrin (d.f.=1, χ=128.85, p<0.0001).
Figure 1.
Gene cluster analysis (p<0.001; fold change > ±1.5) for control and dieldrin-treated microarrays. The color represents intensity of the single dye channel with red being high probe intensity and green being low probe intensity, relative to row (gene) average for mean centered gene data.
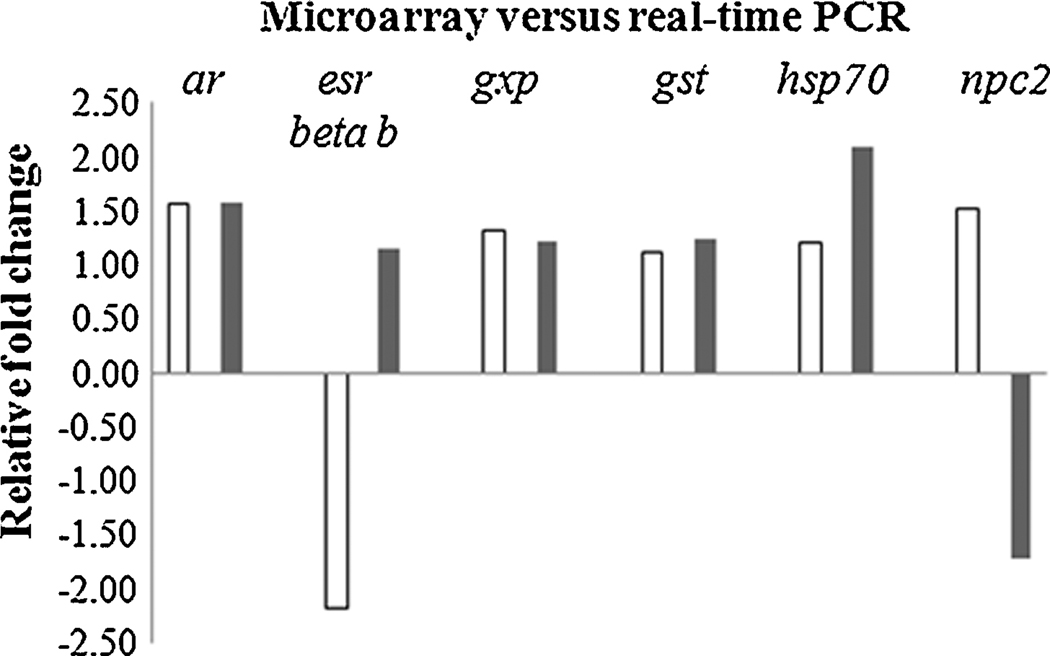
Gene expression was also examined in the hypothalamus of female LMB using real-time PCR for select genes that included ar, esr-beta b, gpx, gst, hsp70, and npc2 (Figure 2). In general, the magnitude and direction were comparable between real-time PCR and microarray data. Relatively small fold changes in gene expression (less than two fold) have been previously observed in neuroendocrine regions of the fish brain (Martyniuk et al., 2006; 2007) and can be an obstacle for the validation of microarray data by real-time PCR. The discrepancy in the direction of expression of the npc2 gene between real-time PCR data and microarray data has also been observed in a second independent experiment with dieldrin (Martyniuk, unpub. data), suggesting that the oligonucleotide probe on the LMB microarray may not be entirely specific for the amplicon that is quantified with real-time PCR. In the case of esr-beta b mRNA, the probe on the microarray was specific to this ER isoform and the primers used in the real-time PCR reaction are specific for esr-beta b (Sabo-Attwood et al., 2004). Therefore the reason for the discrepancy in esr-beta b mRNA between methods in this study is currently difficult to ascertain but may be due to low expression levels of this transcript in the teleostean neuroendocrine brain.
Figure 2.
Relative fold changes for genes analyzed by microarray (white bars; n=4) and real time PCR (black bars; n=6) in the female hypothalamus. Abbreviations are as follows; androgen receptor (ar), estrogen receptor beta b (esr-beta b), glutathione peroxidase (gpx), glutathione-S-transferase (gst), heat shock protein 70 (hsp70), and Niemann-Pick Disease type C2 (npc2).
The major strength of this study is derived from the multiple bioinformatics approaches using gene ontology and pathway analysis to provide a more universal description of gene expression changes occurring in the hypothalamus. These approaches provide valuable information on global genomic processes and are becoming the preferred methods to analyze large genomic datasets compared to focusing on specific transcripts. Functional enrichment analysis was performed for biological processes, molecular functions, and cellular components for transcripts regulated at p<0.05 to gain additional insight into global processes affected by dieldrin. Dieldrin regulated transcripts were involved in DNA repair (GO:0006281), transcription (GO:0006350), regulation of transcription, DNA dependent (GO:0006355), and the ubiquitin cycle (GO:0006512) (Table 3). Gene ontology terms for molecular function that were significantly over-represented in our regulated gene dataset included catalytic activity (GO:0003824), serine-type endopeptidase activity (GO:0004252), and zinc ion binding (GO:0008270). Transcripts for proteins located in the nucleus (GO:0005634) were highly over-represented in the GO category for cellular components, and also included intracellular junction (GO:0005911) and anaphase-promoting complex (GO:0005680).
Table 3.
Functional enrichment analysis for biological process, molecular function, a nd cellular component of dieldrin-mediated transcripts (p<0.05). Babelomics was used to identify gene ontology terms that were significantly over-represented on the LMB microarray after dieldrin treatment.
| GO term | GO term classification | GO identifier | p-value |
|---|---|---|---|
| Biological Process | Ubiquitin Cycle | GO:0006512 | p<0.001 |
| Complement activation, classical pathway | GO:0006958 | p<0.001 | |
| Regulation of transcription, DNA dependent | GO:0006355 | 0.011 | |
| Transcription | GO:0006350 | 0.013 | |
| DNA Repair | GO:0006281 | 0.021 | |
| Cell communication | GO:0007154 | 0.025 | |
| Intracellular Signaling Cascade | GO:0007242 | 0.026 | |
| Lipid transport | GO:0006869 | 0.029 | |
| I-kappaB kinase/NF-kappaB cascade | GO:0007249 | 0.037 | |
| DNA methylation | GO:0006306 | 0.039 | |
| Blood coagulation | GO:0007596 | 0.039 | |
| Protein amino acid phosphorylation | GO:0006468 | 0.041 | |
| Exocytosis | GO:0006887 | 0.042 | |
| Molecular Function | Catalytic activity | GO:0003824 | p<0.001 |
| Serine-type endopeptidase activity | GO:0004252 | p<0.001 | |
| Zinc ion binding | GO:0008270 | 0.014 | |
| DNA binding | GO:0003677 | 0.017 | |
| Nucleic acid binding | GO:0003676 | 0.021 | |
| Kinase activity | GO:0016301 | 0.022 | |
| 3'-5' cyclic-nucleotide phosphodiesterase activity | GO:0004114 | 0.038 | |
| Cellular Component | Nucleus | GO:0005634 | p<0.001 |
| Extracellular region | GO:0005576 | 0.028 | |
| Intercellular junction | GO:0005911 | 0.036 | |
| Mediator complex | GO:0000119 | 0.04 | |
| Anaphase-promoting complex | GO:0005680 | 0.051 |
When investigating further those transcripts highly regulated (p<0.01) and involved in the ubiquitin-proteasome pathway, it was discovered that approximately 86% showed an increase in mRNA steady state levels, suggesting there is a significant induction of protein degradation pathways after exposure of LMB to dieldrin (d.f.= 1; χ = 11.63; p<0.001) (Table 4). Note that many ubiquitin-proteasome related transcripts were not included in the pathway analysis because the gene probe did not meet the requirements that included 1) p<0.001 2) fold change > ±1.5 and 3) human homolog identified (see below). The criteria were used to reduce the complexity of the pathway analysis and to focus on the most likely gene targets affected by dieldrin. However, many of these transcripts are also implicated in the same diseases identified in the pathway analysis (data not shown). There were two genes (traf-interacting protein and checkpoint with forkhead and ring finger domains) that had probes on the microarray that showed significant but opposite fold changes. Each probe was specific for a different region of the transcript and may be detecting changes in other gene family members or closely related transcripts. In these two cases, the probe with the highest probability of being significant is given in the table. The molecular data and bioinformatics approaches investigating global gene patterns indicate that the hypothalamus is responding to stress.
Table 4.
All transcripts regulated b y dieldrin that are involved in the ubiquitin-proteasome pathway (p<0.01). There were significantly more genes induced (19) than down-regulated (3) after treatment to dieldrin.
| NR_Accession | Gene Name | p-value | Fold change |
|---|---|---|---|
| AAW31810 | myc binding protein 2 | 0.0094 | −1.42 |
| CAD38880 | hect carboxyl terminus domain and RCC1-like domain 2 | 0.0027 | −1.39 |
| AAH45309 | ubiquitin-conjugating enzyme E2G 1 | 0.0034 | −1.32 |
| XP_699224 | ring finger protein 127 | 0.0072 | 1.16 |
| CAG12771 | ubiquitin specific peptidase 16 | 0.0005 | 1.18 |
| CAF99126 | neuralized homolog | 0.0055 | 1.22 |
| AAL75965 | f-box and leucine-rich repeat protein 14 | 0.0006 | 1.38 |
| CAG02474 | anaphase promoting complex subunit 7 | 0.0013 | 1.40 |
| AAH44553 | ubiquitin specific peptidase 14 | 0.0019 | 1.43 |
| CAF95617 | u3 small nucleolar RNA-associated protein 18 homolog | 0.0046 | 1.43 |
| AAH44088 | ccr4-not transcription complex, subunit 4 | 0.0038 | 1.46 |
| AAS45170 | bloodthirsty | 0.0033 | 1.46 |
| XP_694020 | thyroid hormone receptor interactor 12 | 0.0004 | 1.47 |
| CAF94074 | baculoviral iap repeat-containing protein 6 | <0.0001 | 1.49 |
| CAA09084 | traf-interacting protein | 0.0007 | 1.53 |
| CAG03056 | nuclear protein localization 4 | <0.0001 | 1.55 |
| CAG12225 | ubiquitin-binding protein homolog | 0.0004 | 1.60 |
| AAH45970 | proteasome subunit, alpha type, 4 | 0.0002 | 1.60 |
| XP_584914 | f-box protein 11 | 0.0057 | 1.65 |
| CAH89580 | checkpoint with forkhead and ring finger domains | 0.0017 | 1.79 |
| CAG02915 | ring finger protein 121 | 0.0083 | 1.85 |
| XP_691280 | tripartite motif-containing 35 isoform 1 | 0.0020 | 1.99 |
3.3 Pathway analysis of differentially regulated transcripts
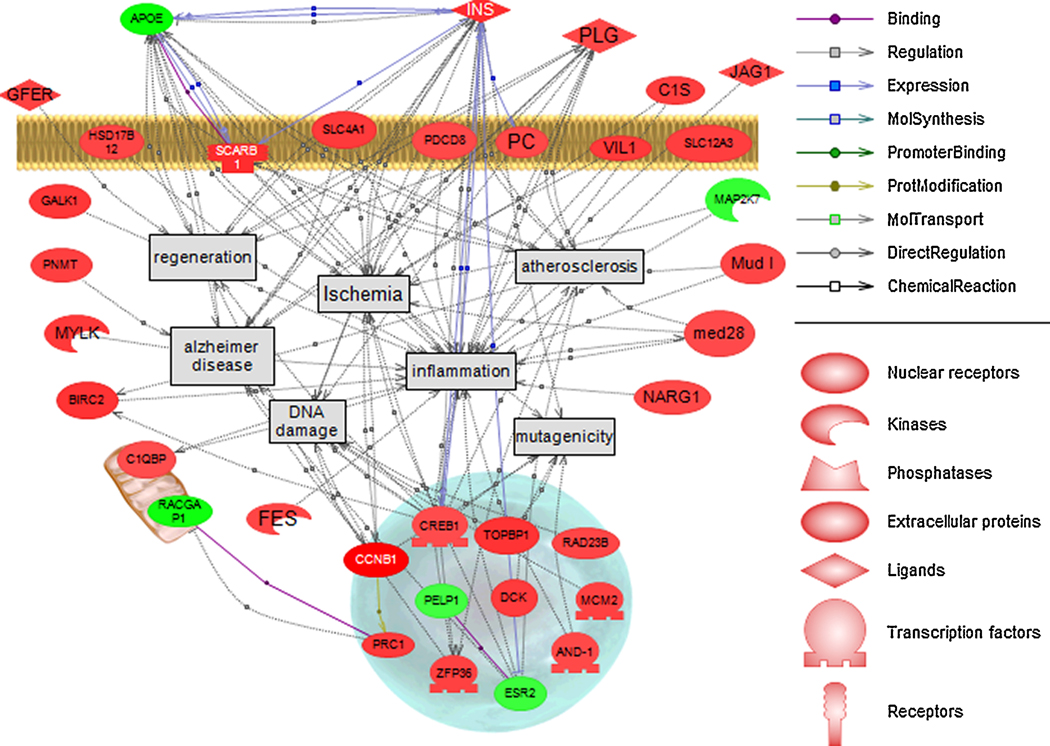
Human homologs were assigned to 179 of 204 or 88% of LMB genes (p<0.001; fold change > ±1.5) and human homologs were used to identify cell processes and diseases associated with transcripts regulated by dieldrin. The gene network identified ischemia, atherosclerosis, inflammation, regeneration, mutagenicity, DNA damage, and Alzheimer’s disease (AD) (Figure 3). Other diseases and cell processes identified in the analysis but not shown in the pathway included different forms of cancer (e.g. breast, liver, leukemia, melanoma, neuroblastoma, lung, and colon), diabetes mellitus, obesity, and blood pressure. Transcripts that were highly significantly regulated (p<0.001) were largely increased in mRNA levels compared to being decreased (d.f.=1, χ = 139.4, p<0.001). Transcripts were located in a variety of cellular compartments that included the nucleus, cytoplasm, plasma membrane, golgi apparatus, and mitochondria. The pathway analysis supports the functional enrichment analysis in that the tissue is responding to a stressor and both processes of DNA damage and DNA repair are occurring in the hypothalamus.
Figure 3.
Pathway analysis of disease associations in the hypothalamus (genes regulated at p<0.001; fold change > ±1.5). Green indicates a down-regulation in the gene and red indicates an induction in gene expression. Each connection has two or more references supporting the association or relationship. Note that dieldrin-mediated genes are located in a variety of cellular compartments and are predominantly induced after treatment.
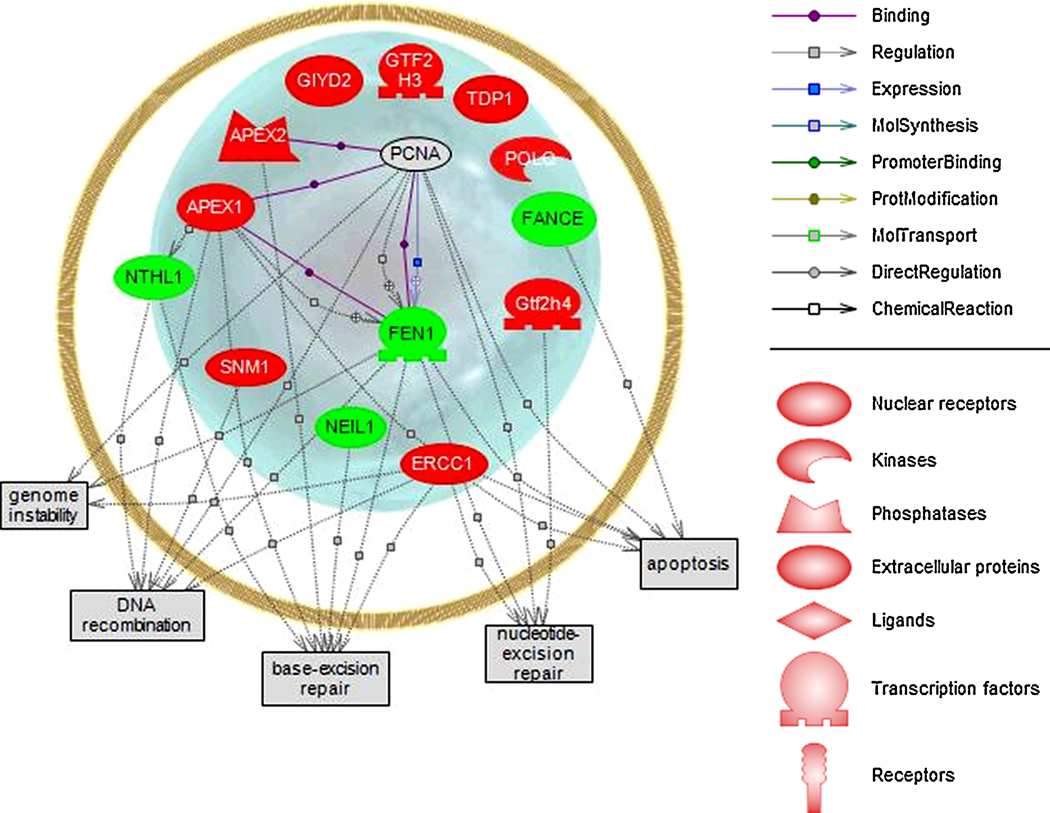
We furthered queried relationships between transcripts (p<0.05) involved in the biological processes of DNA repair (Figure 4). As expected, all transcripts involved in DNA repair were confined to the nucleus and were involved in cell processes that indicate stress in the hypothalamus (apoptosis, nucleotide excision repair, base excision repair, DNA recombination, and genome instability). The program built pathways considering the shortest number of connections among selected entities. Interestingly, proliferating cell nuclear antigen mRNA was not regulated by dieldrin but was identified as directly binding both apex1 and apex2 (apex nuclease 1 and 2, DNA cross-link repair; both induced with dieldrin) as well as fen1 (flap structure-specific endonuclease 1; reduced with dieldrin).
Figure 4.
Pathway analysis of transcripts (p<0.05) involved in the biological process of DNA repair. Green indicates a down-regulation in the gene and red indicates an induction in gene expression. Proliferating cell nuclear antigen had many direct relationships with apurinic/apyrimidinic endonucleases (mRNA increased) and flap structure-specific endonuclease 1 (mRNA decreased) but was not identified in this study as being regulated by dieldrin.
4. Discussion
4.1. The effect of dieldrin on GABA levels in the CNS is tissue dependent
Females had significantly more GABA in the telencephalon compared to the cerebellum which is consistent with the distribution of GABA producing cells within the adult teleostean CNS. Riboprobe staining for both gad1 and gad2 mRNA, the two enzyme isoforms that produce GABA from glutamate, is more abundant and dense in the telencephalon and hypothalamus of goldfish when compared to the cerebellum (Martyniuk et al., 2007). In the same study, enzyme activity of total GAD in the hypothalamus has been shown to be approximately 2–5 times higher than in the cerebellum. Therefore, differences in GABA levels across LMB brain tissues correspond to the distribution of GABA producing cells in the teleostean CNS.
GABA levels in the female hypothalamus and cerebellum were moderately increased by in vivo dieldrin treatment. Increasing GABA production in the CNS may be suggestive of a compensatory mechanism to GABA-A antagonism. In rat, a daily dieldrin injection of 0.25 LD50 significantly lowered levels of glutamic acid and glutamine but increased GABA, glycine, lysine, serine, alanine, and histidine (Ahmed et al., 1986). The present study suggests that GABA concentrations are potentially affected in a region specific manner by dieldrin. However, time-course studies in discrete brain regions are required to better evaluate the effects of dieldrin on GABA levels.
There were no significant cha nges in DA or DOPAC levels in male and female hypothalamus, telencephalon, and cerebellum. Similarly, mice injected with a single subcutaneous injection of dieldrin (40 and 80 mg/kg) after 7 days showed no significant changes in levels of striatal dopamine (DA) and DOPAC (Thiffault et al., 2001). However, in some studies, dieldrin has also been shown to deplete DA production in mammals in vivo. Mice injected with 30 mg/kg dieldrin showed an initial decrease in striatal DA after 6 h followed by an increase in DA above baseline after 24 and 72 h (Sava et al., 2007). Striatal DOPAC was decreased after 6 and 24 h and returned to baseline level at 72 h. In the same study, slow subcutaneous infusion of dieldrin at an estimated cumulative dose of 50 mg/kg into male mice resulted in a significant 20–30% increase in DA after two weeks but no change in DOPAC levels. In another study, male mice injected with 0.3, 1, or 3 mg/kg dieldrin every three days for 30 days exhibited a near 30% reduction in DOPAC in the striatum after treatment to 1 or 3 mg/kg dieldrin (Hatcher et al., 2007). It is plausible that our treatment was not of sufficient length to observe changes in the catecholamine pathway. Select biological processes that are affected by dieldrin, for example cell communication and kinase/phosphatase activity are potential early genomic responses that may correlate to later changes in neurotransmitter levels in the CNS, however, this must be further evaluated with additional studies.
4.2. Dieldrin induces the ubiquitin-proteasome pathway at the genomic level
Many induced transcripts were involved in the ubiquitin-proteasome pathway. The ubiquitin-proteasome pathway has many biological roles, including protein degradation that involves covalent attachment of ubiquitin molecules to targeted proteins and subsequent degradation b y the 26S proteasome complex. In addition to removing mis-folded proteins, this pathway is also involved in protein trafficking, cell signaling, and cell homeostasis. Dieldrin-mediated transcripts included ubiquitin-conjugating enzyme E2G 1 (ube2g1), ring finger protein 121 (rnf121) and ring finger protein 127 (rnf127), hect carboxyl terminus domain and RCC1-like domain 2 (herc2), and ubiquitin specific peptidase 14 (usp14) and 16 (usp16). Protein ubiquitination is required as a signal for 26S proteasome dependent protein degradation. There are three primary enzymes needed for the addition of ubiquitin to the targeted protein; E1 (ubiquitin activating enzyme), E2 (ubiquitin conjugating enzyme), and E3 (ubiquitin ligase). E3 ubiquitin ligases include various subtypes such as HECT type E3s, U-box type E3s, single RING-finger type E3s, and multi subunit RING-finger E3s that contain a range of protein binding motifs. Of interest, disrupted ubiquitin-mediated processes are closely associated with human neurological disorders, for example PD (Marín et al., 2004; Dawson, 2006). PD includes symptoms such as tremors, impaired fine motor dexterity, and changes in gait that are the result of the degeneration and loss of dopaminergic neurons in the substantia nigra. Parkin, for example, is a well known marker for PD and is an E3 ubiquitin ligase that binds to ubiquitin-conjugating enzymes (E2s) through its RING-IBR-RING motif (Takahashi et al., 2003). It is hypothesized that environmental exposure to pesticides may augment disruptions in the protein degradation pathway (Sun et al., 2007) and there is evidence that dieldrin is modulating this pathway in the LMB hypothalamus.
Pathway analysis also identified AD as a disease state associated with many of the dieldrin-mediated transcripts. There is evidence in mammals that the ubiquitin-proteasome pathway is also involved in many other human neurological disorders, including Alzheimer's disease (AD) (Hegde and Upadhya, 2007). AD is a result of senile plaques that form dense fibrous clumps deposited outside neurons due to abnormally folded beta-amyloid and tau proteins. Alpha-synuclein protein aggregates to form insoluble fibrils in amyloids that are associated with Lewy bodies that are present in patients with AD as well as PD. Sun et al. (2005) demonstrated in the rat mesencephalic dopaminergic cell line (N27) that dieldrin impaired ubiquitin-proteasome function additively with alpha-synuclein. GABA can play a role in the etiology of human neurological disorders and can be involved in the production of amyloids (Marcade et al., 2008). In addition, stimulation of GABA-A receptor in the mammalian CNS by muscimol has been shown to reduce amyloid beta protein-induced neurotoxicity (Lee et al., 2005). To our knowledge, it is not known whether antagonizing the GABA-A receptor (i.e. via dieldrin) increases neurotoxicity due to amyloid beta protein in the vertebrate CNS. It is important to note that this study merely points out similarities between dieldrin neurotoxicity and human neurological diseases and acknowledges that mechanisms leading to neurodegeneration are integrated and complex. Nevertheless, this study provides evidence that transcripts involved in the ubiquitin-proteasome pathway are preferentially induced by in vivo dieldrin treatment in the LMB hypothalamus.
4.3. Dieldrin alters genes involved in DNA repair
Functional enrichment and pathway analysis identified DNA repair mechanisms and transcripts involved in DNA damage and apoptosis as being affected by dieldrin. DNA repair mechanisms include base-excision and nucleotide excision repair processes. These findings suggest that the hypothalamus m ay be increasing protective mechanisms (e.g., maintaining cell homeostasis) while at the same time undergoing dieldrin-induced apoptotic processes. Neuronal cell death underlies human neurodegenerative diseases and is thought to be the result of accumulated oxidative stress (Mancuso et al., 2006; Zhou et al., 2008). In LMB, a number of isoforms for glutathione-S-transferase were induced by dieldrin (p<0.05), indicative of an oxidative stress response in the hypothalamus. Anantharam et al. (2007) performed microarray analysis on N27 cells exposed to 100 µM H2O2, a potent compound that generates reactive oxygen species (ROS), after 4 h and identified transcripts that had kinase/phosphatase activity and were Ras/Rab related GTPases. These processes are also involved in apoptotic and ubiquitin–proteasome pathways. In the present study, over represented molecular function GO categories included kinase activity and 3’,5’-cyclic-nucleotide phosphodiesterase activity (GO:0004114). Genes that code proteins with kinase activity included scyl3 protein (scyl3), cell cycle related kinase, p21-activated kinase 7 (pak7), deoxycytidine kinase 2 (dck2), phosphoinositide-3-kinase (pik3), and ras-related GTP-binding protein RAB10 (rab10). Therefore, there appears to be active signaling in the hypothalamus via kinases. Furthermore, in the hypothalamus of LMB, there may be an oxidative stress response as there are common gene families regulated by both H2O2 and dieldrin.
The transcriptomics data presented here are also consistent with mammalian neurophysiological data that demonstrates that dieldrin results in increased apoptosis in the CNS. There is both in vitro and in vivo evidence that dieldrin induces apoptotic pathways in neural tissues and cell types. For example, incubation of dopaminergic PC12 cells with 30–1000 µM dieldrin for 1 h induced ROS resulted in apoptotic cell death and increased DNA fragmentation (Kitazawa et al., 2001). Kanthasamy et al. (2008) exposed N27 cells to 30, 100 or 300 µM dieldrin for 15 min – 3 h and reported increased cytosolic cytochrome c (cytc) and caspase-3 (casp3) activity, both well-characterized cellular responses to stress. Dopaminergic cells treated with dieldrin also show decreased survival of approximately 40% in 48 h resulting from increased formation of ROS, chromatin condensation, DNA fragmentation, and apoptosis (Chun et al., 2001). In the study by Hatcher et al. (2007), male mice injected with 0.3, 1, or 3 mg/kg dieldrin every three days for 30 days showed increased oxidative stress in the striatum. This was based on data that the redox potential of glutathione was significantly elevated in the 3 mg/kg dieldrin-treated group over control animals and the total levels of glutathione were decreased by approximately 50%. In the LMB, steady state mRNA changes in transcripts such as programmed cell death 11 (pdcd11), bcl2-associated athanogene 2 (bag2), flap structure-specific endonuclease 1 (fen1), and fanconi anemia, complementation group E (fance) suggests that apoptotic pathways are also induced in the hypothalamus in response to dieldrin.
Functional enrichment analysis identified DNA repair as a biological process significantly affected by dieldrin. Pathway analysis also revealed that many of the regulated transcripts are genes involved in cell processes such as regeneration and DNA damage. Impaired ability to repair damaged DNA is thought to be a factor underlying neurodegenerative diseases including AD, PD, and amyotrophic lateral sclerosis (Martin, 2008). Regulated genes that were identified as being involved in DNA repair mechanisms included excision repair complementation group 1 and 8 (ercc1 and ercc8), apex nuclease 1 and 2 (apex1 and apex2), DNA cross-link repair, nei endonuclease VIII-like 3, and fanconi anemia - complementation group E and F (fance and fancf). Many of these enzymes function to repair oxidized, deaminated, or alkylated DNA bases through base excision, synthesis, and ligation. Dieldrin has been previously shown to induce DNA repair mechanisms in mammals. Male mice i.p. injected with either 6 or 30 mg/kg dieldrin and treated between 6–72 h showed significant increases in the activity of 8-oxoguanine DNA glycosylase, an enzyme that repairs oxidatively damaged guanosine nucleotides (Sava et al., 2007). In the LMB hypothalamus, there appears to be a dynamic genomic response that includes both processes of DNA damage and repair/regeneration mechanisms.
4.4. Evidence that dieldrin alters inflammation pathways in the brain
In addition to DNA damage and regeneration, pathway analysis supports the hypothesis that dieldrin alters the mRNA levels of genes involved in neuroinflammation. Inflammatory responses in the brain have also been hypothesized as a mechanism contributing to PD. Nagatsu and Sawada (2007) investigated postmortem brains and found that those with idiopathic PD showed evidence of a pro-inflammatory response. Individuals with idiopathic PD had increased levels of cytokines such as TNF-alpha, IL-1 beta, IL-2, IL-4, and IL-6. It is suggested in the study that the increase in cytokines may be part of the pathogenesis of PD and this may lead to accelerated apoptotic death of dopaminergic neurons. In support of an association between dieldrin and inflammation, mice injected with 50 µM dieldrin into air pouches for 6 or 12 h showed a significant increase in the number of total leukocytes 6 h post-injection, suggesting that dieldrin induces neutrophilic inflammation in vivo (Pelletier et al., 2001). In the same study, neutrophils incubated for 24 h with increasing concentrations of dieldrin exhibited a significant increase in IL-8 production from exposure to 50 µM dieldrin. In the present study, transcripts such as coagulation factor XI (f11), toll-interleukin-1 receptor interacting protein II (tlr2), alpha-2-macroglobulin (a2m), and small inducible cytokine subfamily E member 1 (scye1) were induced in the LMB hypothalamus, suggesting that an elevated inflammatory response may also underlie dieldrin neurotoxicity.
4.5. Dieldrin and genes involved in steroid signaling
There was some evidence that both ar and esr-beta b mRNA steady state levels were affected by dieldrin in the hypothalamus after acute injection. There is data suggesting that dieldrin also directly binds steroid hormone receptors, potentially affecting vertebrate reproduction. Dieldrin weakly activates human ER alpha recombinant protein (IC50 = 106 uM) as determined by inhibition binding experiments using tritium-labeled steroid ligands (Scippo et al., 2004). Moreover, dieldrin along with structurally related OCPs endrin, chlordane, aldrin, and endosulfan are considered weak retinoid × receptor (RAR) analogues and are able to activate Rarβ and Rarγ (Lemaire et al., 2005). In LMB, reproduction is impaired after dieldrin treatment. LMB fed pellets treated with dieldrin (0.4 and 0.81 µg/g) for 5 days per week for 120 days had altered gene transcript levels for estrogen receptor (esr) isoforms and androgen receptor (ar) and also showed disrupted steroid synthesis/metabolism (Garcia-Reyero et al., 2006). In the study, exposure of LMB to 0.4 µg/g dieldrin decreased esr-beta b and esr-beta a mRNA in the gonads of both the sexes, decreased esr-alpha and ar mRNA in males, and increased ar mRNA in females. Sex steroid production was also affected in dieldrin-fed LMB, as the production of the major sex hormones 11-ketotestosterone and 17β-estradiol (E2) was decreased in both males and females (Garcia-Reyero et al., 2006). Dieldrin appears to induce transcriptional responses in the LMB gonad which is also associated with altered steroid production.
Noteworthy is that the hypothalamic-pituitary-gonad (HPG) reproductive axis is tightly regulated by steroid feedback mechanisms and it is not known whether reproductive impairment in LMB is due to neurotoxic effects of dieldrin in the CNS or because of altered steroid receptor signaling along the HPG axis. GABA, in addition to other amino acid neurotransmitters, is a key stimulator of luteinizing hormone release from the pituitary in teleost fish (Trudeau et al., 2000). Therefore, GABA antagonism by dieldrin in the CNS may potentially alter steroid receptor expression in the brain and affect GABA-stimulated LH release, resulting in adverse effects in the gonads. Both hypotheses are plausible, and impaired reproduction observed in teleosts by dieldrin may be the result of both dieldrin-mediated neurotoxicity and altered steroid receptor signaling due to ER binding.
4.5. Conclusions
Using genomics and multiple bioinformatics approaches, this study identified dieldrin-mediated genes involved in the ubiquitin-proteasome pathway, DNA damage and repair, apoptosis, and inflammation. There is increasing epidemiological evidence suggesting that exposure to pesticides in the environment may contribute to increased risk of neurodegenerative diseases (Di Monte et al., 2002; Di Monte, 2003; Sun et al., 2007; Drechsel and Patel, 2008). Kanthasamy et al. (2005) reviewed data from the literature about the fate, distribution, localization, and exposure to dieldrin and provided a comprehensive summary on epidemiological evidence supporting the association between dieldrin and PD. In humans, significantly higher concentrations of dieldrin have been detected in the brains of patients that had advanced stages of PD compared to individuals that did not have PD (Fleming, et al., 1994; Corrigan et al., 2000). We caution that the data presented here is not causative for human neurodegenerative diseases and does not suggest fish exhibit neurodegenerative disease symptoms that parallel human disease. There are complex signaling cascades underlying the progression of neurodegeneration and it is plausible that similar molecular cascades presented here between dieldrin neurotoxicity and neurodegeneration may be common responses to ongoing injury in the CNS. This is supported by the data suggesting DNA repair, DNA damage, and regeneration mechanisms are occurring in the hypothalamus.
Although genetics, age, lifestyle, and sex are strong determinants for the onset of neurodegenerative diseases, each of these contributing factors may be exacerbated with chemical exposure over time. It is not well understood how environmental exposures interfere with GABA signaling and whether or not this is associated with increased risks to neurotoxicity and neurodegeneration. In addition to reducing dopamine (DA) levels, GABA does appear to play a role in disease models of PD because of interactions between GABA and the DA system (Tepper and Lee, 2007), and therefore pesticides that impact the GABA system may have implications for human health. Additional efforts in human health should be allocated to the GABAergic system and pesticides that effect GABA signaling, as GABA receptor modulation may result in cellular responses at the genomic level that are associated with neurodegeneration.
Supplementary Material
Acknowledgements
The authors gratefully acknowledge K. Childress for handling and rearing of LMB at the Center for Environmental and Human Toxicology Aquatic Facilities. We thank G. Ostrow for assistance with the scanning of microarrays and R.J. Griffitt for assistance with microarray data analysis. We thank MC Croteau for helpful comments on the manuscript. This research was funded by a National Institutes of Health Pathway to Independence Award granted to C.J.M (K99 ES016767-01A1) and by the Superfund Basic Research Program from the National Institute of Environmental Health Sciences to N.D. and D.B. (RO1 ES015449).
Abbreviations for pathway studios
Abbreviation nomenclature and rules follow that of mammalian proteins.
- APOE
Apolipoprotein E
- APEX 1
Apurinic/apyrimidinic endonuclease 1
- APEX 2
Apurinic/apyrimidinic endonuclease 2
- BIRC2
baculoviral IAP repeat-containing 2
- CREB1
cAMP responsive element binding protein 1
- C1S
complement component 1, s subcomponent
- C1QBP
complement component 1, q subcomponent binding protein
- Mud1
complement component factor H
- CCNB1
cyclin B1
- DCK
deoxycytidine kinase
- SNM1
DNA cross-link repair 1A, PSO2 homolog
- ESR2
estrogen receptor 2 beta
- ERCC1
Excision repair cross-complementing rodent repair deficiency, complementation group 1
- FANCE
Fanconi anemia, complementation group E
- Fen1
Flap structure-specific endonuclease 1
- GALK1
galactokinase 1
- GFER
growth factor, erv1 homolog (S. cerevisiae)
- GTF2H3
General transcription factor IIH, polypeptide 3
- GIYD2; Gtf2h4
General transcription factor II H, polypeptide 4
- HSD17B12
GIY-YIG domain containing 2, hydroxysteroid (17-beta) dehydrogenase 12
- INS2
insulin 2
- JAG
jagged 1
- mitotin (S. cerevisiae) MCM2
MCM2 minichromosome maintenance deficient 2
- med28
mediator of RNA polymerase II transcription, subunit 28 homolog (yeast)
- MAP2K7
mitogen activated protein kinase kinase 7
- NARG1
NMDA receptor-regulated gene 1 (predicted)
- NEIL1
phenylethanolamine-N-methyltransferase, Nei endonuclease VIII-like 1 (E. coli)
- NTHL1; PNMT
Nth (endonuclease III)-like 1
- PLG
plasminogen
- PDCD8
programmed cell death 8
- POLQ
Polymerase (DNA directed), theta
- PCNA
Proliferating cell nuclear antigen
- TDP1
Tyrosyl-DNA phosphodiesterase 1
- PRC1
protein regulator of cytokinesis 1 (predicted)
- PC
pyruvate carboxylase
- RAD23B
RAD23b homolog (S. cerevisiae)
- RACGAP1
Rac GTPase-activating protein 1 (predicted)
- member 1SCARB1
scavenger receptor class B
- TOPBP1
similar to mKIAA0259 protein (predicted)
- MYLK
similar to neuronal myosin light chain kinase 1
- PELP1
similar to Proline, glutamic acid and leucine rich protein 1
- FES
similar to tyrosine kinase Fps/Fes (predicted)
- SLC4A1
solute carrier family 4, member 1
- SLC12A3
solute carrier family 12, member 3
- VIL1
villin 1 (predicted)
- AND-1
WD repeat and HMG-box DNA binding protein 1 (predicted)
- ZFP36
zinc finger protein 36
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest: There are no conflicts of interest to disclose.
References
- Ahmed NA, Rawi SM, el-Behary MH. Effect of dieldrin injection on the level of certain amino acids and some enzymes in rat brain. Comp Biochem Physiol C. 1986;85:437–442. doi: 10.1016/0742-8413(86)90222-7. [DOI] [PubMed] [Google Scholar]
- Al-Shahrour F, Mínguez P, Tárraga J, Montaner D, Alloza E, Vaquerizas JM, Conde L, Blaschke C, Vera J, Dopazo J. BABELOMICS: a systems biology perspective in the functional annotation of genome-scale experiments. Nucleic Acids Res. 2006;34:W472–W476. doi: 10.1093/nar/gkl172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Al-Shahrour F, Minguez P, Vaquerizas JM, Conde L, Dopazo J. Babelomics: a suite of web-tools for functional annotation and analysis of group of genes in high-throughput experiments. Nucleic Acids Res. 2005;33:W460–W464. doi: 10.1093/nar/gki456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anantharam V, Lehrmann E, Kanthasamy A, Yang Y, Banerjee P, Becker KG, Freed WJ, Kanthasamy AG. Microarray analysis of oxidative stress regulated genes in mesencephalic dopaminergic neuronal cells: relevance to oxidative damage in Parkinson's disease. Neurochem Int. 2007;50:834–847. doi: 10.1016/j.neuint.2007.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bloomquist JR. Intrinsic lethality of chloride-channel-directed insecticides and convulsants in mammals. Toxicol Lett. 1992;60:289–298. doi: 10.1016/0378-4274(92)90287-t. [DOI] [PubMed] [Google Scholar]
- Carr RL, Couch TA, Liu J, Coats JR, Chambers JE. The interaction of chlorinated alicyclic insecticides with brain GABA(A) receptors in channel catfish (Ictalurus punctatus) J Toxicol Environ Health A. 1999;56:543–553. doi: 10.1080/00984109909350176. [DOI] [PubMed] [Google Scholar]
- Chun HS, Gibson GE, DeGiorgio LA, Zhang H, Kidd VJ, Son JH. Dopaminergic cell death induced by MPP(+), oxidant and specific neurotoxicants shares the common molecular mechanism. J Neurochem. 2001;76:1010–1021. doi: 10.1046/j.1471-4159.2001.00096.x. [DOI] [PubMed] [Google Scholar]
- Conesa A, Götz S, García-Gómez JM, Terol J, Talón M, Robles M. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21:3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
- Corrigan FM, Wienburg CL, Shore RF, Daniel SE, Mann D. Organochlorine insecticides in substantia nigra in Parkinson's disease. J Toxicol Environ Health A. 2000;59:229–234. doi: 10.1080/009841000156907. [DOI] [PubMed] [Google Scholar]
- Dawson TM. Parkin and defective ubiquitination in Parkinson's disease. J Neural Transm Suppl. 2006;70:209–213. doi: 10.1007/978-3-211-45295-0_32. [DOI] [PubMed] [Google Scholar]
- Di Monte DA. The environment and Parkinson's disease: is the nigrostriatal system preferentially targeted by neurotoxins? Lancet Neurol. 2003;2:531–538. doi: 10.1016/s1474-4422(03)00501-5. [DOI] [PubMed] [Google Scholar]
- Di Monte DA, Lavasani M, Manning-Bog AB. Environmental factors in Parkinson's disease. Neurotoxicology. 2002;23:487–502. doi: 10.1016/s0161-813x(02)00099-2. [DOI] [PubMed] [Google Scholar]
- Drechsel DA, Patel M. Role of reactive oxygen species in the neurotoxicity of environmental agents implicated in Parkinson's disease. Free Radic Biol Med. 2008;44:1873–1886. doi: 10.1016/j.freeradbiomed.2008.02.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95:14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fleming L, Mann JB, Bean J, Briggle T, Sanchez-Ramos JR. Parkinson's disease and brain levels of organochlorine pesticides. Ann Neurol. 1994;36:100–103. doi: 10.1002/ana.410360119. [DOI] [PubMed] [Google Scholar]
- Gant DB, Eldefrawi ME, Eldefrawi AT. Cyclodiene insecticides inhibit GABAA receptor-regulated chloride transport. Toxicol Appl Pharmacol. 1987;88:313–321. doi: 10.1016/0041-008x(87)90206-7. [DOI] [PubMed] [Google Scholar]
- Garcia-Reyero N, Barber DS, Gross TS, Johnson KG, Sepúlveda MS, Szabo NJ, Denslow ND. Dietary exposure of largemouth bass to OCPs changes expression of genes important for reproduction. Aquat Toxicol. 2006;78:358–369. doi: 10.1016/j.aquatox.2006.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garcia-Reyero N, Griffitt RJ, Liu L, Kroll KJ, Farmerie WG, Barber DS, Denslow ND. Construction of a robust microarray from a non-model species largemouth bass, Micropterus salmoides (Lacepede), using pyrosequencing technology. J Fish Biol. 2008;72:2354–2376. doi: 10.1111/j.1095-8649.2008.01904.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gesto M, Tintos A, Soengas JL, Míguez JM. Effects of acute and prolonged naphthalene exposure on brain monoaminergic neutotransmitters in rainbow trout (Oncorhynchus mykiss) Comp Biochem Physiol C. 2006;144:173–183. doi: 10.1016/j.cbpc.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Hatcher JM, Richardson JR, Guillot TS, McCormack AL, Di Monte DA, Jones DP, Pennell KD, Miller GW. Dieldrin exposure induces oxidative damage in the mouse nigrostriatal dopamine system. Exp Neurol. 2007;204:619–630. doi: 10.1016/j.expneurol.2006.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hegde AN, Upadhya SC. The ubiquitin-proteasome pathway in health and disease of the nervous system. Trends Neurosci. 2007;30:587–595. doi: 10.1016/j.tins.2007.08.005. [DOI] [PubMed] [Google Scholar]
- Ikeda T, Nagata K, Shono T, Narahashi T. Dieldrin and picrotoxinin modulation of GABA(A) receptor single channels. Neuroreport. 1998;9:3189–3195. [PubMed] [Google Scholar]
- Jorgenson JL. Aldrin and dieldrin: a review of research on their production, environmental deposition and fate, bioaccumulation, toxicology, and epidemiology in the United States. Environ Health Perspect. 2001;109:113–139. doi: 10.1289/ehp.01109s1113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanthasamy AG, Kitazawa M, Kanthasamy A, Anantharam V. Dieldrin-induced neurotoxicity: relevance to Parkinson's disease pathogenesis. Neurotoxicology. 2005;26:701–719. doi: 10.1016/j.neuro.2004.07.010. [DOI] [PubMed] [Google Scholar]
- Kanthasamy AG, Kitazawa M, Yang Y, Anantharam V, Kanthasamy A. Environmental neurotoxin dieldrin induces apoptosis via caspase-3-dependent proteolytic activation of protein kinase C delta (PKCdelta): Implications for neurodegeneration in Parkinson's disease. Mol Brain. 2008;1:12. doi: 10.1186/1756-6606-1-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitazawa M, Anantharam V, Kanthasamy AG. Dieldrin-induced oxidative stress and neurochemical changes contribute to apoptopic cell death in dopaminergic cells. Free Radic Biol Med. 2001;31:1473–1485. doi: 10.1016/s0891-5849(01)00726-2. [DOI] [PubMed] [Google Scholar]
- Kitazawa M, Anantharam V, Kanthasamy A, Kanthasamy AG. Dieldrin promotes proteolytic cleavage of poly(ADP-ribose) polymerase and apoptosis in dopaminergic cells: protective effect of mitochondrial anti-apoptotic protein Bcl-2. Neurotoxicology. 2004;25:589–598. doi: 10.1016/j.neuro.2003.09.014. [DOI] [PubMed] [Google Scholar]
- Lamai SL, Warner GF, Walker CH. Effects of dieldrin on life stages of the African catfish, Clarias gariepinus (Burchell) Ecotoxicol Environ Saf. 1999;42:22–29. doi: 10.1006/eesa.1998.1723. [DOI] [PubMed] [Google Scholar]
- Lemaire G, Balaguer P, Michel S, Rahmani R. Activation of retinoic acid receptor-dependent transcription by organochlorine pesticides. Toxicol Appl Pharmacol. 2005;202:38–49. doi: 10.1016/j.taap.2004.06.004. [DOI] [PubMed] [Google Scholar]
- Lee BY, Ban JY, Seong YH. Chronic stimulation of GABAA receptor with muscimol reduces amyloid beta protein (25–35)-induced neurotoxicity in cultured rat cortical cells. Neurosci Res. 2005;52:347–356. doi: 10.1016/j.neures.2005.04.008. [DOI] [PubMed] [Google Scholar]
- Marcade M, Bourdin J, Loiseau N, Peillon H, Rayer A, Drouin D, Schweighoffer F, Désiré L. Etazolate, a neuroprotective drug linking GABA(A) receptor pharmacology to amyloid precursor protein processing. J Neurochem. 2008;106:392–404. doi: 10.1111/j.1471-4159.2008.05396.x. [DOI] [PubMed] [Google Scholar]
- Marín I, Lucas JI, Gradilla AC, Ferrús A. Parkin and relatives: the RBR family of ubiquitin ligases. Physiol Genomics. 2004;17:253–263. doi: 10.1152/physiolgenomics.00226.2003. [DOI] [PubMed] [Google Scholar]
- Martin LJ. DNA damage and repair: relevance to mechanisms of neurodegeneration. J Neuropathol Exp Neurol. 2008;67:377–387. doi: 10.1097/NEN.0b013e31816ff780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martyniuk CJ, Xiong H, Crump K, Chiu S, Sardana R, Nadler A, Gerrie ER, Xia X, Trudeau VL. Gene expression profiling in the neuroendocrine brain of male goldfish (Carassius auratus) exposed to 17alpha-ethinylestradiol. Physiol Genomics. 2006;27:328–336. doi: 10.1152/physiolgenomics.00090.2006. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Gerrie ER, Popesku JT, Ekker M, Trudeau VL. Microarray analysis in the zebrafish (Danio rerio) liver and telencephalon after exposure to low concentration of 17alpha-ethinylestradiol. Aquat Toxicol. 2007;84:38–49. doi: 10.1016/j.aquatox.2007.05.012. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Awad R, Hurley R, Finger TE, Trudeau VL. Glutamic acid decarboxylase 65, 67, and GABA-transaminase mRNA expression and total enzyme activity in the goldfish (Carassius auratus) brain. Brain Res. 2007;1147:154–166. doi: 10.1016/j.brainres.2007.02.010. [DOI] [PubMed] [Google Scholar]
- Martyniuk CJ, Kroll KJ, Porak WF, Steward C, Grier HJ, Denslow ND. Seasonal relationship between gonadotropin, growth hormone, and estrogen receptor mRNA expression in the pituitary gland of largemouth bass. Gen Comp Endocrinol. 2009;163:306–317. doi: 10.1016/j.ygcen.2009.04.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso M, Coppede F, Migliore L, Siciliano G, Murri L. Mitochondrial dysfunction, oxidative stress and neurodegeneration. J Alzheimers Dis. 2006;10:59–73. doi: 10.3233/jad-2006-10110. [DOI] [PubMed] [Google Scholar]
- Nagatsu T, Sawada M. Biochemistry of postmortem brains in Parkinson's disease: historical overview and future prospects. J Neural Transm Suppl. 2007;72:113–120. doi: 10.1007/978-3-211-73574-9_14. [DOI] [PubMed] [Google Scholar]
- Nikitin A, Egorov S, Daraselia N, Mazo I. Pathway studio—the analysis and navigation of molecular networks. Bioinformatics. 2003;19:2155–2157. doi: 10.1093/bioinformatics/btg290. [DOI] [PubMed] [Google Scholar]
- Pedrajas JR, Peinado J, López-Barea J. Oxidative stress in fish exposed to model xenobiotics. Oxidatively modified forms of Cu,Zn-superoxide dismutase as potential biomarkers. Chem Biol Interact. 1995;98:267–282. doi: 10.1016/0009-2797(95)03651-2. [DOI] [PubMed] [Google Scholar]
- Peinado JM, McManus KT, Myers RD. Rapid method for micro-analysis of endogenous amino acid neurotransmitters in brain perfusates in the rat by isocratic HPLC-EC. J Neurosci Methods. 1986;18:269–276. doi: 10.1016/0165-0270(86)90013-0. [DOI] [PubMed] [Google Scholar]
- Pelletier M, Roberge CJ, Gauthier M, Vandal K, Tessier PA, Girard D. Activation of human neutrophils in vitro and dieldrin-induced neutrophilic inflammation in vivo. J Leukoc Biol. 2001;70:367–373. [PubMed] [Google Scholar]
- Rozen S, Skaletsky HJ. Primer3 on the W WW for general users and for biologist programmers. In: Krawetz S, Misener S, editors. Bioinformatics Methods and Protocols: Methods in Molecular Biology. Totowa, NJ: Humana Press; 2000. pp. 365–386. [DOI] [PubMed] [Google Scholar]
- Sabo-Attwood T, Kroll KJ, Denslow ND. Differential expression of largemouth bass (Micropterus salmoides) estrogen receptor isotypes alpha, beta, and gamma by estradiol. Mol Cell Endocrinol. 2004;218:107–118. doi: 10.1016/j.mce.2003.12.007. [DOI] [PubMed] [Google Scholar]
- Saldanha AJ. Java Treeview – extensible visualization of microarray data. Bioinformatics. 2004;20:3246–3248. doi: 10.1093/bioinformatics/bth349. [DOI] [PubMed] [Google Scholar]
- Satyanarayan S, Ramakant, Satyanarayan A. Bioaccumulation studies of organochlorinated pesticides in tissues of Cyprinus carpio. J Environ Sci Health B. 2005;40:397–412. doi: 10.1081/pfc-200047572. [DOI] [PubMed] [Google Scholar]
- Sava V, Velasquez A, Song S, Sanchez-Ramos J. Dieldrin elicits a widespread DNA repair and antioxidative response in mouse brain. J Biochem Mol Toxicol. 2007;21:125–135. doi: 10.1002/jbt.20165. [DOI] [PubMed] [Google Scholar]
- Scippo ML, Argiris C, Van De Weerdt C, Muller M, Willemsen P, Martial J, Maghuin-Rogister G. Recombinant human estrogen, androgen and progesterone receptors for detection of potential endocrine disruptors. Anal Bioanal Chem. 2004;378:664–669. doi: 10.1007/s00216-003-2251-0. [DOI] [PubMed] [Google Scholar]
- Sun F, Anantharam V, Latchoumycandane C, Kanthasamy A, Kanthasamy AG. Dieldrin induces ubiquitin-proteasome dysfunction in alpha-synuclein overexpressing dopaminergic neuronal cells and enhances susceptibility to apoptotic cell death. J Pharmacol Exp Ther. 2005;315:69–79. doi: 10.1124/jpet.105.084632. [DOI] [PubMed] [Google Scholar]
- Sun F, Kanthasamy A, Anantharam V, Kanthasamy AG. Environmental neurotoxic chemicals-induced ubiquitin proteasome system dysfunction in the pathogenesis and progression of Parkinson's disease. Pharmacol Ther. 2007;114:327–344. doi: 10.1016/j.pharmthera.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Takahashi R, Imai Y, Hattori N, Mizuno Y. Parkin and endoplasmic reticulum stress. Ann N Y Acad Sci. 2003;991:101–106. doi: 10.1111/j.1749-6632.2003.tb07467.x. [DOI] [PubMed] [Google Scholar]
- Tepper JM, Lee CR. GABAergic control of substantia nigra dopaminergic neurons. Prog Brain Res. 2007;160:189–208. doi: 10.1016/S0079-6123(06)60011-3. [DOI] [PubMed] [Google Scholar]
- Thiffault C, Langston WJ, Di Monte DA. Acute exposure to organochlorine pesticides does not affect striatal dopamine in mice. Neurotox Res. 2001;3:537–343. doi: 10.1007/BF03033209. [DOI] [PubMed] [Google Scholar]
- Ton C, Lin Y, Willett C. Zebrafish as a model for developmental neurotoxicity testing. Birth Defects Res A Clin Mol Teratol. 2006;76:553–567. doi: 10.1002/bdra.20281. [DOI] [PubMed] [Google Scholar]
- Trudeau VL, Spanswick D, Fraser EJ, Larivière K, Crump D, Chiu S, MacMillan M, Schulz RW. The role of amino acid neurotransmitters in the regulation of pituitary gonadotropin release in fish. Biochem Cell Biol. 2000;78:241–259. [PubMed] [Google Scholar]
- Wang XF, Li S, Chou AP, Bronstein JM. Inhibitory effects of pesticides on proteasome activity: implication in Parkinson's disease. Neurobiol Dis. 2006;23:198–205. doi: 10.1016/j.nbd.2006.02.012. [DOI] [PubMed] [Google Scholar]
- Wu YP, Mizukami H, Matsuda J, Saito Y, Proia RL, Suzuki K. Apoptosis accompanied by up-regulation of TNF-alpha death pathway genes in the brain of Niemann-Pick type C disease. Mol Genet Metab. 2005;84:9–17. doi: 10.1016/j.ymgme.2004.08.017. [DOI] [PubMed] [Google Scholar]
- Vetillard A, Benanni S, Saligaut C, Jego P, Bailhache T. Localization of tyrosine hydroxylase and its messenger RNA in the brain of rainbow trout by immunocytochemistry and in situ hybridization. J Comp Neurol. 2002;449:374–389. doi: 10.1002/cne.10296. [DOI] [PubMed] [Google Scholar]
- Zhou C, Huang Y, Przedborski S. Oxidative stress in Parkinson's disease: a mechanism of pathogenic and therapeutic significance. Ann N Y Acad Sci. 2008;1147:93–104. doi: 10.1196/annals.1427.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.