Abstract
Bacillus cereus ATCC 14579 converts the C-terminal 14 residues of a 52-mer prepeptide into a related set of eight variants of the thiocillin subclass of thiazolyl peptide antibiotics by a cascade of posttranslational modifications that alter 13 of those 14 residues. We have introduced prepeptide gene variants into a knockout strain to conduct an alanine scan of all 14 progenitor residues, as well as a serine scan of the six cysteine residues that are converted to thiazoles in the mature natural product. No mature scaffolds were detected for the S1A and S10A mutants, consistent with their roles as the source of the pyridine core. In both the alanine and serine scans, only one substitution mutant failed to produce a mature scaffold: cysteine 11. Cysteine to serine mutants gave mixture of dehydrations, aromatizations, and unaltered alcohol side chains depending on position. Overall, substitutions that altered the trithiazolylpyridine core or reduced the conformational rigidity of the 26-membered macrocyclic loop led to loss of antibiotic activity. In total, 21 peptide mutants were cultured, from which production of 107 compounds was observed and 94 compounds, representing 17 structural mutants were assayed for antibiotic activity. High-resolution NMR solution structures were determined for one mutant and one wild type compound. These structures demonstrate that the tight conformational rigidity of the natural product is severely disrupted by loss of even a single heterocycle, perhaps accounting for the attendant loss of activity in such mutants.
Introduction
Micrococcin P1 (MP1), the founding member of a class of >80 peptide antibiotics, known as the thiazolyl peptides was discovered in 1948 and shown 2 decades later to inhibit the translocation step of ribosomal protein synthesis. MP1 thus exhibits potent activity against methicillin resistant Staphylococcus aureus (MRSA) and other gram-positive bacteria.1–4 More recently, crystal structures have helped to distinguish MP1 from other thiazolyl peptides by characterization of its unique binding mode;5 completion of its first total synthesis has further awakened interest in the medicinal chemistry of MP1 in particular.6,7
The naturally occurring thiazolyl peptides can be closely subdivided into five groups based on the central thiazolylpyridine core, the macrocyclic loop size, and other structural features.1 Representative compounds are illustrated in figure 1.4,8–28 The 26-membered macrocycles, MP1, thiostrepton, and nosiheptide have all been shown to bind directly to the large ribosomal subunit in the region that interacts with elongation factor G.5,29 In contrast, GE-2270A, equipped with a larger, 29-membered ring, binds elongation factor Tu (EF-Tu), preventing formation of the ternary complex and impairing delivery of an incoming amino acyl tRNA.30–32 Berninamycin contains a highly oxygenated 35-membered ring around a thiazolyl-oxazolylpyridine core and reportedly acts on the 23S rRNA in a manner similar to MP1 and Nosiheptide.33
Figure 1.
Examples of thiazolyl peptide antibiotics: Micrococcin P1 (26-member macrocycle), Nosiheptide (26-member macrocycle + 13-member bridge), Thiostrepton (26-member macrocycle + 27-member B-ring), GE2270A (29-member macrocycle), and Berninamycin A (35-member macrocycle).
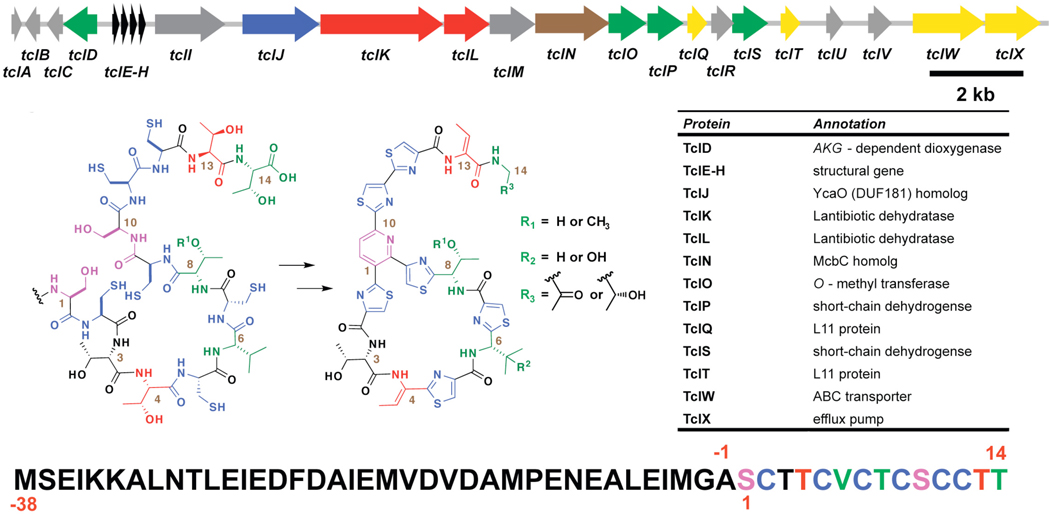
There has been much consideration as to whether these highly modified structures are assembled post-translationally or, perhaps, nonribosomally.34–39 However, recent reports have disclosed that GE2270A, thiomuracin,32 nosiheptide,40 thiostrepton,41,42 and the thiocillins (of which MP1 is the founding member),42,43 are all derived from 50–60 residue prepeptides, synthesized on the ribosome. The sequences of the mature products are derived from the C-termini of these microbial prepeptides. Thus, in the thiocillin producer, Bacillus cereus ATCC 14579, four copies of the structural gene, tclE-H, encode a 52-residue peptide with a 38 amino acid leader sequence, which is cleaved during posttranslational maturation of the final 14 residues (numbered as serine 1 through threonine 14) into the natural product (figure 2). The maturation pathway is mediated by eight to nine adjacently encoded enzymes, including two distinct lantibiotic dehydratases,38,39,44–46 TclK and L, a cyclodehydratase and associated desaturase, similar to heterocyclizing enzymes in the Microcin B17 synthase complex,36,47–49 TclJ and N, respectively, and a putative aza-Diels-Alderase, responsible for setting the central pyridine from two serines present in the prepeptide.50–53 TclQ and TclT encode copies of ribosomal protein L11; these proteins likely affect host resistance by mimicking natural substrate L11 in binding and inactivating the completed natural product until it can be exported, either by the ABC transporter, tclW or the efflux pump tclX. There are a number of tailoring enzymes also present in the gene cluster that affect three seemingly stochastic sets of modifications at residues 6, 8, and 14: an iron-dependent dioxygenase, which may hydroxylate valine 6, an O-methyl transferase, which can methylate the side chain of threonine 8, and two short-chain dehydrogenases, which likely affect β-oxidation/decarboxylation and subsequent reduction at threonine 14. Cumulatively, in B. cereus, these last three modifications yield eight distinct “thiocillin” compounds, including MP143.
Figure 2.
Gene cluster from B. cereus ATCC 14579 responsible for posttranslational biosynthesis of the thiocillins from a 14 residue prepeptide with a 38 residue leader sequence43.
We recently reported preparation of a double-crossover knockout of the tclE-H structural gene in B. cereus strain 14579.54 In this system we were able to introduce plasmid-based variants of the structural gene via Campbell integration, thereby allowing the bacterium to produce new Thiocillin-like antibiotics. We herein describe the application of this gene replacement strategy to a complete alanine scan of the fourteen residues (serine 1-threonine 14; figure 2) that give rise to the mature antibiotic. Additionally, the presence in the gene cluster of homologs of both the lantibiotic dehydratases and the Microcin B17-like cyclodehydratases led us to include a serine scan at the positions of the 6 cysteines that yield the thiazole rings. These scans allow assessment of the biosynthetic assembly line from two perspectives: first, a biosynthetic assessment of the posttranslational cascade’s enzymatic machinery, by demonstrating which positions permit alterations to the amino acid substituent while still allowing completion of the antibiotic biosynthesis. Second, they offer an assessment of retained biological activity, by providing assayable quantities of modified scaffolds for determination of any changes in their antibiotic profile. Although total synthesis of thiazolyl peptides has been rapidly advancing, there have been no reports of structure activity relationships of either the tetracyclic core or the rigidifying elements in the macrocycles, the most characteristic components of this class of antibiotics.7,55–61 Given the complexity of the substrates and the intricacy of the tailoring events (up to fourteen steps in thiocillin maturation), this prepetide replacement strategy appears a practical and rapid means to access a large amount of biosynthetic and biochemical intelligence. Moreover, there are many insights that can be gained only from work with a fully operable biosynthetic system, capable of producing mature compounds.
Results and Discussion
Methods for generation and characterization of mutant antibiotics
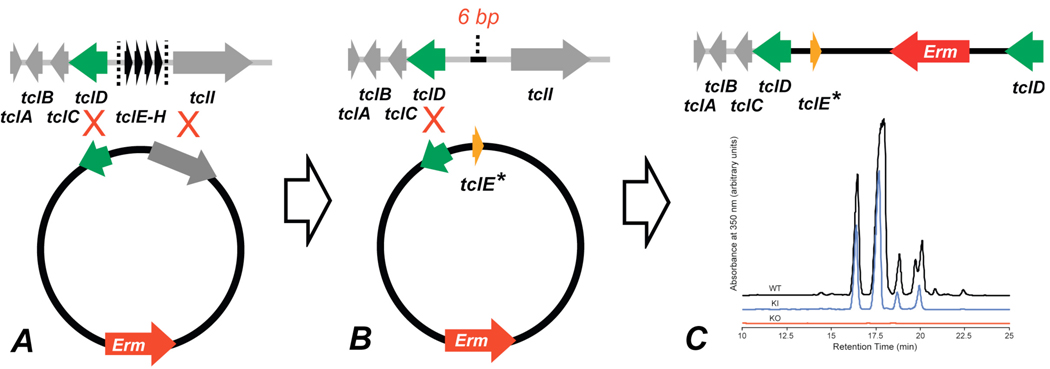
Clean removal of the four copies of the structural gene, tclE-H, was effected by means of homologous recombination with a plasmid baring two areas of homology (Figure 4A).54 Upon excision only a six base pair scar remained. Additionally, isolates from fermentations of the knockout strain were devoid of compound of any kind within the typical range of elution of the natural products and within the limits of detection of our instrumentation. In the work described below, this knockout strain was combined with knock-in plasmids containing a single area of homology in the tclD gene and a single variant copy of the 156bp structural gene, tclE, as well as an erythromycin resistance cassette. New variants were fermented for 72 hrs and compounds isolated by methanol extraction from the pellet (see Supporting Information). Compounds were further purified by reverse-phase HPLC (RP-HPLC; Figure 4C).
Figure 4.
Schematic representation of knockout/knock-in strategy. (A) Excision of four structural gene copies via homologous recombination with a plasmid containing two areas of homology. (B) Knockout strain (exhibiting a 6 base pair scar from the initial knockout) is combined with a second plasmid baring a single area of homology and a single variant copy of the structural gene TclE. (C) Product variant strain is cultured and thiocillins isolated (pictured are comparative HPLC traces of isolates from the wild-type, the knockout, and a wild-type rescue).
From the stochastic hydroxylation at valine 6, O-methylation at threonine 8, and ketone reduction at residue 14, up to eight variants can be generated from the wildtype 52-mer prepropeptide43. Similar combinations of scaffold variants were anticipated in the mutants and multiple products are the norm from such fermentations as seen in figure 6. For the wild type scaffold, we validated that four of the thiocillin variants (Micrococcin P1, Micrococcin P2, YM-266183, and Thiocillin I) had equivalent submicromolar antibiotic potency against B subtilis and Staph aureus strains, suggesting the two fold variants at residues 6, 8, and 14 are not consequential for ribosome binding. For many initial antibiotic assays of the mutants we therefore used pooled fractions. Where further purification proved necessary for determination of specific component antibiotic activity, this could be accomplished with extended gradients on RP-HPLC.
Figure 6.
Schematic representation of thiocillin SAR. Substitution of residues backed by red panels result in loss of antibiotic production, while those backed in yellow sustain production but lose activity. Substitutions at positions illustrated in green allow production of active antibiotic analogues.
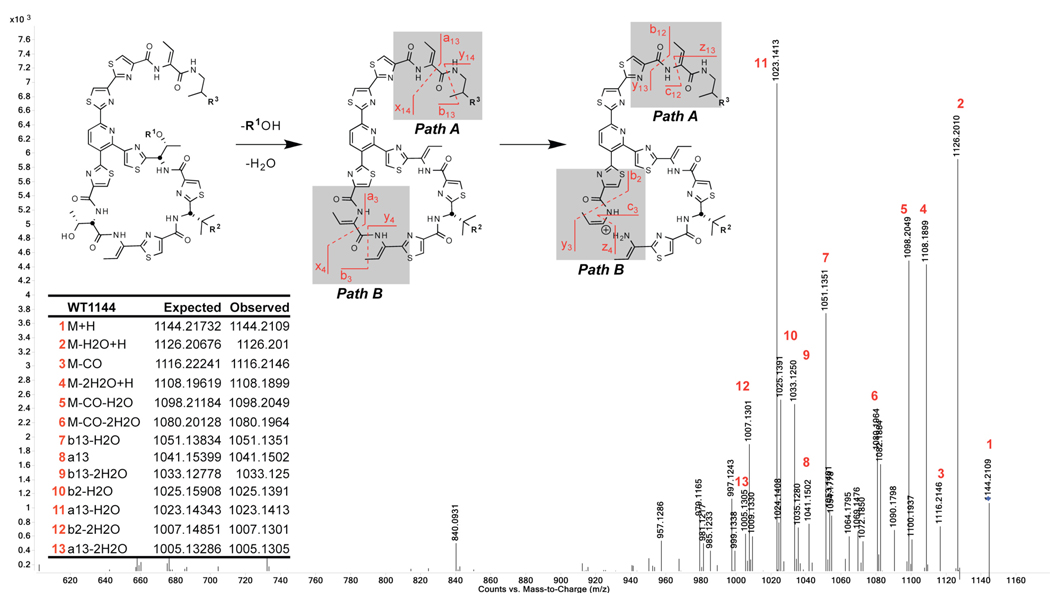
Additionally, due to the complex mixtures of products obtained for most mutant fermentations, the primary means of compound characterization (beyond the variant structural gene sequences) were hi-res LC/MS and MS/MS on an Accurate Mass LC-QTOF 6520 (Agilent Technologies). Essential to these means of characterization were the distinct absorbance of the conjugated trithiazolylpyridine core at 350 nm and the unique fragmentation pattern at intermediate collision energies (illustrated for Micrococcin P2 in Figure 5). Due to the highly modified, constrained secondary structure of the thiocillins, ions from two fragmentation pathways can be reliably discerned in MS/MS spectra: A) at the C-terminus and B) at the internal residues, Thr3 and Thr4. These patterns were very consistent and functioned for almost all mutants examined.
Figure 5.
Common MS/MS fragmentation pathways observed for Thiocillin variants: (A) C-terminus and (B) between threonines 3 and 4 of the macrocycle subunit. Exempletive mass spectrum of Micrococcin P2 with labeled fragment peaks.
Complete alanine scan
As described above, site-specific mutants were generated via Campbell integration of mutant tclE plasmids into B. cereus tclΔE-H. In our first round of transformations, previously reported54, we targeted conservative changes at residues 3,4,6, and 8 in the macrocycle, as well as 13, located on the tail; that set of gene replacements contained alanine substitutions at residues 3, 6, 8, and 13 only. The work reported here began by supplementing this partial scan with substitutions at residues 4 and 14. These six residues are not involved in setting the trithiazolylpyridine core in the mature scaffold, and therefore, alanine substitutions were expected to minimally disrupt biosynthesis. HPLC traces of the UV absorption (350 nm) of methanolic extracts are illustrated in figure 7 and minimum inhibitory concentrations (MICs) from serial dilution liquid culture assays are listed in table 1. All 6 mutants produced well (>1 mg/L) under our standard growth and extraction conditions.
Figure 7.
Anticipated product states of mutant serine side-chains: a) unmodified alcohol, b) dehydroAlanine (dhA), c) oxazoline, and d) oxazole. Sp2 centers and non-rotatable bonds are illustrated in red.
Table 1.
Summary of production, modification, and antibiotic activity of B. cereus thiazole alanine scan mutants.
| Mutant | Compounds Observed |
Additional Modifications |
Production (mg/L) |
MIC (µg/mL)a |
|||
|---|---|---|---|---|---|---|---|
| 168b | COLc | MW2d | |||||
| 1 | S1A | no production | N/A | N/A | N/A | N/A | N/A |
| 2 | C2A | 2 | -- | 0.4 | >8 | >8 | >8 |
| 3 | T3A | 6 | -- | 2.4 | >8 | >8 | >8 |
| 4 | T4A | 7 | -- | 7.4 | >8 | >8 | >8 |
| 5 | C5A | 4 | -- | 0.2 | nt | nt | nt |
| 6 | V6A | 6 | Ala-hydroxylation | 1.5 | 0.13 | 0.06 | 0.06 |
| 7 | C7A | 5 | -- | 2.0 | >8 | >8 | >8 |
| 8 | T8A | 4 | -- | 2.2 | 1 | 0.25 | 0.25 |
| 9 | C9A | 3 | -- | 3.0 | >8 | >8 | >8 |
| 10 | S10A | no production | N/A | N/A | N/A | N/A | N/A |
| 11 | C11A | no production | N/A | N/A | N/A | N/A | N/A |
| 12 | C12A | 5 | -- | N/A | nt | nt | nt |
| 13 | T13A | 7 | -- | 14.0 | 0.13 | 0.06 | 0.06 |
| 14 | T14A | 10 | C-term truncation | 0.4 | >100 | 0.5 | 0.25 |
| 15 | WTf | 6 | -- | 10.0 | 0.5 | 0.06 | 0.13 |
Minimum inhibitory concentrations determined by overnight culture in 96-well plate format.
Bacillus subtilus 168.
Methicillin-resistant Staphylococcus aureus COL.
Methicillin-resistant Staphylococcus aureus MW2.
nt = not tested.
WT = wild type.
Somewhat surprisingly, the V6A mutant provided evidence that, tclD, the iron-dependent valine 6 hydroxylase, was promiscuous. On the native scaffold tclD hydroxylates Cβ, an isopropyl carbon, to yield a tertiary alcohol. The majority of observed compounds from the alanine-6 mutant were hydroxylated on the methyl group to yield the primary alcohol. Although this modification effectively introduces a serine at valine 6, subsequent dehydration, either in the form of the dehydroalanine or as an oxazoline was not observed.
As further evidence of combinatorial outcomes from the posttranslational cascade, at the C-terminus, of the molecule, the T14A mutant bore two alterations in addition to the single amino acid substitution: 1) dehydration of threonine 13 to the dehydrobutyrine side chain was partially inhibited (4 of the 6 largest compounds in the LC/MS display unmodified threonines at this position); 2) minor amounts of the C-terminal residue are cleaved, presumably by carboxypeptidases in the cell during maturation (see SI for LC/MS results). Although the latter modification results in a C-terminal threonine, neither decarboxylation, nor dehydration, nor further truncation could be discerned in these isolates.
To test the timing of this truncation versus C-terminal modification, a ΔT14 deletion mutant was prepared and its product compounds isolated. In this case, the major products contained a C-terminal dehydrobutyrate, however, minor amounts of the unmodified C-terminal residue were also present. More revealing was the observation of products resulting from oxidation/decarboxylation, similar to the natural compounds (see SI for LC/MS results). Combined with the results from the T14A mutant this suggests that the oxidation/decarboxylation occurs early in thiocillin biosynthesis and likely acts to prevent C-terminal proteolytic degradation of the completed antibiotic.
In terms of antibiotic activity, we have noted previously that substitutions at threonines 3 and 4 nearly abolished activity, while those at residues 6 and 8 sustained near wild-type levels54. This result correlates with x-ray structure of MP1 bound to the bacterial 50S ribosomal subunit, which shows residues 3 and 4 turned in towards the binding cleft, in position to make contacts with protein L11.5 In contrast, the opposite edge of the macrocycle, where residues 6 and 8 are located, is essentially solvent exposed in this crystal structure. Residues 3 and 4 are highly conserved throughout thiazolyl peptides bearing a 26-membered ring, including both those with a central piperidine or didehydropiperidine ring, such as thiostrepton, and those with an additional bridging element, like nosiheptide and thiazomycin. Thus, although the biosynthetic machinery can readily accommodate alternative amino acids at these positions, the sequence has specifically evolved as a binding motif for the bacterial 50S ribosomal subunit, which would be retained on account of its antibiotic activity.
Alanine Scan of cysteine and serine residues
A second round of alanine substitutions focused on the eight residues that are converted to seven heterocycles (one pyridine and six thiazoles) (table 1, entries 1, 2, 5, 7, and 9–12). Production levels were substantially diminished relative to the previous set of scan substitutions and some of the anticipated compounds in this set were not detectably produced. Feeding studies have previously shown that serines 1 and 10 are used to construct the central pyridine ring, likely by dual dehydrations and a presumed subsequent [4+2] cycloaddition.62,63 Fundamentally disrupting closure of the pyridine core by introducing alanines in place of these key serines should prevent formation of the natural product scaffold. In agreement, S1A and S10A mutants did not produce observable, thiocillin-like molecules.
Somewhat less predictably, products were not observed in isolates from the C11A mutant. In the mature thiocillin scaffold, cysteine 11 has been converted to a thiazole; this heterocycle could be important for pyridine formation, either as an electron sink for olefin activation in a dehydroAla intermediate or for active site recognition of enzymes later in the pathway. In contrast, disruption mutants of the other two pyridine thiazole substituents, C2A and C9A, did not prevent scaffold maturation, producing 3 and 4 compounds respectively, each of which had a bithiazolylpyridine rather than a trithiazolylpyridine core. Of the remaining thiazole rings found in the macrocycle, C7A produced strongly, while C5A gave compounds insufficient for antibiotic assays. Similarly, the C12A mutant provided minimal amounts of compound.
Three sets of products from thiazole deletion mutants were tested for antibiotic activity (C2A, C7A, and C9A, table 1, entries 2, 7, and 9); in all three cases, loss of activity exceeded 100-fold. These results suggest that the global conformation of the macrocyle is important for inhibition of translation elongation at the ribosome. The loss of the rigidity contributed by a single thiazole, attached to the pyridine core or constraining the macrocyclic loop conformation, disrupts or diminishes that activity. Similar to the two-residue motif at positions 3 and 4 of the macrocycle, the structure of the 26-membered macrocycle is highly conserved in members of the subgroup (SI table). Of this subgroup of thiazolyl peptides, the only known representatives containing less than four fully aromatized heterocycles (e.g. from cysteines 2,5,7, and 9 in the thiocillins) in the macrocyclic ring are those related to thiostrepton and its semi-oxidized aliphatic core; many of those have a second macrocyclic ring which may impose an additional set of conformational constraints to enforce an active architecture to those scaffolds.
In sum, the alanine scan defines a metabolic SAR pattern that can be formulated as depicted in figure 6. Serines 1 and 10 and thiazole 11 all appear essential to post-translational modification and replacement results in loss of compound production. Meanwhile, replacement of the four internal thiazoles, at cysteines 2, 5, 7, and 9, as well as the two conserved threonines, threonine 3 and 4, allows for compound production, while abolishing antibiotic activity. In contrast, positions 6, 8, 12, 13, and 14, on the back half and tail of the molecule, are relatively permissive to substitution.
Cysteine to serine scan
Mutation of each of the six cysteine residues to serines offers complementary information and insight compared to the cysteine to alanine mutations. In addition to an outcome where the serine side chain is unchanged (equivalent to the cysteine to alanine substitution), three other outcomes are possible for posttranslational modification of each of these six serine side chains. One possible route (figure 7) involves processing down the lantibiotic pathway, involving transient phosphorylation and subsequent elimination of inorganic phosphate by either TclK or TclL to yield a dehydro-alanine (dhA) (a to b, figure 7). In the natural product, this pathway is responsible both for initial modification of serines 1 and 10 en route to the pyridine core and for terminal transformation of threonines 4 and 13 into dehydrobutyrines (dhbs). The second route involves processing down the cyclodehydration pathway to heterocycle formation by Tcls J and N. TclJ is presumed to effect the cyclodehydration of cysteines to thiazolines and by analogy here of serines to oxazolines which could persist (a to c, figure 7) or else serve as dehydrogenation substrates for the presumed flavin-dependent desaturase TclN to convert to heteroaromatic oxazoles (c to d, figure 7). As illustrated in figure 6, these four distinct product states from the serine scan introduce different amounts of rigidity in terms of the sp2 character and the rotational constraint they contribute to the maturing peptide backbone.
HPLC traces of extracts from the six thiazole-serine substitutions are illustrated in figure 9 and minimal inhibitory concentrations (MICs) presented in table 2. Depicted alongside the methanolic extracts of the other compounds are isolates from the cell free media of C5S and C7S. This was washed over a C18 SPE cartridge and then eluted with 50% aqueous acetonitrile; the same procedure had been performed for wild-type extracts and mutants for the alanine scan with little or no added material isolated. However, in instances of the C5S and C7S transformants, the majority of products were found in the media. In light of the stark effects of substitutions at threonine 3, a T3S mutant was also included in the serine scan.
Figure 9.
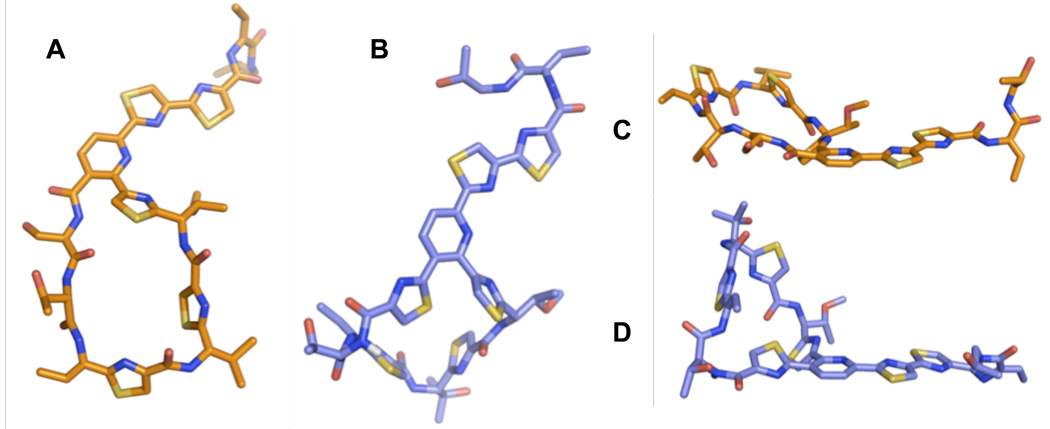
NMR solution phase structures (DMSO-d6) of a C2S-alcohol mutant (A and C) and wild-type thiocillin III (B and D). The amide proton of the “bent” amide in thiocillin III is illustrated in white.
Table 2.
Summary of production, modification, and antibiotic activity of B. cereus thiazole serine scan mutants.
| Mutant | Compounds Observed |
Ratioa | Production (mg/L) |
MIC (µg/mL)a |
|||
|---|---|---|---|---|---|---|---|
| 168b | COLc | MW2d | |||||
| 1 | C2S | 4 | 58:42:0 | 1.9 | >8 | >8 | >8 |
| 2 | T3S | 4 | 100:0:0 | 3.0 | 1 | 0.25 | 0.13 |
| 3 | C5S | 6 | 100:0:0 | 1.3 | >8 | >8 | >8 |
| 4 | C7S | 7 | 77:23:0 | 1.8 | >8 | >8 | >8 |
| 5 | C9S | 31e | 69:22:9 | 0.8 | nt | nt | nt |
| 6 | C11S | no production | N/A | N/A | N/A | N/A | N/A |
| 7 | C12S | 6 | 57:27:16 | 0.4 | 0.5 | 0.25 | 0.25 |
| 8 | WTf | 6 | N/A | 10.0 | 0.5 | 0.06 | 0.13 |
Unmodified:Dehydrated:Oxazole.
Minimum inhibitory concentrations determined by overnight culture in 96-well plate format.
Bacillus subtilus 168.
Methicillin-resistant Staphylococcus aureus COL.
Methicillin-resistant Staphylococcus aureus MW2.
nt = not tested.
WT = wild type.
Variant gave complex mixtures of multiple products (see text/SI)
Levels of production are similar to or less than those of the comparable alanine mutants. Once again, the C11 substitution failed to provide compound, further emphasizing the importance of this residue in posttranslational processing to the mature natural products. Of the mutants that produced isolatable quantities of compounds, all were further analyzed by MS/MS to determine ratios of the individual product states at the newly introduced serine residues. These ratios are based on integration of observed ions in the hi-res mass spectra of compound mixtures and assume that all compounds ionize similarly.
In isolates from all six producers, the major products exhibited an unmodified serine alcohol side chain, with fully formed oxazoles observable only in C9S and C12S mutants.64 In our prior studies on the Microcin B17 pathway, while the McbBCD complex was witnessed to act on both Gly-Ser and Gly-Cys sequence fragments, oxazole formation was almost 2-log slower than thiazole formation.36,65 Although the in vivo system presented here is more complex than homogeneously purified MccB17 synthase, it may be that a similarly slow substrate control is at work in the YcaO homologue TclJ, which could account for the majority of unmodified serine residues in this set of scans. Nonetheless, the ratio unmodified:modified (either dehydrated only or dehydrated and oxidized) is higher at residues adjacent to the heteroaromatic core: C2S, C9S, and C12S, which is conjugated via its biaryl partner at residue 11. Presumably this regioselectivity pattern for cyclodehydration is due to greater substrate affinity of TclJ and N for residues around this core region and may also indicate an ordered sequence of thiazole/oxazole ring formation versus pyridine. In principle, the T3S and hydroxylated V6A (see above) mutants should be susceptible to the same modifications as the six thiazole substitution serines. However, neither oxazole formation, nor any form of dehydration were detected at these serine residues.
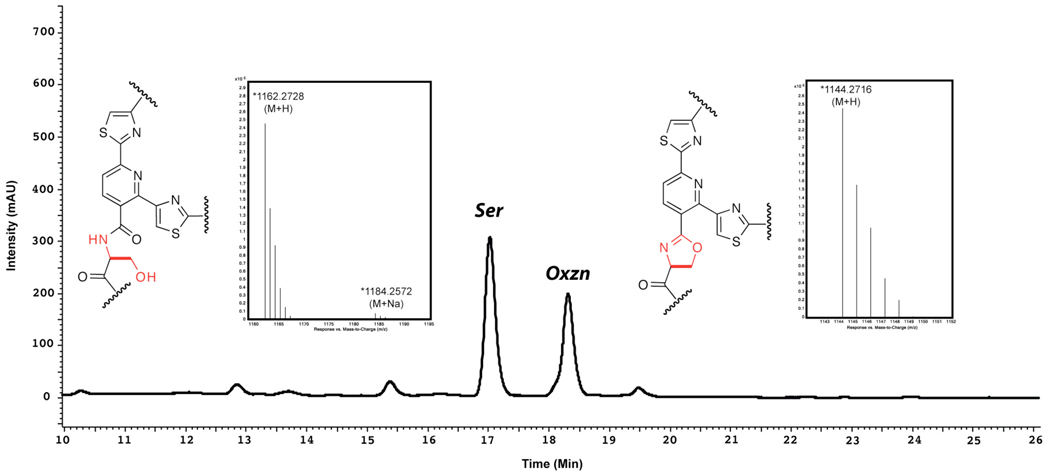
Although the low level of production and the large number of differing compounds made isolation and structural determination of individual products challenging, the C2S mutant gave sufficient quantities of primarily two compounds, enabling separation and identification. In this case, the structures of the two isolated compounds were assigned by combination NMR and MS analysis as the C2-alcohol and oxazoline analogs of thiocilin III as shown in figure 8.66 The 1HNMR spectrum of the oxazoline derivative showed a mixture of two rotamers which began to coalesce on heating; this high degree of rigidity is reflected in the substrates’ stability against autooxidation and in the degradation observed on attempts at chemical oxidation with DBU and bromotrichloromethane. Nevertheless, the core-forming cycloaddition was able to be processed on this derivative.
Figure 8.
HPLC traces (350 nm) of methanolic extracts from 3 day growths of B. cereus C2S mutant. Structures of core regions as determined by LC/MS, MS/MS, and 1D and 2D NMR are depicted adjacent to the individually isolated peaks.
In sum, there are four possible outcomes for each of the six cysteine to serine mutants: unchanged alcohol, dhA, oxazoline, and oxazole for a total of 24 possible variants. Since these could then be subject to stochastic two fold further modifications at the side chains of residues 6,8, and 14, in principle there could be up to 192 variants of the six serine mutants for serines 2, 5, 7, 9, 11, and 12. We have observed 46 of those possible compounds to date, at five of the six positions.
To evaluate antibiotic activity, the C2S derivatives were tested separately, whereas each of the other serine scan mutants was tested as a mixture. As with the alanine scan, all mutants with disruption of the heterocycles arising from residues 2, 5, 7, and 9 (within the macrocyclic substructure) had a greater than 100-fold loss of activity. Despite the presence of the partially cyclized residue in the C2S oxazoline, this derivative also appeared essentially inactive in our assays. Additionally, the C7S mutant products were tested at elevated concentrations to allow treatment with quantities of dehydrated compounds equivalent to wild type MICs, yet still proved inactive. In contrast, the combined products from the C12S mutant were in range of the wild type, suggesting that bithiazole formation on the thiocillin tail is not essential for antibiotic activity. Bithiazoles are known DNA intercalators and the thiazolyl peptide tails are seen to largely associate with the 23S rRNA side of the 50S binding cleft. Still, a number of thiazolyl peptide antibiotics do not contain this motif, clearly to no substantial detriment in activity. Lastly, whereas T3A, T3D, and T3K had all been essentially devoid of activity, the T3S mutant antibiotic activity proved comparable to wild-type thiocillins, confirming the importance of a hydrogen bond donor at this residue of the molecule.54
Conformational analysis of NMR structures
Given the loss of activity in thiazole disruption mutants at cysteines 2, 5, 7, and 9, we endeavored to analyze the structural basis of this change by comparing solution state NMR-derived structures from a wild-type thiocillin and at least one thiazole disruption mutant. As yet, there exists no 3D small-molecule structure of a thiocillin or other unconstrained 26-membered thiazolyl peptides.67 Thus, 2D 1H-1H COSY, ROSY, TOCSY, 13C-edited HSQC, and HMBC were used to obtain complete proton and carbon resonance assignments for homogeneously purified thiocillin II and a C2S analogue bearing an unmodified serine residue. Preliminary folds, based on the chemical structures and basic NMR information describing the relative orientation of aromatic ring systems, were generated with ACD/Chem, energy minimized with MMplus force field using Hypercube Hyperchem 8.0 and diversified by using restrained simulated annealing in XplorNIH. Identified distance restraints were set to 6.5 Angstrom in general due to the nature of the 2D ROSY experiment. In total, 14 non-sequential, non-intra-residual distance constrains between 5 protons were identified and J couplings were included as additional constraints for the simulated annealing. All topology files, .par and .psf, essential for a simulated annealing in xplorNIH were generated with XPLO2D (Uppsala Software Factory). The more common prodrug-algorithm (Dundee University) could not be used due to the size of thiocillin and C2S. For each of the two structures 150 models were calculated and 10 structures with the lowest global energy values were picked for the structural bundle. After the simulated annealing, an energy minimization and a short molecular dynamics simulation of 0.5 ns were performed in an MMplus force field using Hyperchem 8.0. Both structures represent the highest populated, lowest energy fold for thiocillin and C2S. All NMR data were processed and analyzed with Mestrec Mnova 5.3.1 and PerchNMR PE.
In the case of wild type, our calculations gave a structure in reasonable accord with those of free nocathiacin and nosiheptide, with several notable distinctions.16,68 Although bonds to ortho-substituents of the pyridine ring are predictably aromatic, sterics demand that thiazoles 2 and 9 be turned aside, as observed in a number of the reported structures. In our calculated structure, this requires that the macrocycle be deformed at a single amide bond, such that the carbonyl oxygen and amide N-H are not precisely anti-periplanar. Structures incorporating this inflection at any given amide bond in the macrocycle all represented discrete minima. However, the 1H NMR spectrum of thiocillin II displays an attenuated chemical shift for the proton on the N-terminus of residue 3, suggesting that the greater sp3 character lies on the nitrogen of threonine 3. The relevant proton is illustrated in white in figure 9. The geometry about this amide bond contributes to the relatively rigid, compact conformation of thiocillin II. The macrocylce itself is curled into a conformational “lip” on the pro-S side of the trithiazolylpyridine, which is the side oriented towards protein L11 in the crystal structure by Harms et al.5 Somewhat more revealing in terms of the observed SAR, the structure determined for the serine-substituted molecule does not contain this “lip.” Indeed, the C2S structure is much flatter and more flexible than the wild type, with two distinct conformers making substantial contributions to the structural bundle, representing the entire range between both extrema. Presumably, the energy required to contort the open C2S structure into a conformation similar to that of the wild type, with its bent amide, is very high.
Conclusions
We have evaluated the capacity of the eight enzymes in the Bacillus cereus thiocillin gene cluster to posttranslationally process single residue variants of the TclE prepeptide. Of particular interest are the two serines that are condensed to the core pyridine and each of the six cysteines that become thiazoles. We have examined the consequences of serine to alanine mutations and changes of each cysteine to either an alanine or serine. The cysteine to alanine changes delete one thiazole at a time in the three subsections of the mature thiocillin scaffold and examine in vivo if these changes can be tolerated for posttranslational bithiazolylpyridine framework construction, and then, if so, what is the separate and consequent effect on antibiotic activity.
The cysteine to serine changes introduce a higher level of complexity and probe additional facets of the posttranslational machinery that builds the mature antibiotic core. Serine residues could in principle stay unmodified and much of the starting alcohol side chain is seen in the five S2, S5, S7, S9, and S12 variants, and in this sense these approximate the cysteine to alanine changes. More intriguingly the cysteine to serine changes probe the partition of serine side chains down either the lantibiotic type processing pathway to a dehydroalanine or down the heterocycle forming pathway (Figure 7). Both product residues would contribute additional rigidity to the macrocyle, but differ in removal of a Cα stereocenter (dhA) or introduction of an endocyclic double bond (oxazoline). Seen this way, the two elements each represent a half step toward the native structural support. Although serines 1 and 10 are presumed to be processed by the lantibiotic post-translational route on way to the central pyridine ring, the serines 2,7,9,12 can be shunted to the oxazoline/oxazole route, without any detectable partitioning to dhA resiudes. This may ultimately give clues to timing, regiospecificity, and competition between the lantibiotic type phosphorylation/elimination post-translational modifications and the cyclodehydration and heteroaromatization type enzymes in the thiocillin pathway, but also in such related scaffolds as the berninamycin antibiotic. It is clear that conversion of serine to oxazoline and then oxazole is substantially slower and more inefficient than processing of cysteines to thiazoles, where no thiazoline intermediates accumulate. Meanwhile serines introduced at residues 3 and 6 go unmodified.
The three cysteines, 2, 9, and 11, that buttress the central pyridine in the mature thiocillin scaffold can be differentiated from cysteines 5 and 7, which sit in the 26-atom macrocyclic loop and contribute to formation of the conformational “lip” observed in the wild type structure. Pyridine frameworks where either cysteine 2 or 9 are substituted with alanine or serine can be generated, suggesting that the central pyridine ring can be condensed from two putative dhAs (from phosphorylation/Pi elimination at serines 1 and 10), when either thiazole 2 or 9 is missing. However, cysteine 11 and/or the attendant thiazole cannot be replaced with either alanine or serine, suggesting a unique role for this thiazole, among the pyridine substituents. That role could be in setting a conformation where serines 1 and 10 can be processed to the pyridine ring stage, including perhaps participation in the net [4+2] aza cycloaddition process. Resolution of the role of thiazole 11 will await studies with purified enzymes Tcl J, K, L, and N to determine timing of dehydroalanine and dehydrobutyrine formation versus thiazole formation in assembly of the trithiazolylpyridine core.
Assessment of the antibiotic activity indicates that while thiazole 2 or 9 can be dispensed with for building a bithiazolylpyridine core, the resultant mature structural variant has lost at least 100-fold potency as antibiotics. Evaluation of the conformational effects of loss of one of those two thiazoles was undertaken by the construction of NMR-based models, comparing wildtype Thiocillin II with a C2S variant, bearing a pendant alcohol, indicating a very different conformer preference that may lie at the heart of loss of binding to the 50S ribosomal subunit and antibiotic activity. The apparent importance of this native conformation to antibiotic activity underscores the recently reported results of assays employing thiazolyl peptide fragments.69,70
This study is an initial effort to define the requirements for each of the six thiazoles for in vivo conversion by the producer bacterium to the mature thiocillin scaffold by single replacement to Ala or Ser. Concomitant assessment of degree of abrogation of antibiotic activity, presumably reflects disruption of binding to the target site on bacterial 50S ribosomal subunits. It reveals the limits on the ability of B. cereus to engineer oxazole for thiazole replacements and focuses subsequent efforts on the special role of Cys11 in posttranslational maturation of the final antibiotic scaffold. The 20 single mutations in the tclE gene, all 14 residues to Ala and also the six Cys to Ser, when expressed in the B. cereus host, lead to maturation of 16 thiazolyl peptide scaffolds. The S1A and S10A mutants cannot close the pyridine ring and the C11A and C11S mutants also yield no product. The other sixteen prepropeptide mutants get processed to 67 C to A variant and 46 C to S variant scaffolds.
Supplementary Material
Figure 3.
Mechanisms of (A) dehydrobutyrine formation via phosphorylation/elimination as effected by lantibiotic dehydratases; (B) thiazoline formation via cyclodehydration/oxidation and subsequent dehydrogenation to thiazole as with the MccB17 synthase complex; and (C) pyridine formation via putative enzyme-catalyzed [4+2] cycloaddition.
Acknowledgements
We thank Jonathan Swoboda and Jenny O’Neill for guidance with MIC assays. This work was supported by NIH NIAID Grant 20011 and NERCE Grant NIAID U54 AI057 159 (C.T.W.) Reagents were prepared with the assistance of the NERCE Biomolecule Production Core (NIAID U54 AI057 159). In particular, we thank Lauren Perry and Robin Ross (both of NERCE) for invaluable aid with batch fermentations of the individual variants. AK acknowledges the DOE and LANS for a JR Oppenheimer fellowship at the Los Alamos National Laboratory. AAB is supported by an NIH National Cancer Institute Postdoctoral Fellowship (CA136283).
Footnotes
Supporting Information Available: Supporting figures and tables, experimental procedures, and spectral data. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Bagley MC, Dale JW, Merritt EA, Xiong X. Chem Rev. 2005;105:685–714. doi: 10.1021/cr0300441. [DOI] [PubMed] [Google Scholar]
- 2.Hughes RA, Moody CJ. Angew Chem Int Ed Engl. 2007;46:7930–7954. doi: 10.1002/anie.200700728. [DOI] [PubMed] [Google Scholar]
- 3.Pestka S, Brot N. J Biol Chem. 1971;246:7715–7722. [PubMed] [Google Scholar]
- 4.Su TL. Br J Exp Pathol. 1948;29:473–481. [PMC free article] [PubMed] [Google Scholar]
- 5.Harms JM, Wilson DN, Schluenzen F, Connell SR, Stachelhaus T, Zaborowska Z, Spahn CM, Fucini P. Mol Cell. 2008;30:26–38. doi: 10.1016/j.molcel.2008.01.009. [DOI] [PubMed] [Google Scholar]
- 6.Ciufolini MA, Shen Y-C. Organic Letters. 1999;1:1843–1846. doi: 10.1021/ol991115e. [DOI] [PubMed] [Google Scholar]
- 7.Lefranc D, Ciufolini MA. Angew Chem Int Ed Engl. 2009;48:4198–4201. doi: 10.1002/anie.200900621. [DOI] [PubMed] [Google Scholar]
- 8.Bycroft BW, Gowland MS. J. Chem. Soc., Chem. Commun. 1978:256–258. [Google Scholar]
- 9.Fenet B, Pierre F, Cundliffe E, Ciufolini MA. Tetrahedron Lett. 2002;43:2367–2370. [Google Scholar]
- 10.Benazet F, Cartier M, Florent J, Godard C, Jung G, Lunel J, Mancy D, Pascal C, Renaut J, et al. Experientia. 1980;36:414–416. doi: 10.1007/BF01975121. [DOI] [PubMed] [Google Scholar]
- 11.Depaire H, Thomas JP, Brun A, Hull WE, Olesker A, Lukacs G. Tetrahedron Lett. 1977:1401–1402. [Google Scholar]
- 12.Depaire H, Thomas JP, Brun A, Lukacs G. Tetrahedron Lett. 1977:1395–1396. [Google Scholar]
- 13.Depaire H, Thomas JP, Brun A, Olesker A, Lukacs G. Tetrahedron Lett. 1977:1403–1406. [Google Scholar]
- 14.Depaire H, Thomas JP, Brun A, Olesker A, Lukacs G. Tetrahedron Lett. 1977:1397–1400. [Google Scholar]
- 15.Pascard C, Ducruix A, Lunel J, Prange T. J. Am. Chem. Soc. 1977;99:6418–6423. doi: 10.1021/ja00461a039. [DOI] [PubMed] [Google Scholar]
- 16.Prange T, Ducruix A, Pascard C, Lunel J. Nature (London) 1977;265:189–190. doi: 10.1038/265189a0. [DOI] [PubMed] [Google Scholar]
- 17.Anderson B, Hodgkin DC, Viswamitra MA. Nature. 1970;225:233–235. doi: 10.1038/225233a0. [DOI] [PubMed] [Google Scholar]
- 18.Hensens OD, Albers-Schonberg G. J. Antibiot. 1983;36:832–845. doi: 10.7164/antibiotics.36.832. [DOI] [PubMed] [Google Scholar]
- 19.Trejo WH, Dean LD, Pluscec J, Meyers E, Brown WE. J. Antibiot. 1977;30:639–643. doi: 10.7164/antibiotics.30.639. [DOI] [PubMed] [Google Scholar]
- 20.Colombo L, Tavecchia P, Selva E, Gallo GG, Zerilli LF. Org. Mass Spectrom. 1992;27:219–225. [Google Scholar]
- 21.Kettenring J, Colombo L, Ferrari P, Tavecchia P, Nebuloni M, Vekey K, Gallo GG, Selva E. J. Antibiot. 1991;44:702–715. doi: 10.7164/antibiotics.44.702. [DOI] [PubMed] [Google Scholar]
- 22.Selva E, Beretta G, Montanini N, Saddler GS, Gastaldo L, Ferrari P, Lorenzetti R, Landini P, Ripamonti F, et al. J. Antibiot. 1991;44:693–701. doi: 10.7164/antibiotics.44.693. [DOI] [PubMed] [Google Scholar]
- 23.Tavecchia P, Gentili P, Kurz M, Sottani C, Bonfichi R, Lociuro S, Selva E. J Antibiot (Tokyo) 1994;47:1564–1567. doi: 10.7164/antibiotics.47.1564. [DOI] [PubMed] [Google Scholar]
- 24.Tavecchia P, Gentili P, Kurz M, Sottani C, Bonfichi R, Selva E, Lociuro S, Restelli E, Ciabatti R. Tetrahedron. 1995;51:4867–4890. [Google Scholar]
- 25.Abe H, Kushida K, Shiobara Y, Kodama M. Tetrahedron Lett. 1988;29:1401–1404. [Google Scholar]
- 26.Lau RCM, Rinehart KL. J. Antibiot. 1994;47:1466–1472. doi: 10.7164/antibiotics.47.1466. [DOI] [PubMed] [Google Scholar]
- 27.Liesch JM, Millington DS, Pandey RC, Rinehart KL., Jr J. Am. Chem. Soc. 1976;98:8237–8249. doi: 10.1021/ja00441a058. [DOI] [PubMed] [Google Scholar]
- 28.Liesch JM, Rinehart KL., Jr J. Am. Chem. Soc. 1977;99:1645–1646. doi: 10.1021/ja00447a061. [DOI] [PubMed] [Google Scholar]
- 29.Cameron DM, Thompson J, March PE, Dahlberg AE. J Mol Biol. 2002;319:27–35. doi: 10.1016/S0022-2836(02)00235-8. [DOI] [PubMed] [Google Scholar]
- 30.Heffron SE, Jurnak F. Biochemistry. 2000;39:37–45. doi: 10.1021/bi9913597. [DOI] [PubMed] [Google Scholar]
- 31.Parmeggiani A, Krab IM, Okamura S, Nielsen RC, Nyborg J, Nissen P. Biochemistry. 2006;45:6846–6857. doi: 10.1021/bi0525122. [DOI] [PubMed] [Google Scholar]
- 32.Morris RP, Leeds JA, Naegeli HU, Oberer L, Memmert K, Weber E, LaMarche MJ, Parker CN, Burrer N, Esterow S, Hein AE, Schmitt EK, Krastel P. J Am Chem Soc. 2009;131:5946–5955. doi: 10.1021/ja900488a. [DOI] [PubMed] [Google Scholar]
- 33.Thompson J, Cundcliffe E, Stark MJR. J. Gen. Microbiol. 1982;128:875–884. doi: 10.1099/00221287-128-4-875. [DOI] [PubMed] [Google Scholar]
- 34.Carnio MC, Stachelhaus T, Francis KP, Scherer S. Eur J Biochem. 2001;268:6390–6401. doi: 10.1046/j.0014-2956.2001.02591.x. [DOI] [PubMed] [Google Scholar]
- 35.Donia MS, Hathaway BJ, Sudek S, Haygood MG, Rosovitz MJ, Ravel J, Schmidt EW. Nat Chem Biol. 2006;2:729–735. doi: 10.1038/nchembio829. [DOI] [PubMed] [Google Scholar]
- 36.Li YM, Milne JC, Madison LL, Kolter R, Walsh CT. Science. 1996;274:1188–1193. doi: 10.1126/science.274.5290.1188. [DOI] [PubMed] [Google Scholar]
- 37.McIntosh JA, Donia MS, Schmidt EW. Nat Prod Rep. 2009;26:537–559. doi: 10.1039/b714132g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Willey JM, van der Donk WA. Annu Rev Microbiol. 2007;61:477–501. doi: 10.1146/annurev.micro.61.080706.093501. [DOI] [PubMed] [Google Scholar]
- 39.Xie L, Miller LM, Chatterjee C, Averin O, Kelleher NL, van der Donk WA. Science. 2004;303:679–681. doi: 10.1126/science.1092600. [DOI] [PubMed] [Google Scholar]
- 40.Yu Y, Duan L, Zhang Q, Liao R, Ding Y, Pan H, Wendt-Pienkowski E, Tang G, Shen B, Liu W. ACS Chem Biol. 2009;4:855–864. doi: 10.1021/cb900133x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kelly WL, Pan L, Li C. J Am Chem Soc. 2009;131:4327–4334. doi: 10.1021/ja807890a. [DOI] [PubMed] [Google Scholar]
- 42.Liao R, Duan L, Lei C, Pan H, Ding Y, Zhang Q, Chen D, Shen B, Yu Y, Liu W. Chem Biol. 2009;16:141–147. doi: 10.1016/j.chembiol.2009.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brown LC, Acker MG, Clardy J, Walsh CT, Fischbach MA. Proc Natl Acad Sci U S A. 2009;106:2549–2553. doi: 10.1073/pnas.0900008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chatterjee C, Miller LM, Leung YL, Xie L, Yi M, Kelleher NL, van der Donk WA. J Am Chem Soc. 2005;127:15332–15333. doi: 10.1021/ja0543043. [DOI] [PubMed] [Google Scholar]
- 45.Li B, Yu JP, Brunzelle JS, Moll GN, van der Donk WA, Nair SK. Science. 2006;311:1464–1467. doi: 10.1126/science.1121422. [DOI] [PubMed] [Google Scholar]
- 46.Patton GC, van der Donk WA. Curr Opin Microbiol. 2005;8:543–551. doi: 10.1016/j.mib.2005.08.008. [DOI] [PubMed] [Google Scholar]
- 47.Milne JC, Eliot AC, Kelleher NL, Walsh CT. Biochemistry. 1998;37:13250–13261. doi: 10.1021/bi980996e. [DOI] [PubMed] [Google Scholar]
- 48.Milne JC, Roy RS, Eliot AC, Kelleher NL, Wokhlu A, Nickels B, Walsh CT. Biochemistry. 1999;38:4768–4781. doi: 10.1021/bi982975q. [DOI] [PubMed] [Google Scholar]
- 49.Roy RS, Gehring AM, Milne JC, Belshaw PJ, Walsh CT. Nat. Prod. Rep. 1999;16:249–263. doi: 10.1039/a806930a. [DOI] [PubMed] [Google Scholar]
- 50.Auclair K, Sutherland A, Kennedy J, Witter DJ, Van den Heever JP, Hutchinson CR, Vederas JC. J. Am. Chem. Soc. 2000;122:11519–11520. [Google Scholar]
- 51.Watanabe K, Oikawa H, Yagi K, Ohashi S, Mie T, Ichihara A, Honma M. J. Biochem. 2000;127:467–473. doi: 10.1093/oxfordjournals.jbchem.a022629. [DOI] [PubMed] [Google Scholar]
- 52.Kelly WL. Org. Biomol. Chem. 2008;6:4483–4493. doi: 10.1039/b814552k. [DOI] [PubMed] [Google Scholar]
- 53.Romesberg FE, Spiller B, Schultz PG, Stevens RC. Science (Washington, D. C.) 1998;279:1929–1933. doi: 10.1126/science.279.5358.1929. [DOI] [PubMed] [Google Scholar]
- 54.Acker MG, Bowers AA, Walsh CT. J. Am. Chem. Soc. 2009;131:17563–17565. doi: 10.1021/ja908777t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bagley MC, Bashford KE, Hesketh CL, Moody CJ. J. Am. Chem. Soc. 2000;122:3301–3313. [Google Scholar]
- 56.Hughes RA, Thompson SP, Alcaraz L, Moody CJ. Chem. Commun. (Cambridge, U. K.) 2004:946–948. doi: 10.1039/b401580k. [DOI] [PubMed] [Google Scholar]
- 57.Moody CJ, Bagley MC. Chem. Commun. (Cambridge) 1998:2049–2050. [Google Scholar]
- 58.Muller HM, Delgado O, Bach T. Angew Chem Int Ed Engl. 2007;46:4771–4774. doi: 10.1002/anie.200700684. [DOI] [PubMed] [Google Scholar]
- 59.Nicolaou KC, Safina BS, Zak M, Estrada AA, Lee SH. Angew. Chem., Int. Ed. 2004;43:5087–5092. doi: 10.1002/anie.200461340. [DOI] [PubMed] [Google Scholar]
- 60.Nicolaou KC, Zak M, Safina BS, Lee SH, Estrada AA. Angew. Chem., Int. Ed. 2004;43:5092–5097. doi: 10.1002/anie.200461341. [DOI] [PubMed] [Google Scholar]
- 61.Nicolaou KC, Zou B, Dethe DH, Li DB, Chen DY. Angew Chem Int Ed Engl. 2006;45:7786–7792. doi: 10.1002/anie.200602798. [DOI] [PubMed] [Google Scholar]
- 62.Mocek U, Beale JM, Floss HG. J. Antibiot. 1989;42:1649–1652. doi: 10.7164/antibiotics.42.1649. [DOI] [PubMed] [Google Scholar]
- 63.Mocek U, Knaggs AR, Tsuchiya R, Nguyen T, Beale JM, Floss HG. Journal of the American Chemical Society. 1993;115:7557–7568. [Google Scholar]
- 64.The C9S mutant bares additional note. For this mutant, two sets of products were observed in the LC-MS spectra. These sets of products had constituents of equivalent masses, but different retention time (see Supporting Information). It can be imagined that, if the substituted serine at amino acid 9 were to be a substrate for the lantibiotic-type dehydratases, a prepetide so modified would present a choice of dienophiles for the putative aza-Diels-Alder reaction: one resulting from dehydration of serine 9 and another from dehydration of serine 10. The first would result in a smaller, 23-membered macrocycle, while the second would provide the standard 26-membered constituent. Compounds resulting from further post-translational modification of these two different sets of products would have equivalent masses and therefore be essentially indistinguishable in the LC-MS spectra. At present, due to the extremely low level of production in this strain, there is insufficient data to determine, whether such a bi-functional cycloaddition substrate is in fact being produced.
- 65.Belshaw PJ, Roy RS, Kelleher NL, Walsh CT. Chem Biol. 1998;5:373–384. doi: 10.1016/s1074-5521(98)90071-0. [DOI] [PubMed] [Google Scholar]
- 66.Additional evidence of the oxazoline present in the structure of the dehydrated compound was provided by conversion to the parent alcohol compound under conditions of acidic hydrolysis. This method of qualitative analysis worked well with the purified dehydro-C2S compound, but could not be extended cleanly to the more complicated and inseparable mixtures described for the C7S, C9S and C12S mutants.
- 67.Constantine KL, Mueller L, Huang S, Abid S, Lam KS, Li W, Leet JE. J. Am. Chem. Soc. 2002;124:7284–7285. doi: 10.1021/ja026249t. [DOI] [PubMed] [Google Scholar]
- 68.Bond CS, Shaw MP, Alphey MS, Hunter WN. Acta Crystallogr., Sect. D: Biol. Crystallogr. 2001;D57:755–758. doi: 10.1107/s0907444901003134. [DOI] [PubMed] [Google Scholar]
- 69.Nicolaou KC, Zak M, Rahimipour S, Estrada AA, Lee SH, O'Brate A, Giannakakou P, Ghadiri MR. Journal of the American Chemical Society. 2005;127:15042–15044. doi: 10.1021/ja0552803. [DOI] [PubMed] [Google Scholar]
- 70.Starosta AL, Qin H, Mikolajka A, Leung GYC, Schwinghammer K, Nicolaou KC, Chen DYK, Cooperman BS, Wilson DN. Chemistry and Biology. 2009;16:1087–1096. doi: 10.1016/j.chembiol.2009.09.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.