Abstract
In subjects with schizophrenia, impairments in working memory are associated with dysfunction of the dorsolateral prefrontal cortex (DLPFC). This dysfunction appears to be due, at least in part, to abnormalities in γ-aminobutyric acid (GABA)-mediated inhibitory circuitry. To test the hypothesis that altered GABA-mediated circuitry in the DLPFC of subjects with schizophrenia reflects expression changes of genes that encode selective presynaptic and postsynaptic components of GABA neurotransmission, we conducted a systematic expression analysis of GABA-related transcripts in the DLPFC of 14 pairs of schizophrenia and age-, sex- and post-mortem interval-matched control subjects using a customized DNA microarray with enhanced sensitivity and specificity. Subjects with schizophrenia exhibited expression deficits in GABA-related transcripts encoding (1) presynaptic regulators of GABA neurotransmission (67 kDa isoform of glutamic acid decarboxylase (GAD67) and GABA transporter 1), (2) neuropeptides (somatostatin (SST), neuropeptide Y (NPY) and cholecystokinin (CCK)) and (3) GABAA receptor subunits (α1, α4, β3, γ2 and δ). Real-time qPCR and/or in situ hybridization confirmed the deficits for six representative transcripts tested in the same pairs and in an extended cohort, respectively. In contrast, GAD67, SST and α1 subunit mRNA levels, as assessed by in situ hybridization, were not altered in the DLPFC of monkeys chronically exposed to antipsychotic medications. These findings suggest that schizophrenia is associated with alterations in inhibitory inputs from SST/NPY-containing and CCK-containing subpopulations of GABA neurons and in the signaling via certain GABAA receptors that mediate synaptic (phasic) or extrasynaptic (tonic) inhibition. In concert with previous findings, these data suggest that working memory dysfunction in schizophrenia is mediated by altered GABA neurotransmission in certain DLPFC microcircuits.
Keywords: microarray, quantitative PCR, in situ hybridization, GAD, neuropeptides, GABAA receptor
Introduction
A common and core clinical feature of schizophrenia is the impairment of certain cognitive functions, such as working memory, that are mediated by the dorsolateral prefrontal cortex (DLPFC).1–3 The dysfunction of this brain region appears to reflect, at least in part, disturbances in inhibitory circuitry mediated by γ-aminobutyric acid (GABA)-containing interneurons.4 In nonhuman primates, normal working memory function depends on GABA-mediated circuitry in the DLPFC5,6 and decreased levels of the mRNAs encoding the 67 kDa isoform of glutamic acid decarboxylase (GAD67), an enzyme that synthesizes GABA, and GABA transporter 1 (GAT1), a presynaptic transporter for the reuptake of synaptically released GABA, have been replicated in multiple post-mortem studies of schizophrenia.7–13 Indeed, an analysis of all post-mortem studies of schizophrenia conducted in specimens from the Stanley Neuropathology Consortium revealed that three genes expressed in GABA neurons (reelin, parvalbumin (PV) and GAD67) had the most abnormal transcript and protein levels in schizophrenia.14
At the cellular level, the density of neurons with detectable levels of GAD67 mRNA was significantly decreased in schizophrenia subjects,7,9 whereas in neurons with detectable levels of GAD67 mRNA, the expression level per neuron did not differ from control values.9 These observations suggest that the majority of DLPFC GABA neurons express normal levels of GAD67 mRNA in subjects with schizophrenia, but approximately 25–35% of GABA neurons lack detectable levels of this transcript. Furthermore, the affected subpopulation includes the GABA neurons that express the calcium-binding protein PV, whereas those that express calretinin (CR) appear to be unaffected.15 However, abnormalities in PV neurons alone may not completely account for the deficits in expression of GAD67 mRNA since such changes were also observed in cortical layers I, II and V, where relatively few PV-containing GABA neurons are located15,16 and where no changes in PV mRNA expression were found.15 Thus, other subpopulations of GABA neurons present in these layers, such as those that express the calcium-binding protein calbindin16 and/or the neuropeptides somatostatin (SST)17 or cholecystokinin (CCK),18 may be affected and transcripts that are selectively expressed in these subpopulations may be altered in the DLPFC of subjects with schizophrenia.
In addition, GABAA receptor expression in the DLPFC appears to be abnormal in subjects with schizophrenia. For example, increased muscimolbinding in pyramidal neuron cell bodies19,20 and increased GABAA receptor α2 subunits in the axon initial segments of pyramidal neurons21 might represent compensatory receptor upregulation in response to decreased GABA release from GABA neurons, especially those that express PV.4 However, the reports of decreased mRNA levels for the GABAA receptor γ2 and δ subunits22,23 suggest that the downregulation of GABAA receptors containing these subunits might also contribute to disturbances in DLPFC inhibitory circuitry in schizophrenia.
Based on these findings, we hypothesized that altered DLPFC GABA-mediated circuitry in schizophrenia reflects expression changes of genes that encode selective pre- and postsynaptic components of GABA neurotransmission. In order to test this hypothesis, we analyzed the expression pattern (that is, the transcriptome) of a large number of GABA-related transcripts using a customized DNA microarray platform with enhanced sensitivity and specificity. Findings for selected transcripts were verified by real-time quantitative polymerase chain reaction (qPCR) and by in situ hybridization. In situ hybridization studies were also conducted in monkeys chronically exposed to haloperidol or olanzapine.
Materials and methods
Human subjects
Brain specimens in the Brain Tissue Donation Program at the University of Pittsburgh Medical Center were obtained during autopsies conducted at the Allegheny County Coroner’s Office (Pittsburgh, PA, USA) after consent was obtained from the next of kin. For DNA microarray and qPCR studies, 14 pairs of schizophrenia and control subjects matched for sex, and as closely as possible for age and post-mortem interval (PMI), were used in this study (Table 1).
Table 1.
Characteristics of subjects for microarray and qPCR studies
| Control subjects |
Schizophrenia subjects |
||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Pair | Case | Sex/ race |
Age | PMI | RIN | Cause of death | Case | DSM IV diagnosis | Sex/ race |
Age | PMI | RIN | Cause of death |
| 1 | 592 | M/B | 41 | 22.1 | 9 | ASCVD | 533 | Chronic undifferentiated schizophrenia |
M/W | 40 | 29.1 | 8.4 | Accidental asphyxiation |
| 2 | 567 | F/W | 46 | 15.0 | 8.9 | Mitral valve prolapse |
537 | Schizoaffective disorder+ |
F/W | 37 | 14.5 | 8.6 | Suicide by hanging |
| 3 | 516 | M/B | 20 | 14.0 | 8.4 | Gun shot wound to chest |
547 | Schizoaffective disorderB,V |
M/B | 27 | 16.5 | 7.4 | Heat stroke |
| 4 | 630 | M/W | 65 | 21.2 | 9 | ASCVD | 566 | Chronic undifferentiated schizophreniaa, B |
M/W | 63 | 18.3 | 8 | ASCVD |
| 5 | 604 | M/W | 39 | 19.3 | 8.6 | HPCAD | 581 | Chronic paranoid schizophreniab,c,B,V |
M/W | 46 | 28.1 | 7.9 | Accidental combined drug overdose |
| 6 | 546 | F/W | 37 | 23.5 | 8.6 | ASCVD | 587 | Chronic undifferentiated schizophreniaa,B |
F/B | 38 | 17.8 | 9 | Myocardial hypertrophy |
| 7 | 1047 | M/W | 43 | 12.4 | 9 | ASCVD | 722 | Undifferentiated schizophreniad |
M/B | 45 | 9.1 | 9.2 | |
| Gastrointestinal hemorrhage |
|||||||||||||
| 8 | 551 | M/W | 61 | 16.4 | 8.3 | Cardiac tamponade |
625 | Chronic disorganized schizophreniae |
M/B | 49 | 23.5 | 7.6 | ASCVD |
| 9 | 685 | M/W | 56 | 14.5 | 8.1 | HPCAD | 622 | Chronic undifferentiated schizophrenia+ |
M/W | 58 | 18.9 | 7.4 | Right MCA infarction |
| 10 | 681 | M/W | 51 | 11.6 | 8.9 | Hypertrophic cardiomyopathy |
640 | Chronic paranoid Schizophrenia |
M/W | 49 | 5.2 | 8.4 | Pulmonary embolism |
| 11 | 806 | M/W | 57 | 24.0 | 7.8 | Pulmonary thromboembolism |
665 | Chronic paranoid schizophreniab |
M/B | 59 | 28.1 | 9.2 | Intestinal hemorrhage |
| 12 | 822 | M/B | 28 | 25.3 | 8.5 | ASCVD | 787 | Schizoaffective disorderf | M/B | 27 | 19.2 | 8.4 | Suicide by gun shot |
| 13 | 727 | M/B | 19 | 7.0 | 9.2 | Trauma | 829 | Schizoaffective disorderb,d,+,B,V |
M/W | 25 | 5.0 | 9.3 | Suicide by drug overdose |
| 14 | 871 | M/W | 28 | 16.5 | 8.5 | Trauma | 878 | Disorganized schizophreniab,V |
M/W | 33 | 10.8 | 8.9 | Myocardial fibrosis |
Abbreviations: ASCVD, atherosclerotic coronary vascular disease; DSM IV, Diagnosis and Statistical Manual of Mental Disorders, fourth edition; HPCAD, hypoplastic coronary artery disease; PMI, post-mortem interval (in hours); qPCR, real-time quantitative polymerase chain reaction; RIN, RNA integrity number.
Schizophrenic subjects off medications at time of death.
Schizophrenia subjects on benzodiazepines at the time of death.
Schizophrenia subjects on valproic acid at the time of death.
Alcohol abuse, in remission at time of death.
Alcohol dependence, current at time of death.
Other substance abuse, current at time of death.
Other substance abuse, in remission at time of death.
Alcohol abuse, current at time of death.
Other substance dependence, current at time of death.
The mean age, PMI, brain pH, RNA integrity number (RIN, see below) and tissue storage time were virtually identical in the two groups (Table 2). For in situ hybridization studies, we used 23 subject pairs composed of 13 of these 14 pairs (except pair 7, Table 1), plus 10 pairs described in cohort 2 (pairs 1–4, 6, 7 and 9–12) of a previously published study.13 These subject pairs were also matched for sex, and the mean values for age, PMI, brain pH, RIN and tissue storage time were nearly identical for the two groups (Table 2).
Table 2.
Summary of subject characteristics for each study
| Parameter | Microarray/qPCR |
In situ hybridization |
||
|---|---|---|---|---|
| Control | Schizophrenia | Control | Schizophrenia | |
| Number | 14 | 14 | 23 | 23 |
| Sex | 12 M, 2 F | 12 M, 2 F | 17 M, 6 F | 17 M, 6 F |
| Race | 10 W, 4 B | 8W,6B | 18 W, 5 B | 15 W, 8 B |
| Age (years) | 42.2 (14.8) | 42.6 (12.3) | 48.0 (15.5) | 47.9 (14.1) |
| PMI (h) | 17.3 (5.4) | 17.4 (8.0) | 18.0 (5.5) | 17.8 (9.3) |
| Brain pH | 6.88 (0.21) | 6.88 (0.20) | 6.87 (0.22) | 6.81 (0.33) |
| RIN | 8.6 (0.4) | 8.4 (0.7) | 8.7 (0.4) | 8.4 (0.7) |
| Storage time (months at −80°C) | 67.4 (24.8) | 73.7 (18.2) | 80.0 (23.6) | 84.3 (23.7) |
Abbreviations: PMI, post-mortem interval; qPCR, real-time quantitative polymerase chain reaction; RIN, RNA integrity number.
Values are mean (±s.d.).
An independent committee of experienced research clinicians made consensus DSM IV (Diagnosis and Statistical Manual of Mental Disorders, fourth edition, 1994) diagnoses for each subject on the basis of medical records and the results of structured interviews conducted with family members of the deceased as described previously.24 All procedures were approved by the University of Pittsburgh’s Committee for the Oversight of Research Involving the Dead and Institutional Review Board for Biomedical Research.
Tissue preparation
The right hemisphere of each brain was blocked coronally, immediately frozen and stored at −80°C as described previously.9 Cryostat sections (20 µm) from the anterior-posterior level corresponding to the middle portion of the superior frontal sulcus were cut serially and collected into tubes containing Trizol reagent (Invitrogen, Carlsbad, CA, USA) for RNA isolation, or mounted on Super frost plus glass slides (VWR International, West Chester, PA, USA) for Nisslstaining or in situ hybridization. The location of DLPFC area 9 was determined from the Nissl-stained sections using cytoarchitectonic criteria as described previously.9 Total RNA was isolated from Trizol homogenates of sections, cleaned by RNeasy columns (Qiagen, Valencia, CA, USA) and RNA integrity was assessed by measuring RIN25 using the Bioanalyzer 2100 (Agilent Technologies, Walbronn, Germany). For all subjects used in this study, RIN was ≥7.0.
DNA microarray
We utilized a customized DNA microarray (Nimble-Gen systems, Madison, WI, USA) containing probes for 85 GABA-related transcripts. The GABA-related transcripts included (1) 11 transcripts selectively expressed in subsets of GABA neurons; (2) 11 transcripts whose protein products are involved in the synthesis, release, uptake or degradation of GABA; (3) 23 transcripts encoding GABA receptor subunits and related proteins and (4) 40 transcripts whose protein products regulate the function of GABA neurons (Supplementary Table 1). For each transcript, five single-stranded 60-mer DNA probes were synthesized by in situ maskless photolithographic printing. Each of these five independent probes, which were nonoverlapping and biased toward the poly-A tail, were synthesized four times in each of five blocks, resulting in 100 measures per transcript per array (Figure 1a). Probes for more than 800 control transcripts were also printed in each array for global normalization.26
Figure 1.
DNA microarray experiment. Expression levels of each mRNA were assessed by five nonoverlapping 60-mer DNA probes, each of which was synthesized four times in each of five blocks, resulting in 100 independent probe measurement areas per transcript per array (a). Pair-wise comparisons of the expression levels of both mitochondrial ATP synthase F0 subunit 6 (ATP6) and somatostatin (SST) mRNAs are shown in (b) and (c), respectively. For each mRNA, the log2-transformed schizophrenia to control subject ratios are shown for each of the five probes across the 14 subject pairs. The color of each symbol represents data from an individual probe and the relative position of each probe is indicated in panel (a). Horizontal bars at right indicate the mean signal differences between the control and schizophrenia groups determined by each probe. The corresponding fold changes relative to the matched control are shown on the right axis.
Total RNA (7 µg) was reverse-transcribed into double-stranded cDNA using cDNA SuperScript Custom Kit (Invitrogen, Carlsbad, CA, USA) and T7 promoter oligo dT primer. cRNAwas transcribed from cDNA by T7 polymerase in the presence of biotinylated nucleotides (Enzo Life Sciences, Farmingdale, NY, USA) using MEGAscript T7 kit (Ambion, Austin, TX, USA), fragmented into 50–200 bases and hybridized to the custom DNA microarray. Hybridized cRNA was detected by Cy3-streptavidin (Amersham Biosciences, Piscataway, NJ, USA). Scanned microarray images were segmented and analyzed by a customized proprietary system (NimbleGen systems, Madison, WI, USA). Determination of signal levels and scaling were performed using Robust Multi-array Average.26
For each subject, arrays were run in duplicate, with each duplicate analyzing a sample from a new reverse transcription and cRNA synthesis. The signal levels were averaged across the two runs. For scale linearity, signal levels were log2-transformed and differential expression was established using average log2 ratio (ALR) between control and schizophrenia groups (|ALR| = 1 corresponds to a twofold increase or decrease, |ALR| = 0.585 represents a 1.5-fold change and |ALR| = 0.263 depicts a 1.2-fold alteration in expression).
Signal levels determined for each probe were considered to be different between the schizophrenia and control groups if |ALR| > 0.263, and if the results of a paired t-test for comparing expression levels were P < 0.05. The usage of paired t-test reflects the pairing of control and schizophrenia subjects based on sex, age and PMI, each of which may have linear13 and/or nonlinear27 effects on sample conditions including transcript levels. However, the usage of nonpaired t-tests resulted in exactly same number of probes that met the combined criteria for 9 out of the 10 transcripts (except for GABAA receptor β3 subunit, Table 3). We did not perform Bonferroni’s correction, because of the risk of eliminating the vast majority of true expression differences and producing a large increase in false-negative errors.28 Instead, we employed the combination of magnitude of difference and statistical significance criteria in order to reduce false-positive findings and to eliminate statistically significant, but very small, expression changes likely to have a marginal biological effect.28–30 When multiple probes met these criteria for a given transcript, the data from the probe detecting the largest |ALR| were reported and used for further analyses.
Table 3.
Transcripts with expression differences between control and schizophrenia subjects by microarray
| Transcript (abbreviation) | Number of probesa |
||||
|---|---|---|---|---|---|
| Accession | P <0.05 | |ALR| > 0.263 P < 0.05 | ALRb | Fold reduction | |
| somatostatin (SST) | NM_001048 | 4 | 4 | −0.665 | 1.59 |
| GABA A receptor, alpha 1 (GABAA α1) | NM_000806 | 1 | 1 | −0.521 | 1.44 |
| GABA A receptor, delta (GABAA δ) | NM_000815 | 3 | 3 | −0.512 | 1.43 |
| neuropeptide Y (NPY) | NM_000905 | 3 | 3 | −0.462 | 1.38 |
| cholecystokinin (CCK) | NM_000729 | 2 | 2 | −0.462 | 1.38 |
| membrane GABA transporter 1 (GAT1) | NM_003042 | 2 | 1 | −0.458 | 1.37 |
| glutamate decarboxylase 67kDa (GAD67) | NM_000817 | 1 | 1 | −0.411 | 1.33 |
| GABA A receptor, gamma 2 (GABAA γ2) | NM_000816 | 3 | 2 | −0.387 | 1.31 |
| GABA A receptor, alpha 4 (GABAA α4) | NM_000809 | 2 | 1 | −0.317 | 1.25 |
| GABA A receptor, beta 3 (GABAA β3) | NM_000814 | 2 | 2 | −0.312 | 1.24 |
Transcripts are listed in the order of magnitude of change.
Number of probes meeting statistical significance and the predetermined combined criteria for magnitude of difference plus statistical significance.
Average log2 schizophrenia/control across 14 pairs. If multiple probes met the combined criteria, the probe with the largest change is reported.
Cluster analysis was performed on log2-transformed signal levels across the 28 subjects using Euclidean distance in Genes@Work developed by IBM.31,32 Correlations among transcript expression changes across the 14 subject pairs were assessed by Pearson’s correlation analysis of log2-transformed schizophrenia to control signal ratios using Bonferroni’s correction.
The effects of potential confounding factors, such as alcohol dependence and/or abuse, treatment with benzodiazepines and/or valproic acid, diagnosis of schizoaffective disorder or the manner of death, on expression changes of each transcript were assessed by comparing log2-transformed schizophrenia to control signal ratios between subject pairs, with or without the presence of these factors in the subjects with schizophrenia at the time of death, using Student’s t-test.
Real-time qPCR
For six of the GABA-related transcripts that met these microarray criteria for altered expression, real-time qPCR analyses30 were performed on DLPFC samples from the same cohort of subjects. Using 50 ng of total RNA, cDNA synthesis by oligo dT primer and SuperScript II reverse transcriptase (Invitrogen) was conducted. All primer pairs used (Supplementary Table 2) exhibited the high amplification efficacy (> 95%) in the standard curve analysis and specific single products in dissociation curve analysis. After primer validation, the comparative threshold cyle (CT) measurement was performed for quantification using SYBR Green I dye (Applied Biosystems, Foster city, CA, USA) and ABI PRISM 7000 (Applied Biosystems) according to the manufacturer’s instructions. Measurement was performed with two independent reverse transcriptions and four replicates for each reverse transcription. Mitochondrial ATP synthase F0 subunit 6 (ATP6) was used as a normalizing transcript30 because all five probes for this transcript showed the most similar and consistent signal levels across all control and schizophrenia subjects in the microarray analysis (Figure 1b). The dCT (CT for target transcripts − CT for ATP6), indicating the relative expression level of the target transcript to ATP6, was used for comparison by paired t-tests. The usage of paired t-test corresponds to the pairing of control and schizophrenia subjects based on sex, age and PMI. However, nonpaired t-tests resulted in the same statistical conclusions for all transcripts. We did not perform Bonferroni’s correction, because this experiment was performed for the confirmation of the decreased expression levels detected by microarray for each transcript and thus, the probability of false-positive error is considered very low.
In situ hybridization
Histological assessment of gene expression was performed by in situ hybridization for SST and CCK mRNAs in 23 subject pairs. In each subject, three sections evenly spaced at 400 µm were used for the assessment of each mRNA. Sections from all subjects were processed together in a given run, and three runs were performed for each mRNA. Templates for synthesis of riboprobes were obtained by PCR. Specific primer sets amplified a 356 bp fragment for SST mRNA and a 394 bp fragment for CCK mRNA, corresponding to bases 112–467 of the human SST mRNA (NM_001048) and bases 192–585 of the human CCK mRNA (NM_000729). Nucleotide sequencing revealed 100% homologies between the amplified fragments and the previously reported sequences. Amplified fragments were subcloned into the plasmid pSTBlue-1 (Novagen, Madison, WI, USA) and antisense and sense probes were transcribed in vitro in the presence of 35S-CTP (Amersham Biosciences, Piscataway, NJ, USA) using T7 or SP6 RNA polymerase.
Hybridization was performed as described previously.13,15 Following fixation with 4% paraformaldehyde in phosphate-buffered saline, the sections were acetylated, dehydrated through a graded ethanol series and defatted in chloroform for 10 min. The sections were then hybridized with 35S-labeled riboprobes (1 × 107 dpm/ml) in a hybridization buffer at 56°C for 16 h. The sections were washed in a solution containing 0.3 M NaCl, 20 mM Tris-HCl (pH 8.0), 1 mM ethylenediaminetetraacetic acid (pH 8.0) and 50% formamide at 63°C, treated with RNase A (20 µg/ml) at 37°C and washed in 0.1X SSC at 67°C. Sections were then dehydrated through a graded ethanol series, airdried and exposed to BioMax MR film (Kodak, Rochester, NY, USA). After exposure to film, sections were coated with NTB2 emulsion (Kodak) diluted 2:1 with water, exposed at 4°C for 18 and 10 days for SST and CCK mRNAs, respectively, developed with D-19 (Kodak) and counterstained with cresyl violet.
Quantification was performed without knowledge of subject diagnosis by random coding of the sections. Transilluminated autoradiographic film images were captured by a video camera under precisely controlled conditions, digitized and analyzed using a Microcomputer Imaging Device (MCID) system (Imaging Research Inc., London, ON, Canada). Images of adjacent sections stained with cresyl violet were also captured and superimposed onto the autoradiographic images to draw contours of the full thickness of the cortex. Optical density was measured within the contours and expressed as nCi/g of tissue by reference to radioactive Carbon-14 standards (ARC Inc., St Louis, MO, USA) exposed on the same film. All cortical optical density measures were corrected by subtracting background measures in the white matter. For each mRNA, the density measures from three sections were averaged before the statistical analysis was conducted. Two analysis of covariance (ANCOVA) models33 were performed to examine the differences in mRNA expression levels. The first model had diagnostic group as a main effect and subject pair as a blocking factor. Because density measures for each mRNA were not normalized, potential differences in RNA integrity were accounted for by including RIN and tissue storage time as covariates in this model.25 The second model, instead of using subject pair as a blocking factor, included all pairing factors (age, sex and PMI) as covariates in addition to RIN and storage time. Because these two ANCOVAs produced similar results for both SST and CCK mRNAs, only the results from the first model are reported. The correlations among mRNA expression changes determined by microarray, qPCR and in situ hybridization were assessed by Pearson’s correlation analysis.
Antipsychotic-exposed monkeys
As described previously,34 experimentally naive, male macaque monkeys (Macaca fasicularis) (LABS of Virginia, Yemassee, SC, USA), 4.5–5.3 years of age, were chronically exposed to twice daily oral doses of haloperidol, olanzapine or placebo (n = 6 monkeys per group) for 17–27 months. The doses of haloperidol and olanzapine were gradually increased until the monkeys were receiving 24–28mg of haloperidol or 11.0–13.2mg of olanzapine per day. The final trough drug plasma levels were within the range associated with clinical efficacy in humans (~1.5 ng/ml for haloperidol and ~15 ng/ml for olanzapine).34 Matched by terminal body weight, monkeys were euthanized in triads (one animal from each group) on the same day. Brains were rapidly removed, and the right frontal lobe was cut into coronal blocks, frozen in isopentane on dry ice and stored at −80°C. Serial coronal sections (16 µm) were cut from the slabs containing the anterior one-third of the principal sulcus and mounted on glass slides. Two sections evenly spaced at 224 µm were hybridized with antisense RNA probes against human GAD67,13 SST, or GABAA receptor α1 subunit mRNAs as described above. For synthesis of a GABAA α1 subunit probe, a 587 bp fragment, corresponding to bases 846–1432 of the human GABAA α1 subunit mRNA (NM_000806), was amplified by PCR and its sequence verified. The optical density for each mRNA was measured in the gray matter of the DLPFC bordered by the cingulate and principal sulci and corrected by subtracting the white matter density measures. Density values of two sections were averaged for each mRNA. The effects of drug exposure on mRNA expression levels were evaluated by a one-way analysis of variance (ANOVA). All studies were carried out in accordance with the NIH Guide for the Care and Use of Laboratory Animals and were approved by the University of Pittsburgh Institutional Animal Care and Use Committee.
Results
Altered expression of GABA-related transcripts
In our customized microarray analysis, five nonover-lapping 60-mer sequences were selected as probes for each transcript (Figure 1a). The expression differences across the 14 subject pairs detected by 5 different probes for SST mRNA, which showed one of the largest decreases in the schizophrenia subjects, are shown in Figure 1c. In general, each of the five probes revealed a similar pattern of expression change within a subject pair. However, the magnitudes of the detected differences varied across the five probes, and one probe for SST mRNA did not meet the combined criteria for decreased expression in schizophrenia. In contrast, the variance among the five probes was much smaller both within and across subject pairs for ATP6 mRNA, a control transcript that showed no expression difference between subject groups (Figure 1b).
The GABA-related transcripts, for which at least one probe met the combined criteria for a difference between schizophrenia and control groups, are listed in Table 3. These 10 transcripts fell into three categories (Figure 2). The first category involved transcripts encoding presynaptic molecules that regulate GABA neurotransmission, namely GAD67 and GAT1. The second category consisted of transcripts encoding neuropeptides, including SST, neuropeptide Y (NPY) and CCK, each of which is expressed and used as a neuromodulator by a subset of GABA neurons. The third category corresponded to different GABAA receptor subunits, including α1, α4, β3, γ2 and δ. As shown in Table 3, SST mRNA exhibited the most robust decrease in the schizophrenia group with four probes meeting the criteria and one of them detecting ALR = −0.67, which corresponds to a 1.6-fold decrease in mRNA level. For each of these 10 GABA-related transcripts, expression levels were decreased in the schizophrenia subject in at least 10 of the 14 subject pairs (Figure 2).
Figure 2.
Altered expression in schizophrenia of 10 GABA-related transcripts as revealed by DNA microarray analyses. Transcripts are classified into three functional groups. The log2-transformed schizophrenia to control subject ratios are shown across 14 subject pairs. The corresponding fold changes are shown on the right axis. For transcripts in which multiple probes met the combined criteria of a difference between control and schizophrenia subjects, the data from the probe that detected the largest average change are presented.
In order to verify the decreased expression in schizophrenia of six GABA-related transcripts representing the three categories, the expression levels of GAD67, SST, NPY, CCK, GABAA α1 and GABAA δ mRNAs were quantified by real-time qPCR in the same 14 subject pairs. We found significant mean decreases in the expression of GAD67 (12%, t26 = −1.8, P = 0.049), SST (44%, t26 = −3.3, P = 0.003), NPY (28%, t26 = − 2.9, P = 0.007), GABAA α1 (42%, t26 = −8.6, P <0.001) and GABAA δ (25%, t26 = −2.6, P = 0.010) mRNAs in the schizophrenia group (Figure 3). For each of these mRNAs, the pattern of pair-wise expression changes in −ddCT (equivalent to log2 ratio of schizophrenia to control levels)35 corresponded well with the pattern shown by the microarray across 14 pairs (compare Figures 2 and 3). In fact, the expression differences detected by the microarray and qPCR analyses were highly correlated across the 14 subject pairs for each of these five transcripts (r = 0.71, P = 0.005 for GAD67 mRNA; r = 0.79, P = 0.001 for SST mRNA; r = 0.71, P = 0.005 for NPY mRNA; r = 0.53, P = 0.053 for GABAA α1 mRNA and r = 0.58, P = 0.029 for GABAA δ mRNA) (Figure 4). For CCK mRNA, in contrast to the microarray findings, qPCR showed a trend for an increase in subjects with schizophrenia (t26 = 1.4, P = 0.095).
Figure 3.
Real-time quantitative PCR verification of the expression changes for six transcripts. For each subject, dCT was obtained by subtracting CT of ATP mRNA from CT of the target mRNA and then, in each pair, ddCT was obtained by subtracting dCT of the control subject from dCT of the schizophrenia subject. The within-pair expression changes are presented in −ddCT for each of the 14 subject pairs and the mean (±s.d.) −ddCT for all subject pairs is shown in the far right of each graph. The corresponding fold and percentage changes are shown on the right axis. The results of paired t-test comparisons of dCT between control and schizophrenia subjects are shown at the top of each graph.
Figure 4.
Correlations among expression changes assessed by DNA microarray, qPCR and in situ hybridization. For GAD67 (left column) and SST (right column) mRNAs, log2-transformed within-pair differences in mRNA levels determined by microarray are plotted against −ddCT determined by qPCR (upper row) and percentage within-pair differences determined by in situ hybridization are plotted against those calculated from −ddCTs (lower row).
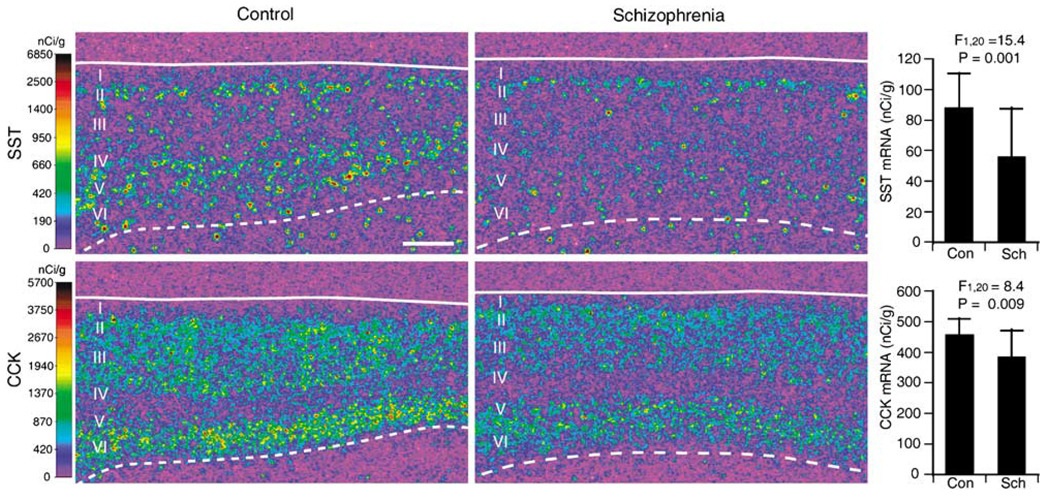
As a further step of verification, and in order to adjudicate the conflicting findings between the microarray and qPCR analyses for CCK mRNA, we conducted quantitative in situ hybridization for SST and CCK mRNAs using 23 subject pairs including 13 pairs (except for pair 7) used in the microarray and qPCR analyses. In DLPFC area 9 of control subjects, SST and CCK mRNAs exhibited distinct laminar patterns of expression (Figure 5). SST mRNA expression was high in layers II and superficial III and in layers IV, V and superficial VI, moderate in deep layer III and deep layer VI and low in layer I. CCK mRNA expression was high in layers II–III and deep layer V and layer VI and low in layers I, IV and superficial V. Microscopic observation of emulsion-coated slides revealed silver grains clustered over Nissl-stained neuronal nuclei for both mRNAs (data not shown). These expression patterns of SST and CCK mRNAs are consistent with previous studies in the human neocortex.36,37 No signal above background was found in sections hybridized with sense RNA probes. The expression levels of SST and CCK mRNAs, as measured in the full thickness of the gray matter, were significantly decreased by 36% (F1,20 = 15.4, P = 0.001) and 15% (F1,20 = 8.4, P = 0.009), respectively, in the schizophrenia group compared to the control group (Figure 5). In addition, our previous study detected a significant 23% decrease (F1,20 = 17.5, P < 0.001) in GAD67 mRNA levels in the schizophrenia subjects in these same 23 subject pairs.13 The percentage expression changes measured by in situ hybridization and qPCR were highly correlated for GAD67 (r = 0.89, P < 0.001) and SST (r = 0.90, P <0.001) mRNAs across the 13 subject pairs common to both studies (Figure 4).
Figure 5.
The expression of SST and CCK mRNAs detected by in situ hybridization. Representative autoradiograms illustrate the expression of SST (upper panels) and CCK (lower panels) mRNAs in DLPFC area 9 of a control subject (left panels) and an age-, sex- and PMI-matched subject with schizophrenia (right panels). The densities of hybridization signals are presented in a pseudocolor manner according to the calibration scales (left). Solid and broken lines indicate the pial surface and the border between gray matter and white matter, respectively. Scale bar = 1 mm. Graphs at the right show the mean (±s.d.) expression levels of SST (upper) and CCK (lower) mRNAs in the control and schizophrenia groups.
Relationship among transcript expression changes
In order to assess the relationships among the expression of 10 GABA-related transcripts with decreased expression levels in schizophrenia, we performed a hierarchical cluster analysis of their expression data obtained by microarray across all 28 subjects. A dendrogram generated by this analysis indicated two major groups (Supplementary Figure). The first group included GAD67, GAT1, SST, NPY and CCK, each of which is expressed by either all or subsets of GABA neurons. The second group consisted of four GABAA receptor subunits, including α1, α4, γ2 and δ. GABAA receptor β3 subunits did not cluster with any of the other transcripts. As the cluster analysis suggested strong coregulation of transcripts within each of the two groups, we next assessed the relationships among their expression changes in schizophrenia by performing secondary correlation analyses across the 14 subject pairs. In the first group, after Bonferroni’s correction, we observed significantly correlated expression changes for GAD67 and SST (r = 0.71, P < 0.005), GAD67 and NPY (r = 0.72, P < 0.004), GAD67 and CCK (r = 0.84, P < 0.001) and SST and NPY (r = 0.81, P < 0.001). Among GABAA receptor subunits, there were significant correlations between α1 and γ2 (r = 0.84, P < 0.001), α1 and δ (r = 0.81, P < 0.001) and γ2 and δ (r = 0.85, P < 0.001).
Potential influence of other factors on GABA-related transcript expression
In order to evaluate the potential effect of long-term exposure to typical or atypical antipsychotic medications, we examined mRNAs representative of the three categories of changed GABA-related transcripts (that is GAD67, SST and GABAA α1) in the DLPFC of monkeys chronically exposed to placebo, haloperidol or olanzapine (n = 6 per group).34 The expression patterns of GAD67, SST and GABAA α1 mRNAs, as revealed by in situ hybridization, did not differ across the groups of monkeys (Figure 6). One-way ANOVA confirmed the absence of an effect of drug exposure on the expression levels of GAD67 (F2,15 = 0.46, P = 0.64), SST (F2,15 = 1.67, P = 0.22) or GABAA α1 (F2,15 = 0.12, P = 0.89) mRNAs.
Figure 6.
The effect of chronic exposure to antipsychotic medications on the expression of GAD67, SST and GABAA receptor α1 subunit mRNAs in the monkey DLPFC. Representative autoradiograms illustrate the expression of GAD67 (upper panels), SST (middle panels) and GABAA α1 (lower panels) mRNAs in the DLPFC of a control (left panels), a haloperidol-exposed (center panels) and an olanzapine-exposed (right panels) monkey. The densities of hybridization signals are presented in a pseudocolor manner according to the calibration scales (left). Solid and broken lines indicate the pial surface and the border between gray matter and white matter, respectively. PS and CS stand for the principal and cingulate sulci, respectively. Scale bar = 1mm. Graphs to the right show the mean (±s.d.) expression levels of GAD67 (upper), SST (middle) and GABAA α1 (lower) mRNAs in the placebo-, haloperidol- and olanzapine-exposed monkey groups.
Because substances such as alcohol, benzodiazepines or valproic acid affect GABA neurotransmission, we assessed whether the usage of these substances by subjects with schizophrenia at the time of death had any effect on the expression changes of the 10 GABA-related transcripts revealed by microarray. For each of these transcripts, no significant differences in log2-tranformed schizophrenia to control signal ratio were detected between pairs with (n = 5) or without (n = 9) alcohol abuse and/or dependence in the schizophrenia subjects, or between pairs with (n = 6) or without (n = 8) the usage of benzodiazepines and/or valproic acid by the schizophrenia subjects (Supplementary Table 3). In addition, no significant differences were observed between pairs with (n = 4) or without (n = 10) the diagnosis of schizoaffective disorder, or between pairs with (n = 3) or without (n = 11) death by suicide in the schizophrenia subjects, for each of the 10 GABA-related transcripts (Supplementary Table 3).
Discussion
In this study, a customized microarray platform revealed expression deficits of multiple GABA-related transcripts in the DLPFC of subjects with schizophrenia. The protein products encoded by these transcripts can be classified into three groups: (1) presynaptic regulators of GABA neurotransmission (GAD67 and GAT1), (2) neuropeptides (SST, NPY, CCK) and (3) GABAA receptor subunits (α1, α4, β3, γ2 and δ). These data, together with previous findings (for review, see Lewis4 and Akbarian and Huang38), provide convergent evidence for altered inhibitory inputs from certain subtypes of GABA neurons via specific GABAA receptors to both the dendritic and perisomatic domains of pyramidal neurons in the DLPFC of subjects with schizophrenia.
These gene expression deficits appear to be related to the disease process of schizophrenia, or at least not a consequence of other factors commonly associated with the illness. The following lines of evidence indicate that these gene expression changes are not attributable to treatment of schizophrenia with anti-psychotics. First, we have previously shown that the expression of GAD67 and GAT1 mRNAs was unaltered in the DLPFC of monkeys chronically exposed to haloperidol and benztropine.9,12 Second, long-term exposure of monkeys to typical (haloperidol) or atypical (olanzapine) antipsychotics did not alter mRNA levels for GAD67, SST or GABAA α1 subunit in the DLPFC (Figure 6). Third, the three subjects with schizophrenia (pairs 2, 9, 13), who were not receiving antipsychotics at the time of death, showed decreased expression for GABA-related transcripts in both microarray and qPCR studies (Figures 2 and 3).
In addition, it is also unlikely that the observed pattern of GABA-related transcript expression changes is caused solely by usage of substances that influence GABA neurotransmission, such as alcohol, benzodiazepines and valproic acid, because we did not observe a significant difference in expression changes for any of the 10 GABA-related transcripts between the pairs with or without comorbid alcohol abuse and/or dependence in the schizophrenia subjects, or between the pairs with or without the use of benzodiazepines and/or valproic acid by schizophrenia subjects, at the time of death (Supplementary Table 3).
Finally, the GABA-related transcript expression changes did not seem to be associated with a diagnosis of schizoaffective disorder or suicide, because the changes did not differ as a function of the presence of these factors in the subjects with schizophrenia (Supplementary Table 3).
Schizophrenia subjects in a subset of the pairs appear to have larger decreases in GABA-related transcript levels than in the other pairs (Figure 2). However, future studies using larger cohorts are necessary to assess if decreased GABA-related transcript expression is specific to a subset of subjects with schizophrenia.
The customized microarray platform used in this study has several advantages compared to previous microarray studies of the DLPFC of subjects with schizophrenia.11,23,39,40 First, the opportunity to select transcripts of interest enabled us to focus on a large number of GABA-related transcripts. Second, the use of five nonoverlapping 60-mer probes provided enhanced specificity and sensitivity for detecting target transcripts and differences in their tissue concentrations.41 Third, the virtual absence of false-positive observations in our data set was demonstrated by the findings that (1) different probes for a given transcript never met the criteria for a difference in opposite directions; (2) among the four transcripts with only one probe meeting the combined criteria for decreased expression in schizophrenia, two transcripts had an additional probe that detected a statistically significant decrease (Table 3); (3) decreased expression levels detected by microarray were verified with real-time qPCR and/or in situ hybridization for all transcripts tested and (4) no expression differences were detected for mRNAs encoding GAD65, CR, or TrkC, whose expression levels were previously reported to be unaltered in the DLPFC of subjects with schizophrenia by other methods.10,13,15 However, false negative errors, which are common in microarray data sets,42,43 are still possible. For example, we did not replicate previously reported alterations for some transcripts, such as PV, BDNF and reelin mRNAs.13–15,44 Therefore, the negative data, without verification by other methods, should not be considered as definitive proof of unaltered transcript expression.
For CCK mRNA, our real-time qPCR failed to confirm the microarray observation of decreased expression. However, we accepted the microarray data for the following reasons. First, in the microarray analysis, two nonoverlapping probes met the combined criteria for decreased expression in schizophrenia subjects (Table 3). Second, in situ hybridization study detected a significant decrease in area 9 of the extended cohort of 23 pairs of control and matched schizophrenia subjects (Figure 5). Third, previous studies reported significant decreases in CCK peptide45 and mRNA levels46 in other frontal areas of schizophrenia subjects. Finally, there may be a limitation in detecting gene expression changes with relatively small magnitudes (<20%) by real-time qPCR.47 These findings demonstrate the value of using multiple methods to uncover true transcript expression differences.28
Our observation of reduced levels of GAD67 and GAT1 mRNAs in the DLPFC of the same subjects with schizophrenia and the correlation between these changes across pairs (r = 0.62, P < 0.018), although not significant after Bonferroni’s correction, are consistent with previous studies indicating that both the synthesis and presynaptic reuptake of GABA are reduced in the subset of GABA neurons that express PV.4,12,15,48 Because a primary reduction in GAT1 does not induce changes in the levels of GAD,49 the downregulation of GAT1, which prolongs the activity of synaptically released GABA,50 is likely to be a compensatory response to decreased GABA synthesis in these neurons.4
The highly significant correlations among the gene expression changes for GAD67, SST and NPY suggest that GAD67 mRNA expression is also decreased in another subset of GABA neurons that express both SST and NPY. In the cortex, SST is expressed by the majority of calbindin-containing GABA neurons, a separate population from those that express PV or CR,16,51,52 and a subset of SST-containing neurons largely overlaps with the majority of NPY-containing neurons.51,53 The localization of SST- and NPY-containing neurons predominantly in layers II and V51,53 (Figure 5) may account for the deficits in GAD67 mRNA expression in these layers, which could not be explained by the expression deficits in PV-containing neurons.9 Because SST- and NPY-containing neurons selectively target distal dendrites of pyramidal neu-rons,53–56 these coordinated gene expression changes suggest that GABA neurotransmission is altered at the dendritic domain of pyramidal neurons in the DLPFC of subjects with schizophrenia. Furthermore, given the functions of SST and NPY as inhibitory neuromodulators,57 their gene expression deficits indicate the presence of additional mechanisms affecting inhibitory regulation of DLPFC circuitry in schizophrenia.
CCK is heavily expressed in GABA neurons that do not contain PV or SST18,58 and is expressed at low levels in some pyramidal neurons.59 The clustering of CCK with GAD67 and their highly correlated expression changes suggest a deficit in GABA synthesis in CCK-containing GABA neurons. The axon terminals of CCK-positive basket neurons converge with those from PV-containing neurons on the perisomatic domain of pyramidal neurons.58 Thus, alterations in GABA regulation on this domain of pyramidal neurons appear to involve at least two subpopulations of GABA neurons in the DLPFC of subjects with schizophrenia.
GABAA receptors containing the α1 and γ2 subunits are enriched in postsynaptic sites where they mediate phasic inhibition.60,61 In contrast, GABAA receptors containing the δ subunit, which is often coassembled with the α4 subunit in the forebrain, are selectively localized to extrasynaptic sites.60–62 These extra-synaptic receptors, which have a high sensitivity to GABA and thus can be activated by ambient GABA molecules in extracellular space, mediate tonic inhibition which reduces the effects of synaptic inputs over time.62,63 Given the predominant localization of the α1, γ2 and δ subunits to dendrites,64,65 the highly correlated expression deficits for these transcripts suggest coordinated downregulation of GABAA receptors mediating phasic and tonic inhibition in the dendritic domain of DLPFC pyramidal neurons in schizophrenia. Because this downregulation is unlikely to be a compensatory response to reduced GABA release, it may represent a primary process in the illness. Consistent with this interpretation, a recent genetic study reported that variants in the GABAA receptor α1 subunit gene were associated with both schizophrenia and altered expression levels of GABAA receptor subunit mRNAs.66 Furthermore, a targeted deletion of the GABAA receptor α1 subunit gene in mice resulted in altered cortical expression of other GABAA receptor subunits, such as increased α2 and decreased γ2 protein levels, as well as decreased levels of SST mRNA.67,68 Together, these findings suggest that the deficit in α1 subunit expression could be an upstream event to other GABA-related gene expression changes in the DLPFC of subjects with schizophrenia.
Although decreased α1 subunit mRNA expression was detected by both microarray and qPCR in this study, two other studies reported increases in the mRNA levels in the DLPFC of subjects with schizophrenia.8,69 This discrepancy might be due to the fact that control and schizophrenia groups were not well matched for factors, such as age, sex and PMI, in these studies. Consistent with this interpretation, another study using 12 pairs of age-, sex- and PMI-matched subjects reported a trend decrease in α1 mRNA levels in schizophrenia.70 However, future studies of independent cohorts are needed to resolve the differences across studies. Previous studies also reported increased muscimol binding in the DLPFC of schizophrenia subjects that was most prominent in pyramidal neuron cell bodies.19,20 However, because muscimol recognizes GABA binding sites in all types of GABAA receptors, decreased expression of some GABAA receptor subunits could be masked by increased expression of other subunits. Indeed, an upregulation of α2 subunit immunoreactivity in the axon initial segment of pyramidal neurons21 and increased α2 mRNA expression in pyramidal neurons71 were observed in the DLPFC of subjects with schizophrenia.
Although establishing the functional significance of our observations requires further verification at the protein level, the results of this study, in concert with previous findings, provide a clearer picture of the nature of altered GABA neurotransmission in the DLPFC of subjects with schizophrenia (Figure 7). Previous studies demonstrated mRNA expression deficits in PV-containing, but not in CR-containing, GABA neurons in the DLPFC of subjects with schizophrenia.9,12,15 These changes were associated with the downregulation of GAT1 in the presynaptic terminals of PV-containing chandelier neurons48 and the upregulation of GABAA receptor α2 subunit in the postsynaptic axon initial segments of pyramidal neurons (Figure 7, lower enlarged square).21 Our current study suggests alterations in GABA neuro-transmission provided by two additional subpopulations of DLPFC GABA neurons: SST- and NPY-containing neurons and CCK-containing basket neurons, which predominately target the distal dendrites and cell bodies of pyramidal neurons, respectively. Furthermore, gene expression deficits for α1 and γ2 GABAA receptor subunits and for δ and α4 subunits suggest decreased synaptic (phasic) and extrasynaptic (tonic) inhibition, respectively, in pyramidal neuron dendrites (Figure 7, upper enlarged square). GABA-mediated regulation at the dendritic domain of pyramidal neurons is important for the selection and integration of excitatory inputs from different cortical and subcortical areas, whereas GABA inputs at the perisomatic domain, including the axon initial segment and cell body, are critical for control of the timing and synchronization of pyramidal neuron firing.72,73 Therefore, the findings summarized in Figure 7 suggest altered GABA-mediated regulation of both inputs to and outputs from DLPFC pyramidal neurons in subjects with schizophrenia. These alterations are certain to affect information processing in DLPFC circuitry and thus are likely to be major contributors to working memory impairments in schizophrenia.
Figure 7.
Schematic summary of alterations in GABA-mediated circuitry in the DLPFC of subjects with schizophrenia. Altered GABA neurotransmission by PV-containing neurons (green) is indicated by gene expression deficits in these neurons and associated changes in their synapses, a decrease in GAT1 expression in their terminals and an upregulation of GABAA receptor α2 subunit at the axon initial segments of pyramidal neurons (lower enlarged square). Decreased expression of both SST and NPY mRNAs indicates alterations in SST and/or NPY-containing neurons (blue) that target the distal dendrites of pyramidal neurons. These changes appear to be accompanied by a down-regulation of GABAA receptor subunits, including the α1 and γ2 subunits present in receptors that mediate synaptic (phasic) inhibition and the α4 and δ subunits present in receptors that mediate extrasynaptic (tonic) inhibition (upper enlarged square), in dendrites of pyramidal neurons. Decreased CCK mRNA levels indicate an alteration of CCK-containing large basket neurons (purple) that represent a separate source of perisomatic inhibition from PV-containing neurons. Gene expression in CR-containing GABA neurons (red) does not seem to be altered. Other neurons, such as PV-containing basket neurons, are not shown because the nature of their involvement in schizophrenia is unclear. G, generic GABA neuron; P, pyramidal neuron; I–IV, layers of the DLPFC.
Supplementary Material
Acknowledgments
This work was supported by NIH grants MH43784 (DAL), MH45156 (DAL), MH067234 (KM) and MH070786 (KM), by an investigator-initiated grant from Eli Lilly (DAL) and by a NARSAD Young Investigator Award (TH). We thank Ms Mary L Brady and Ms Jimette Gilmartin for their assistance in preparing figures and the manuscript.
Footnotes
Supplementary Information accompanies the paper on the Molecular Psychiatry Web site (http://www.nature.com/mp)
References
- 1.Weinberger DR, Berman KF, Zec RF. Physiologic dysfunction of dorsolateral prefrontal cortex in schizophrenia. I. Regional cerebral blood flow evidence. Arch Gen Psychiatry. 1986;43:114–124. doi: 10.1001/archpsyc.1986.01800020020004. [DOI] [PubMed] [Google Scholar]
- 2.Goldman-Rakic PS. Working memory dysfunction in schizophrenia. J Neuropsychiatry Clin Neurosci. 1994;6:348–357. doi: 10.1176/jnp.6.4.348. [DOI] [PubMed] [Google Scholar]
- 3.Elvevåg B, Goldberg TE. Cognitive impairment in schizophrenia is the core of the disorder. Crit Rev Neurobiol. 2000;14:1–21. [PubMed] [Google Scholar]
- 4.Lewis DA, Hashimoto T, Volk DW. Cortical inhibitory neurons and schizophrenia. Nat Rev Neurosci. 2005;6:312–324. doi: 10.1038/nrn1648. [DOI] [PubMed] [Google Scholar]
- 5.Sawaguchi T, Matsumura M, Kubota K. Delayed response deficits produced by local injection of bicuculline into the dorsolateral prefrontal cortex in Japanese macaque monkeys. Exp Brain Res. 1989;75:457–469. doi: 10.1007/BF00249897. [DOI] [PubMed] [Google Scholar]
- 6.Rao SG, Williams GV, Goldman-Rakic PS. Destruction and creation of spatial tuning by disinhibition: GABAA blockade of prefrontal cortical neurons engaged by working memory. J Neurosci. 2000;20:485–494. doi: 10.1523/JNEUROSCI.20-01-00485.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Akbarian S, Kim JJ, Potkin SG, Hagman JO, Tafazzoli A, Bunney WE, Jr, et al. Gene expression for glutamic acid decarboxylase is reduced without loss of neurons in prefrontal cortex of schizophrenics. Arch Gen Psychiatry. 1995;52:258–266. doi: 10.1001/archpsyc.1995.03950160008002. [DOI] [PubMed] [Google Scholar]
- 8.Ohnuma T, Augood SJ, Arai H, McKenna PJ, Emson PC. Measurement of GABAergic parameters in the prefrontal cortex in schizophrenia: focus on GABA content, GABAA receptor a-1 subunit messenger RNA and human GABA transporter-1 (HGAT-1) messenger RNA expression. Neuroscience. 1999;93:441–448. doi: 10.1016/s0306-4522(99)00189-x. [DOI] [PubMed] [Google Scholar]
- 9.Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. Decreased glutamic acid decarboxylase67 messenger RNA expression in a subset of prefrontal cortical gamma-aminobutyric acid neurons in subjects with schizophrenia. Arch Gen Psychiatry. 2000;57:237–245. doi: 10.1001/archpsyc.57.3.237. [DOI] [PubMed] [Google Scholar]
- 10.Guidotti A, Auta J, Davis JM, Gerevini VD, Dwivedi Y, Grayson DR, et al. Decrease in reelin and glutamic acid decarboxylase67 (GAD67) expression in schizophrenia and bipolar disorder. Arch Gen Psychiatry. 2000;57:1061–1069. doi: 10.1001/archpsyc.57.11.1061. [DOI] [PubMed] [Google Scholar]
- 11.Mirnics K, Middleton FA, Marquez A, Lewis DA, Levitt P. Molecular characterization of schizophrenia viewed by microarray analysis of gene expression in prefrontal cortex. Neuron. 2000;28:53–67. doi: 10.1016/s0896-6273(00)00085-4. [DOI] [PubMed] [Google Scholar]
- 12.Volk DW, Austin MC, Pierri JN, Sampson AR, Lewis DA. GABA transporter-1 mRNA in the prefrontal cortex in schizophrenia: decreased expression in a subset of neurons. Am J Psychiatry. 2001;158:256–265. doi: 10.1176/appi.ajp.158.2.256. [DOI] [PubMed] [Google Scholar]
- 13.Hashimoto T, Bergen SE, Nguyen QL, Xu B, Monteggia LM, Pierri JN, et al. Relationship of brain-derived neurotrophic factor and its receptor TrkB to altered inhibitory prefrontal circuitry in schizophrenia. J Neurosci. 2005;25:372–383. doi: 10.1523/JNEUROSCI.4035-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Torrey EF, Barci BM, Webster MJ, Bartko JJ, Meador-Woodruff JH, Knable MB. Neurochemical markers for schizophrenia, bipolar disorder, and major depression in postmortem brains. Biol Psychiatry. 2005;57:252–260. doi: 10.1016/j.biopsych.2004.10.019. [DOI] [PubMed] [Google Scholar]
- 15.Hashimoto T, Volk DW, Eggan SM, Mirnics K, Pierri JN, Sun Z, et al. Gene expression deficits in a subclass of GABA neurons in the prefrontal cortex of subjects with schizophrenia. J Neurosci. 2003;23:6315–6326. doi: 10.1523/JNEUROSCI.23-15-06315.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Condé F, Lund JS, Jacobowitz DM, Baimbridge KG, Lewis DA. Local circuit neurons immunoreactive for calretinin, calbindin D-28k, or parvalbumin in monkey prefrontal cortex: distribution and morphology. J Comp Neurol. 1994;341:95–116. doi: 10.1002/cne.903410109. [DOI] [PubMed] [Google Scholar]
- 17.Lewis DA, Campbell MJ, Morrison JH. An immunohistochemical characterization of somatostatin-28 and somatostatin-28 (1–12) in monkey prefrontal cortex. J Comp Neurol. 1986;248:1–18. doi: 10.1002/cne.902480102. [DOI] [PubMed] [Google Scholar]
- 18.Lund JS, Lewis DA. Local circuit neurons of developing and mature macaque prefrontal cortex: Golgi and immunocytochemical characteristics. J Comp Neurol. 1993;328:282–312. doi: 10.1002/cne.903280209. [DOI] [PubMed] [Google Scholar]
- 19.Hanada S, Mita T, Nishino N, Tanaka C. 3H]Muscimol binding sites increased in autopsied brains of chronic schizophrenics. Life Sci. 1987;40:239–266. doi: 10.1016/0024-3205(87)90341-9. [DOI] [PubMed] [Google Scholar]
- 20.Benes FM, Vincent SL, Marie A, Khan Y. Up-regulation of GABA-A receptor binding on neurons of the prefrontal cortex in schizophrenic subjects. Neuroscience. 1996;75:1021–1031. doi: 10.1016/0306-4522(96)00328-4. [DOI] [PubMed] [Google Scholar]
- 21.Volk DW, Pierri JN, Fritschy J-M, Auh S, Sampson AR, Lewis DA. Reciprocal alterations in pre- and postsynaptic inhibitory markers at chandelier cell inputs to pyramidal neurons in schizophrenia. Cereb Cortex. 2002;12:1063–1070. doi: 10.1093/cercor/12.10.1063. [DOI] [PubMed] [Google Scholar]
- 22.Huntsman MM, Tran BV, Potkin SG, Bunney WE, Jones EG. Altered ratios of alternatively spliced long and short gamma 2 subunit mRNAs of the gamma-amino butyrate type A receptor in prefrontal cortex of schizophrenics. Proc Natl Acad Sci USA. 1998;95:15066–15071. doi: 10.1073/pnas.95.25.15066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Vawter MP, Crook JM, Hyde TM, Kleinman JE, Weinberger DR, Becker KG, et al. Microarray analysis of gene expression in the prefrontal cortex in schizophrenia: a preliminary study. Schizophr Res. 2002;58:11–20. doi: 10.1016/s0920-9964(01)00377-2. [DOI] [PubMed] [Google Scholar]
- 24.Glantz LA, Lewis DA. Decreased dendritic spine density on prefrontal cortical pyramidal neurons in schizophrenia. Arch Gen Psychiatry. 2000;57:65–73. doi: 10.1001/archpsyc.57.1.65. [DOI] [PubMed] [Google Scholar]
- 25.Imbeaud S, Graudens E, Boulanger V, Barlet X, Zaborski P, Eveno E, et al. Towards standardization of RNA quality assessment using user-independent classifiers of microcapillary electrophoresis traces. Nucleic Acids Res. 2005;33:1–12. doi: 10.1093/nar/gni054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Irizarry RA, Hobbs B, Collin F, Beazer-Barclay YD, Antonellis KJ, Scherf U, et al. Exploration, normalization, and summaries of high density oligonucleotide array probe level data. Biostatistics. 2003;4:249–264. doi: 10.1093/biostatistics/4.2.249. [DOI] [PubMed] [Google Scholar]
- 27.Hayes TL, Cameron JL, Fernstrom JD, Lewis DA. A comparative analysis of the distribution of prosomatostatin-derived peptides in human and monkey neocortex. J Comp Neurol. 1991;303:584–599. doi: 10.1002/cne.903030406. [DOI] [PubMed] [Google Scholar]
- 28.Mirnics K, Pevsner J. Progress in the use of microarray technology to study the neurobiology of disease. Nat Neurosci. 2004;7:434–439. doi: 10.1038/nn1230. [DOI] [PubMed] [Google Scholar]
- 29.Unger T, Korade Z, Lazarov O, Terrano D, Sisodia SS, Mirnics K. True and false discovery in DNA microarray experiments: transcriptome changes in the hippocampus of presenilin 1 mutant mice. Methods. 2005;37:261–273. doi: 10.1016/j.ymeth.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 30.Glorioso C, Sabatini M, Unger T, Hashimoto T, Monteggia LM, Lewis DA, et al. Specificity and timing of neocortical transcriptome changes in response to BDNF gene ablation during embryogenesis or adulthood. Mol Psychiatry. 2006;11:633–648. doi: 10.1038/sj.mp.4001835. [DOI] [PubMed] [Google Scholar]
- 31.Lepre J, Rice JJ, Tu Y, Stolovitzky G. Genes@Work: an efficient algorithm for pattern discovery and multivariate feature selection in gene expression data. Bioinformatics. 2004;20:1033–1044. doi: 10.1093/bioinformatics/bth035. [DOI] [PubMed] [Google Scholar]
- 32.Stolovitzky GA, Kundaje A, Held GA, Duggar KH, Haudenschild CD, Zhou D, et al. Statistical analysis of MPSS measurements: application to the study of LPS-activated macrophage gene expression. Proc Natl Acad Sci USA. 2005;102:1402–1407. doi: 10.1073/pnas.0406555102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Neter J, Kutner MH, Nachtsheim CJ, Wasserman W. Applied Linear Statistical Models. 4th edn. McGraw-Hill: Boston; 1996. [Google Scholar]
- 34.Dorph-Petersen K-A, Pierri JN, Perel JM, Sun Z, Sampson AR, Lewis DA. The influence of chronic exposure to antipsychotic medications on brain size before and after tissue fixation: a comparison of haloperidol and olanzapine in macaque monkeys. Neuropsychopharm. 2005;30:1649–1661. doi: 10.1038/sj.npp.1300710. [DOI] [PubMed] [Google Scholar]
- 35.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 36.Dournaud P, Cervera-Pierot P, Hirsch E, Javoy-Agid F, Kordon C, Agid Y, et al. Somatostatin messenger RNA-containing neurons in Alzheimer’s disease: an in situ hybridization study in hippocampus, parahippocampal cortex and frontal cortex. Neuroscience. 1994;61:755–764. doi: 10.1016/0306-4522(94)90399-9. [DOI] [PubMed] [Google Scholar]
- 37.Bachus SE, Hyde TM, Herman MM, Egan MF, Kleinman JE. Abnormal cholesystokinin mRNA levels in entorhinal cortex of schizophrenics. J Psychiatry Res. 1997;31:233–256. doi: 10.1016/s0022-3956(96)00041-6. [DOI] [PubMed] [Google Scholar]
- 38.Akbarian S, Huang HS. Molecular and cellular mechanisms of altered GAD1/GAD67 expression in schizophrenia and related disorders. Brain Res Brain Res Rev. 2006;52:293–304. doi: 10.1016/j.brainresrev.2006.04.001. [DOI] [PubMed] [Google Scholar]
- 39.Hakak Y, Walker JR, Li C, Wong WH, Davis KL, Buxbaum JD, et al. Genome-wide expression analysis reveals dysregulation of myeli-nation-related genes in chronic schizophrenia. Proc Natl Acad Sci USA. 2001;98:4746–4751. doi: 10.1073/pnas.081071198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mimmack ML, Ryan M, Baba H, Navarro-Ruiz J, Iritani S, Faull RL, et al. Gene expression analysis in schizophrenia: reproducible up-regulation of several members of the apolipoprotein L family located in a high-susceptibility locus for schizophrenia on chromosome 22. Proc Natl Acad Sci USA. 2002;99:4680–4685. doi: 10.1073/pnas.032069099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hughes TR, Mao M, Jones AR, Burchard J, Marton MJ, Shannon KW, et al. Expression profiling using microarrays fabricated by an ink-jet oligonucleotide synthesizer. Nat Biotechnol. 2001;19:342–347. doi: 10.1038/86730. [DOI] [PubMed] [Google Scholar]
- 42.Hollingshead D, Lewis DA, Mirnics K. Platform influence on DNA microarray data in postmortem brain research. Neurobiol Dis. 2005;18:649–655. doi: 10.1016/j.nbd.2004.10.020. [DOI] [PubMed] [Google Scholar]
- 43.Mirnics K, Levitt P, Lewis DA. Critical appraisal of DNA microarrays in psychiatric genomics. Biol Psychiatry. 2006;60:163–176. doi: 10.1016/j.biopsych.2006.02.003. [DOI] [PubMed] [Google Scholar]
- 44.Weickert CS, Hyde TM, Lipska BK, Herman MM, Weinberger DR, Kleinman JE. Reduced brain-derived neurotrophic factor in prefrontal cortex of patients with schizophrenia. Mol Psychiatry. 2003;8:592–610. doi: 10.1038/sj.mp.4001308. [DOI] [PubMed] [Google Scholar]
- 45.Gabriel SM, Davidson M, Haroutunian V, Powchik P, Bierer LM, Purohit DP, et al. Neuropeptide deficits in schizophrenia vs. Alzheimer’s disease cerebral cortex. Biol Psychiatry. 1996;39:82–91. doi: 10.1016/0006-3223(95)00066-6. [DOI] [PubMed] [Google Scholar]
- 46.Virgo L, Humphries C, Mortimer A, Barnes T, Hirsch SR, de Belleroche J. Cholecystokinin messenger RNA deficit in frontal and temporal cerebral cortex in schizophrenia. Biol Psychiatry. 1995;37:694–701. doi: 10.1016/0006-3223(94)00206-I. [DOI] [PubMed] [Google Scholar]
- 47.Ding C, Cantor CR. Quantitative analysis of nucleic acids - the last few years of progress. J Biochem Mol Biol. 2004;37:1–10. doi: 10.5483/bmbrep.2004.37.1.001. [DOI] [PubMed] [Google Scholar]
- 48.Pierri JN, Chaudry AS, Woo T-U, Lewis DA. Alterations in chandelier neuron axon terminals in the prefrontal cortex of schizophrenic subjects. Am J Psychiatry. 1999;156:1709–1719. doi: 10.1176/ajp.156.11.1709. [DOI] [PubMed] [Google Scholar]
- 49.Jensen K, Chiu CS, Sokolova I, Lester HA, Mody I. GABA transporter-1 (GAT1)-deficient mice: differential tonic activation of GABAA versus GABAB receptors in the hippocampus. J Neurophysiol. 2003;90:2690–2701. doi: 10.1152/jn.00240.2003. [DOI] [PubMed] [Google Scholar]
- 50.Overstreet LS, Westbrook GL. Synapse density regulates independence at unitary inhibitory synapses. J Neurosci. 2003;23:2618–2626. doi: 10.1523/JNEUROSCI.23-07-02618.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kubota Y, Hattori R, Yui Y. Three distinct subpopulations of GABAergic neurons in rat frontal agranular cortex. Brain Res. 1994;649:159–173. doi: 10.1016/0006-8993(94)91060-x. [DOI] [PubMed] [Google Scholar]
- 52.González-Albo MC, Elston GN, DeFelipe J. The human temporal cortex: characterization of neurons expressing nitric oxide synthase, neuropeptides and calcium-binding proteins, and their glutamate receptor subunit profiles. Cereb Cortex. 2001;11:1170–1181. doi: 10.1093/cercor/11.12.1170. [DOI] [PubMed] [Google Scholar]
- 53.Hendry SHC, Jones EG, Emson PC. Morphology, distribution, and synaptic relations of somatostatin- and neuropeptide Y-immuno-reactive neurons in rat and monkey neocortex. J Neurosci. 1984;4:2497–2517. doi: 10.1523/JNEUROSCI.04-10-02497.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.DeLima AD, Morrison JH. Ultrastructural analysis of somatostatin-immunoreactive neurons and synapses in the temporal and occipital cortex of the macaque monkey. J Comp Neurol. 1989;283:212–227. doi: 10.1002/cne.902830205. [DOI] [PubMed] [Google Scholar]
- 55.Kawaguchi Y, Kubota Y. Physiological and morphological identification of somatostatin- or vasoactive intestinal polypeptide-containing cells among GABAergic cell subtypes in rat frontal cortex. J Neurosci. 1996;16:2701–2715. doi: 10.1523/JNEUROSCI.16-08-02701.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Melchitzky DS, Lewis DA. Synaptic targets of somatostatin-labeled axon terminals in monkey prefrontal cortex. Soc Neurosci Abstr. 2005;675:6. doi: 10.1016/j.neuroscience.2004.08.046. [DOI] [PubMed] [Google Scholar]
- 57.Baraban SC, Tallent MK. Interneuron diversity series: interneuronal neuropeptides - endogenous regulators of neuronal excitability. Trends Neurosci. 2004;27:135–142. doi: 10.1016/j.tins.2004.01.008. [DOI] [PubMed] [Google Scholar]
- 58.Kawaguchi Y, Kondo S. Parvalbumin, somatostatin and cholecystokinin as chemical markers for specific GABAergic interneuron types in the rat frontal cortex. J Neurocytol. 2002;31:277–287. doi: 10.1023/a:1024126110356. [DOI] [PubMed] [Google Scholar]
- 59.Schiffmann SN, Vanderhaeghen JJ. Distribution of cells containing mRNA encoding cholecystokinin in the rat central nervous system. J Comp Neurol. 1991;304:219–233. doi: 10.1002/cne.903040206. [DOI] [PubMed] [Google Scholar]
- 60.Nusser Z, Sieghart W, Somogyi P. Segregation of different GABAA receptors to synaptic and extrasynaptic membranes of cerebellar granule cells. J Neurosci. 1998;18:1693–1703. doi: 10.1523/JNEUROSCI.18-05-01693.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mangan PS, Sun C, Carpenter M, Goodkin HP, Sieghart W, Kapur J. Cultured hippocampal pyramidal neurons express two kinds of GABAA receptors. Mol Pharmacol. 2005;67:775–788. doi: 10.1124/mol.104.007385. [DOI] [PubMed] [Google Scholar]
- 62.Wei W, Zhang N, Peng Z, Houser CR, Mody I. Perisynaptic localization of delta subunit-containing GABA(A) receptors and their activation by GABA spillover in the mouse dentate gyrus. J Neurosci. 2003;23:10650–10661. doi: 10.1523/JNEUROSCI.23-33-10650.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Farrant M, Nusser Z. Variations on an inhibitory theme: phasic and tonic activation of GABA(A) receptors. Nat Rev Neurosci. 2005;6:215–229. doi: 10.1038/nrn1625. [DOI] [PubMed] [Google Scholar]
- 64.Hendry SHC, Huntsman MM, Viñuela A, Mohler H, de Blas AL, Jones EG. GABAA receptor subunit immunoreactivity in primate visual cortex: distribution in macaques and humans and regulation by visual input in adulthood. J Neurosci. 1994;14:2383–2401. doi: 10.1523/JNEUROSCI.14-04-02383.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Fritschy J-M, Mohler H. GABAA-receptor heterogeneity in the adult rat brain: differential regional and cellular distribution of seven major subunits. J Comp Neurol. 1995;359:154–194. doi: 10.1002/cne.903590111. [DOI] [PubMed] [Google Scholar]
- 66.Petryshen TL, Middleton FA, Tahl AR, Rockwell GN, Purcell S, Aldinger KA, et al. Genetic investigation of chromosome 5q GABAA receptor subunit genes in schizophrenia. Mol Psychiatry. 2005;10:1074–1088. doi: 10.1038/sj.mp.4001739. 1057. [DOI] [PubMed] [Google Scholar]
- 67.Kralic JE, Korpi ER, O’Buckley TK, Homanics GE, Morrow AL. Molecular and pharmacological characterization of GABA(A) receptor alpha1 subunit knockout mice. J Pharmacol Exp Ther. 2002;302:1037–1045. doi: 10.1124/jpet.102.036665. [DOI] [PubMed] [Google Scholar]
- 68.Ponomarev I, Maiya R, Harnett MT, Schafer GL, Ryabinin AE, Blednov YA, et al. Transcriptional signatures of cellular plasticity in mice lacking the alpha1 subunit of GABAA receptors. J Neurosci. 2006;26:5673–5683. doi: 10.1523/JNEUROSCI.0860-06.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Impagnatiello F, Guidotti AR, Pesold C, Dwivedi Y, Caruncho H, Pisu MG, et al. A decrease of reelin expression as a putative vulnerability factor in schizophrenia. Proc Natl Acad Sci USA. 1998;95:15718–15723. doi: 10.1073/pnas.95.26.15718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Akbarian S, Huntsman MS, Kim JJ, Tafazzoli A, Potkin SG, Bunney WE, Jr, et al. GABAA receptor subunit gene expression in human prefrontal cortex: comparison of schizophrenics and controls. Cereb Cortex. 1995;5:550–560. doi: 10.1093/cercor/5.6.550. [DOI] [PubMed] [Google Scholar]
- 71.Kim AM, Matzilevich DA, Walsh JP, Benes FM, Woo TW. Parvalbumin-containing neurons and disturbances of prefrontal cortical circuitry in schizophrenia. Soc Neurosci Abstr. 2005;912:1. [Google Scholar]
- 72.Markram H, Toledo-Rodriguez M, Wang Y, Gupta A, Silberberg G, Wu C. Interneurons of the neocortical inhibitory system. Nat Rev Neurosci. 2004;5:793–807. doi: 10.1038/nrn1519. [DOI] [PubMed] [Google Scholar]
- 73.Somogyi P, Klausberger T. Defined types of cortical interneurone structure space and spike timing in the hippocampus. J Physiol. 2005;562:9–26. doi: 10.1113/jphysiol.2004.078915. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.