Abstract
Differences in immunological response among vaccine recipients are determined both by their genetic differences and environmental factors. Knowledge of genetic determinants of immunological response to a vaccine can be used to design a vaccine that circumvents immunogenetic restrictions. The currently available vaccine for typhoid is a pure polysaccharide vaccine, immune response to which is T-cell independent. Little is known about whether genetic variation among vaccinees associates with variation in their antibody response to a polysaccharide vaccine. We conducted a study on 1,000 individuals resident in an area at high-risk for typhoid; vaccinated them with the typhoid vaccine, measured their antibody response to the vaccine, assayed >2,000 curated SNPs chosen from 283 genes that are known to participate in immune-response; and analyzed these data using a strategy to (a) minimize the statistical problems associated with testing of multiple hypotheses, and (b) internally cross-validate inferences, using a half-sample design, with little loss of statistical power. The first stage analysis, using the first half-sample, identified 54 SNPs in 43 genes to be significantly associated with immune response. In the second-stage, these inferences were cross-validated using the second half-sample. First-stage results of only 8 SNPs (out of 54) in 7 genes (out of 43) were cross-validated. We tested additional SNPs in these 7 genes, and found 8 more SNPs to be significantly associated. Haplotypes constructed with these SNPs in these 7 genes also showed significant association. These 7 genes are DEFB1, TLR1, IL1RL1, CTLA4, MAPK8, CD86 and IL17D. The overall picture that has emerged from this study is that (a) immune response to polysaccharide antigens is qualitatively different from that to protein antigens, and (b) polymorphisms in genes involved in polysaccharide recognition, signal transduction, inhibition of T-cell proliferation, pro-inflammatory signaling and eventual production of antimicrobial peptides are associated with antibody response to the polysaccharide vaccine for typhoid.
Electronic supplementary material
The online version of this article (doi:10.1007/s11568-010-9134-1) contains supplementary material, which is available to authorized users.
Keywords: Polysaccharide antigen, Antibody, Immune system, Half-sample design, Single nucleotide polymorphism, Association, Non-cognate T-cell
Introduction
Vaccination is one of the most effective public health tools available to prevent and control the spread of infectious diseases. However, the immune response induced by a vaccine often varies between individuals, implying that a vaccine may not afford equal protection to all vaccinees. The extent of variation in immune response has been extensively studied for some vaccines. Primary failure, as assessed by post-vaccination antibody levels, occurs in a significant proportion of vaccinees; for example, 2–10% for measles vaccine (Poland 1998; Poland et al. 1999), 5–20% for hepatitis B vaccine (Milich and Leroux-Roels 2003; Zuckerman 1996), and 23–40% for typhoid vaccine (Gupta et al. 2008; Sur et al. 2009).
Immunological mechanisms that are responsible for protection against pathogens are often unknown. Measuring clinical or microbiological protection conferred by a vaccine is best done by an artificial or a natural challenge, which is rarely possible for ethical or practical reasons. Vaccine responses are, therefore, commonly monitored by measuring humoral antibody responses and sometimes cell-mediated immune responses. Differences in antibody response (AR) among vaccinees are partly due to environmental factors, such as presence of maternal antibodies, nutrition—in particular vitamin A deficiency—or other infections, and partly due to genetic differences among the vaccinees (Kimman et al. 2007; Poland and Jacobson 1998; van Lovern et al. 2001). Twin studies have shown high and statistically significant heritability for AR to measles (89%), mumps (39%), rubella (46%), hepatitis B (61–77%), oral polio (60%), tetanus (44–64%), diphtheria (49%), Haemophilus influenzae type B [Hib] (51%), and other vaccines (Kimman et al. 2007). Past linkage and association studies conducted to study genomics of vaccine-induced AR have revealed many interesting results. Polymorphic variants in HLA genes have been found to be associated or linked with AR to hepatitis B vaccine (Kruskall et al. 1992), influenza vaccine (Gelder et al. 2002; Poland et al. 2008a), measles vaccine (Poland 1998; Poland et al. 2008b), and BCG tuberculosis vaccine (van Eden et al. 1983), among others. However, non-MHC or non-HLA genes—IL1B, IL4R, IL6, IL10 and TNF—have also been found to be independently associated with response to hepatitis B vaccine (Wang et al. 2004). Interestingly, although measles, mumps and rubella vaccines are all attenuated live vaccines that are administered simultaneously, the responses induced by these vaccines are influenced differently by host genetics (Kimman et al. 2007).
Vaccine immunogenicity is determined in part by the chemical and physical nature of microbial antigens and adjuvants, and also by the genetic make-up of vaccine recipients (Pulendran et al. 2001). The rationale and value of so-called “immunogenetic profiling” has been reviewed (Jin and Wang 2003); knowledge of key immunogenetic associations can be used to design a vaccine that circumvents immunogenetic restrictions. Most available vaccines comprise microbial or added protein components. Insight into the impact of genetic variation in vaccinees on AR has also been obtained in some detail (Kimman et al. 2007) for these vaccines.
Another class of vaccines is polysaccharide (PS) vaccines. PS vaccines are a unique type of inactivated subunit vaccine composed of long chains of sugar molecules that make up the surface capsule of certain bacteria, such as Salmonella enterica serotype typhi (Salmonella typhi). Currently, pure PS vaccines that are available include pneumococcal, meningococcal and typhoid vaccines. The essential feature of immune response to a pure PS vaccine is that it is typically T-cell independent, which means that a PS vaccine stimulates B-cells and non-cognate T-cells without the assistance of cognate T cells (Vos et al. 2000; Snapper 2006). Little is known about the role of genetic variation in vaccinees on antibody response to PS vaccines.
Worldwide, typhoid fever affects 17 million people annually, causing 600,000 deaths. It is caused by S. typhi that is an obligate parasite and has no natural reservoir outside of human. It is a gram-negative enteric bacillus and multi-organ parasite that inhabits the lymphatic tissues of small intestine, liver, spleen and bloodstream of infected individuals. Over 100 strains of this parasite—that differ in levels of virulence—have been isolated. It enters uninfected individuals via the fecal-oral route from infected individuals. Therefore, individuals living under unhygienic conditions—particularly in areas with open sewage, public latrines and unclean drinking water—are most vulnerable to typhoid infection and therefore in the greatest need of vaccination. Salmonella typhimurium, that causes murine typhoid, causes enteritis in humans characterized by self-limited fever and diarrhea and, in some cases, dysentery; symptoms that are rarely observed with S. typhi infection. S. typhi has an outer capsule, Vi polysaccharide, that is absent in S. typhimurium. Vi is a polymer of α-1,4-galacturonic acid with an N-acetyl at position C-2 and variable O-acetylation at C-3 (Szu et al. 1991). The expression of this molecule correlates with virulence, resistance to phagocytosis and resistance to complement-mediated killing (Looney and Steigbigel 1986). Therefore, the Vi PS has been used as a vaccine to protect individuals from S. typhi infection. The objective of this study was to quantify the extent of variation in antibody response to a widely used Vi PS vaccine for typhoid and to discover associations of polymorphisms in candidate genes of vaccinees with antibody response. To our knowledge, this is the first large-scale study on genomics of immune response of a polysaccharide vaccine.
Materials and methods
Institutional ethical approvals were taken from all collaborating institutions before initiation of this study. Written informed consent was taken from each recruited participant. Approval of the Drug Controller General of India was taken before administration of the vaccine to the study participants. Dispatch of aliquots of serum samples collected in this study to the collaborating laboratories in the USA was approved by the Health Ministry Screening Committee, Government of India.
Study participants
Individuals (n = 1,000), unrelated at least to the first-cousin level, aged 12 years or older, inhabiting a socio-economically depressed locality of Kolkata (formerly, Calcutta), India, were recruited into this study. The residents belonged to two maritally isolated, religious groups, Muslim and Hindu. The Muslims of this locality are mostly religious converts, during the last 100 years, to Islam from Hinduism. The individuals recruited into this study are demographically (age-group, gender and religion) representative of the entire locality, since care was taken to recruit into age × gender × religion subgroups by a probability proportional to size sampling scheme, with population sizes of subgroups having been determined by us through an initial complete-enumeration demographic survey of the locality (comprising ~9,950 individuals). The following exclusionary criteria were used, based on self-report, to recruit an individual into this study: that (a) she/he had not had fever lasting for more than three consecutive days in the past 1 year, and (b) she was not pregnant or lactating.
Vaccination and collection of blood samples
Each study participant was injected, intramuscularly, with a 0.5 ml single-dose injectable vaccine, containing 25 μg of the cell surface Vi polysaccharide extracted from S. typhi Ty2 strain with the excipients sodium chloride, sodium dihydrogen phosphate dihydrate, disodium phosphate dihydrate, phenol and water for injection. This vaccine is marketed in India as Typherix® (manufactured by GlaxoSmithKline, Inc.), and is widely used in Asia, Africa and South America, and approved for use elsewhere, including the European Union and the USA.
From each study participant, a blood sample was collected immediately prior to vaccination. Blood samples were also collected 3- and 28-days post-vaccination. Serum and DNA were isolated from these samples, using standard protocols and QiaGen columns.
Antibody response assay
A bead assay was performed using the Bio-Plex (Bio-Rad, Hercules, CA) platform. The Vi PS antigen (supplied by Fina Biosolutions, Rockville, MD, USA) was conjugated to Bio Rad beads [COOH (028)] at a concentration 8.33 × 106 beads/ml in a total volume of 1.50 ml. About 50 μl of human serum was prepared at various dilutions in human serum diluent (PBS, 1% W/V, 5% V/V goat serum, 0.05% Tween 20 V/V and 0.1% Kathon V/V). Day-0 serum was diluted 1:50, 1:400 and 1:3,200; Day-28 serum diluted 1:200, 1:1,600 and 1:12,800 in human serum diluent before incubation with the Vi beads for 3 h. After washing the beads, the bound human IgG was detected by goat anti-human IgG-RPE (1:5,000 in human serum diluent) by incubation for 30 min. Following the final wash steps, the beads were analyzed using the Bio-Plex instrument. A pooled serum with a high anti-Vi IgG response was used to create a standard curve that was assigned an arbirtrary value of 200 ELISA units (EU) per ml. The responses in the individual subjects were compared to the standard curve to calculate anti-Vi IgG EU/ml in the Day-0 and Day-28 serum.
Candidate genes and SNPs
We selected 283 autosomal genes from immunological pathways as candidates for this association study. SNPs from these genes, including about 2 and 1 kb upstream and downstream regions respectively, were chosen from the HapMap database (http://www.hapmap.org). The choice of SNPs was done using a statistical protocol that took into account differences in LD patterns and allele frequencies in the four HapMap populations. The statistical protocol provided a ranking of the “potential informativeness” of SNPs within each gene cataloged in the HapMap database, from which a subset of maximally informative SNPs was chosen. Further details are provided in Supplementary Text S1 and the list of genes and SNPs are provided in Table 1 and Table S1. Genotyping was done using Sequenom (55.7% of assayed SNPs) and Illumina (remaining 44.3% of SNPs) platforms.
Table 1.
Numbers of genes in various functional classes included in this study
| GO categorya | Molecular function | No. of genes |
|---|---|---|
| GO:0004871 | Signal transducer activity | 86 |
| GO:0005102 | Receptor binding | 44 |
| GO:0001664 | G-protein-coupled receptor binding | 41 |
| GO:0003676 | Nucleic acid binding | 21 |
| GO:0000166 | Nucleotide binding | 19 |
| GO:0001584 | Rhodopsin-like receptor activity | 16 |
| GO:0005488 | Binding | 12 |
| GO:0003824 | Catalytic activity | 7 |
| GO:0004857 | Enzyme inhibitor activity | 6 |
| GO:0003823 | Antigen binding | 3 |
| GO:0001871 | Pattern binding | 3 |
| GO:0001530 | Lipopolysaccharide binding | 2 |
| GO:0008009 | Chemokine activity | 1 |
| GO:0001948 | Glycoprotein binding | 1 |
| GO:0015457 | Auxiliary transport protein activity | 1 |
| GO:0032393 | MHC class I receptor activity | 1 |
| GO:0003774 | Motor activity | 1 |
| GO:0032395 | MHC class II receptor activity | 1 |
| GO:0001532 | Interleukin-21 receptor activity | 1 |
| GO:0005125 | Cytokine activity | 1 |
| GO:0000287 | Magnesium ion binding | 1 |
| GO:0005048 | Signal sequence binding | 1 |
| GO:0005515 | Protein binding | 1 |
| GO:0001540 | Beta-amyloid binding | 1 |
| GO:0005215 | Transporter activity | 1 |
| GO:0003712 | Transcription cofactor activity | 1 |
| Miscellaneous (GO category unavailable; information obtained from UNIPROTb database) | Mediation of inflammation and angiogenesis | 1 |
| Peptide binding | 1 | |
| Antimicrobial activity | 3 | |
| T-cell activation | 1 | |
| Defense response with MHC | 1 | |
| Regulation of inflammatory responses | 1 | |
| Putative immune function (Gene: HCG9; lies within MHC class I region) | 1 | |
| Total | 283 | |
aGO (Gene Ontology) category was obtained from gene2go file of NCBI Entrez Gene database (ftp://ftp.ncbi.nlm.nih.gov/gene/DATA/)
Statistical analysis
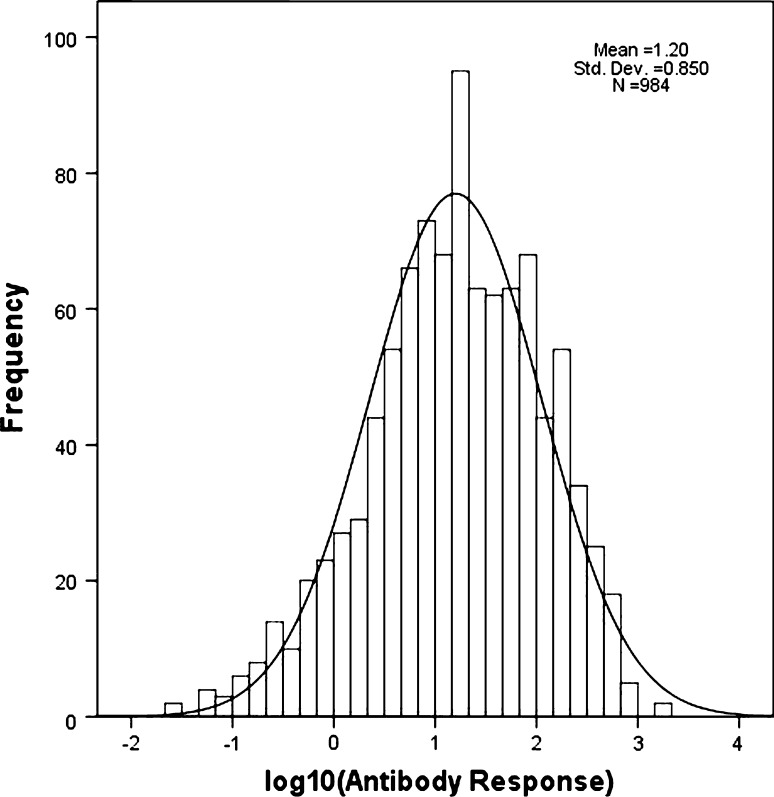
Antibody response (AR) to vaccination was measured as the difference between 28-day post-vaccination and pre-vaccination antibody levels (EU/ml). Values of AR were log10-transformed (logAR) to induce normality to the frequency distribution (Fig. 1) that is essential for using standard parametric statistical methods for hypothesis testing. Curation of genotype data included removal of (a) loci with <90% call rate (8.9% of loci), (b) non-polymorphic loci with MAF <0.05 (24.7% of loci with call rate >90%), (c) significantly deviant from HWE (4.2% of polymorphic loci), (d) individuals with <75% of loci with valid genotype calls, (e) individuals on whom antibody response could not be properly assayed (8 of 1,000 individuals), and (f) individuals showing cryptic relationships (>80% of loci with identical genotypes; 6 pairs of individuals satisfied this criterion, one individual from each pair was randomly chosen for final analysis). After data-curation, the final data set comprised genotypes of 984 individuals at 2,040 SNPs in 283 genes. Table 1 provides the numbers of genes in different classes of molecular function; the complete list of genes, SNPs selected from these genes and other relevant information are provided in Supplementary Table S1. Analysis of Variance (ANOVA) was performed to test equality of mean values of logAR among subgroups of vaccinees.
Fig. 1.
Frequency distribution of log10-transformed values of antibody response among vaccinees
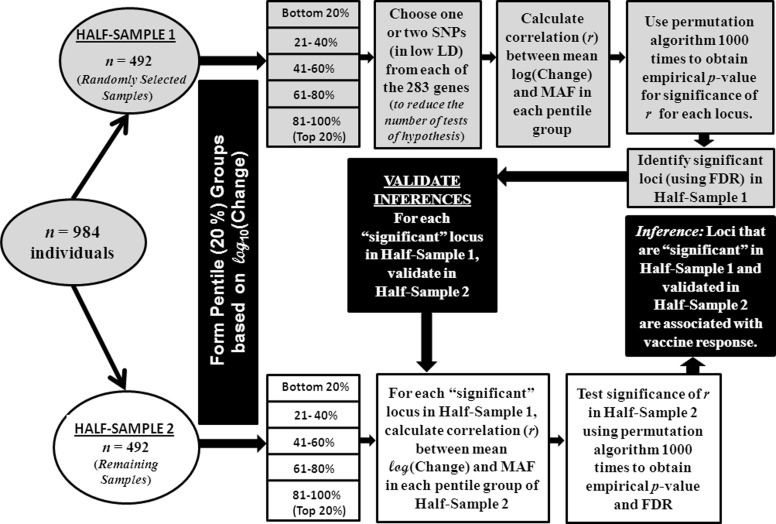
In conducting detailed statistical analyses of these data, we deviated from “standard practice” (Wellcome Trust Case Control Consortium 2007; Wellcome Trust Case Control Consortium et al. 2007; Burgner et al. 2009; Samani et al. 2007; Baranska et al. 2008; Hennig et al. 2008) and adopted a novel strategy. First, we did not classify the vaccinees as “responders” and “non-responders”, because this would involve defining an arbitrary threshold on values of antibody response to enable classification. Instead, we sorted the vaccinees by ascending order of their antibody response and grouped the vaccinees into 5 pentile (20 percentile) groups. Second, we posited that if indeed an allele at a locus was positively (or, negatively) associated with antibody response, then its frequency among vaccinees should monotonically increase (or, decrease) from those belonging to the lowest pentile group of antibody response to those belonging to the highest pentile group. In other words, the correlation coefficient between the minor allele frequency (MAF) at the locus and mean AR of vaccinees belonging to the pentile groups should be statistically significant. Third, in genomic studies involving a large number of associated loci, correction for multiple testing—especially if the loci are in significant linkage disequilibrium (LD); that is, non-independent—remains a major statistical problem (Conneely and Boehnke 2007; Han et al. 2009). To circumvent this, for single-locus association analysis, we reduced the number of tests by selecting from each gene only the highly informative and minimally associated SNPs, initially selecting the SNP located at the most 5′ end of the genomic region covered for the gene. This strategy resulted in the choice of 469 SNPs from the genes under consideration. (The data on the remaining loci were unused in the single-locus association analysis, but were used for haplotype association analysis, as explained later.) Fourth, cross-validation of findings of an association study is often a problem. In addition, replication of a study similar to the present one is particularly difficult because it entails the ethical choice of a population in which individuals will benefit from the vaccination to be provided. If the results are cross-validated in a different population, then in the event of discordance of results with the initial study it is difficult to ascertain whether population differences were the cause of the observed discordance. To avoid these vagaries, we generated an internal cross-validation sample, by randomly splitting the 984 vaccinees into two half-samples, of ~500 vaccinees in each half-sample (with ~100 vaccinees in each of the 5 pentile groups based on antibody response within each half sample). While this strategy in which ~500, instead of ~1,000, vaccinees are considered in the initial association analysis results in some loss of statistical power, our simulation studies showed that the loss of power is only 3–14% depending on the value of the correlation coefficient between MAF and mean AR (results not shown). The reason why the loss of statistical power is small is because a sample of size ~500 individuals (included in each half-sample) is large. The small loss of statistical power is more than offset by the gain in ability to cross-validate results using the same population sample. The strategy is presented as a flow-chart in Fig. 2.
Fig. 2.
Flow-chart describing the strategy used in the conduct of statistical data analyses
Test of significance of the correlation coefficient (r) between MAF and mean antibody response was carried out by a permutation test. Genotypes of individual vaccinees at a locus were randomly permuted and the correlation coefficient was computed in this permuted sample. This procedure was repeated 1,000 times, to produce a histogram of correlation coefficients under the null hypothesis r = 0. This histogram was then used to compute the P-value associated with the observed r at this locus. After generating the P-values for all loci, the FDR procedure (Benjamini and Hochberg 1995) was used to identify statistically significant values of r at the 5% level of significance.
We have also revalidated our inferences using the ‘standard’ procedure for association analysis, in which the null hypothesis tested is that the mean logAR values among the genotype classes are equal. This null hypothesis is tested using ANOVA. All significant allelic associations with logAR detected using the procedures described above were also validated using genotype data using PLINK (Purcell et al. 2007), Version 1.06 (http://www.pngu.mgh.harvard.edu/~purcell/plink/).
To summarize our analytical strategy, we first tested—in a random sample of 50% of our data (half-sample 1)—whether the minor allele frequency at a locus increases (or decreases) with increasing antibody response among groups of vaccinees using correlation analysis, and data-permutation and false discovery rate procedures for assessing significance of the estimated correlation coefficient. For those SNPs that showed significant correlation, we cross-validated the results in the remaining 50% of our data (half-sample 2), using the same methodology (correlation, data-permutation and FDR analyses). Finally, those SNPs for which inferences could be cross-validated, we revalidated the inferences using a ‘standard’ approach of testing equality of mean antibody-response among genotypes by ANOVA.
Haplotypes were inferred using PHASE (Stephens et al. 2001; http://www.stat.washington.edu/stephens/phase.html). Haplotype association analysis was performed using the permutation algorithm described above for single SNP markers.
Results
Demographic characteristics of vaccinees
Among the vaccinees (n = 984), 62.7% were Muslim and 37.3% were Hindu; 46.4% were male and 53.6% were female. The mean age of the vaccinees was 33.7 ± 0.4 years. The male vaccinees (mean age = 34.7 years) were, on the average, 2 years older than the female vaccinees (mean age = 32.7 years); this difference was statistically significant (P = 0.015). However, the mean ages of the vaccinees did not differ significantly by religion (mean age of Muslim and Hindu vaccinees were, respectively, 33.6 and 33.8 years; P = 0.76).
Pre- and post-vaccination antibody levels and antibody response
Mean antibody levels (in EU/ml) prior to vaccination were low, but were high on the 28th day post-vaccination (Table 2). The antibody levels pre- or post-vaccination were not significantly influenced by gender or religion.
Table 2.
Pre- and post-vaccination antibody levels (EU/ml) among vaccinees and their antibody response, classified by gender and religion
| Pre-vaccination | Post-vaccination | Antibody response | |
|---|---|---|---|
| Gender | |||
| Male | 1.77 ± 0.67 | 61.34 ± 5.10 | 59.57 ± 5.00 |
| Female | 1.38 ± 0.27 | 77.45 ± 7.03 | 76.06 ± 6.98 |
| P-value* | 0.185 | 0.088 | 0.100 |
| Religion | |||
| Muslim | 1.78 ± 0.53 | 77.48 ± 6.29 | 75.70 ± 6.23 |
| Hindu | 1.21 ± 0.23 | 57.34 ± 5.51 | 56.14 ± 5.42 |
| P-value* | 0.582 | 0.245 | 0.302 |
* P-values were calculated after log10-transformation of the data to induce normality of the frequency distributions
Enormous variation in both pre- and 28-day post-vaccination antibody levels was observed. Table 3 provides the descriptive statistics for the three variables (Days-0 and 28 antibody levels, and antibody response (AR) = Difference between Day-28 and Day-0 antibody levels) among vaccinees belonging to the five pentile groups based on Day-0 antibody levels. Irrespective of the pre-vaccination antibody level, vaccinees showed considerable variation in post-vaccination levels or antibody response (see also Supplementary Figure S1). The correlation between pre- and post-vaccination antibody levels was low (0.188), but significant (P < 0.0005). Among the vaccinees, the observed differences in the logAR (Fig. 1) could not be explained by variation in age (P = 0.915), or by gender (P = 0.100) or religious group (P = 0.302) differences (Table 2) of the vaccinees. Therefore, we did not stratify the vaccinees by age, gender or religious group for further statistical analyses.
Table 3.
Ranges and descriptive statistics of pre- and post-vaccination levels and antibody response (AR) among vaccinees belonging to the five pentile groups based on pre-vaccination levels
| Day-0 pentile group | Variable | Mean (EU/ml) | SD (EU/ml) | Minimum (EU/ml) | Maximum (EU/ml) |
|---|---|---|---|---|---|
| 1 (n = 196) | Day-0 | 0.026 | 0.014 | 0.000 | 0.050 |
| Day-28 | 7.218 | 10.065 | 0.045 | 68.173 | |
| AR | 7.193 | 10.064 | 0.031 | 68.136 | |
| 2 (n = 197) | Day-0 | 0.081 | 0.020 | 0.050 | 0.120 |
| Day-28 | 21.637 | 56.840 | 0.108 | 691.270 | |
| AR | 21.556 | 56.839 | 0.056 | 691.211 | |
| 3 (n = 197) | Day-0 | 0.210 | 0.060 | 0.121 | 0.328 |
| Day-28 | 40.696 | 66.614 | 0.201 | 514.707 | |
| AR | 40.486 | 66.604 | 0.067 | 514.521 | |
| 4 (n = 197) | Day-0 | 0.607 | 0.208 | 0.329 | 1.079 |
| Day-28 | 74.254 | 96.472 | 0.698 | 717.317 | |
| AR | 73.647 | 96.440 | 0.294 | 716.404 | |
| 5 (n = 197) | Day-0 | 6.891 | 23.264 | 1.084 | 293.561 |
| Day-28 | 205.719 | 234.984 | 2.769 | 1,777.819 | |
| AR | 198.827 | 234.098 | 0.341 | 1,775.567 |
Association analysis
After data-curation, data were available on 984 individuals pertaining to 2,040 SNPs in 283 genes. Of these 2,040 SNPs, the difference in the MAFs between Muslim and Hindu vaccinees was not statistically significant at the 5% level for any locus, after correcting for multiple-testing (Supplementary Figure S2). Thus, even though the vaccinees belonged to two maritally isolated religious groups, there was no genetic stratification or sub-structuring among the vaccinees. The vaccinees were randomly grouped into two half-samples; the demographic characteristics of the vaccinees in these two half-samples were similar and statistically non-significant (Supplementary Table S2 and Supplementary Figure S3). Vaccinees belonging to each half-sample were grouped into 5 pentile groups based on their AR values; that is, groups defined on the basis of AR values sorted in ascending order, with each group comprising approximately 20% of the total sample. The mean values of AR among vaccinees belonging to the pentile groups within half-samples are presented in Table 4. Between the two half-samples, while the mean values of AR were not statistically significant for the first three pentile groups, there were significant differences for the fourth and fifth pentile groups. These differences arose from a few individuals who showed high AR in either of the two half-samples (Supplementary Figure S3), as is evident from the large standard deviations of AR for these two pentile groups. While the increase in mean AR from the first to the third pentile groups is gradual, there is a sharp increase from third to the fourth pentile group and also from the fourth to the fifth pentile group. The mean values of logAR and the MAF for each locus were computed for vaccinees belonging to each pentile group within each half-sample. The differences in logAR between consecutive pentile groups were statistically significant, within each half-sample and for the total sample. The differences in MAFs between half-samples for the 5 pentile groups are presented in Supplementary Figure S4; none of the differences, corrected for multiple-testing, was statistically significant at the 5% level. To partially circumvent the statistical problems associated with multiple-testing, a subset of 469 (out of 2,040) ‘highly informative and minimally associated’ SNPs was chosen, with at least one SNP in each of the 283 genes (see Materials and Methods section for further details), for an initial association analysis. The correlation coefficient (r) between MAF and mean logAR of vaccinees belonging to the pentile groups of half-sample 1 was computed for each of the 469 loci selected for the association analysis. Of these 469 SNPs, only 54 (11.5%) SNPs in 43 genes showed significant correlation (P-values were estimated by data-permutation), at the 5% level (using multiple-testing correction by FDR method), between MAF and mean logAR of vaccinees belonging to the pentile groups (Table 5).
Table 4.
Mean ± SD antibody response (EU/ml) among individuals belonging to various pentile classes
| Half-sample | Pentile class (n in each half-sample) | ||||
|---|---|---|---|---|---|
| 1 (n = 98) | 2 (n = 98) | 3 (n = 98) | 4 (n = 98) | 5 (n = 100) | |
| 1 | 1.28 ± 1.00 | 6.50 ± 1.97 | 17.11 ± 4.46 | 63.80 ± 26.66 | 308.44 ± 252.31 |
| 2 | 1.39 ± 0.97 | 6.49 ± 2.24 | 18.08 ± 4.92 | 47.95 ± 12.52 | 205.31 ± 160.59 |
| P-value | 0.450 | 0.983 | 0.150 | <0.0009 | 0.001 |
Table 5.
List of 54 SNPs in 43 genes, along with the corresponding correlation coefficient between minor allele frequency and antibody response, that showed significant association with antibody response in half-sample 1
| Chromosome no. | Gene | SNP rs # | Corr. coeff. (r) | Chromosome no. | Gene | SNP rs # | Corr. coeff. (r) |
|---|---|---|---|---|---|---|---|
| 1 | TNFRSF8 | 12736809 | −0.94 | 4 | IRF2 | 12512614 | −0.96 |
| 1 | TNFRSF1B | 505844 | −0.97 | 6 | BAT2 | 2736172 | −0.91 |
| 1 | IL12RB2 | 7555183 | −0.99 | 6 | HLA-DOB | 2859579 | −0.99 |
| 1 | TGFBR3 | 1017956 | −0.94 | 6 | IL22RA2 | 13197049 | 0.92 |
| 1 | XCL1 | 6700487 | 0.91 | 6 | CCR6 | 1571878 | −0.96 |
| 1 | PTGS2 | 2745557 | 0.93 | 8 | DEFB1 | 2978873 | 0.98 |
| 1 | IRF6 | 2073486 | −0.93 | 8 | DEFB1 | 2977772 | 0.91 |
| 1 | NLRP3 | 12565738 | −0.92 | 8 | IKBKB | 3747811 | −0.95 |
| 1 | NLRP3 | 10754558 | 0.98 | 9 | PAX5 | 7031673 | −0.91 |
| 2 | IL1R2 | 13022757 | −0.93 | 10 | IL2RA | 10905668 | 0.98 |
| 2 | IL1R2 | 4850993 | −0.95 | 10 | ITGB1 | 10827164 | −0.95 |
| 2 | IL1R2 | 3218848 | 0.98 | 10 | CXCL12 | 10900030 | 0.96 |
| 2 | IL1R2 | 2110562 | −0.92 | 10 | MAPK8 | 12358297 | −0.95 |
| 2 | IL1RL1 | 10208293 | 0.95 | 10 | CHUK | 2230804 | −0.91 |
| 2 | IL18R1 | 1362348 | 0.92 | 11 | CD59 | 3181274 | −0.94 |
| 2 | CTLA4 | 231779 | −0.99 | 11 | CD44 | 112762 | −0.97 |
| 3 | IL5RA | 2290610 | −0.98 | 11 | IL10RA | 4252306 | 0.93 |
| 3 | CMTM8 | 4132830 | −0.96 | 11 | TIRAP | 8177352 | 0.98 |
| 3 | CMTM7 | 17029530 | −0.97 | 13 | IL17D | 1888001 | 0.94 |
| 3 | CD80 | 491407 | −1.00 | 13 | TNFRSF19 | 7338328 | −0.98 |
| 3 | CD80 | 2228017 | −0.97 | 16 | IL4R | 4787423 | 0.96 |
| 3 | CD80 | 2222631 | −0.93 | 17 | CSF3 | 2227321 | −0.95 |
| 3 | CD80 | 16829988 | −0.99 | 17 | MAP2K6 | 12946388 | −0.95 |
| 3 | CD86 | 2681411 | 0.97 | 21 | ITGB2 | 235326 | 0.99 |
| 3 | CD86 | 17203439 | 0.95 | 22 | MAPK1 | 743409 | −0.93 |
| 4 | TLR1 | 5743572 | 0.93 | 22 | MAPK1 | 2283792 | −0.98 |
| 4 | CXCL13 | 171388 | −0.94 | 22 | MAPK1 | 3788332 | −0.94 |
Cross-validation analysis
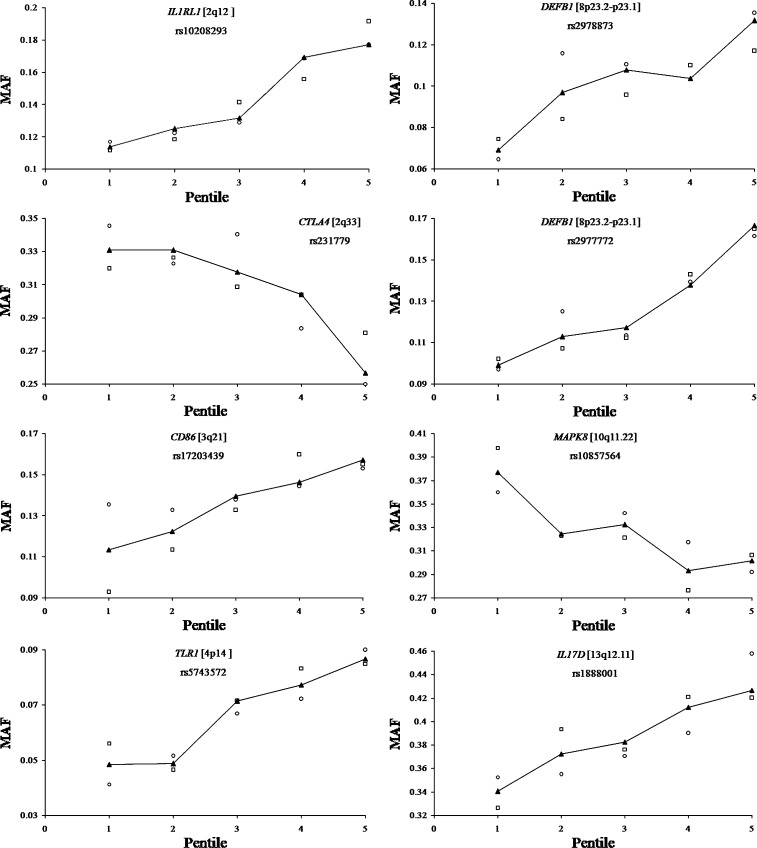
The inference that 54 SNPs were significantly associated with AR was cross-validated using the data on vaccinees belonging to half-sample 2—in which, as expected, the demographic characteristics were similar to those of half-sample 1(Supplementary Table S2 and Supplementary Figure S3). The same procedure of assessing the significance of relationship between logAR and MAF was used for each of the 54 SNPs considered for cross-validation. Of these 54 SNPs, significant association was found only for 8 SNPs—all intronic—in 7 genes (Table 6). Figure 3 shows that there is a near monotonic increase (for 6 SNPs) or decrease (for 2 SNPs) of MAF with increasing AR.
Table 6.
Details of 8 SNPs in 7 Genes that showed significant cross-validated association with antibody response, and of other assayed SNPs in those genes that showed significant association in both half-samples
| Gene | Chromosome no. | Total no. of SNPs assayeda | No. of “significant” SNPs | Details of “significant” SNPs | r In half-sample 1 (n = 492)d | r In half-sample 2 (n = 492)d | P-value using PLINK in the total samplee (n = 984) | ||
|---|---|---|---|---|---|---|---|---|---|
| rs # | nt Position | SNP location within gene (mRNA transcript no. in NCBI database) | |||||||
| IL1RL1 | 2 | 13 | 3 | 4142132b | 102303914 | Intron 1 (NM_016232.4) | 0.96 | 0.94 | 0.011 |
| 1997466b | 102317899 | Intron 1 (NM_016232.4) | 0.91 | 0.94 | 0.016 | ||||
| 10208293 c | 102332742 | Intron 10 (NM_016232.4) | 0.95 | 0.91 | 0.002 | ||||
| CTLA4 | 2 | 1 | 1 | 231779 c | 204442732 | Intron 1 (NM_005214.3) | −0.99 | −0.93 | 0.028 |
| CD86 | 3 | 17 | 1 | 17203439 c | 123278196 | Intron 1 (NM_175862.3) | 0.95 | 0.97 | 0.016 |
| TLR1 | 4 | 24 | 2 | 55815313b | 38481645 | Intron 2 (NM_003263.3) | 0.95 | 0.94 | 0.022 |
| 5743572 c | 38481786 | Intron 2 (NM_003263.3) | 0.98 | 0.93 | 0.050 | ||||
| DEFB1 | 8 | 33 | 7 | 2978873 c | 6717373 | Intron 1 (NM_005218.3) | 0.98 | 0.98 | 0.016 |
| 2977780b | 6717513 | Intron 1 (NM_005218.3) | 0.95 | 0.95 | 0.018 | ||||
| 2980928b | 6717829 | Intron 1 (NM_005218.3) | 0.95 | 0.95 | 0.018 | ||||
| 2978872b | 6718140 | Intron 1 (NM_005218.3) | 0.93 | 0.98 | 0.008 | ||||
| 2951854b | 6718836 | Intron 1 (NM_005218.3) | 0.92 | 0.94 | 0.028 | ||||
| 2980923b | 6720467 | Intron 1 (NM_005218.3) | 0.95 | 0.93 | 0.015 | ||||
| 2977772 c | 6720606 | Intron 1 (NM_005218.3) | 0.91 | 0.94 | 0.013 | ||||
| MAPK8 | 10 | 7 | 1 | 10857564 c | 49282578 | Intron 1 (NM_002750.2) | −0.95 | −0.96 | 0.030 |
| IL17D | 13 | 6 | 1 | 1888001 c | 20183906 | Intron 2 (NM_138284.1) | 0.94 | 0.91 | 0.018 |
aIncluding assayed SNPs in 2 kb regions upstream and downstream of the gene
bAssociation significant in both Half-samples (was not included in the initial analysis of 469 SNPs)
cAssociation found to be significant in Half-Sample 1 and cross-validated in Half-Sample 2 (i.e., included in the initial analysis of 469 SNPs)
d P-values corresponding to these correlation coefficients are all <0.005, and with FDR < 0.05
eEssentially each P-value corresponds to a test of equality of mean values of logAR among individuals belonging to the three genotypes at the locus under consideration
Fig. 3.
Increase/decrease in minor allele frequency (MAF) from the lowest to the highest pentile group of antibody response (open circles pertain to half-sample 1, open squares to half-sample 2 and solid triangles to the complete data)
Significance of the other assayed SNPs in genes showing cross-validated association
To avoid vagaries of multiple-testing, for the association analysis we selected a subset of assayed SNPs that were not in strong LD, and identified 8 associated SNPs in 7 genes (Table 6). We next sought to examine whether any of the remaining assayed SNPs in these 7 genes also showed significant association. In the cross-validated gene, CTLA4, only one SNP was selected for genotype assay, since the size of the gene is small. In the remaining 6 genes (Table 6), 100 SNPs were assayed (Table S1), of which 7 were found to be significantly associated with AR and were cross-validated. Among the additional 93 assayed SNPs in these 6 genes, 8 showed significant association (Table 6). These 8 SNPs were also intronic. Quantitative trait (antibody response) association analysis with genotypes was performed for these 16 SNPs; that is, we tested the null hypothesis that the mean values of logAR among vaccinees belonging to the three genotypes in the complete data set (half samples 1 and 2 pooled) were equal. This was carried out by ANOVA using PLINK (http://www.pngu.mgh.harvard.edu/~purcell/plink/); all differences were found to be significant (Table 6) at the 5% level, implying that mean antibody responses were significantly different among genotypes.
Haplotype analysis
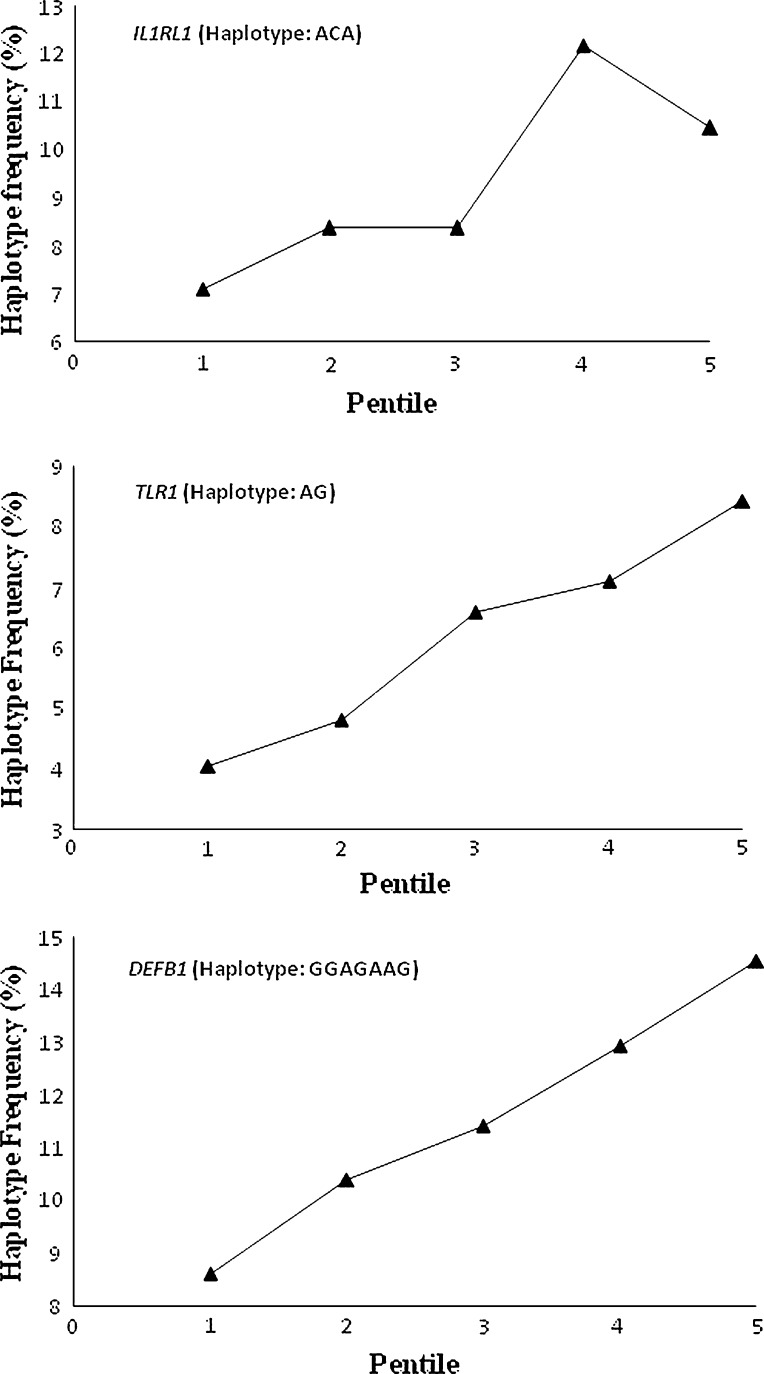
With the exception of CTLA4, for each of the remaining 6 genes (IL1RL1, CD86, TLR1, DEFB1, MAPK8 and IL17D) multiple SNPs were assayed, but were not included in the initial analyses to reduce the number of tests. In these 6 genes, we carried out association analyses with all the assayed SNPs, and found some of these additional SNPs to be significantly associated with AR. The additional SNPs that were significantly associated belonged to 3 genes—IL1RL1, TLR1 and DEFB1 (Table 6). For these 3 genes in which multiple SNPs showed significant association with antibody response, we estimated the frequency of the haplotype comprising minor alleles at the SNP loci that were found to be significantly associated (Table 6). For each of these genes, the signs of the correlation coefficient between MAF and mean logAR of pentile groups for all SNPs that were significantly associated were identical (data not shown). We then estimated the strength of association between the frequencies of these haplotypes and mean logAR of vaccinees belonging to the pentile groups using the total sample (n = 984). Essentially, the nature of and strength (significant at 5% level) of relationship remained unaltered (Fig. 4) from that found in the cross-validation analysis (Fig. 3). Thus, haplotypes based on SNPs in these genes also showed significant association with AR.
Fig. 4.
Increase/decrease in haplotype frequency from the lowest to the highest pentile group of antibody response in the total sample
Discussion
Salmonella typhi, that causes typhoid fever, is encapsulated by a polysaccharide (PS), Vi PS. The expression of Vi PS correlates with virulence and resistance to nonoxidative killing and phagocytosis (Looney and Steigbigel 1986). Infection with encapsulated bacteria can be prevented by vaccination with capsular polysaccharides. A Vi PS vaccine for typhoid is marketed and used in many countries, including India. Since the transmission of the typhoid pathogen is through the oral-fecal route, typically the areas of endemicity of typhoid are also areas where there is low level of personal and public hygiene. Although it is in such areas where a vaccination program against typhoid is likely to have a major public-health impact, residents of such areas are often exposed to S. typhi and other co-infections prior to vaccination. Indeed, several studies have reported that 19–58% percent of subjects that live in endemic areas involved in Vi polysaccharide vaccine studies develop protective levels (1 μg/ml) of anti-Vi antibodies without vaccination (Klugman et al. 1996; Keddy et al. 1999; Panchanathan et al. 2001). These observations suggest that natural, environmental exposure to S. typhi has the potential to induce protective levels of serum, anti-Vi antibodies. Post-vaccination immune response is likely to be affected by prior exposure. Participants in this study were chosen from a high-risk area for typhoid infection. Using appropriate criteria, we excluded individuals who may have been infected with S. typhi at least a year prior to their recruitment in this study, thereby minimizing, to the extent feasible, the confounding effect of immune response due to prior exposure with that due to vaccination. Our data have revealed that post-vaccination antibody levels do not strongly correlate with pre-vaccination levels. This indicates that the vaccine can induce a strong antibody response even if a vaccinee has a high pre-vaccination level of antibody and the vice versa.
Immune response to polysaccharide antigens is qualitatively different from that to protein antigens. Typically, it is T-cell independent. T-cell independent antigens are not consistently immunogenic in children <2 years of age (Chelvarajan et al. 2005). The Vi polysaccharide vaccine represents a T-independent antigen that does not generate antigen-specific CD4+ T cell help. Without CD4+ T cell help, long-term memory B cells are not generated. This explains the need to revaccinate with the Vi polysaccharide vaccine every 2–3 years (Whitaker et al. 2009). However, T-cell-independent type 2 (TI-2) antigens, such as Vi PS, can also stimulate the mature B cells to induce antibody production through non-cognate T-cells (Obukhanych and Nussenzweig 2006) and antibody response to Vi PS vaccine has been shown to be correlated with the number of CD4+ T lymphocytes (Kroon et al. 1999).
Despite their T-cell independent nature, some polysaccharide antigens have been reported to activate dendritic cells and other cell types. When dendritic cells were treated with the polysaccharide from Ganoderma lucidum there was enhanced cell surface expression of CD80, CD86, CD83, CD40, CD54, and HLA-DR (Kroon et al. 1999). It is also known that blood dendritic cells interact with marginal zone (MZ) B cells to initiate TI-2 immune responses as polysaccharide antigens localize preferentially to these (MZ) B cells found in spleen (Achtman et al. 2009; Klouwenberg and Bont 2008). The splenic marginal zone (MZ) has been implicated as a source of circulating IgM memory B cells (Kreutzmann et al. 2003). These findings reveal a larger role for polysaccharides in immune recognition than currently appreciated.
Inter-individual variability in immune response to peptide vaccines has been documented and a number of polymorphic loci in genes related to both innate and adaptive immune systems have been found to be associated with antibody response to vaccines (Sur et al. 2009). However, little is known about genomics of immune response for polysaccharide vaccines. To our knowledge, this is the first large-scale study—involving a large number of vaccinated individuals (~1,000) and a large number of polymorphic loci (>2,000)—on genomics of immune response to a polysaccharide vaccine to prevent S. typhi infection. Our strategy for data analyses was chosen to minimize the possibility of false positive discovery and, with a minimal loss of statistical power, enabled internal cross-validation of results through a half-sample analytical design.
Of the 283 genes considered as candidates for this study of immune response, we have found 16 SNPs in 7 genes to show significant allelic, genotypic and haplotypic associations with immune response to the Vi PS vaccine. These 7 genes are β-defensin 1 (DEFB1) with 7 associated SNPs; toll-like receptor 1 (TLR1) and interleukin 1 receptor-like 1 (IL1RL1) each with 2 associated SNPs; cytotoxic T-lymphocyte-associated antigen 4 (CTLA4), mitogen-activated protein kinase 8 (MAPK8), cluster of differentiation 86—a molecule expressed on antigen-presenting cells that provide costimulatory signals necessary for T cell activation and survival (CD86) and interleukin 17D (IL17D) each with 1 associated SNP.
Though the role of T lymphocytes in the regulation of the antibody production to PS is obscure, anti-PS response involves non-cognate activation of TCR-nonspecific T cells by implicating a role for endogenous CD40-CD40L interactions in the human antibody response (Snapper et al. 2001). It is known that B7 family of co-stimulatory molecules, CD86 along with CD80, present on the antigen-presenting cells (APCs), interact with CD28/CTLA4 receptors on T cells to provide major non-cognate co-stimulatory signals (Khan et al. 2007). Therefore, it is not surprising that CD86 and CTLA4 genes showed significant association with immune response to Vi PS vaccine in the present study, indicative of a non-cognate T-cell dependence. TLR1 showed association with the host response to the vaccine. Downregulation of TLR-mediated immune responses through diminished TLR-mediated cell signaling or expression is an important immune evasion mechanism in some bacterial pathogens (Alvarez 2005; Babu et al. 2009). Data from experiments with human colonic tissue explants indicate that the Vi antigen reduces TLR-mediated IL-8 signaling, thereby evading TLR-mediated host response triggering neutrophil infiltration in the intestinal mucosa (Wilson et al. 2008). However, it is not clear if Vi polysaccharide is recognized by TLR1 or if the TLR1 gene influences host responses to co-infections that then indirectly influences host response to vaccination with Vi PS. All toll-like receptors have a toll-interleukin 1 domain that is responsible for signal transduction; IL1RL1—a gene that we have found to be significantly associated with AR induced by the PS vaccine—participates in this signaling. MAP kinases are involved in a wide variety of cellular processes such as proliferation, differentiation, transcription regulation and development; therefore, the association of a SNP in MAPK8 with antibody response is not unexpected. There are reports demonstrating that the MAP kinase pathway may be one of the targets that is modulated by Vi to dampen early inflammatory responses in Vi polysaccharide treated intestinal epithelial cells (Caco-2; Sharma and Qadri 2004). This may explain why MAPK8 was found to be associated with Vi polysaccharide antibody response. The involvement of a MAP kinase and IL17D, that stimulates the production of other cytokines, can result in pro-inflammatory signals that can induce upregulation of DEFB1 (O’Neil et al. 1999). We have found multiple SNPs in DEFB1 to be significantly associated with Vi polysaccharide vaccine response. Thus, the overall picture that emerges from our findings is that polymorphisms in genes involved in polysaccharide recognition, signal transduction, inhibition of T-cell proliferation, pro-inflammatory signaling and eventual production of antimicrobial peptides are associated with antibody response to the Vi polysaccharide vaccine for typhoid. However, just as co-infection with parasites (Harris et al. 2009) or other infectious agents (Sugata et al. 2008; Lin et al. 2006) can influence immunological response to a pathogen, it is possible, that antibody response to vaccination with Vi polysaccharide may be affected by co-infections or prior exposure. While it is difficult to rule out the involvement of other genes in modulation of immune response to the Vi polysaccharide vaccine, the 7 genes that we have found to be associated with host antibody response to the polysaccharide vaccine provide a foundation for elucidating the functional mechanism of the products of these genes in host response to the polysaccharide vaccine. However, we emphasize that the evaluation of genes that influence host antibody response to the Vi polysaccharide vaccine is complicated by the living conditions (i.e., environment) of the study participants and the lack of knowledge regarding the exact microbial exposure (S. typhi or other infectious agents) of each study participant. It is desirable that similar studies be performed on participants residing in hygienic areas that lack natural exposure to S. typhi, and with access to good quality water and hygienic sewer services, although residents of such areas are unlikely to require vaccination for typhoid.
Electronic supplementary material
Acknowledgments
We are grateful to (a) members of the TCG-ISI Centre for Population Genomics, particularly, Bijan Bairagya, Biplab Dey, Uposoma Dey, Ardhendu Endow, Souvik Mukherjee, Sonia Poddar, Priya Sengupta and Debabrata Sutradhar; (b) members of The Centre for Genomic Application; (c) Shaun M. Kirwan for performance of the antibody response assays; and, (d) investigators, phlebotomists and clinicians who helped in participant recruitment, vaccination, biospecimen collection and monitoring of adverse events. This work was supported by NIAID, National Institutes of Health, U.S.A.; Contract No. HHSN200400067C.
References
- Achtman AH, Hopken UE, Bernert C, Lipp M. CCR7-deficient mice develop atypically persistent germinal centers in response to thymus-independent type 2 Antigens. J Leukoc Biol. 2009;85:409–417. doi: 10.1189/jlb.0308162. [DOI] [PubMed] [Google Scholar]
- Alvarez JI. Inhibition of toll like receptor immune responses by microbial pathogens. Front Biosci. 2005;10:582–587. doi: 10.2741/1554. [DOI] [PubMed] [Google Scholar]
- Babu S, Bhat SQ, Kumar NP, Anuradha R, Kumaran P et al (2009) Attenuation of toll-like receptor expression and function in latent tuberculosis by coexistent filarial infection with restoration following antifilarial chemotherapy. PLoS Negl Trop Dis 3. Available: http://www.plosntds.org/article/info%3Adoi%2F10.1371%2Fjournal.pntd.0000489. Accessed 9 Sep 2009 [DOI] [PMC free article] [PubMed]
- Baranska M, Van Amelsvoort L, Birindelli S, Fustinoni S, Corsini E, et al. Association of pesticide exposure, vaccination response, and interleukin-1 gene polymorphisms. Hum Exp Toxicol. 2008;27:709–713. doi: 10.1177/0960327108100002. [DOI] [PubMed] [Google Scholar]
- Benjamini Y, Hochberg Y. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J R Stat Soc Ser B. 1995;57:289–300. [Google Scholar]
- Burgner D, Davila S, Breunis WB, Ng SB, Li Y et al (2009) A genome-wide association study identifies novel and functionally related susceptibility loci for Kawasaki disease. PLoS Genet 5. Available: http://www.plosgenetics.org/article/info:doi/10.1371/journal.pgen.1000319. Accessed 9 Sep 2009 [DOI] [PMC free article] [PubMed]
- Chelvarajan RL, Collins SM, Van Willigen JM, Bondada S. The unresponsiveness of aged mice to polysaccharide antigens is a result of a defect in macrophage function. J Leukoc Biol. 2005;77:503–512. doi: 10.1189/jlb.0804449. [DOI] [PubMed] [Google Scholar]
- Conneely KN, Boehnke M. So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. Am J Hum Genet. 2007;81:1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gelder CM, Lambkin R, Hart KW, Fleming D, Williams OM, et al. Associations between human leukocyte antigens and nonresponsiveness to influenza vaccine. J Infect Dis. 2002;185:114–117. doi: 10.1086/338014. [DOI] [PubMed] [Google Scholar]
- Gupta D, Faridi MM, Aggarwal A, Kaur I. Seroprevalence of anti-Vi antibodies and immunogenicity of Typhim Vi vaccine in children. Hum Vaccin. 2008;4:305–308. doi: 10.4161/hv.4.4.5824. [DOI] [PubMed] [Google Scholar]
- Han B, Kang HM, Eskin E (2009) Rapid and accurate multiple testing correction and power estimation for millions of correlated markers. PLoS Genet 5. Available: http://www.plosgenetics.org/article/info:doi%2F10.1371%2Fjournal.pgen.1000456%3Bjsessionid=252FD069666D094C2E2295EF4C4FDA28. Accessed 9 Sep 2009 [DOI] [PMC free article] [PubMed]
- Harris JB, Podolsky MJ, Bhuiyan TR, Chowdhury F, Khan AI et al (2009) Immunologic responses to vibrio cholerae in patients co-infected with intestinal parasites in Bangladesh. PLoS Negl Trop Dis 3. Available: http://www.plosntds.org/article/info:doi/10.1371/journal.pntd.0000403. Accessed 9 Sep 2009 [DOI] [PMC free article] [PubMed]
- Hennig BJ, Fielding K, Broxholme J, Diatta M, Mendy M et al (2008) Host Genetic factors and vaccine-induced immunity to Hepatitis B virus infection. PloS ONE 3. Available: http://www.plosone.org/article/info:doi/10.1371/journal.pone.0001898. Accessed 9 Sep 2009 [DOI] [PMC free article] [PubMed]
- Jin P, Wang E. Polymorphism in clinical immunology—from HLA typing to immunogenetic profiling. J Transl Med. 2003;1:8. doi: 10.1186/1479-5876-1-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keddy KH, Klugman KP, Hansford CF, Blondeau C, Bouveret le Cam NN. Persistence of antibodies to the Salmonella typhi Vi capsular polysaccharide vaccine in South African school children ten years after immunization. Vaccine. 1999;17:110–113. doi: 10.1016/S0264-410X(98)00160-1. [DOI] [PubMed] [Google Scholar]
- Khan N, Ghousunnissa S, Jegadeeswaran SKM, Thiagarajan D, Hasnain SE, et al. Anti-B7–1/B7–2 antibody elicits innate-effector responses in macrophages through NF-kB-dependent pathway. Int Immunol. 2007;19:477–486. doi: 10.1093/intimm/dxm012. [DOI] [PubMed] [Google Scholar]
- Kimman TG, Vandebriel RJ, Hoebee B. Genetic variation in the response to vaccination. Community Genet. 2007;10:201–217. doi: 10.1159/000106559. [DOI] [PubMed] [Google Scholar]
- Klouwenberg PK, Bont L (2008) Neonatal and infantile immune responses to encapsulated bacteria and conjugate vaccines. Clin DevImmunol 628963. Available: http://www.umcutrecht.nl/NR/rdonlyres/7DF815ED-779A-4CFC-8209-6A981438FECD/11720/PeterKleinKouwenbergenLouisBont2008.pdf. Accessed 9 Sep 2009 [DOI] [PMC free article] [PubMed]
- Klugman KP, Koornhof HJ, Robbins JB, Le Cam NN. Immunogenicity, efficacy and serological correlate of protection of Salmonella typhi Vi capsular polysaccharide vaccine three years after immunization. Vaccine. 1996;14:435–438. doi: 10.1016/0264-410X(95)00186-5. [DOI] [PubMed] [Google Scholar]
- Kreutzmann S, Manuela M, Weber H, Germing U, Tournilhac O, et al. Human immunoglobulin M memory B cells controlling Streptococcus pneumoniae infections are generated in the spleen. J Exp Med. 2003;197:939–945. doi: 10.1084/jem.20022020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroon FP, van Dissel JT, Ravensbergen E, Nibbering PH, van Furth R. Impaired antibody response after immunization of HIV-infected individuals with the polysaccharide vaccine against Salmonella typhi (Typhim-Vi®) Vaccine. 1999;17:2941–2945. doi: 10.1016/S0264-410X(99)00167-X. [DOI] [PubMed] [Google Scholar]
- Kruskall MS, Alper CA, Awdeh Z, Yunis EJ, Marcus-Bagley D. The immune response to hepatitis B vaccine in humans: inheritance patterns in families. J Exp Med. 1992;175:495–502. doi: 10.1084/jem.175.2.495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin CH, Hsieh CC, Chen SJ, Wu TC, Chung RL, et al. The diagnostic value of serum interleukins 6 and 8 in children with acute gastroenteritiss. J Pediatr Gastroenterol Nutr. 2006;43:25–29. doi: 10.1097/01.mpg.0000235764.30743.5b. [DOI] [PubMed] [Google Scholar]
- Looney RJ, Steigbigel RT. Role of the Vi antigen of Salmonella typhi in resistance to host defense in vitro. J Lab Clin Med. 1986;108:506–516. [PubMed] [Google Scholar]
- Milich DR, Leroux-Roels GG. Immunogenetics of the response to HBsAg vaccination. Autoimmun Rev. 2003;2:248–257. doi: 10.1016/S1568-9972(03)00031-4. [DOI] [PubMed] [Google Scholar]
- O’Neil DA, Porter EM, Elewaut D, Anderson GM, Eckmann L, et al. Expression and regulation of the human b-defensins hBD1 and hBD2 in intestinal epithelium. J Immunol. 1999;163:6718–6724. [PubMed] [Google Scholar]
- Obukhanych TV, Nussenzweig MC. T-independent type II immune responses generate memory B cells. J Exp Med. 2006;203:305–310. doi: 10.1084/jem.20052036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panchanathan V, Kumar S, Yeap W, Devi S, Ismail R, Sarijan S, et al. Comparison of safety and immunogenicity of a Vi polysaccharide typhoid vaccine with a whole-cell killed vaccine in Malaysian Air Force recruits. Bull World Health Organ. 2001;79:811–817. [PMC free article] [PubMed] [Google Scholar]
- Poland GA. Variability in immune response to pathogens: using measles vaccine to probe immunogenetic determinants of response. Am J Hum Genet. 1998;62:215–220. doi: 10.1086/301736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland GA, Jacobson RM. The genetic basis for variation in antibody responses to vaccines. Curr Opin Pediatr. 1998;10:208–215. doi: 10.1097/00008480-199804000-00017. [DOI] [PubMed] [Google Scholar]
- Poland GA, Jacobson RM, Colbourne SA, Thampy AM, Lipsky JJ, et al. Measles antibody seroprevalence rates among immunized Inuit, Innu and Caucasian subjects. Vaccine. 1999;17:1525–1531. doi: 10.1016/S0264-410X(98)00362-4. [DOI] [PubMed] [Google Scholar]
- Poland GA, Ovsyannikova IG, Jacobson RM. Immunogenetics of seasonal influenza vaccine response. Vaccine. 2008;26:D35–D40. doi: 10.1016/j.vaccine.2008.07.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poland GA, Ovsyannikova IG, Jacobson RM. Vaccine immunogenetics: bedside to bench to population. Vaccine. 2008;26:6183–6188. doi: 10.1016/j.vaccine.2008.06.057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pulendran B, Kumar P, Cutler CW, Mohamadzadeh M, Van Dyke T, et al. Lipopolysaccharides from distinct pathogens induce different classes of immune responses in vivo. J Immunol. 2001;167:5067–5076. doi: 10.4049/jimmunol.167.9.5067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MAR, et al. PLINK: a toolset for wholegenome association and population-based linkage analysis. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samani NJ, Erdmann J, Hall AS, Hengstenberg C, Mangino M, et al. Genomewide association analysis of coronary artery disease. N Engl J Med. 2007;357:443–453. doi: 10.1056/NEJMoa072366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma A, Qadri A. Vi polysaccharide of Salmonella typhi targets the prohibitin family of molecules in intestinal epithelial cells and suppresses early inflammatory responses. Proc Natl Acad Sci USA. 2004;101:17492–17497. doi: 10.1073/pnas.0407536101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snapper CM. Differential regulation of protein- and polysaccharide-specific Ig isotype production in vivo in response to intact Streptococcus pneumoniae. Curr Protein Pept Sci. 2006;7:295–305. doi: 10.2174/138920306778017972. [DOI] [PubMed] [Google Scholar]
- Snapper CM, Shen Y, Khan AQ, Colino J, Zelazowski P, et al. Distinct types of T-cell help for the induction of a humoral immune response to Streptococcus pneumoniae. Trends Immunol. 2001;22:308–311. doi: 10.1016/S1471-4906(01)01926-3. [DOI] [PubMed] [Google Scholar]
- Stephens M, Smith N, Donnelly P. A new statistical method for haplotype reconstruction from population data. Am J Hum Genet. 2001;68:978–989. doi: 10.1086/319501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugata K, Taniguchi K, Yui A, Miyake F, Suga S, et al. Analysis of rotavirus antigenemia and extraintestinal manifestations in children with rotavirus gastroenteritis. Pediatrics. 2008;122:392–397. doi: 10.1542/peds.2007-2290. [DOI] [PubMed] [Google Scholar]
- Sur D, Ochiai RL, Bhattacharya SK, Ganguly NK, Ali M, et al. A cluster-randomized effectiveness trial of Vi typhoid vaccine in India. N Engl J Med. 2009;361:403–405. doi: 10.1056/NEJMoa0807521. [DOI] [PubMed] [Google Scholar]
- Szu SC, Li XR, Stone AL, Robbins JB. Relation between structure and immunologic properties of the Vi capsular polysaccharide. Infect Immun. 1991;59:4555–4561. doi: 10.1128/iai.59.12.4555-4561.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Eden W, De Bries RRP, Stanford JL, Rook GAW. HLA-DR3 associated genetic control of response to multiple skin tests with new tuberculins. Clin Exp Immunol. 1983;52:287–292. [PMC free article] [PubMed] [Google Scholar]
- van Lovern H, Van Amsterdam JGC, Vandebriel RJ, Kimman TG, Rumke HC, et al. Vaccine-induced antibody responses as parameters of the influence of endogenous and environmental factors. Environ Health Perspect. 2001;109:757–764. doi: 10.2307/3454816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vos Q, Lees A, Wu ZQ, Snapper CM, Mond JJ. B-cell activation by Tcell-independent type 2 antigens as an integral part of the humoral immune response to pathogenic microorganisms. Immunol Rev. 2000;176:154–170. doi: 10.1034/j.1600-065X.2000.00607.x. [DOI] [PubMed] [Google Scholar]
- Wang C, Tang J, Song W, Lobashevsky E, Wilson CM, et al. HLA and cytokine gene polymorphisms are independently associated with responses to hepatitis B vaccination. Hepatology. 2004;39:978–988. doi: 10.1002/hep.20142. [DOI] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium Genome-wide association study of 14, 000 cases of seven common diseases and 3, 000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wellcome Trust Case Control Consortium. Australo-Anglo-American Spondylitis Consortium (TASC) Burton PR, Clayton DG, Cardon LR, Craddock N, et al. Association scan of 14,500 nonsynonymous SNPs in four diseases identifies autoimmunity variants. Nat Genet. 2007;39:1329–1337. doi: 10.1038/ng.2007.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitaker JA, Franco-Paredes C, del Rio C, Edupuganti S. Rethinking typhoid fever vaccines: implications for travelers and people living in highly endemic areas. J Travel Med. 2009;16:46–52. doi: 10.1111/j.1708-8305.2008.00273.x. [DOI] [PubMed] [Google Scholar]
- Wilson RP, Raffatellu M, Chessa D, Winter SE, Tükel C, et al. The Vi-capsule prevents Toll-like receptor 4 recognition of Salmonella. Cell Microbiol. 2008;10:876–890. doi: 10.1111/j.1462-5822.2007.01090.x. [DOI] [PubMed] [Google Scholar]
- Zuckerman JN. Nonresponse to hepatitis B vaccines and the kinetics of anti-HBs production. J Med Virol. 1996;50:283–288. doi: 10.1002/(SICI)1096-9071(199612)50:4<283::AID-JMV1>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.