Abstract
Acyl-CoA:glycerol-3-phosphate acyltransferase (GPAT) catalyzes the first step during de novo synthesis of glycerolipids. Mammals have at least four GPAT isoforms. Here we report the further characterization of the two recently identified microsomal GPAT3 and GPAT4. Both enzymes are highly expressed in adipose tissues. However, while GPAT3 is highly (∼60-fold) induced during adipocyte differentiation, GPAT4 induction is only modest (∼5-fold), leading to a lower abundance of GPAT4 mRNA in adipocytes. While overexpression of GPAT3 and GPAT4 in either insect or mammalian cells results in a comparable increase of GPAT activity, shRNA-mediated knockdown of GPAT3, but not GPAT4, in 3T3-L1 adipocytes led to a significant decrease in GPAT activity, a profound inhibition of lipid accumulation, and a lack of expression of several adipogenic markers during adipocyte differentiation. These data suggest that GPAT3 may encode the major GPAT isoform in adipocytes and play an important role in adipogenesis. Furthermore, we have shown that both GPAT3 and GPAT4 are phosphorylated by insulin at Ser and Thr residues, leading to increased GPAT activity that is sensitive to wortmannin. Our results reveal a link between the lipogenic effects of insulin and microsomal GPAT3 and GPAT4, implying their importance in glycerolipid biosynthesis.
Keywords: AGPAT6, acyltransferase, triacylglycerol, phospholipid
Acyl-CoA:glycerol-3-phosphate acyltransferase (GPAT, EC 2.3.1.15) catalyzes the acyl-CoA–dependent acylation of glycerol-3-phosphate (G3P), the first step of the de novo triacylglycerol (TAG) and phospholipid biosynthesis. The product of this reaction, lysophosphatidic acid (LPA), is further converted to phosphatidic acid (PA) by 1-acylglycerol-3-phosphate acyltransferase (AGPAT) or LPA acyltansferase (LPAAT). Dephosphorylation of PA by the Mg2+-dependent phosphatidic acid phophatase (encoded by the lipin gene) produces diacylglycerol (DAG), a precursor for both TAG and phospholipids. While these enzymatic activities in the TAG and phospholipid biosynthetic pathway were characterized in the 1950s, the genetic complements of these enzymes have remained unknown until very recently. Mainly due to the availability of large amounts of sequence information as well as advanced bioinformatics tools, great strides have been made to identify “missing genes” encoding these enzymes, including several novel GPAT isoforms (1–4).
GPAT has been suggested to be the rate-limiting enzyme in the TAG biosynthesis pathway because of its low specific activity and position as the initiating enzyme. It has been known for many years that multiple GPAT isoforms exist: a mitochondrial NEM-resistant isoform is primarily found in liver, while a microsomal NEM-sensitive isoform is the major isoform in most other tissues. GPAT1, the first mitochondrial GPAT isoform, was cloned in 1993 (5); GPAT2, a second mitochondrial isoform, has been identified more recently based on residual mitochondrial GPAT activity in GPAT1-deficient mice as well as its sequence homology to GPAT1 (2, 6). Using a combination of bioinformatic and transcriptional profiling approaches, we recently reported the first identification of a gene encoding GPAT3, a microsomal NEM-sensitive GPAT (1). However, despite an abundant expression level of GPAT3 mRNA in adipose tissue, the expression of GPAT3 transcripts in liver is notably low, suggesting the presence of other GPAT isoforms accounting for liver GPAT activity.
We and others noted that GPAT3 is closely related (69% and 80% amino acid identity across the whole molecule and within the acyltransferase domain, respectively) to AGPAT6, an orphan member of the glycerophospholipid acyltransferase superfamily with high expression level in liver (1, 7). Two recent studies independently showed that AGPAT6 encodes a second microsomal GPAT, now named GPAT4 (3, 4). First, both studies demonstrated that GPAT4 overexpression conferred NEM-sensitive GPAT activity in mammalian cells. In addition, AGPAT6−/− mice, which had been generated independently using a gene-trap approach, showed significantly decreased microsomal GPAT activity in liver and mammary gland as well as defects in milk triglyceride production, clearly demonstrating the importance of GPAT4/AGPAT6 in contributing to GPAT activity in these tissues (3, 4). AGPAT6−/− mice also exhibit decreased epididymal and inguinal fat and an almost complete absence of subcutaneous adipose tissue after 4 months of age (8). The AGPAT6-null mice are also resistant to genetic and diet-induced obesity (7, 8). However, microsomal GPAT activity was unchanged in adipose tissue of AGPAT6−/− mice (4), and embryonic fibroblast cells from AGPAT6−/− mice can undergo normal differentiation to adipocytes (8), suggesting other GPAT isoforms than GPAT4 contribute to GPAT activity in adipose tissue. The mechanisms underlying the profound subdermal lipodystrophy in AGPAT6−/− mice are currently unclear.
Here we investigate the relative roles of GPAT3 and GPAT4 in adipose tissue by directly comparing expression, hormonal regulation, and function using 3T3-L1 adipocytes as a model. Our data indicate that GPAT3 is the major microsomal GPAT isoform in both adipose tissue and 3T3-L1 adipocytes. We unexpectedly discovered a role for GPAT3, but not GPAT4, in adipocyte differentiation, adding this gene to the growing list of triglyceride biosynthetic genes important for adipocyte differentiation. Our data also revealed a regulatory role of insulin in modulating GPAT3 and GPAT4 activity by phosporylation.
MATERIALS AND METHODS
Materials
Unless stated otherwise, all lipids and acyl-CoAs were purchased from Avanti Polar Lipids (Alabaster, AL) or Sigma (St. Louis, MO). [14C]glycerol 3-phosphate and [14C]acyl-CoAs were obtained from American Radiolabeled Chemicals, Inc. (St. Louis, MO). Silica TLC plates (60 A) were from Whatman (Florham Park, NJ). All other materials or chemicals were obtained from either Sigma or Invitrogen (Carlsbad, CA).
Cloning and expression of cDNAs
The human GPAT4 with a N-terminus in-frame FLAG epitope (MDYKDDDDL, tag underlined) was cloned into pPCRscript Amp SK(+) vector (Stratagene, La Jolla, CA) by PCR amplification from human leukocytes cDNA libraries (Cloentech, CA). The cDNA insert was subsequently cloned into the EcoRV and NotI sites of mammalian expression vector pcDNA3.1(−)/Hygro, or into the same sites of pFastBacTM1 vector to generate recombinant baculovirus for insect cell expression using the Bac-to-Bac baculovirus expression system according to the manufacturer's instruction (Invitrogen). In our attempt to clone an untagged mouse GPAT4, a cDNA clone encoding a truncated GPAT4 protein (aa 1-204, termed GPAT4-N204× in the present study) was identified by sequencing. The cDNA was similarly engineered into mammalian expression vector pcDNA3.1(−)/Hygro. The human DGAT1, GPAT1, and GPAT3 clones with or without FLAG tag were generated as previously described (1, 9, 10). Expression of recombinant proteins in mammalian HEK293 cells or insect Sf-9 cells was performed as described (1). Protein expression levels were visualized by Coomassie blue staining or Western analysis using anti-FLAG antibody.
GPAT activity assays
GPAT activity assay was performed as described (1), using [14C]glycerol 3-phosphate (G3P) and lauroyl-CoA as substrates. Briefly, the assay was conducted in 75 mM Tris HCl, pH 7.5, 4 mM MgCl2, 1 mg/ml fatty acid free BSA with 150 µM [14C]G3P (55 mCi/mmol) and 50 µM acyl-CoA for 15 min at room temperature in the presence of 50 or 100 µg of total lysate protein. Under some circumstances, the assay was performed under similar conditions with G3P and [14C]palmitoyl-CoA as substrates. Lipids were extracted by using chloroform:methanol (2:1, v/v), dried, and separated by TLC with chloroform:methanol:water (65:25:4, v/v) followed by exposure to a PhosphorImager screen. Where indicated, cell lysates were incubated with or without 0.4 µM NEM for 15 min on ice before the initiation of reaction.
Transcriptional profiling and quantitative RT-PCR analysis
Undifferentiated and differentiated 3T3-L1 adipocytes, tissues from normal, 8- to 12-week-old male C57BL/6J mice, as well as tissues from 10-week-old male ob/ob and age-matched wild-type control mice, were obtained as described (1). For PPARγ agonist treatment, 10-week-old male ob/ob mice were gavaged once per day with 15 mg/kg rosiglitazone or vehicle control for 21 days. RNA was extracted from cells and tissues using Trizol (Invitrogen) and purified using the RNeasy kit, including DNaseI treatment (Qiagen). RNA from human tissues was obtained from Clontech (Mountain View, CA). Taqman real-time quantitative PCR (Q-PCR) was performed by using an ABI Prism 7900 sequence detector (Applied Biosystems, Foster City, CA) with 18S as an internal control as described (1). Under some circumstances, we used plasmid DNAs containing GPAT3 or GPAT4 cDNA to quantitate tissue mRNA abundance. Gene-specific primers and probes were obtained from Applied Biosystems. Relative expression was determined by the Ct method (Applied Biosystems).
ShRNA-mediated knockdown of GPAT3 and GPAT4 in 3T3L1 adipocytes
A set of different 29-nt short hairpin RNA (shRNA) constructs targeting GPAT3 or GPAT4 mRNA was obtained from OriGene Technologies Inc. The gene-specific shRNA cassettes were designed with a pattern of sense-loop-antisense using loop sequence TCAAGAG and inserted into the BamH1 and HindIII cloning sites of the mammalian expression vector pGFP-V-RS containing a puromycin-selection marker. The sense oligonucleotides were GPAT3-sh1: 5′- GATCAGCTTGCTGATTATAGGAACTACAC; GPAT3-sh2: 5′- GCAGACCTGGCGGTGAAGCTCCTGTCCAC; GPAT4 sh1:5′-CACGTTCACTTAATGTGCTACCGTATCTG; GPAT4 sh2: 5′-ATCGCCACCTGGTGGCTAAGAGGCTGACT. All constructs were sequenced to confirm the correct insertion. The recombinant retroviral constructs were transfected into PT67 packaging cell line with Lipofectamine 2000 (Invitrogen) by following the KnockoutTM RNAi systems User Manual from Clontech. Two days after transfection, retrovirus-containing medium was collected to infect 3T3-L1 cells. Puromycin was used to select stable 3T3-L1 cell lines that express targeting shRNAs. The selected 3T3-L1 cells were maintained in puromycin-containing medium for differentiation assays.
3T3-L1 preadipocyte differentiation assay
3T3-L1 cells expressing control shRNA or targeted shRNAs were grown in DMEM supplemented with 10% (v/v) FBS, 100 U/ml penicillin, 100 mg/ml streptomycin, 2 mM L-glutamine, and 2 µg/ml puromycin. Two days after the confluence (defined as day 0), the cells were refreshed with DMEM containing 10% FBS, 2 µg/ml insulin, 0.25 µM dexamethasone and 0.5 mM IBMX. The cells were cultured in this medium for 4 days, then the medium was switched to DMEM containing 10% FBS and 2 µg/ml insulin for 2 days, followed by another 2-day incubation with medium containing only 10% FBS. Neutral lipid accumulation in differentiated cells was determined by Oil Red-O staining after a fixation with 3.7% formaldehyde.
Immunoprecipitation and detection of protein phosphorylation
HepG2 cells were transfected with empty vector, Flag-hGPAT3, or Flag-hGPAT4, respectively. Forty-eight h after transfection, cells were treated with 50 nM insulin (Sigma) for 10 min and lysed with 1% Triton X-100 in ice-cold PBS. Under certain circumstances, cells were treated with 100 nM wortamannin (Sigma) for 2 h before addition of insulin. Cell lysates were incubated with anti-Flag antibody M2 (Sigma) and protein A/G agarose (Santa Cruz Biotechnologies, Santa Cruz, CA) at 4°C for 16 h. Then, the agarose beads were washed with PBS plus 1% Triton X-100, and proteins were eluted with SDS loading buffer. Samples were separated by SDS-PAGE and blotted with anti-Flag, anti-pTyr (Millipore, Billerica, MA), and anti-pSer (Santa Cruz) antibodies, respectively. To monitor insulin signaling, cell lysates were also separated and blotted with anti-pAkt antibody (Cell Signaling Technology, Danvers, MA).
Statistical analysis
Statistical significance was determined by Student's t-test.
RESULTS
Phylogenetic tree of the acyltransferase family
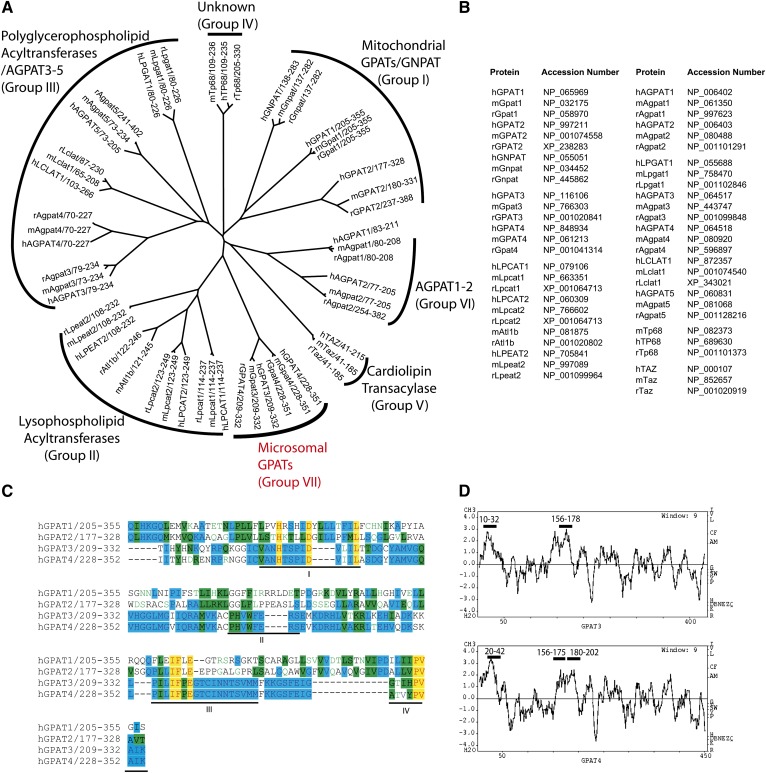
It was previously found that several glycerolipid acyltransferases (e.g., mitochondrial GPAT1, LPAAT1, and LPAAT2) in TAG and/or phospholipid biosynthetic pathway contain a conserved domain characteristic of four conserved motifs (11). These enzymes belong to a superfamily of glycerophospholipid acyltransferases, members of which all share the pfam01553 acyltransferase domain as seen in NCBI database. A phylogenetic analysis of the conserved domain within mouse (m), rat (r), and human (h) orthologs divides the family into several subfamilies (the numbers of subfamilies given below are arbitrary and may not follow the order in the tree in Fig. 1A). The mitochondrial GPAT1, GPAT2, and a dihydroxyacetone-phosphate acyltransferase (GNPAT) belong to the same subfamily. A second subfamily contains three newly discovered lysophospholipid acyltransferases (LPCAT1, LPCAT2/LysoPAFAT, and LPEAT2) as well as an orphan member called Acyltransferase-like 1b (Atl1b) that appears to be present in mouse and rat only. The third subgroup contains the two recently identified polyglycerophospholipid acyltransferases LPGAT1 and LCLAT1, as well as several orphan acyltransferases named AGPAT3, AGPAT4, and AGPAT5. AGPAT3 was recently reported to have both AGPAT and lysophosphatidylinositol acyltransferase (LPIAT) activities with a preference for arachidonoyl-CoA as a donor (12). Tafazzin, a transacylase important for cardiolipin remodeling by operating an acyl-CoA–independent transacylation reaction, and a protein with unknown function called transmembrane protein 68 (TP68) are remotely related to other members and fall into subgroups 4 and 5, respectively. The two proteins with defined AGPAT activity, AGPAT1 and AGPAT2, share high homology and form subgroup 6. Microsomal GPAT3 and GPAT4 are closely related and form a separate subgroup 7. Notably, GPAT3 and GPAT4 do not share appreciable homology with mitochondrial GPAT1 and GPAT2 (less than 20% of amino acid identity even in the predicted glycerolipid acyltransferase domain). The accession numbers for the proteins shown are listed in Fig. 1B. All four GPAT isoforms identified so far are characterized by four classical sequence motifs (I-IV, underlined in Fig. 1C). A fifth motif, which is predicted to be required for enzymatic activity, was noted in some, but not all, family members (7, 13). The hydrophobicity profile between GPAT3 and GPAT4 appears to be very similar (Fig. 1D) and suggests 2 and 3 transmembrane domains, respectively, in GPAT3 and GPAT4 (Fig. 1D, as indicated by bold bars above the corresponding amino acids sequence). Despite the high degree of homology, the two genes are localized in different chromosomes: human GPAT3 resides at 4q21.23; and human GPAT4 is localized at 8p11.21.
Fig. 1.
Phylogenetic alignment of the pfam01553 glycerophospholipid acyltransferases family and sequence analysis of GPATs. A: Phylogenetic tree showing the relatedness of protein sequences of mouse (m), rat (r), and human (h) glycerophospholipid acyltransferases within the region containing the conserved pfam01553 domain. The alignment was performed with ClustalW, and the tree was created using TreeView program. B: Protein accession numbers of glycerophospholipid acyltransferase family members shown in A. C: Sequence alignment of human GPAT1-4. Amino acids that are identical are shadowed in yellow; those that are similar are shadowed in green and/or blue. D: Kyte-Doolittle hydrophobicity profile of human GPAT3 and GPAT4. The predicted transmembrane domains are indicated by bold bars above the corresponding amino acids sequences.
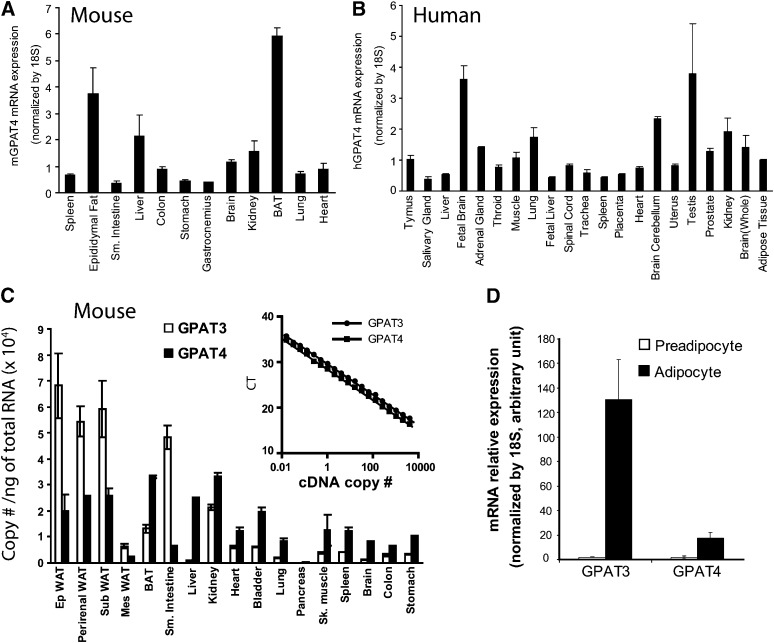
Tissue distribution and regulation of GPAT3 and GPAT4 mRNA
We previously reported the tissue distribution pattern of GPAT3 transcripts (14). In the present study, we set out to examine the mRNA expression of GPAT4 across the similar panel of mouse and human tissues by using quantitative PCR with 18S as internal control. Within the set of tissues examined, mouse GPAT4 was highly expressed in brown adipose tissue, epididymal fat, and liver, followed by kidney, heart, and brain (Fig. 2A). Expression of GPAT4, albeit at lower levels, was also detected in other mouse tissues tested (Fig. 2A). In humans, GPAT4 mRNA was widely expressed in many tissues tested, with the highest expression levels in testis, brain, kidney, and lung (Fig. 2B). Significant levels of human GPAT4 mRNA were also found in adipose tissue and liver (Fig. 2B). As GPAT3 mRNA expression level in both human and mouse liver is notably low, GPAT4 may encode the major microsomal GPAT activity in this tissue. Our data also suggest a more ubiquitous expression pattern of GPAT4 (Fig. 2A, B) compared with GPAT3 (14). To better assess the abundance of GPAT3 and GPAT4 mRNA in mouse tissues, we used plasmids to quantitate amplification for each probe set, which yields almost identical amplification on a molar basis (Fig. 2C, inset). The standard curves generated from each probe set were then used to calculate the copy number of mRNA per ng of total RNA tested. Again, as shown in Fig. 2C, both GPAT3 and GPAT4 mRNA are highly expressed in mouse adipose tissues. However, it appears that GPAT3 transcripts are more abundant than GPAT4 in white adipose tissues examined (including epididymal, perirenal, mensenteric, and subcutaneous fat pad), whereas GPAT4 mRNA expression levels are higher in brown adipose tissue. There are more copies of GPAT3 transcripts than GPAT4 in small intestine (Fig. 2C). GPAT4 is a predominant microsomal GPAT isoform at the mRNA level in liver, kidney, heart, lung, and many other mouse tissues examined (Fig. 2C).
Fig. 2.
Tissue distribution regulation of GPAT3 and GPAT4 mRNA detected by Q-PCR. Tissue distribution of mouse (A) and human (B) GPAT4 mRNA. C: Comparison of GPAT3 and GPAT4 mRNA expression in mouse tissues as determined by Q-PCR analysis. Inset graphs are standard curves generated using GPAT3 and GPAT4 cDNA-containing plasmids and were used to determine copy numbers/ng RNA. D: Induction of GPAT3 and GPAT4 mRNA during 3T3-L1 cells differentiation. Data are expressed as mean ± SD (n = 4). CT, cycle time; GPAT, acyl-CoA:glycerol-3-phosphate acyltransferase; Q-PCR, quantitative PCR.
GPAT3 mRNA was found to be dramatically upregulated during the differentiation of 3T3-L1 adipocytes (∼70-fold) (1). This was confirmed in the present study (Fig. 2D). As GPAT4 is also highly expressed in adipose tissue, we examined its mRNA expression level during the differentiation of 3T3-L1 preadipocytes to adipocytes. Q-PCR analysis revealed that GPAT4 is moderately induced during adipogenesis (∼9-fold). Probably due to its robust induction during the adipogenic program, the level of GPAT3 mRNA in fully differentiated 3T3-L1 adipocytes was significantly higher (∼4-fold) compared with the level of GPAT4 mRNA (Fig. 2D). We previously reported that GPAT3 was upregulated by rosiglitazone (Rosi, a potent PPARγ agonist) treatment, and its expression level was reciprocally regulated in white adipose tissue and liver in ob/ob mice (1). Using the same samples, we found that GPAT4 mRNA levels remain unchanged in white adipose tissue and liver of both ob/ob mice and mice treated with rosiglitazone (supplementary Fig. I), suggesting that GPAT4, unlike GPAT3, is not a target gene for PPARγ activation.
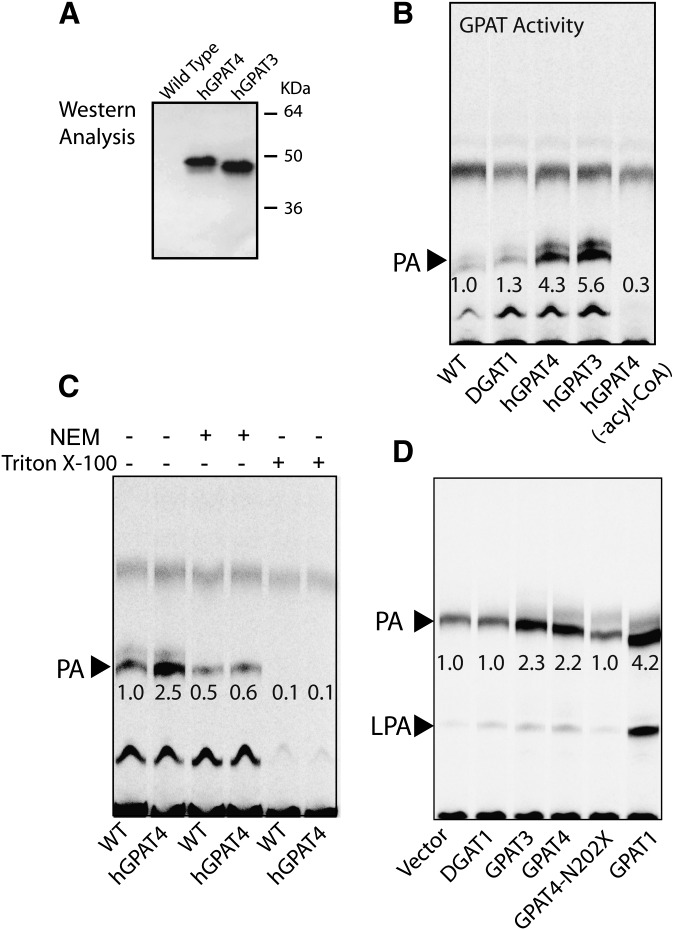
Characterization of GPAT4 activity
To compare the enzymatic activity of GPAT3 and GPAT4, we expressed both proteins, each tagged with an FLAG epitope, in Sf-9 cells using a baculovirus system, and performed the GPAT activity assay in lysates prepared from infected cells. Western analysis with anti-Flag antibody confirmed comparable expression of GPAT3 and GPAT4 proteins, both migrating on SDS-PAGE to predicted positions (∼50 kDa) (Fig. 3A). The expression level of GPAT3 and GPAT4 protein in infected cells was also shown to be similar (Fig. 3A). Baculoviral expression of hGPAT4 and hGPAT3 in Sf9 cells resulted in a profound increase in GPAT activity. As shown in Fig. 3B, the formation of PA, a major end-product of GPAT activity in the presence of cell lysates, was enhanced 4.3- and 5.6-fold, respectively, by hGPAT4 and hGPAT3 overexpression, compared with cells infected with wild-type virus. In the absence of exogenously added acyl-CoA, PA formation was minimal, indicating its specificity as a marker for GPAT activity. As expected, overexpression of DGAT1 in Sf-9 cells did not cause an appreciable increase in GPAT activity (Fig. 4B, lane 2 versus 1). GPAT3 activity has been shown to be sensitive to inactivation by NEM treatment (1). Similar to GPAT3, the increased GPAT activity conferred by GPAT4 overexpression in Sf-9 cells was almost completely abolished by NEM treatment (Fig. 3C, lanes 3, 4 versus lanes 1, 2). Membrane proteins vary by their sensitivity to detergents, while several members of the acyltransferase family, such as DGAT2 and MGAT2, are readily inactivated by detergents, including Triton X-100 and CHAPS; the MBOAT family member DGAT1 is resistant to detergent treatment (15, 16). GPAT4 activity in cell lysates was sensitive to detergent treatment, as indicated by a dramatic decrease in GPAT activity after a preincubation with Triton X-100 (Fig. 3C, lane 6 versus 2).
Fig. 3.
GPAT activity of GPAT3 and GPAT4 expressed in Sf-9 or mammalian cells. A: Western analysis showing expression of GPAT3 and GPAT4 in Sf-9 cells. B: TLC analysis for GPAT activity in Sf9 cells infected with wild-type virus or virus containing hDGAT1, hGPAT4, or hGPAT3 cDNA. C: GPAT activity conferred by GPAT4-overexpression in Sf-9 cells was sensitive to inactivation by NEM and Triton X-100. D: TLC analysis for GPAT activity in mammalian HEK293T cells transfected with empty vector or vectors containing cDNAs corresponding to hDGAT1, hGPAT3, hGPAT4, truncated hGPAT4 encoding N-terminal 202aa, or hGPAT1. The embedded numbers represent the relative levels of formed radiolabeled PA (indicated by an arrow). The fast-migrating band above PA (B and C) may represent a product derived from G3P by endogenous acyl-CoA-independent enzyme(s), possibly phosphatidylglycerol phosphate or phosphatidylglycerol. The identity of the slow-running band below PA (B and C) is unknown. Data are representative of at least two independent experiments with similar results. GPAT, acyl-CoA:glycerol-3-phosphate acyltransferase; LPA, lysophosphatidic acid; NEM, N-ethylmaleimide; PA, phosphatidic acid; WT, wild type.
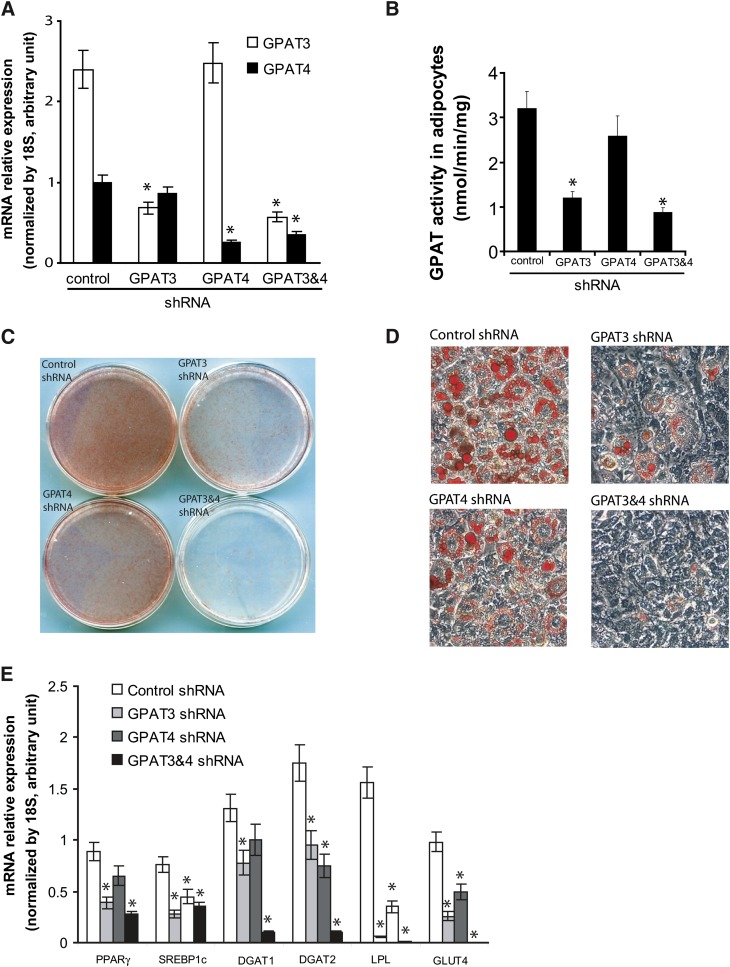
Fig. 4.
Knockdown of GPAT3, but not GPAT4, in 3T3-L1 cells impaired their differentiation to mature adipocytes. A: Ectopic expression of shRNAs specific for GPAT3 or GPAT4 resulted in significant and selective reduction of GPAT3 and GPAT4 mRNA, respectively, in 3T3-L1 adipocytes. Combined expression of both shRNAs led to a significant reduction of both GPAT3 and GPAT4 mRNA. B: GPAT activity in differentiated 3T3-L1 adipocytes expressing different shRNAs. C, D: Oil red-O staining of lipid accumulation in 3T3-L1 adipocytes expressing control shRNA, GPAT3 shRNA, GPAT4 shRNA, or both GPAT3 and GPAT4 shRNAs. Generation of 3T3-L1 cells expressing shRNAs and induction of the cells to differentiated mature adipocytes are described under “Materials and Methods.” Eight days after addition of differentiation agents, cells were fixed and stained with Oil Red-O, and images from either the dishes (C) or a microscopic view of the dishes (D) are shown. E: Expression profile of genes involved in adipogenesis and lipogenesis in differentiated 3T3-L1 adipocytes stably expressing different shRNAs. Values represent the mean of at least three independent measurements ± SD. *P < 0.05 versus control shRNA. DGAT, acyl-CoA:diacylglycerol acyltransferase; GPAT, acyl-CoA:glycerol-3-phosphate acyltransferase; GLUT4, glucose transporter type 4; PPARγ, peroxisome proliferators-activated receptor gamma.
Similar to GPAT3, GPAT4 overexpressed in mammalian HEK293 cells also conferred GPAT activity. As shown in Fig. 4D, overexpression of GPAT3, GPAT4, or mtGPAT1 increased GPAT activity by 2.3-, 2.2-, or 4.2-fold compared with cells transfected with empty vector or DGAT1 cDNA. In contrast, a truncated GPAT4 protein with an elimination of C-terminal acyltransferase domain no longer exhibited GPAT activity (Fig. 3D, lane 5 versus 4). Collectively, these data demonstrate that GPAT4 encodes a protein that displays all the properties characteristic of a microsomal GPAT and has enzymatic activity similar to GPAT3.
GPAT3, but not GPAT4, is critical for the differentiation of 3T3-L1 preadipocytes to adipocytes
GPAT3 and GPAT4 are both highly expressed in adipose tissue. As the major enzymatic activity catalyzing the initial step in de novo triglyceride synthesis, microsomal GPAT was predicted to play a critical role in adipogenesis (17). Knowing the existence of two different microsomal GPAT isoforms, GPAT3 and GPAT4, we felt that it would be important to determine which isoform(s) are required for adipogenesis. To this end, we introduced shRNAs specific for GPAT3 or GPAT4 into 3T3-L1 cells using a retrovirus-mediated approach. A nonspecific shRNA targeting luciferase was used as a control. Q-PCR analysis of GPAT3 and GPAT4 mRNA after differentiation revealed that infection of GPAT3 or GPAT4 shRNA specifically decreased the levels of GPAT3 or GPAT4 mRNA by 70% or 75%, respectively (Fig. 4A). Infection of cells with both GPAT3 and GPAT4 shRNAs led to reduction of both transcripts more than 70% (Fig. 4A). Enzymatic analysis indicates a significant decrease (∼60%) in GPAT activity in 3T3-L1 adipoctes expressing GPAT3 shRNA, whereas knockdown of GPAT4 to a similar level only led to a slight reduction in GPAT activity (Fig. 4B). Oil red-O staining of lipid droplets in differentiated 3T3-L1 adipocytes showed that knockdown of GPAT3 significantly impaired lipid accumulation (Fig. 4C, D). In contrast, the effect of GPAT4 knockdown on adipogenesis of 3T3-L1 cells is only marginal (Fig. 4C, D). Knockdown of both GPAT3 and GPAT4 almost completely inhibited lipid droplet formation (Fig. 4C, D).
To explore the mechanisms by which knockdown of GPAT3 regulates adipocyte differentiation, we performed Q-PCR analysis on a variety of genes important for adipogenesis and lipogenesis, including PPARγ, SREBP1c, GLUT4, DGAT1, DGAT2, and LPL. Expression of PPARγ and SREBP1c, which act as master transcriptional factors for adipocyte differentiation and adipogenesis, were significantly suppressed in adipocytes with knockdown of GPAT3 alone and a combinative knockdown of both GPAT3 and GPAT4 (Fig. 4E). Knockdown of GPAT4 alone yielded a less profound reduction in expression of these two genes (Fig. 4E). As a result, expression levels of genes in glucose and fatty acid uptake (e.g., GLUT4, LPL) and lipogenesis (e.g., DGAT1, DGAT2) were decreased in GPAT3 knockdown cells (Fig. 4E). Thus, knockdown of GPAT3 may not only decrease triglyceride synthesis but impair the process of adipogenic programming. Note that knockdown of GPAT4 also led to a significant, albeit less pronounced (compared with GPAT3 knockdown), decrease in the expression of genes in promoting lipogenesis.
Insulin stimulates the phosphorylation of GPAT3 and GPAT4 and upregulates their activity
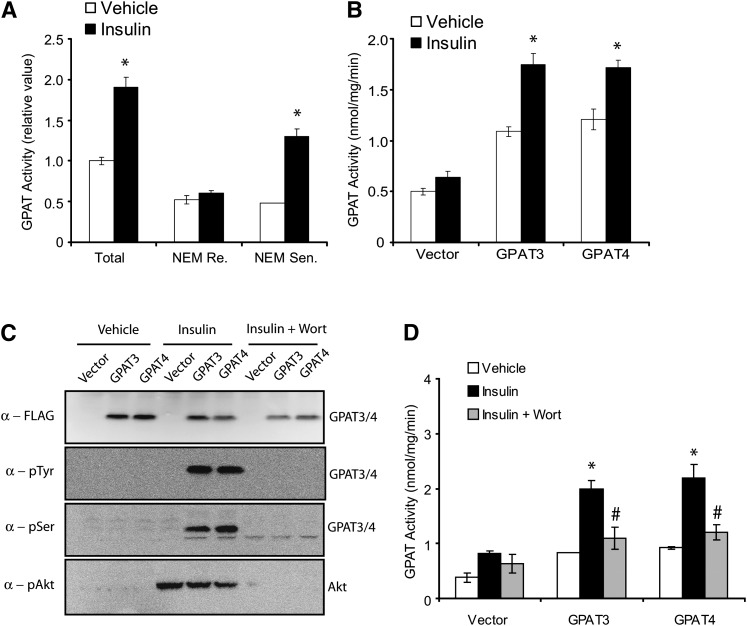
It was previously reported that microsomal GPAT activity is enhanced by insulin, in concert with its lipogenesis-stimulating function. To determine whether an increase in enzymatic activity is due to an insulin-stimulating phosphorylation event on GPAT3 and/or GPAT4, we sought to examine the effects of insulin on phosphorylation and activity of GPAT3 and GPAT4. We first confirmed that in differentiated 3T3-L1 adipocytes, insulin treatment resulted in a significant increase in total GPAT activity that was mainly due to an increase in the NEM-sensitive microsomal GPAT activity (Fig. 5A) (18, 19). We next examined the effects of insulin on FLAG-tagged GPAT3 or GPAT4 overexpressed in HepG2 cells. As expected, overexpression of either GPAT3 or GPAT4 caused a 2- to 3-fold increase in GPAT activity (Fig. 5B, white bars). While insulin had little effect on endogenous GPAT activity in HepG2 cells, it significantly increased the GPAT activity in GPAT3- or GPAT4-overexpressing cells (Fig. 5B, black bars), suggesting that the activity of both GPAT3 and GPAT4 can be enhanced by insulin. It has been well documented that the actions of insulin are mediated by phosphorylation cascades, which eventually lead to transcriptional or posttranscriptional regulation (e.g, phosphorylation) of target proteins. To further investigate whether insulin treatment can activate phosphorylation of GPAT3 and/or GPAT4, and consequently, modulate their functions, we used specific anti-p-Tyr and anti-p-Ser antibodies and wortmannin (a specific inhibitor against phosphatidylinositol 3-OH kinase, which blocks insulin signaling). Western analysis on proteins immunoprecipitated by anti-FLAG antibody indicated the expression of recombinant GPAT3 and GPAT4 (Fig. 5C, top panel). Insulin treatment resulted in phosphorylation of both GPAT3 and GPAT4, as visualized by probing with antiphosphotyrosine antibody (anti-p-Tyr) or antiphosphoserine antibody (anti-p-Ser) (Fig. 5C, middle two panels). Incubating cells with wortmannin completely blocked the insulin-stimulated phosphorylation of GPAT3 and GPAT4 (Fig. 5C). As expected, insulin also stimulated the wortmannin-sensitive Akt phosphorylation (Fig. 5C, bottom panel). Furthermore, wortmannin treatment completely abolished the insulin-induced GPAT activity in GPAT3 and GPAT4, suggesting that the enhancing effect of insulin on activity of GPAT3 and GPAT4 depends on its downstream signaling cascades.
Fig. 5.
Insulin stimulated phosphorylation of GPAT3 and GPAT4 and enhanced their activities. A: Insulin treatment increased NEM-sensitive (Sen) microsomal, but not NEM-resistant (Re) mitochondrial GPAT activity in 3T3-L1 adipocytes. Cells were starved (serum removed from medium) for 18 h before insulin addition. *P < 0.05 versus vehicle. B: Insulin treatment enhanced GPAT activity of HepG2 cells transiently transfected with GPAT3 or GPAT4. *P < 0.05 versus vehicle. C: Insulin stimulated wortmannin (Wort)-sensitive phosphorylation of GPAT3 and GPAT4. Lysates prepared from HepG2 cells transiently transfected with FLAG-tagged GPAT3 or GPAT4 cDNA were immunoprecipitated with anti-FLAG antibody and subjected to Western analysis by phosphospecific antibodies to Tyr or Ser residues. The membrane was also blotted with anti-pAkt antibody to monitor the general insulin-stimulated signaling cascades. D: Wortmannin treatment abolished the insulin-stimulated increase in GPAT activity in HepG2 cells overexpressing GPAT3 or GPAT4. *P < 0.05 versus vehicle; #P < 0.05 versus insulin. GPAT activity was measured with G3P and [14C]palmitoyl-CoA as substrates. The vehicle used is serum-free medium (A and B) or 0.1% DMSO in serum-free medium (C and D). All data are expressed as mean ± SD (n = 3–4). GPAT, acyl-CoA:glycerol-3-phosphate acyltransferase; Ser, Serine; Tyr, Tyrosin.
DISCUSSION
We and others have previously identified two closely related orphan members of the glycerolipid acyltransferase family as ER-associated microsomal GPATs, GPAT3 and GPAT4 (1, 3, 4). Here we further examined the role of these two microsomal GPAT isoforms in regulating glycerolipid metabolism as well as their regulation by insulin or PPARγ. Our data strongly argue that GPAT3 plays an important role in controlling triglyceride biosynthesis in adipocytes. First, detailed analysis of transcript abundance by Q-PCR indicates that there are more copies of GPAT3 transcripts than GPAT4 in both a number of different white adipose tissue depots and differentiated 3T3-L1 cells, suggesting that GPAT3 is a major microsomal GPAT isoform in adipocytes. Furthermore, targeted knockdown of GPAT3 in 3T3-L1 cells significantly decreased GPAT activity and impaired the adipocyte differentiation, whereas knockdown of GPAT4 only had marginal effects, suggesting a more important role of GPAT3 during the conversion of 3T3-L1 to adipocytes. Consistent with our results, the total and NEM-sensitive GPAT activities in gonadal fat from GPAT4−/− mice were reported to remain unchanged compared with wild-type mice, and embryonic fibroblast cells from GPAT4−/− mice can undergo normal differentiation to adipocytes (4, 8). However, the GPAT4−/− mice are resistant to diet-induced obesity and have a substantial reduction of subdermal adipose tissue when they are over 4 months old (8), suggesting an indispensable role of GPAT4 in regulating adipogenesis in vivo. How can we reconcile this dissociation between mice and the cell-based model? One possible explanation is that the lack of subdermal fat in GPAT4-deficient mice could be secondary to the increased energy expenditure in these mice (4). In addition, differentiation of 3T3-L1 cells to adipocytes in vitro is induced by the three drugs within a relatively short period of time (∼7 days) and may not be able to fully recapitulate the adipogenesis induced by diet in vivo. Nevertheless, findings from this study and previous work highlight the complexity of in vivo triglyceride metabolism and can be used as entry point to further understand the role of GPAT3 and GPAT4 in physiological settings.
In contrast to GPAT3 as a major microsomal GPAT isoform in WAT, GPAT4 may encode the primary NEM-sensitive GPAT activity in liver, brown adipose tissue (BAT), and mammary gland. Our Q-PCR data indicate that the expression level of GPAT4 is much higher than GPAT3 in liver and BAT. Consistent with these findings, studies using tissues lysates from GPAT4−/− mice liver revealed that total and NEM-sensitive GPAT activities were reduced by ∼50% and ∼65%, respectively, resulting in 45% less triglyceride content in this organ (4). GPAT activities from BAT were also similarly reduced (4). The content of triglyceride and diglyceride in milk of lactating GPAT4−/− mice was reduced by 90%, probably due to a ∼90% decrease in total GPAT activity in mammary epithelial cells (3, 7). Thus, the two microsomal GPATs appear to exert their function as an initiating enzyme in glycerolipid synthesis in a tissue-selective manner. It is tempting to speculate that one reason for having multiple GPAT isoforms with differential tissue distribution is to facilitate tissue-specific transcriptional regulation. GPAT3 may be more susceptible to be induced by exogenous stimulus and thus represent an inducible isoform of GPAT activity, whereas GPAT4 may be a constitutive isoform. In support of this notion, induction of GPAT3 is at least ten times higher than GPAT4 during the differentiation of 3T3-L1 cells to adipocytes. In addition, GPAT3 is strongly regulated by PPARγ activation, genetically-induced obesity, and nutritional status, while GPAT4 expression is not affected under those conditions (1) (this study). This is reminiscent of other enzymes that form important, biologically active metabolites. For instance, cyclooxygenase (COX) is an enzyme responsible for formation of prostanoids, important mediators for inflammation. There are two active form of COX: COX-1 and COX-2. COX-1 is considered a constitutive enzyme, while COX-2 is an inducible enzyme, becoming abundant in activated inflammatory cells (20).
The impairment on adipocyte differentiation by silencing GPAT3 revealed in the current study is reminiscent of that observed by knockdown of acylglycerol 3-phosphate acyltransferase-2 (AGPAT2) (21), an enzyme catalyzing the second LPA-to-PA converting step in de novo triglyceride biosynthesis. Similarly, suppression of calcium- independent intracellular PLA2β and γ, two other enzymes controlling abundance of cellular PA and LPA by phospholipids remodeling pathway, also prevented adipocyte differentiation (22). Remarkably, PPARγ, C/EBPα, and/or SREBPc, the key transcriptional regulators in the differentiation of adipocytes, are suppressed in all of these studies when levels of LPA- or PA-synthesizing enzymes were reduced. These findings all support the notion that LPA and PA can serve as critical factors in initiating adipogenic program, independent of their roles as triglyceride synthetic precursors. Consistent with this notion, the role of LPA, PA, and their downstream products, such as phospholipids, in regulating cell differentiation and growth has been well documented (23, 24). Our transcriptional profiling data indicate that both GPAT3 and GPAT4 transcripts are expressed at high levels in monocytes, macrophage, and certain myeloid cells (J. Cao et al., unpublished observations); it may not be an exaggeration to speculate that GPAT3 and/or GPAT4 are involved in producing increased levels of LPA and PA in activated inflammatory cells. More recently, Tang et al. reported that overexpression of GPAT3 in HEK293T cells induced mTOR-dependent p70S6K phosphorylation, implying its potential role in mTOR signaling and cancer therapy (25). Further investigations are needed to uncover the pathophysiological functions of these molecules in inflammation and cancer.
Early characterization of GPAT activities in primary tissues and cells showed that both mitochondrial and microsomal GPAT activities are enhanced by insulin, in concert with its lipogenenesis-stimulating function (18, 26, 27). We confirmed these earlier findings in differentiated 3T3-L1 adipocytes (Fig. 5A). With the clones of two microsomal GPATs in hand, we further showed that insulin stimulated the phosphorylation of GPAT3 and GPAT4 and increased their activities. Such effects of insulin on GPAT3 and GPAT4 are sensitive to inactivation by wortmannin, suggesting a dependence on phosphorylation cascades in the insulin signaling pathway. Predicting potential phosphorylation sites with two most popular bioinformatic tools [Scansite (http://scansite.mit.edu) and NetPhos 2.0 Server (http://www.cbs.dtu.dk/services/NetPhos/)] revealed a number of potential Tyrosine (Tyr), Serine (Ser), or Threonine (The) phosphorylation sites in both hGPAT3 and hGPAT4 amino acid sequence (data not shown). Indeed, searching the phosphosites at PhosphoSitePlus (www.phosphosite.org) revealed the presence of three putative phosphorylation sites S68, S77, and Y423 in human GPAT3, which was supported by experimental evidence. No phosphorylation site in hGPAT4 is recorded on the Web site. To identify and confirm the insulin-stimulated phosphorylation sites within GPAT3 and GPAT4 experimentally, further mass spectrometric and/or mutagenic analysis is warranted. Other enzymes in de novo triglyceride synthesis have also been shown to be regulated by insulin-stimulated phosphorylation. For instance, insulin controls the phosphorylation of lipin, the phosphatidic acid phosphatase converting PA to DAG, leading to altered subcellular localization (28, 29).
All four GPAT isoforms identified so far are members of the pfam01553 glycerophospholipid acyltransferase family. Additional genes encoding GPAT activity may exist. For instance, a recent study using GPAT1 knockout mice suggested the existence of a fifth GPAT isoform in heart which is NEM-resistant and localized on mitochondria (30). Given the fact that none of the orphan members share appreciable homology with the identified GPAT1–4 (Fig. 1A), it is unlikely to define additional GPATs out of the pfam01553 family. Instead, new GPAT isoforms may emerge from structurally distinct protein families, members of which recognize substrates that are structurally related to glycerol 3-phosphate. One possible candidate family is the recently discovered membrane-bound-O-acyltransferase (MBOAT) family. Members in this family can recognize a variety of diverse substrates, including neutral lipids (e.g., cholesterol, diacylglycerol, wax) (10, 31); lysophospholipid (e.g., LPC, LPI, and LPE) (32, 33); and peptide (e.g., ghrelin, hedgehog, and Wnt) (34–36). It will be interesting to see whether some enzymes functioning as lysophospholipid acyltransferase can acylate glycerol 3-phosphate.
CONCLUSION
We have examined the tissue distribution, insulin-stimulated phosphorylation, and involvement in adipocyte biology of GPAT3 and GPAT4, the two recently identified microsomal GPATs. Our data suggest that GPAT3 and GPAT4 may play different roles in glycerolipid metabolism: GPAT3 is a critical regulator for lipid accumulation during adipocyte differentiation, whereas GPAT4 may be more responsible for constitutive glycerolipid metabolism. In addition, our results demonstrated that the function of GPAT3 and GPAT4 is controlled by insulin-mediated phosphorylation, implying their importance in lipogenesis. Finally, as enzymes in triglyceride synthesis (e.g., DGAT1) have emerged as important therapeutic targets for treating obesity, type 2 diabetes, and dyslipidemia, molecular identification of GPAT3 and GPAT4—and the subsequent characterization of these enzymes at pathophysiological levels—will help us evaluate them as targets for nutrient modification or intervention for metabolic diseases.
Supplementary Material
Footnotes
Abbreviations:
- AGPAT
- acyl-CoA:acylglycerol-3-phosphate acyltransferase
- BAT
- brown adipose tissue
- C/EBPα
- CCAAT/enhancer binding protein alpha
- DAG
- diacylglycerol
- DGAT
- acyl-CoA:diacylglycerol acyltransferase
- ER
- endoplasmic reticulum
- G3P
- glycerol-3-phosphate
- GPAT
- acyl-CoA:glycerol-3-phosphate acyltransferase
- LPAAT
- lysophosphatidic acid acyltansferase
- NEM
- N-ethylmaleimide
- PPARγ
- peroxisome proliferators-activated receptor gamma
- Q-PCR
- quantitative PCR
- SREBP1c
- sterol regulatory element binding protein 1c
- TAG
- triacylglycerol
- WAT
- white adipose tissue
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of one figure.
REFERENCES
- 1.Cao J., Li J. L., Li D., Tobin J. F., Gimeno R. E. 2006. Molecular identification of microsomal acyl-CoA:glycerol-3-phosphate acyltransferase, a key enzyme in de novo triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 103: 19695–19700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang S., Lee D. P., Gong N., Schwerbrock N. M., Mashek D. G., Gonzalez-Baro M. R., Stapleton C., Li L. O., Lewin T. M., Coleman R. A. 2007. Cloning and functional characterization of a novel mitochondrial N-ethylmaleimide-sensitive glycerol-3-phosphate acyltransferase (GPAT2). Arch. Biochem. Biophys. 465: 347–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chen Y. Q., Kuo M. S., Li S., Bui H. H., Peake D. A., Sanders P. E., Thibodeaux S. J., Chu S., Qian Y. W., Zhao Y., et al. 2008. AGPAT6 is a novel microsomal glycerol-3-phosphate acyltransferase. J. Biol. Chem. 283: 10048–10057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nagle C. A., Vergnes L., Dejong H., Wang S., Lewin T. M., Reue K., Coleman R. A. 2008. Identification of a novel sn-glycerol-3-phosphate acyltransferase isoform, GPAT4, as the enzyme deficient in Agpat6−/− mice. J. Lipid Res. 49: 823–831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yet S. F., Lee S., Hahm Y. T., Sul H. S. 1993. Expression and identification of p90 as the murine mitochondrial glycerol-3- phosphate acyltransferase. Biochemistry. 32: 9486–9491. [DOI] [PubMed] [Google Scholar]
- 6.Lewin T. M., Schwerbrock N. M., Lee D. P., Coleman R. A. 2004. Identification of a new glycerol-3-phosphate acyltransferase isoenzyme, mtGPAT2, in mitochondria. J. Biol. Chem. 279: 13488–13495. [DOI] [PubMed] [Google Scholar]
- 7.Beigneux A. P., Vergnes L., Qiao X., Quatela S., Davis R., Watkins S. M., Coleman R. A., Walzem R. L., Philips M., Reue K., et al. 2006. Agpat6–a novel lipid biosynthetic gene required for triacylglycerol production in mammary epithelium. J. Lipid Res. 47: 734–744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Vergnes L., Beigneux A. P., Davis R., Watkins S. M., Young S. G., Reue K. 2006. Agpat6 deficiency causes subdermal lipodystrophy and resistance to obesity. J. Lipid Res. 47: 745–754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cao J., Liu Y., Lockwood J., Burn P., Shi Y. 2004. A novel cardiolipin-remodeling pathway revealed by a gene encoding an endoplasmic reticulum-associated acyl-CoA:lysocardiolipin acyltransferase (ALCAT1) in mouse. J. Biol. Chem. 279: 31727–31734. [DOI] [PubMed] [Google Scholar]
- 10.Cases S., Smith S. J., Zheng Y. W., Myers H. M., Lear S. R., Sande E., Novak S., Collins C., Welch C. B., Lusis A. J., et al. 1998. Identification of a gene encoding an acyl CoA:diacylglycerol acyltransferase, a key enzyme in triacylglycerol synthesis. Proc. Natl. Acad. Sci. USA. 95: 13018–13023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lewin T. M., Wang P., Coleman R. A. 1999. Analysis of amino acid motifs diagnostic for the sn-glycerol-3-phosphate acyltransferase reaction. Biochemistry. 38: 5764–5771. [DOI] [PubMed] [Google Scholar]
- 12.Yuki K., Shindou H., Hishikawa D., Shimizu T. 2009. Characterization of mouse lysophosphatidic acid acyltransferase 3: an enzyme with dual functions in the testis. J. Lipid Res. 50: 860–869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Agarwal A. K., Arioglu E., De Almeida S., Akkoc N., Taylor S. I., Bowcock A. M., Barnes R. I., Garg A. 2002. AGPAT2 is mutated in congenital generalized lipodystrophy linked to chromosome 9q34. Nat. Genet. 31: 21–23. [DOI] [PubMed] [Google Scholar]
- 14.Cao J., Shan D., Revett T., Li D., Wu L., Liu W., Tobin J. F., Gimeno R. E. 2008. Molecular identification of a novel mammalian brain isoform of acyl-CoA:lysophospholipid acyltransferase with prominent ethanolamine lysophospholipid acylating activity, LPEAT2. J. Biol. Chem. 283: 19049–19057. [DOI] [PubMed] [Google Scholar]
- 15.Cao J., Burn P., Shi Y. 2003. Properties of the mouse intestinal acyl-CoA:monoacylglycerol acyltransferase, MGAT2. J. Biol. Chem. 278: 25657–25663. [DOI] [PubMed] [Google Scholar]
- 16.Cao J., Cheng L., Shi Y. 2007. Catalytic properties of MGAT3, a putative triacylgycerol synthase. J. Lipid Res. 48: 583–591. [DOI] [PubMed] [Google Scholar]
- 17.Coleman R. A., Reed B. C., Mackall J. C., Student A. K., Lane M. D., Bell R. M. 1978. Selective changes in microsomal enzymes of triacylglycerol phosphatidylcholine, and phosphatidylethanolamine biosynthesis during differentiation of 3T3–L1 preadipocytes. J. Biol. Chem. 253: 7256–7261. [PubMed] [Google Scholar]
- 18.Farese R. V., Standaert M. L., Yamada K., Huang L. C., Zhang C., Cooper D. R., Wang Z., Yang Y., Suzuki S., Toyota T., et al. 1994. Insulin-induced activation of glycerol-3-phosphate acyltransferase by a chiro-inositol-containing insulin mediator is defective in adipocytes of insulin-resistant, type II diabetic, Goto-Kakizaki rats. Proc. Natl. Acad. Sci. USA. 91: 11040–11044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Saggerson E. D., Carpenter C. A. 1987. Effects of streptozotocin-diabetes and insulin administration in vivo or in vitro on the activities of five enzymes in the adipose-tissue triacylglycerol-synthesis pathway. Biochem. J. 243: 289–292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mitchell J. A., Warner T. D. 2006. COX isoforms in the cardiovascular system: understanding the activities of non-steroidal anti-inflammatory drugs. Nat. Rev. Drug Discov. 5: 75–86. [DOI] [PubMed] [Google Scholar]
- 21.Gale S. E., Frolov A., Han X., Bickel P. E., Cao L., Bowcock A., Schaffer J. E., Ory D. S. 2006. A regulatory role for 1-acylglycerol-3-phosphate-O-acyltransferase 2 in adipocyte differentiation. J. Biol. Chem. 281: 11082–11089. [DOI] [PubMed] [Google Scholar]
- 22.Su X., Mancuso D. J., Bickel P. E., Jenkins C. M., Gross R. W. 2004. Small interfering RNA knockdown of calcium-independent phospholipases A2 beta or gamma inhibits the hormone-induced differentiation of 3T3–L1 preadipocytes. J. Biol. Chem. 279: 21740–21748. [DOI] [PubMed] [Google Scholar]
- 23.Goetzl E. J., Lee H., Dolezalova H., Kalli K. R., Conover C. A., Hu Y. L., Azuma T., Stossel T. P., Karliner J. S., Jaffe R. B. 2000. Mechanisms of lysolipid phosphate effects on cellular survival and proliferation. Ann. N. Y. Acad. Sci. 905: 177–187. [DOI] [PubMed] [Google Scholar]
- 24.Moolenaar W. H. 1995. Lysophosphatidic acid, a multifunctional phospholipid messenger. J. Biol. Chem. 270: 12949–12952. [DOI] [PubMed] [Google Scholar]
- 25.Tang W., Yuan J., Chen X., Gu X., Luo K., Li J., Wan B., Wang Y., Yu L. 2006. Identification of a novel human lysophosphatidic acid acyltransferase, LPAAT-theta, which activates mTOR pathway. J. Biochem. Mol. Biol. 39: 626–635. [DOI] [PubMed] [Google Scholar]
- 26.Vila M. C., Farese R. V. 1991. Insulin rapidly increases glycerol-3-phosphate-acyltransferase activity in rat adipocytes. Arch. Biochem. Biophys. 284: 366–368. [DOI] [PubMed] [Google Scholar]
- 27.Vila M. C., Milligan G., Standaert M. L., Farese R. V. 1990. Insulin activates glycerol-3-phosphate acyltransferase (de novo phosphatidic acid synthesis) through a phospholipid-derived mediator. Apparent involvement of Gi alpha and activation of a phospholipase C. Biochemistry. 29: 8735–8740. [DOI] [PubMed] [Google Scholar]
- 28.Harris T. E., Huffman T. A., Chi A., Shabanowitz J., Hunt D. F., Kumar A., Lawrence J. C., Jr 2007. Insulin controls subcellular localization and multisite phosphorylation of the phosphatidic acid phosphatase, lipin 1. J. Biol. Chem. 282: 277–286. [DOI] [PubMed] [Google Scholar]
- 29.Huffman T. A., Mothe-Satney I., Lawrence J. C., Jr 2002. Insulin-stimulated phosphorylation of lipin mediated by the mammalian target of rapamycin. Proc. Natl. Acad. Sci. USA. 99: 1047– 1052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lewin T. M., de Jong H., Schwerbrock N. J., Hammond L. E., Watkins S. M., Combs T. P., Coleman R. A. 2008. Mice deficient in mitochondrial glycerol-3-phosphate acyltransferase-1 have diminished myocardial triacylglycerol accumulation during lipogenic diet and altered phospholipid fatty acid composition. Biochim. Biophys. Acta. 1781: 352–358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chang C. C., Huh H. Y., Cadigan K. M., Chang T. Y. 1993. Molecular cloning and functional expression of human acyl- coenzyme A:cholesterol acyltransferase cDNA in mutant Chinese hamster ovary cells. J. Biol. Chem. 268: 20747–20755. [PubMed] [Google Scholar]
- 32.Hishikawa D., Shindou H., Kobayashi S., Nakanishi H., Taguchi R., Shimizu T. 2008. Discovery of a lysophospholipid acyltransferase family essential for membrane asymmetry and diversity. Proc. Natl. Acad. Sci. USA. 105: 2830–2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zhao Y., Chen Y. Q., Bonacci T. M., Bredt D. S., Li S., Bensch W. R., Moller D. E., Kowala M., Konrad R. J., Cao G. 2008. Identification and characterization of a major liver lysophosphatidylcholine acyltransferase. J. Biol. Chem. 283: 8258–8265. [DOI] [PubMed] [Google Scholar]
- 34.Chang S. C., Magee A. I. 2009. Acyltransferases for secreted signalling proteins. [Review] Mol. Membr. Biol. 26: 104–113. [DOI] [PubMed] [Google Scholar]
- 35.Gutierrez J. A., Solenberg P. J., Perkins D. R., Willency J. A., Knierman M. D., Jin Z., Witcher D. R., Luo S., Onyia J. E., Hale J. E. 2008. Ghrelin octanoylation mediated by an orphan lipid transferase. Proc. Natl. Acad. Sci. USA. 105: 6320–6325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang J., Brown M. S., Liang G., Grishin N. V., Goldstein J. L. 2008. Identification of the acyltransferase that octanoylates ghrelin, an appetite-stimulating peptide hormone. Cell. 132: 387–396. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.