Abstract
Placental fatty acid transport and metabolism are important for proper growth and development of the feto-placental unit. The nuclear receptors, liver X receptors α and β (LXRα and LXRβ), are key regulators of lipid metabolism in many tissues, but little is known about their role in fatty acid transport and metabolism in placenta. The current study investigates the LXR-mediated regulation of long-chain acyl-CoA synthetase 3 (ACSL3) and its functions in human placental trophoblast cells. We demonstrate that activation of LXR increases ACSL3 expression, acyl-CoA synthetase activity, and fatty acid uptake in human tropholast cells. Silencing of ACSL3 in these cells attenuates the LXR-mediated increase in acyl-CoA synthetase activity. Furthermore, we show that ACSL3 is directly regulated by LXR through a conserved LXR responsive element in the ACSL3 promoter. Our results suggest that LXR plays a regulatory role in fatty acid metabolism by direct regulation of ACSL3 in human placental trophoblast cells.
Keywords: BeWo cell, fatty acid uptake, acyl-CoA synthetase
The participation of fatty acids in most metabolic pathways, including β-oxidation and biosynthesis of complex lipids (such as triacylglycerols and phospholipids), requires their activation by addition of a CoA group. Mammals express three related families of proteins able to activate long chain fatty acids: the long-chain acyl-CoA synthetases (ACSL), the fatty acid transport proteins (FATP), and the acyl-CoA synthetase bubblegum (ACSBG) family (1–5). Five genes in the ACSL family have been identified based on sequence homology. They are named ACSL1 and ACSL3 to ACSL6 and differ in their tissue and intracellular distribution. The distinct intracellular location of acyl-CoA synthetases has been hypothesized to channel fatty acids to different metabolic fates by activating the fatty acids at different subcellular compartments (6). ACSL3 is localized on lipid droplets or endoplasmic reticulum (ER) membranes, suggesting that this enzyme is likely involved in lipid synthesis (7, 8). The ACSL3 is highly expressed in prostate, skeletal muscle, testis, heart, and placenta (9). Besides activation of fatty acids, a function in transport of fatty acids has been indicated for the FATPs and members of the ACSL family. Forced expression of mammalian ACSL1, ACSL4, and ACSL6 in yeast cells lacking native long-chain acyl-CoA activity lead to enhanced fatty acid uptake (10). ACSL3 has been associated with the biosynthesis of neutral lipids in hepatocytes (8); however, no information is available regarding its role on activation and uptake of fatty acids by human placental trophoblasts.
LXRα and LXRβ are nuclear receptors that are activated by oxysterols, which are oxidized cholesterol derivatives (11). LXR regulates expression of multiple genes involved in efflux, transport, and excretion of cholesterol, the fatty acid biosynthetic pathway and lipoprotein metabolism in a number of tissues [reviewed in (12, 13)]. Furthermore, the recent identification of the fatty acid transporter protein (FAT/CD36) as a direct LXR target gene (14) points to a role for LXR in the regulation of fatty acid transport.
The placental transport of fatty acids is important for proper growth and development of the fetus (15). The placenta is the key organ for the transfer of fatty acids and other nutrients from the mother to the fetus. Placenta may also be involved in the dyslipidemia associated with pregnancy disorders, as observed in preeclampsia (16) and gestational diabetes (17). The placenta governs the fetal supply of fatty acids via two processes: transport and metabolism. The transport is carried out through highly specialized trophoblast cells that form a barrier between maternal and fetal circulation (18). Increased knowledge of the molecular mechanisms involved in this fatty acid transport is important to better understand and possibly prevent adverse fetal development.
The functional significance of LXR in placenta is largely uncharacterized. The few studies published on LXR in placenta indicate diverse roles of LXR, such as placentation and trophoblast invasion (19); inhibition of human chorionic gonadotropin secretion; and cholesterol transport and biosynthesis of lipids (20–22). These findings suggest that LXR may be important in human placentation as well as feto-placental lipid transport and metabolism.
To extend our knowledge on the roles of LXR in placental lipid metabolism, we first searched for LXR target genes in primary human third-trimester trophoblasts. As human placental choriocarcinoma (BeWo) cells have been used extensively as a human placental trophoblast cell model to study fatty acid uptake and metabolism (23, 24), we utilized this cell line for investigation of LXR-mediated regulation of fatty acid metabolism and transport. We report that ligand activation of LXR stimulates acyl-CoA synthetase activity and fatty acid uptake through increased ACSL3 expression in these cells. In addition we demonstrate that ACSL3 is a new direct LXR target gene.
MATERIALS AND METHODS
Materials
T0901317 and LCPUFA were obtained from Cayman Chemicals (Ann Arbor, MI). GW3965, 9-cis retinoic acid (9-cis RA), oleic acid (OA), and oligonucleotides were obtained from Sigma (St Louis, MO). [1-14C] eicosapentaenoic acid (EPA; specific activity 55.0 mCi/mmol) and [1-14C] docosahexaenoic acid (DHA; specific activity 55.5 mCi/mmol) were obtained from American Radiolabeled Chemical (St. Louis, MO). [1-14C] OA (specific activity 54.6 mCi/mmol) and [1-14C] arachidonic acid (ARA; specific activity 56 mCi/mmol) were obtained from Perkin Elmer (Waltham, MA). Cell culture plastic ware was obtained from Becton Dickinson (Franklin Lakes, NJ). Other chemicals and solvents were obtained from Sigma.
Cell lines, primary trophoblasts, and placenta
HTR-8/SVneo cells, which were kindly provided by Dr. Charles H. Graham, Canada, were maintained in RPMI-1640 (25). BeWo cells (American Type Culture Collection, Manassas, VA) were maintained in Ham's F12 medium. JAR cells and COS-1 cells (American Type Culture Collection) were maintained in RPMI-1640 and DMEM, respectively (20, 26). Cell culture media were supplemented with 10% fetal calf serum (FCS), 2 mM L-glutamine, and 1% antibiotics (50 U/ml penicillin and 50 μg/ml streptomycin), except for HTR-8/SVneo cells, where the medium was supplemented with 5% FCS. Cell cultures were incubated at 37°C in 5% CO2.
Placenta biopsies and placentas used for isolation of primary human trophoblasts were obtained from term placentas delivered by caesarean section after uncomplicated pregnancies. Primary human cytotrophoblasts were isolated as described previously (26) with modifications (27). Following isolation, the cytotophoblasts were plated out and maintained in P35 culture dishes (3 × 106 cells/dish) in culture medium (50:50 DMEM:Ham's F12) supplemented with 10% FCS, 2 mM L-glutamine, and 1% antibiotics (50 U/ml penicillin and 50 μg/ml streptomycin) at 37°C in 5% CO. Medium was changed every day. Placenta tissues from six healthy women were snap-frozen in liquid nitrogen before storage at −70°C following caesarean section. The placenta study was approved by the Regional Committee of Medical Research Ethics in Eastern Norway and written consent was obtained from each patient.
Quantitative RT-PCR
For gene expression analysis, total RNA was isolated from human cells and human placenta tissue using ABI 6100 (Applied Biosystems, Foster City, CA) according to the manufacturer's instruction.
For quantitative RT-PCR (qRT-PCR), cDNAs were synthesized from extracted total RNA using high-capacity cDNA Reverse Transcription kit and analyzed using TaqMan Gene Expression Master Mix and the 7900HT Real-Time PCR System (Applied Biosystems). The assay efficiency is according to Applied Biosystems 100% (+/−10%). All genes were analyzed with the same threshold. All assays used are listed in Table 1.
TABLE 1.
Gene assays
| Gene Name | TaqMan Assay Number |
|---|---|
| ACSL1 | Hs00960561_m1 |
| ACSL3 | Hs00244853_m1 |
| ACSL4 | Hs00244871_m1 |
| ACSL5 | Hs00212106_m1 |
| ACSL6 | Hs00362960_m1 |
| FATP1 | Hs01587917_m1 |
| FATP2 | Hs00186324_m1 |
| FATP3 | Hs00225680_m1 |
| FATP4 | Hs00192700_m1 |
| FATP6 | Hs00204034_m1 |
| ACSBG1 | Hs00209500_m1 |
| ACSBG2 | Hs00229479_m1 |
| SREBP-1 | Hs01088691_m1 |
| CD36 | Hs00169627_m1 |
| TBP | Hs99999910_m1 |
| Glyceraldehyde 3-phosphate dehydrogenase | Hs99999905_m1 |
| Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | Hs00237047_m1 |
| B2M | Hs99999907_m1 |
Abbreviations: ACSBG, acyl-CoA synthetase bubblegum; ACSL, long-chain acyl-CoA synthetase; B2M, beta-2-microglobulin; FATP, fatty acid transport protein; SREBP-1, sterol-regulatory element binding protein-1; TBP, TATA box binding protein.
When comparing expression in placenta and BeWo cells, four different endogenous controls were tested. Only B2G was equally expressed in BeWo cells and placenta and was used as the endogenous control in the experiment. The relative RNA expression levels were calculated using the comparative Ct method.
Microarray
Primary human trophoblast cells isolated from three independent placentas were kept in culture for 24 h, after which the cells were stimulated with 1 μM T0901317 or the same amount of vehicle (DMSO) for 24 h before harvesting. The RNA expression profile was obtained using Affymetrix one-cycle gene expression protocol and Affymetrix Human Genome U133 Plus 2.0 arrays. Arrays were scanned using GeneChip ® Scanner 3000 7G (Affymetrix). Image analysis was performed using GeneChip ® Operating Software 1.3 (Affymetrix). The r-package “simpleaffy” from www.bioconductor.org was used to assess the quality of the arrays. Log2 ratios were calculated for every gene comparing the control and stimulation intensities using Microarray Analyze Suite normalized intensities.
Cloning of the ACSL3 promoter and generation of expression vectors
The human and mouse ACSL3 promoter sequences spanning 5000 base pairs (bp) upstream and 3000 bp downstream from the transcription start site were analyzed for potential LXR responsive elements (LXRE) as described before (28). Species homology was determined by BLAST2 alignment (29). The full-length human ACSL3 reporter construct [ACSL3 (−2189/+213)LUC] was generated as described previously (28) using pPCR-Script vector (Stratagene, La Jolla, CA) prior to insertion into the BglII site in the pGL3-Basic luciferase reporter vector (Promega, Madison, WI). Primer sequences were 5′-ACSL3-promoter; 5′-ATA AGATCTGCTCGGCGATTTGAGATAATTGA-3′ and 3′-ACSL3-promoter; 5′-ATAAGATCTAATGCGAGGGGACGAACGTAT-3′. ACSL3-(−125/+213)-LUC was generated by restriction cutting of the full-length reporter with XmaI (New England Biolabs, Ipswich, MA). The [ACSL3-(−2189/+213 LXRE-mut)-LUC] was generated by site-specific mutagenesis using Pfu Turbo DNA polymerase as described previously (28).
Full-length cDNAs encoding the open reading frame of human LXRα (30), human LXRβ (31), human retinoid X receptor (RXR)α (32), and a multicloning site (MCS; Met-KpnI-HindIII-SacI-SpeI: ATG-GGTACC-AAGCTT-GAGCTC-ACTAGT) were PCR-amplified and cloned into the pDONR-221 P4r-P3r vector (Invitrogen). The generated pENTR vectors were recombined into a novel pcDNA3-attP4r-attP3r-DEST vector (generated from pcDNA3; Invitrogen) using LR recombinase, resulting in pcDNA3-hLXRα, pcDNA3-hLXRβ, pcDNA3-hRXRα, and pcDNA3-MCS (empty) expression vectors. Primers and vector maps are available upon request.
Transfection and luciferase assay
For transient transfection experiments, BeWo cells were seeded at a density of 6 × 104 cells/well in 24-well plates in growth medium with serum. The next day, cells were transfected with reporter constructs (300 ng), internal control (pTK Renilla luciferase, 100 ng), and expression vectors (150 ng) or corresponding empty vector (pcDNA3-MCS) using 2 μg/well of LipofectAMINE 2000 reagent diluted in OptiMEM-I as described by the manufacturer (Invitrogen). OptiMEM-I was replaced with normal growth medium containing serum supplemented with vehicle (DMSO, 0.2% final concentration) with/without agonists, and incubated at 37°C for 24 h. Cells were harvested in 100 μl Passive Lysis Buffer (Promega). Luciferase activity was measured using the Dual-Luciferase® Reporter Assay System (Promega) with a TD-20/20 Luminometer (Turner Designs, Sunnyvale, CA).
Electrophoretic mobility shift assay
The hRXRα, hLXRα, and hLXRβ proteins were in vitro translated according to Dalen et al (28). In brief, the pCMX-hRXRα, pCMX-hLXRα, or pCMX-hLXRβ expression vectors, described in the cloning section above, were used separately in a TNT-T7 quick coupled transcription/translation system (Promega). The resulting in vitro translated proteins were mixed (hLXRα with hRXRα; hLXRα with hRXRα) and used in electrophoretic mobility shift assays (EMSA) for size identification of the complexes. For preparation of nuclear extracts, COS-1 cells were transfected in 10-cm dishes with 4 µg of expression vector and 8 µl of LipofectAMINE2000 (Invitrogen) in 3.5 ml of OptiMem (Gibco, Invitrogen) for 5 h, followed by 24-h incubation in 12 ml of DMEM (Sigma) containing serum and antibiotics according to Dalen et al (28). Nuclear extracts were isolated using CelLytic NuCLEAR Extraction Kit (Sigma), according to the manufacturer's instruction with 2× EDTA-free Complete Proteinase Inhibitor Cocktail (Roche Applied Sciences) added. The double-stranded wild-type oligonucleotides and oligoes specifically mutated in the two half-sites used in EMSA were (only one strand is shown): h-ACSL3 LXRE 5′-CTCCGCGGAATGACCCGTAGTAACCTCGCCCCGCC-3′ and h-ACSL3 LXRE-mut 5′-CTCCGCGGAATGAGGCGTAGTAAGCTCGCCCCGCC-3′. Underlined letters indicate the LXRE element and bold letters indicate mutated nucleotides. The double-stranded oligoes were radiolabeled and probes were purified, followed by binding reactions and separation of the protein-DNA complexes from free probes as described (28). For supershift assays, 1 µL mouse monoclonal hLXRα antibody (R and D Systems: PP-K8607-00, Perseus Proteomics) was preincubated with nuclear LXRα protein for 45 min at 4°C.
ACSL3 siRNA
Duplexes of siRNA sequences (Applied Biosystems) targeting human ACSL3 were (ACSL3 siRNA 1 ID# s4999) sense 5′-GAGUAGUUUUCAACGUAAUtt-3′ and antisense 5′-AUUACGUUGAAAACUACUCat-3′; (ACSL3 siRNA 2 ID#s 4997) sense 5′- GGAACUAACUGAACUAGCUtt-3′ and antisense 5′- AGCUAGUUCAGUUAGUUAGUUCCtt-3′; and (ACSL3 siRNA 3 ID#s4998) sense 5′- CAUUAUUGCUGGUAUAACUtt-3′ and antisense 5′-AGUUAUACCAGCAAUAAUGtt-3′. The sequence for the negative control siRNA was (part #4390843) sense 5′-UAACGACGCGACGACGUAAtt-3′ and antisense 5′-UUACGUCGUCGCGUCGUUAtt-3′. Reverse transfection was employed on BeWo cells using LipofectamineTM RNAiMAX Reagent (Invitrogen) according to the manufacturer's instructions. Briefly, RNAi duplexes (10 pmol/well) were complexed with LipofectamineTM RNAiMAX Reagent (3 μl/well) 350 μl OPTI-MEM in 12-well plates. The trypsinated BeWo cells diluted to 80.000 cells/well in 1.5 ml growth medium (Ham's F12 containing serum but not antibiotics) were added to each well. 4 h after the transfection, cells were supplemented with agonists and incubated for 48 h before initiation of acyl-CoA synthetase activity assays. OA uptake was performed 48 h after transfection with siRNA.
Western analysis
Cells were seeded in 12-well plates and treated as described in the ACSL3 siRNA experiment. Cells from three wells were pooled and harvested in 200 μl lysis buffer (33), and protein concentrations measured by BC Assay (Interchim). Proteins (20 μg) from whole cell extracts were separated by a 10% Tris-HCl SDS-PAGE (Criterion™ Precast Gel, Bio-Rad) at 150 V for 2 h and transferred to a nitrocellulose membrane (Hybond-C Extra, Amersham Biosciences) by electro transfer (33). Membranes were incubated with mouse anti-human ACSL3 (Abnova, #H00002181-B01; 1:1000) followed by incubation with secondary antibody [horseradish peroxidase conjugated rabbit anti-mouse antibody (#ab6728; 1:10000)]. Bound antibody was detected using enhanced chemiluminescence (SuperSignal West Dura Extended Substrate, Thermo Sceientific or ECLplus, Amersham Biosciences) and visualized with Hyperfilm MP (Amersham Biosciences). Membranes were stripped and incubated with primary antibody against mouse anti-β-actin (Sigma, #A5441; 1:10000).
Acyl-CoA synthetase activity
BeWo cells grown in medium containing serum were prestimulated with 9-cis RA (1 μM) and/or GW3965 (1 μM) and/or the same amount of vehicle. Acyl-CoA synthetase activity was measured in BeWo cell protein-lysates (5 μg) as described previously (34) with modifications (35). OA (50 µM trace-labeled with [14C] OA 2 µCi/100 μl) was used as substrate.
Fatty acid uptake
Fatty acid uptake was measured as described previously (36) with some modifications. Briefly, fatty acids (100 μM) were complexed with fat-free BSA (BSA; 100 μM, Sigma), in serum-free F-12 HAM medium at a 1:1 fatty acid: BSA molar ratio (23). Radiolabeled fatty acid was applied directly to serum-free medium prior to addition to the cells. BeWo cells (∼80% confluent) were washed once with serum-free medium before incubation for 3 h with the radioactive fatty acid (100 μM) solution (37°C) in the presence of vehicle (DMSO, the same amount used as for the treatment) or triacsin C (10 μM). The reaction was stopped by the addition of ice-cold 0.5% BSA in PBS. Cells were washed twice with room-temperature 0.5% BSA in PBS and twice with PBS to remove any surface bound fatty acids. Further, the cells were lysed and harvested in 0.1 M NaOH and used for scintillation counting. Results were expressed as nmol fatty acid/μg protein. Stimulation with agonists was performed 48 h before the fatty acid uptake study.
Statistics
Data are presented as mean ± SEM. Statistical analyses were performed using two-tailed unpaired student's t-test with equal variance not assumed, unless stated otherwise.
RESULTS
The ACSL3 transcript is induced by synthetic LXR agonists in primary trophoblast cells and trophoblast cell lines
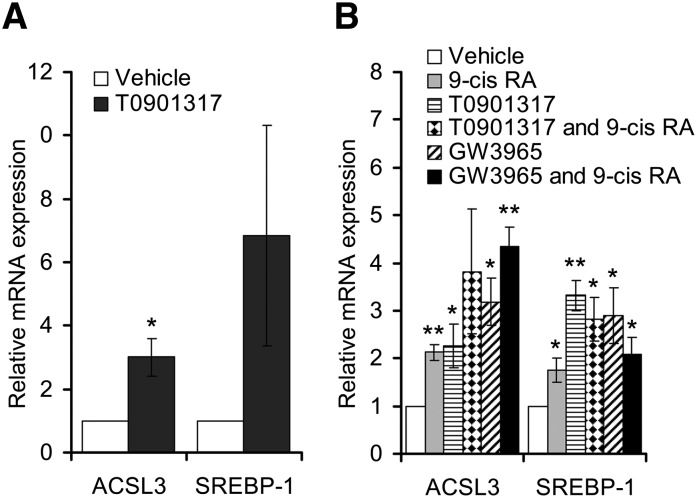
We have previously shown an increased biosynthesis of triacylglycerol following LXR activation in BeWo cells (20). To explore further the role of LXR in placental lipid metabolism, LXR target genes were investigated by performing microarray analyses after stimulating cultured primary human trophoblast cells for 24 h with the synthetic LXR agonist T0901317 (data not shown). Among the regulated genes, we noted an increased expression of ACSL3 mRNA. As expected, established lipogenic LXR target genes such as SREBP-1 (37) and FAS (38) were also induced. A 3-fold induction of the ACSL3 transcript was confirmed by qRT-PCR (Fig. 1A). A similar magnitude of induction was also observed upon LXR stimulation in the cultured human placental BeWo cells (Fig. 1B) and in two other placental cell lines, JAR and HTR-8/neo (data not shown). Both the LXR agonists, GW3965 and T0901317, induced expression of ACSL3 in BeWo cells to a similar extent (Fig. 1B). Because GW3965 is a more specific LXR agonist than T0901317 (39), we henceforth used this agonist for the remaining cell experiments.
Fig. 1.
Upregulation of ACSL3 mRNA expression by synthetic LXR agonists in primary human trophoblasts and BeWo cells. A: Primary trophoblasts were incubated in triplicate with vehicle (white bars) or T0901317 (1 μM, black bars) for 24 h in three independent placenta isolations. B: BeWo cells were incubated with vehicle (white bars), 9-cis RA (1 μM, gray bars), T0901317 (1 μM, bars with small dots), both agonists (bars with big dots), GW3965 (1 μM, striped bars), or GW3965 and 9-cis RA together (black bars) for 48 h. ACSL3 and SREBP-1 expression were analyzed by qRT-PCR and normalized to TBP. Each experiment was performed in triplicate. Data shown represents the means of three independent experiments performed in triplicate (n = 3) ± SEM, relative to control. *P < 0.05 and **P < 0.01 relative to control. ACSL, long-chain acyl-CoA synthetase; BeWo, human placental choriocarcinoma; LXR, liver X receptor; qRT-PCR, quantitative reverse transcription-PCR; SREBP-1, sterol-regulatory element binding protein-1; TBP, TATA box binding protein.
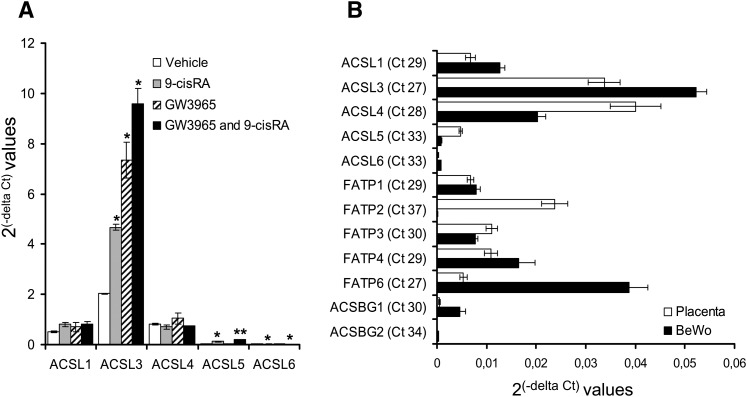
ACSL3, 5, and 6 are induced by LXR agonist, but only ACSL3 is highly expressed in BeWo cells
Acyl-CoA synthetase activity of the ACSL, FATP, and ACSBG families has been described previously (40, 41). Therefore, we investigated the effect of LXR activation on the expression of all the above-mentioned genes in BeWo cells. Among these genes only expression of ACSL3, 5, and 6 were induced upon stimulation with LXR agonist (Fig. 2A and data not shown). Judged by Ct values, ACSL3 and FATP6 had the highest basal expression levels in BeWo cells (Fig. 2B), whereas the expression of ACSL5 and 6 was almost negligible compared with that of ACSL3 (Fig. 2B). Similarly, judged by Ct values, ACSL3, ACSL4, and FATP2 had the highest basal expression in placenta (Fig. 2B). We did not analyze expression of FATP5 as it is not present in human trophoblasts (42). Taken together, these data clearly demonstrate that ACSL3 is the only transcript among the ACSL, FATP, and ACSBG families that was both highly expressed in BeWo cells, and its expression was significantly increased by LXR agonist.
Fig. 2.
Expression of ACSL3, 5, and 6 are increased with synthetic LXR agonist, but only ACSL3 is predominantly expressed in BeWo cells. A: BeWo cells were incubated with vehicle (white bars), 9-cis RA (1 μM, gray bars), GW3965 (1 μM, striped bars), or both agonists together (black bars) for 48 h. Total RNA was analyzed for gene expression of the five ACSL isoforms by qRT-PCR normalized to TBP. The results are presented as means of three independent experiments performed in triplicate (n = 3) ± SEM relative to control. *P < 0.05 and **P < 0.01 B: Basal expression of ACSL, FATP, and ACSBG family members in BeWo cells (white bars) and placenta (black bars) normalized to B2M. The results are represented as 2(-delta Ct) for the means of three independent BeWo cell experiments performed in triplicate (n = 3) and for the means of six independent placenta isolations performed in triplicate (n = 6) ± SEM. Ct values are indicated for placental tissue. ACSBG, acyl-CoA synthetase bubblegum; ACSL, long-chain acyl-CoA synthetase; B2M, beta-2-microglobulin; BeWo, human placental choriocarcinoma; FATP, fatty acid transport protein; LXR, liver X receptor; qRT-PCR, quantitative reverse transcription-PCR; SREBP-1, sterol-regulatory element binding protein-1; TBP, TATA box binding protein.
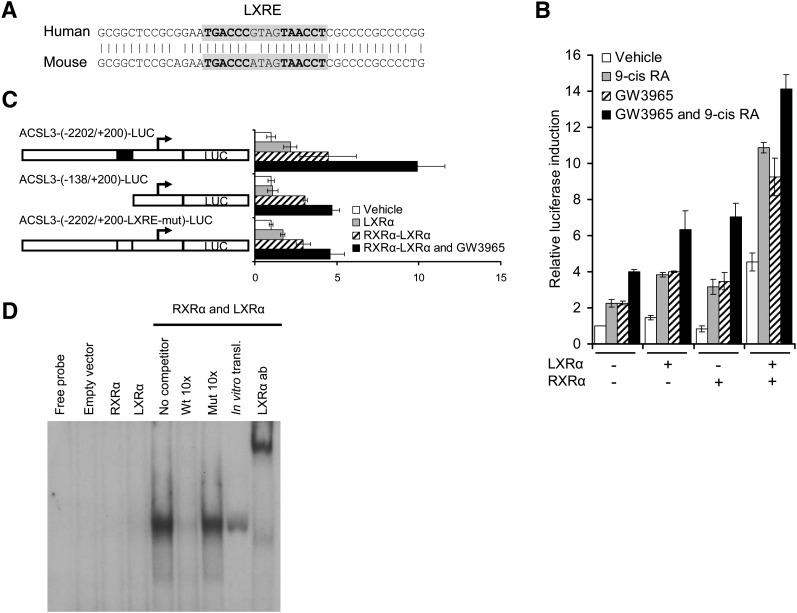
ACSL3 is a direct LXR target gene
To locate the potential LXR responsive region, we performed a theoretical analysis of the ACSL3 promoter for LXR/RXR heterodimer binding sites. A potential LXRE containing a direct repeat with a four-nucleotide spacer (DR-4) was identified 163 bp upstream of the transcription start site (9). The DR-4 element was conserved between the human and mouse ACSL3 promoters (Fig. 3A). To explore the physiological relevance of this element, we cloned a fragment of the ACSL3 promoter (−2202 bp/+200 bp) into the pGL3-basic luciferase reporter vector [hACSL3-(−2202/+200)-LUC]. This construct was transiently transfected into BeWo cells alone or cotransfected with LXRα and RXRα expression vectors in the presence of LXR and/or RXR agonists. A 4-fold upregulation was observed in presence of both agonists in absence of cotransfected LXR and RXR expression vectors (Fig. 3B). This demonstrates that the ligands are able to stimulate transcription of the ACSL3 reporter through activation of endogenously expressed RXR and LXRs. Cotransfection with LXRα and RXRα expression vectors increased reporter activity further, with a maximal 14-fold induction after cotransfection with both expression vectors in the presence of both agonists. Essentially the same results were obtained when cells were cotransfected with RXRα and LXRβ expression vectors (results not shown).
Fig. 3.
ACSL3 is a direct LXR target gene. A: Alignment of the mouse and human LXRE elements in the ACSL3 promoters. B: BeWo cells were transiently transfected with ACSL3-(−2202/+200)-LUC reporter and cotransfected with pLR (internal control), pcDNA3-MCS, pcDNA3-hRXRα, and/or pcDNA3-hLXRα expression vectors as indicated. After transfection, cells were incubated for 24 h with medium containing vehicle (0.2% DMSO, white bars), 9-cisRA (1 μM, gray bars), GW3965 (1 μM, striped bars), or both agonists (black bars). The data is a representative experiment of two independent experiments performed in triplicate (n = 3) ± SEM. C: Transient transfection with ACSL3-(−2202/+200)-LUC, ACSL3-(−138/+200)-LUC, and ACSL3-(−2202/+200-LXRE-mut)-LUC reporters. The cells were cotransfected with pcDNA3-MCS (empty vector, white bars), pcDNA3-hRXRα (gray bars), or pcDNA3-hLXRα and pcDNA3-hRXRα (striped and black bars). After transfection, cells were incubated in medium containing vehicle (0.1% DMSO, white, gray, and striped bars) or GW3965 (1 μM, black bars). The data is a representative experiment of four independent experiments performed in triplicate (n = 3) ± SEM. D: EMSA of nuclear extracts (2 µg) isolated from COS-1 cells transfected with either pcDNA3-hRXRα or pcDNA3-hLXRα expression vectors individually form a strong specific complex with the ACSL3 LXRE only when these extracts are combined (lane 5). Identical results were found for in vitro translated RXRα and LXRα proteins (lane 8). The competition experiments were performed using unlabeled LXRE (lanes 6) and LXRE-mutated oligonucleotides (lanes 7) as competitors in 10-fold molar excess. The upper array indicates the super-shift with LXRα antibody binding to the LXRα-RXRα-LXRE complex (lane 9). ACSL, long-chain acyl-CoA synthetase; BeWo, human placental choriocarcinoma; EMSA, electrophoretic mobility shift assay; LXR, liver X receptor; LXRE, liver X receptor responsive element; RXR, retinoid X receptor.
Next, we generated a truncated version of the ACSL3 reporter construct lacking the LXRE [hACSL3-(−138 bp/+200 bp)-LUC] and a full-length reporter construct containing a mutated form of the LXRE (ACSL3-[(−2202/+200)-LXRE-mut]-LUC), and compared the reporter activity of these constructs against the ACSL3-(−2202/+200)-LUC construct. BeWo cells were transfected with the reporters, cotransfected with expression vectors for LXRα and/or RXRα, and stimulated with LXR agonist. A 10-fold induction was observed for the ACSL3-(−2202/+200)-LUC reporter, but a significantly weaker 5-fold induction was observed for the ACSL3-(−138 bp/+200 bp)-LUC and ACSL3-[(−2202/+200)-LXRE-mut]-LUC reporters. This suggests that the two latter constructs lack one functional LXRE element that is required for full ACSL3 reporter activity. The residual induction that remains in the ACSL3-(−138 bp/+200bp)-LUC and ACSL3-[(−2202/+200)-LXRE-mut]-LUC reporters suggests that the ACSL3 −138/+200 promoter region contains additional sequences that respond to activation of RXR and/or LXR. We were unable to find other potential LXREs in this region using theoretical promoter analysis (data not shown), which suggests that this residual effect might be secondarily mediated through other transcription factors.
To test the ability of LXRα and RXRα proteins to bind to the identified LXRE, we performed EMSA with a 35-mer oligonucleotide containing the ACSL3 LXRE element or a mutated version of the LXRE. As expected, we observed LXRα/RXRα heterodimer binding to the oligonucleotide containing the wild-type (wt) LXRE, but not to the mutated LXRE (Fig. 3D). Similar results were observed using protein extracts containing LXRβ and RXRα proteins (data not shown). The complex sizes were confirmed using in vitro translated proteins. Addition of an antibody against LXRα resulted in a supershift, confirming the specific binding of an LXRα/RXRα heterodimer to the LXRE. Together, these data indicate that the ACSL3 mRNA transcript is induced by LXR agonists through direct binding of an LXRα/RXRα heterodimer to an LXRE in the human ACSL3 promoter, located 163 bp upstream of the transcription start site.
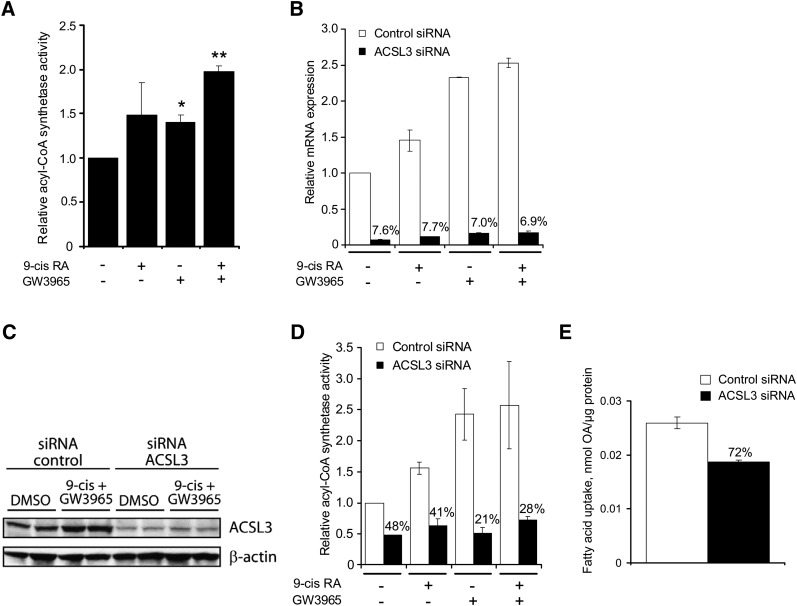
LXR agonist increases total acyl-CoA synthetase activity
After establishing ACSL3 as a direct LXR target gene, we examined whether the acyl-CoA synthetase activity was increased following LXR activation. OA was used as substrate, as ACSL3 is known to have a considerable oleoyl-CoA synthetase activity (1). Incubation of BeWo cells with LXR agonist for 48 h resulted in a significant increase in acyl-CoA synthetase activity, with a maximal 2-fold increase following incubation of both LXR and RXR agonists (Fig. 4A).
Fig. 4.
LXR activation increases acyl-CoA synthetase activity in BeWo cells, and ACSL3 silencing prevents increased acyl-CoA synthetase activity. BeWo cells were either treated directly or reverse-transfected with ACSL3 siRNA or negative control siRNA and incubated for 48 h in medium supplemented with vehicle (0.2% DMSO), 9-cis RA (1 μM), and/or GW3965 (1 μM). A: In cell lysates harvested from BeWo cells prestimulated with vehicle, 9-cis RA (1 μM), and/or GW3965 (1 μM) for 48 h. Acyl-CoA synthetase activity was analyzed in BeWo cell lysates by incubating the lysates in the presence of 100 μM [14C]OA for 8 min at 37°C. Generated [14C]OA-CoA was extracted, and the radioactivity was determined in a scintillation counter. Each experiment was performed in triplicate. The results are the means of three independent experiments performed in triplicate (n = 3) ± SEM. *P < 0.05 and **P < 0.01 relative to control (one tailed t-test). B: Total RNA was extracted and analyzed using qRT-PCR. ACSL3 mRNA expression (normalized to TBP) was compared in cells transfected with ACSL3 siRNA1 (black bar) or negative control siRNA (white bar). The results are the means of two independent experiments performed in duplicates (n = 2) ± SEM. Reduction in expression (in %) relative to negative control siRNA is indicated. C: ACSL3 and β-actin protein expression in cells transfected with control siRNA or ACSL3 siRNA. Each lane contains cells pooled from three wells (n = 2). The experiment was repeated once with similar results. D: Acyl-CoA synthetase activity in cell lysates from cells transfected with ACSL3 siRNA (black bar) or a negative control siRNA (white bar). The results are the means of three independent experiments performed in triplicate (n = 3) ± SEM. Reduction in expression (in %) relative to negative control siRNA is indicated. E: OA uptake in cells transfected with ACSL3 siRNA (black bar) or a negative control siRNA (white bar). A representative experiment of four independent experiments performed in triplicate is shown (n = 3) ± SEM. OA uptake (in %) relative to negative control siRNA is indicated. ACSL, long-chain acyl-CoA synthetase; BeWo, human placental choriocarcinoma; LXR, liver X receptor; OA, oleic acid; qRT-PCR, quantitative reverse transcription-PCR; siRNA, small interfering RNA.
Silencing of ACSL3 attenuated the LXR-mediated increase in total acyl-CoA synthetase activity in BeWo cells
To determine the impact of ACSL3 on the observed increased acyl-CoA synthetase activity following LXR activation, we silenced ACSL3 in BeWo cells using siRNA. Three different ACSL3 siRNA duplexes targeting three different regions of the ACSL3 mRNA were tested. All the tested siRNAs reduced the basal ACSL3 mRNA transcript by ∼90%, with the highest reduction using ACSL3 siRNA1 (data not shown). ACSL3 siRNA1 was selected for further experiments (ACSL3 siRNA). When we silenced ACSL3 in BeWo cells and subsequently stimulated these cells with LXR and/or RXR agonists for 48 h, there was an ∼90% reduction in ACSL3 mRNA expression levels following both ACSL3 silencing and agonist stimulation compared with the negative control siRNA (Fig. 4B). To determine if the protein levels were changed, we analyzed ACSL3 protein content in control- and ACSL3 siRNA-silenced BeWo cells treated with vehicle or 9-cis RA and GW3965 using Western analysis (Fig. 4C). Ligand stimulation increased ACSL3 protein levels in cells receiving control siRNA, whereas a consistent lower level was found in cells receiving ASCL3 siRNA. Together, these results demonstrate that the mRNA and protein levels of the ACSL3 transcript were reduced in cells treated with ACSL3 siRNA.
We then went on to examine the consequence of reduced expression of ACSL3 transcript on acyl-CoA synthetase activity. The acyl-CoA synthetase activity following LXR and RXR activation increased 2.5-fold in cells transfected with control siRNA, whereas no difference in activity was found in cells transfected with ACSL3 siRNA (Fig. 4D). Intriguingly, silencing of ACSL3 alone resulted in ∼50% reduction in the total acyl-CoA synthetase activity in BeWo cells (Fig. 4D).
Activation of free FAs into acyl-CoA is required for downstream metabolism of externally provided or de novo synthesized FAs. To determine whether ASCL3-mediated acyl-CoA synthetase activity is important for basal fatty acid uptake, we performed OA uptake in ACSL3-silenced BeWo cells (Fig. 4E). The basal uptake of OA in BeWo cells transfected with ACSL3 siRNA was reduced compared with control siRNA treated cells.
LXR agonist increases fatty acid uptake in BeWo cells
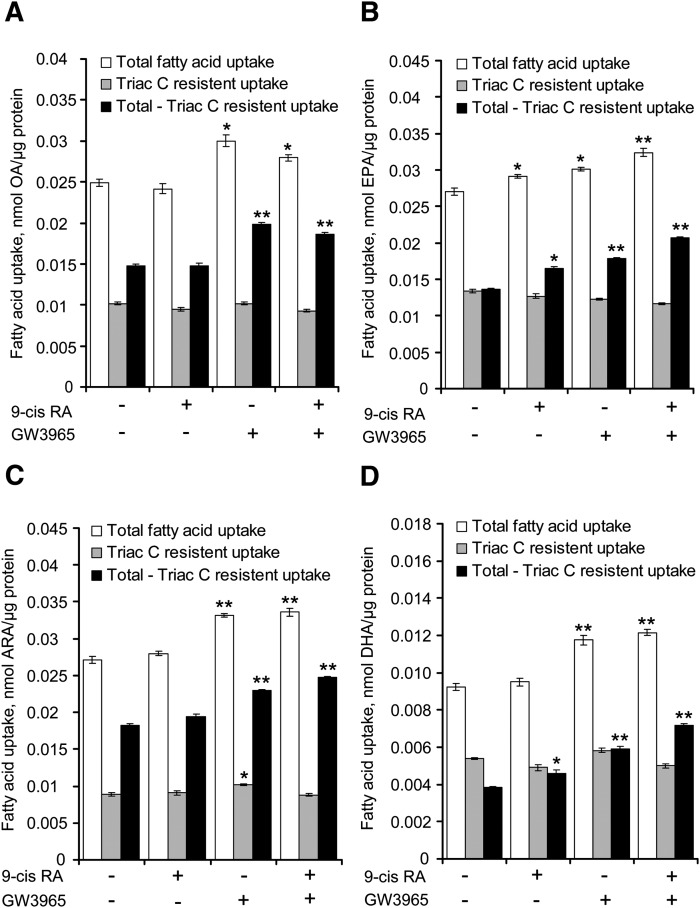
As ACSL3 was shown to affect the fatty acid uptake, we investigated if the LXR-mediated increase in ACSL3 expression and total acyl-CoA synthetase activity also increased the fatty acid uptake in BeWo cells. The cells were prestimulated with vehicle, LXR, and/or RXR agonist for 48 h, and subsequently, uptake of radiolabeled fatty acids was examined as indicated in Fig. 5. Uptake of OA, EPA, DHA, and ARA was significantly increased following LXR agonist stimulation of BeWo cells.
Fig. 5.
LXR activation increases uptake of OA, EPA, ARA, and DHA in BeWo cells. BeWo cells were prestimulated with vehicle, 9-cis RA (1 μM) and/or GW3965 (1 μM) for 48 h. The fatty acid uptake was measured 3 h after the addition of [14C] fatty acids (100 μM) in the presence of vehicle (white bars) or triacsin C (gray bars). The black bars represent total fatty acid uptake (white bars) minus the triacsin C resistant uptake (gray bars). The results are presented as calculated fatty acid uptake (in nmol) normalized to protein (in μg) per well. A: Uptake of [14C]OA. B: Uptake of [14C]EPA. C: Uptake of [14C]ARA. D: Uptake of [14C]DHA. The data are representative experiments of two to four independent experiments performed in triplicate (n = 3) ± SEM. *P < 0.05 and **P < 0.01 relative to control. ARA, arachidonic acid; BeWo, human placental choriocarcinoma; DHA, docosahexaenoic acid; EPA, eicosapentaenoic acid; LXR, liver X receptor; OA, oleic acid.
In parallel, we examined if the increased acyl-CoA synthetase activity following LXR activation accounted for the observed increase in total fatty acid uptake by incubating the cells in the presence of Triacsin C. Triacsin C inhibits the acyl-CoA synthetase activity of the ACSL 1, 3, and 4 proteins, but not the ACSL5 and 6 proteins (43, 44). As ACSL3 is the only acyl-CoA synthetase family member that is both significantly expressed and induced by activation of LXRs, the use of this inhibitor permitted us to separate the activity of ACSL3 from those of the weakly expressed ACSL5 and 6. Inhibition of acyl-CoA synthetase activity with Triacsin C inhibited cellular fatty acid uptake ∼50% for all tested fatty acids. Intriguingly, stimulation with RXR and/or LXR agonists had no effect on the fatty acid uptake in cells treated with Triacsin C, in marked contrast to the increased uptake observed in cells cultured in the absence of Triacsin C (Fig. 5, compare white and gray bars).
To estimate the uptake accounted for by LXR-mediated increased expression of ACSL3, we subtracted the fatty acid uptake in cells treated with Triacsin C (Triac C resistant uptake) from the uptake in untreated cells (total fatty acid uptake). The Triacsin C sensitive uptake (Total − Triac C resistant uptake) in cells receiving LXR and RXR agonists increased ∼46% for DHA, 34% for EPA, 27% for ARA, and 21% for OA compared with control (Fig. 5).
DISCUSSION
LXR regulates cellular lipid metabolism at various levels. The participation of LXRs in cholesterol transport and lipid uptake from lipoproteins is well documented (45, 46), but little information is available regarding their role in cellular fatty acid activation and fatty acid uptake per se. This article demonstrates that activation of LXR increases acyl-CoA synthetase activity through transcriptional activation of ACSL3 expression. Our experiments, using an acyl-CoA synthetase inhibitor (triacsin C) and silencing of ACSL3, suggest that ACSL3 is involved in the LXR-mediated increased acyl-CoA synthetase activity in BeWo cells. Furthermore, our experiments show an increased uptake of EPA, ARA, DHA, and OA in BeWo cells following LXR activation, which suggests that activation of LXRs has the ability to promote uptake of fatty acids in trophoblast cells. To our knowledge, this is the first demonstration of the involvement of LXR in the uptake of fatty acids by placental trophoblasts. Increased uptake of palmitic acid into myotubes stimulated with LXR agonists have been reported previously (47).
In this article we identified ACSL3 as a novel LXR target gene in placental trophoblast cells. Our data show that ligand activation of LXR stimulated ACSL3 expression in primary human trophoblasts and three human trophoblast cell lines. We also observed induction of ASCL3 mRNA in liver, skeletal muscle, heart, and adipose tissue in mice receiving a synthetic LXR agonist (unpublished observations). Thus, the LXR-mediated increase in ACSL3 expression is likely to occur in multiple species and tissues.
Ligand activation of LXRs induces transcription of the ACSL3 gene through binding of an LXR/RXR heterodimer to a characterized LXRE located upstream in the proximal human ASCL3 promoter. LXR is, however, able to induce the generated ACSL3 reporters in absence of the characterized LXRE element but at significant reduced efficiency. This suggests that the promoter contains additional transcriptional binding sites that respond to activation of LXRs. We were unable to identify additional DR-4 like LXREs in the −138 bp/+200 bp region, which argues against a second LXRE in this region. We speculate that the alternative regulation is due to LXR-mediated increased transcription of another transcription factor that binds in the proximal ASCL3 promoter region. Possible candidates are the known LXR target genes SREBP-1c (48) and carbohydrate responsive element binding protein (49), but peroxisome prolifertaor-activated receptor γ (PPARγ) and small heterodimer partner (SHP) could also be involved in this transcriptional regulation of ACSL3. A recent report demonstrated an interrelationship between LXRα, SREBP-1c, PPARγ, and SHP in the transcriptional regulation of glucokinase gene expression in liver (50).
LXRs are known to directly regulate target genes, such as FAS, ACC, and SREBP-1c, in liver (38, 48, 51). Due to the ability of LXRs to change the expression of these genes, LXRs are believed to be key regulators of de novo fatty acid synthesis. Increased intracellular FFAs, as result of increased de novo fatty acid synthesis or increased uptake of fatty acids, increases the risk for intracellular lipotoxicity, while further metabolism into complex lipids may reduce the lipotoxicity (52). LXR activation increases accumulation of triacylglycerol (TAG) in liver, muscle cells, and adipocytes (53–55), an effect which thus far has been explained by stimulation of lipogenesis (38, 48, 51). It remains to be thoroughly investigated if LXR-mediated upregulation of ACSL3 plays a role in this process. ACSL3 is reported to be important for secretion of the lipoprotein VLDL from hepatocytes by incorporation of fatty acids into phosphatidylcholine (56). LXRs are shown to increase the production and secretion of TAG-rich VLDL particles in the liver for TAG export to peripheral tissues (57). Therefore, a direct LXR-mediated increased hepatic expression of ACSL3 could contribute to the increased circulating TAG observed by systemic LXR activation in mice (53).
Despite the pivotal function of acyl-CoA synthetase in lipid metabolism, little information is available on this activity in placental cells. Our siRNA studies suggest that the acyl-CoA synthetase activity of ACSL3 accounts for a major part of long chain fatty acid activation in placental trophoblast cells, as it accounted for 50% of total oleoyl-CoA synthetase activity in BeWo cells. According to our expression data, the remaining activity is presumably accounted for mainly by the abundantly expressed ACSL1, ACSL4, FATP4, and FATP6 genes. All of these proteins are reported to have acyl-CoA synthetase activity with substrate preference for oleic acid as well as for other long or very long chain fatty acids (41, 58). In addition, some of the residual activity can be mediated by traces of ASCL3 protein remaining after siRNA knock down of the gene. Additional silencing studies targeting each of these genes are necessary to determine their individual contribution to acyl-CoA synthetase activity in trophoblast cells.
Growing evidence supports a role for members of the ACSL family in cellular fatty acid uptake (10). This was first shown in yeast, where forced expression of mammalian ACSL1, ACSL4, and ACSL6 in cells lacking native long chain acyl-CoA activity increased total fatty acid uptake (10). The importance of the acyl-CoA synthetase activity for fatty acid uptake is additionally supported by mutation studies where the acyl-CoA synthetase activity domain in FATP4 resulted in reduction of fatty acid uptake in COS-7 cells (59). These results are in line with those of Finstad et al. (60) reporting that uptake of EPA correlated with acyl-CoA synthetase activity and ACSL3 expression levels in two leukemia cell lines. The hypothesis is that the fatty acids are trapped in the cells due to their conversion into membrane-impermeable CoA thioesters directly after their transport across the plasma membrane (10). Our analyses of ACSL3 in BeWo cells revealed a remarkable correlation among ACSL3 expression, acyl-CoA synthetase activity, and fatty acid uptake, indicating that ACSL3 expression is required for efficient fatty acid uptake in trophoblast cells. Further, the correlation among LXR-mediated induction of ACSL3 transcript, acyl-CoA synthetase activity, and fatty acid uptake suggest that ACSL3 is important for LXR- mediated increased uptake and subsequent activation of fatty acid in BeWo cells.
The membrane-bound FATPs, FABPpm and CD36, and intracellular FABPs have so far been believed to be the main proteins involved in long chain fatty acid uptake in placental trophoblast cells [reviewed in (61)]. Our expression analysis detected high expression of several FATPs in BeWo cells, rendering it likely that they contribute to the fatty acid uptake in these cells. However, our studies suggest that ACSL3 is important for activation and uptake of fatty acids in BeWo cells. We showed that approximately half of the oleic acid uptake in BeWo cells is triacsin C sensitive, where ACSL3 likely accounts for the majority of this triacsin C sensitive uptake, as the other triacsin C sensitive proteins that potentially could be involved were expressed at lower levels (ACSL5, ACSL6, and members of the FATP family). A comparable expression level of ACSL3 in BeWo cells and placenta tissue opens the possibility that ACSL3 activity might be as important for fatty acid transport and metabolism in human trophoblasts as in our cultured BeWo cells.
CONCLUSION
Our data shed new light on the important functional relationship between LXRs and ACSL3 in placenta, and they suggest that the fatty acid activation function of ACSL3 contributes to the uptake of fatty acids in trophoblast cells.
Acknowledgments
The authors are grateful to Aud Jørgensen and Borghild Arntsen for their technical assistance. We are also grateful to Nina K Harsem for providing placental tissues and Nina K Harsem and Kari Anne R. Tobin for assistance in isolating placental trophoblast cells.
Footnotes
Abbreviations:
- ACSBG
- acyl-CoA synthetase bubblegum
- ACSL
- long-chain acyl-CoA synthetase
- ARA
- arachidonic acid
- B2M
- beta-2-microglobulin
- BeWo
- human placental choriocarcinoma
- DHA
- docosahexaenoic acid
- DR-4
- direct repeat-4
- EMSA
- electrophoretic mobility shift assay
- EPA
- eicosapentaenoic acid
- FAS
- fatty acid synthase
- FATP
- fatty acid transport protein
- LCPUFA
- long chain polyunsaturated fatty acid
- LXR
- liver X receptor
- LXRE
- LXR responsive element
- OA
- oleic acid
- oxLDL
- oxidized low density lipoproteins
- PPARγ
- peroxisome prolifertaor-activated receptor γ
- qRT-PCR
- quantitative RT-PCR
- RXR
- retinoid X receptor
- SHP
- small heterodimer partner
- siRNA
- small interfering RNA
- SREBP-1
- sterol-regulatory element binding protein-1
- TBP
- TATA box binding protein
- wt
- wild type
This work was supported by grants from the Medical Faculty, University of Oslo; Norwegian Research Council; the Johan Throne Holst Foundation; the Novo Nordic Foundation; European Union (EU) Sixth Framework Programme for Research and Technological Development (FP6); Consortium for Research into Nuclear Receptors in Development and Aging; South-Eastern Norway Regional Health Authority; and the Norwegian Sudden Infant Death Association.
REFERENCES
- 1.Fujino T., Kang M. J., Suzuki H., Iijima H., Yamamoto T. 1996. Molecular characterization and expression of rat acyl-CoA synthetase 3. J. Biol. Chem. 271: 16748–16752. [DOI] [PubMed] [Google Scholar]
- 2.Hall A. M., Smith A. J., Bernlohr D. A. 2003. Characterization of the Acyl-CoA synthetase activity of purified murine fatty acid transport protein 1. J. Biol. Chem. 278: 43008–43013. [DOI] [PubMed] [Google Scholar]
- 3.Hall A. M., Wiczer B. M., Herrmann T., Stremmel W., Bernlohr D. A. 2005. Enzymatic properties of purified murine fatty acid transport protein 4 and analysis of acyl-CoA synthetase activities in tissues from FATP4 null mice. J. Biol. Chem. 280: 11948–11954. [DOI] [PubMed] [Google Scholar]
- 4.Oikawa E., Iijima H., Suzuki T., Sasano H., Sato H., Kamataki A., Nagura H., Kang M. J., Fujino T., Suzuki H., et al. 1998. A novel acyl-CoA synthetase, ACS5, expressed in intestinal epithelial cells and proliferating preadipocytes. J. Biochem. 124: 679–685. [DOI] [PubMed] [Google Scholar]
- 5.Pei Z., Oey N. A., Zuidervaart M. M., Jia Z., Li Y., Steinberg S. J., Smith K. D., Watkins P. A. 2003. The acyl-CoA synthetase “bubblegum” (lipidosin): further characterization and role in neuronal fatty acid beta-oxidation. J. Biol. Chem. 278: 47070–47078. [DOI] [PubMed] [Google Scholar]
- 6.Lewin T. M., Kim J. H., Granger D. A., Vance J. E., Coleman R. A. 2001. Acyl-CoA synthetase isoforms 1, 4, and 5 are present in different subcellular membranes in rat liver and can be inhibited independently. J. Biol. Chem. 276: 24674–24679. [DOI] [PubMed] [Google Scholar]
- 7.Brasaemle D. L., Dolios G., Shapiro L., Wang R. 2004. Proteomic analysis of proteins associated with lipid droplets of basal and lipolytically stimulated 3T3–L1 adipocytes. J. Biol. Chem. 279: 46835–46842. [DOI] [PubMed] [Google Scholar]
- 8.Fujimoto Y., Itabe H., Kinoshita T., Homma K. J., Onoduka J., Mori M., Yamaguchi S., Makita M., Higashi Y., Yamashita A., et al. 2007. Involvement of ACSL in local synthesis of neutral lipids in cytoplasmic lipid droplets in human hepatocyte HuH7. J. Lipid Res. 48: 1280–1292. [DOI] [PubMed] [Google Scholar]
- 9.Minekura H., Kang M. J., Inagaki Y., Suzuki H., Sato H., Fujino T., Yamamoto T. T. 2001. Genomic organization and transcription units of the human acyl-CoA synthetase 3 gene. Gene. 278: 185–192. [DOI] [PubMed] [Google Scholar]
- 10.Tong F., Black P. N., Coleman R. A., Dirusso C. C. 2006. Fatty acid transport by vectorial acylation in mammals: roles played by different isoforms of rat long-chain acyl-CoA synthetases. Arch. Biochem. Biophys. 447: 46–52. [DOI] [PubMed] [Google Scholar]
- 11.Janowski B. A., Willy P. J., Devi T. R., Falck J. R., Mangelsdorf D. J. 1996. An oxysterol signalling pathway mediated by the nuclear receptor LXR alpha. Nature. 383: 728–731. [DOI] [PubMed] [Google Scholar]
- 12.Ulven S. M., Dalen K. T., Gustafsson J. A., Nebb H. I. 2005. LXR is crucial in lipid metabolism. Prostaglandins Leukot. Essent. Fatty Acids. 73: 59–63. [DOI] [PubMed] [Google Scholar]
- 13.Tontonoz P., Mangelsdorf D. J. 2003. Liver X receptor signaling pathways in cardiovascular disease. Mol. Endocrinol. 17: 985–993. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J., Febbraio M., Wada T., Zhai Y., Kuruba R., He J., Lee J. H., Khadem S., Ren S., Li S., et al. 2008. Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology. 134: 556–567. [DOI] [PubMed] [Google Scholar]
- 15.Innis S. M. 2005. Essential fatty acid transfer and fetal development. Placenta. 26(Suppl A): S70–S75. [DOI] [PubMed] [Google Scholar]
- 16.von Versen-Hoynck F., Rajakumar A., Parrott M. S., Powers R. W. 2009. Leptin affects system A amino acid transport activity in the human placenta: evidence for STAT3 dependent mechanisms. Placenta. 30: 361–367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Butte N. F. 2000. Carbohydrate and lipid metabolism in pregnancy: normal compared with gestational diabetes mellitus. Am. J. Clin. Nutr. 71: 1256S–1261S. [DOI] [PubMed] [Google Scholar]
- 18.Dutta-Roy A. K. 2000. Transport mechanisms for long-chain polyunsaturated fatty acids in the human placenta. Am. J. Clin. Nutr. 71: 315S–322S. [DOI] [PubMed] [Google Scholar]
- 19.Pavan L., Hermouet A., Tsatsaris V., Therond P., Sawamura T., Evain-Brion D., Fournier T. 2004. Lipids from oxidized low-density lipoprotein modulate human trophoblast invasion: involvement of nuclear liver X receptors. Endocrinology. 145: 4583–4591. [DOI] [PubMed] [Google Scholar]
- 20.Weedon-Fekjaer M. S., Duttaroy A. K., Nebb H. I. 2005. Liver X receptors mediate inhibition of hCG secretion in a human placental trophoblast cell line. Placenta. 26: 721–728. [DOI] [PubMed] [Google Scholar]
- 21.Burke K. T., Colvin P. L., Myatt L., Graf G. A., Schroeder F., Woollett L. A. 2009. Transport of maternal cholesterol to the fetus is affected by maternal plasma cholesterol concentrations in the golden Syrian hamster. J. Lipid Res. 50: 1146–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lindegaard M. L., Wassif C. A., Vaisman B., Amar M., Wasmuth E. V., Shamburek R., Nielsen L. B., Remaley A. T., Porter F. D. 2008. Characterization of placental cholesterol transport: ABCA1 is a potential target for in utero therapy of Smith-Lemli-Opitz syndrome. Hum. Mol. Genet. 17: 3806–3813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Campbell F. M., Clohessy A. M., Gordon M. J., Page K. R., Dutta-Roy A. K. 1997. Uptake of long chain fatty acids by human placental choriocarcinoma (BeWo) cells: role of plasma membrane fatty acid-binding protein. J. Lipid Res. 38: 2558–2568. [PubMed] [Google Scholar]
- 24.Tobin K. A., Johnsen G. M., Staff A. C., Duttaroy A. K. 2009. Long-chain polyunsaturated fatty acid transport across human placental choriocarcinoma (BeWo) cells. Placenta. 30: 41–47. [DOI] [PubMed] [Google Scholar]
- 25.Graham C. H., Hawley T. S., Hawley R. G., MacDougall J. R., Kerbel R. S., Khoo N., Lala P. K. 1993. Establishment and characterization of first trimester human trophoblast cells with extended lifespan. Exp. Cell Res. 206: 204–211. [DOI] [PubMed] [Google Scholar]
- 26.Kliman H. J., Nestler J. E., Sermasi E., Sanger J. M., Strauss J. F., III 1986. Purification, characterization, and in vitro differentiation of cytotrophoblasts from human term placentae. Endocrinology. 118: 1567–1582. [DOI] [PubMed] [Google Scholar]
- 27.Tobin K. A., Harsem N. K., Dalen K. T., Staff A. C., Nebb H. I., Duttaroy A. K. 2006. Regulation of ADRP expression by long-chain polyunsaturated fatty acids in BeWo cells, a human placental choriocarcinoma cell line. J. Lipid Res. 47: 815–823. [DOI] [PubMed] [Google Scholar]
- 28.Dalen K. T., Ulven S. M., Bamberg K., Gustafsson J. A., Nebb H. I. 2003. Expression of the insulin-responsive glucose transporter GLUT4 in adipocytes is dependent on liver X receptor alpha. J. Biol. Chem. 278: 48283–48291. [DOI] [PubMed] [Google Scholar]
- 29.Altschul S. F., Madden T. L., Schaffer A. A., Zhang J., Zhang Z., Miller W., Lipman D. J. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 25: 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Willy P. J., Umesono K., Ong E. S., Evans R. M., Heyman R. A., Mangelsdorf D. J. 1995. LXR, a nuclear receptor that defines a distinct retinoid response pathway. Genes Dev. 9: 1033–1045. [DOI] [PubMed] [Google Scholar]
- 31.Song C., Kokontis J. M., Hiipakka R. A., Liao S. 1994. Ubiquitous receptor: a receptor that modulates gene activation by retinoic acid and thyroid hormone receptors. Proc. Natl. Acad. Sci. USA. 91: 10809–10813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mangelsdorf D. J., Ong E. S., Dyck J. A., Evans R. M. 1990. Nuclear receptor that identifies a novel retinoic acid response pathway. Nature. 345: 224–229. [DOI] [PubMed] [Google Scholar]
- 33.Dalen K. T., Ulven S. M., Arntsen B. M., Solaas K., Nebb H. I. 2006. PPARalpha activators and fasting induce the expression of adipose differentiation-related protein in liver. J. Lipid Res. 47: 931–943. [DOI] [PubMed] [Google Scholar]
- 34.Wang Y. L., Guo W., Zang Y., Yaney G. C., Vallega G., Getty-Kaushik L., Pilch P., Kandror K., Corkey B. E. 2004. Acyl coenzyme a synthetase regulation: putative role in long-chain acyl coenzyme a partitioning. Obes. Res. 12: 1781–1788. [DOI] [PubMed] [Google Scholar]
- 35.Parkes H. A., Preston E., Wilks D., Ballesteros M., Carpenter L., Wood L., Kraegen E. W., Furler S. M., Cooney G. J. 2006. Overexpression of acyl-CoA synthetase-1 increases lipid deposition in hepatic (HepG2) cells and rodent liver in vivo. Am. J. Physiol. Endocrinol. Metab. 291: E737–E744. [DOI] [PubMed] [Google Scholar]
- 36.Duttaroy A. K., Jorgensen A. 2005. Insulin and leptin do not affect fatty acid uptake and metabolism in human placental choriocarcinoma (BeWo) cells. Prostaglandins Leukot. Essent. Fatty Acids. 72: 403–408. [DOI] [PubMed] [Google Scholar]
- 37.Yoshikawa T., Shimano H., Amemiya-Kudo M., Yahagi N., Hasty A. H., Matsuzaka T., Okazaki H., Tamura Y., Iizuka Y., Ohashi K., et al. 2001. Identification of liver X receptor-retinoid X receptor as an activator of the sterol regulatory element-binding protein 1c gene promoter. Mol. Cell. Biol. 21: 2991–3000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Joseph S. B., Laffitte B. A., Patel P. H., Watson M. A., Matsukuma K. E., Walczak R., Collins J. L., Osborne T. F., Tontonoz P. 2002. Direct and indirect mechanisms for regulation of fatty acid synthase gene expression by liver X receptors. J. Biol. Chem. 277: 11019–11025. [DOI] [PubMed] [Google Scholar]
- 39.Mitro N., Vargas L., Romeo R., Koder A., Saez E. 2007. T0901317 is a potent PXR ligand: implications for the biology ascribed to LXR. FEBS Lett. 581: 1721–1726. [DOI] [PubMed] [Google Scholar]
- 40.Pei Z., Jia Z., Watkins P. A. 2006. The second member of the human and murine bubblegum family is a testis- and brainstem-specific acyl-CoA synthetase. J. Biol. Chem. 281: 6632–6641. [DOI] [PubMed] [Google Scholar]
- 41.Soupene E., Kuypers F. A. 2008. Mammalian long-chain acyl-CoA synthetases. Exp. Biol. Med. (Maywood). 233: 507–521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schaiff W. T., Bildirici I., Cheong M., Chern P. L., Nelson D. M., Sadovsky Y. 2005. Peroxisome proliferator-activated receptor-gamma and retinoid X receptor signaling regulate fatty acid uptake by primary human placental trophoblasts. J. Clin. Endocrinol. Metab. 90: 4267–4275. [DOI] [PubMed] [Google Scholar]
- 43.Kim J. H., Lewin T. M., Coleman R. A. 2001. Expression and characterization of recombinant rat Acyl-CoA synthetases 1, 4, and 5. Selective inhibition by triacsin C and thiazolidinediones. J. Biol. Chem. 276: 24667–24673. [DOI] [PubMed] [Google Scholar]
- 44.Van Horn C. G., Caviglia J. M., Li L. O., Wang S., Granger D. A., Coleman R. A. 2005. Characterization of recombinant long-chain rat acyl-CoA synthetase isoforms 3 and 6: identification of a novel variant of isoform 6. Biochemistry. 44: 1635–1642. [DOI] [PubMed] [Google Scholar]
- 45.Ishimoto K., Tachibana K., Sumitomo M., Omote S., Hanano I., Yamasaki D., Watanabe Y., Tanaka T., Hamakubo T., Sakai J., et al. 2006. Identification of human low-density lipoprotein receptor as a novel target gene regulated by liver X receptor alpha. FEBS Lett. 580: 4929–4933. [DOI] [PubMed] [Google Scholar]
- 46.Zhang Y., Repa J. J., Gauthier K., Mangelsdorf D. J. 2001. Regulation of lipoprotein lipase by the oxysterol receptors, LXRalpha and LXRbeta. J. Biol. Chem. 276: 43018–43024. [DOI] [PubMed] [Google Scholar]
- 47.Kase E. T., Wensaas A. J., Aas V., Hojlund K., Levin K., Thoresen G. H., Beck-Nielsen H., Rustan A. C., Gaster M. 2005. Skeletal muscle lipid accumulation in type 2 diabetes may involve the liver X receptor pathway. Diabetes. 54: 1108–1115. [DOI] [PubMed] [Google Scholar]
- 48.Repa J. J., Liang G., Ou J., Bashmakov Y., Lobaccaro J. M., Shimomura I., Shan B., Brown M. S., Goldstein J. L., Mangelsdorf D. J. 2000. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 14: 2819–2830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Cha J. Y., Repa J. J. 2007. The liver X receptor (LXR) and hepatic lipogenesis. The carbohydrate-response element-binding protein is a target gene of LXR. J. Biol. Chem. 282: 743–751. [DOI] [PubMed] [Google Scholar]
- 50.Kim T. H., Kim H., Park J. M., Im S. S., Bae J. S., Kim M. Y., Yoon H. G., Cha J. Y., Kim K. S., Ahn Y. H. 2009. Interrelationship between liver X receptor alpha, sterol regulatory element-binding protein-1c, peroxisome proliferator-activated receptor gamma, and small heterodimer partner in the transcriptional regulation of glucokinase gene expression in liver. J. Biol. Chem. 284: 15071–15083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Talukdar S., Hillgartner F. B. 2006. The mechanism mediating the activation of acetyl-coenzyme A carboxylase-alpha gene transcription by the liver X receptor agonist T0–901317. J. Lipid Res. 47: 2451–2461. [DOI] [PubMed] [Google Scholar]
- 52.Yamaguchi K., Yang L., McCall S., Huang J., Yu X. X., Pandey S. K., Bhanot S., Monia B. P., Li Y. X., Diehl A. M. 2007. Inhibiting triglyceride synthesis improves hepatic steatosis but exacerbates liver damage and fibrosis in obese mice with nonalcoholic steatohepatitis. Hepatology. 45: 1366–1374. [DOI] [PubMed] [Google Scholar]
- 53.Schultz J. R., Tu H., Luk A., Repa J. J., Medina J. C., Li L., Schwendner S., Wang S., Thoolen M., Mangelsdorf D. J., et al. 2000. Role of LXRs in control of lipogenesis. Genes Dev. 14: 2831–2838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Cozzone D., Debard C., Dif N., Ricard N., Disse E., Vouillarmet J., Rabasa-Lhoret R., Laville M., Pruneau D., Rieusset J., et al. 2006. Activation of liver X receptors promotes lipid accumulation but does not alter insulin action in human skeletal muscle cells. Diabetologia. 49: 990–999. [DOI] [PubMed] [Google Scholar]
- 55.Juvet L. K., Andresen S. M., Schuster G. U., Dalen K. T., Tobin K. A., Hollung K., Haugen F., Jacinto S., Ulven S. M., Bamberg K., et al. 2003. On the role of liver X receptors in lipid accumulation in adipocytes. Mol. Endocrinol. 17: 172–182. [DOI] [PubMed] [Google Scholar]
- 56.Yao H., Ye J. 2008. Long chain acyl-CoA synthetase 3-mediated phosphatidylcholine synthesis is required for assembly of very low density lipoproteins in human hepatoma Huh7 cells. J. Biol. Chem. 283: 849–854. [DOI] [PubMed] [Google Scholar]
- 57.Grefhorst A., Elzinga B. M., Voshol P. J., Plosch T., Kok T., Bloks V. W., van der Sluijs F. H., Havekes L. M., Romijn J. A., Verkade H. J., et al. 2002. Stimulation of lipogenesis by pharmacological activation of the liver X receptor leads to production of large, triglyceride-rich very low density lipoprotein particles. J. Biol. Chem. 277: 34182–34190. [DOI] [PubMed] [Google Scholar]
- 58.Watkins P. A. 2008. Very-long-chain acyl-CoA synthetases. J. Biol. Chem. 283: 1773–1777. [DOI] [PubMed] [Google Scholar]
- 59.Milger K., Herrmann T., Becker C., Gotthardt D., Zickwolf J., Ehehalt R., Watkins P. A., Stremmel W., Fullekrug J. 2006. Cellular uptake of fatty acids driven by the ER-localized acyl-CoA synthetase FATP4. J. Cell Sci. 119: 4678–4688. [DOI] [PubMed] [Google Scholar]
- 60.Finstad H. S., Dyrendal H., Myhrstad M. C., Heimli H., Drevon C. A. 2000. Uptake and activation of eicosapentaenoic acid are related to accumulation of triacylglycerol in Ramos cells dying from apoptosis. J. Lipid Res. 41: 554–563. [PubMed] [Google Scholar]
- 61.Duttaroy A. K. 2009. Transport of fatty acids across the human placenta: a review. Prog. Lipid Res. 48: 52–61. [DOI] [PubMed] [Google Scholar]