Abstract
African ancestry individuals have a more favorable lipoprotein profile than Caucasians, although the mechanisms for these differences remain unclear. We measured fasting serum lipoproteins and genotyped 768 tagging or potentially functional single nucleotide polymorphisms (SNPs) across 33 candidate gene regions in 401 Afro-Caribbeans older than 18 years belonging to 7 multi-generational pedigrees (mean family size 51, range 21–113, 3,426 relative pairs). All lipoproteins were significantly heritable (P < 0.05). Gender-specific analysis showed that heritability for triglycerides was much higher (P < 0.01) in women than in men (women, 0.62 ± 0.18, P < 0.01; men, 0.13 ± 0.17, P > 0.10), but the heritability for LDL cholesterol (LDL-C) was higher (P < 0.05) in men than in women (men, 0.79 ± 0.21, P < 0.01; women, 0.39 ± 0.12, P < 0.01). The top 14 SNPs that passed the false discovery rate threshold in the families were then tested for replication in an independent population-based sample of 1,750 Afro-Caribbean men aged 40+ years. Our results revealed significant associations for three SNPs in two genes (rs5929 and rs6511720 in LDLR and rs7517090 in PCSK9) and LDL-C in both the family study and in the replication study. Our findings suggest that LDLR and PCSK9 variants may contribute to a variation in LDL-C among African ancestry individuals. Future sequencing and functional studies of these loci may advance our understanding of genetic factors contributing to LDL-C in African ancestry populations.
Lipoprotein abnormalities, characterized by elevated levels of LDL cholesterol (LDL-C) and triglycerides (TRIG) and low levels of HDL cholesterol (HDL-C), have a central role in the development of atherosclerotic coronary heart disease (CHD). A recent meta-analysis, including 3,000 individuals with CHD-related deaths, showed that HDL-C and LDL-C are independently associated with CHD risk (1). There is also considerable evidence that high levels of TRIG are an additional, independent risk factor for CHD (2, 3), although this is still controversial (4).
Individuals of African ancestry have a more favorable lipoprotein profile than Caucasians, characterized by lower levels of TRIG and higher levels of HDL-C (5–8). The mechanisms responsible for these ethnic differences remain to be defined. In particular, the differences in TRIG levels are independent of the greater degree of obesity among individuals of African ancestry and several other risk factors and appear to be consistent across African populations in different environments (9), indicating a possible role of genetic factors. Although genetic factors are important in determining lipoprotein levels, little data exists regarding the importance of heredity and specific genetic factors in determining lipoprotein levels in populations of African ancestry, especially outside the US, and the findings from previous studies in African-Americans may not necessarily apply to other African ancestry populations. Recently, several genome-wide association studies identified a number of loci contributing to inter-individual variation in lipoprotein levels (10, 11). However, the majority of these studies were restricted to Caucasian populations. Given the ethnic differences in lifestyle and environmental factors, as well as in genetic background, it is important to examine genes related to lipoprotein metabolism in different ethnic groups. Therefore, we examined the heritability of fasting, serum levels of HDL-C, LDL-C, and TRIG and systematically screened for association with 33 positional and biological candidate genes in large, multigenerational families of African ancestry.
METHODS
The Tobago Family Health Study sample
The Tobago Family Health Study was designed to better understand the role of inheritance, lifestyle, and body weight and composition in the etiology of several common chronic diseases in a population of African ancestry. The population on the Caribbean island of Tobago was largely settled in the late 1700s during the transatlantic slave trade. The first official British records in 1770 enumerated 268 Whites and 3,110 African slaves on the island. There were 15,470 African slaves on the island in 1819 (12). The British occupied Tobago permanently in 1814 and slavery was officially abolished in 1833 (12). The most recent census indicates that there are approximately 54,000 inhabitants on the island. According to the 1990 census data based on self-report, the population of Tobago was 92% African descent, 4.5% mixed, 2% East Indian, 0.4% White, and 1% other (13). We confirmed with molecular markers that the Afro-Caribbean population of Tobago has a low level of non-African admixture (6%) (14) compared with the more genetically heterogeneous African-American population that has much higher degree of non-African admixture (17–23.9%) (15–17).
Probands for the Tobago Family Health Study were identified from an ongoing population-based prostate cancer screening study (18). To be eligible, a proband had to be Afro-Caribbean, have had a spouse who was willing to participate in the study, and have at least six living offspring and/or siblings aged 18+ years who were residing in Tobago. Because we were interested in establishing a community-based sample of families, probands and their family members were recruited without regard to their health status. To date, 401 individuals aged 18–103 years (mean age 43 yrs) belonging to 7 multigenerational families (mean family size 51 individuals) of West African ancestry have been recruited. Among the families, we have the following relationships: 361 parent-offspring, 495 full siblings, 101 grandparent-grandchildren, 1,137 avuncular, 61 half-sibs, and 1,380 cousins (3,535 relative pairs). Written informed consent was obtained from every participant using forms and procedures approved by the Tobago Division of Health and Social Services and University of Pittsburgh Institutional Review Boards.
Replication sample
Using an independent population-based cohort of 1,750 Afro-Caribbean men who live in the same geographic region as the family study, we attempted to replicate the associations with single nucleotide polymorphisms (SNPs) that passed the false discovery rate (FDR) with α = 0.2. The replication sample was a randomly selected subset of a larger study of body composition among 2,500 men aged 40 and older (mean age 59 years) on the island of Tobago (19), with the comprehensive data on anthropometric measures, demographic information, medical history, lifestyle information, and fasting lipoproteins. Men were recruited by word of mouth, via health care workers at the hospital, health centers, or private physicians as well as local advertising by poster, flyers, and public service announcements. Written informed consent was obtained using forms approved by the Institutional Review Boards of the University of Pittsburgh and the Tobago Ministry of Health.
Data collection
Information on lifestyle habits [current smoking (yes/no), current alcohol intake (more than one drink per week, yes/no), walking (minutes per week), and TV viewing (hrs/week)], medical conditions, medication use, and reproductive characteristics in women (age at menarche, menopause, parity, oral contraceptive use) were assessed using standardized interviewer-administered questionnaires that were reviewed with participants in the study clinic. We recorded information on walking, because it is the predominant physical activity form on the island. Height was measured to the nearest 0.1 cm using a wall-mounted stadiometer. Weight was recorded to the nearest 0.1 kg without shoes on a balance-beam scale.
Clinical and metabolic variables
All biochemical assays were performed in the Heinz Nutrition Laboratory at the University of Pittsburgh's Graduate School of Public Health, which has met the accuracy and precision standards of the Centers for Disease Control and Prevention and is CLIA accredited. Serum was prepared after morning, fasting phlebotomy and stored at −70°C until assay. HDL-C was determined using the selective heparin/manganese chloride precipitation method, interassay CV 2.1% (20). LDL-C was calculated by means of the Friedewald equation. TRIG were determined enzymatically using the procedure of Bucolo and David, interassay CV 1.7% (21).
Gene and SNP selection
We used a positional and biological candidate gene approach to prioritize genes. A positional candidate gene approach identified genes that lie under linkage peaks for lipid traits based on four studies that performed genome wide scans to identify loci that may affect lipoprotein metabolism in African ancestry populations: Hypertension Genetic Epidemiology Network (22), HERITAGE Family Study (23, 24), African-American Diabetes Mellitus Study (25), and Genetics of NIDDM Study (26). The strongest linkage signals reported were: 5q33 for LDL-C (25), 19p13 and 19q13 for TRIG (24, 26), 1q25 and 7p21 for HDL-C (25), and 21q22 for LDL-C, HDL-C, and TRIG (22). The genes included: 19q13.2 Cluster (ApoE, ApoC1, ApoC2, and ApoC4), ABCG1 (ATP-binding Cassette, Subfamily G, Member 1), GYS1 (glycogen synthase 1), HSD11B1 [hydroxysteroid (11-β) dehydrogenase 1], LDLR (Low-density Lipoprotein Receptor), LXRβ (liver X receptor β), RETN (resistin), SOAT1 (sterol O-acyltransferase 1), and USF2 (upstream stimulatory factor 2).
A biological candidate gene approach included known physiologically defined genes for lipid metabolism that were selected on the basis of published evidence. Many of these genes have not been previously studied in African ancestry populations. Some genes were identified from animal studies and have not been investigated comprehensively in humans. The biological candidate genes included: 11q23.3 Cluster (ApoA1, ApoA4, ApoA5, and ApoC3), ACDC (Adiponectin), AdipoR1 (Adiponectin Receptor 1), AdipoR2 (Adiponectin Receptor 2), ApoA2 (Apolipoprotein A2), ApoB (Apolipoprotein B), LCAT (Lecithin-cholesterol Acyltransferase), DGAT2 (Acyl-CoA:diacylglycerol acyltransferase 2), FABP4 (fatty acid binding protein 4), FOXC2 (forkhead transcription factor C2), GTF3A (general transcription factor IIIa), HNF4A (Hepatic nuclear factor 4 α), LIPE (hormone-sensitive lipase), LPIN1 (lipin), LIPG (Endothelial Lipase), LPL (Lipoprotein Lipase), MTP (microsomal triglyceride transfer protein), PBEF1 (Visfatin), PCSK9 (Proprotein Convertase Subtilisin Kexin 9), PLIN (perilipin), PPARA (Peroxisome Proliferator-activated Receptor Alpha), PPARG (Peroxisome Proliferator-activated Receptor Gamma), SCARB1 (Scavenger Receptor Class B Type I), and SREBF2 (Sterol regulatory element binding transcription factor 2).
A reference panel of SNPs that spanned 5 kb upstream of the transcriptional start site and 5 kb downstream of the 3′ region of each gene or gene cluster was identified from the International HapMap database (www.hapmap.org) (Phase II, Yoruba population of Ibadan, Nigeria). We used the program HClust to select tag SNPs with a minor-allele frequency (MAF) ≥5% that predicted the remaining SNPs with an r2 ≥ 0.8 (27). In addition, potentially functional SNPs that were either nonsynonymous coding variants, predicted to alter a putative transcription factor binding site in the promoter region, or a putative exon splice enhancer with MAF ≥ 5% in an African reference population were selected for genotyping using the dbSNP database (Build 125) and the PupaSNP (28).
Genotyping in the Tobago Family Health Study
Genotyping of 768 SNPs was performed on genomic DNA using the Illumina GoldenGate Custom assay system. Eight blind duplicates were run and a 100% reproducibility rate was observed. Of the 768 SNPs attempted, 91 SNPs were dropped before statistical analysis, because genotypes were not able to be determined (N = 39), they were monomorphic or had a MAF <1% (N = 48), or they did not conform to the expectations of Hardy-Weinberg equilibrium in unrelated individuals (N = 4, P < 0.005). The 677 remaining SNPs were used in statistical analysis.
Genotyping of SNPs for replication in the Tobago population-based study of men
Fourteen SNPs that passed the FDR (rs5929, rs7517090, rs8030806, rs389261, rs6511720, rs3746575, rs11608456, rs11890442, rs2569540, rs11574739, rs1028583, rs3212198, rs5927, and rs6073435) were genotyped using genomic DNA with the fluorogenic 5′-nuclease TaqMan allelic discrimination assay system (Applied Biosystems, Foster City, CA). The assays were performed under standard conditions on a 7900HT real-time PCR instrument with probes and reagents purchased from Applied Biosystems. The best proxy SNP (rs6031593) was used in place of rs3746575 in the population study (D'=0.92, r2 = 0.72 HapMap Database, phase II release 24) due to rs3746575 assay failure. rs389261 (in the 19q13.2 Cluster) could not be genotyped by TaqMan, and the TaqMan Genotyping Assay for rs11608456 (in the AdipoR2) was not able to be manufactured (no proxies were identified for this SNP; the highest r2 was 0.61, HapMap Database, phase II release 24). All successfully genotyped SNPs conformed to the expectations of Hardy-Weinberg equilibrium (χ2 test of allele frequencies: P > 0.01). The average genotyping completeness rate was 95.8% and the average genotyping consensus rate among the >5% blind replicate samples was 99.2%.
Statistical analyses
First, the distributions of all the traits were assessed for nonnormality, and data were transformed before statistical analysis to reduce nonnormality. Subsequently, all outliers (±3.5 SD) were removed for each trait, and no more than four values were removed for a single variable. We calculated site-specific allele frequencies by gene counting and tested for departures from Hardy-Weinberg equilibrium using a goodness–of-fit statistic. Pairwise estimates of linkage disequilibrium were measured as D' and r2 from the diploid data (29). Quantitative genetic methods were used to model the total variation in all phenotypic parameters as a function of the mean trait value [additive genetic effects, heritability residual (h2r)], effects attributed by the measured covariates, and the uncertain variation due to residual genetic and unmeasured environmental impact plus random errors. We also tested whether the heritabilities in phenotypes differed between men and women (i.e., h2r men = h2r women) by comparing the difference between the heritability estimates in the two sexes with the estimated variance of the difference. Briefly, we expanded the basic variance component model to allow the genetic variances in male and female to differ (30). Then we use the likelihood ratio test to compare the basic versus the expanded model. All analyses were performed using the SOLAR software package (Solar, version 2.1.4; Southwest Foundation for Biomedical Research, San Antonio, TX) (31) under the variance components analytical framework, which accounts for the nonindependence among family members. Covariates of probable importance for lipoprotein metabolism, including age, gender, body mass index (BMI), current smoking, current alcohol intake, minutes walking per week, postmenopausal status, parity, age at menarche, and oral contraceptive use, were included in all models. Based on participants' self-report, none of the participants reported using lipid-lowering medication. After assessment of covariates, association analyses were performed to compare mean trait levels by genotype assuming an additive model.
We used the FDR with α = 0.2 to adjust for multiple hypothesis testing. The FDR method controls the expected proportion of false-positives among all positive results over multiple studies (14). The FDR can be regarded as a post hoc maximizing procedure and is more powerful than multiple-comparison procedures based on the family-wise error rate. SNPs with P-values more significant than the expected FDR distribution with α = 0.2 were considered significant.
Using an independent population-based sample of African ancestry men from the same geographic region, we attempted to replicate associations of LDL-C and TRIG with 14 SNPs that passed the FDR in the family study. The single SNPs were tested for their association with the lipoproteins assuming an additive model. Linear regression was used to test for association between the number of rare alleles and the levels of lipoproteins. All models were adjusted for age, BMI, minutes walking per week, current smoking, and current alcohol intake. Analyses were performed using SAS version 9.1 (SAS Institute, Cary, NC).
RESULTS
Subject characteristics
Mean age of the family study participants was approximately 43 years and ranged from 18 to 103 years (Table 1). Participants were predominantly women (60.3%). The prevalence of obesity was particularly high among women (∼43%). More men than women smoked (11% vs. 1%) and drank alcohol (29% vs. 9%) on a regular basis. Approximately one-third of women were postmenopausal and one-third used oral contraceptives. BMI was significantly greater in women, but waist circumference was similar in men and women. The mean levels of TRIG, HDL-C, and LDL-C were 88.6 ± 46.4 mg/dl, 40.3 ± 12.8 mg/dl and 132.4 ± 42.4 mg/dl, respectively, and all lipoprotein levels were similar in men and women (Table 1).
TABLE 1.
General characteristics of the Afro-Caribbean participants in the Tobago Family Health Study
| Total (N = 471) | Men (N = 187) | Women (N = 284) | |
|---|---|---|---|
| Age (years) | 42.7 ± 16.8 | 42.8 ± 16.9 | 42.6 ± 16.8 |
| Anthropometric | |||
| BMI (kg/m2) | 28.3 ± 6.4 | 26.7 ± 4.9 | 29.4 ± 7.0* |
| Waist circumference (cm) | 89.9 ± 15.4 | 90.1 ± 12.4 | 89.7 ± 17.2 |
| Lifestyle | |||
| Alcohol use (>1 drink/week) (%) | 13.2 | 28.9 | 2.8* |
| Current smoking (%) | 4.9 | 11.4 | 0.7* |
| TV viewing (h/week) | 15.9 ± 8.0 | 16.6 ± 8.5 | 15.5 ± 7.7 |
| Time spent walking per week (min/week) | 47.9 ± 96.7 | 52.6 ± 72.7 | 44.8 ± 109.8 |
| Reproductive | |||
| Use of oral contraceptives (%) | N/A | N/A | 33.1 |
| Ever pregnant (%) | N/A | N/A | 76.7 |
| Number of pregnancies | N/A | N/A | 3.5 ± 2.6 |
| Postmenopausal status (%) | N/A | N/A | 31.6 |
| Lipoprotein levels | |||
| TRIG (mg/dl) | 89 ± 46 | 92 ± 50 | 86.2 ± 43.6 |
| HDL-C (mg/dl) | 40 ± 13 | 41 ± 12 | 39.6 ± 13.2 |
| LDL-C (mg/dl) | 133 ± 42 | 130 ± 40 | 134 ± 43 |
| Medical conditions | |||
| Obesitya (%) | 33.5 | 19.8 | 42.6* |
| Diabetesb (%) | 15.4 | 12.4 | 17.2 |
| Hypertensionc (%) | 28.6 | 30.4 | 27.4 |
Values are unadjusted means (SEM). * P-value for gender difference <0.001.
BMI ≥ 30 kg/m2.
Fasting glucose of ≥126 mg/dl and/or use of insulin/hypoglycemic medications.
Systolic blood pressure of ≥140 mmHg and/or diastolic blood pressure of ≥90 mmHg or currently taking antihypertensive medications.
Heritability analyses
h2r, the proportion of variance due to additive genetic effects, was estimated after removing the variation attributable to significant covariates. All lipid and lipoproteins were significantly heritable (h2r ± SE; TRIG, 0.28 ± 0.11; HDL-C, 0.48 ± 0.11; and LDL-C, 0.44 ± 0.11; P < 0.05 for all). Significant covariates accounted for 24% of the total phenotypic variation for TRIG (age, gender, BMI, menopause, parity, and current oral contraceptive medication intake were significant covariates), 6% of the total phenotypic variation for HDL-C (BMI and current alcohol intake were significant covariates), and 18% of the total phenotypic variation for LDL-C (age, BMI, and menopause were significant covariates).
Gender-stratified analysis showed that the residual heritability for TRIG was much higher (P < 0.01) in women than in men (women, 0.62 ± 0.18, P < 0.01; men, 0.13 ± 0.17, P > 0.10). In contrast, the residual heritability for LDL-C was higher (P < 0.05) in men than in women (men, 0.79 ± 0.21, P < 0.01; women, 0.39 ± 0.12, P < 0.01). Heritability of HDL-C tended to be higher in men but the difference was not significantly different (P = 0.20) (men, 0.51 ± 0.18, P < 0.01; women, 0.33 ± 0.14, P < 0.01).
Family-based association analyses
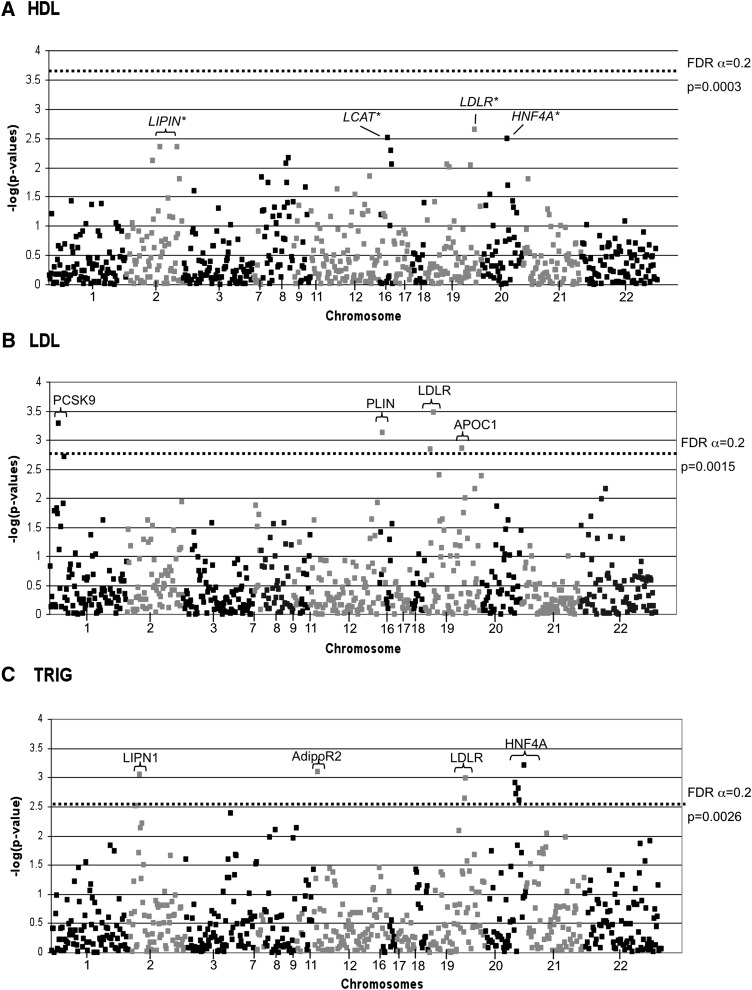
No SNPs passed the FDR cutoff of 0.2 for associations with HDL-C in the families (Fig. 1A). However, SNPs in the LDLR, LCAT, HNF4A, and LIPN1 showed the most suggestive nominal associations with HDL-C (supplementary Table I and Fig. 1A). Using an FDR cutoff of 0.20, five SNPs in four genes (LDLR, PCSK9, PLIN, and APOC1) passed the P-value threshold (P = 0.0015) for associations with LDL-C levels, and nine SNPs in four genes (HNF4A, ADIPOR2, LIPN1, and LDLR) passed the P-value threshold (P = 0.0026) for associations with TRIG levels in the families, after adjustment for potential covariates (Supplementary Tables II and III; Fig. 1B, C).
Fig. 1.
Association results are presented for HDL (A), LDL (B), and TRIG (C). The -log of the P-value is presented on the y axis and SNPs are presented across the x axis in order of chromosome position. The dashed lines in B and C represent the P-value for significant SNPs and gene symbols are presented for those SNPs. No SNPs in HDL were associated beyond the FDR threshold, and the dashed line in A represents the level of significance, which would have been needed. Top HDL findings are indicated in A.
Minor alleles of two SNPs (rs5929 and rs6511720) in LDLR were associated with lower LDL-C in the families (Table 2). The minor allele of PCSK9 rs7517090 was also associated with lower LDL-C (Table 2). On the other hand, minor alleles of APOC1 rs389261 and PLIN rs8030806 were associated with greater LDL-C (Table 2). Three additional SNPs in both LDLR and 19q cluster and seven additional SNPs in PCSK9 showed nominal associations with LDL-C (P < 0.05), but no additional SNPs in PLIN showed nominal associations with LDL-C (supplementary Table II). Each of the five SNPs accounted for 1.5–5.2% of the total phenotypic variability in LDL-C (Table 2). The minor allele frequencies for all significant SNPs in the LDLR, PCSK9, APOC1, and PLIN were comparable to those reported in African ancestry individuals from the Yoruba population of Ibadan, Nigeria (www.hapmap.org). Interestingly, the LDLR rs6511720 has shown the strongest association with HDL-C (nominal P-value = 0.0022), but no other SNPs in the LDLR gene were associated with HDL-C.
TABLE 2.
Adjusted mean (± SE) LDL-C levels for SNPs that showed significant associations with LDL-C after adjusting for multiple comparisons
| Adjusted Means ± SE |
Variation in LDL-C Explained by SNP | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Name | SNP rs no. | Chromosome | Position (bp)* | Study | 1/1 | 1/2 | 2/2 | Additive P-value | |
| LDLR | rs5929 | 19 | 11087800 | Family | 134 ± 2.3 | 124 ± 4.3 | 99 ± 14.7 | 4.6% | 0.00033 |
| Replication | 134 ± 1.1 | 127 ± 2.2 | 128 ± 7.6 | 0.5% | 0.004 | ||||
| LDLR | rs6511720 | 19 | 11063306 | Family | 136 ± 2.2 | 117 ± 4.3 | 93 ± 13.3 | 4.8% | 0.0014 |
| Replication | 135 ± 1.1 | 127 ± 2.0 | 124 ± 6.6 | 0.77% | 0.0002 | ||||
| PCSK9 | rs7517090 | 1 | 55287544 | Family | 138 ± 2.4 | 120 ± 3.4 | 98 ± 10.6 | 5.2% | 0.0005 |
| Replication | 134 ± 1.2 | 132 ± 1.7 | 121 ± 5.0 | 0.9% | 0.05 | ||||
| PLIN | rs8030806 | 15 | 88005145 | Family | 128 ± 2.4 | 137 ± 3.8 | 168 ± 12.8 | 1.5% | 0.0007 |
| Replication | 133 ± 1.1 | 133 ± 2.0 | 118 ± 7.5 | 1.1% | 0.44 | ||||
| 19q13.2 | rs389261 | 19 | 50112183 | Family | 124 ± 2.8 | 137 ± 3.1 | 140 ± 6.7 | 3.2% | 0.0013 |
| Replication | ND | ND | ND | ND | ND | ||||
1, major allele; 2, minor allele. Bold indicates significant P-value. ND, Not determined due to assay failure.
* Chromosomal position was based on the National Center for Biotechnology Information dbSNP, build 36.2.
A significant association with TRIG was observed for the five SNPs in HNF4A, one SNP in AdipoR2 and LPIN1, and two SNPs in LDLR in the families. Nine additional SNPs in HNF4A, two additional SNPs in AdopoR2, five additional SNPs in LPIN1, and six additional SNPs in LDLR were nominally significant (P < 0.05; supplementary Table III). The minor allele frequencies (MAF) for all SNPs were comparable to those reported in African ancestry subjects from the Yoruba population of Ibadan, Nigeria (www.hapmap.org). Each of these 14 SNPs accounted for 3.5–6.7% of the total phenotypic variability in TRIG levels. The major alleles of all significant HNF4A SNPs were associated with lower TRIG levels.
Population-based replication analyses
Twelve out of 14 SNPs were successfully genotyped in an independent population-based sample of 1,750 African ancestry men, participants of the large body composition study among 2,500 men from the same geographic region as the family study. Our replication results revealed significant associations for three successfully genotyped SNPs (in LDLR and PCSK9), which were associated with LDL-C level in families, but not for rs8030806 in PLIN (Table 2). However, no association with TRIG levels was replicated for eight successfully genotyped significant SNPs (Table 3).
TABLE 3.
Adjusted mean (± SE) TRIG levels for SNPs that showed significant associations with TRIG after adjusting for multiple comparisons
| Adjusted Means ± SE |
Variation in TRIG Explained by SNP | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Gene Name | SNP rs no. | Chromosome | Position (bp)* | Study Population | 1/1 | 1/2 | 2/2 | Additive P-value | |
| HNF4A | rs3746575a | 20 | 42491510 | Family | 82 ± 4.3 | 93 ± 3.3 | 114 ± 6.6 | 6.7% | 0.0006 |
| Population-based | 111 ± 2.6 | 112 ± 2.0 | 109 ± 2.9 | 0.21 | 0.57 | ||||
| HNF4A | rs11574739 | 20 | 42476364 | Family | 88 ± 3.1 | 94 ± 4.3 | 146 ± 11.7 | 5.7% | 0.0012 |
| Population-based | 112 ± 1.7 | 110 ± 2.5 | 110 ± 6.9 | 0.16 | 0.47 | ||||
| HNF4A | rs1028583 | 20 | 42484175 | Family | 85 ± 4.3 | 91 ± 3.4 | 112 ± 6.4 | 4.5% | 0.0015 |
| Population-based | 111 ± 2.6 | 111 ± 1.9 | 110 ± 2.9 | 0.011 | 0.96 | ||||
| HNF4A | rs3212198 | 20 | 42477776 | Family | 84 ± 3.9 | 93 ± 3.6 | 112 ± 6.5 | 5.1% | 0.0019 |
| Population-based | 112 ± 2.3 | 110 ± 2.0 | 112 ± 3.2 | 0.03% | 0.78 | ||||
| HNF4A | rs6073435 | 20 | 42487002 | Family | 87 ± 3.7 | 92 ± 3.6 | 119 ± 7.6 | 4.3% | 0.0025 |
| Population-based | 113 ± 2.2 | 109 ± 2.0 | 114 ± 3.4 | 0.11% | 0.87 | ||||
| LDLR | rs2569540 | 19 | 11099239 | Family | 106 ± 4.7 | 90 ± 3.6 | 83 ± 5.1 | 3.6% | 0.001 |
| Population-based | 114 ± 2.5 | 114 ± 20.0 | 111 ± 3.19 | 0.38 | 0.20 | ||||
| LDLR | rs5927 | 19 | 11094941 | Family | 86 ± 3.7 | 95 ± 3.9 | 108 ± 7.2 | 3.5% | 0.0023 |
| Population-based | 111 ± 2.0 | 112 ± 2.1 | 108 ± 4.3 | 0.28% | 0.76 | ||||
| AdipoR2 | rs11608456 | 12 | 1701455 | Family | 97 ± 2.9 | 79 ± 5.3 | 70 ± 18.3 | 3.8% | 0.0007 |
| Population-based | ND | ND | ND | ND | ND | ||||
| LPIN1 | rs11890442 | 2 | 11828454 | Family | 85 ± 3.2 | 98 ± 4.3 | 119 ± 7.6 | 6.1% | 0.0009 |
| Population-based | 111 ± 2.0 | 110 ± 2.1 | 118 ± 4.1 | 0.61 | 0.31 | ||||
1, Major allele; 2, minor allele.
In the population-based Afro-Caribbean replication study rs6031593 was genotyped as a proxy SNP for rs3746575 genotyped in the family study. Bold indicates significant P-value. ND, Not determined due to assay failure.
* Chromosomal position was based on the National Center for Biotechnology Information dbSNP, build 36.2.
DISCUSSION
The current study examined the heritability of lipoprotein phenotypes in a well-characterized collection of extended multi-generational families of West African ancestry on the Caribbean island of Tobago. This relatively isolated and homogeneous population has a low level of non-African ancestry as determined by population ancestry informative molecular markers (32). We found that all lipoprotein traits were significantly heritable, with heritability estimates ranging from 28% to 48%. Significant demographic, lifestyle, reproductive, and medical factors accounted for only 6–24% of the total phenotypic variation in lipoprotein traits. Heritability estimates obtained in our study are in the range of those previously reported in African-Americans [TRIG: 0.14–0.41 (22, 26, 33); LDL-C: 0.39–0.55 (22, 26); HDL-C: 0.38–0.65 (26, 33)]. Our results also suggest that among women, genes may have a much stronger influence on TRIG levels but less strong influence on LDL-C levels than in men. This difference could be related to gender differences in the ability to metabolize fat, interactions with genes on the sex chromosomes, effects related to female reproduction or differences arising from gender-specific hormonal factors, or other environmental exposures, such as diet.
We observed very low levels of TRIG in this Afro-Caribbean population. This finding is consistent with the results from other studies in African ancestry populations (6). On the other hand, we observed lower HDL-C levels than those reported in other studies of African ancestry populations (6). Lower HDL-C observed in our sample might be due to nutritional factors. Also, obesity is strongly associated with low HDL-C (34). The prevalence of overweight and obesity were high in our families, especially among women, which could be an additional explanation for the observed low HDL-C levels. Excessive adiposity and accumulation of body fat is strongly associated with a low concentration, adverse distribution pattern, and abnormal metabolism of HDL particles (35). However, TRIG levels were not influenced by the high prevalence of obesity as TRIG levels were low even among women.
A second objective of our analysis was to determine the association of lipoprotein variation with tagging SNPs spanning 33 candidate gene regions. We used a pathway-driven approach to select positional and biological candidate genes, the majority of which have not yet been investigated comprehensively in an African ancestry population. We successfully replicated an association between LDL-C and two SNPs in the LDLR gene (rs5929 and rs6511720). rs6511720 has also been associated with LDL-C in a large genome-wide association study of lipoproteins (11). Notably, using an independent sample, we replicated the LDLR SNP, rs5929, which showed the strongest association with LDL-C levels in the Tobago Family Study. Although this SNP was not previously reported to be associated with LDL-C, it has a very low frequency in populations of European ancestry (MAF = 0.8%), which could be a potential explanation why it has not been identified in previous population studies of European ancestry individuals. In silico analysis using FASTSNP (36) (http://fastsnp.ibms.sinica.edu.tw/) indicated a possible impact of rs5929, located within a coding region, on splicing regulation. The LDLR SNPs, rs2304182 and rs2738464, which were nominally associated with LDL-C in our study, were previously associated with LDL-C in the Multi-Ethnic Study of Atherosclerosis (37) and the Framingham Offspring Study (38).
A SNP in PCSK9, rs7517090, was associated with LDL-C in the families and was replicated in the population-based sample. Minor alleles of an intronic SNP (rs7517090) in PCSK9 were associated with decreased LDL-C levels in both the family- and population-based Afro-Caribbean samples. This SNP has not been previously associated with LDL-C, but it is absent in populations of European and Asian ancestries (MAF = 0%), whereas in the current sample and in the Yoruba population from HapMap, the MAF is 13.9–19.9%. PCSK9 has recently emerged as a potential target for reducing LDL-C levels, as genetic variation in this gene strongly contributes to variation in LDL-C, particularly among African-Americans (39). PCSK9 post-transcriptionally downregulates the LDLR in the liver and thereby controls the level of LDL-C (40). For example, two rare nonsense mutations in PCSK9 with a combined frequency of 2% in African ancestry individuals were associated with a 28% reduction in mean LDL-C and an 88% reduction in CHD risk (41). Although several variants of PCSK9 have been identified so far, their effect on PCSK9 activity has not been determined.
We report for the first time a potential association of multiple common variants in a transcription factor gene, hepatic nuclear factor 4-α (HNF4A), with decreased TRIG levels in African ancestry population. However, we were unable to replicate these findings in an independent sample of men. Taking into consideration that we observed significant gender differences in heritability estimates for TRIGs and in obesity levels, future studies should test for similar HNF4A associations in an independent sample of African ancestry women and those with a wider range in body weight. Nonetheless, similar to our findings, another family study showed that rs3212198 was associated with TRIG in families of European and Mexican ancestry (42).
Our study has several potential limitations. We only genotyped tagging SNPs with a MAF ≥ 0.05 from the Yoruban population in HapMap. It is plausible that SNPs with a minor allele frequency of 1–5%, rare variants with MAF < 1%, such as those in PCSK9 from previous studies in African-Americans, which we did not genotype, and structural variants also influence lipoproteins in African ancestry populations. Additionally, because we only studied individuals of Afro-Caribbean ancestry, our findings may not be generalizable to populations of other ethnicities. Although none of our participants reported using lipid-lowering medications, some under-reporting of lipid-lowering medication may have occurred. Furthermore, the small number of pedigrees in our analysis may have influenced our heritability estimates and association results. However, previous studies (43, 44) have shown that extended multi-generational pedigrees, such as those in the present study, may provide more precise heritability estimates and may be more powerful than nuclear pedigrees or sib-pairs in detecting and locating disease loci and with fewer false positives. Therefore, our multigenerational families, which contained over 3,000 relative pairs, should have been sufficient to provide a robust estimate of heritability. Finally, the African ancestry population of Tobago appears to have a low level of non-African admixture (6%), but we did not genotype individual ancestry-informative markers in the family or population samples. However, the MAF of all significant SNPs were similar to those reported in the Yoruba population. Our study also has notable strengths, including its inclusion of very large, multi-generational pedigrees of African ancestry from a relatively isolated and homogeneous population with a low level of non-African ancestry. We also assessed a broad array of potential covariates for lipid and lipoprotein levels. Finally, to directly replicate the most promising SNP associations, we genotyped an independent population-based sample derived from the same geographic region and with the same genetic background as our families, which increases the validity and generalizability of our findings.
In conclusion, this study suggests that genetic factors are a significant source of inter-individual differences in fasting lipid and lipoprotein levels among Afro-Caribbeans and that genes may have much stronger influence on the distribution of TRIGs in women than in men of African ancestry. We have confirmed that LDLR and PCSK9 are strong candidate genes for LDL-C in African ancestry individuals. Our study also suggests a potentially novel association between common HNF4A variants and TRIG levels in Afro-Caribbean families. Future sequencing and functional studies of these loci may advance our understanding of the genetic factors contributing to the low LDL-C and TRIG phenotypes in African ancestry populations.
Supplementary Material
Acknowledgments
The authors would also like to thank the participants of both studies and all supporting staff.
Footnotes
Abbreviations:
- BMI
- body mass index
- CHD
- coronary heart disease
- FDR
- false discovery rate
- HDL-C
- HDL cholesterol
- h2r
- residual heritability
- LDL-C
- LDL cholesterol
- MAF
- minor-allele frequency
- SNP
- single nucleotide polymorphism
- TRIG
- triglyceride
Dr. Miljkovic was supported for this project by the Individual National Research Service Award for postdoctoral training from the National Heart, Lung, and Blood Institute (grant F32 HL083641) and is currently supported by the Mentored Research Scientist Development Award from the National Institute of Diabetes and Digestive and Kidney Diseases (grant K01DK083029). Dr. Yerges-Armstrong was supported by the National Institute on Aging (training grant T32AG00181). Genotyping services were provided by the Johns Hopkins University under federal contract number (N01HV48195) from the National Heart, Lung, and Blood Institute and by Seattle SNPs's Program for Genomic Applications from the National Heart, Lung, and Blood Institute (U01 HL66682 and UO1 HL66642). The Tobago Family Health Study was supported, in part, by funding or in-kind services from the Division of Health and Social Services and Tobago House of Assembly, by grants R03-AR050107 and R01-AR049747 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases. The Tobago Population-based Health Study was supported by grant R01AR049747 from the National Institute of Arthritis and Musculoskeletal and Skin Diseases. Its contents are solely the responsibility of the authors and do not necessarily represent the official views of the National Institutes of Health or other granting agencies.
The online version of this article (available at http://www.jlr.org) contains supplementary data in the form of three tables.
REFERENCES
- 1.Lewington S., Whitlock G., Clarke R., Sherliker P., Emberson J., Halsey J., Qizilbash N., Peto R., Collins R. 2007. Blood cholesterol and vascular mortality by age, sex, and blood pressure: a meta-analysis of individual data from 61 prospective studies with 55,000 vascular deaths. Lancet. 370: 1829–1839. [DOI] [PubMed] [Google Scholar]
- 2.Nordestgaard B. G., Benn M., Schnohr P., Tybjaerg-Hansen A. 2007. Nonfasting triglycerides and risk of myocardial infarction, ischemic heart disease, and death in men and women. JAMA. 298: 299–308. [DOI] [PubMed] [Google Scholar]
- 3.Bansal S., Buring J. E., Rifai N., Mora S., Sacks F. M., Ridker P. M. 2007. Fasting compared with nonfasting triglycerides and risk of cardiovascular events in women. JAMA. 298: 309–316. [DOI] [PubMed] [Google Scholar]
- 4.Ginsberg H. N. 2002. New perspectives on atherogenesis: role of abnormal triglyceride-rich lipoprotein metabolism. Circulation. 106: 2137–2142. [DOI] [PubMed] [Google Scholar]
- 5.Miljkovic-Gacic I., Bunker C. H., Ferrell R. E., Kammerer C. M., Evans R. W., Patrick A. L., Kuller L. H. 2006. Lipoprotein subclass and particle size differences in Afro-Caribbeans, African Americans, and white Americans: associations with hepatic lipase gene variation. Metabolism. 55: 96–102. [DOI] [PubMed] [Google Scholar]
- 6.Brown S. A., Hutchinson R., Morrisett J., Boerwinkle E., Davis C. E., Gotto A. M., Jr., Patsch W. 1993. Plasma lipid, lipoprotein cholesterol, and apoprotein distributions in selected US communities. The Atherosclerosis Risk in Communities (ARIC) Study. Arterioscler. Thromb. 13: 1139–1158. [DOI] [PubMed] [Google Scholar]
- 7.Zoratti R. 1998. A review on ethnic differences in plasma triglycerides and high-density-lipoprotein cholesterol: is the lipid pattern the key factor for the low coronary heart disease rate in people of African origin? Eur. J. Epidemiol. 14: 9–21. [DOI] [PubMed] [Google Scholar]
- 8.Webber L. S., Srinivasan S. R., Wattigney W. A., Berenson G. S. 1991. Tracking of serum lipids and lipoproteins from childhood to adulthood. The Bogalusa Heart Study. Am. J. Epidemiol. 133: 884–899. [DOI] [PubMed] [Google Scholar]
- 9.Howard B. V., Criqui M. H., Curb J. D., Rodabough R., Safford M. M., Santoro N., Wilson A. C., Wylie-Rosett J. 2003. Risk factor clustering in the insulin resistance syndrome and its relationship to cardiovascular disease in postmenopausal white, black, hispanic, and Asian/Pacific Islander women. Metabolism. 52: 362–371. [DOI] [PubMed] [Google Scholar]
- 10.Kathiresan S., Musunuru K., Orho-Melander M. 2008. Defining the spectrum of alleles that contribute to blood lipid concentrations in humans. Curr. Opin. Lipidol. 19: 122–127. [DOI] [PubMed] [Google Scholar]
- 11.Willer C. J., Sanna S., Jackson A. U., Scuteri A., Bonnycastle L. L., Clarke R., Heath S. C., Timpson N. J., Najjar S. S., Stringham H. M., et al. 2008. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat. Genet. 40: 161–169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Douglas A. 2003. Tobago: Melancholy Isle. Volume Three 1807–1898. Westindiana, St. Anns, Republic of Trinidad & Tobago. [Google Scholar]
- 13.Tobago C. 1993. Population and Housing Census. Volume 11. Demographic Report: Age Structure. Religion. Ethnic Group. Education. Central Statistical Office, Office of the Prime Minister, Port of Spain, Republic of Trinidad and Tobago. [Google Scholar]
- 14.Miljkovic-Gacic I., Ferrell R. E., Patrick A. L., Kammerer C. M., Bunker C. H. 2005. Estimates of African, European and Native American ancestry in Afro-Caribbean men on the island of Tobago. Hum. Hered. 60: 129–133. [DOI] [PubMed] [Google Scholar]
- 15.Reiner A. P., Ziv E., Lind D. L., Nievergelt C. M., Schork N. J., Cummings S. R., Phong A., Burchard E. G., Harris T. B., Psaty B. M., et al. 2005. Population structure, admixture, and aging-related phenotypes in African American adults: the Cardiovascular Health Study. Am. J. Hum. Genet. 76: 463–477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Parra E. J., Marcini A., Akey J., Martinson J., Batzer M. A., Cooper R., Forrester T., Allison D. B., Deka R., Ferrell R. E., et al. 1998. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 63: 1839–1851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Parra E. J., Kittles R. A., Argyropoulos G., Pfaff C. L., Hiester K., Bonilla C., Sylvester N., Parrish-Gause D., Garvey W. T., Jin L., et al. 2001. Ancestral proportions and admixture dynamics in geographically defined African Americans living in South Carolina. Am. J. Phys. Anthropol. 114: 18–29. [DOI] [PubMed] [Google Scholar]
- 18.Bunker C. H., Patrick A. L., Konety B. R., Dhir R., Brufsky A. M., Vivas C. A., Becich M. J., Trump D. L., Kuller L. H. 2002. High prevalence of screening-detected prostate cancer among Afro-Caribbeans: the Tobago Prostate Cancer Survey. Cancer Epidemiol. Biomarkers Prev. 11: 726–729. [PubMed] [Google Scholar]
- 19.Hill D. D., Cauley J. A., Sheu Y., Bunker C. H., Patrick A. L., Baker C. E., Beckles G. L., Wheeler V. W., Zmuda J. M. 2008. Correlates of bone mineral density in men of African ancestry: the Tobago bone health study. Osteoporos. Int. 19: 227–234. [DOI] [PubMed] [Google Scholar]
- 20.Warnick G. R., Albers J. J. 1978. A comprehensive evaluation of the heparin-manganese precipitation procedure for estimating high density lipoprotein cholesterol. J. Lipid Res. 19: 65–76. [PubMed] [Google Scholar]
- 21.Bucolo G., David H. 1973. Quantitative determination of serum triglycerides by the use of enzymes. Clin. Chem. 19: 476–482. [PubMed] [Google Scholar]
- 22.North K. E., Miller M. B., Coon H., Martin L. J., Peacock J. M., Arnett D., Zhang B., Province M., Oberman A., Blangero J., et al. 2005. Evidence for a gene influencing fasting LDL cholesterol and triglyceride levels on chromosome 21q. Atherosclerosis. 179: 119–125. [DOI] [PubMed] [Google Scholar]
- 23.Feitosa M. F., Borecki I. B., Rankinen T., Rice T., Despres J. P., Chagnon Y. C., Gagnon J., Leon A. S., Skinner J. S., Bouchard C., et al. 2005. Evidence of QTLs on chromosomes 1q42 and 8q24 for LDL-cholesterol and apoB levels in the HERITAGE family study. J. Lipid Res. 46: 281–286. [DOI] [PubMed] [Google Scholar]
- 24.Feitosa M. F., Rice T., North K. E., Kraja A., Rankinen T., Leon A. S., Skinner J. S., Blangero J., Bouchard C., Rao D. C. 2006. Pleiotropic QTL on chromosome 19q13 for triglycerides and adiposity: the HERITAGE Family Study. Atherosclerosis. 185: 426–432. [DOI] [PubMed] [Google Scholar]
- 25.Adeyemo A. A., Johnson T., Acheampong J., Oli J., Okafor G., Amoah A., Owusu S., Agyenim-Boateng K., Eghan B. A., Jr., Abbiyesuku F., et al. 2005. A genome wide quantitative trait linkage analysis for serum lipids in type 2 diabetes in an African population. Atherosclerosis. 181: 389–397. [DOI] [PubMed] [Google Scholar]
- 26.Malhotra A., Wolford J. K. 2005. Analysis of quantitative lipid traits in the genetics of NIDDM (GENNID) study. Diabetes. 54: 3007–3014. [DOI] [PubMed] [Google Scholar]
- 27.Rinaldo A., Bacanu S. A., Devlin B., Sonpar V., Wasserman L., Roeder K. 2005. Characterization of multilocus linkage disequilibrium. Genet. Epidemiol. 28: 193–206. [DOI] [PubMed] [Google Scholar]
- 28.Conde L., Vaquerizas J. M., Ferrer-Costa C., de la Cruz X., Orozco M., Dopazo J. 2005. PupasView: a visual tool for selecting suitable SNPs, with putative pathological effect in genes, for genotyping purposes. Nucleic Acids Res. 33: W501–W505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Devlin B., Risch N. 1995. A comparison of linkage disequilibrium measures for fine-scale mapping. Genomics. 29: 311–322. [DOI] [PubMed] [Google Scholar]
- 30.Brown L. B., Streeten E. A., Shuldiner A. R., Almasy L. A., Peyser P. A., Mitchell B. D. 2004. Assessment of sex-specific genetic and environmental effects on bone mineral density. Genet. Epidemiol. 27: 153–161. [DOI] [PubMed] [Google Scholar]
- 31.Almasy L., Blangero J. 1998. Multipoint quantitative-trait linkage analysis in general pedigrees. Am. J. Hum. Genet. 62: 1198–1211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Benjamini Y., Hochberg Y. 1995. Controlling the false discovery rate: a practical and powerful approach to multiple testing. J. R. Stat. Soc. B. 57: 289–300. [Google Scholar]
- 33.Kullo I. J., Ding K., Boerwinkle E., Turner S. T., de Andrade M. 2006. Quantitative trait loci influencing low density lipoprotein particle size in African Americans. J. Lipid Res. 47: 1457–1462. [DOI] [PubMed] [Google Scholar]
- 34.Ko G. T., Chan J. C., Cockram C. S. 1998. The association between dyslipidaemia and obesity in Chinese men after adjustment for insulin resistance. Atherosclerosis. 138: 153–161. [DOI] [PubMed] [Google Scholar]
- 35.Despres J. P., Moorjani S., Lupien P. J., Tremblay A., Nadeau A., Bouchard C. 1990. Regional distribution of body fat, plasma lipoproteins, and cardiovascular disease. Arteriosclerosis. 10: 497–511. [DOI] [PubMed] [Google Scholar]
- 36.Yuan H. Y., Chiou J. J., Tseng W. H., Liu C. H., Liu C. K., Lin Y. J., Wang H. H., Yao A., Chen Y. T., Hsu C. N. 2006. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 34: W635–W641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Post W., Mychaleckyj J. C., Raynor L., Taylor K. D., Guo X., Watson K. E., Hedrick C., Polak J., Tsai M., Rich S. S., et al. 2007. Variants in the LDL receptor gene are associated with LDL cholesterol in the Multi-Ethnic Study of Atherosclerosis. Presented at the annual meeting of The American Society of Human Genetics, October 23–27, 2007, San Diego, CA. [Google Scholar]
- 38.Estus S., Zhu H., Zou F., Lok J., Younkin S., Manning A. K., Grear K. E., Ling I. F., Tucker H. M., Simpson J. F., et al. 2007. LDLR polymorphisms, cholesterol and Alzheimers disease. Presented at the annual meeting of The American Society of Human Genetics, October 23–27, 2007, San Diego, CA. [Google Scholar]
- 39.Cohen J., Pertsemlidis A., Kotowski I. K., Graham R., Garcia C. K., Hobbs H. H. 2005. Low LDL cholesterol in individuals of African descent resulting from frequent nonsense mutations in PCSK9. Nat. Genet. 37: 161–165. [DOI] [PubMed] [Google Scholar]
- 40.Benjannet S., Rhainds D., Essalmani R., Mayne J., Wickham L., Jin W., Asselin M. C., Hamelin J., Varret M., Allard D., et al. 2004. NARC-1/PCSK9 and its natural mutants: zymogen cleavage and effects on the low density lipoprotein (LDL) receptor and LDL cholesterol. J. Biol. Chem. 279: 48865–48875. [DOI] [PubMed] [Google Scholar]
- 41.Cohen J. C., Boerwinkle E., Mosley T. H., Jr., Hobbs H. H. 2006. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N. Engl. J. Med. 354: 1264–1272. [DOI] [PubMed] [Google Scholar]
- 42.Weissglas-Volkov D., Huertas-Vazquez A., Suviolahti E., Lee J., Plaisier C., Canizales-Quinteros S., Tusie-Luna T., Aguilar-Salinas C., Taskinen M. R., Pajukanta P. 2006. Common hepatic nuclear factor-4alpha variants are associated with high serum lipid levels and the metabolic syndrome. Diabetes. 55: 1970–1977. [DOI] [PubMed] [Google Scholar]
- 43.Slate J., Pemberton J. M., Visscher P. M. 1999. Power to detect QTL in a free-living polygynous population. Heredity. 83: 327–336. [DOI] [PubMed] [Google Scholar]
- 44.Wijsman E. M., Amos C. I. 1997. Genetic analysis of simulated oligogenic traits in nuclear and extended pedigrees: summary of GAW10 contributions. Genet. Epidemiol. 14: 719–735. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.