The crystal structure of the phosphatase domain of Sts-1 at pH 4.6 complexed with sulfate has been determined to 1.35 Å resolution.
Keywords: T-cell receptor, Sts-1, phosphoglycerate mutases, phosphatases, signaling proteins, sulfate
Abstract
The suppressor of T-cell signaling (Sts) proteins are multidomain proteins that negatively regulate the signaling of membrane-bound receptors, including the T-cell receptor (TCR) and the epidermal growth-factor receptor (EGFR). They contain at their C-terminus a 2H-phosphatase homology (PGM) domain that is responsible for their protein tyrosine phosphatase activity. Here, the crystal structure of the phosphatase domain of Sts-1, Sts-1PGM, was determined at pH 4.6. The asymmetric unit contains two independent molecules and each active site is occupied by a sulfate ion. Each sulfate is located at the phosphate-binding site and makes similar interactions with the catalytic residues. The structure suggests an explanation for the lower Michaelis–Menten constants at acidic pH.
1. Introduction
The 2H-phosphatase superfamily of enzymes, which is also referred to as the phosphoglycerate mutase (PGM) superfamily, is a large family of enzymes that includes the diverse acid phosphatases (AcPs), cofactor-dependent phosphoglycerate mutase (dPGM), fructose 2,6-bisphosphatase (F2,6BPase), the Sts proteins (Mikhailik et al., 2007 ▶) and many others. It is so called because the majority of its members are phosphatases that use two catalytic histidine residues to dephosphorylate substrates (Rigden, 2008 ▶; Jedrzejas, 2000 ▶). The first His is the nucleophilic residue during catalysis and belongs to a conserved ‘RHGE’ signature motif. The second His and two conserved Arg residues are scattered in the primary sequence, complete the active site and give it a basic potential character that attracts and stabilizes phosphorylated substrates. Outside of the His and Arg residues, the primary sequences of these enzymes show little conservation. Despite the low sequence homology, the overall tertiary fold is globally maintained; more importantly, the structure of the active site is highly conserved. The diversity among family members is reflected in their substrate diversity and specificity: substrates range from small phosphorylated molecules to phosphoproteins (Sts) and some enzymes are very specific (F2,6BPase) while others are promiscuous (AcPs).
Recently, we have determined the crystal structure of the 2H-phosphatase domain of mouse Sts-1, Sts-1PGM, and of its homologue Sts-2 alone or in complex with phosphate and phosphate mimics. Using site-directed mutagenesis, we characterized their phosphatase activities (Mikhailik et al., 2007 ▶; Chen et al., 2008 ▶; Chen, Jakoncic, Carpino et al., 2009 ▶; Carpino et al., 2009 ▶; Chen, Jakoncic, Parker et al., 2009 ▶). We showed that His380 (Sts-1 numbering), His565, Arg369 and Arg462 are catalytic residues that are part of the active site, which also includes Arg383 and Glu490. In particular, we showed that Sts-1 possesses a protein tyrosine phosphatase (PTP) activity that is severely reduced by the mutation of any of the residues mentioned above. This PTP activity is important for the dephosphorylation of cytosolic and membrane proteins such as ZAP-70 and the epidermal growth-factor receptor (EGFR; Mikhailik et al., 2007 ▶; Raguz et al., 2007 ▶). Despite the 45% sequence identity and the conservation of catalytic residues between the Sts homologues, Sts-2PGM has a relatively weaker activity compared with Sts-1 (Chen et al., 2008 ▶; Chen, Jakoncic, Carpin et al., 2009 ▶).
To shed light on how Sts-1PGM interacts with its phosphorylated substrates, we carried out a series of experiments in which substrates were cocrystallized or soaked into crystals of wild-type or catalytically impaired mutants of Sts-1PGM. However, our attempts at observing any substrate in the active site failed. Since the various Sts-1 crystals were grown at pH values of 6.5–7.0 and since the catalytic activity at these pH values is substantial (Y. Chen & N. Nassar, unpublished work), we argued that the phosphorylated substrates were hydrolyzed by Sts-1 in the crystalline lattice or prior to formation of the crystalline lattice. The idea was then to grow Sts-1 crystals at a low pH, where the catalytic activity of Sts-1 is significantly reduced while its affinity for phosphorylated substrates is improved (Y. Chen & N. Nassar, unpublished work). Here, we describe the crystallization and crystal structure of Sts-1PGM obtained at pH 4.6 and in complex with a sulfate ion.
2. Materials and methods
2.1. Expression and purification
The mouse Sts-1PGM domain (residues 369–640) was cloned as an N-terminally His-tagged protein in the pProEX-HTb vector (Life Technologies), expressed in Escherichia coli BL21-CodonPlus strain (Stratagene) and purified to homogeneity as described in Kleinman et al. (2006 ▶). Purified Sts-1PGM as assessed by SDS–PAGE was concentrated by centrifugation using Amicon Ultra Centrifugal filters (10 kDa molecular-weight cutoff) to ∼23 mg ml−1 as measured by the Bradford protein assay (Bradford, 1976 ▶), shock-frozen in liquid nitrogen and stored at 193 K until use.
2.2. Crystallization and data collection
Crystallization screening of Sts-1PGM was performed at 293 K using the hanging-drop vapour-diffusion method. Protein aliquots were thawed on ice and incubated with fresh dithiothreitol (DTT) at a final concentration of 2 mM prior to use. Thin needle-like crystals were obtained when Sts-1PGM (in 20 mM Tris–HCl pH 8.0, 150 mM NaCl, 5 mM β-mercaptoethanol and 2 mM DTT) was mixed with an equal amount of reservoir solution. The reservoir solution consisted of 22–25%(w/v) polyethylene glycol 2000 monomethylether (PEG 2000 MME, Fluka), 0.2 M ammonium sulfate and 0.1 M sodium acetate pH 4.6–5.5. These crystals were improved by macroseeding, in which the PEG 2000 MME concentration and protein:reservoir ratio were optimized. While performing seeding we noticed that when exposing the crystallization drop to air for a minute or so lozenge-like crystals appeared at the air–drop interface. Those lozenges were seeded without further washing or manipulation into freshly prepared drops. The best crystals were obtained when the seeded drop was made by mixing 2 µl protein solution and 6–9 µl reservoir solution at 22% PEG 2000 MME. The lozenges grew to 0.1 × 0.1 × 0.05 mm in size 24–48 h after seeding. Cryoprotection was achieved by increasing the PEG concentration to 30% and adding 10% ethylene glycol. Crystals were mounted on nylon loops (Hampton Research, Inc.) before flash-cooling in liquid nitrogen.
One diffraction data set was collected from a single crystal to 1.35 Å resolution using a 2k × 2k ADSC Q270 CCD detector on beamline X6A at the National Synchrotron Light Source (NSLS; Brookhaven National Laboratory). All intensities were indexed, processed and scaled with the HKL-2000 package (Otwinowski & Minor, 1997 ▶).
2.3. Structure determination
The diffraction data had clear systematic absences in the h00, 0k0 and 00l reflections that were consistent with the orthorhombic space group P212121. The unit-cell parameters were a = 63.0, b = 80.0, c = 105.4 Å, α = β = γ = 90°, corresponding to a unit-cell volume of 531 216 Å3. The unit-cell parameters are compatible with the presence of two Sts-1PGM monomers in the asymmetric unit, corresponding to a Matthews coefficient V M (Matthews, 1968 ▶) of 2.21 Å3 Da−1 and a solvent content of 44%.
Although the anomalous signal of the sulfur was clear from the data (see Fig. 1 ▶), molecular-replacement phases were found by solving the rotation and translation functions in the program MOLREP (Vagin & Teplyakov, 1997 ▶) from the CCP4 package (Collaborative Computational Project, Number 4, 1994 ▶) using default parameters and chain A of the apo Sts-1PGM coordinates (PDB code 2h0q; Mikhailik et al., 2007 ▶) as a search model. The molecular replacement ultimately confirmed the space group as P212121. Two independent solutions belonging to two unrelated dimers (A and B) were found and the biologically active homodimers were formed by applying the space-group symmetry operations such that molecule A formed a dimer with a symmetry-related copy of molecule B. The crystallographic residual R work was 0.483 at the end of this step. Refinement against the 1.35 Å diffraction data was performed in the program REFMAC5 (Murshudov et al., 1997 ▶) using the maximum-likelihood target function. Model visualization and rebuilding against electron-density maps as well as the addition of water molecules were performed in the program Coot (Emsley & Cowtan, 2004 ▶). Stereochemistry and structure quality were checked using the program PROCHECK (Laskowski et al., 1993 ▶). The final crystallographic residuals R work and R free (calculated by randomly selecting 5% of the data; Brünger, 1992 ▶) were 16.4% and 19.1%, respectively (Table 1 ▶).
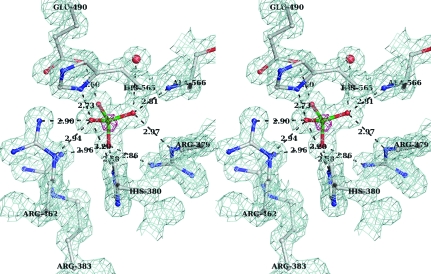
Figure 1.
Stereo representation of the final (2F o − F c) electron-density map (blue mesh) calculated to 1.35 Å resolution and contoured at 1σ around the sulfate molecule. Superposed is the anomalous difference electron-density map (pink mesh) shown for the same region contoured at 4σ. The hydrogen-bonding network between the sulfate moiety and the protein is shown as dotted lines. Amino acids are shown in ball-and-stick representation and labeled. Distances (Å) between H-atom donor and acceptor are shown.
Table 1. Data-collection and refinement statistics.
Values in parentheses are for the last resolution bin.
| Data collection | |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 62.97, b = 80.05, c = 105.45 |
| Resolution (Å) | 20.0–1.35 (1.37–1.35) |
| Rmerge (%) | 5.1 (30.3) |
| 〈I〉/〈σ(I)〉 | 32.5 (3.3) |
| Completeness (%) | 98.3 (83.5) |
| No. of reflections | 669717 |
| No. of unique reflections | 115649 (4846) |
| Multiplicity | 5.8 |
| Mosaicity (°) | 0.24 |
| Refinement statistics | |
| Resolution (Å) | 20.0–1.35 (1.39–1.35) |
| Rwork (%) | 16.2 (26.4) |
| Rfree (%) | 19.1 (31.2) |
| No. of scatterers | 4998 |
| Protein | 4403 |
| Sulfate | 10 |
| Water | 585 |
| Average B factors (Å2) | 13.8 |
| Protein atoms | 12.5 |
| Sulfate ions | 7.9 |
| Water molecules | 24.0 |
| R.m.s.d. from ideal geometry | |
| Bond lengths (Å) | 0.008 |
| Bond angles (°) | 1.297 |
| Ramachandran statistics (%) | |
| Most favored | 93.0 |
| Additionally allowed | 6.6 |
| Generously allowed | 0.4 |
| Outliers | 0.0 |
| PDB code | 3mbk |
3. Results
3.1. Crystallization
We recently showed using the nonspecific substrate para-nitrophenyl phosphate (pNPP) that the Michaelis–Menten constants k cat and K m of the 2H-phosphatase domain Sts-1PGM are significantly reduced at acidic pH values (C. Chen & N. Nassar, unpublished work), suggesting that at acidic pH Sts-1PGM interacts with its substrates without hydrolyzing them. It thus seems logical to search for crystallization conditions of Sts-1PGM in complex with its phosphorylated substrates at acidic pH values in order to obtain an atomic picture of how this domain binds its substrate(s). As a first step, we wanted to check for structural changes that might take place owing to crystallization at acidic pH values. We therefore designed crystallization experiments in which the molecular weight of the PEG used as precipitant and the nature of the salt used as an additive at 0.2 M were varied while the pH was restricted to 4.6–5.5 using sodium acetate as a buffer. While Sts-1PGM crystallizes in space group C2 in PEG 8000 with 0.3 M magnesium acetate or chloride as an additive at pH 6.5–7.0, needle-like crystals were obtained with PEG 2000 MME and ammonium sulfate at pH 4.6. Seeding was employed to increase the size of the needles. During seeding, lozenge-like crystals appeared after the drop was exposed to air. The lozenge-shaped crystals were optimized by seeding, varying the PEG concentration in the reservoir and the composition of the drop. The latter parameter turned out to be crucial to obtaining large single crystals. The best crystals were obtained when protein and reservoir were mixed in a ratio of 1:3 to 1:4.5 and immediately seeded.
3.2. Overall structure
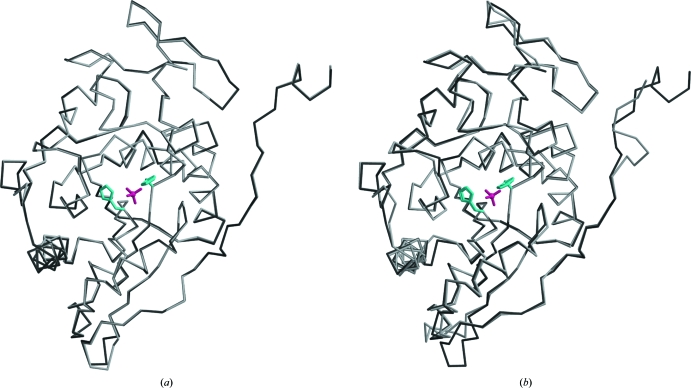
The higher resolution of the diffraction data at pH 4.6 allowed us to build residues C-terminal to the model obtained at pH 6.5 (PDB code 2h0q), which were missing in the C2 crystals, and to improve the quality of the model. However, density for the first and last four residues of the construct was missing. In both copies, the 264 residues of the native protein show clear density apart from the Leu605–Glu607 loop and the side chains of several solvent-exposed lysines. The two molecules in the asymmetric unit superpose with a root-mean-square deviation (r.m.s.d.) of 0.223 Å calculated on Cα positions. The largest deviations occur in the loops Leu529–Ala530 and Leu605–Gly609. These two loops are solvent-exposed and are not involved in catalysis. The low r.m.s.d. value between the two molecules in the asymmetric unit suggests that overall they do not differ significantly (Fig. 2 ▶ a). The C2 and P212121 models also superpose well, with an r.m.s.d. of 0.65 Å calculated on 260 Cα positions between their respective A chains. As shown in Fig. 2 ▶(b), the positions of the catalytic residues are well conserved in the two structures, suggesting that the structure is not affected by the crystallization pH and that the observed decrease in catalytic activity at lower pH values is not a consequence of structural rearrangement of the active site but is likely to arise from the protonation of key catalytic residues.
Figure 2.
(a) Superposition of the two Sts-1PGM monomers that populate the asymmetric unit in the present structure solved at pH 4.6. (b) Superposition of the A chains of Sts-1PGM solved at pH 6.5 (light gray; PDB code 2h0q) and at pH 4.6 (dark gray). The sulfate ion is shown in pink and the side chains of the catalytic His380 and His565 are shown in cyan in ball-and-stick representation to highlight the active site.
The Sts-1PGM monomer has an overall fold characteristic of the 2H-phosphatase superfamily. The core of the enzyme is a seven-stranded β-sheet surrounded by eight α-helices and several 310-helices. At the C-terminus, an eighth strand followed by a 310-helix separate from the core of one monomer and are inserted against the core of the second monomer in the dimer. The dimer interface buries 2296 Å2 of accessible surface area and is comparable in size to the dimer interface of Sts-2PGM (Chen, Jakoncic, Parker et al., 2009 ▶).
3.3. The active site
The active sites at pH 4.6 and 7.0 are remarkably similar. From the early stages of the refinement, a nonprotein electron density consistent with a symmetrical molecule of tetrahedral arrangement such as a phosphate or sulfate was present in the active site of each Sts-1PGM monomer. The anomalous difference electron-density map calculated with phases derived from the final model and ΔF ano as coefficients and displayed at 4σ shows that a heavy atom occupies the centre of the tetrahedron (pink density in Fig. 1 ▶). The vertices of the tetrahedron are within hydrogen-bonding distance of the guanidinium groups of several arginines and are therefore occupied by H-atom acceptors. Given that Sts-1PGM was purified in phosphate-free buffers and given that the crystals were grown in the presence of 0.2 M ammonium sulfate, it is likely that this density is that of a sulfate molecule. Furthermore, bond lengths and anomalous signal peaks corroborate the presence of SO4 2−.
Fig. 1 ▶ depicts as dotted lines the extensive network of hydrogen-bond interactions between the sulfate molecule and the active site. The side chains of His380, His565, Arg379, Arg383, Arg462 and Glu490 as well as the main-chain amino group of Ala566 interact with the four O atoms. The distance between the S atom and N∊2 of the nucleophilic His380 is 3.3 Å, which is large to mimic a nucleophilic attack. Instead, His380 N∊2 is 2.6 Å from one sulfate O atom. This short distance implies a strong hydrogen-bond interaction between these two atoms. Since the sulfate is deprotonated and negatively charged at pH 4.6, N∊2 has to be protonated for the hydrogen-bond interaction to occur. Together with the observation that Nδ1 is also protonated, our data imply that His380 is protonated on both N atoms in the present structure. Another key catalytic residue is Glu490. The carboxylate group of Glu490 is 2.8 Å from one sulfate O atom, suggesting that Glu490 must be protonated for the hydrogen bond to occur. Glu490 can be protonated under the acidic crystallization conditions since the pK a of a glutamate side chain is ∼4.6. A protonated Glu490 is also consistent with its role as a proton donor to the leaving group during catalysis (Mikhailik et al., 2007 ▶).
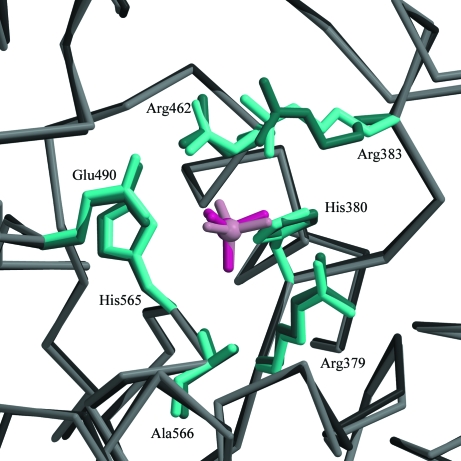
When compared with the structure of Sts-1PGM in complex with phosphate (Mikhailik et al., 2007 ▶), the sulfate makes similar interactions with the active site and occupies exactly the same position (Fig. 3 ▶). The distances of the hydrogen-bond interactions made by the two anions with the protein are similar on average. For example, the interactions made by the sulfate with Arg379, His380, Glu490 and Ala566 are 0.2 Å shorter on average and the interactions made with Arg383 and His565 are 0.2 Å longer on average than those made by the phosphate. This 0.2 Å difference in hydrogen-bond distances is insignificant given the difference in the data resolution used to refine the structures (1.35 Å for the sulfate versus 2.7 Å for the phosphate) and the difference in the error on the coordinates between the two structures.
Figure 3.
Structural alignment in the active site of sulfate-bound (dark pink) and phosphate-bound (light pink) Sts-1PGM. The side chains of conserved catalytic residues are also highlighted.
4. Discussion
Protein tyrosine phosphatases (PTPs) are essential components of signal transduction pathways that regulate the duration and the intensity of the signal to be propagated by dephosphorylating target proteins. Whereas the manner in which Cys-based phosphatases such as PTP-1B interact with their substrates is known (Jia et al., 1995 ▶), how His-based Sts phosphatases recognize their substrates and how their specificity is determined remain unknown. We have solved the crystal structure of Sts-1PGM at pH 4.6. The rationale behind this work is that at acidic pH the k cat and K m of Sts-1PGM for the nonspecific substrate pNPP are significantly reduced. Thus, capturing a complex between Sts-1PGM and a substrate for structural studies should be achievable at acidic pH. The overall structure of the enzyme is identical to the structure previously determined at physiological pH despite the difference in the crystallization space group. Each active site in the asymmetric unit is occupied by one sulfate ion that sits in the phosphate-binding site. The sulfate and phosphate ions make similar interactions with the protein, suggesting that the sulfate is a good phosphate mimic in this case. Although the protonation of His380 has no physiological relevance, the tight hydrogen bond this histidine makes with the sulfate at acidic pH values might stabilize the EP complex, resulting in a low k cat. In addition, a protonated N∊2 cannot carry out a nucleophilic attack on the P atom of a substrate, which would explain the low k cat value experimentally observed at acidic pH values.
Supplementary Material
PDB reference: phosphatase domain of Sts-1, 3mbk
Acknowledgments
We thank Vivan Stojanoff for help during data collection on X6A. Research in NN’s laboratory was supported in part by grants from the NIH (CA-115611) and DOD (NF060060). Research carried out at the X6A beamline, National Synchrotron Light Source, Brookhaven National Laboratory is supported by the US Department of Energy under contract No. DE-AC02-98CH10886. X6A is funded by NIH/NIGMS under agreement Y1 GM-0080-03.
References
- Bradford, M. M. (1976). Anal. Biochem.72, 248–254. [DOI] [PubMed]
- Brünger, A. T. (1992). Nature (London), 355, 472–475. [DOI] [PubMed]
- Carpino, N., Chen, Y., Nassar, N. & Oh, H.-W. (2009). Mol. Immunol.46, 3224–3231. [DOI] [PMC free article] [PubMed]
- Chen, Y., Jakoncic, J., Carpino, N. & Nassar, N. (2009). Biochemistry, 48, 1681–1690. [DOI] [PMC free article] [PubMed]
- Chen, Y., Jakoncic, J., Parker, K. A., Carpino, N. & Nassar, N. (2009). Biochemistry, 48, 8129–8135. [DOI] [PMC free article] [PubMed]
- Chen, Y., Jakoncic, J., Wang, J., Zheng, X., Carpino, N. & Nassar, N. (2008). Biochemistry, 47, 12135–12145. [DOI] [PMC free article] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Jedrzejas, M. J. (2000). Prog. Biophys. Mol. Biol.73, 263–287. [DOI] [PubMed]
- Jia, Z., Barford, D., Flint, A. J. & Tonks, N. K. (1995). Science, 268, 1754–1758. [DOI] [PubMed]
- Kleinman, H., Ford, B., Keller, J., Carpino, N. & Nassar, N. (2006). Acta Cryst. F62, 218–220. [DOI] [PMC free article] [PubMed]
- Laskowski, R. A., Moss, D. S. & Thornton, J. M. (1993). J. Mol. Biol.231, 1049–1067. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Mikhailik, A., Ford, B., Keller, J., Chen, Y., Nassar, N. & Carpino, N. (2007). Mol. Cell, 27, 486–497. [DOI] [PMC free article] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Otwinowski, Z. & Minor, D. (1997). Methods Enzymol.276, 307–326. [DOI] [PubMed]
- Raguz, J., Wagner, S., Dikic, I. & Hoeller, D. (2007). FEBS Lett.581, 4767–4772. [DOI] [PubMed]
- Rigden, D. J. (2008). Biochem. J.409, 333–348. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
PDB reference: phosphatase domain of Sts-1, 3mbk