Deuterated type III antifreeze protein specifically hydrogen reverse-labelled in the methyl groups of leucine and valine residues has been expressed, purified and crystallized. Preliminary neutron data collection showed diffraction to 1.80 Å resolution from a 0.23 mm3 crystal.
Keywords: ocean pout type III antifreeze protein, neutron diffraction, reverse labelling, deuteration
Abstract
Antifreeze proteins (AFPs) are found in different species from polar, alpine and subarctic regions, where they serve to inhibit ice-crystal growth by adsorption to ice surfaces. Recombinant North Atlantic ocean pout (Macrozoarces americanus) AFP has been used as a model protein to develop protocols for amino-acid-specific hydrogen reverse-labelling of methyl groups in leucine and valine residues using Escherichia coli high-density cell cultures supplemented with the amino-acid precursor α-ketoisovalerate. Here, the successful methyl protonation (methyl reverse-labelling) of leucine and valine residues in AFP is reported. Methyl-protonated AFP was expressed in inclusion bodies, refolded in deuterated buffer and purified by cation-exchange chromatography. Crystals were grown in D2O buffer by the sitting-drop method. Preliminary neutron Laue diffraction at 293 K using LADI-III at ILL showed in a few 24 h exposures a very low background and clear small spots up to a resolution of 1.80 Å from a crystal of dimensions 1.60 × 0.38 × 0.38 mm corresponding to a volume of 0.23 mm3.
1. Introduction
The methyl-protonation approach described in this paper is part of a Human Frontier Science Program (HFSP) funded project aiming to develop direct methods for phase determination in neutron protein crystallography (Hauptman & Langs, 2003 ▶; Xu et al., 2005 ▶). In X-ray crystallography, the computational process for the determination of the shape and atomic arrangement of the molecules requires the measurement of differences in diffraction patterns arising either from the introduction of heavy atoms into the native protein crystals (multiple isomorphous replacement method) or from the presence of atoms with strong anomalous dispersion (multi-wavelength or single-wavelength anomalous dispersion methods). Suitable derivatives can be prepared by soaking native crystals in solutions containing atoms of heavy elements such as mercury (Sun & Radaev, 2002 ▶) or by using genetic engineering to introduce Se atoms into the protein molecule (Usón & Sheldrick, 1999 ▶). The goal of the HFSP project is to develop new methods that use neutron radiation. An important difference between X-ray and neutron diffraction involves the scattering from H atoms (Blakeley, 2009 ▶). The two hydrogen isotopes protium and deuterium scatter X-rays the same way, but neutrons are scattered differently. This difference can be used as the basis for making ideal derivatives that provide the information needed to solve protein structures. Our project aims to design practical methods for exploiting the differential neutron scattering of the hydrogen isotopes and to develop technology for deuterating selected parts of protein molecules. We have used North Atlantic ocean pout (Macrozoarces americanus) antifreeze protein (AFP) as a model protein for this purpose. Perdeuterated AFP has recently been crystallized in the ILL–EMBL Deuteration Laboratory (Petit-Haertlein et al., 2009 ▶) and a neutron crystallographic study is in progress.
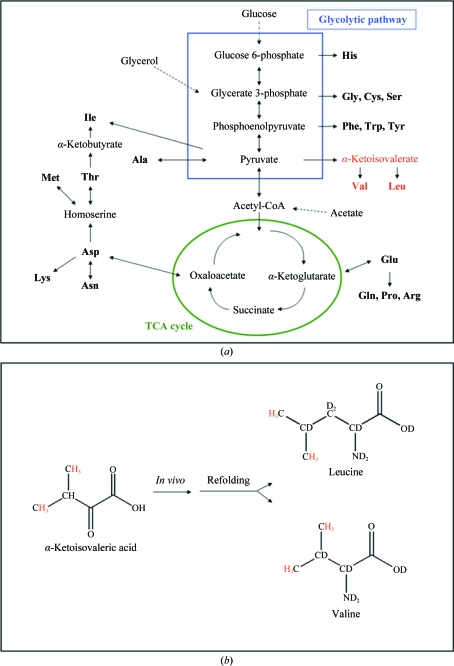
We used type III AFP as a model protein to develop protocols for the incorporation of methyl-protonated valine and leucine residues into perdeuterated proteins for the above-described applications in neutron crystallography. Similar isotopic labelling schemes are available for NMR spectroscopy of high-molecular-weight proteins (Tugarinov et al., 2006 ▶). These labelling strategies are based on the use of selectively protonated α-keto acid precursors in deuterated minimal medium, enabling specific incorporation of 13CH3 groups in selected methyl sites (for metabolic pathways and methyl protonation, see Fig. 1 ▶). Using this approach, the level of incorporation is very high without detectable scrambling of protons. Addition of α-ketoisovalerate at a concentration higher than 100 mg per litre of culture has been shown to give near-complete labelling of leucine and valine methyl groups (Goto et al., 1999 ▶). An alternative approach for the incorporation of protonated amino acids into perdeuterated proteins relies on the availability of suitable auxotrophic host strains to obtain high levels of incorporation of the protonated label and the addition of deuterated amino acids to reduce isotope scrambling (Waugh, 1996 ▶).
Figure 1.
Amino-acid-specific methyl protonation. (a) Schematic illustration of the major metabolic pathways involved in amino-acid biosynthesis in E. coli. (b) Residue-specific methyl protonation of valine and leucine via the precursor α-ketoisovalerate.
Recently, Weiss and coworkers reported methyl protonation to introduce (1H-δ methyl)-leucine and (1H-γ methyl)-valine into deuterated rubredoxin from Pyrococcus furiosus (Weiss et al., 2008 ▶). According to the results of several trials, direct phasing methods did not work for the rubredoxin data sets. The most likely reason for this failure was the poor completeness of the data (David Langs, private communication). Following this experience, special care was taken with the AFP data sets, which are more complete. Direct phasing methods are currently being tried on these data sets. In contrast to the labelling approach of Weiss and coworkers, which was carried out in flask cultures, the labelling in our case was carried out in high-cell-density cultures. Our protocol for the labelling approach has been validated by mass spectrometry and neutron crystallography.
2. Experimental methods
2.1. Expression and purification of deuterated AFP with methyl-protonated valine and leucine residues (AFP D VMP + LMP)
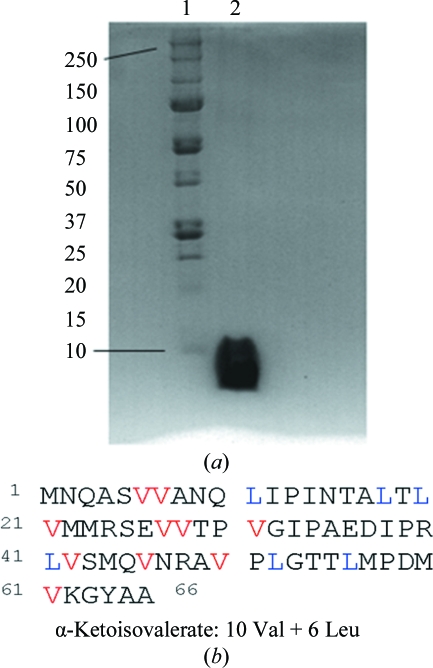
The synthetic gene of the type III antifreeze protein AFP (isoform HPLC12) from the North Atlantic ocean pout M. americanus was subcloned into a pET-28a (Novagen) vector that confers kanamycin resistance. The sequence adjusted to the protein sequence of PDB entry 1hg7 (Antson et al., 2001 ▶) is shown in Fig. 2 ▶(b). Methyl-protonated valine and leucine residues are indicated in colour.
Figure 2.
Expression and labelling of AFP. (a) 12% SDS–Tris-tricine polyacrylamide gel electrophoresis. The standard protein bands (kDa) of Precision Plus pre-stained molecular-weight standards (Bio-Rad) are indicated in lane 1. 25 µg purified AFP D VMP + LMP is shown in lane 2. (b) Primary sequence of AFP. Methyl-protonated valine residues are shown in red and leucine residues in blue.
Recombinant AFP D VMP + LMP was obtained by expression in Escherichia coli BL21 (DE3) cells at the ILL–EMBL Deuteration Facility in Grenoble, France, as previously described for perdeuterated AFP (Petit-Haertlein et al., 2009 ▶), with some modifications mainly concerning the addition of the precursor α-ketoisovalerate and a shortening of the induction time to avoid nonspecific labelling (metabolic isotopic H-scrambling). Briefly, cells were grown in deuterated minimal medium containing 25 mg l−1 kanamycin. Perdeuterated d 8-glycerol (Euriso-top, France) was used as a carbon source. Mineral salts were dried out in a rotary evaporator (Heidolph) at 333 K and labile protons were exchanged for deuterons by dissolving in a minimal volume of D2O and re-drying. Adaptation of BL21 (DE3) cells to deuterated minimal medium was achieved by an adaptation process on minimal medium agar plates (Artero et al., 2005 ▶). 1.5 l deuterated medium was inoculated with a 100 ml preculture of adapted cells in a 3 l fermenter (Labfors, Infors). During the batch and fed-batch phases, the pH was adjusted to 6.8 by addition of NaOD (Euriso-top, France) and the temperature was adjusted to 303 K. The fed-batch phase was initiated when the optical density at 600 nm (OD600) reached a value of 3.7. d 8-Glycerol was added to the culture to keep the growth rate stable during fermentation. 1 h before induction, 1.2 g l−1 α-ketoisovalerate (Sigma–Aldrich) was added to the fermenter culture. The preparation of (3-2H)-ketoisovalerate by alkaline treatment in D2O buffer (Goto et al., 1999 ▶) could be skipped: AFP was initially expressed in its deuterated form with valine and leucine residues hydrogen reverse-labelled in their methyl and Cβ (valine) and Cγ (leucine) positions. AFP refolding in an alkaline deuterated buffer, in which only the methyl H atoms of valines and leucines resist deuterium exchange, generated the H-methyl reverse-labelled AFP. When the OD600 reached a value of 13.7, antifreeze overexpression was induced by the addition of 1 mM IPTG and incubation was continued for only 6 h to avoid isotopic H-scrambling. Cells (56 g wet weight) were then harvested and stored at 193 K.
10 g frozen cell paste was resuspended in 10 ml 8 M guanidine hydrochloride in D2O, stirred on ice for 1 h and sonicated to increase the yield of soluble protein. After centrifugation at 19 000 rev min−1 for 20 min at 277 K, the supernatant containing the soluble proteins was added dropwise to 50 ml ice-cold refolding buffer (50 mM K2PO4, 100 mM NaCl in D2O pD 11.1). The pD value of the deuterated buffers was measured with an H2O-calibrated pH meter. The conversion of pH to pD was accomplished by adding a constant of 0.4 (Kręzel & Bal, 2004 ▶). The AFP solution was then dialysed against 300 ml 50 mM sodium acetate pD 4.1. Insoluble material was removed by centrifugation and the supernatant was filtered through a 0.2 µM membrane. The pD of the protein solution was lowered to 3.1 using citric acid and the antifreeze protein was purified on a 5 ml SP Sepharose column (GE Healthcare). This step removes contaminating proteins; AFP D VMP + LMP is recovered in the flowthrough. Protein purity was assessed by SDS–PAGE (see Fig. 2 ▶ a). AFP D VMP + LMP was concentrated on 5K Amicon Ultra centrifugal filter units (Millipore) to a final concentration of 10 mg ml−1 in 50 mM sodium acetate pD 5.2.
2.2. Determination of deuteration level
The molecular weight of AFP D VMP + LMP and its deuteration level were determined by MALDI mass spectrometry. Laser desorption/ionization mass-spectrometric analysis was performed using a Perseptive Biosystems (Framingham, Massachusetts, USA) Voyager Elite XL time-of-flight mass spectrometer operating with a pulsed nitrogen laser at 337 nm. External calibration was performed with a calibration mixture of three standards from the Applied Biosystems Corporation with m/z values of 5734.59, 11 674.48 and 16 952.56. Samples of concentrated native AFP diluted in 0.1% trifluoroacetic acid (Sigma–Aldrich), with a final concentration of about 10 mM, were mixed with an equal volume of a saturated solution of sinapinic acid (Fluka) prepared in a 50%(v/v) solution of acetonitrile/aqueous 0.3% trifluoroacetic acid directly on the stainless-steel sample plate and air-dried prior to analysis. The values expressed are average masses and correspond to the [M + H]+ ion.
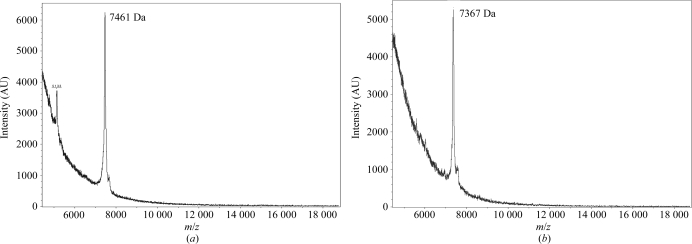
The efficiency of methyl-protonation of valine and leucine residues in AFP was determined by comparing the mass of perdeuterated AFP with the mass of the methyl-protonated variant (AFP D VMP + LMP). For mass-spectrometric analysis both the perdeuterated and the methyl-protonated AFPs were refolded and purified under the same conditions. The measured mass takes the 115 exchangeable H atoms into account since for technical reasons the mass of AFP was determined in hydrogenated solutions. The primary sequence of AFP is shown in Fig. 2 ▶(b), with its ten valine and six leucine residues indicated in colour. Complete methyl-protonation of these 16 residues should lead to a mass decrease of 96 Da. Fig. 3 ▶ shows the mass spectra of both AFP variants, with a measured value of 7461 Da (average of ten measurements) for AFP D and 7367 Da (average of ten measurements) for AFP D VMP + LMP. The measured mass difference (94 Da) is very close to the expected difference.
Figure 3.
MALDI mass spectrum of AFP. (a) MALDI mass spectrum of AFP D prepared in H2O. (b) MALDI mass spectra of AFP D VMP + LMP prepared in H2O. The mass difference (94 Da) is very close to the expected value of 96 for D–H exchange on 32 methyl groups of valine or leucine residues.
2.3. Crystallization
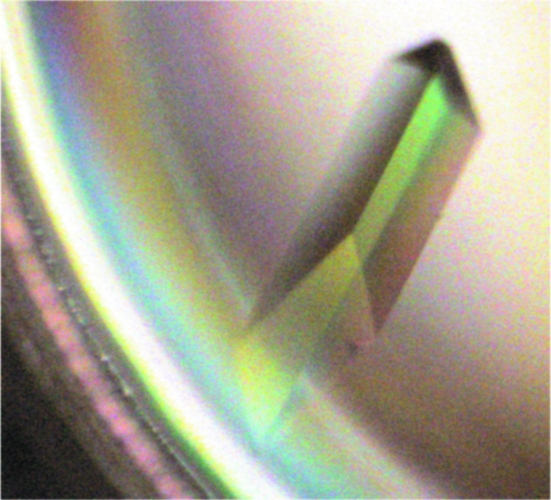
Crystallization conditions were adapted from the crystallization of perdeuterated AFP (Petit-Haertlein et al., 2009 ▶). AFP D VMP + LMP was concentrated to 10 mg ml−1 in 50 mM sodium acetate pD 5.2. Crystallization was performed by the sitting-drop vapour-diffusion method at 285 K. Drops containing 20 µl protein solution and 30 µl precipitant buffer (2.1 M ammonium sulfate, 9% d 8-glycerol) were equilibrated against 1 ml precipitant buffer in the reservoir. After 35 d, a crystal with a volume of 0.042 mm3 (1.4 × 0.2 × 0.15 mm) as shown in Fig. 4 ▶ was fished out, rinsed in 2 µl of a pre-equilibrated drop and then placed in a 48 µl sitting drop at 285 K. The final volume of 0.23 mm3 (1.60 × 0.38 × 0.38 mm) was reached after a few weeks.
Figure 4.
Picture of an AFP D VMP + LMP crystal taken under polarized light. The protein crystallizes in space group P212121, with unit-cell parameters a = 32.7, b = 39.1, c = 46.5 Å at 293 K.
2.4. Neutron quasi-Laue data collection
Neutron quasi-Laue data were collected at 293 K on the LADI-III beamline installed on cold neutron guide H142 at the Institut Laue–Langevin. Using the crystal of AFP D VMP + LMP with a volume of 0.23 mm3, diffraction data were collected to 1.8 Å resolution. As is typical for a Laue experiment, the crystal was held stationary at a different ϕ setting for each exposure, with an angular interval between settings of 7°. Initially, 13 contiguous images were collected using an exposure time of 24 h per image in order to collect the high-resolution data (see Fig. 5 ▶), followed by a low-resolution pass using an exposure time of 2 h per image. Next, the crystal orientation was modified and both the high- and low-resolution passes were repeated. Finally, the crystal orientation was modified again and a further eight images were collected, such that the complete data set comprised 60 images with an average exposure time of 710 min per image. The neutron Laue data images were processed using the Daresbury Laboratory LAUE suite program LAUEGEN, which was modified to account for the cylindrical geometry of the detector (Campbell et al., 1998 ▶). The program LSCALE (Arzt et al., 1999 ▶) was used to determine the wavelength-normalization curve using the intensities of symmetry-equivalent reflections measured at different wavelengths and to apply wavelength-normalization calculations to the observed data. The data were then scaled and merged in SCALA (Collaborative Computational Project, Number 4, 1994 ▶). Data-reduction statistics are summarized in Table 1 ▶.
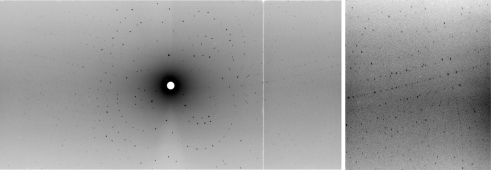
Figure 5.
Neutron Laue diffraction pattern. Left: a neutron Laue diffraction pattern from the 0.23 mm3 crystal of AFP D VMP + LMP collected on the LADI-III beamline at the Institut Laue–Langevin. Right: close up of a high-resolution region.
Table 1. Neutron quasi-Laue data-collection statistics for the AFP D VMP + LMP crystal with a volume of 0.23 mm3 .
Values in parentheses are for the highest resolution shell.
| Source and beamline | Institut Laue–Langevin, LADI-III |
| Wavelength (Å) | 3.25–4.1 |
| No. of images | 60 |
| Oscillation angle (°) | 7 |
| Average exposure time per image (min) | 710 |
| Space group | P212121 |
| Unit-cell parameters (Å) | a = 32.7, b = 39.1, c = 46.5 |
| Resolution range (Å) | 46.52–1.80 (1.90–1.80) |
| No. of observations | 57643 (2085) |
| No. of unique reflections | 5194 (591) |
| Completeness (%) | 89.3 (71.4) |
| Rmerge† | 0.136 (0.169) |
| Rp.i.m.‡ | 0.035 (0.085) |
| Mean I/σ(I) | 14.0 (6.6) |
| Multiplicity | 11.1 (3.5) |
R
merge = 
 , where Ii(hkl) is the intensity of the ith observation of reflection hkl.
, where Ii(hkl) is the intensity of the ith observation of reflection hkl.
The precision-indicating merging statistic R
p.i.m. = 
 .
.
3. Conclusions
A protocol for the incorporation of methyl-protonated valine and leucine residues into perdeuterated proteins in high-cell-density cultures for neutron crystallography applications has been developed. Similar isotopic labelling schemes are available to produce samples for NMR spectroscopy of high-molecular-weight proteins in flask cultures (Tugarinov et al., 2006 ▶). The labelling strategies are based on the use of selectively protonated α-keto acid precursors in deuterated minimal medium, enabling specific incorporation of CH3 groups in selected methyl sites (valine and leucine). Using this approach, the level of incorporation is very high without detectable scrambling of protons. Isotopically labelled α-keto acids are commercially available. Sodium salts of α-ketoisovaleric acids are available in a protonated form at position 3 and can be quantitatively exchanged to 3-2H at high pH in D2O. Alternatively, position 3 can be quantitatively exchanged to 3-2H after incorporation into leucine and valine residues if the recombinant protein resists high alkaline pD solutions (AFP refolding buffer). For high-cell-density cultures the α-keto acid concentration was increased by a factor of ten. Mass spectrometry allowed validation of our labelling approach. Direct evidence of incorporation of methyl-protonated valine and leucine residues has been obtained by neutron crystallography on LADI-III with a ‘radically small’ crystal volume of 0.23 mm3.
Acknowledgments
We thank Bernard Dublet and Eric Forest (Institut de Biologie Structurale, Grenoble, France) for the mass-spectrometric analysis, Susana Teixeira and Jean-Baptiste Artero from the ILL for technical advice and François Dauvergne for technical support on LADI-III. This work was supported by Human Frontier Science Program grant RGP0021/2006-C and benefitted from the activities of the DLAB consortium funded by the European Union under contract HPRI-2001-50065 and from United Kingdom Engineering and Physical Sciences Research Council (EPSRC)-funded activity within the ILL–EMBL Deuteration Laboratory under grant GR/R99393/01. EIH is a member of the ‘Carrera del Investigador’ Conicet-Argentina. This work was supported by the Centre National de la Recherche Scientifique (CNRS), by the Institut National de la Santé et de la Recherche Médicale and the Hôpital Universitaire de Strasbourg (HUS).
References
- Antson, A. A., Smith, D. J., Roper, D. I., Lewis, S., Caves, L. S. D., Verma, C. S., Buckley, S. L., Lillford, P. J. & Hubbard, R. E. (2001). J. Mol. Biol.305, 875–889. [DOI] [PubMed]
- Artero, J.-B., Härtlein, M., McSweeney, S. & Timmins, P. (2005). Acta Cryst. D61, 1541–1549. [DOI] [PubMed]
- Arzt, S., Campbell, J. W., Harding, M. M., Hao, Q. & Helliwell, J. R. (1999). J. Appl. Cryst.32, 554–562.
- Blakeley M. P. (2009). Crystallogr. Rev.15, 157–218.
- Campbell, J. W., Hao, Q., Harding, M. M., Nguti, N. D. & Wilkinson, C. (1998). J. Appl. Cryst.31, 496–502.
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Goto, N. K., Gardner, K. H., Mueller, G. A., Willis, R. C. & Kay, L. E. (1999). J. Biomol. NMR, 13, 369–374. [DOI] [PubMed]
- Hauptman, H. A. & Langs, D. A. (2003). Acta Cryst. A59, 250–254. [DOI] [PubMed]
- Kręzel, A. & Bal, W. (2004). J. Inorg. Biochem.98, 161–166. [DOI] [PubMed]
- Petit-Haertlein, I., Blakeley, M. P., Howard, E., Hazemann, I., Mitschler, A., Haertlein, M. & Podjarny, A. (2009). Acta Cryst. F65, 406–409. [DOI] [PMC free article] [PubMed]
- Sun, P. D. & Radaev, S. (2002). Acta Cryst. D58, 1099–1103. [DOI] [PubMed]
- Tugarinov, V., Kanelis, V. & Kay, L. E. (2006). Nature Protoc.1, 749–754. [DOI] [PubMed]
- Usón, I. & Sheldrick, G. M. (1999). Curr. Opin. Struct. Biol.9, 643–648. [DOI] [PubMed]
- Waugh, D. S. (1996). J. Biomol. NMR, 8, 184–192. [DOI] [PubMed]
- Weiss, K. L., Meilleur, F., Blakeley, M. P. & Myles, D. A. A. (2008). Acta Cryst. F64, 537–540. [DOI] [PMC free article] [PubMed]
- Xu, H., Hauptman, H. A. & Langs, D. A. (2005). Hydrogen and Hydration-Sensitive Structural Biology, pp. 197–205. Tokyo: KubaPro Co. Ltd.