The glycosylated fungal haem-thiolate peroxygenase AaPII was crystallized in three different forms from a charge-heterogeneous protein sample. Structure solution of the 45 kDa protein was achieved with SAD utilizing the haem iron and the structure was determined at 2.2 Å resolution.
Keywords: aromatic peroxygenases, haloperoxidases, haem thiolates, fungal enzymes, glycoproteins, charge heterogeneity, anomalous scattering
Abstract
Some litter-decaying fungi secrete haem-thiolate peroxygenases that oxidize numerous organic compounds and therefore have a high potential for applications such as the detoxification of recalcitrant organic waste and chemical synthesis. Like P450 enzymes, they transfer oxygen functionalities to aromatic and aliphatic substrates. However, in contrast to this class of enzymes, they only require H2O2 for activity. Furthermore, they exhibit halogenation activity, as in the well characterized fungal chloroperoxidase, and display ether-cleavage activity. The major form of a highly glycosylated peroxygenase was produced from Agrocybe aegerita culture media, purified to apparent SDS homogeneity and crystallized under three different pH conditions. One crystal form containing two molecules per asymmetric unit was solved at 2.2 Å resolution by SAD using the anomalous signal of the haem iron. Subsequently, two other crystal forms with four molecules per asymmetric unit were determined at 2.3 and 2.6 Å resolution by molecular replacement.
1. Introduction
Aromatic peroxygenases (APOs) are extracellular haem-thiolate proteins that are produced by soil-litter and wood-colonizing fungi (Pecyna et al., 2009 ▶). The best-studied APO proteins are from agaric basidiomycetes (colloquially called mushrooms) such as Agrocybe aegerita and Coprinellus radians (Hofrichter & Ullrich, 2006 ▶; Anh et al., 2007 ▶). The actual function of APOs is not yet fully understood, but owing to their enormous versatility and secretion into the fungal micro-environment (as ‘digestive’ enzymes), APOs may be involved in the degradation and detoxification of recalcitrant organic materials. Among other functions, APOs are capable of transferring oxygen functionalities to aromatic substrates (e.g. naphthalene and toluene) and in this way resemble P450 enzymes (Ullrich & Hofrichter, 2005 ▶; Anh et al., 2007 ▶). However, unlike the latter, which are intracellular monooxygenases that require NAD(P)H as an electron donor as well as specific electron-transfer proteins (flavin reductase), APOs work outside the fungal cells (hyphae) and only require H2O2 for function (Ullrich & Hofrichter, 2007 ▶). In addition to aromatic oxygenation, APOs catalyze the peroxide-driven hydroxylation of alkyl side chains, the cleavage of ethers and the N- and S-oxidation of heterocycles (Kluge et al., 2009 ▶; Ullrich et al., 2008 ▶; Aranda et al., 2009 ▶; Kinne et al., 2009 ▶). The enzyme also shows halogenating activity; however, this is less pronounced than in fungal chloroperoxidase (CPO) and is restricted to the nonspecific bromination of activated substrates (Ullrich & Hofrichter, 2005 ▶). APO proteins are usually secreted as several isozymes and isoforms which differ with respect to their isoelectric points (Ullrich et al., 2009 ▶). They can be produced, for example using wild-type strains of A. aegerita, in stirred-tank bioreactors in yields of up to 30 mg APO protein per litre (Ullrich et al., 2004 ▶). The major form of APO from A. aegerita, AaPII (A. aegerita peroxygenase isozyme II), consists of 328 amino acids and shares 29% sequence identity with CPO in the N-terminal half, while there is no homology for the remaining peptide. No sequence homology to any other peroxidase or P450 enzyme could be found. AaPII is highly glycosylated, with about 20% carbohydrate content, and has six potential N-glycosylation sites (Pecyna et al., 2009 ▶). Here, we report the crystallization and structure determination of AaPII. Using a mixture of two AaPII isoforms, several crystallization conditions within the pH range 4.6–8.5 yielded suitable single crystals belonging to orthorhombic space groups. The crystals contained two or four molecules per asymmetric unit and diffracted to about 2 Å resolution. Diffraction data were collected at the ESRF synchrotron, including anomalous data at the Fe edge. The structure of the crystals obtained at pH 8.5, which contained two molecules per asymmetric unit, was solved by SAD using the anomalous signal of the haem iron and preliminarily refined at 2.2 Å resolution. Subsequently, the structures of two other crystal forms obtained at pH 4.6 and 5.6, both with four molecules in the asymmetric unit, were determined using the molecular-replacement (MR) technique at resolutions of 2.3 and 2.6 Å, respectively.
2. Materials and methods
2.1. Culture conditions, protein purification and enzyme assay
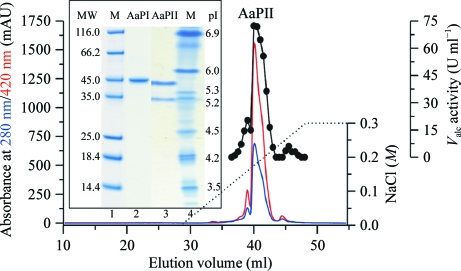
Pre-cultivation of A. aegerita (deposited in the German collection of microorganisms and cell cultures; DSM No. 22459) was performed in 3%(w/v) soybean-flour medium (Hensel Voll-Soja, Schoenenberger GmbH, Magstadt, Germany) in agitated Erlenmeyer flasks at 297 K. Enzyme production was carried out in a 5 l stirred-tank bioreactor (Biostat B, Braun Biotech International GmbH, Melsungen, Germany) containing the same medium (Ullrich et al., 2004 ▶). The culture liquid was harvested at the maximum activity level of about 1500 U l−1. The filtrated enzyme-containing liquid was concentrated 60-fold by two steps of ultrafiltration using a tangential flow cassette system (Omega Minisette, 10 kDa cutoff, Pall Corporation, Hauppauge, New York, USA) and a 150 ml stirred cell system (10 kDa cutoff polyethersulfone membrane, Pall Life Sciences, Dreieich, Germany). The crude AaP preparation was further purified by three steps of fast protein liquid chromatography (FPLC) using an ÄKTA system (GE Healthcare Europe GmbH, Freiburg, Germany) equipped with SP Sepharose, Mono Q and Mono S columns. Separation was carried out using sodium acetate (10 mM, pH 4.75–6.5) as the solvent and increasing sodium chloride gradients (0–0.3 or 0–1.0 M) for protein elution (Fig. 1 ▶). Elution was monitored at 420 nm (haem) and 280 nm (total protein). Fractions with AaP activity were pooled, concentrated, dialyzed against 10 mM sodium acetate buffer and stored at 277 K. The AaPII fraction is one of the most abundant isoenzymes produced by the fungus. After these three steps AaPII was purified to a homogenous molecular weight but still split into two different pI values of 5.6 and 5.2 (Fig. 1 ▶, inset). Peroxygenase activity was measured by following the oxidation of veratryl alcohol to veratraldehyde (∊310 = 9.3 mM −1 cm−1) in 50 mM sodium citrate–phosphate buffer pH 7 (Ullrich et al., 2004 ▶). A subsequent improvement of the FPLC techniques including chromatofocusing led to fractions with a homogenous pI (Ullrich et al., 2009 ▶).
Figure 1.
FPLC elution profile of the last step of purification on the strong cation exchanger Mono S. Inset: SDS–PAGE and isoelectric focusing PAGE of the pooled AaP fraction II. Lane 1, SDS–PAGE with molecular-weight markers (kDa); lane 2, SDS–PAGE of purified AaPII; lane 3, IEF gel of purified AaPII showing two isoforms; lane 4, IEF gel of pI markers.
2.2. SDS–PAGE, analytical IEF and protein determination
To determine the purity of the protein, AaP fraction II was electrophoretically analyzed using a Mini-Cell electrophoresis system (XCell Sure Lock, Invitrogen, Karlsruhe, Germany) and 10% NuPAGE Novex Bis-Tris precast gels as well as Novex pre-cast IEF gels (pH 3–7, Invitrogen). Electrophoretic separation was carried out according to the manufacturer’s protocol. Protein bands were visualized with the Colloidal Blue Staining Kit from Invitrogen. The protein concentration was determined by the method of Bradford using the Roti-Nanoquant Protein Assay Kit (Roth, Karlsruhe, Germany) with bovine serum albumin H4 (Roth) as the standard.
2.3. Crystallization
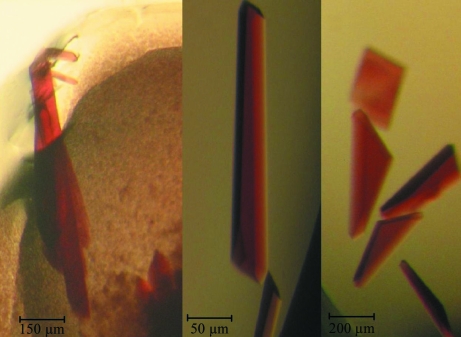
Purified AaPII, still containing the two isoforms with pI 5.6 and 5.2, was used at concentrations of 5, 10 and 20 mg ml−1 in 10 mM sodium acetate pH 6.0 for initial crystallization screens applying the Crystal Screen 1 (Hampton Research, USA) and Screen 6 (Jena Bioscience, Germany) kits. The hanging-drop vapour-diffusion technique was utilized in 24-well plates by mixing 2 µl protein solution and 2 µl reservoir solution. The drops were allowed to equilibrate against 1 ml reservoir solution at 292 K. After refinement of initial crystallization conditions, larger drops of 10 µl were set up with the hanging-drop technique and 20 µl drops were set up with the sitting-drop technique. Using ammonium sulfate as the precipitant, a total of three suitable crystal forms were obtained at different pH values (Fig. 2 ▶) and were termed AaPIIpH46, AaPIIpH56 and AaPIIpH85 in the following.
Figure 2.
Crystals of (a) AaPIIpH85, (b) AaPIIpH56 and (c) AaPIIpH46. The largest crystal of AaPIIpH85 has a length of 1.2 mm.
2.4. Data collection and processing
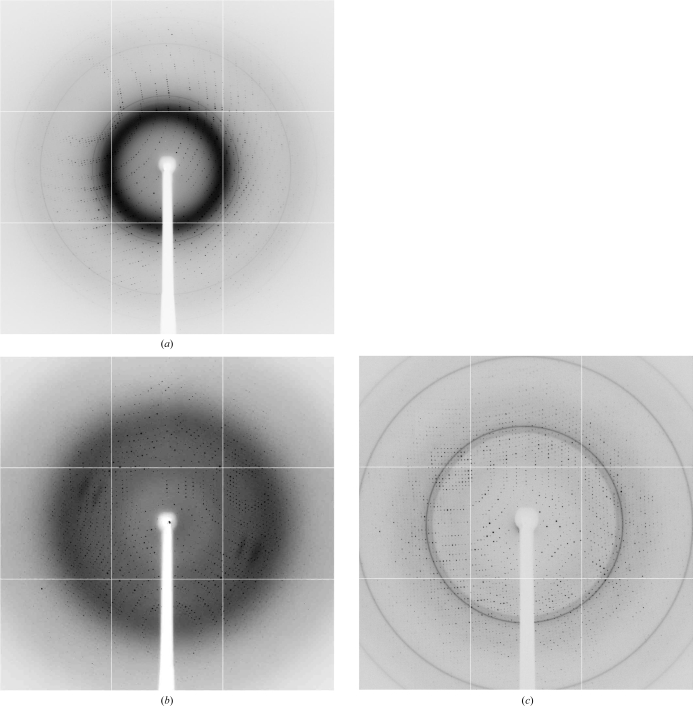
Prior to data collection at cryotemperatures, AaPIIpH85 and AaPIIpH46 were treated with Paratone-N oil and PFO (both from Hampton Research, USA), respectively, and flash-cooled in liquid nitrogen. AaPIIpH56 did not require additional cryoprotectant as the crystallization solution contained 20% glycerol (Table 1 ▶). Native as well as MAD X-ray diffraction data were collected on the macromolecular crystallographic beamline ID23-1 at the ESRF (Grenoble, France). The beamline was equipped with a MiniKappa goniometer, a sample changer and a remote semi-automatic sample-alignment device. Diffraction data were recorded on a Q315R ADSC CCD detector and processed and scaled with the XDS program package (Kabsch, 2010a ▶,b ▶). All AaPII crystal forms initially diffracted to 1.8–2.3 Å resolution (Fig. 3 ▶), but rapidly lost resolution during data collection. Details of the data-collection and processing statistics are given in Tables 2 ▶ and 3 ▶.
Table 1. Conditions of AaPII crystallization.
AaPIIpH85, AaPIIpH56 and AaPIIpH46 are the crystal forms obtained at pH 8.5, pH 5.6 and pH 4.6, respectively.
| Crystal | AaPIIpH85 | AaPIIpH56 | AaPIIpH46 |
|---|---|---|---|
| Precipitant | 2.0 M ammonium sulfate | 2.4 M ammonium sulfate | 2.0 M ammonium sulfate |
| Buffer | 100 mM Tris–HCl pH 8.5 | 100 mM sodium citrate pH 5.6 | 200 mM sodium acetate pH 4.6 |
| Salt/additive | None | 20%(v/v) glycerol | None |
| Protein concentration (mg ml−1) | 10 | 10 | 5 |
| Cryoprotectant | Paratone-N | None | PFO |
Figure 3.
Diffraction patterns of (a) AaPIIpH85, (b) AaPIIpH56 and (c) AaPIIpH46 crystals. The edges of the frames correspond to 1.8, 2.2 and 2.3 Å, respectively.
Table 2. Crystal characteristics, native data-collection and processing statistics of AaPII crystals.
Values in parentheses refer to the highest resolution shell.
| Crystal | AaPIIpH85 | AaPIIpH56 | AaPIIpH46 |
|---|---|---|---|
| Space group | P212121 | P21212 | P21212 |
| Unit-cell parameters (Å) | |||
| a | 77.29 | 113.17 | 112.75 |
| b | 95.03 | 144.65 | 144.88 |
| c | 127.20 | 135.13 | 134.46 |
| No. of molecules per asymmetric unit | 2 | 4 | 4 |
| X-ray source | ID23 at ESRF | ID23 at ESRF | ID23 at ESRF |
| Wavelength (Å) | 0.9724 | 1.0720 | 0.9760 |
| Temperature (K) | 100 | 100 | 100 |
| No. of crystals | 1 | 1 | 1 |
| Resolution (Å) | 47.51–2.19 (2.25–2.19) | 49.09–2.60 (2.66–2.60) | 48.94–2.31 (2.37–2.31) |
| Total No. of reflections | 345058 (17639) | 436897 (28588) | 465760 (32925) |
| No. of unique reflections | 47667 (3057) | 68909 (4793) | 97006 (6821) |
| Completeness (%) | 98.0 (86.7) | 99.5 (95.1) | 99.4 (95.7) |
| I/σ(I) | 16.9 (6.0) | 13.8 (4.8) | 15.4 (5.5) |
| Rmerge† (%) | 7.4 (39.8) | 11.5 (45.1) | 6.5 (22.0) |
As defined by Diederichs & Karplus (1997 ▶).
Table 3. Data-collection, processing and phasing statistics of AaPIIpH85 data used for SAD structure solution.
The data set was produced from the same crystal as in Table 2 ▶. Values in parentheses are for the highest resolution shell.
| Crystal | AaPIIpH85 |
|---|---|
| Unit-cell parameters (Å) | |
| a | 77.30 |
| b | 95.15 |
| c | 127.20 |
| Oscillation range (°) | 0.25 |
| No. of frames | 600 |
| Wavelength (Å) | 1.7385 |
| Resolution (Å) | 47.51–3.0 (3.18–3.0) |
| Total No. of reflections (Friedels counted separately) | 108188 (12585) |
| No. of unique reflections (Friedels counted separately) | 35924 (5567) |
| Completeness (%) | 98.5 (94.6) |
| I/σ(I) | 21.9 (10.81) |
| Rmeas† (%) | 5.1 (9.4) |
| Internal correlation coefficient of anomalous signal‡ (%) | 73 (12) |
| Phasing power§ (ano) | 0.994 |
| FOM§ (centric/acentric) | 0.052/0.312 |
| FOM after density modification to 2.52 Å resolution | 0.881 |
2.5. Molecular-replacement experiments
Prior to MAD data collection with synchrotron radiation, native data were collected using a rotating-anode generator and structure solution was attempted by molecular replacement using the known structure of CPO as the search model (PDB code 1cpo; Sundaramoorthy et al., 1995 ▶). From the Matthews coefficient, it was assumed that AaPIIpH46 and AaPIIpH56 contained four to six molecules per asymmetric unit, while for AaPIIpH85 crystals two molecules would be the most likely case. Patterson maps calculated by the molecular-replacement program MOLREP (Vagin & Teplyakov, 1997 ▶) from the CCP4 suite of programs (Collaborative Computational Project, Number 4, 1994 ▶) revealed pseudo-translation of the AaPIIpH46 and AaPIIpH56 crystals. Moreover, it became evident from self-rotation functions (MOLREP) that in these crystal forms twofold noncrystallographic symmetry axes are almost parallel to a crystallographic symmetry axis or are close to a mirror plane of the rotation function, complicating the structure solution even further. Therefore, we directed our efforts towards the AaPIIpH85 crystals, which contain only two molecules per asymmetric unit. Numerous runs with the programs MOLREP and Phaser (McCoy et al., 2005 ▶) from the CCP4 package were calculated by systematically changing various parameters such as resolution and radius of integration and using several truncated and/or modified fragments of the search molecule. Nevertheless, all those attempts failed.
2.6. Structure determination by SAD
The crystals of AaPIIpH85 were chosen for structure determination with the SAD technique since they (i) diffract to the highest resolution, (ii) contain only two molecules per asymmetric unit and (iii) do not exhibit pseudo-symmetry.
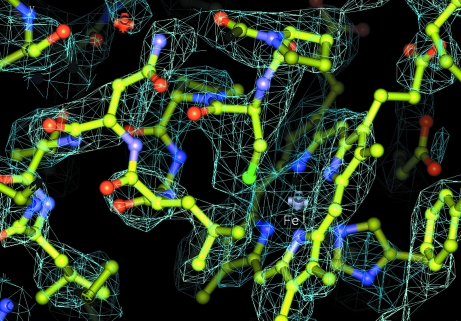
Data sets corresponding to peak, high-energy remote and inflection wavelengths were collected and processed. However, owing to radiation damage only the first 600 (of 720) frames of the peak wavelength (Table 3 ▶) were found to show a useful anomalous signal. Consequently, this data set was used for structure solution. As the first step, a two-site substructure was obtained using SHELXD (Sheldrick, 2008 ▶; Schneider & Sheldrick, 2002 ▶) with data to 4 Å resolution. This cutoff was chosen because beyond it the internal correlation of the anomalous signal fell below 30%. As the composition of the asymmetric unit was initially unknown, a two-site solution could indicate one protein molecule containing one Fe and one Mn atom or two molecules each containing only the haem iron. Owing to the low resolution and weak anomalous signal, the structure could not be solved with SHELXE; however, the correct hand could be established. The heavy-atom sites were further refined in autoSHARP (Vonrhein et al., 2007 ▶; Table 3 ▶). SOLOMON (Abrahams & Leslie, 1996 ▶) density-modification statistics (calculated within autoSHARP) indicated the presence of two complete molecules. It was not possible to trace the resulting electron density (calculated at 3 Å resolution) with ARP/wARP (Langer et al., 2008 ▶). However, further density modification up to 2.52 Å resolution within autoSHARP (Table 3 ▶) resulted in an interpretable map (Fig. 4 ▶). With subsequent model building in Buccaneer (Cowtan, 2006 ▶) and refinement in REFMAC5 (Skubák et al., 2004 ▶), which are part of the CCP4 program suite, a partial structural model was obtained. Finally, the model was completed manually in Coot (Emsley & Cowtan, 2004 ▶) and further refined in REFMAC5.
Figure 4.
Electron-density map at 2.52 Å resolution calculated after density modification in autoSHARP. The region of the map shows the haem (the iron is labelled) and its close environment at the proximal side comprising a helical segment containing the Fe-ligating cysteine.
3. Results and discussion
Initially, only rather small and very thin AaPII crystals were obtained using condition Nos. 32 and 47 of Hampton Research Crystal Screen (Jancarik & Kim, 1991 ▶), both of which contained 2.0 M ammonium sulfate. Since no other conditions yielded crystals, it was assumed that ammonium sulfate was a good precipitant. In particular, all PEG-based conditions left the drops clear. Subsequently, the ammonium sulfate-based Screen 6 from Jena Bioscience was tested and showed several conditions that were promising for crystallization with 1.2–2.2 M ammonium sulfate in the pH range 4.6–8.5. These conditions were refined and single crystals (Fig. 2 ▶) that were suitable for further X-ray experiments were obtained from three of them. The final conditions are given in Table 1 ▶. Although the AaPIIpH46 and AaPIIpH56 crystals are essentially isomorphous, structure determination was attempted for both since the crystals were obtained at different pH values. AaPII exhibits a broad pH spectrum for activity, which would be interesting to investigate structurally. Moreover, it was not initially evident whether or not the different crystal forms contained the same isozyme or isoforms or a mixture of them. Similar to other fungal enzymes, e.g. lignin peroxidase and laccases, AaPII is highly glycosylated. These enzymes often exhibit charge heterogeneity caused by variations of these glycosylation sites, which usually results in problems in obtaining single crystals of suitable quality or at all. This charge heterogeneity, or in other words the presence of isoforms, can be detected by the presence of several peaks in analytical IEF gels of seemingly pure protein samples. In the case of the first structure determination of a fungal laccase with a full complement of all four essential coppers, single crystals could only be obtained after purification of the enzyme to apparent charge homogeneity (Piontek et al., 2002 ▶; Antorini et al., 2002 ▶). Although our AaPII protein samples consisted of two isoforms, single crystals of high quality could readily be obtained.
Solution of the phase problem was initially attempted by molecular replacement, but this method failed. Considering the low sequence homology of the only available search model, CPO, this method was unlikely to succeed. Furthermore, CPO lacks a C-terminal peptide of 72 amino acids owing to two proteolytic cleavages (Sundaramoorthy et al., 1995 ▶), resulting in a mature protein of only 299 amino acids. For comparison, AaPII comprises a total of 328 amino acids.
Finally, the AaPIIpH85 crystal structure solution was approached using the MAD technique. Its structure solution presented a challenging case because two molecules of AaPII per asymmetric unit represent a molecular mass of about 90 kDa. Based on its relatively high sequence homology to an Mn-binding motif in CPO, it was initially assumed that AaPII (Pecyna et al., 2009 ▶) also contains an occupied Mn-binding site (Sundaramoorthy et al., 1995 ▶; Kühnel et al., 2006 ▶). However, no anomalous signal for an Mn atom was detected. Although the presence of a second anomalous scatterer, e.g. Mn, would have greatly facilitated structure solution, the structure could be solved with only two anomalous scatterers: the two haem irons. Nowadays, easy access to synchrotrons with their tuneable X-ray radiation facilitates the structure solution of proteins containing a natural anomalous scatterer such as, for example, transition metals in redox enzymes. The case of AaPII demonstrates that only one anomalous scatterer, albeit in combination with phase-modification procedures, is sufficient to solve the structure of a 45 kDa protein.
Using the electron density calculated with SAD phases at 3.0 Å resolution and subsequent density modification to 2.52 Å resolution, an initial protein model could be obtained comprising residues 8–328 of subunit A and residues 5–324 of subunit B. The two structural models correspond to the gene sequence of apo1 (Pecyna et al., 2009 ▶). After manual building and preliminary refinement at 2.2 Å resolution all N-glycosylation sites could be identified, confirming the assignment of the sequence. With one AaPII model in hand, we subsequently solved the other two crystal structures by MR. Inspection of electron-density maps revealed that the protein sequences of the four subunits in AaPIIpH46 and in AaPIIpH56 also correspond to the gene sequence of apo1 (Pecyna et al., 2009 ▶). Further refinement is currently in progress. Whether or not this will reveal the existence of two isoforms in the crystals corresponding to the two peaks in the isoelectric focusing gels remains an open question at this stage.
The recently discovered peroxidase/peroxygenase from A. aegerita belongs to the haem-thiolate proteins, which include physiologically important enzymes such as cytochrome P450, nitric oxide synthase and chloroperoxidase. The latter was the only known haem-thiolate peroxidase for more than 40 years and only in 2004 was a second enzyme of this type (AaP) described. As in P450s, both enzymes (CPO and AaP) show a characteristic shift of the Soret band towards 450 nm after reduction with dithionite and flushing with carbon monoxide (Hofrichter & Ullrich, 2006 ▶), which is thought to be caused by the axial haem ligand, which is a cysteine. Though CPO and AaP have strong to moderate chlorinating and brominating activities, respectively, it is questionable whether these activities are of physiological relevance. In particular, AaP, which only oxidizes bromide (and in turn brominates substrates such as phenol), must have another function since bromide, unlike chloride, is a rare anion in terrestrial environments. In this context, the oxygen-transfer potential of AaP, which includes epoxidations, hydroxylations and ether bond cleavages of/in little-activated substrates, is of particular interest (Ullrich & Hofrichter, 2007 ▶; Kinne et al., 2009 ▶). CPO has also been found to transfer peroxide oxygen to substrate molecules (Manoj & Hager, 2008 ▶); however, this catalytic feature is limited to activated carbon or heteroatoms (e.g. the double bond in styrene or the sulfur in thioanisole) and does not include aromatic substrates (Aranda et al., 2009 ▶). Nevertheless, both enzymes represent versatile biocatalysts with remarkable catalytic properties, which makes them promising for applications in organic synthesis (Hofrichter & Ullrich, 2006 ▶; Manoj & Hager, 2008 ▶). With the structure determination of this new enzyme, we have a tool in hand to explore its catalytic mechanism and proceed with structure–function studies.
Acknowledgments
We gratefully acknowledge the opportunity to collect diffraction data on the synchrotron beamline ID23-1 at the ESRF and thank the staff for their technical assistance and beamline support. This project was financed by a grant to KP and MH from the Commission of the European Communities within a project of the Sixth European Framework Programme (BIORENEW contract NMP2-CT-2006-026456). Dr Ignacio Fita (IBMB/CSIC, Barcelona, Spain) is thanked for giving us the opportunity to collect preliminary diffraction data from AaPII crystals on a rotating-anode generator in his laboratory.
References
- Abrahams, J. P. & Leslie, A. G. W. (1996). Acta Cryst. D52, 30–42. [DOI] [PubMed]
- Anh, D. H., Ullrich, R., Benndorf, D., Svatos, A., Muck, A. & Hofrichter, M. (2007). Appl. Environ. Microbiol.73, 5477–5485. [DOI] [PMC free article] [PubMed]
- Antorini, M., Herpoël-Gimbert, I., Choinowski, T., Sigoillot, J. C., Asther, M., Winterhalter, K. & Piontek, K. (2002). Biochim. Biophys. Acta, 1594, 109–114. [DOI] [PubMed]
- Aranda, E., Kinne, M., Kluge, M., Ullrich, R. & Hofrichter, M. (2009). Appl. Microbiol. Biotechnol.82, 1057–1066. [DOI] [PubMed]
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Cowtan, K. (2006). Acta Cryst. D62, 1002–1011. [DOI] [PubMed]
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Emsley, P. & Cowtan, K. (2004). Acta Cryst. D60, 2126–2132. [DOI] [PubMed]
- Hofrichter, M. & Ullrich, R. (2006). Appl. Microbiol. Biotechnol.71, 276–288. [DOI] [PubMed]
- Jancarik, J. & Kim, S.-H. (1991). J. Appl. Cryst.24, 409–411.
- Kabsch, W. (2010a). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Kabsch, W. (2010b). Acta Cryst. D66, 133–144. [DOI] [PMC free article] [PubMed]
- Kinne, M., Poraj-Kobielska, M., Ralph, S. A., Ullrich, R., Hofrichter, M. & Hammel, K. E. (2009). J. Biol. Chem.284, 29343–29349. [DOI] [PMC free article] [PubMed]
- Kluge, M., Ullrich, R., Dolge, C., Scheibner, K. & Hofrichter, M. (2009). Appl. Microbiol. Biotechnol.81, 1071–1076. [DOI] [PubMed]
- Kühnel, K., Blankenfeldt, W., Terner, J. & Schlichting, I. (2006). J. Biol. Chem.281, 23990–23998. [DOI] [PubMed]
- Langer, G. G., Cohen, S. X., Perrakis, A. & Lamzin, V. S. (2008). Nature Protoc.3, 1171–1179. [DOI] [PMC free article] [PubMed]
- Manoj, K. M. & Hager, L. P. (2008). Biochemistry, 47, 2997–3003. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Storoni, L. C. & Read, R. J. (2005). Acta Cryst. D61, 458–464. [DOI] [PubMed]
- Pecyna, M. J., Ullrich, R., Bittner, B., Clemens, A., Scheibner, K., Schubert, R. & Hofrichter, M. (2009). Appl. Microbiol. Biotechnol.84, 885–897. [DOI] [PubMed]
- Piontek, K., Antorini, M. & Choinowski, T. (2002). J. Biol. Chem.277, 37663–37669. [DOI] [PubMed]
- Schneider, T. R. & Sheldrick, G. M. (2002). Acta Cryst. D58, 1772–1779. [DOI] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Skubák, P., Murshudov, G. N. & Pannu, N. S. (2004). Acta Cryst. D60, 2196–2201. [DOI] [PubMed]
- Sundaramoorthy, M., Terner, J. & Poulos, T. L. (1995). Structure, 3, 1367–1377. [DOI] [PubMed]
- Ullrich, R., Dolge, C., Kluge, M. & Hofrichter, M. (2008). FEBS Lett.582, 4100–4106. [DOI] [PubMed]
- Ullrich, R. & Hofrichter, M. (2005). FEBS Lett.579, 6247–6250. [DOI] [PubMed]
- Ullrich, R. & Hofrichter, M. (2007). Cell. Mol. Life Sci.64, 271–293. [DOI] [PMC free article] [PubMed]
- Ullrich, R., Liers, C., Schimpke, S. & Hofrichter, M. (2009). Biotechnol. J.4, 1619–1626. [DOI] [PubMed]
- Ullrich, R., Nüske, J., Scheibner, K., Spantzel, J. & Hofrichter, M. (2004). Appl. Environ. Microbiol.70, 4575–4581. [DOI] [PMC free article] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. (2007). Methods Mol. Biol.364, 215–230. [DOI] [PubMed]