Thermostable recombinant L2 lipase from thermophilic Bacillus sp. L2 has been crystallized by using counter-diffusion method and diffracted to 2.7 Å resolution. The crystal belongs to the primitive orthorhombic space group P212121 with unit-cell parameters a = 87.44, b = 94.90, c = 126.46 Å.
Keywords: lipases, L2 lipase, Bacillus sp. L2, thermostable lipase, counter-diffusion method
Abstract
Purified thermostable recombinant L2 lipase from Bacillus sp. L2 was crystallized by the counter-diffusion method using 20% PEG 6000, 50 mM MES pH 6.5 and 50 mM NaCl as precipitant. X-ray diffraction data were collected to 2.7 Å resolution using an in-house Bruker X8 PROTEUM single-crystal diffractometer system. The crystal belonged to the primitive orthorhombic space group P212121, with unit-cell parameters a = 87.44, b = 94.90, c = 126.46 Å. The asymmetric unit contained one single molecule of protein, with a Matthews coefficient (V M) of 2.85 Å3 Da−1 and a solvent content of 57%.
1. Introduction
Lipases are well known as a physiologically and commercially important group of enzymes that have many industrial applications such as in organic chemical processing, detergent formulation, synthesis of biosurfactants, the agrochemical industry, paper manufacture, nutrition, cosmetics and pharmaceutical processing (Jaeger & Eggert, 2002 ▶; Sharma et al., 2001 ▶). Thermostable lipases from both mesophiles and thermophiles are currently gaining much attention in industry. These thermostable lipases are suitable for harsh industrial processes that commonly require high-temperature operation. By conducting biotechnological processes at elevated temperatures, the risk of contamination by mesophiles can be reduced.
Thermostable L2 lipase from Bacillus sp. L2 is highly active over a wide range of temperature (328–353 K) and pH (6–9) and has an optimum temperature as high as 343 K (Shariff et al., 2007 ▶). Its stability at high temperature and alkaline pH, and broad substrate specificity make it a novel potential additive in detergent formulation for more effective oil-stain removal at high temperatures. However, the structure of this interesting enzyme has not been determined. In this paper, we describe the crystallization of L2 lipase and the collection of X-ray diffraction data to 2.7 Å resolution.
2. Materials and methods
2.1. Protein expression and purification
Escherichia coli M15[pREP4] harbouring pQE-30/L2 was grown in LB medium supplemented with 50 µg ml−1 ampicillin and 25 µg ml−1 kanamycin on a rotary shaker (200 rev min−1) at 310 K. The culture was induced with 1 mM IPTG for 2 h. The cells were harvested by centrifugation and resuspended in binding buffer A (20 mM sodium phosphate, 500 mM NaCl, 20 mM imidazole pH 7.4) before sonication (Branson 250 sonifier; output 2, duty cycle 30, 2 min) and clarified by centrifugation (10 000 rev min−1, 30 min). The clarified lysate was collected and applied onto an Ni-Sepharose 6 Fast Flow affinity-chromatography column (XK 16/20, Amersham Bioscience, USA) equilibrated with binding buffer A. After elution with a linear gradient of imidazole from 20 to 500 mM and 0–500 mM NaCl, the fraction containing mature L2 lipase was desalted by dialysis against 50 mM Tris–HCl, 20 mM NaCl and 5% glycerol (buffer B) overnight. The sample was then applied onto a Super Q Toyopearl 650 S (Tosoh) column equilibrated with 50 mM Tris–HCl pH 9.0. After elution with a linear gradient of 0–300 mM NaCl, the fraction containing mature L2 lipase was desalted by dialysis against buffer B overnight. The homogeneity and identity of the purified sample were assessed using SDS–PAGE (Laemmli, 1970 ▶) and native PAGE. The sample was then concentrated to 3 mg ml−1 by ultrafiltration (Amicon Ultra-15, 10 kDa cutoff).
2.2. Crystallization
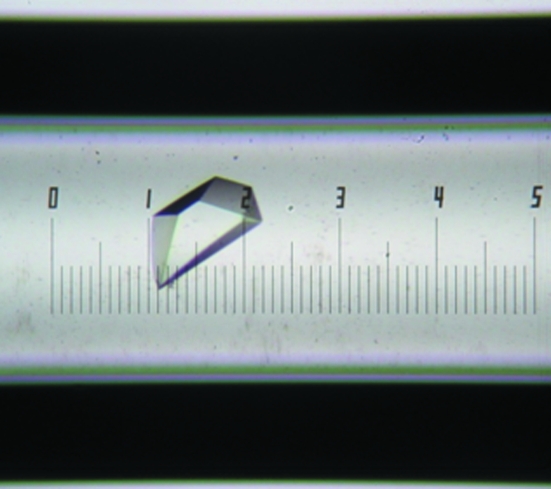
Preliminary crystal screening was performed by both the hanging-drop and sitting-drop methods using commercially available screening kits from Hampton Research (Crystal Screen, Crystal Screen 2, PEG/Ion Screen and Index Screen). Typically, 1 µl protein solution (2 mg ml−1) was mixed with reservoir solution in a 1:1 ratio and the resulting drop was equilibrated against reservoir solution at 293 K. From the series of crystallization screens tested, formulation No. 22 [0.1% MES pH 6.5, 12%(w/v) PEG 20 000] of Crystal Screen 2 conferred the best reproducible crystal formation. For optimization purposes, this formulation was used in the counter-diffusion crystallization method using the Crystal-Tube Kit (Confocal Science Inc.). Good-quality crystals were obtained after two weeks of incubation at 293 K using 20% PEG 6000, 50 mM MES pH 6.5 and 50 mM NaCl. The approximate dimensions of the crystal were 0.05 × 0.12 × 0.02 mm (Fig. 1 ▶).
Figure 1.
L2 lipase crystals obtained in a capillary using 20% PEG 6000, 50 mM MES pH 6.5, 50 mM NaCl. The approximate dimensions of the crystal are 0.05 × 0.12 × 0.02 mm.
2.3. X-ray data collection and processing
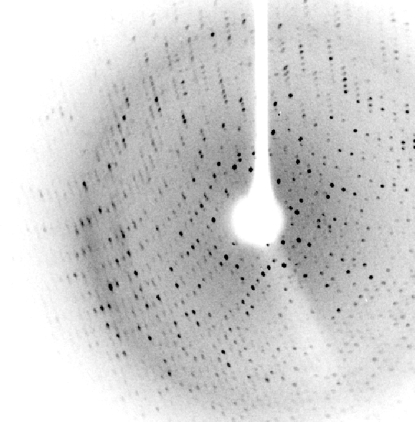
Crystals were mounted in Hampton loops using 30%(v/v) glycerol as a cryoprotectant and flash-cooled to 100 K in a stream of liquid nitrogen. Data were collected in-house on a Bruker X8 PROTEUM single-crystal diffractometer system with a MICROSTAR microfocus rotating-anode generator and a PLATINUM135 CCD detector placed at a distance of 6 cm. The exposure time was 60 s for the low-resolution pass and 120 s for high-resolution data collection. The best crystal diffracted to 2.7 Å resolution (Fig. 2 ▶). The data were indexed with PROTEUM, integrated with SAINT and scaled with SADABS (all from Bruker). After scaling, statistics were obtained and space-group determination was performed with XPREP (Bruker). In light of the statistics, the data were cut at 2.9 Å.
Figure 2.
X-ray diffraction pattern of an L2 lipase crystal.
3. Results and discussion
Recombinant L2 lipase was expressed in E. coli and purified with a molecular weight of 43 kDa. A single crystal of L2 lipase diffracted to 2.7 Å resolution and belonged to the primitive orthorhombic space group P212121, with unit-cell parameters a = 87.44, b = 94.90, c = 126.46 Å. Data-collection statistics are summarized in Table 1 ▶. A total of 167 801 reflections were merged into 44 888 unique reflections. Specific volume calculations using the molecular weight and the unit-cell parameters indicated that the crystal contained one protein molecule per asymmetric unit, with a Matthews coefficient (V M) of 2.85 Å3 Da−1 (Matthews, 1968 ▶) and a solvent content of 57%.
Table 1. Data-collection statistics for L2 lipase.
Values in parentheses are for the highest resolution shell.
| Crystal data | |
| Space group | P212121 |
| Unit-cell parameters (Å) | |
| a | 87.44 |
| b | 94.90 |
| c | 126.46 |
| Data processing | |
| Temperature (K) | 100 |
| Wavelength (Å) | 1.54 |
| Resolution (Å) | 15.0–2.7 (3.00–2.90) |
| No. of diffraction images | 1090 |
| Oscillation angle per frame (°) | 0.3 |
| Unique data | 44888 |
| Redundancy (%) | 4.05 (4.23) |
| Data completeness (%) | 99.50 (98.6) |
| Mean I/σ(I) | 15.43 (4.92) |
| Molecules per asymmetric unit | 1 |
| Matthews coefficient (Å3 Da−1) | 2.85 |
| Solvent content (%) | 57 |
| Rmerge (%) | 9.5 (19.8) |
Comparison with reports from the literature showed that this is the only thermostable lipase solved at high resolution that belongs to the orthorhombic space group P212121; other thermostable lipases such as L1 lipase, T1 lipase and P1 lipase all belong to the monoclinic space group C2 (Jeong et al., 2001 ▶; Matsumura et al., 2008 ▶; Sinchaikul et al., 2001 ▶). The change in the space group of L2 lipase occurs despite it only differing from the other thermostable lipases by 2–5 amino acids. In addition, it is the only thermostable lipase crystal to be successfully crystallized by the counter-diffusion crystallization method.
A preliminary attempt to solve the structure of L2 lipase by using the lipase from Geobacillus zalihae T1 (PDB code 2dsn; Matsumura et al., 2007 ▶), which has high homology, as a search model showed promising results. Complete structure determination and refinement studies are still in progress.
Acknowledgments
We thank Dr Marianna Biadene for her contribution in helping us with data collection and fruitful discussions. This research was supported by the Ministry of Science, Technology and Innovation, Malaysia (09-05-MGI-GMB001).
References
- Jaeger, K. & Eggert, T. (2002). Curr. Opin. Biotechnol.13, 390–397. [DOI] [PubMed]
- Jeong, S.-T., Kim, H.-K., Kim, S.-J., Pan, J.-G., Oh, T.-K. & Ryu, S.-E. (2001). Acta Cryst. D57, 1300–1302. [DOI] [PubMed]
- Laemmli, U. K. (1970). Nature (London), 227, 680–685. [DOI] [PubMed]
- Matsumura, H., Yamamoto, T., Leow, T. C., Mori, T., Salleh, A. B., Basri, M. Inoue, T., Kai, Y. & Rahman, R. N. Z. A. (2008). Proteins, 70, 592–598. [DOI] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- Shariff, F. M. S., Leow, T. C., Salleh, A. B., Basri, M. & Rahman, R. N. Z. R. A. (2007). J. Basic Microbiol.47, 406–412. [DOI] [PubMed]
- Sharma, R., Chisti, Y. & Banerjee, U. C. (2001). Biotechnol. Adv.19, 627–662. [DOI] [PubMed]
- Sinchaikul, S., Sookkheo, B., Phutrakul, S., Pan, F.-M. & Chen, S.-T. (2001). Protein Expr. Purif.22, 388–398. [DOI] [PubMed]