GIMAP2, a guanine nucleotide-binding protein belonging to the GIMAP family, has been crystallized in various nucleotide-loaded states.
Keywords: GTPase of immunity-associated proteins, GIMAP2, IMAP2
Abstract
GTPases of immunity-associated proteins (GIMAPs) are important regulators of T-cell death and survival. Here, the crystallization and data collection of three GIMAP2 constructs in various nucleotide-loaded states is described. Selenomethionine-substituted carboxy-terminally truncated GIMAP2 (amino-acid residues 1–260; GIMAP21–260) in the nucleotide-free form crystallized in space group P212121 and the crystals diffracted X-rays to 1.5 Å resolution. The phase problem was solved using the single anomalous dispersion (SAD) protocol. GDP-bound GIMAP221–260 and GDP-bound GIMAP21–234 crystallized in space group P212121 and the crystals diffracted X-rays to 2.9 and 1.7 Å resolution, respectively. GTP-bound GIMAP21–234 crystallized in space group C2221 and the crystals diffracted to 1.9 Å resolution. These results will allow a detailed structural analysis of GIMAP2, which will provide insight into the architecture and function of the GIMAP family.
1. Introduction
GIMAPs [GTPases of immunity-associated proteins, also known as immunity-associated nucleotide-binding proteins (IANs) or IMAPs] are a GTPase family that are exclusively found in vertebrates and plants (for a review, see Nitta & Takahama, 2007 ▶). The seven human GIMAPs are clustered on chromosome 7 and are composed of an amino-terminal guanine-nucleotide binding domain (G-domain) followed by a varying C-terminal extension of 50–110 amino acids in length. In GIMAP4, GIMAP5 and GIMAP7 this extension shows a high propensity to form a coiled-coil structure. GIMAP1, GIMAP2 and GIMAP5 additionally possess one or two additional predicted transmembrane (TM) helices at the C-terminus. An exception is GIMAP8, which consists of three consecutive G-domains, each followed by a short extension. The closest homologues of the GIMAP family for which structural information is available are the Toc GTPases in plants, which have approximately 18% sequence identity (Sun et al., 2002 ▶).
GIMAPs are mainly expressed in the tissues of the immune system, such as the spleen, thymus and lymph nodes. Within these tissues, they are present in T cells and B cells (Krucken et al., 2004 ▶; Nitta et al., 2006 ▶). The biobreeding diabetes-prone (BBDP) rat strain, which is devoid of functional GIMAP5 owing to a frameshift mutation, shows a severe reduction in the number of peripheral T cells (lymphopaenia) concomitant with an increased rate of apoptosis of the remaining cells (MacMurray et al., 2002 ▶; Michalkiewicz et al., 2004 ▶; Pandarpurkar et al., 2003 ▶). Furthermore, BBDP rats develop spontaneous autoimmune diabetes. Similar to BBDP rats, GIMAP5-knockout mice show a significant loss of peripheral T cells and a complete absence of natural killer cells (Schulteis et al., 2008 ▶). These mice die after approximately 15 weeks owing to liver failure.
In mouse thymocytes, significant up-regulation of GIMAP3, GIMAP4 and GIMAP5 was demonstrated at the stage of positive selection. Overexpression of GIMAP4 increased the number of apoptotic CD4/CD8 double-positive thymocytes. Knockdown of GIMAP5 interfered with thymocyte development between the double-negative and double-positive stages. In the same study, the interaction of GIMAP3 and GIMAP5 with the anti-apoptotic proteins Bcl-2 and Bcl-xL and with the pro-apoptotic Bax was reported, as well as selective interaction of GIMAP4 with Bax (Nitta et al., 2006 ▶).
Taken together, these results suggest that GIMAPs play a critical role in the development of T lymphocytes, especially in the regulation of apoptosis during selection processes in the thymus. In order to analyze the structure and mechanism of GIMAPs, we purified and crystallized three human GIMAP2 constructs in different nucleotide-loaded states. Complemented with biochemical and cell-biological experiments, these studies will shed light on the mechanism and function of GIMAPs in lymphocyte development.
2. Materials and methods
2.1. Cloning and protein preparation
The cDNA coding for human GIMAP2 was obtained from Geneservice (Cambridge, England). GIMAP2 constructs coding for amino acids 1–260 (GIMAP21–260) and 21–260 (GIMAP221–260) were designed that lacked the two predicted TM helices (residues 261–337) of wild-type GIMAP2. A third construct encompassing amino acids 1–234 (GIMAP21–234) was devoid of the C-terminal extension and corresponded to the GIMAP2 G-domain. Constructs were PCR-amplified using the following oligonucleotide primers: 5′-GGCGGATCCATGGACCAAAATGAACACAGTCACTG-3′ (forward, BamHI restriction site indicated in bold) and 5′-GGCCTCGAGCTACTATAACCTAGGAGTCTTGCGTTC-3′ (reverse, XhoI restriction site indicated in bold) for GIMAP21–260, 5′-GGCGGAT CCTCTGAGCTGAGAATCATCCTGGTG-3′ (forward, BamHI restriction site indicated in bold) for GIMAP221–260 and 5′-GGCCTCGAGCTACTATTCATCTGATCCCACAGGTCCAC-3′ (reverse, XhoI restriction site indicated in bold) for GIMAP21–234. Inserts were cloned in the pGEX6P1 expression vector (GE Healthcare, München, Germany) using standard restriction-enzyme cloning methods to generate GST-fusion constructs. 10 l TB modified medium (Carl Roth, Karlsruhe, Germany) supplemented with 50 µg ml−1 ampicillin and 34 µg ml−1 chloramphenicol was inoculated in a 1:100 ratio with an overnight Escherichia coli Rosetta (DE3) culture in LB medium containing the expression constructs. Cells were grown with shaking to an OD600 of 0.5 at 310 K. Bacterial cultures were cooled to 291 K, protein expression was induced by the addition of 40 µM isopropyl β-d-1-thiogalactopyranoside (IPTG) and cultures were grown for another 20 h. Bacteria were collected by centrifugation at 5000 rev min−1 for 7 min and pellets were resuspended in 30 ml lysis buffer per litre of bacterial culture containing 50 mM HEPES pH 7.5, 500 mM NaCl, 2.5 mM dithiothreitol (DTT), 100 µM Pefabloc SC, 1 µg ml−1 DNAse I and 1 mM magnesium chloride. The cell suspension was stored at 233 K.
2.2. Expression of selenomethionine-labelled protein
To produce SeMet-labelled protein, M9 minimal medium [6 g l−1 Na2HPO4, 3 g l−1 KH2PO4, 1 g l−1 NH4Cl, 0.5 g l−1 NaCl, 1 mM MgSO4, 0.4%(w/v) glucose, 0.1 g l−1 riboflavin, 0.1 g l−1 niacin amide, 0.01 g l−1 pyridoxine, 0.1 g l−1 thiamine] was inoculated with an overnight LB culture of the respective expression clone at a 1:100 dilution. Cells were grown to an OD600 of 0.5 at 310 K. An amino-acid supplement (l-lysine, l-phenylalanine and l-threonine to a final concentration of 100 mg l−1, and l-isoleucine, l-leucine, l-valine and l-SeMet to a final concentration of 50 mg l−1) was added to inhibit endogenous methionine biosynthesis and start SeMet incorporation (Doublié, 1997 ▶). The bacterial culture was cooled to 291 K 15 min after addition. Protein expression was induced by the addition of 40 µM IPTG. Further growth and harvesting of the cells was conducted in the same way as for the native protein.
2.3. Protein purification
All purification procedures were carried out on ice or at 277 K. A bacterial suspension from a 10 l culture was mechanically ruptured using a microfluidizer (Microfluidics, Newton, USA) for at least two passes. The soluble fraction of the lysate was prepared by centrifugation at 50 000g for 45 min at 277 K. The supernatant was filtered using a filter pore size of 0.2 µm and applied onto a chromatography column loaded with 15 ml reduced glutathione-Sepharose beads (GSH-Sepharose; GE Healthcare, München, Germany) equilibrated with five column volumes (CV) of buffer I (50 mM HEPES pH 7.5, 500 mM NaCl, 2.5 mM DTT). The column was washed with buffer I until E 280 reached baseline. The column was then washed with 3 CV buffer II (50 mM HEPES pH 7.5, 500 mM NaCl, 2.5 mM DTT, 10 mM EDTA) to remove protein-bound Mg2+ and nucleotide. The column was washed again with 3 CV buffer I to remove EDTA. GSH-Sepharose beads were resuspended in 1 CV buffer I and transferred to a Falcon tube. 1 mg GST-PreScission protease (GE Healthcare, München, Germany) was added to cleave the amino-terminal GST tag. Five amino acids, Gly-Pro-Leu-Gly-Ser, remained as a cloning artifact at the N-terminus after proteolytic cleavage. The suspension was incubated for 15 min on a roller and for a further 20 h at 277 K without agitation. The column was re-packed and cleaved GIMAP2 was collected by eluting with buffer I until E 280 reached baseline. After concentrating the sample to 2 ml, the protein was applied onto a Superdex 75 16/60 gel-filtration column (GE Healthcare, München, Germany) equilibrated with 10 mM HEPES pH 7.5, 150 mM NaCl, 2.5 mM DTT. The peak fractions containing the protein of interest were pooled and concentrated to approximately 35 mg ml−1. Protein concentration was determined by measuring the absorption at 280 nm using an calculated extinction coefficient of 28 420 M −1 cm−1 (Gasteiger et al., 2005 ▶). The protein was flash-frozen in liquid nitrogen in 25 µl aliquots and stored at 193 K. SeMet-labelled protein was purified in the same way as native protein. For all constructs, the yield of purified native protein as well as that of SeMet-labelled protein was approximately 1 mg per litre of bacterial culture.
2.4. Crystallization
Before crystallization, the proteins were diluted to 15 mg ml−1 with 10 mM HEPES pH 7.5, 150 mM NaCl, 2.5 mM DTT. Magnesium chloride and nucleotides of the highest purity (Jena Bioscience, Jena, Germany), where applicable, were added to a final concentration of 2 mM. We observed that even in its original buffer GIMAP2 showed heavy precipitation after 1 d incubation at room temperature. Consequently, all further crystallization experiments were conducted at 277 K. Since commercial screens did not yield any initial crystallization conditions, a variety of precipitants were tested for their compatibility with GIMAP2 crystal formation. These crystallization attempts were performed in 24-well hanging-drop vapour-diffusion setups. The reservoir contained 0.5 ml solution and crystallization drops were composed of 1 µl protein solution plus 1 µl reservoir solution. Single crystals of nucleotide-free GIMAP21–260 grew in conditions containing 11–19% 2-propanol. After further refinement, the final crystals used for data collection were grown in 11–12% 2-propanol and 15% glycerol. Crystals grew to dimensions of approximately 200 × 50 × 50 µm and could be directly cryocooled. SeMet-derivatized crystals were prepared in the same way.
GDP-loaded GIMAP221–260 crystallized in 19% 2-propanol. Crystals (approximately 100 ×10 × 10 µm) were prepared for cryocooling by brief transfer to a solution containing 10 mM HEPES pH 7.5, 150 mM NaCl, 2.5 mM DTT, 2 mM magnesium chloride, 2 mM GDP, 19% 2-propanol and 20% glycerol.
GDP-loaded GIMAP21–234 crystallized in 12.5% 2-propanol and 30% glycerol and could be cryocooled without the addition of further cryoprotectant. Crystals of GTP-loaded GIMAP21–234 could be obtained by performing an additive screen (Hampton Research, Aliso Viejo, California, USA). The final crystallization condition contained 6% 2-propanol and 10 mM ATP. Single needles (approximately 100 × 10 × 10 µm) were cryocooled after brief transfer to a solution containing 10 mM HEPES pH 7.5, 150 mM NaCl, 2.5 mM DTT, 2 mM magnesium chloride, 2 mM GTP, 6% 2-propanol, 10 mM ATP and 25% glycerol. Notably, GIMAPs contain a canonical G-domain including the guanine-specificity motif G4 and are not known to bind ATP. All crystals were cryocooled by plunging them into liquid nitrogen.
2.5. Data collection
Data sets were recorded from single crystals using the rotation method with a ϕ increment of 1° at a temperature of 100 K. Beamline 14.2 at BESSY II, Berlin, Germany was used with a MAR 165 CCD detector, with the exception of the GIMAP21–234–GTP needles, for which a data set was recorded using the microfocus setup at BESSY II beamline 14.1, which is equipped with a Rayonics MX-225 CCD detector. Initial indexing and determination of an optimal data-collection strategy was performed using MOSFLM (Leslie, 2006 ▶). A fluorescence scan of SeMet-derivatized nucleotide-free GIMAP21–260 crystals was conducted using a Bruker AXS/Roentec X-ray fluorescence detector. The recorded intensities were integrated with the program XDS (Kabsch, 2010 ▶). Molecular-replacement trials were carried out using the programs MOLREP (Vagin & Teplyakov, 1997 ▶) and Phaser (McCoy et al., 2007 ▶) via the CCP4 graphical interface v.6.0.2 (Collaborative Computational Project, Number 4, 1994 ▶).
3. Results and discussion
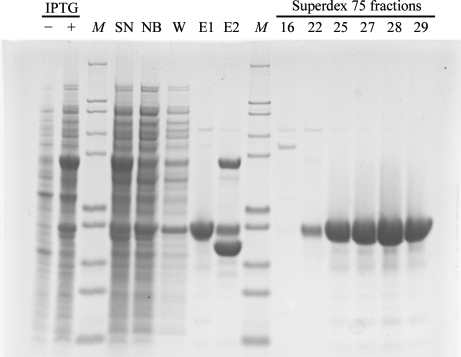
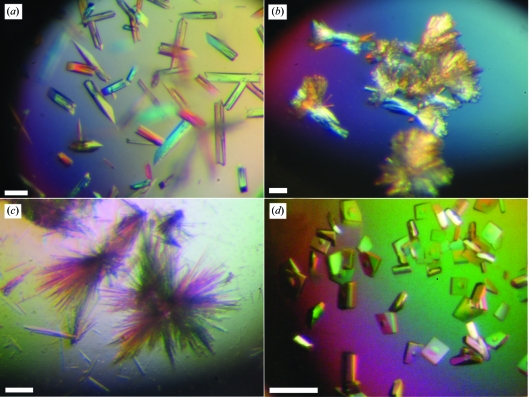
Three GIMAP2 constructs were purified to homogeneity (Fig. 1 ▶). The use of 2-propanol as a precipitant favoured the crystallization of all of the GIMAP constructs in the presence of various nucleotides. Crystals of nucleotide-free GIMAP21–260 lacking the two predicted TM helices appeared after 1 h and grew to their final dimensions overnight (Fig. 2 ▶ a). They diffracted X-rays to 1.7 Å resolution and belonged to the orthorhombic space group P212121, with unit-cell parameters a = 57.4, b = 60.8, c = 72.0 Å, α = β = γ = 90°.
Figure 1.
Typical purification procedure of GIMAP2 constructs using GIMAP21–260 as an example. 4–12% SDS–PAGE of various samples taken during purification. −/+ IPTG, whole-cell bacteria lysates before and after induction; M, molecular-weight markers, bands correspond to 200, 116, 97, 66, 55, 36.5, 31, 21.5, 14 and 6 kDa (from top to bottom); SN, supernatant of bacterial lysates after ultracentrifugation; NB, nonbound material after GSH-Sepharose column; W, wash fraction of the same column; E1, eluate after protease cleavage containing the GIMAP construct; E2, GSH elution showing GST and the GST-tagged protease.
Figure 2.
GIMAP2 crystals. (a) Crystals of nucleotide-free GIMAP21–260. (b) Needle clusters of GDP-loaded GIMAP221–260. (c) Crystals of GTP-bound GIMAP21–234. (d) Crystals of GDP-loaded GIMAP21–234. The scale bars correspond to 100 µm.
Initial attempts to solve the structure by molecular replacement using the distantly related Toc34 GTPase from pea as the template failed. Therefore, SeMet-derivatized protein was prepared and crystallized in the absence of nucleotide. Using an X-ray fluorescence scan, the peak energy was determined to be 12 655.69 eV, with f′ and f′′ values of 5.14 and −7.58, respectively. The data set collected at peak energy showed anomalous signal to high resolution. SeMet-substituted crystals were isomorphous to the native crystals but diffracted X-rays to higher resolution. Consequently, a low-energy remote data set was collected from a second derivatized crystal to a resolution of 1.5 Å. A single anomalous dispersion (SAD) protocol was applied to solve the phase problem, as implemented in the program autoSHARP (Vonrhein et al., 2007 ▶). All seven selenium sites were found using SHELXD (Sheldrick, 2008 ▶). 218 of 260 amino-acid residues were automatically placed by phenix.autobuild (Adams et al., 2010 ▶) and could be refined against the high-resolution data set to initial R work and R free factors of 24% and 26%, respectively, using the program REFMAC5 (Murshudov et al., 1997 ▶). Further model building and refinement are in progress.
Since all crystallization attempts for this construct failed in the presence of GDP and GTP, we generated a second GIMAP2 construct in which the first 20 residues were deleted. Bioinformatics analysis using the RONN server (Yang et al., 2005 ▶) suggested that these residues were disordered. GIMAP221–260–GDP crystallized as clusters of small needles using 2-propanol as a precipitant (Fig. 2 ▶ b). Single needles were broken off from these clusters and used for data collection. The crystals diffracted X-rays to a maximal resolution of 2.9 Å and belonged to space group P212121, with unit-cell parameters a = 62.7, b = 76.5, c = 101.6 Å, α = β = γ = 90°. Analysis of the Matthews coefficient V M (Table 1 ▶) suggested the presence of two GIMAP2 molecules in the asymmetric unit. This was confirmed by molecular replacement using the program MOLREP with the preliminary nucleotide-free GIMAP2 coordinates as a search model. Despite extensive screening efforts, GTP-loaded GIMAP221–260 could not be crystallized.
Table 1. Data-collection statistics.
Values in parentheses are for the highest resolution shell.
| SeMet GIMAP21-260 | |||||
|---|---|---|---|---|---|
| Peak | Low-energy remote | GIMAP221–260 | GIMAP21–234 | GIMAP21–234 | |
| Nucleotide | GDP | GDP | GTP | ||
| Beamline | BESSY 14.2 | BESSY 14.2 | BESSY 14.2 | BESSY 14.2 | BESSY 14.1 |
| Wavelength (Å) | 0.9797 | 0.9849 | 0.9184 | 0.9184 | 0.9184 |
| Space group | P212121 | P212121 | P212121 | P212121 | C2221 |
| Unit-cell parameters | |||||
| a (Å) | 57.4 | 57.4 | 62.7 | 42.9 | 58.0 |
| b (Å) | 60.8 | 60.8 | 76.5 | 61.8 | 69.2 |
| c (Å) | 72.0 | 72.0 | 101.6 | 185.3 | 116.6 |
| α = β = γ (°) | 90 | 90 | 90 | 90 | 90 |
| No. of molecules in asymmetric unit | 1 | 1 | 2 | 2 | 1 |
| VM† (Å3 Da−1) | 2.15 | 2.15 | 2.26 | 2.35 | 2.22 |
| Resolution (Å) | 50–1.75 (1.80–1.75) | 50–1.50 (1.59–1.50) | 50–2.89 (3.07–2.89) | 50–1.70 (1.80–1.70) | 50–1.90 (2.00–1.90) |
| No. of measured reflections | 183804 (11439) | 179638 (28189) | 45534 (6795) | 224594 (32614) | 95706 (11945) |
| No. of unique reflections | 48628 (3489) | 40986 (6487) | 11194 (1690) | 54984 (8607) | 18848 (2939) |
| Rmeas‡ (%) | 6.8 (50.0) | 7.8 (59.7) | 22.0 (62.3) | 6.6 (57.9) | 6.8 (52.6) |
| I/σ(I) | 14.6 (3.2) | 14.7 (3.6) | 8.5 (3.5) | 15.8 (3.0) | 18.1 (3.0) |
| Completeness (%) | 99.1 (95.9) | 99.7 (99.1) | 98.2 (95.8) | 99.2 (98.3) | 99.7 (98.8) |
| Overall B factor (Wilson) (Å2) | 26.9 | 22.3 | 35.0 | 27.1 | 32.9 |
| Anomalous phasing power | 1.8 | ||||
| Figure of merit (acentric/centric) | 0.435/0.133 | ||||
A third construct was prepared that lacked the C-terminal extension (residues 235–260) to aid crystallization. GIMAP21–234 formed spherulites in the presence of GTP and 2-propanol. Using an additive screen, we were able to grow crystals that could be mounted for data collection (Fig. 2 ▶ c). The crystals diffracted X-rays to a maximal resolution of 1.9 Å and belonged to the orthorhombic space group C2221, with unit-cell parameters a = 58, b = 69.2, c = 116.6 Å, α = β = γ = 90°.
This construct was also crystallized in the GDP-loaded state in the presence of glycerol and 2-propanol (Fig. 2 ▶ d). The crystals diffracted to 1.7 Å resolution and belonged to space group P212121, with unit-cell parameters a = 42.9, b = 61.8, c = 185.3 Å, α = β = γ = 90°. Preliminary molecular-replacement analysis of these data again confirmed the presence of two GIMAP2 molecules in the asymmetric unit. All X-ray data and phasing statistics are summarized in Table 1 ▶. The structures of GDP-loaded and GTP-loaded GIMAP2 will be solved by molecular replacement using the nucleotide-free structure as a search model. Structure analysis of all of the crystals described in this paper is expected to yield new insights into the architecture and function of this G-protein family.
Acknowledgments
The authors would like to thank the macromolecular crystallography group at BESSY II, Berlin, especially Uwe Mueller, Jörg Schulze and Georg Zocher, for assistance at the beamlines. Furthermore, we would like to thank Sabine Werner for excellent technical assistance and Jürgen J. Müller and Katja Fälber for critical reading of the manuscript.
References
- Adams, P. D. et al. (2010). Acta Cryst. D66, 213–221.
- Collaborative Computational Project, Number 4 (1994). Acta Cryst. D50, 760–763.
- Diederichs, K. & Karplus, P. A. (1997). Nature Struct. Biol.4, 269–275. [DOI] [PubMed]
- Doublié, S. (1997). Methods Enzymol.276, 523–530. [PubMed]
- Gasteiger, E., Hoogland, C., Gattiker, A., Duvaud, S., Wilkins, M. R., Appel, R. D. & Bairoch, A. (2005). The Proteomics Protocols Handbook, edited by J. M. Walker, pp. 571–607. Totowa: Humana Press.
- Kabsch, W. (2010). Acta Cryst. D66, 125–132. [DOI] [PMC free article] [PubMed]
- Krucken, J., Schroetel, R. M., Muller, I. U., Saidani, N., Marinovski, P., Benten, W. P., Stamm, O. & Wunderlich, F. (2004). Gene, 341, 291–304. [DOI] [PubMed]
- Leslie, A. G. W. (2006). Acta Cryst. D62, 48–57. [DOI] [PubMed]
- MacMurray, A. J. et al. (2002). Genome Res.12, 1029–1039. [DOI] [PMC free article] [PubMed]
- Matthews, B. W. (1968). J. Mol. Biol.33, 491–497. [DOI] [PubMed]
- McCoy, A. J., Grosse-Kunstleve, R. W., Adams, P. D., Winn, M. D., Storoni, L. C. & Read, R. J. (2007). J. Appl. Cryst.40, 658–674. [DOI] [PMC free article] [PubMed]
- Michalkiewicz, M., Michalkiewicz, T., Ettinger, R. A., Rutledge, E. A., Fuller, J. M., Moralejo, D. H., Van, Y. B., MacMurray, A. J., Kwitek, A. E., Jacob, H. J., Lander, E. S. & Lernmark, A. (2004). Physiol. Genomics, 19, 228–232. [DOI] [PubMed]
- Murshudov, G. N., Vagin, A. A. & Dodson, E. J. (1997). Acta Cryst. D53, 240–255. [DOI] [PubMed]
- Nitta, T., Nasreen, M., Seike, T., Goji, A., Ohigashi, I., Miyazaki, T., Ohta, T., Kanno, M. & Takahama, Y. (2006). PLoS Biol.4, e103. [DOI] [PMC free article] [PubMed]
- Nitta, T. & Takahama, Y. (2007). Trends Immunol.28, 58–65. [DOI] [PubMed]
- Pandarpurkar, M., Wilson-Fritch, L., Corvera, S., Markholst, H., Hornum, L., Greiner, D. L., Mordes, J. P., Rossini, A. A. & Bortell, R. (2003). Proc. Natl Acad. Sci. USA, 100, 10382–10387. [DOI] [PMC free article] [PubMed]
- Schulteis, R. D. et al. (2008). Blood, 112, 4905–4914. [DOI] [PMC free article] [PubMed]
- Sheldrick, G. M. (2008). Acta Cryst. A64, 112–122. [DOI] [PubMed]
- Sun, Y.-J., Forouhar, F., Li, H.-M., Tu, S.-L., Yeh, Y.-H., Kao, S., Shr, H.-L., Chou, C.-C., Chen, C. & Hsiao, C.-D. (2002). Nature Struct. Biol.9, 95–100. [DOI] [PubMed]
- Vagin, A. & Teplyakov, A. (1997). J. Appl. Cryst.30, 1022–1025.
- Vonrhein, C., Blanc, E., Roversi, P. & Bricogne, G. (2007). Methods Mol. Biol.364, 215–230. [DOI] [PubMed]
- Yang, Z. R., Thomson, R., McNeil, P. & Esnouf, R. M. (2005). Bioinformatics, 21, 3369–3376. [DOI] [PubMed]