Abstract
Background
Chromosome copy gain, loss and LOH involving most chromosomes have been reported in many cancers, but less is known about chromosome instability in premalignant conditions. 17p LOH and DNA content abnormalities have been previously reported to predict progression from Barrett’s esophagus (BE) to esophageal adenocarcinoma (EA). Here, we evaluated genome-wide chromosomal instability in multiple stages of BE and EA in whole biopsies.
Methods
42 patients were selected to represent different stages of progression from BE to EA. Whole BE or EA biopsies were minced, aliquots processed for flow cytometry and genotyped with a paired constitutive control for each patient using 33,423 SNPs.
Results
Copy gains, losses, and LOH increased in frequency and size between early and late stage BE (p<0.001), with SNP abnormalities increasing from <2% to >30% in early and late stages, respectively. A set of statistically significant events were unique to either early, late or both stages, including previously reported and novel abnormalities. The total number of SNP alterations was highly correlated with DNA content aneuploidy and was sensitive and specific to identify patients with concurrent EA (empirical ROC AUC=0.91).
Conclusions
With the exception of 9p LOH, most copy gains, losses, and LOH detected in early stages of BE were smaller than those detected in later stages, and few chromosomal events were common in all stages of progression. Measures of chromosomal instability can be quantified in whole biopsies using SNP-based genotyping and have potential to be an integrated platform for cancer risk stratification in BE.
Keywords: Barrett’s esophagus, Esophageal cancer, Chromosome copy number loss, gain, LOH, Aneuploid, Wavelets, Biomarkers, SNP array, Beadchip
Introduction
Genomic instability has been hypothesized to play a crucial role in development of cancer for decades(1), and there is substantial evidence to support this hypothesis. Numerous studies have investigated genome-wide copy number changes and LOH(2–5) in EA and other cancers, including renal cell(6), colon(7, 8), cervical(9), pancreas(10), lung(11, 12), and glioblastoma(13). These studies have reported that the cancer genome acquires extensive chromosomal changes during neoplastic progression from a normal cell to cancer. Many of these alterations confer a selective advantage for the cancers (“drivers”) relative to normal cells, but others appear to be hitchhikers that are evolutionarily neutral (“passengers”)(14). Few studies have utilized SNP-based technologies to evaluate genome-wide copy number and LOH in premalignant stages of neoplastic progression compared to those in cancer(15–17). Characterization of premalignant conditions is important to understand the biology of neoplastic progression as well as to identify potential targets for cancer prevention and to develop methods for cancer risk stratification or early diagnosis of an asymptomatic cancer.
The incidence of esophageal adenocarcinoma (EA) is rising more rapidly than any other cancer in the US(18, 19). EA is typically detected at an advanced stage in which it is rapidly fatal with mortality greater than 80%(20, 21). Barrett’s esophagus (BE) is a condition that develops in a subset of patients with chronic gastroesophageal reflux disease in which the normal squamous epithelium of the esophagus is replaced by intestinal metaplasia(22). Although BE is the only known precursor of EA, most patients with BE die of causes unrelated to BE or EA(23, 24). Further, recent evidence suggests that in most cases BE appears to be a successful adaptation to a harsh intraesophageal environment with chronic acid and bile reflux associated with erosion, ulceration, reactive oxygen species and oxidative damage(25–28). Thus, there is a need for biomarkers that can distinguish common, benign BE patients for whom surveillance can be safely prolonged, rare cases that will progress and therefore merit careful surveillance or intervention to prevent EA, and patients who require early detection because they have already developed a small EA that cannot be visualized endoscopically.
BE is one of the best models of human intraepithelial neoplasia because the premalignant epithelium can be safely visualized and biopsied so that genomic changes can be compared in different stages of neoplastic evolution and then studied longitudinally by endoscopic biopsy surveillance for EA risk management(29, 30). Genetic progression models and longitudinal studies of oral leukoplakia have also shown successes in using somatic genetic biomarkers for risk stratification in head and neck cancer(31–35). This is in contrast to many other premalignant conditions that are typically removed when detected, such as colonic adenomas, or are unable to be systematically sampled for biopsy examination over time because of limited accessibility or potential clinical complications. Studies of BE indicate that inactivation of CDKN2A by loss of heterozygosity (LOH), methylation and/or mutation is selected as an early event that predisposes to large clonal expansions in the BE segment(36–38). Inactivation of TP53 by mutation and LOH is subsequently selected, typically in a CDKN2A deficient background, and predisposes to progression to increased 4N fractions (G2/tetraploidy), aneuploidy and EA(14, 29, 32–34, 38–42).
We reported recently in a 10-year prospective cohort study that a panel of chromosome instability biomarkers, including LOH spanning the CDKN2A and TP53 loci on chromosome arms 9p and 17p, and DNA content abnormalities (increased 4N, and aneuploidy) identified a high-risk patient subset that had a 79% five-year cumulative incidence of EA and a low-risk subset with a 0% cumulative EA incidence to nearly eight years of follow-up after their baseline endoscopic biopsy surveillance(43). This and other studies support the hypothesis that progressive chromosomal instability and clonal evolution drive progression from a benign premalignant state to cancer(29, 32–34, 42, 44). However, it has been difficult to combine these biomarkers into a single platform for clinical use because various and complex sample processing methods are required, including flow cytometric cell sorting and multiple research assays for LOH detection. Further, regions of LOH evaluated in these studies were based on low density microsatellite polymorphisms(45–47). In addition, genome-wide studies of chromosomal instability in EA have identified common regions of copy gain, loss, copy neutral LOH, and a small number of homozygous deletions(2), but these events have not been quantified in early stages of progression in BE.
Here, we evaluate genome-wide copy gain, loss, LOH and assess SNP-based quantification of aneuploidy in a cross-sectional study of patients representing different stages of progression from early BE metaplasia to advanced EA using paired samples with Illumina multisample 33K SNP BeadChips. Selection of patients and biopsies in this study was guided by previous extensive characterization of the Seattle Barrett’s esophagus cohort, which allowed more precise estimation of risk based on molecular characterization and more endoscopic follow-up data than are typically available. Chromosome alterations including genome-wide copy gain, loss, and LOH were evaluated and compared across the progression stages using whole, unpurified biopsies. Overall genomic instability was quantified relative to DNA content aneuploidy in the same biopsy to assess feasibility of using SNP-arrays as a common platform for biomarkers of EA risk.
Material and Methods
Study participants and biological samples
Participants were enrolled in the Seattle Barrett’s Esophagus Study, which was approved by the Human Subjects Division of the University of Washington in 1983 and renewed annually thereafter with reciprocity from the Fred Hutchinson Cancer Research Center (FHCRC) Institutional Review Board from 1993 to 2001. Since 2001, the study has been approved by the FHCRC IRB with reciprocity from the University of Washington Human Subjects Division.
Mucosal biopsies used in this cross sectional study were obtained from 42 patients using previously defined protocols for endoscopic mucosal biopsy and evaluation of surgical resection specimens(48, 49). A constitutive control sample from gastric tissue was collected for each of the 42 participants and used as the reference for paired LOH and copy number analysis. The 42 participants each had histologically diagnosed specialized intestinal metaplasia and represented four different stages of neoplastic progression in Barrett’s esophagus defined by
[BE Early] 12 surveillance endoscopic biopsies from 12 participants with Barrett’s esophagus who never developed DNA content abnormalities (aneuploidy or 4N fraction >6%), high-grade dysplasia, or EA during any follow-up endoscopies (mean 118.3 months, range 59.2 to 182.6 months) and in whom no TP53 mutation or 17p LOH was detected at baseline (M:F=1:11, mean age 58);
[BE instability] 12 endoscopic biopsies from 12 participants with Barrett’s esophagus who developed a DNA content abnormality (aneuploidy and/or increased 4N) during surveillance (mean 104.5 months, range 42.2 to 208.3 months) but in whom no TP53 mutation or 17p LOH was detected at baseline (M:F=3:9, mean age 63). During this follow-up period, 6 of 12 participants had a diagnosis of high-grade dysplasia and one participant developed cancer after 14 years. DNA content abnormalities and high-grade dysplasia are known risk factor for progression to EA(48);
[Advanced BE] 10 Barrett’s mucosal biopsies within premalignant Barrett’s epithelium from 10 surgical resections in participants with microscopic EAs, including nine intramucosal and one with superficial submucosal invasion detected during endoscopic surveillance (mean BE surveillance 21.1 months, range 4.2 to 42.3 months, M:F=2:8, mean age 63) and
[EA] 8 large esophageal adenocarcinoma specimens averaging 4.8 cm in diameter (range 1.5 cm to 9 cm) from the surgical resection specimens of 8 patients (M:F=0:8, mean age 75).
DNA content flow cytometry to determine ploidy of BE/EA biopsies
Whole biopsy specimens and gastric controls, including fresh/frozen endoscopic biopsies (n = 48) or surgical resection specimens (n=36), were processed. For the BE and EA samples, whole endoscopic biopsies (n=24) or surgical resection specimens (n=18) were minced in buffer used for preparing tissue for flow cytometry (146 mM NaCl, 10mM Tris Base (pH 7.5), 1 mM CaCl2, 0.5 mM MgSO4, 0.05% Bovine serum albumin (BSA), 21 mM MgCl2, 0.2% Igepal CA-630 Sigma I3021 as previously described(50). Approximately 90ul of unfixed nuclei were saturated with DAPI (5 µg/ml, Accurate Chemical, Westbury, NY) and evaluated for DNA content on a Cytopeia inFlux flow cytometer (Seattle, WA). Listmode files were analyzed using Multicycle (Phoenix Flow Systems, San Diego, CA) as described previously(51, 52).
SNP-based copy gain, loss, and LOH using Illumina Infinium technology
DNA from the remaining minced tissue of each whole biopsy was extracted using standard Puregene DNA Isolation Kit as recommended by the manufacturer (Gentra Systems, Inc. Minneapolis, MN), and quantitated with the Quant-iT™ PicoGreen® dsDNA Assay Kit following manufacturers protocol (Invitrogen) on the CytoFluor II plate reader. DNA for SNP arrays was not purified by flow cytometry because this study is designed to identify abnormalities that ultimately may be assessed in whole biopsies without further purification in a clinical setting. Each sample was genotyped using a Illumina 12-sample Human Hap300_Pool10 Beadchip targeting 33,423 SNP loci (33K SNP array) from the HumanHap300 BeadChip and processed at Illumina using standard multi-sample Infinium protocols and reagents according to manufacturer’s instructions. DNA (average 223.8ng input) was amplified, enzymatically fragmented, precipitated, resuspended in hybridization buffer, denatured, and hybridized to the BeadChip overnight at 48°C. After the overnight incubation, the BeadChip was washed, primer extended, and stained on a Tecan Genesis/EVO robot using a Tecan GenePaint slide processing system. After staining, BeadChips were washed and coated, then scanned using Illumina’s BeadArray Reader. The probe intensity data were extracted and analyzed using Illumina's BeadStudio 3.1 software.
Genotyping data consists of two channel intensity data corresponding to the two alleles. Data is generated as rectangular coordinates of the raw A versus raw B allele intensities. After normalization with BeadArray Reader, the genotyping data were transformed to a polar coordinate plot of normalized intensity R = Xnorm + Ynorm and allelic intensity ratio theta=(2/pi)* arctan (Ynorm / Xnorm), where Xnorm and Ynorm represent transformed normalized signals from alleles A and B for a particular locus. For each patient, a pair of normal gastric (reference) and a BE/EA sample (subject) was processed in paired mode, using default reference clustering. The B allele frequency parameter, based upon the theta values, is calculated for each locus as described previously (53).
Statistical Methods
We examined the MA plots (signal intensity vs. signal ratio plot) for each sample and found no bias. Paired constitutive controls for each patient were used to ensure that all abnormalities detected were somatic lesions and not constitutive copy number variations that could arise when using a single reference sample for all patients(54, 55). Paired sample analysis of BE or EA samples (defined as “sub”) paired with the control gastric sample (defined as “ref”) from each patient generated two parameters, the log-ratio log2(Rsub/Rref) and |dAlleleFreq sub-ref| that were used for copy number and LOH detection(53). The |dAlleleFreq sub-ref| represents the absolute value of the difference in allele frequency between the subject (BE or EA) and reference (paired constitutive control) samples. Since both samples are from the same patient, the difference of allele frequency at any given locus should be approximately zero except in regions exhibiting somatic chromosomal anomalies such as copy number differences or LOH. Two stages of data analysis were performed in this study. The first identified copy gain, copy loss, and LOH for each chromosome arm. The second tested whether or not the frequency of a gain, loss or LOH identified in a given segment along the chromosome arm reached statistical significance across patients.
Specifically, we defined the normal baseline copy number (2N) on each chromosome using |dAlleleFreq sub-ref|. After wavelet processing (see below) of |dAlleleFreq sub-ref|, regions with signal values including at least 10% contiguous SNPs on either chromosome arm with a |dAlleleFreq sub-ref| equal to zero were identified. These regions were used to set the baseline chromosome copy number (adjusted Log2(Rsub/Rref) to zero and was applied to specifically to that chromosome). If a contiguous region of 10% of SNPs could not be found on a chromosome, then no adjustment was made for a baseline copy number. A wavelet method was used to smooth both copy number Log2(Rsub/Rref) and |dAlleleFreq sub-ref| signal values(56, 57). Daubechies3 wavelets were used in this study, and array signal was decomposed to the second level with the wavelets and reconstructed following dynamic thresholding (depending on the signal to noise ratio for that sample). For a specific constitutive genotype “AB”, after above wavelet procedure processing (including thresholding), probes with Log2(Rsub/Rref) signal values that deviated from zero were identified as either copy gain (allelic imbalance, e.g. AAB or balanced, e.g. AABB) or loss (deletion or homozygous deletion). Probes with |dAlleleFreq sub-ref| which deviated from zero after wavelet processing including thresholding were identified to be LOH (allelic imbalance, e.g A and AAB or copy neutral LOH, e.g.AA). Infrequently, there were a few noncontiguous probe signal levels that were more than four standard deviations from the mean signal of a chromosome arm. Since some of those signal values could be smoothed out by wavelets, those signal values were included as lesions for that patient and subjected to consistency testing in the second stage of analysis. A Kruskal-Wallis test was used to compare the median number of lesions and sizes of lesions in each group (Figure 1 and 2). For the size of events, we defined a gain, loss or LOH segment as continuous until there were at least two adjacent probes without that lesion. Thus, the boundaries of a lesion segment were defined by at least two or more probes without the lesion (Figure 2). Poisson regression was used to test the frequency of the lesion in each group (Figure 2).
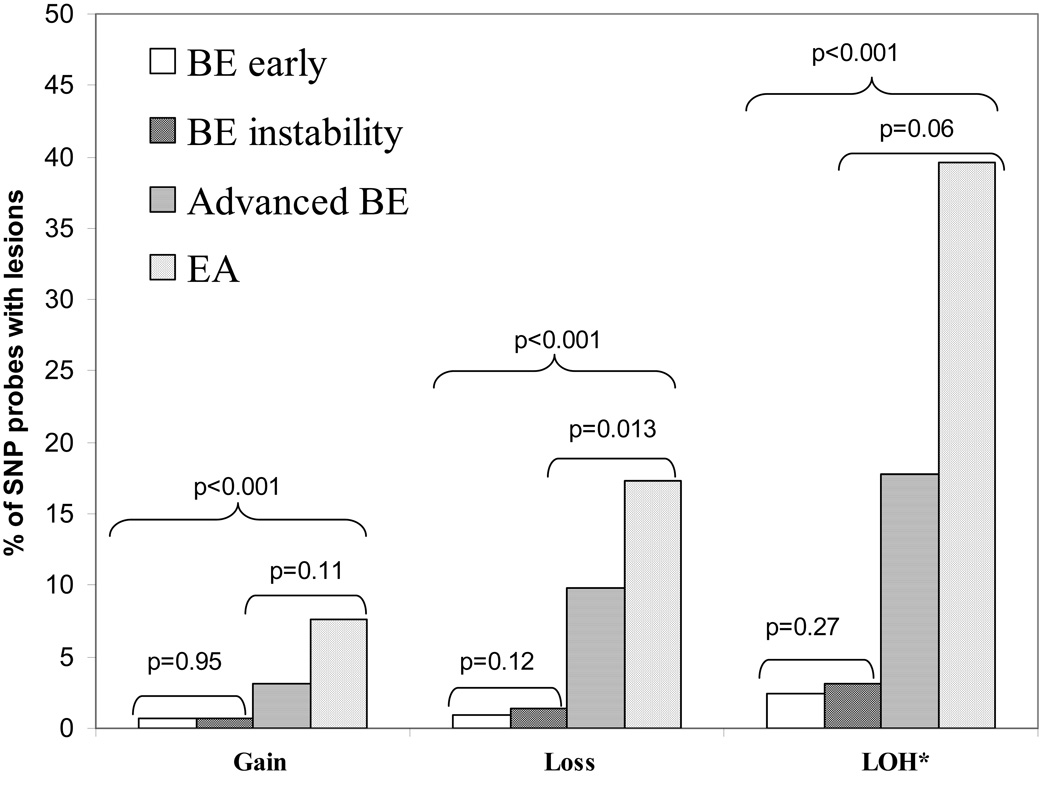
Figure 1.
Percent of probes across the genome with copy gain, loss and LOH in different stages of neoplastic progression measured in the 33K SNP array. * % SNP probes with LOH was calculated using only those SNPs that were heterozygous in the paired constitutive control.
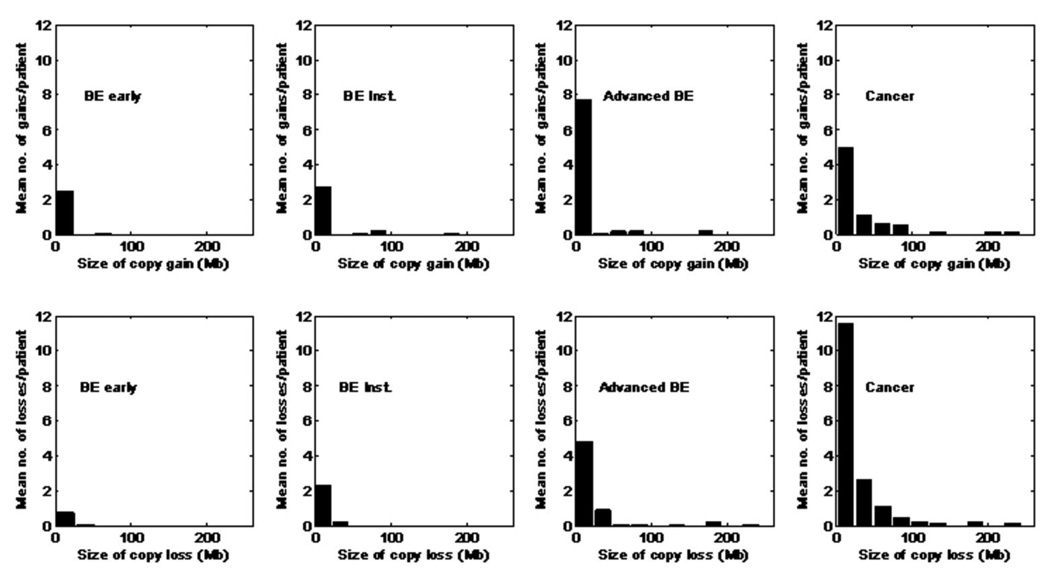
Figure 2.
Gain and loss size distribution of the four progression stages.
In the second stage of analysis the significance of frequencies of lesions across the patients was tested. Copy number gain, loss, and LOH, were tested separately along the chromosome arms. Specifically for each of the three types of lesions, every chromosome arm was divided into 0.5Mb windows, although the last window may be smaller than 0.5Mb. If one or more lesions were included in a window in the first stage of the analysis, then one event was counted for that window. If no lesion was called in the first stage, the window was given a value of zero. Fisher’s exact test was used to test the number of events across the patients of a comparison group. We compared the lesion frequency within each window in both the early and late stage separately, relative to the null hypothesis of no lesion in a window. The p-values of these comparisons were adjusted to have a false positive discovery number of 1 or less(58). Starting and ending positions of any given lesion on a chromosome may be different across multiple patients. Therefore, to test whether a lesion was common among patients, we used the window method as a practical approach, considering sample size and SNP informativity rates. The 0.5Mb window size was determined by considering the specific array probe density (coverage of the genome) and SNP informativity (heterozygosity) rate for LOH detection(59). Larger windows have higher probability of containing an informative SNP, but the resolution for detection is lower; whereas a smaller window will have higher resolution but may not contain informative SNPs. Using an average SNP informativity rate of 0.26 in dbSNP and considering the multi-sample Illumina 33K BeadChip density, we selected a 0.5Mb window size in order to have 0.8 LOH detection power. We evaluated window sizes from 0.1–1.0 Mb and obtained essentially identical results. Empirical Receiver Operating Curves were generated according to previously described methods(60). All the analyses were carried out in Matlab (version 7.1, The Mathworks Inc.) and Statistical Analysis System (SAS version 9.1, SAS Institute Inc.)
Results
Molecular and pathology criteria defining stages of progression
The 42 pairs of biopsies evaluated in this study were obtained from 42 patients including 6 women and 36 men of mean age 64.3 years (range 43 to 93). Table 1 depicts the participant selection criteria with regard to molecular and pathology measures for each stage of progression. Patients were classified using previously evaluated flow-cytometrically purified biopsies (mean 9.64 biopsies per patient) for 17p LOH and TP53 mutation (at baseline only), and DNA content abnormalities by flow cytometry as described previously(29, 43).
Table 1.
Molecular and pathologic stages of progression.
| Patient Characteristics at Time of Sampling |
EA Risk Biomarkers* in Follow-up |
||
|---|---|---|---|
| “Early stage” | BE early (N=12) |
• No biomarker abnormalities* | • 0/12 develop DNA content abnormality • 0/12 develop high-grade dysplasia |
|
Endoscopic biopsies from Barrett’s epithelium |
BE instability (N=12) |
• No biomarker abnormalities* | • 12/12 develop DNA content abnormality • 6/12 with diagnosis of high-grade dysplasia |
| “Late stage” | Advanced BE (N=10) |
•17pLOH,TP53 mutation • Aneuploidy • Pre-operative endoscopic diagnosis of small, early cancer; samples from |
Not applicable |
|
Surgical resection specimens | |||
| EA tumor (N=8) |
•17pLOH,TP53 mutation •Aneuploidy • Samples from large EA tumor mass |
Not applicable | |
Biomarkers= 17pLOH (spanning TP53), TP53 mutation, DNA content abnormality (Aneuploidy and/or Increased 4N), high-grade dysplasia
Genome-wide chromosome instability in different stages of neoplastic progression
Genome-wide chromosome instability was evaluated in patient samples representing the four different stages of neoplastic progression described in Table 1. Figure 1 shows the average percent of SNPs with copy gain or loss abnormalities, and percentage of informative SNPs with allelic imbalance due to copy gain, or loss of heterozygosity [LOH] including copy loss and copy neutral events using paired constitutive controls (see methods). The number of SNPs with copy gain, loss, and LOH show a highly statistically significant trend of increasing number of altered SNPs across each of the four progression stages (p<0.001). In early stages of progression, very few SNPs have copy number or LOH abnormalities (Figure 1). In late stages of BE progression, the frequency of SNP alterations increased dramatically (Figure 1). Direct comparisons of number of SNP probes with gain, loss and LOH between the early stages of progression (BE early vs. BE instability groups), and also between the late stages of BE (advanced BE and EA groups) did not reach p=0.01 significance level, given that the statistical power of the tests are small when the sample size of each group is not large.
To distinguish between a high a number of small chromosomal events versus a small number of larger events, for example 100 regions each including 10 SNPs, compared to 10 regions each including 100 SNPs, we assessed the size of altered chromosomal copy number regions. Figure 2 shows the distribution of copy gain and loss segment sizes in the four stages of BE progression.
There was a trend for increased copy gain segment size with progression stages (p<0.01). However, copy gain frequency and segment size did not reach statistical significance between early BE and BE instability stages, given the samples sizes of comparison groups (p=0.65, p=0.70 respectively, Figure 2). Similarly, the frequency and segment size of copy gain also did not reach statistical significance at p=0.01 between advanced BE and EA stages (p=0.84, p=0.04, respectively). However, when the combined early stages (BE early and BE instability) were compared to the combined late stages (advanced BE and EA), the frequency and segment size of copy gain were both significantly different (p<0.001).
There was a statistically significant difference in the number of regions of copy loss between the BE early and BE instability groups (p=0.005, Figure 2). However, many of these regions were based on loss of a single SNP. Neither the total number of SNPs with loss (p=0.12, Figure 1) nor the median sizes of loss (p=0.20, Figure 2) were different in the two groups. Similarly, neither frequency nor segment size of copy loss reached a p=0.01 level of statistical significance between advanced BE and EA (p=0.04, p=0.23, respectively) given the sample size. However, when the combined early stages were compared to the combined late stages, frequency and segment size of copy loss were both significantly different (p<0.001).
SNP distribution patterns along the chromosome and the heterozygosity rate of specific SNPs affect the ability to call LOH segment sizes precisely(59). Therefore, we plotted the distribution of LOH using allele frequency differences between paired constitutive controls and BE/EA samples (|dAlleleFreq sub-ref|) for each informative probe displayed by chromosome in each sample (Figure 3).
Figure 3.
A. Distribution of informative SNPs with LOH (black bars) on chromosomes 1 to 5 (rows, oriented p-arm at the bottom and q-arm at the top of each chromosome) in individual samples from 42 patients (columns). Regions with no black bars represent either non-informative SNPs or no LOH. Patients are grouped along the x-axis into the four stages of neoplastic progression and the numbers along the x axis refer to individual patients.
B. Genome wide LOH map of chromosomes 6 to 12 for individual samples (n=42). The organization is the same as in Figure 3A.
C. Genome wide LOH map of chromosomes 13 to 22 of individual samples (n=42). The organization is the same is in Figure 3A.
Somatic copy gain, loss and LOH lesions might confer a selective advantage to the clone. Alternatively, a lesion may be the result of random, non-selected events arising during neoplastic evolution, especially in later stages of progression after inactivation of TP53. Therefore, we examined the frequency of each lesion across patients. The numbers of patients in each of the four stages are small and the results from Figures 1 and 2 indicate that BE early and BE instability groups have statistically similar magnitudes (albeit not the same) of copy gain, loss, and LOH frequencies and sizes relative to each other as compared with advanced BE and EA groups. Therefore, patients from the BE early and BE instability groups were combined into a single early stage group in order to get more power for lesion detection. Also, advanced BE and EA have similar (albeit not the same) magnitude of instability relative to each other as compared to the two early groups. Therefore, the advanced BE and EA patients were combined into a late stage group and evaluated separately. Statistically significant levels of chromosome copy change and LOH events among patients are shown for early stage (supplemental Figure 1A, 1B) and late stage (supplemental Figure 1C, 1D). A complete list of statistically significant abnormalities identified in each 0.5Mb window detected in early and late stages of progression are presented in supplement materials (Table S1 and Table S2).
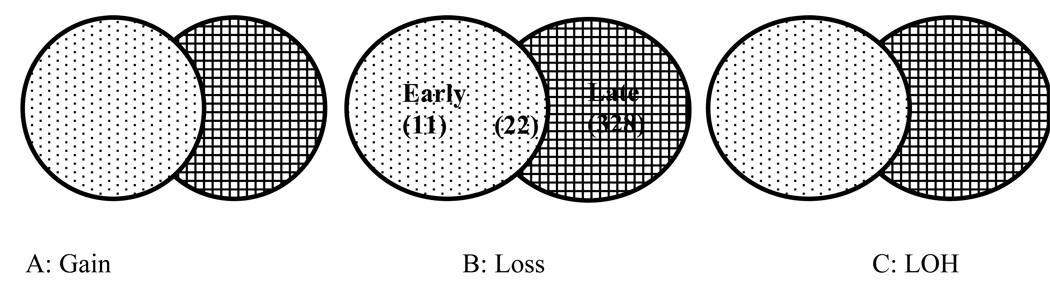
We also evaluated the relationship between the significant alterations identified independently in genome-wide analysis of early (BE early and BE instability) and late (Advanced BE and EA) stages of progression. Figure 4 illustrates the relationships among the significant copy gain (Figure 4A), copy loss (4B) and LOH (4C) windows. Of the 56 0.5Mb windows identified with copy gain in late stages, only 2 of 56 (4%) also reached statistical significance in early stages of progression. Of the windows identified as reaching statistical significance in late stages, 22 of 350 (6%) copy loss and 71 of 878 (8%) LOH events also reached statistical significance in early stages of progression. The specific lesions and locations that were common in both early and late stages are shown in Table 2. For the patients in this study, the average rate of informative SNPs on this array is 0.33. Thus, there is a significant probability for small segments with copy loss to have no informative SNPs available for LOH analysis. The LOH events listed in Table 2 that did not reach statistical significance in the small regions of copy loss on chromosomes 1, 4, 10, 11, 12, 18, and 20 were mainly due to lack of informative SNPs.
Figure 4.
Number of 0.5MB windows with chromosome lesions that reached statistical significance across patients in Early (BE early and BE instability) and Late (advanced BE and EA) stages of neoplastic progression. Light dots: The number of 0.5 MB windows that reached statistical significance in early stage (BE early and BE instability) samples only but were not significant in late stage samples. Intersection of dots and cross hatched: The number of events that reached statistical significance both in early stage (BE early and BE instability) and late stage (advanced BE and EA) samples. Cross hatched: The number of events that reached statistical significance in late stage (advanced BE and EA) samples only but were not significant in early stages.
Table 2.
Lesions that reached statistical significance in both early and late stages of progression.
| Lesion type | [Chromosome] (location and range in ± 0.5MB) |
|---|---|
| Gain | [8] (145.5); [10] (73.5) |
| Loss |
[1] (186.6); [3] (59.8~60.8); [4] (33.9); [9] (9.0~12.1, 20.5~25.0, 28.5~30.5, 44.5); [10] (68.0); [11] (38.5); [12] (28.3); [16] (77.3); [18] (64.9); [20] (22.0) |
| LOH | [3] (59.8 ~ 60.8); [9] (0.5 ~ 38.1); [16] (77.3) |
Comparison of genome-wide SNP-based LOH to flow cytometric DNA content aneuploidy in unpurified, whole biopsies
DNA content abnormalities as measured by flow cytometry have been reported to be useful for cancer risk stratification in BE(48, 61, 62). To test the ability of a SNP platform to detect populations of cells with abnormal DNA content, we minced each biopsy, divided the homogenate in half, and processed half using the multi-sample Illumina 33K SNP arrays and the other half by DNA content flow cytometry using our standard protocol and analysis as described in methods. Multiple genetic mechanisms can generate allelic imbalance resulting in LOH measured by allele frequency differences between paired constitutive controls and BE/EA samples (|dAlleleFreq sub-ref|)| including copy gain in a heterogenous population, copy loss and copy neutral LOH(55, 63). Although small regions of LOH may not be detected if there are insufficient informative SNPs, genome-wide chromosomal instability arising in a population of cells would be detected by the LOH signal of a SNP-based platform, with the exception of balanced copy number alterations such as a pure tetraploid population or pure population with homozygous deletions. We compared genome-wide LOH results obtained in unpurified biopsies to DNA content flow cytometry for aneuploidy detection in the same biopsies (Table 3). Aneuploidy was detected by flow cytometry in 11 of 42 biopsies. A threshold of ≥ 1000 SNPs with LOH throughout the genome (gw-LOH), which represents about 9% of the average number of informative heterozygous SNPs on the 33K platform, provided the optimal sensitivity and specificity for discriminating flow-cytometric diploid and aneuploid samples. 10 of 11 (~91%) of the aneuploid populations that were detected by flow cytometry had ≥ 1000 SNPs with LOH. The one exception was the only near-diploid aneuploid sample in the study. This sample had a flow-cytometric DNA content of 2.3N. Patients with near-diploid aneuploid populations are at lower risk of progressing to EA(61). There were 7 samples with ≥ 1000 SNPs with LOH but which were diploid by flow cytometry. Six of seven samples were from patients with EA or advanced BE. Thus, gw-LOH has potential to be a surrogate for flow cytometric DNA content aneuploidy.
Table 3.
Comparison of SNP-based genome wide LOH relative to flow cytometric DNA content in unpurified, whole biopsies.
6/7 patients were advanced BE or EA.
2.3N near-diploid aneuploid by flow-cytometry
The relationship between genome wide LOH (33K BeadChip) and stages of progression
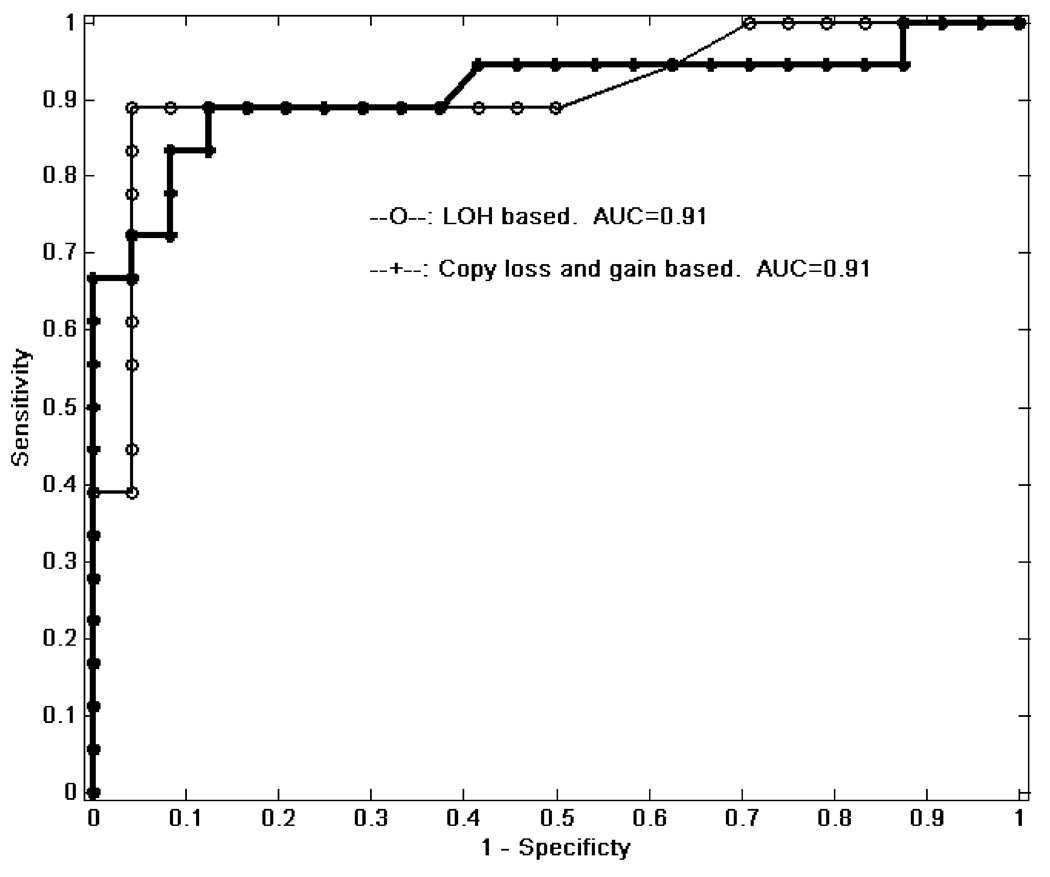
We also evaluated the sensitivity and specificity of genome wide LOH in whole biopsies using the 33k array for distinguishing patients with or without concurrent cancer. The total number of informative SNPs with LOH (LOHTotal) across all 22 chromosomes was summed for each patient. The mean LOHTotal for early stages (BE early and BE-instability) was 456 (standard error = 123) compared to 3445 (standard error = 587) for advanced stages (advanced BE and EA). If LOHTotal was used to distinguish between early and late stages, we obtained an AUC of 0.91 as shown in the empirical ROC curves with the raw data in Figure 5. The results were essentially identical using paired samples for copy gain and loss.
Figure 5.
ROC curve for separating advanced stages (advanced BE and EA) vs. early stages (BE or BE instability) using total genome wide LOH (AUC=0.91) or copy gain and loss (AUC=0.91).
Finally, we compared genome wide LOH measurements to DNA content flow cytometric aneuploidy for sample classification of early and late stages of progression (Table 4). The results are promising for SNP-based assessment of aneuploidy for cancer risk in whole, unpurified biopsies.
Table 4.
Comparison of DNA content flow cytometry alone and genome wide LOH for discriminating progression stages.
| Tissue type | Diploid | Aneuploid | gw-LOH (<1000) |
gw-LOH (≥1000) |
|---|---|---|---|---|
| BE-Early or BE-instability |
24 | 0 | 23 | 1 |
| Advanced BE or EA |
7 | 11 | 2 | 16 |
Discussion
A previous chromosome instability biomarker panel for cancer risk stratification in Barrett’s esophagus required Ki67/DNA content multi-parameter flow cytometric cell sorting for enrichment of proliferating epithelial cells, a panel of STR polymorphisms on chromosomes 9 and 17 for LOH analysis, and DNA content flow cytometric assessment of tetraploidy and aneuploidy (43). Although this chromosome instability panel accurately identified patients at high- and low risk for progression to EA, it required a constellation of technologies that are difficult to perform outside of research centers. Here, we report that 9p and 17p LOH could be detected in whole, unprocessed biopsies, and that genome-wide SNP based measures of LOH and copy number performed as well or better than DNA content flow cytometry for detection of aneuploidy, providing a significant advance towards validating the biomarkers in a clinically compatible, SNP DNA array platform. A similar platform might also be useful in assessing cancer risk in oral leukoplakia, based on the previous panel of 9p and 3p LOH, TP53 abnormalities and chromosome polysomy (43). We used SNP DNA arrays on neoplastic and paired constitutive control samples from each individual to provide better resolution for smaller genomic events, improve signal-to-noise ratios, and differentiate constitutive copy number variation from somatic genetic events that arise during neoplastic progression. Since DNA arrays can measure LOH, copy number, and known mutations as well as DNA methylation, in which bisulfite treatment produces a C→T SNP at CpG sites, this technology may provide a common platform to compare these biomarkers in validation studies to efficiently select those markers most appropriate for patient management.
In this study, we demonstrated feasibility for SNP-based assessment of somatic copy number gain, loss and LOH in patients at different stages of neoplastic progression as well as a genome-wide measure of aneuploidy. Validation of a common biomarker platform as well as validation of reliable markers for risk stratification in BE will require one or more large cohort studies. The ideal cohort study would include patients at all levels of EA risk and the biomarker(s) would be evaluated on prospectively collected samples. Testing of multiple biopsies, for example, one biopsy every two cm, from each endoscopic procedure for a given patient will likely increase reliability because neoplastic clones occupy variably sized regions of a Barrett’s segment. Well designed cohort studies using a common platform to detect LOH, copy change, and DNA methylation abnormalities should be able to identify a small set of unbiased, reliable markers for BE risk stratification. Validated biomarkers from the cohort studies could be translated into a less expansive, clinically compatible SNP analysis platform, such as custom GoldenGate, VeraCode BeadExpress, or Pyrosequencing assays, for routine, large-scale sample testing in the clinical setting. New tools such as massively parallel sequencing technologies are likely to significantly advance our understanding of the underlying somatic-genetic mechanisms in cancer, including detection of tandem duplications, balanced translocations or inversions, and discovery of as yet unidentified critical genes. However, these technologies are not yet feasible for large scale cohort studies or clinical application.
Using paired constitutive controls, we consistently detected recurrent copy gains, losses and LOH in unpurified biopsies even in early benign BE that showed no evidence of progression to EA for a mean of 10 years. At advanced stages of progression, the neoplastic genome contained frequent and extensive regions of copy gain, loss and LOH, consistent with a recent report characterizing EA using high-density SNP arrays(2). To characterize chromosomal instability during neoplastic progression and identify regions that may contain genes useful for cancer prevention, cancer risk stratification and early detection, we examined chromosomal abnormalities found only in patients at early stages of progression, those common in patients at both early and late stages and those only detected in patients at late stages of progression. Only a small number of abnormalities were frequent in early stages of progression, and with the single exception of chromosome arm 9p, the sizes of chromosome abnormalities were small at early stages compared to late stages. Of the significant 0.5Mb windows with copy gain, loss and LOH identified in late stages of progression, only 4%, 6%, and 8%, respectively, also reached statistical significance in early stages of progression. Chromosome arm 9p was unusual in the sense that it acquired large regions of copy loss and LOH spanning most of the arm at early stages of progression. Within these large 9pLOH events we found three statistically significant, distinct regions of copy loss spanning 9.0~12.1, 20.5~25.0, 28.5~30.5 Mb, with the most frequent being potential homozygous deletions at the CDKN2A/ARF locus, which have been reported previously in multiple cancer types(2, 64–67). CDKN2A abnormalities have been associated with large clonal expansions in BE as well as other conditions (14, 36, 37, 68), and may be selected in BE as part of an adaptation to a harsh acid reflux environment (25–28). Additionally, chromosome 9 is highly structurally polymorphic with more than 1000 annotated genes and has evolved many intra- and interchromosomal duplications(69, 70). Two other common abnormalities were significant in early stage BE including copy loss spanning the FHIT (3p, 59.8 ~60.6 Mb) and WWOX (16q, 77.3 Mb) loci, detected in paired samples in 46% and 24% of early stage patients. These regions have been extensively studied in EA and other cancer types (71–82), and recently Nancarrow et al reported these regions as the two most common regions of homozygous deletions in EA, detected in 74% and 35%, respectively(2). This is consistent with the patients with advanced BE and EA in the present study in whom we detected FHIT and WWOX copy loss in 78% and 56%, respectively, in the late stage biopsies. Other relatively small lesions that contain multiple genes reached statistical significance among patients at early stages such as copy gain on 8q24.3 and 10q22.1.
The paradox of specific chromosomal abnormalities that appear to be selected both in early and late stages of progression is that these events occur too frequently in early stages to be sufficient for development of EA because the rate of progression to EA is very low in most persons with BE(43, 48, 61, 83–86). This suggests several possibilities. First, these early, high-frequency events might be necessary but not sufficient for progression. Second, the frequent, early events might be selected as part of the adaptation to gastroesophageal reflux, but have relatively little involvement in progression to EA, and could appear in late stages as hitchhikers (“passengers”) on selected events that drive progression to EA. Third, they might represent regions susceptible to chromosome damage such as fragile sites(87) that could then hitchhike on selected abnormalities such as an epigenetically modified progenitor cell population(88). Regardless of mechanism, patients with early stage BE appear to have a low risk of progressing to, or dying of, EA(23, 24, 43, 48, 61, 83–86, 89, 90). Effective identification of high risk patients will be required to guide cancer prevention strategies in BE, focusing on patients who may benefit from the intervention because their risk of cancer is high, and reassuring low-risk patients who are unlikely to benefit from an intervention targeting only the BE mucosa.
In this manuscript, we also report that frequent loss or LOH involving whole chromosome arms were observed in late stages of progression, including 3p, 5, 9p, 13, 17p and 18q (Figures 1 to 3 and supplemental Figure 1, Table S2), many of which have recently been reported and reviewed (55), as well as in earlier low density STR allelotypes(45–47, 91). Large recurring regions of copy gain, loss and LOH may provide reproducible assessment of aneuploidy by higher density SNP platforms and this could be combined with an array assessment of more localized regions of copy gain, loss or LOH in a common platform to assess chromosomal instability for risk stratification in BE and other conditions. We also confirmed 17p status in the SNP arrays compared to previous STR results in 38 of 42 patients (90%) even though we did not use the same biopsies in the current study and at least two of the discordant cases could be proven to be due to sampling limitations because the samples also lacked aneuploid populations that were present in the original biopsies (data not shown).
Our study has limitations, including the relatively small sample size in each of the participant categories, and validation of our results will require large cohort studies. However, our results provide important insight into the level of chromosomal instability relative to stages of neoplastic progression in BE. In addition, supervised statistical methods could be used for sample classification, but we chose not to prematurely exclude or include markers from the present study because of the limited sample sizes and probe densities. Further, although the current SNP platforms are potentially technically limited in detecting a perfect 4N tetraploid population, a large, pure tetraploid clone may be rare in an evolving neoplastic population of cells. Despite these potential limitations, this study succeeded in proof of concept that LOH, copy number and aneuploidy can be assessed in unprocessed whole biopsies from Barrett’s esophagus using a SNP array. By assessing chromosome instability in premalignant and malignant stages of neoplastic progression, the results can guide future hypothesis-driven research to better evaluate genomic abnormalities for cancer risk stratification, early detection and identification of candidate targets for cancer prevention.
In summary, our results indicate that SNP array assessment of genome-wide chromosomal changes that develop during neoplastic progression provide promise for improved EA risk stratification in patients with BE, and this hypothesis, as well as others based on DNA methylation, can be efficiently tested in cohort studies, using prospectively accumulated biospecimens (EDRN phase 4)(92). This single-platform assessment of chromosome instability biomarkers might also be used to identify patients at high risk of developing EA for cancer prevention trials.
Supplementary Material
Acknowledgements
This study was funded by NIH P01CA91955 and CCM was partially supported by the Commonwealth Universal Research Enhancement Program at the Pennsylvania Department of Health and the Pew Charitable Trust.
We thank David Cowan for database management, Christine Karlsen for patient care coordination, Valerie Cerera for flow-cytometry. Illumina, BeadArray, and Infinium are registered trademarks or trademarks of Illumina, Inc.
References
- 1.Nowell PC. The clonal evolution of tumor cell populations. Science. 1976;194:23–28. doi: 10.1126/science.959840. [DOI] [PubMed] [Google Scholar]
- 2.Nancarrow DJ, Handoko HY, Smithers BM, et al. Genome-wide copy number analysis in esophageal adenocarcinoma using high-density single-nucleotide polymorphism arrays. Cancer Res. 2008;68:4163–4172. doi: 10.1158/0008-5472.CAN-07-6710. [DOI] [PubMed] [Google Scholar]
- 3.Croft J, Parry EM, Jenkins GJ, et al. Analysis of the premalignant stages of Barrett's oesophagus through to adenocarcinoma by comparative genomic hybridization. Eur J Gastroenterol Hepatol. 2002;14:1179–1186. doi: 10.1097/00042737-200211000-00004. [DOI] [PubMed] [Google Scholar]
- 4.El-Rifai W, Frierson HF, Jr, Moskaluk CA, et al. Genetic differences between adenocarcinomas arising in Barrett's esophagus and gastric mucosa. Gastroenterology. 2001;121:592–598. doi: 10.1053/gast.2001.27215. [DOI] [PubMed] [Google Scholar]
- 5.Moskaluk CA, Hu J, Perlman EJ. Comparative genomic hybridization of esophageal and gastroesophageal adenocarcinomas shows consensus areas of DNA gain and loss. Genes Chromosomes Cancer. 1998;22:305–311. [PubMed] [Google Scholar]
- 6.Cifola I, Spinelli R, Beltrame L, et al. Genome-wide screening of copy number alterations and LOH events in renal cell carcinomas and integration with gene expression profile. Mol Cancer. 2008;7:6. doi: 10.1186/1476-4598-7-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gaasenbeek M, Howarth K, Rowan AJ, et al. Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex changes and multiple forms of chromosomal instability in colorectal cancers. Cancer Res. 2006;66:3471–3479. doi: 10.1158/0008-5472.CAN-05-3285. [DOI] [PubMed] [Google Scholar]
- 8.Oosting J, Lips EH, van Eijk R, et al. High-resolution copy number analysis of paraffin-embedded archival tissue using SNP BeadArrays. Genome Res. 2007;17:368–376. doi: 10.1101/gr.5686107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kloth JN, Oosting J, van Wezel T, et al. Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer. BMC Genomics. 2007;8:53. doi: 10.1186/1471-2164-8-53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Harada T, Chelala C, Bhakta V, et al. Genome-wide DNA copy number analysis in pancreatic cancer using high-density single nucleotide polymorphism arrays. Oncogene. 2008;27:1951–1960. doi: 10.1038/sj.onc.1210832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lindblad-Toh K, Tanenbaum DM, Daly MJ, et al. Loss-of-heterozygosity analysis of small-cell lung carcinomas using single-nucleotide polymorphism arrays. Nat Biotechnol. 2000;18:1001–1005. doi: 10.1038/79269. [DOI] [PubMed] [Google Scholar]
- 12.Weir BA, Woo MS, Getz G, et al. Characterizing the cancer genome in lung adenocarcinoma. Nature. 2007;450:893–898. doi: 10.1038/nature06358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lo KC, Bailey D, Burkhardt T, et al. Comprehensive analysis of loss of heterozygosity events in glioblastoma using the 100K SNP mapping arrays and comparison with copy number abnormalities defined by BAC array comparative genomic hybridization. Genes Chromosomes Cancer. 2008;47:221–237. doi: 10.1002/gcc.20524. [DOI] [PubMed] [Google Scholar]
- 14.Maley CC, Galipeau PC, Li X, et al. The combination of genetic instability and clonal expansion predicts progression to esophageal adenocarcinoma. Cancer Res. 2004;64:7629–7633. doi: 10.1158/0008-5472.CAN-04-1738. [DOI] [PubMed] [Google Scholar]
- 15.Mei R, Galipeau PC, Prass C, et al. Genome-wide detection of allelic imbalance using human SNPs and high-density DNA arrays. Genome Res. 2000;10:1126–1137. doi: 10.1101/gr.10.8.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuziak T, Jeong J, Majewski T, et al. High-resolution whole-organ mapping with SNPs and its significance to early events of carcinogenesis. Lab Invest. 2005;85:689–701. doi: 10.1038/labinvest.3700270. [DOI] [PubMed] [Google Scholar]
- 17.Lai LA, Paulson TG, Li X, et al. Increasing genomic instability during premalignant neoplastic progression revealed through high resolution array-CGH. Genes Chromosomes Cancer. 2007;46:532–542. doi: 10.1002/gcc.20435. [DOI] [PubMed] [Google Scholar]
- 18.Pohl H, Welch HG. The role of overdiagnosis and reclassification in the marked increase of esophageal adenocarcinoma incidence. J Natl Cancer Inst. 2005;97:142–146. doi: 10.1093/jnci/dji024. [DOI] [PubMed] [Google Scholar]
- 19.Holmes RS, Vaughan TL. Epidemiology and pathogenesis of esophageal cancer. Sem Rad Oncology. 2006 doi: 10.1016/j.semradonc.2006.09.003. In press. [DOI] [PubMed] [Google Scholar]
- 20.Farrow DC, Vaughan TL. Determinants of survival following the diagnosis of esophageal adenocarcinoma (United States) Cancer Causes Control. 1996;7:322–327. doi: 10.1007/BF00052937. [DOI] [PubMed] [Google Scholar]
- 21.Brown LM, Devesa S. Epidemiologic trends in esophageal and gastric cancer in the United States. Surg Oncol Clin N Am. 2002;11:235–256. doi: 10.1016/s1055-3207(02)00002-9. [DOI] [PubMed] [Google Scholar]
- 22.Wang KK, Sampliner RE. Updated guidelines 2008 for the diagnosis, surveillance and therapy of Barrett's esophagus. Am J Gastroenterol. 2008;103:788–797. doi: 10.1111/j.1572-0241.2008.01835.x. [DOI] [PubMed] [Google Scholar]
- 23.Anderson LA, Murray LJ, Murphy SJ, et al. Mortality in Barrett's oesophagus: results from a population based study. Gut. 2003;52:1081–1084. doi: 10.1136/gut.52.8.1081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Moayyedi P, Akhtar-Danesh N, Talley NJ, Jankowski J. Mortality risks associated with Barrett's oesophagus: authors' reply. Aliment Pharmacol Ther. 2008;27:853–854. doi: 10.1111/j.1365-2036.2007.03582.x. [DOI] [PubMed] [Google Scholar]
- 25.Dixon J, Strugala V, Griffin SM, et al. Esophageal mucin: an adherent mucus gel barrier is absent in the normal esophagus but present in columnar-lined Barrett's esophagus. Am J Gastroenterol. 2001;96:2575–2583. doi: 10.1111/j.1572-0241.2001.04159.x. [DOI] [PubMed] [Google Scholar]
- 26.Jovov B, Van Itallie CM, Shaheen NJ, et al. Claudin-18: a dominant tight junction protein in Barrett's esophagus and likely contributor to its acid resistance. Am J Physiol Gastrointest Liver Physiol. 2007;293:G1106–G1113. doi: 10.1152/ajpgi.00158.2007. [DOI] [PubMed] [Google Scholar]
- 27.Tobey NA, Argote CM, Vanegas XC, Barlow W, Orlando RC. Electrical parameters and ion species for active transport in human esophageal stratified squamous epithelium and Barrett's specialized columnar epithelium. Am J Physiol Gastrointest Liver Physiol. 2007;293:G264–G270. doi: 10.1152/ajpgi.00047.2007. [DOI] [PubMed] [Google Scholar]
- 28.Ostrowski J, Mikula M, Karczmarski J, et al. Molecular defense mechanisms of Barrett's metaplasia estimated by an integrative genomics. J Mol Med. 2007;85:733–743. doi: 10.1007/s00109-007-0176-3. [DOI] [PubMed] [Google Scholar]
- 29.Barrett MT, Sanchez CA, Prevo LJ, et al. Evolution of neoplastic cell lineages in Barrett oesophagus. Nat Genet. 1999;22:106–109. doi: 10.1038/8816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Maley CC, Galipeau PC, Finley JC, et al. Genetic clonal diversity predicts progression to esophageal adenocarcinoma. Nat Genet. 2006;38:468–473. doi: 10.1038/ng1768. [DOI] [PubMed] [Google Scholar]
- 31.Califano J, van der Riet P, Westra W, et al. Genetic progression model for head and neck cancer: implications for field cancerization. Cancer Res. 1996;56:2488–2492. [PubMed] [Google Scholar]
- 32.Califano J, Westra WH, Meininger G, et al. Genetic progression and clonal relationship of recurrent premalignant head and neck lesions. Clin Cancer Res. 2000;6:347–352. [PubMed] [Google Scholar]
- 33.Lee JJ, Hong WK, Hittelman WN, et al. Predicting cancer development in oral leukoplakia: ten years of translational research. Clin Cancer Res. 2000;6:1702–1710. [PubMed] [Google Scholar]
- 34.Rosin MP, Cheng X, Poh C, et al. Use of allelic loss to predict malignant risk for low-grade oral epithelial dysplasia. Clin Cancer Res. 2000;6:357–362. [PubMed] [Google Scholar]
- 35.Mao L, Lee JS, Fan YH, et al. Frequent microsatellite alterations at chromosomes 9p21 and 3p14 in oral premalignant lesions and their value in cancer risk assessment. Nat Med. 1996;2:682–685. doi: 10.1038/nm0696-682. [DOI] [PubMed] [Google Scholar]
- 36.Wong DJ, Paulson TG, Prevo LJ, et al. p16 INK4a lesions are common, early abnormalities that undergo clonal expansion in Barrett's metaplastic epithelium. Cancer Res. 2001;61:8284–8289. [PubMed] [Google Scholar]
- 37.Eads CA, Lord RV, Wickramasinghe K, et al. Epigenetic patterns in the progression of esophageal adenocarcinoma. Cancer Res. 2001;61:3410–3418. [PubMed] [Google Scholar]
- 38.Maley CC, Galipeau PC, Li X, et al. Selectively advantageous mutations and hitchhikers in neoplasms: p16 lesions are selected in Barrett's esophagus. Cancer Res. 2004;64:3414–3427. doi: 10.1158/0008-5472.CAN-03-3249. [DOI] [PubMed] [Google Scholar]
- 39.Galipeau PC, Cowan DS, Sanchez CA, et al. 17p (p53) allelic losses, 4N (G2/tetraploid) populations, and progression to aneuploidy in Barrett's esophagus. Proc Natl Acad Sci U S A. 1996;93:7081–7084. doi: 10.1073/pnas.93.14.7081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Reid BJ, Prevo LJ, Galipeau PC, et al. Predictors of progression in Barrett's esophagus II: baseline 17p (p53) loss of heterozygosity identifies a patient subset at increased risk for neoplastic progression. Am J Gastroenterol. 2001;96:2839–2848. doi: 10.1111/j.1572-0241.2001.04236.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dolan K, Morris AI, Gosney JR, Field JK, Sutton R. Loss of heterozygosity on chromosome 17p predicts neoplastic progression in Barrett's esophagus. J Gastroenterol Hepatol. 2003;18:683–689. doi: 10.1046/j.1440-1746.2003.03048.x. [DOI] [PubMed] [Google Scholar]
- 42.Pan H, Califano J, Ponte JF, et al. Loss of heterozygosity patterns provide fingerprints for genetic heterogeneity in multistep cancer progression of tobacco smoke-induced non-small cell lung cancer. Cancer Res. 2005;65:1664–1669. doi: 10.1158/0008-5472.CAN-04-3297. [DOI] [PubMed] [Google Scholar]
- 43.Galipeau PC, Li X, Blount PL, et al. NSAIDs modulate CDKN2A, TP53, and DNA content risk for future esophageal adenocarcinoma. PLoS Med. 2007;4:e67. doi: 10.1371/journal.pmed.0040067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.McDermott KM, Zhang J, Holst CR, et al. p16(INK4a) prevents centrosome dysfunction and genomic instability in primary cells. PLoS Biol. 2006;4:e51. doi: 10.1371/journal.pbio.0040051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Barrett MT, Sanchez CA, Galipeau PC, et al. Allelic loss of 9p21 and mutation of the CDKN2/p16 gene develop as early lesions during neoplastic progression in Barrett's esophagus. Oncogene. 1996;13:1867–1873. [PubMed] [Google Scholar]
- 46.Hammoud ZT, Kaleem Z, Cooper JD, et al. Allelotype analysis of esophageal adenocarcinomas: evidence for the involvement of sequences on the long arm of chromosome 4. Cancer Res. 1996;56:4499–4502. [PubMed] [Google Scholar]
- 47.Dolan K, Garde J, Gosney J, et al. Allelotype analysis of oesophageal adenocarcinoma: loss of heterozygosity occurs at multiple sites. Br J Cancer. 1998;78:950–957. doi: 10.1038/bjc.1998.607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Reid BJ, Levine DS, Longton G, Blount PL, Rabinovitch PS. Predictors of progression to cancer in Barrett's esophagus: baseline histology and flow cytometry identify low- and high-risk patient subsets. Am J Gastroenterol. 2000;95:1669–1676. doi: 10.1111/j.1572-0241.2000.02196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Rabinovitch PS, Reid BJ, Haggitt RC, Norwood TH, Rubin CE. Progression to cancer in Barrett's esophagus is associated with genomic instability. Lab Invest. 1989;60:65–71. [PubMed] [Google Scholar]
- 50.Paulson TG, Galipeau PC, Reid BJ. Loss of heterozygosity analysis using whole genome amplification, cell sorting, and fluorescence-based PCR. Genome Res. 1999;9:482–491. [PMC free article] [PubMed] [Google Scholar]
- 51.Reid BJ, Haggitt RC, Rubin CE, Rabinovitch PS. Barrett's esophagus. Correlation between flow cytometry and histology in detection of patients at risk for adenocarcinoma. Gastroenterology. 1987;93:1–11. [PubMed] [Google Scholar]
- 52.Blount PL, Galipeau PC, Sanchez CA, et al. 17p allelic losses in diploid cells of patients with Barrett's esophagus who develop aneuploidy. Cancer Res. 1994;54:2292–2295. [PubMed] [Google Scholar]
- 53.Peiffer DA, Le JM, Steemers FJ, et al. High-resolution genomic profiling of chromosomal aberrations using Infinium whole-genome genotyping. Genome Res. 2006;16:1136–1148. doi: 10.1101/gr.5402306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Redon R, Ishikawa S, Fitch KR, et al. Global variation in copy number in the human genome. Nature. 2006;444:444–454. doi: 10.1038/nature05329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Nancarrow DJ, Handoko HY, Stark MS, Whiteman DC, Hayward NK. SiDCoN: a tool to aid scoring of DNA copy number changes in SNP chip data. PLoS ONE. 2007;2:e1093. doi: 10.1371/journal.pone.0001093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hsu L, Self SG, Grove D, et al. Denoising array-based comparative genomic hybridization data using wavelets. Biostatistics. 2005;6:211–226. doi: 10.1093/biostatistics/kxi004. [DOI] [PubMed] [Google Scholar]
- 57.Burrus CS, Gopinath RA, Guo H. Introduction to Wavelets and Wavelet Transforms. Prentice Hall; 1998. [Google Scholar]
- 58.Storey JD. A direct approach to false discovery rates. Journal of the Royal Statistical Society, Series B. 2002;64:479–498. [Google Scholar]
- 59.Li X, Self SG, Galipeau PC, Paulson TG, Reid BJ. Direct inference of SNP heterozygosity rates and resolution of LOH detection. PLoS Comput Biol. 2007;3:e244. doi: 10.1371/journal.pcbi.0030244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pepe MS. The statistical evaluation of medical tests for classification and prediction. Oxford University Press; 2003. p. 320. [Google Scholar]
- 61.Rabinovitch PS, Longton G, Blount PL, Levine DS, Reid BJ. Predictors of progression in Barrett's esophagus III: baseline flow cytometric variables. Am J Gastroenterol. 2001;96:3071–3083. doi: 10.1111/j.1572-0241.2001.05261.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Teodori L, Gohde W, Persiani M, et al. DNA/protein flow cytometry as a predictive marker of malignancy in dysplasia-free Barrett's esophagus: thirteenyear follow-up study on a cohort of patients. Cytometry. 1998;34:257–263. doi: 10.1002/(sici)1097-0320(19981215)34:6<257::aid-cyto3>3.0.co;2-s. [DOI] [PubMed] [Google Scholar]
- 63.Wongsurawat VJ, Finley JC, Galipeau PC, et al. Genetic mechanisms of TP53 loss of heterozygosity in Barrett's esophagus: implications for biomarker validation. Cancer Epidemiol Biomarkers Prev. 2006;15:509–516. doi: 10.1158/1055-9965.EPI-05-0246. [DOI] [PubMed] [Google Scholar]
- 64.Florl AR, Schulz WA. Peculiar structure and location of 9p21 homozygous deletion breakpoints in human cancer cells. Genes Chromosomes Cancer. 2003;37:141–148. doi: 10.1002/gcc.10192. [DOI] [PubMed] [Google Scholar]
- 65.Cairns P, Polascik TJ, Eby Y, et al. Frequency of homozygous deletion at p16/CDKN2 in primary human tumours. Nat Genet. 1995;11:210–212. doi: 10.1038/ng1095-210. [DOI] [PubMed] [Google Scholar]
- 66.Kabat GC, Ng SK, Wynder EL. Tobacco, alcohol intake, and diet in relation to adenocarcinoma of the esophagus and gastric cardia. Cancer Causes Control. 1993;4:123–132. doi: 10.1007/BF00053153. [DOI] [PubMed] [Google Scholar]
- 67.Kasahara T, Bilim V, Hara N, Takahashi K, Tomita Y. Homozygous deletions of the INK4a/ARF locus in renal cell cancer. Anticancer Res. 2006;26:4299–4305. [PubMed] [Google Scholar]
- 68.Califano J, Leong PL, Koch WM, et al. Second esophageal tumors in patients with head and neck squamous cell carcinoma: an assessment of clonal relationships. Clin Cancer Res. 1999;5:1862–1867. [PubMed] [Google Scholar]
- 69.Humphray SJ, Oliver K, Hunt AR, et al. DNA sequence and analysis of human chromosome 9. Nature. 2004;429:369–374. doi: 10.1038/nature02465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Park JP, Wojiski SA, Spellman RA, Rhodes CH, Mohandas TK. Human chromosome 9 pericentric homologies: implications for chromosome 9 heteromorphisms. Cytogenet Cell Genet. 1998;82:192–194. doi: 10.1159/000015097. [DOI] [PubMed] [Google Scholar]
- 71.Iliopoulos D, Guler G, Han SY, et al. Roles of FHIT and WWOX fragile genes in cancer. Cancer Lett. 2006;232:27–36. doi: 10.1016/j.canlet.2005.06.048. [DOI] [PubMed] [Google Scholar]
- 72.Lisitsyn NA, Lisitsina NM, Dalbagni G, et al. Comparative genomic analysis of tumors: detection of DNA losses and amplification. Proc Natl Acad Sci U S A. 1995;92:151–155. doi: 10.1073/pnas.92.1.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Albrecht B, Hausmann M, Zitzelsberger H, et al. Array-based comparative genomic hybridization for the detection of DNA sequence copy number changes in Barrett's adenocarcinoma. J Pathol. 2004;203:780–788. doi: 10.1002/path.1576. [DOI] [PubMed] [Google Scholar]
- 74.Mueller J, Werner M, Siewert JR. Malignant progression in Barrett's esophagus: pathology and molecular biology. Recent Results Cancer Res. 2000;155:29–41. doi: 10.1007/978-3-642-59600-1_3. [DOI] [PubMed] [Google Scholar]
- 75.van Dekken H, Vissers CJ, Tilanus HW, Tanke HJ, Rosenberg C. Clonal analysis of a case of multifocal oesophageal (Barrett's) adenocarcinoma by comparative genomic hybridization. J Pathol. 1999;188:263–266. doi: 10.1002/(SICI)1096-9896(199907)188:3<263::AID-PATH374>3.0.CO;2-Y. [DOI] [PubMed] [Google Scholar]
- 76.Michael D, Beer DG, Wilke CW, Miller DE, Glover TW. Frequent deletions of FHIT and FRA3B in Barrett's metaplasia and esophageal adenocarcinomas. Oncogene. 1997;15:1653–1659. doi: 10.1038/sj.onc.1201330. [DOI] [PubMed] [Google Scholar]
- 77.Kuroki T, Trapasso F, Shiraishi T, et al. Genetic alterations of the tumor suppressor gene WWOX in esophageal squamous cell carcinoma. Cancer Res. 2002;62:2258–2260. [PubMed] [Google Scholar]
- 78.Kuroki T, Yendamuri S, Trapasso F, et al. The tumor suppressor gene WWOX at FRA16D is involved in pancreatic carcinogenesis. Clin Cancer Res. 2004;10:2459–2465. doi: 10.1158/1078-0432.ccr-03-0096. [DOI] [PubMed] [Google Scholar]
- 79.Aqeilan RI, Kuroki T, Pekarsky Y, et al. Loss of WWOX expression in gastric carcinoma. Clin Cancer Res. 2004;10:3053–3058. doi: 10.1158/1078-0432.ccr-03-0594. [DOI] [PubMed] [Google Scholar]
- 80.Aqeilan RI, Donati V, Gaudio E, et al. Association of Wwox with ErbB4 in breast cancer. Cancer Res. 2007;67:9330–9336. doi: 10.1158/0008-5472.CAN-07-2147. [DOI] [PubMed] [Google Scholar]
- 81.Watson JE, Doggett NA, Albertson DG, et al. Integration of high-resolution array comparative genomic hybridization analysis of chromosome 16q with expression array data refines common regions of loss at 16q23-qter and identifies underlying candidate tumor suppressor genes in prostate cancer. Oncogene. 2004;23:3487–3494. doi: 10.1038/sj.onc.1207474. [DOI] [PubMed] [Google Scholar]
- 82.Pimenta FJ, Cordeiro GT, Pimenta LG, et al. Molecular alterations in the tumor suppressor gene WWOX in oral leukoplakias. Oral Oncol. 2007 doi: 10.1016/j.oraloncology.2007.08.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Thomas T, Abrams KR, De Caestecker JS, Robinson RJ. Meta analysis: Cancer risk in Barrett's oesophagus. Aliment Pharmacol Ther. 2007;26:1465–1477. doi: 10.1111/j.1365-2036.2007.03528.x. [DOI] [PubMed] [Google Scholar]
- 84.Dulai GS, Shekelle PG, Jensen DM, et al. Dysplasia and risk of further neoplastic progression in a regional Veterans Administration Barrett's cohort. Am J Gastroenterol. 2005;100:775–783. doi: 10.1111/j.1572-0241.2005.41300.x. [DOI] [PubMed] [Google Scholar]
- 85.Hage M, et al. 5-aminolevulinic acid photodynamic therapy versus argon plasma coagulation for ablation of Barrett's esophagus: a randomised trial. Gut. 2004;53:785–790. doi: 10.1136/gut.2003.028860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Sharma P, Falk GW, Weston AP, et al. Dysplasia and cancer in a large multicenter cohort of patients with Barrett's esophagus. Clin Gastroenterol Hepatol. 2006;4:566–572. doi: 10.1016/j.cgh.2006.03.001. [DOI] [PubMed] [Google Scholar]
- 87.Wang YH. Chromatin structure of human chromosomal fragile sites. Cancer Lett. 2006;232:70–78. doi: 10.1016/j.canlet.2005.07.040. [DOI] [PubMed] [Google Scholar]
- 88.Feinberg AP, Ohlsson R, Henikoff S. The epigenetic progenitor origin of human cancer. Nat Rev Genet. 2006;7:21–33. doi: 10.1038/nrg1748. [DOI] [PubMed] [Google Scholar]
- 89.van der Burgh A, Dees J, Hop WC, van Blankenstein M. Oesophageal cancer is an uncommon cause of death in patients with Barrett's oesophagus. Gut. 1996;39:5–8. doi: 10.1136/gut.39.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Solaymani-Dodaran M, Logan RF, West J, Card T. Mortality associated with Barrett's esophagus and gastroesophageal reflux disease diagnoses-a population-based cohort study. Am J Gastroenterol. 2005;100:2616–2621. doi: 10.1111/j.1572-0241.2005.00340.x. [DOI] [PubMed] [Google Scholar]
- 91.Jenkins GJ, Doak SH, Parry JM, et al. Genetic pathways involved in the progression of Barrett's metaplasia to adenocarcinoma. Br J Surg. 2002;89:824–837. doi: 10.1046/j.1365-2168.2002.02107.x. [DOI] [PubMed] [Google Scholar]
- 92.Sullivan Pepe M, Etzioni R, Feng Z, et al. Phases of biomarker development for early detection of cancer. J Natl Cancer Inst. 2001;93:1054–1061. doi: 10.1093/jnci/93.14.1054. [DOI] [PubMed] [Google Scholar]
- 93.Taylor J, Tibshirani R, Efron B. The 'miss rate' for the analysis of gene expression data. Biostatistics. 2005;6:111–117. doi: 10.1093/biostatistics/kxh021. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.