Abstract
This review describes the family of intrinsically disordered proteins, members of which fail to form rigid 3-D structures under physiological conditions, either along their entire lengths or only in localized regions. Instead, these intriguing proteins/regions exist as dynamic ensembles within which atom positions and backbone Ramachandran angles exhibit extreme temporal fluctuations without specific equilibrium values. Many of these intrinsically disordered proteins are known to carry out important biological functions which, in fact, depend on the absence of specific 3-D structure. The existence of such proteins does not fit the prevailing structure-function paradigm, which states that unique 3-D structure is a prerequisite to function. Thus, the protein structure-function paradigm has to be expanded to include intrinsically disordered proteins and alternative relationships among protein sequence, structure, and function. This shift in the paradigm represents a major breakthrough for biochemistry, biophysics and molecular biology, as it opens new levels of understanding with regard to the complex life of proteins. This review will try to answer the following questions: How were intrinsically disordered proteins discovered? Why don't these proteins fold? What is so special about intrinsic disorder? What are the functional advantages of disordered proteins/regions? What is the functional repertoire of these proteins? What are the relationships between intrinsically disordered proteins and human diseases?
1. Introduction
Proteins are the major components of the living cell. They play crucial roles in the maintenance of life, and their dysfunctions are known to cause development of different pathological conditions. Although proteins possess an almost endless variety of biological functions, one class of them, known as enzymes, biological catalysts, attracted the major attention of researchers in the early days of protein science. A catalyst is a material or substance that speeds up a chemical or biochemical reaction. Without the catalyst, such a reaction would have occurred anyway but at a much slower rate. Importantly, the catalyst is never used up in the reaction – there is always the same amount at the start and the end of the reaction.
Historically, a long-standing belief has been that the specific functionality of a given protein is determined by its unique 3-D structure. The primary origin of this structure-function paradigm is the “lock and key” hypothesis formulated in 1894 by Emil Fischer to explain the astonishing specificity of the enzymatic hydrolysis of glucoside multimers by different types of similar enzymes, where one enzyme could hydrolyze α- but not β-glycosidic bonds, and another could hydrolyze β- but not α-glycosidic bonds [1]. Based on these observations Fischer [1] wrote (as translated in [2]) “To use a picture, I would like to say that enzyme and glucoside have to fit to each other like a lock and key in order to exert a chemical effect on each other.” In this analogy, the lock is the enzyme, the key-hole is the active site of enzyme, and the key is the substrate. Similar to the situation for which only the correctly shaped key opens a particular lock, it has been hypothesized that only the correctly shaped/sized substrate (key) could fit into the key-hole (active site) of the particular lock (enzyme).
For a long period of time, the validity of “lock and key” model and its associated sequence-structure-function paradigm was unquestioned, especially after the crystal structures of proteins started to be solved by X-ray diffraction. In fact, the first determined 3-D structure of an enzyme, lysozyme, for which a bound inhibitor was co-crystallized with the protein, immediately showed that the precise locations of certain amino acid side chains is almost certainly what facilitates catalysis [3]. Since the first reports on X-ray crystallographic structures at atomic resolution for myoglobin [4, 5] and lysozyme [3], more than 61,575 protein structures have been deposited into the Protein Data Bank [6] as of November 17, 2009, most of which have been determined by X-ray diffraction but also with a small percentage of which have been determined by the newer methods based on NMR spectroscopy. These structures, especially those determined by X-ray crystallography, seemed to continue to reinforce a static view of functional protein structure, with the enzyme active site being considered to be a rigid and sturdy lock, providing an exact fit to only one substrate (key).
In reality, not all proteins are structured throughout their entire lengths. Instead, many proteins are in fact highly flexible or structurally disordered, and dozens of examples of functional yet disordered regions have been reported based on X-ray structure determination studies or based on the characterization of protein structure by other biophysical techniques [7-21]. For example, many proteins in the Protein Data Bank (PDB) have portions of their sequences missing from the determined structures (so-called missing electron density) [22, 23]. A common reason for missing electron density is that the unobserved atom, side chain, residue, or region fails to scatter X-rays coherently due to variation in position from one protein to the next, e.g. the unobserved atoms are flexible or disordered.
For several examples, comparison of the results of the X-ray and NMR analyses of the same protein has revealed that solution and crystal structures can be quite different, with solution structures being much more flexible [24, 25]. Evidently the interactions in the crystal lattice reduce protein flexibility and, in some cases, have even been observed to induce disorder to order transitions in functionally important protein regions. Thus, NMR spectroscopy is able to directly confirm the flexibility of protein segments that are missing in crystallographic experiments and can sometimes indicate flexible regions that have become rigid due to crystal contacts [26-28].
Unstructured proteins and unstructured regions can be characterized by a variety of additional biophysical and biochemical methods such as small angle X-ray scattering, Raman optical activity, circular dichroism, and protease sensitivity to name a few. Indeed, more than 20 different methods have been focused on disordered protein regions with each giving different pieces of information about the unstructured state [29-31]. When time and money permit, unstructured proteins should be studied by multiple biophysical methods in order to gain a fuller understanding of their characteristics [29-31].
Some proteins represent a big challenge for protein crystallographers because of their flexible and very dynamic nature. Myelin basic protein (MBP) exemplifies these troublemakers [32]. One exhaustive series of attempts to crystallize MBP for X-ray diffraction has been reported, where the authors tried 4,600 different crystallization conditions but were unable to induce crystallization of MBP [33]. Based on these observations the myelin basic protein has been suggested to belong to the category on “uncrystallizable” proteins. It can be safely assumed that many other unsuccessful crystallization attempts for numerous other proteins have not been reported, since negative results are generally assumed to be unsuitable for publication. In the case of MBP, several additional studies suggest that this protein lacks fixed 3D structure, existing instead as in intrinsically disordered ensemble, which in turn have been suggested to provide the basis for its multifunctionality [34]. Well-structured proteins often fail to crystallize, so not every crystallization failure should be ascribed to structural disorder. Nevertheless, we wonder how many crystallization failures denote these multifunctional yet unstructured proteins.
The importance of flexible structure for some proteins emerged from studies on protein folding. In fact, it has been pointed out that partially structured folding intermediates (such as the molten globule [35-41] and the pre-molten globule [37, 42-44]), which preserve some main elements of native secondary structure and their approximate mutual positions in 3-D space, but differ from the rigid globular state by looser packing of side chains and by the dramatic increase in the mobility of loops and ends of chain, are apparently ideal for some protein functions. The pre-molten globule is much more compact than the random coil but is less compact and has less secondary structure as compared to the molten globule (see below for additional discussion). By adjusting the solvent conditions, many proteins can be made to exist as stable, artificially induced, molten globules or as stable pre-molten globules, suggesting that these forms are not always transient folding intermediates [37, 38, 42-46].
Molten globule formation is likely driven by hydrophobic collapse, but with insufficiently tight side chain packing to form stable structures [38]. Pre-molten globule formation, on the other hand, evidently arises due to water being a poor solvent for polypeptides [42, 47-49] (see below for further discussion of this structural form). Recent studies on model homopolymer amino acids shed additional light on the concept that collapse can be driven by water not being a good solvent for proteins. Despite their lack of hydrophobic side chains, both polyglycine and polyglutamine form collapsed forms lacking appreciable secondary structure, likely because water is a poor solvent for both of these polymers [49-52]. Given the hydrophilic nature of polyglutamine, these results suggest that collapse from water being a poor solvent is likely to be a general phenomenon for proteins that lack a significant net charge. Both of these homopolymers contain dynamic, fluctuating structures that involve rapidly exchanging hydrogen bonds. While these homopolymers and the pre-molten globule state may share the property of arising from water being a poor solvent, the latter form contains much more secondary structure than the former, probably due in part to the presence of hydrophobic side chains. Further comparisons of various model homopolymers with different pre-molten globule proteins are needed to better understand their similarities and differences.
Some proteins exist as stable molten globules or as stable pre-molten globules, suggesting that for these proteins such partially folded forms can be associated with function (e.g., see [53-58]). Indeed, molten globules have been suggested to be involved in a number of physiological processes [37, 59, 60] such as interaction with chaperones [61], protein insertion into membranes [62, 63] and interaction with ligands (summarized in [64, 65]). Although functionality has been attributed to the molten globule- or pre-molten globule-like conformations for the examples cited above, the major emphasis still remains focused on the concept that these partially folded structures represent kinetic folding intermediates trapped by chaperones just after the protein biosynthesis but before proteins become completely fold [37, 59, 60], or appear as a result of point mutations preventing polypeptides from complete folding [37, 66]. Some other proteins (such as pore-forming domains of some toxins, or proteins that act as carriers of large hydrophobic ligands) were assumed to have originally a rigid structure but were forced somehow to denature to fulfill their functions [38, 60].
Many proteins with flexible structures have been discovered one-by-one. Some of these proteins were observed as atypical cases of polyfunctional proteins (e.g., serum albumin [67]), or polypeptides with unusual amino acid compositions (e.g., prothymosine α [7-9]), or proteins involved in the binding of large partners (RNA, DNA, proteins, and heme, e.g., histones [10], ribosomal proteins [11], myoglobin [12] and cytochrome c [13, 14]) or in the binding of large numbers of small partners (e.g., osteocalcin [15]). For some of these highly flexible proteins the increased conformational flexibility was even suggested to be of functional significance, with these data indicating that sometimes proteins do not need to be rigid to be functional.
From the 1980s onwards, a number of researchers pointed out that lack of structure or flexibility can be important for biological function. Huber and Bennett [16] pointed out that missing regions of electron density of several proteins likely carried out important functions. Several papers in the late 1980s (reviewed by Sigler [17]) suggested that several important transcription factors carry out function without specific structure, requiring instead the existence of rather ill-defined “acid blobs or negative noodles.” To describe the open and relatively mobile conformation of the caseins, which allows rapid and extensive degradation of these proteins to smaller peptides by proteolytic enzymes, Holt and Sawyer suggested the term “rheomorphic protein” (meaning flowing shape) [18] and proposed later that the rheomorphism of the casein phosphoproteins is important for the protection of the mammary gland against pathological calcification during lactation by allowing the protein to combine rapidly with nuclei of calcium phosphate to form stable calcium phosphate nanoclusters [19, 20]. In a similar time frame, Pontius extended his earlier work to suggest that unstructured proteins could have an advantage for certain types of molecular interactions [21]. Based on the observations that tau protein in solution resembled a Gaussian polymer being characterized by the lack of detectable secondary structure and compact folded conformation, together with the facts that this protein exhibited the following properties: 1. a high conformational flexibility similar to that of denatured protein; 2. a high resistance to heat and acid treatment without losing its ability to promote microtubule formation; 3. a rod-like or highly extended appearance in the electron microscope; and 4. a binding of tau to microtubules that was not defined by clearly identifiable residues, but rather was distributed over many weakly interacting sites within the C-terminal half, tau was regarded as a “natively denatured” protein [68]. In 1995 study, Gast et al. [8] pointed out that prothymosin α, an acidic protein with an unusual amino acid composition, is characterized by a high evolutionary conservation and wide tissue distribution, yet this protein adopts a random coil-like conformation under physiological conditions in vitro. These authors also raised an important question: “Whether this is a rare or a hitherto-overlooked but widespread phenomenon in the field of macromolecular polypeptides?” [8]. A year later, similar conformational behavior was described for another biologically important protein, α-synuclein (also known as the non-Aβ component of Alzheimer's disease amyloid precursor protein, NACP), which was shown to possess high stability to heat denaturation, a highly charged amino acid sequence, a “random coil” structure as demonstrated by CD, an abnormally high Stokes radius, and an abnormal SDS binding leading to unusual mobility on SDS–PAGE [69]. The authors also have pointed out that since similar diagnostic properties were earlier reported for several other proteins, all of them should be combined in a new class of “natively unfolded proteins” [69].
Despite the significant number of important experimental results described for these unstructured proteins, the concept that these proteins form an important and novel structure-function class simply failed to take hold. Part of the problem apparently was that the information about flexible yet functional proteins was scattered in the literature, and so the concept of biological function originating from conformational flexibility was re-discovered many times and given many different names [7-21, 53-58, 67-69]. As a result, for a very long time each “non-traditional” protein with highly unusual structural properties and/or strange conformational behavior was typically considered to be a rare exception to the general rule that the function requires rigid 3-D structure. Also, these disordered proteins contradicted the widely-accepted protein structure-function paradigm. Perhaps especially due to this reason, the number of these proteins was assumed without evidence to be insignificantly small. Therefore, the tipping point for a concept change did not occur, and general questions about biological roles of disordered proteins were not being asked.
The situation has begun to change since the mid to late 1990s due significantly to the efforts of four research groups that came to the important conclusion that naturally flexible proteins, instead of being just rare exceptions, represent a very broad class of proteins [70-73]. Interestingly, this important conclusion was reached at about the same time independently by four groups of investigators who emphasized rather different approaches, namely bioinformatics [72, 74-88], NMR spectroscopy [70, 89, 90], protein folding/misfolding [9, 64, 71, 91-95], and protein structural characterization [73]. The work of these four groups of course was strongly influenced by, and depended significantly upon, the many specific examples described by previous workers [7-21, 53-58, 67-69] but differed from previous efforts in that the lack of structure itself became the focus of attention with special efforts directed towards understanding the differences in function and mechanism between structured and unstructured proteins.
By now, many proteins have been shown to lack rigid 3-D structure under physiological conditions in vitro, existing instead as dynamic ensembles of interconverting structures. These proteins have been given various names including rheomorphic [18], intrinsically disordered [72], natively denatured [68], natively unfolded [69, 71], intrinsically unstructured [70, 73], mostly unstructured [55], and natively disordered [29]). Each of these terms has advantages and limitations. Not on this list of names is flexible, which is commonly applied to unstructured proteins but which evidently has not been suggested as a class name. Disordered proteins and regions are certainly highly flexible [96], but the word “flexible” has been used to describe many types of backbone and side chain mobility important for function [96], for example the motions in regions of high B-factor [97]. This general use of the word “flexible” does not make it such a good choice as a general descriptor for these ill-structured proteins. The term rheomorphic seems appropriate for extended random coils but perhaps not for molten globules, nor for collapsed random coils. The terms intrinsically unstructured and natively unfolded may be also be suitable for extended random coils and even those that are collapsed, but these terms don't seem to appropriately describe proteins that form transient or stable secondary structure. The term disorder suffers because of its negative connotation and its possible confusion with a pathological state, yet, on the other hand, disorder can be used for proteins like the molten globule that form substantial secondary structure but that nevertheless are highly dynamic and non-uniform. For this last reason, herein we will call these proteins “intrinsically disordered” (ID).
By “intrinsic disorder” we mean that the protein exists as a structural ensemble, either at the secondary or at the tertiary level. In other words, in contrast to structured or ordered proteins whose 3-D structure is relatively stable with Ramachandran angles that vary slightly around their equilibrium positions but with occasional cooperative conformational switches, intrinsically disordered proteins or regions exist as dynamic ensembles in which the atom positions and backbone Ramachandran angles vary significantly over time with no specific equilibrium values, and these ensembles typically undergo non-cooperative conformational changes. Both extended (random coil-like) regions with perhaps some secondary structure and collapsed (partially folded or molten globule-like and pre-molten globule-like) domains with poorly packed side chains are included in our view of intrinsic disorder [72, 92].
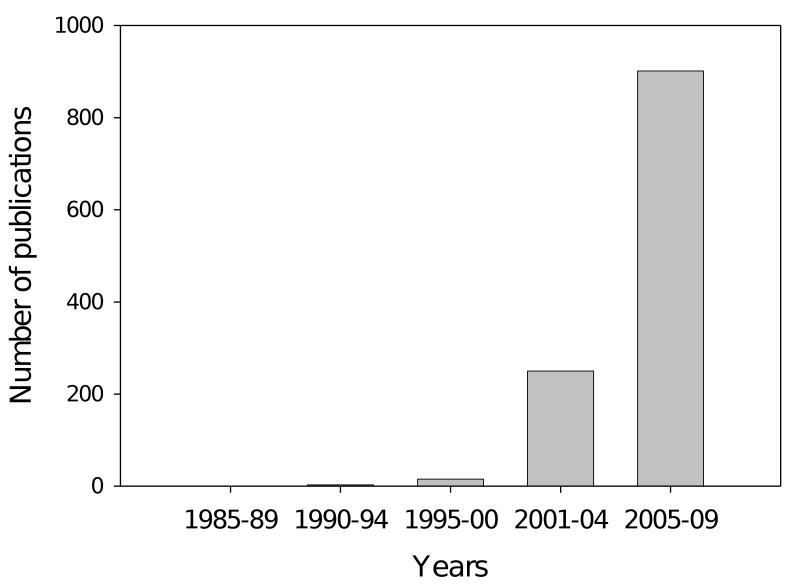
Since publication of key studies and reviews describing this new concept [53-58, 70-73, 98, 99], the literature on these proteins is virtually exploding (see Figure 1). Bioinformatics studies indicate that about 25 to 30% of eukaryotic proteins are mostly disordered [100], that more than half of eukaryotic proteins have long regions of disorder [85, 100], and that more than 70% of signaling proteins have long disordered regions [101].
Figure 1. Time-dependent increase in the number of PubMed hits dealing with ID proteins.
The following keywords have been used to perform this search: intrinsically disordered, natively unfolded, intrinsically unstructured, intrinsically unfolded and intrinsically flexible.
Now it is recognized that ID is a very abundant phenomenon. In fact, many proteins were shown to contain regions of disorder or even to be entirely disordered. Uversky et al. compiled a list of 91 disordered proteins characterized by NMR, circular dichroism or other biophysical techniques [71]. A subsequent search of X-ray crystal structures and the literature have expanded this list to more than 200 proteins that contain disordered regions of 30 consecutive residues or longer as characterized by X-ray crystallography, proteolytic digestion or other physical analyses such as NMR or circular dichroism [72]. The commonness of intrinsic disorder was estimated by predicting disorder for whole genomes, including both known and putative protein sequences (see below for the discussion of the disorder predictors). Such predictions have been published for 31 genomes that span the 3 kingdoms. The percentage of sequences in each genome with segments predicted to have ≥ 40 consecutive disordered residues was used to gain an overview of proteomic disorder. For so many consecutive predictions of disorder, the false positive error rate was estimated from ordered proteins to be less than 0.5% of the segments of 40 and less than 6% of the fully ordered proteins [72, 85]. The eukaryotes exhibited more disorder by this measures than either the prokaryotes or the archaea, with C. elegans; A. thaliana; S. cerevisiae; and D. melanogaster predicted to have 52-67% of their proteins with such long predicted regions of disorder, while bacteria and archaea were predicted to have 16-45% and 26-51% of their proteins with such long disorder regions, respectively [85, 100]. The increased amount of disorder in the eukaryota is very likely related to the increase in cellular signaling in the eukaryota [72, 85, 100]. The functional repertoire and advantages of intrinsic disorder will be discussed below.
2. The protein non-folding problem
2.1. Why ID proteins do not fold
Similar to the “normal” protein for which it has been shown that the correct folding into its relatively rigid biologically active conformation is determined by its amino acid sequence, the absence of rigid structure in the “non-traditional” ID proteins may also be somehow encoded in the specific features of their amino acid sequences. In fact, some of the ID proteins have been discovered due their unusual amino acid sequence compositions and the absence of regular structure in these proteins has been explained by the specific features of their amino acid sequences including the presence of numerous uncompensated charged groups (often negative); i.e., a high net charge at neutral pH, arising from the extreme pI values in such proteins [8, 69, 102], and a low content of hydrophobic amino acid residues [8, 102]. Interestingly, the first predictor of intrinsic disorder was developed by R. J. P. Williams based on the abnormally high ratio of the number of charged residues divided by the number of hydrophobic residues for the two ID proteins [98]. Although this predictor was used to separate just two ID proteins from a small set of ordered proteins, this paper is significant as being the first indication that ID proteins have amino acid compositions that differ substantially from those of proteins with 3-D structure. Subsequent investigation suggests, however, that the predictor developed by R.J.P. Williams does not work well in general [103].
Later, the use of charge and hydropathy to distinguish structured and ID protein was rediscovered with two important differences: first, normailized net charge, not total number of charged residues, was used; second, normalized hydrophobicity using the Kyte and Doolittle scale [104], not total number of hydrophobic residues, was used [71]. For this second charge-hydropathy approach, 275 natively folded and 91 natively unfolded proteins (i.e., proteins which at physiologic conditions have been reported to have the NMR chemical shifts of a random-coil, and/or lack significant ordered secondary structure (as determined by CD or FTIR), and/or show hydrodynamic dimensions close to those typical of an unfolded polypeptide chain) have been assembled from the literature searches. From the comparison of these datasets it has been concluded that the combination of low mean hydrophobicity and relatively high net charge represents an important prerequisite for the absence of compact structure in proteins under physiological conditions [71].
The above observation was used to develop a charge-hydropathy (CH) plot method of analysis that distinguishes ordered and disordered proteins based only on their net charges and hydropathies [71]. Figure 2A represents the original CH-plot and shows that natively unfolded proteins are specifically localized within a specific region of C-H phase space. Furthermore, ID and ordered proteins can be separated by a linear boundary, above which a polypeptide chain with a given mean net charge will most probably be unfolded [71].
Figure 2. Peculiarities of amino acid composition of ID proteins.
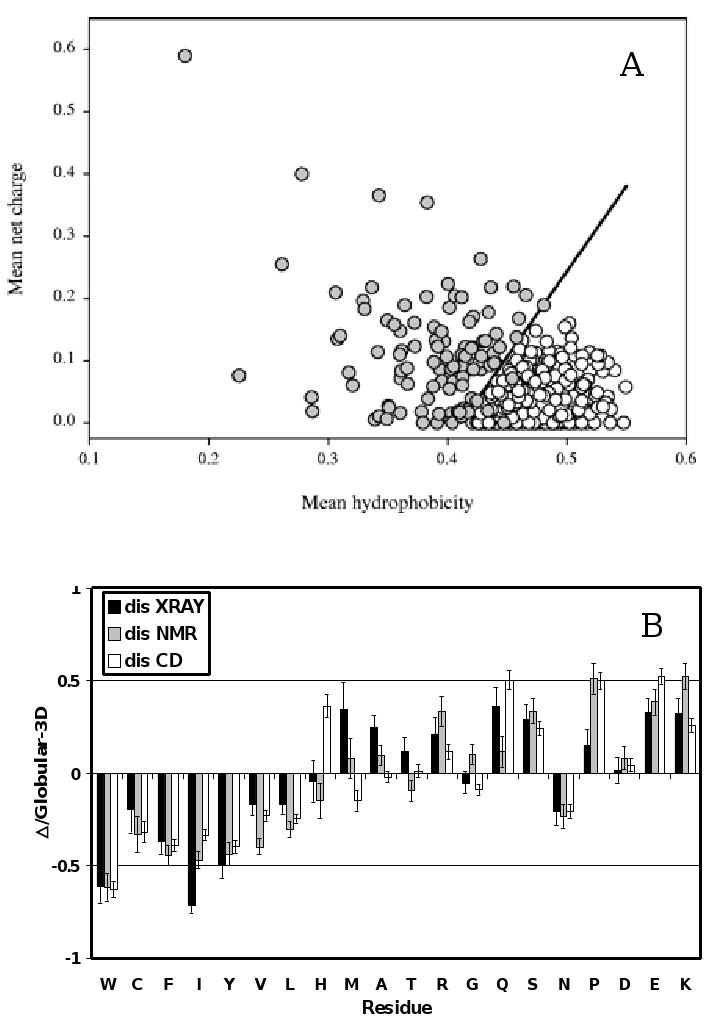
A. Comparison of the mean net charge and the mean hydrophobicity for a set of 275 folded (open circles) and 91 natively unfolded proteins (gray circles). The solid line represents the border between intrinsically unstructured and native proteins (see text). B. Order/Disorder composition profile. Comparisons of amino acid compositions of ordered protein with each of three databases of disordered protein. The ordinates are (% amino acid in disordered dataset - % amino acid in ordered dataset) / (% amino acid in ordered dataset) = Δ/globular_3D. The residues are ordered according to the Vihinen's flexibility scale [105]. Names of each database indicate how the disordered regions were identified. Negative values indicate that the disordered database has less than order, positive indicates more than order.
From the physical viewpoint, such a combination of low hydrophobicity with high net charge as a prerequisite for intrinsic unfoldedness makes perfect sense: high net charge leads to charge-charge repulsion, and low hydrophobicity means less driving force for protein compaction. In other words, these features are characteristic for ID proteins with the coil-like (or close to coil-like) structures. This can explain why R.J.P. Williams original approach, which used a measure of total charge rather than net charge, does not work nearly so well as CH-plot method mentioned above. Obviously, such highly disordered proteins represent only a small subset of the ID protein realm.
More detailed analysis was elaborated to gain additional information on the compositional difference between ordered and ID proteins. Comparison of a non-redundant set of ordered proteins with several datasets of disorder (where proteins were grouped based on different techniques, X-ray crystallography, NMR and CD, used to identify disorder) revealed that disordered regions share at least some common sequence features over many proteins [74, 75]. These differences in amino acid compositions are visualized in Figure 2B. Here, the relative content of each amino acid in a given disordered dataset has been expressed as (Disordered–Ordered)/(Ordered). Thus, negative peaks correspond to the amino acids in which the disordered segments are depleted compared with the ordered ones, and positive peaks indicate the amino acids in which ID regions are enriched [72]. The arrangement of the amino acids from least to most flexible was based on the scale established by Vihinen et al. [105]. This scale was defined by the average residue B-factors of the backbone atoms for 92 unrelated proteins. Figure 2B shows that the disordered proteins are significantly depleted in bulky hydrophobic (Ile, Leu, and Val) and aromatic amino acid residues (Trp, Tyr, and Phe), which would normally form the hydrophobic core of a folded globular protein, and also possess low content of Cys and Asn residues. The depletion of ID protein in Cys is also crucial as this amino acid residue is known to have a significant contribution to the protein conformation stability via the disulfide bond formation or being involved in coordination of different prosthetic groups. In fact, since the thiolate anion is one of the strongest biological nucleophiles, the thiol group of cysteine is one of the most reactive functional groups found in proteins, participating in a range of different redox reactions that do not directly involve, but can be coupled to, electron transfer [106]. Cellular SH groups are implicated in the coordination of metal ions and the defense against oxidants, and the reversible formation of disulfide bonds is involved in regulation of enzyme activity, signal transduction, transcriptional activity, and protein folding [107]. Obviously, Cys has quite different order-disorder promoting properties in its oxidized (disulfide-bonded) or bound form, and the reduced/unbound form, where the disulfide bond formation and coordination of different prosthetic groups strongly favors stable ordered structure.
The depleted residues, Trp, Tyr, Phe, Ile, Leu, Val, Cys and Asn were proposed to be called order-promoting amino acids. On the other hand, ID proteins were shown to be substantially enriched in polar, disorder-promoting, amino acids: Ala, Arg, Gly, Gln, Ser, Glu, and Lys and also in the hydrophobic, but structure braking Pro [72, 83, 84, 108, 109]. Note that these biases in the amino acid compositions of ID proteins are also consistent with the low overall hydrophobicity and high net charge characteristic of the natively unfolded proteins (see above). The concepts of compositional profiling discussed above have being implemented in a form of the Compositional Profiler, a web-based exploratory data mining tool for discovery and visualization of amino acid composition differences [109].
In addition to amino-acid composition, the disordered segments have also been compared with the ordered ones by various attributes such as hydropathy, net charge, flexibility index, helix propensities, strand propensities, and compositions for groups of amino acids such as W + Y + F (aromaticity). As a result, 265 property-based attribute scales [83] and more than 6,000 composition-based attributes (e.g., all possible combinations having one to four amino acids in the group) have been compared [110]. It has been established that ten of these attributes, including 14 Å contact number, hydropathy, flexibility, β-sheet propensity, coordination number, R+E+S+P, bulkiness, C+F+Y+W, volume, and net charge, provide fairly good discrimination between order and disorder [72]. Later, 517 amino acid scales (including a variety of hydrophobicity scales, different measures of side chain bulkiness, polarity, volume, compositional attributes, the frequency of each single amino acid and so on) were analyzed to construct a new amino acid attribute, e.g. a novel amino acid scale that discriminates between order and disorder [111]. This scale out-performed the other 517 amino acid scales for the discrimination of order and disorder and provided a new ranking for the tendencies of the amino acid residue to promote order or disorder (from order promoting to disorder promoting): W, F, Y, I, M, L, V, N, C, T, A, G, R, D, H, Q, K, S, E, P [111].
Thus, the ID proteins differ dramatically from the ordered proteins in their amino acid sequences. These differences were used to develop different predictors of intrinsic disorder.
2.2. How to predict ID protein from amino acid sequence
In 1997, the first Predictor Of Natural Disordered Regions (PONDR®) was developed. This predictor used 10 of the above-mentioned sequence attributes [77]. PONDR® operates from primary sequence data alone, using the nonlinear models (feed-forward neural networks) as the basis for the order/disorder discrimination [77]. The reason for developing this predictor was to test whether intrinsic disorder arises from the amino acid sequence. If disorder is predictable from sequence with accuracies better than expected by chance, then evidently such regions of sequence have the information to specify lack of structure.
While working on PONDR® development, datasets of ordered and disordered protein sequence segments as characterized by X-ray diffraction [77], by NMR [75, 86], by X-ray diffraction partitioned by location [87], or by homology [78, 86] were assembled. Balanced datasets were used to train neural network predictors with various inputs. Predictions of order/disorder on out-of-sample, balanced datasets (e.g. using 5-cross validation), produced accuracies in the range of 70-84% (Table 1). Accuracies on individual proteins can vary by 10% from the averaged values. The relatively high prediction accuracies strongly support the use of amino acid sequence to predict disorder as an element of native protein structure and support the hypothesis that disorder is encoded by the amino acid sequence.
Table 1.
Accuracy and improvement of neural network predictors of natural disordered regions (PONDRs®)
| Name | Training Set | # Disordered Residues | Accuracies % | |
|---|---|---|---|---|
| Ordera | Disorderb | |||
| XL1 | 7 X-ray | 502 | 71 | 47 |
| VL1 | 7 NMR, 8 X-ray | 1,366 | 83 | 45 |
| XL-XT | VL1 plus XTc | 71 | 59 | |
| VL2 | 53 X-ray, 35 NMR, 52 CD |
17,978 | 76 | 65 |
| VL3d | 54 X-ray, 40 NMR, 58 CD |
22,434 | 84 | 59 |
| VSL1e | 230 long DRf 983 short DR Ordered regions |
25,958 9,632 354,169 |
83 | 79 |
| VSL2g | 230 long DR 983 short DR Ordered regions |
25,958 9,632 354,169 |
81 | 82 |
O_PDB_S25
Combined dis_X-ray, dis_NMR and dis_CD
XT is a joint name for the N-terminus (XN), and the C-terminus (XC) predictors, which were trained using x-ray crystallographic data, where the terminal disordered regions were 5 or more amino acids in length.
Besides the addition of a few more chains, substantial cleaning of the training databases was carried out between VL2 and VL3. Several incorrectly labeled chains were identified and fixed and order/disorder boundaries were adjusted in a few other proteins
The VSL1 predictor combines two predictors optimized for long (>30 residues) and short (≤30 residues) disordered regions, respectively, using weights generated by a third meta-predictor. The attributes used include amino acid frequencies, sequence complexity, ratio of net charge / hydrophobicity, averaged flexibility, and averaged PSI-BLAST profiles calculated over symmetric input windows.
Disordered region
VSL2 is a slightly improved version of VSL1 predictor. The training data for VSL2 were slightly different: 8 ambiguous sequences were removed; His-tags were not used in training, short DR of 1-3 residues were not used in training. Also, linear SVM instead of logistic regression was used for VSL2 version (Kang Peng, personal communication).
Note: Both VSL1 and VSL2 take advantage of length dependencies
Since the time of its first introduction, PONDR® has undergone dramatic development and several versions of the predictors with the increased accuracy and reliability are currently available. Recently, the prediction accuracy of the PONDR family of disorder predictors has been considerably increased using the greatly expanded database of disordered proteins and improved computational techniques [112, 113]. This includes PONDR® VL-XT, VL3, and VSL1 predictors, access to which is provided by Molecular Kinetics, Inc. (at http://www.pondr.com/).
Table 1 shows that to achieve balanced accuracies of order and disorder prediction, we were able to improve disorder prediction only slightly while losing accuracy on order evaluation (e.g., compare data for VSL1 and VSL2). This observation is quite interesting and suggests that this effect likely arises from ordered-like fragments located within the disordered regions: the order prediction accuracy is lost when these types of sequences are considered to be disordered. Table 1 also shows that the accuracy of prediction of ordered residues continues to be better as compared to disordered residues.
The current view on this asymmetry is that regions structurally characterized as disordered often contain local regions with a strong tendency to become ordered – and these do become ordered when the correct binding partner comes along (e.g., see for early NMR studies [53-58]). Many examples of this type of behavior are discussed below. In fact, we noticed some time ago that predictions of order in regions characterized to be disordered often identify potential binding sites [88]. Based on these observations an algorithm has been elaborated [114] that identifies regions having a propensity for of α-helix-forming molecular recognition elements (α-MoREs) based on a discriminant function that indicates such regions while giving a low false-positive error rate on a collection of structured proteins (see below for more details). The MoRE segments have been renamed molecular recognition features (MoRFs) [115], and the algorithm for their prediction improved by increasing the sizes of the training sets [116].
Since publication of first ID predictors, numerous researchers have designed many algorithms to predict disordered proteins utilizing specific biochemical properties and biased amino acid compositions of ID proteins and using various prediction ideas and different computing techniques. Many of these predictors can be accessed via public servers (see Table 2). A recent review of algorithms for intrinsic disorder prediction revealed that since the first predictors were published, more than 50 predictors of disorder have been developed [103]. Furthermore, in this review, the basic concepts of various prediction methods of intrinsically disordered proteins were summarized, the strength and shortcomings of many of the disorder predictors were analyzed, and difficulties and the directions of future development of intrinsically disordered protein prediction techniques were discussed [103].
Table 2.
Protein disorder predictors
PONDR® is a family of ID predictors, which includes VL-XT, VL3,
Formally, SEG is not a disorder predictor. It is an indicator of low sequence complexity regions
As a recognition of the increased interest to the phenomenon of intrinsic protein disorder, starting from 2004 disorder prediction has been included as part of the biennial Critical Assessment of Techniques for Protein Structure Prediction (called CASP). Although only four groups participated in CASP5 disorder prediction experiments, CASP8 attracted 25 groups [117]. The ability to predict disorder from sequence with really high accuracy in a truly blind experiment adds confidence in the various results obtained by disorder predictions [118]. The rapid increase in the number of disordered predictors indicated in the recent review [103] is due in large measure to the popularization of disorder prediction in the CASP experiments (see Figure 3).
Figure 3. Time-dependent increase in the total number of IDP predictors.
The list of predictors includes: the first suggested predictor of IDPs [378]; the first formal predictor of IDPs [379]; predictor of ID in calcineurin family [380]; CH-Plot [71]; CDF [85]; PONDR® VL-XT [84]; GlobPlot [381]; DisEMBL [382]; DISOPRED [383]; flavors of protein disorder [122]; NORSp [384]; predictor by using reduced amino acid alphabet [385]; DISOPRED2 [307]; DRIPPRED [386]; FoldUnfold [387, 388]; Softberry (http://www.softberry.com); VaZyMolO [389]; PONDR® VL3-E [113]; IUPred [301, 390]; FoldIndex [391]; RONN [392]; DISpro [393]; PONDR® VSL1 [394]; CDF [100]; combined CDF/CH-Plot predictor [100]; α-MoRF [114]; Prelink [395]; PONDR® VSL2 [396]; Spritz [397]; DisPSSMP [398]; IUP predictor [399]; disorder prediction in calmodulin partners [233]; Decision trees [268]; Wiggle [400]; iPDA [401]; PrDOS [402]; SGT [403]; Ucon [404]; α-MoRF II [116]; composition profiler [109]; POODLE-L [405]; POODLE-S [406]; POODLE-W [403]; NORSnet [404]; OnD-CRF [407]; predictor by using bayesian multinomial classifier [408]; DISOclust [409]; Top-IDP [111]; DPROT [410]; hierarchical classifier [411]; MetaPrDOS [412]; MeDor [413]; Draai [414]; CDF-ALL [415]; IUPforest-L [416].
While more than 50 order-disorder predictors have been published [103], here we will discuss just one of these, developed by Rost and co-workers [119], to illustrate that the problem of intrinsic disorder prediction has been examined in very distinct ways. This study was initiated by the observation of long, irregular sequences, or “loopy regions,” that are visible in x-ray crystal and NMR structures. Since such regions are observed in X-ray structures, they don't fit our definition of intrinsic disorder. However, such loopy regions typically have no internally buried regions and thus probably exist as disordered regions that become ordered by either crystal contacts or by interactions with the surfaces of globular protein domains. The observed loopy regions were not used as a training set, however, but rather to motivate the development of the algorithm. The resulting Rost algorithm was based upon the absence of predicted secondary structure and the presence of predicted solvent accessibility in fairly long regions. This algorithm is used to indicate the presence of such “loopy” proteins, which were also called regions of “no regular secondary structure” (NORS) [119]. The NORS indicator as developed gave a low false positive prediction rate on globular, ordered protein, but its performance has not been evaluated on a set of proteins known to be disordered. Comparisons between prediction results for the NORS algorithm and results for PONDR indicate that these predictors, in fact, overlap significantly, but are by no means coincident (unpublished observations).
All of the predictors developed so far use as input the protein amino acid sequence and its attributes. The attributes are allied in different combinations and applied to classify each residue within a local sequence region (or in some cases to classify the entire protein) as either ordered or disordered. Different approaches are used to weigh and combine the various features. Predictors of disorder are based on various computational approaches, including, analytical algebraic functions, linear least squares, logistic regression, neural networks, and support vector machines. Finally, it is necessary to emphasize that even although modern predictors of intrinsic disorder use different (and in some cases very sophisticated) computational approaches, they are mostly based on the concepts elaborated in the pioneering computational studies mentioned above [71, 72, 77, 83] or are based on derivatives of these concepts.
Comparing several predictors on an individual protein of interest or on a protein dataset often provides additional insight regarding the predicted disorder if any exists. This is illustrated by a recent study, in which two distinct methods for using amino acid sequences to predict which proteins are likely to be mostly disordered were been compared [100]. These two binary predictors of whole protein structure or disorder are the cumulative distribution function (CDF) analysis and the charge-hydropathy (CH) plot. The CDF is based on PONDR® VL-XT, which predicts the order-disorder class for every residue in a protein [85, 100]. CDF analysis summarizes these per-residue predictions by plotting PONDR® scores against their cumulative frequency, which allows ordered and disordered proteins to be distinguished based on the distribution of prediction scores. The second binary predictor of whole protein order-disorder classification is the charge-hydropathy plots [71], in which ordered and disordered proteins being plotted in charge-hydropathy space can be separated to a significant degree by a linear boundary as discussed above in some detail.
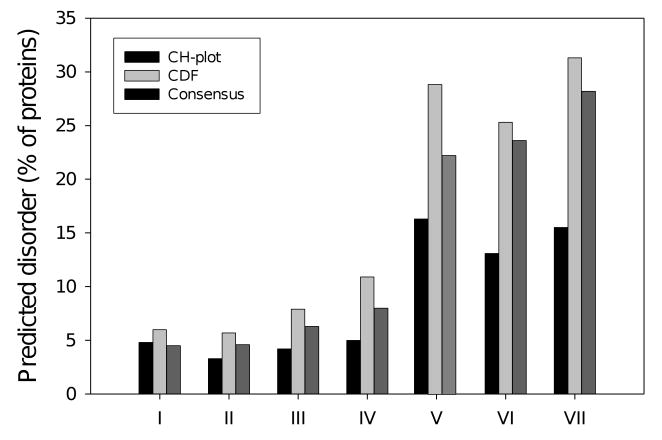
Interestingly, CDF analysis predicts a much higher frequency of disorder in sequence databases as compared to CH-plot discrimination [114]. However, the vast majority of disordered proteins predicted by charge-hydropathy discrimination were also predicted by CDF analysis. These findings are not a big surprise, as CH-plot analysis discriminates protein using only two attributes, mean net charge and mean hydrophobicity, whereas PONDR® VL-XT (and consequently the resulting CDF) is a neural network, which is a nonlinear classifier, trained to distinguish order and disorder based on a relatively large feature space (including average coordination number, amino acid compositions (aromatic and charged residues), and net charge). Thus, CH feature space can be considered as a subset of PONDR® VL-XT feature space [100]. Importantly, these findings may be physically interpretable in terms of different types of disorder, collapsed (molten globule-like) and extended (pre-molten globule- and coil-like). Under this consideration, the CH-plot classification discriminates proteins with the extended disorder from a set of globular conformations (molten globule-like or rigid well-structured proteins) and proteins predicted to be disordered by the CH-plot approach are likely to belong to the extended disorder class. On the other hand, PONDR®-based approaches can discriminate all disordered conformations (coil-like, pre-molten globules and molten globules) from rigid well-folded proteins, suggesting that CH classification is roughly a subset of PONDR® VL-XT, in both predictions of disorder and feature space [100]. Based on this reasoning, several interesting conclusions have been reached. It has been suggested that if a protein is predicted to be disordered by both CH and CDF, then, it is likely to be in the extended disorder class. However, a protein predicted to be disordered by CDF but predicted to be ordered by CH-plot might have properties consistent with a dynamic, collapsed chain; i.e., it is likely to be in the native molten globule class. Finally, proteins predicted to be ordered by both algorithms are of course likely to be in the well-structured class [100]. Importantly, the fact that CDF analysis predicts about 2-fold higher frequency of disorder in sequence databases than CH-plot classification suggests that approximately half of disordered proteins in different proteomes possess extended disorder, whereas another half represents proteins with the collapsed disorder [100].
The discussed above difference in the sensitivity of CDF and CH-plot predictors to different levels of overall disorderedness was utilized in CDF-CH-plot analysis [120]. To illustrate the basic concepts of this approach Figure 4C shows the results of separation for two example proteins in the CH-CDF phase space. Here, each spot corresponds to a single protein and its coordinates are calculated as a distance of this protein from the boundary in the corresponding CH-plot (Y-coordinate) (see Figure 4B) and an averaged distance of the corresponding CDF curve from the boundary (X-coordinate) (see Figure 4A). Positive and negative Y values correspond to proteins which, according to CH-plot analysis, are predicted to be natively unfolded or compact, respectively. Whereas positive and negative X values are attributed to proteins that, by the CDF analysis, are predicted to be ordered or intrinsically disordered, respectively. Therefore, this plot has four quadrants: pink quadrant (-, -) contains proteins predicted to be disordered by CDF, but compact by CH-plot (i.e., proteins with molten globule-like properties); red quadrant (-,+) includes proteins predicted to be disordered by both methods (i.e., random coils and pre-molten globules); blue quadrant (+,-) contains ordered proteins; violet quadrant (+,+) includes proteins predicted to be disordered by CH-plot, but ordered by the CDF analysis [120]. Application of this approach to the whole genomes revealed that ∼30% protein in mouse (Figure 5) and human proteomes (data not shown) potentially possess the properties of native molten globules.
Figure 4. Binary predictors of intrinsic disorder.
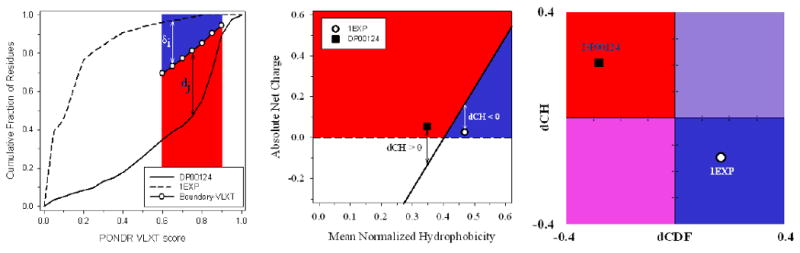
A. CDF analysis. Dashed curve located above the boundary represents CDF curve of ordered protein, whereas solid line located below the boundary corresponds to the CDF curve of IDP. Here, δi and dj (where i and j range from 1 to 7) are attributed to the ordered and disordered protein, respectively, and represent the distances of points at the CDF curve from the corresponding boundary points. The averaged distance of a given CDF curve from a boundary line is calculated as or . B. CH-plot analysis. Black square located above the boundary corresponds to ordered protein, open circle located below the boundary represents disordered protein. C. CH-CDF analysis. Black square corresponds to disordered protein DP00124, whereas open circle represents ordered protein 1EXP. X-coordinates were calculated as averaged distances of corresponding CDF curves from a boundary (positive dCDF distance corresponds to a protein predicted to be ordered by CDF analysis, negative dCDF distance corresponds to a protein predicted to be disordered by CDF analysis, see plot A). Y-coordinates were obtained as distances from spots corresponding to proteins to boundary. Positive and negative dCH distances correspond to protein predicted by CH-plot to be disordered or ordered, respectively, see plot B.
Figure 5. CH-CDF plot for mice proteins.
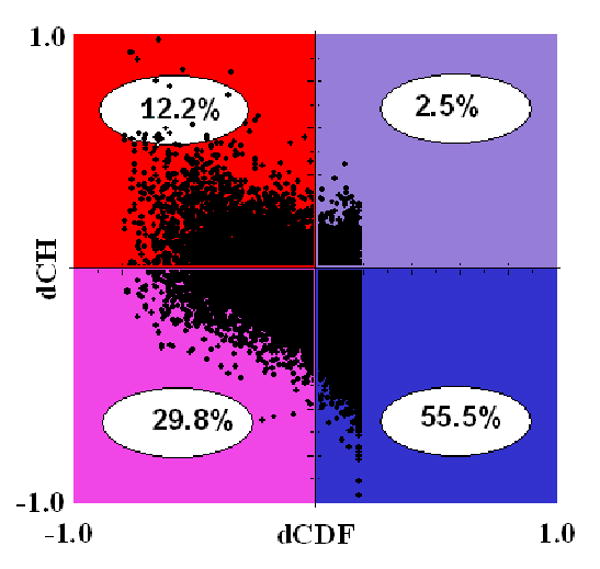
The principles of this computational tool are described in the Approach section. Quadrants contain differently disordered proteins: red quadrant contains extended IDPs (predicted to be disordered by CDF and CH-plot analysis), pink quadrant contains native molten globules (predicted to be disordered by CDF and ordered by CH-plot), the blue quadrant contains globular proteins (predicted to be ordered by both CDF and CH-plot analyses), whereas the violet quadrant contains proteins predicted to be ordered by CDF and disordered by CH-plot.
The interpretation that proteins in the (-.-) quadrant are likely to be molten globules needs further exploration. For example, a protein with distinct structured and disordered domains might fall into the (-,-) quadrant and not be a native molten globule at all. A protein that is a molten globule as a monomer might form a structured complex. This association could be with itself, with another protein, or with a non-protein ligand. Indeed a number of proteins are known to be ill-structured as monomers but form structured dimers [121]. Similarly, many proteins are ill-structured in the apo-forms but become structured after the specific ligand binding [64, 65]. Thus, we are in the process of investigating collections of proteins that map to the 4 quadrants of Figure 5 to better understand the extent to which the various proteins conform to the simple concepts described above.
As it has been already mentioned, protein disorder is a multi-faced phenomenon; i.e., disordered proteins, being mobile, flexible, and dynamic, might have very different structural features, which range from collapsed molten globule-like conformation to extended coil-like state. It has been suggested that, just as an ordered protein is comprised of different types of secondary structure (α-helices, β-strands, β-turns, 310-helices and others), ID protein can also be made up of distinguishable types of disorder [122]. To check this hypothesis, a unique partitioning algorithm based on the differential prediction accuracies has been developed [122]. This algorithm used the notion that a specialized predictor built on a given disorder flavor should have significantly higher same-flavor accuracy than other-flavor predictors or than a global predictor applied to the same given flavor. Application of this partitioning algorithm to known disordered proteins identified three distinctive flavors of disorder, arbitrarily called V, C, and S [122]. Importantly, the flavor-specific disordered proteins have been shown to be distinguishable not only by their amino acid compositions, but also by disordered sequence locations, and biological functions. Based on these observations, it was proposed that specific flavor-function relationships do exist and thus it is possible (in principle) to identify the functions of disordered regions from their amino acid sequences alone, without any need for specific structural knowledge [122].
2.3. What is so special about ID proteins
ID proteins have dynamic structures that interconvert on a number of timescales and have been shown to have many similarities to non-native states of “normal” globular proteins, which may exist in at least four different conformations: native (ordered), molten globule, pre-molten globule, and coil-like [42-44, 123]. Using this analogy, it has been established that ID proteins and regions under physiological conditions in vitro might contain collapsed-disorder (i.e., where ID is present in a form of molten globules) and extended-disorder (i.e., regions where ID is present in a form of random coil or pre-molten globule) [29, 42, 72]. This is illustrated by Figure 6, which schematically represents the three types of disorder mentioned above, molten globule, pre-molten globule and coil for a model polypeptide chain of 100 residues long. A model structure of a 100 residues-long globular protein is shown for comparison. Figure clearly shows that there is a dramatic difference in the hydrodynamic volumes occupied by a polypeptide chain in these four conformations. These differences further increase for a longer polypeptide chain.
Figure 6. Illustrative examples of ID proteins.
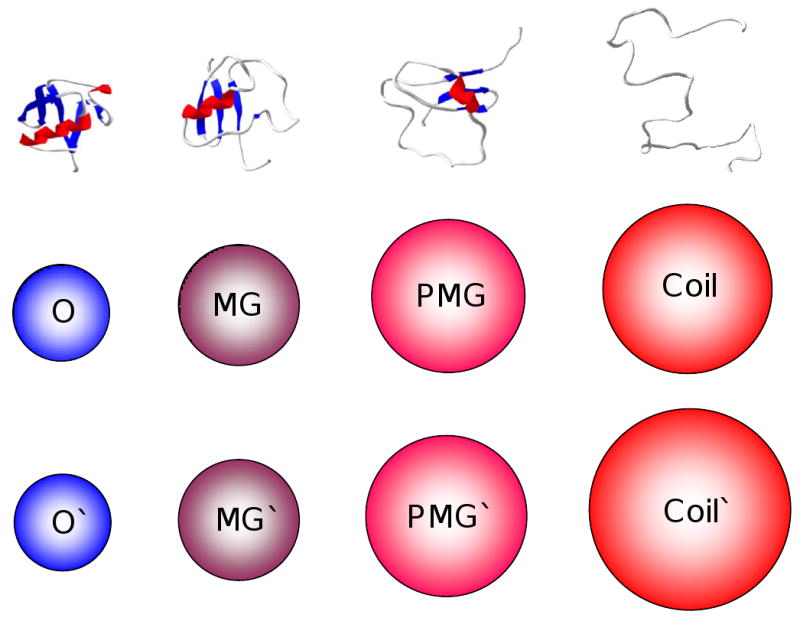
Top line: Collapsed (molten globule-like, MG) disorder; Extended (pre-molten globule-like, PMG) disorder; (coil-like, coil) disorder. Ordered globular protein of same length is also shown for comparison. Figure represents model structures of a 100 residue-long polypeptide chain. Middle line: Relative hydrodynamic volumes occupied by a 100 residue-long polypeptide chain in these four conformations. Bottom line: Relative hydrodynamic volumes occupied by a 500 residue-long polypeptide chain in these four conformations. Spheres in the middle and bottom lines show an increase in the hydrodynamic volume relative to the volume of the corresponding ordered protein.
2.3.1. Collapsed disorder
The structural properties of the molten globule (which was originally described as universal folding intermediate of globular proteins) are well known and have been systematized in number of reviews (e.g., see [38] and references therein). The protein molecule in this intermediate state has no (or has only a trace of) rigid cooperatively melted tertiary structure. However, it is characterized not only by the well-developed secondary structure, but also by the presence of some topology, i.e., relatively fixed mutual positioning of the secondary structure elements [124-131]. A considerable increase in the accessibility of a protein molecule to proteases was noted as a specific property of the molten globule [132-136]. The transformation into this intermediate state is accompanied by a considerable increase in the affinity of a protein molecule to the hydrophobic fluorescence probes (such as 8-anilinonaphthalene-1-sulfonate, ANS), and this behavior should be considered as a characteristic property of the molten globule state [137]. The averaged value for the increase in the hydrodynamic radius in the molten globule state compared with the native state is no more than 15%, which corresponds to volume increase of ∼50% [37, 38, 42, 92, 93]. Small-angle X-ray scattering analysis has revealed that the protein molecule in this partially folded state has a globular structure typical of ordered globular proteins [138-142].
2.3.2. Extended disorder
A significant number of sequences encodes for the extendedly disordered proteins that are characterized by low sequence complexity. Are these proteins random coils, or do they possess residual or transient structure? If they have residual or transient structure, how should they be classified? Based on the analysis of the available literature, it has been concluded that such proteins do not possess uniform structural properties, as expected for members of a single thermodynamic entity. In fact, they may be divided into two structurally different groups, intrinsic coils and intrinsic pre-molten globules [42, 92, 93]. Proteins from the first group have hydrodynamic dimensions typical of considerably unfolded polypeptide chain in poor solvent (see below), and do not possess any (or almost not any) ordered secondary structure. Proteins from the second group are more compact (see below), and exhibit some amount of residual secondary structure. However, they are still less dense than native globular or molten globule proteins [42, 92, 93]. Other properties of extendedly disordered proteins can be derived from structural features described below for unfolded and pre-molten globular conformations of globular proteins.
Obviously, intrinsically disordered proteins with very high net charges are expected to be more extended and behave more similar to random coils (i.e., similar to conformations adopted by proteins in the denaturant guanidinium hydrochloride). The validity of this hypothesis was recently illustrated via the analysis of the set of nucleoporins (Nups) containing long natively unfolded domains with phenylalanine-glycine repeats (FG domains). These Nups constitute a gate of the nuclear pore complex (NPC), where the FG domains form a malleable network of disordered polypeptides, which selects and size-discriminates against diffusing macromolecules [143]. In this study, most nucleoporin FG domains were shown to adopt collapsed molten-globular configurations and were characterized by a low content of charged amino acids. Others adopted more extended, coil-like conformations, were structurally more dynamic, and were characterized by a high content of charged amino acids. Many nucleoporins contained both types of structures in a biphasic distribution along their polypeptide chain. For example, the N-terminus of Nsp1 (AA 1-172; Nsp1n), had a low charged-AA content of 2% and adopted molten globular configurations, whereas the remainder of the Nsp1 FG domain (AA 173-603; Nsp1m) had a charged AA content of 36% and adopted extended-coil configurations [143].
The unfolded protein is essentially never a true random coil. In fact, the existence of significant residual structure in the unfolded globular protein has been described even under the most severe denaturing conditions, such as high concentrations of strong denaturants [144-147]. Thus, coil-like ID proteins are not completely random, but are characterized by the presence of some residual (and highly flexible) structure. This fact is very important for the functioning of these proteins (see below).
The structural peculiarities of a polypeptide chain in the pre-molten globule state are briefly outlined below. The protein molecule in this state has no rigid tertiary structure. It is characterized by a considerable secondary structure, although much less pronounced than that of the native or the molten globule protein (protein in the pre-molten globule state has ∼50% of the native secondary structure, whereas in the molten globule state the corresponding value is noticeably higher). The protein molecule in the pre-molten globule state is considerably less compact than in the molten globule or native states, but it is still more compact than the random coil (its hydrodynamic volume in the molten globule, the pre-molten globule and the unfolded states, in comparison with that of the native state, increases 1.5, ∼3 and ∼12 times, respectively, see Figure 6).
The protein molecule in the pre-molten globule state can effectively interact with the hydrophobic fluorescent probe ANS, though essentially weaker than in the molten globule state. This means that at least part of the hydrophobic clusters of polypeptide chain accessible to the solvent is already formed in the pre-molten globule state [37, 38, 43, 44, 148]. Despite this ability to interact with ANS, the pre-molten globule state the protein molecule does not exhibit globular structure [141, 142, 148]. The last observation indicates that the pre-molten globule probably represents a “squeezed” and partially ordered form of the coil [42, 47, 48, 142]. Importantly, local structural elements of these squeezed coils may occupy native-like positions [43, 44, 142]. This fact is of functional importance too.
2.3.3. Conformational behavior
Because of the ID proteins possess strong biases in their amino acid compositions one might expect that this will be reflected in their conformational behavior. Recently, the peculiarities of the responses of the ID proteins to changes in their environment were systemized in a comprehensive review [149]. As summarized in [149], the conformational behavior of IDPs is typically characterized by a low cooperativity (or the complete lack thereof) during denaturant-induced unfolding of any structure that might exist, by the lack of measurable excess heat absorption peak(s) characteristic for the melting of ordered proteins, by a gain of structure in response to heat and changes in pH, by the ability to gain structure in the presence of various counter ions, osmolytes, membranes and binding partners, and by different, protein-specific responses to macromolecular crowding [149].
An increase in temperature often induces the partial folding of intrinsically unstructured proteins (i.e., proteins with extended disorder), rather than the unfolding that is typically observed for globular proteins. The effects of elevated temperature may be attributed to the increased strength of the hydrophobic interaction at higher temperatures, leading to a stronger hydrophobic driving force for partial folding [92, 93, 149].
For a number of extendedly-disordered proteins it has been shown that a decrease (or increase) in pH induces partial folding of intrinsically unordered proteins due to the minimization of their large net charge present at neutral pH, thereby decreasing charge/charge intramolecular repulsion and permitting hydrophobic-driven collapse to the partially-folded conformation [92, 93, 149].
Importantly, this high temperature and extreme pH stability of ID proteins can be used isolate them from cell extracts. For example, disordered proteins with extended disorder can be separated from ordered proteins by their intrinsic indifference to denaturing conditions that originates from the lack of tertiary and secondary structure. In other words, ID proteins can be isolated from ordered ones as proteins which “survived” (i.e., remained soluble) harsh denaturing conditions usually leading to the precipitation of ordered proteins, including extensive heating/boiling or incubation in the presence of trichloroacetic acid (TCA), or perchloric acid (PCA) [150, 151] [152, 153].
2.4. How to identify and structurally characterize ID proteins
Computational analyses clearly show that the ID proteins and proteins with long ID regions are highly abundant in nature. Several experimental approaches are sensitive to the intrinsic disorder of a given protein or its part and therefore have been used to provide structural information on ID proteins. Ironically, the choice of suitable techniques for the characterization of disordered proteins is based on the experience retrieved from the studies on “traditional” ordered proteins. In fact, almost all experimentally validated ID proteins were discovered and structurally characterized by techniques elaborated for the analysis of structure and self-organization of ordered proteins, where the information on the presence of intrinsic disorder was typically retrieved from the absence of a signal characteristic for the ordered protein. These studies clearly showed that in many ways ID proteins resemble denatured states of well-structured proteins.
The unique 3-D structure of a globular protein is stabilized by non-covalent interactions (conformational forces) of different types, such as hydrogen bonds, hydrophobic forces, electrostatic interactions, van der Waals interactions, etc. Being different in their physical bases, these forces are known to respond differently to changes in the protein environment. In fact, some of the forces are either weakened or even completely eliminated under particular conditions, whereas other conformational forces remain unchanged or are even intensified under the same conditions. This gives raise to the formation of various partially folded conformations with properties intermediate between those of the well-ordered and the completely unfolded states.
By analogy with different conformations of globular proteins, intrinsically disordered proteins can be divided into different groups, depending on the amount of disorder they possess. This gives several structurally different classes of intrinsic disorder: native molten globules, native pre-molten globules and native coils. As these conformations possess defined structural differences along with increasing amounts of disorder, they may be discriminated from one another by several physicochemical methods [29, 30, 154, 155]. Some of the most widely used techniques for identification of intrinsic disorder in proteins are briefly outlined below.
X-ray crystallography defines missing electron density in many protein structures, which may correspond to disordered region(s). The increased flexibility of atoms in such a region leads to the non-coherent X-ray scattering, making them unobserved [72, 156-158]. Since structured domains can wobble on flexible hinges and also be unobserved, long unobserved regions are not always disordered.
Heteronuclear multidimensional NMR is an extremely powerful technique for protein 3D-structure determination in solution and for the characterization of protein dynamics. Recent advances in this technology have allowed the complete assignment of resonances for several unfolded and partially folded proteins, as well as the disordered fragments of folded proteins [70, 90, 155, 159-161]. These methods can also provide direct measurement of the mobility of unstructured regions.
There are two types of optically active chromophores in proteins, side groups of aromatic amino acid residues and peptide bonds [162, 163]. Circular dichroism (CD) spectra in the near ultraviolet region (250-350 nm), also called the aromatic region, reflect the symmetry of the environment of aromatic amino acid residues and, consequently, are characteristic of protein tertiary structure. The lack of rigid tertiary structure in a protein containing aromatic residues may be easily detected by the simplified near-UV CD spectrum with low intensity.
Diminishing of ordered secondary structure may be detected by several spectroscopic techniques including far-UV CD [162-168], optical rotary dispersion (ORD) [169], Fourier transform infra-red spectroscopy (FTIR) [71], Raman optical activity [170], and deep-UV resonance Raman spectroscopy [171, 172].
Hydrodynamic parameters obtained from techniques such as gel-filtration, viscometry, SAXS, SANS, sedimentation, dynamic and static light scattering may help in determining whether a protein is compact or unfolded. The unfolding of a protein molecule results in an essential increase in its hydrodynamic volume. For instance, there is a well documented 15-20% increase in the hydrodynamic radius of globular proteins upon their transformation into the molten globule state, while the hydrodynamic volume of the pre-molten globule is even larger (see Figure 6). Furthermore, native and unfolded conformations of globular proteins possess very different molecular mass dependencies of their hydrodynamic radii, RS [42, 47, 48, 92, 93, 142, 173, 174]. As a result, ID proteins will have an increased hydrodynamic volume relative to ordered proteins of similar molecular mass, leading to an increase in their apparent molecular mass. For example, for the spheres shown in Figure 6 this increase is translated in the following numbers. For a 100 residues-long polypeptide with a real molecular mass of 11 kDa, the apparent molecular mass of a molten globular conformation estimated from the hydrodynamic data (e.g. gel-filtration) is 16 kDa, for the native pre-molten globule this number is 29 kDa, whereas for the native coil the column will give ∼40 kDa. For a 500 residue-long ID protein with the real molecular mass of 55 kDa these numbers are: ∼90 kDa, 180 kDa and 375 kDa if the protein is a native molten globule, a native pre-molten globule or a native coil, respectively.
Another very important structural parameter is the degree of globularity, which reflects the presence or absence of a tightly packed core in a protein molecule. This information may be extracted from the analysis of small angle X-ray scattering (SAXS) data in form of a Kratky plot, which is a plot of I(S)*S2 versus S, with I(S) being the scattering intensity, and S being the scattering vector given by 2sinθ/λ, where θ is the scattering angle, and λ is the wavelength of X-ray. The shape of the Kratky plot curve is sensitive to the conformational state of the scattering protein molecules [140, 141, 175]. A scattering curve in the Kratky coordinates has a characteristic maximum for globular proteins in either their native or molten globule states (i.e., states with globular structure). However, if a protein is completely unfolded or in a pre-molten globule conformation (i.e., with no globular structure), such a maximum is absent [95, 140, 141].
Additional knowledge on the intramolecular mobility and compactness of a protein may be extracted from the analysis of various fluorescence characteristics. This includes FRET, shape and position of the intrinsic fluorescence spectrum, fluorescence anisotropy and lifetime, accessibility of the chromophore groups to external quenchers, and steady state and time-resolved parameters of the fluorescent dyes. Overall, these techniques add important information on the conformational dynamics of a polypeptide. As discussed above, the ability of a partially folded polypeptide chain to interact with hydrophobic fluorescent probes, such as ANS [137], is a very useful property which can be used for identification of the ID proteins.
Increased proteolytic degradation in vitro of intrinsically disordered proteins indirectly confirms by their increased flexibility [72, 133-136, 176].
Immunochemical methods may also be applied toward the elucidation of protein disorder. The immunoglobulins obtained against a given protein may be specific for different levels of macromolecule: the primary structure [177, 178], the secondary structure [179], or the tertiary structure [177, 178]. In the latter case, the antigenic determinants may reside on either the neighbouring residues in the chain (loops) [177, 178] or on spatially distant residues [179]. Furthermore, it has been shown that antibodies in the immune serum may possess a high affinity to the internal elements of an antigen [179]. Thus, antibodies may be successfully used to study the structural changes, which a protein-immunogen undergoes upon changes of the experimental conditions. For example, antibodies obtained against the Ca2+-saturated F1-fragment of prothrombin did not interact with the calcium-free apo-form of this protein [180]. An analogous effect also was observed in the case of osteocalcin [181].
Intrinsic disorder may be detected by the analysis of protein conformational stability. For example, the presence or absence of a cooperative transition on the calorimetric melting curve for a given protein is a simple and convenient criterion indicating the presence or absence of a rigid tertiary structure [38, 154, 182]. Furthermore, it has been shown that the steepness of urea- or GdmCl-induced unfolding curves depends strongly on whether a given protein has a rigid tertiary structure (i.e., it is native) or is already denatured and exists as a molten globule [149, 183, 184]. To extend this type of analysis, the values of Δνeff (which is the difference in the number of denaturant molecules “bound” to one protein molecule in its two states) should be determined. Then this quantity should be compared to the and values corresponding to the native to coil and molten globule to coil transitions in globular protein of a given molecular mass, respectively [183, 184].
Finally, unique electrophoretic mobility of ID proteins should be mentioned. Electrophoresis is mostly applied either to determine the molecular mass of proteins or to elucidate the charge difference and/or form of the macromolecule. However, it has been pointed out that due to their unique amino acid compositions, ID proteins bind less sodium dodecyl sulphate (SDS) than “normal” proteins [73, 185]. As a result, they possess abnormal mobility in SDS polyacrylamide gel electrophoresis experiments and their apparent molecular masses determined by this technique are often 1.2-1.8 times higher than real one calculated from sequence data or measured by mass spectrometry (for example, see [185]).
In addition to the traditional techniques sensitive to the lack of ordered structure in a given protein, some novel experimental approaches have been elaborated to characterize ID proteins both in vitro and in vivo (see above, [53-58]). One of the very promising approaches to analyze ID proteins in their natural environments (i.e., within cells) is in-cell NMR spectroscopy [186-189]. This method is based on the notion that most nuclei in natural substances are NMR-inactive and hence not detectable by NMR methods. Therefore, the isotope-effect can be exploited as a selective visualization filter, where a molecule of interest in which NMR-inactive nuclei are substituted with NMR-active stable isotopes suddenly becomes ‘visible’ to the spectroscopic eye [186-189].
Several ID proteins have been investigated using the in vivo NMR analysis in prokaryotic cells. Examples include FglM [190] and α-synuclein [191, 192]. FglM a 97-residue polypeptide from Salmonella typhimurium, which regulates flagellar synthesis by binding to the transcription factor δ28 [193]. Although unbound FlgM is mostly unstructured, its C-terminal half can form a transient α-helix [194]. Interaction with δ28 in vitro leads to the partial folding of this C-terminal domain, which is manifested by the disappearance of a set of C-terminal NMR resonance signals [195]. Since inside the living E. coli cells the same set of NMR resonance signals were absent, it has been concluded that in cellular environment a structural rearrangement took place in the FlgM C-terminal domain that was similar to the one observed with δ28 [190]. Importantly, the N-terminal half of FglM remained unfolded even in the over-crowded cellular environment [190]. The in vivo NMR analysis of another ID protein, α-synuclein, revealed that this protein remained soluble, monomeric and, in contrast to FglM completely unfolded inside live E. coli cells [191, 192]. Although this technique is very attractive being potentially able to provide detailed information on the IDP structure and dynamics inside the cell, significant precaution should be taken while performing the in vivo NMR analysis. In fact, two in-cell NMR reports describing the in-cell dynamics of another IDP, apo-cytochrome b5, were recently retracted [196, 197] because protein leakage from the cells led to misleading data [198].
In-cell NMR analysis of target proteins in eukaryotic cells is a rather new addition to the rapidly growing field of high-resolution in vivo NMR studies. Recently, the conformational in vivo properties of the human Tau protein inside the Xenpus laevis oocytes were analyzed by the in-cell NMR. Although this study revealed that intracellular crowding did not induce dramatic conformational rearrangements of this ID protein, the in-cell NMR spectra displayed several resonance signals that strongly suggested that residues of Tau became post-translationally phosphorylated by endogenous Xenopus kinases and other spectral features suggested that a noticeable portion of intracellular was shown to be bound to endogenous microtubules [199].
Utilization of various single-molecule techniques represents another interesting development in the field of structural characterization of ID proteins. Since the conformational landscape of an ID protein does not have a single, highly stable ordered structure, being characterized by a set of marginally-stable interconverting conformations, whose equilibrium is driven by the depths and profiles of their energy minima and by the effect of the environment upon them, the capability of resolving the properties of individual protein molecules and quantify subpopulations is particularly crucial for ID proteins. Two single-molecule approaches have been reported in studies of the conformational properties of ID proteins, single molecule fluorescence resonance energy transfer (SM-FRET) [200] and atomic force microscopy-based single molecule force spectroscopy (SMFS) [201, 202].
SM-FRET trajectories contain detailed information about conformational motions associated with, for example, protein-protein or protein-DNA interactions. For example, single molecule spectroscopy revealed that protein-protein interactions related to cell signaling [203] or in the DNA damage recognition [204] are characterized by the large-amplitude fluorescence intensity fluctuations at a time-scale ranging over several orders of magnitude, from milliseconds to sub-seconds, suggesting that these interactions involve highly flexible intrinsically disordered structures [205, 206].
Recently, SM-FRET was used to analyze the shape of α-synuclein bound to detergent micelles and lipid vesicles [207]. By strategically placing maleimide donor (Alexa 488) and acceptor (Alexa 594) in different regions of the protein it has been shown that α-synuclein formed a bend helix when bound to highly curved SDS micelles and existed as an elongated helix interacting with more physiological 100 nm diameter lipid vesicles [207].
SMFS has been found to be a very useful tool to gain insights on the conformational equilibria of monomeric ID proteins, providing crucial information on the conformational heterogeneity of monomeric α-synuclein [202]. In fact, SMFS analysis revealed that the conformational equilibrium of this classical ID protein included three main classes of conformations, such as disordered and “α-like” structures, and that the relative abundance of various conformers was modulated by changes in the environment, mutations and pharmacological strategies [201, 202]. Based on these observations it was suggested that SMFS can explore the full conformational space of an ID protein at the single-molecule level, detecting even poorly populated conformers and measuring their distribution in a variety of biologically important conditions [202]. Interestingly, the application of another “non-traditional” technique, the electrospray ionization mass spectrometry analysis of protein ion charge state distributions, supported the existence of the conformational heterogeneity of α-synuclein, and revealed that this ID protein samples four distinct conformational states, ranging from a highly structured one to a random coil [208].
Recently, to visualize individual molecules in solution under physiological conditions, a high-speed atomic force microscopy (AFM), where the successive AFM images were captured at rates of 5–17 frames per second, was utilized [209]. To illustrate the power of this approach, a highly mobile protein was visualized by high-speen AFM. In this study, the protein studied is named FACT (facilitates chromatin transcription), which is a highly conserved eukaryotic heterodimer consisting of structure-specific recognition protein-1 (SSRP1) and the protein SPT16, which is crucial for transcript elongation through nucleosomes by RNA polymerase II [210]. The high-speed AFM analysis revealed the existence of two undulating and wobbling tail-like segments of different lengths that protrude from the main body of FACT [209]. In addition to providing the first direct visualization of the high intrinsic mobility of an IDP, this approach revealed unique mechanical properties of the tail domains, which appeared to be in conformations that were more relaxed than random coils. Based on these exciting data the authors concluded that the high-speed AFM represents a novel imaging technology that can be utilized for visualization and characterization of flexible intrinsically disordered region in other important proteins at the single-molecule level [209].
Since the information on the presence of intrinsic disorder in a given protein is typically retrieved from the absence of a signal characteristic for the ordered protein, misidentification and misclassification of ID proteins are still very common. Obviously, the simultaneous application of several techniques mentioned above to a given protein provides the most unambiguous evidence of its disordered features. The size of this journal review is too limited to provide in depth analysis of various experimental tools for structural characterization of ID proteins. Recently, a comprehensive volume dedicated to this issue was published [31] and we address the interested readers to this book.
2.5. What is the functional repertoire of ID proteins
2.5.1. ID proteins and their functions
As already discussed, intrinsic disorder is very common in nature. Figure 7 further illustrates this conclusion showing that intrinsic disorder is abundant at the proteome level, and many proteins were not only predicted to have long disordered regions, but to be disordered along their entire lengths. This high natural abundance of ID proteins clearly suggests that, although intrinsically disordered proteins fail to form fixed 3D-structures under physiological conditions, they likely carry out important biological functions, which has been confirmed by several comprehensive studies [70-73, 80, 90, 92, 93, 101, 211-213]. Furthermore, sites of posttranslational modifications (acetylation, hydroxylation, ubiquitination, methylation, phosphorylation, etc.) and sites of regulatory proteolytic attack are frequently associated with regions of intrinsic disorder [82].
Figure 7. Predicted abundance of mostly disordered proteins in several proteomes.
I, Y. pestis; II, E. coli; III, A. fulgidus; IV, M. thermoautotrophicum; V, S. cerevisiae; VI, A. thaliana; VII, M. musculus. Analysis was performed by three predictors of mostly disordered proteins: the charge-hydropathy (CH) plot, the cumulative distribution function (CDF) of PONDR® VL-XT score, and a consensus predictor that combines the CH-plot and CDF predictors. The main point is that eukaryotes appear to contain far more intrinsic disorder as compared to prokaryotes. This amount of predicted disorder has important functional consequences, and so proteomic experiments need to be redesigned to recognize and explore intrinsically disordered proteins.
Among the various functions found for disordered regions, even superficial analysis of “natively unfolded” proteins revealed that many of them undergo disorder-to-order transitions when stabilized by binding with specific targets [71]. In fact, for the majority of proteins described in that study, the existence of ligand-induced folding was established. Examples include induced structure formation upon binding with DNA (or RNA) for protamines, Max protein, high mobility group proteins HMG-14 and HMG-17, osteonectine, SDRD protein, chromatogranins A and B, Δ131Δ fragment of SNase, and histone H1. Other examples include the folding of cytochrome c in the presence of heme, the folding of ostecalcine induced by cations, secondary structure formation in parathyroid hormone related protein induced by membrane association, structure formation in glucocorticoid receptor brought about by association with trimethylamine N-oxide, folding of histidine-rich protein II induced by heme; and structure formation and compaction of prothymosin-α mediated by zinc [71]. Therefore, among the major functions of these unstructured, intrinsically disordered proteins are nucleic acid binding, metal ion binding, heme binding and interaction with membrane bilayers [71]. In addition to mentioned disorder-to-order-transition, there are many pre-structured or pre-organized motifs in IDPs that bind to target proteins [53-58].
More than 150 proteins have been identified in early studies as containing functional disordered regions, or being completely disordered, yet performing vital cellular roles [72, 80]. Twenty-eight separate functions were assigned for these disordered regions, including molecular recognition via binding to other proteins, or to nucleic acids [80, 81]. An alternative view is that functional disorder fits into at least five broad classes based on their mode of action [73]. These broad classes, with illustrative examples, are outlined in Table 3. Based on the thorough analysis of available literature data protein and RNA chaperones were added to this list [214].
Table 3. Functional classes of disorder.
Based on data reported in [73]
| Class | Example | Function |
|---|---|---|
| Entropic chains | Microtubule-associated protein 2 | Entropic bristle, spacing in microtubule architecture |
| Effectors | 4EBP1, 2, 3 | Inhibitor of translation initiation |
| Scavengers | Caseins | Inhibition of calcium precipitation in milk |
| Assemblers | Caldesmon | Actin polymerization |
| Display sites | CREB transactivator domain | Regulation by phosphorylation |
Recently, a novel computational tool was elaborated for the evaluation of a correlation between the functional annotations in the Swiss-Prot database and the predicted intrinsic disorder [215-217]. The approach is based on the hypothesis that if a function described by a given keyword relies on intrinsic disorder, then the keyword-associated protein would be expected to have a greater level of predicted disorder compared to the protein randomly chosen from the Swiss-Prot. To test this hypothesis, functional keywords associated with 20 or more proteins in Swiss-Prot were found and corresponding keyword-associated datasets of proteins were assembled. For each keyword-associated set, one thousand length-matching and number-matching sets of random proteins were drawn from Swiss Prot. Order-disorder predictions were carried out for the keyword-associated sets and for the matching random sets. If a function described by a given keyword were carried out by a long region of disordered protein, one would expect the keyword-associated set to have a greater amount of predicted disorder compared to the matching random sets. The keyword-associated set would be expected to have less prediction of disorder compared to the random sets if the keyword-associated function were carried out by structured protein. Given the predictions for the function-associated and matching random sets, it is possible to calculate the p-values, where a p-value > 0.95 suggests a disorder-associated function, a p-value < 0.05 suggests an order-associated function, and intermediate p-values are ambiguous [215-217]. The application of this tool revealed that out of 710 Swiss-Prot keywords, 310 functional keywords were associated with ordered proteins, 238 functional keywords were attributed to disordered proteins, and the remainder 162 keywords yield ambiguity in the likely function-structure associations [215-217].
Obviously, for all these ID proteins, the traditional structure-function paradigm is insufficient, suggesting that a more comprehensive view of the protein structure/function relationships is needed. In fact, a new paradigm was offered [72, 80, 82] to elaborate the sequence-to-structure-to-function scheme in a way that includes the novel functions of disordered proteins (Figure 8). The complex data supporting this revised view were summarized in The Protein Trinity Hypothesis [82], which suggested that native proteins can be in one of three states, the solid-like ordered state, the liquid-like collapsed-disordered state or the gas-like extended-disordered state. Function is then viewed to arise from any one of the three states or from transitions between them. This model was subsequently expanded to include a fourth state (pre-molten globule) and transitions between all four states [92].
Figure 8. Involvement of intrinsic disorder in protein function.
Note that the classical structure-function paradigm cannot describe many of the function proteins perform.
Some important activities ascribed to ID proteins do not directly involve coupled binding and folding, but rather are dependent on the flexibility, pliability and plasticity of the backbone. We are calling these “entropic chain activities”, as they rely entirely on an extended random-coil conformation of a polypeptide that has to maintain constant motion during functioning. Discovering new entropic chain activities and estimating the fraction of ID regions involved in such activities are both intriguing problems [72].
One illustrative example of such entropic chain activities is provided by the voltage gated potassium channel. This tetrameric integramembrane protein cycles among three states: closed (sensitive to voltage), open, and inactive (insensitive to voltage). In the ball and chain mechanism for ion channel inactivation, a highly flexible “chain” carries out a random search until the “ball” plugs the open channel (Figure 9, top panel). Portions of this figure are based on findings of Antz et al. [218]. For simplicity, only four of the proposed ten states are shown [219]. The inactivation depends on a binding interaction between the channel opening and the “ball”.
Figure 9. Example of an entropic clock. Top panel.
Simplified model of a Shaker-type voltage-gated K+ ion channel (blue) with ‘ball and chain’ timing mechanism. The ‘ball and chain’ is comprised of an inactivation, or ball, domain (yellow) that is tethered to the pore assembly by a disordered chain (red) of ∼ 60 residues. For simplicity, only four of the proposed ten states are shown. The cytoplasmic side of the assembly is oriented downward. A. Closed state prior to membrane depolarization. Note that conformational changes of the pore have sealed the channel and a positive charge on the cytoplasmic side of the pore assembly excludes binding of the ball domain. B. Open state following membrane depolarization. C. After depolarization, the cytoplasmic side of the pore opening assumes a negative charge that facilitates interaction with the positively charged ball domain. D. Inactivation of the channel occurs when the ball domain occludes the pore. The transition from C to D does not involve charge migration and can be modeled as a random walk of the ball domain towards the pore opening. (Portions of figure based on Antz et al. [218]). Bottom panel: Schematic presentation of the ‘chain’ length-dependent timing of channel inactivation. Different lengths of the ‘chain’ region of N-terminal domain result in different rates of channel inactivation [220, 221], where shorter ‘chain’ causes a more rapid inactivation (A), whereas a longer ‘chain’ produces slower inactivation (B). Modified from [225].
The time of opening is also a crucially important. This “time of being open” function depends directly on the length and flexibility of the disordered “chain.” An extended disordered region functions as one component of an entropic clock found in some ion channels. Charge migrations within the tetrameric pore proteins are associated with the majority of state changes of voltage-gated K+ ion channels [219]. However, the timing of the inactivation step is determined by the time it takes for a mobile domain to find and block the channel. The movement of the mobile domain is restricted by a tether composed of ∼ 60 disordered residues [220, 221] (Figure 9, bottom panel). The timing of channel inactivation is a function of the length of the disordered tether.
Since ion channels serve to modulate the excitability of nerve cells, their malfunction can have substantial impact on human health. Mutations in the human homolog of the Shaker K+ channel (KCNA1) can lead to myokymia, partial epilepsy, or episodic ataxia [222, 223]. The ball and chain model was originally developed from experiments showing that protease digestion caused the open state to remain open, and then adding back the trypsin-released peptide led to channel inactivation. Recent NMR data provides direct confirmation of the flexibility of the “chain” region. If channel closure were to involve a random search by a flexible chain, the time of closure would be inversely proportional to the square of the length of the chain [224]. Genetic engineering of the Shaker K+ channel was used to construct channels chains of various lengths. The inactivation times for the chains followed the expected dependence on length. Shorter chains speeded up inactivation and longer chains slowed inactivation [220]. Taken together these data strongly support the ball and chain mechanism.
Finally, in one study sperm ion channels were found to have apparently disordered regions of different length in different primates. Furthermore, the underlying mutations over time were consistent with positive selection [225]. Thus, while the time of being open function is evidently not related directly to protein structure, this function is still subject to natural selection, thus providing very strong evidence of its importance.
Variety of complex and highly coordinated interactions of proteins represents the mechanistic foundation of the organism's physiology and function. This coordination is frequently achieved via recognition of specific and unique identification regions. For proteins, these identification regions are frequently located inside the ID regions [53-58, 213, 226]. Therefore, many ID regions are involved in regulation, recognition, signaling and control pathways where high-specificity/low-affinity interactions with multiple partners are necessary prerequisites. In this way, the functional diversity provided by disordered regions complements those of ordered protein regions.
2.5.2. ID proteins as hubs in protein signaling networks
Many diverse systems may best be described as networks with complex topologies, which is often assumed to be either completely regular or completely random [227]. An important feature of the random networks (such as the highway system) is that, despite the random placement of links, most nodes have approximately same number of links in the resulting systems [227, 228]. In a random network, the nodes follow a Poisson distribution with a bell shape. Nodes that have significantly more or fewer links than the average are difficult to find in such a system [228]. On the other hand, certain other networks, known as “scale-free” or “small-world” networks, are different: they have hubs, with many connections, and ends, that aren't connected to anything but a hub (if highways were like this, there would be a lot of dead-ends) [229, 230]. Scale-free networks combine the local clustering of connections characteristic of regular networks with occasional long-range connections between clusters, as can be expected to occur in random networks. Thus, some nodes have a very large number of connections to other nodes, whereas most nodes have just a few [228]. The popular nodes, called hubs, can have hundreds, thousands or even millions of links depending on the type of network being described. It has been emphasized that from this perspective, the network appears to have no scale [228]. For such scale-free networks the distance between nodes follows a power-law distribution [231]. Examples of scale-free networks include the author-collaboration network, the airline routes, the World Wide Web, the metabolic network, the protein domain network, and, apparently, the protein signaling networks inside cells (reviewed in [230]).
A recent proposal is that α-synuclein, p53 and many other ID proteins that interact with large numbers of distinct partners form hubs in the scale-free protein-protein interaction network inferred for the cell [213, 226]. To test the roles of disorder in the specific case of protein-protein interaction networks, we first collected a set of structurally characterized hub proteins [213]. Several hub proteins were found to be disordered from one end to the other, and yet to be capable of binding large numbers of partners. Other hubs contained both ordered and disordered regions. For these hubs, many, but not all, of the interactions mapped to the regions of disorder. Two highly structured hubs were found. For both of these structured hubs, 14-3-3 and calmodulin, the binding regions of their partner proteins were found to be intrinsically disordered [232, 233].
Overall, this initial study suggested two primary mechanisms by which disorder is utilized in protein-protein interaction networks, namely one disordered region binding to many partners and many disordered region binding to one partner. Several groups have tested these ideas further via comprehensive bioinformatics studies on collections of hub proteins, and the results of these studies supported the hypothesis that hub proteins commonly use disordered regions to bind to multiple partners [234-239]. These bioinformatics studies include further refinement of the analysis with the suggestion that disorder is very commonly used for regions that bind sequentially to multiple partners (so called “date hubs” [238]).
2.5.3. ID in scaffold proteins
Scaffold proteins represent a subclass of hub proteins that typically have a modest number of interacting partners. Scaffolds are often found at the central parts of functional complexes where they interact with most of their partners at the same time and therefore act as party hubs [235]. Some scaffolds selectively bring together specific proteins within a signaling pathway and provide selective spatial orientation and temporal coordination to facilitate and promote interactions among interacting proteins. Scaffolding can influence the specificity and kinetics of signaling interactions. Scaffolds can bind simultaneously to multiple participants in a particular pathway and facilitate and/or modify the specificity of pathway interactions [240]. Scaffolds may act on individual proteins by changing their conformation and thus their activity and on interaction partners by providing proximity and spatial orientation [240]. Some scaffolds create focal points for spatial and temporal coordination of enzymatic activity of kinases and phosphatases.
Modulation of the phosphorylation state of downstream members of signal transduction pathways is a primary mode of action for many scaffold proteins. Compartmentalization is provided by the fact that the activity of bound members is directed towards neighboring substrates that may or may not be bound to the scaffold. Enzymes may be activated or inhibited upon association with the scaffold. Associations are dynamic and may serve to coordinate the responses among pathways. Scaffolds contain several domains for protein-protein interaction. Furthermore, scaffold proteins can play a role in modulating the activation of alternative pathways by promoting interactions between various signaling proteins [241].
In order to understand the role of ID for scaffolding functions, several well-characterized scaffold proteins with structurally and functionally characterized ID regions were analyzed [241]. Based on the analysis of these several famous scaffolds, including axin, breast cancer type 1 susceptibility protein (BRCA1), A-kinase anchoring proteins AKAP79 and AKAP250, microtubule-associated protein 2 (MAP2), titin and several others, large ID regions appear to be crucial for successful scaffold function. These signaling scaffold proteins utilize the various features of highly flexible ID regions to obtain more functionality from less structure [241].
The more function from less structure conclusion was further supported by a recent study on structural properties of the CASK-interactive protein 1 [242], which is a post-synaptic density protein in mammalian neurons where it acts as a specific scaffold interacting with many important proteins including κ-casein (CASK), stathmin-3, synaptotagmin, neurexin-2, septin-4, neural cell adhesion molecule L1 (L1CAM), SH2/SH3 adaptor protein NCK-alpha (NCK1), and several others. Using a set of bioinformatics tools, CD spectroscopy, wide-line and 1H-NMR spectroscopy, limited proteolysis and gel filtration chromatography, the entire C-terminal proline-rich region of 800 amino acids of CASK-interactive protein1 exhibits the set of characteristics associated with being intrinsically disordered [242]. Furthermore, the authors extended their finding of a high level of ID in CASK-interactive protein 1 by assembling a set of 74 scaffold proteins and predicting their disorder by three different algorithms. A very high fraction of the residues was found to fall into local disorder, and ordered domains of these scaffold proteins were shown to be connected by linker regions which were mostly disordered. Thus, the usual design of a scaffold protein includes a set of short globular domains (∼80 amino acids on average) connected by longer linker regions (∼150 residues on average) with crucial binding functions [242].
2.5.4. The functional advantages of ID proteins/regions
Importantly, even sturdy lock holes (i.e., protein active sites) have been shown to be rather flexible. In fact, as early as in 1958 it has been recognized that some enzymes could act on rather differently shaped substrates, suggesting that a degree of flexibility would be needed to fit the different substrates and thereby to be functional. To explain these ideas, a modification of the “lock and key” model called the “induced fit” theory was proposed by Koshland [243]. According to this theory and its subsequent modifications/interpretations, the enzyme is partially flexible and the substrate does not simply bind to the active site, but it has to bring about changes to the shape of the active site to activate the enzyme and make the reaction possible. Substantial experimental evidence has been accumulated to support this view for many different enzymes. For example, the existence of functional flexibility within the active site has been demonstrated by X-ray crystallographic analysis of E. coli dihydrofolate reductase liganded with different cofactors and substrates. In fact, Sawaya and Kraut have analyzed crystal structures of different forms of this protein, including the holo-enzyme, the Michaelis complex, the ternary product complex, the tetrahydrofolate binary complex as well as the tetrahydrofolate-NADPH complex. These structures can be used to reconstruct a 2.1 Å resolution movie, depicting the sequence of events during the catalytic cycle, which showed that the enzyme adopts different conformational substates while complexed with different ligands, suggesting that the process of enzymatic catalysis might be accompanied by significant conformational changes [244].
Signaling and regulation are proposed to be among the most important functions of ID proteins/regions [101]. Qualitatively, it seems reasonable that highly mobile proteins would provide a better basis for signaling and recognition. For example, disordered regions can bind partners with both high specificity and low affinity [245]. This means that the regulatory interactions can be specific and also can be easily dispersed. Obviously this represents a keystone of signaling – turning a signal off is as important as turning it on [72].
Another crucial property of ID proteins for their function in signaling networks is binding diversity; i.e., their ability to partner with many other proteins and other ligands, such as nucleic acids [89]. This opens the possibility for one regulatory region or one regulatory protein to bind to many different partners. A protein that binds to multiple partners might be expected to be crucial for a number of different biological processes and therefore might be especially important for the survival of the cell. In agreement with this idea, proteins that make multiple interactions are more likely to lead to lethality if deleted [246].
There are several other reasons of why ID proteins might be superior for certain tasks compared to their ordered counterparts. This includes, but is not limited to: binding commonality in which multiple, distinct sequences recognize a common binding site (with perhaps different folds in the various complexed ID proteins) [176]; the ability to form large interaction surfaces as the disordered region wraps-up [247] or surrounds its partner [248]; faster rates of association by reducing dependence on orientation factors and by enlarging target sizes [21]; and faster rates of dissociation by unzippering mechanisms [72].
An interesting consequence of the capability of ID proteins to interact with different binding partners is their polymorphism in bound state; i.e., an ID protein (or ID region) can have completely different geometries in the rigidified structures induced by associating with its partner, depending on the nature of the bound partner. Crystallographic studies on glycogen synthase kinase 3β (GSK3β), a Ser/Thr protein kinase and its interactions with FRAT1 and axin provide an illustrative example of these polymorphic bound states [249]. Figure 10 shows that a sharp turn breaks the structure of FRAT peptide into two distinct and separate α-helical segments, whereas the axin peptide is bound as a single unbroken α-helix with a turn and an irregular tail [249]. Overall, despite the fact that the primary binding sites for axin and FRAT on GSK3β have been found to overlap substantially in the crystal structures, so that their binding is mutually exclusive, the GSK3β-interacting regions of these two proteins were shown to possess negligible sequence similarity [249]. Furthermore, even although both bound peptides are primarily helical, their detailed structures and interactions with GSK3β have substantial differences (Figure 10). The ability of GSK3β to bind two different proteins with high specificity via the same binding site is mediated by the conformational plasticity of the 285–299 loop. In fact, in crystals of unbound GSK3β, this loop is highly mobile and poorly defined. However as shown in Figure 10 this loop folds quite differently when it accommodates the two different protein ligands. While some residues in this versatile binding site in GSK3β are involved in interactions with both axin and FRAT, others are involved uniquely with one or the other [249].
Figure 10. Polymorphism in the bound state.
Comparison of axin and FRAT binding to GSK3β. The binding sites for the axin (383–401) peptide and FRAT (197–222) peptides are co- localized in the C-terminal domain of GSK3β. However, the two peptides have no sequence homology, have different conformations in their bound state, and possess different sets of interactions with GSK3β.
Another interesting example of the phenomenon discussed above is the polymorphism in bound state of p53 protein. There are four domains in p53: the unfolded N-terminal translational activation domain, the structured central DNA binding domain, and the unstructured C-terminal tetramerization and regulatory domain. At the transactivation region, p53 interacts with TFIID, TFIIH, Mdm2, RPA, CBP/p300 and CSN5/Jab1 among many other proteins [250]. At the C-terminal domain, it interacts with GSK3β, PARP-1, TAF1, TRRAP, hGcn5, TAF, 14-3-3, S100B(ββ) and many other proteins [250]. Figure 11 summarizes currently available information on p53 interactions and structure [251]. Some of these interactions are mapped to regions of the p53 sequence together with the order/disorder tendencies of p53 as revealed by PONDR® VL-XT. In PONDR plot, segments with scores above 0.5 correspond to the disordered regions, whereas those below 0.5 correspond to the ordered regions/binding sites.
Figure 11. p53 interaction with different binding partners illustrate peculiarities of one-to-many signaling.
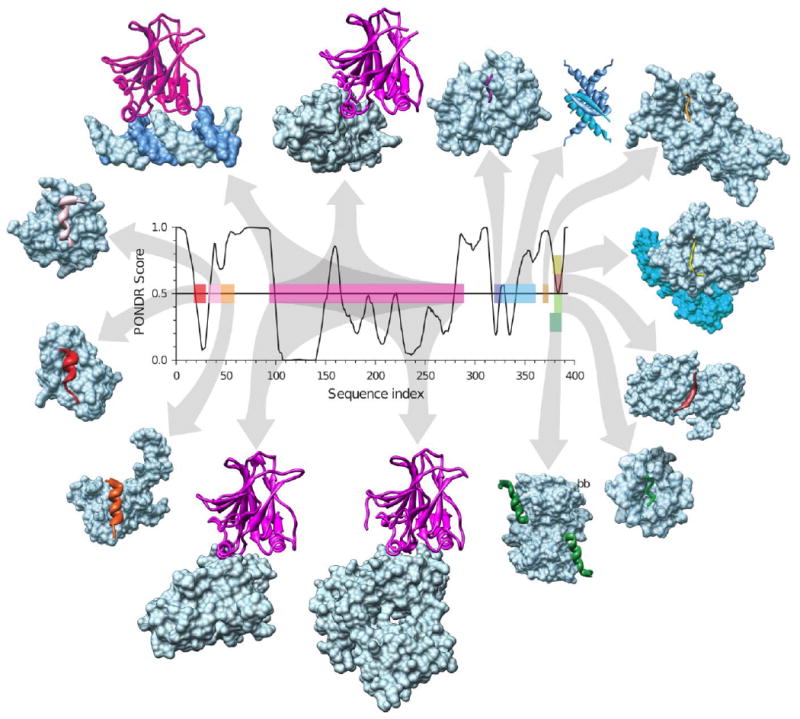
A structure versus disorder prediction on the p53 amino acid sequence is shown in the center of the figure (up = disorder, down = order) along with the structures of various regions of p53 bound to fourteen different partners. The predicted central region of structure with the predicted amino and carbonyl termini as being disordered have been confirmed experimentally for p53. The various regions of p53 are color coded to show their structures in the complex and to map the binding segments to the amino acid sequence. Starting with the p53-DNA complex (top, left, magenta protein, blue DNA), and moving in a clockwise direction, the Protein Data Bank IDs and partner names are given as follows for the fourteen complexes: (1tsr – DNA), (1gzh – 53BP1), (1q2d – gcn5), (3sak – p53 (tetrametization domain)), (1xqh – set9), (1h26 – cyclinA), (1ma3 – sirtuin), (1jsp – CBP bromo domain), (1dt7 – s100ββ), (2h1l – sv40 Large T antigen), (1ycs – 53BP2), (2gs0 – PH), (1ycr – MDM2), and (2b3g – rpa70).
This presentation reveals interesting correlations of the peculiarities of disorder prediction with the well-understood domains of p53. These correlations are indicated in order from amino terminal to carboxyl terminal along the sequence correlating with a clockwise arrangement for the molecular structures. Two first downward spikes in the PONDR plot are located within the transcription activation domain that binds to Tfb1, Mdm2 and Rpa70. A long prediction of mostly ordered structure (residues 100-280) matches the domain that binds to DNA. This large, centrally located DNA binding domain is predicted and observed to be structured, both when it is bound to DNA (upper left, note the DNA molecule as a partner) and when it binds in a similar fashion to 3 different protein partners (one above the prediction curve and two below, all are indicated by the magenta color for the similarly folded p53 central domain). A sharp downward spike doublet (residues 315-360) overlaps with the tetramerization domain. A sharp downward spike at the C-terminus of p53 matches the negative regulatory domain that binds to Cyclin A, Sirtuin, CBP and the S100ββ dimer [250]. Furthermore, a short fragment from a linker connecting the tetramerization and the negative regulatory domain interacts with SET9. Finally, another short fragment that corresponds to the first sharp downward spike of the tetramerization domain binds to tGcn5. Binding regions of the transactivation and negative regulatory domains were shown to undergo coupled folding and binding resulting from their interaction with corresponding binding partners; i.e., they represent illustrative examples various molecular recognition features (MoRFs) (analysed in details in [251]).
Figure 11 clearly shows that many complexes are formed that involve the disordered regions of p53. An interesting aspect of these many partnerships is that, for each interaction, typically only a short region of p53 becomes structured upon binding. For one particular complex (Upper Center, light blue color), one region of disorder self-associates to form a dimer (with nearly all of the buried residues in the dimer interface), and this dimer further aggregates into a tetramer. Thus, this association involves the coupled binding and folding of a disordered region. For another set of four complexes (Right side, colors of light yellow, red, and light and dark green), the same short segment near the p53 C-terminus binds to four different partners. Because this segment is unstructured to begin with, it can adopt different conformations when binding to the different partners. For this particular example, the disordered segment adopts an α-helix, a β-strand, and two different coils upon binding with its four different partners [251].
Summarizing, among various advantages of intrinsic lack of structure and function-related disorder-to-order transitions are [70, 72, 74, 79, 84, 241, 252]:
Decoupled specificity and strength of binding provides for high-specificity-low-affinity interactions. Pre-formed recognition elements and α-MoRFs can contribute to this phenomenon. A continuum of binding strength likely exists. Binding regions containing predominant preformed structure contribute more free energy to binding and those regions with little or no preformed structure contribute little free energy to binding;
Increased speed of interaction due to greater capture radius and the ability to spatially search through interaction space;
Strengthened encounter complex allows for less stringent spatial orientation requirements;
Efficient regulation via rapid degradation;
Increased interaction (surface) area per residue;
A single disordered region may bind to several structurally diverse partners;
Many distinct (structured) proteins may bind a single disordered region;
Intrinsic disorder provides ability to overcome steric restrictions, enabling larger interaction surfaces in protein-protein and protein-ligand complexes than those obtained with rigid partners;
Unstructured regions fold to specific bound conformations according to the template provided by structured partners;
Efficient regulation via posttranslational modification; i.e., phosphorylation, methylation, ubiquitination, SUMOylation, etc.;
Ease of regulation/redirection and production of otherwise diverse forms by alternative splicing;
The possibility of overlapping binding sites due to extended linear conformation;
Diverse evolutionary rates with some ID proteins being highly conserved and other ID proteins possessing high evolutionary rates. The latter ones can evolve into sophisticated and complex interaction centers (scaffolds) that can be easily tailored to the needs of divergent organisms;
Flexibility that allows masking (or not) of interaction sites or that allows interaction between bound partners;
2.5.5. Functional importance of disorder-to-order transitions
Molecular recognition and binding functions of ID proteins are proposed to be within three broad classes: effector, scavenger, and assembler [73]. In each of these cases, a significant disorder-to-order transition occurs upon binding to the target molecule. Some illustrative examples of this coupled folding and binding include the following: cation binding to α-synuclein [253]; self-association of an inhibitory subunit of phosphodiesterase [94]; formation of a functioning ribosome [254] from disordered ribosomal proteins [11]; zinc-induced folding of the DNA-binding domain of the 1,25-dihydroxyvitamin D3 receptor [255]; the SNARE complex formation from the disordered components, Snc1 and Sec9 [256]; interaction of intrinsically disordered caldesmon with its binding partner calmodulin [257].
Many other protein-protein and protein-nucleic acid interactions involve coupled folding and binding of at least one of the partners [70-73, 80, 90, 92, 93, 101, 211-213, 226]. Obviously, the amino acid compositions and the structures of such natively disordered proteins enable a very large accessible surface area (ASA) in the absence of binding partner(s), whereas the binding-induced disorder-to-order transition is accompanied by a dramatic decrease in ASA and also by the concomitant release of a large number of water molecules. In agreement with the latter aspect of coupled binding and folding, the binding of lac repressor to its regulatory site is accompanied by the release of ∼260 water molecules, as estimated by the osmotic stress method introduced by Parsegian and co-workers [258-260], which is based on the measurement of the change in the number of water molecules present in system compartments that are inaccessible to neutral solutes [261]. Thus, the lack of ability to decrease ASA intramolecularly (i.e., to fold) is compensated in many ID proteins by their outstanding capability to do so upon interaction with their natural partners.
The function-associated conformational changes and disorder-to-order transitions may be brought about by alterations in environmental or cellular conditions. The importance of this mechanism originates from the following simple reasoning: a large decrease in conformation entropy, which accompanies disorder-to-order transition, uncouples specificity from binding strength. This phenomenon enables highly specific interactions to be easily reversible, which is beneficial for cells, especially in the inducible responses typically involved in signaling and regulation. In 2000 and 2002, NMR analysis revealed the existence of specific “pre-existing”, “pre-organized”, “structural pre-ordering”, and “preformed” structural elements in several IDPs. These studies not only experimentally demonstrated existence of IDPs, but also showed that pre-existing (or pre-organized, pre-ordering, preformed) elements in IDPs are actively involved in target binding [53-58]. A recent computational study of such binding illustrated that the disordered partner contains a “conformational preference” for the structure it will take upon binding, and that these so-called “preformed elements” tend to be helices [262]. This research validates previous findings for individual protein-protein interactions, such as p27Kip1 [53, 263] and p53 [55], both of which have disordered regions with significant helical character that form α-helices upon binding to their partners.
Prediction of the sequence locations of regions that undergo disorder-to-order transitions has been accomplished [114]. This work was derived from observations that PONDR® VL-XT sometimes gives short regions of predicted order bounded by regions of predicted disorder for several known binding sites [88]. This was first noticed in the 4E binding protein one (4EBP1), which NMR studies had shown to be completely disordered [264]. However, a short stretch of 4EBP1 undergoes a disorder-to-order transition upon binding to eukaryotic translation initiation factor 4E [265]. The activity of eukaryotic translation initiation factor 4E is inhibited by this binding. The structure of this complex is shown in Figure 12A, whereas the PONDR® VL-XT prediction for 4EBP1 is shown in Figure 12B. Note that there is a sharp dip in the PONDR® VL-XT score in the area of the binding region. This drop is flanked by long regions of predicted disorder.
Figure 12. Examples of binding regions and their positions relative to PONDR®predicted order.
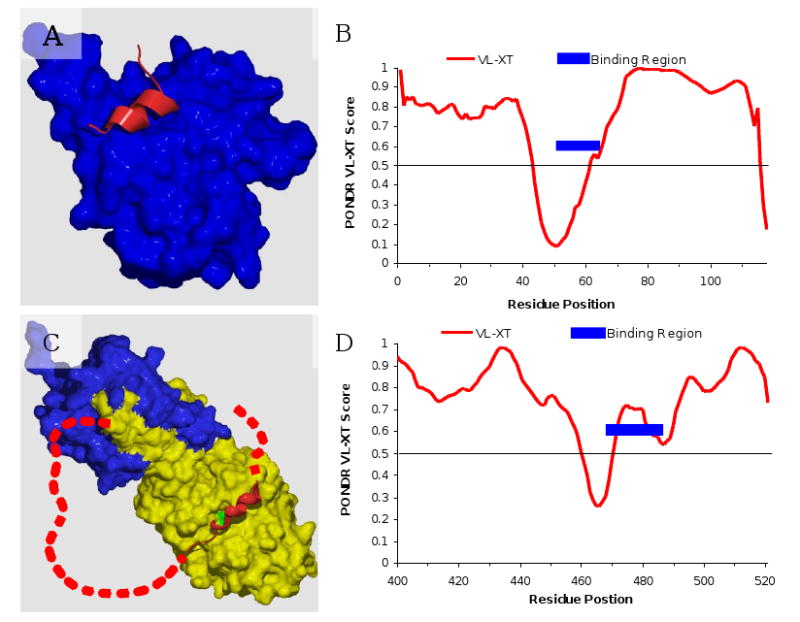
A. Eukaryotic initiation factor (blue) and the binding region of 4EBP1 (red). B. The PONDR® VL-XT prediction for 4EBP1 with the binding region designated (blue bar). C. The B (blue) and A (yellow) subunits of calcineurin and the autoinhibitory region of the A subunit (red helix) in the midst of observed disordered sequence (red dashes). D. The PONDR® VL-XT prediction for the last 121 amino acid residues of the A subunit with the autoinhibitory region indicated (blue bar). Modified from [114].
A similar situation exists in a case of calcineurin, a calcium-calmodulin dependent protein phosphatase, which is important in many signaling pathways. The crystal structure of calcineurin is shown in Figure 12C [266]. The A subunit's catalytic domain is followed by a stretch of 95 residues invisible in the x-ray crystal, a short (18 residue) helix spanning the autoinhibitory domain, and another stretch of 35 residues missing from the X-ray crystal [266]. The PONDR® VL-XT prediction of most of this region is shown in Figure 12D, which illustrates that the region of predicted order overlaps with the autoinhibitory helix.
Another interesting example is the E. coli ribonuclease RNase E protein. Using a combination of several biophysical techniques, Luisi and colleagues determined that the C-terminal half of the RNase E, which is involved in multiple protein-protein interactions, is intrinsically disordered. Application of PONDR® VL-XT gives four sharp downward spikes in disordered C-terminal half of this protein, which were labeled A, B, C, and D in the graph showing the prediction [267]. Synthetic peptides corresponding to these regions were tested for their binding to various partners of RNase E. Fragment C bound enolase, and fragment D bound PNPase. Peptide B was shown to contribute to RNA binding. Finally, peptide A became involved in a coiled-coil. Given the functional importance of the peptides A, B, C, and D, the authors named the corresponding dips in the PONDR VL-XT plots “regions of increased structural propensity” (RISPs). In this nomenclature, the peptides could be renamed RISP A, B, C, and D, respectively.
The crystal structure of the RISP C/enolase complex was subsequently determined, and RISP C was found to exist mostly as an α-helix [268]. These findings clearly show that short regions of predicted order bounded by extended regions of predicted disorder in PONDR® VL-XT plots can be used to identify binding sites that involved disorder-to-order transitions upon complex formation.
These findings by Luisi and co-workers are consistent with work briefly mentioned above indicating that downward spikes or dips in PONDR VL-XT plots commonly indicate binding regions [75, 114, 116]. Here we will discuss this work in more detail.
Based on many additional observations similar to those given above for 4E BP1, the calcineurin autoinhibitory domain, and RNase E, we proposed the existence of particular protein structural elements or features that mediate the binding events of initially disordered regions. This element consists of a short region that undergoes coupled binding and folding within a longer region of disorder. Originally, these features were termed “molecular recognition elements,” (MoREs). Later, they were renamed Molecular Recognition Features (MoRFs), to emphasize their conformational transition as they “morph” from disorder to order. An algorithm has been recently elaborated [114] that identifies regions having a propensity for of α-helix-forming molecular recognition features (α-MoRFs) based on a discriminant function that indicates such regions while giving a low false-positive error rate on a collection of structured proteins. Application of this predictor to databases of genomics and functionally annotated proteins indicates that α-MoREs are likely to be play important roles protein-protein interactions involved in signaling events [114].
Later, the MoRF prediction algorithms were improved by (1) including additional α-MoRF examples and their cross species homologues in the positive training set; (2) extracting monomer structure chains from PDB as the negative training set; (3) including attributes from recently developed disorder predictors, secondary structure predictions, and amino acid indices as attributes; and (4) constructing neural network based predictors and performing validation [116]. The original MoRF algorithm was trained on a small number of α-MoRF examples (14 regions from 12 proteins). Over 50 MoRF regions from PDB plus one cross species homologue of each structure-based example were included in the new positive training set. Over 1500 attributes, including disorder predictions, secondary structure predictions and amino acid indices were evaluated by conditional probability method. The top attributes, including VSL2 and VL3 disorder predictions and other physicobiological propensities, were used to develop the feed forward neural networks. Since the first stage of the stacked predictor architecture was the identification of potential MoRF regions from disorder prediction profiles, an appropriate disorder predictor should be chosen which would consistently identifies known binding regions as “dips” – or short regions of predicted order within longer regions of predicted disorder – in the profiles of predicted disorder probability. To this end, the performance of disorder predictors such as PONDR® VLXT, PONDR® VL3, PONDR® VL2, PONDR® VSL2B, PONDR® VSL2P, PONDR® VL3BA, RONN, IUPred, DisPro, DRIPPRED, and DISOPRED on a set of ID proteins with known binding regions was analyzed. Although many predictors gave similar general disorder/order predictions for the target proteins, PONDR® VLXT was more sensitive for features associated with regions potentially undergoing disorder-to-order transition than other predictors and therefore it was selected for the identification of potential MoRFs for the first stage of the prediction algorithm [116]. Combination of all these efforts produced a novel, highly accurate MoRF identifying tool, the sensitivity, specificity and accuracy of which were 0.87 ± 0.10, 0.87 ± 0.11, and 0.87 ± 0.08 over 10-cross validation, respectively [116].
Systematic studies of PDB entries revealed that protein complexes deposited in this database often comprise a short peptide bound to a larger globular protein. Analysis of literature data showed that some of these short peptides, being specifically folded (in a form of α-helix, β-hairpin, β-strand, polyproline II helix, or irregular structure, etc.) within their complexes with globular partners, are intrinsically disordered prior the corresponding complex formation. Thus, all these regions can be considered as illustrative members of the subset of protein-protein interactions involving disorder-to-order transitions during the complex formation, known as MoRFs. In our recent study, 2,512 polypeptide chains were extracted from PDB, which satisfied the prerequisite of being short (lengths ≤ 70 residues) and bound to a globular partner (with chains ≥ 100 residues). As the next step, all sequences having any occurrences of ‘X’ and ‘Z’ and being shorter than 10 residues were removed. This preprocessing of data resulted in a dataset comprising of ∼1,200 chains (∼55,000 residues and an average chain length of 33.5 residues). This was followed up by the removal of redundancy amongst these ∼1200 chains. The final dataset included 372 non-redundant protein chains (9,093 residues). The secondary structure assignment showed that, 27% of this dataset consisted of α-helical residues, 12% were β-sheet residues and approximately 48% of the residues had an irregular conformation. The remaining 13% of the residues were found to be disordered as they were characterized by missing coordinate information in their respective PDB files [115]. We believe that these collections of experimentally proven α-, β-, and ι-MoRFs represent useful datasets that can be used to find sequence attributes for discriminating these different MoRFs from each other and from ordered proteins.
One interesting observation on the disorder-based prediction of these binding sites is that MoRFs that form helix or sheet upon binding to their partners usually exhibit dips in their corresponding PONDR VL-XT plots, whereas regions that bind as irregular structure often exhibit high-value but featureless curves in their corresponding PONDR VL-XT plots [269]. Additional work is in progress to determine whether the binding regions that acquire helix or sheet structure are systematically correlated with dips in the PONDR plots while binding regions that lack structure are systematically correlated with the absence of such dips.
Other predictors of disorder are also beginning to be used to identify potential binding sites in disordered regions and these binding sites are being named “ANCHORs” [270]. An interesting aspect of this work is the proposal that long regions of disorder apparently contain localized ANCHORs that correspond to more tightly binding subregions within the longer disordered region. Perhaps longer binding disordered regions also have MoRF subregions, and perhaps there is an overlap between ANCHORs and MoRFs. Further work will test these possibilities.
Rather than disorder-based approaches, other workers are using motif-based approaches for identifying possible binding sites involving short linear regions of proteins, either eukaryotic linear motifs (ELMs) [271, 272] or short liner motifs (SLiMs) [273, 274]. One study shows considerable coincidence between binding sites found by MoRFs and those found by either motif-based methods [275], suggesting that ELMs, SLiMs and MoRFs are all similar, and also suggesting that ELMs and SLiMs occur mostly in disordered regions. This conclusion was further supported by a recent study of the evolutionary conservation of SLiMs recognized by SH2, SH3 and Ser/Thr kinase domains in both ordered and disordered protein regions was systematically analyzed. This analysis revealed that SLiMs were more conserved in disordered regions rather than in ordered regions. This correlation between SLiM conservation and disorder prediction demonstrated that functional SLiMs recognized by SH2, SH3 and Ser/Thr kinase domain occurred more often in disordered as compared to structured regions of proteins [276].
3. Intrinsic disorder and alternative splicing
Alternative splicing (AS) is a process by which two or more mature mRNAs are produced from a single precursor pre-mRNA by the inclusion or omission of different segments [277, 278]. The “exons” are joined to form the mRNA and the “introns” are left out [279]. But so far, AS of mRNA has been commonly observed only in multicellular eukaryotes [280], including plants, apicompexans, diatoms, amoebae, animals and fungi (reviewed in [281]). Genes shared among animals, fungi and plants show high levels of alternative splicing, suggesting that some type of alterntive splicing may have been already present in the unicellular ancestor of these groups, and further suggesting that alternative splicing was likely co-opted to help solve problems associated with the evolution of multicellularity [281].
For humans and other mammals, multiple proteins are often produced from a single gene since 40 – 60% the genes yield proteins via the AS mechanism [282-284] with more recent studies giving even higher estimates for the percentages of alternatively spliced genes. AS very likely provides an important mechanism for enhancing protein diversity in multicellular eukaryotes [285]. AS has affects on a diversity of protein functions such as protein-protein interactions, ligand binding, and enzymatic activity [286-288]. Therefore it comes as no surprise that abnormal AS has been associated with numerous human diseases, examples being myotonic dystrophy [289], Axoospermia [290], Alzheimer's disease [291], Parkinson's disease [292, 293], and cancer [294].
Removal of a piece of sequence from a structured protein would often lead to dysfunctional protein folding, most often causing loss of function (sometimes, however, the AS isoform of structured protein can maintain function, albeit typically with a reduction in activity). Why, then, is the AS phenomenon so common in nature? The analysis of the effect of AS on structured proteins revealed AS-induced alterations are generally of small size, are usually located on the protein surface, and are most often located in coil regions [295]. Given the small sizes and locations of the changes resulting from alternative splicing, the different splice variants were predicted to fold into the same overall structures, with only slight structural perturbations that could be functionally important [295, 296].
The structural implications given above are interesting, but only a small fraction of alternative splicing events have been mapped to structured proteins. Since 40% to 60% of mammalian (human) genes are estimated to undergo alternative splicing, and since there are several thousand mammalian proteins in PDB [157], we would expect to find several thousand examples to study. So far, however, despite exhaustive searches of PDB, only 20 examples have been reported [295]. Based on the failure to find a significant number of examples of alternative splicing that map to regions of structure, it was hypothesized that the protein folding problems discussed above would be solved for different isoforms if the alternatively spliced regions of mRNA were to code for regions of ID protein. If AS were to map to ID regions, both multiple and long splice variants would be allowed because structural perturbation would not be a problem.
To test this hypothesis a collection of human proteins with structurally characterized regions of order and disorder was built and an exhaustive search on alternative splicing for all of these proteins was performed. This generated a set of 46 human proteins with 75 alternatively spliced segments all of which were located in structurally characterized regions [297]. Importantly, of these 75 alternatively spliced regions of RNA, 43 (57%) coded for entirely disordered protein regions, 18 (24%) coded for regions containing both ordered and disordered subregions (with the splice boundaries very often in, or very near to, the disordered regions), and just 14 (19%) coded for fully structured regions [297]. Next, to increase the number of examples, a collection of SwissProt human proteins labeled as having AS isoforms were identified. This approach generated a set of 558 proteins with 1,266 regions that are absent from one isoform due to AS. Disorder/order propensities of these AS proteins and regions were predicted together with the disorder/order propensities of the 46 structurally characterized proteins and for their 75 regions that were affected by alternative splicing. This analysis revealed an excellent correlation between predictions and observations of disorder in the 46 structurally characterized proteins. For the 1,266 regions from SwissProt, the predicted abundance of disorder closely matched the corresponding predictions for the 75 with known structure. These data strongly suggest that AS occurs mostly in regions of RNA that code for disordered protein [297]. Recently another AS dataset was developed by assembling UniProt information irrespective of organism. This dataset contains 15,678 proteins with 36,320 AS regions. Disorder predictions on these AS fragments gave results nearly identical to those obtained for the smaller set of 1,266 AS regions mentioned above. Overall these data provide strong indication that AS regions of mRNA code for intrinsically disordered regions much more often than for structured regions.
These findings have crucial functional implementations. Since disorder plays various roles in protein functions and in protein-protein interaction networks, modification of such functions could be readily accomplished by AS within disordered regions. Thus, a linkage between AS and signaling by disordered regions provides a novel and plausible mechanism that could underlie and support cell differentiation, which ultimately gave rise to multicellular organisms in nature [297].
4. Controlled chaos: On the tight regulation of ID proteins in the living cells
ID proteins are real, abundant, diversified, and vital. Functions of ID proteins are mostly complementary to the catalytic activities of ordered proteins [70-72, 80-82, 92, 93, 101, 108, 211, 213, 215-217, 226, 298-300]. Many disorder-related functions (e.g., signaling, control, regulation and recognition) appear to be incompatible with well-defined, stable 3-D structures [70-73, 82, 92, 93, 108, 211, 213, 215-217, 226, 299-301]. Intrinsic disorder is assumed to provide several functional advantages including increased interaction surface area, structural plasticity to interact with several targets, high specificity for given partners combined with high kon and koff rates that enable rapid association with the partner without an excessive binding strength, the ability to fold upon binding and accessible post-translational modification sites. Structurally, ID proteins range from completely unstructured polypeptides (native coils, that resemble the highly unfolded states of globular proteins) to extended partially structured forms (native pre-molten globules) or even to compact disordered ensembles that may contain significant secondary structure (native molten globules) [42, 53-58, 72, 92, 93]. These proteins are highly abundant in nature (∼55% of eukatyotic proteins are predicted to contain at least one disordered region that is at least 40 amino acids in length [72] and are often associated with human diseases [302-304].
The highly dynamic nature of ID proteins points towards chaos. However, the evolutionary persistence of these highly dynamic proteins, their unique functionality and their involvement in all the major cellular processes provides evidence that this chaos is tightly controlled [305]. To answer the question on how are these proteins governed and regulated inside the cell Gsponer et al. conducted a detailed study focused at the intricate mechanisms of the ID protein regulation [306]. To this end, all the Saccharomyces cerevisiae proteins were grouped into three classes using one of the available disorder predictors, DisoPred2 [307]: (i) 1971 highly ordered proteins with just 0 – 10% of their residues predicted to be disordered; (ii) 2711 moderately disordered proteins with 10 – 30% of their residues predicted to be disordered; and (iii) 2020 highly disordered proteins containing 30 – 100% of their residues predicted to be disordered. Then, the correlations between intrinsic disorder and the various regulation steps of protein synthesis and degradation were evaluated.
To examine the transcription of genes encoding ID proteins and ordered proteins, the transcriptional rates and the degradation rates of the corresponding transcripts were compared [306]. This analysis revealed that the transcriptional rates of mRNAs encoding ID proteins and ordered proteins were comparable. However the ID protein-encoding transcripts were generally less abundant than transcripts encoding ordered proteins due to the increased decay rates of the former set (see Figure 13).
Figure 13. Mechanisms of IDP regulation inside the cell.
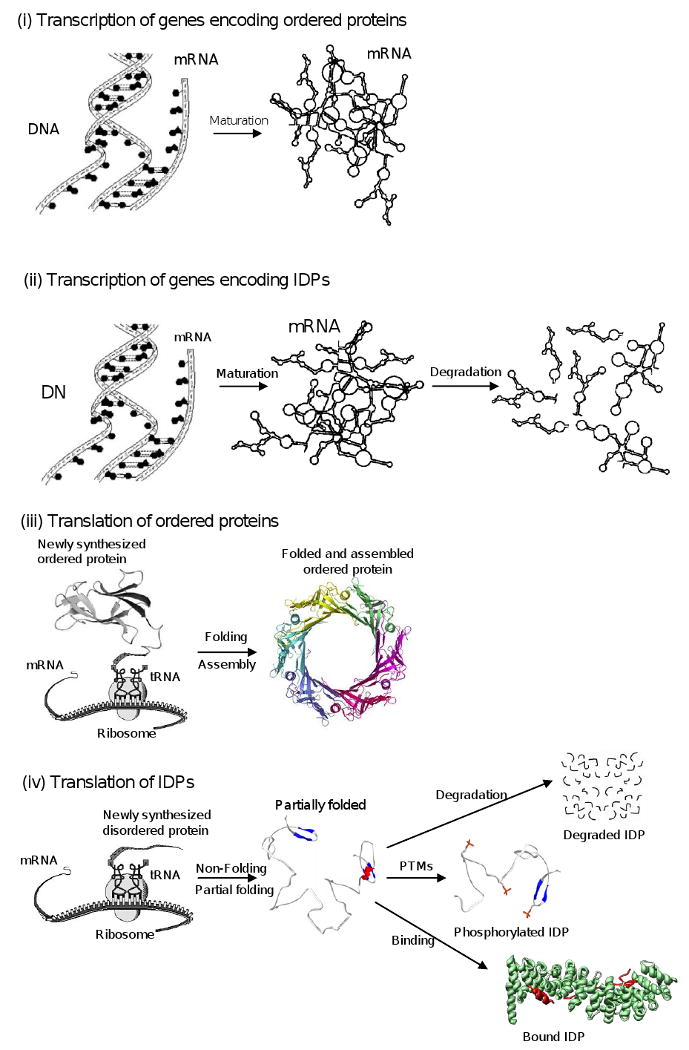
Regulation of ordered proteins (i) and IDPs (ii) at the transcriptional level. mRNAs encoding ordered proteins and IDPs are transcribed with comparable rates; however, IDP-encoding mRNAs are subjected to faster degradation. Therefore, the pool of the IDP-encoding mRNAs is significantly smaller than the number of mRNAs encoding the ordered proteins. Regulation of ordered proteins (iii) and IDPs (iv) at the translational level. The biosynthesis of ordered proteins is noticeably faster than that of IDPs. When synthesized, IDPs are either subjected to fast degradation, to various posttranslational modifications, PTMs, (including phosphorylation as shown in the plot), or to binding with specific partners. As a result of slow transcription and fast degradation, the overall level of IDPs inside the cells is lower and their half-lives are generally shorter than those of ordered proteins. However, some IDPs can be present at high quantities and/or for long periods of time due to either specific PTMs or due to the interactions with some specific factors.
The existence of tight regulation of the IDP abundance was also established at the protein level. In fact, ID proteins were shown to be less abundant than ordered proteins due to the lower rate of protein synthesis and shorter protein half-lives (see Figure 13). As the abundance and half-life in a cell of certain proteins can be further modulated via their post-translational modifications such as phosphorylation [308], the experimentally determined yeast kinase-substrate network was analyzed next. The ID proteins were shown to be substrates of twice as many kinases as were the ordered proteins. Furthermore, the vast majority of kinases whose substrates were ID proteins were either regulated in a cell-cycle dependent manner, or were activated upon exposure to particular stimuli or stress [306]. Therefore, post-translational modifications may not only serve as an important mechanism for the fine-tuning of ID protein functions, but possibly these modifications may also be necessary to tune the ID protein availability under the different cellular conditions. In addition to Saccharomyces cerevisiae, similar regulation trends were also found in Schizosaccharomyces pombe and Homo sapiens [306]. Based on these observations it has been concluded that both unicellular and multicellular organisms appear to use similar mechanisms for regulation of the ID protein availability.
Overall, this study clearly demonstrated that there is an evolutionarily conserved tight control of synthesis and clearance of most ID proteins. This tight control is directly related to the major roles of ID proteins in signaling, where it is crucial for a given protein to be available in appropriate amounts and not to be present longer than needed [306].
Although the abundance of many ID proteins may be under strict control as discussed above, some ID proteins could be present in cells in large amounts or/and for long periods of time due to either specific post-translational modifications or via interactions with other factors. These events could promote changes in cellular localization of ID proteins or protect them from the degradation machinery [72, 216, 308-310]. Therefore, the chaos seemingly introduced into the protein world by the discovery of ID proteins is under the tight control [305].
In an independent study, a global scale relationship between the predicted fraction of protein disorder and RNA and protein expression in E. coli was analyzed [311]. It has been shown that fraction of protein disorder were positively correlated with both measured RNA expression levels of E. coli genes in three different growth media (LB rich medium and N-C- minimal media supplemented with glycerol as a carbon source and either ammonium chloride or arginine as a nitrogen source) and with predicted abundance levels of E. coli proteins. When a subset of 216 E. coli proteins that are known to be essential for the survival and growth of this bacterium were analyzed, the correlation between protein disorder and expression level became even more evident. In fact, essential proteins had on average a much higher fraction of disorder (0.36), had a higher number of proteins classified as completely disordered (19% vs. 2% for E. coli proteome), and were expressed at a higher level in all three media than an average E. coli gene [311].
To better understand the function-disorder relationship for highly expressed E. coli proteins, manual literature mining was carried out for a group of proteins that had high levels of predicted intrinsic disorder, revealing that the disorder predictions matched well with the experimentally elucidated regions of protein flexibility and disorder [311]. A direct link between protein disorder and protein level in E. coli cells could also result because the disordered proteins may carry out essential control and regulation functions that are needed to respond to the various environmental conditions. Another possibility is that ID proteins might undergo more rapid degradation compared to structured proteins, which cells can counter by increasing mRNA levels of the corresponding genes. In this case, higher synthesis and degradation rates could make the levels of these proteins very sensitive to the environment, with slight changes in either production or degradation leading to significant shifts in protein levels [311].
Even more support for the tight control of ID proteins inside the cell has come from the analysis of cellular regulation of so-called “vulnerable” proteins [312]. The integrity of the soluble protein functional structures is maintained in part by a precise network of hydrogen bonds linking the backbone amide and carbonyl groups. In a well-ordered protein, hydrogen bonds are shielded from water attack, preventing backbone hydration and the total or partial denaturation of the soluble structure under physiological conditions [313, 314]. Since soluble protein structures may be more or less vulnerable to water attack depending on their packing quality, a structural attribute, protein vulnerability, was introduced as the ratio of solvent-exposed backbone hydrogen bonds (which represent local weaknesses of the structure) to the overall number of hydrogen bonds [312].
Vulnerability can be related to protein intrinsic disorder as the inability of a particular protein fold to protect intramolecular hydrogen bonds from water attack may result in backbone hydration leading to local or global unfolding. Since binding of a partner can help to exclude water molecules from the microenvironment of the preformed bonds, a vulnerable soluble structure gains extra protection of its backbone hydrogen bonds through the complex formation [313].
To understand the role of structure vulnerability in transcriptome organization, the relationship between the structural vulnerability of a protein and the extent of co-expression of genes encoding its binding partners was analyzed. This study revealed that structural vulnerability can be considered as a determinant of transcriptome organization across tissues and temporal phases [312]. Finally, by interrelating vulnerability, disorder propensity and co-expression patterns, the role of protein intrinsic disorder in transcriptome organization was confirmed, since the correlation between the extent of intrinsic disorder of the most disordered domain in an interacting pair and the expression correlation of the two genes encoding the respective interacting domains was evident [312].
5. D2 concept: Disorder in disorders
5.1. What is the relationship between ID proteins and human diseases?
Because ID proteins play crucial roles in numerous biological processes, many of these proteins are implicated in human disease. For example, several human diseases originate from the deposition of stable, ordered, filamentous protein aggregates, commonly referred to as amyloid fibrils. In each of these pathological states, a specific protein or protein fragment changes from its natural soluble form into insoluble fibrils, which accumulate in a variety of organs and tissues [315-321]. More than 20 different proteins are known so far to be involved in these diseases. These proteins are unrelated in terms of sequence or starting structure.
Several ID proteins are found among the amyloidogenic proteins and are clearly associated with the development of neurodegenerative diseases [321, 322]. An incomplete list of disorders associated with ID proteins includes Alzheimer's disease (deposition of amyloid-β, tau-protein, α-synuclein fragment NAC [323-326]; Niemann-Pick disease type C, subacute sclerosing panencephalitis, argyrophilic grain disease, myotonic dystrophy, and motor neuron disease with neurofibrillary tangles (accumulation of tau-protein in form of neurofibrillary tangles [325]); Down's syndrome (nonfilamentous amyloid-β deposits [327]); Parkinson's disease, dementia with Lewy body, diffuse Lewy body disease, Lewy body variant of Alzheimer's disease, multiple system atrophy and Hallervorden-Spatz disease (deposition of α-synuclein in a form of Lewy body, or Lewy neuritis [328]); prion diseases (deposition of PrPSC [329]); and a family of polyQ diseases, which are a group of neurodegenerative disorders caused by expansion of GAC trinucleotide repeats coding for PolyQ in the gene products [330]. Furthermore, most mutations in rigid globular proteins associated with accelerated fibrillation and protein deposition diseases have been shown to destabilize the native structure, increasing the steady-state concentration of partially folded (disordered) conformers [315-321].
The maladies given above have been called conformational diseases, as they are characterized by the conformational changes, misfolding and aggregation of an underlying protein. However, there is another side to this coin: protein functionality. In fact, many of the proteins associated with the conformational disorders are also involved in recognition, regulation and cell signaling. For example, functions ascribed to α-synuclein, a protein involved in several neurodegenerative disorders, include binding fatty acids and metal ions; regulation of certain enzymes, transporters, and neurotransmitter vesicles; and regulation of neuronal survival (reviewed in [328]). Overall, there are about 50 proteins and ligands that were shown to physically interact and/or co-localize with this protein. Furthermore, α-synuclein has amazing structural plasticity and adopts a series of different monomeric, oligomeric and insoluble conformations (reviewed in [331]). The choice between these conformations is determined by the peculiarities of the protein environment, assuming that α-synuclein has an exceptional ability to fold in a template-dependent manner. Based on these observations, we hypothesize that the development of the conformational diseases may originate from the misidentification, misregulation and missignaling, accompanied by misfolding. In other words, mutations and/or changes in the environment may result in protein confusion, for which its ID becomes lost, thus reducing its capability to recognize proper binding partners and leading to the formation of nonfunctional and deadly aggregates.
Recent analysis of so-called polyglytamine diseases gives support to this hypothesis [332]. Polyglytamine diseases are a specific group of hereditary neurodegeneration caused by expansion of CAG triplet repeats in an exon of disease genes which leads to the production of a disease protein containing an expanded polyglutamine, polyQ, stretch. Nine neurodegenerative disorders, including Kennedy's disease, Huntington's diseases, spinocerebellar atrophy-1, -2, -3, -6, 7, 17, and dentatorubral pallidoluysian atrophy are known to belong to this class of diseases [333-336]. In most polyQ diseases, expansion to over 40 repeats leads to the onset [336].
Molecular processes such as the unfolded protein response, protein transport, synaptic transmission and transcription are all implicated in the pathology of polyQ diseases [332]. Importantly, more than 20 transcription-related factors have been reported to interact with pathological polyQ proteins. Furthermore, these interactions were shown to repress the transcription, leading finally to the neuronal dysfunction and death (reviewed in [332]). These results suggest that polyQ diseases represent kind of transcriptional disorder [332], supporting our misidentification hypothesis for at least some of the conformational disorders.
So far, three computational/bioinformatics approaches have been elaborated to estimate the abundance of IDPs in various pathological conditions. The first approach is based on the assembly of specific datasets of proteins associated with a given disease and the computational analysis of these datasets using a number of disorder predictors [101, 120, 302, 337-339]. In essence, this is an analysis of individual proteins extended to a set of independent proteins. A second approach utilized the diseasome, a network of genetic diseases where the related proteins are interlinked within one disease and between different diseases [340]. A third approach is based on the evaluation of the association between a particular protein function (including the disease-specific functional keywords) with the level of intrinsic disorder in a set of proteins known to carry out this function [215-217]. These three approaches are briefly described below, whereas the results of their application are presented in the subsequent section.
For the first time, the dataset analysis approach was used in 2002 when it was found that 79% of cancer-associated and 66% of cell-signaling proteins contain predicted regions of disorder of 30 residues or longer [101]. In contrast, only 13% of a set of proteins with well-defined ordered structures contained such long regions of predicted disorder. For this study, cancer-associated proteins were defined as those human proteins in Swiss-Prot containing the keyword “oncogene” (this included anti- and proto-oncogenes) or containing the word “tumor” in the description field. In experimental studies, the presence of disorder has been directly observed in several cancer-associated proteins, including p53 [55], p57kip2 [341], Bcl-XL and Bcl-2 [342], c-Fos [343], and most recently, a thyroid cancer associated protein, TC-1 [344]. Following a similar analytical model, a dataset of 487 proteins related to cardiovascular disease (CVD) was collected and analyzed [338]. On average, CVD-related proteins were found to be highly disordered. The percentage of proteins with 30 or more consecutive disordered residues was 61% for CVD-associated proteins. Many proteins were predicted to be wholly disordered, with 101 proteins from the CVD dataset predicted to have a total of almost 200 specific disorder-based binding motifs (thus about 2 binding sites per protein), α-MoRFs [338]. Finally, the dataset analysis revealed that in addition to being abundant in cancer- and CVD-related proteins, intrinsic disorder is commonly found in such maladies as neurodegenerative diseases and diabetes [302, 339].
The human diseasome systematically links the human disease phenome (which includes 1,284 human genetic diseases, 867 of which had at least one link to other diseases, and 516 diseases formed a giant component) with the human disease genome (which contains 1,777 disease genes of which 1,377 were shown to be connected to other disease genes, and 903 genes belonged to a giant cluster) [345]. The abundance of intrinsic disorder in human diseasome was evaluated using a set of computational tools such as PONDR® VSL2, CDF-analysis, CH-plot, and α-MoRF prediction [340]. These analyses uncovered an unfoldome associated with human genetic diseases and revealed that intrinsic disorder is common in proteins associated with many human genetic diseases. Also different disease classes were shown to vary in the IDP contents of their associated proteins and α-MoRFs were found to be very common in the diseasome. Indeed, α-MoRF abundance correlated with the intrinsic disorder level. Finally, some disease classes were shown to have a significant fraction of genes affected by alternative splicing, and the alternatively spliced regions in the corresponding proteins were predicted to be highly disordered and in some diseases to contain a significant number of MoRFs [340].
The studies on correlation of ID with various functional key-words [215-217] revealed that many diseases show strong correlations with proteins predicted to be disordered. Contrary to this, no disease-associated proteins were found to be strongly correlated with absence of disorder [216]. Among disease-related Swiss-Prot keywords strongly associated with ID were oncoproteins, malaria, trypanosomiasis, human immunodeficiency virus (HIV) and acquired immunodeficiency syndrome (AIDS), deafness, obesity, cardiovascular disease, diabetes mellitus, albinism, and prion [216]. In agreement with this bioinformatics analysis, at least one illustrative, experimentally validated example of functional disorder or order was found for the vast majority of functional keywords related to diseases [216].
Summarizing, intrinsic disorder is highly abundant among proteins associated with various human diseases likely because of the importance of signaling to wide range of disorders. Since ID proteins are very common in various diseases, the “disorder in disorders” or D2 concept was introduced to summarize work in this area [302] and concepts of the disease-related unfoldome and unfoldomics were developed [304].
5.2. ID proteins as novel drug targets
Molecular recognition is the most fundamental process in biology and underlies essentially every process crucial to life. How one DNA strand recognizes another, how a protein recognizes a particular locus on a DNA molecule, how a single-stranded DNA binding protein recognizes ssDNA without regard to local sequence, how one protein recognizes another, how a protein recognizes a ligand, how an enzyme recognizes its substrate, and how a drug molecule specifically recognizes its target represent some of the well-studied examples of this phenomenon. Both our basic understanding of life and our ability to derive practical benefit from our understanding, such as the discovery of new drug molecules, depend on the mechanisms of molecular recognition. Molecular recognition is to life as the quantum mechanics of the hydrogen atom is to the periodic table and chemistry.
With regard to molecular recognition by proteins, for more than 100 years thinking has been dominated by the lock and key concept according to which the specific functionality of a given protein is predetermined by the unique spatial positioning of amino acid side chains and prosthetic groups, predestinated, in turn, through a defined three-dimensional (3D) structure. This concept has represented one of the cornerstones in protein biology, chemistry and physics. This model has prevailed for more than a century, both creating and shaping the universe of modern protein science.
As discussed above and elsewhere, the functions of ID proteins and regions may arise from the specific disordered form, from inter-conversion of disordered species, or from transitions between disordered and ordered conformations. Very often ID proteins/regions are involved in regulation, signaling and control pathways, where binding to multiple partners and high-specificity/low-affinity interactions play crucial roles [300]. Since many proteins associated with various human diseases are either completely disordered or contain long disordered regions [302, 304], and since some of these disease-related ID proteins/regions are involved in recognition, regulation and signaling, these proteins/regions clearly represent novel potential drug targets.
The possibility of interrupting the action of disease-associated proteins (including via the modulation of protein-protein interactions) presents an extremely attractive objective for the development of new drugs [346, 347]. The rational design of enzyme inhibitors depends on the classical view of protein function, which states that three-dimensional structure is an obligatory prerequisite for function. While this approach has led to many successful drug molecules that target enzymatic domains, this approach has influenced thinking with regard to all types of protein functions, even those functions that depend on ID proteins and regions such as the protein-protein interactions described above. Due to the failure to recognize the important roles of disorder in protein function, current and evolving methods of drug discovery suffer from an overly rigid view of protein function.
Structure-aided design techniques have been modified and extended for the discovery of molecules that function as inhibitors of protein-protein interactions. Computational evaluations can be used to search a protein's interaction surface for sites that will potentially bind small molecules [348], provided that a reliable 3-D structure is available. Techniques can then be applied to design small molecules that will bind to these surfaces and simultaneously prevent hydrophobic collapse of the targeted protein-protein interaction. The low affinity of these leads for their targets often limits the biological relevance of these molecules, because high affinity is typically needed for successful drug molecules. The higher the affinity, the lower the dose needed to cause effect. Lower doses can help to reduce the chance of side effects that might arise from alternative, low affinity interactions. For these and numerous other reasons, the search for drug molecules that act by disrupting protein-protein interactions has been mostly unsuccessful, with a contributing factor being that many of the approaches were borrowed directly from techniques used for the discovery of enzyme inhibitors [349].
The difficulty of designing drugs that target protein-protein interaction surfaces may result from the inherent contradiction between the character of structured protein-protein interfaces and the nature of small molecule binding. As accessed from available crystal structures, the majority of small molecule effectors greater than 300 Da bind a contiguous epitope of five or more residues [350]. In contrast, many interfaces between two ordered proteins are much more complex, with epitopes consisting of discontinuous or a combination of multiple contiguous epitopes [351]. The divergent natures of these two binding systems can help to explain why attempts to find small molecule inhibitors of protein-protein interactions have so far met with limited success.
A new approach that resolves the inherent contradiction in discovering drugs that target protein-protein interactions would accelerate the creation of new drugs that act by inhibiting specific interconnections of protein signaling and regulation networks. One such alternative approach would be to target protein-protein interactions that utilize disordered regions in proteins. Disordered proteins often bind their partners with a relatively short length of contiguous residues, which become ordered upon binding [88, 114, 116]. Two general concepts for targeting interactions involving ID proteins and regions can be envisioned. If one partner is disordered and the second is structured, one can target the binding site on the structured partner. This would be similar to traditional drug discovery approaches. Alternatively, one can target possible binding sites on the ID protein or region directly, especially if both partners lack fixed structure. An interesting twist of this second disorder-based approach for drug discovery is that targeting disordered regions can be described as inducing structure to prevent function. By now, finding molecules that bind to structured partners of disordered proteins and finding molecules that bind directly to specific sites in disordered regions have both identified promising leads for the drug discovery process. Each of these alternative approaches will be discussed in turn.
Promising molecules have been found that act by binding to the structured protein Mdm2 and thereby block the binding of a disordered region of p53. This is an illustration of the first of the two alternative approaches mentioned in the preceding paragraph. To give some background regarding how this discovery was made, we will first discuss the p53 molecule and the studies on this molecule that led to the novel approach for drug discovery.
The p53 tumor suppressor protein is at the center of a large signaling network. This protein regulates expression of genes involved in numerous cellular processes, including cell cycle progression, apoptosis induction, DNA repair, as well as others involved in responding to cellular stress [250]. When p53 function is lost, either directly through mutation or indirectly through several other mechanisms, the cell often undergoes cancerous transformation [352, 353]. Cancers showing mutations in p53 are found in colon, lung, esophagus, breast, liver, brain, reticuloendothelial tissues and hemopoietic tissues [352].
When activated, p53 accumulates in the nucleus and binds to specific DNA sequences [353, 354]. The p53 molecule induces or inhibits over 150 genes, including p21, GADD45, MDM2, IGFBP3, and BAX [354]. The overall structure of p53 consists of three primary domains: an amino-terminal transactivation region, a central DNA binding domain, and a carboxy-terminal tetramerization and regulatory region. The transactivation region of p53 interacts with TFIID, TFIIH, MDM2, RPA, CBP/p300 and CSN5/Jab1 among other proteins [250]. In addition, the p53 protein is post-translationally modified in many ways and these modifications affect its interaction with various protein partners. The majority of the known modifications are phosphorylations, but there are also multiple acetylations and even one SUMOylation [250]. All of the modification sites except two (at residues 20 and 320) are found in regions predicted to be disordered by PONDR® [251]. All of this information taken together clearly illustrates that p53 is a critical protein within the cell.
The p53 protein is regulated by several different mechanisms, but the one being discussed in detail here is the inhibition of its activity by binding to Mdm2, an E3 ubiquitin ligase. Mdm2 associates with a short stretch of p53, residues 13-29 (see Figure 14A). This region of p53 is within the transactivation domain, thus p53 cannot activate or inhibit other genes when Mdm2 is bound. Mdm2 binding leads to ubiquitination of p53 and thus targets it for destruction. Mdm2 also contains a nuclear export signal that causes p53 to be transported out of the nucleus.
Figure 14. Druggable p53–Mdm2 interaction.
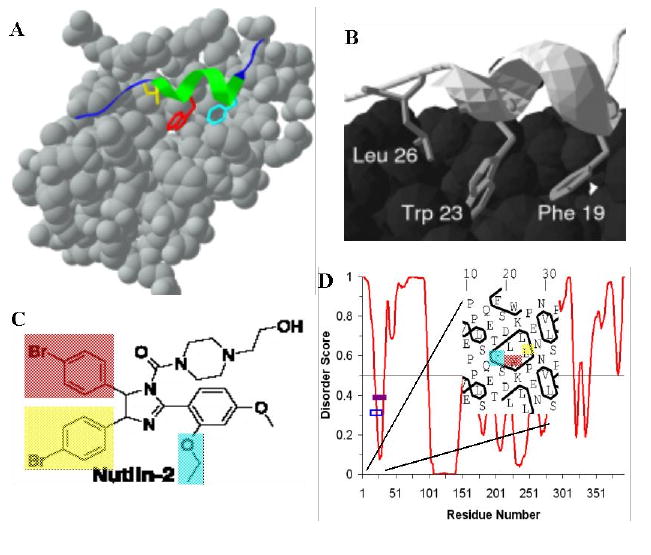
Protein disorder features and small molecule design. The p53 peptide (in color) bound to Mdm2 (PDB 1YCR, in gray scale) is shown in (A). Close-up view of p53 (ribbon) bound to Mdm2 (globular). The side chains of p53's crucial residues for the interaction (Phe 19, Trp 23, Leu 26) are shown (B). Notice that residues Phe19, Trp23 and Leu26 of p53 are pointing into the Mdm2 binding pocket. By comparison, the small molecule nutlin-2, designed to mimic the side chains of the residues from p53 is shown in (C). The PONDR® VL-XT plot of p53 is shown in (D), which indicates that this fragment of p53 might undergo disorder-to-order transition upon binding to Mdm2. The purple bar represents the predicted α-MoRF region (α-helical molecular recognition feature) [114, 116], the hollow box represents the determined binding region, which shows a good agreement between the two. Hydrophobic cluster analysis of binding region is shown. Figure is modified from [364].
X-ray crystallographic studies of the p53-Mdm2 complex reveal that the Mdm2 binding region of p53 forms a helical structure that binds into a deep groove on the surface of Mdm2 (see Figure 14A) [355]. On the other hand, NMR studies of p53 show that the unbound N-terminal region lacks fixed structure, although it does possess an amphipathic helix that forms secondary structure part of the time [55] and therefore represents an illustrative example of the α-MoRF concept. This amphipathic helix observed part of the time in the unbound state is the same helix that binds to Mdm2. A close examination of the interface between the p53 and Mdm2 proteins reveals that Phe19, Trp23, and Leu26 of p53 are the major contributors to the interaction, with the side chains of these three amino acids pointing down into a crevice on the Mdm2 surface (Figure 14B).
Because of the apparent simplicity of the interface, as well as the importance of the p53-Mdm2 interaction, this protein-protein interaction has been investigated as a possible drug target by many researchers. Several peptide inhibitors of the interaction have been created [356-359]. These peptides were all derived from the region of p53 that binds to Mdm2. Additionally all successful peptide inhibitors contained the three crucial residues involved in the interaction, Phe19, Trp23, and Leu26 [360].
In addition to the peptide inhibitors, several small, drug-like molecules have been found to block the p53-Mdm2 interaction [360-363]. While some of these were natural products, others were from a class of cis-imidazolines called “Nutlins”. These latter molecules increased the level of p53 in cancer cell lines. This drastically decreased the viability of these cells, causing most of them to undergo apoptosis. When one of the Nutlin compounds was given orally to mice, researchers saw a 90% inhibition of tumor growth compared to the control. The structure of Nutlin-2 (Figure 14C) was shown to mimic the crucial residues of p53, with two bromophenyl groups fitting into Mdm2 in the same pockets as Trp23 and Leu26, and an ethyl-ether side chain filling the spot normally taken by Phe19 [361-363].
Remarkably, the disorder prediction for p53 using PONDR® VL-XT software showed a sharp downward spike (near residue 25 in Figure 14D) indicating predicted ordered region near the N-terminus of the protein. Furthermore, the α-MoRF identifier was able to recognize the region of p53 that binds to Mdm2 as a region of molecular recognition [251]. These data suggest that the disordered sides of druggable protein-protein interactions are predictable. This successful nutlin story marks the potential beginning of a new era, the signaling-modulation era, in targeting drugs to protein-protein interactions. Importantly, this druggable p53-Mdm2 interaction involves a disorder-to-order transition. Principles of such a transition are generally understood and therefore can use to find similar drug targets [364].
In addition to nutlins, seven other promising drug molecules have been found that act by blocking protein-protein interactions [346, 347]. While protein disorder is not mentioned in any of the papers describing how a small molecule can block protein-protein interactions, the disorder-based analysis revealed that four of these interactions involve one structured partner and one disordered partner, with 3 of the 4 disordered segments becoming helix upon binding (see Figure 15). Therefore, the p53 – Mdm2 complex is not the only member of this class currently known to be blocked by a small drug-like molecule. We fully expect many more examples to appear shortly, and we also expect some of these examples to lead to useful drug molecules. Our previous bioinformatics studies suggest that p53-Mdm2-like interactions are likely to be extremely common [114, 116]. We therefore predict that this type of interaction will lead eventually to a cornucopia of new drug targets that operate by blocking disorder-based protein-protein interactions.
Figure 15. IDPs as drug targets.

Protein-protein interactions involving α-helical or β-strand portion of the partners are used to design small molecules for cancer drugs. A. A ribbon diagram of complex of Bcl-xL and BAK fragment was regenerated from PDB 1BXL. Small molecules were designed based on the 20-residue helix of BAK to inhibit the interaction. B. A ribbon diagram of complex of MDM2 and P53 fragment was regenerated from PDB 1YCR. Small molecule inhibitors were designed based on the structure of the helical fragment of P53. C. A ribbon diagram of complex of IL-2 receptor α and IL-2 was regenerated from PDB 1Z92. Small molecules were designed based on the α-helix portion of IL-2 that interacts with the receptor. D. A ribbon diagram of complex of β-catenin and T cell factor was regenerated from PDB 1G3J. The structure of β-catenin is consisted of 12 tri-helical repeats (except the repeat 7, which just has two helical units). Small molecules from a natural-product library were screened and a couple of inhibitors were found. However, the binding sites for the small molecule inhibitors were not clear. E. A ribbon diagram of complex of XIAP and Smac fragment was regenerated from PDB 1G3F. Small molecule inhibitors were designed based on the β-strand fragment (AVPIAQKSE) of Smac.
For the examples given above, the drug molecules mimic a critical region of the disordered partner (which folds upon binding) and compete with this region for its binding site on the structured partner. We argue that these druggable sites are likely to operate by the coupled binding and folding mechanism and utilize interaction sites that are small enough and compact enough to be easily mimicked by small molecules. We have developed methods for predicting such binding sites in disordered regions [349] and have elaborated the bioinformatics tools to identify which disordered binding regions can be easily mimicked by small molecules [364]. Overall, these examples represent the first approach mentioned above of how disorder can lead to drug discovery.
The second approach mentioned above is the targeting of small molecules to the disordered regions of proteins. Drugs targeting these regions will likely function through inducing the disordered region to form an ordered structure that is unlike its structure in its complex with its binding partner, thereby preventing the protein-protein interaction.
The principles of small molecule binding to disordered regions have not been well studied, but sequence specific, small molecule binding to short peptides was observed more than a decade ago [365]. This work has been followed up with more than 80 articles that cite this publication, but none of these follow-up articles have made the connection to drug discovery via to binding to disordered proteins.
Rather than searching specifically for small molecules that bind to ID proteins or regions, several laboratories found such molecules via an indirect approach. These several laboratories discovered small molecules, Myc-Max compounds, which inhibit the interaction between c-Myc and its obligate heterodimerization partner, Max [366-376]. Later it was discovered that these inhibitors bind to unstructured protein [375, 377]. Before discussing the role of disorder in these studies, we will first provide some biological background for these studies.
Deregulation of the c-Myc transcription factor is involved in many types of cancer, making this oncoprotein an attractive target for drug discovery. In order to bind DNA, regulated target gene expression, and function in most biological contexts, c-Myc must dimerize with Max, which lacks a transactivation segment. The interaction regions of both Max and c-Myc are disordered as monomers. They undergo mutual coupled binding and folding when their zipper domains interact to form a helical coiled coil [377].
One approach to c-Myc inhibition has been to disrupt the formation of this dimeric complex. In a search for effective inhibitors of the c-Myc-Max interactions, high throughput screening led to the discovery of 7 inhibitors [368, 373]. These molecules were subsequently shown to bind to one of three discrete sites within the 85-residue bHLHZip domain of c-Myc. These three sites are located within a region of c-Myc that is disordered before it binds to Max.
These binding sites are composed of short contiguous stretches of amino acids that can selectively and independently bind the small molecules. Inhibitor binding induces only local conformational changes, preserves the overall disorder of c-Myc, and inhibits dimerization with Max. Furthermore, the binding of multiple inhibitors to c-Myc was shown to occur simultaneously and independently on the three independent sites. Based on these observations it has been concluded that a rational and generic approach to the inhibition of protein-protein interactions involving ID proteins may therefore be possible through the targeting of ID sequence [377].
Ideally, a drug that targets a given protein-protein interaction should be tissue specific. Although some proteins are unique for a given tissue, many more proteins have very wide distribution, being present in several tissues and organs. How can one develop tissue-specific drugs targeting such abundant proteins?
Often, tissue specificity for many of the abundant proteins is achieved via the alternative splicing of the corresponding pre-mRNAs, which generates two or more protein isoforms from a single gene. Estimates indicate that between 35 and 60% of human genes yield protein isoforms by means of alternatively spliced mRNA [282]. The added protein diversity from alternative splicing is thought to be important for tissue-specific signaling and regulatory networks in the multicellular organisms. Recently, it has been established that the regions of alternative splicing in proteins are enriched in intrinsic disorder [297]. Since disorder is frequently utilized in protein binding regions, having alternative splicing of pre-mRNA coupled to regions of protein disorder was proposed to lead to tissue-specific signaling and regulatory diversity [297]. Therefore, associating alternative splicing with protein disorder enables the time- and tissue-specific modulation of protein function. These findings open a unique opportunity to develop tissue-specific drugs modulating the function of a given ID protein/region (with a unique profile of disorder distribution) in a target tissue and not affecting the functionality of this same protein (with different disorder distribution profile) in other tissues.
6. Conclusions
Intrinsically disordered (ID) proteins are widespread and represent a distinct protein tribe, with disorder being an important structural element that exists at various levels of protein structure. Such ID proteins are commonly involved in recognition, regulation and cell signaling functions and have biophysical characteristics that are well disposed for this role. They are much more common in eukaryota in comparison to prokaryota and archaea, reflecting the greater need for disorder-associated signaling and regulation in nucleated cells. Changes in the environment and/or mutation(s) would be expected to affect the normal function of the ID proteins, leading to misidentification and missignaling. This, in turn, can result in misfolding and aggregation, which are known to be associated with the pathogenesis of numerous disease states. Finally, disorder-based signaling represents a new opportunity to be exploited via the drug discovery process.
Acknowledgments
We thank Zoran Obradovic, Chris Oldfield, Bin Xue, Pedro Romero, Marc Cortese, and Ya-Yue Van for their continuing support and collaborations in the field of the ID proteins studies. This work was supported in part by the grants R01 LM007688-01A1 (to A.K.D and V.N.U.) and GM071714-01A2 (to A.K.D and V.N.U.) from the National Institutes of Health and the Program of the Russian Academy of Sciences for the “Molecular and cellular biology” (to V. N. U.). We gratefully acknowledge the support of the IUPUI Signature Centers Initiative.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Fischer E. Einfluss der configuration auf die wirkung der enzyme. Ber Dt Chem Ges. 1894;27:2985–2993. [Google Scholar]
- 2.Lemieux UR, Spohr U. How Emil Fischer was led to the lock and key concept for enzyme specificity. Adv Carbohydrate Chem Biochem. 1994;50:1–20. [PubMed] [Google Scholar]
- 3.Blake CC, Koenig DF, Mair GA, North AC, Phillips DC, Sarma VR. Structure of hen egg-white lysozyme. A three-dimensional Fourier synthesis at 2 Angstrom resolution. Nature. 1965;206:757–761. doi: 10.1038/206757a0. [DOI] [PubMed] [Google Scholar]
- 4.Kendrew JC, Bodo G, Dintzis HM, Parrish RG, Wyckoff H, Phillips DC. A three-dimensional model of the myoglobin molecule obtained by x-ray analysis. Nature. 1958;181:662–666. doi: 10.1038/181662a0. [DOI] [PubMed] [Google Scholar]
- 5.Kendrew JC, Dickerson RE, Stranberg BE, H RJ, Davies DR, Phillips DC, Shore VC. Structure of myoglobin: a three-dimensional Fourier synthesis at 2 Å resolution. Nature. 1960;185:422–427. doi: 10.1038/185422a0. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J Mol Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 7.Watts JD, Cary PD, Sautiere P, Crane-Robinson C. Thymosins: both nuclear and cytoplasmic proteins. Eur J Biochem. 1990;192:643–651. doi: 10.1111/j.1432-1033.1990.tb19271.x. [DOI] [PubMed] [Google Scholar]
- 8.Gast K, Damaschun H, Eckert K, Schulze-Forster K, Maurer HR, Muller-Frohne M, Zirwer D, Czarnecki J, Damaschun G. Prothymosin alpha: a biologically active protein with random coil conformation. Biochemistry. 1995;34:13211–13218. doi: 10.1021/bi00040a037. [DOI] [PubMed] [Google Scholar]
- 9.Uversky VN, Gillespie JR, Millett IS, Khodyakova AV, Vasiliev AM, Chernovskaya TV, Vasilenko RN, Kozlovskaya GD, Dolgikh DA, Fink AL, Doniach S, Abramov VM. Natively unfolded human prothymosin alpha adopts partially folded collapsed conformation at acidic pH. Biochemistry. 1999;38:15009–15016. doi: 10.1021/bi990752+. [DOI] [PubMed] [Google Scholar]
- 10.Boublik M, Bradbury EM, Crane-Robinson C, Johns EW. An investigation of the conformational changes of histone F2b by high resolution nuclear magnetic resonance. Eur J Biochem. 1970;17:151–159. doi: 10.1111/j.1432-1033.1970.tb01147.x. [DOI] [PubMed] [Google Scholar]
- 11.Venyaminov SY, Gudkov AT, Gogia ZV, Tumanova LG. Absorption and circular dichroism spectra of individual proteins from Escherichia coli ribosomes, Pushchino, Russia. 1981 [Google Scholar]
- 12.Breslow E, Beychok S, Hardman KD, Gurd FR. Relative conformations of sperm whale metmyoglobin and apomyoglobin in solution. J Biol Chem. 1965;240:304–309. [PubMed] [Google Scholar]
- 13.Stellwagen E, Rysavy R, Babul G. The conformation of horse heart apocytochrome c. J Biol Chem. 1972;247:8074–8077. [PubMed] [Google Scholar]
- 14.Fisher WR, Taniuchi H, Anfinsen CB. On the role of heme in the formation of the structure of cytochrome c. J Biol Chem. 1973;248:3188–3195. [PubMed] [Google Scholar]
- 15.Isbell DT, Du S, Schroering AG, Colombo G, Shelling JG. Metal ion binding to dog osteocalcin studied by 1H NMR spectroscopy. Biochemistry. 1993;32:11352–11362. doi: 10.1021/bi00093a012. [DOI] [PubMed] [Google Scholar]
- 16.Huber R, Bennett WS., Jr Functional significance of flexibility in proteins. Biopolymers. 1983;22:261–279. doi: 10.1002/bip.360220136. [DOI] [PubMed] [Google Scholar]
- 17.Sigler PB. Transcriptional activation. Acid blobs and negative noodles. Nature. 1988;333:210–212. doi: 10.1038/333210a0. [DOI] [PubMed] [Google Scholar]
- 18.Holt C, Sawyer L. Caseins as rheomorphic proteins: interpretation of primary and secondary structures of the αS1-, β- and κ-caseins. J Chem Soc Faraday Trans. 1993;89:2683–2692. [Google Scholar]
- 19.Holt C, Wahlgren NM, Drakenberg T. Ability of a beta-casein phosphopeptide to modulate the precipitation of calcium phosphate by forming amorphous dicalcium phosphate nanoclusters. Biochem J. 1996;314(Pt3):1035–1039. doi: 10.1042/bj3141035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Holt C, Timmins PA, Errington N, Leaver J. A core-shell model of calcium phosphate nanoclusters stabilized by beta-casein phosphopeptides, derived from sedimentation equilibrium and small-angle X-ray and neutron-scattering measurements. Eur J Biochem. 1998;252:73–78. doi: 10.1046/j.1432-1327.1998.2520073.x. [DOI] [PubMed] [Google Scholar]
- 21.Pontius BW. Close encounters: why unstructured, polymeric domains can increase rates of specific macromolecular association. Trends Biochem Sci. 1993;18:181–186. doi: 10.1016/0968-0004(93)90111-y. [DOI] [PubMed] [Google Scholar]
- 22.Bloomer AC, Champness JN, Bricogne G, Staden R, Klug A. Protein disk of tobacco mosaic virus at 2.8 A resolution showing the interactions within and between subunits. Nature. 1978;276:362–368. doi: 10.1038/276362a0. [DOI] [PubMed] [Google Scholar]
- 23.Bode W, Schwager P, Huber R. The transition of bovine trypsinogen to a trypsin-like state upon strong ligand binding. The refined crystal structures of the bovine trypsinogen-pancreatic trypsin inhibitor complex and of its ternary complex with Ile-Val at 1.9 A resolution. J Mol Biol. 1978;118:99–112. doi: 10.1016/0022-2836(78)90246-2. [DOI] [PubMed] [Google Scholar]
- 24.Lian LY. NMR structural studies of glutathione S-transferase. Cell Mol Life Sci. 1998;54:359–362. doi: 10.1007/s000180050164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sidote DJ, Hoffman DW. NMR structure of an archaeal homologue of ribonuclease P protein Rpp29. Biochemistry. 2003;42:13541–13550. doi: 10.1021/bi030170z. [DOI] [PubMed] [Google Scholar]
- 26.Mizutani H, Saraboji K, Malathy Sony SM, Ponnuswamy MN, Kumarevel T, Krishna Swamy BS, Simanshu DK, Murthy MR, Kunishima N. Systematic study on crystal-contact engineering of diphthine synthase: influence of mutations at crystal-packing regions on X-ray diffraction quality. Acta Crystallogr D Biol Crystallogr. 2008;64:1020–1033. doi: 10.1107/S0907444908023019. [DOI] [PubMed] [Google Scholar]
- 27.Kobe B, Guncar G, Buchholz R, Huber T, Maco B, Cowieson N, Martin JL, Marfori M, Forwood JK. Crystallography and protein-protein interactions: biological interfaces and crystal contacts. Biochem Soc Trans. 2008;36:1438–1441. doi: 10.1042/BST0361438. [DOI] [PubMed] [Google Scholar]
- 28.Bahadur RP, Zacharias M. The interface of protein-protein complexes: analysis of contacts and prediction of interactions. Cell Mol Life Sci. 2008;65:1059–1072. doi: 10.1007/s00018-007-7451-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Daughdrill GW, Pielak GJ, Uversky VN, Cortese MS, Dunker AK. Natively disordered proteins. In: Buchner J, Kiefhaber T, editors. Handbook of Protein Folding, Wiley-VCH. Verlag GmbH & Co.; Weinheim, Germany: 2005. pp. 271–353. [Google Scholar]
- 30.Receveur-Brechot V, Bourhis JM, Uversky VN, Canard B, Longhi S. Assessing protein disorder and induced folding. Proteins. 2006;62:24–45. doi: 10.1002/prot.20750. [DOI] [PubMed] [Google Scholar]
- 31.Longhi S, Uverskym VN, editors. Instrumental Analysis of Intrinsically Disordered Proteins: Assessing Structure and Conformation. John Wiley & Sons, Inc; Hoboken, New Jersey, USA: 2010. [Google Scholar]
- 32.Sedzik J, Kirschner DA. Is myelin basic protein crystallizable? Neurochem Res. 1992;17:157–166. doi: 10.1007/BF00966794. [DOI] [PubMed] [Google Scholar]
- 33.Harauz G, Ishiyama N, Hill CM, Bates IR, Libich DS, Fares C. Myelin basic protein-diverse conformational states of an intrinsically unstructured protein and its roles in myelin assembly and multiple sclerosis. Micron. 2004;35:503–542. doi: 10.1016/j.micron.2004.04.005. [DOI] [PubMed] [Google Scholar]
- 34.Harauz G, Ladizhansky V, Boggs JM. Structural polymorphism and multifunctionality of myelin basic protein. Biochemistry. 2009;48:8094–8104. doi: 10.1021/bi901005f. [DOI] [PubMed] [Google Scholar]
- 35.Dolgikh DA, Abaturov LV, Bolotina IA, Brazhnikov EV, Bychkova VE, Gilmanshin RI, Lebedev Yu O, Semisotnov GV, Tiktopulo EI, Ptitsyn OB, et al. Compact state of a protein molecule with pronounced small-scale mobility: bovine alpha-lactalbumin. Eur Biophys J. 1985;13:109–121. doi: 10.1007/BF00256531. [DOI] [PubMed] [Google Scholar]
- 36.Dolgikh DA, Gilmanshin RI, Brazhnikov EV, Bychkova VE, Semisotnov GV, Venyaminov S, Ptitsyn OB. Alpha-Lactalbumin: compact state with fluctuating tertiary structure? FEBS Lett. 1981;136:311–315. doi: 10.1016/0014-5793(81)80642-4. [DOI] [PubMed] [Google Scholar]
- 37.Ptitsyn OB, Bychkova VE, Uversky VN. Kinetic and equilibrium folding intermediates. Philos Trans R Soc Lond B Biol Sci. 1995;348:35–41. doi: 10.1098/rstb.1995.0043. [DOI] [PubMed] [Google Scholar]
- 38.Ptitsyn OB. Molten globule and protein folding. Adv Protein Chem. 1995;47:83–229. doi: 10.1016/s0065-3233(08)60546-x. [DOI] [PubMed] [Google Scholar]
- 39.Arai M, Kuwajima K. Role of the molten globule state in protein folding. Adv Protein Chem. 2000;53:209–282. doi: 10.1016/s0065-3233(00)53005-8. [DOI] [PubMed] [Google Scholar]
- 40.Kuwajima K. The molten globule state as a clue for understanding the folding and cooperativity of globular-protein structure. Proteins. 1989;6:87–103. doi: 10.1002/prot.340060202. [DOI] [PubMed] [Google Scholar]
- 41.Kuwajima K. The molten globule state of alpha-lactalbumin. Faseb J. 1996;10:102–109. doi: 10.1096/fasebj.10.1.8566530. [DOI] [PubMed] [Google Scholar]
- 42.Uversky VN. Protein folding revisited. A polypeptide chain at the folding-misfolding-nonfolding cross-roads: which way to go? Cell Mol Life Sci. 2003;60:1852–1871. doi: 10.1007/s00018-003-3096-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Uversky VN, Ptitsyn OB. Further evidence on the equilibrium “pre-molten globule state”: four-state guanidinium chloride-induced unfolding of carbonic anhydrase B at low temperature. J Mol Biol. 1996;255:215–228. doi: 10.1006/jmbi.1996.0018. [DOI] [PubMed] [Google Scholar]
- 44.Uversky VN, Ptitsyn OB. “Partly folded” state, a new equilibrium state of protein molecules: four-state guanidinium chloride-induced unfolding of beta-lactamase at low temperature. Biochemistry. 1994;33:2782–2791. doi: 10.1021/bi00176a006. [DOI] [PubMed] [Google Scholar]
- 45.Kim PS, Baldwin RL. Specific intermediates in the folding reactions of small proteins and the mechanism of protein folding. Annu Rev Biochem. 1982;51:459–489. doi: 10.1146/annurev.bi.51.070182.002331. [DOI] [PubMed] [Google Scholar]
- 46.Kim PS, Baldwin RL. Intermediates in the folding reactions of small proteins. Annu Rev Biochem. 1990;59:631–660. doi: 10.1146/annurev.bi.59.070190.003215. [DOI] [PubMed] [Google Scholar]
- 47.Tcherkasskaya O, Uversky VN. Polymeric aspects of protein folding: a brief overview. Protein Pept Lett. 2003;10:239–245. doi: 10.2174/0929866033478988. [DOI] [PubMed] [Google Scholar]
- 48.Tcherkasskaya O, Davidson EA, Uversky VN. Biophysical constraints for protein structure prediction. J Proteome Res. 2003;2:37–42. doi: 10.1021/pr025552q. [DOI] [PubMed] [Google Scholar]
- 49.Crick SL, Jayaraman M, Frieden C, Wetzel R, Pappu RV. Fluorescence correlation spectroscopy shows that monomeric polyglutamine molecules form collapsed structures in aqueous solutions. Proc Natl Acad Sci U S A. 2006;103:16764–16769. doi: 10.1073/pnas.0608175103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vitalis A, Wang X, Pappu RV. Atomistic simulations of the effects of polyglutamine chain length and solvent quality on conformational equilibria and spontaneous homodimerization. J Mol Biol. 2008;384:279–297. doi: 10.1016/j.jmb.2008.09.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vitalis A, Wang X, Pappu RV. Quantitative characterization of intrinsic disorder in polyglutamine: insights from analysis based on polymer theories. Biophys J. 2007;93:1923–1937. doi: 10.1529/biophysj.107.110080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wang X, Vitalis A, Wyczalkowski MA, Pappu RV. Characterizing the conformational ensemble of monomeric polyglutamine. Proteins. 2006;63:297–311. doi: 10.1002/prot.20761. [DOI] [PubMed] [Google Scholar]
- 53.Bienkiewicz EA, Adkins JN, Lumb KJ. Functional consequences of preorganized helical structure in the intrinsically disordered cell-cycle inhibitor p27(Kip1) Biochemistry. 2002;41:752–759. doi: 10.1021/bi015763t. [DOI] [PubMed] [Google Scholar]
- 54.Chi SW, Kim DH, Lee SH, Chang I, Han KH. Pre-structured motifs in the natively unstructured preS1 surface antigen of hepatitis B virus. Protein Sci. 2007;16:2108–2117. doi: 10.1110/ps.072983507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 56.Ramelot TA, Gentile LN, Nicholson LK. Transient structure of the amyloid precursor protein cytoplasmic tail indicates preordering of structure for binding to cytosolic factors. Biochemistry. 2000;39:2714–2725. doi: 10.1021/bi992580m. [DOI] [PubMed] [Google Scholar]
- 57.Sayers EW, Gerstner RB, Draper DE, Torchia DA. Structural preordering in the N-terminal region of ribosomal protein S4 revealed by heteronuclear NMR spectroscopy. Biochemistry. 2000;39:13602–13613. doi: 10.1021/bi0013391. [DOI] [PubMed] [Google Scholar]
- 58.Zitzewitz JA, Ibarra-Molero B, Fishel DR, Terry KL, Matthews CR. Preformed secondary structure drives the association reaction of GCN4-p1, a model coiled-coil system. J Mol Biol. 2000;296:1105–1116. doi: 10.1006/jmbi.2000.3507. [DOI] [PubMed] [Google Scholar]
- 59.Bychkova VE, Pain RH, Ptitsyn OB. The ‘molten globule’ state is involved in the translocation of proteins across membranes? FEBS Lett. 1988;238:231–234. doi: 10.1016/0014-5793(88)80485-x. [DOI] [PubMed] [Google Scholar]
- 60.Bychkova VE, Ptitsyn OB. The molten globule in vitro and in vivo. Chemtracts Biochem Molec Biol. 1993;4:133–163. [Google Scholar]
- 61.Martin J, Langer T, Boteva R, Schramel A, Horwich AL, Hartl FU. Chaperonin-mediated protein folding at the surface of groEL through a ‘molten globule’-like intermediate. Nature. 1991;352:36–42. doi: 10.1038/352036a0. [DOI] [PubMed] [Google Scholar]
- 62.van der Goot FG, Gonzalez-Manas JM, Lakey JH, Pattus F. A ‘molten-globule’ membrane-insertion intermediate of the pore-forming domain of colicin A. Nature. 1991;354:408–410. doi: 10.1038/354408a0. [DOI] [PubMed] [Google Scholar]
- 63.van der Goot FG, Lakey JH, Pattus F. The molten globule intermediate for protein insertion or translocation through membranes. Trends Cell Biol. 1992;2:343–348. [PubMed] [Google Scholar]
- 64.Uversky VN, Narizhneva NV. Effect of natural ligands on the structural properties and conformational stability of proteins. Biochemistry (Mosc) 1998;63:420–433. [PubMed] [Google Scholar]
- 65.Uversky VN. A rigidifying union: The role of ligands in protein structure and stability. In: Pandalai SG, editor. Recent Research Developments in Biophysics & Biochemistry. Vol. 3. Transworld Research Network; Kerala, India: 2003. pp. 711–745. [Google Scholar]
- 66.Bychkova VE, Ptitsyn OB. Folding intermediates are involved in genetic diseases? FEBS Lett. 1995;359:6–8. doi: 10.1016/0014-5793(95)00004-s. [DOI] [PubMed] [Google Scholar]
- 67.Karush F. Heterogeneity of the binding sites of bovine serum albumin. J Am Chem Soc. 1950;72:2705–2713. [Google Scholar]
- 68.Schweers O, Schonbrunn-Hanebeck E, Marx A, Mandelkow E. Structural studies of tau protein and Alzheimer paired helical filaments show no evidence for beta-structure. J Biol Chem. 1994;269:24290–24297. [PubMed] [Google Scholar]
- 69.Weinreb PH, Zhen W, Poon AW, Conway KA, Lansbury PT., Jr NACP, a protein implicated in Alzheimer's disease and learning, is natively unfolded. Biochemistry. 1996;35:13709–13715. doi: 10.1021/bi961799n. [DOI] [PubMed] [Google Scholar]
- 70.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 71.Uversky VN, Gillespie JR, Fink AL. Why are “natively unfolded” proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 72.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 73.Tompa P. Intrinsically unstructured proteins. Trends Biochem Sci. 2002;27:527–533. doi: 10.1016/s0968-0004(02)02169-2. [DOI] [PubMed] [Google Scholar]
- 74.Dunker AK, Garner E, Guilliot S, Romero P, Albrecht K, Hart J, Obradovic Z, Kissinger C, Villafranca JE. Protein disorder and the evolution of molecular recognition: theory, predictions and observations. Pac Symp Biocomput. 1998:473–484. [PubMed] [Google Scholar]
- 75.Garner E, Cannon P, Romero P, Obradovic Z, Dunker AK. Predicting Disordered Regions from Amino Acid Sequence: Common Themes Despite Differing Structural Characterization. Genome Inform Ser Workshop Genome Inform. 1998;9:201–213. [PubMed] [Google Scholar]
- 76.Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Garner E, Guilliot S, Dunker AK. Thousands of proteins likely to have long disordered regions. Pac Symp Biocomput. 1998:437–448. [PubMed] [Google Scholar]
- 77.Romero P, Obradovic Z, Kissinger CR, Villafranca JE, Dunker AK. Identifying disordered regions in proteins from amino acid sequences. IEEE Int Conf Neural Netw. 1997;1:90–95. [Google Scholar]
- 78.Romero, Obradovic, Dunker K. Sequence Data Analysis for Long Disordered Regions Prediction in the Calcineurin Family. Genome Inform Ser Workshop Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 79.Dunker AK, Obradovic Z, Romero P, Kissinger C, Villafranca E. On the importance of being disordered. PDB Newsletter. 1997;81:3–5. [Google Scholar]
- 80.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 81.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 82.Dunker AK, Obradovic Z. The protein trinity--linking function and disorder. Nat Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 83.Williams RM, Obradovic Z, Mathura V, Braun W, Garner EC, Young J, Takayama S, Brown CJ, Dunker AK. The protein non-folding problem: amino acid determinants of intrinsic order and disorder. Pac Symp Biocomput. 2001:89–100. doi: 10.1142/9789814447362_0010. [DOI] [PubMed] [Google Scholar]
- 84.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 85.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 86.Romero P, Obradovic Z, Dunker AK. Intelligent data analysis for protein disorder prediction. Artif Intel Rev. 2000;14:447–484. [Google Scholar]
- 87.Li X, Romero P, Rani M, Dunker AK, Obradovic Z. Predicting Protein Disorder for N-, C-, and Internal Regions. Genome Inform Ser Workshop Genome Inform. 1999;10:30–40. [PubMed] [Google Scholar]
- 88.Garner E, Romero P, Dunker AK, Brown C, Obradovic Z. Predicting Binding Regions within Disordered Proteins. Genome Inform Ser Workshop Genome Inform. 1999;10:41–50. [PubMed] [Google Scholar]
- 89.Kriwacki RW, Hengst L, Tennant L, Reed SI, Wright PE. Structural studies of p21Waf1/Cip1/Sdi1 in the free and Cdk2-bound state: conformational disorder mediates binding diversity. Proc Natl Acad Sci U S A. 1996;93:11504–11509. doi: 10.1073/pnas.93.21.11504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Dyson HJ, Wright PE. Coupling of folding and binding for unstructured proteins. Curr Opin Struct Biol. 2002;12:54–60. doi: 10.1016/s0959-440x(02)00289-0. [DOI] [PubMed] [Google Scholar]
- 91.Uversky VN, Gillespie JR, Millett IS, Khodyakova AV, Vasilenko RN, Vasiliev AM, Rodionov IL, Kozlovskaya GD, Dolgikh DA, Fink AL, Doniach S, Permyakov EA, Abramov VM. Zn(2+)-mediated structure formation and compaction of the “natively unfolded” human prothymosin alpha. Biochem Biophys Res Commun. 2000;267:663–668. doi: 10.1006/bbrc.1999.2013. [DOI] [PubMed] [Google Scholar]
- 92.Uversky VN. Natively unfolded proteins: a point where biology waits for physics. Protein Sci. 2002;11:739–756. doi: 10.1110/ps.4210102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Uversky VN. What does it mean to be natively unfolded? Eur J Biochem. 2002;269:2–12. doi: 10.1046/j.0014-2956.2001.02649.x. [DOI] [PubMed] [Google Scholar]
- 94.Uversky VN, Permyakov SE, Zagranichny VE, Rodionov IL, Fink AL, Cherskaya AM, Wasserman LA, Permyakov EA. Effect of zinc and temperature on the conformation of the gamma subunit of retinal phosphodiesterase: a natively unfolded protein. J Proteome Res. 2002;1:149–159. doi: 10.1021/pr0155127. [DOI] [PubMed] [Google Scholar]
- 95.Uversky VN, Li J, Fink AL. Evidence for a partially folded intermediate in alpha-synuclein fibril formation. J Biol Chem. 2001;276:10737–10744. doi: 10.1074/jbc.M010907200. [DOI] [PubMed] [Google Scholar]
- 96.Teilum K, Olsen JG, Kragelund BB. Functional aspects of protein flexibility. Cell Mol Life Sci. 2009;66:2231–2247. doi: 10.1007/s00018-009-0014-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Radivojac P, Obradovic Z, Smith DK, Zhu G, Vucetic S, Brown CJ, Lawson JD, Dunker AK. Protein flexibility and intrinsic disorder. Protein Sci. 2004;13:71–80. doi: 10.1110/ps.03128904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Williams RJ. The conformational mobility of proteins and its functional significance. Biochem Soc Trans. 1978;6:1123–1126. doi: 10.1042/bst0061123. [DOI] [PubMed] [Google Scholar]
- 99.Namba K. Roles of partly unfolded conformations in macromolecular self-assembly. Genes Cells. 2001;6:1–12. doi: 10.1046/j.1365-2443.2001.00384.x. [DOI] [PubMed] [Google Scholar]
- 100.Oldfield CJ, Cheng Y, Cortese MS, Brown CJ, Uversky VN, Dunker AK. Comparing and combining predictors of mostly disordered proteins. Biochemistry. 2005;44:1989–2000. doi: 10.1021/bi047993o. [DOI] [PubMed] [Google Scholar]
- 101.Iakoucheva LM, Brown CJ, Lawson JD, Obradovic Z, Dunker AK. Intrinsic disorder in cell-signaling and cancer-associated proteins. J Mol Biol. 2002;323:573–584. doi: 10.1016/s0022-2836(02)00969-5. [DOI] [PubMed] [Google Scholar]
- 102.Hemmings HC, Jr, Nairn AC, Aswad DW, Greengard P. DARPP-32, a dopamine- and adenosine 3′:5′-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. II. Purification and characterization of the phosphoprotein from bovine caudate nucleus. J Neurosci. 1984;4:99–110. doi: 10.1523/JNEUROSCI.04-01-00099.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 104.Kyte J, Doolittle RF. A simple method for displaying the hydropathic character of a protein. J Mol Biol. 1982;157:105–132. doi: 10.1016/0022-2836(82)90515-0. [DOI] [PubMed] [Google Scholar]
- 105.Vihinen M, Torkkila E, Riikonen P. Accuracy of protein flexibility predictions. Proteins. 1994;19:141–149. doi: 10.1002/prot.340190207. [DOI] [PubMed] [Google Scholar]
- 106.Jacob C, Giles GI, Giles NM, Sies H. Sulfur and selenium: the role of oxidation state in protein structure and function. Angew Chem Int Ed Engl. 2003;42:4742–4758. doi: 10.1002/anie.200300573. [DOI] [PubMed] [Google Scholar]
- 107.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr Opin Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 108.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Vacic V, Uversky VN, Dunker AK, Lonardi S. Composition Profiler: a tool for discovery and visualization of amino acid composition differences. BMC Bioinformatics. 2007;8:211. doi: 10.1186/1471-2105-8-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Li X, Obradovic Z, Brown CJ, Garner EC, Dunker AK. Comparing predictors of disordered protein. Genome Inform Ser Workshop Genome Inform. 2000;11:172–184. [PubMed] [Google Scholar]
- 111.Campen A, Williams RM, Brown CJ, Meng J, Uversky VN, Dunker AK. TOP-IDP-scale: a new amino acid scale measuring propensity for intrinsic disorder. Protein Pept Lett. 2008;15:956–963. doi: 10.2174/092986608785849164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Vucetic S, Radivojac P, Obradovic Z, Brown CJ, Dunker AK. Methods for improving protein disorder prediction. IEEE Int. Conf. Neural Netw; Washington, DC. 2001. [Google Scholar]
- 113.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 114.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005 doi: 10.1021/bi050736e. In press. [DOI] [PubMed] [Google Scholar]
- 115.Mohan A, Oldfield CJ, Radivojac P, Vacic V, Cortese MS, Dunker AK, Uversky VN. Analysis of molecular recognition features (MoRFs) J Mol Biol. 2006;362:1043–1059. doi: 10.1016/j.jmb.2006.07.087. [DOI] [PubMed] [Google Scholar]
- 116.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining alpha-helix-forming molecular recognition features with cross species sequence alignments. Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Noivirt-Brik O, Prilusky J, Sussman JL. Assessment of disorder predictions in CASP8. Proteins. 2009;77 9:210–216. doi: 10.1002/prot.22586. [DOI] [PubMed] [Google Scholar]
- 118.Melamud E, Moult J. Evaluation of disorder predictions in CASP5. Proteins. 2003;53 6:561–565. doi: 10.1002/prot.10533. [DOI] [PubMed] [Google Scholar]
- 119.Liu J, Tan H, Rost B. Loopy proteins appear conserved in evolution. J Mol Biol. 2002;322:53–64. doi: 10.1016/s0022-2836(02)00736-2. [DOI] [PubMed] [Google Scholar]
- 120.Mohan A, Sullivan WJ, Jr, Radivojac P, Dunker AK, Uversky VN. Intrinsic disorder in pathogenic and non-pathogenic microbes: discovering and analyzing the unfoldomes of early-branching eukaryotes. Mol Biosyst. 2008;4:328–340. doi: 10.1039/b719168e. [DOI] [PubMed] [Google Scholar]
- 121.Gunasekaran K, Tsai CJ, Nussinov R. Analysis of ordered and disordered protein complexes reveals structural features discriminating between stable and unstable monomers. J Mol Biol. 2004;341:1327–1341. doi: 10.1016/j.jmb.2004.07.002. [DOI] [PubMed] [Google Scholar]
- 122.Vucetic S, Brown CJ, Dunker AK, Obradovic Z. Flavors of protein disorder. Proteins. 2003;52:573–584. doi: 10.1002/prot.10437. [DOI] [PubMed] [Google Scholar]
- 123.Fink AL. Compact intermediate states in protein folding. Annu Rev Biophys Biomol Struct. 1995;24:495–522. doi: 10.1146/annurev.bb.24.060195.002431. [DOI] [PubMed] [Google Scholar]
- 124.Baum J, Dobson CM, Evans PA, Hanley C. Characterization of a partly folded protein by NMR methods: studies on the molten globule state of guinea pig alpha-lactalbumin. Biochemistry. 1989;28:7–13. doi: 10.1021/bi00427a002. [DOI] [PubMed] [Google Scholar]
- 125.Bushnell GW, Louie GV, Brayer GD. High-resolution three-dimensional structure of horse heart cytochrome c. J Mol Biol. 1990;214:585–595. doi: 10.1016/0022-2836(90)90200-6. [DOI] [PubMed] [Google Scholar]
- 126.Chyan CL, Wormald C, Dobson CM, Evans PA, Baum J. Structure and stability of the molten globule state of guinea-pig alpha-lactalbumin: a hydrogen exchange study. Biochemistry. 1993;32:5681–5691. doi: 10.1021/bi00072a025. [DOI] [PubMed] [Google Scholar]
- 127.Jeng MF, Englander SW, Elove GA, Wand AJ, Roder H. Structural description of acid-denatured cytochrome c by hydrogen exchange and 2D NMR. Biochemistry. 1990;29:10433–10437. doi: 10.1021/bi00498a001. [DOI] [PubMed] [Google Scholar]
- 128.Wu LC, Laub PB, Elove GA, Carey J, Roder H. A noncovalent peptide complex as a model for an early folding intermediate of cytochrome c. Biochemistry. 1993;32:10271–10276. doi: 10.1021/bi00089a050. [DOI] [PubMed] [Google Scholar]
- 129.Eliezer D, Yao J, Dyson HJ, Wright PE. Structural and dynamic characterization of partially folded states of apomyoglobin and implications for protein folding. Nat Struct Biol. 1998;5:148–155. doi: 10.1038/nsb0298-148. [DOI] [PubMed] [Google Scholar]
- 130.Bose HS, Whittal RM, Baldwin MA, Miller WL. The active form of the steroidogenic acute regulatory protein, StAR, appears to be a molten globule. Proc Natl Acad Sci U S A. 1999;96:7250–7255. doi: 10.1073/pnas.96.13.7250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bracken C. NMR spin relaxation methods for characterization of disorder and folding in proteins. J Mol Graph Model. 2001;19:3–12. doi: 10.1016/s1093-3263(00)00136-4. [DOI] [PubMed] [Google Scholar]
- 132.Merrill AR, Cohen FS, Cramer WA. On the nature of the structural change of the colicin E1 channel peptide necessary for its translocation-competent state. Biochemistry. 1990;29:5829–5836. doi: 10.1021/bi00476a026. [DOI] [PubMed] [Google Scholar]
- 133.Fontana A, Fassina G, Vita C, Dalzoppo D, Zamai M, Zambonin M. Correlation between sites of limited proteolysis and segmental mobility in thermolysin. Biochemistry. 1986;25:1847–1851. doi: 10.1021/bi00356a001. [DOI] [PubMed] [Google Scholar]
- 134.Fontana A, Polverino de Laureto P, De Phillips V. Molecular aspects of proteolysis of globular proteins. In: van den Tweel W, Harder A, Buitelear M, editors. Protein Stability and Stabilization. Elsevier Science; Amsterdam, The Netherlands: 1993. pp. 101–110. [Google Scholar]
- 135.Fontana A, Zambonin M, Polverino de Laureto P, De Filippis V, Clementi A, Scaramella E. Probing the conformational state of apomyoglobin by limited proteolysis. J Mol Biol. 1997;266:223–230. doi: 10.1006/jmbi.1996.0787. [DOI] [PubMed] [Google Scholar]
- 136.Fontana A, Polverino de Laureto P, De Filippis V, Scaramella E, Zambonin M. Probing the partly folded states of proteins by limited proteolysis. Fold Des. 1997;2:R17–26. doi: 10.1016/S1359-0278(97)00010-2. [DOI] [PubMed] [Google Scholar]
- 137.Semisotnov GV, Rodionova NA, Razgulyaev OI, Uversky VN, Gripas AF, Gilmanshin RI. Study of the “molten globule” intermediate state in protein folding by a hydrophobic fluorescent probe. Biopolymers. 1991;31:119–128. doi: 10.1002/bip.360310111. [DOI] [PubMed] [Google Scholar]
- 138.Eliezer D, Chiba K, Tsuruta H, Doniach S, Hodgson KO, Kihara H. Evidence of an associative intermediate on the myoglobin refolding pathway. Biophys J. 1993;65:912–917. doi: 10.1016/S0006-3495(93)81124-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Kataoka M, Kuwajima K, Tokunaga F, Goto Y. Structural characterization of the molten globule of alpha-lactalbumin by solution X-ray scattering. Protein Sci. 1997;6:422–430. doi: 10.1002/pro.5560060219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Semisotnov GV, Kihara H, Kotova NV, Kimura K, Amemiya Y, Wakabayashi K, Serdyuk IN, Timchenko AA, Chiba K, Nikaido K, Ikura T, Kuwajima K. Protein globularization during folding. A study by synchrotron small-angle X-ray scattering. J Mol Biol. 1996;262:559–574. doi: 10.1006/jmbi.1996.0535. [DOI] [PubMed] [Google Scholar]
- 141.Uversky VN, Karnoup AS, Segel DJ, Seshadri S, Doniach S, Fink AL. Anion-induced folding of Staphylococcal nuclease: characterization of multiple equilibrium partially folded intermediates. J Mol Biol. 1998;278:879–894. doi: 10.1006/jmbi.1998.1741. [DOI] [PubMed] [Google Scholar]
- 142.Tcherkasskaya O, Uversky VN. Denatured collapsed states in protein folding: example of apomyoglobin. Proteins. 2001;44:244–254. doi: 10.1002/prot.1089. [DOI] [PubMed] [Google Scholar]
- 143.Yamada Y, Calestagne-Morelli A, Uversky VN, Lau EY, Krishnan VV, Phillips JL, Newsam S, Colvin ME, Rexach MF. Distinct categories of natively unfolded structures with separate functions in FG nucleoporins. Cell Cycle. 2010 doi: 10.1074/mcp.M000035-MCP201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Hammarstrom P, Carlsson U. Is the unfolded state the Rosetta Stone of the protein folding problem? Biochem Biophys Res Commun. 2000;276:393–398. doi: 10.1006/bbrc.2000.3360. [DOI] [PubMed] [Google Scholar]
- 145.Shortle D. The expanded denatured state: an ensemble of conformations trapped in a locally encoded topological space. Adv Protein Chem. 2002;62:1–23. doi: 10.1016/s0065-3233(02)62003-0. [DOI] [PubMed] [Google Scholar]
- 146.Smith LJ, Fiebig KM, Schwalbe H, Dobson CM. The concept of a random coil. Residual structure in peptides and denatured proteins. Fold Des. 1996;1:R95–106. doi: 10.1016/S1359-0278(96)00046-6. [DOI] [PubMed] [Google Scholar]
- 147.Shimizu S, Chan HS. Origins of protein denatured state compactness and hydrophobic clustering in aqueous urea: inferences from nonpolar potentials of mean force. Proteins. 2002;49:560–566. doi: 10.1002/prot.10263. [DOI] [PubMed] [Google Scholar]
- 148.Uverskii VN. How many molten globules states exist? Biofizika. 1998;43:416–421. [PubMed] [Google Scholar]
- 149.Uversky VN. Intrinsically Disordered Proteins and Their Environment: Effects of Strong Denaturants, Temperature, pH, Counter Ions, Membranes, Binding Partners, Osmolytes, and Macromolecular Crowding. Protein J. 2009 doi: 10.1007/s10930-009-9201-4. [DOI] [PubMed] [Google Scholar]
- 150.Cortese MS, Baird JP, Uversky VN, Dunker AK. Uncovering the unfoldome: enriching cell extracts for unstructured proteins by Acid treatment. J Proteome Res. 2005;4:1610–1618. doi: 10.1021/pr050119c. [DOI] [PubMed] [Google Scholar]
- 151.Csizmok V, Szollosi E, Friedrich P, Tompa P. A novel two-dimensional electrophoresis technique for the identification of intrinsically unstructured proteins. Mol Cell Proteomics. 2006;5:265–273. doi: 10.1074/mcp.M500181-MCP200. [DOI] [PubMed] [Google Scholar]
- 152.Galea CA, High AA, Obenauer JC, Mishra A, Park CG, Punta M, Schlessinger A, Ma J, Rost B, Slaughter CA, Kriwacki RW. Large-scale analysis of thermostable, mammalian proteins provides insights into the intrinsically disordered proteome. J Proteome Res. 2009;8:211–226. doi: 10.1021/pr800308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Galea CA, Pagala VR, Obenauer JC, Park CG, Slaughter CA, Kriwacki RW. Proteomic studies of the intrinsically unstructured mammalian proteome. J Proteome Res. 2006;5:2839–2848. doi: 10.1021/pr060328c. [DOI] [PubMed] [Google Scholar]
- 154.Uversky VN. A multiparametric approach to studies of self-organization of globular proteins. Biochemistry (Mosc) 1999;64:250–266. [PubMed] [Google Scholar]
- 155.Eliezer D. Biophysical characterization of intrinsically disordered proteins. Curr Opin Struct Biol. 2009;19:23–30. doi: 10.1016/j.sbi.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Bhalla J, Storchan GB, MacCarthy CM, Uversky VN, Tcherkasskaya O. Local flexibility in molecular function paradigm. Mol Cell Proteomics. 2006;5:1212–1223. doi: 10.1074/mcp.M500315-MCP200. [DOI] [PubMed] [Google Scholar]
- 157.Le Gall T, Romero PR, Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in the Protein Data Bank. J Biomol Struct Dyn. 2007;24:325–342. doi: 10.1080/07391102.2007.10507123. [DOI] [PubMed] [Google Scholar]
- 158.Mohan A, Uversky VN, Radivojac P. Influence of sequence changes and environment on intrinsically disordered proteins. PLoS Comput Biol. 2009;5:e1000497. doi: 10.1371/journal.pcbi.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Eliezer D. Characterizing residual structure in disordered protein States using nuclear magnetic resonance. Methods Mol Biol. 2007;350:49–67. doi: 10.1385/1-59745-189-4:49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Mittag T, Forman-Kay JD. Atomic-level characterization of disordered protein ensembles. Curr Opin Struct Biol. 2007;17:3–14. doi: 10.1016/j.sbi.2007.01.009. [DOI] [PubMed] [Google Scholar]
- 161.Jensen MR, Markwick PR, Meier S, Griesinger C, Zweckstetter M, Grzesiek S, Bernado P, Blackledge M. Quantitative determination of the conformational properties of partially folded and intrinsically disordered proteins using NMR dipolar couplings. Structure. 2009;17:1169–1185. doi: 10.1016/j.str.2009.08.001. [DOI] [PubMed] [Google Scholar]
- 162.Adler AJ, Greenfield NJ, Fasman GD. Circular dichroism and optical rotatory dispersion of proteins and polypeptides. Methods Enzymol. 1973;27:675–735. doi: 10.1016/s0076-6879(73)27030-1. [DOI] [PubMed] [Google Scholar]
- 163.Fasman GD. Circular Dichroism and the Conformational Analysis of Biomolecules. Plenem Press; New York: 1996. [Google Scholar]
- 164.Provencher SW, Glockner J. Estimation of globular protein secondary structure from circular dichroism. Biochemistry. 1981;20:33–37. doi: 10.1021/bi00504a006. [DOI] [PubMed] [Google Scholar]
- 165.Johnson WC., Jr Secondary structure of proteins through circular dichroism spectroscopy. Annu Rev Biophys Biophys Chem. 1988;17:145–166. doi: 10.1146/annurev.bb.17.060188.001045. [DOI] [PubMed] [Google Scholar]
- 166.Woody RW. Circular dichroism. Methods Enzymol. 1995;246:34–71. doi: 10.1016/0076-6879(95)46006-3. [DOI] [PubMed] [Google Scholar]
- 167.Kelly SM, Price NC. The application of circular dichroism to studies of protein folding and unfolding. Biochim Biophys Acta. 1997;1338:161–185. doi: 10.1016/s0167-4838(96)00190-2. [DOI] [PubMed] [Google Scholar]
- 168.Vassilenko KS, Uversky VN. Native-like secondary structure of molten globules. Biochim Biophys Acta. 2002;1594:168–177. doi: 10.1016/s0167-4838(01)00303-x. [DOI] [PubMed] [Google Scholar]
- 169.Chen E, Kumita JR, Woolley GA, Kliger DS. The kinetics of helix unfolding of an azobenzene cross-linked peptide probed by nanosecond time-resolved optical rotatory dispersion. J Am Chem Soc. 2003;125:12443–12449. doi: 10.1021/ja030277+. [DOI] [PubMed] [Google Scholar]
- 170.Smyth E, Syme CD, Blanch EW, Hecht L, Vasak M, Barron LD. Solution structure of native proteins with irregular folds from Raman optical activity. Biopolymers. 2001;58:138–151. doi: 10.1002/1097-0282(200102)58:2<138::AID-BIP30>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 171.Xu M, Ermolenkov VV, He W, Uversky VN, Fredriksen L, Lednev IK. Lysozyme fibrillation: deep UV Raman spectroscopic characterization of protein structural transformation. Biopolymers. 2005;79:58–61. doi: 10.1002/bip.20330. [DOI] [PubMed] [Google Scholar]
- 172.Xu M, Ermolenkov VV, Uversky VN, Lednev IK. Hen egg white lysozyme fibrillation: a deep-UV resonance Raman spectroscopic study. J Biophotonics. 2008;1:215–229. doi: 10.1002/jbio.200710013. [DOI] [PubMed] [Google Scholar]
- 173.Tanford C. Protein denaturation. Adv Protein Chem. 1968;23:121–282. doi: 10.1016/s0065-3233(08)60401-5. [DOI] [PubMed] [Google Scholar]
- 174.Uversky VN. Use of fast protein size-exclusion liquid chromatography to study the unfolding of proteins which denature through the molten globule. Biochemistry. 1993;32:13288–13298. doi: 10.1021/bi00211a042. [DOI] [PubMed] [Google Scholar]
- 175.Glatter O, Kratky O. Small Angle X-ray Scattering. London, England: 1982. [DOI] [PubMed] [Google Scholar]
- 176.Iakoucheva LM, Kimzey AL, Masselon CD, Bruce JE, Garner EC, Brown CJ, Dunker AK, Smith RD, Ackerman EJ. Identification of intrinsic order and disorder in the DNA repair protein XPA. Protein Sci. 2001;10:560–571. doi: 10.1110/ps.29401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Amit AG, Mariuzza RA, Phillips SE, Poljak RJ. Three-dimensional structure of an antigen-antibody complex at 6 A resolution. Nature. 1985;313:156–158. doi: 10.1038/313156a0. [DOI] [PubMed] [Google Scholar]
- 178.Wilson IA, Haft DH, Getzoff ED, Tainer JA, Lerner RA, Brenner S. Identical short peptide sequences in unrelated proteins can have different conformations: a testing ground for theories of immune recognition. Proc Natl Acad Sci U S A. 1985;82:5255–5259. doi: 10.1073/pnas.82.16.5255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Fujio H, Takagaki Y, Ha YM, Doi EM, Soebandrio A, Sakato N. Native and non-native conformation-specific antibodies directed to the loop region of hen egg-white lysozyme. J Biochem. 1985;98:949–962. doi: 10.1093/oxfordjournals.jbchem.a135375. [DOI] [PubMed] [Google Scholar]
- 180.Furie B, Furie BC. Conformation-specific antibodies as probes of the gamma-carboxyglutamic acid-rich region of bovine prothrombin. Studies of metal-induced structural changes. J Biol Chem. 1979;254:9766–9771. [PubMed] [Google Scholar]
- 181.Delmas PD, Stenner DD, Romberg RW, Riggs BL, Mann KG. Immunochemical studies of conformational alterations in bone gamma-carboxyglutamic acid containing protein. Biochemistry. 1984;23:4720–4725. doi: 10.1021/bi00315a030. [DOI] [PubMed] [Google Scholar]
- 182.Privalov PL. Stability of proteins: small globular proteins. Adv Protein Chem. 1979;33:167–241. doi: 10.1016/s0065-3233(08)60460-x. [DOI] [PubMed] [Google Scholar]
- 183.Ptitsyn OB, Uversky VN. The molten globule is a third thermodynamical state of protein molecules. FEBS Lett. 1994;341:15–18. doi: 10.1016/0014-5793(94)80231-9. [DOI] [PubMed] [Google Scholar]
- 184.Uversky VN, Ptitsyn OB. All-or-none solvent-induced transitions between native, molten globule and unfolded states in globular proteins. Fold Des. 1996;1:117–122. doi: 10.1016/S1359-0278(96)00020-X. [DOI] [PubMed] [Google Scholar]
- 185.Iakoucheva LM, Kimzey AL, Masselon CD, Smith RD, Dunker AK, Ackerman EJ. Aberrant mobility phenomena of the DNA repair protein XPA. Protein Sci. 2001;10:1353–1362. doi: 10.1110/ps.ps.40101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186.Serber Z, Corsini L, Durst F, Dotsch V. In-cell NMR spectroscopy. Methods Enzymol. 2005;394:17–41. doi: 10.1016/S0076-6879(05)94002-0. [DOI] [PubMed] [Google Scholar]
- 187.Serber Z, Dotsch V. In-cell NMR spectroscopy. Biochemistry. 2001;40:14317–14323. doi: 10.1021/bi011751w. [DOI] [PubMed] [Google Scholar]
- 188.Serber Z, Keatinge-Clay AT, Ledwidge R, Kelly AE, Miller SM, Dotsch V. High-resolution macromolecular NMR spectroscopy inside living cells. J Am Chem Soc. 2001;123:2446–2447. doi: 10.1021/ja0057528. [DOI] [PubMed] [Google Scholar]
- 189.Serber Z, Selenko P, Hansel R, Reckel S, Lohr F, Ferrell JE, Jr, Wagner G, Dotsch V. Investigating macromolecules inside cultured and injected cells by in-cell NMR spectroscopy. Nat Protoc. 2006;1:2701–2709. doi: 10.1038/nprot.2006.181. [DOI] [PubMed] [Google Scholar]
- 190.Dedmon MM, Patel CN, Young GB, Pielak GJ. FlgM gains structure in living cells. Proc Natl Acad Sci U S A. 2002;99:12681–12684. doi: 10.1073/pnas.202331299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 191.Li C, Charlton LM, Lakkavaram A, Seagle C, Wang G, Young GB, Macdonald JM, Pielak GJ. Differential dynamical effects of macromolecular crowding on an intrinsically disordered protein and a globular protein: implications for in-cell NMR spectroscopy. J Am Chem Soc. 2008;130:6310–6311. doi: 10.1021/ja801020z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.McNulty BC, Young GB, Pielak GJ. Macromolecular crowding in the Escherichia coli periplasm maintains alpha-synuclein disorder. J Mol Biol. 2006;355:893–897. doi: 10.1016/j.jmb.2005.11.033. [DOI] [PubMed] [Google Scholar]
- 193.Hughes KT, Gillen KL, Semon MJ, Karlinsey JE. Sensing structural intermediates in bacterial flagellar assembly by export of a negative regulator. Science. 1993;262:1277–1280. doi: 10.1126/science.8235660. [DOI] [PubMed] [Google Scholar]
- 194.Daughdrill GW, Hanely LJ, Dahlquist FW. The C-terminal half of the anti-sigma factor FlgM contains a dynamic equilibrium solution structure favoring helical conformations. Biochemistry. 1998;37:1076–1082. doi: 10.1021/bi971952t. [DOI] [PubMed] [Google Scholar]
- 195.Daughdrill GW, Chadsey MS, Karlinsey JE, Hughes KT, Dahlquist FW. The C-terminal half of the anti-sigma factor, FlgM, becomes structured when bound to its target, sigma 28. Nat Struct Biol. 1997;4:285–291. doi: 10.1038/nsb0497-285. [DOI] [PubMed] [Google Scholar]
- 196.Bryant JE, Lecomte JT, Lee AL, Young GB, Pielak GJ. Protein dynamics in living cells. Biochemistry. 2005;44:9275–9279. doi: 10.1021/bi050786j. [DOI] [PubMed] [Google Scholar]
- 197.Bryant JE, Lecomte JT, Lee AL, Young GB, Pielak GJ. Cytosol has a small effect on protein backbone dynamics. Biochemistry. 2006;45:10085–10091. doi: 10.1021/bi060547b. [DOI] [PubMed] [Google Scholar]
- 198.Pielak GJ. Retraction. Biochemistry. 2007;46:8206. doi: 10.1021/bi700744h. [DOI] [PubMed] [Google Scholar]
- 199.Bodart JF, Wieruszeski JM, Amniai L, Leroy A, Landrieu I, Rousseau-Lescuyer A, Vilain JP, Lippens G. NMR observation of Tau in Xenopus oocytes. J Magn Reson. 2008;192:252–257. doi: 10.1016/j.jmr.2008.03.006. [DOI] [PubMed] [Google Scholar]
- 200.Mukhopadhyay S, Krishnan R, Lemke EA, Lindquist S, Deniz AA. A natively unfolded yeast prion monomer adopts an ensemble of collapsed and rapidly fluctuating structures. Proc Natl Acad Sci U S A. 2007;104:2649–2654. doi: 10.1073/pnas.0611503104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Brucale M, Sandal M, Di Maio S, Rampioni A, Tessari I, Tosatto L, Bisaglia M, Bubacco L, Samori B. Pathogenic mutations shift the equilibria of alpha-synuclein single molecules towards structured conformers. Chembiochem. 2009;10:176–183. doi: 10.1002/cbic.200800581. [DOI] [PubMed] [Google Scholar]
- 202.Sandal M, Valle F, Tessari I, Mammi S, Bergantino E, Musiani F, Brucale M, Bubacco L, Samori B. Conformational equilibria in monomeric alpha-synuclein at the single-molecule level. PLoS Biol. 2008;6:e6. doi: 10.1371/journal.pbio.0060006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 203.Tan H, Nalbant P, Toutchkine A, Hu D, Vorpagel ER, Hahn KM, Lu HP. Single-molecule study of protein-protein interaction dynamics in a cell signaling system. J Phys Chem B. 2004;108:737–744. [Google Scholar]
- 204.Lu HP, Iakoucheva LM, Ackerman EJ. Single-molecule conformational dynamics of fluctuating noncovalent DNA-protein interactions in DNA damage recognition. J Am Chem Soc. 2001;123:9184–9185. doi: 10.1021/ja0058942. [DOI] [PubMed] [Google Scholar]
- 205.Lu HP. Probing single-molecule protein conformational dynamics. Acc Chem Res. 2005;38:557–565. doi: 10.1021/ar0401451. [DOI] [PubMed] [Google Scholar]
- 206.Lu HP. Single-molecule study of protein-protein and protein-DNA interaction dynamics. Methods Mol Biol. 2005;305:385–414. doi: 10.1385/1-59259-912-5:385. [DOI] [PubMed] [Google Scholar]
- 207.Trexler A, Rhoades E. Synuclein binds large unilamellar vesicles as an extended helix. Biochemistry. 2009 doi: 10.1021/bi900114z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 208.Frimpong A, Abzalimov RR, Uversky VN, Kaltashov IA. Characterization of intrinsically disordered proteins with electrospray ionization mass spectrometry: Conformational heterogeneity of alpha-synuclein. Proteins: Structure, Function, and Bioinformatics. 2010 doi: 10.1002/prot.22604. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 209.Miyagi A, Tsunaka Y, Uchihashi T, Mayanagi K, Hirose S, Morikawa K, Ando T. Visualization of intrinsically disordered regions of proteins by high-speed atomic force microscopy. Chemphyschem. 2008;9:1859–1866. doi: 10.1002/cphc.200800210. [DOI] [PubMed] [Google Scholar]
- 210.Belotserkovskaya R, Oh S, Bondarenko VA, Orphanides G, Studitsky VM, Reinberg D. FACT facilitates transcription-dependent nucleosome alteration. Science. 2003;301:1090–1093. doi: 10.1126/science.1085703. [DOI] [PubMed] [Google Scholar]
- 211.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 212.Fink AL. Natively unfolded proteins. Curr Opin Struct Biol. 2005;15:35–41. doi: 10.1016/j.sbi.2005.01.002. [DOI] [PubMed] [Google Scholar]
- 213.Dunker AK, Cortese MS, Romero P, Iakoucheva LM, Uversky VN. Flexible nets: The roles of intrinsic disorder in protein interaction networks. FEBS Journal. 2005;272:5129–5148. doi: 10.1111/j.1742-4658.2005.04948.x. [DOI] [PubMed] [Google Scholar]
- 214.Tompa P, Csermely P. The role of structural disorder in the function of RNA and protein chaperones. FASEB J. 2004;18:1169–1175. doi: 10.1096/fj.04-1584rev. [DOI] [PubMed] [Google Scholar]
- 215.Vucetic S, Xie H, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 2. Cellular components, domains, technical terms, developmental processes, and coding sequence diversities correlated with long disordered regions. J Proteome Res. 2007;6:1899–1916. doi: 10.1021/pr060393m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Functional anthology of intrinsic disorder. 3. Ligands, post-translational modifications, and diseases associated with intrinsically disordered proteins. J Proteome Res. 2007;6:1917–1932. doi: 10.1021/pr060394e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Xie H, Vucetic S, Iakoucheva LM, Oldfield CJ, Dunker AK, Uversky VN, Obradovic Z. Functional anthology of intrinsic disorder. 1. Biological processes and functions of proteins with long disordered regions. J Proteome Res. 2007;6:1882–1898. doi: 10.1021/pr060392u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 218.Antz C, Geyer M, Fakler B, Schott MK, Guy HR, Frank R, Ruppersberg JP, Kalbitzer HR. NMR structure of inactivation gates from mammalian voltage-dependent potassium channels. Nature. 1997;385:272–275. doi: 10.1038/385272a0. [DOI] [PubMed] [Google Scholar]
- 219.Armstrong CM, Bezanilla F. Inactivation of the sodium channel. II. Gating current experiments. J Gen Physiol. 1977;70:567–590. doi: 10.1085/jgp.70.5.567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hoshi T, Zagotta WN, Aldrich RW. Biophysical and molecular mechanisms of Shaker potassium channel inactivation. Science. 1990;250:533–538. doi: 10.1126/science.2122519. [DOI] [PubMed] [Google Scholar]
- 221.Zagotta WN, Hoshi T, Aldrich RW. Restoration of inactivation in mutants of Shaker potassium channels by a peptide derived from ShB. Science. 1990;250:568–571. doi: 10.1126/science.2122520. [DOI] [PubMed] [Google Scholar]
- 222.Herson PS, Virk M, Rustay NR, Bond CT, Crabbe JC, Adelman JP, Maylie J. A mouse model of episodic ataxia type-1. Nat Neurosci. 2003;6:378–383. doi: 10.1038/nn1025. [DOI] [PubMed] [Google Scholar]
- 223.Lerche H, Jurkat-Rott K, Lehmann-Horn F. Ion channels and epilepsy. Am J Med Genet. 2001;106:146–159. doi: 10.1002/ajmg.1582. [DOI] [PubMed] [Google Scholar]
- 224.Liebovitch LS, Selector LY, Kline RP. Statistical properties predicted by the ball and chain model of channel inactivation. Biophys J. 1992;63:1579–1585. doi: 10.1016/S0006-3495(92)81732-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Podlaha O, Zhang J. Positive selection on protein-length in the evolution of a primate sperm ion channel. Proc Natl Acad Sci U S A. 2003;100:12241–12246. doi: 10.1073/pnas.2033555100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 226.Uversky VN, Oldfield CJ, Dunker AK. Showing your ID: intrinsic disorder as an ID for recognition, regulation and cell signaling. J Mol Recognit. 2005;18:343–384. doi: 10.1002/jmr.747. [DOI] [PubMed] [Google Scholar]
- 227.Erdös P, Rényi A. On the evolution of random graphs. Publ Math Inst Hung Acad Sci. 1960;5:17–61. [Google Scholar]
- 228.Barabasi AL, Bonabeau E. Scale-free networks. Sci Am. 2003;288:60–69. doi: 10.1038/scientificamerican0503-60. [DOI] [PubMed] [Google Scholar]
- 229.Watts DJ, Strogatz SH. Collective dynamics of ‘small-world’ networks. Nature. 1998;393:440–442. doi: 10.1038/30918. [DOI] [PubMed] [Google Scholar]
- 230.Goh KI, Oh E, Jeong H, Kahng B, Kim D. Classification of scale-free networks. Proc Natl Acad Sci U S A. 2002;99:12583–12588. doi: 10.1073/pnas.202301299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Barabasi AL, Albert R. Emergence of scaling in random networks. Science. 1999;286:509–512. doi: 10.1126/science.286.5439.509. [DOI] [PubMed] [Google Scholar]
- 232.Bustos DM, Iglesias AA. Intrinsic disorder is a key characteristic in partners that bind 14-3-3 proteins. Proteins. 2006;63:35–42. doi: 10.1002/prot.20888. [DOI] [PubMed] [Google Scholar]
- 233.Radivojac P, Vucetic S, O'Connor TR, Uversky VN, Obradovic Z, Dunker AK. Calmodulin signaling: analysis and prediction of a disorder-dependent molecular recognition. Proteins. 2006;63:398–410. doi: 10.1002/prot.20873. [DOI] [PubMed] [Google Scholar]
- 234.Patil A, Nakamura H. Disordered domains and high surface charge confer hubs with the ability to interact with multiple proteins in interaction networks. FEBS Lett. 2006;580:2041–2045. doi: 10.1016/j.febslet.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 235.Ekman D, Light S, Bjorklund AK, Elofsson A. What properties characterize the hub proteins of the protein-protein interaction network of Saccharomyces cerevisiae? Genome Biol. 2006;7:R45. doi: 10.1186/gb-2006-7-6-r45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 236.Haynes C, Oldfield CJ, Ji F, Klitgord N, Cusick ME, Radivojac P, Uversky VN, Vidal M, Iakoucheva LM. Intrinsic disorder is a common feature of hub proteins from four eukaryotic interactomes. PLoS Comput Biol. 2006;2:e100. doi: 10.1371/journal.pcbi.0020100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Dosztanyi Z, Chen J, Dunker AK, Simon I, Tompa P. Disorder and sequence repeats in hub proteins and their implications for network evolution. J Proteome Res. 2006;5:2985–2995. doi: 10.1021/pr060171o. [DOI] [PubMed] [Google Scholar]
- 238.Singh GP, Dash D. Intrinsic disorder in yeast transcriptional regulatory network. Proteins. 2007;68:602–605. doi: 10.1002/prot.21497. [DOI] [PubMed] [Google Scholar]
- 239.Singh GP, Ganapathi M, Dash D. Role of intrinsic disorder in transient interactions of hub proteins. Proteins. 2007;66:761–765. doi: 10.1002/prot.21281. [DOI] [PubMed] [Google Scholar]
- 240.Liu W, Rui H, Wang J, Lin S, He Y, Chen M, Li Q, Ye Z, Zhang S, Chan SC, Chen YG, Han J, Lin SC. Axin is a scaffold protein in TGF-beta signaling that promotes degradation of Smad7 by Arkadia. EMBO J. 2006;25:1646–1658. doi: 10.1038/sj.emboj.7601057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Cortese MS, Uversky VN, Dunker AK. Intrinsic disorder in scaffold proteins: getting more from less. Prog Biophys Mol Biol. 2008;98:85–106. doi: 10.1016/j.pbiomolbio.2008.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 242.Balazs A, Csizmok V, Buday L, Rakacs M, Kiss R, Bokor M, Udupa R, Tompa K, Tompa P. High levels of structural disorder in scaffold proteins as exemplified by a novel neuronal protein, CASK-interactive protein1. FEBS J. 2009;276:3744–3756. doi: 10.1111/j.1742-4658.2009.07090.x. [DOI] [PubMed] [Google Scholar]
- 243.Koshland DE., Jr Application of a theory of enzyme specificity to protein synthesis. Proc Natl Acad Sci USA. 1958;44:98–104. doi: 10.1073/pnas.44.2.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 244.Sawaya MR, Kraut J. Loop and subdomain movements in the mechanism of Escherichia coli dihydrofolate reductase: crystallographic evidence. Biochemistry. 1997;36:586–603. doi: 10.1021/bi962337c. [DOI] [PubMed] [Google Scholar]
- 245.Schulz GE. Nucleotide Binding Proteins. In: Balaban M, editor. Molecular Mechanism of Biological Recognition. Elsevier/North-Holland Biomedical Press; New York: 1979. pp. 79–94. [Google Scholar]
- 246.Jeong H, Mason SP, Barabasi AL, Oltvai ZN. Lethality and centrality in protein networks. Nature. 2001;411:41–42. doi: 10.1038/35075138. [DOI] [PubMed] [Google Scholar]
- 247.Choo Y, Schwabe JW. All wrapped up. Nat Struct Biol. 1998;5:253–255. doi: 10.1038/nsb0498-253. [DOI] [PubMed] [Google Scholar]
- 248.Meador WE, Means AR, Quiocho FA. Target enzyme recognition by calmodulin: 2.4 A structure of a calmodulin-peptide complex. Science. 1992;257:1251–1255. doi: 10.1126/science.1519061. [DOI] [PubMed] [Google Scholar]
- 249.Dajani R, Fraser E, Roe SM, Yeo M, Good VM, Thompson V, Dale TC, Pearl LH. Structural basis for recruitment of glycogen synthase kinase 3beta to the axin-APC scaffold complex. Embo J. 2003;22:494–501. doi: 10.1093/emboj/cdg068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 250.Anderson CW, Appella E. Signaling to the p53 tumor suppressor through pathways activated by genotoxic and nongenotoxic stress. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling. Academic Press; New York: 2004. pp. 237–247. [Google Scholar]
- 251.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2008;9 1:S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 252.Brown CJ, Takayama S, Campen AM, Vise P, Marshall TW, Oldfield CJ, Williams CJ, Dunker AK. Evolutionary rate heterogeneity in proteins with long disordered regions. J Mol Evol. 2002;55:104–110. doi: 10.1007/s00239-001-2309-6. [DOI] [PubMed] [Google Scholar]
- 253.Uversky VN, Li J, Fink AL. Metal-triggered structural transformations, aggregation, and fibrillation of human alpha-synuclein. A possible molecular NK between Parkinson's disease and heavy metal exposure. J Biol Chem. 2001;276:44284–44296. doi: 10.1074/jbc.M105343200. [DOI] [PubMed] [Google Scholar]
- 254.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 255.Craig TA, Veenstra TD, Naylor S, Tomlinson AJ, Johnson KL, Macura S, Juranic N, Kumar R. Zinc binding properties of the DNA binding domain of the 1,25-dihydroxyvitamin D3 receptor. Biochemistry. 1997;36:10482–10491. doi: 10.1021/bi970561b. [DOI] [PubMed] [Google Scholar]
- 256.Rice LM, Brennwald P, Brunger AT. Formation of a yeast SNARE complex is accompanied by significant structural changes. FEBS Lett. 1997;415:49–55. doi: 10.1016/s0014-5793(97)01091-0. [DOI] [PubMed] [Google Scholar]
- 257.Permyakov SE, Millett IS, Doniach S, Permyakov EA, Uversky VN. Natively unfolded C-terminal domain of caldesmon remains substantially unstructured after the effective binding to calmodulin. Proteins. 2003;53:855–862. doi: 10.1002/prot.10481. [DOI] [PubMed] [Google Scholar]
- 258.Parsegian VA, Rand RP, Fuller NL, Rau DC. Osmotic stress for the direct measurement of intermolecular forces. Methods Enzymol. 1986;127:400–416. doi: 10.1016/0076-6879(86)27032-9. [DOI] [PubMed] [Google Scholar]
- 259.Parsegian VA, Rand RP, Rau DC. Macromolecules and water: probing with osmotic stress. Methods Enzymol. 1995;259:43–94. doi: 10.1016/0076-6879(95)59039-0. [DOI] [PubMed] [Google Scholar]
- 260.Parsegian VA, Rand RP, Rau DC. Osmotic stress, crowding, preferential hydration, and binding: A comparison of perspectives. Proc Natl Acad Sci U S A. 2000;97:3987–3992. doi: 10.1073/pnas.97.8.3987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Fried MG, Stickle DF, Smirnakis KV, Adams C, MacDonald D, Lu P. Role of hydration in the binding of lac repressor to DNA. J Biol Chem. 2002;277:50676–50682. doi: 10.1074/jbc.M208540200. [DOI] [PubMed] [Google Scholar]
- 262.Fuxreiter M, Simon I, Friedrich P, Tompa P. Preformed structural elements feature in partner recognition by intrinsically unstructured proteins. J Mol Biol. 2004;338:1015–1026. doi: 10.1016/j.jmb.2004.03.017. [DOI] [PubMed] [Google Scholar]
- 263.Lacy ER, Filippov I, Lewis WS, Otieno S, Xiao L, Weiss S, Hengst L, Kriwacki RW. p27 binds cyclin-CDK complexes through a sequential mechanism involving binding-induced protein folding. Nat Struct Mol Biol. 2004;11:358–364. doi: 10.1038/nsmb746. [DOI] [PubMed] [Google Scholar]
- 264.Fletcher CM, Wagner G. The interaction of eIF4E with 4E-BP1 is an induced fit to a completely disordered protein. Protein Sci. 1998;7:1639–1642. doi: 10.1002/pro.5560070720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 265.Mader S, Lee H, Pause A, Sonenberg N. The translation initiation factor eIF-4E binds to a common motif shared by the translation factor eIF-4 gamma and the translational repressors 4E-binding proteins. Mol Cell Biol. 1995;15:4990–4997. doi: 10.1128/mcb.15.9.4990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 266.Kissinger CR, Parge HE, Knighton DR, Lewis CT, Pelletier LA, Tempczyk A, Kalish VJ, Tucker KD, Showalter RE, Moomaw EW, et al. Crystal structures of human calcineurin and the human FKBP12-FK506-calcineurin complex. Nature. 1995;378:641–644. doi: 10.1038/378641a0. [DOI] [PubMed] [Google Scholar]
- 267.Callaghan AJ, Aurikko JP, Ilag LL, Gunter Grossmann J, Chandran V, Kuhnel K, Poljak L, Carpousis AJ, Robinson CV, Symmons MF, Luisi BF. Studies of the RNA degradosome-organizing domain of the Escherichia coli ribonuclease RNase E. J Mol Biol. 2004;340:965–979. doi: 10.1016/j.jmb.2004.05.046. [DOI] [PubMed] [Google Scholar]
- 268.Chandran V, Luisi BF. Recognition of enolase in the Escherichia coli RNA degradosome. J Mol Biol. 2006;358:8–15. doi: 10.1016/j.jmb.2006.02.012. [DOI] [PubMed] [Google Scholar]
- 269.Dunker AK. Another window into disordered protein function. Structure. 2007;15:1026–1028. doi: 10.1016/j.str.2007.08.001. [DOI] [PubMed] [Google Scholar]
- 270.Dosztanyi Z, Meszaros B, Simon I. ANCHOR: web server for predicting protein binding regions in disordered proteins. Bioinformatics. 2009;25:2745–2746. doi: 10.1093/bioinformatics/btp518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 271.Puntervoll P, Linding R, Gemund C, Chabanis-Davidson S, Mattingsdal M, Cameron S, Martin DM, Ausiello G, Brannetti B, Costantini A, Ferre F, Maselli V, Via A, Cesareni G, Diella F, Superti-Furga G, Wyrwicz L, Ramu C, McGuigan C, Gudavalli R, Letunic I, Bork P, Rychlewski L, Kuster B, Helmer-Citterich M, Hunter WN, Aasland R, Gibson TJ. ELM server: A new resource for investigating short functional sites in modular eukaryotic proteins. Nucleic Acids Res. 2003;31:3625–3630. doi: 10.1093/nar/gkg545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 272.Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, Seiler M, Davey NE, Haslam N, Weatheritt RJ, Budd A, Hughes T, Pas J, Rychlewski L, Trave G, Aasland R, Helmer-Citterich M, Linding R, Gibson TJ. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 273.Davey NE, Edwards RJ, Shields DC. The SLiMDisc server: short, linear motif discovery in proteins. Nucleic Acids Res. 2007;35:W455–459. doi: 10.1093/nar/gkm400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Edwards RJ, Davey NE, Shields DC. SLiMFinder: a probabilistic method for identifying over-represented, convergently evolved, short linear motifs in proteins. PLoS One. 2007;2:e967. doi: 10.1371/journal.pone.0000967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 275.Fuxreiter M, Tompa P, Simon I. Local structural disorder imparts plasticity on linear motifs. Bioinformatics. 2007;23:950–956. doi: 10.1093/bioinformatics/btm035. [DOI] [PubMed] [Google Scholar]
- 276.Ren S, Uversky VN, Chen Z, Dunker AK, Obradovic Z. Short Linear Motifs recognized by SH2, SH3 and Ser/Thr Kinase domains are conserved in disordered protein regions. BMC Genomics. 2008;9 2:S26. doi: 10.1186/1471-2164-9-S2-S26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Sambrook J. Adenovirus amazes at Cold Spring Harbor. Nature. 1977;268:101–104. doi: 10.1038/268101a0. [DOI] [PubMed] [Google Scholar]
- 278.Black DL. Mechanisms of alternative pre-messenger RNA splicing. Annu Rev Biochem. 2003;72:291–336. doi: 10.1146/annurev.biochem.72.121801.161720. [DOI] [PubMed] [Google Scholar]
- 279.Gilbert W. Why genes in pieces? Nature. 1978;271:501. doi: 10.1038/271501a0. [DOI] [PubMed] [Google Scholar]
- 280.Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5:773–782. doi: 10.1038/nrg1451. [DOI] [PubMed] [Google Scholar]
- 281.Irimia M, Rukov JL, Penny D, Roy SW. Functional and evolutionary analysis of alternatively spliced genes is consistent with an early eukaryotic origin of alternative splicing. BMC Evol Biol. 2007;7:188. doi: 10.1186/1471-2148-7-188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Stamm S, Ben-Ari S, Rafalska I, Tang Y, Zhang Z, Toiber D, Thanaraj TA, Soreq H. Function of alternative splicing. Gene. 2005;344:1–20. doi: 10.1016/j.gene.2004.10.022. [DOI] [PubMed] [Google Scholar]
- 283.Brett D, Hanke J, Lehmann G, Haase S, Delbruck S, Krueger S, Reich J, Bork P. EST comparison indicates 38% of human mRNAs contain possible alternative splice forms. FEBS Lett. 2000;474:83–86. doi: 10.1016/s0014-5793(00)01581-7. [DOI] [PubMed] [Google Scholar]
- 284.Johnson JM, Castle J, Garrett-Engele P, Kan Z, Loerch PM, Armour CD, Santos R, Schadt EE, Stoughton R, Shoemaker DD. Genome-wide survey of human alternative pre-mRNA splicing with exon junction microarrays. Science. 2003;302:2141–2144. doi: 10.1126/science.1090100. [DOI] [PubMed] [Google Scholar]
- 285.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 286.Minneman KP. Splice variants of G protein-coupled receptors. Molecular interventions. 2001;1:108–116. [PubMed] [Google Scholar]
- 287.Thai TH, Kearney JF. Distinct and opposite activities of human terminal deoxynucleotidyltransferase splice variants. J Immunol. 2004;173:4009–4019. doi: 10.4049/jimmunol.173.6.4009. [DOI] [PubMed] [Google Scholar]
- 288.Scheper W, Zwart R, Baas F. Alternative splicing in the N-terminus of Alzheimer's presenilin 1. Neurogenetics. 2004;5:223–227. doi: 10.1007/s10048-004-0195-y. [DOI] [PubMed] [Google Scholar]
- 289.Roberts R, Timchenko NA, Miller JW, Reddy S, Caskey CT, Swanson MS, Timchenko LT. Altered phosphorylation and intracellular distribution of a (CUG)n triplet repeat RNA-binding protein in patients with myotonic dystrophy and in myotonin protein kinase knockout mice. Proc Natl Acad Sci U S A. 1997;94:13221–13226. doi: 10.1073/pnas.94.24.13221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 290.Ma K, Inglis JD, Sharkey A, Bickmore WA, Hill RE, Prosser EJ, Speed RM, Thomson EJ, Jobling M, Taylor K, et al. A Y chromosome gene family with RNA-binding protein homology: candidates for the azoospermia factor AZF controlling human spermatogenesis. Cell. 1993;75:1287–1295. doi: 10.1016/0092-8674(93)90616-x. [DOI] [PubMed] [Google Scholar]
- 291.Lovestone S, Reynolds CH, Latimer D, Davis DR, Anderton BH, Gallo JM, Hanger D, Mulot S, Marquardt B, Stabel S, et al. Alzheimer's disease-like phosphorylation of the microtubule-associated protein tau by glycogen synthase kinase-3 in transfected mammalian cells. Curr Biol. 1994;4:1077–1086. doi: 10.1016/s0960-9822(00)00246-3. [DOI] [PubMed] [Google Scholar]
- 292.Beyer K, Domingo-Sabat M, Humbert J, Carrato C, Ferrer I, Ariza A. Differential expression of alpha-synuclein, parkin, and synphilin-1 isoforms in Lewy body disease. Neurogenetics. 2008;9:163–172. doi: 10.1007/s10048-008-0124-6. [DOI] [PubMed] [Google Scholar]
- 293.Beyer K, Domingo-Sabat M, Lao JI, Carrato C, Ferrer I, Ariza A. Identification and characterization of a new alpha-synuclein isoform and its role in Lewy body diseases. Neurogenetics. 2008;9:15–23. doi: 10.1007/s10048-007-0106-0. [DOI] [PubMed] [Google Scholar]
- 294.Venables JP. Aberrant and alternative splicing in cancer. Cancer Res. 2004;64:7647–7654. doi: 10.1158/0008-5472.CAN-04-1910. [DOI] [PubMed] [Google Scholar]
- 295.Wang P, Yan B, Guo JT, Hicks C, Xu Y. Structural genomics analysis of alternative splicing and application to isoform structure modeling. Proc Natl Acad Sci U S A. 2005;102:18920–18925. doi: 10.1073/pnas.0506770102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 296.Furnham N, Ruffle S, Southan C. Splice variants: a homology modeling approach. Proteins. 2004;54:596–608. doi: 10.1002/prot.10568. [DOI] [PubMed] [Google Scholar]
- 297.Romero PR, Zaidi S, Fang YY, Uversky VN, Radivojac P, Oldfield CJ, Cortese MS, Sickmeier M, LeGall T, Obradovic Z, Dunker AK. Alternative splicing in concert with protein intrinsic disorder enables increased functional diversity in multicellular organisms. Proc Natl Acad Sci U S A. 2006;103:8390–8395. doi: 10.1073/pnas.0507916103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 298.Dunker AK, Uversky VN. Signal transduction via unstructured protein conduits. Nat Chem Biol. 2008;4:229–230. doi: 10.1038/nchembio0408-229. [DOI] [PubMed] [Google Scholar]
- 299.Dunker AK, Silman I, Uversky VN, Sussman JL. Function and structure of inherently disordered proteins. Curr Opin Struct Biol. 2008;18:756–764. doi: 10.1016/j.sbi.2008.10.002. [DOI] [PubMed] [Google Scholar]
- 300.Dunker AK, Oldfield CJ, Meng J, Romero P, Yang JY, Chen JW, Vacic V, Obradovic Z, Uversky VN. The unfoldomics decade: an update on intrinsically disordered proteins. BMC Genomics. 2008;9 2:S1. doi: 10.1186/1471-2164-9-S2-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 301.Dosztanyi Z, Csizmok V, Tompa P, Simon I. IUPred: web server for the prediction of intrinsically unstructured regions of proteins based on estimated energy content. Bioinformatics. 2005;21:3433–3434. doi: 10.1093/bioinformatics/bti541. [DOI] [PubMed] [Google Scholar]
- 302.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 303.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Protein disorder in the human diseasome: unfoldomics of human genetic diseases. BMC Genomics. 2009;10 1:S12. doi: 10.1186/1471-2164-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 304.Uversky VN, Oldfield CJ, Midic U, Xie H, Xue B, Vucetic S, Iakoucheva LM, Obradovic Z, Dunker AK. Unfoldomics of human diseases: linking protein intrinsic disorder with diseases. BMC Genomics. 2009;10 1:S7. doi: 10.1186/1471-2164-10-S1-S7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 305.Uversky VN, Dunker AK. Biochemistry. Controlled chaos. Science. 2008;322:1340–1341. doi: 10.1126/science.1167453. [DOI] [PubMed] [Google Scholar]
- 306.Gsponer J, Futschik ME, Teichmann SA, Babu MM. Tight regulation of unstructured proteins: from transcript synthesis to protein degradation. Science. 2008;322:1365–1368. doi: 10.1126/science.1163581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 307.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 308.Grimmler M, Wang Y, Mund T, Cilensek Z, Keidel EM, Waddell MB, Jakel H, Kullmann M, Kriwacki RW, Hengst L. Cdk-inhibitory activity and stability of p27Kip1 are directly regulated by oncogenic tyrosine kinases. Cell. 2007;128:269–280. doi: 10.1016/j.cell.2006.11.047. [DOI] [PubMed] [Google Scholar]
- 309.Iakoucheva LM, Radivojac P, Brown CJ, O'Connor TR, Sikes JG, Obradovic Z, Dunker AK. The importance of intrinsic disorder for protein phosphorylation. Nucleic Acids Res. 2004;32:1037–1049. doi: 10.1093/nar/gkh253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 310.Tompa P. The interplay between structure and function in intrinsically unstructured proteins. FEBS Lett. 2005;579:3346–3354. doi: 10.1016/j.febslet.2005.03.072. [DOI] [PubMed] [Google Scholar]
- 311.Paliy O, Gargac SM, Cheng Y, Uversky VN, Dunker AK. Protein disorder is positively correlated with gene expression in Escherichia coli. J Proteome Res. 2008;7:2234–2245. doi: 10.1021/pr800055r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 312.Chen J, Liang H, Fernandez A. Protein structure protection commits gene expression patterns. Genome Biol. 2008;9:R107. doi: 10.1186/gb-2008-9-7-r107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 313.Fernandez A, Scheraga HA. Insufficiently dehydrated hydrogen bonds as determinants of protein interactions. Proc Natl Acad Sci U S A. 2003;100:113–118. doi: 10.1073/pnas.0136888100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 314.Fernandez A. Keeping dry and crossing membranes. Nat Biotechnol. 2004;22:1081–1084. doi: 10.1038/nbt0904-1081. [DOI] [PubMed] [Google Scholar]
- 315.Kelly JW. The alternative conformations of amyloidogenic proteins and their multi-step assembly pathways. Curr Opin Struct Biol. 1998;8:101–106. doi: 10.1016/s0959-440x(98)80016-x. [DOI] [PubMed] [Google Scholar]
- 316.Dobson CM. Protein misfolding, evolution and disease. Trends Biochem Sci. 1999;24:329–332. doi: 10.1016/s0968-0004(99)01445-0. [DOI] [PubMed] [Google Scholar]
- 317.Bellotti V, Mangione P, Stoppini M. Biological activity and pathological implications of misfolded proteins. Cell Mol Life Sci. 1999;55:977–991. doi: 10.1007/s000180050348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 318.Uversky VN, Talapatra A, Gillespie JR, Fink AL. Protein deposits as the molecular basis of amyloidosis. I. Systemic amyloidoses. Med Sci Monitor. 1999;5:1001–1012. [Google Scholar]
- 319.Uversky VN, Talapatra A, Gillespie JR, Fink AL. Protein deposits as the molecular basis of amyloidosis. II. Localized amyloidosis and neurodegenerative disordres. Med Sci Monitor. 1999;5:1238–1254. [Google Scholar]
- 320.Rochet JC, Lansbury PT., Jr Amyloid fibrillogenesis: themes and variations. Curr Opin Struct Biol. 2000;10:60–68. doi: 10.1016/s0959-440x(99)00049-4. [DOI] [PubMed] [Google Scholar]
- 321.Uversky VN, Fink AL. Conformational constraints for amyloid fibrillation: the importance of being unfolded. Biochim Biophys Acta. 2004;1698:131–153. doi: 10.1016/j.bbapap.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 322.Uversky VN, Fink AL. Pathways to amyloid fibril formation: Partially folded intermediates in fibrillation of unfolded proteins. In: Sipe JD, editor. Amyloid Proteins: The Beta Pleated Sheet Conformation and Disease. Wiley-VCH, Verlag GmbH & Co. KGaA; Weinheim, Germany: 2005. pp. 247–265. [Google Scholar]
- 323.Glenner GG, Wong CW. Alzheimer's disease and Down's syndrome: sharing of a unique cerebrovascular amyloid fibril protein. Biochem Biophys Res Commun. 1984;122:1131–1135. doi: 10.1016/0006-291x(84)91209-9. [DOI] [PubMed] [Google Scholar]
- 324.Masters CL, Multhaup G, Simms G, Pottgiesser J, Martins RN, Beyreuther K. Neuronal origin of a cerebral amyloid: neurofibrillary tangles of Alzheimer's disease contain the same protein as the amyloid of plaque cores and blood vessels. Embo J. 1985;4:2757–2763. doi: 10.1002/j.1460-2075.1985.tb04000.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 325.Lee VM, Balin BJ, Otvos L, Jr, Trojanowski JQ. A68: a major subunit of paired helical filaments and derivatized forms of normal Tau. Science. 1991;251:675–678. doi: 10.1126/science.1899488. [DOI] [PubMed] [Google Scholar]
- 326.Ueda K, Fukushima H, Masliah E, Xia Y, Iwai A, Yoshimoto M, Otero DA, Kondo J, Ihara Y, Saitoh T. Molecular cloning of cDNA encoding an unrecognized component of amyloid in Alzheimer disease. Proc Natl Acad Sci U S A. 1993;90:11282–11286. doi: 10.1073/pnas.90.23.11282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 327.Wisniewski KE, Dalton AJ, McLachlan C, Wen GY, Wisniewski HM. Alzheimer's disease in Down's syndrome: clinicopathologic studies. Neurology. 1985;35:957–961. doi: 10.1212/wnl.35.7.957. [DOI] [PubMed] [Google Scholar]
- 328.Dev KK, Hofele K, Barbieri S, Buchman VL, van der Putten H. Part II: alpha-synuclein and its molecular pathophysiological role in neurodegenerative disease. Neuropharmacology. 2003;45:14–44. doi: 10.1016/s0028-3908(03)00140-0. [DOI] [PubMed] [Google Scholar]
- 329.Prusiner SB. Shattuck lecture--neurodegenerative diseases and prions. N Engl J Med. 2001;344:1516–1526. doi: 10.1056/NEJM200105173442006. [DOI] [PubMed] [Google Scholar]
- 330.Zoghbi HY, Orr HT. Polyglutamine diseases: protein cleavage and aggregation. Curr Opin Neurobiol. 1999;9:566–570. doi: 10.1016/S0959-4388(99)00013-6. [DOI] [PubMed] [Google Scholar]
- 331.Uversky VN. A protein-chameleon: conformational plasticity of alpha-synuclein, a disordered protein involved in neurodegenerative disorders. J Biomol Struct Dyn. 2003;21:211–234. doi: 10.1080/07391102.2003.10506918. [DOI] [PubMed] [Google Scholar]
- 332.Okazawa H. Polyglutamine diseases: a transcription disorder? Cell Mol Life Sci. 2003;60:1427–1439. doi: 10.1007/s00018-003-3013-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 333.Cummings CJ, Zoghbi HY. Fourteen and counting: unraveling trinucleotide repeat diseases. Hum Mol Genet. 2000;9:909–916. doi: 10.1093/hmg/9.6.909. [DOI] [PubMed] [Google Scholar]
- 334.Gusella JF, MacDonald ME. Molecular genetics: unmasking polyglutamine triggers in neurodegenerative disease. Nat Rev Neurosci. 2000;1:109–115. doi: 10.1038/35039051. [DOI] [PubMed] [Google Scholar]
- 335.Orr HT. Beyond the Qs in the polyglutamine diseases. Genes Dev. 2001;15:925–932. doi: 10.1101/gad.888401. [DOI] [PubMed] [Google Scholar]
- 336.Fischbeck KH. Polyglutamine expansion neurodegenerative disease. Brain Res Bull. 2001;56:161–163. doi: 10.1016/s0361-9230(01)00577-9. [DOI] [PubMed] [Google Scholar]
- 337.Uversky VN, Roman A, Oldfield CJ, Dunker AK. Protein intrinsic disorder and human papillomaviruses: increased amount of disorder in E6 and E7 oncoproteins from high risk HPVs. J Proteome Res. 2006;5:1829–1842. doi: 10.1021/pr0602388. [DOI] [PubMed] [Google Scholar]
- 338.Cheng Y, LeGall T, Oldfield CJ, Dunker AK, Uversky VN. Abundance of intrinsic disorder in protein associated with cardiovascular disease. Biochemistry. 2006;45:10448–10460. doi: 10.1021/bi060981d. [DOI] [PubMed] [Google Scholar]
- 339.Uversky VN. Intrinsic disorder in proteins associated with neurodegenerative diseases. Front Biosci. 2009;14:5188–5238. doi: 10.2741/3594. [DOI] [PubMed] [Google Scholar]
- 340.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Protein disorder in the human diseasome: Unfoldomics of human genetic diseases. PLoS Computational Biology. 2008 doi: 10.1186/1471-2164-10-S1-S12. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 341.Adkins JN, Lumb KJ. Intrinsic structural disorder and sequence features of the cell cycle inhibitor p57Kip2. Proteins. 2002;46:1–7. doi: 10.1002/prot.10018. [DOI] [PubMed] [Google Scholar]
- 342.Chang BS, Minn AJ, Muchmore SW, Fesik SW, Thompson CB. Identification of a novel regulatory domain in Bcl-X(L) and Bcl-2. Embo J. 1997;16:968–977. doi: 10.1093/emboj/16.5.968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 343.Campbell KM, Terrell AR, Laybourn PJ, Lumb KJ. Intrinsic structural disorder of the C-terminal activation domain from the bZIP transcription factor Fos. Biochemistry. 2000;39:2708–2713. doi: 10.1021/bi9923555. [DOI] [PubMed] [Google Scholar]
- 344.Sunde M, McGrath KC, Young L, Matthews JM, Chua EL, Mackay JP, Death AK. TC-1 is a novel tumorigenic and natively disordered protein associated with thyroid cancer. Cancer Res. 2004;64:2766–2773. doi: 10.1158/0008-5472.can-03-2093. [DOI] [PubMed] [Google Scholar]
- 345.Goh KI, Cusick ME, Valle D, Childs B, Vidal M, Barabasi AL. The human disease network. Proc Natl Acad Sci U S A. 2007;104:8685–8690. doi: 10.1073/pnas.0701361104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 346.Arkin MR, Wells JA. Small-molecule inhibitors of protein-protein interactions: progressing towards the dream. Nat Rev Drug Discov. 2004;3:301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 347.Arkin M. Protein-protein interactions and cancer: small molecules going in for the kill. Curr Opin Chem Biol. 2005;9:317–324. doi: 10.1016/j.cbpa.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 348.Liang J, Edelsbrunner H, Woodward C. Anatomy of protein pockets and cavities: measurement of binding site geometry and implications for ligand design. Protein Sci. 1998;7:1884–1897. doi: 10.1002/pro.5560070905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 349.Cochran AG. Antagonists of protein-protein interactions. Chem Biol. 2000;7:R85–94. doi: 10.1016/s1074-5521(00)00106-x. [DOI] [PubMed] [Google Scholar]
- 350.Rodi DJ, Agoston GE, Manon R, Lapcevich R, Green SJ, Makowski L. Identification of small molecule binding sites within proteins using phage display technology. Comb Chem High Throughput Screen. 2001;4:553–572. doi: 10.2174/1386207013330779. [DOI] [PubMed] [Google Scholar]
- 351.Blundell TL, Burke DF, Chirgadze D, Dhanaraj V, Hyvonen M, Innis CA, Parisini E, Pellegrini L, Sayed M, Sibanda BL. Protein-protein interactions in receptor activation and intracellular signalling. Biol Chem. 2000;381:955–959. doi: 10.1515/BC.2000.117. [DOI] [PubMed] [Google Scholar]
- 352.Hollstein M, Sidransky D, Vogelstein B, Harris CC. p53 mutations in human cancers. Science. 1991;253:49–53. doi: 10.1126/science.1905840. [DOI] [PubMed] [Google Scholar]
- 353.Balint EE, Vousden KH. Activation and activities of the p53 tumour suppressor protein. Br J Cancer. 2001;85:1813–1823. doi: 10.1054/bjoc.2001.2128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 354.Zhao R, Gish K, Murphy M, Yin Y, Notterman D, Hoffman WH, Tom E, Mack DH, Levine AJ. Analysis of p53-regulated gene expression patterns using oligonucleotide arrays. Genes Dev. 2000;14:981–993. [PMC free article] [PubMed] [Google Scholar]
- 355.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 356.Bottger A, Bottger V, Sparks A, Liu WL, Howard SF, Lane DP. Design of a synthetic Mdm2-binding mini protein that activates the p53 response in vivo. Curr Biol. 1997;7:860–869. doi: 10.1016/s0960-9822(06)00374-5. [DOI] [PubMed] [Google Scholar]
- 357.Wasylyk C, Salvi R, Argentini M, Dureuil C, Delumeau I, Abecassis J, Debussche L, Wasylyk B. p53 mediated death of cells overexpressing MDM2 by an inhibitor of MDM2 interaction with p53. Oncogene. 1999;18:1921–1934. doi: 10.1038/sj.onc.1202528. [DOI] [PubMed] [Google Scholar]
- 358.Chene P, Fuchs J, Bohn J, Garcia-Echeverria C, Furet P, Fabbro D. A small synthetic peptide, which inhibits the p53-hdm2 interaction, stimulates the p53 pathway in tumour cell lines. J Mol Biol. 2000;299:245–253. doi: 10.1006/jmbi.2000.3738. [DOI] [PubMed] [Google Scholar]
- 359.Garcia-Echeverria C, Chene P, Blommers MJ, Furet P. Discovery of potent antagonists of the interaction between human double minute 2 and tumor suppressor p53. J Med Chem. 2000;43:3205–3208. doi: 10.1021/jm990966p. [DOI] [PubMed] [Google Scholar]
- 360.Chene P. Inhibition of the p53-MDM2 interaction: targeting a protein-protein interface. Mol Cancer Res. 2004;2:20–28. [PubMed] [Google Scholar]
- 361.Klein C, Vassilev LT. Targeting the p53-MDM2 interaction to treat cancer. Br J Cancer. 2004;91:1415–1419. doi: 10.1038/sj.bjc.6602164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 362.Vassilev LT. Small-molecule antagonists of p53-MDM2 binding: research tools and potential therapeutics. Cell Cycle. 2004;3:419–421. [PubMed] [Google Scholar]
- 363.Vassilev LT, Vu BT, Graves B, Carvajal D, Podlaski F, Filipovic Z, Kong N, Kammlott U, Lukacs C, Klein C, Fotouhi N, Liu EA. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science. 2004;303:844–848. doi: 10.1126/science.1092472. [DOI] [PubMed] [Google Scholar]
- 364.Cheng Y, LeGall T, Oldfield CJ, Mueller JP, Van YY, Romero P, Cortese MS, Uversky VN, Dunker AK. Rational drug design via intrinsically disordered protein. Trends Biotechnol. 2006;24:435–442. doi: 10.1016/j.tibtech.2006.07.005. [DOI] [PubMed] [Google Scholar]
- 365.Chen CT, Wagner H, Still WC. Fluorescent, sequence-selective peptide detection by synthetic small molecules. Science. 1998;279:851–853. doi: 10.1126/science.279.5352.851. [DOI] [PubMed] [Google Scholar]
- 366.Pescarolo MP, Bagnasco L, Malacarne D, Melchiori A, Valente P, Millo E, Bruno S, Basso S, Parodi S. A retro-inverso peptide homologous to helix 1 of c-Myc is a potent and specific inhibitor of proliferation in different cellular systems. FASEB J. 2001;15:31–33. doi: 10.1096/fj.00-0422fje. [DOI] [PubMed] [Google Scholar]
- 367.Berg T, Cohen SB, Desharnais J, Sonderegger C, Maslyar DJ, Goldberg J, Boger DL, Vogt PK. Small-molecule antagonists of Myc/Max dimerization inhibit Myc-induced transformation of chicken embryo fibroblasts. Proc Natl Acad Sci U S A. 2002;99:3830–3835. doi: 10.1073/pnas.062036999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 368.Yin X, Giap C, Lazo JS, Prochownik EV. Low molecular weight inhibitors of Myc-Max interaction and function. Oncogene. 2003;22:6151–6159. doi: 10.1038/sj.onc.1206641. [DOI] [PubMed] [Google Scholar]
- 369.Kiessling A, Sperl B, Hollis A, Eick D, Berg T. Selective inhibition of c-Myc/Max dimerization and DNA binding by small molecules. Chem Biol. 2006;13:745–751. doi: 10.1016/j.chembiol.2006.05.011. [DOI] [PubMed] [Google Scholar]
- 370.Mo H, Henriksson M. Identification of small molecules that induce apoptosis in a Myc-dependent manner and inhibit Myc-driven transformation. Proc Natl Acad Sci U S A. 2006;103:6344–6349. doi: 10.1073/pnas.0601418103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 371.Xu Y, Shi J, Yamamoto N, Moss JA, Vogt PK, Janda KD. A credit-card library approach for disrupting protein-protein interactions. Bioorg Med Chem. 2006;14:2660–2673. doi: 10.1016/j.bmc.2005.11.052. [DOI] [PubMed] [Google Scholar]
- 372.Bagnasco L, Tortolina L, Biasotti B, Castagnino N, Ponassi R, Tomati V, Nieddu E, Stier G, Malacarne D, Parodi S. Inhibition of a protein-protein interaction between INI1 and c-Myc by small peptidomimetic molecules inspired by Helix-1 of c-Myc: identification of a new target of potential antineoplastic interest. FASEB J. 2007;21:1256–1263. doi: 10.1096/fj.06-7082com. [DOI] [PubMed] [Google Scholar]
- 373.Wang H, Hammoudeh DI, Follis AV, Reese BE, Lazo JS, Metallo SJ, Prochownik EV. Improved low molecular weight Myc-Max inhibitors. Mol Cancer Ther. 2007;6:2399–2408. doi: 10.1158/1535-7163.MCT-07-0005. [DOI] [PubMed] [Google Scholar]
- 374.Follis AV, Hammoudeh DI, Daab AT, Metallo SJ. Small-molecule perturbation of competing interactions between c-Myc and Max. Bioorg Med Chem Lett. 2009;19:807–810. doi: 10.1016/j.bmcl.2008.12.025. [DOI] [PubMed] [Google Scholar]
- 375.Follis AV, Hammoudeh DI, Wang H, Prochownik EV, Metallo SJ. Structural rationale for the coupled binding and unfolding of the c-Myc oncoprotein by small molecules. Chem Biol. 2008;15:1149–1155. doi: 10.1016/j.chembiol.2008.09.011. [DOI] [PubMed] [Google Scholar]
- 376.Mustata G, Follis AV, Hammoudeh DI, Metallo SJ, Wang H, Prochownik EV, Lazo JS, Bahar I. Discovery of novel myc-max heterodimer disruptors with a three-dimensional pharmacophore model. J Med Chem. 2009;52:1247–1250. doi: 10.1021/jm801278g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 377.Hammoudeh DI, Follis AV, Prochownik EV, Metallo SJ. Multiple independent binding sites for small-molecule inhibitors on the oncoprotein c-Myc. J Am Chem Soc. 2009;131:7390–7401. doi: 10.1021/ja900616b. [DOI] [PubMed] [Google Scholar]
- 378.Williams RJ. The conformation properties of proteins in solution. Biol Rev Camb Philos Soc. 1979;54:389–437. doi: 10.1111/j.1469-185x.1979.tb00843.x. [DOI] [PubMed] [Google Scholar]
- 379.Romero P, Obradovic Z, Kissinger C, Villafranca JE, Dunker AK. Identifying disordered regions in proteins from amino acid sequence. Proc IEEE Int Conf Neural Networks. 1997;1:90–95. [Google Scholar]
- 380.Romero P, Obradovic Z, Dunker AK. Sequence Data Analysis for Long Disordered Regions Prediction in the Calcineurin Family. Genome Inform Ser Workshop Genome Inform. 1997;8:110–124. [PubMed] [Google Scholar]
- 381.Linding R, Russell RB, Neduva V, Gibson TJ. GlobPlot: Exploring protein sequences for globularity and disorder. Nucleic Acids Res. 2003;31:3701–3708. doi: 10.1093/nar/gkg519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 382.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure (Camb) 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 383.Jones DT, Ward JJ. Prediction of disordered regions in proteins from position specific score matrices. Proteins. 2003;53:573–578. doi: 10.1002/prot.10528. [DOI] [PubMed] [Google Scholar]
- 384.Liu J, Rost B. NORSp: Predictions of long regions without regular secondary structure. Nucleic Acids Res. 2003;31:3833–3835. doi: 10.1093/nar/gkg515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 385.Weathers EA, Paulaitis ME, Woolf TB, Hoh JH. Reduced amino acid alphabet is sufficient to accurately recognize intrinsically disordered protein. FEBS Lett. 2004;576:348–352. doi: 10.1016/j.febslet.2004.09.036. [DOI] [PubMed] [Google Scholar]
- 386.MacCallum RM. Order/disorder prediction with self organizing maps. CASP6 online paper [Google Scholar]
- 387.Garbuzynskiy SO, Lobanov MY, Galzitskaya OV. To be folded or to be unfolded. Protein Sci. 2004;13:2871–2877. doi: 10.1110/ps.04881304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 388.Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. FoldUnfold: web server for the prediction of disordered regions in protein chain. Bioinformatics. 2006;22:2948–2949. doi: 10.1093/bioinformatics/btl504. [DOI] [PubMed] [Google Scholar]
- 389.Ferron F, Rancurel C, Longhi S, Cambillau C, Henrissat B, Canard B. VaZyMolO: a tool to define and classify modularity in viral proteins. J Gen Virol. 2005;86:743–749. doi: 10.1099/vir.0.80590-0. [DOI] [PubMed] [Google Scholar]
- 390.Dosztányi Z, Csizmók V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 391.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 392.Yang ZR, Thomson R, McNeil P, Esnouf RM. RONN: the bio-basis function neural network technique applied to the detection of natively disordered regions in proteins. Bioinformatics. 2005;21:3369–3376. doi: 10.1093/bioinformatics/bti534. [DOI] [PubMed] [Google Scholar]
- 393.Cheng J, Sweredoski MJ, Baldi P. Accurate prediction of protein disordered regions by mining protein structure data. Data Mining and Knowledge Discovery. 2005;11:213–222. [Google Scholar]
- 394.Obradovic Z, Peng K, Vucetic S, Radivojac P, Dunker AK. Exploiting heterogeneous sequence properties improves prediction of protein disorder. Proteins. 2005;61:176–182. doi: 10.1002/prot.20735. [DOI] [PubMed] [Google Scholar]
- 395.Coeytaux K, Poupon A. Prediction of unfolded segments in a protein sequence based on amino acid composition. Bioinformatics. 2005;21:1891–1900. doi: 10.1093/bioinformatics/bti266. [DOI] [PubMed] [Google Scholar]
- 396.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. BMC Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 397.Vullo A, Bortolami O, Pollastri G, Tosatto SC. Spritz: a server for the prediction of intrinsically disordered regions in protein sequences using kernel machines. Nucleic Acids Res. 2006;34:164–168. doi: 10.1093/nar/gkl166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 398.Su CT, Chen CY, Ou YY. Protein disorder prediction by condensed PSSM considering propensity for order or disorder. BMC Bioinformatics. 2006;7:319. doi: 10.1186/1471-2105-7-319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 399.Yang MQ, Yang JY. IUP: Intrinsically Unstructured Protein predictor - A software tool for analyzing polypeptide sequences. IEEE BIBE. 2006;2006:1–13. [Google Scholar]
- 400.Gu J, Gribskov M, Bourne PE. Wiggle-predicting functionally flexible regions from primary sequence. PLoS Comput Biol. 2006;2:e90. doi: 10.1371/journal.pcbi.0020090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 401.Su CT, Chen CY, Hsu CM. iPDA: integrated protein disorder analyzer. Nucleic Acids Res. 2007;35:465–472. doi: 10.1093/nar/gkm353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 402.Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:460–464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 403.Shimizu K, Muraoka Y, Hirose S, Tomii K, Noguchi T. Predicting mostly disordered proteins by using structure-unknown protein data. BMC Bioinformatics. 2007;8:78. doi: 10.1186/1471-2105-8-78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 404.Schlessinger A, Punta M, Rost B. Natively unstructured regions in proteins identified from contact predictions. Bioinformatics. 2007;23:2376–2384. doi: 10.1093/bioinformatics/btm349. [DOI] [PubMed] [Google Scholar]
- 405.Hirose S, Shimizu K, Kanai S, Kuroda Y, Noguchi T. POODLE-L: a two-level SVM prediction system for reliably predicting long disordered regions. Bioinformatics. 2007;23:2046–2053. doi: 10.1093/bioinformatics/btm302. [DOI] [PubMed] [Google Scholar]
- 406.Shimizu K, Hirose S, Noguchi T. POODLE-S: web application for predicting protein disorder by using physicochemical features and reduced amino acid set of a position-specific scoring matrix. Bioinformatics. 2007;23:2337–2338. doi: 10.1093/bioinformatics/btm330. [DOI] [PubMed] [Google Scholar]
- 407.Wang L, Sauer UH. OnD-CRF: predicting order and disorder in proteins using conditional random fields. Bioinformatics. 2008;24:1401–1402. doi: 10.1093/bioinformatics/btn132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 408.Bulashevska A, Eils R. Using bayesian multinomial classifier to predict where a given protein sequence is intrinsically disordered. J Theor Biol. 2008;254:799–803. doi: 10.1016/j.jtbi.2008.05.040. [DOI] [PubMed] [Google Scholar]
- 409.McGuffin LJ. Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics. 2008;24:1798–1804. doi: 10.1093/bioinformatics/btn326. [DOI] [PubMed] [Google Scholar]
- 410.Sethi D, Garg A, Raghava GP. DPROT: prediction of disordered proteins using evolutionary information. Amino Acids. 2008;35:599–605. doi: 10.1007/s00726-008-0085-y. [DOI] [PubMed] [Google Scholar]
- 411.Yang JY, Yang MQ. Identification of Intrinsically Unstructured Proteins using hierarchical classifier. Int J Data Min BioinformInt. 2008;2:121–133. doi: 10.1504/ijdmb.2008.019093. [DOI] [PubMed] [Google Scholar]
- 412.Ishida T, Kinoshita K. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics. 2008;24:1344–1348. doi: 10.1093/bioinformatics/btn195. [DOI] [PubMed] [Google Scholar]
- 413.Lieutaud P, Canard B, Longhi S. MeDor: a metaserver for predicting protein disorder. BMC Genomics. 2008;9 2:S25. doi: 10.1186/1471-2164-9-S2-S25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 414.Han P, Zhang X, Feng ZP. Predicting disordered regions in proteins using the profiles of amino acid indices. BMC Bioinformatics. 2009;10 1:S42. doi: 10.1186/1471-2105-10-S1-S42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 415.Xue B, Oldfield CJ, Dunker AK, Uversky VN. CDF it all: Consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Letters. 2009 doi: 10.1016/j.febslet.2009.03.070. In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 416.Han P, Zhang X, Norton RS, Feng ZP. Large-scale prediction of long disordered regions in proteins using random forests. BMC Bioinformatics. 2009;10:8. doi: 10.1186/1471-2105-10-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 417.Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. Expected packing density allows prediction of both amyloidogenic and disordered regions in protein chains. J Phys. 2007;19 doi: 10.1371/journal.pcbi.0020177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 418.Schlessinger A, Punta M, Yachdav G, Kajan L, Rost B. Improved disorder prediction by combination of orthogonal approaches. PLoS ONE. 2009;4:e4433. doi: 10.1371/journal.pone.0004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 419.Wootton JC. Non-globular domains in protein sequences: automated segmentation using complexity measures. Comput Chem. 1994;18:269–285. doi: 10.1016/0097-8485(94)85023-2. [DOI] [PubMed] [Google Scholar]