Abstract
Spastin, the most common locus for mutations in hereditary spastic paraplegias1, and katanin are related microtubule-severing AAA ATPases2–6 involved in constructing neuronal7–10 and noncentrosomal7,11 microtubule arrays and in segregating chromosomes12,13. The mechanism by which spastin and katanin break and destabilize microtubules is unknown, in part owing to the lack of structural information on these enzymes. Here we report the X-ray crystal structure of the Drosophila spastin AAA domain and provide a model for the active spastin hexamer generated using small-angle X-ray scattering combined with atomic docking. The spastin hexamer forms a ring with a prominent central pore and six radiating arms that may dock onto the microtubule. Helices unique to the microtubule-severing AAA ATPases surround the entrances to the pore on either side of the ring, and three highly conserved loops line the pore lumen. Mutagenesis reveals essential roles for these structural elements in the severing reaction. Peptide and antibody inhibition experiments further show that spastin may dismantle microtubules by recognizing specific features in the carboxy-terminal tail of tubulin. Collectively, our data support a model in which spastin pulls the C terminus of tubulin through its central pore, generating a mechanical force that destabilizes tubulin–tubulin interactions within the microtubule lattice. Our work also provides insights into the structural defects in spastin that arise from mutations identified in hereditary spastic paraplegia patients.
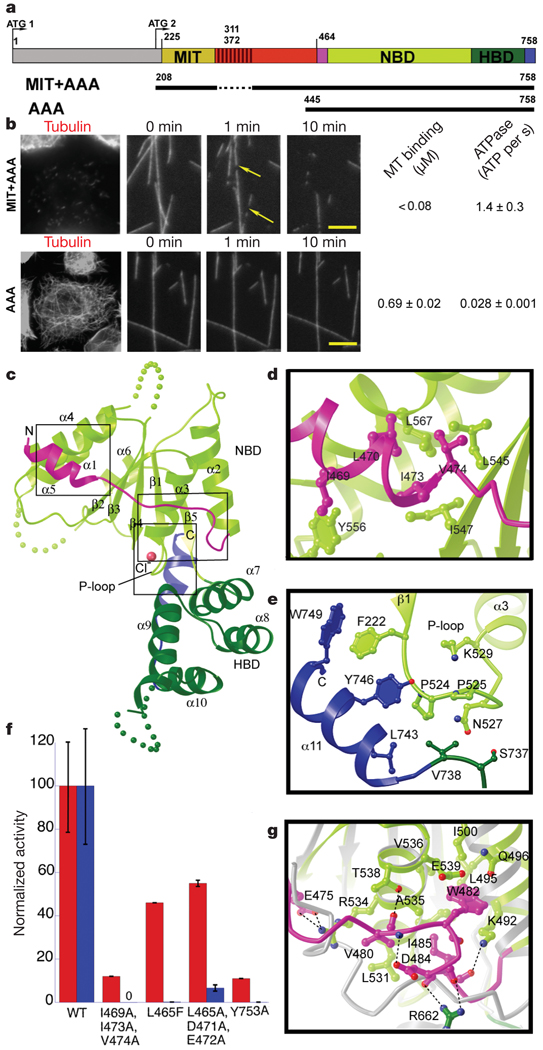
Drosophila spastin is composed of an amino-terminal domain, a microtubule-interacting and -trafficking (MIT) domain that alone binds weakly to microtubules3, a poorly conserved linker element, and a carboxy-terminal AAA ATPase domain (Fig. 1a). The N-terminal region is not required for severing, because a MIT–AAA construct lacking this region robustly severs microtubules (Fig. 1a, b)3–5, has an ATPase rate similar to the full-length protein4 and displays tight microtubule binding (Fig. 1b). The N-terminal region also may not be expressed in all spastin isoforms (see Supplementary Information). A segment of the poorly conserved linker (residues 390–442) is also not essential for robust microtubule-severing in vivo; however, truncation of the linker to <40 residues abolishes severing but not microtubule binding (Supplementary Fig. 1). The AAA construct has weak severing, ATPase and microtubule-binding activities compared with a longer construct containing the AAA and MIT domains (Fig. 1b and Supplementary Fig. 1). These results differ from a recent study14 that concluded that the MIT domain is not involved in microtubule-severing.
Figure 1. X-ray structure of the nucleotide-free AAA domain of spastin.
a, Domain structure of Drosophila spastin: grey, N-terminal domain; red, linker (exon 4, absent in the shorter isoform of spastin used in this study, is hatched); and the AAA domain (coloured according to the X-ray structure). NBD, nucleotide-binding domain; HBD, four-helix bundle domain. Two potential start codons (ATG) are shown (see Supplementary Methods for discussion). The N-terminal boundary of the AAA domain is based on our X-ray structure and differs from that of ref. 14. A segment of the structurally important N-terminal helix of the AAA domain is within what the authors of ref. 14 define as a microtubule-binding domain. The MIT + AAA and AAA constructs are shown schematically below. b, Left, MIT + AAA disassembles the microtubule network when transfected in Drosophila S2 cells and when added to microtubules in vitro, but AAA has no detectable activity at the same concentration (0.15 µM). (Weak severing is observed at higher concentrations, Supplementary Fig. 1.) Arrows indicate breaks in microtubules. Scale bar, 5 µm. Right, microtubule (MT)-binding and ATPase activities of MIT + AAA and AAA. Microtubule-binding affinity was determined for the Walker B E583Q mutant, which is a stable hexamer and is inactive in severing. c, Ribbon representation of the spastin AAA domain crystal structure. N-terminal helix/loop, magenta; NBD, light green; HBD, dark green; C-terminal helix, blue. The pink sphere depicts a chloride ion. d, Conserved hydrophobic interactions between the N-terminal helix and the main body of the NBD. e, Conserved interactions between the C-terminal helix and the P loop. f, ATPase (red) and microtubule-severing (blue) rates of N- and C-terminal helix mutants. Error bars represent standard errors of the mean (see Methods). WT, wild type. g, Detail of the superposition of spastin and ATP-bound NSF structures15, showing contacts that keep the N-terminal flap of monomeric spastin (magenta) in an open conformation, unable to stabilize the nucleotide or interact with the neighbouring protomer. Spastin is colour-coded as in panel c. NSF is in grey. Dashed lines, hydrogen bonds.
We solved the X-ray structure of the nucleotide-free, monomeric AAA domain of Drosophila spastin (residues 464–758) at 2.7Å resolution (Rfree = 28.7%; Supplementary Information). Similar to other AAA proteins, the enzymatic core of spastin contains a central α/β nucleotide-binding domain (NBD) and a smaller four-helix bundle domain (HBD). A marked feature of the spastin structure is its open nucleotide pocket, which explains the absence of a bound nucleotide, despite the presence of 0.5 mM adenosine 5’-O-(3-thiotriphosphate) (ATPγS) in the crystallization solution. Comparison of our nucleotide-free spastin structure with the ATP-bound structure of N-ethylmaleimide-sensitive fusion protein (NSF) (an AAA protein involved in membrane fusion15) reveals that an extended loop involved in nucleotide contact and protomer–protomer interactions in NSF (Supplementary Fig. 2) is pulled away from the nucleotide pocket in spastin by the packing of the linchpin Trp 482 in a conserved hydrophobic pocket (Fig. 1g). The pocket for Trp 482 is sub-optimal, compatible with movement of the tryptophan and rearrangement of the flap on ATP-induced hexamerization or/and substrate binding.
Uniquely among known AAA structures, spastin has two helices (N-terminal α1 and C-terminal α11) that embrace the NBD (Fig. 1c and Supplementary Fig. 3). The amphipathic N-terminal helix is anchored to the body of the NBD by interdigitating hydrophobic residues (Leu 470/Ile 473/Val 474; Fig. 1d). Mutation of these residues to alanine reduced ATPase activity by ~90% and abolished microtubule-severing, while preserving microtubule binding (Fig. 1f and Supplementary Fig. 4). Mutation of invariant Leu 567 located at the helix α1–NBD interface causes hereditary spastic paraplegias (HSP)16. Solvent-exposed residues on the N-terminal helix also have important roles; L465F and the triple mutant L465A/D471A/E472A markedly decreased microtubule-severing (Fig. 1f) without significantly affecting the ATPase. The C-terminal helix, which is also present in the closely related enzyme VPS4 (vacuolar sorting protein 4)17, and part of the preceding conserved linker wrap around the phosphate-binding loop (P loop) of the NBD (Fig. 1e). Mutation of the highly conserved Tyr 753 at the end of the C-terminal helix to alanine effectively inactivated the enzyme (Fig. 1f), whereas a Y753F mutation still showed severing activity in vivo (Supplementary Fig. 4). Thus, our structural and mutational analyses indicate that helices α1 and α11 of spastin have important roles in allosteric control of the ATP-binding site and possibly in substrate binding (discussed below).
We next obtained structural information on a hexameric spastin construct using small-angle X-ray scattering (SAXS). A Caenorhabditis elegans MIT–AAA construct was used because it is monodisperse at the concentrations (> 5 mg ml−1) required for collecting high-quality SAXS data. Compared to Drosophila spastin, C.elegans spastin lacks an N-terminal domain and has a shorter linker between the MIT and AAA domains; nonetheless, it displays microtubule-severing activity18 (Supplementary Fig. 5a). We first examined the oligomeric state of C. elegans spastin in its nucleotide-free (apo) and ATP-bound states by static multi-angle light scattering. To create a stable, noncycling ATP-bound state, we prepared a well-described AAA mutation that blocks nucleotide hydrolysis (E278Q; E583Q in Drosophila spastin). Static light scattering revealed that the apoenzyme exists in equilibrium between a monomeric and a weak dimeric state, whereas ATP-bound spastin is a hexamer, a quaternary structure adopted by many AAA proteins19 (Supplementary Fig. 5). Unlike many AAA ATPases, but similar to katanin20, spastin exists mostly as a monomer at submicromolar concentrations, even in the presence of ATP (data not shown).
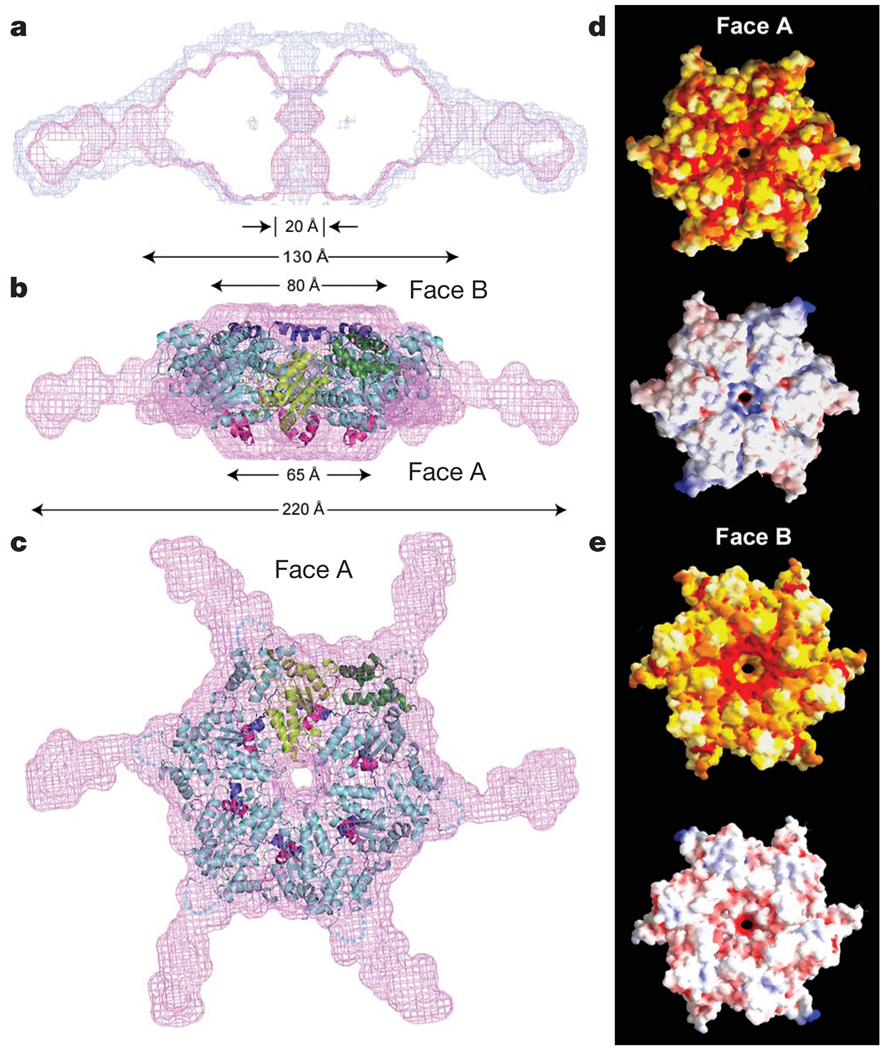
SAXS data were used to generate low-resolution ab initio21 models of three-dimensional arrangements of scattering centres that provide the shape of the molecular envelope of the hexamer (Methods; Fig. 2 and Supplementary Fig. 6). The models from seven independent ab initio simulations were aligned, averaged and filtered on the basis of occupancy to obtain a most probable model. The close agreement between the total volume enclosed by the superposition of the individual runs (the composite structure) and the most probable density map (the filtered structure) indicates the robustness of the ab initio reconstructions. The filtered structure shows a central ring with a double trapezoid cross-section (130Å × 65Å), a ~20-Å-diameter central pore, and slender arms radiating ~50Å outward and extending towards one face of the ring (Fig. 2). The clear reconstruction of the arms also indicates that the linker, although unlikely to be rigid, adopts some defined structure and is not completely disordered. Shortening the linker to <40 residues disables microtubule-severing (but not microtubule-binding, Supplementary Fig. 1), suggesting some length and/or sequence requirement for this region. The asymmetric position of the arms defines a polarity to the overall structure (two faces, herein termed face A and B). We generated an atomic model for the AAA hexameric core of spastin by superimposing our nucleotide-free spastin monomer X-ray structure onto the crystal structure of the NSF hexamer. This model was docked into the SAXS reconstruction with the N- and C-terminal helices on faces A and B, respectively (Figs 2b,c; for details about the fit, see Supplementary Fig. 6 and Supplementary Information).
Figure 2. Model of active, hexameric spastin from light and small-angle X-ray scattering.
a, Ab initio SAXS reconstructions21 of C. elegans spastin (MIT + AAA; residues 15–452, ATP-hydrolysis-deficient E278Q mutant). Shown is a cross-section through the filtered (magenta) and composite (blue) SAXS envelopes. The composite structure consists of the aligned, superimposed and summed models from seven independent simulations, whereas the filtered model corresponds to the most probable density map. b, c, Fit of a spastin hexameric model into the SAXS reconstruction (equatorial (b) and axial (c) views). In the absence of an atomic model of the MIT + linker, its precise location within the envelope is uncertain. Maximal diameter is given at various heights of the structure. c shows face A of the hexamer. For details, see Methods. Colour-coding for spastin is as in Fig. 1c. d, e, Surface properties of face A (d) and face B (e). Top image of d and e, solvent-accessible surface of the spastin hexamer model, colour-coded for amino acid similarity as in Supplementary Fig. 3 (white, 40% identity, to dark red, 100% identity, among spastin and katanins). Bottom, solvent-accessible surface of the spastin hexamer model, colour-coded for electrostatic potential (red, negative; blue, positive, ranging from −12 kT to 12 kT).
Several AAA proteins (for example, the bacterial proteins ClpX, ClpA and ClpB) remodel their substrates by threading the end of the polypeptide chain through a central pore in their rings19,22,23. The microtubule-severing activities of spastin and katanin depend on the ~20-residue disordered and negatively charged C-terminal tails of tubulin3,6, suggesting an analogous mechanism for spastin and katanin. In support of this model, we found that a 23-mer peptide corresponding to the C-terminal tail of β-tubulin inhibited microtubule-severing by ~70% at 0.5 mM, whereas a randomized (scrambled) peptide of identical amino acid composition or an α-tubulin peptide that contains the C-terminal tyrosine (α-Tyr peptide) did not show detectable effects (Fig. 3a). The large concentration of peptide needed to observe inhibition is not surprising given the high local concentration of tubulin tails encountered by microtubule-bound spastin. Involvement of the β-tubulin tail is consistent with genetic data showing that a charge-reversal mutation in this region suppresses the lethality of ectopic katanin activity24. We also found that an antibody that recognizes exposed glutamate residues on the C-terminal tails of tubulin (detyrosinated α-tubulin with a final C-terminal glutamate as well as β-tubulin and polyglutamylated tubulin) completely inhibited spastin-mediated severing. In contrast, a ‘Tyr’ antibody that recognizes β-tubulin with a C-terminal tyrosine25 (~50% of brain tubulin26,27) did not inhibit severing, even though the antibody binds to microtubules (Fig. 3b and Supplementary Fig. 7b). Although we did not detect a robust inhibitory effect of a detyrosinated α-tubulin peptide, an antibody that recognizes the tail of Glu– α-tubulin27 partially inhibited severing (Supplementary Fig. 7c). Collectively, these in vitro data support a model in which spastin interacts with the acidic tubulin C-terminal peptide during the severing reaction and may recognize specific features of the C-terminal peptide.
Figure 3. Role of the tubulin C terminus and the spastin pore in microtubule-severing.
a, Effects of tubulin C-terminal peptides on microtubule-severing in vitro. Addition of α-Tyr peptide had no detectable effect on severing rates, whereas a β-tubulin C-terminal peptide reduced the severing rate. A scrambled peptide had no detectable effect (see Methods). Scale bar, 5 µm. b, Antibodies (Ab) recognizing Glu–α-tubulin, β-tubulin and polyglutamylated tubulin (anti-‘Glu’) inhibit spastin-mediated severing completely at 1:2 antibody:tubulin molar ratio (the same level of protection was seen even 30 min after spastin addition), whereas antibodies recognizing Tyr– α-tubulin (anti-Tyr) did not protect against severing, even at a 5:1 antibody:tubulin molar ratio (antibody binding to these microtubules demonstrated in Supplementary Fig. 7b). c, Left, conservation of the three pore loops. Loop 1 residues are conserved in all AAA ATPases; loops 2 and 3 are specific to the spastin subfamily. Effects on microtubule-severing of mutations in pore loop residues are shown on top of the alignment: red, inactive; black, severely crippled; green, active. Asterisks denote disease mutations. The effects of mutations generally decrease in severity from the pore entrance to the exit, with loop 1 being the least permissive to substitutions. Right, positions of pore loops (labelled 1, 2 and 3) in the spastin hexamer, in a cross-sectional view of the pore. Side-chains for residues K555, Y556 and D559 as well as residues 592–596 are not visible in the electron density maps and are presumed to be disordered. d, Left, ATPase (red) and microtubule-severing (blue) rates for selected mutants. Error bars represent standard errors of the mean (see Methods). Right, molecular surface of the hexameric spastin model showing in yellow the location of residues on face A that impair severing. Loop residues that impair severing are shown in red.
To explore this model further, we examined the roles of three solvent-exposed loops within the pore that are highly conserved among spastins and katanins (Fig. 3c). Mutations in pore loop 1 of Drosophila spastin, which has been shown to be important for the substrate-remodelling activity of several other AAA proteins22,23,28, abolished severing (Figs 3c, d) but preserved microtubule binding (Supplementary Table 2 and Supplementary Fig. 8). After submission of this work, similar results were obtained in ref. 14. Mutations of solvent-exposed residues in pore loops 2 and 3 also completely inhibited or severely crippled the enzyme (Figs 3c, d). However, the disease mutant S589Y retains some activity, suggesting neurons are susceptible to disease with partial spastin activity. Mutations of surface residues leading to the pore (Fig. 3d) also markedly affected the activity of spastin (for example, L465F and L465A/D471A/E472A in Fig. 1f, and K562A and K562R in Supplementary Fig. 4).
In conclusion, the combination of X-ray crystallography, SAXS ab initio reconstructions and structure-guided mutagenesis provides the first structural information on microtubule-severing proteins and allows us to propose a molecular model for spastin-mediated severing (Fig. 4a). Owing to their similar domain organization and high sequence similarity, this model probably pertains to katanin as well. We propose that face A of the spastin AAA ring docks onto the microtubule, placing the positively charged N-terminal pore entrance in contact with the negatively charged C terminus of tubulin. The translocation from face A to face B would correspond to the direction of substrate translocation proposed for the distantly related AAA ATPases ClpX, ClpA and ClpB22,28,29. The linker and MIT domains extending from the ring would make additional contacts with the microtubule, thus increasing microtubule avidity and potentially stabilizing the hexamer on the microtubule20. On the basis of our affinity measurements, only a subset of the six arms is likely to make strong binding interactions with the microtubule (Fig. 4a).
Figure 4. Proposed mechanism of severing by spastin and effects of disease mutations.
a, Proposed mechanism for microtubule-severing by spastin. The spastin AAA core is shown in cyan with pore loops 1, 2 and 3 highlighted in red and numbered in the figure. The MIT domains are shown as gold ovals. The valency of the interaction of the MIT domains with the microtubule is unknown. On the basis of affinity measurements, it is likely that not all MIT domains are engaged with the microtubule (the potentially unengaged MIT domain is shown hatched). The tubulin heterodimers forming the microtubule are shown in green as a ribbon representation, whereas the C-terminal tubulin tails are shown in red cartoon representation. b, Left, molecular surface of spastin (face A). One protomer is shown in a ribbon representation and residues mutated in HSP patients are shown as violet spheres. Right, in addition to mapping to the pore loops (S589Y, R601L, P631L), disease mutations can interfere with ATP binding (F522C, N527K, K529R) and protomer–protomer interactions (D697N, R704Q, R641C, R601L, P631L). G511R maps to a loop on face A where it could destabilize protomer–protomer interactions and/or the microtubule-binding interface (Supplementary Fig. 4).
We propose that the tubulin polypeptide is threaded through the pore, perhaps driven by nucleotide-driven conformational changes of the pore loops. However, spastin may not need to completely translocate the tubulin polypeptide substrate, but instead just grip the C-terminal tubulin tail and exert mechanical ‘tugs’ that might partially unfold tubulin or locally destabilize protomer–protomer interactions, leading to catastrophic breakdown of the microtubule lattice. It also remains possible that the MIT domains could participate in this nucleotide-driven process by binding and ‘feeding’ the C-terminal tails to the pore. Further biophysical characterization will be needed to decipher the structural details of substrate recognition and mechanical force production. Our data also suggest that spastin may selectively recognize post-translationally modified tubulins (‘Glu’ tubulins) that are part of stable microtubules. Consistent with this idea, loss of spastin in Drosophila results in the accumulation of stabilized polyglutamylated tubulin in neurons8 and spastin knockout mice show axonal swellings enriched in detyrosinated, stable microtubules30. Our structure also provides the first glimpse into how spastin disease mutations contribute to spastin dysfunction and disease, most of which we suggest are involved in destabilizing protomer–protomer interactions, microtubule- or ATP-binding (Fig. 4b and Supplementary Fig. 4); in such cases, spastin-linked HSP is probably caused by haploinsufficiency and not a dominant negative effect. Further elucidation of the mechanistic details of how spastin interacts with particular tubulin isoforms and post-translational modifications and leads to microtubule destabilization may provide insight into the origin of spastin paraplegias and potential treatments for this disease.
METHODS SUMMARY
Crystallographic statistics can be found in Supplementary Table 1.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgements
We thank C. Ralston for access to beamlines at the Advanced Light Source (Lawrence Berkeley National Laboratory), G. Hura for assistance during the SAXS experiments and data processing, N. Zhang for assistance with molecular biology, D. Southword for advice with the static multi-angle scattering experiments, T. Huckaba for the anti-Glu α-tubulin antibody, and H. Bourne and A. Ferre-D’Amare for support and critical reading of the manuscript. R.D.V. is a Howard Hughes Medical Institute investigator. A.R.-M. has received support from the Damon Runyon Cancer Research Foundation, the NIH and the Burroughs Wellcome Fund.
Footnotes
Supplementary Information is linked to the online version of the paper at www.nature.com/nature.
Author Information Atomic coordinates and structure factor amplitudes have been deposited in the Protein Data Bank under the accession number 3B9P.
Reprints and permissions information is available at www.nature.com/reprints.
References
- 1.Hazan J, et al. Spastin, a new AAA protein, is altered in the most frequent form of autosomal dominant spastic paraplegia. Nature Genet. 1999;23:296–303. doi: 10.1038/15472. [DOI] [PubMed] [Google Scholar]
- 2.Frickey T, Lupas AN. Phylogenetic analysis of AAA proteins. J. Struct. Biol. 2004;146:2–10. doi: 10.1016/j.jsb.2003.11.020. [DOI] [PubMed] [Google Scholar]
- 3.Roll-Mecak A, Vale RD. The Drosophila homologue of the hereditary spastic paraplegia protein, spastin, severs and disassembles microtubules. Curr. Biol. 2005;15:650–655. doi: 10.1016/j.cub.2005.02.029. [DOI] [PubMed] [Google Scholar]
- 4.Evans KJ, Gomes ER, Reisenweber SM, Gundersen GG, Lauring BP. Linking axonal degeneration to microtubule remodeling by Spastin-mediated microtubule severing. J. Cell Biol. 2005;168:599–606. doi: 10.1083/jcb.200409058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Salinas S, et al. Human spastin has multiple microtubule-related functions. J. Neurochem. 2005;95:1411–1420. doi: 10.1111/j.1471-4159.2005.03472.x. [DOI] [PubMed] [Google Scholar]
- 6.McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 7.Roll-Mecak A, Vale RD. Making more microtubules by severing: a common theme of noncentrosomal microtubule arrays? J. Cell Biol. 2006;175:849–851. doi: 10.1083/jcb.200611149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Trotta N, Orso G, Rossetto MG, Daga A, Broadie K. The hereditary spastic paraplegia gene, spastin, regulates microtubule stability to modulate synaptic structure and function. Curr. Biol. 2004;14:1135–1147. doi: 10.1016/j.cub.2004.06.058. [DOI] [PubMed] [Google Scholar]
- 9.Sherwood NT, Sun Q, Xue M, Zhang B, Zinn K. Drosophila Spastin regulates synaptic microtubule networks and is required for normal motor function. PLoS Biol. 2004;2:e429. doi: 10.1371/journal.pbio.0020429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wood JD, et al. The microtubule-severing protein Spastin is essential for axon outgrowth in the zebrafish embryo. Hum. Mol. Genet. 2006;15:2763–2771. doi: 10.1093/hmg/ddl212. [DOI] [PubMed] [Google Scholar]
- 11.Burk DH, Liu B, Zhong R, Morrison WH, Ye ZH. A katanin-like protein regulates normal cell wall biosynthesis and cell elongation. Plant Cell. 2001;13:807–827. [PMC free article] [PubMed] [Google Scholar]
- 12.Srayko M, Buster DW, Bazirgan OA, McNally FJ, Mains PE. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 2000;14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang D, Rogers GC, Buster DW, Sharp DJ. Three microtubule severing enzymes contribute to the “Pacman-flux” machinery that moves chromosomes. J. Cell Biol. 2007;177:231–242. doi: 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.White SR, Evans KJ, Lary J, Cole JL, Lauring B. Recognition of C-terminal amino acids in tubulin by pore loops in Spastin is important for microtubule severing. J. Cell Biol. 2007;176:995–1005. doi: 10.1083/jcb.200610072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yu RC, Hanson PI, Jahn R, Brunger AT. Structure of the ATP-dependent oligomerization domain of N-ethylmaleimide sensitive factor complexed with ATP. Nature Struct. Biol. 1998;5:803–811. doi: 10.1038/1843. [DOI] [PubMed] [Google Scholar]
- 16.Fonknechten N, et al. Spectrum of SPG4 mutations in autosomal dominant spastic paraplegia. Hum. Mol. Genet. 2000;9:637–644. doi: 10.1093/hmg/9.4.637. [DOI] [PubMed] [Google Scholar]
- 17.Scott A, et al. Structural and mechanistic studies of VPS4 proteins. EMBO J. 2005;24:3658–3669. doi: 10.1038/sj.emboj.7600818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Matsushita-Ishiodori Y, Yamanaka K, Ogura T. The C. elegans homologue of the spastic paraplegia protein, spastin, disassembles microtubules. Biochem. Biophys. Res. Commun. 2007;359:157–162. doi: 10.1016/j.bbrc.2007.05.086. [DOI] [PubMed] [Google Scholar]
- 19.Sauer RT, et al. Sculpting the proteome with AAA(+) proteases and disassembly machines. Cell. 2004;119:9–18. doi: 10.1016/j.cell.2004.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hartman JJ, Vale RD. Microtubule disassembly by ATP-dependent oligomerization of the AAA enzyme katanin. Science. 1999;286:782–785. doi: 10.1126/science.286.5440.782. [DOI] [PubMed] [Google Scholar]
- 21.Svergun DI, Petoukhov MV, Koch MH. Determination of domain structure of proteins from X-ray solution scattering. Biophys. J. 2001;80:2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hinnerwisch J, Fenton WA, Furtak KJ, Farr GW, Horwich AL. Loops in the central channel of ClpA chaperone mediate protein binding, unfolding, and translocation. Cell. 2005;121:1029–1041. doi: 10.1016/j.cell.2005.04.012. [DOI] [PubMed] [Google Scholar]
- 23.Schlieker C, et al. Substrate recognition by the AAA+ chaperone ClpB. Nature Struct. Mol. Biol. 2004;11:607–615. doi: 10.1038/nsmb787. [DOI] [PubMed] [Google Scholar]
- 24.Lu C, Srayko M, Mains PE. The Caenorhabditis elegans microtubule-severing complex MEI-1/MEI-2 katanin interacts differently with two superficially redundant beta-tubulin isotypes. Mol. Biol. Cell. 2004;15:142–150. doi: 10.1091/mbc.E03-06-0418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wehland J, Willingham MC, Sandoval IV. A rat monoclonal antibody reacting specifically with the tyrosylated form of alpha-tubulin. I. Biochemical characterization, effects on microtubule polymerization in vitro, and microtubule polymerization and organization in vivo. J. Cell Biol. 1983;97:1467–1475. doi: 10.1083/jcb.97.5.1467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rodriguez JA, Borisy GG. Tyrosination state of free tubulin subunits and tubulin disassembled from microtubules of rat brain tissue. Biochem. Biophys. Res. Commun. 1979;89:893–899. doi: 10.1016/0006-291x(79)91862-x. [DOI] [PubMed] [Google Scholar]
- 27.Gundersen GG, Kalnoski MH, Bulinski JC. Distinct populations of microtubules: tyrosinated and nontyrosinated alpha tubulin are distributed differently in vivo. Cell. 1984;38:779–789. doi: 10.1016/0092-8674(84)90273-3. [DOI] [PubMed] [Google Scholar]
- 28.Siddiqui SM, Sauer RT, Baker TA. Role of the processing pore of the ClpX AAA+ ATPase in the recognition and engagement of specific protein substrates. Genes Dev. 2004;18:369–374. doi: 10.1101/gad.1170304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee S, Choi JM, Tsai FT. Visualizing the ATPase cycle in a protein disaggregating machine: structural basis for substrate binding by ClpB. Mol. Cell. 2007;25:261–271. doi: 10.1016/j.molcel.2007.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Tarrade A, et al. A mutation of spastin is responsible for swellings and impairment of transport in a region of axon characterized by changes in microtubule composition. Hum. Mol. Genet. 2006;15:3544–3558. doi: 10.1093/hmg/ddl431. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.