Abstract
Protein intrinsic disorder is becoming increasingly recognized in proteomics research. While lacking structure, many regions of disorder have been associated with biological function. There are many different experimental methods for characterizing intrinsically disordered proteins and regions; nevertheless, the prediction of intrinsic disorder from amino acid sequence remains a useful strategy especially for many large-scale proteomics investigations. Here we introduced a consensus artificial neural network (ANN) prediction method, which was developed by combining the outputs of several individual disorder predictors. By eight-fold cross-validation, this meta-predictor, called PONDR-FIT, was found to improve the prediction accuracy over a range of 3 to 20% with an average of 11% compared to the single predictors, depending on the datasets being used. Analysis of the errors shows that the worst accuracy still occurs for short disordered regions with less than ten residues, as well as for the residues close to order/disorder boundaries. Increased understanding of the underlying mechanism by which such meta-predictors give improved predictions will likely promote the further development of protein disorder predictors. The access to PONDR-FIT is available at www.disprot.org.
Keywords: natively unfolded, intrinsically unstructured, intrinsically disordered, highly flexible, highly dynamic, structurally disordered, predictor, PONDR
1. Introduction
Many proteins or partial regions of proteins lack stable and well-defined three-dimensional structures under physiological conditions in vitro [1–4]. These proteins and regions are often called intrinsically disordered proteins (IDPs) or intrinsically disordered regions (IDRs) among many other names [5–6]. The phenomena of disordered protein or residues have been extensively observed in nature. About 70% of PDB structures have some disordered residues, while about 25% have IDRs > 10 residues in length [7]. The fraction of proteins in PDB with IDRs increases substantially if only eukaryotic proteins are considered. Also, many PDB structures represent only fragments of the full-length protein sequences, many protein structures involve complexes with various-sized ligands, and the process of crystallization can induce IDRs to fold into structure, so the actual disordered content in proteins from PDB is very likely much higher than the amount estimated from missing electron density.
The first proteomics study focusing on intrinsically disordered proteins revealed that these proteins are typically missed due to their low hydrophobicity when standard methods are used [8]. Subsequently, more targeted methods have been developed [9–12]. These systematic studies revealed not only the abundance of IDRs, but also the direct linkage between IDRs and many diseases.
Although IDPs and IDRs don’t have specified 3D structures, many are nevertheless actively involved in a diverse array of biological functions [1] typically involving signaling, recognition and regulation [1, 13–15]. Further studies revealed that disordered proteins are also extensively associated with human diseases, such as cancer, cardiovascular diseases, amyloidoses, neurodegenerative diseases, diabetes, etc [16]. The sequence-to-structure-to-function paradigm does not seem suitable for IDPs [1]. Instead, the functions of disordered proteins arise from multiple inter-converting conformations. Thus, locating the disordered residues along the sequences of various proteins and identifying their movements will be important for further understanding the mechanisms behind the functions of IDPs and IDRs.
One approach that has been important in the study of IDPs and IDRs has been the use of disorder predictors. This method is extremely powerful in terms of time and cost to the study of disordered proteins, compared to traditional experimental methods, at the proteome level [8–10, 17]. Starting with small collections of IDPs and IDRs determined by various methods, various researchers have built predictors that use amino acid sequence as inputs and that give structure (order) or disorder as outputs. More than 50 disorder predictors have been developed by now [18]. These include the early PONDR series [19–21], DisEMBL [22], DISOPRED [23], and NORSnet [24], which used artificial neural networks; PONDR®VSL2 [21], DISOPRED2 [25], and POODLE [26], which employed support vector machines; and DISPro [27], which used a combination of neural networks and Bayesian methods. Decision tree based methods have also been applied to this problem [28]. Several additional predictors take advantage of template sequences to predict disordered residues, such as PrDOS [29] and DISOclust [30]. Several physics-based methods have also been developed, including the charge-hydropathy plot [3], IUPred [31], Ucon[32], and FoldIndex[33]. Specific amino acid scales have also provided the basis of predictors such as FoldUnfold [34] and TopIDP [35].
In addition to differences in the computational method being used, various predictors have also selected different combinations of input features[18]. Commonly used features include the following: amino acid sequence [22, 25], amino acid composition [19–21, 26], sequence complexity often measured as Shannon’s entropy [19–21, 26], position specific scoring matrices [20–21, 23–24], predicted secondary structure [24, 27], and predicted accessible surface area[24, 27].
The prediction results from a collection of predictors can be used as the inputs for another predictor, called a meta-predictor. This approach often results in improvement in prediction accuracy evidently because the different predictors have information arising from different sequence features, different prediction models, and different training sets. This approach has been previously applied to the prediction of both secondary and tertiary structure of proteins [36–38]. In summary, altogether more than 50 predictors of intrinsic protein disorder have been described [18].
With regard to disordered proteins, PONDRs VLXT, VSL2, and VL3 are all meta-predictors, because they are constructed from combinations of individual predictors. More recently, two additional meta-predictors, metaPrDOS [39] and MD [40], were developed for the specific purpose of improving disorder prediction accuracy. The former is a combination of PrDOS, DISpro, DISPROT(VSL2P), DISOPRED2, POODLE-S, IUPred, and DisEMBL, and the latter is composed of NORSnet, PROFbval [41], Ucon, and DISOPRED2. These two meta-predictors accomplished their stated purpose in that both showed significantly improved performance compared to any one of their individual component predictors.
Here we report the development of PONDR-FIT, assembled by combining PONDR-VLXT, PONDR-VSL2, PONDR-VL3, FoldIndex, IUPred, and TopIDP. This meta-predictor represents a completely new combination of predictors not used in any previous meta-predictor. We show that PONDR-FIT is comparable to the other meta-predictors with significantly improved accuracy in the aggregate as compared to its individual component predictors. In addition, our study of the errors made by this predictor identifies the remaining problems that are impeding the further improvement of disorder prediction.
2. Methods
2.1. Datasets
Two groups of datasets are used in this study. Each group contains experimentally verified disordered and structured residues. The first group is composed from the previously generated fully ordered dataset (FOD) and fully disordered dataset (FDD) [42]. By definition, proteins in FOD have only structured residues while proteins in FDD have only disordered residues. The second group is a recently enlarged partially disordered dataset (PDD) prepared by the method of Oldfield, et al. [8], which contains both structured and disordered residues in the same protein with the structured residues comprising PDDS subset and the disordered residues forming PDDM subset. The predictor was trained on both groups of datasets to compare the influence of dataset on the accuracy of predictor.
FOD has 554 chains and 113,895 residues. This dataset was selected from PDB database of July 20, 2008 by choosing only single-chain and non-membrane structures obtained by x-ray crystallography and characterized by unit cells with primitive space groups. Structures having ligands or disulfide bonds and structures with missing residues were removed from the dataset. The remaining sequences were clustered by using BLASTCLUST from NCBI (http://ncbi.nlm.nih.gov/BLAST) to put sequences with at least 25% sequence identity into a cluster. The longest sequence of each cluster was included in the final FOD.
FDD was assembled from DisProt [43], version 4.5 of July 17, 2008 by selecting only sequences that contain no structured residues. The final dataset has 84 protein chains and 17,420 disordered residues.
PDD was selected from PDB x-ray structures with resolution higher than 3.0 angstroms, by choosing single chain proteins with no ligands or partners. The sequences were clustered by using BLASTCLUST with a sequence identity cutoff of 30%. The longest sequence in each cluster was selected for inclusion. The resulting set of sequences was further filtered by keeping only those sequences that have 20 or more consecutive disordered residues as identified by xml2pdb [44]. Choosing 20 disordered residues as the cutoff value was a compromise. By choosing 10 as the cutoff, there are 1116 sequences, and by choosing 30, there are only 361 sequences. With 20 disordered residues as the cutoff, there are 647 sequences with a total of 230,314 residues, in which 16011 disordered residues are located within 1376 disordered regions of length 20 or longer. This dataset was further divided into structured and disordered subsets, with the structured residues comprising PDDS and the disordered residues forming PDDM.
2.2. Individual disorder predictors
The individual predictors used in the analysis are PONDR® VLXT [19], PONDR® VL3 [20], PONDR® VSL2 [21], IUPred [31], FoldIndex [33], and TopIDP [45]. All of these predictors use the primary sequence as the input and give an individual score output, one for each amino acid in the sequence, indicating each residue’s likelihood of being structured or disordered.
PONDR® VLXT applies three different neural networks, one for each terminal region and one for the internal region of the sequence. Each neural network is trained by a specific dataset containing only the amino acid residues of that specific region. The final prediction result uses the individual predictors in their respective regions. The transition from one predictor to another is accomplished by computing the average scores of the two predictors for a short region of overlap at the boundary between the two regions. The input features of neural networks include selected compositions and profiles from the primary sequences. PONDR® VLXT may underestimate the occurrence of long disordered regions in proteins. However, this method has significant advantages in finding potential binding sites [46–47].
PONDR® VL3 employs ten neural networks and selects the final prediction by simple majority voting. The input features of these predictors are various sequence profiles. This predictor has higher accuracy in predicting longer disordered regions.
PONDR® VSL2 is a combination of neural network predictors for both short and long disordered regions. A length limit of 30 residues divides short and long disordered regions. Each individual predictor is trained by the dataset containing sequences of that specific length. And the final prediction is a weighted average determined by a second layer predictor. PONDR® VSL2 applies not only the sequence profile, but also the result of sequence alignments from PSI-blast and secondary structure prediction from PHD and PSI-pred. This predictor is so far the most accurate predictor in the PONDR family.
IUPred assumes that globular proteins have larger numbers of effective inter-residue interactions (negative free energy) than disordered proteins due to the different types of amino acids involved in possible residue contacts. Based on this idea, a composition-based pair-wise interaction matrix was shown to give values similar to those obtained from a structure-based interaction matrix. Structured and disordered proteins were compared by this approach, with the structured proteins found to have a significantly lower free energy estimate, thus giving a means to predict whether a protein is structured or disordered using amino acid sequence as input.
FoldIndex is a method developed from charge-hydrophobicity plots by rearranging the terms in the basic equation and by adding the technique of sliding windows. The charge-hydrophobicity plot was designed to determine if a protein is disordered or not. By applying a sliding window of 21 amino acids centered at a specific residue, the position of this segment on charge-hydrophobicity plot can be calculated, and the distance of this position away from the boundary line is taken as an indication whether the central residue is disordered or not.
TopIDP is a numerical scale giving the order-disorder propensity for each amino acid. This scale was determined by maximizing the differences in conditional probabilities for structured versus disordered regions of proteins for the central residues in windows of 21 residues.
2.3. Performance evaluation
We applied the two main criteria used in CASP6 [48] and CASP7 [49] to evaluate the prediction performance. One criterion is the area under the Receiver Operating Characteristic (ROC) curve. The second criterion is the accuracy of correct prediction rates averaged over both ordered and disordered samples.
For plotting ROC curves, four individual quantities need to be determined in advance, i.e. TP (true positive), FN (false negative), TN (true negative), and FP (false positive). These four quantities can be further formulated into: Sensitivity (Sens) = TP / (TP + FN), and Specificity (Spec) = TN / (TN + FP). Since the prediction output here is always a real number, by setting different threshold values of the disordered status, the values of sensitivity and specificity will change accordingly. By taking (1-specificity) as×axis, and sensitivity as y axis, all the data pairs corresponding to the minimal threshold value to the maximal threshold value will make a continuous curve. This is the ROC curve, the area under this curve is a reliable indication for the quality of the prediction. The value of AUC is between 0 and 1, the larger the area, the better the predictor. Errors of this analysis are estimated by bootstrapping, where a subset constituting 90% of the total sample is randomly selected from the dataset at each bootstrapping step and the analysis is repeated multiple times. A total of 200 bootstrapping steps is used to give an average and errors for the prediction.
For simplicity, the average of sensitivity and specificity is also a useful indicator: Accuracy (ACC) = (Sensitivity + Specificity) / 2. This quantity can directly reflect the balance of prediction accuracy between structured and disordered residues. No weighting is used in this estimate, even if the number of structured and disordered residues is very substantially unbalanced.
2.4. Consensus prediction and 8-fold cross-validation
The consensus predictor (PONDR-FIT) is a combination of all six of the above predictors. For each residue along the sequence, the prediction results of the six individual predictors on a sliding window of 21 residues centered at that residue, were fed into a single layer artificial neural network (ANN) with 20 hidden units. Hence, the total number of input features is 126. The single output at the output layer is the disorder score of the meta-predictor for that centered residue.
We trained the artificial neural network by 8-fold cross-validation on FOD and FDD. Both FOD and FDD were equally divided into 8 groups. Each time, seven groups were used for training and control, the other for independent testing. To avoid over-fitting, during each training process, the seven groups were further split into two classes: the first one, with 90% of the samples, was used for training; the remaining 10% was used for selecting the optimized parameters. The final performance was evaluated by the average of 8 independent testing sets. The artificial neural network was also trained on the PDD dataset. In this circumstance, both the ordered and disordered residues in PDD were split into 8 groups, respectively. The other procedures are the same as those used with FOD and FDD.
3. Results
We selected six individual predictors for the construction of a composite predictor. In brief, this set of 6 individual predictors was selected because they use different predictive approaches, because they emphasize different features of the sequence, and because they all give acceptable accuracies as individual predictors. Apparently, any number out of 6 individual predictors can be combined together to build up the meta-predictor. There are 6, 15, 20, and 15 different combinations containing 5, 4, 3 and 2 out of the 6 individual predictors, respectively. Hence, there are 6 + 15 + 20 + 15 = 56 distinct ways of building up the meta-predictors by taking different combinations of 6 individual predictors. Of course, each predictor can be used individually and all 6 predictors can be combined, giving 7 more predictors for a total of 63 different predictors from the overall set of 6 predictors.
The first issue is whether the various meta-predictors containing more individual predictors are more accurate than the predictors having fewer individual predictors. Understanding this issue is important for the development of meta-predictors. To carry out these comparisons, we used a common machine learning model, namely a simple artificial neural network (ANN) using the outputs from the individual predictors as inputs and using a single hidden layer. We used 8-fold cross validation to give accuracies (ACC), and we estimated the areas under the curves (AUCs) of receiver operating characteristic (ROC) curves to analyze the results as described in Materials and Methods.
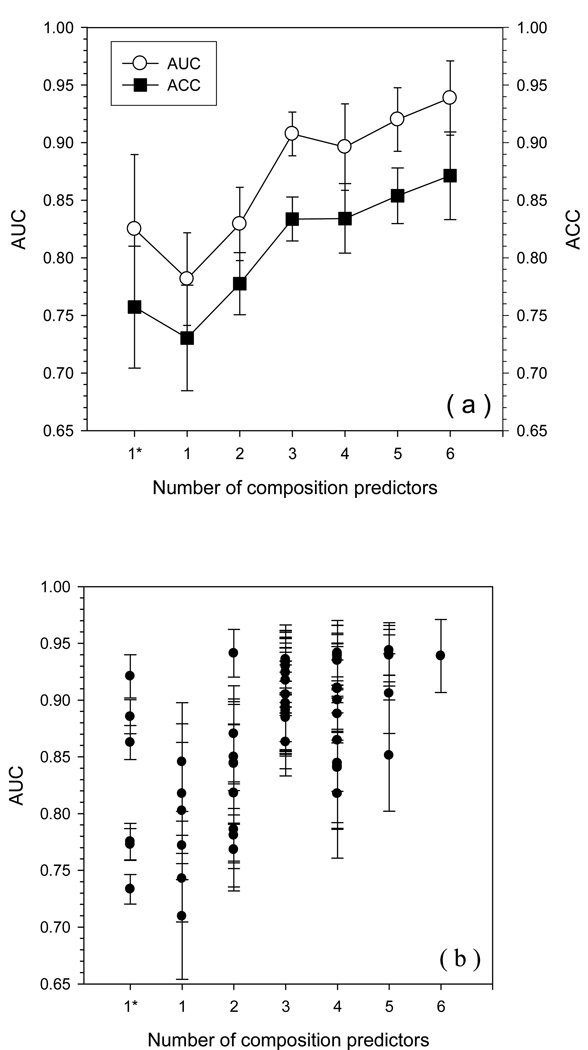
We tested all 63 predictors by 8-fold cross validation on the fully ordered dataset (FOD) and on the fully disordered dataset (FDD) to give both ACC and AUC values (Fig. 1). From the results shown in Fig.1 (a), the performance evaluated by the AUC values parallels that assessed by the ACC values. Overall, the general prediction performance improves as the number of individual predictors increases inside the composite predictor.
Fig. 1.
Prediction accuracy of all possible composite predictors from 6 individual predictors on FOD/FDD. All composite predictors with the same number of individual predictors are classified into the same group. 1* indicates the AUC (ACC) value of each individual predictor from their original predictions and the error bar is calculated by bootstrapping. All other error bars are from 8-fold cross-validation. The y-axis is the AUC (or ACC) value. The x-axis is the number of individual predictors used in the combination. Panel (a) shows the averaged performance for each group with the same number of individual predictors. The open circles represent AUC values while filled squares represent ACC values. Panel (b) shows the prediction performance of each possible composite predictor. Each filled circle represents one type of combination.
Due to the relatively large standard error within each group, the prediction performances of the various composite predictors were compared one-by-one, in order to determine which subsets give better predictions. These results are shown in Fig.1 (b). For the datasets of FOD and FDD, the most accurate individual predictor is PONDR®VL3 which achieved an AUC of 0.92. One composite predictor with 2 individual predictors surpasses PONDR®VL3 by around 0.02, which is (VL3 + TopIDP). Three composite predictors composed of 4 individual predictors also achieved the same accuracy. These three composite predictors are (VSL2 + VL3 + FoldIndex + TopIDP), (VSL2 + VL3 + FoldIndex + TopIDP), and (VLXT + VL3 + IUPred + TopIDP). Two composite predictors with 5 individual predictors reached the level of 0.94. They are (VSL2 + VL3 + FoldIndex + IUPred + TopIDP) and (VLXT + VSL2 + VL3 + IUPred + TopIDP). The 6-component composite predictor obtained a similar accuracy with an AUC of 0.94. Apparently, for the FOD and FDD datasets, all of the highest performing composite predictors have both VL3 and TopIDP in their compositions. A sequential analysis about the influence of number of components on the accuracy, which was previously applied in PsiPred [50], is very interesting. 6-component meta-predictor has the highest accuracy. Compared to this 6-component-predictor, 5-component-predictor without ToPIDP has the least accuracy among all other 5-component-predictors. In this least-accurate 5-component predictor, one individual-predictor can be moved out to produce another 4-component predictor. There are 5 different 4-component predictors by choosing various individual predictors. Among these five predictors, the predictor without VL3 has the lowest accuracy. By repeating this process, the “steepest descent line” is TopIDP-VL3-IUPred-VSL2-VLXT-FoldIndex. Similarly, by selecting the predictor which causes the minimal loss of accuracy at each step of reducing the number of component predictors, we may have the “slowest descent line” which is FoldIndex-VL3-VSL2-VLXT- TopIDP-IUPred. This process can also be implemented in a reversed order by which we start at a single predictor and gradually add individual predictor into the meta-predictor. For individual predictors which may contribute the highest accuracy increment to previous meta-predictor, we had a “steepest ascent line” which is VL3-TopIDP-VSL2-IUPred-FoldIndex-VLXT. By choosing individual predictors which presents the smallest improvement of accuracy, we got the “slowest ascent line” FoldIndex-TopIDP-VLXT-VSL2-IUPred-VL3. Apparently, the order of predictors in steepest descent line and steepest ascent line is almost the same with each other.
The next question, which is also important for assembling the meta-predictor, is which individual predictors should be used? Although observations discussed above support VL3 and TopIDP as components of the final predictor, it would be advantageous to check the performance of each individual predictor on different data, because the results from Fig.1 are based on the optimization and analysis on the data from FOD and FDD only. All the sequences in these two datasets are fully disordered or ordered and thus have long consecutive disorder. That is likely one reason that PONDR-VL3 plays such an important role in the accuracy improvement. To gain additional insight, a comparison of the performance of the 6 individual predictors on various datasets was carried out (Table 1). The accuracy of PONDR-VL3 drops dramatically when the PDD dataset is substituted for the FOD/FDD datasets, while PONDR-VSL2 has a high and stable prediction regardless of which dataset is used. Thus, in the final meta-predictor, for the purpose of eliminating the influence arising from the choice of data and the method of prediction, we will use all 6 predictors to form the meta-predictor that we are calling PONDR-FIT. As shown in Table 1, the prediction performance of the meta-predictor is only 2~3% higher than the best of its individual composition predictors, in terms of the ACC values. However, the best individual predictor is quite different for the two sets of data, while PONDR-FIT is the best overall for both sets of data.
Table 1.
Prediction accuracy of PONDR-FIT and its individual composition predictors in various datasets.
| FOD/FDD | PDD (PDDS/PDDM) | |||
|---|---|---|---|---|
| ACC | AUC | ACC | AUC | |
| PONDR-VLXT | 70.3 ± 0.4% | 0.77 ± 0.04 | 67.2 ± 0.7% | 0.74 ± 0.03 |
| PONDR-VSL2 | 78.5 ± 0.3% | 0.86 ± 0.03 | 74.6 ± 0.6% | 0.82 ± 0.03 |
| PONDR-VL3 | 84.1 ± 0.4% | 0.92 ± 0.03 | 70.1 ± 0.6% | 0.76 ± 0.03 |
| FoldIndex | 67.5 ± 0.4% | 0.73 ± 0.04 | 62.3 ± 0.7% | 0.68 ± 0.04 |
| IUPred | 80.5 ± 0.3% | 0.89 ± 0.02 | 67.2 ± 0.5% | 0.74 ± 0.02 |
| TopIDP | 73.5 ± 0.4% | 0.78 ± 0.04 | 53.5 ± 0.6% | 0.54 ± 0.04 |
| PONDR-FIT | 87.1 ± 3.3% | 0.94 ± 0.03 | 76.6 ± 0.3% | 0.85 ± 0.01 |
Because the amino acid compositions are different among various datasets, the differing prediction accuracies for the different datasets are not strange at all. Fig. 2 shows the composition difference among FOD, FDD, PDDM, and PDDS. From the composition profiles, the composition of FOD is clearly similar to that of PDDS; however, the compositions of FDD and PDDM are not very close to each other at all. For quantifying the differences between them, we further applied the summed Kullback-Leibler divergence [51] to estimate the overall similarities between different datasets as shown in Table 2. Datasets with KL divergence less than 0.01 are regarded as similar datasets [52], while the others are clearly distinguishable datasets. By this criterion, FOD and PDDS are very similar to each other, while FDD and PDDM are obviously distinct. Hence, predictors trained on FOD and FDD, may fail to obtain similar prediction accuracy when tests are carried out on PDDS/PDDM. A further comparison is illustrated in Table 3. The meta-predictor trained on FOD and FDD gives very poor accuracy when applied to the PDD. However, a meta-predictor trained on PDD achieved quite good accuracy on PDD.
Fig. 2.
Relative compositions of (a) PDDS and (b) PDDM, compared to FOD and FDD. The black bars are calculated by (CP - CFOD) / CFOD, while the slashed bars are calculated from (CP - CFDD) / CFDD, where Cp is the composition of one type of amino acid in either PDDS or PDDM. All data were generated by bootstrapping.
Table 2.
Summed Kullback-Leibler divergence among four datasets.
| FOD | PDDS | FDD | PDDM | |
|---|---|---|---|---|
| FOD | 0 | 0.008 | 0.075 | 0.114 |
| PDDS | 0 | 0.073 | 0.129 | |
| FDD | 0 | 0.033 | ||
| PDDM | 0 |
Table 3.
Prediction accuracy of PONDR-FIT on naturally-distinct datasets.
| Optimization Datasets | Validation Datasets | ACC | AUC |
|---|---|---|---|
| FOD / FDD | PDDS / PDDM | 63.4 ± 0.4% | 0.677 ± 0.001 |
| PDDM / PDDS | FOD / FDD | 76.0 ± 2.6% | 0.831 ± 0.029 |
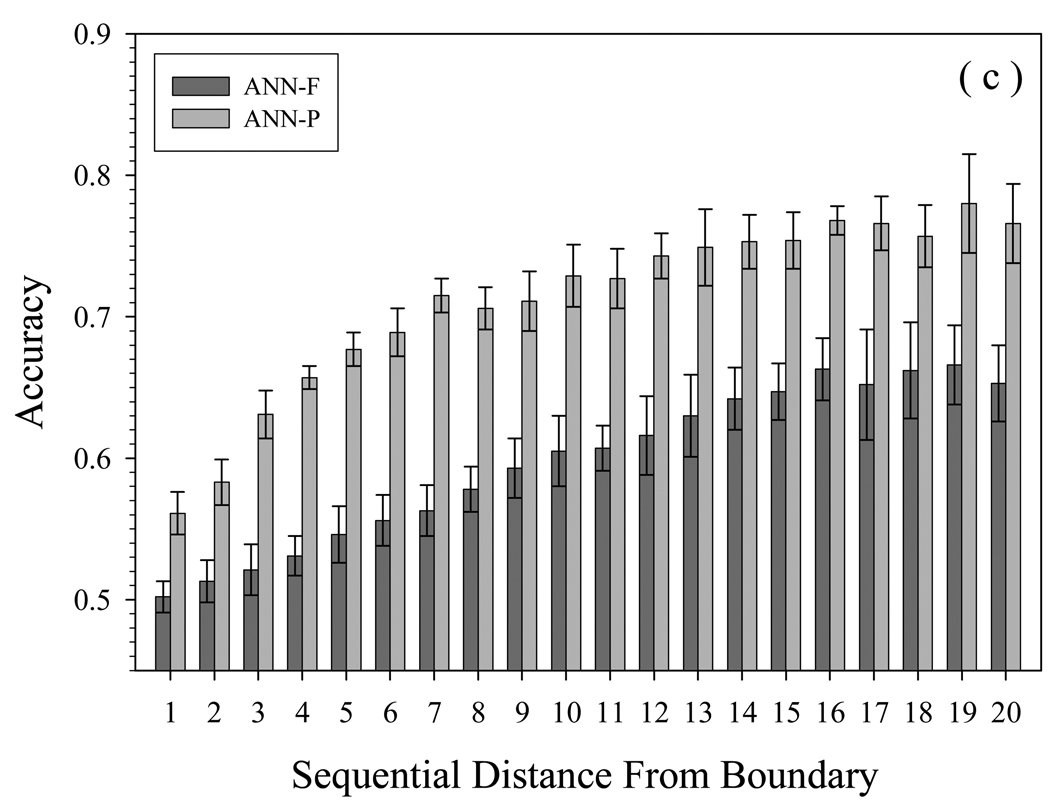
Why does the accuracy on PDD drop so quickly when shifting the training sets from PDD to FOD/FDD? The prediction errors of different categories are presented in Fig. 3. From Fig. 3(a), for each residue type, the accuracy obtained by training on FOD/FDD is less than that when training on PDD. The highest accuracy was achieved for histidine and methionine, residues with intermediate tendency to be present in disordered segments. Other residues of intermediate tendency for disorder were predicted less accurately, with slight further accuracy decreases for disorder-prone residues, followed by additional accuracy decreases for order-prone residues. Fig. 3(b) shows the effect of IDR length on prediction accuracy. Neural networks trained using FOD/FDD never achieve the accuracy of PDD-trained neural networks, for any IDR length. For short IDRs (<10AA), the accuracy is the lowest, giving values less than 0.6. PDD-trained networks are significantly more accurate predictors of disorder with short IDRs (<20 residues), the accuracy is consistently over 0.7. For very long IDRs (>70AA), the PDD-trained predictor is on average slightly better than predictor trained using FOD/FDD. Fig.3(c) shows the variation of prediction accuracy with distance of the predicted IDR from the boundary between structured and disordered residues. With the PDD-trained neural networks, the influence of the boundary is eliminated after a distance of only 7 residues, again showing their superiority for predictions of short IDRs.
Fig. 3.
Prediction accuracy of PDD tested on different neural networks, by (a) residue type, (b) length of IDR, and (c) position away from boundary of structured and disordered residues. Y-axes show the accuracy of 8-fold cross validation by neural networks, after training on FOD/FDD (ANN-F) or PDDS/PDDM (ANN-P). The order of amino acids in (a) is their tendency to be present in disordered segments. In (b), the x-axis shows IDRs arranged by length, in 5-residue increments. In (c), the x-axis is sequential distance, which is the number of residues separating the beginning of the IDR from the boundary between structured and disordered segments. The composition ratio in (a) and (b) is the percentage of that residue or IDR in all samples.
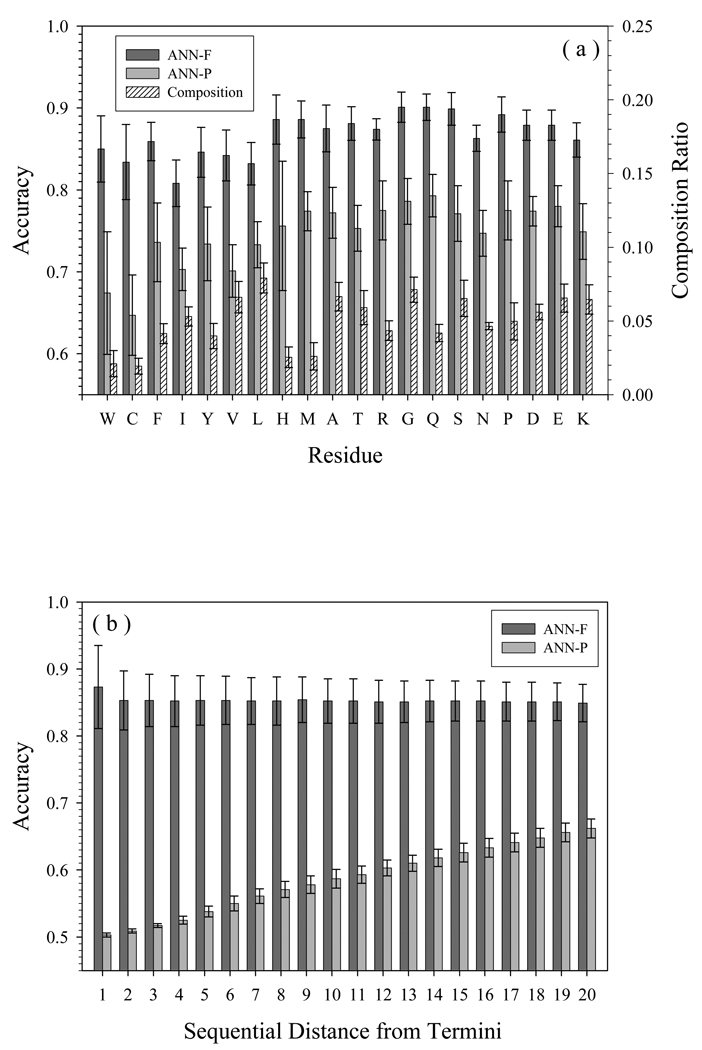
For the purpose of comparing the accuracy difference of various predictors on various dataset, the accuracy dependence on residue types and on distance from termini in FOD/FDD is presented in Fig. 4(a) and (b), respectively. Here, the accuracy dependence on length of segment was ignored because all the sequences in FOD/FDD datasets are either fully structured or fully disordered, and the majority of them are longer than 100 residues which is the up-limit of Fig. 3(b). As shown in Fig. 4(a), the residue dependence of FOD/FDD-trained predictor is marginal compared to PDD-trained predictor. However, structure-prone residues have less accuracy than disorder-prone residues. For each type of residues, FOD/FDD-trained predictor is much more accurate than PDD-trained predictor.
Fig. 4.
Prediction accuracy of FOD/FDD on different neural networks, by (a) residue type and (b) distance from termini. Y-axes show the accuracy of 8-fold cross validation by neural networks, after training on FOD/FDD (ANN-F) or PDDS/PDDM (ANN-P). The order of amino acids in (a) is their tendency to be present in disordered segments. The composition ratio in (a) is the percentage of that residue in all samples.
As to the boundary effect, since proteins in FOD/FDD datasets are either fully structured or fully disordered, we can only take both N- and C-termini as the boundary. The influence of boundary is shown in Fig. 4(b). FOD/FDD-trained predictor has almost no boundary effect, while PDD-trained predictor is very sensitive to the distance from boundary. Since the FOD/FDD trained network is biased to reply that all residues are either disordered or ordered, the bars in Fig. 4(b) are the same length: this predictor is insensitive to the locations of residues. On the other hand, as there are no obvious restrictions or biases for the PDD trained network, this predictor is sensitive to the locations, and therefore can be useful not only to predict PDD, but also FOD/FDD.
Although we separated them as different factors affecting the prediction accuracy, the boundary effect and compositional profile may be the two different sides of the same phenomenon. All the machine learning methods employ the sliding window to generate the input. The location distant from the boundary decides the ratio of structure-prone and disorder-prone residues. Therefore, the concepts of location and composition may be strongly correlated under these circumstances.
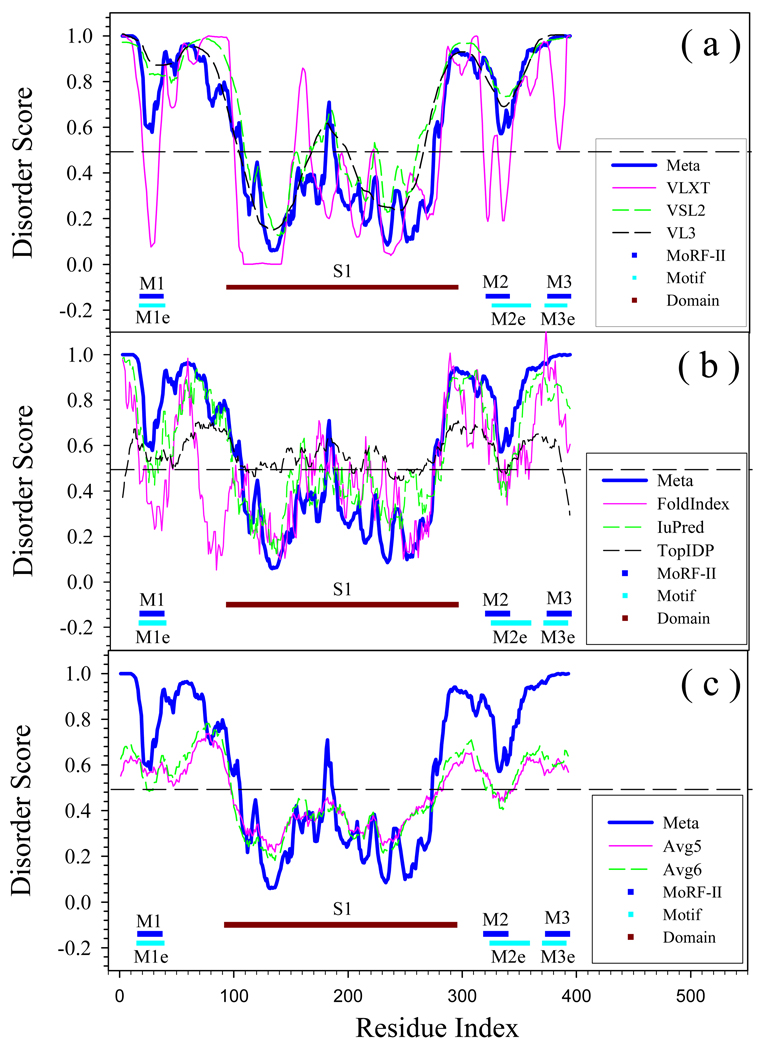
Figs. 5 – 9 show the results of this new prediction scheme applied to human p53 (Fig. 5), human cyclic-AMP-response-element-binding protein (CREB) (Fig. 6), human CREB binding protein (CBP) (Fig. 7), an α-helical transmembrane (TM) protein (Fig. 8) and a β-structural TM protein (Fig. 9), compared to the results of these proteins analysis by various individual predictors. Fig. 5(a) compares PONDR-FIT, PONDR®VLXT, PONDR®VSL2, and PONDR®VL3. Fig. 5(b) shows the difference of PONDR-FIT from FoldIndex, IUPred, and TopIDP. The last inset Fig. 5(c) compares PONDR-FIT to two average values from 6 individual predictors. It is clear that in Fig. 5(a), the general trend of PONDR-FIT is very similar to VL3 and VSL2.
Fig. 5.
Prediction of intrinsically disordered residues in Human P53 (SwissProt ID: P04637) by ANN-P (PONDR-FIT) and its 6 individual component predictors. Short horizontal lines indicated by M1, M2, and M3, are three MoRF regions predicted by MoRF-II predictor. M1e (3DAC [83]), M2e (1AIE [84]), and M3e (1MA3 [59]) are experimentally identified binding motifs. Brown line S1 (2BIQ [85]) is crystallized DNA-binding domain. The dashed lines at 0.5 of Y-axis are threshold lines for disordered/structured residues. Residues with a score above this line are predicted disordered, and residues with a score below 0.5 are predicted to be ordered. Meta, VLXT, VSL2, and VL3 in (a) correspond to the prediction from PONDR-FIT, PONDR®VLXT, PONDR®VSL2, PONDR®VL3. While in (b) FoldIndex, IUPred, and TopIDP are the compared predictors. In (c), Avg5 is the mathematical average of five individual, omitting PONDR®VLXT; Avg6 is the mathematical average of all six individual predictors.
Fig.9.
Prediction of intrinsically disordered residues in a β-structural transmembrane protein, outer membrane protein G from E. coli (Swissprot id: P76045). The transmembrane regions are shown by black lines. Information about these regions is extracted from SwissProt. Structured regions are shown by brown horizontal lines. Brakes between these brown lines correspond to regions with missing electron density (2F1C, [91]). Plot (a) Represents predictions by PONDR-FIT, PONDR®VLXT, PONDR®VSL2, PONDR®VL3; Plot (b) represents data for PONDR-FIT, FoldIndex, IUPred, and TopIDP; Plot (c) compares PONDR-FIT with Avg5 and Avg6.
Fig.6.
Prediction of intrinsically disordered residues in human CREB protein CREB1 (SwissProt ID: P16220) by ANN-P (PONDR-FIT) and its 6 individual component predictors. Dark blue horizontal line is a MoRF region predicted by MoRF-II predictor. There are no intrinsically ordered domains or regions in this protein. However, one segment of this protein, KID domain, forms a crystallizable complex with the KIX domain of CREB binding protein (1KDX, [69]). This segment is indicated as M1e and is shown as light blue line. Red bar corresponds to the DNA-binding domain, whereas yellow bar indicated the dimerization domain. Plot (a) Represents predictions by PONDR-FIT, PONDR®VLXT, PONDR®VSL2, PONDR®VL3; Plot (b) represents data for PONDR-FIT, FoldIndex, IUPred, and TopIDP; Plot (c) compares PONDR-FIT with Avg5 and Avg6.
Fig. 7.
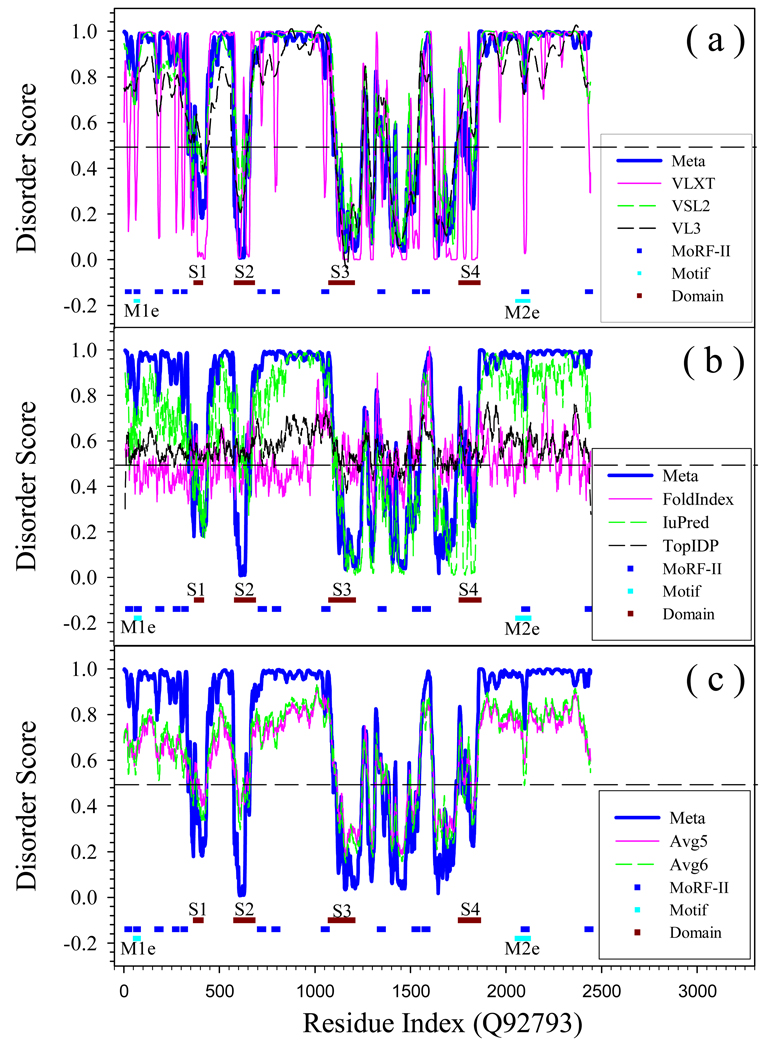
Prediction of intrinsically disordered residues in human CREB binding protein (SwissProt ID: Q92793) by ANN-P (PONDR-FIT) and its 6 individual component predictors. Short blue horizontal lines are MoRF regions predicted by MoRF-II predictor. M1e (NCBD) and M2e (1ZOQ [86], NRID) are verified binding motifs [5]. These are shown by light blue horizontal lines. Experimentally validated structured domain are represented by brown horizontal lines: S1 (1LIQ [87], TAZ1); S2 (1SB0 [88], KIX); S3 (3DWY [89], Bromo); and S4 (2KJE [77], TAZ2). Plot (a) Represents predictions by PONDR-FIT, PONDR®VLXT, PONDR®VSL2, PONDR®VL3; Plot (b) represents data for PONDR-FIT, FoldIndex, IUPred, and TopIDP; Plot (c) compares PONDR-FIT with Avg5 and Avg6.
Fig.8.
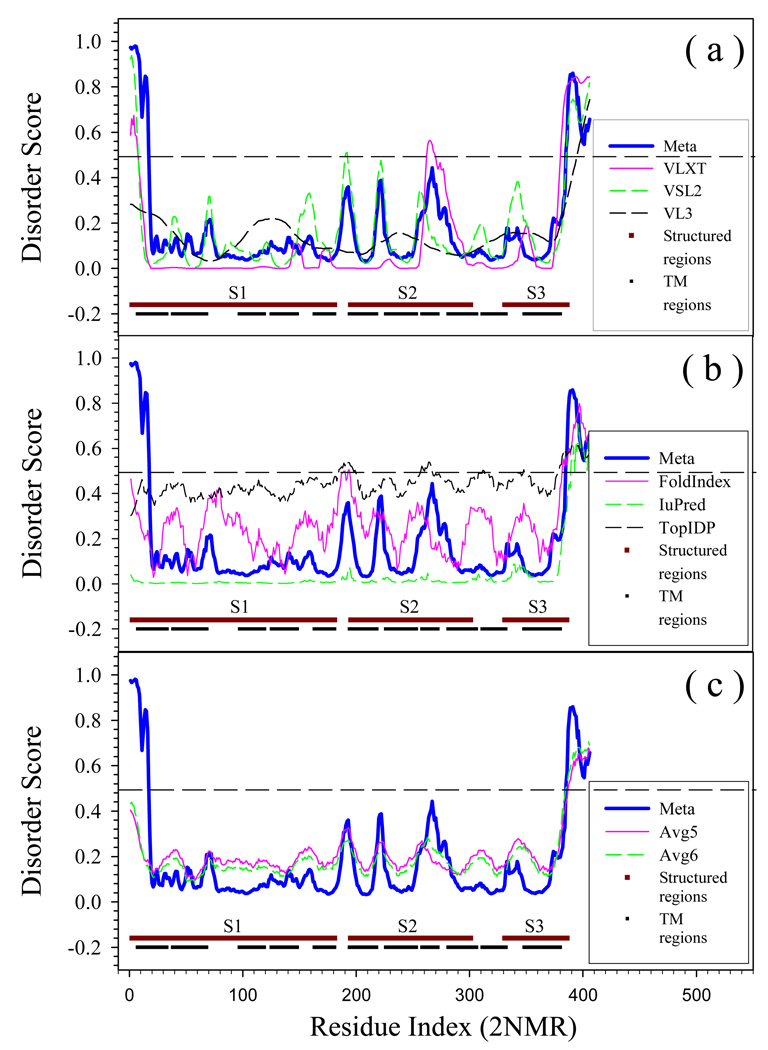
Prediction of intrinsically disordered residues in an α-helical transmembrane protein, ammonia channel from E.coli (Swissprot ID: P69681). The trans-membrane regions are shown by black lines. Information about these regions is extracted from SwissProt. Structured regions are shown by brown horizontal lines. Brakes between these brown lines correspond to regions with missing electron density (2NMR, [90]). Plot (a) Represents predictions by PONDR-FIT, PONDR®VLXT, PONDR®VSL2, PONDR®VL3; Plot (b) represents data for PONDR-FIT, FoldIndex, IUPred, and TopIDP; Plot (c) compares PONDR-FIT with Avg5 and Avg6.
In Fig. 5(b), FoldIndex is very different from PONDR-FIT in both N and C terminal segments, whereas TopIDP shows the strongest effect of termini. Fig. 5(b) shows numerous differences in the various prediction profiles, reflecting the fundamental differences between the PONDR family of predictors and other predictors. It is also interesting in Fig. 5(c), that although the averaged values can imitate the trend of disordered states along the sequence, they may fail to identify biologically important regions. For example, PONDR-FIT clearly shows that two segments, AA20-AA30 and AA330-AA340, have higher tendency of being structured and are flanked by disordered regions, another segment AA180-AA190 is a disordered region connecting two structured domains at both sides. All these three regions have sharp transition from one disordered status to the other. However, in the curves shown by averaged values, the sharp transitions disappeared or became rather smooth.
The most obvious differences between PONDR-FIT and VLXT are four dips found by VLXT near residues 30, 320, 340 and 390, as well as a peak near residue 160. These VLXT dips may correspond to MoRF regions, which are structure-prone segments in disordered regions [46]. In VLXT prediction, MoRF appears as a dip falling down from high-disordered score to lowdisordered score. Although MoRF itself has the tendency of being structured, it is flanked by disordered regions and is very likely to be disordered under the physiological conditions. This assumption has been well tested by several other predictors (VSL2, VL3, and PONDR-FIT) and in other studies [15]. MoRF plays important roles in molecular recognition and signaling. Actually, above mentioned four dips were predicted to be three MoRFs as indicated by M1, M2, and M3 in Fig.5. Importantly, these predicted MoRFs coincide with structurally characterized regions of p53 responsible for protein-protein interactions (see regions indicated M1e, M2e and M3e). In fact, X-ray crystallographic studies of the p53-Mdm2 complex reveal that the N-terminal region of p53 that binds Mdm2 (indicated as M1e region in Fig. 5) forms a helical structure that is situated in a deep groove on the surface of Mdm2 [53], NMR studies show that the unbound N-terminal region lacked fixed structure, although a region within it does form of an amphipathic helix some fraction of the time [54]. In addition to Mdm2, the transactivation region of p53 interacts with TFIID, TFIIH, RPA, CBP/p300 and CSN5/Jab1 [55]. M2e/M2 region (see Fig. 5) corresponds to the p53 tetramerization domain. While this domain is structured, the structure is acquired upon the formation of the complex [56].
At the C-terminal domain, p53 interacts with GSK3β, PARP-1, TAF1, TRRAP, hGcn5, TAF, 14-3-3, S100ββ and many other proteins [57]. Our recent computational analysis [56] of structures currently in the RCSB Protein Data Bank shows that the same C-terminal disorder region of p53 (residues 374 to 388, indicated as M3e region in Fig. 5) forms three major secondary structure types in the bound state depending on which partner it binds: an α-helix when associating with S100ββ[58], a β-sheet with sirtuin [59], an irregular structure with CBP [60], and a different irregular structure with cyclin A2 [61]. Interaction of this fragment with S100ββ occurs in a Ca2+-dependent manner [62]. In the absence of S100ββ, this p53 peptide exists as a random coil as determined by NMR. However, much of this C-terminal peptide adopts a helical conformation when bound to Ca2+ loaded S100ββ [58].
Finally, the region marked S1 (see Fig. 5) represents a well-folded DNA-binding domain of p53. This domain mediates interactions between p53 and 3 endogenous partners, DNA [63], 53BP1 [64], and 53BP2 [65], and p53 and one exogenous partner, namely, the large-T antigen (LTag) from simian virus 40 [66].
Altogether, data shown in Fig. 5 suggests that PONDR-FIT lacks the ability to find all the functionally important residues represented by MoRFs, being able to detect a MoRF (M1), a tetramertization domain (M2) and a structured DNA-binding domain (S1). Comparison of predictions by PONDR-FIT and individual disorder predictors for several proteins shows the same trend, where PONDR-FIT is able to visualize some of the disorder-based functional sites.
Fig. 6 represents the results of the disorder analysis in human cyclic-AMP-response-element- binding protein (CREB), which contains the kinase-inducible transcriptional-activation domain (KID), one of the most well-characterized examples of the coupled folding and binding processes [5]. In fact, the KID polypeptide is intrinsically disordered, both as an isolated peptide and in full-length CREB [67] [68], but it folds to form a pair of orthogonal helices on binding to its target partner, the KID-binding (KIX) domain of CREB-binding protein (CBP) [69]. CREB is an important transcription factor involved in learning and memory [70–71]. The key steps involved in CREB-mediated gene transcription include dimerization, binding at the cAMP response element (CRE), a specialized stretch of DNA that contains the consensus nucleotide sequence TGACGTCA, and phosphorylation of a single serine residue by calmodulin-dependent kinases [72–73]. Phosphorylation of CREB activates a cascade of events that involves recruitment of associated proteins such as CREB-binding protein (CBP) and assembly of a larger transcriptional complex [71]. CREB consists of an NH2-terminal activation domain (residues 1-253) and a smaller, COOH-terminal domain (residues 284-341), which includes bZIP DNA-binding (residues 284-305) and dimerization (residues 311-332) domains. The activation domain of CREB contains two glutamine-rich regions (Q1, residues 11-88, and Q2M, residues 166-283), which are important for basal activity. The kinase-inducible domain (KID, residues 101-160) includes several phosphorylation sites and confers the phosphorylation-induced activity to CREB [68]. Fig. 6 shows that the CREB dimerization domain is detected by PONDR-FIT as a peculiar double-dip minimum in the disorder score distribution. Note that the tetramerization domain of p53 is characterized by the same double-dip appearance in the PONDR-FIT plot (see Fig. 5). Although the region corresponding to the KID domain is characterized by the overall high disorder scores, this fragment has several local minima which roughly coincide with location of α-helices induced by interaction with KIX domain of CBP (residues 120-129 and 133-141). Notably, the DNA-binding domain of CREB is “invisible” to all predictors, which is not a surprise since this region is highly charged containing 55% of arginines and lysines.
Fig.7 illustrates the disorder distribution in an important partner of CREB, a modular transcriptional co-activator, CREB-binding protein (CBP). Of the 2,442 amino acids of the human CBP, more than 50% are in the intrinsically disordered regions [5]. CBP has long mostly disordered tails (residues 1-346 and 1855-2442) and is characterized by the beads-on-the-string structure, where independently folded domains or groups of them are connected by long disordered linkers. The mentioned independently folded domains are the transcriptional-adaptor zinc-finger-1 (TAZ1) domain (residues 347-439), the KID-binding (KIX) domain (residues 587-673), the Bromo domain (residues 1081-1197), the plant homeodomain (PHD, residues 1224-1317), the histone acetyltransferase (HAT) domain (residues 1318-1693), the zinc-binding domain near the dystrophin WW domain (ZZ, residues 1694-1751), and the transcriptional-adaptor zinc-finger-2 (TAZ2) domain (residues 1763-1854). The tertiary structure of four of these folded domains, TAZ1, PHD, ZZ, and TAZ2, is stabilized by zinc binding [5], and 3D structures of the TAZ1 [74], KIX [69], Bromo [60], ZZ [75], and TAZ2 [76–77]) domains have been determined by NMR. In addition to folded domains, CBP contains the disordered nuclear-receptor-interaction domain (NRID, residues 61-75) and the unstructured nuclear-receptor co-activator-binding domain (NCBD, residues 2058-2116) also known as the IRF-3 binding domain (IBiD). NCBD is highly helical, but possesses properties of the native molten globule [78]. Once again, Fig. 7 shows that PONDR-FIT is able to “see” many of the functional domains of this protein.
Finally, Figs. 8 and 9 represents the results of disorder analysis in two transmembrane (TM) proteins, an α-helical TM protein, ammonia channel from E.coli (Fig. 8), and a β-structural TM protein, outer membrane protein G from E. coli (Fig. 9). Since the TM proteins are very different from globular and disordered proteins [52], and since TM proteins were excluded from the training datasets, we did not expect that PONDR-FIT can be directly applied to TM proteins. However, Figs. 8 and 9 clearly show that this predictor works reasonably well on these difficult targets too. In fact, the majority of regions with missing electron density (indicated in Figs. 8 and 9 as regions between structured regions shown by brown lines) are located in regions either predicted by PONDR-FIT as disordered or as regions with increased propensity for disorder. Furthermore, the majority of trans-membrane regions are located in fragments of these proteins predicted to be ordered by PONDR-FIT.
4. Discussion
Disorder prediction will remain useful for proteomics studies, either when used in combination with experiments [11–12] or when used as stand-alone values over entire genomes [25, 79]. We are in the process of using PONDR-FIT to carry out disorder predictions across all model organisms. The results will be published and posted on our Database of Protein Disorder (www.disprot.org) [43].
All combinations of the individual predictors were tried to determine the effects of the various individual predictors on the results obtained (Fig. 1). As expected, in general the meta-predictor combinations containing high-accuracy individual predictors also have high accuracy and the achievable accuracy of a meta-predictor depends on the combination of individual predictors. Taking into consideration that each predictor classifies samples in the high-dimensional phase space, combination of various predictors results in a new division of the phase space with integrated dimensions. The training process of meta-predictor is an optimization process in the phase space for new classifier. Obviously, not every new division will be better than individual ones. On the other side, with integration of new dimensions, improvement can be expected. This is exactly the results observed in Fig.1. Even though the improvement has been achieved in most cases, an overall ceiling was observed for all the composite predictors. The best performance of the composite predictors is only ~2% higher than the best of the individual predictor in that combination. The similar phenomenon with improvement of 2~3% was also observed in another two meta-predictors, metaPrDOS [39] and MD [40]. According to previous estimates, the possible highest prediction accuracy in terms of sensitivity for disordered residues in a general dataset is around 85% [17, 80]. Compared with the value of accuracy in Table 1, more improvement can be expected in the future.
Prediction of disorder depends on many factors, including the relationship if any between the training sets used to develop the predictor and the testing sets used to estimate prediction accuracy. These various factors lead to uncertainties when attempts are made to compare the accuracies of different disorder predictors in different publications because the data used for training and testing are not standardized. Table 1, in which two types of data were used, illustrates the resulting uncertainties with regard to comparing prediction performance among the various predictors and their combinations. These results show that the performances change markedly when different data are used.
One would like to compare the accuracies of the two previous meta-predictors, META-Disorder, and metaPrDOS, and the meta-predictor described here, PONDR-FIT, but this is difficult due to the various uncertainties described briefly above. Here we would like propose a mechanism for making such comparisons more reliable. Note that all three of these meta-predictors employed IUPred as one of the components. Thus, one way to compare the accuracies would be to use the results from IUPred as in internal standard. By this approach, the accuracy improvement of the meta-predictor as compared to IUPred becomes a indicator measure to compare the performances of the three meta-predictors.
The only measure used in all three studies is the AUC, so we will restrict comparisons to this value. In terms of AUC, the META-Disorder predictor improved the accuracy to 0.80 compared to IUPred at 0.76, while metaPrDOS achieved 0.90 compared to IUPred at 0.83. Thus, their AUC improvements over IUPred are 5.2% and 8.4%, respectively, for META-Disorder and metaPrDOS. Here we found that, compared to IUPred, PONDR-FIT improved the AUC by 5.3% for the FOD/FDD data and by 14.9% for the PDD datasets. The smaller improvement was observed for the FOD/FDD data because the accuracy was already very high for this data, and these results demonstrate that the details of the dataset being used limit even this method of comparing results. However, if we assume that the PDD data is closer to the data used to train and test the previous meta-predictors, then the 14.9% improvement brought about by PONDER-FIT suggests that this latest meta-predictor does very well compared to the 5.2% improvement by META-Disorder and the 8.4% by metaPrDOS.
The error analysis in Table 3 and Fig.3 has potential new applications. As shown by Table 3, neural networks trained by PDD achieved the similar accuracy on partially disordered sequences as on fully disordered sequences. Meanwhile, neural networks trained by FOD/FDD are much better for long disordered sequences than partially ones. Fig.3 shows the errors under various conditions. The bottle necks of current disorder prediction are in boundary regions and short IDRs.
In prediction of secondary structure, interactions between the segment of interest and the rest of the protein can override the structural tendencies of the segment of interest and thereby reduce prediction accuracy [81]. We suspect that prediction of disorder for short regions of disorder and at boundary regions becomes less accurate because short regions of disorder and boundary regions are more likely to be affected by attractions or repulsions relative to the nearby structured protein. That is, we speculate that interactions between the segment of interest and the nearby structured protein can induce either structure or disorder in the segment of interest, thereby reducing the prediction accuracy. We made one attempt to improve predictions at boundaries with some success [82], but that study was probably limited by both insufficient data and poor models that failed to properly account for the effects of the possible non-local interactions. The reduced accuracy of predictions of short disordered regions and at boundary regions suggests that focused study on these problems could bring about important improvements in predictor accuracies.
Comparison in Figs. 5-9 between meta-prediction and averaged values of prediction shows another side of the importance of meta-prediction. The averaged values correspond to a different refinement scheme – simple majority voting. For a simple binary output, this majority voting may not lose too much information. However, for real value predictions, the complex method like neural networks is clearly superior in providing prediction information that better correlates with experimentally determined details regarding structure and disorder. .
Acknowledgements
This work was supported in part by the grants R01 LM007688-01A1 (to A.K.D and V.N.U.), R01 GM071714-01A2 (to A.K.D and V.N.U.), and R01 GM73784 (to R.L.D.) from the National Institute of Health, the grant EF 0849803 from the National Science Foundation, and the Program of the Russian Academy of Sciences for the “Molecular and Cellular Biology” (to V.N.U.). We gratefully acknowledge the support of the IUPUI Signature Centers Initiative.
Abbreviations
- IDP
Intrinsically Disordered Protein
- IDR
Intrinsically Disordered Region
- PDD
Partially Disordered Dataset
- PDDS
Structured residues in PDD
- PDDM
residues with Missing electron density in PDD
- FOD
Fully Ordered Dataset
- FDD
Fully Disordered Dataset
- ANN
Artificial Neural Networks
- ANN-P
ANN trained by PDD
- ANN-F
ANN trained by FOD/FDD
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Wright PE, Dyson HJ. Intrinsically unstructured proteins: re-assessing the protein structure-function paradigm. J Mol Biol. 1999;293:321–331. doi: 10.1006/jmbi.1999.3110. [DOI] [PubMed] [Google Scholar]
- 2.Dunker AK, Lawson JD, Brown CJ, Williams RM, Romero P, Oh JS, Oldfield CJ, Campen AM, Ratliff CM, Hipps KW, Ausio J, Nissen MS, Reeves R, Kang C, Kissinger CR, Bailey RW, Griswold MD, Chiu W, Garner EC, Obradovic Z. Intrinsically disordered protein. J Mol Graph Model. 2001;19:26–59. doi: 10.1016/s1093-3263(00)00138-8. [DOI] [PubMed] [Google Scholar]
- 3.Uversky VN, Gillespie JR, Fink AL. Why are "natively unfolded" proteins unstructured under physiologic conditions? Proteins. 2000;41:415–427. doi: 10.1002/1097-0134(20001115)41:3<415::aid-prot130>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 4.Tompa P. The functional benefits of protein disorder. Journal of Molecular Structure-Theochem. 2003;666:361–371. [Google Scholar]
- 5.Dyson HJ, Wright PE. Intrinsically unstructured proteins and their functions. Nat Rev Mol Cell Biol. 2005;6:197–208. doi: 10.1038/nrm1589. [DOI] [PubMed] [Google Scholar]
- 6.Dunker AK, Obradovic Z. The protein trinity--linking function and disorder. Nat Biotechnol. 2001;19:805–806. doi: 10.1038/nbt0901-805. [DOI] [PubMed] [Google Scholar]
- 7.Obradovic Z, Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK. Predicting intrinsic disorder from amino acid sequence. Proteins. 2003;53 Suppl 6:566–572. doi: 10.1002/prot.10532. [DOI] [PubMed] [Google Scholar]
- 8.Cortese MS, Baird JP, Uversky VN, Dunker AK. Uncovering the unfoldome: enriching cell extracts for unstructured proteins by acid treatment. J Proteome Res. 2005;4:1610–1618. doi: 10.1021/pr050119c. [DOI] [PubMed] [Google Scholar]
- 9.Csizmok V, Dosztanyi Z, Simon I, Tompa P. Towards proteomic approaches for the identification of structural disorder. Curr Protein Pept Sci. 2007;8:173–179. doi: 10.2174/138920307780363479. [DOI] [PubMed] [Google Scholar]
- 10.Midic U, Oldfield CJ, Dunker AK, Obradovic Z, Uversky VN. Protein disorder in the human diseasome: unfoldomics of human genetic diseases. BMC Genomics. 2009;10 Suppl 1:S12. doi: 10.1186/1471-2164-10-S1-S12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Galea CA, Pagala VR, Obenauer JC, Park CG, Slaughter CA, Kriwacki RW. Proteomic studies of the intrinsically unstructured mammalian proteome. J Proteome Res. 2006;5:2839–2848. doi: 10.1021/pr060328c. [DOI] [PubMed] [Google Scholar]
- 12.Galea CA, High AA, Obenauer JC, Mishra A, Park CG, Punta M, Schlessinger A, Ma J, Rost B, Slaughter CA, Kriwacki RW. Large-scale analysis of thermostable, mammalian proteins provides insights into the intrinsically disordered proteome. J Proteome Res. 2009;8:211–226. doi: 10.1021/pr800308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunker AK, Brown CJ, Obradovic Z. Identification and functions of usefully disordered proteins. Adv Protein Chem. 2002;62:25–49. doi: 10.1016/s0065-3233(02)62004-2. [DOI] [PubMed] [Google Scholar]
- 14.Dunker AK, Brown CJ, Lawson JD, Iakoucheva LM, Obradovic Z. Intrinsic disorder and protein function. Biochemistry. 2002;41:6573–6582. doi: 10.1021/bi012159+. [DOI] [PubMed] [Google Scholar]
- 15.Minezaki Y, Homma K, Kinjo AR, Nishikawa K. Human transcription factors contain a high fraction of intrinsically disordered regions essential for transcriptional regulation. J Mol Biol. 2006;359:1137–1149. doi: 10.1016/j.jmb.2006.04.016. [DOI] [PubMed] [Google Scholar]
- 16.Uversky VN, Oldfield CJ, Dunker AK. Intrinsically disordered proteins in human diseases: introducing the D2 concept. Annu Rev Biophys. 2008;37:215–246. doi: 10.1146/annurev.biophys.37.032807.125924. [DOI] [PubMed] [Google Scholar]
- 17.Radivojac P, Iakoucheva LM, Oldfield CJ, Obradovic Z, Uversky VN, Dunker AK. Intrinsic disorder and functional proteomics. Biophys J. 2007;92:1439–1456. doi: 10.1529/biophysj.106.094045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.He B, Wang K, Liu Y, Xue B, Uversky VN, Dunker AK. Predicting intrinsic disorder in proteins: an overview. Cell Res. 2009;19:929–949. doi: 10.1038/cr.2009.87. [DOI] [PubMed] [Google Scholar]
- 19.Romero P, Obradovic Z, Li X, Garner EC, Brown CJ, Dunker AK. Sequence complexity of disordered protein. Proteins. 2001;42:38–48. doi: 10.1002/1097-0134(20010101)42:1<38::aid-prot50>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 20.Peng K, Vucetic S, Radivojac P, Brown CJ, Dunker AK, Obradovic Z. Optimizing long intrinsic disorder predictors with protein evolutionary information. J Bioinform Comput Biol. 2005;3:35–60. doi: 10.1142/s0219720005000886. [DOI] [PubMed] [Google Scholar]
- 21.Peng K, Radivojac P, Vucetic S, Dunker AK, Obradovic Z. Length-dependent prediction of protein intrinsic disorder. Bmc Bioinformatics. 2006;7:208. doi: 10.1186/1471-2105-7-208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Jones DT, Ward JJ. Prediction of disordered regions in proteins from position specific score matrices. Proteins. 2003;53 Suppl 6:573–578. doi: 10.1002/prot.10528. [DOI] [PubMed] [Google Scholar]
- 24.Schlessinger A, Liu J, Rost B. Natively unstructured loops differ from other loops. PLoS Comput Biol. 2007;3:e140. doi: 10.1371/journal.pcbi.0030140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ward JJ, Sodhi JS, McGuffin LJ, Buxton BF, Jones DT. Prediction and functional analysis of native disorder in proteins from the three kingdoms of life. J Mol Biol. 2004;337:635–645. doi: 10.1016/j.jmb.2004.02.002. [DOI] [PubMed] [Google Scholar]
- 26.Shimizu K, Hirose S, Noguchi T. POODLE-S: web application for predicting protein disorder by using physicochemical features and reduced amino acid set of a position-specific scoring matrix. Bioinformatics. 2007;23:2337–2338. doi: 10.1093/bioinformatics/btm330. [DOI] [PubMed] [Google Scholar]
- 27.Cheng J, Sweredoski MJ, Baldi P. Accurate prediction of protein disordered regions by mining protein structure data. Data Mining Knowl. Disc. 2005;11:213–222. [Google Scholar]
- 28.Han P, Zhang X, Norton RS, Feng ZP. Predicting disordered regions in proteins based on decision trees of reduced amino acid composition. J Comput Biol. 2006;13:1723–1734. doi: 10.1089/cmb.2006.13.1723. [DOI] [PubMed] [Google Scholar]
- 29.Ishida T, Kinoshita K. PrDOS: prediction of disordered protein regions from amino acid sequence. Nucleic Acids Res. 2007;35:W460–W464. doi: 10.1093/nar/gkm363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McGuffin LJ. Intrinsic disorder prediction from the analysis of multiple protein fold recognition models. Bioinformatics. 2008;24:1798–1804. doi: 10.1093/bioinformatics/btn326. [DOI] [PubMed] [Google Scholar]
- 31.Dosztanyi Z, Csizmok V, Tompa P, Simon I. The pairwise energy content estimated from amino acid composition discriminates between folded and intrinsically unstructured proteins. J Mol Biol. 2005;347:827–839. doi: 10.1016/j.jmb.2005.01.071. [DOI] [PubMed] [Google Scholar]
- 32.Schlessinger A, Punta M, Rost B. Natively unstructured regions in proteins identified from contact predictions. Bioinformatics. 2007;23:2376–2384. doi: 10.1093/bioinformatics/btm349. [DOI] [PubMed] [Google Scholar]
- 33.Prilusky J, Felder CE, Zeev-Ben-Mordehai T, Rydberg EH, Man O, Beckmann JS, Silman I, Sussman JL. FoldIndex: a simple tool to predict whether a given protein sequence is intrinsically unfolded. Bioinformatics. 2005;21:3435–3438. doi: 10.1093/bioinformatics/bti537. [DOI] [PubMed] [Google Scholar]
- 34.Galzitskaya OV, Garbuzynskiy SO, Lobanov MY. FoldUnfold: web server for the prediction of disordered regions in protein chain. Bioinformatics. 2006;22:2948–2949. doi: 10.1093/bioinformatics/btl504. [DOI] [PubMed] [Google Scholar]
- 35.Campen A, Williams RM, Brown CJ, Meng J, Uversky VN, Dunker AK. TOP-IDP-scale: a new amino acid scale measuring propensity for intrinsic disorder. Protein Pept Lett. 2008;15:956–963. doi: 10.2174/092986608785849164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rychlewski L, Fischer D. LiveBench-8: the large-scale, continuous assessment of automated protein structure prediction. Protein Sci. 2005;14:240–245. doi: 10.1110/ps.04888805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.von Grotthuss M, Pas J, Wyrwicz L, Ginalski K, Rychlewski L. Application of 3D-Jury, GRDB, and Verify3D in fold recognition. Proteins. 2003;53 Suppl 6:418–423. doi: 10.1002/prot.10547. [DOI] [PubMed] [Google Scholar]
- 38.Fischer D. 3D-SHOTGUN: a novel, cooperative, fold-recognition meta-predictor. Proteins. 2003;51:434–441. doi: 10.1002/prot.10357. [DOI] [PubMed] [Google Scholar]
- 39.Ishida T, Kinoshita K. Prediction of disordered regions in proteins based on the meta approach. Bioinformatics. 2008;24:1344–1348. doi: 10.1093/bioinformatics/btn195. [DOI] [PubMed] [Google Scholar]
- 40.Schlessinger A, Punta M, Yachdav G, Kajan L, Rost B. Improved disorder prediction by combination of orthogonal approaches. PLoS ONE. 2009;4:e4433. doi: 10.1371/journal.pone.0004433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Moran O, Roessle MW, Mariuzza RA, Dimasi N. Structural features of the full-length adaptor protein GADS in solution determined using small-angle X-ray scattering. Biophys J. 2008;94:1766–1772. doi: 10.1529/biophysj.107.116590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Xue B, Oldfield CJ, Dunker AK, Uversky VN. CDF it all: Consensus prediction of intrinsically disordered proteins based on various cumulative distribution functions. FEBS Lett. 2009 doi: 10.1016/j.febslet.2009.03.070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sickmeier M, Hamilton JA, LeGall T, Vacic V, Cortese MS, Tantos A, Szabo B, Tompa P, Chen J, Uversky VN, Obradovic Z, Dunker AK. DisProt: the Database of Disordered Proteins. Nucleic Acids Res. 2007;35:D786–D793. doi: 10.1093/nar/gkl893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.xml2pdb. Roland Dunbrack: http://dunbrack.fccc.edu/xml2pdb.php.
- 45.Campen A, Williams MR, Brown JC, Meng J, Uversky V, Dunker AK. TOPIDP-Scale: A new amino acid scale measuring propensity for intrinsic disorder. Protein & Peptide Lett. 2008 doi: 10.2174/092986608785849164. Accepted. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Oldfield CJ, Cheng Y, Cortese MS, Romero P, Uversky VN, Dunker AK. Coupled folding and binding with alpha-helix-forming molecular recognition elements. Biochemistry. 2005;44:12454–12470. doi: 10.1021/bi050736e. [DOI] [PubMed] [Google Scholar]
- 47.Cheng Y, Oldfield CJ, Meng J, Romero P, Uversky VN, Dunker AK. Mining a-Helic-Forming Molecular Recognition Features with Cross Species Sequence Alignments. Biochemistry. 2007;46:13468–13477. doi: 10.1021/bi7012273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jin Y, Dunbrack RL., Jr Assessment of disorder predictions in CASP6. Proteins. 2005;61 Suppl 7:167–175. doi: 10.1002/prot.20734. [DOI] [PubMed] [Google Scholar]
- 49.Bordoli L, Kiefer F, Schwede T. Assessment of disorder predictions in CASP7. Proteins. 2007;69 Suppl 8:129–136. doi: 10.1002/prot.21671. [DOI] [PubMed] [Google Scholar]
- 50.McGuffin LJ, Bryson K, Jones DT. The PSIPRED protein structure prediction server. Bioinformatics. 2000;16:404–405. doi: 10.1093/bioinformatics/16.4.404. [DOI] [PubMed] [Google Scholar]
- 51.Lin JH. Divergence Measures Based on the Shannon Entropy. Ieee Transactions on Information Theory. 1991;37:145–151. [Google Scholar]
- 52.Xue B, Li L, Meroueh SO, Uversky VN, Dunker AK. Analysis of structured and intrinsically disordered regions of transmembrane proteins. Mol Biosyst. 2009 doi: 10.1039/B905913J. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Kussie PH, Gorina S, Marechal V, Elenbaas B, Moreau J, Levine AJ, Pavletich NP. Structure of the MDM2 oncoprotein bound to the p53 tumor suppressor transactivation domain. Science. 1996;274:948–953. doi: 10.1126/science.274.5289.948. [DOI] [PubMed] [Google Scholar]
- 54.Lee H, Mok KH, Muhandiram R, Park KH, Suk JE, Kim DH, Chang J, Sung YC, Choi KY, Han KH. Local structural elements in the mostly unstructured transcriptional activation domain of human p53. J Biol Chem. 2000;275:29426–29432. doi: 10.1074/jbc.M003107200. [DOI] [PubMed] [Google Scholar]
- 55.Anderson CW, Appella E. Signaling to the p53 tumor suppressor through pathways activated by genotoxic and nongenotoxic stress. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling. New York: Academic Press; 2004. pp. 237–247. [Google Scholar]
- 56.Oldfield CJ, Meng J, Yang JY, Yang MQ, Uversky VN, Dunker AK. Flexible nets: disorder and induced fit in the associations of p53 and 14-3-3 with their partners. BMC Genomics. 2007;9:S1. doi: 10.1186/1471-2164-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Anderson CW, Appella E. Signaling to the p53 tumor suppressor through pathways activated by genotoxic and nongenotoxic stress. In: Bradshaw RA, Dennis EA, editors. Handbook of Cell Signaling. New York: Academic Press; 2003. pp. 237–247. [Google Scholar]
- 58.Rustandi RR, Baldisseri DM, Weber DJ. Structure of the negative regulatory domain of p53 bound to S100B(betabeta) Nat Struct Biol. 2000;7:570–574. doi: 10.1038/76797. [DOI] [PubMed] [Google Scholar]
- 59.Avalos JL, Celic I, Muhammad S, Cosgrove MS, Boeke JD, Wolberger C. Structure of a Sir2 enzyme bound to an acetylated p53 peptide. Mol Cell. 2002;10:523–535. doi: 10.1016/s1097-2765(02)00628-7. [DOI] [PubMed] [Google Scholar]
- 60.Mujtaba S, He Y, Zeng L, Yan S, Plotnikova O, Sachchidanand, Sanchez R, Zeleznik-Le NJ, Ronai Z, Zhou MM. Structural mechanism of the bromodomain of the coactivator CBP in p53 transcriptional activation. Mol Cell. 2004;13:251–263. doi: 10.1016/s1097-2765(03)00528-8. [DOI] [PubMed] [Google Scholar]
- 61.Lowe ED, Tews I, Cheng KY, Brown NR, Gul S, Noble ME, Gamblin SJ, Johnson LN. Specificity determinants of recruitment peptides bound to phospho-CDK2/cyclin A. Biochemistry. 2002;41:15625–15634. doi: 10.1021/bi0268910. [DOI] [PubMed] [Google Scholar]
- 62.Rustandi RR, Drohat AC, Baldisseri DM, Wilder PT, Weber DJ. The Ca(2+)-dependent interaction of S100B(beta beta) with a peptide derived from p53. Biochemistry. 1998;37:1951–1960. doi: 10.1021/bi972701n. [DOI] [PubMed] [Google Scholar]
- 63.Cho Y, Gorina S, Jeffrey PD, Pavletich NP. Crystal structure of a p53 tumor suppressor-DNA complex: understanding tumorigenic mutations. Science. 1994;265:346–355. doi: 10.1126/science.8023157. [DOI] [PubMed] [Google Scholar]
- 64.Joo WS, Jeffrey PD, Cantor SB, Finnin MS, Livingston DM, Pavletich NP. Structure of the 53BP1 BRCT region bound to p53 and its comparison to the Brca1 BRCT structure. Genes Dev. 2002;16:583–593. doi: 10.1101/gad.959202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gorina S, Pavletich NP. Structure of the p53 tumor suppressor bound to the ankyrin and SH3 domains of 53BP2. Science. 1996;274:1001–1005. doi: 10.1126/science.274.5289.1001. [DOI] [PubMed] [Google Scholar]
- 66.Lilyestrom W, Klein MG, Zhang R, Joachimiak A, Chen XS. Crystal structure of SV40 large T-antigen bound to p53: interplay between a viral oncoprotein and a cellular tumor suppressor. Genes Dev. 2006;20:2373–2382. doi: 10.1101/gad.1456306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Radhakrishnan I, Perez-Alvarado GC, Dyson HJ, Wright PE. Conformational preferences in the Ser133-phosphorylated and non-phosphorylated forms of the kinase inducible transactivation domain of CREB. FEBS Lett. 1998;430:317–322. doi: 10.1016/s0014-5793(98)00680-2. [DOI] [PubMed] [Google Scholar]
- 68.Richards JP, Bachinger HP, Goodman RH, Brennan RG. Analysis of the structural properties of cAMP-responsive element-binding protein (CREB) and phosphorylated CREB. J Biol Chem. 1996;271:13716–13723. doi: 10.1074/jbc.271.23.13716. [DOI] [PubMed] [Google Scholar]
- 69.Radhakrishnan I, Perez-Alvarado GC, Parker D, Dyson HJ, Montminy MR, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of CREB: a model for activator:coactivator interactions. Cell. 1997;91:741–752. doi: 10.1016/s0092-8674(00)80463-8. [DOI] [PubMed] [Google Scholar]
- 70.Yin JC, Tully T. CREB and the formation of long-term memory. Curr Opin Neurobiol. 1996;6:264–268. doi: 10.1016/s0959-4388(96)80082-1. [DOI] [PubMed] [Google Scholar]
- 71.Carlezon WA, Jr, Duman RS, Nestler EJ. The many faces of CREB. Trends Neurosci. 2005;28:436–445. doi: 10.1016/j.tins.2005.06.005. [DOI] [PubMed] [Google Scholar]
- 72.Shaywitz AJ, Greenberg ME. CREB: a stimulus-induced transcription factor activated by a diverse array of extracellular signals. Annu Rev Biochem. 1999;68:821–861. doi: 10.1146/annurev.biochem.68.1.821. [DOI] [PubMed] [Google Scholar]
- 73.Mayr B, Montminy M. Transcriptional regulation by the phosphorylation-dependent factor CREB. Nat Rev Mol Cell Biol. 2001;2:599–609. doi: 10.1038/35085068. [DOI] [PubMed] [Google Scholar]
- 74.Dames SA, Martinez-Yamout M, De Guzman RN, Dyson HJ, Wright PE. Structural basis for Hif-1 alpha /CBP recognition in the cellular hypoxic response. Proc Natl Acad Sci U S A. 2002;99:5271–5276. doi: 10.1073/pnas.082121399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ponting CP, Blake DJ, Davies KE, Kendrick-Jones J, Winder SJ. ZZ and TAZ: new putative zinc fingers in dystrophin and other proteins. Trends Biochem Sci. 1996;21:11–13. [PubMed] [Google Scholar]
- 76.De Guzman RN, Liu HY, Martinez-Yamout M, Dyson HJ, Wright PE. Solution structure of the TAZ2 (CH3) domain of the transcriptional adaptor protein CBP. J Mol Biol. 2000 303;:243–253. doi: 10.1006/jmbi.2000.4141. [DOI] [PubMed] [Google Scholar]
- 77.Ferreon JC, Martinez-Yamout MA, Dyson HJ, Wright PE. Structural basis for subversion of cellular control mechanisms by the adenoviral E1A oncoprotein. Proc Natl Acad Sci U S A. 2009;106:13260–13265. doi: 10.1073/pnas.0906770106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Demarest SJ, Deechongkit S, Dyson HJ, Evans RM, Wright PE. Packing, specificity, and mutability at the binding interface between the p160 coactivator and CREB-binding protein. Protein Sci. 2004;13:203–210. doi: 10.1110/ps.03366504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Dunker AK, Obradovic Z, Romero P, Garner EC, Brown CJ. Intrinsic protein disorder in complete genomes. Genome Inform Ser Workshop Genome Inform. 2000;11:161–171. [PubMed] [Google Scholar]
- 80.Mohan A, Uversky VN, Radivojac P. Influence of sequence changes and environment on intrinsically disordered proteins. PLoS Comput Biol. 2009;5:e1000497. doi: 10.1371/journal.pcbi.1000497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kihara D. The effect of long-range interactions on the secondary structure formation of proteins. Protein Sci. 2005;14:1955–1963. [Google Scholar]
- 82.Radivojac P, Obradovic Z, Brown CJ, Dunker AK. Prediction of boundaries between intrinsically ordered and disordered protein regions. Pac Symp Biocomput. 2003:216–227. [PubMed] [Google Scholar]
- 83.Popowicz GM, Czarna A, Holak TA. Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain. Cell Cycle. 2008;7:2441–2443. doi: 10.4161/cc.6365. [DOI] [PubMed] [Google Scholar]
- 84.Mittl PR, Chene P, Grutter MG. Crystallization and structure solution of p53 (residues 326–356) by molecular replacement using an NMR model as template. Acta Crystallogr D Biol Crystallogr. 1998;54:86–89. doi: 10.1107/s0907444997006550. [DOI] [PubMed] [Google Scholar]
- 85.Joerger AC, Ang HC, Veprintsev DB, Blair CM, Fersht AR. Structures of p53 cancer mutants and mechanism of rescue by second-site suppressor mutations. J Biol Chem. 2005;280:16030–16037. doi: 10.1074/jbc.M500179200. [DOI] [PubMed] [Google Scholar]
- 86.Qin BY, Liu C, Srinath H, Lam SS, Correia JJ, Derynck R, Lin K. Crystal structure of IRF-3 in complex with CBP. Structure. 2005;13:1269–1277. doi: 10.1016/j.str.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 87.Sharpe BK, Matthews JM, Kwan AH, Newton A, Gell DA, Crossley M, Mackay JP. A new zinc binding fold underlines the versatility of zinc binding modules in protein evolution. Structure. 2002;10:639–648. doi: 10.1016/s0969-2126(02)00757-8. [DOI] [PubMed] [Google Scholar]
- 88.Zor T, De Guzman RN, Dyson HJ, Wright PE. Solution structure of the KIX domain of CBP bound to the transactivation domain of c-Myb. J Mol Biol. 2004;337:521–534. doi: 10.1016/j.jmb.2004.01.038. [DOI] [PubMed] [Google Scholar]
- 89.Filippakopoulos P, Picaud S, Fedorov O, Karim R, Pike ACW, Von Delft F, Arrowsmith CH, Edwards AM, Wickstroem M, Bountra C, Knapp S. Crystal Structure of the Bromodomain of Human CREBBP. in, To be published. [Google Scholar]
- 90.Javelle A, Lupo D, Zheng L, Li XD, Winkler FK, Merrick M. An unusual twin-his arrangement in the pore of ammonia channels is essential for substrate conductance. J Biol Chem. 2006;281:39492–39498. doi: 10.1074/jbc.M608325200. [DOI] [PubMed] [Google Scholar]
- 91.Subbarao GV, van den Berg B. Crystal structure of the monomeric porin OmpG. J Mol Biol. 2006;360:750–759. doi: 10.1016/j.jmb.2006.05.045. [DOI] [PubMed] [Google Scholar]