Abstract
BCL6 protects germinal center (GC) B cells against DNA damage–induced apoptosis during somatic hypermutation and class-switch recombination. Although expression of BCL6 was not found in early IL-7–dependent B cell precursors, we report that IL-7Rα–Stat5 signaling negatively regulates BCL6. Upon productive VH-DJH gene rearrangement and expression of a μ heavy chain, however, activation of pre–B cell receptor signaling strongly induces BCL6 expression, whereas IL-7Rα–Stat5 signaling is attenuated. At the transition from IL-7–dependent to –independent stages of B cell development, BCL6 is activated, reaches expression levels resembling those in GC B cells, and protects pre–B cells from DNA damage–induced apoptosis during immunoglobulin (Ig) light chain gene recombination. In the absence of BCL6, DNA breaks during Ig light chain gene rearrangement lead to excessive up-regulation of Arf and p53. As a consequence, the pool of new bone marrow immature B cells is markedly reduced in size and clonal diversity. We conclude that negative regulation of Arf by BCL6 is required for pre–B cell self-renewal and the formation of a diverse polyclonal B cell repertoire.
BCL6 functions as a transcriptional repressor (Chang et al., 1996; Seyfert et al., 1996; Shaffer et al., 2000) in normal and malignant germinal center (GC) B cells, and belongs to the BTB/POZ (Bric-à-brac, tramtrack, broad complex/Pox virus zinc finger) zinc finger family of proteins. In diffuse large B cell lymphomas (DLBCLs), BCL6 is frequently translocated into the Ig heavy or light chain loci (e.g., t(3;14)(q27;q32); Ye et al., 1993). During normal B cell development, BCL6 expression was only found in GC B cells (Cattoretti et al., 1995; Allman et al., 1996), in which BCL6 is critical for survival and proliferation. In the absence of BCL6, GC formation is abrogated (Dent et al., 1997; Ye et al., 1997). This is mainly attributed to the central negative regulatory effect of BCL6 on DNA damage response genes in GC B cells (Ranuncolo et al., 2007). Through somatic hypermutation and DNA double-strand break (DSB) events resulting from class-switch recombination in GCs combined with replication errors owing to a high proliferation rate, GC B cells are exposed to a high level of DNA damage stress (Schlissel et al., 2006; Liu et al., 2008). Therefore, the ability of BCL6 to suppress DNA damage response and checkpoint genes (Shaffer et al., 2000; Shvarts et al., 2002; Phan and Dalla-Favera, 2004; Phan et al., 2005, Ranuncolo et al., 2008) as well as the DNA damage sensor ATR (Ranuncolo et al., 2007) is essential for GC B cell proliferation and survival.
Extensive DNA damage not only occurs in GCs but also during early B cell development in the bone marrow (Schlissel et al., 2006). However, previous studies focused on the function of BCL6 within GCs, and a role of BCL6 in early B cell development was not examined in detail. Non-GC B cells, such as pre–B cells, sustain DNA damage owing to DNA DSBs during V(D)J recombination and replication errors linked to their high proliferation rate. In pre–B cells, DNA DSBs during V(D)J recombination first target one DH and JH and then multiple VH segments. This is followed by Vκ-Jκ gene rearrangement and potentially multiple additional rearrangements targeting the κ-deleting element (KDE) and a Jκ-Cκ intron element or an upstream Vκ segment, before Vλ and Jλ gene segments are ultimately rearranged (Rolink et al., 1993). V(D)J recombination–induced DSBs activate a multifunctional expression program (Bredemeyer et al., 2008), including proapoptotic pathways (Guidos et al., 1996) that can promote cell death unless survival pathways are concomitantly activated. It is currently unclear through which mechanisms pre–B cells are protected against extensive DNA damage stress caused by DSB events during V(D)J recombination at three chromosomal regions (VH-DJH at 14q32; Vκ-Jκ, KDE-Jκ intron, and KDE-Vκ at 2p12; and Vλ−Jλ at 22q11). Searching for factors that could protect pre–B cells from DNA damage–induced apoptosis, we established two in vitro systems for inducible pre–B cell differentiation and Ig κ and λ light chain gene rearrangement (Rolink et al., 1993; Muljo and Schlissel, 2003; Klein et al., 2005). These cell-culture systems allow for a detailed analysis of the differentiation of large cycling pre–BII cells (i.e., fraction C’; Rolink et al., 1995; Hardy and Hayakawa, 2001) to small resting pre–BII cells (i.e., fraction D) and ultimately to the immature B cell stage (i.e., fraction E). In this paper, we demonstrate that BCL6 function is critical for pre–B cell survival at the transition from IL-7–dependent to –independent stages of B cell development.
RESULTS
BCL6 is up-regulated during Ig light chain gene rearrangement
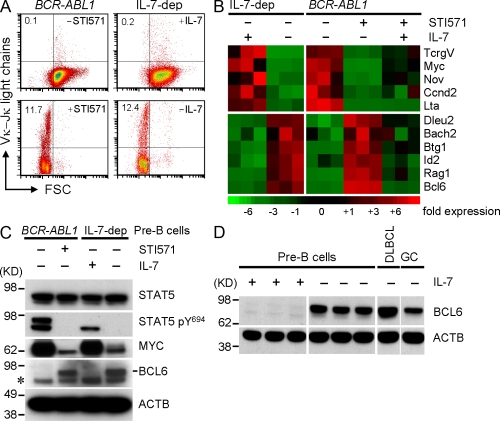
To initiate Vκ-Jκ and Vλ-Jλ gene rearrangements in pre–B cells and to study factors that protect pre–B cells against DSB-mediated DNA damage stress during these recombination events, we used two cell-culture systems that were previously described by us and others (Rolink et al., 1993; Muljo and Schlissel, 2003; Klein et al., 2005). Although a recent study demonstrated that IL-7–Stat5 signaling prevents premature Ig light chain rearrangement in pro–B cells (Malin et al., 2010), withdrawal of IL-7 from pre–B cell cultures strongly induces differentiation and Ig light chain gene rearrangement (Rolink et al., 1993). More recently, pharmacological inhibition of v-ABL1 and BCR-ABL1 in transformed pre–B cells was demonstrated to have the same effect (Muljo and Schlissel, 2003; Klein et al., 2005). In this study, we confirmed that withdrawal of IL-7 from IL-7–dependent mouse pre–B cells and inhibition of BCR-ABL1 kinase activity in BCR-ABL1–transformed pre–B cells induced the transition from large cycling pre–B cells to small resting pre–B cells, with subsequent expression of κ light chains on the cell surface (Fig. 1 A). Inhibition of ABL1 kinase activity in transformed pre–B cells recapitulates the pre–B to immature B cell transition, including sequential initiation of DSB events at the Vκ, KDE, and Vλ gene segments (Muljo and Schlissel, 2003; Klein et al., 2005). These experiments were performed using IL-7–dependent and BCR-ABL1–transformed pre–B cells from SLP65-deficient mice. In the presence of IL-7 or upon transformation by BCR-ABL1, SLP65-deficient B cell precursors give rise to large cycling pre–BII cells, whereas IL-7–dependent and BCR-ABL1–transformed SLP65 wild-type cells are arrested at the pro–B/pre–BI cell stage of development. To search for factors that are up-regulated in response to induced Ig light chain gene rearrangement in pre–B cells as potential protective factors against DSB-induced DNA damage stress, we performed Affymetrix GeneChip analyses of IL-7–dependent and BCR-ABL1–transformed pre–B cells before and after inhibition of IL-7 and BCR-ABL1 kinase signaling (Fig. 1 B). IL-7 withdrawal and BCR-ABL1 kinase inhibition by STI571 (10 µmol/liter STI571 for 16 h) resulted in very similar gene expression changes (Fig. 1 B). Some of these gene expression changes can be attributed to dephosphorylation of STAT5-Y694 (Walker et al., 2007), at which both the IL-7 and (BCR-) ABL1 signaling pathways converge (Banerjee and Rothman, 1998; Fig. 1 C). When genes were sorted based on fold increase after induced differentiation, BCL6 ranked first in the analysis (Fig. 1 B). Of note, the MYC protooncogene was among the genes on the opposite extreme of this analysis. Silencing of MYC and de novo expression of BCL6 upon inhibition of IL-7 or BCR-ABL1 signaling was confirmed at the protein level by Western blot analysis and correlated with STAT5 dephosphorylation at Y694 (Fig. 1 C). BCL6 is expressed at very high levels in GC B cells and serves a critical role in GC B cell survival (Dent et al., 1997; Ye et al., 1997; Phan and Dalla-Favera, 2004). Likewise, BCL6 functions as a protooncogene in DLBCL cells, where it is often expressed at very high levels owing to the BCL6-IGH translocation (t(3;14)(q27;q32); Ye et al., 1993). For these reasons, we studied BCL6 protein levels in pre–B cells upon IL-7 withdrawal as compared with GC B cells and DLBCLs by Western blotting (Fig. 1 D). Of note, withdrawal of IL-7 resulted in dramatic up-regulation of BCL6 protein expression, which reached levels comparable to both DLBCLs and GC B cells.
Figure 1.
Regulation of BCL6 during inducible pre–B cell differentiation. (A) IL-7–dependent and BCR-ABL1–transformed pre–B cells were induced to differentiate by withdrawal of 10 ng/ml IL-7 and ABL1 kinase inhibition (2 µmol/liter STI571), respectively. Cell size (FSC) and κ light chain surface expression were monitored by flow cytometry (n = 5). Numbers indicate percentages. (B) To identify genes that are differentially regulated during induced pre–B cell differentiation, we studied pre–B cells stimulated to differentiate in a microarray analysis. Genes were sorted based on the ratio of gene expression values observed upon withdrawal of IL-7 from IL-7–dependent pre–B cells. (C) Likewise, protein lysates from pre–B cells in the presence or absence of induced differentiation (treatment with 10 µmol/liter STI571 or withdrawal of IL-7 for 24 h) were analyzed by Western blotting using antibodies against STAT5, phosphorylated STAT5 at Y694, BCL6 (clone N3), MYC, and an ACTB antibody as loading control (n = 6). The asterisk denotes a nonspecific band that is consistently observed with the N3 BCL6 antibody. Of note, BCR-ABL1 kinase signaling results in stronger STAT5 tyrosine phosphorylation at Y694 and detection of two phosphoproteins compared with IL-7–dependent STAT5 phosphorylation, where only one band is detected (see D). (D) To directly compare BCL6 protein expression levels in pre–B cells upon IL-7 withdrawal and DLBCL cells and tonsillar GC B cells, Western blot analyses were repeated with cell lysates from these cell populations (cell lysates from three IL-7 withdrawal experiments). FSC, forward scatter.
The balance between MYC and BCL6 regulates Vκ-Jκ light chain gene recombination
To investigate whether these in vitro observations are relevant for mechanisms of pre–B cell differentiation in vivo, we measured the mRNA levels of BCL6 and MYC at various stages of B lymphopoiesis isolated from primary mouse bone marrow (Fig. S1). To this end, hematopoietic progenitor cells (HPCs; c-kit+ Sca-1+), pro–B cells (c-kit+ B220+; fractions B and C), large pre–BII cells (CD25+ B220low; fraction C’), and small pre–BII cells (fraction D), as well as immature B cells (B220low IgM+; fraction E), were isolated and studied by quantitative RT-PCR for BCL6 and MYC mRNA levels (Fig. 2 A). In most subsets of early B lymphopoiesis, MYC mRNA levels were significantly higher when compared with BCL6. In particular, the ratio of BCL6/MYC mRNA levels was very low in large cycling pre–BII cells (fraction C’; ratio = 0.1). Strikingly, however, this ratio increased by >150-fold in small resting pre–BII cells (fraction D; ratio = 18). Given the role of MYC in cell-cycle regulation, lower mRNA levels of MYC in small resting compared with large cycling pre–BII cells were expected. The finding of a 20-fold up-regulation of BCL6 in small resting compared with large cycling pre–BII cells is novel albeit consistent with the cytokine context of pre–B cells at this differentiation stage: pre–BII cells transitioning from the large cycling to the small resting stage down-regulate expression of the IL-7 receptor (Hardy and Hayakawa, 2001; Johnson et al., 2008). Withdrawal of IL-7 receptor signaling results in strong de novo expression of BCL6, as demonstrated in Fig. 1, which correlates with dephosphorylation of STAT5 and down-regulation of MYC (Fig. 1 C). The inverse relationship of BCL6 and MYC expression is consistent with our finding that BCL6 is recruited and directly binds to the MYC promoter in BCR-ABL1–transformed pre–B cells (Fig. 2 B), suggesting that BCL6 functions as a transcriptional repressor of MYC. In addition to BCL6, expression levels of MYC are also negatively regulated by STI571 itself, i.e., via dephosphorylation of STAT5.
Figure 2.
The balance between MYC and BCL6 regulates Vκ-Jκ light chain gene recombination. (A) HPCs, pro–B cells (fractions B and C), large cycling pre–BII cells (fraction C’), small resting pre–BII cells (fraction D), and immature B cells (fraction E) were sorted from normal mouse bone marrow (n = 3) and subjected to quantitative RT-PCR to measure mRNA levels of BCL6 and MYC relative to Hprt. Mean values ± SD of three experiments are given. Numbers in the bar chart denote the ratios of BCL6 versus MYC mRNA levels. (B) Recruitment of BCL6 to the MYC promoter was identified by ChIP-on-chip analysis and confirmed by single locus QChIP analysis. ChIP-on-chip analysis was performed for three BCR-ABL1–transformed pre–B cell lines (each two replicates) under control conditions or after treatment with 10 µmol/liter STI571 for 24 h. Two replicate experiments for one BCR-ABL1–transformed pre–B ALL cell line are shown. (C and D) IL-7–dependent (C) and BCR-ABL1–transformed (D) pre–B cells were induced to differentiate by IL-7 withdrawal or inhibition of BCR-ABL1 kinase activity. To test the function of MYC and BCL6 during induced pre–B cell differentiation, pre–B cells were transduced with retroviral vectors encoding BCL6-IRES-GFP, MYC-IRES-GFP, or an IRES-GFP empty vector control. Percentages of κ light chain+ GFP+ cells are indicated (mean values of three experiments ± SD). (E) IL-7–dependent bone marrow pre–B cells from MYCfl/fl and MYCfl/fl × mx-Cre mice were transduced with retroviral vectors encoding BCL6-IRES-GFP or an IRES-GFP (empty vector control) and cultured in the presence of 10 ng/ml IL-7. 2,500 U/ml IFN-β was added to all cultures to induce expression of Cre and MYC deletion in Mx-Cre × MYCfl/fl but not MYCfl/fl pre–B cells (mean values ± SD; n = 3).
To study the functional significance of concomitant up-regulation of BCL6 and down-regulation of MYC during induced pre–B cell differentiation and Ig light chain gene recombination, we performed gain- and loss-of-function experiments (Fig. 2, C–E). In one set of experiments, IL-7– and BCR-ABL1–driven pre–B cells were transduced with retroviral BCL6 and MYC vectors or a GFP empty vector control. Upon induced pre–B cell differentiation, the pool of κ light chain–expressing cells was significantly increased by BCL6 overexpression (Fig. 2, C and D). It should be noted that overexpression of BCL6 likely results in a competitive disadvantage of the BCL6-GFP–transduced cells. As shown in transformed pre–B cells (Fig. 2 B) and, previously, in DLBCLs (Ci et al., 2009), BCL6 functions as a transcriptional repressor of MYC and may, hence, contribute to the quiescent phenotype of small resting pre–BII cells (BCL6high MYClow) as opposed to large cycling pre–BII cells (BCL6low MYChigh; Fig. 2 A).
In contrast, overexpression of MYC reduced the frequency of κ light chain+ cells as compared with GFP empty vector controls in BCR-ABL1–transformed pre–B cells (treatment with STI571; Fig. 2 D). Also, IL-7–dependent pre–B cells showed a trend toward MYC-induced reduction of κ light chain+ cells, which, however, was not statistically significant. Collectively, these findings suggest that BCL6 and MYC have opposite effects on the regulation of pre–B cell differentiation. To test this possibility in a genetic experiment, we combined retroviral overexpression of BCL6 with conditional deletion of MYC in IL-7–dependent pre–B cells from MYCfl/fl mice (Fig. 2 E). Overexpression of BCL6 alone and conditional deletion of MYC alone already had a statistically significant, albeit subtle, effect on MYCfl/fl pre–B cells in the presence of IL-7. Strikingly, however, combined overexpression of BCL6 with Cre-mediated deletion of MYC strongly promoted differentiation of pre–B cells into κ light chain–producing immature B cells, even in the presence of IL-7 (Fig. 2 E). The results demonstrate that the balance between BCL6 and MYC determines the large cycling (fraction C’) versus small resting (fraction D) phenotypes of pre–BII cells. We conclude that the genetically induced inversion of the BCL6/MYC ratio in pre–BII cells in this experiment is required and sufficient to recapitulate the effect of IL-7 withdrawal on pre–B cell differentiation.
Pre–B cell receptor activation induces expression of BCL6 via down-regulation of IL-7 responsiveness
In Fig. 1 C, we observed that up-regulation of BCL6 correlated with dephosphorylation of STAT5 Y694 upon withdrawal of IL-7 receptor signaling. This finding suggests that active STAT5 (phosphorylated at Y694) functions as a negative regulator of BCL6 expression downstream of the IL-7 receptor. To mechanistically address this possibility, IL-7–dependent pre–B cells were transduced with a retroviral vector encoding a constitutively active STAT5 mutant (Onishi et al., 1998) or an empty vector control. Withdrawal of IL-7 resulted in both dephosphorylation of STAT5 and up-regulation of BCL6 in pre–B cells that were transduced with an empty vector. In contrast, pre–B cells transduced with the constitutively active STAT5 mutant maintained some STAT5 phosphorylation even after IL-7 withdrawal, and up-regulation of BCL6 was greatly attenuated (Fig. 3 A). This finding demonstrates that active STAT5 functions as a negative regulator downstream of the IL-7 receptor. To verify that withdrawal of IL-7 not only results in BCL6 up-regulation in long-term IL-7–dependent pre–B cell cultures, we used a neutralizing anti–IL-7 antibody to inactivate IL-7 receptor signaling in freshly ex vivo–isolated (B220+ MACS) bone marrow B cell precursors (Fig. 3 B). Although both populations strongly up-regulated BCL6 in response to anti–IL-7 antibody treatment, the antibody had a stronger effect in long-term pre–B cell cultures than in freshly isolated ex vivo B220+ bone marrow cells. This difference likely reflects a higher degree of enrichment for IL-7–responsive cells in long-term IL-7 pre–B cell cultures than in B220+ bone marrow cells (Fig. 3 B).
Figure 3.
Pre–B cell receptor activation induces expression of BCL6 via down-regulation of IL-7 responsiveness. IL-7–dependent pre–B cells were transduced with a retroviral vector encoding a constitutively active STAT5 mutant (STAT5-CA-GFP; Onishi et al., 1998) or an empty vector control (GFP). (A) GFP-expressing transduced cells were sorted, subjected to IL-7 withdrawal, and analyzed by Western blotting for expression of BCL6 and tyrosine phosphorylation of STAT5 using β-actin as a loading control (n = 3). (B) Mouse B cell precursors were isolated by B220+ MACS from freshly harvested bone marrow cells. Freshly isolated bone marrow B cell precursors and IL-7–dependent pre–B cell cultures were treated with 10 µg/ml of a neutralizing anti–IL-7 antibody overnight and were subjected to Western blot analysis (three experiments were performed). (C) Bone marrow B cell precursors from RAG2−/− tTA/μ chain–transgenic mice are unable to express an endogenous μ chain but carry a functionally prerearranged μ chain under control of tetO sequences (Hess et al., 2001). These mice express a tTA under control of endogenous μ chain regulatory elements, and withdrawal of tetracycline results in activation of μ chain expression (routinely performed quality control). The effect of tetracycline-inducible activation of μ chain expression on BCL6 expression and STAT5 tyrosine phosphorylation was determined by Western blotting (n = 3; right). (D) IL-7–dependent pre–B cells lacking the pre–B cell receptor–related linker molecule SLP65 were transduced with retroviral expression vectors encoding either SLP65-GFP or GFP alone. Surface expression levels of IL-7Rα chain were measured by flow cytometry (the experiment was performed twice). The histogram shows the IL-7Rα levels in transduced GFP+ (green) and untransduced (gray) cells.
Although these findings establish that IL-7Rα–STAT5 signaling negatively regulates BCL6 expression in pre–BII cells, it is not clear how BCL6 expression can be activated in these cells. To address this question, we used a model in which inducible de novo synthesis of the pre–B cell receptor recapitulates the transition of pro–B cells (fractions B and C) into large cycling pre–BII cells (after 2 d; fraction C’) and then small resting pre–BII cells (after 4 d; fraction D): pro–B cells from RAG2−/− tetracycline-controlled transactivator (tTA)/μ chain–transgenic mice (Hess et al., 2001) are unable to express an endogenous μ chain because of a lack of RAG2-dependent V(D)J recombination. Although these pro–B cells carry their endogenous Ig heavy chain loci in germline configuration, they also harbor a functionally prerearranged μ chain under the control of tetracycline operator (tetO) sequences. In addition, RAG2−/− tTA/μ chain–transgenic mice express a tTA under control of endogenous μ chain regulatory elements. We verified induction of μ chain expression in RAG2−/− tTA/μ chain–transgenic IL-7–dependent pro–B cell cultures by flow cytometry (Fig. 3 C). In agreement with previously published data (Schuh et al., 2008), we observed that induction of μ chain expression in RAG2−/− tTA/μ chain–transgenic pro–B cells results in down-regulation of CD43 and up-regulation of CD25, which is consistent with a pro–B to pre–B cell transition (Hardy and Hayakawa, 2001). 2 d after induction of μ chain expression, the cells had assumed a large cycling phenotype (fraction C’), followed by cell-cycle exit and reduction of cell size (small resting pre–BII cell; fraction D; Trageser et al., 2009). A Western blot analysis performed with cells that were harvested after 4 d of μ chain induction showed strong up-regulation of BCL6 together with near complete dephosphorylation of STAT5 (Fig. 3 C). These findings demonstrate that the transition from IL-7Rα–STAT5 signaling in large cycling pre–BII cells (fraction C’) to BCL6 expression in small cycling pre–BII cells (fraction D) is initiated by the pre–B cell receptor. The finding of pre–B cell receptor–mediated down-regulation of IL-7Rα–STAT5 signaling is consistent with a recent study, which demonstrated that IL-7Rα–STAT5 signaling prevents premature Ig light chain gene recombination in pro–B cells (Malin et al., 2010). Experiments to delineate the details of pre–B cell receptor–dependent activation of BCL6 are currently under way. In this context it is noteworthy that reconstitution of the pre–B cell receptor linker molecule SLP65 in SLP65−/− pre–B cells results in down-regulation of IL-7Rα surface expression (Fig. 3 D) and reduced IL-7 responsiveness (Schebesta et al., 2002). In addition, a recent study showed that pre–B cell receptor–dependent activation of SLP65 leads to dephosphorylation of STAT5 via inhibition of JAK3 (Nakayama et al., 2009). We conclude that BCL6 activation in B cell precursors is naturally confined to the pre–B cell compartment and is induced after de novo synthesis of the pre–B cell receptor based on productively rearranged VH-DJH gene segments. We propose that down-regulation of IL-7 responsiveness represents an important aspect through which pre–B cell receptor signaling induces expression of BCL6 in small resting pre–BII cells.
BCL6 is required for normal polyclonal B lymphopoiesis
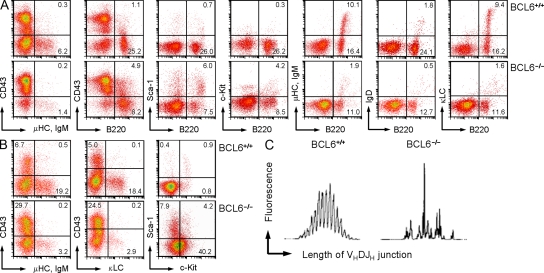
To investigate the physiological relevance of BCL6 during early B cell development, we performed a detailed flow cytometry analysis of bone marrow samples from age-matched BCL6−/− and BCL6+/+ mice (n = 4). Although the total number of B220+ B cell lineage cells was only slightly reduced in BCL6−/− compared with wild-type bone marrow, the frequency of IgM+ and κ light chain+ immature B cells was significantly diminished (Fig. 4, A and B; statistical analysis in Table S1). Compared with wild-type cells, c-kit+ B220+ (fraction B) and CD43+ B220+ (fraction C) pro–B cell populations were slightly expanded in BCL6−/− bone marrow. In agreement with previous studies (Dent et al., 1997; Ye et al., 1997), κ light chain–expressing immature B cells (fraction E) were detected in BCL6−/− bone marrow, but their frequency was approximately fivefold lower than in wild-type mice (P = 0.03; Table S1).
Figure 4.
Normal polyclonal B lymphopoiesis requires BCL6 survival signaling in late pre–B cells. Bone marrow mononuclear cells from BCL6+/+ and BCL6−/− mice were analyzed by flow cytometry using the indicated antibody combinations (numbers indicated percentages). (A) Bone marrow cells were analyzed using a FSC/SSC gate together with propidium iodide exclusion (viable lymphocytes). (B) Cells were analyzed using a B220+ gate. Four mice were studied in each group, and a detailed statistical analysis including absolute cell numbers is presented in Table S1. (C) To examine the clonal composition of κ+ immature B cells (fraction E), B220+ IgD− κ+ cells were sorted and analyzed by Ig spectratyping. Spectratyping analysis separates Ig gene rearrangements based on the length of their VH-DJH junction, and the height (fluorescence intensity) of each size peak indicates its relative representation within the B cell population analyzed. Individual size peaks are typically separated by 3 bp, reflecting the reading frame in functional VH-DJH gene rearrangements. In total, four pairs of BCL6+/+ and BCL6−/− bone marrow–derived immature B cell samples were analyzed by spectratyping. FSC, forward scatter; SSC, side scatter.
To study the clonal composition of IgM+ κ+ bone marrow immature B cells (fraction E) that developed in the absence of BCL6, we sorted 8,000 B220+ IgD− κ+ bone marrow cells from both BCL6−/− and BCL6+/+ bone marrow and analyzed them by spectratype analysis. Although wild-type B220+ IgD− κ+ bone marrow immature B cells exhibit a broad polyclonal repertoire, the profile of BCL6−/− bone marrow immature B cells was oligoclonal (Fig. 4 C). These findings are based on sorted immature B cells from four pairs of BCL6+/+ and BCL6−/− mice, and suggest that BCL6 is required for normal polyclonal B cell production. To verify the clonal composition of the primary B cell repertoire in BCL6+/+ and BCL6−/− mice, we performed a detailed sequence analysis for two pairs of mice where immature B cells from bone marrow and mature B cells from spleens were available for cell sorting from the same animals (Fig. 5). Interestingly, in two BCL6−/− mice, spectratyping identified clonal expansions with the same peak size (i.e., the same length of the VH-DJH junction; Fig. 5 A) that were amplified from both the bone marrow and the spleen of these mice. This finding raises the question of whether the clonal expansions among bone marrow immature B cells and mature splenic B cells in BCL6−/− mice are derived from the same dominant clone.
Figure 5.
BCL6 is required for the development of a diverse primary B cell repertoire. (A) For two pairs of BCL6+/+ and BCL6−/− mice (I and II), in addition to bone marrow samples spleen tissue was also available. Immature B cells (fraction E) from bone marrow and mature B cells from spleens (fraction F) were sorted and subjected to spectratyping analysis to study length diversity of VH-DJH junctions. In two BCL6−/− mice, clonal expansions were identified that occurred both in the bone marrow and in the spleen with the same peak size, i.e., the same length of the VH-DJH junction. Two such clonal expansions were found in BCL6−/− mouse I (I.1 and I.2) and one expansion was found in BCL6−/− mouse II (II.1). (B) To test whether these expansions with the same length of the VH-DJH junction indeed belong to one clone, we performed a detailed sequence analysis of these VH-DJH junctions. Alignments of VH, D, and JH gene segments are shown as well as N/P nucleotides in the junctional regions. Internal heptamers are highlighted in red, and sites at which the junctional homology with the parental VH-DJH rearrangement ends are indicated by arrows.
To test this possibility, we performed a comprehensive sequence analysis of VH-DJH junctions in bone marrow immature B cells and splenic mature B cells from BCL6+/+ and BCL6−/− mice, respectively (Fig. 5 B and Table S2). Consistent with the finding of a restricted B cell repertoire in BCL6−/− mice (Fig. 4 C and Fig. 5 A), we amplified multiple clonally related sequences from BCL6−/− but not from BCL6+/+ mice (available from GenBank/EMBL/DDBJ under accession nos. FN652762, FN652763, FN652764, FN652765, FN652766, FN652767, FN652768, FN652769, FN652770, FN652771, FN652772, FN652773, FN652774, FN652775, FN652776, FN652777, and FN652778; Table S2). In addition, clonal expansions within the bone marrow immature B cell and mature splenic B cell pool in BCL6−/− mice carried the same VH-DJH junction and, hence, share the same clonal origin (Fig. 5 B). Two large clones were amplified from the bone marrow and spleen of BCL6−/− mouse #I, and one clonal expansion in the bone marrow of BCL6−/− mouse #II was also recovered from the spleen. These findings further illustrate that even though the size of a peripheral B cell pool may be normal in BCL6−/− mice (Dent et al., 1997; Ye et al., 1997), the primary B cell repertoire in BCL6−/− mice is restricted to a small number of dominant clones.
A more detailed analysis of VH-DJH junctions revealed that many other seemingly unique VH-DJH rearrangements amplified from BCL6−/− mature splenic B cells in fact harbor the same D-JH junction (gray shading in Table S2) but have rearranged different VH gene segments. These clones are derived from one parental bone marrow immature B cell clone but subsequently diversified in the spleen by VH replacement, i.e., by substitution of the initially rearranged VH segment by a VH segment located upstream. VH replacement (Reth et al., 1986) involves the recombination signal sequence (RSS) of a nonrearranged VH gene and a cryptic RSS (i.e., an isolated heptamer) within the preexisting VHDJH joint. The cryptic RSS (highlighted in red in Fig. 5 B) is located at the 3′ end of the majority of VH gene segments in mice and humans (Radic and Zouali, 1996). The spectratyping profiles shown in Fig. 5 A seem to suggest that the extreme oligoclonality observed among BCL6−/− bone marrow immature B cells is alleviated to some degree within the pool of BCL6−/− mature splenic B cells. It is likely, however, that many of these additional clones have diversified by VH gene replacement, share the same D-JH junction, and are, hence, derived from a very small number of immature B cells that have left the bone marrow. We conclude that the ability of small resting pre–BII cells to up-regulate BCL6 is critical for the development of a diverse primary B cell repertoire.
BCL6 is required for self-renewal signaling at the pre–B cell stage
Previous work demonstrated that a fraction of BCL6−/− mice suffers from chronic inflammation (Dent et al., 1997; Ye et al., 1997), which may affect the composition of the B cell progenitor populations within the bone marrow. In the animals we analyzed in this study, no signs of inflammatory disease were noted. In addition, we tested the ability of BCL6−/− pre–B cells to differentiate under cell-culture conditions, which excludes potential side effects from inflammation (Fig. S2). We first incubated bone marrow samples from BCL6+/+ and BCL6−/− mice in the presence of 10 ng/ml IL-7, which selectively induces proliferation of bone marrow pre–B cells. In the presence of high IL-7 concentrations, the majority of the bone marrow HPCs die, whereas B cell lineage cells vigorously proliferate (Fig. S2 A). We verified that the IL-7–dependent and BCR-ABL1–transformed cells used are indeed pre–B cells based on positive intracellular staining for µ chain and CD25 expression (Fig. S2 B). IL-7–dependent pre–B cell proliferation was robustly induced in bone marrow samples from BCL6+/+ mice, and pre–B cell cultures can be expanded for up to 2 mo until pre–B cell cultures become senescent (unpublished data). In striking contrast, BCL6−/− pre–B cells consistently failed to propagate ex vivo even in the presence of 10 ng/ml IL-7. Although wild-type pre–B cells start to vigorously expand on day 3, no viable B cell precursors can be detected in IL-7 cultures from BCL6−/− bone marrow (Fig. S2 A). Because BCL6−/− pre–B cells failed to differentiate and died upon IL-7 withdrawal even after short-term culture (Fig. S2 A), we studied differentiation in BCR-ABL1–transformed pre–B cells, as described in Fig. 1 A. Compared with wild-type pre–B cells, transformation of fresh BCL6−/− pre–B cells was significantly delayed, cells proliferated at a reduced rate, and in some instances transformation failed entirely (unpublished data). However, we found after successful BCR-ABL1 transformation of BCL6+/+ and BCL6−/− cells that inhibition of BCR-ABL1 kinase activity induced differentiation into κ+ immature B cells in 3% of wild-type pre–B cells within 3 d, whereas no significant differentiation but markedly enhanced cell death was observed in BCL6−/− pre–B cells (Fig. S3 A).
BCL6 protects pre–B cells against apoptosis during Vκ-Jκ recombination
We next compared V(D)J recombinase activity in BCL6−/− and wild-type pre–B cells before and after induced differentiation. To this end, we transduced BCR-ABL1–transformed pre–B cells with a retrovirus carrying an inverted GFP cassette flanked by RSS as in Vκ and Jκ gene segments. Upon inversion-mediated recombination of the RSS-flanked GFP, the GFP cassette will be in the correct orientation and expressed from the retroviral long terminal repeat (Fig. S3 B). Equal numbers of BCL6−/− and wild-type pre–B cells were transduced and subjected to selection for antibiotic resistance encoded by the RSS-GFP reporter construct (Wossning et al., 2006). We detected spontaneous V(D)J recombinase activity in both untreated BCR-ABL1–transformed BCL6+/+ and BCL6−/− pre–B cells, and the frequency of cells with recombinase activity was higher in the wild-type cells (∼20% compared with ∼2%; P < 0.01; Fig. S3 B). When BCR-ABL1–transformed pre–B cells were stimulated to differentiate by BCR-ABL1 kinase inhibition, the frequency of cells undergoing RSS-dependent recombination events increased in both BCL6−/− and wild-type pre–B cells. RSS-targeted recombination events were followed by de novo expression of κ light chains in wild-type pre–B cells. Strikingly, however, the frequency of κ+ cells did not significantly increase in STI571-treated BCL6−/− pre–B cells despite recombination activity in these cells (Fig. S3 B). This difference between BCL6−/− and BCL6+/+ pre–B cells could indicate that BCL6 is required for targeting of the V(D)J recombinase to the Ig κ light chain locus. Alternatively, BCL6 may have a role in the intracellular processing of κ light chains and their pairing with Ig heavy chains. Finally, Vκ-Jκ rearranging pre–B cells might undergo apoptosis in the absence of BCL6 before κ light chains can be expressed on the cell surface. To directly test the latter hypothesis, we measured the percentage of apoptotic cells (annexin V+) in BCL6+/+ and BCL6−/− pre–B cells with (RSS-GFP+) and without (RSS-GFP−) indication of V(D)J recombinase activity (Fig. S3 C). If lack of κ light chain expression on BCL6−/− pre–B cells was indeed linked to apoptosis during Vκ-Jκ rearrangement, one would expect that RSS-GFP+ cells were preferentially apoptotic in the case of BCL6−/− but not BCL6+/+ cells. Induction of differentiation by STI571 treatment resulted in a strong increase of V(D)J recombinase activity, both in BCL6+/+ and BCL6−/− pre–B cells (Fig. S3 C). Among BCL6+/+ cells, only a minority of V(D)J recombining pre–B cells (RSS-GFP+) was apoptotic in the presence (24% of all RSS-GFP+ cells) or absence (13% of RSS-GFP+ cells) of STI571-treatment. In contrast, however, no enrichment of viable cells was observed among BCL6−/− pre–B cells that underwent V(D)J recombination: approximately one half of BCL6−/− V(D)J recombining pre–B cells (RSS-GFP+) was apoptotic in the presence and absence of STI571 treatment (Fig. S3 C). In summary, STI571 treatment induced increased V(D)J recombinase activity and increased cell death both in BCL6+/+ and BCL6−/− cells. However, the outcome of STI571-induced V(D)J recombination activity differs between BCL6+/+ and BCL6−/− cells: in BCL6+/+ cells, STI571-induced recombinase activity results in higher output of κ+ cells. In BCL6−/− cells, STI571-induced recombination results in increased cell death (Fig. S3, B and C).
These findings suggest that DNA DSB events during Vκ-Jκ gene rearrangement are directly linked to apoptosis in BCL6−/− pre–B cells. To test this possibility, we induced differentiation in IL-7–dependent B cell precursors carrying a homozygous deletion of the RAG1 gene, which is required for V(D)J recombination. Upon withdrawal of IL-7, RAG1−/− B cell precursors undergo phenotypic changes consistent with differentiation, as in wild-type pre–B cells (Fig. S4), but fail to introduce DNA DSBs at Vκ- and Jκ genes. To test the role of BCL6 in this system, we used a peptidomimetic inhibitor of BCL6 (retro-inverso BCL6 peptide inhibitor [RI-BPI]; Cerchietti et al., 2009), which was recently developed for the treatment of GC-derived B cell lymphoma (Polo et al., 2004). Withdrawal of IL-7 had a similar effect in both RAG1−/− and wild-type B cell precursors, and induced phenotypic differentiation (down-regulation of CD43 and up-regulation of CD25) in most cells but apoptosis only in a small fraction (Fig. S4). Concomitant withdrawal of IL-7 and peptidomimetic inhibition of BCL6 increased the fraction of apoptotic cells in both Rag1−/− and wild-type B cell precursors. Although >50% of RAG1−/− B cell precursors survived simultaneous inhibition of IL-7 and BCL6 signaling, almost all cells in which DNA DSBs could be introduced during induced Vκ-Jκ gene rearrangement underwent apoptosis (annexin V+; Fig. S4). Likewise, concomitant inhibition of IL-7 and BCL6 compromised the survival of differentiating (CD25+ CD43−) subclones, unless introduction of DNA DSBs during V(D)J recombinase activity was prevented in RAG1−/− B cell precursors (Fig. S4). These striking differences indicate that BCL6 function in differentiating pre–B cells is mainly needed to counteract the apoptotic DNA damage response induced by DNA DSBs during Ig light chain gene rearrangement.
BCL6 is required for transcriptional repression of DNA damage response genes during Ig light chain recombination
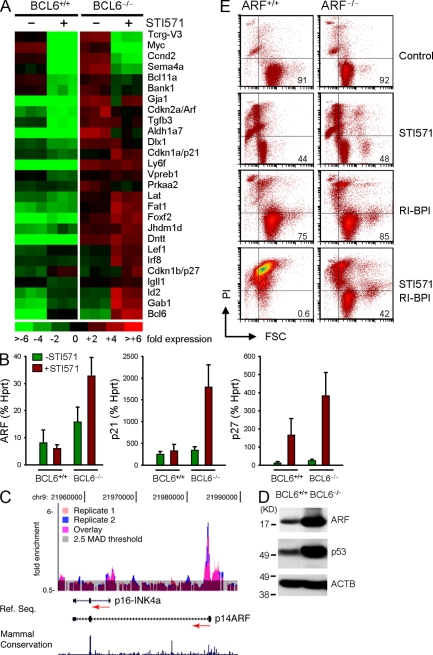
To elucidate the mechanism of BCL6-mediated pre–B cell survival signaling, we next investigated the gene expression pattern in BCL6−/− and BCL6+/+ pre–B cells (Fig. 6 A). Importantly, well-known cell-cycle checkpoint regulators (CDKN1A/p21, CDKN1B/p27, and CDKN2A/Arf) and inducers of cellular senescence (CDKN2A/Arf) are among the targets of BCL6-mediated transcriptional repression (Fig. 6, A and B), which directly supports a central role of BCL6 in pre–B cell self-renewal signaling and protection against DNA damage–induced apoptosis (e.g., during Vκ-Jκ gene rearrangement). These differences could be confirmed by quantitative RT-PCR for ARF, p21, and p27 (highlighted in Fig. 6, A and B). In the absence of BCL6, p21, p27, and ARF showed excessive up-regulation after STI571 treatment, and Arf mRNA levels were increased in BCL6-deficient pre–B cells even in the absence of induced differentiation. Although p21 and p27 were previously identified as target genes of BCL6-mediated transcriptional repression (Shaffer et al., 2000; Phan and Dalla-Favera, 2004; Phan et al., 2005), transcriptional silencing of ARF by BCL6 represents a novel finding. To determine whether BCL6 could bind to the ARF locus, we performed BCL6 chromatin immunoprecipitation (ChIP)–on-chip using a genomic microarray tiling across the entire CDKN2A locus (Fig. 6 C) in the human B cell lymphoma cell line OCI-Ly1. The CDKN2A gene contains two start exons (1α and 1β) that give rise to two alternative transcripts, p14ARF and p16INK4A. The ChIP-on-chip result demonstrated that BCL6 was strongly bound to sequences proximal to the p14ARF transcriptional start site (Fig. 6 C). ChIP-on-chip also showed that BCL6 also bound, albeit weakly, to sequences related to p16INK4a (Fig. 6 C). BCL6 binding to the p14ARF promoter was confirmed by performing additional quantitative ChIP (QChIP) assays using primers to amplify this site. BCL6 single-locus QChIP for CDKN2A/p14ARF showed 0.74 ± 0.2% of input for BCL6 compared with 0.03 ± 0.01% of input using a β-actin antibody as negative control (n = 3; P = 0.02).
Figure 6.
BCL6 promotes pre–B cell survival by negative regulation of ARF. (A) BCR-ABL1–transformed BCL6+/+ and BCL6−/− pre–B cells were induced to differentiate (10 µmol/liter STI571) and studied by microarray analysis. Genes were sorted based on the ratio of gene expression values in STI571-treated BCL6+/+ compared with BCL6−/− pre–B cells. (B) Differences in mRNA levels of checkpoint genes (ARF, p21, and p27) were verified by quantitative RT-PCR using HPRT as a reference (mean values ± SD; n = 3). (C) Recruitment of BCL6 to the genomic region of the CDKN2A locus (ARF) was verified by ChIP-on-chip analysis and confirmed by single locus QChIP analysis (n = 3). BCL6–DNA complexes were immunoprecipitated using an anti-BCL6 polyclonal antibody. ChIP products and their respective input genomic fragments were amplified by ligation-mediated PCR. QChIP was performed again at this stage for selected positive control loci to verify that the enrichment ratios were retained (not depicted). The genomic products of two biological ChIP replicates were labeled with Cy5 (for ChIP products) and Cy3 (for input) and cohybridized on genomic tiling arrays including the genomic region of the CDKN2A locus (Chr9:21,950629–22,003329). After hybridization, the relative enrichment for each probe was calculated as the signal ratio of ChIP to input. Peaks of enrichment for BCL6 relative to input were uploaded as custom tracks into the University of California, Santa Cruz genome browser and graphically represented as histograms. Peaks involving five or more oligonucleotide probes above threshold (2.5-fold SD above average enrichment) were considered positive. (D) To determine whether recruitment of BCL6 to the Arf (CDKN2A) promoter affects expression levels of proteins in the Arf–p53 pathway, BCR-ABL1–transformed BCL6+/+ and BCL6−/− pre–B cells were analyzed by Western blotting. (E) BCR-ABL1–transformed pre–B cells from ARF+/+ and ARF−/− mice were induced to differentiate (+STI571) or cultured under control conditions in the presence or absence of 5 µmol/liter of a peptidomimetic BCL6 inhibitor (RI-BPI; Cerchietti et al., 2009). Cell size (FSC) and viability (propidium iodide uptake) were measured by flow cytometry after 1 and 3 d (n = 3). Percentages of viable cells are indicated for each condition. FSC, forward scatter; PI, propidium iodide.
BCL6 promotes pre–B cell survival by negative regulation of ARF
Consistent with the identification of CDKN2A (ARF) as a direct transcriptional target of BCL6, we found that protein levels of both Arf and its downstream effector p53 are excessively increased in BCR-ABL1–transformed BCL6−/− pre–B cells (Fig. 6 D). To investigate the functional significance of transcriptional repression of ARF by BCL6, we studied the role of ARF in a genetic loss-of-function model. We tested whether pre–B cell apoptosis resulting from inhibition of BCL6 is in part mediated by excessive activation of Arf. To this end, we induced differentiation in wild-type and Arf-deficient BCR-ABL1 pre–B cells by treatment with STI571 in the presence or absence of concomitant BCL6 inhibition (RI-BPI; Fig. 6 E). Induced differentiation (STI571) or inhibition of BCL6 (RI-BPI) alone had little effect on the viability of both wild-type and ARF−/− pre–B cells. Simultaneous induction of differentiation and inhibition of BCL6, however, induced apoptosis in the vast majority of wild-type pre–B cells (viability < 1%). In contrast, a significant portion of ARF−/− pre–B cells survived concomitant induction of differentiation and inhibition of BCL6 (viability > 40%; Fig. 6 E). We conclude that de novo expression of BCL6 at the transition from IL-7–dependent to –independent stages of B cell development is required to promote pre–B cell survival by negative regulation of Arf.
DISCUSSION
Based on our observations, we propose the following scenario of BCL6-mediated survival signaling at the transition from IL-7–dependent to –independent stages of B cell development (Fig. S5). Large cycling pre–BII cells express high levels of the IL-7 receptor (Fig. 3 D; Hardy and Hayakawa, 2001; Johnson et al., 2008), ligation of which induces tyrosine phosphorylation of JAK1, JAK3, and STAT5 (Palmer et al., 2008). STAT5 phosphorylation at Y694 leads to transcriptional suppression of BCL6 (Fig. 1 C and Fig. 3 A; Walker et al., 2007) and transcriptional activation of MYC, which in turn promotes cell-cycle progression via CCND2 (Bouchard et al., 2001). A recent study demonstrated that IL-7–STAT5 signaling in these cells actively prevents Ig light chain gene recombination (Malin et al., 2010). The Ig light chain loci IL-7–STAT5–dependent pro– and large cycling pre–BII cells are in germline configuration, and the level of DNA damage (e.g., DSB) is low (Fig. S5, left). Activation of pre–B cell receptor signaling in large cycling pre–BII cells, however, induces down-regulation of the IL-7 receptor (Fig. 3 D), down-regulation of IL-7 responsiveness (Schebesta et al., 2002; Johnson et al., 2008), and ultimately dephosphorylation of STAT5 (Fig. 3 C). Dephosphorylation of STAT5 may either result from reduced IL-7 responsiveness (Schebesta et al., 2002; Johnson et al., 2008) or interference of the pre–B cell receptor signaling molecule SLP65 with JAK3–STAT5 signal transduction (Nakayama et al., 2009). Termination of IL-7Rα–JAK1–STAT5 signaling leads to down-regulation of MYC-CCND2 and, hence, loss of the “large cycling” phenotype of pre–BII cells. At the same time, loss of IL-7Rα–JAK1–STAT5 signaling allows transcriptional activation of BCL6, which leads to further transcriptional repression of MYC-CCND2 (Fig. 2 B; Ci et al., 2009). The identification of MYC and CCND2 as transcriptional targets of BCL6 can explain that expression of MYC and BCL6 are mutually exclusive at the transition from large cycling pre–BII cells (MYC+ BCL6−) to small resting pre–BII cells (MYC− BCL6+).
After productive VH-DJH recombination, pre–B cell receptor signaling induces up-regulation of IRF4/8 and other factors needed for Ig light chain gene rearrangement (Schebesta et al., 2002; Johnson et al., 2008). In consequence, de novo expression of BCL6 upon IL-7 withdrawal coincides with initiation of Ig light chain gene rearrangement. Recombination events at Ig light chain loci (Vκ-Jκ, KDE-Jκ intron, and KDE-Vκ at 2p12; Vλ-Jλ at 22q11) involve multiple DNA DSBs and induce activation of DNA damage response genes and checkpoint genes, including ARF, p21, and p27, which are all negatively regulated by BCL6 (Fig. 6, A–D). In the absence of BCL6, Ig light chain gene rearrangement leads to excessive activation of DNA damage response and checkpoint genes (ARF, p53, p21, and p27; Fig. 6), and only a few small resting pre–BII cells survive the transition from IL-7–dependent to –independent B cell development. Based on the timing of its expression and its ability to curb an excessive DNA damage response during light chain gene recombination, BCL6 plays a central role in the generation of a diverse primary B cell repertoire.
MATERIALS AND METHODS
BCL6−/−, MYCfl/fl × mx-Cre, ARF (CDKN2A)−/−, SLP65−/−, and RAG1−/− mice.
Bone marrow from BCL6−/− mice (a gift from R. Dalla-Favera, Columbia University, New York, NY; Ye et al., 1997) was harvested, and bone marrow cells were cultured either in the presence of 10 ng IL-7/ml in RetroNectin-coated (Takara Bio Inc.) Petri dishes or retrovirally transformed by BCR-ABL1 as described in In vitro pre–B cell differentiation assays. Likewise, bone marrow B cell precursors from MYCfl/fl × mx-Cre and MYCfl/fl mice (Baena et al., 2005) were cultured in the presence of 10 ng/ml IL-7 on RetroNectin. Deletion of Myc was induced by IFN-β–mediated (2,500 U/ml; PBL Interferon Source) induction of mx-Cre activation. SLP65−/− pre–B cells were extracted from bone marrow of SLP65−/− mice (Jumaa et al., 1999). RAG1−/− and CDKN2A−/− mice were purchased from the Jackson Laboratory. All pre–B cells derived from bone marrow of mice were maintained in IMDM (Invitrogen) with GlutaMAX containing 20% fetal bovine serum, 100 IU/ml penicillin, 100 µg/ml streptomycin, 50 µM 2-mercaptoethanol, and 10 ng/ml of recombinant mouse IL-7 (PeproTech) at 37°C in a humidified incubator with 5% CO2. All mouse experiments were subject to approval by the Children’s Hospital Los Angeles Institutional Animal Care and Use Committee.
RAG2−/− tTA/μ chain–transgenic mice.
RAG2−/− tTA/μ chain–transgenic mice (Hess et al., 2001) are unable to express an endogenous μ chain because of a lack of RAG2-dependent V(D)J recombination but carry a functionally prerearranged μ chain under control of tetO sequences in the germline. In addition, RAG2−/− tTA/μ chain–transgenic mice express a tTA under control of endogenous μ chain regulatory elements.
In vitro pre–B cell differentiation assays.
Polyclonal pre–B cells were either propagated in the presence of 10 ng/ml IL-7 or transformed using a murine stem cell virus (MSCV) retrovirus encoding BCR-ABL1-IRES-GFP (Pear et al., 1998) or BCR-ABL1-IRES-Neo. Induction of differentiation was either induced by withdrawal of IL-7 (Rolink et al., 1993) or inhibition of BCR-ABL1 kinase activity using 2 µmol/liter STI571. STI571 (imatinib) was provided by Novartis. After 3 d of IL-7 withdrawal or STI571 treatment, successful induction of differentiation was verified by flow cytometry analysis of κ light chain surface expression.
Neutralization of IL-7 activity in ex vivo–isolated bone marrow B cell precursors.
Bone marrow was isolated from C57B/6J mice, and B cell lineage cells were isolated using B220+ MACS beads (Miltenyi Biotec). Viability and purity of isolated B cell lineage cells was verified by flow cytometry. B220+ cells were incubated overnight on OP9 stroma cells with 1 ng/ml IL-7 in the presence or absence of 10 µg/ml of a neutralizing anti–mouse IL-7 antibody (goat polyclonal IgG; R&D Systems). After 12 h, cells were harvested and protein lysates were analyzed by Western blotting for expression of BCL6 using β-actin as loading control.
RI-BPI.
Homodimerization of the BCL6 BTB domain forms a lateral groove motif, which is required to recruit the silencing mediator of retinoid and thyroid receptors (SMRT) and the nuclear receptor corepressor (N-CoR) to form a BCL6 repression complex (Ahmad et al., 2003). Amino acid side chains protruding into this groove make extensive contact with an 18-residue BCL6 binding domain (BBD) peptide that is conserved between SMRT and N-CoR. The BCL6 lateral groove residues that contact N-CoR and SMRT are unique and are not present in other BTB domain proteins. A recombinant peptide containing the SMRT BBD along with a cell penetrating the TAT domain was able to block interaction of BCL6 with SMRT and N-CoR (Polo et al., 2004). Based on this initial work, a retro-inverso/fusogenic peptidomimetic molecule with superior potency and stability was developed (Cerchietti et al., 2009) and used for the BCL6 inhibition cell-culture experiments described. RI-BPI represents a retro-inverso TAT-BBD-fusogenic peptide (Cerchietti et al., 2009) and was synthesized by Biosynthesis, Inc. and stored lyophilized at −20°C until reconstituted with sterile water immediately before use. The purity determined by HPLC-MS was 95% or higher.
Retroviral transduction.
Transfections of MSCV-based retroviral constructs encoding BCL6-IRES-GFP, MYC-IRES-GFP, SLP65-IRES-GFP, BCR-ABL1-IRES-GFP, BCR-ABL1-IRES-Neo, or Neo and GFP empty vector controls were performed using Lipofectamine 2000 (Invitrogen) with Opti-MEM media (Invitrogen). Retroviral supernatant was produced by cotransfecting 293FT cells with the plasmids pHIT60 (gag-pol) and pHIT123 (ecotropic env; provided by D.B. Kohn, University of California, Los Angeles, Los Angeles, CA). Cultivation was performed in high glucose DMEM (Invitrogen) with GlutaMAX containing 10% fetal bovine serum, 100 IU/ml penicillin, 100 µg/ml streptomycin, 25 mM Hepes, 1 mM sodium pyruvate, and 0.1 mM of nonessential amino acids. Regular media were replaced after 16 h by growth media containing 10 mM sodium butyrate. After 8 h of incubation, the media was changed back to regular growth media. 24 h later, the virus supernatant were harvested, filtered through a 0.45-µm filter, and loaded by centrifugation (2,000 g for 90 min at 32°C) two times on 50 µg/ml RetroNectin-coated nontissue 6-well plates. 2–3 × 106 pre–B cells were transduced per well by centrifugation at 500 g for 30 min and maintained overnight at 37°C with 5% CO2 before transferring into culture flasks.
V(D)J recombination reporter assay.
For construction of the recombination reporter plasmid, a PCR fragment including both 5′ and 3′ RSS amplified from the vector pJH288 was ligated into the retroviral expression vector pMOWS containing a puromycin resistance gene for selection (Wossning et al., 2006). GFP cDNA was ligated in between the RSS in reverse orientation. BCL6−/− and wild-type BCR-ABL1+ pre–B cells were transduced with the RSS-GFP reporter retrovirus. 3 d after transduction, transduced cells were selected in the presence of 1 µg/ml puromycin for 5 d. At this time, no viable cells were detected in a nontransduced parallel culture.
Quantitative RT-PCR.
Total RNA from cells was extracted using the RNeasy isolation kit from QIAGEN. cDNA was generated using a poly(dT) oligonucleotide and SuperScript III Reverse Transcriptase (Invitrogen). Quantitative real-time PCR was performed with the SYBRGreenER mix (Invitrogen) and a real-time PCR system (ABI7900HT; Applied Biosystems) according to standard PCR conditions. Primers for quantitative RT-PCR are listed in Table S3.
Western blotting.
Cells were lysed in CelLytic buffer (Sigma-Aldrich) supplemented with 1% protease inhibitor cocktail (Thermo Fisher Scientific). 10 µg of protein mixture per sample was separated on NuPAGE (Invitrogen) 4–12% Bis-Tris gradient gels and transferred on polyvinylidene fluoride membranes (Immobilion; Millipore). For the detection of mouse and human proteins by Western blotting, primary antibodies were used together with an immunodetection system (WesternBreeze; Invitrogen). The following antibodies were used: human BCL6 (clones D8 and N3; Santa Cruz Biotechnology, Inc.), mouse BCL6 (rabbit polyclonal; Cell Signaling Technology), ARF (4C6/4; Cell Signaling Technology), p53 (1C12; Cell Signaling Technology), global STAT5 (3H7; Cell Signaling Technology), and phospho-Y694 STAT5 (14H2; Cell Signaling Technology). Antibodies against β-actin were used as a loading control (H4; Santa Cruz Biotechnology, Inc.).
Flow cytometry.
Antibodies against mouse CD19 (1D3), B220 (RA3-6B2), c-kit (2B8), CD25 (7D4), CD43 (S7), IgM or μ chain (R6-60.2 and II/41), IgD (11–26), and κ light chains (187.1) as well as respective isotype controls were purchased from BD. Anti–mouse Sca-1 antibody (clone 177228) was obtained from R&D Systems and anti–mouse IL-7Rα (A7R34) was obtained from eBioscience. For apoptosis analyses, annexin V, propidium iodide, and 7-AAD were used (BD).
Affymetrix GeneChip analysis.
Total RNA from cells used for microarray was isolated by RNeasy purification. Biotinylated cRNA was generated and fragmented according to the Affymetrix protocol and hybridized to 430 mouse microarrays (Affymetrix). After scanning (GeneChip Scanner 3000 7G; Affymetrix) the GeneChip arrays, the generated data files were imported to BRB Array Tool (available at http://linus.nci.nih.gov/BRB-ArrayTools.html) and processed using the robust multi-array average algorithm for normalization and summarization. To determine relative signal intensities, the ratio of intensity for each sample in a probe set was calculated by normalizing to the mean value of grouped samples. Ratios were exported in Gene Cluster and visualized as a heatmap with Java Treeview. Microarray data are available from the Gene Expression Omnibus under accession nos. GSE21664 (IL-7 withdrawal from pre–B cells) and GSE20987 (comparison of BCL6+/+ and BCL6−/− BCR-ABL1–transformed pre–B cells).
ChIP on chip.
Human B cell lymphoma OCI-Ly1 cells were fixed and washed, and chromatin was sheared to an average length of 600 bp. BCL6–DNA complexes were immunoprecipitated using an anti-BCL6 polyclonal antibody (N3). Enrichment of known target genes was validated by quantitative real-time PCR. ChIP products and their respective input genomic fragments were amplified by ligation-mediated PCR as previously reported (Ranuncolo et al., 2007). QChIP was performed again at this stage for selected positive control loci to verify that the enrichment ratios were retained. The genomic products of two biological ChIP replicates were labeled with Cy5 (for ChIP products) and Cy3 (for input), and cohybridized on custom-designed genomic tiling arrays generated by NimbleGen Systems Inc. These high-density tiling arrays contain 50-residue oligonucleotides with an average overlap of 25 bases, omitting repetitive elements. Included in the arrays were the genomic region of the CDKN2A locus (Chr9:21950629–22003329) according to the human genome May 2004 assembly. After hybridization, the relative enrichment for each probe was calculated as the signal ratio of ChIP to input. Peaks of enrichment for BCL6 relative to input were captured with a 5-probe sliding window, and the results were uploaded as custom tracks into the University of California, Santa Cruz genome browser and graphically represented as histograms. The cutoff threshold is defined as 2.5 times the SD above the average relative enrichment on the entire array. Peaks involving five or more oligonucleotide probes above this threshold were considered positive hits.
QChIP.
3 × 107 cells were used for ChIP. Cells were double cross-linked with 1.5 mmol/liter EGS (Thermo Fisher Scientific) for 30 min, followed by 1% formaldehyde (Sigma-Aldrich) for 10 min at room temperature. They were lysed (1% Triton X-100, 150 mmol/liter NaCl, 20 mmol/liter Tris, pH 8, 0 and 1 mmol/liter EDTA) and sonicated using a sonicator (Branson Digital Sonifier 450) for 40 min. Cells were immunoprecipitated with rabbit polyclonal antibody to BCL6 (N3) and rabbit polyclonal antibody to β-actin (H-196; Santa Cruz Biotechnology, Inc.) as a control. Eluted DNA was quantified by PCR (Opticon Engine 2; MJ Research) with the SYBR Green Kit (QIAGEN). Primers to confirm binding of BCL6 to the ARF (CDKN2A) promoter region are given in Table S3. Serial dilutions of input DNA were performed to assess the relative amount of product that was enriched after immunoprecipitation.
Clonality analysis and spectratyping of B cell populations.
VH-DJH gene rearrangements from B cell populations were amplified using PCR primers specific for the J558 VH region gene together with a primer specific for the Cμ constant region gene. Using a FAM-conjugated Cμ constant region or a JH gene-specific primer in a run-off reaction, PCR products were labeled and subsequently analyzed on a capillary sequencer (ABI3100; Applied Biosystems) by fragment-length analysis. Sequences of primers used are given in Table S3.
Online supplemental material.
Fig. S1 shows our sorting strategy for HPCs, pro–B and pre–BI cells, large cycling pre–BII cells (fraction C’), small resting pre–BII cells (fraction D), and immature B cells from the bone marrow of BCL6+/+ and BCL6−/− mice. Fig. S2 shows survival and growth of IL-7–dependent pre–B cells from bone marrow of BCL6+/+ and BCL6−/− mice under cell-culture conditions and cytoplasmic stainings for μ chain expression in IL-7–dependent and BCR-ABL1–transformed pre–B cells from SLP65-deficient mice. Fig. S3 depicts induction of κ light chain expression on BCR-ABL1–transformed pre–B cells from BCL6+/+ and BCL6−/− mice. In addition, transformed BCL6+/+ and BCL6−/− pre–B cells were transduced with a reporter construct for V(D)J recombinase activity. STI571-induced V(D)J recombinase activity was assayed along with κ light chain expression and apoptosis (annexin V) by flow cytometry. Fig. S4 shows differentiation and survival of RAG1+/+ and RAG1−/− pre–B cells upon IL-7 withdrawal and concomitant BCL6 inhibition. Fig. S5 presents a proposed scenario for the role of BCL6 during the transition from IL-7–dependent to –independent stages of B cell development. Table S1 presents a quantitative analysis of flow cytometry studies of BCL6+/+ and BCL6−/− B cell precursor populations. Table S2 presents a detailed sequence analysis of junctional regions within VH-DJH rearrangements in κ+ immature B cells from the bone marrow and spleen of BCL6+/+ and BCL6−/− mice. Table S3 lists sequences of oligonucleotide primers used for clonality and spectratyping analyses, quantitative RT-PCR, and QChIP. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20091299/DC1.
Acknowledgments
We would like to thank R. Dalla-Favera for sharing BCL6−/− mice generated in his laboratory and wild-type controls with us. We would like to thank M.R. Lieber and L. Hennighausen for critical discussions.
This work was supported by grants from the National Institutes of Health/National Cancer Institute (R01CA104348 to A.M. Melnick, 5R01CA085573 to B.H. Ye, and R01CA137060, R01CA139032, and R21CA152497 to M. Müschen), Translational Research Program grants from the Leukemia and Lymphoma Society (6132-09 and 6097-10), the V Foundation for Cancer Research (M. Müschen), the William Laurence and Blanche Hughes Foundation and a Stand Up to Cancer–American Association for Cancer Research Innovative Research Grant (IRG00909 to M. Müschen), and a grant from the Deutsche Forschungsgemeinschaft (MU1616/5-1). A.M. Melnick and M. Müschen are Scholars of the Leukemia and Lymphoma Society.
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- ChIP
- chromatin immunoprecipitation
- DLBCL
- diffuse large B cell lymphoma
- DSB
- double-strand break
- GC
- germinal center
- HPC
- hematopoietic progenitor cell
- KDE
- κ-deleting element
- QChIP
- quantitative ChIP
- RI-BPI
- retro-inverso BCL6 peptide inhibitor
- RSS
- recombination signal sequence
- tetO
- tetracycline operator
- tTA
- tetracycline-controlled transactivator
References
- Ahmad K.F., Melnick A., Lax S., Bouchard D., Liu J., Kiang C.L., Mayer S., Takahashi S., Licht J.D., Privé G.G. 2003. Mechanism of SMRT corepressor recruitment by the BCL6 BTB domain. Mol. Cell. 12:1551–1564 10.1016/S1097-2765(03)00454-4 [DOI] [PubMed] [Google Scholar]
- Allman D., Jain A., Dent A., Maile R.R., Selvaggi T., Kehry M.R., Staudt L.M. 1996. BCL-6 expression during B-cell activation. Blood. 87:5257–5268 [PubMed] [Google Scholar]
- Baena E., Gandarillas A., Vallespinós M., Zanet J., Bachs O., Redondo C., Fabregat I., Martinez-A C., de Alborán I.M. 2005. c-Myc regulates cell size and ploidy but is not essential for postnatal proliferation in liver. Proc. Natl. Acad. Sci. USA. 102:7286–7291 10.1073/pnas.0409260102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banerjee A., Rothman P. 1998. IL-7 reconstitutes multiple aspects of v-Abl-mediated signaling. J. Immunol. 161:4611–4617 [PubMed] [Google Scholar]
- Bouchard C., Dittrich O., Kiermaier A., Dohmann K., Menkel A., Eilers M., Lüscher B. 2001. Regulation of cyclin D2 gene expression by the Myc/Max/Mad network: Myc-dependent TRRAP recruitment and histone acetylation at the cyclin D2 promoter. Genes Dev. 15:2042–2047 10.1101/gad.907901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bredemeyer A.L., Helmink B.A., Innes C.L., Calderon B., McGinnis L.M., Mahowald G.K., Gapud E.J., Walker L.M., Collins J.B., Weaver B.K., et al. 2008. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 456:819–823 10.1038/nature07392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cattoretti G., Chang C.C., Cechova K., Zhang J., Ye B.H., Falini B., Louie D.C., Offit K., Chaganti R.S., Dalla-Favera R. 1995. BCL-6 protein is expressed in germinal-center B cells. Blood. 86:45–53 [PubMed] [Google Scholar]
- Cerchietti L.C., Yang S.N., Shaknovich R., Hatzi K., Polo J.M., Chadburn A., Dowdy S.F., Melnick A. 2009. A peptomimetic inhibitor of BCL6 with potent antilymphoma effects in vitro and in vivo. Blood. 113:3397–3405 10.1182/blood-2008-07-168773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang C.C., Ye B.H., Chaganti R.S., Dalla-Favera R. 1996. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc. Natl. Acad. Sci. USA. 93:6947–6952 10.1073/pnas.93.14.6947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ci W., Polo J.M., Cerchietti L., Shaknovich R., Wang L., Yang S.N., Ye K., Farinha P., Horsman D.E., Gascoyne R.D., et al. 2009. The BCL6 transcriptional program features repression of multiple oncogenes in primary B cells and is deregulated in DLBCL. Blood. 113:5536–5548 10.1182/blood-2008-12-193037 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent A.L., Shaffer A.L., Yu X., Allman D., Staudt L.M. 1997. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 276:589–592 10.1126/science.276.5312.589 [DOI] [PubMed] [Google Scholar]
- Guidos C.J., Williams C.J., Grandal I., Knowles G., Huang M.T., Danska J.S. 1996. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes Dev. 10:2038–2054 10.1101/gad.10.16.2038 [DOI] [PubMed] [Google Scholar]
- Hardy R.R., Hayakawa K. 2001. B cell development pathways. Annu. Rev. Immunol. 19:595–621 10.1146/annurev.immunol.19.1.595 [DOI] [PubMed] [Google Scholar]
- Hess J., Werner A., Wirth T., Melchers F., Jäck H.M., Winkler T.H. 2001. Induction of pre-B cell proliferation after de novo synthesis of the pre-B cell receptor. Proc. Natl. Acad. Sci. USA. 98:1745–1750 10.1073/pnas.041492098 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson K., Hashimshony T., Sawai C.M., Pongubala J.M., Skok J.A., Aifantis I., Singh H. 2008. Regulation of immunoglobulin light-chain recombination by the transcription factor IRF-4 and the attenuation of interleukin-7 signaling. Immunity. 28:335–345 10.1016/j.immuni.2007.12.019 [DOI] [PubMed] [Google Scholar]
- Jumaa H., Wollscheid B., Mitterer M., Wienands J., Reth M., Nielsen P.J. 1999. Abnormal development and function of B lymphocytes in mice deficient for the signaling adaptor protein SLP-65. Immunity. 11:547–554 10.1016/S1074-7613(00)80130-2 [DOI] [PubMed] [Google Scholar]
- Klein F., Feldhahn N., Mooster J.L., Sprangers M., Hofmann W.K., Wernet P., Wartenberg M., Müschen M. 2005. Tracing the pre-B to immature B cell transition in human leukemia cells reveals a coordinated sequence of primary and secondary IGK gene rearrangement, IGK deletion, and IGL gene rearrangement. J. Immunol. 174:367–375 [DOI] [PubMed] [Google Scholar]
- Liu M., Duke J.L., Richter D.J., Vinuesa C.G., Goodnow C.C., Kleinstein S.H., Schatz D.G. 2008. Two levels of protection for the B cell genome during somatic hypermutation. Nature. 451:841–845 10.1038/nature06547 [DOI] [PubMed] [Google Scholar]
- Malin S., McManus S., Cobaleda C., Novatchkova M., Delogu A., Bouillet P., Strasser A., Busslinger M. 2010. Role of STAT5 in controlling cell survival and immunoglobulin gene recombination during pro-B cell development. Nat. Immunol. 11:171–179 10.1038/ni.1827 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muljo S.A., Schlissel M.S. 2003. A small molecule Abl kinase inhibitor induces differentiation of Abelson virus-transformed pre-B cell lines. Nat. Immunol. 4:31–37 10.1038/ni870 [DOI] [PubMed] [Google Scholar]
- Nakayama J., Yamamoto M., Hayashi K., Satoh H., Bundo K., Kubo M., Goitsuka R., Farrar M.A., Kitamura D. 2009. BLNK suppresses pre-B-cell leukemogenesis through inhibition of JAK3. Blood. 113:1483–1492 10.1182/blood-2008-07-166355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onishi M., Nosaka T., Misawa K., Mui A.L., Gorman D., McMahon M., Miyajima A., Kitamura T. 1998. Identification and characterization of a constitutively active STAT5 mutant that promotes cell proliferation. Mol. Cell. Biol. 18:3871–3879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Palmer M.J., Mahajan V.S., Trajman L.C., Irvine D.J., Lauffenburger D.A., Chen J. 2008. Interleukin-7 receptor signaling network: an integrated systems perspective. Cell. Mol. Immunol. 5:79–89 10.1038/cmi.2008.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pear W.S., Miller J.P., Xu L., Pui J.C., Soffer B., Quackenbush R.C., Pendergast A.M., Bronson R., Aster J.C., Scott M.L., Baltimore D. 1998. Efficient and rapid induction of a chronic myelogenous leukemia-like myeloproliferative disease in mice receiving P210 bcr/abl-transduced bone marrow. Blood. 92:3780–3792 [PubMed] [Google Scholar]
- Phan R.T., Dalla-Favera R. 2004. The BCL6 proto-oncogene suppresses p53 expression in germinal-centre B cells. Nature. 432:635–639 10.1038/nature03147 [DOI] [PubMed] [Google Scholar]
- Phan R.T., Saito M., Basso K., Niu H., Dalla-Favera R. 2005. BCL6 interacts with the transcription factor Miz-1 to suppress the cyclin-dependent kinase inhibitor p21 and cell cycle arrest in germinal center B cells. Nat. Immunol. 6:1054–1060 10.1038/ni1245 [DOI] [PubMed] [Google Scholar]
- Polo J.M., Dell’Oso T., Ranuncolo S.M., Cerchietti L., Beck D., Da Silva G.F., Prive G.G., Licht J.D., Melnick A. 2004. Specific peptide interference reveals BCL6 transcriptional and oncogenic mechanisms in B-cell lymphoma cells. Nat. Med. 10:1329–1335 10.1038/nm1134 [DOI] [PubMed] [Google Scholar]
- Radic M.Z., Zouali M. 1996. Receptor editing, immune diversification, and self-tolerance. Immunity. 5:505–511 10.1016/S1074-7613(00)80266-6 [DOI] [PubMed] [Google Scholar]
- Ranuncolo S.M., Polo J.M., Dierov J., Singer M., Kuo T., Greally J., Green R., Carroll M., Melnick A. 2007. Bcl-6 mediates the germinal center B cell phenotype and lymphomagenesis through transcriptional repression of the DNA-damage sensor ATR. Nat. Immunol. 8:705–714 10.1038/ni1478 [DOI] [PubMed] [Google Scholar]
- Ranuncolo S.M., Polo J.M., Melnick A. 2008. BCL6 represses CHEK1 and suppresses DNA damage pathways in normal and malignant B-cells. Blood Cells Mol. Dis. 41:95–99 10.1016/j.bcmd.2008.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reth M., Gehrmann P., Petrac E., Wiese P. 1986. A novel VH to VHDJH joining mechanism in heavy-chain-negative (null) pre-B cells results in heavy-chain production. Nature. 322:840–842 10.1038/322840a0 [DOI] [PubMed] [Google Scholar]
- Rolink A., Grawunder U., Haasner D., Strasser A., Melchers F. 1993. Immature surface Ig+ B cells can continue to rearrange κ and λ L chain gene loci. J. Exp. Med. 178:1263–1270 10.1084/jem.178.4.1263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolink A., Ghia P., Grawunder U., Haasner D., Karasuyama H., Kalberer C., Winkler T., Melchers F. 1995. In-vitro analyses of mechanisms of B-cell development. Semin. Immunol. 7:155–167 10.1016/1044-5323(95)90043-8 [DOI] [PubMed] [Google Scholar]
- Schebesta M., Pfeffer P.L., Busslinger M. 2002. Control of pre-BCR signaling by Pax5-dependent activation of the BLNK gene. Immunity. 17:473–485 10.1016/S1074-7613(02)00418-1 [DOI] [PubMed] [Google Scholar]
- Schlissel M.S., Kaffer C.R., Curry J.D. 2006. Leukemia and lymphoma: a cost of doing business for adaptive immunity. Genes Dev. 20:1539–1544 10.1101/gad.1446506 [DOI] [PubMed] [Google Scholar]
- Schuh W., Meister S., Herrmann K., Bradl H., Jäck H.M. 2008. Transcriptome analysis in primary B lymphoid precursors following induction of the pre-B cell receptor. Mol. Immunol. 45:362–375 10.1016/j.molimm.2007.06.154 [DOI] [PubMed] [Google Scholar]
- Seyfert V.L., Allman D., He Y., Staudt L.M. 1996. Transcriptional repression by the proto-oncogene BCL-6. Oncogene. 12:2331–2342 [PubMed] [Google Scholar]
- Shaffer A.L., Yu X., He Y., Boldrick J., Chan E.P., Staudt L.M. 2000. BCL-6 represses genes that function in lymphocyte differentiation, inflammation, and cell cycle control. Immunity. 13:199–212 10.1016/S1074-7613(00)00020-0 [DOI] [PubMed] [Google Scholar]
- Shvarts A., Brummelkamp T.R., Scheeren F., Koh E., Daley G.Q., Spits H., Bernards R. 2002. A senescence rescue screen identifies BCL6 as an inhibitor of anti-proliferative p19(ARF)-p53 signaling. Genes Dev. 16:681–686 10.1101/gad.929302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trageser D., Iacobucci I., Nahar R., Duy C., von Levetzow G., Klemm L., Park E., Schuh W., Gruber T., Herzog S., et al. 2009. Pre–B cell receptor–mediated cell cycle arrest in Philadelphia chromosome–positive acute lymphoblastic leukemia requires IKAROS function. J. Exp. Med. 206:1739–1753 10.1084/jem.20090004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker S.R., Nelson E.A., Frank D.A. 2007. STAT5 represses BCL6 expression by binding to a regulatory region frequently mutated in lymphomas. Oncogene. 26:224–233 10.1038/sj.onc.1209775 [DOI] [PubMed] [Google Scholar]
- Wossning T., Herzog S., Köhler F., Meixlsperger S., Kulathu Y., Mittler G., Abe A., Fuchs U., Borkhardt A., Jumaa H. 2006. Deregulated Syk inhibits differentiation and induces growth factor–independent proliferation of pre–B cells. J. Exp. Med. 203:2829–2840 10.1084/jem.20060967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye B.H., Lista F., Lo Coco F., Knowles D.M., Offit K., Chaganti R.S., Dalla-Favera R. 1993. Alterations of a zinc finger-encoding gene, BCL-6, in diffuse large-cell lymphoma. Science. 262:747–750 10.1126/science.8235596 [DOI] [PubMed] [Google Scholar]
- Ye B.H., Cattoretti G., Shen Q., Zhang J., Hawe N., de Waard R., Leung C., Nouri-Shirazi M., Orazi A., Chaganti R.S., et al. 1997. The BCL-6 proto-oncogene controls germinal-centre formation and Th2-type inflammation. Nat. Genet. 16:161–170 10.1038/ng0697-161 [DOI] [PubMed] [Google Scholar]