Abstract
Human BDCA3+ dendritic cells (DCs) were suggested to be homologous to mouse CD8α+ DCs. We demonstrate that human BDCA3+ DCs are more efficient than their BDCA1+ counterparts or plasmacytoid DCs (pDCs) in cross-presenting antigen and activating CD8+ T cells, which is similar to mouse CD8α+ DCs as compared with CD11b+ DCs or pDCs, although with more moderate differences between human DC subsets. Yet, no specific marker was known to be shared between homologous DC subsets across species. We found that XC chemokine receptor 1 (XCR1) is specifically expressed and active in mouse CD8α+, human BDCA3+, and sheep CD26+ DCs and is conserved across species. The mRNA encoding the XCR1 ligand chemokine (C motif) ligand 1 (XCL1) is selectively expressed in natural killer (NK) and CD8+ T lymphocytes at steady-state and is enhanced upon activation. Moreover, the Xcl1 mRNA is selectively expressed at high levels in central memory compared with naive CD8+ T lymphocytes. Finally, XCR1−/− mice have decreased early CD8+ T cell responses to Listeria monocytogenes infection, which is associated with higher bacterial loads early in infection. Therefore, XCR1 constitutes the first conserved specific marker for cell subsets homologous to mouse CD8α+ DCs in higher vertebrates and promotes their ability to activate early CD8+ T cell defenses against an intracellular pathogenic bacteria.
DCs are central to immune defenses in mammals. In mice, three subsets of DCs are resident of lymphoid organs (Crozat et al., 2010). Plasmacytoid DCs (pDCs) are professional producers of IFN-α and -β, contributing to immune defenses against viruses (Baranek et al., 2009). CD11b+ DCs preferentially prime CD4+ T cells and promote humoral immunity (Carter et al., 2006; Dudziak et al., 2007). CD8α+ DCs are endowed with a unique efficiency in priming CD8+ T cells and in cross-presenting exogenous antigens (Carter et al., 2006; Dudziak et al., 2007). CD8α+ DCs are required for the natural induction of strong CD8+ T cell responses against tumors (Hildner et al., 2008; Sancho et al., 2008) or West Nile virus (Hildner et al., 2008). Specific delivery of vaccine antigens to CD8α+ DCs is especially efficient for vaccination against intracellular pathogens or tumors (Bonifaz et al., 2004; Nchinda et al., 2008). Therefore, identification of human DC subsets functionally homologous to mouse CD8α+ DCs (CD8α+-type DCs) should be a major step forward for the design of innovative vaccination or immunotherapeutic strategies against cancer or infections (Crozat et al., 2010). So far, no conserved marker has been identified to specifically and unambiguously define CD8α+-type DCs in several mammalian species. Although human BDCA3+ and mouse CD8α+ DCs express the C-type lectin CLEC9A, which is known to be involved in cross-presentation in mice, this marker is also found on some human CD14+ monocytes and on mouse pDCs (Caminschi et al., 2008; Huysamen et al., 2008; Sancho et al., 2008). It is of note that no orthologue of CLEC9A has been identified yet in non mammalian vertebrate species.
We have recently performed comparative genomics studies of human, mouse, and sheep DC subsets to help identify potential homologies between these cell types across mammalian species (Robbins et al., 2008; unpublished data). We found that human blood BDCA3+ DCs share a specific gene signature with mouse CD8α+ DCs and proposed that they could be human professional cross-presenting DCs (Robbins et al., 2008; Crozat et al., 2010). In this paper, we demonstrate that human BDCA3+ DCs are more potent than their BDCA1+ counterparts or than pDCs for CD8+ T cell activation through antigen cross-presentation. We established elsewhere that sheep lymph CD26+ DCs (Epardaud et al., 2004) are also equivalents to mouse CD8α+ DCs based on gene expression and functions such as superior efficacy for presentation of soluble antigen to CD8+ T cells (unpublished data). In this paper, we identify the XC chemokine receptor 1 (XCR1) as the first universal marker specifically expressed by the CD8α+-type DCs from three different mammalian species: ovine CD26+ DCs, mouse CD8α+ DCs, and human BDCA3+ DCs. We show that the Xcr1 gene is present and well conserved in all higher vertebrates from reptiles to human. The ligand of XCR1, chemokine (C motif) ligand 1 (XCL1), is specifically expressed by activated NK and CD8+ T cells in mouse and human. We show that Xcl1 mRNA is stored selectively in memory CD8+ T cells, allowing them to rapidly produce high levels of this chemokine upon stimulation. Finally, we show that XCR1−/− mice have decreased CD8+ T cell responses to Listeria monocytogenes (Lm) associated with higher bacterial loads early after infection. Overall, our study strongly suggests an important and conserved role in mammals for XCR1 in the cross talk between NK or CD8+ T cells and CD8α+-type DCs, identifying this molecule as a novel tool to survey and target the DCs endowed with the best cross-presentation capacity across species.
RESULTS
Human BDCA3+ DCs are functional homologues of mouse CD8α+ DCs for cross-presentation
To assess the cross-presentation capacity of human BDCA3+ DCs, we purified these cells together with other types of DCs. BDCA1+ DCs, BDCA3+ DCs, and pDCs were magnetically enriched from PBMCs by negative selection and then positively sorted by flow cytometry as depicted in Fig. 1 A. The purity of sorted cells was >95% as observed by postsorting FACS reanalysis. Sorted HLA-A2+ DC subsets were cultured with apoptotic uninfected, or chronically HIV-infected, H9 cells in the presence of maturation stimuli, poly I:C for BDCA3+ DCs, LPS or poly I:C for BDCA1+ DCs, and R848 for pDCs. They were then cultured with an HIV-1 Pol-specific CD8+ T cell line. To avoid viral replication, experiments were performed in the presence of Saquinavir, an HIV protease inhibitor (Hoeffel et al., 2007). BDCA3+ DCs induced 40% more IFN-γ ELISPOTs than poly I:C–stimulated BDCA1+ DCs or R848-stimulated pDCs and 60% more as compared with LPS-stimulated BDCA1+ DCs (Fig. 1 B). Therefore, as expected from their gene expression profile (Robbins et al., 2008; Crozat et al., 2010), human BDCA3+ DCs had higher cross-presentation capacities than other human blood DC subsets and represent functional homologues of mouse CD8α+ DCs for cross-presentation.
Figure 1.
Human blood BDCA3+ DCs are endowed with a higher cross-presentation ability than BDCA1+ DCs and pDCs. (A) Sorting of DC populations from PBMCs. First, CD3 and CD14-depleted PBMCs were gated based on SSC-W and SSC-A to exclude cellular doublets and on FSC-A and SSC-A. Next, CD19−HLA-DR+ events and CD14−CD16− events were sequentially selected. HLA-DR+CD123hi events, corresponding to pDCs, were then sorted and the remaining events falling in the HLA-DR+CD123lo/− were distinguished for BDCA1+ DC and BDCA3+ DC sorting. Sorting gates are delineated by red lines. (B) Cross-presentation assay with human DCs. Purified BDCA1+ DCs, BDCA3+ DCs, and pDCs were cultured with apoptotic cells at a 1:1 ratio for 16 h in the presence of maturation signal, 100 ng/ml LPS, 10 µg/ml poly I:C, or 10 µM R848 as indicated. After loading, DCs were extensively washed and used (5,000/well) in a 16-h IFN-γ ELISPOT assay with HIV-specific CD8+ T cells lines as effectors (15,000/well). Data are presented as the mean and range of duplicate wells for one representative experiment of at least two.
XCR1 is selectively expressed by mouse CD8α+, human BDCA3+, and sheep CD26+ DCs
Based on gene chip analyses, the expression of the XCR1 gene is a selective marker for both human BDCA3+ DCs and mouse CD8α+ DCs when compared with a large panel of cell types, including other DC subsets, a variety of other leukocytes and of nonhematopoietic cells, and all types of tissue (Fig. 2 A). This contrasts with the markers currently used to identify these DC subsets, which are more broadly expressed as clearly evident for BDCA3 in human or CADM1 in human and mouse (Fig. 2 A). Further extending this striking convergent observation in mouse and human DCs, we found that in a distant mammalian species, i.e., sheep, the XCR1 gene was also selectively expressed in the CD8α+-like skin lymph CD26+ DC (Fig. 2 B). Thus, among the genes differentially expressed between DC subsets in a conserved manner across the three mammalian species examined, the XCR1 gene stands out as the most discriminating marker for mouse spleen CD8α+ DCs, human blood BDCA3+ DCs, and sheep lymph CD26+ DCs (Fig. 2, A and B). This is also the case for tonsil BDCA3+ DCs (Fig. 2 A, orange circle). Xcr1 mRNA was also selectively expressed in CD11c+SIRPα−CD24+ cells from BM FLT3-L cultures (Fig. 3 A), which correspond to a subset of in vitro–derived DCs equivalent to spleen CD8α+ DCs which can be referred to as eCD8α+ DCs (Naik et al., 2005).
Figure 2.
Xcr1 gene expression by different subsets of immune cells isolated from mice, human, and sheep. (A) Microarray analysis of the expression of the XCR1, THBD (BDCA3), CADM1, and LILRA4 (BDCA2) human genes and of the Xcr1, CD8a, Cadm1, and Siglech mouse genes in 96 different cell types or tissues, in human (top) and mouse (bottom), respectively. The human data were retrieved from the GEO database, normal tissues and cell types from the GSE7307 dataset, PBMC-derived macrophages from GSE4883, monocyte-derived DCs from GSE7509, monocyte-derived macrophages from GSM213500, and alveolar macrophages from GSE2125, and blood and tonsil DC subsets were retrieved from the E-TABM-34 dataset (Lindstedt et al., 2005) of the EBI ArrayExpress database. The data for the other leukocyte subsets directly isolated from normal human blood were described previously (Du et al., 2006; Robbins et al., 2008) and can be downloaded from http://www-microarrays.u-strasbg.fr/files/datasetsE.php. The data for the mouse were downloaded from the BioGPS public database (http://biogps.gnf.org). Green circles, pDCs (dark, blood; light, tonsil); red circles, mouse spleen CD8α+ DCs and human blood BDCA3+ DCs; orange circles, human tonsil BDCA3+ DCs; blue circles, mouse spleen CD11b+ DCs or human BDCA1+ DCs (dark, blood; light, tonsil); brown circles, mouse spleen; yellow circles, mouse lymph nodes; gray, all other cell types and tissues. Results are expressed as mean and SD for at least three independent values for most human data points. (B) Xcr1 expression determined by real-time PCR on sorted mouse spleen DC subsets and sheep leukocytes. Mouse DCs were defined as lin−CD11c+Bst2+ for pDCs, lin−CD11c+Bst2−CD8α+ for CD8α+ DCs, and CD8α− for CD11b+ DCs. Results are representative of at least two independent experiments. Sheep skin lymph CD1b+CD26+ and CD1b+CD26− DCs were sorted by flow cytometry to >99% purity. Sheep CD4+ T, CD8+ T, NK, and B cells were purified from afferent lymph, and CD14+ monocytes from blood, by flow cytometry sorting to >95% purity. Results are mean ± SEM of triplicate real-time RT-PCR reactions and they are representative of two different sheep for lymphocytes and of three different sheep for DCs. (C) β-galactosidase expression in splenocytes from XCR1+/+ and XCR1−/− mice. β-galactosidase expression was assessed by FDG staining. The results are shown from one mouse representative of at least eight animals studied in four independent experiments.
Figure 3.
XCR1 expression and functions on mouse DC subsets from BM FLT3-L cultures. (A) DC subsets from FLT3-L BM cultures from C57BL/6Ncrl mice were sorted and examined by real-time PCR for expression of the Xcr1 gene. (B) Transwell assay on DC subsets derived from FLT3-L cultures of XCR1+/− and XCR1−/− BM cells. Results are representative of two experiments and expressed as mean ± SEM from duplicate wells for each data point. eCD8α+: FLT3-L BM-DC equivalents to spleen CD8α+ DCs and defined as SIRPα−CD24+CD11c+ cells. eCD11b+: FLT3-L DC equivalents to spleen CD11b+ DCs and defined as SIRPα+CD24−CD11c+ cells. pDC: FLT3-L BM-pDC defined as CD11clowSiglec-H+ cells.
To assess XCR1 protein expression pattern on mouse leukocytes, we resorted to a reporter/knockout mouse strain expressing the β-galactosidase enzyme under the control of the Xcr1 promoter. When total spleen leukocytes were isolated from XCR1−/− mice and stained for β-galactosidase activity using fluorescein di-β-d-galactopyranoside (FDG) as a substrate, >80% of the cells that stained positive with FDG were CD8α+ DCs, and >75% of CD8α+ DCs were FDG+ (Fig. 2 C). Thus, Xcr1 was confirmed to be expressed selectively by the majority of mouse CD8α+ DCs. Altogether, these results showed that XCR1 is a conserved and very discriminating marker for the identification of mouse, ovine, and human CD8α+-type DCs.
XCR1 protein is active on mouse CD8α+, human BDCA3+, and sheep CD26+ DCs
We then performed transwell migration assays with recombinant XCL1 and mixtures of the three subsets of mouse spleen DCs (Fig. 4 A) or their in vitro equivalents derived in BM FLT3-L cultures (Fig. 3 B). In both instances, XCL1 induced the specific migration of CD8α+-type DCs. Moreover, only XCR1+/+ and XCR1+/− CD8α+ or eCD8α+ DCs were responsive to XCL1 (Fig. 3 B and Fig. 4 A), but not their XCR1−/− counterparts, demonstrating that XCR1 is required for DC responses to XCL1. We then performed transwell assays with enriched DC preparations from human blood and sheep lymph. Only human BDCA3+ DCs and sheep CD26+ DCs, and none of the other leukocyte subsets tested, were found to migrate in response to XCL1 (Fig. 4 A). Therefore, XCR1 is required for DC responses to XCL1, and its engagement induces the selective motility of mouse, ovine, and human CD8α+-type DCs.
Figure 4.
XCR1 is functionally active on mouse CD8α+, human BDCA3+, and sheep CD26+ DCs, and Xcl1 mRNA is stored in quiescent NK cells and memory CD8+ T lymphocytes. Transwell migration assay on enriched human blood DCs or lymphocytes, sheep lymph CD26+ versus CD26− DCs, and splenic DCs from XCR1−/− and C57BL/6NCrl (XCR1+/+) mice. Results are representative of at least two independent experiments for each species and expressed as mean ± SEM from duplicate wells for each data point. (B) Expression of the XCL1 gene in human and mouse cell types and tissues, based on the same gene chips data as used in Fig. 1 A, with the following additions: black triangles, NK cells; purple triangles, resting peripheral CD8+ T cells; purple diamond, anti-CD3 activated human T cells; purple square, mouse CD8+ thymocytes; violet square, mouse CD4+ thymocytes. Results are expressed as mean and SD for at least three independent values for most human data points. (C) Expression of the XCL1 gene in sheep leukocytes as assessed by real-time PCR on the same lymph or blood cells as shown in Fig. 2. Results are mean ± SEM of triplicate real-time RT-PCR reactions, and they are representative of two different sheep for lymphocytes and of three different sheep for DCs. (D) Results of Xcl1 gene expression in mouse CD8+ T cell subsets. Xcl1 expression was measured by real-time PCR on sorted naive or TIM, or antiviral TCM CD8+ T cell subsets. Expression of Ccl5 and Ifng were also evaluated as controls, as the genes are expressed to higher levels in memory CD8+ T cells (Walzer et al., 2003). Results are represented as mean ± SD for mean values from duplicate real-time PCR reactions performed on mRNAs from naive T cells or from TIM from three individual mice each, and from TCM from four individual pools of seven mice.
Xcl1 mRNA is stored in quiescent NK cells and memory CD8+ T lymphocytes
XCL1 is produced and secreted specifically by activated mouse and human NK and CD8+ T cells (Müller et al., 1995; Dorner et al., 2002), whereas no XCL1 protein is detected by intracellular staining in quiescent mouse NK cells (Dorner et al., 2002). The Xcl1 gene is specifically expressed in NK and CD8+ T cells already at steady state not only in mouse and human (Fig. 4 B) but also in sheep (Fig. 4 C). Moreover, similar to what we previously reported for CCL5 (Walzer et al., 2003), we show that XCL1 mRNA is selectively expressed to high levels in antiviral central memory CD8+ T lymphocytes (TCMs) and, to a lesser extent, in memory T cells induced by sterile inflammatory conditions (inflammatory memory CD8+ T cells [TIMs]; Fig. 4 D). Cytokine secretion profiling of the mouse CD8+ T cell subsets demonstrated a rapid and higher level secretion of XCL1 by TIM after antigen-specific reactivation in vitro compared with naive CD8+ T cells (unpublished data). The selective expression of Xcl1 in mouse TCM was confirmed independently by mining published mouse microarray datasets retrieved through the GEO database (Sarkar et al., 2008; Fig. S1 A). Therefore, XCL1 mRNA is specifically expressed at steady state in NK and, to a lesser extent, CD8+ T cells, further induced in these cell types upon activation, including during viral infections in mouse and human (Fig. S1; Hyrcza et al., 2007; Sarkar et al., 2008), and stored in TCM. Hence, XCR1 expression is likely integral to the unique potency of CD8α+-type DCs for interaction with NK and CD8+ T cells.
Xcr1 is conserved in vertebrates and may be an ancient marker for CD8α+-type DCs.
Sequences similar to the Xcr1 locus are available from many mammals including all the species with a full genome assembly. Only one gene per species was found, located in the same genomic region as several CC chemokine receptors (CCRs). A unique Xcr1 sequence was also identified from genomes of birds and the Anole lizard. Several sequences similar to Xcr1 were present in teleost fish, such as zebrafish and stickleback, but only one in others like Fugu. The Xcr1 sequence is well conserved in all eutherian species examined (>75% identity with the human sequence), in marsupials (∼70% identity), in birds and reptiles (60–65% identity), and in fish (50–60% identity; Fig. S2). These sequences were aligned with those of several human CCRs, and phylogenetic analyses were performed using different methods. Trees reconstructed were congruent and demonstrated that the Xcr1-like sequences found in the different vertebrate taxa are true orthologues of the mammalian Xcr1 gene (Fig. 5 A and Fig. S2), which was confirmed by synteny (Fig. 5, B and C). The multiple genes found in fish probably derive from the complex history of fish genomes that have been marked by large duplication events, fast evolution, and extensive genome rearrangements (Ravi and Venkatesh, 2008). It is remarkable that both stickleback Xcr1 co-orthologues are closely linked to paralogues of the marker FYCO1, which is also encoded in the close neighborhood of mammalian Xcr1 (Fig. 5 B-C). This conserved synteny indicates that the stickleback Xcr1-like genes indeed constitute valid co-orthologues of the mammalian receptor. Hence, XCR1 is a promising candidate marker to identify, isolate, study, and possibly target CD8α+-type DCs in all vertebrates.
Figure 5.
Xcr1 evolution in vertebrates. (A). Neighbor-joining (NJ) phylogenetic analysis. Xcr1 sequences from selected vertebrate species were extracted from the Ensembl databases and aligned with relevant human CCR sequences using ClustalW. The evolutionary history was inferred using the NJ method. The bootstrap consensus tree inferred from 1,000 replicates is taken to represent the evolutionary history of the taxa analyzed. The percentages of replicate trees in which the associated taxa clustered together in the bootstrap test are shown next to the branches. The tree is drawn to scale, with branch lengths in the same units as those of the evolutionary distances used to infer the phylogenetic tree. The evolutionary distances were computed using the Poisson correction method and are in the units of the number of amino acid substitutions per site. All positions containing gaps and missing data were eliminated from the dataset (complete deletion option). There were a total of 267 positions in the final dataset. Phylogenetic analyses were conducted in MEGA4 (Tamura et al., 2007). Abbreviations for species are: HS, Homo sapiens (Ensembl ID: ENSG00000173578); BT, Bos taurus (ENSBTAG00000020019); MM, Mus musculus (ENSMUSG00000060509); FR, Fugu rubripes (ENSTRUG00000012394); GG, Gallus gallus (ENSGALG00000011735); MD, Monodelphis domestica (ENSMODG00000024025); TG, Taeniopygia guttata (ENSTGUG00000004516); DR, Danio rerio (ENSDARG00000054847, ENSDART00000077124, and ENSDART00000074943); and GA, Gasterosteus aculeatus (ENSGACG00000011742 and ENSGACG00000017177). Human CCR IDs are as follows: CX3CR1 (ENSG00000168329); CCR8 (ENSG00000179934); CCR4 (ENSG00000183813); CCR1 (ENSG00000163823); CCR3 (ENSG00000183625); CCR2 (ENSG00000121807); and CCR5 (ENSG00000160791). (B) Map of the human genes in the vicinity of the XCR1 gene. (C) Conserved syntenies retrieved in different vertebrates in the neighborhood of Xcr1. Abbreviations for species are the same as in A. The locations of the genes are given as the number of the corresponding chromosome (for mammals and birds) or linkage group (for teleost), followed by a colon and the position of the sequence in base pairs on the chromosome/group.
XCR1 promotes induction of CD8+ T cell responses and control of bacterial load in vivo early during Lm infection
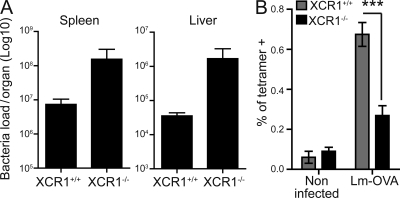
CD8α+ DCs, but not CD11b+ DCs or pDCs, isolated from mice infected with Lm present bacterial antigens for the priming of naive CD8+ T cells (Belz et al., 2005). Treatment of Lm-infected mice with pertussis toxin compromises the later activation of adoptively transferred antibacterial CD8+ T cells (Aoshi et al., 2008). Pertussis toxin is a nonspecific inhibitor of G proteins that is widely used to inhibit chemokine receptor signaling. Thus, it was suggested that a chemokine receptor expressed on DCs is required to enable them to induce CD8+ T cell responses to Lm infection. We reasoned that XCR1 could contribute to this process. Therefore, we challenged XCR1−/− and XCR1+/+ mice with Lm to evaluate the impact of XCR1 deficiency on the control of the bacteria load and on the induction of CD8+ T cell responses (Fig. 6). As compared with XCR1+/+ mice, XCR1−/− animals infected with WT Lm displayed a log increase in their bacteria loads measured in livers and spleens at day 3 after inoculation (Fig. 6 A). There was still a trend toward higher bacterial loads in the spleen and liver of XCR1−/− mice at day 4 after inoculation, although it was weaker than at day 3 (unpublished data). Because anti-Lm CD8+ T cell responses are readily detectable at 3–4 d after infection (Khanna et al., 2007), their alteration in XCR1−/− animals could contribute to the higher bacterial replication in these animals. To compare the levels of anti-Lm CD8+ T cell responses between XCR1−/− and XCR1+/+ mice, they were infected with a recombinant Lm strain expressing the ovalbumin antigen (Lm-OVA), and anti-OVA CD8+ T cells were enumerated by MHC class I tetramer staining at day 4 after inoculation. XCR1−/− mice exhibited significantly fewer anti-Lm CD8+ T cells than XCR1+/+ animals (Fig. 6 B). Thus, these data suggest that XCR1 activity in CD8α+ DCs allows them to more efficiently induce antigen-specific CD8+ T cell responses and, hence, promote bacteria control early after infection with Lm.
Figure 6.
XCR1 deficiency affects Lm control and antibacterial CD8+ T cell responses early after infection. (A) Bacterial loads in the spleen and liver of XCR1−/− versus XCR1+/+ mice at day 3 after infection. Results are represented as mean ± SEM for three animals per group and from one representative experiment out of two. (B) Frequency of anti-OVA(SIINFEKL) tetramer+ cells within total spleen CD8+ T lymphocytes from Lm-OVA–infected mice at day 4 after infection. Results are represented as mean ± SEM from two independent experiments pooled for statistical analysis. ***, P < 0.005 using an unpaired Mann-Whitney non parametric test, eight mice per group.
DISCUSSION
The demonstration that human blood BDCA3+ DCs isolated ex vivo are more potent than their BDCA1+ counterparts or pDCs for antigen cross-presentation and CD8+ T cell activation fulfills our prediction, based on our comparative genomics studies, that human BDCA3+ DCs represent, in this respect, a functional homologue of mouse CD8α+ DCs and an interesting target for the design of innovative vaccination or immunotherapeutic strategies against infections by intracellular pathogens or cancer (Robbins et al., 2008; Crozat et al., 2010). More generally, our results demonstrate a common specialization of mouse CD8α+, human BDCA3+, and sheep CD26+ DCs for CD8+ T cell activation through cross-presentation of exogenous antigens and identical high selective expression of XCR1 by these cells that is consistent with the role of this molecule in the promotion of the interactions between CD8α+-like DCs and CD8+ T cells. These results establish a definitive proof of concept that validates our comparative genomics approach to hunt for conserved functions based on conserved molecular pathways for DC subsets across several mammalian species (Robbins et al., 2008; Crozat et al., 2010).This work exemplifies how critical knowledge can be gained to understand important immunological processes by using comparisons between species as a tool and by developing tight collaborations between teams working with different animal models. In particular, this work illustrates the utility of nonprimate animal models for understanding human immunology, provided that a rational approach is followed to help focusing on conserved biological processes and molecular functions.
Based on gene chip analyses, we previously reported that mouse CD8α+ DCs specifically expressed the Xcr1 gene, to high levels, whereas none of the other leukocyte subsets examined did. Indeed, in the Supplemental material of our paper (Robbins et al., 2008), we reported Xcr1 as the third most specific gene for mouse CD8α+ DCs in our analyses. This result was confirmed and significantly extended by Dorner et al. (2009). Indeed, these authors additionally reported that XCR1 engagement induces calcium mobilization specifically in CD8α+ DCs and that XCL1 responses are required for the induction of optimal primary CD8+ T cell responses by CD8α+ DCs in response to soluble model antigens in vitro and in vivo. No functional ProbeSet specific for the human XCR1 gene had been identified on the Affymetrix Human Genome U133 Plus 2.0 Array until very recently. XCR1 was, therefore, not reported by us (Robbins et al., 2008) or by Dorner et al. (2009) to be specifically expressed in human BDCA3+ DCs. However, by mapping on the human genome all individual orphan ProbeSets that were giving a high and specific signal only in human BDCA3+ DCs as compared with other blood leukocyte subsets, we identified that the 1561226_at ProbeSet mapped to the human XCR1 gene, which was later confirmed by the November 2009 update of the Ensembl genome browser. Hence, we demonstrate in this paper, for the first time, to the best of our knowledge, a conserved specific expression and activity of XCR1 on CD8α+-type DCs derived in vitro in mouse BM FLT3-L cultures or isolated ex vivo from human and sheep. In our hands, XCL1 did not induce any maturation of DCs, either alone or in combination with IFN-γ, and did not seem to affect TLR-induced maturation of DCs in vitro (unpublished data). We confirm the specific expression of XCR1 ligands in NK cells and CD8+ T lymphocytes in mouse and human and extend it to sheep. We show that Xcl1 mRNA is stored in quiescent NK cells and antiviral memory CD8+ T lymphocytes. We show that XCR1 promotes the induction of CD8+ T cell responses and the control of bacterial load in vivo early during Lm infection. Our phylogenetic analyses show the presence of a unique and well conserved XCR1 orthologue in all higher vertebrates from reptiles to human. Altogether, these observations strongly suggest that the XCL1→XCR1 chemotactic pathway constitutes one of the critical and conserved molecular bases promoting selective and efficient cross talk between CD8α+-type DCs and CD8+ T lymphocytes as well as NK cells.
In summary, our results demonstrate a functional homology between mouse CD8α+, sheep CD26+, and human BDCA3+ DCs and identify XCR1 as the best candidate to date for the identification of CD8α+-type DCs in all vertebrates, with a very narrow expression pattern highly restricted to these cell types. In addition, we show that, contrary to the molecules classically used to identify CD8α+-like DCs, XCR1 is not a mere phenotypical marker of these cells but actually contributes to confer on them their exquisite ability for CD8+ T cell activation. Thus, our study brings significant advances toward the generation of a unifying phenotypic and functional view of mammalian DC subsets, with major implications both for basic and applied immunology. Our findings open a way for the general survey by immunohistofluorescence of the anatomical location of CD8α+-type DCs and for their specific targeting in vivo for vaccination against intracellular pathogens or tumors threatening human or domestic animal health.
MATERIALS AND METHODS
Cell purification and culture for human DC subset cross-presentation assay.
For each experiment, >500 × 106 PBMCs were isolated by Ficoll-Hypaque density gradient centrifugation (GE Healthcare) from peripheral blood cytapheresis residues obtained from normal healthy volunteer donors through the Etablissement Français du Sang according to the French ethics committee on human experimentations, within a convention with Institut National de la Santé et de la Recherche Médicale. Primary DCs (BDCA1+ DCs, BDCA3+ DCs, and pDCs) were isolated from PBMCs depleted of CD3+ and CD14+ cells using CD3+ and CD14+ MACS microbeads and an AutoMACS (Miltenyi Biotec). Enriched cells were labeled and DCs were sorted on a FACSAria flow cytometer (BD) as events negative for CD14, CD16, and CD19 and positive for HLA-DR and BDCA1 for BDCA1+ DCs, HLA-DR and BDCA3 for BDCA3+ DCs, or HLA-DR and CD123 for pDCs. The following antibodies were used: HLA-DR-PerCp (G46-6), CD14-PE-Cy7 (M5E2), CD16-APC-H7 (3G8; all obtained from BD), CD19-ECD (J3-119; Beckman Coulter), BDCA1-Pacific Blue (L161; BioLegend), BDCA3-APC (AD5-14H12), and CD123-PE (AC145; both obtained from Miltenyi Biotec). Cell sorting gave a purity >95%. CD8+ primary T cell lines specific for the HLA-A2–restricted HIV-Pol476-84 epitope were generated as previously described (Hoeffel et al., 2007) using PBMCs from HLA-A2+ HIV+ individuals from a cohort study with the approval of Cochin Hospital’s ethics committee. They were cultured in the presence of 1 µM Pol476-484 in complete medium (CM; RPMI 1640–GlutaMAX containing 100 U/ml penicillin, 100 mg/ml streptomycin, 1% nonessential amino acids, 1 mM sodium pyruvate, and 10 mM Hepes buffer; Life Technologies) + 5% human serum (BioWest) and maintained for 4 wk at 0.7 × 106–106 cells/ml, adding 10 U/ml IL-2 (Roche) twice a week. Uninfected (American Type Culture Collection; HTB-176) or chronically infected (American Type Culture Collection; CRL-8543) HLA-A2− H9 cells were UV-irradiated (400 mJ/cm2 at 312 nm) to induce their apoptosis and incubated at 106 cells/ml for 2 h in CM + 10% FCS (Invitrogen). Purified BDCA-1+ DCs, BDCA-3+ DCs, and pDCs were cultured with apoptotic cells at a 1:1 ratio for 16 h in the presence of a maturation signal, 100 ng/ml LPS (Sigma-Aldrich), or 10 µg/ml poly I:C (InvivoGen) or 10 µM R848 (InvivoGen) in CM + 5% human AB serum. After loading, DCs were extensively washed and used (5,000/well) in a 16-h IFN-γ ELISPOT assay with HIV-specific CD8+ T cells lines as effectors (15,000/well). All cultures were performed in the presence of 1 µM Saquinavir (Roche), an HIV-protease inhibitor, to avoid HIV replication (Hoeffel et al., 2007).
Animals.
C57BL/6Ncrl mice were provided by Charles River. XCR1−/− mice (B6.129P2-Xcr1<tm1Dgen>/J mice; MGI:3604538) were generated by Deltagen. Frozen XCR1+/− embryos were provided by The Jackson Laboratory for strain reviviscence in the Centre d’Immunologie de Marseille-Luminy (CIML). In these mice, the β-galactosidase is expressed under the Xcr1 promoter and in place of Xcr1, which permits a detection of the natural expression pattern of Xcr1. All mice were kept under pathogen-free conditions in the CIML animal care facility. Mouse experiments were conducted in accordance with institutional guidelines for animal care and use (French Provence Ethical Committee Protocol no. 04/2005 and US Office of Laboratory Animal Welfare Assurance A5665-01). Sheep were cannulated at the Center of Research in Interventional Imaging, Institut National de La Recherche Agronomique-Assistance Publique Hôpitaux de Paris (Jouy en Josas, France) for collecting prescapular skin afferent lymph as previously described (Schwartz-Cornil et al., 2006), and the sheep were housed in the Domestic Animal Experimental Unit INRA (Paris-Sud ethic commitee no. 08002A, Jouy en Josas, France).
Memory CD8+ T cell generation.
TIMs were generated by intraperitoneal injection of 50 nmol influenza A nucleoprotein NP366-374 in PBS in naive thymectomized transgenic F5 mice (Mbitikon-Kobo et al., 2009). Antiviral TCMs were generated by immunizing F5 mice with a recombinant vaccinia virus including the NP68 epitope as previously described (Cottalorda et al., 2009). Spleens and lymph nodes were collected 6 wk after infection for cell preparation as described in the next section.
Cell preparation, staining, and cell sorting for real-time PCR or transwell migration assay.
Mouse splenocytes were prepared and DC subsets sorted as described previously (Robbins et al., 2008). CD8+ T cells purified from mouse spleen and lymph nodes were stained for surface expression of CD44, CD8, and F5 TCR (Walzer et al., 2002). Naive CD8+ T cells, TIM, and antiviral TCM were sorted by flow cytometry as CD8+/CD44low, CD8+/CD44int, and CD8+/CD44high/F5 TCR+, respectively. β-Galactosidase enzyme activity in splenocytes from XCR1−/− mice was revealed using FDG as a substrate according to the manufacturer’s instructions (Invitrogen). In brief, FDG was diluted in warm distilled water at 2 mM, and cells were mixed 1:1 with the substrate. After an incubation of one minute at 37°C, cells were placed on ice, and the enzymatic reaction was terminated by adding ice-cold PBS. Cells were stained with a marker of viability and promptly acquired by flow cytometry. Sheep CD4+ T, CD8+ T, NK, and B cells were enriched from afferent lymph, and monocytes were enriched from blood. Sheep leukocytes were stained with mouse mAbs against CD1b (clone TH97A; Parsons et al., 1991), CD26 (CC69; Gliddon and Howard, 2002), CD8α (CACT80C; MacHugh and Sopp, 1991), CD4 (ST4; Hopkins, 1991), CD14 (CAM36A; Berthon and Hopkins, 1996), and NKp46 (AKS4). AKS4 was produced against bovine NKp46 according to a previously described protocol (Storset et al., 2004), and it cross reacts with ovine NK cells defined as CD16+/CD14− blood leukocytes (Elhmouzi-Younes et al., 2010). NKp46 has been shown to be the best discriminating and conserved marker for NK cell identification in mammals (Storset et al., 2004; Walzer et al., 2007). The DU2-104 mAb is used to characterize all B cells in sheep (Press et al., 1993). Sheep cells were reacted with 2 µg/ml of primary antibodies, stained with fluorochrom-conjugated purified Ig directed to specific mouse isotypes, and sorted by FACS with >95% purity.
Microarray and real-time PCR experiments.
The generation and analysis of microarray data for mouse and human DC subsets were described elsewhere (Robbins et al., 2008; Zucchini et al., 2008). All datasets used are public and their references for download from databases are given in the legends of figures. A compendium of human hgu133plus2 Affymetrix.CEL files was established, using Bioconductor (release 2.5) in the R statistical environment (version 2.10.1). This consisted of the quality control assessment using the AffyPLM package (Bolstad et al., 2004; e.g., visual examination of hybridization chip images, relative log expression, and normalized unscaled standard errors plots) and the robust multichip analysis normalization (Irizarry et al., 2003) with the Affy package. Normalized log2 scaled values were then averaged among replicate arrays for each probe, and standard deviations were calculated. For real-time PCR experiments, total RNA was prepared with either the RNA NOW reagent (Biogentex) for mouse CD8+ T cells or the RNeasy Isolation kit (QIAGEN) for mouse DCs. RNA was reverse transcribed into cDNA with the QuantiTect reverse transcription kit (QIAGEN), and the cDNA was analyzed by real-time PCR using the Power SYBR green PCR master mix (Applied Biosystems). Primers were as follows: mouse Xcr1, 5′-CTCAGCCTTGTGGGTAACAGC-3′ and 5′-ACAGGCAGTAGACAGGAGAAC-3′; mouse Ccl5, 5′-GCACCTGCCTCACCATATGG-3′ and 5′-AGCACTTGCTGCTGGTGTAG-3′; mouse Ifng, 5′-CCAGCGCCAAGCATTCAATGAG-3′ and 5′-GACAATCTCTTCCCCACCCCG-3′; mouse ubiquitin, 5′-AAGAATTCAGATCGGATGACACACT-3′ and 5′-GCCACTTGGAGGTTGACACTTT-3′; sheep XCR1, 5′-TGCCATCTTCCACAAGGTGTT-3′ and 5′-ACGGAGGCGAGGAACCA-3′; sheep XCL1, 5′-TGAGCCAGAGCAAGCCTACA-3′ and 5′-TCACTACCCAGTCAGGGTCACA-3′; and sheep GAPDH, 5′-CACCATCTTCCAGGAGCGAG-3′ and 5′-CCAGCATCACCCCACTTGAT-3′. Relative gene expression was calculated using the ΔΔCt method with b-actin for mouse DCs, ubiquitin for mouse CD8+ T cells, and GAPDH for sheep leukocytes as the endogenous control housekeeping genes.
Transwell migration assay.
Mouse DCs were obtained by density gradient enrichment from splenocytes or derived in vitro from BM Flt3-L cultures as previously described (Sjölin et al., 2006). Human DCs were enriched using the human Blood Dendritic Cell Isolation kit II (Miltenyi Biotec). Sheep DCs were enriched by density gradient. Cells were resuspended in migration medium (0.5% BSA in RPMI) and distributed in the upper chamber of a 24 Transwell system insert (5-µm pore size; Corning). The lower chamber was filled with migration medium alone or containing 300 ng/ml of mouse recombinant XCL1 (R&D Systems) for mouse cells, 500 ng/ml of human recombinant XCL1 (R&D Systems) for human cells, and 100 ng/ml of recombinant mouse XCL1 for sheep cells. Cells were incubated for 3–4 h at 37°C in 5% CO2. A constant number of polybead polystyrene microspheres (Polyscience) was added in each lower chamber before harvesting migrated cells. Migrated cells were then stained and analyzed by flow cytometry. A constant event of microspheres for each well was acquired to evaluate the absolute number of cells recovered after migration. The percentage of migrated cells was calculated by the following formula for each subset: [number of cells having migrated in response to XCL-1 − number of cells having migrated spontaneously]/[total number of input cells] × 100. All experiments were performed with duplicate wells.
Multianalyte profiling in supernatant from mouse CD8+ T cells.
2 × 105 sorted naive CD8+ T cells and TIM were stimulated for 6 h with the NP68 peptide at a concentration of 10 nM in 200 µl of medium. Supernatants were analyzed using Rodent MAP 1.0 (Rules Based Medicine).
Infections with Lm.
The Lm WT strain 10403S and the Lm strain that had been engineered to express OVA (Lm-OVA; gift from Grégoire Lauvau, Albert Einstein College of Medicine, Bronx, NY) were prepared from clones grown from organs of infected mice. Stocks of bacteria were kept frozen at −80°C. For infections, a sample of the frozen stock was grown to a logarithmic phase (OD600 = 0.05–0.15) in broth heart infusion (BHI) medium (Sigma-Aldrich), diluted in PBS, and injected into the retroorbital vein. Bacterial loads in organs were evaluated after infection of mice with one LD50 of WT Lm (5 × 104 bacteria). CD8+ T cell responses were measured in animals infected with 0.1 LD50 of Lm-OVA (5 × 103 bacteria). The dose of Lm injected was confirmed by CFU counting of the inoculum. To measure bacterial titers, spleens and livers were harvested at day 3 and dissociated on nylon meshes in 5 ml of dissociation buffer (0.2% NP-40). Serial dilutions were performed and plated onto BHI agar plates. For analysis of Lm-specific CD8+ T cell responses at day 4, OVA-specific CD8+ T cells were readily detected using OVA-iTag-H-2Kb-SIINFEKL PE-conjugated MHC class I tetramers (Beckman Coulter).
Phylogenetic analysis.
12 representatives of XCR1 sequences were retrieved from Ensembl genome assemblies of relevant vertebrate species. No good candidate for XCR1 counterpart was found in other phyla. Chromosomal locations were obtained from Ensembl (November 2009 release). Conserved syntenies were searched manually in Ensembl and with the Genomicus tool (http://www.dyogen.ens.fr/genomicus/cgi-bin/search.pl). The amino acid sequences of XCR1 domain were aligned using ClustalW and Mega4 programs. Any site at which the alignment introduces a gap in any sequence was excluded, and a comparable set of sites was therefore used for the phylogenetic analyses. The phylogenetic analysis was made using two methods: NJ and parsimony using the MEGA 4 program (Tamura et al., 2007).
Online supplemental material.
Fig. S1 documents the expression pattern of XCL1 in mouse NK cells, mouse TCM, and human CD8+ T cells during viral infections. Fig. S2 shows the alignment of XCR1 protein sequences and the Maximum Parsimony phylogenetic analysis of Xcr1 evolution in vertebrates. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20100223/DC1.
Acknowledgments
We thank Michel Bonneau (Centre de Recherche en Imagerie Interventionnelle, Jouy en Josas, France) for the sheep surgery, Céline Urien for excellent technical support, the Domestic Animal Experimental Unit (UCEA, Institut National de La Recherche Agronomique, Jouy en Josas, France) for sheep care, the Centre d’Immunologie de Marseille-Luminy (CIML) staff of the mouse care facilities and the flow cytometry core facility for assistance, and Grégoire Lauvau (Albert Einstein College of Medicine, New York, USA) for providing the Lm strains. We thank Lene Vimeux (Centre National de la Recherche Scientifique [CNRS] UMR 8104) for generating T cell lines, and Antonio Cosma, Sabrina Guenounou, Thibault Andrieu, and Roger Le Grand (flow cytometry cell sorting core facility of the Commissariat à l’Énergie Atomique [CEA] in Fontenay aux Roses, France). We thank Bernard Charley and Bernard Malissen for critical reading of the manuscript.
This work was supported by Association pour la recherche sur le cancer (postdoctoral fellowship to K. Crozat and research grants to M. Dalod and to J. Marvel), CNRS (postdoctoral fellowship to R. Guiton), Agence nationale de recherches sur le SIDA et les hépatites virales (fellowship to C-A. Dutertre), Institut National de la Santé et de la Recherche Médicale (Jeune Chercheur fellowship to V. Feuillet), Ligue nationale contre le cancer (fellowship to E. Ventre), institutional grants to CIML, and the Agence nationale de la recherche program Genanimal 2006 (research grant to I. Schwartz-Cornil).
The authors declare no competing financial interests.
Footnotes
Abbreviations used:
- CCR
- CC chemokine receptor
- FDG
- fluorescein di-β-d-galactopyranoside
- Lm
- Listeria monocytogenes
- NJ
- neighbor joining
- pDC
- plasmacytoid DC
- TCM
- central memory CD8+ T lymphocyte
- TIM
- inflammatory memory CD8+ T cell
- XCL1
- chemokine (C motif) ligand 1
- XCR1
- XC chemokine receptor 1
References
- Aoshi T., Zinselmeyer B.H., Konjufca V., Lynch J.N., Zhang X., Koide Y., Miller M.J. 2008. Bacterial entry to the splenic white pulp initiates antigen presentation to CD8+ T cells. Immunity. 29:476–486 10.1016/j.immuni.2008.06.013 [DOI] [PubMed] [Google Scholar]
- Baranek T., Zucchini N., Dalod M. 2009. Plasmacytoid dendritic cells and the control of herpesvirus infections. Viruses. 1:383–419 10.3390/v1030383 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Belz G.T., Shortman K., Bevan M.J., Heath W.R. 2005. CD8alpha+ dendritic cells selectively present MHC class I-restricted noncytolytic viral and intracellular bacterial antigens in vivo. J. Immunol. 175:196–200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berthon P., Hopkins J. 1996. Ruminant cluster CD14. Vet. Immunol. Immunopathol. 52:245–248 10.1016/0165-2427(96)05568-7 [DOI] [PubMed] [Google Scholar]
- Bolstad B.M., Collin F., Simpson K.M., Irizarry R.A., Speed T.P. 2004. Experimental design and low-level analysis of microarray data. Int. Rev. Neurobiol. 60:25–58 10.1016/S0074-7742(04)60002-X [DOI] [PubMed] [Google Scholar]
- Bonifaz L.C., Bonnyay D.P., Charalambous A., Darguste D.I., Fujii S., Soares H., Brimnes M.K., Moltedo B., Moran T.M., Steinman R.M. 2004. In vivo targeting of antigens to maturing dendritic cells via the DEC-205 receptor improves T cell vaccination. J. Exp. Med. 199:815–824 10.1084/jem.20032220 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminschi I., Proietto A.I., Ahmet F., Kitsoulis S., Shin Teh J., Lo J.C., Rizzitelli A., Wu L., Vremec D., van Dommelen S.L., et al. 2008. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood. 112:3264–3273 10.1182/blood-2008-05-155176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carter R.W., Thompson C., Reid D.M., Wong S.Y., Tough D.F. 2006. Preferential induction of CD4+ T cell responses through in vivo targeting of antigen to dendritic cell-associated C-type lectin-1. J. Immunol. 177:2276–2284 [DOI] [PubMed] [Google Scholar]
- Cottalorda A., Mercier B.C., Mbitikon-Kobo F.M., Arpin C., Teoh D.Y., McMichael A., Marvel J., Bonnefoy-Bérard N. 2009. TLR2 engagement on memory CD8(+) T cells improves their cytokine-mediated proliferation and IFN-gamma secretion in the absence of Ag. Eur. J. Immunol. 39:2673–2681 10.1002/eji.200939627 [DOI] [PubMed] [Google Scholar]
- Crozat K., Guiton R., Guilliams M., Henri S., Baranek T., Schwartz-Cornil I., Malissen B., Dalod M. 2010. Comparative genomics as a tool to reveal functional equivalences between human and mouse dendritic cell subsets. Immunol. Rev. 234:177–198 10.1111/j.0105-2896.2009.00868.x [DOI] [PubMed] [Google Scholar]
- Dorner B.G., Scheffold A., Rolph M.S., Huser M.B., Kaufmann S.H., Radbruch A., Flesch I.E., Kroczek R.A. 2002. MIP-1alpha, MIP-1beta, RANTES, and ATAC/lymphotactin function together with IFN-gamma as type 1 cytokines. Proc. Natl. Acad. Sci. USA. 99:6181–6186 10.1073/pnas.092141999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner B.G., Dorner M.B., Zhou X., Opitz C., Mora A., Güttler S., Hutloff A., Mages H.W., Ranke K., Schaefer M., et al. 2009. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity. 31:823–833 10.1016/j.immuni.2009.08.027 [DOI] [PubMed] [Google Scholar]
- Du X., Tang Y., Xu H., Lit L., Walker W., Ashwood P., Gregg J.P., Sharp F.R. 2006. Genomic profiles for human peripheral blood T cells, B cells, natural killer cells, monocytes, and polymorphonuclear cells: comparisons to ischemic stroke, migraine, and Tourette syndrome. Genomics. 87:693–703 10.1016/j.ygeno.2006.02.003 [DOI] [PubMed] [Google Scholar]
- Dudziak D., Kamphorst A.O., Heidkamp G.F., Buchholz V.R., Trumpfheller C., Yamazaki S., Cheong C., Liu K., Lee H.W., Park C.G., et al. 2007. Differential antigen processing by dendritic cell subsets in vivo. Science. 315:107–111 10.1126/science.1136080 [DOI] [PubMed] [Google Scholar]
- Elhmouzi-Younes J., Boysen P., Pende D., Storset A.K., Le Vern Y., Laurent F., Drouet F. 2010. Ovine CD16+/CD14- blood lymphocytes present all the major characteristics of natural killer cells. Vet. Res. 41:4 10.1051/vetres/2009052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epardaud M., Bonneau M., Payot F., Cordier C., Mégret J., Howard C., Schwartz-Cornil I. 2004. Enrichment for a CD26hi SIRP- subset in lymph dendritic cells from the upper aero-digestive tract. J. Leukoc. Biol. 76:553–561 10.1189/jlb.0404223 [DOI] [PubMed] [Google Scholar]
- Gliddon D.R., Howard C.J. 2002. CD26 is expressed on a restricted subpopulation of dendritic cells in vivo. Eur. J. Immunol. 32:1472–1481 [DOI] [PubMed] [Google Scholar]
- Hildner K., Edelson B.T., Purtha W.E., Diamond M., Matsushita H., Kohyama M., Calderon B., Schraml B.U., Unanue E.R., Diamond M.S., et al. 2008. Batf3 deficiency reveals a critical role for CD8alpha+ dendritic cells in cytotoxic T cell immunity. Science. 322:1097–1100 10.1126/science.1164206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoeffel G., Ripoche A.C., Matheoud D., Nascimbeni M., Escriou N., Lebon P., Heshmati F., Guillet J.G., Gannagé M., Caillat-Zucman S., et al. 2007. Antigen crosspresentation by human plasmacytoid dendritic cells. Immunity. 27:481–492 10.1016/j.immuni.2007.07.021 [DOI] [PubMed] [Google Scholar]
- Hopkins J. 1991. Workshop studies on the ovine CD4 homologue. Vet. Immunol. Immunopathol. 27:101–102 10.1016/0165-2427(91)90087-S [DOI] [PubMed] [Google Scholar]
- Huysamen C., Willment J.A., Dennehy K.M., Brown G.D. 2008. CLEC9A is a novel activation C-type lectin-like receptor expressed on BDCA3+ dendritic cells and a subset of monocytes. J. Biol. Chem. 283:16693–16701 10.1074/jbc.M709923200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyrcza M.D., Kovacs C., Loutfy M., Halpenny R., Heisler L., Yang S., Wilkins O., Ostrowski M., Der S.D. 2007. Distinct transcriptional profiles in ex vivo CD4+ and CD8+ T cells are established early in human immunodeficiency virus type 1 infection and are characterized by a chronic interferon response as well as extensive transcriptional changes in CD8+ T cells. J. Virol. 81:3477–3486 10.1128/JVI.01552-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khanna K.M., McNamara J.T., Lefrançois L. 2007. In situ imaging of the endogenous CD8 T cell response to infection. Science. 318:116–120 10.1126/science.1146291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lindstedt M., Lundberg K., Borrebaeck C.A. 2005. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J. Immunol. 175:4839–4846 [DOI] [PubMed] [Google Scholar]
- MacHugh N.D., Sopp P. 1991. Individual antigens of cattle. Bovine CD8 (BoCD8). Vet. Immunol. Immunopathol. 27:65–69 10.1016/0165-2427(91)90081-M [DOI] [PubMed] [Google Scholar]
- Mbitikon-Kobo F.M., Vocanson M., Michallet M.C., Tomkowiak M., Cottalorda A., Angelov G.S., Coupet C.A., Djebali S., Marçais A., Dubois B., et al. 2009. Characterization of a CD44/CD122int memory CD8 T cell subset generated under sterile inflammatory conditions. J. Immunol. 182:3846–3854 10.4049/jimmunol.0802438 [DOI] [PubMed] [Google Scholar]
- Müller S., Dorner B., Korthaüer U., Mages H.W., D’Apuzzo M., Senger G., Kroczek R.A. 1995. Cloning of ATAC, an activation-induced, chemokine-related molecule exclusively expressed in CD8+ T lymphocytes. Eur. J. Immunol. 25:1744–1748 10.1002/eji.1830250638 [DOI] [PubMed] [Google Scholar]
- Naik S.H., Proietto A.I., Wilson N.S., Dakic A., Schnorrer P., Fuchsberger M., Lahoud M.H., O’Keeffe M., Shao Q.X., Chen W.F., et al. 2005. Cutting edge: generation of splenic CD8+ and CD8- dendritic cell equivalents in Fms-like tyrosine kinase 3 ligand bone marrow cultures. J. Immunol. 174:6592–6597 [DOI] [PubMed] [Google Scholar]
- Nchinda G., Kuroiwa J., Oks M., Trumpfheller C., Park C.G., Huang Y., Hannaman D., Schlesinger S.J., Mizenina O., Nussenzweig M.C., et al. 2008. The efficacy of DNA vaccination is enhanced in mice by targeting the encoded protein to dendritic cells. J. Clin. Invest. 118:1427–1436 10.1172/JCI34224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons K.R., Howard C.J., Sopp P. 1991. Immunohistology of workshop monoclonal antibodies to the bovine homologue of CD1. Vet. Immunol. Immunopathol. 27:201–206 10.1016/0165-2427(91)90101-H [DOI] [PubMed] [Google Scholar]
- Press C.M., Hein W.R., Landsverk T. 1993. Ontogeny of leucocyte populations in the spleen of fetal lambs with emphasis on the early prominence of B cells. Immunology. 80:598–604 [PMC free article] [PubMed] [Google Scholar]
- Ravi V., Venkatesh B. 2008. Rapidly evolving fish genomes and teleost diversity. Curr. Opin. Genet. Dev. 18:544–550 10.1016/j.gde.2008.11.001 [DOI] [PubMed] [Google Scholar]
- Robbins S.H., Walzer T., Dembélé D., Thibault C., Defays A., Bessou G., Xu H., Vivier E., Sellars M., Pierre P., et al. 2008. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9:R17 10.1186/gb-2008-9-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sancho D., Mourão-Sá D., Joffre O.P., Schulz O., Rogers N.C., Pennington D.J., Carlyle J.R., Reis e Sousa C. 2008. Tumor therapy in mice via antigen targeting to a novel, DC-restricted C-type lectin. J. Clin. Invest. 118:2098–2110 10.1172/JCI34584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sarkar S., Kalia V., Haining W.N., Konieczny B.T., Subramaniam S., Ahmed R. 2008. Functional and genomic profiling of effector CD8 T cell subsets with distinct memory fates. J. Exp. Med. 205:625–640 10.1084/jem.20071641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz-Cornil I., Epardaud M., Bonneau M. 2006. Cervical duct cannulation in sheep for collection of afferent lymph dendritic cells from head tissues. Nat. Protoc. 1:874–879 10.1038/nprot.2006.147 [DOI] [PubMed] [Google Scholar]
- Sjölin H., Robbins S.H., Bessou G., Hidmark A., Tomasello E., Johansson M., Hall H., Charifi F., Karlsson Hedestam G.B., Biron C.A., et al. 2006. DAP12 signaling regulates plasmacytoid dendritic cell homeostasis and down-modulates their function during viral infection. J. Immunol. 177:2908–2916 [DOI] [PubMed] [Google Scholar]
- Storset A.K., Kulberg S., Berg I., Boysen P., Hope J.C., Dissen E. 2004. NKp46 defines a subset of bovine leukocytes with natural killer cell characteristics. Eur. J. Immunol. 34:669–676 10.1002/eji.200324504 [DOI] [PubMed] [Google Scholar]
- Tamura K., Dudley J., Nei M., Kumar S. 2007. MEGA4: Molecular Evolutionary Genetics Analysis (MEGA) software version 4.0. Mol. Biol. Evol. 24:1596–1599 10.1093/molbev/msm092 [DOI] [PubMed] [Google Scholar]
- Walzer T., Arpin C., Beloeil L., Marvel J. 2002. Differential in vivo persistence of two subsets of memory phenotype CD8 T cells defined by CD44 and CD122 expression levels. J. Immunol. 168:2704–2711 [DOI] [PubMed] [Google Scholar]
- Walzer T., Marçais A., Saltel F., Bella C., Jurdic P., Marvel J. 2003. Cutting edge: immediate RANTES secretion by resting memory CD8 T cells following antigenic stimulation. J. Immunol. 170:1615–1619 [DOI] [PubMed] [Google Scholar]
- Walzer T., Bléry M., Chaix J., Fuseri N., Chasson L., Robbins S.H., Jaeger S., André P., Gauthier L., Daniel L., et al. 2007. Identification, activation, and selective in vivo ablation of mouse NK cells via NKp46. Proc. Natl. Acad. Sci. USA. 104:3384–3389 10.1073/pnas.0609692104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zucchini N., Bessou G., Robbins S.H., Chasson L., Raper A., Crocker P.R., Dalod M. 2008. Individual plasmacytoid dendritic cells are major contributors to the production of multiple innate cytokines in an organ-specific manner during viral infection. Int. Immunol. 20:45–56 10.1093/intimm/dxm119 [DOI] [PMC free article] [PubMed] [Google Scholar]