Abstract
In recent years, human dendritic cells (DCs) could be subdivided into CD304+ plasmacytoid DCs (pDCs) and conventional DCs (cDCs), the latter encompassing the CD1c+, CD16+, and CD141+ DC subsets. To date, the low frequency of these DCs in human blood has essentially prevented functional studies defining their specific contribution to antigen presentation. We have established a protocol for an effective isolation of pDC and cDC subsets to high purity. Using this approach, we show that CD141+ DCs are the only cells in human blood that express the chemokine receptor XCR1 and respond to the specific ligand XCL1 by Ca2+ mobilization and potent chemotaxis. More importantly, we demonstrate that CD141+ DCs excel in cross-presentation of soluble or cell-associated antigen to CD8+ T cells when directly compared with CD1c+ DCs, CD16+ DCs, and pDCs from the same donors. Both in their functional XCR1 expression and their effective processing and presentation of exogenous antigen in the context of major histocompatibility complex class I, human CD141+ DCs correspond to mouse CD8+ DCs, a subset known for superior antigen cross-presentation in vivo. These data define CD141+ DCs as professional antigen cross-presenting DCs in the human.
The adaptive immune response is initiated through presentation of antigen to T cells by DCs. In the mouse, DCs can be broadly grouped into plasmacytoid DCs (pDCs) and conventional DCs (cDCs; earlier termed myeloid DCs). Mouse cDCs can be further subdivided into several DC types, which are apparently specialized for optimal antigen uptake, processing, and presentation to T cells in different body compartments (Steinman and Banchereau, 2007; Heath and Carbone, 2009; Segura and Villadangos, 2009). One particular type of antigen presentation is cross-presentation: in this case, extracellular antigen is not classically presented in the context of MHC-II but is instead shunted into the MHC-I presentation pathway (Bevan, 2006; Shen and Rock, 2006; Villadangos et al., 2007). CD8+ T cells can thus be activated by antigens taken up from the extracellular space and then differentiate into cytotoxic T cells. This mechanism is thought to be of major importance for the recognition of viral or bacterial antigens when DCs are not directly infected. In these instances, debris of cells that were infected and have subsequently undergone apoptosis as part of a cellular stress reaction is taken up and cross-presented by specialized DCs. Through this type of processing, the antigenic composition of the pathogen can become visible to the CD8+ T cell immune system. In the mouse, extensive experimentation has demonstrated that within cDCs, CD8+ DCs are the most effective in antigen cross-presentation (den Haan et al., 2000; Iyoda et al., 2002; Schulz and Reis e Sousa, 2002; Heath et al., 2004). Whether mouse pDCs play a significant role in antigen presentation and more so in antigen cross-presentation is controversial (Colonna et al., 2004; Liu, 2005; Villadangos and Young, 2008).
We have recently shown in the mouse system that splenic CD8+ DCs (and their counterparts in other organs) are the only cells in the body expressing XCR1, a chemokine receptor with a unique ligand, XCL1 (Dorner et al., 2009). In vitro, XCL1 induces potent chemotaxis of XCR1+ CD8+ DCs. In vivo, XCL1 secreted by activated CD8+ T cells augments their expansion and differentiation into cytotoxic T cells when the antigen is cross-presented by CD8+ DCs in the context of MHC-I (Dorner et al., 2009). Collectively, these observations indicate that the XCL1–XCR1 communication axis optimizes the cooperation of antigen-specific CD8+ T cells with XCR1+ DCs, which cross-present antigen to them.
Based on our studies in the mouse, we were interested to determine whether human DCs express XCR1. Human DCs have been extensively phenotyped in the past and subdivided again into pDC and into CD1c+ (BDCA-1+), CD16+, and CD141+ (BDCA-3+) cDC subsets (Dzionek et al., 2000; MacDonald et al., 2002; Piccioli et al., 2007; for review see Ju et al., 2010). Meticulous gene expression analyses of all human and mouse DCs have recently revealed a large gene expression program shared by human and mouse pDCs, and also led to the suggestion that human CD141+ DCs correspond to mouse CD8+ DCs (Robbins et al., 2008). In spite of this groundbreaking work on the subdivision of human DCs into subsets, information on the function of human primary DCs remained very scarce, apparently because of the limitations imposed by the very low frequencies of DCs in human blood (CD1c+ DCs, 0.31 ± 0.14% SD; CD16+ DCs, 0.75 ± 0.41%; CD141+ DCs, 0.04 ± 0.03%; pDCs, 0.29 ± 0.08%; n = 8; not depicted). Instead, antigen cross-presentation in the human system was essentially analyzed with DCs derived from monocytes in culture (Fonteneau et al., 2003), a system that may not reflect all of the functional properties of primary DCs.
In the present study, we demonstrate that CD141+ DCs are the only population in human blood that expresses the chemokine receptor XCR1. Human CD141+ DCs react to the chemokine XCL1 by mobilization of intracellular Ca2+ ([Ca2+]i) and by strong chemotaxis in vitro. More importantly, our experiments demonstrate that primary CD141+ DCs excel in cross-presentation of antigen when directly compared with CD1c+ DCs, CD16+ DCs, and pDCs from the same donors. Collectively, these functional data strongly indicate that human CD141+ DCs are the homologue of mouse CD8+ DCs. At the same time, the professional capacity of human CD141+ DCs to cross-present antigen is of major interest in the ongoing quest to develop vaccines capable of inducing antiviral or antitumor cytotoxicity in the human.
RESULTS
Strategy for identification and sorting of human cDC subsets
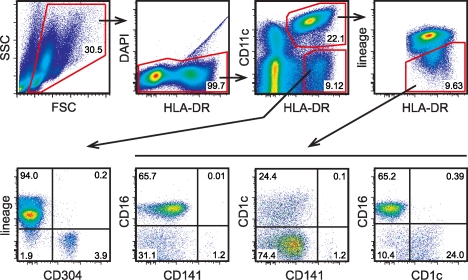
The highly restricted expression of XCR1 in subsets of mouse DCs was suggestive of a similarly restricted expression in the human. Therefore, we first established a reliable process for the identification and sorting of DC subsets from human blood. Although the phenotype of pDCs in human blood could be established unequivocally (Robinson et al., 1999; Dzionek et al., 2000), several differing approaches were undertaken to define human cDCs in the past (Dzionek et al., 2000; MacDonald et al., 2002; Lindstedt et al., 2005; Piccioli et al., 2007). Our strategy was based on the observation that all human blood cDCs express HLA-DR and CD11c (MacDonald et al., 2002), and on the conclusion that HLA-DR+ CD34+ cells should not be regarded as DCs (Piccioli et al., 2007). Using the gating depicted in Fig. 1, we were able to clearly define distinct populations within cDCs, which in total represent ∼1.1% (±0.38% SD) of PBMCs in human blood (not depicted). The cDCs are composed of subsets expressing CD16 (68.2 ± 37.3%), CD1c (28.2 ± 12.7%), and the very small subset expressing CD141 (3.6 ± 2.7%). This gating strategy was the basis for the isolation of all cDC subsets to a very high purity.
Figure 1.
Strategy for defining human DC subsets in the blood. PBMCs from human blood were enriched by density gradient centrifugation and stained for the indicated cell-surface markers (for the mAb used for staining see Materials and methods). Flow sorting (after magnetic cell enrichment) was performed on the principle of the gating strategy shown. This approach allowed us to isolate CD304+ pDCs, CD16+ DCs, CD1c+ DCs, and CD141+ DCs to very high purities. The inset numbers represent the percentage of the gated cells in the respective gating step. FSC, forward scatter; SSC, side scatter.
The chemokine receptor XCR1 is selectively expressed in human CD141+ DCs
To identify cells expressing XCR1 in the human, we set up a highly sensitive and specific quantitative RT-PCR (qRT-PCR), with forward and reverse primers in exons 1 and 3, respectively, and a probe in the noncoding region of exon 3 (F1, R1, and P1; Fig. 2). This assay gave a very low signal with PBMCs (∼300 copies in 100 ng of total RNA; unpublished data). To identify the source of this signal, we isolated various cell types from PBMCs to a very high purity (see Materials and methods) and subjected them to qRT-PCR. No signal was obtained in T cells, B cells, NK cells, granulocytes, monocytes, pDCs, CD1c+ DCs, and CD16+ DCs (Fig. 2 B). In contrast, a strong XCR1 signal (∼700,000 copies in 100 ng of total RNA) was obtained with CD141+ DCs (Fig. 2 B). In addition, we tested monocyte-derived DCs (MoDCs) in culture and found them to be negative (Fig. 2 B). To exclude cell-specific expression of a XCR1 splice variant, we used a primer set within the coding region of XCR1 for the qRT-PCR (F2, R2, and P2; Fig. 2 A) and obtained identical results (not depicted). In all assays, the β2-microglobulin signal was used as a positive control (unpublished data). Collectively, these experiments determined that in human blood, XCR1 is only expressed in CD141+ DCs.
Figure 2.
The chemokine receptor XCR1 is selectively expressed in CD141+ DCs. (A) Organization of the human XCR1 gene (E1, exon 1; E2, exon 2; and E3, exon 3); the coding region is shown in black, and the primer–probe sets used for expression analyses are indicated. (B) qRT-PCR of total RNA from T cells, B cells, NK cells, granulocytes, monocytes, pDCs, CD1c+ DCs, CD16+ DCs, and CD141+ DCs isolated to a very high purity (>97.5%) from PBMCs and tested with primer–probe set 1 (F1, R1, and probe P1). Cultured MoDCs were also tested. Identical results were obtained using primer–probe set 2 (F2, R2, and P2; not depicted). The test systems used allowed the detection of ≥200 copies of XCR1 in cDNA reverse transcribed from 100 ng of total RNA. All PCR analyses were performed with cell subsets from at least two donors. Error bars represent means ± SEM.
XCL1 mobilizes Ca2+ in CD141+ DCs but not in CD1c+ and CD16+ DCs
To test the function of XCR1 in human CD141+ DCs, we isolated this subset as well as the other cDC subsets from human blood (purity in all instances was >98.7%) and exposed them to XCL1. In a single-cell Ca2+ assay, ∼70% of all fura-2–loaded CD141+ DCs responded to XCL1 with a characteristic transient increase in [Ca2+]i indicative of a phosphoinositide hydrolysis–dependent and inositol-1,4,5-trisphosphate–triggered mobilization of Ca2+ from storage organelles. Approximately 20% of the cells responded only to the positive control (a mixture of CCL2, CCL21, CXCL9, and CX3CL1, selected based on the data of Robbins et al. [2008]), and 10% gave no response (Fig. 3). CD1c+ and CD16+ DCs, although alive and reactive, failed to respond to XCL1 under identical conditions (unpublished data). These results indicate that XCR1 is expressed on the cell surface in the majority of CD141+ DCs and can transduce an activation signal upon binding of its chemokine ligand, XCL1.
Figure 3.
XCL1 induces a [Ca2+]i signal in CD141+ DCs. CD141+ DCs were flow sorted to a purity >98.7%, immobilized on poly–l-lysine–coated glass coverslips, and loaded with 2 µM fura-2/AM. Cells were imaged in a monochromator-assisted digital video imaging system and challenged with 1 µg/ml XCL1 as indicated (left arrow). Subsequently, the same cells were challenged again with a mixture of 100 ng/ml CCL2, 200 ng/ml CCL21, 200 ng/ml CXCL9, and 1 ng/ml CX3CL1 used as a positive control (right arrow). The data shown represent [Ca2+]i concentrations of 300 single cells (gray lines) measured in two independent experiments. The mean [Ca2+]i signal averaged over all cells responding to XCL1 is indicated (black line).
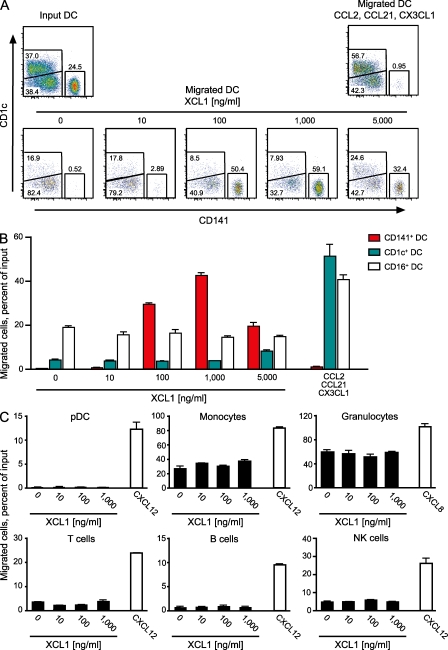
XCL1 selectively chemoattracts CD141+ DCs
To test human XCL1 for chemotaxis, highly purified, flow-sorted DC subtypes were mixed at a ratio of ∼20% CD141+, 40% CD16+, and 40% CD1c+ DCs (Fig. 4 A, Input DC) and assayed in a Transwell system. Without addition of a chemokine, only CD16+ DCs within this mixed population exhibited some background migration (Fig. 4 A). Upon addition of increasing amounts of XCL1, a selective migration of CD141+ DCs was observed (Fig. 4 A). At the optimal concentrations of XCL1 (100–1,000 ng/ml), >40% of all input CD141+ DCs migrated (in one experiment >70%). Checkerboard tests with XCL1 indicated true chemotaxis and not only chemokinesis (unpublished data). CD1c+ and CD16+ DCs responded to various concentrations of XCL1 only by background migration; positive controls with a mixture of the chemokines CCL2, CCL21, and CX3CL1 (Fig. 4 A, Migrated DC) ensured that this unresponsiveness to XCL1 was not caused by a general unresponsiveness of these DC subsets. In the literature, migration of T cells, B cells, NK cells, and granulocytes was repeatedly reported in response to XCL1 (for a compilation of references see Dorner et al., 2009). When we performed analogous Transwell experiments, XCL1 failed to induce migration with any of these cell populations, whereas the respective positive control chemokines (CXCL12 and CXCL8) were effective (Fig. 4 C). Collectively, these experiments established that XCL1 is a chemokine with selective action on CD141+ DCs.
Figure 4.
XCL1 selectively induces chemotaxis in CD141+ DCs. (A) A mixture of highly purified, flow-sorted DC subtypes (20% CD141+, 40% CD16+, and 40% CD1c+ DCs; Input DC) was tested for migration in response to medium alone or to serial dilutions of XCL1 (10–5,000 ng/ml) in a Transwell system. A combination of the chemokines CCL2, CCL21, and CX3CL1 was used as a positive control for the DC subsets (Migrated DC). The absolute numbers of CD141+, CD1c+, and CD16+ DCs in input and migrated cell populations are truly represented in the dot plots, because all cells within a defined volume were included in the analysis in each instance. (B) Proportion of migrated CD1c+, CD16+, and CD141+ DCs in the experiment shown in A. (C) Proportion of migrated pDCs, monocytes, granulocytes, T cells, B cells, and NK cells in response to XCL1 (10–1,000 ng/ml) or the chemokines CXCL12 and CXCL8, which were used as positive controls. For migration assays of B cells, NK cells, and monocytes, PBMCs were magnetically depleted of T cells, and for T cell migration, PBMCs were used directly. For migration assays of granulocytes, whole blood cells were used after erythrocyte lysis with ACK buffer, and pDCs were magnetically enriched from PBMCs with the Plasmacytoid Dendritic Cell Isolation Kit (Miltenyi Biotec). All experiments with DCs were performed three times; all other populations were assayed twice. Error bars represent means ± SEM.
CD141+ DCs efficiently cross-present antigen
In the mouse system, XCR1 is exclusively expressed on a DC subset (bearing the surface marker CD8 in the spleen) known for its superior capacity to cross-present antigen in vivo (Dorner et al., 2009). We therefore decided to directly compare human CD141+ DCs with the other DC subtypes for antigen cross-presentation when purified from the blood of single donors. To this end, CD141+ DCs, CD1c+ DCs, CD16+ DCs, and pDCs from HLA-A*0201 donors were co-cultured with a CD8+ T cell clone recognizing the human CMV (HCMV) peptide NLVPMVATV (pp65495–503) when presented in the context of HLA-A*0201. Addition of recombinant pp65 protein in a soluble form to co-cultures with CD141+ DCs triggered strong IFN-γ secretion by the T cell clone, indicating an efficient intracellular degradation of pp65 and subsequent presentation of the NLVPMVATV peptide on the surface of this DC subset (Fig. 5 A). Addition of recombinant pp65 protein to co-cultures with CD1c+ or CD16+ DCs resulted in clearly lower IFN-γ signals; in the case of pDCs the signal was minimal (Fig. 5 A). Addition of OVA, a protein whose degradation products cannot be recognized by the T cell clone, gave only minimal background signals, as was the case in cultures containing only T cells or DCs (Fig. 5 A). Positive controls with the pp65 peptide NLVPMVATV present in the co-cultures (containing lower numbers of T cells and DCs) gave a comparably strong IFN-γ signal with all cDC subsets but a lower signal with pDCs (Fig. 5 A), although the viability of all DC subtypes at the end of the co-culture period was in the range of 60–70%.
Figure 5.
Capacity of CD141+, CD1c+, and CD16+ DCs to cross-present soluble and cell-associated HCMV pp65 antigen. (A) CD8+ T cell clone 10, specific for the HLA-A*0201–restricted HCMV pp65 peptide NLVPMVATV (pp65495–503), was co-cultured with CD141+ DCs, CD1c+ DCs, CD16+ DCs, or pDCs obtained from the buffy coat of one HLA-A*0201+ blood donation, with 3 µg/ml of recombinant soluble HCMV pp65 added for the entire culture period. The activation of the T cell clone was determined by measuring the concentration of IFN-γ in the supernatants at the termination of culture after 20 h. Negative controls included addition of irrelevant protein OVA (3 µg/ml) and cultures with only the T cell clone or DCs; addition of 1 µg/ml of pp65495–503 peptide to the co-cultures served as a positive control. Shown is one representative experiment out of nine; each experiment was performed with cells from a different donor (all experiments included all cDCs subsets; four of them also included pDCs). (B) CD141+, CD1c+, or CD16+ DCs isolated from leukapheresis PBMCs of one HLA-A*0201+ donor were co-cultured with CD8+ T cell clone 61 at variable ratios (from 1:1 to 1:16) with cell-associated pp65 antigen added for the entire culture period, and IFN-γ was determined in the supernatant after 24 h. Negative controls included irrelevant MART-127–35 peptide, and positive controls included HCMV pp65495–503 peptide (both at 1 µg/ml). Shown are results representative of three experiments with different donors. Error bars represent means ± SEM.
Because cross-presentation is mainly implicated in the uptake of cell debris, we also performed analogous tests with the cell-associated HCMV pp65 protein. CD141+, CD1c+, or CD16+ DCs were co-cultured with a CD8+ T cell clone at variable ratios of 1:1 to 1:16 in the presence of cell material obtained by repeated freeze–thaw cycles of a HCMV pp65 transfectant. These cross-presentation tests gave strong IFN-γ signals with CD141+ DCs, and only low responses with CD1c+ and CD16+ DCs (Fig. 5 B). The signals obtained with the negative control peptide MART-127–35 were always minimal with CD141+ and CD1c+ DCs; the positive controls with the pp65495–503 peptide indicated similar expression of HLA-A*0201 on the surface of these cDC subsets. Collectively, these tests with soluble or cell-associated HCMV pp65 demonstrated a superior capacity of CD141+ DCs to cross-present antigen to CD8+ T cells.
DISCUSSION
The very low frequencies of pDCs and cDCs in human blood strongly impeded functional studies with primary populations in the past. As a result, data on antigen presentation, and in particular on antigen cross-presentation, remained scarce. This limited information also made it difficult to directly compare human cDC subsets with the extensively characterized mouse cDC subtypes. In the mouse, it has been recognized that each DC subset is optimally equipped for the uptake, recognition, and presentation of different antigens or pathogens. One particular mouse cDC subset bearing the CD8 marker in the spleen (CD8+ DCs) very efficiently takes up antigen from the extracellular space and cross-presents it to CD8+ T cells (den Haan et al., 2000; Iyoda et al., 2002; Schulz and Reis e Sousa, 2002; Heath et al., 2004). This professional antigen cross-presentation is regarded as important for the immune defense of intracellular viral and bacterial pathogens, as well as for the eradication of tumor tissue (Lin et al., 2008; Shortman and Heath 2010). We have recently shown that these mouse splenic CD8+ DCs (and their counterparts in other organs) exclusively express the chemokine receptor XCR1 and selectively migrate to its chemokine ligand XCL1 (Dorner et al., 2009).
We now have developed an effective scheme for the isolation of all currently defined human DC subsets, including pDCs (CD304+) and cDCs (CD1c+, CD16+, or CD141+) to a very high purity from single-blood donations. This approach allowed us to search for the human counterpart of the cross-presenting mouse CD8+ DCs. In the past, concordant expression of certain cell-surface molecules on both human CD141+ DCs and mouse CD8+ DCs led to initial suggestions that these two DC subsets may be related (Galibert et al., 2005; Caminschi et al., 2008). By meticulously comparing the gene expression patterns of all known human and mouse DC subsets, a recent study substantiated these suggestions (Robbins et al., 2008). In our current work, we now demonstrate that CD141+ DCs, which represent only ∼3–4% of all DCs in human blood, exclusively express the chemokine receptor XCR1 and selectively respond to the chemokine ligand XCL1 by [Ca2+]i mobilization and chemotaxis. More importantly in terms of biological function, we now show that CD141+ DCs excel in cross-presentation of antigen to CD8+ T cells when directly compared with all other human pDC and cDC subsets in vitro. Collectively, these results define CD141+ DCs as professional antigen–cross-presenting DCs in the human and allow the conclusion that this DC subset is the homologue of mouse CD8+ DCs.
The identification of professional human cross-presenting DCs is an important step forward, because it indicates that the biological concepts on antigen cross-presentation derived from extensive experimentation in the mouse can apparently be transferred to the human. However, many questions remain to be answered. For example, the identification of cross-presenting DCs in the blood raises the issue of whether these cells are also resident in lymphoid tissues and other organs. Our preliminary data clearly point in this direction. In the thymus, we could observe that CD141+ cDCs express XCR1 mRNA, and CD141+ cells obtained from tonsillectomies also gave substantial PCR signals for XCR1. Attempts to locate human CD141+ DCs in lymphoid tissues using histology were confounded by a relatively diffuse expression pattern obtained in areas populated by lymphoid cells and by the dominant expression of CD141 on the endothelium (Esmon et al., 1982). A definitive proof of the presence of human DCs capable of antigen cross-presentation in lymphoid tissues will require further phenotypic dissection of the local DC subtypes combined with antigen presentation studies in vitro.
We are not aware of any data in the literature on the capacity of CD141+ DCs to present or cross-present antigen to T cells. With other primary human DC subsets, functional data on cross-presentation are very scarce. Schnurr et al. (2005) functionally compared primary CD1c+ DCs (cultured overnight with GM-CSF) with primary pDCs (cultured with IL-3) and found only CD1c+ DCs capable of antigen cross-presentation to CD8+ T cells, whereas pDCs were totally ineffective. In contrast, Hoeffel et al. (2007) reported a similar cross-presentation by cDCs and pDCs, and Di Pucchio et al. (2008) also found a comparable cross-presentation capacity of CD1c+ DCs and pDCs. In neither of the cited reports have CD141+ DCs been removed before experimentation, but this technical aspect is unlikely to be the cause for the differing results, because the frequency of CD141+ DCs within the tested DC populations is so low. Instead, other technical details may have influenced the results. Hoeffel et al. (2007), e.g., have used a lipopeptide as a source of antigen (for a short-term assay), whereas Di Pucchio et al. (2008) assessed antigen cross-presentation by quantifying CD8+ T cell proliferation over 6 d. In our experiments, CD141+ DCs were clearly superior to CD1c+ and CD16+ DCs in their capacity to cross-present soluble or cell-associated antigen to CD8+ T cells in a short-term, highly quantitative assay system in which survival of all tested DC subpopulations was similar. In the same setup, pDCs were consistently ineffective.
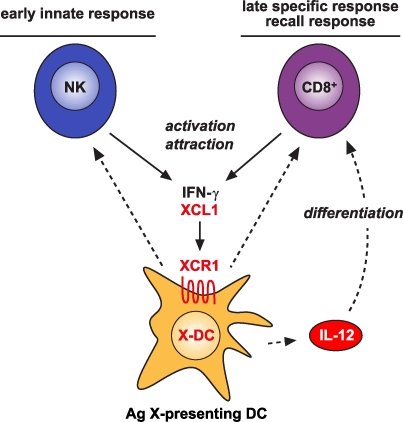
The tight correlation between the expression (and function) of the chemokine receptor XCR1 and the capacity of DCs to cross-present antigen in the mouse and in the human is suggestive of a biological linkage. We have previously demonstrated that XCL1, the only known chemokine ligand for XCR1, is coexpressed with IFN-γ by NK cells and CD8+ T cells upon activation in vitro (Dorner et al., 2002). Further, in the mouse Listeria infection model, XCL1 was secreted by NK cells in the early phase of the infection and by antigen-experienced CD8+ T cells in the recall response (in each instance together with IFN-γ). These observations led to the concept of XCL1 and IFN-γ acting (together with other cytokines) as a functional cytokine unit in the defense of certain infections (Dorner et al., 2002). With the recognition of cross-presenting DCs as the only cells bearing XCR1, this concept can be further refined. Thus, in the early innate response, XCL1 may ensure contact and thus exchange of information between activated NK cells and XCR1-bearing DCs. Later, in the adaptive phase and on reexposure, this cytokine functional unit is handed over to CD8+ T cells and thus ensures proper communication between cross-presenting DCs and CD8+ T cells (Fig. 6), as also demonstrated experimentally (Dorner et al., 2009). The XCL1–XCR1 communication axis thus apparently optimizes the differentiation and function of innate and adaptive cytotoxic cells required for an effective response against certain (intracellular?) pathogens.
Figure 6.
Involvement of the XCL1–XCR1 communication axis in the innate and adaptive cytotoxic responses to cross-presented microbial and tumor antigens. Secretion of the chemokine XCL1 by activated NK cells specifically attracts XCR1-expressing DCs capable of antigen cross-presentation. This ensures an effective communication between these cells in the innate phase of the immune response. In the adaptive phase, secretion of XCL1 by activated CD8+ T cells optimizes the communication with antigen cross-presenting DCs and facilitates the differentiation of CD8+ T cells to cytotoxic cells.
Finally, the identification of human professional cross-presenting DCs provides new avenues for the development of antiviral and antitumor vaccines based on the induction of cytotoxic T cells (Steinman and Banchereau, 2007; Andrews et al., 2008; Caminschi et al., 2009). The involvement of the XCL1–XCR1 communication axis in the innate and adaptive cytotoxic responses against certain infections (and possibly also tumors) makes XCR1 an interesting target for the delivery of prophylactic or therapeutic vaccines to cross-presenting DCs. The selectivity of XCR1 expression on these DCs, unmatched by any other known cell-surface molecule, and the typical internalization of chemokine receptors upon ligand binding further make this targeting route attractive.
MATERIALS AND METHODS
Antibodies and flow cytometry.
Antibodies recognizing CD19 (clone BU12; Flavell et al., 1995), CD3 (OKT3), CD14 (63D3), HLA-DR (L243; all from American Type Culture Collection), CD11c (BU15), CD15 (W6D3), CD16 (3G8), CD45 (HI30), CD56 (HCD56), HLA-A2 (BB7.2; BioLegend), CD1c (AD5-8E7), CD141 (AD5-14H12), and CD304 (AD5-17F6; Miltenyi Biotec) were used. Data were acquired on a flow cytometer (LSR II; BD) and analyzed using FlowJo software (Tree Star, Inc.).
Cell isolation for RNA preparation, fluorometric [Ca2+]i determination, and cross-presentation assays.
PBMCs were prepared by standard Biocoll (Biochrom) density gradient centrifugation of buffy coats from preoperative autologous deposits or of peripheral blood cells obtained from volunteers by leukapheresis. Leukapheresis was based on written informed consent and was approved by the local ethics committee. For preparation of granulocytes, whole blood was subjected to erythrocyte lysis with ACK buffer (155 mM NH4Cl, 10 mM KHCO3, 0.1 mM EDTA) followed by flow sorting (CD45+, CD15+, CD3−, CD14−, CD19−, CD141−) to a purity >99.7%. T cells (CD3+, CD19−, CD141−), B cells (CD19+, HLA-DR+, CD3−, CD141−), NK cells (CD16+, CD56+, CD3−, CD141−, HLA-DR−), and monocytes (CD14+, CD3−, CD19−, CD141−) were flow sorted from PBMCs to a purity >99.4%. CD1c+ DCs (CD1c+, CD11c+, HLA-DR+, CD16−, CD141−, lin−; ≥99.5% pure), CD16+ DCs (CD16+, CD11c+, HLA-DR+, CD1c−, CD141−, lin−; ≥99.2% pure), CD141+ DCs (CD141+, CD11c+, HLA-DR+, CD1c+, CD16−, lin−; ≥97.5% pure), and pDCs (CD304+, HLA-DR+, CD11c−, CD141−, lin−; ≥99.9% pure) were flow sorted from PBMCs after magnetic enrichment with the respective cell isolation kits from Miltenyi Biotec. In all flow sorting experiments, cells were preincubated with 2 mg/ml Endobulin (Baxter Hyland-Immuno Division) for 5 min at 4°C to block unspecific binding of mAbs. Flow sorting of cells was performed on a cell sorter (FACSAria II; BD). For generation of MoDCs, monocytes were enriched from PBMCs with CD14 microbeads and cultured for 5 d in RPMI 1640 medium supplemented with 10% FCS, penicillin, streptomycin, 500 U/ml GM-CSF, and 100 U/ml IL-4.
PCR.
Total RNA was prepared using the High Pure RNA Isolation Kit (Roche). RNA concentration, purity, and integrity were determined on a 2100 Bioanalyzer (Agilent Technologies) and by photometrical reading on a NanoDrop 1000 (Thermo Fisher Scientific). For determination of XCR1 expression, 100 ng of total RNA were reverse transcribed into cDNA with hexamer primers and the AMV Reverse Transcription System (Promega), and analyzed by quantitative PCR using the following primer–probe sets: 5′-TCAAGACGCATGTAAAGAGGTGTAG-3′ (forward primer F1), 5′-GTTGCCTGAGGACTCCATCTG-3′ (reverse primer R1), and 5′-FAM-TGCTCTAAACGTCCCTGCCATCTGGT-TAMRA-3′ (probe P1); and 5′-TTGCCTGTGTGGATCTCCC-3′ (forward primer F2), 5′-CGGTGGATGGTCATGATGG-3′ (reverse primer R2), and 5′-FAM-CATCAGCCTCTACAGCAGCATCTTCTTCCT-TAMRA-3′ (probe P2). Amplification of β2-microglobulin was used as a positive control: 5′-GCCGTGTGAACCATGTGACT-3′ (forward), 5′-CGGCATCTTCAAACCTCCA-3′ (reverse), and 5′-FAM-TAAGTGGGATCGAGACATGTAAGCAGCATC-TAMRA-3′ (probe). To generate standards for mRNA/cDNA copy quantitation, the specific XCR1 gene fragments were amplified and cloned into pJET1.2 vectors using the CloneJET PCR cloning kit (Fermentas). For quantitative PCR, primers were mixed with 10 µl ABsolute QPCR Mix including ROX (ABgene) and 1/10 of the cDNA in a 20-µl PCR reaction. PCR was performed and quantified on a 7500 Fast Real-Time PCR System (Applied Biosystems) with initial enzyme activation at 95°C for 15 min, followed by 40 cycles (95°C for 15 s and 60°C for 1 min).
Fluorometric [Ca2+]i determination.
CD141+, CD1c+, and CD16+ DCs were freshly flow sorted to a purity >98.7%, as described in Cell isolation for RNA preparation… The sorted cell suspensions were washed (200 g for 5 min) and resuspended in Hepes-buffered solution (HBS) containing 128 mM NaCl, 6 mM KCl, 1 mM MgCl2, 1 mM CaCl2, 5.5 mM glucose, 10 mM Hepes, and 0.2% BSA, supplemented with 2 µM fura-2/AM (Invitrogen). Cell suspensions were allowed to settle and adhere on poly–l-lysine–coated glass coverslips at 37°C and 5% CO2 for 30 min in a humidified atmosphere. Coverslips were superfused with HBS, mounted in a bath chamber, and imaged with an inverted microscope (Axiovert 100; Carl Zeiss, Inc.) using a UV-transmissive 10×/0.5 Fluar objective. During and after application of 1,000 ng/ml XCL1 or a chemokine mix (100 ng/ml CCL2, 200 ng/ml CCL21, 200 ng/ml CXCL9, 1 ng/ml CX3CL1) as a positive control, the fura-2/AM fluorescence was sequentially excited with monochromatic light of 340, 358, and 380 nm, and fluorescence emission was detected through a 512-nm-long pass filter with a cooled charge-coupled device camera (TILL-Photonics). Signals arising from the bound and unbound Ca2+ indicator were averaged over regions of interest that covered the area of single immobilized cells. Spectrally overlapping signals were separated and [Ca2+]i was calculated by a spectral fingerprinting method (Lenz et al., 2002). To assess a possible impact of contaminating fluorescence arising from fluorochrome-labeled antibodies, cell suspensions were treated and imaged in a similar way but without fura-2/AM loading. Under these conditions, there were no fluorescence signals discernible when excited at 340, 358, or 380 nm; we thus conclude that the fluorescence signals required no additional preprocessing.
Chemotaxis assay.
For chemotaxis assays, 105 cells suspended in 100 µl of medium (RPMI 1640, 1% BSA, 50 µM β-ME, 100 µg/ml penicillin/streptomycin) were placed into the upper chamber of a Transwell-24 system (6.5-mm diameter, 5-µm pore polycarbonate membrane; Costar; Corning). The lower chamber was filled with chemotaxis medium containing recombinant human XCL1 (R&D Systems) or any of the chemokines CCL2 (100 ng/ml), CCL21 (200 ng/ml), CX3CL1 (1 ng/ml), CXCL12 (200 ng/ml for T cells, B cells, NK cells, and monocytes; 100 ng/ml for pDCs), and CXCL8 (100 ng/ml; all from R&D Systems), and the cells were incubated for 150 min at 37°C in 5% CO2. In the lower chamber, cDCs (CD141+, CD1c+, CD16+), pDCs (CD304+, HLA-DR+, CD11c−, CD3−, CD14−, CD19−, CD56−), T cells (CD3+, CD19−, CD14−), B cells (CD19+, CD3−, CD14), NK cells (CD56+, CD16+, CD3−, CD14−, CD19−), monocytes (CD14+, CD3−, CD19−), and granulocytes (CD15+, CD45+, CD14−, CD19−) were identified by the indicated markers using flow cytometry. The absolute number of input or migrated cells was determined by counting all cells in a defined volume with a flow cytometer, and the percentage of migrated cells was calculated by dividing the number of cells in the lower chamber by the number of input cells (number of migrated cells/number of input cells × 100). All experiments were performed with duplicate wells.
HCMV pp65495–503–specific CD8+ T cell clone.
An HLA-A*0201–restricted, HCMV pp65–specific CD8+ T cell clone was prepared according to the procedure of Fonteneau et al. (2001). In brief, T cells from an HLA-A*0201+ donor were stimulated with autologous mature MoDCs, which were pulsed with pp65495–503. The activated T cells were expanded and cloned by limiting dilution; two of the pp65-specific CD8+ T cell clones (clones 10 and 61) were further expanded on irradiated feeder cells (an EBV line [2 × 104 cells/well] and PBMCs [2 × 105 cells/well], both irradiated with 43 Gy) using 1 µg/ml leucoagglutinin PHA-L (Sigma-Aldrich) and 150 U/ml of recombinant IL-2. Before being used in an assay, the T cell clones were rested for at least 10 d.
Cross-presentation assay.
Soluble recombinant HCMV pp65 (low endotoxin) was obtained from Miltenyi Biotec. Recombinant OVA (Sigma-Aldrich), from which LPS was removed using EndoTrap red (Hyglos), resulting in <0.5 U of endotoxin per milligram of protein as determined by the LAL assay (Charles River), was taken up in the same buffer as recombinant pp65. For cell-associated pp65 antigen, full-length HCMV pp65 cDNA cloned into the pcDNA3.1/myc-His vector (24 µg) was mixed with 24 µl Lipofectamine 2000 in 3 ml of Opti-MEM medium (all from Invitrogen) and used to transfect 2 × 106 HeLa cells (American Type Culture Collection) overnight according to the manufacturer’s instructions. Transfected HeLa cells were trypsinized, adjusted to 2 × 106 cells/ml in RPMI 1640 containing 10% FCS, and subjected to four freeze–thaw cycles. Transfection efficiency was monitored by Western blotting of 5 µg of protein lysate (determined with a BCA Protein Assay; Thermo Fisher Scientific) using HCMV pp65 mAb (CH12; Santa Cruz Biotechnology, Inc.). Cross-presentation of pp65 antigens by DCs was examined as described previously (Fonteneau et al., 2003). In brief, highly purified DC subtypes were co-cultured with a T cell clone in round-bottom 96-well plates in RPMI 1640, 10% FCS (Biochrom), penicillin, and streptomycin in the presence of antigen for 20–24 h. In cultures with recombinant soluble pp65 or OVA proteins (each at 3 µg/ml), T cell clone 10 was co-cultured with DCs (both at 4 × 104 cells/well) in 100 µl of complete medium; for the positive control with pp65495–503 peptide (1 µg/ml), T cell clone 10 and DCs (both at 104 cells/well) were co-cultured in 30 µl of medium. In cultures with cell-associated pp65, T cell clone 61 (5 × 104 cells/well) was co-cultured at variable ratios with DC subsets in a final volume of 150 µl of complete medium in the presence of HeLa pp65 lysate (50 µl) or MART-127–35 peptide AAGIGILTV or pp65495–503 peptide NLVPMVATV (both synthesized in our own facility and used at 1 µg/ml). In all instances, stimulation of the T cell clone was determined by quantifying the amount of IFN-γ secreted into the supernatant. ELISA for IFN-γ was performed according to the manufacturer’s instructions (OptEIA Human IFN-γ ELISA Set; BD), optical density was determined at 450 nm, and the data were evaluated with Revelation software (version 4.21; Dynex Technologies).
Acknowledgments
We are grateful for the advice given by Drs. J. Röck and A. Dzionek. Some cell flow sorts were performed by Dr. D. Kunkel in the FACS facility of the Berlin-Brandenburg Center for Regenerative Therapies.
This work was supported by the Wilhelm Sander Foundation and in part by grants from the German Ministry of Health and the Deutsche Forschungsgemeinschaft (Kr 827/16-1 and TR52 TP B06 to R.A. Kroczek and KL 427/15-1 to P.M. Kloetzel).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- cDC
- conventional DC
- HCMV
- human CMV
- MoDC
- monocyte-derived DC
- pDC
- plasmacytoid DC
- qRT-PCR
- quantitative RT-PCR
References
- Andrews D.M., Maraskovsky E., Smyth M.J. 2008. Cancer vaccines for established cancer: how to make them better? Immunol. Rev. 222:242–255 10.1111/j.1600-065X.2008.00612.x [DOI] [PubMed] [Google Scholar]
- Bevan M.J. 2006. Cross-priming. Nat. Immunol. 7:363–365 10.1038/ni0406-363 [DOI] [PubMed] [Google Scholar]
- Caminschi I., Proietto A.I., Ahmet F., Kitsoulis S., Shin Teh J., Lo J.C., Rizzitelli A., Wu L., Vremec D., van Dommelen S.L., et al. 2008. The dendritic cell subtype-restricted C-type lectin Clec9A is a target for vaccine enhancement. Blood. 112:3264–3273 10.1182/blood-2008-05-155176 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caminschi I., Lahoud M.H., Shortman K. 2009. Enhancing immune responses by targeting antigen to DC. Eur. J. Immunol. 39:931–938 10.1002/eji.200839035 [DOI] [PubMed] [Google Scholar]
- Colonna M., Trinchieri G., Liu Y.J. 2004. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 5:1219–1226 10.1038/ni1141 [DOI] [PubMed] [Google Scholar]
- den Haan J.M., Lehar S.M., Bevan M.J. 2000. CD8+ but not CD8− dendritic cells cross-prime cytotoxic T cells in vivo. J. Exp. Med. 192:1685–1696 10.1084/jem.192.12.1685 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Pucchio T., Chatterjee B., Smed-Sörensen A., Clayton S., Palazzo A., Montes M., Xue Y., Mellman I., Banchereau J., Connolly J.E. 2008. Direct proteasome-independent cross-presentation of viral antigen by plasmacytoid dendritic cells on major histocompatibility complex class I. Nat. Immunol. 9:551–557 10.1038/ni.1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner B.G., Scheffold A., Rolph M.S., Hüser M.B., Kaufmann S.H., Radbruch A., Flesch I.E., Kroczek R.A. 2002. MIP-1α, MIP-1β, RANTES, and ATAC/lymphotactin function together with IFN-γ as type 1 cytokines. Proc. Natl. Acad. Sci. USA. 99:6181–6186 10.1073/pnas.092141999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorner B.G., Dorner M.B., Zhou X., Opitz C., Mora A., Güttler S., Hutloff A., Mages H.W., Ranke K., Schaefer M., et al. 2009. Selective expression of the chemokine receptor XCR1 on cross-presenting dendritic cells determines cooperation with CD8+ T cells. Immunity. 31:823–833 10.1016/j.immuni.2009.08.027 [DOI] [PubMed] [Google Scholar]
- Dzionek A., Fuchs A., Schmidt P., Cremer S., Zysk M., Miltenyi S., Buck D.W., Schmitz J. 2000. BDCA-2, BDCA-3, and BDCA-4: three markers for distinct subsets of dendritic cells in human peripheral blood. J. Immunol. 165:6037–6046 [DOI] [PubMed] [Google Scholar]
- Esmon N.L., Owen W.G., Esmon C.T. 1982. Isolation of a membrane-bound cofactor for thrombin-catalyzed activation of protein C. J. Biol. Chem. 257:859–864 [PubMed] [Google Scholar]
- Flavell D.J., Flavell S.U., Boehm D.A., Emery L., Noss A., Ling N.R., Richardson P.R., Hardie D., Wright D.H. 1995. Preclinical studies with the anti-CD19-saporin immunotoxin BU12-SAPORIN for the treatment of human-B-cell tumours. Br. J. Cancer. 72:1373–1379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fonteneau J.F., Larsson M., Somersan S., Sanders C., Münz C., Kwok W.W., Bhardwaj N., Jotereau F. 2001. Generation of high quantities of viral and tumor-specific human CD4+ and CD8+ T-cell clones using peptide pulsed mature dendritic cells. J. Immunol. Methods. 258:111–126 10.1016/S0022-1759(01)00477-X [DOI] [PubMed] [Google Scholar]
- Fonteneau J.F., Kavanagh D.G., Lirvall M., Sanders C., Cover T.L., Bhardwaj N., Larsson M. 2003. Characterization of the MHC class I cross-presentation pathway for cell-associated antigens by human dendritic cells. Blood. 102:4448–4455 10.1182/blood-2003-06-1801 [DOI] [PubMed] [Google Scholar]
- Galibert L., Diemer G.S., Liu Z., Johnson R.S., Smith J.L., Walzer T., Comeau M.R., Rauch C.T., Wolfson M.F., Sorensen R.A., et al. 2005. Nectin-like protein 2 defines a subset of T-cell zone dendritic cells and is a ligand for class-I-restricted T-cell-associated molecule. J. Biol. Chem. 280:21955–21964 10.1074/jbc.M502095200 [DOI] [PubMed] [Google Scholar]
- Heath W.R., Carbone F.R. 2009. Dendritic cell subsets in primary and secondary T cell responses at body surfaces. Nat. Immunol. 10:1237–1244 10.1038/ni.1822 [DOI] [PubMed] [Google Scholar]
- Heath W.R., Belz G.T., Behrens G.M., Smith C.M., Forehan S.P., Parish I.A., Davey G.M., Wilson N.S., Carbone F.R., Villadangos J.A. 2004. Cross-presentation, dendritic cell subsets, and the generation of immunity to cellular antigens. Immunol. Rev. 199:9–26 10.1111/j.0105-2896.2004.00142.x [DOI] [PubMed] [Google Scholar]
- Hoeffel G., Ripoche A.C., Matheoud D., Nascimbeni M., Escriou N., Lebon P., Heshmati F., Guillet J.G., Gannagé M., Caillat-Zucman S., et al. 2007. Antigen crosspresentation by human plasmacytoid dendritic cells. Immunity. 27:481–492 10.1016/j.immuni.2007.07.021 [DOI] [PubMed] [Google Scholar]
- Iyoda T., Shimoyama S., Liu K., Omatsu Y., Akiyama Y., Maeda Y., Takahara K., Steinman R.M., Inaba K. 2002. The CD8+ dendritic cell subset selectively endocytoses dying cells in culture and in vivo. J. Exp. Med. 195:1289–1302 10.1084/jem.20020161 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ju X., Clark G., Hart D.N.J. 2010. Review of human DC subtypes. Methods Mol. Biol. 595:3–20 10.1007/978-1-60761-421-0_1 [DOI] [PubMed] [Google Scholar]
- Lenz J.C., Reusch H.P., Albrecht N., Schultz G., Schaefer M. 2002. Ca2+-controlled competitive diacylglycerol binding of protein kinase C isoenzymes in living cells. J. Cell Biol. 159:291–302 10.1083/jcb.200203048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin M.L., Zhan Y., Villadangos J.A., Lew A.M. 2008. The cell biology of cross-presentation and the role of dendritic cell subsets. Immunol. Cell Biol. 86:353–362 10.1038/icb.2008.3 [DOI] [PubMed] [Google Scholar]
- Lindstedt M., Lundberg K., Borrebaeck C.A. 2005. Gene family clustering identifies functionally associated subsets of human in vivo blood and tonsillar dendritic cells. J. Immunol. 175:4839–4846 [DOI] [PubMed] [Google Scholar]
- Liu Y.J. 2005. IPC: professional type 1 interferon-producing cells and plasmacytoid dendritic cell precursors. Annu. Rev. Immunol. 23:275–306 [DOI] [PubMed] [Google Scholar]
- MacDonald K.P., Munster D.J., Clark G.J., Dzionek A., Schmitz J., Hart D.N. 2002. Characterization of human blood dendritic cell subsets. Blood. 100:4512–4520 10.1182/blood-2001-11-0097 [DOI] [PubMed] [Google Scholar]
- Piccioli D., Tavarini S., Borgogni E., Steri V., Nuti S., Sammicheli C., Bardelli M., Montagna D., Locatelli F., Wack A. 2007. Functional specialization of human circulating CD16 and CD1c myeloid dendritic-cell subsets. Blood. 109:5371–5379 10.1182/blood-2006-08-038422 [DOI] [PubMed] [Google Scholar]
- Robbins S.H., Walzer T., Dembélé D., Thibault C., Defays A., Bessou G., Xu H., Vivier E., Sellars M., Pierre P., et al. 2008. Novel insights into the relationships between dendritic cell subsets in human and mouse revealed by genome-wide expression profiling. Genome Biol. 9:R17 10.1186/gb-2008-9-1-r17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robinson S.P., Patterson S., English N., Davies D., Knight S.C., Reid C.D. 1999. Human peripheral blood contains two distinct lineages of dendritic cells. Eur. J. Immunol. 29:2769–2778 [DOI] [PubMed] [Google Scholar]
- Schnurr M., Chen Q., Shin A., Chen W., Toy T., Jenderek C., Green S., Miloradovic L., Drane D., Davis I.D., et al. 2005. Tumor antigen processing and presentation depend critically on dendritic cell type and the mode of antigen delivery. Blood. 105:2465–2472 10.1182/blood-2004-08-3105 [DOI] [PubMed] [Google Scholar]
- Schulz O., Reis e Sousa C. 2002. Cross-presentation of cell-associated antigens by CD8α+ dendritic cells is attributable to their ability to internalize dead cells. Immunology. 107:183–189 10.1046/j.1365-2567.2002.01513.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Segura E., Villadangos J.A. 2009. Antigen presentation by dendritic cells in vivo. Curr. Opin. Immunol. 21:105–110 10.1016/j.coi.2009.03.011 [DOI] [PubMed] [Google Scholar]
- Shen L., Rock K.L. 2006. Priming of T cells by exogenous antigen cross-presented on MHC class I molecules. Curr. Opin. Immunol. 18:85–91 10.1016/j.coi.2005.11.003 [DOI] [PubMed] [Google Scholar]
- Shortman K., Heath W.R. 2010. The CD8+ dendritic cell subset. Immunol. Rev. 234:18–31 10.1111/j.0105-2896.2009.00870.x [DOI] [PubMed] [Google Scholar]
- Steinman R.M., Banchereau J. 2007. Taking dendritic cells into medicine. Nature. 449:419–426 10.1038/nature06175 [DOI] [PubMed] [Google Scholar]
- Villadangos J.A., Young L. 2008. Antigen-presentation properties of plasmacytoid dendritic cells. Immunity. 29:352–361 10.1016/j.immuni.2008.09.002 [DOI] [PubMed] [Google Scholar]
- Villadangos J.A., Heath W.R., Carbone F.R. 2007. Outside looking in: the inner workings of the cross-presentation pathway within dendritic cells. Trends Immunol. 28:45–47 10.1016/j.it.2006.12.008 [DOI] [PubMed] [Google Scholar]