Abstract
Small ubiquitin-like modifier (SUMO) modification of proteins (SUMOylation) and deSUMOylation have emerged as important regulatory mechanisms for protein function. SENP1 (SUMO-specific protease) deconjugates SUMOs from modified proteins. We have created SENP1 knockout (KO) mice based on a Cre–loxP system. Global deletion of SENP1 (SENP1 KO) causes anemia and embryonic lethality between embryonic day 13.5 and postnatal day 1, correlating with erythropoiesis defects in the fetal liver. Bone marrow transplantation of SENP1 KO fetal liver cells to irradiated adult recipients confers erythropoiesis defects. Protein analyses show that the GATA1 and GATA1-dependent genes are down-regulated in fetal liver of SENP1 KO mice. This down-regulation correlates with accumulation of a SUMOylated form of GATA1. We further show that SENP1 can directly deSUMOylate GATA1, regulating GATA1-dependent gene expression and erythropoiesis by in vitro assays. Moreover, we demonstrate that GATA1 SUMOylation alters its DNA binding, reducing its recruitment to the GATA1-responsive gene promoter. Collectively, we conclude that SENP1 promotes GATA1 activation and subsequent erythropoiesis by deSUMOylating GATA1.
The small ubiquitin-like modifier (SUMO) can be covalently attached to a large number of proteins through formation of isopeptide bonds with specific lysine residues of target proteins (Müller et al., 2001; Seeler and Dejean, 2003; Gill, 2004). SUMO, like ubiquitin (Pickart, 2001), is covalently attached to substrate proteins via an isopeptide bond between a C-terminal glycine and a lysine residue in the substrate. A consensus SUMO acceptor site has been identified consisting of the sequence ØKXE (Ø is a large hydrophobic amino acid and K is the site of SUMO conjugation). A large number of proteins with important roles in many cellular processes, including gene expression, chromatin structure, signal transduction, and maintenance of the genome, have been found to be SUMOylated (Müller et al., 2001; Gill, 2004). Nuclear proteins, including transcription factors, are a major group of proteins among SUMOylated proteins. These include p53, c-Jun, Sp3, Elk-1, p300 histone acetyltransferase, HDAC (histone deacetylase), PML, HIPKs, and the GATA family proteins. SUMOylation of these proteins has not been generally associated with increased protein degradation. Rather, SUMO modification regulates protein localization, protein–protein or protein–DNA interactions, and/or biochemical activities. The consequence of SUMOylation on protein function is substrate specific.
SUMOylation is a dynamic process that is mediated by activating, conjugating, and ligating enzymes and is readily reversed by a family of SUMO-specific proteases (Li and Hochstrasser, 1999). The enzymatic machinery that adds and removes SUMO is similar to, but distinct from, the well known ubiquitination machinery. The E1-activating and E2-conjugating enzymes involved in SUMOylation are highly related to the ubiquitination system. However, the SUMO E3 ligase that promotes transfer of SUMO from the E2 to specific substrates and the SUMO-specific protease that removes SUMO from specific substrates are different from those involved in ubiquitination and deubiquitination. In contrast to the large number of ubiquitin E3 ligases, only three groups of SUMO E3 ligases have been identified (Gill, 2004): RanBP2, the PIAS proteins, and the polycomb group protein Pc2. Six members of the sentrin-specific protease (SENP) family (SENP1–3 and SENP5–7) have been reported in the mammalian system (Gong et al., 2000; Kim et al., 2000; Nishida et al., 2000; Best et al., 2002; Hang and Dasso, 2002; Kadoya et al., 2002; Zhang et al., 2002; Bailey and O’Hare, 2004). SENP1 is a protease that appears to deconjugate a large number of SUMOylated proteins (Gong et al., 2000). Different members of these SUMO-specific proteases appear to localize in different cellular compartments, where they regulate protein function by modifying protein stability, cellular localization, and protein–protein interactions (Suzuki et al., 1999; Gong et al., 2000; Kim and Iwao, 2000; Best et al., 2002; Kadoya et al., 2002; Zhang et al., 2002; Bailey and O’Hare, 2004). However, the in vivo function of these SUMO-specific proteases is largely unknown.
Erythropoiesis is a complex multistage process that is conserved in human, mouse, and zebrafish systems (Godin and Cumano, 2002; Orkin and Zon, 2008). During mouse development, erythropoiesis first occurs within the blood islands of the embryos. This stage, which is termed primitive, is characterized by the production of nucleated red cells that express embryonic globins. After development of definitive hematopoietic progenitors in the blood islands and the aorta-gonad-mesonephros region, erythropoiesis shifts to the liver around day 10 of gestation, where it remains for the majority of fetal life. This stage is characterized by profound expansion of erythroid progenitors and expression of a definitive adult pattern of globin molecules (Peschle et al., 1985). Erythropoiesis takes place within the BM microenvironment after birth. During definitive erythropoiesis in fetal liver or adult BM, the committed erythroid progenitors first give rise to erythroid burst-forming unit (BFU-e) and then to erythroid CFU (CFU-e) cells, which, in turn, mature into enucleated erythrocytes through multiple steps.
Previous studies have demonstrated that GATA1, a member of the GATA family consisting of six transcription factors (GATA1–GATA6), is essential for normal erythropoiesis. GATA1-deficient embryonic stem (ES) cells are able to contribute to all different tissues in chimeric mice, with the exception of the mature red blood cells (Pevny et al., 1991; Weiss et al., 1994). Expression of GATA1 during embryogenesis appears after GATA2, a GATA family member involved in primitive hematopoiesis (Godin and Cumano, 2002; Orkin and Zon, 2008). However, GATA1 is highly expressed in fetal liver (embryonic day (E) 11–15). Consistently, GATA1-null mouse embryos die from severe anemia between E10.5 and 11.5 (Fujiwara et al., 1996). GATA1 consists of three functional domains: an N-terminal activation domain, the N-terminal zinc finger (N finger), and the C-terminal zinc finger (C finger). GATA1 binds to the DNA consensus sequence (A/T)GATA(A/G) by two zinc finger motifs (Ferreira et al., 2005). The critical role of GATA1 in erythropoiesis is attributed to its activity in driving expression of many erythropoietic genes. These genes include heme biosynthesis enzyme hemoglobins, erythropoietic growth factor receptor EpoR and c-Kit, mitogenic factor c-Myb, and antiapoptotic protein Bcl-XL (Zon et al., 1991; Chiba et al., 1993; Gregory et al., 1999; Lacombe and Mayeux, 1999). Among these factors, EpoR has been well established to be critical for definitive erythropoiesis in mouse fetal liver (Lin et al., 1996; Wu et al., 1995; Lee et al., 2001). Interestingly, the role of GATA1 in erythropoiesis is dose dependent. GATA1 knockdown embryos (GATA1.05), which express only ∼5% of the WT GATA1 levels, also die between E11.5 and 12.5 (Takahashi et al., 1997). Other GATA1 knockdown mice (GATA1 low; McDevitt et al., 1997), which express ∼20% of the WT GATA1 levels, die between E13.5 and 14.5 as a result of ineffective primitive and definitive erythroid differentiation, some are born alive (2% instead of the expected 25%), and a small number survive to adulthood.
The function of GATA1 is tightly regulated by interaction with transcriptional cofactors as well as by several posttranslational modifications including phosphorylation, acetylation, and SUMOylation (Boyes et al., 1998; Cantor and Orkin, 2002; Collavin et al., 2004; Yu et al., 2005; Hernandez-Hernandez et al., 2006; Lamonica et al., 2006; Zhao et al., 2006). However, the in vivo effects of these modifications on GATA1 activity have not been defined. In the present study, we show that global deletion of SENP1 in mice (SENP1 KO) causes erythropoietic defects in fetal liver. We show that the GATA1 activity and its target genes are significantly reduced in fetal liver of SENP1 KO mice as a result of accumulation of SUMOylated GATA1. Mechanistic studies suggest that SENP1 directly deSUMOylates GATA1, regulating GATA1 DNA-binding activity and GATA1-dependent EpoR expression and erythropoiesis. Our study provides a novel mechanism by which GATA1 activity is modulated during erythropoiesis.
RESULTS
Generation of SENP1 KO mice by homologous recombination
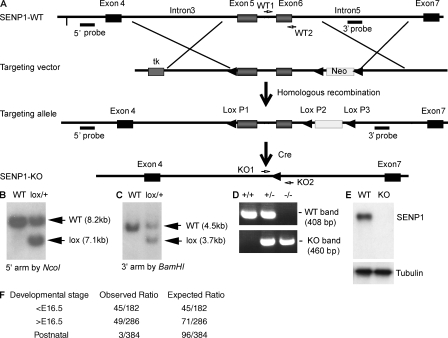
We generated SENP1lox/lox mice based on homologous recombination. We first generated global deletion of SENP1 by mating SENP1+/lox mice with β-actin–Cre mice to obtain SENP1+/− mice, with deletion of both the targeted region (exons 5 and 6) and the Neo gene (Fig. 1, A–C). SENP1+/− mice appeared normal and fertile. We attempted to generate SENP1−/− (SENP1 KO) mice by intercrossing SENP1+/−, and no live SENP1 KO mice were obtained after weaning. We then carefully examined mouse genotyping and phenotypes at E13.5 and 16.5 of embryonic development and on the day of birth (postnatal day [P] 0). Typical genotyping of SENP1+/+ (WT), SENP1+/− (heterozygote), and SENP1−/− (KO) mice are shown in Fig. 1 D. Deletion of SENP1 in SENP1 KO mice was verified by RT-PCR using primers amplifying exons 5–6 and by Western blotting with an anti-SENP1 antibody recognizing the C-terminal domain of SENP1. Neither SENP1 transcripts encoding for exons 5–6 (not depicted) nor SENP1 protein was detected in SENP1 KO E13.5 fetal liver (Fig. 1 E). The ratios of live and dead SENP1 KO embryos were analyzed (Fig. 1 F). Only three SENP1 KO littermates among 384 born pulps survived to 2 wk of age, and each had severe anemia (unpublished data). We focused on E13.5–16.5 to further characterize the SENP1 KO phenotype.
Figure 1.
Generation of SENP1-flox and KO mice. (A) Schematic diagram for strategy to generate SENP1-flox mice. The targeting vector contains intron 3 at the 5′ arm, exons 5/6 in the targeting region, and intron 5 at the 3′ arm. The 5′ probe with NcoI digest and 3′ probe with BamHI digest are indicated. PCR primers for WT (WT1 and WT2) and KO alleles (KO1 and KO2) are shown. (B and C) Screening of ES clones. G418-resistant stable ES clones were obtained after electroporation of the SENP1 targeting vector. Genomic DNA from these clones were extracted and used for screening. Clones were first verified as positive for lox P1, P2, and P3 by PCR with primers. The positive clones with SENP1 were further screened by Southern blotting using the 5′ probe after NcoI digest (B) and 3′ probe after BamHI digest (C). WT and lox alleles with expected sizes are indicated. Two independent SENP1lox/+ ES cell clones were injected into C57BL/6 blastocysts. Chimeras were further bred with C57BL/6 females for germline transmission. (D) Generation of SENP1+/lox and SENP1+/− mice. SENP1+/lox mice were mated with β-actin–Cre mice. Tail genomic DNA was used to determine SENP1 deletion by PCR with KO primers adjacent to 5′ of lox P1 (KO1) and 3′ of lox P3 (KO2) to obtain heterozygous mice with a deletion of both the targeting region (the exons 5/6) and the Neo gene (SENP1+/−). SENP1 KO mice were obtained by intercrossing between the heterozygous (SENP1+/−) male and female. Representative genotyping of +/+, +/−, and −/− mice are shown with WT primers (within the exon 6 as indicated in A) and KO primers. (E) SENP1 deletion in embryos. Deletion of SENP1 in embryos was confirmed by Western blotting with anti-SENP1 antibody. Tubulin was used as a loading control. (F) The observed numbers of SENP1 KO mice during different embryonic and perinatal stages (E9.5–16.5, E16.5–P0, and P1–21). The observed and expected ratios are shown.
Erythropoiesis defects in SENP1 KO fetal liver
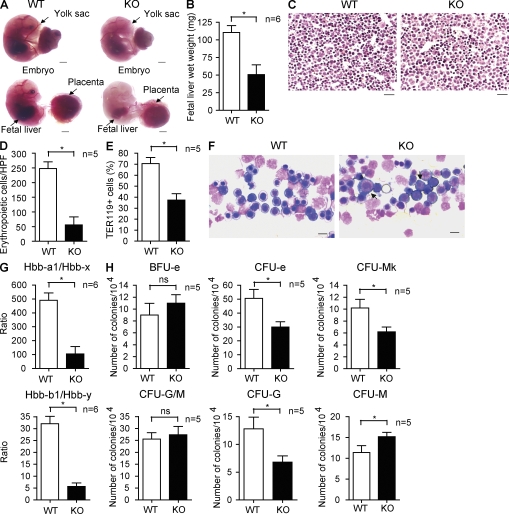
Starting at E13.5, SENP1 KO embryos and fetal livers were paler and smaller than those of WT and heterozygote. Both the SENP1 KO embryo and yolk sac have poor blood vessel development with fewer red blood cells, indicating anemia in SENP1 KO (Fig. 2 A). Erythropoiesis is initiated in the yolk sac at around E8.0 during normal mouse development (primitive erythropoiesis; Orkin and Zon, 2008). Starting around E11 until 1 wk after birth, the fetal liver is the major erythropoietic organ. Fetal liver in SENP1 KO was smaller in size (Fig. 2 A) and lighter in weight (Fig. 2 B). Histological examination of fetal liver sections revealed a marked decrease in the number of morphologically identifiable small-sized erythroid cells (erythroblasts) with condensed chromatin nucleus in SENP1 KO fetal liver (Fig. 2, C and D [quantification]) as well as by FACS with erythrocyte marker Ter119 (Fig. 2 E). Although decreased erythropoietic islands were clearly discernable in SENP1 KO fetal livers, fetal liver touch preparations by May-Giemsa staining analysis showed a disproportionate abundance of immature erythroblasts, such as proerythroblasts, compared with WT mice (Fig. 2 F).
Figure 2.
Erythropoietic defects in fetal liver of SENP1 KO mice. (A) Appearance of WT and SENP1 KO embryos at E16.5. Freshly dissected embryos without staining were photographed. Embryo with yolk sac (top) and without yolk sac (bottom) are shown. Yolk sac, placenta, and fetal liver are indicated. Bars, 200 µM. (B) Wet weight of fetal livers in WT and SENP1 KO embryos (n = 6 each group). Data are mean ± SEM from five fetal livers per group. *, P < 0.01. (C) Histology examination of WT and SENP1 KO E13.5 fetal livers by hematoxylin/eosin staining. Bars, 100 µM. (D) Number of erythropoietic cells per high-power field (40×) was quantified from five sections. *, P < 0.01. (E) Freshly isolated E13.5 WT or SENP1 KO fetal liver cells were incubated in PharmLyse to remove mature red blood cells, and the remaining erythroid progenitor cells were doubly stained with anti-Ter119 followed by flow cytometry (FACS) analysis. The percentage of Ter119+ cells is shown. Data are mean ± SEM from five fetal livers per group. *, P < 0.01. (F) E13.5 fetal liver touch preparations and May-Giemsa staining. Arrows indicate proerythroblasts. Bars, 20 µM. (G) Quantitative RT-PCR analysis of hemoglobin (Hbb) forms in E13.5 WT and SENP1 KO fetal livers. 18S rRNA was used for normalization. Hbb-a1 and Hbb-b1 are adult hemoglobin, whereas Hbb-x and Hbb-y are the corresponding embryonic forms. Data are mean ± SEM from six embryos of each genotype. *, P < 0.01. (H) E13.5 fetal liver cells from WT and SENP1 KO mice were used for colony forming assays of immature BFU-e, CFU-e, CFU-mk, CFU-G/M, and CFU-G or CFU-M. 104 cells were seeded as duplicates from WT and SENP1 KO fetal livers (n = 5 each). *, P < 0.01. Data are mean ± SEM from triplicates of two independent experiments.
To further determine if SENP1 KO mice display defects in erythropoiesis leading to anemia, hemoglobin levels in E13.5 fetal livers from SENP1 WT and SENP1 KO mice were analyzed by quantitative RT-PCR. The SENP1 KO fetal liver showed a marked decrease in adult forms of hemoglobin (Hbb-a1 and Hbb-b1) with a dramatic increase in the corresponding embryonic forms (Hbb-x and Hbb-y; Fig. 2 F, ratios of Hbb-a1/Hbb-x and Hbb-b1/Hbb-y). To determine the stage of erythroid differentiation at which SENP1 KO develop defects, we compared the abilities of fetal liver cells derived from WT and SENP1 KO E13.5 embryos to form CFU-e and more immature BFU-e. SENP1-deletion had no effects on the number of the BFU-e (Fig. 2 G, BFU-e). However, the number of CFU-e showed a significant (40%) decrease in SENP1 KO fetal liver cells (Fig. 2 G, CFU-e). Similar reduction in megakaryocyte CFU (CFU-mk), the closest differentiation pathway with erythropoiesis, was observed in SENP1 KO (Fig. 2 G, CFU-mk). In contrast, granulocyte/monocyte CFU (CFU-G/M) was not significantly altered (Fig. 2 G, CFU-G/M). Interestingly, SENP1 KO showed an increase in CFU-G concomitant with an increase in CFU-M (Fig. 2 G, CFU-G and CFU-M). This in vitro observation is consistent with the in vivo results that monocytes in SENP1 KO transplanted recipient mice are increased compared with WT mice (Fig. 3 C).
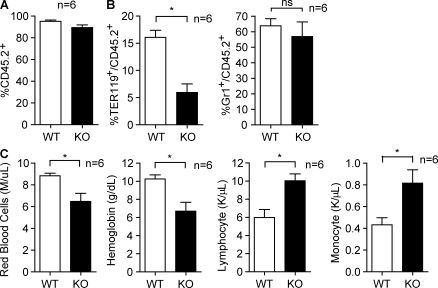
Figure 3.
Erythropoiesis defects in SENP1 KO fetal liver confirmed by BMT. (A) Flow cytometry analysis of engraftment ratio in BM of recipient mice. E13.5 fetal liver cells of WT and SENP KO mice (CD45.2+) were injected into irradiated Pep3B recipients (CD45.1+). Data are mean ± SEM from six mice per group. (B) Flow cytometry analysis in BM of recipient mice. Erythroid cells (TER119+) and granulocytes (Gr1+) were quantified by FACS with respective antibodies. The percentage of each cell type in CD45.2+ population from six mice is shown. Data are mean ± SEM from six mice per group. *, P < 0.01. (C) CBC analysis from peripheral blood of recipient mice. Hemoglobin concentration, numbers of red blood cells, lymphocytes, and monocytes are shown. Data are mean ± SEM from six mice per group. *, P < 0.01. Similar results were observed in additional two independent experiments with the same number of mice in each group.
Transplantation of SENP1 KO fetal liver cells into adult recipient BM confers erythropoiesis defects
To further determine if SENP1 KO erythroid progenitor cells exhibit an intrinsic defect in erythropoiesis, we measured erythropoietic activity of fetal liver cells from WT and SENP1 KO after transplantation into myeloablated adult WT mice. To this end, E13.5 fetal liver cells of WT or SENP KO mice (CD45.2+) were respectively injected into irradiated Pep3B recipients (CD45.1+). The engraftment ratio in BM of recipient mice after fetal liver transplantation was analyzed by flow cytometry analysis using CD45.2 and CD45.1 surface marker after 6 mo. The results suggest that engraftment could reach >95%, and no difference was detected between WT and SENP1 KO fetal liver cells in the reconstitution of recipient BM (Fig. 3 A, percentage of CD45.2+ cells). We confirmed the genotype of the donated fetal liver cells 6 mo after BM transplantation (BMT) by genotyping blood samples with WT or SENP1 KO–specific primers (unpublished data). We then measured precursor cells in BM of recipient mice by flow cytometry with specific antibodies. Erythroid cells (TER-119+/CD45.2+ ratio) were significantly decreased in mice receiving KO versus WT fetal liver cells, whereas there was no significant difference in granulocyte maturation for KO versus WT donors (Gr-1+CD45.2+ ratio; Fig. 3 B). We also performed complete blood counts (CBCs) of the peripheral blood from WT, WT BMT, and KO BMT groups. Consistent with the decreased number of erythroid cells (TER-119+) and megakaryocytes (CD41+) in their BM, SENP1 KO recipients showed decreases in hemoglobin, hematocrit, and number of red blood cell compared with the recipients of WT cells. However, lymphocytes and monocytes were increased (Fig. 3 C). These results suggest that SENP1 KO fetal liver transplantation confers an anemic phenotype. It is of note that the effects of SENP1 deletion on transplanted mice were less than those of fetal liver, suggesting a potential compensatory event in recipient mice. Indeed, we observed enlarged spleens and extramedullary hematopoiesis with increased erythroid precursor clumps, megakaryocytes and other developing myeloid populations in the SENP1 KO transplanted mice (Fig. S1).
Decreased GATA1 activity in SENP1 KO fetal liver and erythroid progenitors
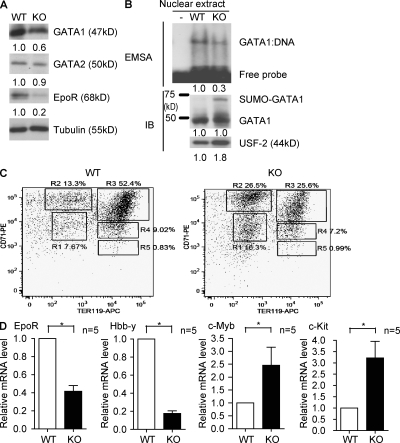
To analyze the molecular determinants of the SENP1 KO erythropoietic phenotype, we examined protein expression of several critical factors regulating erythropoiesis. Consistent with previously published results (Cheng et al., 2007), protein expression of Epo and its upstream regulator HIF1α was reduced in SENP1 KO fetal liver (Fig. S2). Notably, erythropoietic transcriptional factor GATA1 and its downstream target erythropoietin receptor (EpoR) were also reduced in protein expression (Fig. 4 A). GATA2, a transcriptional factor involved in primitive erythropoiesis (Tsai et al., 1994), was not significantly altered by SENP1 deletion. Furthermore, GATA1 activity in fetal liver was reduced (more than threefold) as determined by electrophoretic mobility shift assay (EMSA) using equivalent GATA1 protein input from fetal liver nuclear extracts (Fig. 4 B).
Figure 4.
Decreased GATA1 activity and GATA1-responsive genes in SENP1 KO fetal liver and stage-matched hematopoietic progenitors. (A) Expression of GATA1, GATA2, and EpoR was examined in E13.5 fetal liver of WT and SENP1 KO mice by Western blotting with respective antibodies. The relative protein levels were quantified by taking normalized WT (vs. tubulin) as 1.0. Five other embryos of WT or KO exhibited similar results. (B) GATA1 activity in SENP1 KO fetal liver. Nuclear extracts were isolated from E13.5 WT and SENP1 KO fetal livers (n = 3 each group), and an equal amount of GATA1 proteins from WT and KO were used for the GATA1 DNA binding activity assay by EMSA with a GATA1-specific oligonucleotide probe. A nuclear protein USF2 was used as a loading control. A representative gel from three independent experiments is shown. The relative GATA1 DNA-binding activity and protein levels were quantified by taking WT as 1.0. (C) Representative flow cytometric analysis of erythroid differentiation in vivo. Freshly isolated E13.5 WT or SENP1 KO fetal liver cells were doubly stained with anti–CD71-PE and anti–Ter119-APC. Flow cytometry density plots of all viable fetal liver cells from representative WT and SENP1 KO embryo in the same littermates are shown. Regions R1–R5 define distinct populations of erythroid progenitor cells at different stages of differentiation. Primitive progenitor cells (including mature BFU-Es and CFU-Es) and proerythroblasts are shown in R1, proerythroblasts and early basophilic erythroblasts in R2, early and late basophilic erythroblasts in R3, chromatophilic and orthochromatophilic erythroblasts in R4, and late orthochromatophilic and reticulocytes in R5. Numbers represent the percentage of cells within that region. Two independent experiments are performed. (D) Transcripts for EpoR, Hbb-y, c-Myb, and c-Kit from sorted fetal liver cells from R2 areas were quantified by quantitative RT-PCR and normalized to β-actin. Data are mean ± SEM (n = 5 from each group) from two independent experiments. *, P < 0.01 versus WT group.
To examine if reduction of GATA1-responsive gene expression is not a result of decreased total numbers of erythroid cells in SENP1 KO, we isolated stage-matched erythroid cells by FACS sorting with anti-CD71 and anti-Ter119. Freshly isolated E13.5 WT or SENP1 KO fetal liver cells were doubly stained with anti-CD71 and anti-Ter119. Based on these two surface markers, the proportion of erythroid progenitors at different stages of differentiation, termed R1–R5, can be assessed as described previously (Zhang et al., 2003). In SENP1 KO fetal liver, a twofold increase in number of less mature erythroblasts (R1 and R2) and concomitant twofold decrease of more mature erythroblasts (R3–R5) were detected (Fig. 4 C), which is consistent with increased proerythroblasts in SENP1 KO fetal liver visualized by May-Giemsa staining (Fig. 2 F). We first examined GATA1 expression in these stage-matched erythroid cells. WT and SENP1 KO showed no differences in GATA1 transcripts (stage R1–R3) and protein expression (stage R2–R3; Fig. S3). We then compared GATA1-responsive genes in the R1 and R2 populations isolated from WT and SENP1 KO mice by quantitative RT-PCR. Transcripts for GATA1-inducible genes EpoR and Hbb-y (an embryonic hemoglobin expressed in R1 and R2 populations) were decreased in SENP1 KO cells. In contrast, GATA1-repressive genes, such as c-Myb and c-Kit, were up-regulated in SENP1 KO cells (Fig. 4 D, data from R2 populations). Similar patterns of gene expression were obtained from R1 populations but not in more mature R3 populations (unpublished data). These data suggest that GATA1 activity and its dependent gene expression were reduced in SENP1 KO erythroid progenitors, causing a defect in erythropoietic maturation and subsequent decrease in total number of erythroid cells observed in fetal liver. Decrease of GATA1 expression observed in SENP1 KO fetal liver (Fig. 4 A) is a result of a smaller number of total erythroid cells (Fig. 2, C–E).
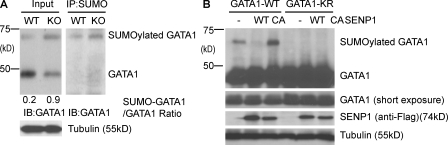
SENP1 directly regulates GATA-1 SUMOylation
SENP1 is an endopeptidase that removes SUMO from conjugated protein targets. We reasoned that deletion of SENP1 leads to GATA1 SUMOylation, reducing GATA1 activity. To test our hypothesis, we first examined if a SUMOylated form of GATA1 could be detected in SENP1 KO fetal liver in the presence of deSUMOylase inhibitor. The SUMOylated form of GATA1 was dramatically increased in SENP1 KO fetal liver, which was confirmed by immunoprecipitation with anti-SUMO followed by Western blotting with anti-GATA1 (Fig. 5 A). To determine if SENP1 directly deSUMOylates GATA1, GATA1 WT, or a SUMO-defective mutant, GATA1-K137R was coexpressed with SENP1 WT or a catalytic inactive form with a C603A mutation (SENP1 CA). As expected, SUMOylated GATA1 could be detected for GATA1 WT but not for the GATA1-K137R mutant. Importantly, SUMOylated GATA1 was diminished by SENP1 WT. In contrast, SENP1 CA increased SUMOylated GATA1 (Fig. 5 B), suggesting that the catalytic inactive form of SENP1 may function as a dominant-negative mutant.
Figure 5.
SENP1 regulates GATA1 SUMOylation. (A) Increased SUMOylated GATA1 in E13.5 SENP1 KO fetal liver. WT and KO fetal livers were dissected in the presence of 1 mM N-ethylmaleimide (deSUMOylase inhibitor) and total homogenates were subjected to Western blotting with anti-GATA1. Tubulin was used as a control. The SUMOylated form of GATA1 was determined by immunoprecipitation with anti-SUMO followed by Western blotting with anti-GATA1. The ratios of SUMO-GATA1/GATA1 were quantified. Data are representative from three pairs of embryos. (B) SENP1 deSUMOylates GATA1. GATA1 WT or GATA1-K137R was cotransfected with SENP1 WT or SENP1 CA in 293T cells as indicated. GATA1 and SENP1 proteins were detected by Western blotting with anti-GATA1 and anti-Flag, respectively. GATA1 conjugated with SUMO is indicated. Experiments were repeated three times.
Critical role of SENP1-mediated GATA1 deSUMOylation in erythropoiesis
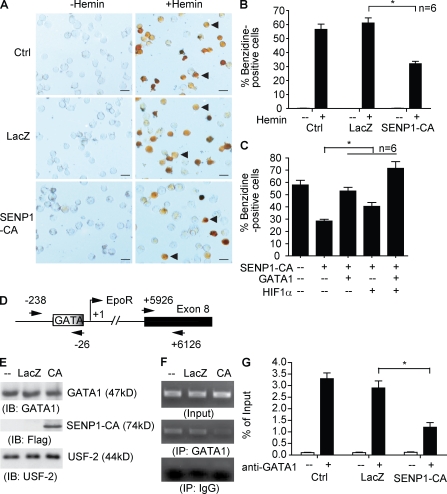
To directly test the role of SENP1-mediated GATA SUMOylation in erythropoiesis, we determined effects of SENP1 CA on erythropoiesis in a human erythroleukemic cell line (HEL), which undergoes erythropoiesis in the presence of hemin (Martin and Papayannopoulou, 1982). HEL cells were infected with adenovirus expressing LacZ or SENP-C603A followed by treatment with hemin for 96 h, and erythropoiesis was measured for hemoglobin-positive cells by benzidine staining. Infection efficiency was >90% as measured by β-galactosidase staining (unpublished data). SENP1 CA significantly blocked hemin-induced erythropoiesis as indicated by a reduced number of benzidine-positive cells (Fig. 6, A and B [quantification]). Similar results were obtained for expression of EpoR and β-hemoglobin by quantitative RT-PCR (unpublished data).
Figure 6.
A critical role of SENP1-mediated GATA1 deSUMOylation in erythropoiesis. (A and B) Overexpression of a catalytic form of SENP1 (C603A) inhibits erythropoiesis. Erythroleukemic cell line HEL cells were either uninfected or infected with adenovirus expressing LacZ or SENP1 CA. Cells were treated with hemin (40 µM) for 96 h and hemoglobin-positive cells by benzidine staining (brown cells indicated by arrowheads in A). Bars, 20 µM. Quantitative results shown in B are mean ± SEM determined from six different areas of each group. *, P < 0.01. Similar results were obtained from additional four experiments. (C) Overexpression of GATA1 or HIF1α rescued SENP1 CA–induced defects in hematopoiesis. HEL cells were infected with adenovirus expressing SENP1 CA in the presence of GATA1 or HIF1α. Erythropoiesis assays were performed as in A and B. Quantitative results shown in C are mean ± SEM determined from six different areas of each group. *, P < 0.01. Experiments were repeated twice. (D–G) Overexpression of SENP1 CA inhibits GATA1 recruitment to the EpoR promoter. (D) Human EpoR promoter region with GATA1 binding site at −45 and the transcriptional site (TSS). A PCR fragment for ChIP assay covering −238 to −26 is indicated. Exon 8 region (+5926 to +6126) was used for a control. HEL cells were infected with adenovirus expressing LacZ or SENP1 CA, followed by treatment with hemin for 96 h as described in A. (E) Nuclear extract was used for Western blotting with anti-GATA1 and anti-Flag (for SENP1 CA). Nuclear protein USF2 was used as a control. (F) Nuclear extracts were subjected to ChIP assay with anti-GATA1 followed by PCR amplification of the EpoR promoter GATA1-binding site and exon 8. An IgG isotype was used as a control. Representative ChIP PCR product of the EpoR promoter GATA1 binding site is shown. (G) Quantitative analyses of the PCR products are presented as ratio of ChIP/Input, and data are mean ± SEM from three independent experiments. *, P < 0.01.
It has been previously reported that erythropoietic deficiency of SENP1 mutant mice is partly a result of inappropriate HIF1α activity (Cheng et al., 2007), and we also observed reduction of HIF1α expression in SENP1 KO fetal liver (Fig. S2). We next determined relative contributions of GATA1 and HIF1α, two erythropoietic transcription factors regulated by SUMOylation, to SENP1-mediated erythropoiesis. To this end, SENP1 CA was coexpressed with GATA1 or HIF1α into HEL cells (Fig. S4 A, protein expression of the transgenes), and erythropoiesis was measured for hemoglobin-positive cells by benzidine staining. Results showed that coexpression of GATA1 or HIF1α partially, and together additively, rescued SENP1 CA–induced hematopoietic defects (Fig. 6 c). These data suggest that both GATA1 and HIF1α contribute to SENP1-mediated erythropoiesis.
Recruitment of GATA1 to the GATA1 site in the EpoR promoter is critical for EpoR expression and subsequent erythropoiesis (Zon et al., 1991). To directly determine the effects of SENP1 CA on GATA1 activity and EpoR transcription in HEL cells, we measured GATA1 binding to the EpoR promoter in SENP1 CA–infected cells by chromatin immunoprecipitation (ChIP) assay. HEL cells were infected and treated with hemin for 96 h, and nuclei were subjected to ChIP with anti-GATA1 followed by a PCR amplification of the GATA1 binding site in the EpoR promoter. An isotype IgG and a region on exon 8 were used as a negative control for immunoprecipitation and PCR amplification, respectively (Fig. 6 D). Expression of SENP1 had no effect on the total level of GATA1 (Fig. 6 E). However, expression of SENP1 CA significantly blunted the binding of GATA1 to the EpoR promoter region (Fig. 6, F and G [quantification], representative ChIP gel). These data strongly support a critical role of SENP1 in controlling GATA1 recruitment to the EpoR promoter and subsequent erythropoiesis.
GATA1 SUMOylation inhibits its DNA binding
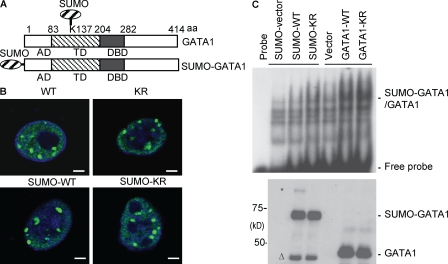
Reduction of GATA1 recruitment to EpoR promoter in SENP1 KO cells could be a result of alterations of SUMOylated GATA1 in protein stability, nuclear localization, and protein and/or DNA-binding activity as demonstrated for other transcriptional factors (Müller et al., 2001; Seeler and Dejean, 2003; Gill, 2004), and we next examined if SUMO conjugation affects these activities of GATA1. SUMOylation, like other posttranslational modification, is a dynamic process. Only a small proportion of a given SUMOylated protein can be detected (Fig. 5), and it has been difficult to determine a suppressive effect of SUMO conjugation on a specific protein. To get around this issue, we used a SUMO fusion strategy, which has been applied to other SUMOylated proteins such as p53 and GATA4 (Wang et al., 2004; Carter et al., 2007). SUMO fusion to the N terminus of GATA1 (Fig. 7 A) had no effects on its expression or its stability as measured for its half-life in the presence of protein synthesis inhibitor cycloheximide (Fig. S5 A). GATA1 WT K137R with or without SUMO fusion showed a similar pattern of nuclear localization (Fig. 7 B; and Fig. S6, A and B [quantification]). However, SUMO fusion completely diminished the DNA binding activity of GATA1 as measured by EMSA using a GATA1 probe (Fig. 7 C).
Figure 7.
GATA1 SUMOylation inhibits its DNA binding. (A) Schematic diagram for GATA1 structural domains and expression construct for SUMO-GATA1 fusion protein (bottom). The natural SUMOylation site on GATA1 (K137) is indicated (top). AD, acidic domain; TD, transactivation domain; DBD, DNA-binding domain. (B) GATA1 WT, GATA1-K137R, SUMO-GATA1 WT, and SUMO-GATA1-KR show a similar punctate staining in the nucleus. Localization of GATA1 in BAEC (which lacks endogenous GATA1) was determined by indirect immunofluorescence microscope with anti-GATA1 followed by an FITC-conjugated anti–rat secondary antibody. The nucleus was counterstained by DAPI (blue). Representative images from 20 cells in each group are shown. Bars, 5 µM. (C) SUMO-GATA1 reduces the DNA binding activity. Various GATA1 constructs were transfected into 293T, and nuclear extracts were subjected to EMSA using a GATA1-specific oligonucleotide probe (top) and Western blotting with anti-GATA1 (bottom). * indicates a SUMOylated form of SUMO-GATA1 and Δ indicates a potential deconjugated/cleaved GATA1. Similar results were obtained from two additional experiments.
Critical role of GATA1 SUMOylation in erythropoiesis
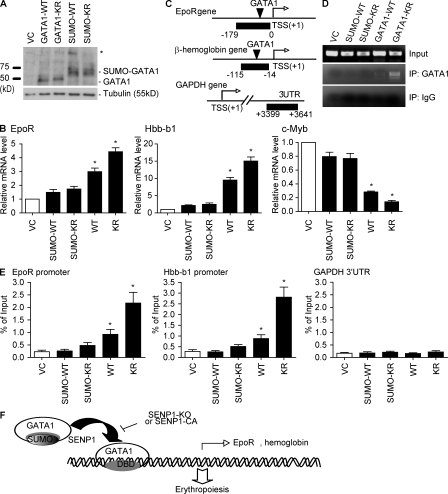
Finally, we determined the functional significance of GATA1 SUMOylation in erythropoiesis. To this end, HEL cells were infected with retrovirus encoding GATA1 WT, GATA1-K137R, SUMO-GATA1, or SUMO-GATA1K137R (Fig. S7 A, expression). Erythropoiesis was measured for hemoglobin-positive cells by benzidine staining as described in Fig. 6, except with no hemin treatment. Expression of GATA1 WT, induced erythropoiesis, and expression of KR strongly induced erythropoiesis, as indicated by a reduced number of benzidine-positive cells. In contrast, SUMO fusion proteins (GATA1 WT and KR) failed to induce erythropoiesis (Fig. S7, B and C [quantification]).
To eliminate any endogenous GATA1 effects, we reconstituted various GATA1 forms into G1E cells, a GATA1-null erythroid precursor cell line defective in erythropoiesis (Welch et al., 2004). G1E cells were infected with retrovirus encoding GATA1 WT, GATA1-K137R, SUMO-GATA1, or SUMO-GATA1K137R. Expression of GATA1 proteins was determined by Western blotting (Fig. 8 A). Transcripts of GATA1 responsiveness were determined by quantitative RT-PCR. Overexpression of GATA1 WT and GATA1-KR strongly enhanced transcription of the GATA1-inducible genes (EpoR and β-hemoglobin) but reduced the GATA1-repressive genes (Myb). In contrast, SUMO conjugation of GATA1 diminished GATA1 responsiveness of these genes (Fig. 8 B).
Figure 8.
Regulation of erythropoiesis by GATA1 SUMOylation. G1E cells were infected with retrovirus encoding GATA1 WT, GATA1-K137R, SUMO-GATA1, or SUMO-GATA1K137R. A retrovirus vector (VC) was a control. (A) Expression of GATA1 was determined by Western blot. * indicates a SUMOylated form of SUMO-GATA1. (B) Transcripts for EpoR, Hbb-b1, and c-Myb from G1E cells were quantified by quantitative RT-PCR and normalized to β-actin 48 h after retroviral infection as indicated. Data are mean ± SEM from three independent experiments. *, P < 0.01. (C–F) SUMO-GATA1 fails to be recruited to the EpoR and Hbb-b1 promoters. The promoter regions with a GATA1 binding site in mouse EpoR and Hbb-b1 genes are shown in C. TSS, the transcriptional site. PCR fragments for ChIP assay are indicated. A 3′ UTR promoter region from the GAPDH gene was used for a control. Nuclei were subjected to ChIP assay as described in Fig. 6. Representative ChIP PCR products of the GATA1-binding site on EpoR promoter are shown in D and quantitative analyses of PCR products (ratio of ChIP/Input) for the three genes are shown in E. Data are mean ± SEM from three independent experiments. *, P < 0.01 versus VC group. (F) Model for a critical role of SENP1 in regulation of GATA1 and GATA1-dependent erythropoiesis. GATA1 is synthesized and SUMOylated by an unknown SUMO ligase to retain in an inactive state. Upon erythropoietic stimuli, deSUMOylated GATA1 binds to the GATA1 sites within a gene promoter, inducing expression of GATA1-dependent erythropoiesis genes including EpoR, hemoglobin, and GATA1 itself. SENP1 KO mice show enhanced GATA1 SUMOylation, which blocks the GATA1 DNA-binding activity and subsequent associations with the promoters of erythropoietic genes, leading to erythropoiesis defects and anemia.
To directly determine if diminished GATA1 responsiveness of gene expression is the result of a blocked recruitment of SUMO-GATA1 to the GATA1-responsive genes, we measured the binding of GATA1 proteins to the EpoR and β-hemoglobin promoters in GATA1-infected G1E cells by ChIP assay. The GAPDH gene promoter was used as a negative control (Fig. 8 C). Similar to Fig. 6, nuclei were subjected to ChIP with anti-GATA1 followed by a PCR amplification of the GATA1 binding site in the EpoR and β-hemoglobin promoters. An isotype IgG was used as a negative control for immunoprecipitation. Consistent with the DNA binding activity and the gene induction of GATA1 proteins, both GATA1 WT and GATA1-K137R bound to the EpoR promoter region, and GATA1-K137R was stronger than GATA1 WT as expected. However, SUMO-GATA1 failed to be recruited to the EpoR and β-hemoglobin promoters (a representative ChIP gel from the EpoR gene is shown in Fig. 8 D with quantification in Fig. 8 E).
DISCUSSION
In this paper, we report that we have created SENP1 KO mice based on a homologous recombination system. The SENP1 KO mouse tissues express neither the SENP1 transcript encoding exons 4–5, as verified by RT-PCR, nor the SENP1 protein, as verified by Western blotting with anti-SENP1 antibody recognizing the C-terminal catalytic domain. There have been two studies on SENP1 mutant mice, and mice from both these studies were derived from ES cell lines with a retroviral vector that had been randomly inserted into the enhancer region or the coding region of the SENP1 gene, causing inactivation of the gene (Yamaguchi et al., 2005; Cheng et al., 2007). Consistent with our SENP1 KO mice, these SENP1 mutant mice show severe fetal anemia. Cheng et al. (2007) have also demonstrated that SENP1, via a direct deSUMOylation of HIF1α, regulates HIF1α stability and HIF1α-dependent genes, including Epo, causing defects of erythropoiesis. Consistent with this finding, we also observed that HIF-1α protein and Epo gene expression were reduced in our SENP1 KO mice and that overexpression of HIF1α could partially rescue a hematopoiesis defect induced by SENP1 deletion. Importantly, we have found that GATA1 is another important target of SENP1 during hematopoiesis (Fig. 8 F).
The evidence for the role of SENP1 in regulation of GATA1 activity is compelling. The quantitative RT-PCR analysis for mRNA, Western blotting for protein expression, and EMSA for the DNA binding activity clearly show that the GATA1 activity is down-regulated in fetal liver of the SENP1 KO mice. We found that a SUMOylated form of GATA1 is accumulated in SENP1 KO mice, correlating with down-regulation of GATA1-dependent genes including EpoR and β-hemoglobin. We further show that SENP1 can directly deSUMOylate GATA1, modulating GATA1-dependent EpoR expression and erythropoiesis by in vitro assays. Gene-targeting and loss-of-function studies have proved that GATA1 plays an essential role in erythropoiesis and megakaryopoiesis (Yamamoto et al., 1990; Weiss and Orkin, 1995). Indeed, SENP1 KO mice show defects in both processes in fetal liver and in the BM with transplanted fetal liver cells. We observed increased lymphocytes and monocytes in SENP1 KO fetal liver–transplanted recipient. This is consistent with the data that GATA1 deficiency could enhance multipotential differentiation ability of erythroid-committed cells (Kitajima et al., 2006). SENP1 KO mice show a milder anemia phenotype than GATA1-deficient mice, which die at E10.5 as a result of severe anemia with arrest of erythroid maturation. This is expected because GATA1 in SENP1 KO cells are not entirely SUMOylated. Therefore, GATA1 activity and GATA1-dependent gene expression is reduced, but not completely eliminated, in SENP1 KO. Therefore, SENP1 KO mice show a similar phenotype to GATA1 knockdown (GATA1.05 and GATA1-low mice, which express only ∼5–20% of the WT GATA1 levels, respectively; McDevitt et al., 1997; Takahashi et al., 1997). GATA1 SUMOylation is balanced by SUMO ligases and SENPs. Further investigations are needed to identify the SUMO ligases responsible for GATA1 SUMOylation and the efficiency of the ligase on GATA1. There are six members of the SENP family (SENP1–3 and SENP5–7), and each SENP has different cellular locations and, likely, has different substrate specificities (Gong et al., 2000; Kim et al., 2000; Nishida et al., 2000; Best et al., 2002; Hang and Dasso, 2002; Kadoya et al., 2002; Zhang et al., 2002; Bailey and O’Hare, 2004). Whether or not other SENPs members could deSUMOylate GATA1 during erythropoiesis still needs to be determined.
The function of GATA1 can be regulated by an array of posttranslational modifications, including phosphorylation and acetylation as well as SUMOylation (Boyes et al., 1998; Cantor and Orkin, 2002; Collavin et al., 2004; Yu et al., 2005; Hernandez-Hernandez et al., 2006; Lamonica et al., 2006; Zhao et al., 2006). Phosphorylation of GATA1 and its role in regulation of GATA1 function has been extensively studied. GATA1 can be strongly phosphorylated at residue Ser26 of the N-terminal acidic activation domain by MAPK and at Ser310 of the DNA-binding domain by Akt in response to growth factors such as Epo (Yu et al., 2005; Zhao et al., 2006). Phosphorylation of GATA1 increases GATA-1–mediated transcription without significantly changing the DNA-binding affinity of GATA1. However, GATA1 acetylation by cofactor p300 enhances its DNA-binding and recruitment to chromatin (Boyes et al., 1998; Hernandez-Hernandez et al., 2006; Lamonica et al., 2006). In contrast, the role of SUMOylation of GATA1 has not been well defined. GATA1 SUMOylation was initially identified based on its interaction with the SUMO E3 ligase PIASy (Collavin et al., 2004). Given the fact that only a small proportion (<5%) of GATA1 can be SUMOylated in cells, an inhibitory effect of SUMOylation on GATA1 cannot be readily detected. We have taken three different approaches to address this issue. First, we delete the SUMO endopeptidase SENP1 or express a dominant-negative form of SENP1 to promote GATA1 SUMOylation. This enhanced GATA1 SUMOylation dramatically represses the DNA binding and the transcriptional activities of GATA1 in erythropoiesis. Second, we use GATA1-null cells to show that a non-SUMOytable GATA1 mutant (K137R) has a higher activity than GATA1 in DNA-binding and induction of erythropoiesis. Finally, we demonstrate that SUMOylated GATA1, in a GATA1-SUMO fusion form, blunts the DNA-binding activity and subsequent recruitment to the EpoR and β-hemoglobin gene promoters and erythropoiesis. Collectively, we conclude that GATA1 SUMOylation inhibits its DNA-binding and erythropoietic activity.
Our study is the first demonstration of a functional significance of GATA1 SUMOylation. The exact mechanism by which SUMO modification inhibits GATA1 DNA binding and the subsequent recruitment to chromatin is currently unclear. It is conceivable that SUMOylation of GATA1, which is the opposite of acetylation, alters GATA1 conformation in a way that is unfavorable for DNA interaction. Whether or not SUMOylation of GATA1 in cells prevents GATA1 acetylation or its interactions with other coactivators are under investigation.
MATERIALS AND METHODS
Targeted inactivation of SENP1 gene by homologous recombination.
The SENP1 targeting arms were isolated from BAC clone identified by screening a Research Genetics (Invitrogen) BAC library using SENP1 cDNA as a probe. The targeting vector was constructed in a pEASY-flox backbone to contain a loxP site inserted upstream of SENP1 exon 5 and a neomycin cassette (Neo) flanked by two loxP sites down stream of exon 6 using standard molecular procedures. The linearized targeting construct was electroporated into 129/C57B/6 ES cells, and the targeted clones were selected with G418 and gancyclovir. Resistant clones were screened for homologous recombination by PCR and confirmed by Southern blot analysis. Two independent SENP1lox ES cell clones were injected into C57BL/6 blastocysts. Chimeras were further bred with C57BL/6 female for germline transmission. SENP1lox/lox mice were mated with β-actin–Cre mice to mediate a recombination in vivo resulting in a complete deletion of SENP1 exon 5 and exon 6 and a frameshift of the downstream catalytic domain.
Histology and immunohistochemistry.
Embryos or fetal tissues were fixed for immunohistochemical and hematoxylin/eosin staining in 4% paraformaldehyde. For histological analysis, tissues were embedded in paraffin, cut into 5-µM sections, and stained with routine hematoxylin/eosin. For immunohistochemical staining, tissue sections were stained with certain antibodies (e.g., anti-GATA1 and anti-EpoR) as described previously (He et al., 2006; Zhang et al., 2008). Bound primary antibodies were detected using avidin-biotin-peroxidase (NovaRed peroxidase substrate kit; Vector Laboratories). Pictures from four random areas of each section and five sections per mouse were taken using a digital camera (Kodak) mounted on a light microscope (Carl Zeiss, Inc.). Images were quantified using the Matlab software (The Math Works, Inc.) as described previously (He et al., 2006; Zhang et al., 2008).
Colony formation assay.
Cells were prepared from murine E13.5 fetal livers. After lysis of mature red blood cells by PharmLyse (BD), cells were mixed with MethoCult M3334 to detect BFU-e or MethoCult M3434 to detect CFU-e, CFU-G, CFU-G, and CFU-G/M assays (STEMCELL Technologies Inc.). Cells were plated in 35-mm dishes and cultured at 37°C with 5% CO2 for 3–8 d. For the CFU-e assay, benzidine-positive CFU-e colonies were scored on day 3. For BFU-e assay, benzidine-positive BFU-e colonies were scored on day 8. For MegaCult-C assays, fetal liver cells were used according to the manufacturer’s protocols (STEMCELL Technologies Inc.). 50 ng/ml human thrombopoietin, 50 ng/ml human IL-11, 10 ng/ml murine IL-3, and 20 ng/ml human IL-6 (PeproTech) were used in these assays. The cultures were incubated at 37°C with 5% CO2 for 6–8 d. Colonies containing more than three megakaryocytes (AchE+ cells) were considered CFU-MKs (Cheng et al., 2009). Duplicate assays were performed for each mouse.
BMT of fetal liver cells.
BMT was performed in lethally irradiated mice (irradiation 2× 5.5 Gy within 3 h) with WT or SENP1 KO fetal liver cells. Fetal liver cells were first isolated from E13.5 embryos of WT or SENP1 KO mice (CD45.2+), and red cells were removed by PharmLyse and subsequent washing with PBS. Cells were counted and 105 cells were injected into the tail vein of the Pep3B recipient (CD45.1+). Successful BMT was controlled by FACS analysis of peripheral blood with anti-CD45.1 and anti-CD45.2 antibodies or by genotyping of SENP1 gene 6 wk after BMT. The mice were then subjected to FACS analysis for specific cell markers or CBC analysis. All animal studies were approved by the institutional animal care and use committees of Yale University.
Flow cytometric analysis of specific cell surface markers.
To analyze erythrocytes, megakaryocytes, and other cell types, the fetal liver cells or BM cells were stained with standard protocols using PE-Ter119, PE-Gr1, PE-CD41a, PE-CD71, or APC-Ter119 (BD). FACS analysis was performed with an LSRII flow cytometer (BD) and FlowJo software (Tree Star, Inc.). Isotype controls were used in each experiment.
Murine peripheral blood counts.
Mice were anesthetized with Methoxyflurane (Medical Developments International) followed by retroorbital bleeding (∼100 µl) using EDTA-treated glass capillary tubes. The blood was evacuated into tubes with 5 µl of 0.2M EDTA to prevent clot formation. The CBC of blood sample was performed using a Hemavet 950FS (Drew Scientific) according to the manufacturer’s protocol.
Plasmids, adenovirus and retrovirus.
Mammalian expression plasmids for SENP1 WT were described previously (Li et al., 2008). Expression plasmid for SENP1-K137R was a gift from E. Yeh (The University of Texas M.D. Anderson Cancer Center, Houston, TX). Expression plasmids GATA1 and K137R mutant were gifts from C. Santoro (Università del Piemonte Orientale, Novara, Italy). Expression plasmid for EpoR (B. Forget, Yale University, New Haven, CT) SUMO-GATA1 fusion plasmid was constructed by cloning GATA1 into pSUMOstar vector (LifeSensors). Adenoviral vectors expressing flag-tagged SENP1-C603A (pAd-SENP1CA) and pAd-GATA1 was constructed as we described for pAd-AIP1 (Zhang et al., 2008). HIF1α adenovirus stock was a gift from F. Giordano (Yale University). All adenovirus was amplified in 293T cells and purified by ultracentrifugation. HEL cells were infected with adenovirus at MOI of 100 for 48–72 h before subsequent assays. Retroviral vector pMY.IRES.EGFP and ecotropic packaging cell line PLAT-E were gifts from T. Kitamura (Tokyo University, Tokyo, Japan). GATA-1 WT, GATA1-K137R, Sumo-GATA1 WT, and Sumo-GATA1-K137R cDNAs were inserted into the multicloning site of pMY.IRES.EGFP. The retrovirus supernatant was obtained by a transient transfection of the retroviral vectors to the PLAT-E packaging cell line using Lipofectamine 2000 (Invitrogen). 106 G1E cells were resuspended in 2 ml retrovirus supernatant with 1 µg polybrene per ml and the infection was done by spinoculation (2 h, 25°C, 2,500 rpm).
Cell culture of G1E and HEL cells.
GATA1-null G1E cell was a gift from M.J. Weiss, (University of Pennsylvania School of Medicine, Philadelphia, PA) and was cultured in IMDM supplemented with 15% fetal calf serum, 1:10,000 monothioglycerol (Sigma-Aldrich), human recombinant erythropoietin (R&D Systems) at 2 U/ml, and kit ligand (PeproTech) at 50 ng/ml and penicillin/streptomycin at 37°C in 5% CO2 in a humidified incubator as previously described (Weiss and Orkin, 1995). HEL cells were cultured in RPMI 1640 medium (Invitrogen) supplemented with 10% fetal calf serum, 2 mM L-glutamine, and penicillin/streptomycin at 37°C in 5% CO2 in a humidified incubator. To induce differentiation, 2 × 105 cells per ml were cultured in the presence of 40 µM hemin (Sigma-Aldrich) for 3 d or as specified.
Immunoprecipitation and immunoblotting.
Cell lysis, protein preparation, and immunoblotting were performed as described previously (Li et al., 2008). For detection of SUMOylated proteins, 1 mM N-ethylmaleimide (deSUMOylase inhibitor) was added to cell lysates. Antibodies against anti-SENP1, GATA1, GATA2, EpoR, USF-2, and tubulin were obtained from Santa Cruz Biotechnology, Inc., anti-HIF1α was obtained from Novus Biologicals, and anti–Flag-POD was obtained from Sigma-Aldrich.
EMSA and ChIP assay.
The double-stranded oligonucleotide containing a GATA consensus site from the EpoR gene promoter was used for EMSA. Preparation of nuclear extracts and EMSA were performed as previously described (Zhang et al., 1995). The ChIP assay was performed exactly as previously described (Letting et al., 2004; Ji et al., 2008) and analyzed by real-time PCR. All the primers are listed in Table S1. Antibodies included anti-GATA1 (ab1963; Abcam), a positive control (anti-RNA polymerase II), and a negative control (normal IgG).
Gene expression analysis.
Total RNA of fetal liver was isolated by using phenol/chloroform and isolated using RNeasy kit with DNase I digestion (QIAGEN). Reverse transcription was done by standard procedure (Super Script First-Strand Synthesis System; Invitrogen) using 1 µg of total RNA. Quantitative real-time PCR was performed by using iQ SYBR Green Supermix on iCycler Real-Time Detection system (Bio-Rad Laboratories). Specific primers for mouse GATA1, GATA2, EpoR, Epo, c-Myb, c-Kit, and various isoforms of hemoglobin genes were used. 18 S ribosomal RNA or β actin was used as internal control. All the primers are listed in Table S1. The relative amount of mRNA normalized with control group as indicated was quantified.
Indirect immunofluorescence confocal microscopy.
Fixation, permeabilization, and staining of cultured BAEC were performed as described previously (Li et al., 2008). Alexa Fluor 488 (green)–conjugated secondary antibodies (Invitrogen) were used. Confocal immunofluorescence microscopy was performed using a confocal microscope (TCS SP5; Leica), and the acquired images were transferred to Photoshop 6.0 (Adobe) to generate the final figures.
Statistical analysis.
All data are expressed as means ± SEM. Statistical significance of differences between different conditions was performed using Prism (GraphPad Software, Inc.) with a two-tailed unpaired Student’s t test. A value of P < 0.01 was considered as statistically significant.
Online supplemental material.
Fig. S1 shows enlarged spleen with extramedullary hematopoiesis in recipient mice with SENP1 KO fetal liver cells. Fig. S2 shows a decreased expression of HIF-1α and Epo in E13.5 of SENP1 KO fetal liver. Fig. S3 shows similar GATA1 expressions in stage-matched hematopoietic progenitors. Fig. S4 shows a rescue of SENP-CA–induced hematopoiesis by overexpression of GATA1 or HIF-1α. Fig. S5 shows no effects of SUMO conjugation on GATA1 protein stability. Fig. S6 shows a similar punctate staining from GATA1 WT and GATA1 mutants. Fig. S7 shows an inability of SUMO-GATA1 to induce erythropoiesis. Table S1 lists primers used in this study. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092215/DC1.
Acknowledgments
This work was supported by National Institutes of Health (NIH) grants HL077357, HL070295, HL65978, and HL085789 to W. Min. Funding was also provided by the Yale Center of Excellence in Molecular Hematology (NIH DK072442).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- BFU-e
- erythroid burst-forming unit
- BMT
- BM transplantation
- CBC
- complete blood count
- CFU-e
- erythroid CFU
- CFU-G/M
- granulocyte/monocyte
- CFU-mk
- megakaryocyte CFU
- ChIP
- chromatin immunoprecipitation
- EMSA
- electrophoretic mobility shift assay
- ES
- embryonic stem
- SENP
- sentrin-specific protease
- SUMO
- small ubiquitin-like modifier
References
- Bailey D., O’Hare P. 2004. Characterization of the localization and proteolytic activity of the SUMO-specific protease, SENP1. J. Biol. Chem. 279:692–703 10.1074/jbc.M306195200 [DOI] [PubMed] [Google Scholar]
- Best J.L., Ganiatsas S., Agarwal S., Changou A., Salomoni P., Shirihai O., Meluh P.B., Pandolfi P.P., Zon L.I. 2002. SUMO-1 protease-1 regulates gene transcription through PML. Mol. Cell. 10:843–855 10.1016/S1097-2765(02)00699-8 [DOI] [PubMed] [Google Scholar]
- Boyes J., Byfield P., Nakatani Y., Ogryzko V. 1998. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 396:594–598 10.1038/25166 [DOI] [PubMed] [Google Scholar]
- Cantor A.B., Orkin S.H. 2002. Transcriptional regulation of erythropoiesis: an affair involving multiple partners. Oncogene. 21:3368–3376 10.1038/sj.onc.1205326 [DOI] [PubMed] [Google Scholar]
- Carter S., Bischof O., Dejean A., Vousden K.H. 2007. C-terminal modifications regulate MDM2 dissociation and nuclear export of p53. Nat. Cell Biol. 9:428–435 10.1038/ncb1562 [DOI] [PubMed] [Google Scholar]
- Cheng J., Kang X., Zhang S., Yeh E.T. 2007. SUMO-specific protease 1 is essential for stabilization of HIF1alpha during hypoxia. Cell. 131:584–595 10.1016/j.cell.2007.08.045 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng E.C., Luo Q., Bruscia E.M., Renda M.J., Troy J.A., Massaro S.A., Tuck D., Schulz V., Mane S.M., Berliner N., et al. 2009. Role for MKL1 in megakaryocytic maturation. Blood. 113:2826–2834 10.1182/blood-2008-09-180596 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiba T., Nagata Y., Kishi A., Sakamaki K., Miyajima A., Yamamoto M., Engel J.D., Todokoro K. 1993. Induction of erythroid-specific gene expression in lymphoid cells. Proc. Natl. Acad. Sci. USA. 90:11593–11597 10.1073/pnas.90.24.11593 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collavin L., Gostissa M., Avolio F., Secco P., Ronchi A., Santoro C., Del Sal G. 2004. Modification of the erythroid transcription factor GATA-1 by SUMO-1. Proc. Natl. Acad. Sci. USA. 101:8870–8875 10.1073/pnas.0308605101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferreira R., Ohneda K., Yamamoto M., Philipsen S. 2005. GATA1 function, a paradigm for transcription factors in hematopoiesis. Mol. Cell. Biol. 25:1215–1227 10.1128/MCB.25.4.1215-1227.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiwara Y., Browne C.P., Cunniff K., Goff S.C., Orkin S.H. 1996. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl. Acad. Sci. USA. 93:12355–12358 10.1073/pnas.93.22.12355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill G. 2004. SUMO and ubiquitin in the nucleus: different functions, similar mechanisms? Genes Dev. 18:2046–2059 10.1101/gad.1214604 [DOI] [PubMed] [Google Scholar]
- Godin I., Cumano A. 2002. The hare and the tortoise: an embryonic haematopoietic race. Nat. Rev. Immunol. 2:593–604 [DOI] [PubMed] [Google Scholar]
- Gong L., Millas S., Maul G.G., Yeh E.T. 2000. Differential regulation of sentrinized proteins by a novel sentrin-specific protease. J. Biol. Chem. 275:3355–3359 10.1074/jbc.275.5.3355 [DOI] [PubMed] [Google Scholar]
- Gregory T., Yu C., Ma A., Orkin S.H., Blobel G.A., Weiss M.J. 1999. GATA-1 and erythropoietin cooperate to promote erythroid cell survival by regulating bcl-xL expression. Blood. 94:87–96 [PubMed] [Google Scholar]
- Hang J., Dasso M. 2002. Association of the human SUMO-1 protease SENP2 with the nuclear pore. J. Biol. Chem. 277:19961–19966 10.1074/jbc.M201799200 [DOI] [PubMed] [Google Scholar]
- He Y., Luo Y., Tang S., Rajantie I., Salven P., Heil M., Zhang R., Luo D., Li X., Chi H., et al. 2006. Critical function of Bmx/Etk in ischemia-mediated arteriogenesis and angiogenesis. J. Clin. Invest. 116:2344–2355 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hernandez-Hernandez A., Ray P., Litos G., Ciro M., Ottolenghi S., Beug H., Boyes J. 2006. Acetylation and MAPK phosphorylation cooperate to regulate the degradation of active GATA-1. EMBO J. 25:3264–3274 10.1038/sj.emboj.7601228 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W., Yang L., Yu L., Yuan J., Hu D., Zhang W., Yang J., Pang Y., Li W., Lu J., et al. 2008. Epigenetic silencing of O6-methylguanine DNA methyltransferase gene in NiS-transformed cells. Carcinogenesis. 29:1267–1275 10.1093/carcin/bgn012 [DOI] [PubMed] [Google Scholar]
- Kadoya T., Yamamoto H., Suzuki T., Yukita A., Fukui A., Michiue T., Asahara T., Tanaka K., Asashima M., Kikuchi A. 2002. Desumoylation activity of Axam, a novel Axin-binding protein, is involved in downregulation of beta-catenin. Mol. Cell. Biol. 22:3803–3819 10.1128/MCB.22.11.3803-3819.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., Iwao H. 2000. Molecular and cellular mechanisms of angiotensin II-mediated cardiovascular and renal diseases. Pharmacol. Rev. 52:11–34 [PubMed] [Google Scholar]
- Kim K.I., Baek S.H., Jeon Y.J., Nishimori S., Suzuki T., Uchida S., Shimbara N., Saitoh H., Tanaka K., Chung C.H. 2000. A new SUMO-1-specific protease, SUSP1, that is highly expressed in reproductive organs. J. Biol. Chem. 275:14102–14106 10.1074/jbc.275.19.14102 [DOI] [PubMed] [Google Scholar]
- Kitajima K., Zheng J., Yen H., Sugiyama D., Nakano T. 2006. Multipotential differentiation ability of GATA-1-null erythroid-committed cells. Genes Dev. 20:654–659 10.1101/gad.1378206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe C., Mayeux P. 1999. The molecular biology of erythropoietin. Nephrol. Dial. Transplant. 14:22–28 10.1093/ndt/14.suppl_2.22 [DOI] [PubMed] [Google Scholar]
- Lamonica J.M., Vakoc C.R., Blobel G.A. 2006. Acetylation of GATA-1 is required for chromatin occupancy. Blood. 108:3736–3738 10.1182/blood-2006-07-032847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee R., Kertesz N., Joseph S.B., Jegalian A., Wu H. 2001. Erythropoietin (Epo) and EpoR expression and 2 waves of erythropoiesis. Blood. 98:1408–1415 10.1182/blood.V98.5.1408 [DOI] [PubMed] [Google Scholar]
- Letting D.L., Chen Y.Y., Rakowski C., Reedy S., Blobel G.A. 2004. Context-dependent regulation of GATA-1 by friend of GATA-1. Proc. Natl. Acad. Sci. USA. 101:476–481 10.1073/pnas.0306315101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S.J., Hochstrasser M. 1999. A new protease required for cell-cycle progression in yeast. Nature. 398:246–251 10.1038/18457 [DOI] [PubMed] [Google Scholar]
- Li X., Luo Y., Yu L., Lin Y., Luo D., Zhang H., He Y., Kim Y.O., Kim Y., Tang S., Min W. 2008. SENP1 mediates TNF-induced desumoylation and cytoplasmic translocation of HIPK1 to enhance ASK1-dependent apoptosis. Cell Death Differ. 15:739–750 10.1038/sj.cdd.4402303 [DOI] [PubMed] [Google Scholar]
- Lin C.S., Lim S.K., D’Agati V., Costantini F. 1996. Differential effects of an erythropoietin receptor gene disruption on primitive and definitive erythropoiesis. Genes Dev. 10:154–164 10.1101/gad.10.2.154 [DOI] [PubMed] [Google Scholar]
- Martin P., Papayannopoulou T. 1982. HEL cells: a new human erythroleukemia cell line with spontaneous and induced globin expression. Science. 216:1233–1235 10.1126/science.6177045 [DOI] [PubMed] [Google Scholar]
- McDevitt M.A., Shivdasani R.A., Fujiwara Y., Yang H., Orkin S.H. 1997. A “knockdown” mutation created by cis-element gene targeting reveals the dependence of erythroid cell maturation on the level of transcription factor GATA-1. Proc. Natl. Acad. Sci. USA. 94:6781–6785 10.1073/pnas.94.13.6781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller S., Hoege C., Pyrowolakis G., Jentsch S. 2001. SUMO, ubiquitin’s mysterious cousin. Nat. Rev. Mol. Cell Biol. 2:202–210 10.1038/35056591 [DOI] [PubMed] [Google Scholar]
- Nishida T., Tanaka H., Yasuda H. 2000. A novel mammalian Smt3-specific isopeptidase 1 (SMT3IP1) localized in the nucleolus at interphase. Eur. J. Biochem. 267:6423–6427 10.1046/j.1432-1327.2000.01729.x [DOI] [PubMed] [Google Scholar]
- Orkin S.H., Zon L.I. 2008. Hematopoiesis: an evolving paradigm for stem cell biology. Cell. 132:631–644 10.1016/j.cell.2008.01.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peschle C., Mavilio F., Carè A., Migliaccio G., Migliaccio A.R., Salvo G., Samoggia P., Petti S., Guerriero R., Marinucci M., et al. 1985. Haemoglobin switching in human embryos: asynchrony of zeta----alpha and epsilon----gamma-globin switches in primitive and definite erythropoietic lineage. Nature. 313:235–238 10.1038/313235a0 [DOI] [PubMed] [Google Scholar]
- Pevny L., Simon M.C., Robertson E., Klein W.H., Tsai S.F., D’Agati V., Orkin S.H., Costantini F. 1991. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 349:257–260 10.1038/349257a0 [DOI] [PubMed] [Google Scholar]
- Pickart C.M. 2001. Mechanisms underlying ubiquitination. Annu. Rev. Biochem. 70:503–533 10.1146/annurev.biochem.70.1.503 [DOI] [PubMed] [Google Scholar]
- Seeler J.S., Dejean A. 2003. Nuclear and unclear functions of SUMO. Nat. Rev. Mol. Cell Biol. 4:690–699 10.1038/nrm1200 [DOI] [PubMed] [Google Scholar]
- Suzuki T., Ichiyama A., Saitoh H., Kawakami T., Omata M., Chung C.H., Kimura M., Shimbara N., Tanaka K. 1999. A new 30-kDa ubiquitin-related SUMO-1 hydrolase from bovine brain. J. Biol. Chem. 274:31131–31134 10.1074/jbc.274.44.31131 [DOI] [PubMed] [Google Scholar]
- Takahashi S., Onodera K., Motohashi H., Suwabe N., Hayashi N., Yanai N., Nabesima Y., Yamamoto M. 1997. Arrest in primitive erythroid cell development caused by promoter-specific disruption of the GATA-1 gene. J. Biol. Chem. 272:12611–12615 10.1074/jbc.272.19.12611 [DOI] [PubMed] [Google Scholar]
- Tsai F.Y., Keller G., Kuo F.C., Weiss M., Chen J., Rosenblatt M., Alt F.W., Orkin S.H. 1994. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature. 371:221–226 10.1038/371221a0 [DOI] [PubMed] [Google Scholar]
- Wang J., Feng X.H., Schwartz R.J. 2004. SUMO-1 modification activated GATA4-dependent cardiogenic gene activity. J. Biol. Chem. 279:49091–49098 10.1074/jbc.M407494200 [DOI] [PubMed] [Google Scholar]
- Weiss M.J., Orkin S.H. 1995. GATA transcription factors: key regulators of hematopoiesis. Exp. Hematol. 23:99–107 [PubMed] [Google Scholar]
- Weiss M.J., Keller G., Orkin S.H. 1994. Novel insights into erythroid development revealed through in vitro differentiation of GATA-1 embryonic stem cells. Genes Dev. 8:1184–1197 10.1101/gad.8.10.1184 [DOI] [PubMed] [Google Scholar]
- Welch J.J., Watts J.A., Vakoc C.R., Yao Y., Wang H., Hardison R.C., Blobel G.A., Chodosh L.A., Weiss M.J. 2004. Global regulation of erythroid gene expression by transcription factor GATA-1. Blood. 104:3136–3147 10.1182/blood-2004-04-1603 [DOI] [PubMed] [Google Scholar]
- Wu H., Liu X., Jaenisch R., Lodish H.F. 1995. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoietin or the erythropoietin receptor. Cell. 83:59–67 10.1016/0092-8674(95)90234-1 [DOI] [PubMed] [Google Scholar]
- Yamaguchi T., Sharma P., Athanasiou M., Kumar A., Yamada S., Kuehn M.R. 2005. Mutation of SENP1/SuPr-2 reveals an essential role for desumoylation in mouse development. Mol. Cell. Biol. 25:5171–5182 10.1128/MCB.25.12.5171-5182.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamamoto M., Ko L.J., Leonard M.W., Beug H., Orkin S.H., Engel J.D. 1990. Activity and tissue-specific expression of the transcription factor NF-E1 multigene family. Genes Dev. 4:1650–1662 10.1101/gad.4.10.1650 [DOI] [PubMed] [Google Scholar]
- Yu Y.L., Chiang Y.J., Chen Y.C., Papetti M., Juo C.G., Skoultchi A.I., Yen J.J. 2005. MAPK-mediated phosphorylation of GATA-1 promotes Bcl-XL expression and cell survival. J. Biol. Chem. 280:29533–29542 10.1074/jbc.M506514200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang R., Min W., Sessa W.C. 1995. Functional analysis of the human endothelial nitric oxide synthase promoter. Sp1 and GATA factors are necessary for basal transcription in endothelial cells. J. Biol. Chem. 270:15320–15326 10.1074/jbc.270.25.15320 [DOI] [PubMed] [Google Scholar]
- Zhang H., Saitoh H., Matunis M.J. 2002. Enzymes of the SUMO modification pathway localize to filaments of the nuclear pore complex. Mol. Cell. Biol. 22:6498–6508 10.1128/MCB.22.18.6498-6508.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Socolovsky M., Gross A.W., Lodish H.F. 2003. Role of Ras signaling in erythroid differentiation of mouse fetal liver cells: functional analysis by a flow cytometry-based novel culture system. Blood. 102:3938–3946 10.1182/blood-2003-05-1479 [DOI] [PubMed] [Google Scholar]
- Zhang H., He Y., Dai S., Xu Z., Luo Y., Wan T., Luo D., Jones D., Tang S., Chen H., et al. 2008. AIP1 functions as an endogenous inhibitor of VEGFR2-mediated signaling and inflammatory angiogenesis in mice. J. Clin. Invest. 118:3904–3916 10.1172/JCI36168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Kitidis C., Fleming M.D., Lodish H.F., Ghaffari S. 2006. Erythropoietin stimulates phosphorylation and activation of GATA-1 via the PI3-kinase/AKT signaling pathway. Blood. 107:907–915 10.1182/blood-2005-06-2516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zon L.I., Youssoufian H., Mather C., Lodish H.F., Orkin S.H. 1991. Activation of the erythropoietin receptor promoter by transcription factor GATA-1. Proc. Natl. Acad. Sci. USA. 88:10638–10641 10.1073/pnas.88.23.10638 [DOI] [PMC free article] [PubMed] [Google Scholar]