Abstract
How proliferative and inhibitory signals integrate to control liver regeneration remains poorly understood. A screen for antiproliferative factors repressed after liver injury identified transducer of ErbB2.1 (Tob1), a member of the PC3/BTG1 family of mito-inhibitory molecules as a target for further evaluation. Tob1 protein decreases after 2/3 hepatectomy in mice secondary to posttranscriptional mechanisms. Deletion of Tob1 increases hepatocyte proliferation and accelerates restoration of liver mass after hepatectomy. Down-regulation of Tob1 is required for normal liver regeneration, and Tob1 controls hepatocyte proliferation in a dose-dependent fashion. Tob1 associates directly with both Caf1 and cyclin-dependent kinase (Cdk) 1 and modulates Cdk1 kinase activity. In addition, Tob1 has significant effects on the transcription of critical cell cycle components, including E2F target genes and genes involved in p53 signaling. We provide direct evidence that levels of an inhibitory factor control the rate of liver regeneration, and we identify Tob1 as a crucial check point molecule that modulates the expression and activity of cell cycle proteins.
Liver regeneration is controlled by a panoply of factors performing diverse functions, including cytokine signaling (suppressor of cytokine signaling 3 [Socs3], IL-6, and Tnf; Feingold et al., 1988; Sakamoto et al., 1999; Zimmers et al., 2003; Riehle et al., 2008), transcription (NF-κb and Stat3; Cressman et al., 1994, 1995), growth factor response (egf [epidermal growth factor] and TGF-β; Russell, 1988; Jones et al., 1995), glucose metabolism (Bucher and Swaffield, 1975), bile acid metabolism (Huang et al., 2006), and platelet-derived signaling involving both extracellular nucleotides (Beldi et al., 2008) and serotonin (Lesurtel et al., 2006). Advances in defining and characterizing these signals leave unanswered the fundamental question of how multiple inputs are integrated into the discrete functional response of hepatocyte proliferation after injury.
Restoration of liver mass requires stimulatory inputs. Inhibition or removal of IL-6, Stat3, Egf, c-Jun, or serotonin, among others, delays liver regeneration (Jones et al., 1995; Sakamoto et al., 1999; Selzner and Clavien, 2000; Lesurtel et al., 2006; Natarajan et al., 2007; Moh et al., 2007). Furthermore, addition of a variety of molecules, including IL-6, HGF (hepatocyte growth factor), and thyroid hormone, enhances liver regeneration (Francavilla et al., 1994; Ogura et al., 2001; Zimmers et al., 2003).
Direct evidence that reduction in inhibitory inputs is critical for restoration of liver mass after injury has been more difficult to obtain, and this remains a highly likely but poorly understood paradigm. Clearly, inhibitory inputs must play some role. For example, both TGF-β and its family member activin inhibit liver regeneration, and the activin antagonist follistatin enhances regeneration (Jakowlew et al., 1991; Schwall et al., 1993; Kogure et al., 1995). These experiments, however, which were performed with pharmacologic doses of the molecules, do not identify the actual mediator. Furthermore, both TGF-β1 and activin βA are increased after partial hepatectomy, making it difficult to conclude that either molecule inhibits regeneration after hepatectomy (Ueda et al., 2003; Takamura et al., 2005; Michalopoulos, 2007). Similarly, although mice with deletions in Socs3 or p21 exhibit enhanced liver regeneration, both are induced after hepatectomy, suggesting they are not direct inhibitors of hepatocyte proliferation (Stepniak et al., 2006; Riehle et al., 2008). Other candidate inhibitors of hepatocyte proliferation, including p27 and CCAAT enhancer binding protein α (C/ebp-α), are difficult to study because of baseline size or metabolic abnormalities in null mice (Flodby et al., 1996; Hayashi et al., 2003).
Constitutive repressors that control the rate of hepatocyte proliferation may be defined by the following characteristics: baseline concentrations that are decreased after hepatectomy, enhancement of hepatocyte proliferation in mice with targeted deletion, and prevention of normal regeneration when maintained at high levels. To find such molecules, we undertook a screen for genes that decrease after partial hepatectomy and might have antimitotic activity.
We then focused on transducer of ErbB2.1 (Tob1) based on our characterization of its expression pattern after hepatectomy and its known antiproliferative functions (Otu et al., 2007). Initially identified by interactions with p185erbB2, proteins in the Tob1 family regulate cell cycle (Matsuda et al., 1996; Yoshida et al., 2000, 2003a). Nuclear localization and phosphorylation are critical for Tob1 function, which occurs via at least two distinct mechanisms (Suzuki et al., 2002; Kawamura-Tsuzuku et al., 2004). The first is to influence cyclin-dependent kinase (Cdk) activity by binding to Caf1, and the second is to modulate mRNA degradation by regulating the first step of the process, deadenylation (Ikematsu et al., 1999; Tucker et al., 2001; Temme et al., 2004; Ezzeddine et al., 2007; Nishida et al., 2007). In this paper, we show that Tob1 fulfills the criteria for a critical constitutive inhibitor of hepatocyte proliferation, and we provide evidence that Tob1 acts as an integrative molecule to control the rate of hepatocyte proliferation after hepatectomy by influencing the activity and expression of cell cycle proteins.
RESULTS
Tob1 mRNA and protein levels decline after partial hepatectomy
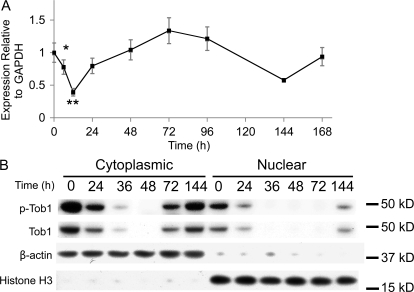
To determine how Tob1 mRNA levels change as liver mass is restored, real-time PCR was performed on regenerating liver after 2/3 hepatectomy in C57B/6 mice (Fig. 1 A). There is high baseline Tob1 mRNA expression in the uninjured liver (time 0). Levels decline acutely but recover by 24 h, reaching baseline by 48 h after hepatectomy. At 6 and 12 h after hepatectomy, these differences were significant (n = 3–9 animals). In contrast, no changes in Tob1 mRNA expression were observed in sham-operated control mice (unpublished data).
Figure 1.
Tob1 mRNA expression and protein levels are decreased after 2/3 hepatectomy. (A) Real-time PCR of Tob1 mRNA in liver samples at various intervals after hepatectomy. *, P = 0.05; **, P = 0.001. Each time point consists of at least three and as many as nine animals. Error bars in this and all other figures are standard error. (B) Western blot analysis of total and phosphorylated Tob1 protein in whole cell and nuclear extracts at various times after 2/3 hepatectomy. β-Actin demonstrated the purity of the cytoplasmic fraction, whereas histone H3 demonstrated the purity of the nuclear fraction. Both serve as loading controls. All blot lanes are the most representative sample from among at least three different animals.
Nuclear localization and phosphorylation state of Tob1 were analyzed separately by Western blotting (Fig. 1 B). In response to 2/3 hepatectomy, cytoplasmic protein levels of both Tob1 and p-Tob1 decrease by 24 h, are nearly undetectable by 36 h, and begin to recover by 72 h, reaching basal levels by 144 h. Nuclear levels of Tob1 and p-Tob1 are generally lower overall. Levels decline by 24 h after hepatectomy and, by 36 h, are nearly undetectable. Recovery begins by 144 h after injury. Purity of cytoplasmic and nuclear fractions was verified by blotting for β-actin (cytoplasmic) and histone H3 (nuclear), which also served as loading controls. No changes in Tob1 protein expression occurred in sham-operated mice (unpublished data). Collectively, our results suggest that Tob1 protein decreases after hepatectomy, primarily via posttranscriptional mechanisms. Because the proportion of Tob1 that is phosphorylated does not change dramatically over time, decreases in levels of p-Tob1 are likely a result of protein degradation and not dephosphorylation.
Liver regeneration after partial hepatectomy is accelerated in Tob1-null mice.
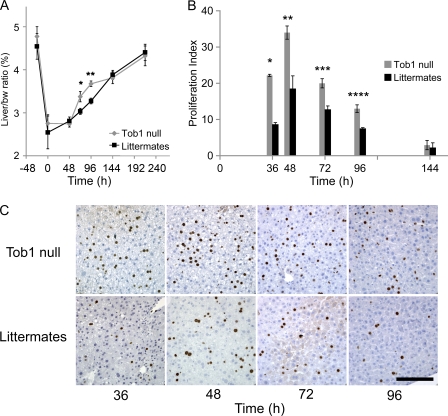
To determine the functional consequences of loss of Tob1, comparisons were made between WT and Tob1-null mice. Western blot analysis confirmed disruption of the Tob1 gene indicated by undetectable levels of Tob1 protein, which is consistent with published results (Yoshida et al., 2000, 2003a; and not depicted). Adult Tob1-null mice and their WT littermates demonstrate similar baseline liver/body weight ratio, exhibit similar minimal baseline hepatocyte proliferation as measured by BrdU labeling, and have similar histology (Fig. 2 A and not depicted). After partial hepatectomy, Tob1-null mice demonstrate a faster recovery of liver/body weight ratio beginning at 72 h and continuing through 96 h (Fig. 2 A; n = 3–6). Liver/body weight ratios are equivalent 9 d after partial hepatectomy when the ratio returns to normal. Tob1-null mice demonstrate more rapid onset of hepatocyte proliferation and higher absolute percentage of proliferating hepatocytes from 36 to 96 h after hepatectomy compared with WT littermates (Fig. 2, B and C; n = 3–6). These results demonstrate that Tob1 functions to decrease the pace of regeneration after hepatectomy.
Figure 2.
Loss of Tob1 enhances liver regeneration and hepatocyte proliferation after hepatectomy. (A) 2/3 hepatectomy was performed on Tob1-null and WT mice. Restoration of liver mass was measured by liver/body weight (bw) ratio at the indicated time points. n = 3–6 per time point. *, P = 0.03; **, P = 0.001. (B and C) BrdU incorporation in the liver at indicated time points after 2/3 hepatectomy. n = 3–6. *, P < 0.001; **, P = 0.008; ***, P = 0.01; ****, P = 0.007. Bar, 150 µM.
Mouse urinary protein (MUP)–Tob1–adeno-associated virus 8 (AAV8) expresses Tob1 protein in the liver
We next investigated whether hepatocyte proliferation rates depend on Tob1 protein levels. To answer this question, it was necessary to develop a system in which Tob1 protein levels could be precisely manipulated. To accomplish this, we assembled an AAV8 with an MUP promoter to drive Tob1 expression in the liver (MUP-Tob1-AAV8; Ho et al., 2008).
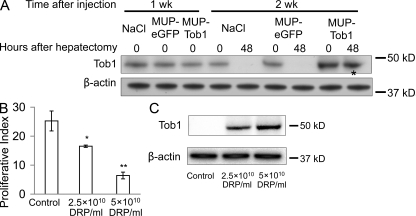
To determine the time course of expression of Tob1 in mice injected with MUP-Tob1-AAV8, Western blotting was performed at different times after injection (Fig. 3 A). 1 wk after injection, there was no difference between Tob1 protein levels in control mice who received no virus (NaCl), a control virus containing enhanced GFP (eGFP; MUP-eGFP-AAV8), or the MUP-Tob1-AAV8. By 2 wk after injection, Tob1 expression in mice receiving MUP-Tob1-AAV8 was robust and above the level of mice receiving no virus or MUP-eGFP-AAV8. As expected from the use of the MUP promoter to drive Tob1 expression, Tob1 protein expression was maintained through 48 h after hepatectomy (Fig. 3 A, *). Tob1 expression was maintained through 3 wk after injection (unpublished data). Each band shown is the most representative band from among three separate animals. These data show that MUP-Tob1-AAV8 efficiently expresses recombinant protein 2 wk after injection. They also demonstrate that expression from the MUP promoter is high enough to overcome any degradation mechanisms after hepatectomy.
Figure 3.
MUP-Tob1-AAV8 virus inhibits hepatocyte proliferation after hepatectomy in a dose-dependent fashion. (A) Western blot of mouse liver after injection of MUP-Tob1-AAV8, MUP-eGFP-AAV8, or NaCl. The asterisk denotes Tob1 expression maintained even 48 h after hepatectomy and 2 wk after injections. (B) Tob1-null mice were infected with indicated doses of MUP-Tob1-AAV8 virus or MUP-eGFP-AAV8 (control). 3 wk later, 2/3 hepatectomy was performed and livers analyzed 48 h after hepatectomy. BrdU labeling is shown. n = 3. *, P = 0.01; **, P < 0.05. (C) Western blot of Tob1 in whole cell lysates. β-Actin is used to demonstrate equal protein loading. All blot lanes are the most representative sample from among at least three different animals.
Tob1 levels control the rate of hepatocyte proliferation after partial hepatectomy
To precisely manipulate Tob1 levels, we performed reconstitution experiments in which Tob1-null mice were infected with MUP-Tob1-AAV8 so that the only source of Tob1 would be the virus. To determine optimal dosing of the virus, we noted that immunofluorescence for eGFP demonstrated that 5 × 1010 DNase-resistant particles (DRPs) of MUP-eGFP-AAV8 produces protein expression in ∼90% of hepatocytes, whereas at 2.5 × 1010 DRP, ∼50% of hepatocytes express (unpublished data). These doses were used for MUP-Tob1-AAV8. As the amount of virus was increased, proliferation rates decreased 48 h after hepatectomy (Fig. 3 B). Mice receiving 5 × 1010 DRP of MUP-Tob1-AAV8 demonstrated lowest proliferation of hepatocytes after hepatectomy, whereas mice with 2.5 × 1010 DRP MUP-Tob1-AAV8 or 5 × 1010 DRP control MUP-eGFP-AAV8 virus exhibited progressively higher levels of hepatocyte proliferation (P < 0.05 between all groups). To verify that Tob1 protein levels decrease in the predicted fashion, Western blotting was performed on liver lysates prepared from specimens at 48 h (Fig. 3 C). 5 × 1010 DRP of virus produced the highest amount of Tob1 protein, whereas 2.5 × 1010 DRP produced a lower level. These results demonstrate that Tob1 levels control the rate of hepatocyte proliferation after hepatectomy in a dose-dependent fashion.
Effect of Loss of Tob1 on kinase activity
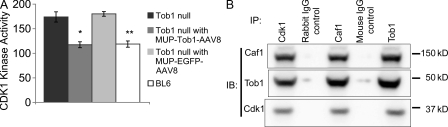
Tob1 associates with a variety of proteins and has multiple functions including a critical role in early G1 progression (Matsuda et al., 1996; Yoshida et al., 2000, 2003a). In particular, Tob1 modulates Cdks via interaction with Caf1 to influence cell cycle progression (Ikematsu et al., 1999; Nishida et al., 2007). To determine whether modulation of Cdk1 activity might explain the effect of Tob1 on liver regeneration, we performed kinase assays in WT mice, Tob1-null mice, and Tob1-null mice with MUP-Tob1-AAV8 or control MUP-eGFP-AAV8 virus. Consistent with this hypothesis, compared with WT mice, hepatic Cdk1 kinase activity was increased in Tob1-null mice. This activity was reduced to normal levels with the introduction of MUP-Tob1-AAV8 to the Tob1-null mice (Fig. 4 A, Tob1 null: 174 ± 17.9; Tob1 null with MUP-Tob1-AAV8: 117.4 ± 13.4; Tob1 null with MUP-eGFP-AAV8: 180.2 ± 13.1; WT C57B/6: 118.75 ± 9.75; n = 4–6, P = 0.035 and P = 0.036). These results suggest that Tob1 modulates cell cycle progression by inhibiting Cdk1 kinase activity.
Figure 4.
Loss of Tob1 results in increases in Cdk1 activity, and Tob1 and Caf1 bind to Cdk1. (A) Cdk1 kinase activity in the liver of Tob1-null mice infected or not with MUP-Tob1-AAV8 or MUP-eGFP-AAV8. n = 4–6. *, P = 0.035; **, P = 0.036. (B) Coimmunoprecipitation of Cdk1, Tob1, and Caf1. IP, immunoprecipitation antibodies; IB, immunoblot antibodies. Mouse IgG control is the negative control. The samples were run on the same gel and each lane is the most representative among three samples.
Tob1 and Caf1 associate with the cyclin–cdk complex
To further elucidate the mechanism of action of Tob1, we tested whether Tob1 and Caf1 associate with the cyclin–cdk complex in the liver. To accomplish this, multiple coimmunoprecipitation experiments were performed. Using antibodies to Cdk1, both Tob1 and Caf1 were precipitated. Similarly, antibodies to Tob1 coimmunoprecipitated both Cdk1 and Caf1. Finally, antibodies to Caf1 coimmunoprecipitated Cdk1 and Tob1 in normal adult liver (Fig. 4 B). Negative controls included both a rabbit and a mouse IgG. Positive control lanes consisted of whole cell lysates (unpublished data). These results demonstrate that all three proteins colocalize in a common complex. Our data provide mechanistic support for the hypothesis that Tob1 modulates Cdk activity by direct association with the cyclin–cdk complex.
Effect of loss of Tob1 on pathways critical for liver regeneration
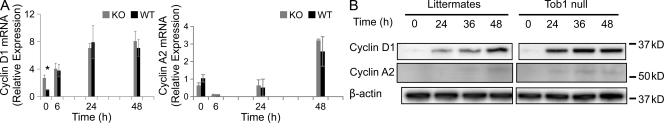
To place Tob1 in the context of the many factors known to be involved in liver regeneration, we performed several studies of the effect of Tob1 deletion on other genes. We first studied how loss of Tob1 affected the cell cycle proteins cyclin D1 and A2. Cyclin D1 is important for initiation of DNA synthesis, whereas cyclin A2 drives the transition to mitosis from S phase (Malumbres and Barbacid, 2009). At baseline, mRNA for cyclin D1 is increased in Tob1-null compared with WT mice (n = 3; Fig. 5 A). Although there are no significant changes in cyclin A2 mRNA expression in the Tob1-null mice compared with WT mice, protein levels of both cyclins increase faster in the Tob1-null mice (Fig. 5 B). In particular, cyclin D1 is increased more strongly by 24 h continuing through 36 h, whereas cyclin A is more robustly expressed by 24 h. These results show that Tob1 inhibits cyclin D1 expression at baseline and acts to delay the rise in both cyclin D1 and A2 necessary for cell division. Therefore, after hepatectomy, decreases in Tob1 would be expected to allow increases in cyclin D1 and A2 that are necessary for normal hepatocyte proliferation.
Figure 5.
Cyclin D1 and A2 expression increase more quickly in Tob1-null mice compared with WT controls. (A) Real-time PCR of cyclin D1 and cyclin A2 mRNA at indicated time points after hepatectomy. Each point represents at least three mice. *, P = 0.012. (B) Cyclin D1 and A2 protein was measured by immunoblot at indicated time points after hepatectomy. β-Actin was the loading control. All lanes are the most representative of three separate animals. Each row was run on the same gel, and the break represents portions of the gel not shown.
We next investigated the effect of loss of Tob1 on cytokine signals Socs3 and IL-6. Both are up-regulated soon after partial hepatectomy and are critical for liver regeneration (Zimmers et al., 2003; Riehle et al., 2008). No change was observed in the mRNA expression of either of these molecules in WT mice versus Tob1-null mice over the initial 48 h after hepatectomy (Fig. S1). Both exhibit an acute rise by 6 h after injury. These results suggest that Tob1 does not act by affecting cytokine transcription.
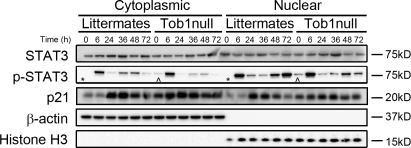
IL-6 is essential for Stat3 activation via phosphorylation and nuclear translocation (Cressman et al., 1994; Zimmers et al., 2003). To determine if Tob1 influences this pathway, we examined Stat3 protein levels, phosphorylation, and nuclear localization in Tob1-null and WT mice. Stat3 protein levels are fairly constant and not dramatically different between Tob1-null and WT littermates, with the exception that nuclear Stat3 is higher in WT mice than in Tob1-null mice at baseline, and nuclear Stat3 increases more robustly in Tob1-null mice than in the WT after hepatectomy (Fig. 6). In contrast, p-Stat3 is present in both cytoplasmic and nuclear fractions of quiescent adult liver from Tob1-null mice but not in WT littermates (Fig. 6, ^ vs. *). In both Tob1 and WT mice, Stat3 undergoes dramatic activation with markedly increased phosphorylation by 6 h after hepatectomy. These results suggest that at baseline, Tob1 helps to suppress hepatocyte proliferation by suppressing Stat3 phosphorylation and therefore its activation.
Figure 6.
Loss of Tob1 affects expression of proteins important for restoration of liver mass after injury. Total and phosphorylated Stat3 and p21 in the liver of Tob1-null mice and WT littermates was measured by immunoblot at indicated time points after hepatectomy. β-Actin and histone H3 were loading controls for the cytoplasmic and nuclear extracts, respectively. Note the presence of p-Stat3 in quiescent adult liver from Tob1-null mice but not in WT littermates (^ vs. *). All samples were run on the same gel and each lane is the most representative among three samples.
Repression of p21 activity is required for hepatocyte proliferation after hepatectomy (Stepniak et al., 2006). To determine the effect of loss of Tob1 on p21, we examined mRNA and protein expression in Tob1-null mice compared with WT littermates. p21 mRNA is induced in both Tob1-null and WT normal adult mice and at 6, 24, and 48 h after hepatectomy (unpublished data). Despite baseline p21 levels being higher in Tob1-null mice, nuclear and cytoplasmic p21 levels increase in a similar fashion in WT and Tob1-null mice (Fig. 6). The presence of Tob1, therefore, serves to suppress p21 levels at baseline.
p27 is a negative regulator of cell cycle, and p27-null mice accelerate DNA replication after partial hepatectomy (Hayashi et al., 2003). To determine whether Tob1 influences p27, mRNA and protein levels were examined. Loss of Tob1 has no effect on p27 mRNA levels after hepatectomy. p27 protein levels are slightly higher at baseline in Tob1-null mice than in the WT (Fig. S2). These results suggest that Tob1 functions to suppress p27 levels at baseline.
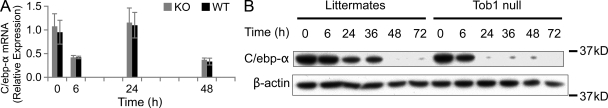
C/ebp-α functions as an inhibitor of hepatocyte proliferation and newborn mice lacking C/ebp-α exhibit increased hepatocyte proliferation (Flodby et al., 1996). There were no differences in mRNA levels between Tob1-null mice and WT mice. In contrast, Tob1-null mice demonstrated accelerated loss of C/ebp-α protein after hepatectomy compared with WT mice (Fig. 7). These results suggest that Tob1 tends to maintain C/ebp-α protein levels via posttranscriptional mechanisms.
Figure 7.
Effect of loss of Tob1 on C/EBP-α. (A) C/EBP-α mRNA was measured by real-time PCR in the liver of Tob1-null and WT littermates at indicated time points after hepatectomy. n = 3. P = NS. (B) C/EBP-α protein was measured by immunoblot. β-Actin was the loading control. All lanes are the most representative of three separate animals.
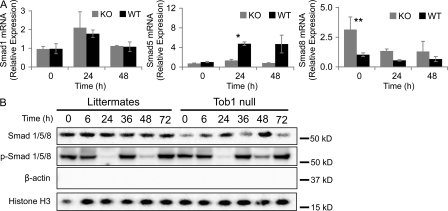
Tob proteins bind to Smad1, 5, and 8, preventing their activity as a transcription factors, and may have a role in their translocation to nuclear bodies (Yoshida et al., 2003b; Ross and Hill, 2008). To determine whether loss of Tob1 affects mRNA or protein levels for these Smads, real-time PCR and Western blotting were performed. Loss of Tob1 has no effect on mRNA for Smad1 (see Fig. 9 A). In contrast, loss of Tob1 blocks the robust induction of Smad5, which occurs in WT mice after 2/3 hepatectomy (n = 3; Fig. 8 A), and loss of Tob1 leads to generally higher levels of Smad8 mRNA after hepatectomy (n = 3). Nuclear Smad protein levels are generally lower in Tob1-null mice. Because Smad signaling occurs via phosphorylation and nuclear localization, a better measure of the action of Tob1 on Smad signaling is to look at activation. In this case, nuclear levels of phosphorylated Smads are remarkably similar between Tob1-null mice and their littermates. Interestingly, Smad activation decreases dramatically by 24 h after hepatectomy in both Tob1-null and WT mice (Fig. 8 B). This data suggests that although Tob1 has effects on Smad mRNA transcription and protein levels, Tob1 does not directly impact Smad activation.
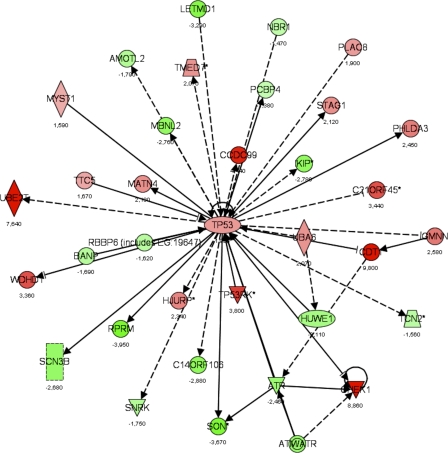
Figure 9.
Gene interaction networks affected by loss of Tob1. A dataset of genes regulated in the Tob1-null mice (LCB > |1.2|), but not in WT mice, was used to determine how loss of Tob1 affects gene expression after 2/3 hepatectomy. From this gene list input, a score was determined that ranks networks according to how relevant they are to the genes in the input dataset (see Results). The score for the p53 network was 34, the negative exponent of the p-value (P = 10−34), and this network is shown here. Each gene is given a heat map color that corresponds to the degree of regulation; reds are increased and greens are decreased, both in proportion to the intensity of the color. Numbers represent fold change. Shapes denote function as follows: trapezoids, transporters; diamonds, enzymes; ovals, transcription regulators; triangles, kinases; rectangles, ion channels; double circles, complex; circles, other. Arrows denote the first molecule acts on the second, and perpendicular lines denote the first inhibits the second. Solid lines are direct interactions, and dashed lines are indirect interactions. Data are from two distinct chips, each with three different animals.
Figure 8.
Loss of Tob1 does not change Smad activation. (A) Smad1, Smad5, and Smad8 mRNA was measured by real-time PCR in the liver of Tob1-null (KO) and WT (WT) mice at different times after hepatectomy (n = 3; *, P = 0.001) and increased Smad8 expression at 24 h (n = 3; **, P = 0.01). Each point represents at least three mice. (B) Total and phosphorylated Smad 1/5/8 protein was measured by immunoblot in nuclear extracts of Tob1-null mice. β-Actin and histone H3 immunoblots demonstrate purity of the nuclear extracts and serve as a loading control. All lanes are the most representative of three separate animals.
Absence of Tob1 affects expression of genes involved in a variety of pathways
To provide further insight into how loss of Tob1 enhances liver regeneration after partial hepatectomy, we performed microarray analysis of samples from WT and Tob1-null mice as normal adults and 24 h after 2/3 hepatectomy. Each experimental point (e.g., WT at 24 h after hepatectomy) used data from two chips, each with a different set of three pooled animals. Each chip was run in duplicate. Affymetrix 2.0 chips containing >45,000 oligonucleotides were used. The full dataset is available from GEO under the accession number GSE21836.
Analyzing the effect of deletion of a specific gene using microarray data can be performed in several ways to answer different questions. To maximize the relevance of the conclusions, we reasoned as follows: Tob1 loss is the critical event in accelerating hepatocyte proliferation and restoration of liver mass. Tob1 levels decrease after hepatectomy in WT mice, so that Tob1-null mice can be considered to have an accelerated loss of Tob1. Because Tob1 has no effect on baseline hepatocyte proliferation and there is a clear effect by 36 h, we chose 24 h to analyze, a time between these points which we reasoned would give enough time for transcriptional differences to become apparent.
To perform this analysis, we constructed a list of genes significantly changed in the WT compared with normal adult liver after hepatectomy and a separate list of genes significantly changed in the Tob1-null mice 24 h after partial hepatectomy compared with uninjured livers from Tob1-null mice. We arranged these in a Venn diagram and chose only genes significantly changed in the Tob1-null and not in the WT mice for further analysis.
We chose a threshold for differential regulation as a lower confidence bound (LCB) > |1.2| (see Irizarry et al., 2003 and Otu et al., 2007 for algorithms and methods). This was chosen because the LCB is a stringent estimate of differential expression and most likely corresponds to an actual difference of >1.2. This level was chosen prospectively as the most accurate choice for reliable differential expression. Using this threshold, there were 616 genes induced in the Tob1-null mice at 24 h after hepatectomy that were not induced in the WT mice at this time point. We note 969 genes are induced in the WT mice that were not induced in the Tob1-null mice after hepatectomy.
To validate this entire dataset, a total of 20 genes were interrogated using real-time PCR. Examining each gene in both WT and Tob1-null mice between 0 and 24 h generated 40 relationships that could be characterized as increased, decreased, or not changed. For the array data, an LCB > |1.2| was considered a change, whereas for the real-time PCR data, a p-value of <0.05 was considered a change. In 34 of these 40 (85%), the real-time data and the array data were concordant (Fig. S3).
We next identified networks differentially regulated in the Tob1-null mice compared with WT mice. This type of analysis is complementary to individual gene information and may be more robust because it examines many genes to identify biological processes. To accomplish this, a score was generated (Ingenuity Systems software) that takes into account the number of genes in the network and the size of the network. It allows a calculation of the approximate fit between each network and the genes from the input dataset, allowing the network to be ranked. The score is a p-value calculation to determine the possibility that the network shown could be found by random chance. Analysis of this set of genes, in which expression changed between 0 and 24 h after hepatectomy in the Tob1-null mice but not in WT mice, allowed identification of pathways affected by loss of Tob1. We limited our analysis to pathways with P < 0.01, given the large number of pathways and genes examined. 20 pathways were identified in this manner. The most significant pathway affected was p53 signaling (Fig. 9), with P < 10−6, followed by insulin-like growth factor 1 signaling, with P = 0.00004. The full dataset can be found in Table S2.
Loss of Tob1 influences transcriptional pathways
To better understand the transcription factors and pathways affected by the loss of Tob1, we used promoter analysis and interaction network toolset (PAINT). This algorithm takes an input set of genes, in our case the genes differentially expressed in the Tob1-null mice, and identifies transcriptional response elements (TREs) in the flanking or known promoter regions of the input gene list that are predicted to bind to known transcription factors. In this way, PAINT helps form a picture of which of a smaller number of genes (the transcription factors) are responsible for the larger number of changes in gene expression patterns.
Using our dataset, this analysis identified several transcription factors expected to bind to these regions, including Ets-1, FoxD3, Cre-BP1, E2F, and c-Jun (Table S1). This is evidence that these transcription factors play a role in many of the gene expression patterns observed in the Tob1-null mice. Because E2F targets are important for many cellular processes, including cell cycle regulation, we then looked at these genes by category according to the gene list generated by Bracken et al. (2004; Fig. S4). To determine which of these are predominately affected by loss of Tob1, our input list included only genes induced in Tob1-null mice but not WT mice at 24 h. Analysis of this list revealed that most were involved in DNA replication. These results suggest that the presence of Tob1 inhibits E2F targets responsible for S phase.
DISCUSSION
We hypothesized that repression of inhibitory signals was a key mechanistic paradigm during liver regeneration. To test this postulate, a screen was undertaken for genes that might be constitutive inhibitors of liver regeneration, i.e., repressed after liver injury. The antiproliferative molecule Tob1, a member of the PC3/BTG1 family of antiproliferative molecules, was identified as potentially fulfilling these requirements.
Our major findings are that Tob1 acts as a constitutive repressor of hepatocyte proliferation after hepatectomy. Tob1 protein levels decrease after hepatectomy, most likely as a result of protein degradation. Decreased Tob1 is necessary for normal liver regeneration, as the rate of hepatocyte proliferation is determined by endogenous Tob1 levels. Tob1 suppresses Cdk1 kinase activity and associates with Caf1 in the Cdk complex. Further antiproliferative activities of Tob1 are likely a result of effects on a variety of cell cycle proteins, including the suppression of cyclin D1 and A2 and inhibition of E2F targets concerned with DNA synthesis.
These results are important for several reasons. Demonstrating that levels of an inhibitory protein control the rate of hepatocyte proliferation after hepatectomy support a novel paradigm in liver regeneration in which inhibitory factors must be actively suppressed for normal regeneration. This model is difficult to prove explicitly without reconstituting the gene in the null mouse. Our AAV-8 construct provides this functionality and allows us to show that the proliferation rate depends directly on Tob1 levels. These results are in contrast to work with, for example, Socs3 and p21 (Stepniak et al., 2006; Riehle et al., 2008). In these cases, although null mice for either demonstrate enhanced liver regeneration, protein levels actually increase after hepatectomy, suggesting that their effect is indirect.
Two major findings point to a mechanism of action for Tob1. The first is that Tob1 binds to Caf1 within a complex with Cdk1. Cdk1 is a critical Cdk activated by cyclin A and then cyclin B to facilitate mitosis (Malumbres and Barbacid, 2009). To determine the nature of this interaction, we examined kinase activity in the Tob1-null mice. Increased kinase activity in Tob1-null mice suggests that Tob1 acts to inhibit kinase activity, delaying cell cycle progression.
A defining aspect of Tob1 is that it seems to be a multifunctional protein involved in a variety of cellular processes. In addition to its effect on kinase activity, we note that loss of Tob1 influences the transcription of multiple genes important for cell cycle progression. Loss of Tob1 accelerates the rise in cyclin D1 and A2 protein levels. In a similar fashion, loss of Tob1 accelerates the rise in E2F targets, preferentially affecting those important for DNA synthesis. Collectively, these results suggest that Tob1 represses the transcription of genes important for cell cycle progression. How Tob1 acts to do this is unclear. It is possible that a wide-ranging effect of Tob1 on RNA degradation might be responsible for effects on these multiple target genes. Alternatively, Tob1 might directly affect a few transcription factors, for example, E2F, with multiple downstream effects.
Other antiproliferative proteins that may regulate cell cycle are influenced by loss of Tob1. These include p21, which suppresses increases in PCNA, cyclins, and cdk2-associated kinase activity, p27, which inhibits cell cycle progression, and C/ebp-α, which interacts with the cyclin–Cdk complex (Albrecht et al., 1998; Greenbaum et al., 1998; Wang et al., 2001; Kossatz et al., 2004; Weymann et al., 2009). Increased levels of p21 and p27 observed in Tob1-null mice suggest that these antiproliferative proteins may be responding to or compensating for a primed hepatocyte to prevent proliferation. Contrast this with the more rapid decrease in C/ebp-α in Tob1-null mice, suggesting that Tob1 helps maintain C/ebp-α. It is tempting to suggest, therefore, that Tob1 and C/ebp-α are regulated along the same molecular signaling pathway, whereas p21 and p27 may be part of a separate pathway.
Dysregulation of p53 signaling identified by array analysis suggests an additional mechanism for Tob1 in cell cycle progression. p53, an important tumor suppressor, can initiate cell cycle arrest during times of stress (Boehme and Blattner, 2009). In addition, there is an additive effect on tumorigenesis when p53 and Tob1 are simultaneously deleted (Yoshida et al., 2003a). Collectively, these and our results place Tob1 as a possible mediator of p53 activity on the cell cycle.
Temporally, major changes in Tob1 protein levels occur after cytokine up-regulation in response to hepatectomy. It is therefore not surprising that loss of Tob1 has minimal effects on mRNA for either IL-6 or Socs3. Our findings that both are increased after hepatectomy are consistent with published data (Sakamoto et al., 1999; Campbell et al., 2001). IL-6 promotes Stat3 activation and Socs3 is part of a negative-feedback loop to inhibit Stat3 activation. We find, consistent with published results, that Stat3 is activated after hepatectomy (Cressman et al., 1994; Zimmers et al., 2003; Riehle et al., 2008). In addition, our finding that Stat3 is activated even in quiescent livers in Tob1-null mice suggests that Tob1 may influence cytokine signaling not by influencing cytokine transcription but rather at the signal transduction level.
Tob1 binds to the downstream effectors of Bmp signaling, including Smad1, 5, and 8, and promotes their localization to the nucleus (specifically the nuclear bodies), which may result in the suppression of their activity. BMP signals are transduced by Smad phosphorylation and nuclear translocation (Feng and Derynck, 2005). Although Smad protein levels and mRNA are altered in Tob1-null mice, are no changes in the level or phosphorylation of Smads in WT versus Tob1-null mice, arguing that Tob1 does not have a direct effect on Smad signaling.
Tob1 affects the transcription of targets of the transcription factors Ets-1, FoxD3, Cre-BP1, E2F, and c-Jun. Ets-1 binds to a portion of the p53 promoter and may regulate its signaling (Baillat et al., 2009). FoxD3 is important for regulating endodermal-specific promoters (Guo et al., 2002). Cre-BP1 forms a homodimer or heterodimer with c-Jun and stimulates CRE-dependent transcription. Identification of both Cre-BP1 and c-Jun TREs in genes dysregulated in Tob1-null mice is particularly interesting because c-Jun, a member of the AP-1 complex, is necessary for liver regeneration by repressing p53/p21 and p38 MAPK (Stepniak et al., 2006). The specific interaction between Tob1 and c-Jun remains unknown but potentially important.
Importantly, our data show that although Tob1 levels control the rate of hepatocyte proliferation, Tob1 does not ultimately control liver size. At baseline, livers in Tob1-null mice are normal size, and by 9 d after hepatectomy, livers in Tob1-null mice again return to normal size. In our hands, hepatocyte proliferation after hepatectomy begins >24 h after surgery and peaks at 48 h. Although there are studies indicating that peak proliferation occurs earlier, our results are consistent with those of others (Lesurtel et al., 2006).
When analyzing our array data, we used a cutoff for differential expression of an LCB of |1.2|. This was chosen as a stringent estimate and the actual difference in expression is likely much higher than this (Ramalho-Santos et al., 2002; Wang et al., 2006). In fact, choice of this cutoff was validated by extensive real-time data showing 85% concordance with the array data. Given the intrinsic limitations of these methods to corroborate each other as a result of alternative splicing, normalization choices, and the difficulty arrays have in detecting low abundance transcripts, this concordance is consistent with other studies (Wang et al., 2006).
In summary, these results suggest a model in which Tob1 antagonizes proliferative signals and directly suppresses cell cycle progression by binding to and modulating Cdk activity and inhibiting transcription of multiple proteins critical for cell cycle progression. These findings demonstrate that an inhibitor of proliferation must be blocked or suppressed for normal liver regeneration. Restoration of liver mass after hepatectomy is therefore best thought of as a balance between stimulatory and inhibitory factors. In human diseases in which liver function and regeneration are suppressed, it may be that understanding this pathway could point to therapeutic modalities.
MATERIALS AND METHODS
Mice.
Tob1-null mice on a C57BL/6 background were a gift from I. Bagchi (University of Illinois at Urbana-Champaign, Champaign, IL). These mice are derived from a line produced and maintained by T. Yamamoto (University of Tokyo, Tokyo, Japan; Yoshida et al., 2000). As described previously, Tob1-deficient mice are predisposed to spontaneous tumor development in the liver and elsewhere (Yoshida et al., 2003a). Viral constructs work best in male mice; thus, all mice used in this study were male between 7 and 10 wk of age. Genotyping was performed as previously described (Yoshida et al., 2003a).
2/3 hepatectomy.
All surgeries were performed according to the National Institutes of Health guidelines for the humane treatment of laboratory animals and with approval of the Harvard Institutional Animal Care and Use Committee. Mice were anesthetized with 60 mg/kg ketamine (Hospira) and 7 mg/kg xylazine (Phoenix Pharmaceuticals) and positioned supine. A transverse incision was made inferior to the xiphoid process, which was excised. The median and left lateral lobes of the liver were eviscerated and ligated. At tissue recovery, mice were anesthetized and weighed. Livers were excised, rinsed, blotted, and weighed. Sections were snap frozen in liquid nitrogen, fixed in 10% neutral buffered formalin, or preserved in RNALater (QIAGEN). Mortality was <5% and not associated with a particular genotype.
AAV8 vector production: subcloning and packaging.
A 2,300-bp murine Tob1 cDNA was obtained from the American Type Culture Collection. The 1,090-bp coding sequence was reframed via PCR to eliminate the untranslated regions and provide new flanking restriction sites and an optimized Kozak sequence. The PCR product was cloned into pAMP, an AAV2 vector plasmid in which expression is controlled by the highly specific MUP promoter element (Ho et al., 2008). An AAV2-MUP-eGFP vector plasmid was constructed as a negative control. Both vectors were cross-packaged in AAV8 via triple plasmid transfection into AAV-293 cells (Agilent Technologies). No helper virus was used. In brief, each AAV vector plasmid, an AAV2/8 rep/cap plasmid providing AAV2 replicase and AAV8 capsid functions, and a third plasmid encoding adenovirus helper functions, pHelper (Agilent Technologies), were cotransfected into 293 cells at a molar ratio of 1:1:1. Cells were harvested 48 h after transfection. Cell pellets were resuspended in DME, and intracellular virus particles released by three consecutive rounds of freeze-thaw, followed by centrifugation at 13,000 rpm for 10 min to remove particulates. Vector stocks were stored at −80°C and titered by real-time PCR using an ABI Prism 7700 Sequence Detection system (Applied Biosystems).
Virus injections.
MUP-Tob1-AAV8, MUP-eGFP-AAV8 (negative control), or vehicle (saline) was administered via tail veins as previously described (Ho et al., 2008). Injections were performed at 200 µl for a total dose of either 5 × 1010 DRP or 2.5 × 1010 DRP. These amounts were chosen based on a linear relationship between MUP-eGFP-AAV8 injection amounts and fluorescence at this dose. 2/3 hepatectomies were performed at the indicated times after virus injections. Excised livers were rinsed in PBS and snap frozen in liquid nitrogen or fixed in 10% neutral buffered formalin.
Immunohistochemistry.
Staining for BrdU (BD) was performed on fixed paraffin-embedded liver sections according to the manufacturer’s instructions (rat ABC Staining System; Santa Cruz Biotechnology, Inc.). Primary antibody was rat anti–mouse BrDU (Santa Cruz Biotechnology, Inc.). Proliferation index was quantified as the percentage of labeled hepatocyte nuclei that stained with BrdU over at least three high-power fields (400×).
mRNA isolation and real-time RT-PCR.
Total mRNA was purified from ∼30 mg of liver tissue preserved in RNALater with the RNeasy Mini kit (QIAGEN). 1 µg mRNA was reverse transcribed to cDNA using a High Capacity cDNA Reverse Transcription kit (Applied Biosystems). An ABI 7700 Sequence Detector (Applied Biosystems) was used for all real-time PCR. cDNA template was diluted 1:5 and amplified using inventoried Taqman gene expression assays (Applied Biosystems) under standard conditions. Gene expression levels were normalized to GAPDH using the comparative CT method. Data were analyzed using ABI Sequence Detector Software (Applied Biosystems).
Protein sample preparation, immunoprecipitation, and Western blot analysis.
Whole cell liver lysates were prepared by homogenizing 50 mg of frozen liver tissue in lysis buffer (50 mM Tris, 100 mM NaCl, 5 mM EDTA, 10% glycerol, and 0.5% NP-40) containing phosphatase inhibitor (Thermo Fisher Scientific) and protease inhibitor cocktail (Roche). Samples were sonicated for 30 s and clarified by centrifugation. Protein concentration was determined with the DC assay kit (Bio-Rad Laboratories).
Nuclear and cytoplasmic fractions were prepared as previously described (Andrews and Faller, 1991), with some modification: liver tissue was homogenized in hypotonic buffer (10 mM Tris, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, and 0.1% NP-40) containing protease inhibitor cocktail and cells were allowed to swell on ice for 10 min. Nuclei were pelleted and the supernatant containing the cytoplasmic fraction cleared. Nuclei were resuspended and lysed with 1% NP-40 and the supernatant collected. Protein concentration was determined with a BCA kit (Thermo Fisher Scientific). Enrichment quality was determined by immunoblotting for β-actin and histone H3. Protein was denatured and separated by SDS-PAGE in 10% Criterion XT Bis-Tris precast gels (Bio-Rad Laboratories) and transferred to nitrocellulose (Millipore). All antibody dilutions were made in 5% milk in TBS buffer. Primary antibodies used were the following: mouse monoclonal anti-Tob1 (Abcam), rabbit anti-Smad1/5/8 (Santa Cruz Biotechnology, Inc.), mouse monoclonal anti-cyclin D1 (Santa Cruz Biotechnology, Inc.), rabbit anti-cyclin A2 (EMD), mouse monoclonal anti-actin (Sigma-Aldrich), goat anti-GAPDH (Santa Cruz Biotechnology, Inc.), rabbit anti-histone H3 (Millipore), mouse monoclonal anti-Cdk1 (Abcam), rabbit anti-phosphoTob1 (Abcam), rabbit anti-Caf1 (Santa Cruz Biotechnology, Inc.), rabbit anti-C/ebp-α, rabbit anti-Stat3, and mouse monoclonal anti-p21 (Santa Cruz Biotechnology, Inc.). HRP-conjugated species-specific secondary antibodies were obtained from eBioscience, Jackson ImmunoResearch Laboratories, Santa Cruz Biotechnology, Inc., or Promega. In some cases, membranes were stripped with stripping buffer (Boston Bioproducts) according to the manufacturer’s instructions, washed extensively, and then reblocked and reprobed.
Coimmunoprecipitation.
Cell lysates were prepared using the NE-PER cytoplasmic and nuclear extraction kit (Thermo Fisher Scientific). Lysate preclearing and red protein G affinity gel bead preparation was accomplished by washing 50 µl EZview red protein G affinity gel beads in 750 µl of iced lysis buffer, followed by centrifugation for 30 s at 8,200 g and washing. Beads were resuspended in 50 µl of cold lysis buffer. 500 µg of lysate was added to the beads, incubated with gentle mixing for 1 h at 4°C, and centrifuged at 8,200 g for 10 min at 4°C, and the supernatant was retained. 5 µg of mouse monoclonal anti-Cdk1 (Santa Cruz Biotechnology, Inc.), rabbit polyclonal anti-Caf1 (Santa Cruz Biotechnology, Inc.), or mouse monoclonal anti-Tob1 (Abcam), or mouse monoclonal isotype control (Abcam) and rabbit isotype control (Abcam) was used. Secondary antibodies were anti–rabbit IgG, HRP-linked antibody (Cell Signaling Technology), and anti–mouse TrueBlot ULTRA: HRP-linked antibody (eBioscience).
Kinase assays.
EZview red protein G beads, using the mouse monoclonal anti-Cdk1 (Santa Cruz Biotechnology, Inc.) antibody as the initial immunoprecipitation antibody, were prepared as per the manufacturer’s instructions. Kinase buffer, 10 mM ATP (Cell Signaling Technology), and Rb biotinylated peptide (Cell Signaling Technology) were combined with the equilibrated beads to a final reaction volume of 50 µl and the final concentration of kinase buffer, 200 µM ATP, and 1.5 µM Rb peptide and incubated at room temperature for 30 min. 50 µl of Stop buffer (50 mM EDTA, pH 8.0) was added and 25 µl/well transferred to a 96-well streptavidin-coated plate (PerkinElmer) with 75 µl ddH2O/well and incubated at room temperature for 60 min. Each well was washed, and 100 µl/well of rabbit polyclonal phospho-Rb antibody (Cell Signaling Technology; 1:1,000 dilution) was added and incubated at room temperature for 120 min. Each well was washed, and 100 µl/well of anti–rabbit HRP-linked antibody (Cell Signaling Technology; 1:1,000 dilution) was added and incubated at room temperature for 30 min. Each well was washed, and 100 µl/well of TMB substrate (Cell Signaling Technology) was added and incubated at room temperature for 15 min. 100 µl/well of STOP solution (Cell Signaling Technology) was added to each well and a SpectraMax M5 microtiter plate reader (MDS Analytical Technologies) used to read the absorbance at 450 nm.
Microarray analysis.
Each mRNA sample consisted of 10 µg of high-quality total mRNA pooled from three animals and was run on an Affymetrix mouse 430 2.0 array as previously described (Otu et al., 2007). Two independent chips, each loaded with a different set of three pooled animals, were examined in duplicate. LCB of fold change was determined as previously described (Otu et al., 2007). Genes with an LCB of >|1.2| were considered dysregulated in the Tob1-null mice. The LCB is a stringent estimate of the LCB of the fold change (Irizarry et al., 2003; Otu et al., 2007).
Promoter analysis.
PAINT (version 3.9; http://www.dbi.tju.edu/dbi/tools/paint) was used to determine TREs in the promoter regions of genes regulated in Tob1-null mice, but not in WT mice, 24 h after hepatectomy as compared with normal adult liver. Promoter regions were retrieved considering 5 kb upstream of the initiation codon. The number of gene identifiers for which upstream sequences were found was 4,264, representing 2,829 unique promoters. The match filter was set to minimize false positives, the TRE core similarity threshold was set at 1.00, and we included TREs found on the complimentary strand. The output file contained a matrix of 159 transcription factors.
Data analysis.
All comparisons were performed using a two-tailed unpaired Student’s t test. All statistical tests used at least three and as many as nine different samples for each time point and genotype. * in the figures denotes statistical significance as defined by P < 0.05. All error bars in the figures are standard error.
Online supplemental material.
Figs. S1 and S2 show mRNA or protein expression patterns after hepatectomy in Tob1-null mice for three proteins known to be important for liver regeneration: IL-6, Socs 3, and p27. Fig. S3 shows real-time PCR validation of 20 genes found to be regulated using gene array analysis after hepatectomy in Tob1-null mice compared with WT mice. Fig. S4 examines categories of E2F targets differentially induced after hepatectomy in Tob1-null mice compared with WT mice. Table S1 shows transcription factors expected to bind to putative promoter regions in the genes differentially induced 24 h after hepatectomy in Tob1-null mice compared with WT mice. Table S2 shows pathways regulated differently after hepatectomy in Tob1-null mice compared with wild-type mice. Online supplemental material is available at http://www.jem.org/cgi/content/full/jem.20092434/DC1.
Acknowledgments
We wish to thank Dr. Michael Sirover and Dr. Xian Li for critical review of the manuscript.
This work was supported in part by National Institutes of Health Grant DK64648 (S.J. Karp), HL007734 (K.J. Ho), the Beth Israel Deaconess Medical Center (S.J. Karp), the Julie Henry Fund at Beth Israel Deaconess Medical Center (S.J. Karp), the Warren Fellowship of Harvard Medical School (K.J. Ho), and the Harvard-Dubai Foundation for Medical Research (H.H. Otu).
The authors have no conflicting financial interests.
Footnotes
Abbreviations used:
- AAV-8
- adeno-associated virus 8
- Cdk
- cyclin-dependent kinase
- C/ebp-α
- CCAAT enhancer binding protein α
- DRP
- DNase-resistant particle
- eGFP
- enhanced GFP
- LCB
- lower confidence bound
- MUP
- mouse urinary protein
- PAINT
- promoter analysis and interaction network toolset
- Socs3
- suppressor of cytokine signaling 3
- Tob1
- transducer of ErbB2.1
- TRE
- transcriptional response element
References
- Albrecht J.H., Poon R.Y., Ahonen C.L., Rieland B.M., Deng C., Crary G.S. 1998. Involvement of p21 and p27 in the regulation of CDK activity and cell cycle progression in the regenerating liver. Oncogene. 16:2141–2150 10.1038/sj.onc.1201728 [DOI] [PubMed] [Google Scholar]
- Andrews N.C., Faller D.V. 1991. A rapid micropreparation technique for extraction of DNA-binding proteins from limiting numbers of mammalian cells. Nucleic Acids Res. 19:2499 10.1093/nar/19.9.2499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baillat D., Laitem C., Leprivier G., Margerin C., Aumercier M. 2009. Ets-1 binds cooperatively to the palindromic Ets-binding sites in the p53 promoter. Biochem. Biophys. Res. Commun. 378:213–217 10.1016/j.bbrc.2008.11.035 [DOI] [PubMed] [Google Scholar]
- Beldi G., Wu Y., Sun X., Imai M., Enjyoji K., Csizmadia E., Candinas D., Erb L., Robson S.C. 2008. Regulated catalysis of extracellular nucleotides by vascular CD39/ENTPD1 is required for liver regeneration. Gastroenterology. 135:1751–1760 10.1053/j.gastro.2008.07.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boehme K.A., Blattner C. 2009. Regulation of p53—insights into a complex process. Crit. Rev. Biochem. Mol. Biol. 44:367–392 10.3109/10409230903401507 [DOI] [PubMed] [Google Scholar]
- Bracken A.P., Ciro M., Cocito A., Helin K. 2004. E2F target genes: unraveling the biology. Trends Biochem. Sci. 29:409–417 10.1016/j.tibs.2004.06.006 [DOI] [PubMed] [Google Scholar]
- Bucher M.L., Swaffield M.N. 1975. Regulation of hepatic regeneration in rats by synergistic action of insulin and glucagon. Proc. Natl. Acad. Sci. USA. 72:1157–1160 10.1073/pnas.72.3.1157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Campbell J.S., Prichard L., Schaper F., Schmitz J., Stephenson-Famy A., Rosenfeld M.E., Argast G.M., Heinrich P.C., Fausto N. 2001. Expression of suppressors of cytokine signaling during liver regeneration. J. Clin. Invest. 107:1285–1292 10.1172/JCI11867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cressman D.E., Greenbaum L.E., Haber B.A., Taub R. 1994. Rapid activation of post-hepatectomy factor/nuclear factor kappa B in hepatocytes, a primary response in the regenerating liver. J. Biol. Chem. 269:30429–30435 [PubMed] [Google Scholar]
- Cressman D.E., Diamond R.H., Taub R. 1995. Rapid activation of the Stat3 transcription complex in liver regeneration. Hepatology. 21:1443–1449 10.1002/hep.1840210531 [DOI] [PubMed] [Google Scholar]
- Ezzeddine N., Chang T.C., Zhu W., Yamashita A., Chen C.Y., Zhong Z., Yamashita Y., Zheng D., Shyu A.B. 2007. Human TOB, an antiproliferative transcription factor, is a poly(A)-binding protein-dependent positive regulator of cytoplasmic mRNA deadenylation. Mol. Cell. Biol. 27:7791–7801 10.1128/MCB.01254-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feingold K.R., Soued M., Grunfeld C. 1988. Tumor necrosis factor stimulates DNA synthesis in the liver of intact rats. Biochem. Biophys. Res. Commun. 153:576–582 10.1016/S0006-291X(88)81134-3 [DOI] [PubMed] [Google Scholar]
- Feng X.H., Derynck R. 2005. Specificity and versatility in tgf-beta signaling through Smads. Annu. Rev. Cell Dev. Biol. 21:659–693 10.1146/annurev.cellbio.21.022404.142018 [DOI] [PubMed] [Google Scholar]
- Flodby P., Barlow C., Kylefjord H., Ahrlund-Richter L., Xanthopoulos K.G. 1996. Increased hepatic cell proliferation and lung abnormalities in mice deficient in CCAAT/enhancer binding protein alpha. J. Biol. Chem. 271:24753–24760 10.1074/jbc.271.40.24753 [DOI] [PubMed] [Google Scholar]
- Francavilla A., Carr B.I., Azzarone A., Polimeno L., Wang Z., Van Thiel D.H., Subbotin V., Prelich J.G., Starzl T.E. 1994. Hepatocyte proliferation and gene expression induced by triiodothyronine in vivo and in vitro. Hepatology. 20:1237–1241 [PubMed] [Google Scholar]
- Greenbaum L.E., Li W., Cressman D.E., Peng Y., Ciliberto G., Poli V., Taub R. 1998. CCAAT enhancer- binding protein beta is required for normal hepatocyte proliferation in mice after partial hepatectomy. J. Clin. Invest. 102:996–1007 10.1172/JCI3135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo Y., Costa R., Ramsey H., Starnes T., Vance G., Robertson K., Kelley M., Reinbold R., Scholer H., Hromas R. 2002. The embryonic stem cell transcription factors Oct-4 and FoxD3 interact to regulate endodermal-specific promoter expression. Proc. Natl. Acad. Sci. USA. 99:3663–3667 10.1073/pnas.062041099 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi E., Yasui A., Oda K., Nagino M., Nimura Y., Nakanishi M., Motoyama N., Ikeda K., Matsuura A. 2003. Loss of p27(Kip1) accelerates DNA replication after partial hepatectomy in mice. J. Surg. Res. 111:196–202 10.1016/S0022-4804(03)00052-0 [DOI] [PubMed] [Google Scholar]
- Ho K.J., Bass C.E., Kroemer A.H., Ma C., Terwilliger E., Karp S.J. 2008. Optimized adeno-associated virus 8 produces hepatocyte-specific Cre-mediated recombination without toxicity or affecting liver regeneration. Am. J. Physiol. Gastrointest. Liver Physiol. 295:G412–G419 10.1152/ajpgi.00590.2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang W., Ma K., Zhang J., Qatanani M., Cuvillier J., Liu J., Dong B., Huang X., Moore D.D. 2006. Nuclear receptor-dependent bile acid signaling is required for normal liver regeneration. Science. 312:233–236 10.1126/science.1121435 [DOI] [PubMed] [Google Scholar]
- Ikematsu N., Yoshida Y., Kawamura-Tsuzuku J., Ohsugi M., Onda M., Hirai M., Fujimoto J., Yamamoto T. 1999. Tob2, a novel anti-proliferative Tob/BTG1 family member, associates with a component of the CCR4 transcriptional regulatory complex capable of binding cyclin-dependent kinases. Oncogene. 18:7432–7441 10.1038/sj.onc.1203193 [DOI] [PubMed] [Google Scholar]
- Irizarry R.A., Bolstad B.M., Collin F., Cope L.M., Hobbs B., Speed T.P. 2003. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 31:e15 10.1093/nar/gng015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakowlew S.B., Mead J.E., Danielpour D., Wu J., Roberts A.B., Fausto N. 1991. Transforming growth factor-beta (TGF-beta) isoforms in rat liver regeneration: messenger RNA expression and activation of latent TGF-beta. Cell Regul. 2:535–548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones D.E., Jr., Tran-Patterson R., Cui D.M., Davin D., Estell K.P., Miller D.M. 1995. Epidermal growth factor secreted from the salivary gland is necessary for liver regeneration. Am. J. Physiol. 268:G872–G878 [DOI] [PubMed] [Google Scholar]
- Kawamura-Tsuzuku J., Suzuki T., Yoshida Y., Yamamoto T. 2004. Nuclear localization of Tob is important for regulation of its antiproliferative activity. Oncogene. 23:6630–6638 10.1038/sj.onc.1207890 [DOI] [PubMed] [Google Scholar]
- Kogure K., Omata W., Kanzaki M., Zhang Y.Q., Yasuda H., Mine T., Kojima I. 1995. A single intraportal administration of follistatin accelerates liver regeneration in partially hepatectomized rats. Gastroenterology. 108:1136–1142 10.1016/0016-5085(95)90212-0 [DOI] [PubMed] [Google Scholar]
- Kossatz U., Dietrich N., Zender L., Buer J., Manns M.P., Malek N.P. 2004. Skp2-dependent degradation of p27kip1 is essential for cell cycle progression. Genes Dev. 18:2602–2607 10.1101/gad.321004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lesurtel M., Graf R., Aleil B., Walther D.J., Tian Y., Jochum W., Gachet C., Bader M., Clavien P.A. 2006. Platelet-derived serotonin mediates liver regeneration. Science. 312:104–107 10.1126/science.1123842 [DOI] [PubMed] [Google Scholar]
- Malumbres M., Barbacid M. 2009. Cell cycle, CDKs and cancer: a changing paradigm. Nat. Rev. Cancer. 9:153–166 10.1038/nrc2602 [DOI] [PubMed] [Google Scholar]
- Matsuda S., Kawamura-Tsuzuku J., Ohsugi M., Yoshida M., Emi M., Nakamura Y., Onda M., Yoshida Y., Nishiyama A., Yamamoto T. 1996. Tob, a novel protein that interacts with p185erbB2, is associated with anti-proliferative activity. Oncogene. 12:705–713 [PubMed] [Google Scholar]
- Michalopoulos G.K. 2007. Liver regeneration. J. Cell. Physiol. 213:286–300 10.1002/jcp.21172 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moh A., Iwamoto Y., Chai G.X., Zhang S.S., Kano A., Yang D.D., Zhang W., Wang J., Jacoby J.J., Gao B., et al. 2007. Role of STAT3 in liver regeneration: survival, DNA synthesis, inflammatory reaction and liver mass recovery. Lab. Invest. 87:1018–1028 10.1038/labinvest.3700630 [DOI] [PubMed] [Google Scholar]
- Natarajan A., Wagner B., Sibilia M. 2007. The EGF receptor is required for efficient liver regeneration. Proc. Natl. Acad. Sci. USA. 104:17081–17086 10.1073/pnas.0704126104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishida K., Horiuchi M., Noda N.N., Takahasi K., Iwasaki N., Minami A., Inagaki F. 2007. Crystallization and preliminary crystallographic analysis of the Tob-hCaf1 complex. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 63:1061–1063 10.1107/S1744309107057466 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogura Y., Hamanoue M., Tanabe G., Mitsue S., Yoshidome S., Nuruki K., Aikou T. 2001. Hepatocyte growth factor promotes liver regeneration and protein synthesis after hepatectomy in cirrhotic rats. Hepatogastroenterology. 48:545–549 [PubMed] [Google Scholar]
- Otu H.H., Naxerova K., Ho K., Can H., Nesbitt N., Libermann T.A., Karp S.J. 2007. Restoration of liver mass after injury requires proliferative and not embryonic transcriptional patterns. J. Biol. Chem. 282:11197–11204 10.1074/jbc.M608441200 [DOI] [PubMed] [Google Scholar]
- Ramalho-Santos M., Yoon S., Matsuzaki Y., Mulligan R.C., Melton D.A. 2002. “Stemness”: transcriptional profiling of embryonic and adult stem cells. Science. 298:597–600 10.1126/science.1072530 [DOI] [PubMed] [Google Scholar]
- Riehle K.J., Campbell J.S., McMahan R.S., Johnson M.M., Beyer R.P., Bammler T.K., Fausto N. 2008. Regulation of liver regeneration and hepatocarcinogenesis by suppressor of cytokine signaling 3. J. Exp. Med. 205:91–103 10.1084/jem.20070820 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross S., Hill C.S. 2008. How the Smads regulate transcription. Int. J. Biochem. Cell Biol. 40:383–408 10.1016/j.biocel.2007.09.006 [DOI] [PubMed] [Google Scholar]
- Russell W.E. 1988. Transforming growth factor beta (TGF-beta) inhibits hepatocyte DNA synthesis independently of EGF binding and EGF receptor autophosphorylation. J. Cell. Physiol. 135:253–261 10.1002/jcp.1041350212 [DOI] [PubMed] [Google Scholar]
- Sakamoto T., Liu Z., Murase N., Ezure T., Yokomuro S., Poli V., Demetris A.J. 1999. Mitosis and apoptosis in the liver of interleukin-6-deficient mice after partial hepatectomy. Hepatology. 29:403–411 10.1002/hep.510290244 [DOI] [PubMed] [Google Scholar]
- Schwall R.H., Robbins K., Jardieu P., Chang L., Lai C., Terrell T.G. 1993. Activin induces cell death in hepatocytes in vivo and in vitro. Hepatology. 18:347–356 [DOI] [PubMed] [Google Scholar]
- Selzner M., Clavien P.A. 2000. Failure of regeneration of the steatotic rat liver: disruption at two different levels in the regeneration pathway. Hepatology. 31:35–42 10.1002/hep.510310108 [DOI] [PubMed] [Google Scholar]
- Stepniak E., Ricci R., Eferl R., Sumara G., Sumara I., Rath M., Hui L., Wagner E.F. 2006. c-Jun/AP-1 controls liver regeneration by repressing p53/p21 and p38 MAPK activity. Genes Dev. 20:2306–2314 10.1101/gad.390506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., K-Tsuzuku J., Ajima R., Nakamura T., Yoshida Y., Yamamoto T. 2002. Phosphorylation of three regulatory serines of Tob by Erk1 and Erk2 is required for Ras-mediated cell proliferation and transformation. Genes Dev. 16:1356–1370 10.1101/gad.962802 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takamura K., Tsuchida K., Miyake H., Tashiro S., Sugino H. 2005. Activin and activin receptor expression changes in liver regeneration in rat. J. Surg. Res. 126:3–11 10.1016/j.jss.2005.01.002 [DOI] [PubMed] [Google Scholar]
- Temme C., Zaessinger S., Meyer S., Simonelig M., Wahle E. 2004. A complex containing the CCR4 and CAF1 proteins is involved in mRNA deadenylation in Drosophila. EMBO J. 23:2862–2871 10.1038/sj.emboj.7600273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tucker M., Valencia-Sanchez M.A., Staples R.R., Chen J., Denis C.L., Parker R. 2001. The transcription factor associated Ccr4 and Caf1 proteins are components of the major cytoplasmic mRNA deadenylase in Saccharomyces cerevisiae. Cell. 104:377–386 10.1016/S0092-8674(01)00225-2 [DOI] [PubMed] [Google Scholar]
- Ueda S., Yamanoi A., Hishikawa Y., Dhar D.K., Tachibana M., Nagasue N. 2003. Transforming growth factor-beta1 released from the spleen exerts a growth inhibitory effect on liver regeneration in rats. Lab. Invest. 83:1595–1603 10.1097/01.LAB.0000095686.10639.C8 [DOI] [PubMed] [Google Scholar]
- Wang H., Iakova P., Wilde M., Welm A., Goode T., Roesler W.J., Timchenko N.A. 2001. C/EBPalpha arrests cell proliferation through direct inhibition of Cdk2 and Cdk4. Mol. Cell. 8:817–828 10.1016/S1097-2765(01)00366-5 [DOI] [PubMed] [Google Scholar]
- Wang Y., Barbacioru C., Hyland F., Xiao W., Hunkapiller K.L., Blake J., Chan F., Gonzalez C., Zhang L., Samaha R.R. 2006. Large scale real-time PCR validation on gene expression measurements from two commercial long-oligonucleotide microarrays. BMC Genomics. 7:59 10.1186/1471-2164-7-59 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weymann A., Hartman E., Gazit V., Wang C., Glauber M., Turmelle Y., Rudnick D.A. 2009. p21 is required for dextrose-mediated inhibition of mouse liver regeneration. Hepatology. 50:207–215 10.1002/hep.22979 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., Tanaka S., Umemori H., Minowa O., Usui M., Ikematsu N., Hosoda E., Imamura T., Kuno J., Yamashita T., et al. 2000. Negative regulation of BMP/Smad signaling by Tob in osteoblasts. Cell. 103:1085–1097 10.1016/S0092-8674(00)00211-7 [DOI] [PubMed] [Google Scholar]
- Yoshida Y., Nakamura T., Komoda M., Satoh H., Suzuki T., Tsuzuku J.K., Miyasaka T., Yoshida E.H., Umemori H., Kunisaki R.K., et al. 2003a. Mice lacking a transcriptional corepressor Tob are predisposed to cancer. Genes Dev. 17:1201–1206 10.1101/gad.1088003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida Y., von Bubnoff A., Ikematsu N., Blitz I.L., Tsuzuku J.K., Yoshida E.H., Umemori H., Miyazono K., Yamamoto T., Cho K.W. 2003b. Tob proteins enhance inhibitory Smad-receptor interactions to repress BMP signaling. Mech. Dev. 120:629–637 10.1016/S0925-4773(03)00020-0 [DOI] [PubMed] [Google Scholar]
- Zimmers T.A., McKillop I.H., Pierce R.H., Yoo J.Y., Koniaris L.G. 2003. Massive liver growth in mice induced by systemic interleukin 6 administration. Hepatology. 38:326–334 10.1053/jhep.2003.50318 [DOI] [PubMed] [Google Scholar]