Abstract
Objectives
We characterized pairwise and higher order patterns of non-nucleoside reverse transcriptase inhibitor (NNRTI)-selected mutations because multiple mutations are usually required for clinically significant resistance to second-generation NNRTIs.
Patients and methods
We analysed viruses from 13 039 individuals with sequences containing at least one of 52 published NNRTI-selected mutations, including 1133 viruses from individuals who received efavirenz but no other NNRTI and 1510 viruses from individuals who received nevirapine but no other NNRTI. Of the 17 reported etravirine resistance-associated mutations (RAMs), Y181C/I/V, L100I, K101P and M230L were considered major based on published in vitro susceptibility data.
Results
Efavirenz preferentially selected for 16 mutations, including L100I (14% versus 0.1%, P < 0.001), K101P (3.3% versus 0.4%, P < 0.001) and M230L (2.8% versus 1.3%, P = 0.004), whereas nevirapine preferentially selected for 12 mutations, including Y181C/I/V (48% versus 6.9%, P < 0.001). Twenty-nine pairs of NNRTI-selected mutations covaried significantly, including Y181C with seven other mutations (A98G, K101E/H, V108I, G190A/S and H221Y), L100I with K103N, and K101P with K103S. Two pairs (Y181C + V179F and Y181C + G190S) were predicted to confer >10-fold decreased etravirine susceptibility. Seventeen percent of sequences had three or more NNRTI-selected mutations, mostly in clusters of covarying mutations. Many clusters had Y181C plus a non-major etravirine RAM; few had more than one major etravirine RAM.
Conclusions
Although major etravirine RAMs rarely occur in combination, 2 of 29 pairs of covarying mutations were associated with >10-fold decreased etravirine susceptibility. Viruses with three or more NNRTI-selected mutations often contained Y181C in combination with one or more minor etravirine RAMs; however, phenotypic and clinical correlates for most of these higher order combinations have not been published.
Keywords: Multidrug resistance, etravirine, antiviral therapy
Introduction
The non-nucleoside reverse transcriptase inhibitors (NNRTIs) bind to a hydrophobic pocket in the reverse transcriptase (RT) enzyme close to, but not contiguous with, the polymerase active site. These compounds inhibit HIV-1 replication allosterically by blocking translocation of the template–primer following nucleotide incorporation, or by displacing the catalytic aspartate residues relative to the polymerase-binding site. Mutations responsible for NNRTI resistance occur in this hydrophobic inhibitor-binding pocket.1 A single mutation in this pocket can lead to high-level resistance to one or more first-generation NNRTIs, including efavirenz and nevirapine. However, two or more mutations are required to cause high-level resistance to etravirine, rilpivirine and other second-generation NNRTIs.2–5 Therefore, we decided to explore the landscape of NNRTI resistance mutations in combinations of two or more, as viruses with these combinations are likely to be most troublesome for NNRTIs with higher genetic barriers to resistance.
We used publicly available HIV-1 RT sequence data from the HIV Drug Resistance Database (HIVDB)6 to identify patterns of mutations associated with efavirenz and nevirapine, the two most commonly used first-generation NNRTIs, and examined pairwise and higher order correlations among NNRTI-selected mutations. Our results provide new insights into the genetic mechanisms of NNRTI resistance and have important implications for the use of second-generation NNRTIs.
Patients and methods
Virus RT sequences
HIV-1 Group M RT sequences were compiled from published studies in the HIVDB. These included a set of 47 350 sequences from 39 687 individuals and a set of 9349 sequences from a large reference laboratory.7 To use no more than a single sequence per individual, we selected the sequence with the greatest number of NNRTI-selected mutations from each of the individuals in the first set and excluded all those (n = 2548) from the second set that differed from each other at <5.0% of their nucleotide positions.
We studied only sequences obtained by direct PCR sequencing of plasma HIV-1 RNA because these are the sequences obtained in clinical settings. Sequences of molecular clones were excluded because of their higher probability of containing PCR errors.8 Sequences obtained from other sources, particularly proviral peripheral blood mononuclear cell DNA, were excluded because they are at higher risk of containing defective genomes such as viruses with G to A hypermutation.9,10
To assess the patterns in which NNRTI-selected mutations occur, we analysed only those sequences containing one or more NNRTI-selected mutations. This enabled us to study sequences obtained from individuals who are likely to be NNRTI experienced, regardless of the availability of a complete treatment history or the possibility of non-adherence.
We analysed 52 NNRTI-selected mutations at 30 positions. These mutations included: (i) 46 non-polymorphic NNRTI-selected mutations at 28 positions (I94L, A98G, L100I, K101E/H/N/P, K102N, K103H/N/S/T, S105T, V106A/M, E138Q, T139R, I178F, V179F, Y181C/I/V, Y188C/H/L, G190A/C/E/Q/S, H221C/Y, K223T, P225H, F227L/Y, M230L, Y232H, L234I, P236L, D237E, K238N/T, V241M, Q242L and Y318F);11 and (ii) several polymorphic mutations that have been associated with decreased susceptibility to one or more NNRTIs (V108I) or are included in the etravirine resistance-associated mutations (RAMs) (V90I, V106I, E138A and V179D/T).4
The complete list of etravirine RAMs was defined as V90I, A98G, L100I, K101E/H/P, V106I, E138A, V179D/F/T, Y181C/I/V, G190A/S and M230L.4 Of these, L100I, K101P, Y181C/I/V and M230L were considered to be major etravirine RAMs based on their marked effect on etravirine susceptibility.4,12–14
Comparison of nevirapine- and efavirenz-selected mutations
We characterized each NNRTI-selected mutation by its frequency within sequences from individuals with a history of having received either efavirenz but no other NNRTI, or nevirapine but no other NNRTI. We then calculated the Pearson χ2 statistic for the association of each NNRTI-selected mutation with efavirenz or nevirapine treatment, based on a 2 × 2 contingency table of the numbers of isolates from patients treated with each drug and the numbers of isolates with and without the mutation. For this analysis only, electrophoretic mixtures were scored as mutations.
Covariation analysis
We used the Jaccard similarity coefficient (J) to identify pairwise correlations among the 52 NNRTI mutations in 13 039 sequences from unique individuals. Covariation analysis was run on the full set of sequences and on the subsets corresponding to sequences from individuals with a history of receiving either efavirenz or nevirapine but no other NNRTI.
We created 2 × 2 contingency tables to indicate the number of times a sequence contained both mutations, only one of the two mutations or neither of the two mutations. Overall, there were 1288 contingency tables representing all possible pairs of mutations except for permutations of mutation pairs at the same position: 1326 (52 choose 2) pairs minus 38 pairs of mutations at the same position. For each comparison, sequences containing an electrophoretic mixture at either of the two positions were excluded because of the inability to determine whether the two NNRTI-selected mutations were present in the same genome.
For a pair of mutations X and Y, the Jaccard similarity coefficient is calculated as J = NXY/(NXY + NX0 + N0Y) where NXY represents the number of sequences containing mutation X and mutation Y, NX0 represents the number of sequences containing X but not Y, and N0Y represents the sequences containing Y but not X. This statistic therefore examines only the sequences containing both X and Y in the context of sequences containing at least one of the mutations, thereby eliminating the heavy weighting given to the double-negative category in such statistical analyses as the χ2 test. This is particularly important so as not to exaggerate the significance of pairs of rare mutations, where the double-negative category is very large.15
To test whether the observed Jaccard similarity coefficients were statistically significant, we calculated the expected value of the Jaccard similarity coefficient (JRAND) for each pair of mutations as if the two mutations (X and Y) occurred independently. JRAND was calculated as the mean Jaccard similarity coefficient after 2000 random rearrangements of the X or Y vector (containing 0 or 1 for the presence or absence of a mutation, respectively). JSE, the standard error of J, was calculated using a jackknifed procedure, which removed one sequence at a time, repeatedly for each sequence.15 The standardized score Z, where Z = (J − JRAND)/JSE, indicates a significant positive association when Z > 1.96 or a significant negative association when Z is less than −1.96, at an unadjusted P < 0.05. The case of NXY = 0 results in a Z-score of negative infinity; Fisher's exact one-tailed test, using the R command fisher.test with a left-tailed alternative hypothesis, was used to approximate the P value for those comparisons with NXY = 0.
Holm's correction was used to control the familywise error rate for multiple pairwise comparisons.16 The P values of the n pairwise comparisons are ranked in ascending order, where p(1) is the smallest (ranked first) and p(i) is the ith ranked. Starting from p(1), one compares each p(i) with π/(n − i + 1) where π is the false discovery rate (in this case, 0.05) and n is the total number of pairwise comparisons, until p(j) > π/(n − j + 1). All correlations between pairs for p(1), … , p(j − 1) are considered statistically significant.
We modelled the distributions of mutations in each sequence using Poissonness plots, following the method of Hoaglin,17 and conducted a parametric bootstrap using a multivariate Poisson simulation to assess the distribution of positive and negative correlations obtained from the Jaccard covariation analysis. This step was important in evaluating the possible biological significance of the negative correlations.
We also examined the extent to which the 52 NNRTI-selected mutations covaried with mutations at 12 major nucleoside reverse transcriptase inhibitor (NRTI) resistance positions, including M41L, K65R, D67N, T69S_SS, K70E, K70R, L74V, L74I, V75M/T, Y115F, Q151M, M184V/I, L210W, T215F and T215Y.18 Sequences that contained at least one NNRTI-selected mutation and at least one of the above NRTI-selected mutations were analysed using the same methods described for assessing covariation within the NNRTI mutation dataset.
Multidimensional scaling
We performed multidimensional scaling on the pairwise association data using a matrix D of dissimilarity coefficients (JD = 1 − J, where J is the Jaccard similarity coefficient) for the 22 mutations found in the significantly positively associated pairs (corrected P < 0.05): A98G, L100I, K101E, K101H, K101P, K102N, K103N, K103S, V106A, V106I, V106M, V108I, V179D, V179F, Y181C, Y188L, G190A, G190S, H221Y, P225H, F227L and K238T. For a list of mutations (X1, X2, … , Xn), multidimensional scaling constructs points in two-dimensional (2-D) space such that the Euclidean distances between these points approximate the entries in the dissimilarity matrix.19 For a given matrix, it computes points X1, X2, … , Xn in 2-D space such that S = Σi,j(dist2(Xi,Xj) − dij2) is minimized where dist(Xi,Xj) is the Euclidean distance between Xi and Xj, and dij is the dissimilarity between Xi and Xj in the matrix D. This was performed using the R function cmdscale (classical multidimensional scaling).
Cluster analysis
We also identified all sequences containing three or more NNRTI-selected mutations and characterized the patterns of mutations by their frequency in the dataset and by whether they were composed of mutations that covaried with one another. Sets of three or more mutations in which each mutation covaried with each of the other members in the set were referred to as clusters or cliques (a term derived from graph theory). A lower threshold for covariation was used for this analysis (uncorrected P < 0.05) than in the pairwise comparison, to facilitate the identification of clusters. As for all previous covariation analyses, we used no more than one sequence per patient.
Results
Viruses and sequences
HIV-1 Group M RT sequences from 13 039 individuals had ≥1 of the 52 study-defined NNRTI-selected mutations. Of these individuals, 2394 were NNRTI naive, 5699 were NNRTI experienced and 4946 had uncertain NNRTI treatment histories. Among the NNRTI-experienced individuals, 1133 had received efavirenz only; 2292 had received nevirapine only (1510 who were treated with nevirapine and 782 women who received a single dose of nevirapine to prevent mother-to-child transmission); and 430 had received both efavirenz and nevirapine. The complete NNRTI treatment history was unavailable for the remaining 1844 NNRTI-experienced individuals. The dataset contained 6977 sequences (54% of the total) with one mutation, 3829 (29%) with two, 1521 (12%) with three, 501 (4%) with four, 172 (1%) with five, 36 (0.3%) with six, 2 with seven, and 1 with eight mutations. Sequences from 10 504 individuals (81%) belonged to subtype B, 747 (6%) to subtype C, 363 (3%) to circulating recombinant form (CRF) 01_AE, 320 (2%) to CRF 02_AG, 210 (2%) to subtype A and 895 (7%) to other subtypes or recombinants.
Comparison of efavirenz- and nevirapine-selected mutations
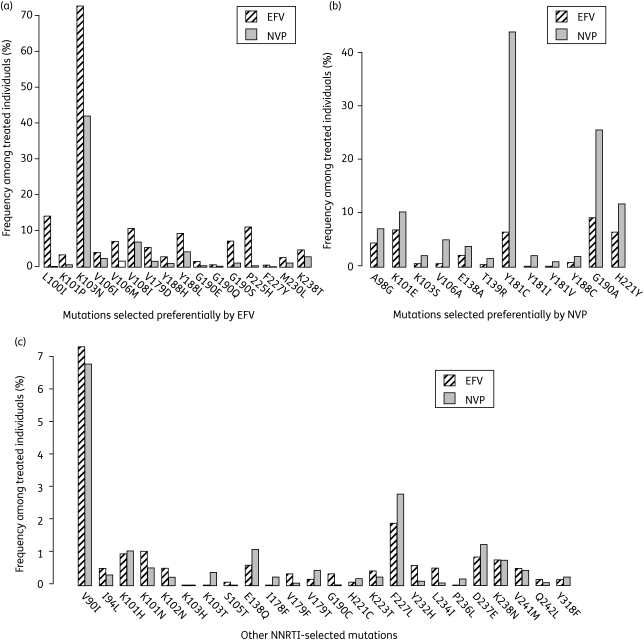
Figure 1 compares the prevalence of each of the 52 NNRTI-selected mutations in the sequences from the 1133 individuals who received efavirenz only and from the 1510 individuals who received nevirapine only. The most common mutations in sequences from efavirenz-experienced individuals were K103N (72% of the sequences), L100I (14%), P225H (11%), V108I (11%), G190A (9.5%), Y188L (9.3%), G190S (7.2%), K101E (7.2%) and V106M (7.0%). The most common mutations in sequences from nevirapine-experienced individuals were Y181C (46% of the sequences), K103N (43%), G190A (26%), H221Y (12%) and K101E (11%). Twenty-eight mutations deviated significantly (P < 0.05) from the distribution that would be expected in the absence of treatment association. Figure 1a shows the 16 mutations that occurred significantly more commonly in efavirenz-exposed individuals (L100I, K101P, K103N, V106I/M, V108I, V179D, Y188H/L, G190E/S/Q, P225H, F227Y, M230L and K238T), and Figure 1b shows the 12 that occurred significantly more commonly in nevirapine-exposed individuals (A98G, K101E, K103S, V106A, E138A, T139R, Y181C/I/V, Y188C, G190A and H221Y). Figure 1c shows the frequencies of the mutations that were not significantly associated with either drug.
Figure 1.
Treatment associations of NNRTI-selected mutations. This figure shows the associations of 52 NNRTI-selected mutations on treatment with efavirenz (EFV) or nevirapine (NVP), based on a Pearson's χ2 analysis of the frequency of the mutation among sequences from individuals treated with EFV or NVP but no other NNRTI. (a) The 16 mutations preferentially selected (P < 0.05) by efavirenz; (b) the 12 mutations preferentially selected by nevirapine; (c) the remaining 24 mutations, which were not significantly associated with either drug.
Among the sequences from 782 women treated with a single dose of nevirapine, the most frequently observed mutations were K103N (28%), Y181C (15%), Y188C (6%), G190A (5%) and E138A (4%). Y188C was the only mutation to occur significantly more frequently among the single-dose nevirapine sequences (6%) than among the full dataset of 1510 nevirapine-experienced individuals (2%).
Sequences from 213 individuals (27%) had at least one etravirine RAM, including 118 (15%) with at least one major etravirine RAM (115 with Y181C, 1 with L100I and 1 with both). No sequence contained >3 etravirine RAMs.
Covariation of NNRTI-selected mutations
To examine the extent of covariation among the NNRTI-selected mutations, we performed separate Jaccard analyses of the complete set of sequences from 13 039 individuals with viruses containing at least one NNRTI-selected mutation, the 1133 sequences from individuals who had received only efavirenz and the 1510 sequences from individuals who had received only nevirapine. Among the 13 039 sequences that comprised the full dataset, 66 of the 1288 possible mutation pairs had positive covariances, and 29 of these covariances were statistically significant after the Holm's adjustment for multiple comparisons (P < 0.05). Table 1 shows the results of this analysis on the 29 pairs of significantly covarying mutations. G190A (n = 9), Y181C (n = 8), V108I (n = 6), K101E (n = 4), H221Y (n = 4) and F227L (n = 4) were the most common mutations among these 29 pairs. Tables S1 to S4 [available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)] show the results of the analysis on all 1288 mutation pairs for the complete dataset, the efavirenz-alone dataset and the nevirapine-alone dataset.
Table 1.
Positively correlated pairs of NNRTI-selected mutations
| Occurrencesb |
Associationc |
|||||
|---|---|---|---|---|---|---|
| Mutation X | Mutation Y | Covariation within EFV/NVP subsetsa | XY | X only | Y only | P |
| Y181C | G190A | EFV | 848 | 1711 | 985 | 1.8E-90 |
| K101E | G190A | EFV, NVP | 445 | 347 | 1367 | 3.6E-75 |
| Y181C | H221Y | EFV, NVP | 430 | 2105 | 264 | 7.2E-56 |
| V108I | H221Y | EFV, NVP | 255 | 743 | 430 | 5.4E-46 |
| L100I | K103N | EFV | 571 | 48 | 4910 | 6.7E-44 |
| V108I | Y181C | NVP | 424 | 589 | 2116 | 2.2E-32 |
| V106A | F227L | NVP | 96 | 66 | 128 | 1.7E-31 |
| K101E | Y181C | EFV | 351 | 444 | 2258 | 3.4E-28 |
| K101E | G190S | EFV, NVP | 133 | 672 | 194 | 1.6E-26 |
| K103N | P225H | EFV | 301 | 5118 | 30 | 1.2E-21 |
| K101H | G190A | NVP | 116 | 58 | 1766 | 2.4E-18 |
| K103S | G190A | NVP | 117 | 76 | 1753 | 1.2E-17 |
| A98G | Y181C | EFV | 319 | 617 | 2311 | 2.3E-14 |
| V106I | Y188L | EFV | 110 | 705 | 579 | 1.1E-11 |
| K103N | K238T | NVP | 219 | 5245 | 69 | 5.1E-11 |
| K101H | Y181C | 91 | 84 | 2573 | 5.2E-9 | |
| A98G | V108I | 139 | 791 | 887 | 1.1E-8 | |
| G190A | H221Y | 174 | 1671 | 524 | 1.2E-8 | |
| A98G | G190A | 215 | 731 | 1657 | 3.3E-8 | |
| G190A | F227L | 79 | 1797 | 144 | 1.0E-7 | |
| V108I | G190A | 221 | 798 | 1606 | 3.4E-7 | |
| V106M | V179D | EFV | 37 | 122 | 567 | 4.5E-7 |
| K101E | V108I | 116 | 679 | 912 | 6.4E-7 | |
| Y181C | G190S | EFV | 120 | 2547 | 224 | 5.8E-6 |
| K101P | K103S | 24 | 207 | 175 | 1.5E-5 | |
| V108I | F227L | 46 | 980 | 175 | 2.1E-5 | |
| H221Y | F227L | 37 | 673 | 186 | 3.3E-5 | |
| V179F | Y181C | 25 | 1 | 2648 | 8.3E-5 | |
| K102N | G190A | 26 | 21 | 1868 | 1.8E-4 | |
aIndicates whether the pair of mutations also covaried significantly (uncorrected P < 0.05) within the subsets of sequences from individuals experienced in efavirenz (EFV) or nevirapine (NVP).
bOccurrences in 13 039 sequences comprising the full dataset; only one isolate per patient was included in the analysis and sequences with mixtures at a given position were excluded from comparisons at that position. XY, isolates containing both mutations; X only, isolates containing mutation X but not mutation Y; Y only, isolates containing mutation Y but not mutation X.
cP values are uncorrected values from the Jaccard covariation analysis. All mutation pairs shown above have correlations that are significant, with a Holm’s correction for multiple pairwise comparisons at P < 0.05.
Of the 29 pairs of mutations that were significantly correlated in the full dataset, 8 pairs covaried (uncorrected P < 0.05) in the efavirenz-alone subset, 5 pairs covaried in the nevirapine-alone subset and 4 pairs covaried in both subsets (Table 1). In 13 of the 29 significantly covarying pairs, both mutations were preferentially selected by the same NNRTI.
The major etravirine RAM Y181C was significantly associated with G190A, H221Y, V108I, A98G, G190S and V179F. L100I was significantly associated with K103N, and K101P was significantly associated with K103S. M230L was most strongly associated with K103N (P = 0.003 uncorrected, not significant following correction for multiple comparisons).
We also found 225 negatively correlated mutation pairs, including 116 that had statistically significant negative correlations after adjusting for multiple comparisons. However, a parametric bootstrap using the multivariate Poisson analysis of a random dataset containing the same number of mutations per sample and the same prevalence of each mutation indicates that a large number of negative correlations is to be expected because our analysis included only isolates containing at least one NNRTI resistance mutation.
Multidimensional scaling
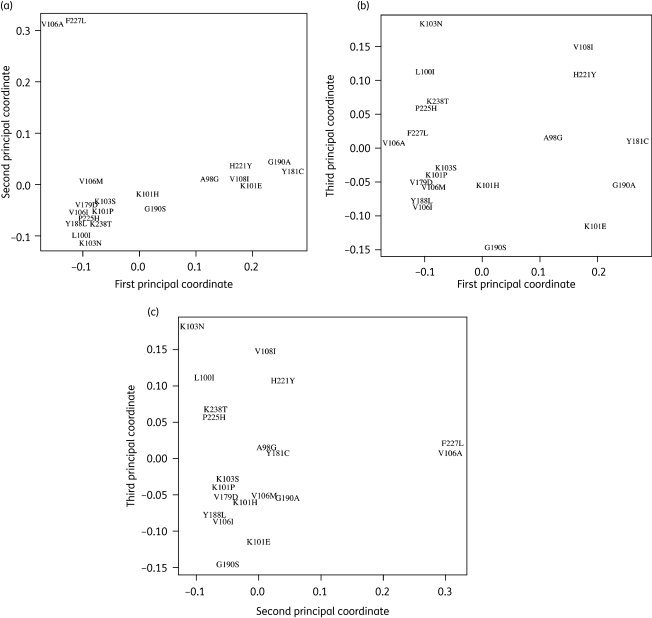
We used the 22 mutations that contribute to ≥1 of the 29 significantly covarying pairs in the full dataset for multidimensional scaling. Figure 2a, b and c shows the mutations on axes representing the first versus second, first versus third and second versus third principal coordinates. The first principal coordinate accounts for 36% of the total inertia and separates the predominantly nevirapine-selected mutations A98G, K101E, V108I, Y181C, G190A and H221Y from the other NNRTI resistance mutations. The second principal coordinate accounts for 25% of the total inertia and separates out V106A and F227L. The third principal component accounts for 15% of the total inertia and separates K103N and its associated mutations (L100I, P225H and K238T) from G190S and Y188L. The remaining principal coordinates did not significantly improve the goodness of fit of the multidimensional scaling and were therefore not plotted. Although V179F covaried with Y181C, it is not shown in Figure 2 because it occurred too uncommonly to be accurately placed by multidimensional scaling.
Figure 2.
Multidimensional scaling of NNRTI-selected mutations. This figure is a 2-D projection of the distances among 21 of the 22 mutations occurring in significantly covarying pairs (corrected P < 0.05, Table 1) in sequences containing at least one NNRTI-selected mutation: (a) compares the first and second principal coordinates; (b) compares the first and third principal coordinates; (c) compares the second and third principal coordinates. The distance between any two mutations is measured by their Jaccard dissimilarity coefficient, JD, where JD is equal to 1 minus the Jaccard similarity coefficient. The R command cmdscale was used to compare the first three principal coordinates from a table of JDs for each pairwise comparison. V179F is not included in this graphic; despite a significant positive correlation with Y181C, the comparison produces a high Jaccard dissimilarity coefficient, causing a misleading placement in multidimensional scaling.
NNRTI–NRTI correlations
To assess the covariation of NNRTI- and NRTI-selected mutations, we performed a Jaccard analysis on sequences from 9564 individuals with viruses containing at least one mutation from each category. After adjusting for multiple comparisons at P < 0.05, we identified 39 positive and 40 negative statistically significant correlations between 52 NNRTI-selected mutations and the 17 major NRTI-selected mutations. The strongest positive correlations were between the NRTI mutations L74V/I and the NNRTI mutations Y181C and L100I. Another notable, though weaker, positive correlation exists between K65R and Y181C. The NNRTI-associated mutation A98G was also strongly positively correlated (corrected P < 0.01) with the thymidine analogue mutations M41L, D67N, L210W and T215F/Y. The full results of this analysis are available in Tables S1 to S4 [available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)].
Higher order clustering of NNRTI mutations
Approximately 17% (n = 2233) of RT sequences with one or more NNRTI-selected mutation contained three or more NNRTI-selected mutations. Table 2 shows the 20 most commonly occurring mutational triplets, of which 14 meet the definition of a mutational cluster, or clique (each mutation covaries with each other mutation in the triplet). The remaining six triplets contain K103N in combination with a pair of covarying mutations. The 20 triplets listed in Table 2 occurred in 48% (n = 1069) of sequences with three or more mutations.
Table 2.
The 20 most common mutational triplets
| Mutation X | Mutation Y | Mutation Z | Occurrences* | Cluster† |
|---|---|---|---|---|
| K101E | Y181C | G190A | 242 | yes |
| V108I | Y181C | G190A | 190 | yes |
| K103N | Y181C | G190A | 180 | |
| V108I | Y181C | H221Y | 165 | yes |
| A98G | Y181C | G190A | 153 | yes |
| Y181C | G190A | H221Y | 141 | yes |
| K103N | Y181C | H221Y | 135 | |
| K103N | V108I | H221Y | 102 | |
| K101E | V108I | Y181C | 96 | yes |
| K101E | V108I | G190A | 89 | yes |
| K103N | V108I | Y181C | 87 | |
| V108I | G190A | H221Y | 79 | yes |
| A98G | V108I | Y181C | 74 | yes |
| A98G | K103N | Y181C | 74 | |
| A98G | K101E | G190A | 71 | yes |
| A98G | K101E | Y181C | 68 | yes |
| K101H | Y181C | G190A | 66 | yes |
| K101E | Y181C | H221Y | 53 | yes |
| A98G | K103N | V108I | 53 | |
| K101E | Y181C | G190S | 47 | yes |
*Occurrences are the number of sequences, from unique patients, that contain mutations X, Y and Z.
†Indicates whether the triplet can be described by a mutational cluster, where each pair of mutations within the triplet is positively correlated with P < 0.05.
In addition to the 14 mutational clusters included in the 20 most commonly occurring triplets, we identified 43 less common clusters. The 57 clusters could be characterized as follows: (i) 3 clusters of five mutations: A98G/K101E/V108I/Y181C/G190A, K101E/V108I/Y181C/G190A/H221Y and K103H/V108I/Y181C/G190A/H221Y, each of which contains within it 5 clusters of four mutations and 10 clusters of three mutations (n = 48); (ii) 3 additional clusters of four mutations: K101H/V108I/Y181C/G190A, K101N/K103H/Y181C/G190A and V108I/G190A/H221Y/F227L, each of which contains 4 clusters of three mutations (n = 15); and (iii) 7 additional clusters of three mutations: K101E/Y181C/G190S, K101H/Y181C/G190S, K103N/E138Q/K238T, S105T/V106A/F227L, V106A/G190A/F227L, V106M/G190A/F227L and V179F/Y181C/G190A. Two clusters of four mutations and 11 clusters of three mutations were present in more than one superset.
Discussion
Prior to the licensing of etravirine in 2008, only a few studies examined the patterns of NNRTI resistance mutations arising following NNRTI treatment failure because failure with one NNRTI generally presaged failure with a second NNRTI.20–24 Since the licensing of etravirine, however, several studies have examined the likelihood of virological response to etravirine among individuals in whom previous NNRTI therapies were unsuccessful.4,25–29 Moreover, the introduction of etravirine has led to the identification of novel NNRTI-selected mutations that often decrease susceptibility to efavirenz and nevirapine as well as to etravirine or other second-generation NNRTIs.2,30,31
In this study, we sought to identify and quantify associations among NNRTI-selected mutations. Our analysis differs from previous covariation analyses15,22 in that we examined an expanded set of mutations—including recently identified non-polymorphic NNRTI-selected mutations10 and the complete list of etravirine RAMs—in a very large number of sequences. In addition, we sought to control for associations that might reflect similar treatment exposure rather than a potential biophysical mechanism. Toward this end, we investigated only sequences containing one or more NNRTI-selected mutations, to minimize the likelihood that a history of NNRTI exposure (NNRTI naive versus NNRTI experienced) was responsible for the association. We then systematically examined the preferential selection of NNRTI resistance mutations by efavirenz or nevirapine and the covariation of mutations within subsets of efavirenz- or nevirapine-experienced individuals, to assess whether pairwise covariations could reflect the selection pressure of the NNRTI received.
Although the viruses that emerge during treatment with efavirenz or nevirapine are often resistant to both drugs, our analysis shows that efavirenz and nevirapine select for NNRTI resistance mutations in very different proportions. For example, nevirapine selects for V106A, Y181C/I/V and G190A significantly more frequently than efavirenz, whereas efavirenz preferentially selects for L100I, K101P, V106M, Y188L, G190S and P225H. These data suggest that patients developing virological failure while receiving nevirapine may be at increased risk of developing virological failure with etravirine because Y181C/I/V—which occurs in 45% of individuals receiving nevirapine—forms the foundation for high-level etravirine resistance.4,12,13 Conversely, although the two efavirenz-selected mutations L100I and K101P are major etravirine RAMs, combined they occur in <20% of patients receiving efavirenz. Nevirapine-associated etravirine RAMs, including Y181C, G190A and E138A, were also among the most commonly observed mutations in sequences from 782 women treated with a single dose of nevirapine.
With the exception of the uncommon mutations Y181I/V, multiple mutations are required for moderate to high-level etravirine resistance (>10-fold reduction in susceptibility).4,26 We identified 29 pairs of significantly covarying NNRTI-selected mutations in viruses from ∼13 000 individuals whose sequences contained at least one NNRTI-selected mutation. Of these 29 pairs, 22 contained at least one etravirine RAM. Nine of these 22 pairs contained major etravirine RAMs: Y181C (n = 7), L100I (n = 1) or K101P (n = 1), but only 2 pairs are predicted to confer moderate to high-level etravirine resistance: Y181C + V179F and Y181C + G190S. Although V179F and G190S do not reduce etravirine susceptibility when they occur individually, they are highly synergistic with Y181C; Y181C + V179F reduces etravirine susceptibility >100-fold and Y181C + G190S reduces etravirine susceptibility ∼20-fold.2,4,12,13
Approximately 17% (n = 2233) of individuals with one or more NNRTI-selected mutations had viruses with at least three NNRTI-selected mutations. Nearly half of these viruses contained at least 1 of 20 common patterns of mutation triplets, suggesting that these patterns of mutations are particularly compatible with enzymatic function and drug resistance. Most of these patterns consisted solely of mutations that covaried with one another (mutational clusters), and the remainder consisted of K103N in combination with one or more pairs of covarying mutations. Y181C was the only major etravirine RAM among the 20 most common mutational triplets. However, eight of these triplets contained 2 of the 17 etravirine RAMs, and six triplets had 3 etravirine RAMs. According to published phenotypic data, only 1 of the 20 triplets is predicted to have high-level resistance to etravirine—K101E, Y181C and G190S—but intermediate resistance to etravirine has previously been associated with the presence of Y181C or K101E plus at least two other NNRTI resistance mutations.2,4,26
In a previous study, Rhee et al.15 found 11 significant pairwise correlations between NNRTI-selected mutations (P < 0.05 corrected), of which two pairs, K103N/Y181C and K103N/V108I, were not positively correlated in our analysis. This is almost certainly because we included only sequences containing one or more NNRTI-selected mutation, reducing the likelihood that the co-occurrence of mutations was confounded by a history of NNRTI exposure. The following nine pairs were still significantly correlated in our analysis: Y181C/G190A, K101E/G190A, Y181C/H221Y, V108I/H221Y, L100I/K103N, V108I/Y181C, V106A/F227L, K101E/Y181C and K103N/P225H. Likewise, Ceccherini-Silberstein et al.22 reported five significant associations among the NNRTI-selected mutations that we studied: Y181C/H221Y, V108I/H221Y, L100I/K103N, K103N/P225H and K103N/K238T. Although we found that four of the pairs described by these studies consist of mutations selected preferentially by nevirapine (K101E, Y181C, G190A and H221Y) and that K103N and its associated mutations are all selected preferentially by efavirenz, we also found that the covariance of every pair was independent of the particular NNRTI received, suggesting a potential biophysical interaction rather than treatment pressure.
In conclusion, viruses from patients with virological failure following nevirapine treatment are more likely to have major etravirine RAMs than viruses from patients who failed treatment with efavirenz. This is because Y181C/I/V occur more commonly with nevirapine than with efavirenz, and because mutations at this position provide the foundation for high-level etravirine resistance. Few patterns among covarying pairs of mutations and the most common mutational triplets are predicted to be associated with moderate to high-level etravirine resistance. However, most patterns contained one or more minor etravirine RAMs in combination with a major etravirine RAM, particularly Y181C. These relatively constrained higher order mutational patterns constitute a potential target for future studies of the phenotypic and clinical significance of etravirine RAMs.
Funding
This work was supported by the National Institutes of Health (AI068581 and AI46148 to E. C. R, S. Y. R. and R. W. S., and R01GM086884 to S. H.).
Transparency declarations
None to declare.
Supplementary data
Tables S1 to S4 are available as Supplementary data at JAC Online (http://jac.oxfordjournals.org/)].
Supplementary Material
References
- 1.Sarafianos SG, Marchand B, Das K, et al. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. doi:10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Azijn H, Tirry I, Vingerhoets J, et al. TMC278, a next-generation nonnucleoside reverse transcriptase inhibitor (NNRTI), active against wild-type and NNRTI-resistant HIV-1. Antimicrob Agents Chemother. 2010;54:718–27. doi: 10.1128/AAC.00986-09. doi:10.1128/AAC.00986-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moyle G, Boffito M, Manhard K, et al. Antiviral activity of RDEA806, a novel HIV non-nucleoside reverse transcriptase inhibitor, in treatment of näive HIV patients. XVII International AIDS Conference; 2008; Mexico City. Geneva, Switzerland: International AIDS Society; Abstract THAB0403. [Google Scholar]
- 4.Vingerhoets J, Tambuyzer L, Azijn H, et al. Resistance profile of etravirine: combined analysis of baseline genotypic and phenotypic data from the randomized, controlled Phase III clinical studies. AIDS. 2010;24:503–14. doi: 10.1097/QAD.0b013e32833677ac. doi:10.1097/QAD.0b013e32833677ac. [DOI] [PubMed] [Google Scholar]
- 5.Zala C, Murphy R, Zhou XJ, et al. IDX899, a novel HIV-1 NNRTI with high barrier to resistance, provides suppression of HIV viral load in treatment-naïve HIV-1-infected subjects. XVII International AIDS Conference; 2008; Mexico City. Geneva, Switzerland: International AIDS Society; Abstract THAB0402. [Google Scholar]
- 6.Rhee SY, Gonzales MJ, Kantor R, et al. Human immunodeficiency virus reverse transcriptase and protease sequence database. Nucleic Acids Res. 2003;31:298–303. doi: 10.1093/nar/gkg100. doi:10.1093/nar/gkg100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bonhoeffer S, Chappey C, Parkin NT, et al. Evidence for positive epistasis in HIV-1. Science. 2004;306:1547–50. doi: 10.1126/science.1101786. doi:10.1126/science.1101786. [DOI] [PubMed] [Google Scholar]
- 8.Learn GH, Jr, Korber BT, Foley B, et al. Maintaining the integrity of human immunodeficiency virus sequence databases. J Virol. 1996;70:5720–30. doi: 10.1128/jvi.70.8.5720-5730.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gifford RJ, Rhee SY, Eriksson N, et al. Sequence editing by Apolipoprotein B RNA-editing catalytic component [corrected] and epidemiological surveillance of transmitted HIV-1 drug resistance. AIDS. 2008;22:717–25. doi: 10.1097/QAD.0b013e3282f5e07a. doi:10.1097/QAD.0b013e3282f5e07a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sanchez G, Xu X, Chermann JC, et al. Accumulation of defective viral genomes in peripheral blood mononuclear cells of human immunodeficiency virus type 1-infected individuals. J Virol. 1997;71:2233–40. doi: 10.1128/jvi.71.3.2233-2240.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Shahriar R, Rhee SY, Liu TF, et al. Nonpolymorphic human immunodeficiency virus type 1 protease and reverse transcriptase treatment-selected mutations. Antimicrob Agents Chemother. 2009;53:4869–78. doi: 10.1128/AAC.00592-09. doi:10.1128/AAC.00592-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vingerhoets J, Azijn H, Fransen E, et al. TMC125 displays a high genetic barrier to the development of resistance: evidence from in vitro selection experiments. J Virol. 2005;79:12773–82. doi: 10.1128/JVI.79.20.12773-12782.2005. doi:10.1128/JVI.79.20.12773-12782.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Das K, Clark AD, Jr, Lewi PJ, et al. Roles of conformational and positional adaptability in structure-based design of TMC125-R165335 (etravirine) and related non-nucleoside reverse transcriptase inhibitors that are highly potent and effective against wild-type and drug-resistant HIV-1 variants. J Med Chem. 2004;47:2550–60. doi: 10.1021/jm030558s. doi:10.1021/jm030558s. [DOI] [PubMed] [Google Scholar]
- 14.Coakley E, Chappey C, Benhamida J, et al. Defining the upper and lower clinical cutoffs for etravirine in the PhenoSense HIV assay. Abstracts of the Sixteenth Conference on Retroviruses and Opportunistic Infections; 2009; Montreal, Quebec, Canada. Alexandria, VA, USA: Foundation for Retrovirology and Human Health; Abstract 687. [Google Scholar]
- 15.Rhee SY, Liu TF, Holmes SP, et al. HIV-1 subtype B protease and reverse transcriptase amino acid covariation. PLoS Comput Biol. 2007;3:e87. doi: 10.1371/journal.pcbi.0030087. doi:10.1371/journal.pcbi.0030087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Holm S. A simple sequentially rejective multiple test procedure. Scand J Stat. 1979;6:65–70. [Google Scholar]
- 17.Hoaglin DC. A poissonness plot. Am Stat. 1980;34:146–9. doi:10.2307/2683871. [Google Scholar]
- 18.Shafer RW, Schapiro JM. HIV-1 drug resistance mutations: an updated framework for the second decade of HAART. AIDS Rev. 2008;10:67–84. [PMC free article] [PubMed] [Google Scholar]
- 19.Mardia KV, Kent JT, Bibby JM. Multivariate Analysis. London: Academic Press; 1979. [Google Scholar]
- 20.Antinori A, Zaccarelli M, Cingolani A, et al. Cross-resistance among nonnucleoside reverse transcriptase inhibitors limits recycling efavirenz after nevirapine failure. AIDS Res Hum Retroviruses. 2002;18:835–8. doi: 10.1089/08892220260190308. doi:10.1089/08892220260190308. [DOI] [PubMed] [Google Scholar]
- 21.Bannister WP, Ruiz L, Cozzi-Lepri A, et al. Comparison of genotypic resistance profiles and virological response between patients starting nevirapine and efavirenz in EuroSIDA. AIDS. 2008;22:367–76. doi: 10.1097/QAD.0b013e3282f3cc35. doi:10.1097/QAD.0b013e3282f3cc35. [DOI] [PubMed] [Google Scholar]
- 22.Ceccherini-Silberstein F, Svicher V, Sing T, et al. Characterization and structural analysis of novel mutations in human immunodeficiency virus type 1 reverse transcriptase involved in the regulation of resistance to nonnucleoside inhibitors. J Virol. 2007;81:11507–19. doi: 10.1128/JVI.00303-07. doi:10.1128/JVI.00303-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deforche K, Camacho RJ, Grossman Z, et al. Bayesian network analyses of resistance pathways against efavirenz and nevirapine. AIDS. 2008;22:2107–15. doi: 10.1097/QAD.0b013e32830fe940. doi:10.1097/QAD.0b013e32830fe940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bacheler L, Jeffrey S, Hanna G, et al. Genotypic correlates of phenotypic resistance to efavirenz in virus isolates from patients failing nonnucleoside reverse transcriptase inhibitor therapy. J Virol. 2001;75:4999–5008. doi: 10.1128/JVI.75.11.4999-5008.2001. doi:10.1128/JVI.75.11.4999-5008.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Llibre JM, Santos JR, Puig T, et al. Prevalence of etravirine-associated mutations in clinical samples with resistance to nevirapine and efavirenz. J Antimicrob Chemother. 2008;62:909–13. doi: 10.1093/jac/dkn297. doi:10.1093/jac/dkn297. [DOI] [PubMed] [Google Scholar]
- 26.Poveda E, de Mendoza C, Pattery T, et al. Phenotypic impact of resistance mutations on etravirine susceptibility in HIV patients with prior failure to nonnucleoside analogues. AIDS. 2008;22:2395–8. doi: 10.1097/QAD.0b013e32831692fb. doi:10.1097/QAD.0b013e32831692fb. [DOI] [PubMed] [Google Scholar]
- 27.Poveda E, Garrido C, de Mendoza C, et al. Prevalence of etravirine (TMC-125) resistance mutations in HIV-infected patients with prior experience of non-nucleoside reverse transcriptase inhibitors. J Antimicrob Chemother. 2007;60:1409–10. doi: 10.1093/jac/dkm372. doi:10.1093/jac/dkm372. [DOI] [PubMed] [Google Scholar]
- 28.Marcelin AG, Flandre P, Descamps D, et al. Factors associated with virological response to etravirine in nonnucleoside reverse transcriptase inhibitor-experienced HIV-1-infected patients. Antimicrob Agents Chemother. 2010;54:72–7. doi: 10.1128/AAC.01051-09. doi:10.1128/AAC.01051-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Poveda E, Anta L, Blanco JL, et al. Etravirine resistance associated mutations in HIV-infected patients failing efavirenz or nevirapine in the Spanish antiretroviral resistance database. AIDS. 2010;24:469–71. doi: 10.1097/QAD.0b013e328331a4b8. doi:10.1097/QAD.0b013e328331a4b8. [DOI] [PubMed] [Google Scholar]
- 30.Maiga AI, Descamps D, Morand-Joubert L, et al. Resistance-associated mutations to etravirine (TMC-125) in antiretroviral-naive patients infected with non-B HIV-1 subtypes. Antimicrob Agents Chemother. 2010;54:728–33. doi: 10.1128/AAC.01335-09. doi:10.1128/AAC.01335-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gatanaga H, Ode H, Hachiya A, et al. Combination of V106I and V179D polymorphic mutations in human immunodeficiency virus type 1 reverse transcriptase confers resistance to efavirenz and nevirapine but not to etravirine. Antimicrob Agents Chemother. 2010;54:1596–602. doi: 10.1128/AAC.01480-09. doi:10.1128/AAC.01480-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.