Abstract
The bacterium Caulobacter crescentus encodes a two-component signalling protein, LovK, that contains an N-terminal photosensory LOV domain coupled to a C-terminal histidine kinase. LovK binds a flavin cofactor, undergoes a reversible photocycle, and displays regulated ATPase and autophosphorylation activity in response to visible light. Femtosecond to nanosecond visible absorption spectroscopy demonstrates congruence between full-length LovK and isolated LOV domains in the mechanism and kinetics of light-dependent cysteinyl-C4(a) adduct formation and rupture, while steady-state absorption and fluorescence line narrowing (FLN) spectroscopies reveal unique features in the electronic structure of the LovK flavin cofactor. In agreement with other sensor histidine kinases, ATP binds specifically to LovK with micromolar affinity. However, ATP binding to the histidine kinase domain of LovK has no apparent effect on global protein structure as assessed by differential Fourier transform infrared (FTIR) spectroscopy. Cysteinyl adduct formation results in only minor changes in the structure of LovK as determined by differential FTIR. This study provides insight into the structural underpinnings of LOV-mediated signal transduction in the context of a full-length histidine kinase. In particular, the data provide evidence for a model in which small changes in the tertiary/quaternary structure of LovK, as triggered by photon detection in the N-terminal LOV sensory domain, are sufficient to regulate histidine kinase activity.
Bacterial cells must detect and adapt to a broad range of chemical and physical signals in their environment to survive. Two-component signalling systems, comprised of sensor histidine kinases and their cognate response regulator proteins, provide the primary sensory/signaling link between changes in the intra- and extracellular environment of the cell and the genetic control circuitry regulating cell adaptation (1). Despite the abundance of annotated two-component signalling genes and their importance in the regulation of microbial biology, our understanding of the functions, interactions, and regulatory mechanisms of two-component proteins remains rudimentary. We know little of the physical and chemical identity of the signals to which sensor histidine kinases respond, nor do we generally understand how signal detection translates into regulation of histidine kinase activity. However, recent biochemical and structural studies on natural and engineered sensor kinases provide an emerging picture of how signal detection by a sensory domain regulates activity of the histidine kinase domain.
Visible light is an environmental signal that has been largely ignored by those who study cell regulation in chemotrophic bacteria. Indeed, until recently there had been little reason to believe that visible light was a relevant regulatory input for most bacterial species. However, genome mining across a range of gram-positive and gram-negative chemotrophs has identified dozens of histidine kinases that contain putative photosensory LOV domains (2-7). Originally defined as the loci for visible photon absorption in the plant photoreceptors known as phototropins (8, 9), photosensory LOV domains are now known to be conserved in varied proteins across the plant, fungal, bacterial, and archaeal kingdoms (2). The genome of the aquatic α-proteobacterium Caulobacter crescentus encodes a cytoplasmic LOV histidine kinase, LovK, that functions as a regulator of cell surface physiology (10). LovK has the spectroscopic hallmarks of a LOV protein, including flavin cofactor binding and a reversible photocycle involving light-dependent formation of a cysteinyl-C4(a) adduct (3, 10). Additionally, the kinase activity of purified LovK is regulated by light (10).
Light is a signal that can be acutely controlled, making it an excellent experimental switch to probe molecular mechanisms of signal detection and transduction. While there is extensive data on the structure and dynamics of individual photosensory LOV domains isolated from their natural protein context (11), the biochemical and biophysical properties of full-length, multi-domain LOV proteins are not well characterized. The simple, two-domain protein, LovK, provides an excellent model to probe the molecular mechanism of LOV-mediated signal transduction on multiple time and length scales in the context of a sensor histidine kinase. We have applied a range of spectroscopic tools to the study of LovK dynamics upon blue light absorption. Here we demonstrate that the mechanism and kinetics of cysteinyl-C4(a) adduct formation and rupture are generally conserved between full-length LovK and isolated LOV domains from other bacterial and plant species. Moreover, we define unique features in the ground-state electronic structure of the FMN cofactor bound to LovK, and describe light-dependent protein structural changes in this photosensory histidine kinase.
Materials and Methods
Protein expression and purification
The LovK sensor histidine kinase (gene number CCNA_00287) was amplified from chromosomal DNA of C. crescentus strain NA1000. This gene was cloned into the EcoRI and HindIII sites of pET28a (Novagen, Madison, WI) to create the overexpression strain FC410 previously described (10). FC410 was grown at 37°C in 4 liters of LB broth and 30 μg/mL of Kanamycin to an OD600 of 0.4 (1 cm path length) at which point the temperature was lowered to 20°C and the cells were induced with 500 μM isopropyl-beta-D-thiogalactopyranoside (IPTG) for 4 hours. Cells were lysed in 20 mM Tris-HCl (pH 8.0), 200 mM NaCl, 1 mM EDTA, 100 mM imidazole. The recombinant His6-LovK protein was purified using Ni2+ Chelating Resin (GE Amersham Pharmacia, Piscataway, NJ). Affinity purified LovK was dialyzed overnight against 20 mM Tris-HCl (pH = 8.0), 20 mM NaCl, 100 mM imidazole and further purified via Q-Sepharose (GE Amersham Pharmacia, Piscataway, NJ) anion exchange. A 50 milliliter volume of purified fractions from Q-Sepharose was concentrated to a volume of 600 μl in a Centriprep 3000 Dalton molecular weight cutoff concentrator (Millipore, Billerica, MA). At this concentration, a portion of LovK forms an unstable precipitate; we spun this concentrated protein sample at 15,000 x g for 10 minutes to produce a clarified, concentrated, and stable sample.
Residues 407-563 of A. sativa phot1 (i.e. the LOV2 domain) were expressed as previously described (12). Briefly, cells were grown at 37 °C to an OD600 of 0.4, induced with 500 μM isopropyl β-d-thiogalactoside, and grown for an additional 14 h after induction at 20 °C. Cells were lysed via sonication in 20 mM Tris-HCl (pH 8.0), 150 mM NaCl. phot1 LOV2 was purified on Ni-NTA resin (Qiagen, Valencia, CA) and protein was concentrated in a Centriprep 3000 Dalton molecular weight cutoff concentrator (Millipore, Billerica, MA).
Steady-state absorption spectroscopy
Steady-state absorption spectra were recorded on a commercial spectrometer (Perkin-Elmer, Groningen, Netherlands) at 25°C. To measure the lit-state spectra, a blue LED (emitting at 470 nm; 5 mW output power) was used for photoconversion at saturating intensity.
Fluorescence Anisotropy Analysis of LovK-ATP-BODYPI binding
Fluorescence anisotropy assays of adenosine 5′-triphosphate, BODIPY® TR 2′-(or-3′)-O-(N-(2-aminoethyl)urethane) (Invitrogen Molecular Probes, Carlsbad, CA) binding to LovK were conducted at 25°C using a Fluoromax-3 fluorimeter (JY-Horiba). We hereafter refer to this fluorescent ATP analog as BODYPI-ATP. Anisotropy at eight LovK concentrations was measured with a constant BODYPI-ATP concentration of 100 nM; the average of two independent sets of measurements is shown. Experimental error at each point was ≈5%. Anisotropy raw data were transformed into fractional saturation according to
Fitting with Hill equation yields the dissociation constant (Kd) and Hill coefficient (n) according to
Femtosecond transient absorption spectroscopy
Prior to the experiment, concentrated LovK was dissolved in 20 mM Tris, 150 mM NaCl, 100 mM imidazole at pH 8.0. For the time-resolved experiments, the absorbance of the samples was adjusted to 0.3 mm−1 at 447 nm. The samples were loaded in a flow system containing a cuvette with a path length of 1 mm, and flowed at a speed of approximately 5 cm/s by means of a peristaltic pump. The flow cuvette was mounted on a shaker that continuously vibrated the cuvette sideways at a frequency of 20 Hz with an amplitude of 1 mm.
Femtosecond transient absorption spectroscopy was carried out with an amplified Ti:sapphire laser system (Coherent Legend USP, operating at 1 kHz) described earlier (13). Part of the output of the amplifier was frequency-doubled in a nonlinear crystal and used as a pump beam. A white-light continuum was generated by focusing the rest of the output of the regenerative amplifier at 800 nm on a 1 mm sapphire crystal, and used as a probe beam. The pump and the probe beam were focused on the sample cuvette at a spot with a diameter of 150 μm. Femtosecond time delays between the pump and probe were controlled by a delay line covering delays up to 4 ns. The pump and probe were polarized parallel. After passing through the sample, the probe beam was dispersed by a polychromator and projected on a 256 element diode array detector.
The time-resolved spectra were analyzed with a global analysis program using a kinetic model consisting of sequentially interconverting species, i.e., 1 → 2 → 3 → … (14), in which the arrows indicate successive monoexponential decays of increasing time constants; these constants can be regarded as the lifetime of each species. Associated with each species are a lifetime and a difference spectrum, denoted the evolution-associated difference spectrum (EADS). The first EADS corresponds to the time-zero difference spectrum. This procedure enables us to visualize clearly the evolution of the (excited) states of the system. The instrument response function was fit to a Gaussian (120 fs full width at half-maximum). To illustrate our observations optimally, we present them in the form of EADS instead of raw time-resolved spectra.
Fluorescence line narrowing (FLN) spectroscopy
Fluorescence Line Narrowing (FLN) spectra were measured on a U1000 double monochromator (Jobin Yvon, Longjumeau, France) equipped with an ultra-sensitive, deep-depleted, front illuminated CCD (Jobin Yvon). For FLN measurements the LovK sample was dissolved in 20 mM Tris, 50 mM NaCl at pH 8.0 and concentrated to an OD of 60 cm−1 using a Centricon spin concentrator (Millipore, Billerica, MA) with a 3000 molecular weight cutoff filter. Prior to measurement, a 5 μL drop of concentrated protein was deposited on a cover slip and mounted on the cryostat tube; samples were maintained at 10 K in a helium flow cryostat (Air Liquide, Sassenage, France). The laser power for FLN experiment was less than 100 microwatts. Excitation at 488.1 nm for FLN was provided by an Innova 100 argon laser (Coherent, Palo Alto, Calif.). The LovK spectrum was obtained at a spectral resolution of 1 cm−1.
Differential FTIR spectroscopy
Infrared difference spectra were recorded on an FTIR spectrometer (IFS 66s Bruker) equipped with a nitrogen cooled photovoltaic MCT detector (20 MHz, KV 100, Kolmar Technologies). A blue LED (emitting at 470 nm; 5 mW output power) was used for photoconversion at saturating intensity. Low-temperature spectra were measured using a Optisat DN cryostat (Oxford) with an ITC4 temperature controller (Oxford) and liquid nitrogen as the coolant. A dark-adapted protein sample was cooled to the set temperature and, after a full equilibration of 15 min, background (FT−1 of I0) and sample (FT−1 of I) interferograms were recorded before and after switching on the blue light, respectively. Background and sample interferograms are the average of 500 and 2000 interferograms, recorded at 4 cm−1 resolution, respectively. After Fourier transform the resultant absorption spectrum represents the light minus dark FTIR difference spectrum. The sample was brought to room temperature between measurements at different temperatures. Two recordings have been averaged for each temperature. The FTIR sample was made with a drop of 2 μL of OD450 nm of about 60 cm−1 (~4 mM LovK in 20 mM Tris/HCl pH 8, 50 mM NaCl) spread between two tightly fixed CaF2 windows without any spacer and greased for tightness.
Results and Discussion
Comparative steady-state absorption spectroscopy of LovK and phot1-LOV2
The steady-state absorption spectra of purified C. crescentus LovK before and after illumination with blue light were measured and compared to absorption spectra of the well-characterized LOV2 domain of Avena sativa phototropin1 (phot1-LOV2). The maximum visible flavin absorption peak of LovK is at 447 nm with vibronic peaks at 474 nm and 426 nm. These bands are attributed to a S0-S1 transition in the isoalloxazine moiety (15) and are qualitatively identical to that measured for phot1-LOV2 (Figure 1A). The near-UV flavin absorption band of LovK, can be attributed to S0-S2 / S0-Sn transitions (16, 17), and is blue-shifted relative to phot1-LOV2. Both LovK and phot1-LOV2 have two peaks in this wavelength range with similar gaps: 371/355 nm and 376/360 nm, respectively (Figure 1A). The light minus dark difference absorption spectra of LovK and phot1-LOV2 show that the absorption peak indicative of cysteinyl-C4(a) adduct formation in the illuminated state (18) is also blue-shifted in LovK (Figures 1B and 1C). The structural and chemical basis of this hypsochromicity is not known, but may be a result of slight differences in the conformation and dynamics of the isoalloxazine moiety, and/or in composition of the residues of the FMN binding pocket in LovK relative to phot1-LOV2.
Figure 1.
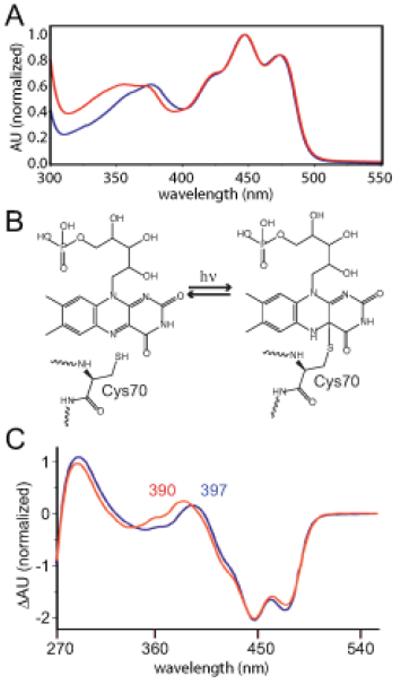
(A) Absorption spectra of C. crescentus LovK (red) and A. sativa phot1-LOV2 (blue). (B) Cartoon illustrating light-dependent adduct formation between LovK residue C70 and C(4a) of the FMN isoalloxazine moiety. (C) Light minus dark difference absorption spectra of C. crescentus LovK (red) and A. sativa phot1-LOV2 (blue). Absorption maxima for the adduct species in LovK (390 nm) and phot1-LOV2 (397 nm) are labelled
Characterization of nucleotide binding to LovK
We utilized fluorescence anisotropy to measure the dissociation constant (Kd) of purified full-length LovK and the fluorescent ATP analog, BODYPI-ATP (see Materials and Methods). BODYPI-ATP was excited at 590 nm and fluorescence anisotropy measured between 618 and 625 nm (λEmmax =621.5 nm) at a resolution of 1 nm. Anisotropy changes of BODIPY-ATP fluorescence expressed as a fractional saturation function of LovK concentration show a typical saturation curve (Figure 2). The Kd of LovK with BODYPI-ATP equals [LovK] at which Y=0.5, according to Y=[LovK]/(Kd+[LovK])
Figure 2.
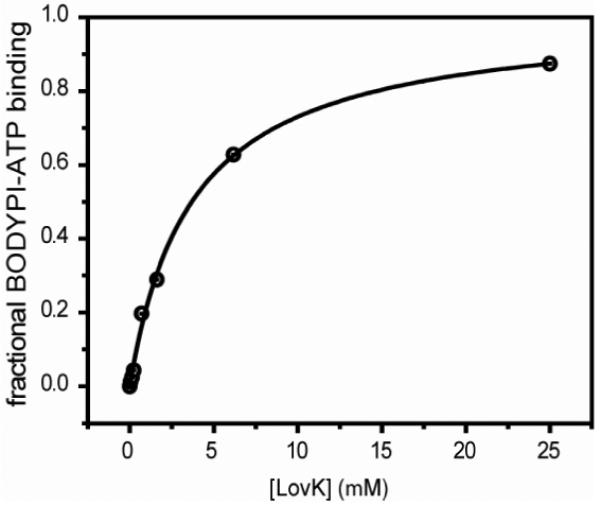
Measuring LovK ATP binding affinity by fluorescence anisotropy. The dependence of signal from the fluorescent ATP analog, BODIPY-ATP, on the concentration of LovK is shown in terms of fraction of LovK bound to BODIPY-ATP. Data are fit to the Hill equation (black line; see Materials and Methods).
BODYPI-ATP binds LovK with a Kd of 3.8 μM and a Hill coefficient of 1.0, evidencing a single binding site for BODYPI-ATP on LovK. Binding competition experiments using an excess of non-fluorescent ATP (1 mM) result in a reversal of the anisotropy changes after 20 min. This indicates that BODYPI-ATP is displaced from LovK due to competition for the same binding site. The measured affinity of LovK for BODYPI-ATP is ≈15-fold tighter than the affinity of the natural ATP nucleotide for the osmosensor histidine kinase, EnvZ, of Escherichia coli (19), ≈50-fold tighter than the affinity of ATP for the E. coli chemotaxis histidine kinase, CheA (20), and ~10-fold tighter than BODYPI-GTP for the STAS domain of full length LOV-YtvA from Bacillus subtilis (21). However, the affinity of BODYPI-ATP for LovK is similar to published affinities of the fluorescent ATP analog 2′(3′)-O-(trinitrophenyl)adenosine 5′-trisphosphate (TNP-ATP) for the CheA and EnvZ histidine kinases of E. coli (19, 22). Thus, the addition of hydrophobic fluorescent moieties via 2′ or 3′ linkage to the nucleotide sugar does not appear to perturb nucleotide binding specificity but confers higher binding affinity with histidine kinases. We did not observe hydrolysis or a change in affinity of BODYPI-ATP with LovK upon illumination with blue light, which is inconsistent with the known light-regulated kinase activity of LovK. However, previous structural work on Thermatoga maritima CheA bound to TNP-ATP (23) suggests that the presence of a 2′/3′-covalent-linked fluorescent moiety such as BODYPI affects the conformation of bound ATP, and thus perturbs the capacity of histidine kinases to hydrolyze the nucleotide γ-phosphate. We could not detect a measurable change in LovK structure upon nucleotide binding by difference Fourier Transform Infrared (FTIR) Spectroscopy (see Materials and Methods).
Dark state recovery of LovK in presence of base catalysts
The rate of thermal recovery from the covalent cysteinyl-C4(a) adduct state back to the non-bonded “dark” state is dramatically enhanced by the presence of low-molecular weight bases such as imidazole or 4-dimethylaminopyridine (DMAP) (24, 25). Although the exact mechanism of base-mediated rate enhancement has not been established, the linear and non-saturable relationship between recovery rate and base concentration (24) suggests a bimolecular reaction involving a direct general base effect on N5 of the flavin cofactor. A previous study has reported that LovK dark recovery from the cysteinyl-C4(a) adduct state (see Figure 1C) is slow in presence of 10 mM imidazole, being best fitted as the sum of two exponential decays with recovery half-times of 1.7 h and 32 h (10). In the absence of imidazole (across a range of buffer and salt conditions) LovK precipitates, and exhibits an apparent dark recovery that is ≈20 times slower. However, this recovery rate could not be accurately measured due to sample scattering. To stabilize the protein, we have measured dark recovery of LovK at 450 nm after full photoconversion of the sample with a blue LED (Emmax = 470 nm) in the presence of 750 mM imidazole and 500 mM DMAP. In agreement with previous data on full-length LovK, a single exponent is insufficient to fit these data (Figure 3). Decay from the adduct state in the presence of 750 mM imidazole can be fitted by two exponents with rates that correspond to half-lives of 3 (30%) and 23 (70%) minutes. LovK recovery is biexponential and further accelerated in presence of 500 mM DMAP, a catalyst that is more basic than imidazole, with rates that correspond to half-lives of 2 (50%) and 10 min (50%). The relative decay rate enhancement by DMAP in LovK is in agreement with previous data on phot1-LOV2 (24).
Figure 3.
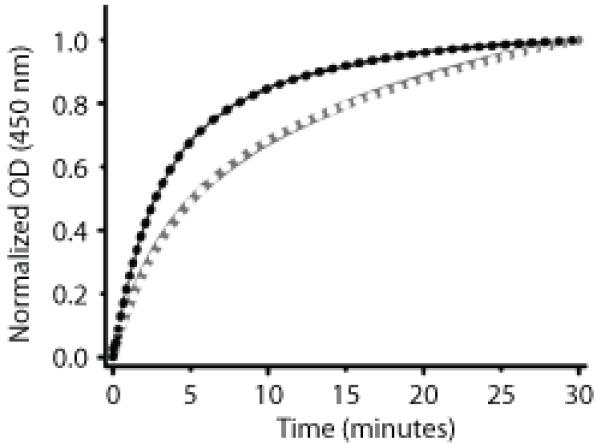
Dark recovery kinetics of LovK from the cysteinyl-C4(a) adduct state in the presence of 500 mM DMAP (black dots) and 800 mM imidazole (white dots). Biexponential fits to the data are shown as a black line (DMAP) and gray line (imidazole).
Conservation of singlet-excited state FMN dynamics in a multi-domain LOV histidine kinase
Femtosecond transient absorption measurements were carried out on LovK at pH 8 at 25°C. The sample was excited at 400 nm and the time-resolved absorbance changes were monitored over a wavelength range of 420 to 705 nm. Global analysis of the time-resolved data demonstrates that two components are required for an adequate description of FMN excitation dynamics between 150 fs and 4 ns. The first component has a lifetime of 2 ns; the second component does not decay on the time scale of our experiment (4 ns). Each component is characterized by an evolution associated difference spectrum (EADS) and an associated lifetime. The resulting EADS and kinetic traces at selected wavelengths are shown in Figure 4. The first EADS can be assigned to the initially created singlet excited state of FMN. It shows a broad bleach band extending from 430 to 500 nm that exhibits vibronic structure near 450 and 475 nm. This negative band corresponds to the loss of FMN ground state absorption upon photoexcitation. The first EADS is further characterized by a pronounced stimulated emission band centred at 534 nm, sharp excited state absorption (ESA) at 505 nm, and broad ESA at wavelengths longer than 620 nm. These spectral features are generally found for the singlet excited state of oxidized flavins, in solution as well as bound to a protein (26-31). The FMN singlet excited state evolves in 2 ns to the second EADS, which does not decay in the 4 ns time window of the experiment. The second EADS is assigned to the FMN triplet state, and is characterized by very broad ESA with two maxima near 515 and 650 nm and a minimum near 550 nm. The ratio (ΔOD450Singlet/ΔOD 450Triplet )−1 reflects the quantum yield of triplet state formation and is estimated to be ≈0.7, similar to that found in phot1 LOV2 domains (12, 32), Chlamydomonas phot LOV1 (33) and Bacillus YtvA (34). The singlet excited-state lifetime of 2 ns and the EADS spectral shape of the singlet and triplet state of FMN bound to LovK agree well with femtosecond transient absorption data collected on isolated LOV2 domains of plant phototropins (12). Ultrafast IR and (time-resolved) FTIR experiments on plant LOV2 domains were consistent with an unprotonated FMN triplet as the primary photoproduct (35-37). The strictly single-exponential decay of ~2 ns observed in LovK, plant phot1 LOV2 and Chlamydomonas phot LOV1 is consistent with an intersystem crossing process on FMN as the primary photochemical reaction, enhanced by the presence of the nearby conserved cysteine (12, 38, 39). This contrasts with the situation in other flavin-binding photoreceptors such as BLUF domains and cryptochromes, where rapid, multiphasic electron transfer processes from aromatic side chains quench the flavin singlet-excited state on the picosecond timescale (27, 28, 40-45). Our time-resolved analysis of LovK thus demonstrates that FMN excited state dynamics are highly congruent in a taxonomically diverse range of isolated LOV domains (12, 32, 34, 46) and in full-length multi-domain LOV proteins, such as LovK.
Figure 4.
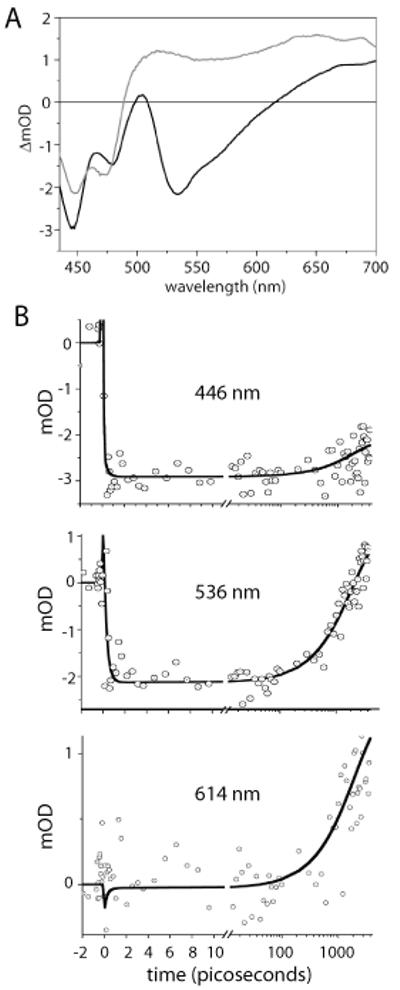
(A) EADS of LovK pumped at 400 nm and monitored from femtoseconds to nanoseconds. FMN singlet excited state (black line) decays to a triplet state (gray line) in 2 ns with a yield of about 70%. (B) Kinetic absorption traces at 446, 536 and 614 nm of LovK pumped at 400 nm; fit in black line.
Probing the ground-state electronic structure of the LovK flavin cofactor by FLN spectroscopy
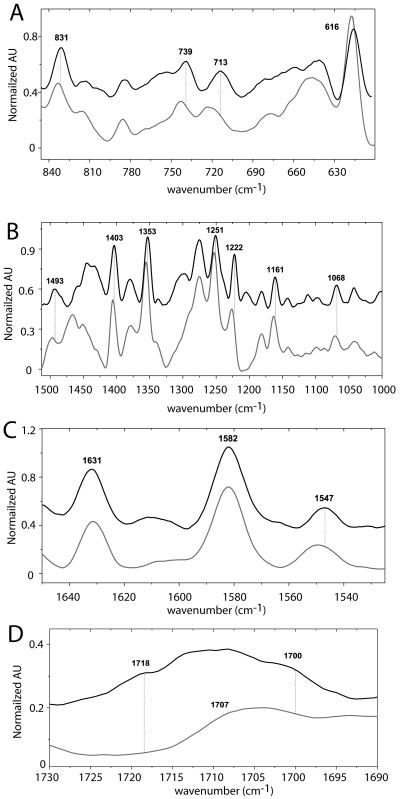
We next characterized vibrational modes of LovK FMN in the dark state. The high level of fluorescence of LovK complicates analysis of the electronic structure of FMN by resonance Raman spectroscopy. Fluorescence line narrowing (FLN) spectroscopy permits measurement of high quality vibrational patterns of inherently fluorescent molecules, such as oxidized FMN bound to LovK (38). The FLN spectrum of LovK was obtained at 10 K using a narrow 488.1 nm excitation. The spectrum of phot1-LOV2 was measured under the same conditions (38) and used for comparison. Whereas visible absorption spectroscopy revealed only small differences between LovK and phot1-LOV2, the FLN vibrational spectra reveal significant differences in the fine structure of FMN (Figure 5). LovK exhibits a unique vibrational signature between 1100 and 600 cm−1: phot1-LOV2 and Adiantum capillus-veneris phy3-LOV2 (38) have very similar band profiles in this region, while the spectrum of LovK is downshifted in this spectral window (Figure 5B). Specifically, bands at 1068, 831, and 616 cm−1 are downshifted by 2 cm−1, and bands at 739 and the 713 cm−1 are downshifted 4 and 8 cm−1, respectively (Figures 5A and 5B). A similar downshift of 5 to 2 cm−1 is also observed in LovK between 1540 and 1100 cm−1 at bands 1161m, 1222s, 1251m, 1353w, 1403m and 1493s cm−1. However bands at 1042, 1181 and 1275 cm−1 overlap in LovK and phot1-LOV2 (Figure 5B). In addition, the 1465-1420 spectral region reveals shifted bands, but the line shape is too complex to conclusively attribute band positions.
Figure 5.
Fluorescence line narrowing (FLN) spectra of LovK (upper black line) and phot1-LOV2 (lower gray line) in different spectral regions (A) 850-610 cm−1, (B) 1510-1000 cm−1, (C) 1650-1525 cm−1 and (D) 1730-1690 cm−1.
Only one of the two C=N stretch bands, assigned to C10a=N1 (38), is distinct. This band is centred at 1547 cm−1 in LovK and 1551 cm−1 in phot1-LOV2 (Figure 5C). LovK exhibits C=C and C4a=N5 frequencies at 1632 cm−1 and 1582 cm−1, respectively, which are similar to phot1-LOV2 but significantly different than phot1-LOV2 C39A. This result indicates the ICT/quinoid character of the FMN in LovK is comparable to phot1-LOV2 (38). In the C=O region (Figure 5D) the signals are very small but bands at 1717 and 1700 cm−1, which appear as shoulders of the band centred at 1710 cm−1, can be detected. We assign the bands at 1718 cm−1 and 1700 cm−1 to two C(4)=O conformers under different hydrogen bonding conditions (47); the broad band at 1710 cm−1 is not a vibrational band but is rather assigned as a residual of the baseline subtraction.
In conclusion, FLN spectroscopy demonstrates that the overall environment of the FMN cofactor binding site is similar in LovK and phot1-LOV2. However, there are clear differences in electronic configuration of the isoalloxazine moiety between these bacterial and plant protein photoreceptors. C=C and C4a=N5 bands are very similar, whereas C10a=N1 and lower stretching frequency modes involving mainly N3, C4, C10a, N10, N1 and C2 (38) show a lower double bond character indicating different electronic density in this region of the isoalloxazine moiety. Below 1000 cm−1 the bands can be assigned to ring deformation, evidencing a different distortion of the isoalloxazine ring of the FMN in LovK.
The effect of cysteinyl-adduct formation on vibrational state of the LovK flavin cofactor
Light minus dark Fourier transform infrared (FTIR) spectra of LovK exhibit the classical bandshift pattern for adduct formation (Figure 6A) (48). Adduct formation bandshifts are not temperature dependent except for 1250-1247 (−) /1302-1306 (+) cm−1 and 1554(−)/1542(+) cm−1. These latter bands have an atypical temperature dependence in which the C=N stretch doublet ratio at 1580-1550 cm−1 is reversed at room temperature (RT) relative to low temperature. The atypically strong temperature dependence of the adduct marker bands at 1582(−) and −1554(−)/1540(+) cm−1 are assigned to stretch modes of C4a=N5 coupled to C10a=N1 in the dark state and C10a=N1 alone in the adduct state. We propose this is due to alternative adduct state conformations at different temperatures. The adduct band at 1540 cm−1 downshifts 2 cm−1 between 150K and RT. A significant contribution of Amide II signal in this spectral region, above 150K, is excluded due to the small structural changes observed at RT.
Figure 6.
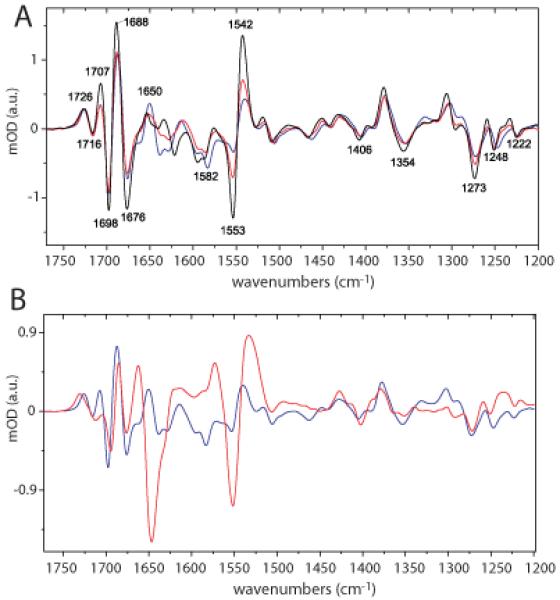
(A) Low temperature FTIR light minus dark spectra recorded at 150 (black), 200 (red) and 300 K (blue). (B) Comparison of light minus dark FTIR spectrum of LovK (blue line) with Phot1-LOV2 (red line) at 300K.
As observed in the Bacillus LOV-STAS protein, YtvA (48), the C=N stretch doublet 1580-1550 cm−1 ratio is approximately 1. In addition, the C10a=N1 stretching frequency of LovK at 1540 cm−1 is upshifted by 6 cm−1 in the adduct state relative to phot1-LOV2 (Figure 6B) evidencing a more localized C10a=N1 bond in the LovK adduct state. The observed differences in C=N stretches in LovK and YtvA relative to phot1-LOV2 may be associated with the blue-shifted absorbance of the adduct species in the bacterial LOV proteins, LovK and YtvA. While the structural and chemical bases of these relative spectral shifts are not known, they may be a result of an altered protein-flavin hydrogen bond network.
Light-dependent changes in LovK protein structure
Differential Fourier transform infrared spectroscopy (FTIR) is a well-established method to monitor changes in the secondary structure of proteins before and after some physical or biochemical perturbation. In the case of LOV domains, FTIR can provide insight into the effects of photon absorption and subsequent cysteinyl-flavin adduct formation on protein structure. Moreover, this method permits one to monitor the temperature dependence of light-dependent changes in LOV secondary structure.
Overall, the FTIR light minus dark spectrum of LovK at 25°C resembles C. reinhardtii phot-LOV2, A. capillus-veneris phy3-LOV1 and B. subtilis YtvA, which exhibit weak protein structural changes upon blue-light illumination (48). The C(4)=O frequency of LovK at 1726 cm−1 and weak β-sheet bands at 1635(−)/1613(+)cm−1 are indicative of small structural changes upon illumination (Figure 6) (49). The C(4)=O band in the lit state at 1726 cm−1 is also consistent with small changes in β-sheet structure; this band is upshifted to 1730 cm−1 in phot1-LOV2 and phy3-LOV2 domains at RT where more extensive light-dependent β-sheet motions occur. The temperature dependence of the β-sheet bands near 1635 cm−1 between 150 and 300 K evidences a model in which the majority of this light-induced change in LovK protein structure takes place outside the LOV domain. Specifically, the C(4)=O band is temperature independent within this temperature range evidencing a similar hydrogen bond strength between C(4)=O and Gln133/Asn112, which is inconsistent with substantial β-sheet motion in the LOV domain of LovK. Unfortunately, this hypothesis could not be explicitly tested by comparison with the isolated LOV domain of LovK due to problems with protein stability at the concentrations needed for FTIR spectroscopy. FTIR data on full-length LovK suggest a tightening of β-sheet involving a relatively small number of amino acids. Moreover, the absence of a strong band near 1645 cm−1 excludes large changes in the structure of α-helix upon illumination, as is observed in Jα of phot1-LOV2 (47) (Figure 6B).
As in A. sativa phot1-LOV2, the C(2)=O stretch is at 1676(−) cm−1 in the dark state while the C(4)=O stretches at 1716 and 1698 (−) cm−1 are both upshifted by 4 cm−1 (Figure 6B). These upshifts are indicative of weaker hydrogen bonding between C(4)=O and Gln133/Asn112 in dark state LovK relative to what is observed between orthologous residues and C(4)=O in phot1-LOV2. The positive band at 1688 cm−1 is upshifted by 3 cm−1 indicating a weaker C(2)=O hydrogen bond to Asn102 in the lit state. Thus Asn102 is either positioned further away from C(2)=O in LovK relative to phot1-LOV2 or is more dynamic/disordered than in phot1-LOV2.
In phy3-LOV1 (50) and the phy3-LOV2 Q1029L mutant (49), the positive band at 1707 cm−1 and corresponding bands at 1695 and 1692 cm−1 are moderately temperature dependent, and are assignable to loop regions of the structure (49-51). LovK, like phy3-LOV1 and the phy3-LOV2 Q1029L mutant, is one of the few LOV proteins that does not show evidence of loop tightening upon adduct formation at RT. Rather, LovK loop bands in the lit state are upshifted by 10-15 cm−1 relative to phy3-LOV domains providing evidence for considerably stronger loop loosening in LovK upon photon absorption and adduct formation than in phy3-LOV1 and phy3-LOV2 Q1029L (49). It is notable that LovK encodes a leucine residue at position 114, which corresponds to a phenylalanine in phy3-LOV2 (F1010) and a leucine in phy3-LOV1. The single amino acid change F1010L in phy3-LOV2 is known to strongly attenuate the large structural rearrangements that occur in phy3-LOV2 in response to adduct formation (52). Notably, mutation of this site in B. subtilis YtvA disrupts communication between the N-terminal LOV domain and the C-terminal STAS domain. The exact role of this residue in LOV signalling has yet to be elucidated, and may vary across protein systems (53).
With the exception of weak β-sheet strengthening upon illumination, the FTIR light minus dark difference spectrum of LovK globally resembles that of the LOV1 domain of the chimeric phytochrome-phototropin protein, phy3 (50). It may be that there are common structural and dynamic features between LOV-mediated signal transduction by the LOV domain of LovK and the LOV1 domain of phy3. The LovK light minus dark spectrum also closely resembles that of YtvA-LOV (48), suggesting a common mode of signal propagation between bacterial LOV-STAS and LOV-histidine kinase proteins that requires only minor changes in the overall structure of the protein. This can be contrasted with plant phototropin LOV2 domains which exhibit larger structural changes in β-sheet and α-helix structure prior to kinase activation (Figure 6B) (54), which eventually propagates to affect structural change in the serine/threonine kinase domain (55). Thus, these data support an emerging picture of heterogeneity in the structural mechanisms through which LOV domains can regulate various output domains.
Acknowledgements
We thank Toh Kee Chua for assistance with data analysis, and Aretha Fiebig for help with TOC figure design. The Avena phot1-LOV2 expression construct was provided by the lab of Kevin Gardner.
Abbreviations
- LOV
light oxygen voltage
- ATP
adenosine triphosphate
- FLN
fluorescence line narrowing
- FTIR
Fourier transform infrared
- IPTG
isopropyl-beta-D-thiogalactopyranoside
- LED
light-emitting diode
- EADS
evolution-associated difference spectrum
- FT
Fourier transform
- CCD
charge-coupled device
- FMN
flavin mononucleotide
- STAS
sulfate transporter anti-sigma factor antagonist domain
Footnotes
M.T.A.A. was supported by the Netherlands Foundation for Scientific Research through the Molecule to Cell Program and the Human Frontier Science Program. E.B.P. was supported by NIH Training Grant 5T32GM007183-34. J.T.M.K. was supported by the Earth and Life Sciences Council of the Netherlands Foundation for Scientific Research via a VIDI grant. S.C. acknowledges support for this project from National Institutes of Health award 1R01GM087353-1, the Arnold and Mabel Beckman Foundation (BYI), and the Mallinckrodt Foundation.
References
- 1.Hoch JA, Silhavy TJ. Two-Component Signal Transduction. ASM Press; Washington, D.C.: 1995. [Google Scholar]
- 2.Crosson S, Rajagopal S, Moffat K. The LOV domain family: Photoresponsive signaling modules coupled to diverse output domains. Biochemistry. 2003;42:2–10. doi: 10.1021/bi026978l. [DOI] [PubMed] [Google Scholar]
- 3.Losi A. The bacterial counterparts of plant phototropins. Photochem Photobiol Sci. 2004;3:566–574. doi: 10.1039/b400728j. [DOI] [PubMed] [Google Scholar]
- 4.Losi A. Flavin-based photoreceptors in bacteria. In: Silva E, Edwards AM, editors. Flavins - Photochemistry and Photobiology. RCS Publishing; Dorchester, UK: 2006. pp. 217–269. [Google Scholar]
- 5.Losi A, Gärtner W. Bacterial bilin- and flavin-binding photoreceptors. Photochem Photobiol Sci. 2008;7:1168–1178. doi: 10.1039/b802472c. [DOI] [PubMed] [Google Scholar]
- 6.Purcell EB, Crosson S. Photoregulation in prokaryotes. Curr Opin Microbiol. 2008;11:168–178. doi: 10.1016/j.mib.2008.02.014. [DOI] [PubMed] [Google Scholar]
- 7.Swartz TE, Tseng TS, Frederickson MA, Paris G, Comerci DJ, Rajashekara G, Kim JG, Mudgett MB, Splitter GA, Ugalde RA, Goldbaum FA, Briggs WR, Bogomolni RA. Blue-light-activated histidine kinases: two-component sensors in bacteria. Science. 2007;317:1090–1093. doi: 10.1126/science.1144306. [DOI] [PubMed] [Google Scholar]
- 8.Christie JM, Reymond P, Powell GK, Bernasconi P, Raibekas AA, Liscum E, Briggs WR. Arabidopsis NPH1: a flavoprotein with the properties of a photoreceptor for phototropism. Science. 1998;282:1698–1701. doi: 10.1126/science.282.5394.1698. [DOI] [PubMed] [Google Scholar]
- 9.Christie JM, Salomon M, Nozue K, Wada M, Briggs WR. LOV (light, oxygen, or voltage) domains of the blue-light photoreceptor phototropin (nph1): binding sites for the chromophore flavin mononucleotide. Proc Natl Acad Sci USA. 1999;96:8779–8783. doi: 10.1073/pnas.96.15.8779. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Purcell EB, Siegal-Gaskins D, Rawling DC, Fiebig A, Crosson S. A photosensory two-component system regulates bacterial cell attachment. Proc Natl Acad Sci USA. 2007;104:18241–18246. doi: 10.1073/pnas.0705887104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Losi A. Flavin-based Blue-light Photosensors: A Photobiophysics Update. Photochem Photobiol. 2007;83:1283–1300. doi: 10.1111/j.1751-1097.2007.00196.x. [DOI] [PubMed] [Google Scholar]
- 12.Kennis JT, Crosson S, Gauden M, van Stokkum IH, Moffat K, van Grondelle R. Primary reactions of the LOV2 domain of phototropin, a plant blue-light photoreceptor. Biochemistry. 2003;42:3385–3392. doi: 10.1021/bi034022k. [DOI] [PubMed] [Google Scholar]
- 13.Berera R, van Grondelle R, Kennis JT. Ultrafast transient absorption spectroscopy: principles and application to photosynthetic systems. Photosynth Res. 2009;101:105–118. doi: 10.1007/s11120-009-9454-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van Stokkum IHM, Scherer T, Brouwer AM, Verhoeven JW. Conformational dynamics of flexibly and semirigidly bridged electron donor-acceptor systems as revealed by spectrotemporal parametrization of fluorescence. J Phys Chem. 1994;98:852–866. [Google Scholar]
- 15.Sun M, Moore TA, Song PS. Molecular luminescence studies of flavins. I. The excited states of flavins. J Am Chem Soc. 1972;94:1730–1740. doi: 10.1021/ja00760a052. [DOI] [PubMed] [Google Scholar]
- 16.Salzmann S, Martinez-Junza V, Zorn B, Braslavsky SE, Mansurova M, Marian CM, Gärtner W. Photophysical properties of structurally and electronically modified flavin derivatives determined by spectroscopy and theoretical calculations. J Phys Chem A. 2009;113:9365–9375. doi: 10.1021/jp905724b. [DOI] [PubMed] [Google Scholar]
- 17.Salzmann S, Silva-Junior MR, Thiel W, Marian CM. Influence of the LOV domain on low-lying excited states of flavin: a combined quantum-mechanics/molecular-mechanics investigation. J Phys Chem B. 2009;113:15610–15618. doi: 10.1021/jp905599k. [DOI] [PubMed] [Google Scholar]
- 18.Salomon M, Christie JM, Knieb E, Lempert U, Briggs WR. Photochemical and mutational analysis of the FMN-binding domains of the plant blue light receptor, Phototropin. Biochemistry. 2000;39:9401–9410. doi: 10.1021/bi000585+. [DOI] [PubMed] [Google Scholar]
- 19.Plesniak L, Horiuchi Y, Sem D, Meinenger D, Stiles L, Shaffer J, Jennings PA, Adams JA. Probing the nucleotide binding domain of the osmoregulator EnvZ using fluorescent nucleotide derivatives. Biochemistry. 2002;41:13876–13882. doi: 10.1021/bi020331j. [DOI] [PubMed] [Google Scholar]
- 20.Surette MG, Levit M, Liu Y, Lukat G, Ninfa EG, Ninfa A, Stock JB. Dimerization is required for the activity of the protein histidine kinase CheA that mediates signal transduction in bacterial chemotaxis. J Biol Chem. 1996;271:939–945. doi: 10.1074/jbc.271.2.939. [DOI] [PubMed] [Google Scholar]
- 21.Buttani V, Gärtner W, Losi A. NTP-binding properties of the blue-light receptor YtvA and effects of the E105L mutation. Eur Biophys J. 2007;36:831–839. doi: 10.1007/s00249-007-0155-1. [DOI] [PubMed] [Google Scholar]
- 22.Stewart RC, VanBruggen R, Ellefson DD, Wolfe AJ. TNP-ATP and TNP-ADP as probes of the nucleotide binding site of CheA, the histidine protein kinase in the chemotaxis signal transduction pathway of Escherichia coli. Biochemistry. 1998;37:12269–12279. doi: 10.1021/bi980970n. [DOI] [PubMed] [Google Scholar]
- 23.Bilwes AM, Quezada CM, Croal LR, Crane BR, Simon MI. Nucleotide binding by the histidine kinase CheA. Nat Struct Biol. 2001;8:353–360. doi: 10.1038/86243. [DOI] [PubMed] [Google Scholar]
- 24.Alexandre MTA, Arents JC, van Grondelle R, Hellingwerf KJ, Kennis JTM. A base-catalyzed mechanism for dark state recovery in the Avena sativa phototropin-1 LOV2 domain. Biochemistry. 2007;46:3129–3137. doi: 10.1021/bi062074e. [DOI] [PubMed] [Google Scholar]
- 25.Zoltowski BD, Vaccaro B, Crane BR. Mechanism-based tuning of a LOV domain photoreceptor. Nat Chem Biol. 2009;5:827–834. doi: 10.1038/nchembio.210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonetti C, Stierl M, Mathes T, van Stokkum IH, Mullen KM, Cohen-Stuart TA, van Grondelle R, Hegemann P, Kennis JT. The role of key amino acids in the photoactivation pathway of the Synechocystis Slr1694 BLUF domain. Biochemistry. 2009;48:11458–11469. doi: 10.1021/bi901196x. [DOI] [PubMed] [Google Scholar]
- 27.Gauden M, Grinstead JS, Laan W, van Stokkum IH, Avila-Perez M, Toh KC, Boelens R, Kaptein R, van Grondelle R, Hellingwerf KJ, Kennis JT. On the role of aromatic side chains in the photoactivation of BLUF domains. Biochemistry. 2007;46:7405–7415. doi: 10.1021/bi7006433. [DOI] [PubMed] [Google Scholar]
- 28.Gauden M, van Stokkum IH, Key JM, Lührs DC, van Grondelle R, Hegemann P, Kennis JT. Hydrogen-bond switching through a radical pair mechanism in a flavin-binding photoreceptor. Proc Natl Acad Sci USA. 2006;103:10895–10900. doi: 10.1073/pnas.0600720103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gauden M, Yeremenko S, Laan W, van Stokkum IH, Ihalainen JA, van Grondelle R, Hellingwerf KJ, Kennis JT. Photocycle of the flavin-binding photoreceptor AppA, a bacterial transcriptional antirepressor of photosynthesis genes. Biochemistry. 2005;44:3653–3662. doi: 10.1021/bi047359a. [DOI] [PubMed] [Google Scholar]
- 30.Stanley RJ, MacFarlane AW. Ultrafast excited state dynamics of oxidized flavins: Direct observations of quenching by purines. J Phys Chem A. 2000;104:6899–6906. [Google Scholar]
- 31.Toh KC, van Stokkum IH, Hendriks J, Alexandre MT, Arents JC, Perez MA, van Grondelle R, Hellingwerf KJ, Kennis JT. On the signaling mechanism and the absence of photoreversibility in the AppA BLUF domain. Biophys J. 2008;95:312–321. doi: 10.1529/biophysj.107.117788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Swartz TE, Corchnoy SB, Christie JM, Lewis JW, Szundi I, Briggs WR, Bogomolni RA. The photocycle of a flavin-binding domain of the blue light photoreceptor phototropin. J Biol Chem. 2001;276:36493–36500. doi: 10.1074/jbc.M103114200. [DOI] [PubMed] [Google Scholar]
- 33.Losi A, Kottke T, Hegemann P. Recording of blue light-induced energy and volume changes within the wild-type and mutated phot-LOV1 domain from Chlamydomonas reinhardtii. Biophys J. 2004;86:1051–1060. doi: 10.1016/S0006-3495(04)74180-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Losi A, Polverini E, Quest B, Gärtner W. First evidence for phototropin-related blue-light receptors in prokaryotes. Biophys J. 2002;82:2627–2634. doi: 10.1016/S0006-3495(02)75604-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Alexandre MT, Domratcheva T, Bonetti C, van Wilderen LJ, van Grondelle R, Groot ML, Hellingwerf KJ, Kennis JT. Primary reactions of the LOV2 domain of phototropin studied with ultrafast mid-infrared spectroscopy and quantum chemistry. Biophys J. 2009;97:227–237. doi: 10.1016/j.bpj.2009.01.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pfeifer A, Majerus T, Zikihara K, Matsuoka D, Tokutomi S, Heberle J, Kottke T. Time-resolved Fourier transform infrared study on photoadduct formation and secondary structural changes within the phototropin LOV domain. Biophys J. 2009;96:1462–1470. doi: 10.1016/j.bpj.2008.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sato Y, Iwata T, Tokutomi S, Kandori H. Reactive cysteine is protonated in the triplet excited state of the LOV2 domain in Adiantum phytochrome3. J Am Chem Soc. 2005;127:1088–1089. doi: 10.1021/ja0436897. [DOI] [PubMed] [Google Scholar]
- 38.Alexandre MT, van Grondelle R, Hellingwerf KJ, Robert B, Kennis JT. Perturbation of the ground-state electronic structure of FMN by the conserved cysteine in phototropin LOV2 domains. Phys Chem Chem Phys. 2008;10:6693–6702. doi: 10.1039/b810040c. [DOI] [PubMed] [Google Scholar]
- 39.Holzer W, Penzkofer A, Fuhrmann M, Hegemann P. Spectroscopic characterization of flavin mononucleotide bound to the LOV1 domain of Phot1 from Chlamydomonas reinhardtii. Photochem Photobiol. 2002;75:479–487. doi: 10.1562/0031-8655(2002)075<0479:scofmb>2.0.co;2. [DOI] [PubMed] [Google Scholar]
- 40.Bonetti C, Mathes T, van Stokkum IH, Mullen KM, Groot ML, van Grondelle R, Hegemann P, Kennis JT. Hydrogen bond switching among flavin and amino acid side chains in the BLUF photoreceptor observed by ultrafast infrared spectroscopy. Biophys J. 2008;95:4790–4802. doi: 10.1529/biophysj.108.139246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brazard J, Usman A, Lacombat F, Ley C, Martin MM, Plaza P, Mony L, Heijde M, Zabulon G, Bowler C. Spectro-temporal characterization of the photoactivation mechanism of two new oxidized cryptochrome/photolyase photoreceptors. J Am Chem Soc. 2010;132:4935–4945. doi: 10.1021/ja1002372. [DOI] [PubMed] [Google Scholar]
- 42.Dragnea V, Waegele M, Balascuta S, Bauer C, Dragnea B. Time-resolved spectroscopic studies of the AppA blue-light receptor BLUF domain from Rhodobacter sphaeroides. Biochemistry. 2005;44:15978–15985. doi: 10.1021/bi050839x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kennis JT, Groot ML. Ultrafast spectroscopy of biological photoreceptors. Curr Opin Struct Biol. 2007;17:623–630. doi: 10.1016/j.sbi.2007.09.006. [DOI] [PubMed] [Google Scholar]
- 44.Shibata Y, Murai Y, Satoh Y, Fukushima Y, Okajima K, Ikeuchi M, Itoh S. Acceleration of electron-transfer-induced fluorescence quenching upon conversion to the signaling state in the blue-light receptor, TePixD, from Thermosynechococcus elongatus. J Phys Chem B. 2009;113:8192–8198. doi: 10.1021/jp901631b. [DOI] [PubMed] [Google Scholar]
- 45.Shirdel J, Zirak P, Penzkofer A, Breitkreuz H, Wolf E. Absorption and fluorescence spectroscopic characterisation of the circadian blue-light photoreceptor cryptochrome from Drosophila melanogaster (dCry) Chem Phys. 2008;352:35–47. [Google Scholar]
- 46.Kottke T, Heberle J, Hehn D, Dick B, Hegemann P. Phot-LOV1: photocycle of a blue-light receptor domain from the green alga Chlamydomonas reinhardtii. Biophys J. 2003;84:1192–1201. doi: 10.1016/S0006-3495(03)74933-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Alexandre MT, van Grondelle R, Hellingwerf KJ, Kennis JT. Conformational heterogeneity and propagation of structural changes in the LOV2/J-alpha domain from Avena sativa phototropin 1 as recorded by temperature-dependent FTIR spectroscopy. Biophys J. 2009;97:238–247. doi: 10.1016/j.bpj.2009.03.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bednarz T, Losi A, Gärtner W, Hegemann P, Heberle J. Functional variations among LOV domains as revealed by FT-IR difference spectroscopy. Photochem Photobiol Sci. 2004;3:575–579. doi: 10.1039/b400976b. [DOI] [PubMed] [Google Scholar]
- 49.Nozaki D, Iwata T, Ishikawa T, Todo T, Tokutomi S, Kandori H. Role of Gln1029 in the photoactivation processes of the LOV2 domain in Adiantum phytochrome3. Biochemistry. 2004;43:8373–8379. doi: 10.1021/bi0494727. [DOI] [PubMed] [Google Scholar]
- 50.Iwata T, Nozaki D, Tokutomi S, Kandori H. Comparative investigation of the LOV1 and LOV2 domains in Adiantum phytochrome3. Biochemistry. 2005;44:7427–7434. doi: 10.1021/bi047281y. [DOI] [PubMed] [Google Scholar]
- 51.Iwata T, Nozaki D, Tokutomi S, Kagawa T, Wada M, Kandori H. Light-induced structural changes in the LOV2 domain of Adiantum phytochrome3 studied by low-temperature FTIR and UV-visible spectroscopy. Biochemistry. 2003;42:8183–8191. doi: 10.1021/bi0345135. [DOI] [PubMed] [Google Scholar]
- 52.Yamamoto A, Iwata T, Tokutomi S, Kandori H. Role of Phe1010 in light-induced structural changes of the neo1-LOV2 domain of Adiantum. Biochemistry. 2008;47:922–928. doi: 10.1021/bi701851v. [DOI] [PubMed] [Google Scholar]
- 53.Tang Y, Cao Z, Livoti E, Krauss U, Jaeger KE, Gärtner W, Losi A. Interdomain signalling in the blue-light sensing and GTP-binding protein YtvA: a mutagenesis study uncovering the importance of specific protein sites. Photochem Photobiol Sci. 2010;9:47–56. doi: 10.1039/b9pp00075e. [DOI] [PubMed] [Google Scholar]
- 54.Harper SM, Christie JM, Gardner KH. Disruption of the LOV-J alpha helix interaction activates phototropin kinase activity. Biochemistry. 2004;43:16184–16192. doi: 10.1021/bi048092i. [DOI] [PubMed] [Google Scholar]
- 55.Pfeifer A, Mathes T, Lu Y, Hegemann P, Kottke T. Blue light induces global and localized conformational changes in the kinase domain of full-length phototropin. Biochemistry. 2010;49:1024–1032. doi: 10.1021/bi9016044. [DOI] [PubMed] [Google Scholar]