Abstract
Objective
To develop an efficient method for single hematopoietic stem cell (HSC) transplantation for high-level hematopoietic engraftment.
Methods
We combined single cell sorting with short-term culture of putative HSCs. Mouse bone marrow cells that had been highly enriched for HSCs were individually deposited into 96 well culture plate and incubated in the presence of mouse c-kit ligand and either mouse interleukin-11 or human recombinant granulocyte colony-stimulating factor. One week later, the resulting clones of cells were individually transplanted into lethally irradiated recipients. We also carried out time course analysis of proliferation of the individual clones. Finally, we used micromanipulation of the paired progenies of the single cells and studied self-renewal and differentiation potentials of HSCs again in combination with transplantation.
Results
There was a correlation between clone size at day 7 of culture and engraftment at two months post-transplantation. Small clones, such as those consisting of fewer than 15 cells, often showed high-level multi-lineage engraftment while clones consisting of 40 or more cells showed very low levels of engraftment. Daily observation of cell divisions of individual clones revealed that some HSCs are the in G0 state for as long as 1 week despite the presence of permissive cytokines. Studies using micromanipulation of paired progenies documented the ability of an HSC to generate two HSCs as well as asymmetric cell divisions.
Conclusions
Single cell sorting combined with short-term culture of individual putative HSCs provides an efficient method for single HSC transplantation. Analyses of the kinetics of individual HSCs provided direct evidence for HSC cell cycle dormancy, self-renewal and expansion.
Keywords: Hematopoietic stem cells, Cell culture, Cell cycle, Self-renewal, Transplantation
Introduction
Controversies abounded in the stem cell plasticity research area for almost a decade, as amply documented in this issue of Experimental Hematology. Numerous reports expressed diametrically opposing views about the ability of stem cells, including hematopoietic stem cells (HSCs), to cross organ/tissue boundaries. Several years ago, we also became interested in the tissue reconstituting abilities of HSCs and, to obtain unequivocal information on the plasticity of HSCs, planned a strategy for single HSC transplantation. Analysis of the tissue constructing ability of HSCs requires an efficient method for single HSC transplantation that yields high-level, multi-lineage engraftment. When we tested the two published methods for single HSC transplantation [1,2], neither provided the efficiency and the nature of engraftment required for our endeavor in our laboratory, perhaps due to subtle differences in the specific sorting conditions. Rather, our efficiencies of single HSC engraftment by direct transplantation of individual single sorted cells were closer to the results described by Smith et al [3]. We, therefore, devised a method combining single cell deposition and short-term (one week) cell culture of putative HSCs. We reasoned that, because HSCs in the steady state bone marrow (BM) are cell cycle dormant, selection of small clones after one-week culture for transplantation would raise the efficiency of generating the desired mice. Using this method, we were able to consistently generate mice showing high-level, multi-lineage hematopoietic engraftment that could last over a year. Availability of the tissues of mice engrafted with a single HSC from transgenic enhanced green fluorescent protein (EGFP) mice enabled us to uncover the significant tissue reconstituting capability of HSCs as described in this issue. In a series of clonal HSC transplantation studies, we discovered that HSCs can generate the connective tissues including various types of fibroblasts/myofibroblasts [4] and adipocytes [5].
The method described in this paper for enrichment for putative HSCs is the initial method we described and incorporates preliminary enrichment based on use of magnetic beads [6]. To improve the enrichment technique, we have subsequently made a number of modifications such as use of Miltenyi microbeads [7] or sorting for side population (SP) cells [7,8] or tip SP cells [2]. Regardless of the specific cell enrichment strategy for putative HSCs, the adoption of short-term culture to identify the cells in cell cycle dormancy was critical for improving the efficiency of obtaining mice with high-level multi-lineage hematopoietic engraftment. For studies of the mechanisms of HSC differentiation, a genealogical approach based on single stem cell transplantation is of critical importance. The clonal HSC engraftment method described here is also likely to be a useful tool toward this aim.
In our pursuit of higher efficiency of clonal transplantation, we carried out daily examination of the proliferation of the individual clones and made a direct observation of cell cycle dormancy and self-renewal of HSCs. Further, micromanipulation of paired putative HSCs and subsequent transplantation of the resulting clones revealed that a single HSC can generate two HSCs. These observations provide not only the scientific rationale for the described clonal engraftment method but also direct evidence for self renewal, expansion and cell cycle dormancy of HSCs that are further elaborated in the Discussion.
Materials
Phycoerythrin (PE)-conjugated D7 (anti-Ly-6A/E [anti-Sca-1]; rat IgG2a), allophycocyanin (APC)-conjugated 2B8 (anti-CD117 [anti-c-kit]; rat IgG2b), fluorescein isothiocyanate (FITC)-conjugated or biotinylated RAM34 (anti-CD34; rat IgG2a), PE-conjugated or biotinylated RB6-8C5 (anti-Ly-6G and Ly-6C [anti-Gr-1]; rat IgG2b), PE-conjugated or biotinylated RA3-6B2 (anti-CD45R/B220; rat IgG2a), PE-conjugated 30-H12 (anti-CD90.2 [anti-Thy-1.2]; rat IgG2b), biotinylated TER-119 (anti-Ly776 [anti-TER-119]; rat IgG2b), biotinylated GK1.5 (anti-L3T4 [anti-CD4]; rat IgG2b), biotinylated 53-6.7 (anti-Ly-2 [anti-CD8a]; rat IgG2a) and PE-conjugated A20 (anti-CD45.1; mouse IgG2a) were purchased from Pharmingen (San Diego, CA). PE-conjugated M1/70.15 (anti-Mac-1; rat IgG2b) were purchased from Caltag Laboratories (Burlingame, CA). Hybridoma 14.8 (anti-B220; rat IgG2b), M1/70.15.11.5 (anti-Mac-1; rat IgG2b), GK1.5 (anti-CD4; rat IgG2b), and 53.6.72 (anti-CD8; rat IgG2b) were purchased from American Type Culture Collection (Rockville, MD). Recombinant mouse c-kit ligand was kindly provided by Kirin Brewery Co., Tokyo, Japan. Recombinant mouse interleukin-11 and recombinant human granulocyte colony-stimulating factor (G-CSF) were purchased from R&D Systems (Minneapolis, MN).
Single HSC Transplantation
Transgenic EGFP mice (C57BL/6-CD45.2 background) [9] were kindly provided by Dr. M. Okabe (Osaka University, Japan). Ten- to 12-week-old EGFP mice were used as donors. Ten- to 14-week-old C57BL/6- CD45.1 mice (Jackson Laboratories, Bar Harbor, ME) were used as irradiated recipients. These two strains of mice were bred and maintained at the Animal Research Facility of the Veterans Affairs Medical Center. All aspects of the animal research have been conducted in accordance with the guidelines set by the Institutional Animal Care and Use Committee of the Department of Veterans Affairs Medical Center. BM cells were flushed from the femurs and tibiae of EGFP mice and pooled. Cells were washed twice with phosphate-buffered saline (PBS) containing 0.1% bovine serum albumin (BSA, Sigma-Aldrich, St. Louis, MO), made into single-cell suspension by repeated pipetting, and filtered through 40-μm nylon mesh. Mononuclear cells (MNCs) were isolated by density gradient centrifugation using Lympholyte M (Cedarlane Laboratories Limited, Ontario, Canada). BM cells binding to anti-Gr-1, anti-Mac-1, anti-B220, anti-CD4, anti-CD8, and anti-TER-119 were removed by using two depletions with immunomagnetic beads (Dynabeads M-450 coupled to sheep anti-rat IgG; DYNAL, Great Neck, NY) to prepare lineage-negative (Lin−) cells. The resulting Lin− cells were stained with PE-conjugated anti-Sca-1, APC-conjugated anti-c-kit, biotin-conjugated anti-CD34 and mouse lineage panel [biotin-conjugated anti-Gr-1, biotin-conjugated anti-mouse CD45R/B220, biotin-conjugated anti-mouse CD3e and biotin-conjugated anti-mouse TER-119 (Becton Dickinson, San Jose, CA)], followed by streptavidin-conjugated PharRed (Pharmingen). After addition of propidium iodide (PI) at a concentration of 1 μg/mL, the cells were washed twice, resuspended in PBS containing 0.1% BSA, and kept on ice until cell sorting. Cell sorting was performed on FACSVantage (Becton Dickinson, San Jose, CA) with appropriate isotype-matched controls. The Lin− cells were first enriched for Sca-1+ c-kit+ cells. Automated deposition of single Lin− Sca-1+ c-kit+ CD34− cells [1] into Corning round-bottom 96-well plates was then performed with Clone-Cyt (Becton Dickinson). Sixty wells in the center of the 96-well plates were used for deposition. Eighteen hours after deposition, the presence of single cells in individual wells was confirmed on an inverted microscope. The plating efficiency ranged 50 to 80% in each experiment.
Single cell culture was carried out in media containing α-modification of Eagle’s medium (αMEM; ICN Biomedicals, Aurora, OH), 20% fetal bovine serum (FBS; Atlanta Biologicals, Norcross, GA), 1% deionized fraction V BSA, 1 × 10−4 mol/L 2-mercaptoethanol (Sigma-Aldrich) and a combination of 10 ng/mL mouse c-kit ligand and either 100 ng/mL of mouse interleukin-11 or human recombinant G-CSF. Earlier, we have observed that interleukin-11 [10] and G-CSF [11], in synergy with interleukin-3 or c-kit ligand [12], supports proliferation of multi-potential hematopoietic progenitors. The 96-well plates were incubated for 1 week at 37°C in a humidified atmosphere with 5% CO2 in air. The individual clonal population of cells thus generated was transplanted into lethally irradiated C57BL/6-Ly5.1 mice (Jackson Laboratories) together with 500 Lin− Sca-1+ c-kit+ CD34+ BM cells from Ly-5.1 mice. The latter type of cells contained only short-term repopulating cells and acted as radio-protective cells [1]. The recipient mice had been prepared with a single 950-cGy dose of total body irradiation using a 4 × 106 V linear accelerator. Because the majority of HSCs are dormant in cell cycle and begin cell divisions a few days after initiation of cell culture, selection of clones consisting of 15 or fewer cells after one-week incubation significantly enhanced the efficiency of generating mice with high level multi-lineage engraftment. The result of our study correlating the size of individual clones and the levels and types of lineage expression at 2 months post-transplantation is presented in Table 1. The size of the clones varied significantly and a correlation existed between the size of clones and engraftment levels. Seven out of 28 mice transplanted with clones in the smallest (1–10 cells) group showed multi-lineage engraftment, while mice transplanted with clones consisting of 40 or more cells showed very low level of engraftment. The majority of the cells in the smallest clones were undifferentiated blasts cells according to staining with May-Grunwald Giemsa (data not shown). No blast cells were detected in wells containing 20 or more cells and macrophages dominated larger colonies. A representative analysis of a clonally engrafted mouse revealing high-level multi-lineage engraftment is presented in Figure 1.
Table 1.
Day 7 cell counts of clones and their engraftment at 2 months post-transplantation.
| Day 7 Cell Count | Engraftment | Multilineage Expression | % Donor Cells |
|---|---|---|---|
| 1–10 | 7/28 | 7/28 | 57.2 – 86.2 |
| 11–21 | 5/15 | 3/15 | 1.2 – 53.5 |
| 21–40 | 2/28 | 2/28 | 19.2, 30.0 |
| 41–80 | 2/47 | 0/47 | 2.0, 4.0 |
| 80+ | 2/6 | 0/6 | 1.0, 4.0 |
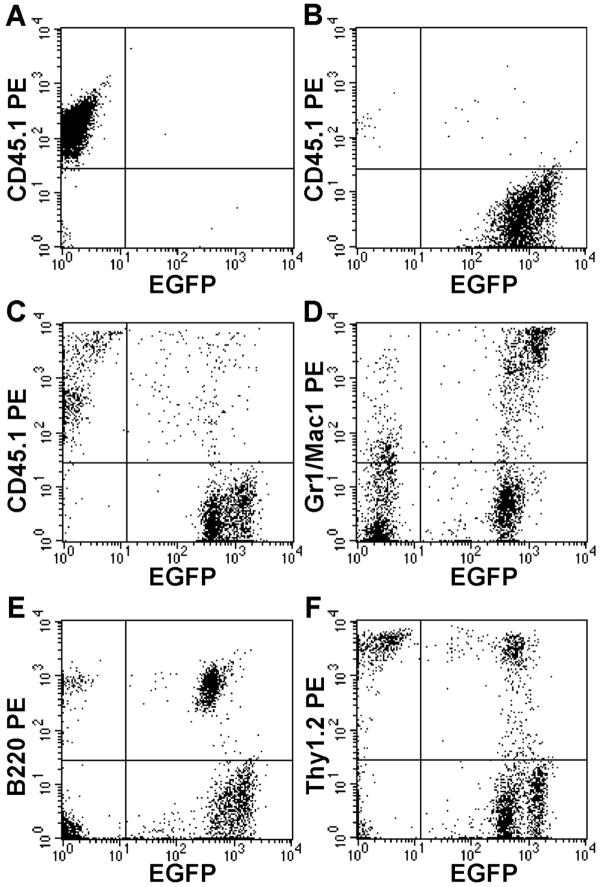
Figure 1.
Multi-lineage engraftment. Representative analysis of peripheral blood nucleated cells from a clonally engrafted mouse 2 months post-transplant shows EGFP+ donor cells representing (C) 70% total cells and 80% of granulocytes-macrophages (D), 85% of B-cells (E) and 50% of T-cells (F); (A–B) controls.
Observation of Cell Cycle Dormancy
The observation that only small clones contain HSCs was consistent with the concept that HSCs in the steady state bone marrow are in the G0 state [13]. In order to document the dormancy of HSCs, we incubated individual putative HSCs in culture with IL-11 and c-kit ligand and daily recorded the size (cell count) of the individual clones by direct microscopic observation of the culture plate. On day 7 of culture, we harvested the content of each well and individually transplanted into lethally irradiated recipients for analysis of the engraftment capability of each clone. In a series of experiments, a total of nineteen mice survived for 2 months or longer post-transplantation. Table 2 presents the day 7 cell count of the individual clones (in a descending order), the day in which doublets were first detected together with the information on the levels and types of the engraftment by individual clones. One cell (Clone #1, Table 2) never divided and, upon transplantation, yielded multi-lineage reconstitution. In agreement with Table 1, high-level, multilineage engraftment was associated with small clones caused by long dormancy. The majority of clones showing multi-lineage engraftment were identified as doublets on day 3. In some clones revealing multi-lineage engraftment, doublets were first detected on as late as days 4, 5 and 7 of culture. These findings strongly suggest that the original HSCs remained in the G0 state for many days despite their exposure to permissive cytokine milieu.
Table 2.
Engraftment characteristics of individual clones*
| Clone | Day 7 Cell Count | Day Doublets Detected | % Donor Cells | Lineage Expression+ |
|---|---|---|---|---|
| 1 | 1 | - | 58.1 | GM, T, B |
| 2 | 2 | 7 | 85.1 | GM, T, B |
| 3 | 3 | 3 | 67.0 | GM, T, B |
| 4 | 3 | 5 | 80.0 | GM, T, B |
| 5 | 6 | 4 | 81.9 | GM, T, B |
| 6 | 6 | 3 | 86.2 | GM, T, B |
| 7 | 8 | 5 | 57.2 | GM, T, B |
| 8 | 9 | 4 | 86.2 | GM, T, B |
| 9 | 11 | 3 | 31.5 | GM, T, B |
| 10 | 12 | 3 | 1.2 | T |
| 11 | 14 | 3 | 53.5 | GM, T, B |
| 12 | 15 | 3 | 4.7 | GM, T |
| 13 | 16 | 3 | 23.4 | GM, T, B |
| 14 | 21 | 2 | 19.2 | GM, T, B |
| 15 | 27 | 3 | 30.0 | GM, T, B |
| 16 | 57 | 3 | 2.0 | T |
| 17 | 80 | 3 | 4.0 | B |
| 18 | 87 | 2 | 4.0 | T, B |
| 19 | 100 | 3 | 1.0 | B |
Analyzed at 2 months post-transplant.
GM: granulocyte/macrophage, B: B-cell, T: T-cell.
Engraftment by Paired Clones
The ability of HSCs to self-renew has been an accepted notion since the inception of the concept of HSCs. Identification of doublets in the time course analysis of the individual clones provided an excellent opportunity to examine this process directly. Accordingly, we next carried out micromanipulation of the doublets for transplantation. As soon as a doublet was detected, the pair of cells were separated and were placed into a new pair of wells using the micromanipulation technique described previously [14,15] and incubated in the same culture media. On day 7 from single cell deposition, the paired sub-clones were individually transplanted into irradiated recipients together with radio-protective cells from Ly-5.1 mice. From a total of seventy-five pairs tested by transplantation, seven pairs showed multi-lineage engraftment in one or both recipients. Detailed information of the individual clones and the engraftment observed from these clones is presented in Table 3. These provide the direct proof for the self-renewal property of HSCs. In two instances (Exps. 1 and 2), both sub-clones of the pair showed multi-lineage engraftment 4–6 months post-transplantation, unequivocally demonstrating that one HSC can generate two HSCs. Analysis of the engraftment seen in Exp. 1 is presented in Figure 2. A total of five experiments documented disparate fates of the pairs. In Exp. 3, both mice survived 4 months, but the engraftment levels were strikingly different. In Exp. 4–7, only one member of the pair showed long-term engraftment and the other died after surviving varying time intervals.
Table 3.
Multi-lineage engraftment from doublets.
| Exp. No. | Day Doublets Detected | Analysis of Engraftment |
||||
|---|---|---|---|---|---|---|
| Mo. Post-Transplant | % Donor Cells* |
|||||
| Total | GM | T | B | |||
| 1 | 3 | 4 | 93 | 88 | 61 | 99 |
| 4 | 87 | 85 | 55 | 99 | ||
| 2 | 4 | 6 | 96 | 88 | 79 | 99 |
| 6 | 89 | 68 | 71 | 99 | ||
| 3 | 4 | 4 | <1.0 | |||
| 4 | 26 | 16 | 20 | 80 | ||
| 4 | 3 | 6 Died (Day 31) | 92 | 75 | 66 | 99 |
| 5 | 3 | 6 Died (Day 48) | 33 | 62 | 20 | 51 |
| 6 | 3 | 2 Died (Day 33) | 65 | 79 | 27 | 85 |
| 7 | 5 | 6 Died (Day 13) | 34 | 30 | 21 | 42 |
Radio-protective cells given in Exps. 1, 2 and 4 and those in Exps. 3, 5 and 6 were 500 and 1,000 Lin− Sca-1+ c-kit+ CD34+ BM cells, respectively. The mouse in Exp. 7 was given 1×105 nucleated BM cells as radio-protective cells.
GM: granulocyte/macrophage, T: T-cell, B: B-cell lineage.
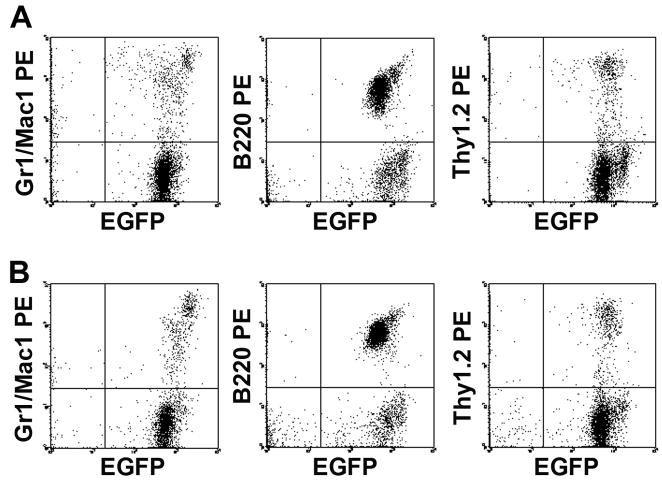
Figure 2.
Flow cytometric analyses at 4 months post-transplantation of hematopoietic engraftment in the pair (A and B) of mice presented as Exp #1 in Table 3. Both show identical, high level engraftment in myeloid (Mac-1/Gr-1-PE), B-cell (B220-PE) and T-cell (Thy-1.2-PE) lineages.
Discussion
The concept of cell cycle dormancy of stem cells was originally proposed by Lajtha [13] almost five decades ago and subsequently was supported by a number of studies using a variety of indirect measurements such as tritiated thymidine “suicide” technique [16], resistance to cell cycle-dependent 5-fluorouracil [17] and serial observation of the growth of individual multipotential blast cell colonies [18]. In the current paper, we used direct analysis of the time of cell divisions of long-term engrafting cells and unequivocally established cell cycle dormancy of HSCs. The majority of putative HSCs remained as single cells for 2 to 6 days in culture despite the presence of cytokines that support proliferation of early progenitors. Our observation is in agreement with the study based on high-resolution video monitoring showing that the duration of the cell cycles of clones containing HSCs were significantly longer than those without HSCs [19]. This observation of cell cycle dormancy of HSCs in culture proved to be of critical value in our studies of differentiation potentials of HSCs. Transplantation of clones consisting of 20 or fewer cells after single cell deposition and one-week culture [6,7,20] led to efficient generation of mice showing high-level multilineage hematopoietic engraftment and greatly facilitated our identification of hitherto unknown potentials of HSCs, i.e. several types of fibroblast/myofibroblasts as summarized in our review [4] and adipocytes [5]. This efficient method for generating mice with clonal HSC engraftment will be useful for documentation of additional tissue reconstituting properties of HSCs.
While self-renewal of HSCs has been an accepted notion for a long time, no definitive experimental approach existed for this concept until the demonstration of hematopoietic reconstitution by single HSCs [1,3]. The paired daughter cell experiment described in this paper enabled direct observation of self-renewal of HSCs. Since at least one of the paired clones derived of a single HSC yielded long-term engraftment, the initial cell division of the original HSCs must have involved self-renewal. Twice in the paired daughter cell experiment, an HSC generated two HSCs, providing direct evidence not only for self-renewal but also expansion of HSCs. Based on serial transplantation, it was calculated that over 8000-fold expansion of HSCs is possible in four transfers [21]. Asymmetric cell division was suggested by a total of five experiments. These results are remarkably similar to those of Ema, et al [22] who also documented expansion of HSCs and asymmetry in engraftment. Together, these observations are consistent with the stochastic mechanism of stem cell self-renewal and commitment of HSCs proposed originally by Till et al [23].
Acknowledgments
This work was supported by the Office of Research and Development, Medical Research Services, Department of Veterans Affairs and grants R01-DK054197 and R01-HL069123 from the National Institutes of Health.
The authors thank Dr. Haiqun Zeng for assistance in FACS sorting, Dr. Amanda C. LaRue for assistance in preparation of this manuscript and the staff members of Department of Radiation Oncology for assistance in irradiation of mice.
Footnotes
Conflict of Interest Disclosure
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Literature Cited
- 1.Osawa M, Hanada K, Hamada H, Nakauchi H. Long-term lymphohematopoietic reconstitution by a single CD34-low/negative hematopoietic stem cell. Science. 1996;273:242–245. doi: 10.1126/science.273.5272.242. [DOI] [PubMed] [Google Scholar]
- 2.Matsuzaki Y, Kinjo K, Mulligan RC, Okano H. Unexpectedly efficient homing capacity of purified murine hematopoietic stem cells. Immunity. 2004;20:87–93. doi: 10.1016/s1074-7613(03)00354-6. [DOI] [PubMed] [Google Scholar]
- 3.Smith LG, Weissman IL, Heimfeld S. Clonal analysis of hematopoietic stem-cell differentiation in vivo. Proc Natl Acad Sci U S A. 1991;88:2788–2792. doi: 10.1073/pnas.88.7.2788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ogawa M, Larue AC, Drake CJ. Hematopoietic origin of fibroblasts/myofibroblasts: its patho-physiological implications. Blood. 2006;108:2893–2896. doi: 10.1182/blood-2006-04-016600. [DOI] [PubMed] [Google Scholar]
- 5.Sera Y, LaRue AC, Moussa O, et al. Hematopoietic stem cell origin of adipocytes. Exp Hematol. 2009;37:1108–1120. doi: 10.1016/j.exphem.2009.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Masuya M, Drake CJ, Fleming PA, et al. Hematopoietic origin of glomerular mesangial cells. Blood. 2003;101:2215–2218. doi: 10.1182/blood-2002-04-1076. [DOI] [PubMed] [Google Scholar]
- 7.Ebihara Y, Masuya M, Larue AC, et al. Hematopoietic origins of fibroblasts: II. In vitro studies of fibroblasts, CFU-F, and fibrocytes. Exp Hematol. 2006;34:219–229. doi: 10.1016/j.exphem.2005.10.008. [DOI] [PubMed] [Google Scholar]
- 8.Goodell MA, Brose K, Paradis G, Conner AS, Mulligan RC. Isolation and functional properties of murine hematopoietic stem cells that are replicating in vivo. J Exp Med. 1996;183:1797–1806. doi: 10.1084/jem.183.4.1797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Okabe M, Ikawa M, Kominami K, Nakanishi T, Nishimune Y. ‘Green mice’ as a source of ubiquitous green cells. FEBS Lett. 1997;407:313–319. doi: 10.1016/s0014-5793(97)00313-x. [DOI] [PubMed] [Google Scholar]
- 10.Musashi M, Yang YC, Paul SR, Clark SC, Sudo T, Ogawa M. Direct and synergistic effects of interleukin 11 on murine hemopoiesis in culture. Proc Natl Acad Sci U S A. 1991;88:765–769. doi: 10.1073/pnas.88.3.765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ikebuchi K, Clark SC, Ihle JN, Souza LM, Ogawa M. Granulocyte colony-stimulating factor enhances interleukin 3-dependent proliferation of multipotential hemopoietic progenitors. Proc Natl Acad Sci U S A. 1988;85:3445–3449. doi: 10.1073/pnas.85.10.3445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tsuji K, Lyman SD, Sudo T, Clark SC, Ogawa M. Enhancement of murine hematopoiesis by synergistic interactions between steel factor (ligand for c-kit), interleukin-11, and other early acting factors in culture. Blood. 1992;79:2855–2860. [PubMed] [Google Scholar]
- 13.Lajtha LG. On the Concept of the Cell Cycle. J Cell Physiol. 1963;62(SUPPL1):143–145. [PubMed] [Google Scholar]
- 14.Suda T, Suda J, Ogawa M. Single-cell origin of mouse hemopoietic colonies expressing multiple lineages in variable combinations. Proc Natl Acad Sci U S A. 1983;80:6689–6693. doi: 10.1073/pnas.80.21.6689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Suda T, Suda J, Ogawa M. Disparate differentiation in mouse hemopoietic colonies derived from paired progenitors. Proc Natl Acad Sci U S A. 1984;81:2520–2524. doi: 10.1073/pnas.81.8.2520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker AJ, McCulloch EA, Siminovitch L, Till JE. The Effect of Differing Demands for Blood Cell Production on DNA Synthesis by Hemopoietic Colony-Forming Cells of Mice. Blood. 1965;26:296–308. [PubMed] [Google Scholar]
- 17.Hodgson GS, Bradley TR. Properties of haematopoietic stem cells surviving 5-fluorouracil treatment: evidence for a pre-CFU-S cell? Nature. 1979;281:381–382. doi: 10.1038/281381a0. [DOI] [PubMed] [Google Scholar]
- 18.Ikebuchi K, Wong GG, Clark SC, Ihle JN, Hirai Y, Ogawa M. Interleukin 6 enhancement of interleukin 3-dependent proliferation of multipotential hemopoietic progenitors. Proc Natl Acad Sci U S A. 1987;84:9035–9039. doi: 10.1073/pnas.84.24.9035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dykstra B, Ramunas J, Kent D, et al. High-resolution video monitoring of hematopoietic stem cells cultured in single-cell arrays identifies new features of self-renewal. Proc Natl Acad Sci U S A. 2006;103:8185–8190. doi: 10.1073/pnas.0602548103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.LaRue AC, Masuya M, Ebihara Y, et al. Hematopoietic origins of fibroblasts: I. In vivo studies of fibroblasts associated with solid tumors. Exp Hematol. 2006;34:208–218. doi: 10.1016/j.exphem.2005.10.009. [DOI] [PubMed] [Google Scholar]
- 21.Iscove NN, Nawa K. Hematopoietic stem cells expand during serial transplantation in vivo without apparent exhaustion. Curr Biol. 1997;7:805–808. doi: 10.1016/s0960-9822(06)00341-1. [DOI] [PubMed] [Google Scholar]
- 22.Ema H, Takano H, Sudo K, Nakauchi H. In vitro self-renewal division of hematopoietic stem cells. J Exp Med. 2000;192:1281–1288. doi: 10.1084/jem.192.9.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Till JE, McCulloch EA, Siminovitch L. A Stochastic Model of Stem Cell Proliferation, Based on the Growth of Spleen Colony-Forming Cells. Proc Natl Acad Sci U S A. 1964;51:29–36. doi: 10.1073/pnas.51.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]