Summary
Protein and protein-lipid interactions, with and within specific areas in the cell membrane, are critical in order to modulate the cell signaling events required to maintain cell functions and viability. Biological bilayers are complex, dynamic platforms, and thus in vivo observations usually need to be preceded by studies on model systems that simplify and discriminate the different factors involved in lipid-protein interactions. Fluorescence microscopy studies using giant unilamellar vesicles (GUVs) as membrane model systems provide a unique methodology to quantify protein binding, interaction and lipid solubilization in artificial bilayers. The large size of lipid domains obtainable on GUVs, together with fluorescence microscopy techniques, provides the possibility to localize and quantify molecular interactions. FCS (Fluorescence Correlation Spectroscopy) can be performed using the GUV model to extract information on mobility and concentration. Two-photon Laurdan GP (Generalized Polarization) reports on local changes in membrane water content (related to membrane fluidity) due to protein binding or lipid removal from a given lipid domain. In this review, we summarize the experimental microscopy methods used to study the interaction of human apolipoprotein A–I (apoA-I) in lipid-free and lipid-bound conformations with bilayers and natural membranes. Results described here help us to understand cholesterol homeostasis, and offer a methodological design suited to different biological systems.
Keywords: rHDL, apoA-I, Fluorescence microscopy, Laurdan GP, GUVs
1. Introduction
Biological bilayers, such as plasma membranes, offer a unique platform in which a dynamic arrangement of lipids regulates the molecular interactions involved in cellular functions. Small changes such as cholesterol content and fluidity could induce growing or coalescence of microdomains that, as a consequence, favor preferential partitioning of proteins involved in cellular signaling events critical to cell viability [1]. In vivo, lipid rafts have been postulated to be aggregates of phospholipids, cholesterol and glycosphingolipids [2–6] associated with important biological processes such as endocytosis, signaling, protein transport, apoptosis, and cytoskeleton organization [7].
To maintain cell viability there exist specific pathways that control membrane composition; one of them is the reverse cholesterol transport (RCT) pathway, the mechanism by which excess cholesterol is removed from peripheral cells [8, 9]. In order to occur efficiently, apolipoproteins as apolipoprotein A–I (apoA-I) must interact with key components at the plasma membrane in a process that is strongly dependent on protein conformation and membrane composition and heterogeneity.
This article reviews the use of fluorescence microscopy methodologies in the analysis of apoA-I and HDL interactions with artificial and natural membranes, as an example of the use of microscopy on this type of system.
2. The RCT cycle: apoA-I and reconstituted HDL particles (rHDL) interaction with membranes
Cholesterol is a key lipid component of biological membranes, and its content and partitioning between raft and more fluid domains can be modified in order to regulate several cellular processes as enzyme activity, signal transduction, opening/closing of channels [10] etc. By the Reverse Cholesterol Transport (RCT), human HDL transports excess cholesterol from peripheral tissues to the liver for excretion into bile and feces [11]. Many steps in the RCT are under investigation as possible therapeutic targets, in order to improve cholesterol efflux and thus reducing cardiovascular risk.
Even though the RCT pathway is not completely understood, it is believed that the first steps involve the interaction of lipid-free or lipid-poor apolipoproteins (for instance apoA-I) with the ATP Binding Cassette transporter ABCA1 at the plasma membrane. This event initiates the efficient removal of cholesterol and phospholipids, promoting the apoA-I rearrangement into disc-shaped particles (preβHDL). This process is unidirectional and requires ATP as an energy source, mediating not only removal of plasma membrane cholesterol but its mobilization from internal pools as well [12]. The preβHDL particles further remove lipids very efficiently from the plasma membrane, probably involving different mechanisms such as acceptors of spontaneously solubilized cholesterol, or specific interaction of apoA-I with other membrane proteins, such as ABCG1 [13]. Lipidated products serve as substrates for plasma Lecithin Chol acyl transferase (LCAT), and other lipid transfer proteins giving rise to circulating, mature HDLs, which are recognized by liver receptors and thus catabolized [14]. Passive diffusional lipid removal by HDL also contributes to ensure the efficiency of cholesterol homeostasis.
In support of the function mentioned for HDL in RCT, a substantial body of evidence agrees that high levels of circulating HDL inversely correlate with the risk of developing cardiovascular disease (CVD) [15]. Nevertheless, the efficiency of HDL to solubilize cholesterol, and/or to interact with key components of the RCT pathway, is strongly dependent on HDL particle stability, size and chemical integrity [16, 17]. HDL lipids and proteins can be subjected to cellular environments that induce oxidation, glycation or other modifications, especially during systemic inflammation, diabetes and chronic renal or coronary heart disease. In these cases, HDL becomes dysfunctional and loses many of its atheroprotective roles such as LCAT activation [18], inhibition of LDL oxidation [19], cholesterol efflux [20], etc. Thus, newer research suggests that the “dynamic flux” of macrophages-derived cholesterol through the RCT pathway is more important to determine risk of CVD than the steady-state concentration [11], and that the quality of the lipoproteins, rather than the quantity, should be considered in order to establish the predictable protective role of HDL [19]
In addition, the efficiency of apoA-I interaction with membranes and lipid removal depends not only on protein conformation but also on the heterogeneity of the membrane, which is believed to play a key role in the regulation of some of the steps involved [21]. Despite their enrichment in cholesterol, lipid rafts do not seem to be essential for lipid efflux mediated by apoA-I, since it was suggested that cholesterol is preferentially acquired from the loosely packed, “non-raft” microdomains [22, 23]. Thus, it was proposed that one of the functions of the ABCA1, should be to induce a redistribution of cholesterol and sphingomyelin from rafts to non-rafts domains in order for them to be accessible to acceptors [21]. These studies support the following hypothesis: “the interaction of apoA-I and HDL with the cellular membrane, and its capacity to remove phospholipids and cholesterol is dependent on the composition and distribution of lipid domains in the plasma membrane”.
By visualizing artificial models, we studied different aspects of these interactions using two-photon fluorescence microscopy. Analogs of the preβHDL containing apoA-I and different molar ratios of lipids have been reconstituted, and interactions with model bilayers and cells were observed and quantified; our results clearly show that protein binding to bilayers is dependent on both protein conformation and membrane composition; we discuss here published and new data and the extrapolation of the in vitro data to in vivo systems with possible implications.
3. What can Microscopy teach us about the apoA-I/HDL –membrane interaction?
a. Models for microscopy studies and fluorescence microscopy techniques
Although most of the lipid models systems available (SUVs, MLVs, LUVs etc) can be used in microscopy studies, the most appropriate systems are planar membranes [24] and Giant Unilamellar Vesicles (GUVs), since they allow spatial resolution and visualization. This work is focused on the use of GUVs as model systems.
GUVs are constructed by the electroformation method published by Angelova et al in 1986 [25] and the methodology published by Pott et al [26, 27] which allows GUVs to be grown at higher salt concentrations required for protein studies in solution.
GUVs can be produced from pure lipids, lipid mixtures, natural lipids extracted from cells, and also from entire membranes containing proteins and lipids. We will refer here to the methodologies and techniques used to study the influence of lipid segregation in the lipid-protein interaction, therefore most of the mixtures used show, under certain conditions of temperature and lipid composition, phase separation visible under the microscope.
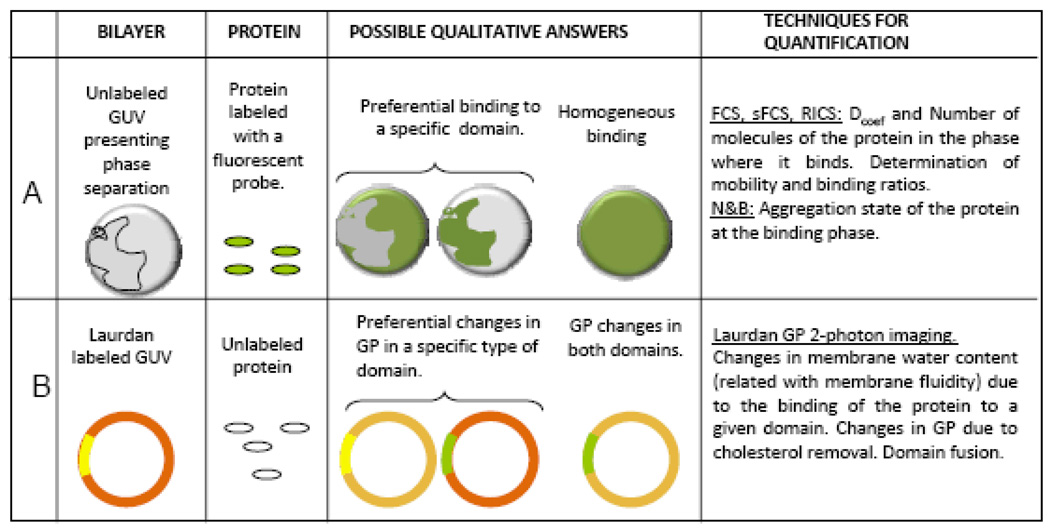
Studies of lipid-protein interaction using systems that show separation of macro domains, address the hypothesis that in vivo membranes are heterogeneous and present segregation of lipid domains. Studies on cells indicate that the major portion of the plasma membrane is in the liquid-ordered state [28]; instead, the raft theory states that heterogeneity in the membrane is due to the existence of areas with ordered packing (lo-phase) in a more fluid (ld) continuous phase. This idea comes from the discovery that glycosphingolipids cluster in the Golgi apparatus before being sorted to the apical surface of polarized epithelial cells [2, 29] and experiments showing that glycosphingolipid clusters tend to be insoluble in Triton X-100 at 4°C, forming detergent resistant membranes (DRM) rich in both cholesterol and glycosylphosphatidyl inositol (GPI)-anchored proteins [30]. Rafts are theoretical structures postulated to exist in cellular membranes of similar compositions and phase (lo) of DRMs in equilibrium with the rest of the membrane in a more fluid phase (ld). Based on the composition of the DRMs, synthetic mixtures called raft-like mixtures are used to reconstitute model membrane systems where lipid phases co-exist [31–35], in order to study the influence of phase segregation on protein-lipid interaction. Figure 1 shows a diagram describing the methodologies and techniques used in these types of studies.
Figure 1.
Schematic representation for the experimental protocols used in the studies of lipid-protein interactions in Giant Unilamellar Vesicle system presenting phase co-existence. [A] Binding experiments are performed adding labeled protein to the chamber containing the GUVs. After the incubation period the protein may preferentially bind to one or both types of domains. Different fluctuation techniques (described in the text) can be used to quantify the binding. Circular objects represent the top view of the GUV presenting lipid domains. [B] Laurdan GP imaging is used to detect changes in water content (related to membrane fluidity) in the lipid bilayer due to the interaction with proteins. After the interaction, the GP changes may occur in the two phases or in one of them, both the GP value and the size of the domain can be quantified and give information of the interaction. The ring shapes represent the GP image of a GUV taken at the equatorial plane and presenting domain separation (two different colors represent two macro-domains with different GP value), this configuration is preferred for GP quantification because all the Laurdan molecules (located parallel to the lipids) are excited by the circular polarized light usually used for excitation.
[A] Protein binding (Figure 1 A): GUVs presenting macro-domain separation can be used to assess the binding of a protein to a specific macro-domain. The protein under study (labeled with a fluorescent molecule) is added to the chamber containing the GUVs and, after the appropriate incubation period, measurements assessing for binding are performed. Several techniques are suited to extract information about binding: the first qualitative answer is given simply by fluorescence intensity measurements: in this case the intensity image can show homogeneous or domain-specific binding and the images will show a sphere (the GUV) totally fluorescent or fluorescent in some specific areas (see Figure 1A, Possible Qualitative Answers). To assess the lipid phase where the protein preferentially binds, a membrane probe such as Laurdan or Prodan can be added to the same GUV after the binding has occurred or, if a dual emission channel system is available, the lipid phase is measured in one channel and the binding is measured in the other channel using fluorescent dyes with good emission spectral separation and the appropriate filters [36–38].
In addition, fluctuation techniques can be used to quantify the binding affinity, including Fluorescent Correlation Spectroscopy (FCS), scanning FCS (sFCS), Raster Image Correlation Spectroscopy (RICS) and Number and Brightness analysis (N&B). FCS [39–43] measures the fluctuations in fluorescent intensity in a small volume generated by a pinhole in confocal microscopes or by the 2-photon excitation process [44–46]. Fluctuations due to diffusion of the molecules in and out of the small volume are detected as a function of time and the Autocorrelation Function (ACF) analysis gives the diffusion coefficient (Dcoef) and the average number of particles in the observation volume (Ň). The fluctuation can also be analyzed using PCH [47], where the probability of detecting photons per sampling time is calculated and thus the number of molecules in the observation volume (N) together with its molecular brightness (β) are extracted. FCS measurements on the co-existing macro-domains of the GUVs can report the mobility (Dcoef) and the number of molecules of bound and free protein allowing the estimation of a the dissociation constant. Scanning FCS [48] gives the same information as point FCS with the big advantage of allowing sampling of several point at the same time, and permits one to discriminate fluctuations coming from the movement of the GUV itself by cross-correlation analysis of points separated in space [49–51]. RICS [52] provides the same dynamic information as FCS as well as information on the spatial correlation between points along the scanning path. As the laser performs the raster scanning movement, it creates a space–time matrix of pixels within the image. The temporal and spatial sampling of the laser beam during the raster scanning is known, that is: the time the laser samples each pixel (pixel dwell time); the time between scan lines (line time) and the time between images (frame time). Therefore, the images contain information on the microsecond time scale for pixels along the horizontal scanning axis, millisecond time scale along the vertical scanning axis or between scan lines, and on the sub-second to second or longer time scale between successive images. This technique can provide maps of the diffusion coefficient and the number of molecules of the bound and free species. Number and Brightness (N&B) analysis is based in moment analysis, for each pixel in an image stack [53, 54]. From the average intensity in each pixel and the variance of the intensity distribution, the number and brightness (aggregation) of mobile particles is determined, thereby providing a new contrast mechanism in the images based on a molecular property. If the protein of interest aggregates when it binds to the membrane, this technique can accurately detect oligomerization [55].
Furthermore, all these techniques performed in a two-channel emission setup allow one to correlate protein binding and membrane phase and to ascertain the protein binding stoichiometry, etc. [56–58]. If the GUVs have been constructed with a fluorescent protein integrated to the bilayer, the same techniques will provide valuable information on motility and aggregation state of the protein immersed in the membrane [27, 59].
[B] Changes in membrane phases (Figure 1B): GUVs presenting lipid segregation can also be a good model to study the properties of lipid bilayers during protein interaction. Valuable information is given by the fluorescent dye Laurdan, used as a membrane probe because of its large excited state dipole moment, which results in its ability to report the extent of water penetration into the bilayer surface due to the dipolar relaxation effect [60]. Water penetration has been correlated with lipid packing and membrane fluidity [61, 62]. The emission spectrum of Laurdan is centered at 440 nm if the membrane is in the gel phase and at 490 nm when it is solubilized in the liquid crystalline phase. The GP value (Generalized Polarization), defined as the difference of intensities at 440 minus 490 nm divided by the sum, measures the emission shift. To calculate the GP value in a two-photon microscope (to avoid photo-bleaching) an excitation wavelength of 780 nm and a two-channel system with the corresponding filters on the emission is used [31, 63].
In a GUV presenting phase separation, the emission of Laurdan will depend on the water content of each phase: the emission from a fluid-ordered phase is blue shifted, as compared with the emission from the fluid-disordered domain. In this way spatial identification of coexisting phases can be realized directly from the GP image. Thus, Laurdan GP imaging [31, 63, 64] can quantify and localize changes in the packing of the lipids in the bilayer due to protein binding, cholesterol removal, etc. GP images are usually taken in the equatorial plane of the GUV [31, 63]; in this configuration all the Laurdan molecules located parallel to the phospholipids can be excited with a circular polarized light (Figure 1B). Under this condition the GP images can be processed and the macro-domains separated and studied independently. The areas of each macro-domain can be quantified to study specific morphological changes associated with the interaction. These methodologies have been used for the study of several proteins [36–38, 55, 65–67] and the apoA-I/HDL system [68–72].
b. ApoA-I interaction with heterogeneous membranes
The idea that the interaction of apoA-I with membranes is favored by bilayer irregularities was suggested in 1978 by Pownall et al [73] following the interaction of apoA-I with multilamellar DMPC liposomes by light scattering. The data showed a decrease in light scattering of the DMPC MLVs after addition of apoA-I (interpreted as lipid solubilization) only at 24.8°C; the authors concluded that this behavior arises from the formation of a “structural determinant” associated with coexisting gel and liquid crystalline phases, and this determinant was necessary for the efficient interaction. Later in 2007, two-photon microscopy studies done by our group showed that this “determinant” indeed exists at the phase transition of DMPC [63]. Figure 2A shows the top view intensity image of a DMPC GUV at 24.5°C (similar image of the already published [63]). Close to the transition temperature of DMPC, the lipids molecules in gel and liquid phase co-exists and there is a large area of defects. The particular bilayer state existing at the transition temperature is characterized as having high stress, which induces volume and shape changes [74] and high permeation to small molecules [75]. This state could allow deeper penetration of some of the amphipatic helices of apoA-I, inducing a conformational rearrangement in the protein in which the hydrophobic faces of the helices come into close contact with the acyl chains of the lipids favoring their removal.
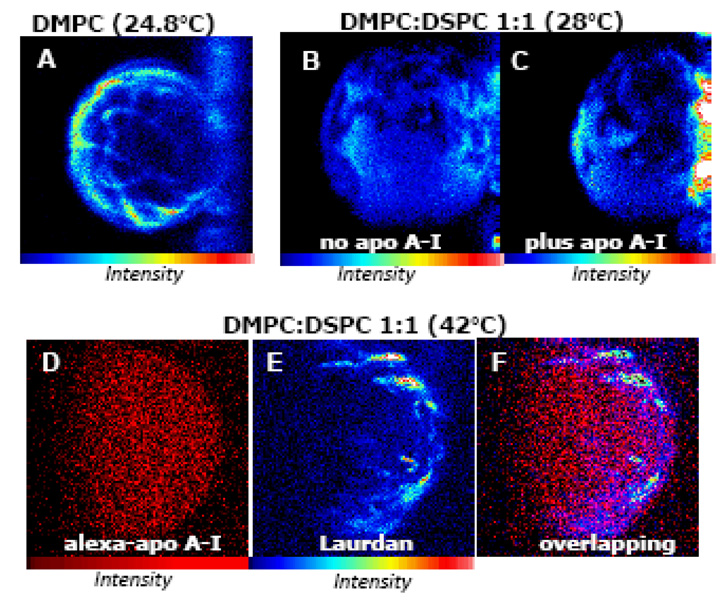
Figure 2. ApoA-I interaction with heterogeneous membranes.
[A] Membrane heterogeneities existing at the transition temperature [63]: Laurdan intensity image (top view) of a DMPC GUV at 24.5°C attached to the platinum wire (structure on the right). [B and C] Lipid solubilization from DMPC:DSPC 1:1 GUV by apoA-I occurs at 28°C [68] as evidenced by the decrease in volume of the GUV as comparing the Laurdan intensity image before (B) and after (C) incubation with apoA-I for 2 h. [D, E and F] ApoA-I binding to heterogeneous bilayers: a GUV made of DMPC:DSPC (0.35:0.65 molar ratio) at 42°C. A target GUV was chosen using the CCD camera and the control image taken before the addition of the labeled protein showed background signal ~150 total counts (image not shown). After 2 hours incubation with Alexa 488-apoA-I the intensity image (D) showed ~ 130,000 total counts defining the shape of the GUV, which indicates protein binding. Next, Laurdan was added to the chamber (final concentration of 0.76 µM) and the same target GUV was imaged (image E) revealing the heterogeneities existing on the membrane (total counts increased 10 times with respect to image D). Figure F corresponds to the overlapping of images D and E showing that binding of the protein does not correlate with the membrane heterogeneities. No changes in the size of the GUV occurred at this temperature after adding apoA-I. Experiments were performed in a two photon microscope previously described [68, 70]. For both probes (Alexa-488 and Laurdan) excitation wavelength of 780 nm was used and the fluorescence emission was observed through a broad band-pass filter from 350 to 600 nm (BG39 filter, Chroma Technology, Brattleboro, VT). Blue-red color scale is used for Laurdan intensity images and red for Alexa 488.
In the interaction of apoA-I with binary mixtures, the efficiency of lipid removal is also dependent on a special membrane condition. Scattering measurements for MLVs of DMPC:DSPC also show a particular temperature at which lipids are solubilized (28°C) [76]. In this regard, we have used DMPC:DSPC 1:1 molar ratio GUVs, and demonstrated that domain segregation is not the important factor for lipid solubilization by apoA-I . In these liposomes, domain coexistence is present from ~ 50 to 24°C but effective lipid solubilization only occurs at ~28°C [68]; this fact is detected by a decrease in the size of the GUV after addition of apoA-I (Figure 2B and 2C) as a consequence of solubilization of both lipids from the binary mixture, but with higher preference for the more fluid component [68].
The preferential binding of apoA-I to areas of different lipid packing was assessed using Alexa-488 labeled apoA-I (alexa488-apoA-I) and GUVs of DMPC:DSPC (0.35:0.65 molar ratio) at 42°C (temperature where phase coexistence exists). A target, unlabeled GUV was localized using a CCD camera and alexa488-apoA-I was added to the chamber to a final protein concentration of 10 µg/ml. After 2h incubation the intensity image (2D) shows the GUV defined by the brightness of labeled apoA-I, indicating the binding of the protein to the bilayer. Next, Laurdan was added to the chamber and the same GUV was imaged. The Laurdan intensity image (Figure 2E) evidences a membrane topology similar to the “structural determinant” associated with coexisting gel and liquid crystalline phases on DMPC (Figure 2A); in this mixture the irregularities can be attributed to the coexistence of the DMPC and DSPC molecules which at 42°C are over and below their transition temperatures respectively (Tm DMPC = 24°C, Tm DSPC = 55°C) (Figure 2E). Figure 2F shows the overlapping of image 2D and 2E evidencing the fact that apoA-I binding is not dependent on the lipid packing.
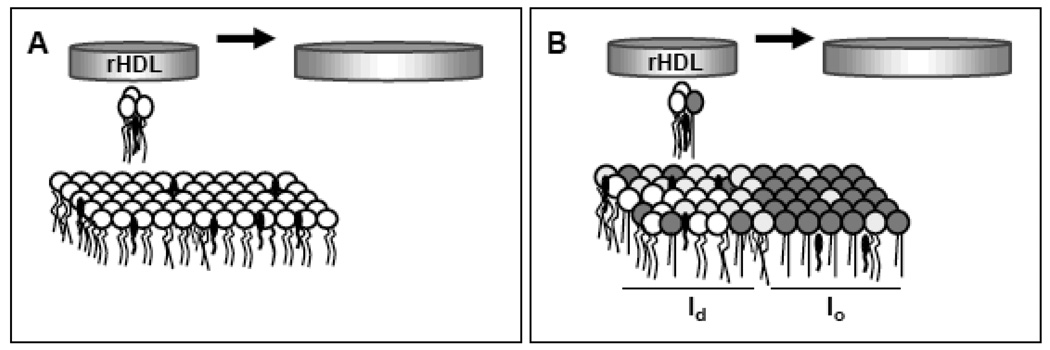
Similar results of homogeneous binding were obtained on POPC:SM:FC GUVs presenting phase separation [72]. Figure 3 shows a diagram with the main conclusions of our published studies on the interaction of lipid-free apoA-I and membranes in vitro [63, 68, 70, 72]: apoA-I is able to bind to the lipids in an homogeneous lipid phase (Figure 3A, [64]) but effective lipid solubilization occurs only under particular conditions: either at the transition temperature of a pure lipid or where the membrane is composed of small fluid domains nucleated within a continuous gel phase (a few degrees above the melting temperature of the more-liquid component in a binary mixture); under these conditions interfacial packing defects are maximal (Figure 3B) [68].
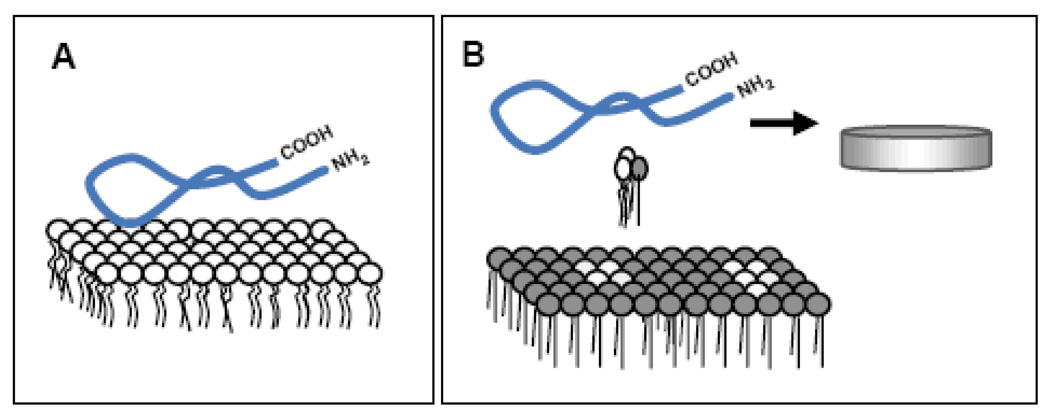
Figure 3.
Scheme of the different interactions of lipid-free ApoA-I with membranes analyzed by our technical approach: [A] Lipid-free apoA-I interaction with homogeneous phospholipid bilayers results in high binding but non efficient lipid removal [70]. [B] Efficient lipid solubilization occurs from bilayers having high interfacial packing defects, with small fluid domains nucleated within a continuous gel phase [68]. In the diagram the blue ribbon represents the lipid-free apoA-I, circles with two legs represent the phospholipids in disordered state (white) and ordered state (gray),
c. Cholesterol efflux and Laurdan GP
ApoA-I constitutes ~ 70% of the protein moiety of HDL and its conformation is highly flexible in order to rearrange in response to changes in HDL lipids during catabolism. Human apoA-I contains a series of highly homologous 11- and 22-residue amphipathic α-helices. These amphipathic α-helices are defined by the arrangement of positively and negatively charged amino acids on the helical polar face as class A and Y. Helix 1 is a Class A helix together with helices 2, and 5–8, which have positively charged amino acids surrounding the non polar face with negative residues clustered at the center of the polar face. Helix 3–4 and 10 are class Y helices, organized similarly but having a positive charge disrupting the negative cluster [77]. Mutants lacking helix 10 show lower rates of cholesterol efflux and recently Davidson et al [78] have shown that helix 10 is critical for promoting optimal cholesterol efflux via the ABCA1 pathway from RAW macrophages. Deletion of helix 1 also reduced lipid binding affinity [79].
As circulating HDL are composed of heterogeneous and dynamic group of particles with different sizes, shapes and compositions, models have been created in order to understand the conformational arrangement of apoA-I involved in lipid homeostasis. Thus, homogeneous lipoprotein complexes homologous to the preβ-HDL particles observed as the first intermediates of cholesterol solubilization [80] have been reconstituted (rHDL) with apoA-I and different amounts of cholesterol and phospholipids. These particles can be obtained by the sodium cholate method [70], or by spontaneous solubilization of lipids at the transition temperature.
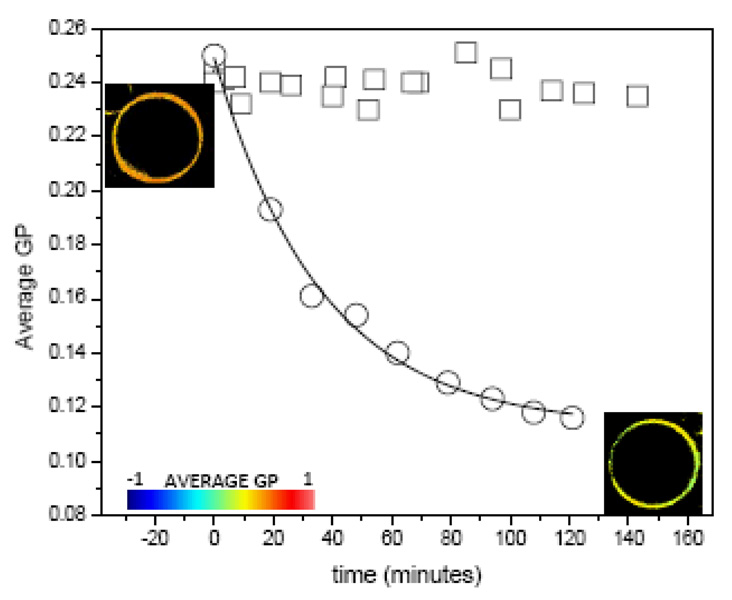
The organization of apoA-I in the rHDL has been reviewed before and three models have been proposed: the picket fence [81], the belt [82] and the hairpin models [79]. Förster Resonance Energy Transfer (FRET) data from our previous studies strongly support the hairpin arrangement of apoA-I in these HDL complexes [83]. The three models agree that helices 10 and 1 are directly involved in the interaction of the HDL particles with the lipids in the bilayer. In order to determine the importance of helix 1 for protein interaction and cholesterol solubilization from membranes, we used an apoA-I mutant, H4@H1, having the putative high-lipid affinity helix 1 replaced with a second copy of a lower lipid-affinity helix 4 [84] but which still has helix 10 in its normal location at the C terminus. The lipid-free mutant exhibited cholesterol efflux capabilities similar to WT on RAW macrophages, however, it exhibited markedly reduced (50%) lipid association characteristics in the DMPC clearance experiments. Reported changes in GP values of GUVs of POPC-32%cholesterol incubated with 78Å rHDL containing Wt apoA-I, demonstrated that rHDL efficiently remove cholesterol from the lipid bilayer [69]. FCS data on the same system showed that the 78Å rHDL interact with the bilayer as independent units (constant number of particles) and that their size increase (Dcoef decrease) to 96Å, results also consistent with lipid removal [70, 85]. Figure 4 shows the comparison of the GP changes observed on POPC-32% GUV incubated with equal concentration of rHDL containing the WT apoA-I and the H4@H1 mutant (with altered helix 1). Changes in the GP values during the incubation with the wild type protein rHDL are consistent with published data [69], however no changes in GP were observed during 2 hours incubation of POPC-32% FC GUVs with the mutant rHDL (Figure 4). The lipid-free H4@H1 exhibited cholesterol efflux capabilities similar to Wt from RAW macrophages, however as expected, it exhibited markedly reduced (~50%) lipid association characteristics in the DMPC clearance experiments. It could be possible then that lipid association is important in order to facilitate passive cholesterol diffusion through the aqueous medium.
Figure 4.
Kinetics of cholesterol removal by particles of 96Å of wild type apoA-I rHDL (circles) [69] and of the mutant H1@H4 (squares) at 36.5°C from POPC-30% cholesterol GUVs. Solid line for the opened circles symbols corresponds to a first order exponential decay fit with a time constant of 39.3+/−0.1. GP images of the GUV at the beginning (top left) and end (bottom right) of the incubation time are also shown and colored according to the GP scale going from −1 to 1.
d- HDL interaction with membranes having co-existing phases
In the process of cholesterol removal by HDL, the extent and direction of the net cholesterol movement will depend on the ratio of efflux to influx and is determined by the properties of the acceptor (the HDL particle) and the donor (the membrane) [86]. Factors reducing the packing density of lipid molecules have been proposed to enhance the rate of cholesterol transfer [87]. Results from “raft-like” mixtures show that HDL particles preferentially remove cholesterol from lipid domains characterized as liquid disordered (ld) [69]. Figure 5 shows the results obtained using Laurdan GP imaging to study the interaction of DOPC:DPPC:FC (1:1:1 molar ratio) and rHDL at 25°C. At this temperature the phase diagram indicates that liquid ordered domains (lo) (rich in DPPC) coexist with ld domains (rich in DOPC), with cholesterol partitioning between the two phases [88]. Laurdan GP can differentiate lo from ld phases and Figure 5B shows them in different colors (lo in orange and ld in light green); rHDL were added to the GUV at 25°C and the GP value of each phase was measured before (Figure 5C) and after (Figure 5D) 1 hour incubation period. The GP value of the ld phase decreased from 0.1 to 0.02, indicating cholesterol removal, while the lo remained unchanged [69]. Contrary to lipid-free apoA-I that requires a high surface of domains coexistence to remove lipids [68], lipid solubilization towards rHDL depends on the accessibility for cholesterol (defined by the particular characteristic of the lipids present at the bilayer). The diagram in Figure 6 shows the possible underlying process of the interaction of rHDL with membranes containing cholesterol. In the POPC:FC (32%) (Figure 6A) the un-saturations present in the acyl chains of POPC introduce kinks that limit the ability of cholesterol to order, to mix homogeneously, to reduce interfacial elasticity [89], and to lower the in-plane elasticity [90]. Cholesterol locates more superficially and thus desorbs better from the membrane to the aqueous interface and transfers to the rHDL. A similar situation is observed in the co-existing ld-phase in the DOPC:DPPC:FC (1:1:1) system. The ld-phase is enriched in the unsaturated phospholipid (DOPC) [88] and the kinks on the acyl chains facilitate the transfer of cholesterol and phospholipids to the rHDL (Figure 6B). On the other hand, the co-existing lo-phase is formed mainly by DPPC, that contains a chain structural motif similar to natural sphingolipids [89]. It has been suggested that cholesterol molecules can locate in this type of bilayer with part of their tail in the adjacent leaflet [91] allowing formation of tail-to-tail dimers [92], and then impeding its desorption (Figure 6B).
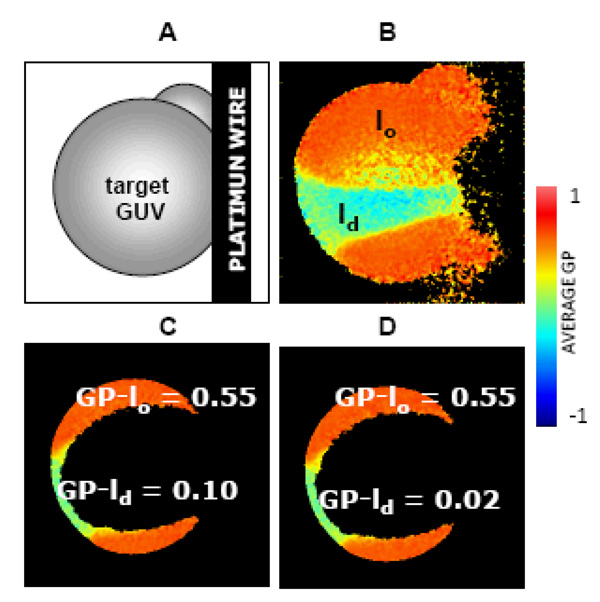
Figure 5. Interaction of rHDL with phase co-existing GUV.
[A] Diagram of the target GUV attached to the Pt wire, [B] GP image of the target GUV (top view) of DOPC:DPPC:FC (1:1:1 molar ratio) at 24.8°C presenting lo (orange) and ld (light green) separated phases, a small GUV on the top right can also be seen on the top right and the discontinuity on the right shows the place of attachment of the GUV to the platinum wire. For GP measurements, a GP image is taken on the center of the same target GUV and the GP values for each phase are showing in the figure at time zero [C] and after 60 minutes incubation with 10ug/ml 96Å rHDL [D] [69]. False color representation according to the palette with GP values going from −1 to 1 is used.
Figure 6.
Scheme of the interaction of lipid-bound apoA-I (rHDL) with membranes analyzed by our technical approach [A] rHDL interact with, and solubilize phospholipids and cholesterol from homogeneous bilayers as independent units and growing in size according to FCS measurements insert reference. [B] If cholesterol is distributed in two phases with different packing (lo/ld), rHDL preferentially remove phospholipids and cholesterol from the more disordered (ld) domain [69]. As in Figure 3, circles with two legs represent the phospholipids in disordered state (white) and ordered state (gray). Black elliptical shape with one leg represents the cholesterol molecules.
e. HDL interaction with cells membranes
Visualization of protein interactions with lipid domains in vivo is difficult due to the fast dynamics of lipid arrangement in membranes and the lack of methodologies to visualize membrane heterogeneity [5]. Thus, the most common methodologies used to study this type of interactions are the isolation of detergent resistant membranes (DRMs) [10] and the use of fluorescent dyes on fixed cells [28]. In the Keystone Symposium on Lipid Rafts and Cell Function in 2006 [93] rafts were defined as ”small (10–200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched domains”. Laurdan GP imaging have been used in the search for cell membrane heterogeneity in erythrocytes [94, 95] and macrophages [96].
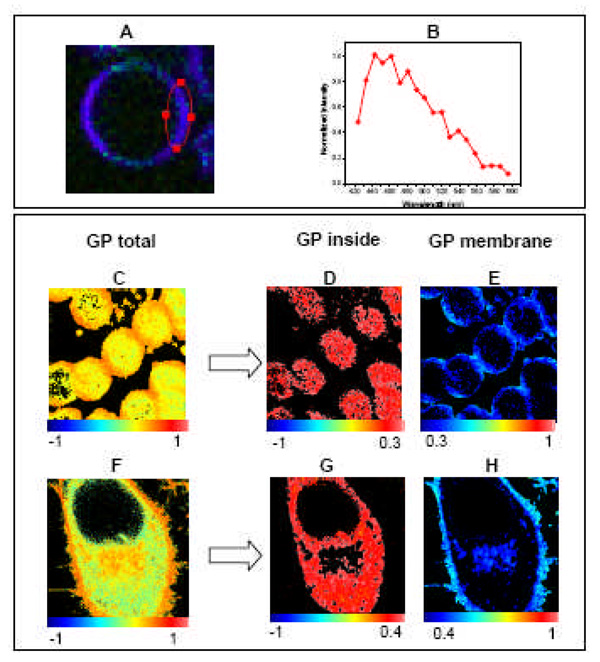
Erythrocytes are well-studied systems. Human erythrocytes were labeled with 1µM Laurdan and Figure 7A shows the spectral image corresponding to the overlap of 19 images taken simultaneously at different emission wavelengths while exciting Laurdan at 780 nm. Figure 7B shows the spectrum corresponding to Laurdan inside the erythrocyte membrane. The spectrum centered at 440 nm indicates that membrane lipids are in an ordered phase in this system. In a microscope, a GP image (Figure 7C) is obtained using two photon excitation at 780 nm, two interference filters in the emission centered at 440 and 490 nm, and applying the GP formula pixel by pixel [31, 63].
Figure 7. GP imaging in alive cells.
Top panel: Human red blood cells were labeled with 1 uM Laurdan for 15 minutes and imaged at 37°C. [A] Laurdan spectral image corresponding to the overlapping of 19 images taken simultaneously at different emission wavelength while exciting Laurdan at 780 nm (taken in a Zeiss Meta 710). [B] Normalized emission spectrum of Laurdan in the erythrocyte membrane taken from the area encircled in red in image A. Bottom panel: Using SimFCS, the pixels in the GP image can be separated in those located inside the cells (low GP) and the ones corresponding to the plasma membrane (high GP). Analysis for Human erythrocytes [C, D and E] and Hela cells [F, G and D] are presented.
The GP image of human erythrocytes in Figure 7C, shows areas in the cells with different GP values. Using the SimFCS program (www.LFD.uci.edu), the pixels with low GP values (−1<GP<0.3) corresponding to the interior of the cells (Figure 7D) can be separated from the pixels with high GP values (0.3 <GP<1) corresponding to the plasma membrane (Figure 7E). The GP image corresponding to the plasma membrane (Figure 7E), shows no visible macro-domain separation in agreement with previous results shown by the Bell’s group [94, 95]. Figure 7F shows the GP image of a nucleated cell (Hela cell) labeled with 1µM Laurdan. The separation of the pixels corresponding to the cytosol (Figure 7G) and the ones from the membrane (Figure 7H) also indicate the lack of macro-domain separation in the membrane of Hela cells (Figure 7H). Interestingly, the nuclear membrane cannot be distinguished from the rest of the cytosol indicating that the nuclear membrane is more fluid than the plasma membrane.
It seems to be an apparent contradiction between the data presented here reporting the lack of macro-domain separation in cell membranes and data reported by Gaus et al [96]. These authors reported the 3D-projection GP image of a macrophage showing extended areas with high GP values localized in knob-like membrane protrusions. The authors discussed that these areas could be either large rafts exclusive of this cell type, or areas enriched in small rafts with high GP. Our results in several nucleated cells show similar macroscopic regions of different GP. Most cells present evaginations, filopodia and, protrusions. In the 3D projection, these areas appear to have consistently higher GPs than in the rest of the cell. When we carefully separate the protrusions from the rest of the plasma membrane we found that pixels belonging to the non-protrusive part of the plasma membrane are relatively homogeneous in GP value without evident separation into macroscopic domains. The fact that macro-domains are not visible by GP imaging in the relatively flat part of the plasma membrane does not exclude the existence of domains smaller than the pixel size. If these domains are also highly mobile [93], other techniques capable of capturing the fast dynamics will be needed to visualize them [5, 50].
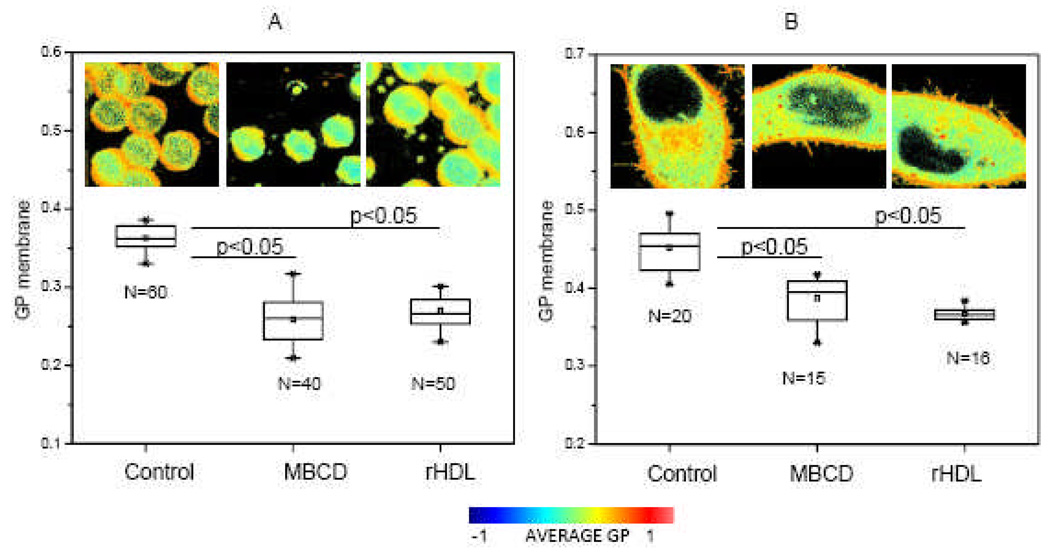
We report here the use of these two cellular systems (erythrocytes and Hela cells) and the GP imaging technique to study cholesterol removal from the plasma membrane by rHDL and methyl beta cyclo dextrines (MβCD). When human erythrocytes are incubated with 3.5 mM MβCD for 2 hours at 37°C approximately 90% of the total cholesterol [97] is removed, inducing a decrease in GP of 0.1 units (Figure 8A). The same decrease in GP is obtained when erythrocytes are incubated with 300 µg/ml rHDL for 2 hours at 37°C. As seen in the images in Figure 8A, we found that MβCD induces changes in shape of the cells (Figure 8A, second image from the left) as previously reported [94]. Instead, the use of rHDL as cholesterol acceptor was less damaging for the cells and changes in shape were not observed (Figure 8A, third image from the left). Incubation of HeLa cells with 10 mM MβCD for 60 min [98] induces the same changes in GP as incubation with 300 µg/ml rHDL for 2h at 37°C. Both types of cells shown in Figure 8 lack (erythrocytes) or have minor (Hela cells) expression of the ABCA1 transporter. Studies done by our group [99] using CHO cells (which express ABCA1), show a similar behavior than the one described for erythrocytes and HeLa. Nevertheless, as cells were not previously loaded with cholesterol, it is possible that ABCA1 was not highly activated under our experimental conditions. Interestingly, working with HeLa overexpressing ABCA1, Zarubica et al [100], reported that over expression of functional ABCA1 triggers not only apoA-I but MβCD- mediated cholesterol efflux. They attributed this fact to a participation of ABCA1 in the redistribution of membrane associated cholesterol into pools readily accessible to external acceptors.
Figure 8. Effect of cholesterol acceptors on the GP membrane of alive cells.
[A] GP membrane values for human erythrocytes in buffer (Control), incubated with MβCD 3.5mM for 120 min, and incubated with 96Å rHDL 300 µg/ml for 2h. Buffer used: 10mM Phosphate, 147 mM NaCl, 3 mM KCl, pH 7.4. [B] GP membrane values for HeLa cells in culture media (Control), incubated with MβCD 10 mM for 60 min and incubated with 96Å rHDL 300µg/ml for 2h. Temperature for all the experiments was 37°C and N corresponds to the number of cells analyzed. ANOVA test was performed to compare the control data and the data after incubation with the cholesterol acceptors. Results show a significant difference with p<0.05. Images presented correspond to the complete GP image. Palette shows the color scale used for all the images.
The fact that GP imaging of the plasma membrane does not show macro-domain separation still leaves open the possibility that domains are smaller than the pixel size. If domains smaller than the pixel size indeed exists in cell membranes, the mechanism behind the decrease in GP value when cholesterol is removed (Figure 8) has to be explained considering that the average membrane GP measured corresponds to:
where: GPld, GPlo and fld , flo are the GP and the fraction (number of pixels) of the ld and lo phase respectively [63]. Thus, the average GP value measured in the membrane is the result of cholesterol equilibrium between the membrane and the internal cholesterol storage, etc. Further studies are needed to understand the cellular mechanism behind these observations in relation to lipid segregation in cell membranes and the connection between the changes in membrane fluidity and cholesterol equilibrium.
Acknowledgements
The work presented here was supported by funds from the National Institutes of Health (Grant PHS 5 P41 RR-03155, P50-GM076516 US) for S.A.S., G.O. and E.G.; the ANPCyT, Argentina, grants N° 14443 and PICT 2106-2008 to M.A.T. and the Australian FABLS N° R108 and International Cooperation (CONICET) to M.A.T. and S.A.S. We acknowledge Dr W. S. Davidson (U. Cincinatti, OH) and Dr. A.M.O.Gomes (Federal University of Rio de Janeiro, Brazil) for the kind donation of the apoA-I mutant H1@H4 and HeLa cells respectively.
Abbreviations
- DMPC
1,2 di-myristoyl phosphatidyl choline
- POPC
1-palmitoyl-2-oleylphosphatidylcholine
- DSPC
1,2 di-stearoyl phosphatidyl choline
- DOPC
1,2 di oleylphosphatidylcholine
- DPPC 1
2-dipalmitoylphatidylcholine
- SM
sphyngomyelin
- MβCD
methyl beta cyclo dextrine
- HDL
High Density Lipoproteins
- SUVs, MLVs, LUVs
small unilamellar, multilamellar and large unilamellar vesicles, respectively
- PCH
Photon Counting Histogram
- FC
free cholesterol
References
- 1.Hao M, Mukherjee S, Maxfield FR. PNAS. 2001;98:13072–13077. doi: 10.1073/pnas.231377398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Simons K, Ikonen E. Nature. 1997;387:569–572. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 3.Lichtenberg D, Goñi FM, Heerklotz H. Trends Biochem. Sci. 2005;30:430–436. doi: 10.1016/j.tibs.2005.06.004. [DOI] [PubMed] [Google Scholar]
- 4.Silva LC, Futerman AH, Prieto M. Biophys. J. 2009;96:3210–3222. doi: 10.1016/j.bpj.2008.12.3923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jacobson K, Mouritsen OG, Anderson RGW. Nat. Cell Biol. 2007;9:7–14. doi: 10.1038/ncb0107-7. [DOI] [PubMed] [Google Scholar]
- 6.Munro S. Cell. 2003;115:377–388. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 7.McIntosh TJ, Vidal A, Simon SA. Biophys. J. 2003;85:1656–1666. doi: 10.1016/S0006-3495(03)74595-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Groen AK, Oude Elferink RP, Verkade HJ, Kuipers F. Ann. Med. 2004;36:135–145. doi: 10.1080/07853890310020635. [DOI] [PubMed] [Google Scholar]
- 9.Nofer J-R, Walter M, Assmann G. Expert Rev. Cardiovasc. Ther. 2005;3:1071–1086. doi: 10.1586/14779072.3.6.1071. [DOI] [PubMed] [Google Scholar]
- 10.Wang X-q, Paller AS. J. Invest. Dermatol. 2006;126:951–953. doi: 10.1038/sj.jid.5700282. [DOI] [PubMed] [Google Scholar]
- 11.Rader DJ, Alexander ET, Weibel GL, Billheimer J, Rothblat GH. J. Lipid Res. 2009;50:S189–S194. doi: 10.1194/jlr.R800088-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamauchi Y, Chang CCY, Hayashi M, Abe-Dohmae S, Reid PC, Chang T-Y, Yokoyama S. J. Lipid Res. 2004;45:1943–1951. doi: 10.1194/jlr.M400264-JLR200. [DOI] [PubMed] [Google Scholar]
- 13.Vaughan AM, Oram JF. J. Lipid Res. 2006;47:2433–2443. doi: 10.1194/jlr.M600218-JLR200. [DOI] [PubMed] [Google Scholar]
- 14.Barter PJ. Atheroscler. Suppl. 2002;3:39–47. doi: 10.1016/s1567-5688(02)00041-7. [DOI] [PubMed] [Google Scholar]
- 15.Gordon DJ, Rifkind BM. N. Engl. J. Med. 1989;321:1311–1316. doi: 10.1056/NEJM198911093211907. [DOI] [PubMed] [Google Scholar]
- 16.Guha M, Gantz DL, Gursky O. J. Lipid Res. 2008;49:1752–1761. doi: 10.1194/jlr.M800106-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cavigiolio G, Shao B, Geier EG, Ren G, Heinecke JW, Oda MN. Biochemistry. 2008;47:4770–4779. doi: 10.1021/bi7023354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shao B, Cavigiolio G, Brot N, Oda MN, Heinecke JW. Proc. Natl. Acad. Sci. USA. 2008;105:12224–12229. doi: 10.1073/pnas.0802025105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Navab M, Anantharamaiah GM, Reddy ST, Van Lenten BJ, Ansell BJ, Fogelman AM. Nat. Clin. Pract. Endocrinol. Metab. 2006;2:504–511. doi: 10.1038/ncpendmet0245. [DOI] [PubMed] [Google Scholar]
- 20.Hansel B, Kontush A, Bonnefont-Rousselot D, Bruckert E, Chapman MJ. Curr. Atheroscler. Rep. 2006;8:501–509. doi: 10.1007/s11883-006-0026-8. [DOI] [PubMed] [Google Scholar]
- 21.Landry YD, Denis M, Nandi S, Bell S, Vaughan AM, Zha X. J. Biol. Chem. 2006;281:36091–36101. doi: 10.1074/jbc.M602247200. [DOI] [PubMed] [Google Scholar]
- 22.Mendez AJ, Lin G, Wade DP, Lawn RM, Oram JF. J. Biol. Chem. 2001;276:3158–3166. doi: 10.1074/jbc.M007717200. [DOI] [PubMed] [Google Scholar]
- 23.Drobnik W, Borsukova H, Böttcher A, Pfeiffer A, Liebisch G, Schütz GL, Schindler H, Schmitz G. Traffic. 2002;3:268–278. doi: 10.1034/j.1600-0854.2002.030404.x. [DOI] [PubMed] [Google Scholar]
- 24.Rosetti CM, Maggio B. Biophys. J. 2007;93:4254–4267. doi: 10.1529/biophysj.107.112441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Angelova MI, Dimitrov DS. Faraday Discuss. Chem. Soc. 1986;81:303–311. [Google Scholar]
- 26.Pott T, Bouvrais H, Méléard P. Chem. Phys. Lipids. 2008;154:115–119. doi: 10.1016/j.chemphyslip.2008.03.008. [DOI] [PubMed] [Google Scholar]
- 27.Montes LR, Alonso A, Goñi FM, Bagatolli LA. Biophys. J. 2007;93:3548–3554. doi: 10.1529/biophysj.107.116228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sun Y, Hao M, Luo Y, Liang C-p, Silver DL, Cheng C, Maxfield FR, Tall AR. J. Biol. Chem. 2003;278:5813–5820. doi: 10.1074/jbc.M208687200. [DOI] [PubMed] [Google Scholar]
- 29.Simons K, van Meer G. Biochemistry. 1988;27:6197–6202. doi: 10.1021/bi00417a001. [DOI] [PubMed] [Google Scholar]
- 30.Brown D. Braz. J. Med. Biol. Res. 1994;27:309–315. [PubMed] [Google Scholar]
- 31.Bagatolli LA, Sanchez SA, Hazlett TL, Gratton E. Methods Enzymol. 2003;360:481–500. doi: 10.1016/s0076-6879(03)60124-2. [DOI] [PubMed] [Google Scholar]
- 32.Veatch SL, Keller SL. Biophys. J. 2003;85:3074–3083. doi: 10.1016/S0006-3495(03)74726-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brown DA. Proc. Natl. Acad. Sci. USA. 2001;98:10517–10518. doi: 10.1073/pnas.191386898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Veatch SL, Keller SL. Phys. Rev. Lett. 2002;89:268101. doi: 10.1103/PhysRevLett.89.268101. [DOI] [PubMed] [Google Scholar]
- 35.Veatch SL, Keller SL. Phys. Rev. Lett. 2005;94:148101. doi: 10.1103/PhysRevLett.94.148101. [DOI] [PubMed] [Google Scholar]
- 36.Sanchez SA, Bagatolli LA, Gratton E, Hazlett TL. Biophys. J. 2002;82:2232–2243. doi: 10.1016/S0006-3495(02)75569-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nicolini C, Baranski J, Schlummer S, Palomo J, Lumbierres-Burgues M, Kahms M, Kuhlmann J, Sánchez SA, Gratton E, Waldmann H, Winter R. J. Am. Chem. Soc. 2006;128:192–201. doi: 10.1021/ja055779x. [DOI] [PubMed] [Google Scholar]
- 38.Bagatolli LA, Binns DD, Jameson JM, Albanesi JP. J. Protein Chem. 2002;21:383–391. doi: 10.1023/a:1021126415320. [DOI] [PubMed] [Google Scholar]
- 39.Berland KM, So PTC, Gratton E. Biophys. J. 1995;68:694–701. doi: 10.1016/S0006-3495(95)80230-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Thompson NL. In: Topics in Fluorescence Spectroscopy. Lakowicz JR, editor. 1991. pp. 337–378. [Google Scholar]
- 41.Chen Y, Muller JD, Berland KM, Gratton E. Methods. 1999;19 doi: 10.1006/meth.1999.0854. 234-152. [DOI] [PubMed] [Google Scholar]
- 42.Thompson NL, Lieto AM, Allen NW. Curr. Opin. Struct. Biol. 2002;12:634–641. doi: 10.1016/s0959-440x(02)00368-8. [DOI] [PubMed] [Google Scholar]
- 43.Levin MK, Carson JH. Differentiation. 2004;72:1–10. doi: 10.1111/j.1432-0436.2004.07201002.x. [DOI] [PubMed] [Google Scholar]
- 44.Masters BR, So PT, Kim KH, Buehler C, Gratton E. Methods Enzymol. 1999;307:513–536. doi: 10.1016/s0076-6879(99)07031-7. [DOI] [PubMed] [Google Scholar]
- 45.Brakenhoff GJ, Muller M, Ghauharali RI. J. Microscopy. 1996;183:140–144. doi: 10.1046/j.1365-2818.1996.870647.x. [DOI] [PubMed] [Google Scholar]
- 46.So PTC, French T, Yu WM, Berland KM, Dong CY, Gratton E. In: Fluorescence Imaging Spectroscopy and Microscopy. Wang XF, Herman B, editors. 1996. pp. 351–374. [Google Scholar]
- 47.Chen Y, Müller JD, So PTC, Gratton E. Biophys. J. 1999;77:553–567. doi: 10.1016/S0006-3495(99)76912-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Berland KM, So PTC, Chen Y, Mantulin WW, Gratton E. Biophys. J. 1996;71:410–420. doi: 10.1016/S0006-3495(96)79242-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ruan Q, Cheng MA, Levi M, Gratton E, Mantulin WW. Biophys. J. 2004;87:1260–1267. doi: 10.1529/biophysj.103.036483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Celli A, Beretta S, Gratton E. Biophys. J. 2008;94:104–116. doi: 10.1529/biophysj.107.105353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Garcia-Marcos A, Sánchez SA, Parada P, Eid JS, Jameson DM, Remacha M, Gratton E, Ballesta JPG. Biophys. J. 2008;94:2884–2890. doi: 10.1529/biophysj.107.121822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Digman M, Sengupta P, Wiseman SP, Brown CM, Horwitz AR, Gratton E. Biophys. J. 2005;88:33–36. doi: 10.1529/biophysj.105.061788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Digman MA, Dalal RB, Horwitz AR, Gratton E. Biophys. J. 2008;94:2320–2332. doi: 10.1529/biophysj.107.114645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Dalal RB, Digman MA, Horwitz AR, Vetri V, Gratton E. Microsc. Res. Tech. 2008;71:69–81. doi: 10.1002/jemt.20526. [DOI] [PubMed] [Google Scholar]
- 55.Krishnan K, Holub O, Gratton E, Clayton AHA, Cody S, Moens PDJ. Biophys. J. 2009;96:5112–5121. doi: 10.1016/j.bpj.2009.03.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Patel RC, Kumar U, Lamb DC, Eid JS, Rocheville M, Grant M, Rani A, Hazlett TL, Patel SC, Gratton E, Patel YC. Proc. Natl. Acad. Sci. USA. 2002;99:3294–3299. doi: 10.1073/pnas.042705099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Digman MA, Wiseman PW, Choi CK, Horwitz AR, Gratton E. Proc. Natl. Acad. Sci. USA. 2009;106:2170–2175. doi: 10.1073/pnas.0806036106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Digman MA, Gratton E. WIREs. Syst. Biol. Med. 2009;1:273–282. doi: 10.1002/wsbm.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Inoue M, Digman MA, Cheng MA, Breusegem SY, Halaihel N, Sorribas V, Mantulin WW, Gratton E, Barry NP, Levi M. J. Biol. Chem. 2004;279:49160–49171. doi: 10.1074/jbc.M408942200. [DOI] [PubMed] [Google Scholar]
- 60.Weber G, Farris FJ. Biochemistry. 1979;18:3075–3078. doi: 10.1021/bi00581a025. [DOI] [PubMed] [Google Scholar]
- 61.Parasassi TS, DeStasio G, Ravagnan G, Rusch R, Gratton E. Biophys. J. 1991;60:179–189. doi: 10.1016/S0006-3495(91)82041-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Parasassi T, Gratton E. J. Fluorescence. 1995;8:365–373. doi: 10.1007/BF00718783. [DOI] [PubMed] [Google Scholar]
- 63.Sánchez SA, Tricerri MA, Gunther G, Gratton E. In: Modern Research and Educational Topics in Microscopy. Applications in Physical/Chemical Sciences. Mendez-Vilas A, Diaz, editors. 2007. pp. 1007–1014. [Google Scholar]
- 64.Parasassi T, Krasnowska EK, Bagatolli LA, Gratton E. J. Fluorescence. 1998;8:365–373. [Google Scholar]
- 65.Henning MF, Sánchez SA, Bakás L. Biochem. Biophys. Res. Commun. 2009;383:22–26. doi: 10.1016/j.bbrc.2009.03.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Janosch S, Nicolini C, Ludolph B, Peters C, Volkert M, Hazlet TL, Gratton E, Waldmann H, Winter R. J. Am. Chem. Soc. 2004;126:7496–7503. doi: 10.1021/ja049922i. [DOI] [PubMed] [Google Scholar]
- 67.Kahya N, Brown D, Schwille P. Biochemistry. 2005;44:7479–7489. doi: 10.1021/bi047429d. [DOI] [PubMed] [Google Scholar]
- 68.Tricerri MA, Toledo JD, Sanchez SA, Hazlett TL, Gratton E, Jonas A, Garda HA. J. Lipid Res. 2005;46:669–678. doi: 10.1194/jlr.M400340-JLR200. [DOI] [PubMed] [Google Scholar]
- 69.Sánchez SA, Tricerri MA, Gratton E. J. Lipid Res. 2007;48:1689–1700. doi: 10.1194/jlr.M600457-JLR200. [DOI] [PubMed] [Google Scholar]
- 70.Tricerri MA, Sanchez SA, Arnulphi C, Durbin DM, Gratton E, Jonas A. J. Lipid Res. 2002;43:187–197. [PubMed] [Google Scholar]
- 71.Puff N, Lamaziere A, Seigneuret M, Trugnan G, Angelova M. Chem. Phys. Lipids. 2005;133:195–202. doi: 10.1016/j.chemphyslip.2004.10.003. [DOI] [PubMed] [Google Scholar]
- 72.Arnulphi C, Sánchez SA, Tricerri MA, Gratton E, Jonas A. Biophys. J. 2005;89:285–295. doi: 10.1529/biophysj.104.047480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Pownall HJ, Massey JB, Kusserow SK, Gotto AM., Jr Biochemistry. 1978;17:1183–1188. doi: 10.1021/bi00600a008. [DOI] [PubMed] [Google Scholar]
- 74.Bagatolli LA, Gratton E. Biophys. J. 1999;77:2090–2101. doi: 10.1016/S0006-3495(99)77050-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Clerc SG, Thompson TE. Biophys.J. 1995;68:2333–2341. doi: 10.1016/S0006-3495(95)80415-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Swaney JB. J. Biol. Chem. 1980;255:8791–8797. [PubMed] [Google Scholar]
- 77.Brouillette CG, Anantharamaiah GM. Biochim. Biophys. Acta. 1995;1256:103–129. doi: 10.1016/0005-2760(95)00018-8. [DOI] [PubMed] [Google Scholar]
- 78.Panagotopulos SE, Witting SR, Horace EM, Hui DY, Maiorano JN, Davidson WS. J. Biol. Chem. 2002;277:39477–39484. doi: 10.1074/jbc.M207005200. [DOI] [PubMed] [Google Scholar]
- 79.Rogers DP, Roberts LM, Lebowitz J, Datta G, Anantharamaiah GM, Engler JA, Brouillette CG. Biochemistry. 1998;37:11714–11725. doi: 10.1021/bi973112k. [DOI] [PubMed] [Google Scholar]
- 80.Jonas A. Methods Enzymol. 1986;128:553–582. doi: 10.1016/0076-6879(86)28092-1. [DOI] [PubMed] [Google Scholar]
- 81.Phillips JC, Wriggers W, Li Z, Jonas A, Schulten K. Biophys. J. 1997;73:2337–2346. doi: 10.1016/S0006-3495(97)78264-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Segrest JP, Jones MK, Klon AE, Sheldahl CJ, Hellinger M, eLoof DH, Harvey SC. J. Biol. Chem. 1999;274:31755–31758. doi: 10.1074/jbc.274.45.31755. [DOI] [PubMed] [Google Scholar]
- 83.Tricerri MA, Agree AKB, Sanchez SA, Bronski J, Jonas A. Biochemistry. 2001;40:5065–5074. doi: 10.1021/bi002815q. [DOI] [PubMed] [Google Scholar]
- 84.Palgunachari MN, Mishra VK, Lund-Katz S, Phillips MC, Adeyeye SO, Alluri S, Anantharamaiah GM, Segrest JP. Arterioscler. Thromb. Vasc. Biol. 1996;16:328–338. doi: 10.1161/01.atv.16.2.328. [DOI] [PubMed] [Google Scholar]
- 85.Cho KH, Durbin DM, Jonas A. J. Lipid Res. 2001;42:379–389. [PubMed] [Google Scholar]
- 86.Rothblat GH, de la Llera-Moya M, Atger V. J. Lipid Res. 1999;40:781–796. [PubMed] [Google Scholar]
- 87.Phillips MC, Gillotte KL, Haynes MP, Johnson WJ, Lund-Katz S, Rothblat GH. Atherosclerosis. 1998;137:13–17. doi: 10.1016/s0021-9150(97)00312-2. [DOI] [PubMed] [Google Scholar]
- 88.Veatch SL, Polozov IV, Gawrisch K, Keller SL. Biophys. J. 2004;86:2910–2922. doi: 10.1016/S0006-3495(04)74342-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Brown RE. J. Cell Sci. 1998;111:1–9. doi: 10.1242/jcs.111.1.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Needham D, McIntosh TJ, Evans E. Biochemistry. 1988;27:4668–4673. doi: 10.1021/bi00413a013. [DOI] [PubMed] [Google Scholar]
- 91.Sankaram MB, Thompson TE. Proc. Natl. Acad. Sci. USA. 1991;88:8686–8690. doi: 10.1073/pnas.88.19.8686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Harris JS, Epps DE, Davio SR, Kezdy FJ. Biochemistry. 1995;34:3851–3857. doi: 10.1021/bi00011a043. [DOI] [PubMed] [Google Scholar]
- 93.Pike LJ. J. Lipid Res. 2006;47:1597–1598. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 94.Stott BM, Vu MP, McLemore CO, Lund MS, Gibbons E, Brueseke TJ, Wilson-Ashworth HA, Bell JD. J. Lipid Res. 2008;49:1202–1215. doi: 10.1194/jlr.M700479-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Smith SK, Farnbach AR, Harris FM, Hawes AC, Jackson LR, Judd AM, Vest RS, Sanchez S, Bell JD. J. Biol. Chem. 2001;276:22732–22741. doi: 10.1074/jbc.M010880200. [DOI] [PubMed] [Google Scholar]
- 96.Gaus K, Gratton E, Kable EP, Jones AS, Gelissen I, Kritharides L, Jessup W. Proc. Natl. Acad. Sci. USA. 2003;100:15554–15559. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Giddings KS, Johnson AE, Tweten RK. Proc. Natl. Acad. Sci. U S A. 2003;100:11315–11320. doi: 10.1073/pnas.2033520100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kansau I, Berger C, Hospital M, Amsellem R, Nicolas V, Servin AL, Bernet-Camard MF. Infect. Immun. 2004;72:3733–3742. doi: 10.1128/IAI.72.7.3733-3742.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Jaureguiberry MS, Tricerri MA, Sanchez SA, Garda HA, Finarelli GS, Gonzalez MC, Rimoldi OJ. J. Membr. Biol. 2010 doi: 10.1007/s00232-010-9245-6. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Plazzo ZAAP, Stöckl M, Trombik T, Hamon Y, Müller P, Pomorski T, Herrmann A, Chimini G. FASEB J. 2009;23:1775–17785. doi: 10.1096/fj.08-122192. [DOI] [PubMed] [Google Scholar]