Abstract
The silent information regulator (Sir2) family proteins are NAD+-dependent deacetylases. Although a few substrates have been identified, functions of the bacteria Sir2-like protein (CobB) still remain unclear. Here the role of CobB on Escherichia coli chemotaxis was investigated. We used Western blotting and mass spectrometry to show that the response regulator CheY is a substrate of CobB. Surface plasmon resonance (SPR) indicated that acetylation affects the interaction between CheY and the flagellar switch protein FliM. The presence of intact flagella in knockout strains ΔcobB, Δacs, Δ(cobB) Δ(acs), Δ(cheA) Δ(cheZ), Δ(cheA) Δ(cheZ) Δ(cobB) and Δ(cheA) Δ(cheZ) Δ(acs) was confirmed by electron microscopy. Genetic analysis of these knockout strains showed that: (i) the ΔcobB mutant exhibited reduced responses to chemotactic stimuli in chemotactic assays, whereas the Δacs mutant was indistinguishable from the parental strain, (ii) CheY from the ΔcobB mutant showed a higher level of acetylation, indicating that CobB can mediate the deacetylation of CheY in vivo, and (iii) deletion of cobB reversed the phenotype of Δ(cheA) Δ(cheZ). Our findings suggest that CobB regulates E. coli chemotaxis by deacetylating CheY. Thus a new function of bacterial cobB was identified and also new insights of regulation of bacterial chemotaxis were provided.
Introduction
Members of the Sir2 family of proteins are nicotinamide adenine dinucleotide (NAD+)-dependent deacetylase enzymes that modulate gene silencing (Rusche et al., 2003), cell cycle regulation (Dryden et al., 2003), fatty acid metabolism (Starai et al., 2002), lifespan extension (Rogina and Helfand, 2004) and apoptosis (Langley et al., 2002). Many organisms contain multiple orthologues of the Sir2 family proteins, and thus it is expected that other substrates under the control of Sir2 proteins will be revealed (Starai et al., 2004). In Salmonella enterica it has been reported that acetylation blocks the adenylating activity of the enzyme acetyl-CoA synthetase (Acs) and activation of the acetylated enzyme requires the activity of the Sir2-like protein CobB (Starai et al., 2002). When the concentration of acetate in the environment is low, Acs is responsible for conversion of acetate to acetyl-CoA (AcCoA) (Starai et al., 2003). However, the physiological roles of CobB in bacteria still remain largely unknown.
Chemotaxis is a mechanism by which bacteria respond to changes in the chemical compositions of their environment, approaching attractants and avoiding repellents. The chemotactic response of bacteria such as Escherichia coli is accomplished by signal transmission between two supramolecular complexes: the receptor complexes and the flagellar–motor complexes. The response regulator of bacterial chemotaxis, CheY, shuttles back and forth between the complexes and transduces the signal from the receptors to the flagella (Silversmith and Bourret, 1999; Sourjik, 2004). CheY can be modulated by two covalent modifications: phosphorylation and acetylation (Baker et al., 2006). CheY is phosphorylated by phosphoryl groups either by its associated autophosphorylating protein kinase, CheA, or by small phosphodonor molecules such as acetyl phosphate (acetyl-P). In fact, the contribution of acetyl-P is minimal because the phosphotransfer rate from CheA to CheY is orders of magnitude faster than the rate from acetyl-P to CheY (Wolfe, 2005). The phosphorylated form, CheY-P, binds to the switch protein FliM much better than does the non-phosphorylated form at the base of the flagellar motor, which then increases the probability of shifting the direction of flagellar rotation from the default direction, counterclockwise (CCW), to clockwise (CW) (Welch et al., 1993). CheY-P dephosphorylates spontaneously, an activity enhanced by another chemotaxis protein, CheZ (Zhao et al., 2002). This dephosphorylation reduces the binding of CheY to the switch (Bren et al., 1996).
The acetylation of CheY is carried out by acetyl-CoA synthetase (Acs, with acetate as the acetyl donor) (Barak et al., 1992) or by autoacetylation (with AcCoA as the acetyl donor) (Barak et al., 2006). Recently, the acetylation of CheY has been detected in vivo, and it was found to result mainly from CheY autoacetylation (Yan et al., 2008). The acetylation sites are six lysine residues – lysines 91, 92, 109, 119, 122 and 126, all clustered at the C-terminus of the protein and localized on the surface that binds FliM, CheZ and CheA (Barak et al., 2004). Although acetylation, like phosphorylation, appears to influence chemotaxis of E. coli (Barak et al., 1998; Ramakrishnan et al., 1998; Barak and Eisenbach, 2001), the role of CheY acetylation in bacterial chemotaxis still remains obscure.
Although CheY was known to undergo acetylation, enzymes responsible for its deacetylation were not fully investigated. Since CobB was shown to be a protein deacetylase, it is possible that CheY might be one of its substrates and so the correlation between CobB and chemotaxis of E. coli was carefully investigated. We speculated that CobB could catalyse CheY deacetylation and hence regulate bacterial chemotaxis. Here, we addressed this speculation very carefully by using biochemical and genetic analysis. To our knowledge, this is the first time to show that CobB regulates E. coli chemotaxis by deacetylating CheY. This broadens our understandings of the physiological roles of Sir2 family protein in bacteria and also provides new insights into the regulation of bacterial chemotaxis.
Results
CobB deacetylates Acs and CheY in vitro
To test whether CobB could deacetylate Acs in E. coli, acetylated Acs (AcAcs) was incubated with CobB in the presence of NAD+. We performed Western blotting analysis to test the acetylation level of Acs using an anti-acetyl-lysine antibody. The result demonstrated that CobB markedly reduces the acetylation level of AcAcs, and that this effect can be alleviated in the presence of the Sir2 inhibitor nicotinamide (NAM) (Fig. S1).
To provide primary evidence that CheY is the substrate of CobB, acetylated CheY (AcCheY) was deacetylated and protein acetylation level was analysed with an anti-acetyl-lysine antibody. As shown in Fig. 1, the acetylation level of AcCheY was not affected in the absence of CobB or NAD+ (lanes 1, 2). In contrast, the acetylation level of AcCheY decreased greatly in the presence of CobB and NAD+ (lane 3). NAM reduced the deacetylation of AcCheY by CobB (lane 4). Additionally, the acetylation level of CobB itself is relatively high, consistent with a recent report that human Sirt2, a member of the Sir2 family, can be acetylated by p300, a protein with protein acetyltransferase activity (Han et al., 2008). These results clearly demonstrate that in E. coli, CobB can deacetylate CheY in vitro.
Fig. 1.
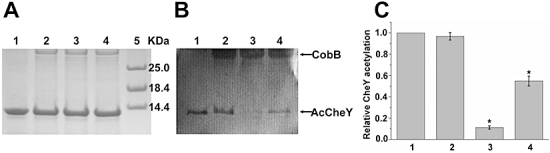
CobB deacetylates AcCheY in vitro. The acetylation levels of all proteins were determined by Western blotting using a specific anti-acetyl-lysine antibody. Experiments were replicated three times, and representative results are shown. A and B. (A) SDS-PAGE of samples. (B) Western blot of the gel in (A). Lane 1, AcCheY + NAD+; lane 2, AcCheY + CobB; lane 3, AcCheY + CobB + NAD+; lane 4, AcCheY + CobB + NAD+ + NAM; lane 5, marker. The concentration of AcCheY and CobB was 15 µM and 3 µM respectively. The reaction was carried out for 6 h at 25°C. C. Average levels of CheY acetylation, quantified from the Western blots of three experiments (mean ± SD) using AlphaView image analysis software and normalized relative to the value obtained in absence of CobB (lane 1). An asterisk indicates a statistically significant difference from lane 1 (P < 0.01; anova analysis).
To identify the site(s) of acetylation and deacetylation, AcCheY and D-CheY (deacetylated AcCheY by CobB) were prepared, enzymatically digested and analysed by LC-MS in a LTQ-Orbitrap mass spectrometer. Lysine-acetylated peptide can be identified as they have a mass increment of 42 Da (Larsen et al., 2006). The acetylated peptide in AcCheY was shown to be K(91).KENIIAAAQAGASGYVVK(109)PFTAATLEEK.L, and had a molecular mass of 2918.55, +42 Da heavier than the equivalent peptide in D-CheY (2876.53). The fragment ion signals reflect the amino acid sequence as read from either the N-terminal (b-ion series) or the C-terminal (y-ion series) direction. As shown in Fig. 2A, the boxed b(3) and b(5) have a mass of 42 Da greater than the corresponding ions in the CobB-treated sample (Fig. 2C). This indicates that the N-terminal residue lysine 91 is acetylated in AcCheY. The boxed b(18)++ and y(11)++ ions in Fig. 2B have a mass of 21 Da greater than the corresponding ions in the Fig. 2C. Subsequent ions in the series b(19–27)++ and y(12–27)++ all showed a mass increased by 21 Da in the acetylated sample. This indicates the presence of an acetyl group on lysine 109 in AcCheY. More detailed data are available in Tables S1 and S2. Thus, these mass spectrometry results reveal that the acetylated residues in CheY are lysine 91 and 109, and that CobB can catalyse CheY deacetylation at the same sites.
Fig. 2.
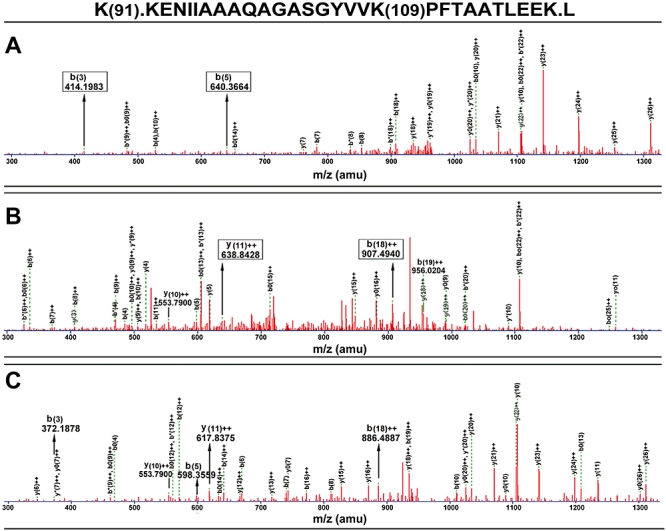
LC-MS/MS analysis confirms that lysine 91 and 109 are acetylated in AcCheY, and can be deacetylated by CobB. A. Tryptic digestion of AcCheY with Lys91 acetylated. The boxed b(3) and b(5) ions have a mass of 42 Da greater than the corresponding ions in the CobB-treated sample. B. Tryptic digestion of AcCheY with Lys109 acetylated. The boxed b(18)++ and y(11)++ ions have a mass of 21 Da greater than the corresponding ions in the CobB-treated sample. C. Tryptic digestion of D-CheY.
Influence of acetylation on the interaction between CheY and FliM
To investigate whether acetylation of CheY affects the interaction between CheY and FliM, surface plasmon resonance (SPR), a sensitive technology to measure the molecular interactions, was employed to study interaction between FliM and CheY. The SPR 3000 system contains a dual-channel measuring cell. The working channel is linked to a sensor chip while the reference channel is linked to the same chip without the immobilized sensor element protein His-FliM. As shown in Fig. 3, interactions between His-FliM and CheY-His (or AcCheY-His) were detected. The response unit (RU) values were proportional to sample concentrations. At the same sample concentration, AcCheY-His exhibited a significantly lower binding signal with His-FliM, compared with CheY-His. The KD values of interactions between His-FliM and AcCheY-His, and between His-FliM and CheY-His are 9.4 and 1.63 µM respectively. These results therefore suggest that acetylation of CheY reduces its binding affinity to FliM.
Fig. 3.
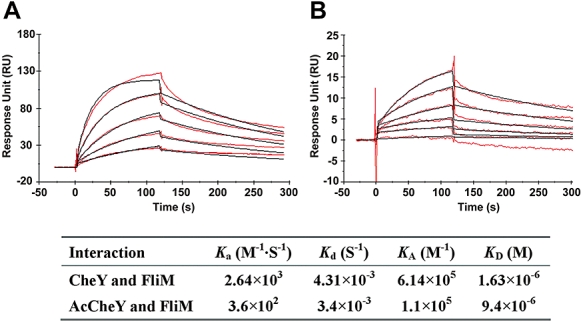
SPR analysis of CheY and AcCheY binding to FliM. Approximately 1120 RU FliM was immobilized on CM5 sensor surface using amine coupling method. This experiment was replicated at least three times. A. Responses of CheY at the concentrations 1–16 µM (from lower to upper). B. Responses of AcCheY at the concentrations 1–32 µM (from lower to upper). The red lines represent protein injections at the indicated concentration. The black lines represent the global fit of the entire data set to 1:1 Langmuir binding model. The inserted table lists the kinetic constants derived from the sensorgrams.
Chemotactic behaviour of ΔcobB, Δacs and Δ(cobB) Δ(acs) mutants
To investigate whether CobB can regulate chemotaxis of E. coli, a ΔcobB mutant (RL001) was constructed and its phenotype was tested. In 2003, Starai et al. reported that cobB mutants have trouble growing on low concentrations of acetate because Acs remains acetylated, which means that Acs remains constitutively off (Starai et al., 2003). Since growth on low concentration of acetate requires Acs (Kumari et al., 1995), cobB mutants grow poorly on low acetate. In our study, 10 mM acetate was used as the lowest concentration to test the ability of RL001 to grow on acetate. Figure S2 shows that the ΔcobB mutant (RL001) grew poorly on low levels of acetate. Thus, this result confirms that the ΔcobB mutant behaves as expected.
In order to test whether flagellum synthesis or function was affected by deletion of cobB, RL001 was observed under the electron microscope. As shown in Fig. S3, RL001 exhibited normal shape and similar numbers of flagella as W3110. Since flagellum assembly in the ΔcobB mutant is normal, we speculated that CobB might affect chemotaxis. To this end, the effect of RL001 on chemotaxis was tested by several distinct chemotaxis assays. Furthermore, in order to rule out the possibility that the effect of CobB on chemotaxis may be caused by its ability to deacetylate Acs, the chemotactic responses of the Δacs mutant (RL002) and Δ(cobB) Δ(acs) (RL0012) mutant were also examined.
In the swarm assay, bacteria migrate outward from the site of inoculation; migration on semi-solid motility plates depends on: the ability to transport and metabolize serine, aspartate and threonine, the ability to assemble flagella, the ability to rotate those flagella, the ability to sense these attractants (serine, aspartate and threonine) gradients (Wolfe and Berg, 1989). On tryptone broth semi-solid agar, the parental strain W3110 formed at least two rings, as did the ΔcobB mutant (Fig. 4A). However, the migration rate of the mutant was slower than that of the parental strain. In contrast, the Δacs mutant was indistinguishable from the W3110 parental strain (Fig. 4B). To confirm that the similarity between them was not strain dependent, strains AJW613 and its Δacs mutant derivative AJW803 were also examined and similar results were observed (Fig. S4). In addition, the Δ(cobB) Δ(acs) mutant showed similar migration rates with the ΔcobB mutant (data not shown).
Fig. 4.
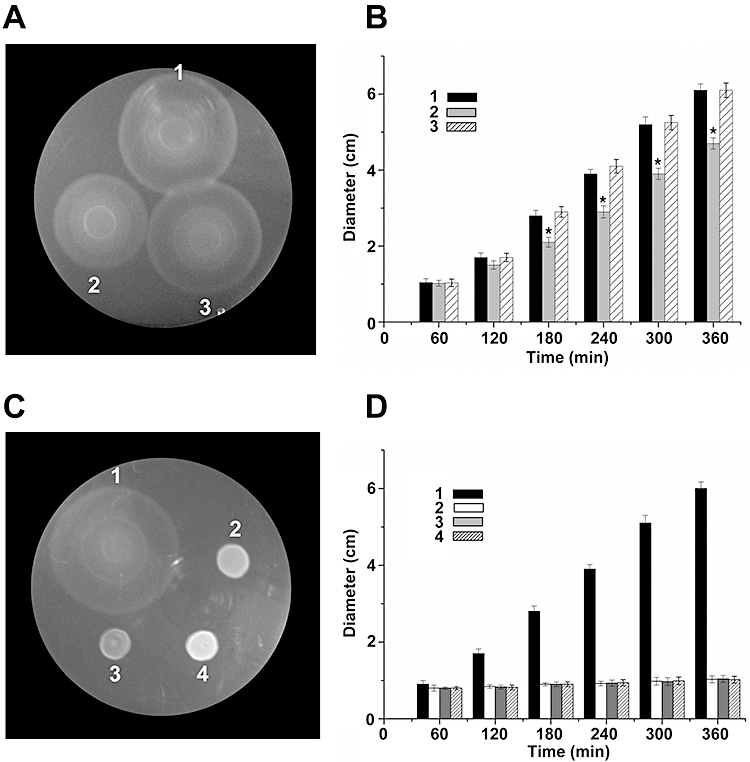
Swarm assays on semi-solid TB plates. Cells were inoculated near the centre of tryptone swarm plates containing 0.2% agar and incubated at 35°C. Experiments were replicated four times, and representative results are shown. A. Swarm ring formation by: (1) wild-type strain W3110, (2) W3110 ΔcobB mutant and (3) W3110 Δacs mutant. Photographs were taken after 3 h. B. Displacement (diameter) of the outermost edge of swarms of strains used in (A). Data are means ± SD from four independent experiments. An asterisk indicates a statistically significant difference from W3110 (P < 0.05; anova analysis). C. Swarm ring formation by: (1) wild-type strain W3110, (2) W3110 Δ(cheA) Δ(cheZ) mutant, (3) W3110 Δ(cheA) Δ(cheZ) Δ(cobB) mutant and (4) W3110 Δ(cheA) Δ(cheZ) Δ(acs) mutant. Photographs were taken after 3.5 h. D. Displacement (diameter) of the outermost edge of swarms of strains used in (C). Data are means ± SD from four independent experiments.
To determine what led to the reduced migration rate for the ΔcobB mutant, we used other assays (plug and capillary assays) that do not depend on the ability of the cells to establish their own gradient, that is the cells do not need to be able to transport and metabolize attractants or repellants. In the plug assay, chemotactically responsive bacteria in semi-solid agar accumulate near a plug containing an attractant or at a distance from a plug containing a repellent (Tso and Adler, 1974). When galactose (50 mM) was used as the attractant, results showed that strain W3110 accumulated normally near the galactose plug, while the accumulation of ΔcobB and Δ(cobB) Δ(acs) mutants near the plug was markedly reduced. However, only a very slight reduction was observed for the Δacs mutant (Fig. S5, upper panel). Likewise, W3110 formed a bacteria-deficient zone around the plug containing the repellent NiSO4 (25 mM), but the repulsion of ΔcobB and Δ(cobB) Δ(acs) mutants was markedly reduced and the response of the Δacs mutant was similar to W3110 (Fig. S5, middle panel). In a modified drop assay in which leucine (100 mM) was used, similar results were observed except that the repellent response of the Δacs mutant seemed to be even more marked than that of W3110 (Fig. S5, lower panel). Quantified results are shown in Fig. 5A.
Fig. 5.
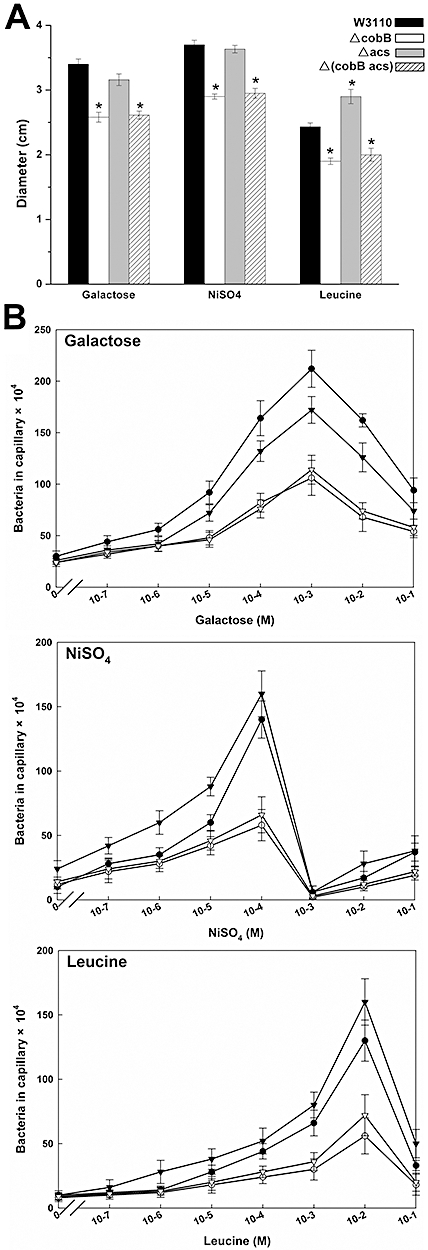
Chemotactic responses of W3110, ΔcobB mutant, Δacs mutant and Δ(cobB) Δ(acs) mutant. A. Plug assays and drop assays. The concentrations of galactose plug, NiSO4 plug and leucine drop were 50 mM, 25 mM and 100 mM respectively. The medium was supplemented with 30 mM acetate. The results were quantified with a ruler. Data are means ± SD from four independent experiments. An asterisk indicates a statistically significant difference from W3110 (P < 0.05; anova analysis). B. Capillary assays. Assays were carried out at 35°C for 1 h. Only concentrations indicated by the symbols were tested. Data are means ± SD from three independent experiments. (▾) W3110; (○) W3110 ΔcobB mutant; (▾) W3110 Δacs mutant; (▿) W3110 Δ(cobB) Δ(acs) mutant.
To further test the above results, capillary assays which do not depend on the ability of the cells to establish their own gradient were also performed. Concentration–response curves for various attractants and repellants are shown in Fig. 5B. The extent of accumulation of W3110 in the capillary increased with the concentration of the galactose attractant. In comparison, the extent of the accumulation of the ΔcobB and Δ(cobB) Δ(acs) mutants was reduced and the response was only about half of the wild-type strain. The response of the Δacs mutant was greater than 80% of W3110. When capillaries containing only motility buffer were inserted into the suspensions of bacteria in which the repellent NiSO4 or leucine were present, typical concentration-dependent accumulations of wild-type cells were observed. Again, for each repellent, the response of ΔcobB and Δ(cobB) Δ(acs) appeared to be markedly reduced in comparison with W3110. In contrast, responses of the Δacs mutant were similar with those of W3110.
These observations suggest that the deficiency in chemotaxis of the ΔcobB mutant is not due to a lack of Acs activity, and thus we conclude that CobB acts on chemotaxis by a mechanism that does not involve Acs. Since CheY is acetylated and CobB is a deacetylase, we speculated that CobB influences chemotaxis by altering the acetylation status of CheY.
Analysis of the in vivo acetylation level of CheY
To provide more direct evidence for CheY deacetylation by CobB in vivo, CheY acetylation levels in strain W3110, and in the ΔcobB mutant (RL001) and Δacs mutant (RL002) were analysed by Western blotting. As shown in Fig. 6A, the concentration of total proteins was similar in the three strains. Polyclonal anti-CheY antibodies were used to monitor CheY expression levels and the results (Fig. 6B) showed there was no difference in CheY expression level between these strains. However, Western blotting using an anti-acetyl-lysine antibody revealed that CheY acetylation level in the ΔcobB mutant was significantly higher than that of W3110, whereas only a slight difference was observed between W3110 and the Δacs mutant (Fig. 6C and D). These data suggest that the absence of CobB results in an obvious increase of in the CheY acetylation level, and further indicates that CobB-mediated deacetylation has an effect on the CheY acetylation, not only in vitro, but also in vivo.
Fig. 6.
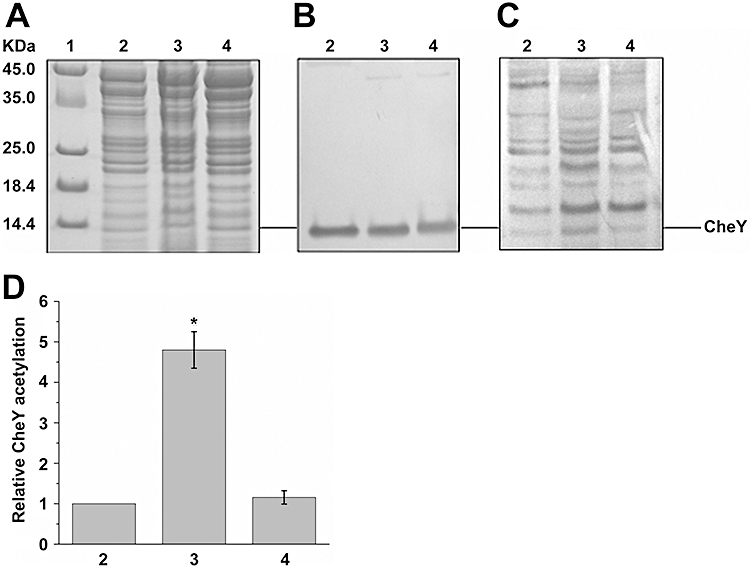
Western blotting analysis of CheY acetylation in vivo. The 30 µg protein lysate was resolved in 12–15% SDS-PAGE and analysed by Western blotting. Experiments were replicated three times, and representative results are shown. A–C. (A) SDS-PAGE of the extracts. (B) Western blot with a CheY antibody. (C) Western blot with an anti-acetyl-lysine antibody. Lane 1, marker; lane 2, W3110; lane 3, W3110 ΔcobB mutant; lane 4, W3110 Δacs mutant. D. Average levels of CheY acetylation, quantified from the Western blots of three experiments (mean ± SD) using AlphaView image analysis software and normalized relative to the value obtained in W3110 (lane 2). An asterisk indicates a statistically significant difference from lane 2 (P < 0.01; anova analysis).
Effects of cobB or acs deletion on the chemotactic behaviour of Δ(cheA) Δ(cheZ) mutant
In 2004, Barak and Eisenbach found that phosphorylation and acetylation of CheY affect with each other: CheA inhibits acetylation, whereas CheZ enhances it. Conversely, Acs enhances phosphorylation and acetate inhibits this enhancement (Barak and Eisenbach, 2004). Therefore, we chose to test the role of the cobB mutant in a cheA cheZ double mutant background. To achieve this, three mutants were constructed, including a cheA cheZ double mutant (RL003), a cheA cheZ cobB triple mutant (RL0031) and a cheA cheZ acs triple mutant (RL0032).
On a tryptone swarm plate, mutant RL003, RL0031 and RL0032 should not form chemotactic bands because they failed to respond to serine, aspartate and threonine chemical attractants. As shown in Fig. 4C and D, they did not form rings on 0.2% agar within 4 h at 35°C, unlike W3110. When the incubation time was prolonged, similar results were obtained.
Plug assay showed that all three strains formed a bacteria-free zone in the presence of a repellent-containing plug (NiSO4), and did not show any response to the attractant galactose (Fig. S6, upper and lower panels). Interestingly, the repulsion of the Δ(cheA) Δ(cheZ) mutant was greater than that of W3110, whereas Δ(cheA) Δ(cheZ) Δ(cobB) showed a similar degree of repulsion as W3110. However, the response of the Δ(cheA) Δ(cheZ) Δ(acs) mutant to the repellent was indistinguishable from the Δ(cheA) Δ(cheZ) mutant. Likewise, results obtained in the leucine-containing drop assay were similar to those of the plug assay (Fig. S6, middle panel). The plug assay was quantified as shown in Fig. 7A.
Fig. 7.
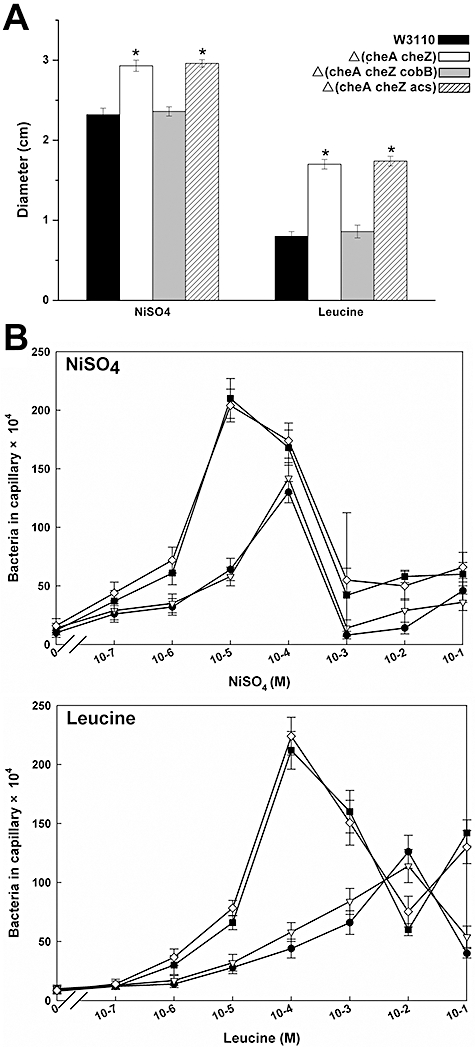
Chemotactic responses of W3110, Δ(cheA) Δ(cheZ), Δ(cheA) Δ(cheZ) Δ(cobB) and Δ(cheA) Δ(cheZ) Δ(acs). A. Plug assays and drop assays. The concentrations of NiSO4 plug, leucine drop and galactose plug were 25 mM, 100 mM and 50 mM respectively. The results were quantified with a ruler. Data are means ± SD from four independent experiments. An asterisk indicates a statistically significant difference from W3110 (P < 0.05; anova analysis). B. Capillary assays. Only the concentrations indicated by the symbols were tested. Data are means ± SD from three independent experiments. (▪) W3110; ( ) W3110 Δ(cheA) Δ(cheZ) mutant; (▿) W3110 Δ(cheA) Δ(cheZ) Δ(cobB) mutant; (◊) W3110 Δ(cheA) Δ(cheZ) Δ(acs) mutant.
) W3110 Δ(cheA) Δ(cheZ) mutant; (▿) W3110 Δ(cheA) Δ(cheZ) Δ(cobB) mutant; (◊) W3110 Δ(cheA) Δ(cheZ) Δ(acs) mutant.
In the capillary assay, the responses of the Δ(cheA) Δ(cheZ) and Δ(cheA) Δ(cheZ) Δ(acs) mutants to the repellents (NiSO4 and leucine) were similar (Fig. 7B), which is consistent with our observations above. Furthermore, these two mutants were repelled by leucine into the capillaries, with a peak response at 10−4 M leucine. In contrast, the peak response of W3110 and the Δ(cheA) Δ(cheZ) Δ(cobB) mutant was about 10−2 M leucine (Fig. 7B). Thus, chemotactic responses of the Δ(cheA) Δ(cheZ) mutant and the Δ(cheA) Δ(cheZ) Δ(acs) mutant towards the repellents appear to be more sensitive and greater compared with those of W3110 and the Δ(cheA) Δ(cheZ) Δ(cobB) mutant.
Taken together, this study provides evidence that CobB does indeed participate in E. coli chemotaxis through deacetylation of CheY. Our results also confirm that the two covalent modifications of CheY, phosphorylation and acetylation, are linked in vivo.
Discussion
In this study, we proposed that CobB regulates E. coli chemotaxis by deacetylating CheY. A new function of bacterial cobB gene was identified and new insights of regulation of bacterial chemotaxis were provided herein. The main findings are discussed below.
AcCheY is a substrate of CobB
Several observations prompted us to speculate that AcCheY could be a substrate of CobB. CheY is known to undergo Acs-mediated acetylation or catalyse its own acetylation. We wondered whether CheY acetylation, like acetylation of histones and eukaryotic transcription factors, might also be reversible. Prior to this report, a deacetylase responsible for CheY deacetylation has not been fully investigated (Barak et al., 2004; 2006). CobB is known to be a NAD+-dependent deacetylase and its eukaryotic orthologues have been shown to regulate many biological processes. Using Western blotting with an anti-acetyl lysine antibody we have shown that AcCheY is deacetylated by CobB. Data from LC-MS/MS analysis showed that lysine residues 91 and 109 in AcCheY are acetylated in the peptide K(91).KENIIAAAQAGASGYVVK(109)PFTAATLEEK.L and no lysine residues were found to be acetylated in the same peptide obtained from D-CheY (Fig. 2). This provides sufficient evidence to support that AcCheY is a substrate of CobB. In addition, previous reports showed six lysine residues 91, 92, 109, 119, 122 and 126 are the main acetylation sites (Barak et al., 2004). The differences between the previous and the current results may be due to the following reasons: (i) we used 67% acetonitrile containing 2.5% trifluoroacetic acid (TFA) for extracting the peptides in comparison with 5% formic acid; thus the peptide extraction may not be complete, (ii) the acetylation of lysine residues 92 and 122 of AcCheY may prevent its cleavage by trypsin, and (iii) because the acetyl (C-term) was not considered as variable modification, the acetylation of lysine 119 was not found.
CheY acetylation affects its interaction with FliM
Although many previous observations (Wolfe et al., 1988; Barak et al., 1998) suggested that CheY acetylation is involved in chemotaxis, the mechanism is still obscure. Structural and functional studies of CheY have revealed that the acetylation sites of CheY are clustered at the C-terminus and appear to be involved in the binding of CheY to FliM (Lee et al., 2001; Dyer and Dahlquist, 2006). In 1998, Ramakrishnan et al. measured AcCheY–FliM binding using a protein cross-linking assay, but found the ability of AcCheY to bind to FliM in vitro was indistinguishable from that of CheY (Ramakrishnan et al., 1998). They favour the possibility that acetylation of CheY affects a post-FliM-binding step. However, the sensitivity of the protein cross-linking method used was low. In fact, SPR analysis, a sensitive technique for measuring molecular interactions, clearly demonstrates that the acetylation of CheY reduces its binding to FliM. (Besides, because the batch of AcCheY is probably heterogeneous and consists of both acetylated and non-acetylated CheY molecules, it is reasonable that the binding values for AcCheY are average values of the at least two populations in this batch. This suggests that the actual KD of AcCheY binding to FliM is even higher.) This observation may be the consequence of one or both of the following hypothetical possibilities. (i) The acetyl groups on CheY may affect the structure of the protein. It has been reported that Lys109 forms a hydrogen bond through its ε-amine with the carboxyl group of Asp57, the phosphorylation site (Volz and Matsumura, 1991). If the ε-amine is acetylated, then hydrogen bond will not be able to form and thus protein structure may be changed. (ii) Based on the BeF3-–CheY–FliM16 complex structure (pdb id 1F4V), the interactions between the activated CheY and the N-terminal FliM peptides are mediated with a combination of three types of non-covalent bonding: hydrophobic interactions, hydrogen bonds and salt bridges (Lee et al., 2001). Among these interactions, the side-chains of K119 and K122 of CheY form stable salt bridges with the side-chains of D12 and N16 of FliM respectively (Fig. S7). Additionally, the salt bridge between K122 and N16 is replaced by a hydrogen bond in the unphosphorylated CheY–FliM16 complex (pdb id 2B1J) (Dyer and Dahlquist, 2006). Interestingly, both K119 and K122 are among the six identified acetylation sites of CheY (Barak et al., 2004). Acetylation on the ε-amine groups of K119 and K122 would neutralize these two residues, and result in breaking the two bonds between CheY and FliM, which obviously would destabilize the overall inter-molecule interaction. This notion is consistent with the observation that AcCheY had a weaker binding affinity with FliM than CheY.
Co-regulation of CheY acetylation and phosphorylation in vivo
Earlier studies demonstrated that the contributions of CheA and CheZ to the phosphorylation level of CheY appear to be important (Mayover et al., 1999). In 2004, Barak and Eisenbach found that CheA strongly inhibits the acetylation of CheY and that CheZ has the opposite effect (Barak and Eisenbach, 2004). In addition, the presence of Acs enhances the phosphorylation level of CheY. Even though this result suggests that CheY acetylation and phosphorylation may be linked, there is a lack of direct evidence for their co-regulation in vivo. Interestingly, we observed that the double deletion mutant Δ(cheA) Δ(cheZ) showed enhanced responses to repellents, but this phenotype could be reversed by further deletion of cobB. This result may be interpreted to mean that the phosphorylation level of CheY changed in the absence of CheA and CheZ, and that this change was affected by the deletion of cobB. These findings strongly suggest that CobB is involved in the E. coli chemotaxis via CheY deacetylation, and that the acetylation and phosphorylation of CheY affect each other in vivo. Further experiments will be necessary to unravel the precise mechanism by which these two modifications are co-regulated.
Potential Role of Acs in acetylating CheY in vivo
Although it has been reported that the acetylation of CheY is carried out by Acs (Barak et al., 1992) or by autoacetylation (Barak et al., 2006), the role of Acs on acetylating CheY in vivo is uncertain. Based on our observations that the Δacs mutant shows similar chemotactic responses to the wild-type strain W3110, three hypothetical situations may be considered: (i) in spite of the fact that Acs can probably transfer acetyl groups to CheY in vitro (Barak et al., 2004), the effect is minor in vivo, (ii) CheY undergoes autoacetylation with acetyl-CoA as the acetyl group (Barak et al., 2006), and the acetylation level is mainly the outcome of autoacetylation in vivo (Yan et al., 2008), and (iii) an unknown acetyltransferase is required for the acetylation of CheY. In any case, Acs has little or no effect on the acetylation of CheY in vivo.
CobB regulates E. coli chemotaxis by deacetylating CheY
Our observations show: (i) AcCheY is a substrate of CobB, (ii) acetylation of CheY affects the binding affinity between CheY and FliM, (iii) E. coliΔcobB has intact flagellum, and (iv) E. coliΔcobB had reduced chemotactic responses in the presence of both attractants and repellents. Taken together, these data strongly suggest CobB regulates E. coli chemotaxis by deacetylating CheY. And this proposition is further supported by the fact that deletion of cobB reverses the effect of a double knockout of cheA and cheZ. Since in vitro experiments have shown that CobB can catalyse the deacetylation of both Acs and CheY, the effect of CobB on chemotaxis may arise from its ability to deacetylate Acs or CheY or both. To distinguish these three possibilities, Δacs and Δ(cobB) Δ(acs) mutants were used as controls. Chemotactic assays showed that the Δacs mutant was indistinguishable from W3110 and the Δ(cobB) Δ(acs) mutant had similar phenotype to ΔcobB mutant (Fig. 5 and Fig. S5). Therefore, we suggest that CobB works through something other than Acs, perhaps via CheY. In comparison with W3110 and the Δacs mutant, the higher level of CheY acetylation in the ΔcobB mutant (Fig. 6) is consistent with the possibility that loss of CobB activity decreases the rate of CheY deacetylation.
Experimental procedures
Materials
AcCoA, NAD+, NAM, synthetic l-leucine, d-galactose and NiSO4 were purchased from Sigma. All chemicals used were of the highest purity available. Anti-acetyl-lysine antibody was obtained from Cell Signaling Technology, and alkaline phosphatase-conjugated goat anti-rabbit antibody was from Sigma. Capillary pipettes (Drummond ‘Microcaps’, 1 µl capacity) were from Fisher Scientific.
Bacterial strains
Escherichia coli strains constructed in this work were derived from strain W3110, a K-12 derivative that is wild type for chemotaxis. All the strains and plasmids used are listed in Table 1. Strains AJW613 and AJW803 were generous gifts of A.J. Wolfe (Loyola University Chicago, Maywood, IL). The gene knockout mutants were constructed with the λ Red recombination system as previously described (Datsenko and Wanner, 2000). The mutants were verified by junction PCR and subsequent sequencing using primers that anneal to the genomic region outside the recombination locus. The double or triple gene knockout mutants were generated using the same procedures, except that the kanamycin resistance gene of the single-gene knockout mutant was first eliminated using the helper plasmid pCP20. All primers used are listed in Table S3.
Table 1.
Bacterial Strains and plasmids used in this study.
| Strain or plasmid | Relevant genotype/phenotype | Reference or source |
|---|---|---|
| E. coli strains | ||
| W3110 | Wild type for chemotaxis; F- λ- IN(rrnD-rrnE)1 rph-1 | Laboratory stock |
| RL001 | W3110 ΔcobB::Km | This study |
| RL002 | W3110 Δacs::Km | This study |
| RL0012 | W3110 ΔcobBΔacs::Km | This study |
| RL003 | W3110 ΔcheAΔcheZ::Km | This study |
| RL0031 | W3110 ΔcheAΔcobBΔcheZ::Km | This study |
| RL0032 | W3110 ΔcheAΔacsΔcheZ::Km | This study |
| AJW613 | Wild type for chemotaxis; ΔlacX74 thi-1 thr-1(Am) leuB6 metF159(Am) rpsL136λlacY | A.J. Wolfe |
| AJW803 | AJW613 Δacs::Km-1 | Kumari et al. (1995) |
| DH5α | Host for plasmid propagation | Laboratory stock |
| BL21(DE3) | Host for protein expression | Laboratory stock |
| AD494(DE3) | Host for protein expression, KmR | Laboratory stock |
| Plasmids | ||
| pKD46 | Red recombinase expression plasmids, ApR | Yale CGSC |
| pKD4 | Template plasmids containing a kanamycin resistance gene flanked by FRT sites, ApR, KmR | Yale CGSC |
| pCP20 | FLP helper plasmid, ApR, CmR | Yale CGSC |
| pET20b-cheY | CheY-His expression plasmids, ApR | This study |
| pTricHis2C-acs | Acs-His expression plasmids, ApR | This study |
| pET32a-cobB | His-CobB expression plasmids, ApR | This study |
| pET28a-fliM | His-FliM expression plasmids, KmR | This study |
Expression and purification of recombinant proteins
CobB, Acs, CheY and FliM encoding genes from E. coli W3110 genome were cloned into bacterial expression vectors (Table 1). Transformed E. coli AD494 (λDE3)/pET32a-cobB, E. coli DH5α/pTrcHis2C-acs, E. coli BL21 (λDE3)/pET20b-cheY and E. coli BL21 (λDE3)/pET28a-fliM were induced with isopropyl-β-d-thiogalactopyranoside (IPTG), lysed and purified by nickel affinity chromatography (detailed primer sequence, expression and purification are described in Supporting information).
In vitro deacetylation assays
To obtain acetylated CheY, we incubated CheY-His with acetyl-CoA and 50 mM Tris-HCl buffer (pH 8.0) for 20 h at 35°C, then separated AcCheY from the low-molecular-mass components by ultrafiltration (Barak et al., 2006). The acetylation level of AcCheY was determined by Western blotting and then mass spectrometry. Deacetylation of AcAcs and AcCheY by His-CobB was performed in 50 mM Tris-HCl buffer (pH 8.0) in the presence or absence of NAD+ (1 mM), and in the presence or absence of NAM (10 mM). Specific protein concentrations and reaction times are indicated in figure legends.
Preparation of anti-CheY antibodies
A polyclonal antibody against CheY was raised by immunization of rabbits with purified CheY protein. CheY antibodies were purified from rabbit serum by affinity chromatography using protein A beads (Beyotime Institute of Biotechnology, Beijing, China) with glycine-HCl as elutant, dialysed against phosphate-buffered saline. The titre of the purified antibody was determined by ELISA.
Analysis of CheY acetylation level in vivo
Strains W3110, RL001 and RL002 were grown to OD590 = 0.4–0.5 in Vogel-Bonner minimal medium supplemented with 30 mM acetate (Vogel and Bonner, 1956). Cells were harvested and lysed as described (Yan et al., 2008). Equal amounts (30 µg of total protein) of the cell-free extracts were subjected to SDS-PAGE. Three gels were used: one was stained with Coomassie blue, and the other two were used for Western blotting with CheY antibody and an anti-acetyl-lysine antibody.
Western blot analysis
The samples were separated by SDS-PAGE (12–15% acrylamide) and then transferred to PVDF membranes using a Bio-Rad SD device (Bio-Rad Laboratories) (20–30 min at 15 V). The membrane was blocked overnight at 4°C in 1× TBST (Tris buffered saline plus 0.1% Tween-20) containing 5% NFDM (non-fat dry milk). Primary rabbit anti-CheY (1:5000) or acetylated-lysine (1:1000) polyclonal antibodies were diluted in TBST/1% NFDM and incubated at 37°C for 2 h. The blot was washed with TBST and incubated with 1:5000 dilution of alkaline phosphatase-conjugated goat anti-rabbit antibody for 1 h at 37°C, then detected according to the manufacturer's instructions.
LC-MS/MS analysis
Prepared digested peptides were analysed with a Finnigan Surveyor HPLC system coupled online with a LTQ-Orbitrap mass spectrometer (Thermo Electron, San Jose, CA) equipped with a nanospray source. Briefly, the peptide mixtures were loaded onto a C18 column (100 µm i.d., 10 cm long, 5 µm resin from Michrom Bioresources, Auburn, CA) using an autosampler. Peptides were eluted with a 0–35% gradient (Buffer A, 0.1% formic acid and 5% ACN; Buffer B, 0.1% formic acid and 95% ACN) over 80 min and detected online in LTQ-Orbitrap mass spectrometer using a data-dependent TOP10 method (Haas et al., 2006) (detailed protein digestion, protein and peptide identification are described in Supporting information).
SPR analysis
Interactions between FliM and CheY, FliM and AcCheY were analysed using BIAcore 3000 (BIAcore AB, Uppsala, Sweden) at 25°C. Approximately 1120 RU His-FliM was immobilized on CM5 sensor chip with an amine-coupling protocol of the BIAcore manual. Before sample measurement, the sensor chip was equilibrated with running buffer at a rate of 40 µl min−1. The running buffer contained 10 mM HEPES (pH 7.4), 200 mM NaCl, 3 mM EDTA and 0.05% (v/v) P20. Samples were injected at different concentrations at a flow rate of 40 µl min−1 for 2 min. After 3–4 min dissociation phase, bound protein was removed with 30 s wash with 10 mM NaOH. No specific binding to a blank flow cell was subtracted to obtain corrected sensorgrams. Equilibrium and kinetic constants were calculated by a global fit to 1:1 Langmuir binding model (BIA evaluation 4.1 software).
Chemotaxis assays
Swarm assays were carried out on TB semi-solid agar plates [0.2% agar containing kanamycin (50 µg ml−1) when needed] as previously described (Adler, 1966). The migration rate was measured as described previously (Wolfe and Berg, 1989). Chemical-in-plug assays were also carried out as described for both attractants and repellents (Tso and Adler, 1974). Briefly, strains were grown in TB and washed twice in motility buffer (10 mM potassium phosphate buffer, pH 7.0; 100 µM EDTA). Bacteria were then homogeneously distributed in semi-solid agar (0.3%), and the stimulus-containing plug (2% agar) was placed at the centre of the plate before solidification of the agar. A modified drop assay was also performed, in which the stimulus (5 µl) was added directly to the centre of the bacteria-containing plate after solidification of the agar (Barak and Eisenbach, 2001). A modified capillary assay for positive chemotaxis was carried out as described (Han and Cooney, 1993). Capillaries containing the attractant were immersed in a suspension of bacteria and incubated for 1 h at 35°C. The number of bacteria in each capillary was determined by inoculating its content on a TB agar plate. If bacteria accumulated in the attractant-contained capillary to a greater extent than in the attractant-free capillary, attraction was considered to have taken place. Capillary assays for negative chemotaxis (chemical-in-pond) were carried out as described (Tso and Adler, 1974).
Acknowledgments
We thank Eric Verdin for providing plasmid pTrcHis2C-acs2 and E. coli Genetic Center (Yale CGSC) for providing plasmids pKD46, pKD4 and pCP20. This project was supported by China Academy of Science (KSCX2-YW-R-164).
Supporting information
Additional supporting information may be found in the online version of this article.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
References
- Adler J. Chemotaxis in bacteria. Science. 1966;153:708–716. doi: 10.1126/science.153.3737.708. [DOI] [PubMed] [Google Scholar]
- Baker MD, Wolanin PM, Stock JB. Signal transduction in bacterial chemotaxis. Bioessays. 2006;28:9–22. doi: 10.1002/bies.20343. [DOI] [PubMed] [Google Scholar]
- Barak R, Eisenbach M. Acetylation of the response regulator, CheY, is involved in bacterial chemotaxis. Mol Microbiol. 2001;40:731–743. doi: 10.1046/j.1365-2958.2001.02425.x. [DOI] [PubMed] [Google Scholar]
- Barak R, Eisenbach M. Co-regulation of acetylation and phosphorylation of CheY, a response regulator in chemotaxis of Escherichia coli. J Mol Biol. 2004;342:375–381. doi: 10.1016/j.jmb.2004.07.021. [DOI] [PubMed] [Google Scholar]
- Barak R, Welch M, Yanovsky A, Oosawa K, Eisenbach M. Acetyladenylate or its derivative acetylates the chemotaxis protein CheY in vitro and increases its activity at the flagellar switch. Biochemistry. 1992;31:10099–10107. doi: 10.1021/bi00156a033. [DOI] [PubMed] [Google Scholar]
- Barak R, Abouhamad WN, Eisenbach M. Both acetate kinase and acetyl coenzyme A synthetase are involved in acetate-stimulated change in the direction of flagellar rotation in Escherichia coli. J Bacteriol. 1998;180:985–988. doi: 10.1128/jb.180.4.985-988.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barak R, Prasad K, Shainskaya A, Wolfe AJ, Eisenbach M. Acetylation of the chemotaxis response regulator CheY by acetyl-CoA synthetase purified from Escherichia coli. J Mol Biol. 2004;342:383–401. doi: 10.1016/j.jmb.2004.07.020. [DOI] [PubMed] [Google Scholar]
- Barak R, Yan J, Shainskaya A, Eisenbach M. The chemotaxis response regulator CheY can catalyze its own acetylation. J Mol Biol. 2006;359:251–265. doi: 10.1016/j.jmb.2006.03.033. [DOI] [PubMed] [Google Scholar]
- Bren A, Welch M, Blat Y, Eisenbach M. Signal termination in bacterial chemotaxis: CheZ mediates dephosphorylation of free rather than switch-bound CheY. Proc Natl Acad Sci USA. 1996;93:10090–10093. doi: 10.1073/pnas.93.19.10090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA, Wanner BL. One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci USA. 2000;97:6640–6645. doi: 10.1073/pnas.120163297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dryden SC, Nahhas FA, Nowak JE, Goustin AS, Tainsky MA. Role for human SIRT2 NAD-dependent deacetylase activity in control of mitotic exit in the cell cycle. Mol Cell Biol. 2003;23:3173–3185. doi: 10.1128/MCB.23.9.3173-3185.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyer CM, Dahlquist FW. Switched or not?: the structure of unphosphorylated CheY bound to the N terminus of FliM. J Bacteriol. 2006;188:7354–7363. doi: 10.1128/JB.00637-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haas W, Faherty BK, Gerber SA, Elias JE, Beausoleil SA, Bakalarski CE, et al. Optimization and use of peptide mass measurement accuracy in shotgun proteomics. Mol Cell Proteomics. 2006;5:1326–1337. doi: 10.1074/mcp.M500339-MCP200. [DOI] [PubMed] [Google Scholar]
- Han G, Cooney JJ. A modified capillary assay for chemotaxis. J Ind Microbiol Biotechnol. 1993;12:396–398. [Google Scholar]
- Han Y, Jin YH, Kim YJ, Kang BY, Choi HJ, Kim DW, et al. Acetylation of Sirt2 by p300 attenuates its deacetylase activity. Biochem Biophys Res Commun. 2008;375:576–580. doi: 10.1016/j.bbrc.2008.08.042. [DOI] [PubMed] [Google Scholar]
- Kumari S, Tishel R, Eisenbach M, Wolfe AJ. Cloning, characterization, and functional expression of acs, the gene which encodes acetyl coenzyme A synthetase in Escherichia coli. J Bacteriol. 1995;177:2878–2886. doi: 10.1128/jb.177.10.2878-2886.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Langley E, Pearson M, Faretta M, Bauer UM, Frye RA, Minucci S, et al. Human SIR2 deacetylates p53 and antagonizes PML/p53-induced cellular senescence. EMBO J. 2002;21:2383–2396. doi: 10.1093/emboj/21.10.2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larsen MR, Trelle MB, Thingholm TE, Jensen ON. Analysis of posttranslational modifications of proteins by tandem mass spectrometry. Biotechniques. 2006;40:790–798. doi: 10.2144/000112201. [DOI] [PubMed] [Google Scholar]
- Lee SY, Cho HS, Pelton JG, Yan D, Henderson RK, King DS, et al. Crystal structure of an activated response regulator bound to its target. Nat Struct Biol. 2001;8:52–56. doi: 10.1038/83053. [DOI] [PubMed] [Google Scholar]
- Mayover TL, Halkides CJ, Stewart RC. Kinetic characterization of CheY phosphorylation reactions: comparison of P-CheA and small-molecule phosphodonors. Biochemistry. 1999;38:2259–2271. doi: 10.1021/bi981707p. [DOI] [PubMed] [Google Scholar]
- Ramakrishnan R, Schuster M, Bourret RB. Acetylation at Lys-92 enhances signaling by the chemotaxis response regulator protein CheY. Proc Natl Acad Sci USA. 1998;95:4918–4923. doi: 10.1073/pnas.95.9.4918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rogina B, Helfand SL. Sir2 mediates longevity in the fly through a pathway related to calorie restriction. Proc Natl Acad Sci USA. 2004;101:15998–16003. doi: 10.1073/pnas.0404184101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- Silversmith RE, Bourret RB. Throwing the switch in bacterial chemotaxis. Trends Microbiol. 1999;7:16–22. doi: 10.1016/s0966-842x(98)01409-7. [DOI] [PubMed] [Google Scholar]
- Sourjik V. Receptor clustering and signal processing in E. coli chemotaxis. Trends Microbiol. 2004;12:569–576. doi: 10.1016/j.tim.2004.10.003. [DOI] [PubMed] [Google Scholar]
- Starai VJ, Celic I, Cole RN, Boeke JD, Escalante-Semerena JC. Sir2-dependent activation of acetyl-CoA synthetase by deacetylation of active lysine. Science. 2002;298:2390–2392. doi: 10.1126/science.1077650. [DOI] [PubMed] [Google Scholar]
- Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. Short-chain fatty acid activation by acyl-coenzyme A synthetases requires SIR2 protein function in Salmonella enterica and Saccharomyces cerevisiae. Genetic. 2003;163:545–555. doi: 10.1093/genetics/163.2.545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Starai VJ, Takahashi H, Boeke JD, Escalante-Semerena JC. A link between transcription and intermediary metabolism: a role for Sir2 in the control of acetyl-coenzyme A synthetase. Curr Opin Microbiol. 2004;7:115–119. doi: 10.1016/j.mib.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Tso WW, Adler J. Negative chemotaxis in Escherichia coli. J Bacteriol. 1974;118:560–576. doi: 10.1128/jb.118.2.560-576.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel HJ, Bonner DM. Acetylornithinase of Escherichia coli: partial purification and some properties. J Biol Chem. 1956;218:97–106. [PubMed] [Google Scholar]
- Volz K, Matsumura P. Crystal structure of Escherichia coli CheY refined at 1.7-A resolution. J Biol Chem. 1991;266:15511–15519. doi: 10.2210/pdb3chy/pdb. [DOI] [PubMed] [Google Scholar]
- Welch M, Oosawa K, Aizawa S, Eisenbach M. Phosphorylation-dependent binding of a signal molecule to the flagellar switch of bacteria. Proc Natl Acad Sci USA. 1993;90:8787–8791. doi: 10.1073/pnas.90.19.8787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ. The acetate switch. Microbiol Mol Biol Rev. 2005;69:12–50. doi: 10.1128/MMBR.69.1.12-50.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Berg HC. Migration of bacteria in semisolid agar. Proc Natl Acad Sci USA. 1989;86:6973–6977. doi: 10.1073/pnas.86.18.6973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolfe AJ, Conley MP, Berg HC. Acetyladenylate plays a role in controlling the direction of flagellar rotation. Proc Natl Acad Sci USA. 1988;85:6711–6715. doi: 10.1073/pnas.85.18.6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan J, Barak R, Liarzi O, Shainskaya A, Eisenbach M. In vivo acetylation of CheY, a response regulator in chemotaxis of Escherichia coli. J Mol Biol. 2008;376:1260–1271. doi: 10.1016/j.jmb.2007.12.070. [DOI] [PubMed] [Google Scholar]
- Zhao R, Collins EJ, Bourret RB, Silversmith RE. Structure and catalytic mechanism of the E. coli chemotaxis phosphatase CheZ. Nat Struct Biol. 2002;9:570–575. doi: 10.1038/nsb816. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


