Abstract
The Caenorhabditis elegans one-cell embryo is a powerful system in which to study microtubule organization because this large cell assembles both meiotic and mitotic spindles within the same cytoplasm over the course of 1 h in a stereotypical manner. The fertilized oocyte assembles two consecutive acentrosomal meiotic spindles that function to reduce the replicated maternal diploid set of chromosomes to a single-copy haploid set. The resulting maternal DNA then unites with the paternal DNA to form a zygotic diploid complement, around which a centrosome-based mitotic spindle forms. The early C. elegans embryo is amenable to live-cell imaging and electron tomography, permitting a detailed structural comparison of the meiotic and mitotic modes of spindle assembly.
Keywords: Mitosis, Meiosis, Microtubules, Caenorhabditis elegans, Spindle, 3D, Electron tomography
Introduction
Meiotic and mitotic spindles are marvels of intracellular architecture. These complex microtubule-based structures comprise a myriad of molecular components that together facilitate the segregation of genetic material within the germline and somatic tissues. By studying the structural features of spindles, we hope to better understand how these higher-order structures form and function in the cell. This review focuses on some of the recent progress made using molecular genetics and electron microscopy to study microtubule organization in both meiotic and mitotic spindles in Caenorhabditis elegans.
The early events in the C. elegans embryo have been well documented (Fig. 1). An oocyte arrested in prophase of meiosis I becomes fertilized in the proximal region of the hermaphrodite gonad while passing through the spermatheca [1]. A signal generated by the sperm promotes maturation of the oocytes just prior to fertilization [2, 3]. Although unfertilized embryos can continue developing until anaphase of meiosis I, fertilization of the oocyte normally occurs within a few minutes after entering the spermatheca, and this is essential for the completion of the meiotic divisions [4–6]. The sperm contributes a haploid set of paternal chromosomes and a pair of centrioles to the oocyte cytoplasm. However, these sperm-derived structures do not directly participate in meiotic spindle assembly.
Fig. 1.
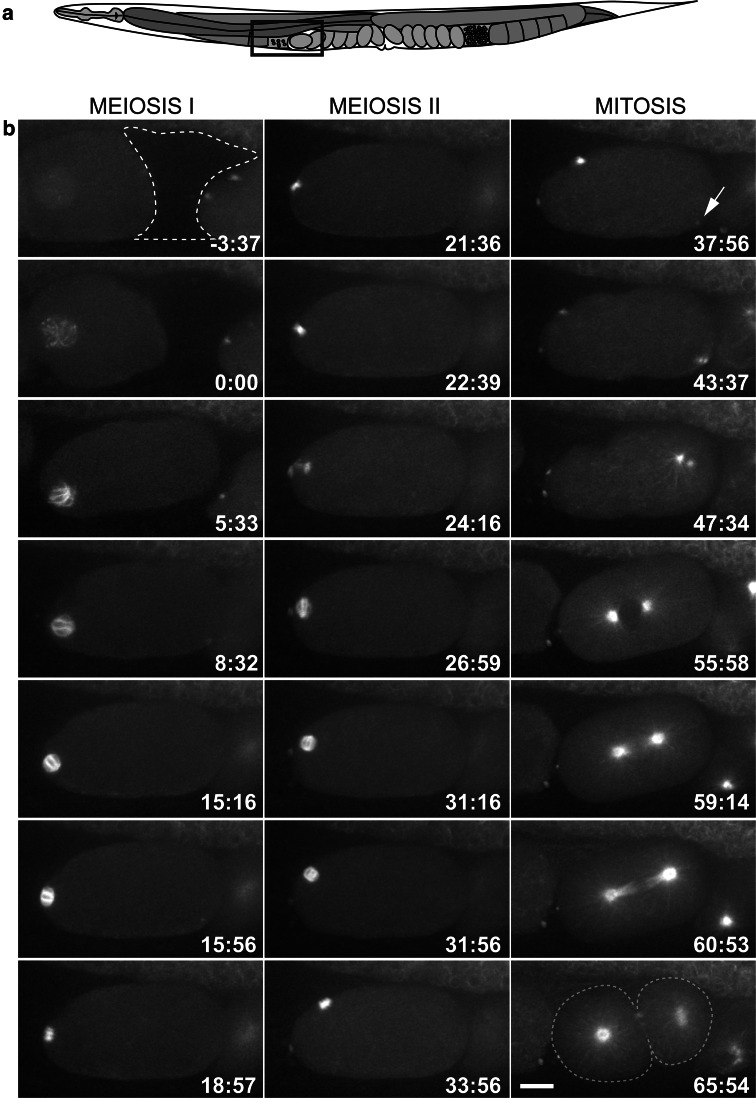
Development of the one-cell C. elegans embryo as viewed by in utero-imaging of a worm expressing GFP::β-tubulin. a Schematic drawing of an adult hermaphrodite. The box depicts the region of the uterus imaged. b Microtubules are observed by fluorescent light microscopy around the oocyte nucleus (t = 0) before it enters the spermatheca (dashed line). Anterior is left in all panels. The oocyte usually enters the spermatheca within 5 min, at which time fertilization occurs and a pointed bipolar array becomes visible. Each round of meiosis takes approximately 20 min. At the end of meiosis II (t = 37:56), the centrosomal microtubules become visible (arrow), and centrosome separation begins soon after. The mitotic spindle forms in the center of the embryo and the spindle skews towards the posterior in anaphase to produce two daughter cells of unequal size. Scale bar is 10 μm in b
Upon fertilization of the oocyte, amorphous arrays of microtubules can be distinguished around the female chromosomes. As the chromosomes condense, this array of microtubules is arranged in an elongated (‘pointed’) bipolar spindle that is about 13 μm long at early metaphase. This meiosis I spindle then shortens to roughly 3–4 μm and adopts a barrel-like shape, and this shortening is accompanied by a rotation of the spindle relative to the surface of the embryo. At anaphase, chromatin is separated to a maximum of 3–4 μm with microtubules arranged between the chromosomes. The second meiotic division then follows a similar pattern [7, 8].
After the completion of meiosis, female and male pro-nuclei form and increase in size. The female pro-nucleus migrates from the anterior hemisphere of the embryo to meet the male pro-nucleus in the posterior. Occasionally both pronuclei reform in the same end of the one-cell embryo; however, it is the sperm position that specifies posterior fate [9–13]. After fertilization, the sperm-delivered centrioles stay associated with the male pro-nucleus and accumulate maternally provided pericentriolar material (PCM) required for the nucleation of microtubules. The microtubules grow out to make contact with the female pro-nucleus and this connection facilitates a dynein-dependent migration and meeting of the two pro-nuclei [14–16]. Continuing with this largely invariant developmental pattern, the two pro-nuclei meet, and the pro-nuclei/centrosome complex migrates to the center of the embryo and rotates. The chromosomes then align on the metaphase plate, and after a few minutes, the centrosomes separate and pull sister chromatids apart in anaphase [17]. A precise displacement of the mitotic spindle towards the posterior at this time positions the plane of cleavage such that the anterior daughter cell is larger than the posterior daughter (for a review, see [18]).
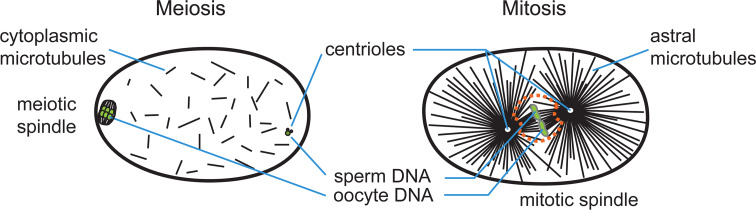
There are two major structural differences between the two female meiotic spindles and the first mitotic spindle in C. elegans (Fig. 2). First, the meiotic spindles are anastral and relatively small (~3–4 μm) at metaphase [7, 8]. In contrast, the metaphase mitotic spindle of the one-cell embryo is large (~14 μm) and has prominent spindle poles with a profusion of astral microtubules [17]. Second, meiotic spindle assembly takes place around chromatin in an acentrosomal environment. In contrast, mitotic spindle assembly in C. elegans is strictly dependent on centrosomes. These structural differences imply that distinct mechanisms determine how microtubules are formed and modulated in these two systems (for reviews, see [19–21]). The early C. elegans embryo assembles the meiotic and mitotic spindles sequentially within the same cytoplasm over the course of 1 h, making it an exceptional model to study both spindle types. Here, we review the structure of microtubules, the modulation of microtubule dynamics, the structure and function of the centrosome, and the mechanisms of acentrosomal and centrosomal spindle assembly in the elegant worm.
Fig. 2.
Schematic comparison of spindle assembly in meiosis and mitosis in the early C. elegans embryo. Chromatin is shown in green. The nuclear envelope (orange dashes) persists in the early stages of mitotic spindle assembly, but disappears by anaphase. Microtubules initially invade the nuclear space through polar fenestrae
Microtubules
Microtubules provide a major structural and mechanical role in the assembly and function of all spindles. Microtubules are fiber-like polymers composed of α/β tubulin-heterodimers that are added during polymerization (growth) or removed during depolymerization (shrinkage) [22, 23]. Most tubulin dimers in solution have GTP bound to both subunits. Hydrolysis of the β-subunit-bound GTP to GDP occurs after polymerization, such that microtubules initially have a GTP-capped growing end, with hydrolysis to GDP triggering depolymerization (reviewed in [24]). A single string of tubulin subunits assembled head-to-tail (α/β, α/β,…) constitutes a microtubule protofilament. Lateral association of protofilaments results in the formation of a 2D sheet that closes to form the hollow microtubule cylinder, the predominant form of polymeric tubulin found in the cell [25, 26]. Conversely, lateral dissociation of protofilaments results in the disassembly of the microtubule lattice [27, 28].
Due to the inherent asymmetry of the α/β tubulin heterodimer, the polymer exhibits a polarity, with a β “plus” and an α “minus” end [29]. This polarity has consequences for microtubule dynamics because heterodimers are preferentially added to or subtracted from the plus end; microtubules grown in vitro display fast-growing plus ends and slow-growing minus ends [22]. Electron microscopy has revealed that the structure of a growing microtubule tip is very different from one that is shrinking. At the plus end of a growing microtubule, the tube is split open along the longitudinal axis, giving a flared 2D sheet at the tip; the open seam closes back into a tube toward the minus end [25, 26]. In contrast, microtubules that are actively shrinking display a striking “ram’s horns” morphology with individual protofilaments peeling away from the fast depolymerizing end [27, 28]. Importantly, these tip morphologies are often diagnostic for microtubule behavior, allowing inferences to be made on the polarity and dynamic state of microtubules within cells.
The intrinsic property of microtubules to undergo stochastic changes between phases of slow growth and fast shrinkage (termed dynamic instability) allows the microtubule cytoskeleton to rapidly adapt to the changing requirements of the cell [22]. Furthermore, dynamic microtubules confer considerable morphological plasticity to the structures that they form within different cell types such as neurons or fibroblasts, or within different cell cycle states, such as interphase or mitosis. Therefore, intracellular factors that control microtubule dynamics will ultimately direct the timing, placement and architecture of higher order structures such as the spindle. We next discuss a few well-studied examples of proteins and protein complexes that regulate microtubule behavior in the cell.
To be or not to be a microtubule: regulating polymer dynamics
Preparation for mitosis requires a significant reorganization of the cytoskeleton. The interphase array of microtubules must be remodeled into a dynamic bipolar mitotic array that can segregate chromatids and position the site of the future cytokinesis furrow [30, 31]. This microtubule reorganization requires changes to individual microtubule dynamics as well as to the rate of microtubule nucleation (for a review on regulators of microtubule dynamics, see [32]).
A number of microtubule stabilizing proteins have been identified through biochemical purification methods, including early efforts [33–38], as well as the more recent mass spectrometry approaches using microtubule pellets from Drosophila embryos [39]. Since their identification, many of these microtubule-associated proteins (MAPs) have turned out to be important regulators of microtubule behavior during spindle assembly (for a review of MAPs, see [40]). One well-studied MAP that promotes microtubule growth is the microtubule polymerase XMAP215. In vitro work suggests that these conserved proteins bind α/β-tubulin heterodimers and assist in supplying them to the growing microtubule end [41–43]. Curiously, in vivo, XMAP215 homologues are most prevalent at spindle poles, but the protein is not often detectable on growing microtubule plus ends further away from the spindle poles. It could be that microtubule plus-end growth requires XMAP215 for polymerization near the surface of the centrosome, but not further away from it. Future work with new probes and advanced imaging techniques may shed light on this seemingly contrary result. In C. elegans, the XMAP215 homologue ZYG-9 (zygote-defective) is required for meiotic and mitotic spindle assembly [44], and for normal microtubule polymerization rates [45].
Although the stabilization of microtubules is clearly important for the assembly of structures such as the spindle, equally important is the controlled removal of tubulin subunits. In contrast to microtubule polymerases, the kinesin-13 family of microtubule depolymerases has been shown to induce microtubule shrinkage [46, 47]. These kinesins do not exhibit traditional motor movement along microtubules, but seem to be specialized for regulating microtubule growth behavior [48–50]. In vitro, MCAK accumulates at both the plus and the minus ends of microtubules. Interestingly, this pattern is mirrored in vivo, with prevalent location of the protein at spindle poles as well as chromosomes during mitosis. One of the roles for kinesin-13 proteins is to correct mis-attached chromatids during spindle assembly [51–56]. When microtubules from the same pole attach to both sister chromatids (syntelic attachment), or when microtubules from opposite poles attach to one chromatid (merotelic attachment), chromosomally located kinesin-13 induces microtubule depolymerization to allow re-attachment of each sister to microtubules from opposite poles. Another role for these kinesins is in regulating microtubule outgrowth from the centrosome [45, 57, 58]. When the C. elegans kinesin-13 member KLP-7 (kinesin-like protein) is depleted, the centrosomes exhibit a twofold increase in the number of astral microtubules [57]. Separate chromatin- and centrosome-located kinesin-13 members are used in Drosophila to modulate the dynamics of microtubules during spindle assembly and the shortening of spindle microtubules in anaphase A [59]. The activities of both microtubule polymerases and depolymerases can be balanced such that microtubule dynamics are tuned to fit the needs of a highly dynamic bipolar spindle array [60].
The centrosome: a primary generator of microtubules
In vitro experiments performed 25 years ago indicated that microtubules can spontaneously polymerize from purified components if the concentration of tubulin is sufficiently high (~14 μM) [22]. However, in many cells microtubules rarely spontaneously polymerize in the cytoplasm, rather they emanate from specific microtubule organizing centers (MTOCs) [61]. Indeed, microtubules polymerize in vitro at a much lower concentration (~3 μM) when centrosomal MTOCs are present [22]. In the early one-cell embryo of C. elegans, one can detect individual microtubules growing in random directions throughout the cytoplasm, in the absence of obvious MTOCs [45]. Spontaneous polymerization of microtubules at this stage is perhaps due to a generous maternal contribution of tubulin present in the early embryonic cytoplasm. However, this population of microtubules completely disappears by metaphase of mitosis, concomitantly with the maturation of the centrosomes, which rapidly take over the role of nucleating microtubules. From a combination of live imaging and electron tomography, we estimate that over 2,000 microtubules (representing both growing and shrinking populations) can emanate from a single centrosome in one-cell C. elegans embryos.
The size, position, composition and function of MTOCs can vary greatly between different cells or different organisms. The major MTOC in most animal cells is the centrosome, composed of a pair of centrioles surrounded by pericentriolar material (PCM). An analogous structure in budding yeast is the spindle pole body (SPB) [62]. However, unlike the spherical centrosome, this multi-layered structure is embedded in the nuclear membrane. Both the SPB and the centrosome provide a nucleating surface from which microtubules emanate. Microtubules growing out from these MTOCs arrange with a typical polarity: microtubule minus ends are anchored to the nucleating surface, and the plus ends grow away from it. In S. cerevisiae, nuclear microtubules grow towards chromosomes from the nuclear SPB surface (the nuclear envelope does not break down during mitosis), whereas cytoplasmic microtubules grow out from the cytoplasm-facing surface [63]. In higher eukaryotes, centrosomal microtubules have the potential to grow out radially in all directions, although different systems display varying levels of symmetry in the microtubule arrays that form.
Centrioles: at the heart of the centrosome
By electron microscopy the centrosome appears as an electron-dense region from which microtubules radiate. At the center of the centrosome is a centriole pair, consisting of a mother centriole and a daughter centriole arranged orthogonally to each other (Fig. 4a). In animal cells the centriole is required for both the formation of centrosomes and cilia/flagella [64]. Each centriole is a cylindrical structure that can range in diameter from 100 to 250 nm, and in length from 100 to 400 nm, depending on the organism (for a review, see [65]). Ultrastructural studies have shown that the embryonic centriole in C. elegans is ‘simple’ in structure, with a central tube surrounded by nine singlet microtubules arranged parallel to the long axis of the tube [17, 66–68]. Centriolar microtubules are composed of α/β-tubulin heterodimers but, unlike their cytoplasmic counterparts, they are extremely stable [69]. The ninefold symmetry of centriolar microtubules is widespread in nature. However, the Drosophila embryo has nine doublet microtubules, and mammalian centrosomes have nine triplet microtubules [65]. The doublet or triplet microtubules are thought to be due, in part, to δ- and ε-tubulin subunits, both of which are absent in C. elegans [70].
Fig. 4.
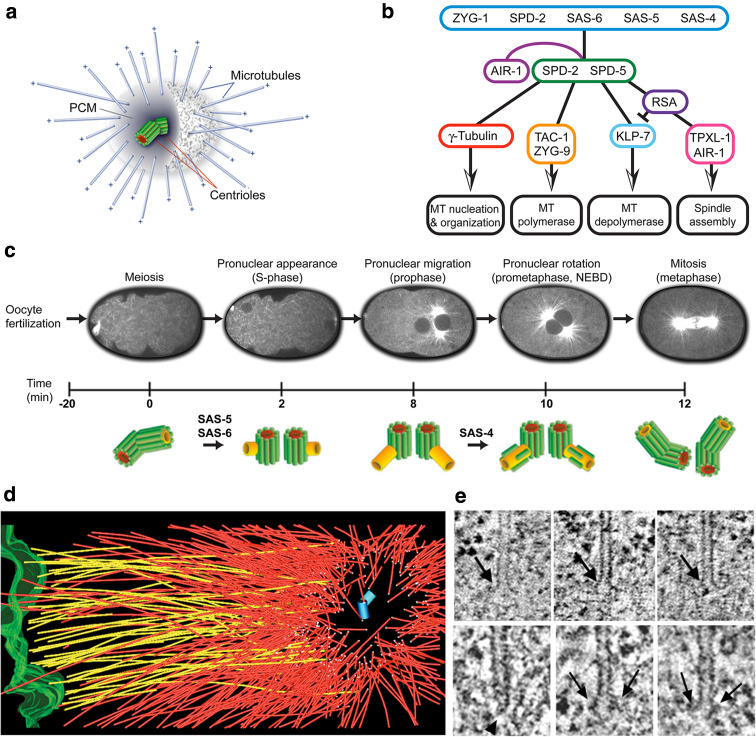
Mitotic spindle organization in the C. elegans embryo. a Schematic representation of the C. elegans centrosome, with a pair of centrioles surrounded by pericentriolar material (PCM) and minus-end nucleated/anchored microtubules. b Functional levels of the centrosome. Cartoon illustrating core proteins responsible for centriole duplication (first level) and PCM assembly (second level). Proteins of the third level influence microtubule nucleation, polymerization and depolymerization. The aurora kinase AIR-1 is involved at multiple steps, in both the maturation of the centrosome and the regulation of downstream effectors. c Ultrastructural pathway of centriole duplication in C. elegans early embryos (modified from [68]). d Partial reconstruction of a metaphase half-spindle. Microtubules that contact the chromosomes (green) are designated kinetochore microtubules (shown in yellow). Other microtubules are displayed in red. Microtubule ends in the spindle pole, surrounding the centriole pair (blue cylinders), are indicated by white spheres. e Kinetochore microtubules have an open, flared plus-end morphology (upper row). About 80% of the microtubules in the centrosome (lower row) have closed, capped minus ends (arrowhead), although some microtubules with open ends (arrows) are observed (modified from [71])
In C. elegans, the mother centriole is positioned at the center of the centrosome, and the daughter centriole is slightly off-center [71]. The daughter centriole does not appear to be necessary for PCM accumulation or organization. For instance, when centriole duplication is blocked, centrosomes containing only a mother centriole still accumulate wild-type levels of PCM, nucleate wild-type numbers of microtubules, and the cell cycle proceeds normally through the first round of mitosis [45, 72].
Genetic and genome-wide RNAi screens have shown that centriole duplication in C. elegans is dependent on at least five components of the centrosome: the conserved protein SPD-2 (spindle defective) [73, 74], the kinase ZYG-1 [67] and the coiled-coil proteins SAS-4 (spindle assembly) [72, 75], SAS-5 [76] and SAS-6 [77, 78] (Fig. 4b). Electron tomography of the C. elegans one-cell-stage embryo through the first mitotic division revealed that daughter centriole assembly begins with the formation and elongation of a hollow tube, which forms in close proximity and orthogonal to the mother centriole. The peripheral assembly of the nine singlet microtubules around the central tube then completes the daughter centriole [68] (Fig. 4c). Coupling of electron tomography with RNAi-based depletions of centriole components revealed that SPD-2 and ZYG-1 are required for the initiation of centriole duplication. In a ‘second wave,’ SAS-6 and SAS-5 are required for central tube formation and elongation, and finally, SAS-4 is required for the assembly of the nine singlet microtubules onto the central tube [68]. Centriolar SAS-4 has been reported to be in a dynamic equilibrium with the cytoplasmic pool until late prophase, when it is stably incorporated in a step that requires γ-tubulin and microtubule assembly [79]. With the exception of SAS-5, counterparts of these centriole proteins have been found in other species [80–89]. Specifically, Chlamydomonas SAS-6 (bld12p) is a component of the pinwheel/cartwheel in the proximal end of the centriole, establishing the ninefold symmetry [87]. Furthermore, the central hub of the cartwheel was not detected in bld12p mutants. Likewise, DSAS-6 organizes a tube-like centriole in Drosophila [85]. An analogous central tube has not been identified yet in human centrioles, at any stage of the duplication process.
From centrioles to centrosomes
Through largely unknown mechanisms, centrioles provide a blueprint for centrosome assembly and centriole duplication. These two functions limit centrosome numbers to two per cell, each of which forms a spindle pole [65, 90]. SPD-2 and SAS-4 localize both to the centriole and to the pericentriolar area. While spd-2(RNAi) results in a centriole duplication defect, it also results in smaller, immature and mis-positioned centrosomes [73, 74]. This suggests that SPD-2 is likely a part of the molecular machinery linking centrioles to PCM recruitment.
Depletion of SAS-4 also results in a centriole duplication defect, which is visible as a monopolar spindle array in each cell of the two-cell-stage embryo [72]. Interestingly, weak sas-4(RNAi) results in asymmetric spindles in the two-cell-stage embryo, such that each half of the spindle is a different length. By electron microscopy these asymmetric spindles also have differently sized centrosomes. The larger centrosome contains a single ‘complete,’ paternally derived centriole, and the smaller centrosome contains a single incomplete centriole with centriolar microtubules missing. Furthermore, these smaller centrosomes with incomplete centrioles were shown by light microscopy to contain less SAS-4 and γ-tubulin. Thus, SAS-4 levels in the cell contribute to centriole structure and PCM size. The effect on PCM seems to be directly related to the centriolar microtubule defect because the paternally derived centrioles have normal PCM recruitment, despite sharing the same cytoplasm with their defective daughter, which has lower levels of PCM [72].
Another link between the centriole and centrosome is the recently described RNA-binding protein SZY-20 (suppressor of ZYG-1) [91]. szy-20 was identified in a screen for genetic suppressors of a temperature-sensitive (ts) mutation in the zyg-1 gene [92]. Depletion of SZY-20 results in larger centrosomes that nucleate more microtubules and an increase in the level of ZYG-1 at the centrosome [91]. This latter result is likely responsible for the suppression of the temperature-sensitive zyg-1 mutant. Interestingly, artificially increasing the size of PCM in vertebrate somatic cells can also stimulate the overproduction of centrioles [93] in a manner reminiscent of de novo centriole assembly in cultured cells [94]. It could be that the PCM itself positively reinforces centriole assembly by bringing the necessary factors closer to the centriole [77]. Centriole enlargement or over-duplication was not observed in the szy-20 mutant [91], suggesting that centriole duplication could be regulated through a number of distinct pathways, depending on the tissue-type or organism.
Centrosome size increases during the course of mitosis. During this centrosome maturation process, additional proteins are recruited to promote robust microtubule outgrowth and assemble the bipolar spindle. Like other PCM components, the worm aurora-A kinase, AIR-1 (Aurora and Ipl1-related), localizes to the centrosome throughout the cell cycle, but its levels increase significantly at the onset of mitosis, reaching a maximum around anaphase [95]. While many PCM components have been identified in various systems, the picture is still unclear as to how the centrosome assembles in terms of the protein-protein interactions and protein modifications involved [96]. In C. elegans, the core PCM components required for the recruitment of all other factors are SPD-2 [73, 74] and SPD-5 [10]. These proteins are required for centrosome growth and maturation. Concomitant with centrosome maturation, many proteins required for spindle assembly accumulate at the centrosome, such as those required for microtubule nucleation, growth and shrinkage.
γ-Tubulin: supporting microtubule outgrowth from the centrosomal surface
The outer surface of the PCM provides a platform for the nucleation of microtubules and a point of attachment for their minus ends [97]. One factor that is thought to promote the nucleation and/or stabilization of microtubule minus ends is γ-tubulin. This protein was identified through work in the fungus Aspergillus and is widely conserved through evolution (for reviews, see [98, 99]). In Aspergillus depletion of γ-tubulin resulted in a reduction in the number and length of microtubules, and had severe effects on mitotic spindle assembly [100]. In all organisms examined so far, γ-tubulin is part of a heteromeric complex that contains two members of the Spc97/Spc98 protein family conserved from Drosophila to humans [101, 102]. Both proteins contain two gamma ring protein (grip) domains, a characteristic shared by many γ-tubulin interacting proteins [101]. Depletion of CeGrip1 leads to the inability to recruit γ-tubulin to the centrosome [103]. This complex, termed the “small” γ-tubulin complex (γ-TuSC), is a subunit of a larger complex referred to as the γ-tubulin ring complex (γ-TuRC) [104, 105]. The γ-TuRC has the shape of an open ring with a diameter of approximately 25 nm, similar to a microtubule. Within this ring each γ-tubulin molecule is thought to be positioned at the end of each protofilament [106–108], although an alternative model has been proposed [109]. The γ-TuRC possesses microtubule capping and nucleating activity in vitro [105, 110]. Electron microscopy studies looking at the structure of microtubules assembled from γ-tubulin in vitro revealed a cap-like structure at microtubule minus ends [106–108]. Similar cone-shaped structures have also been observed at the minus ends of microtubules nucleated from SPBs of budding yeast [63, 111–113] and centrosomes from Drosophila [114] and C. elegans [71].
Although γ-tubulin appears to provide kinetically dominant sites for microtubule nucleation, it is apparently not absolutely necessary for the formation of centrosomal microtubules in vivo. Surprisingly, γ-tubulin-depleted early C. elegans embryos contain extensive microtubule arrays, but fail in chromosome segregation and cytokinesis [103, 115]. In addition, the overall organization of the centrosome appears to be disrupted in tbg-1(RNAi) embryos, such that microtubules extend into the core of the centrosome rather than abutting near the outer surface [103, 115]. Electron tomography of centrosomes in tbg-1(RNAi) embryos reveals that centrioles lose their central position within the centrosome (O’Toole, Srayko and Müller-Reichert, unpublished results). Also the end morphology of the microtubule minus ends is altered. Upon depletion of γ-tubulin, the cone-shaped structure as seen in wild-type embryos is lost, and the majority of the microtubule ends are open as observed by electron tomography (O’Toole, Srayko, and Müller-Reichert, unpublished results). Therefore, centrosomal γ-tubulin could function primarily in anchoring and organizing of microtubules at the centrosome, rather than in their nucleation per se.
Structurally, the nature of the PCM remains one of the biggest mysteries of the centrosome field, and we have only a rudimentary knowledge about this ‘filamentous network’ that can carry out all the functions of the animal spindle pole.
Acentrosomal spindle assembly: female meiosis in C. elegans
Although most animal cells use centrosomes to nucleate microtubules when building the bipolar spindle apparatus, one exception is widespread: the female meiotic spindle [7, 116–118]. The prevalence of acentrosomal spindles in female meiosis and higher plant cells poses the question: are centrosomes absolutely necessary for spindle assembly in mitotic animal cells? A growing number of experiments suggest that alternative non-centrosomal pathways indeed exist in vertebrate cells [119, 120] as well as in Drosophila [82, 121–123]. For C. elegans, it appears that a non-centrosomal pathway is either not present or not sufficient for the first mitotic division, despite its use during female meiosis. The absence of centrosomal MTOCs in female meiosis has a profound impact on microtubule nucleation, spindle assembly and the mechanism of chromosome segregation, yet it does not seem to affect the overall fidelity of the process. We next discuss the loss of centrosomes from the female germline and the consequences of this loss on meiotic spindle assembly.
No centrosomes? No problem! (…at least not for female meiosis)
Centriole elimination resulting in sex-specific dimorphism of the female and male meiotic spindles is prevalent throughout the animal kingdom, suggesting that this system has evolved to ensure that the first mitosis of the zygote proceeds only when both male and female genetic material is present. Centriole elimination from the oocyte rather than the sperm appears to be more common. For many organisms, the centriole plays an important role as a basal body for flagellar production and motility [64, 124]. However, C. elegans sperm are amoeboid and crawl towards the oocyte using the contractility of a unique cytoskeletal polymer known as the MSP (major sperm protein) [125]. Centriole elimination from the oocytes as opposed to the sperm, therefore, could be advantageous, because the acentrosomal spindles lack robust astral microtubules, which might help limit the amount of cytoplasm lost upon polar body extrusion [7].
The process by which centrioles are eliminated from the oocyte cytoplasm is largely mysterious. In C. elegans, centrioles disappear during oocyte maturation, which occurs within the reflexed region of the gonad arm [7, 126]. Although the mechanism is unclear, one gene implicated in centriole elimination is the cyclin-dependent kinase inhibitor cki-2, which negatively regulates the cyclin E-Cdk1 complex [126]. As electron tomography has been used successfully to visualize the structural pathway of centriole assembly in the early C. elegans embryo [68], it is likely that this technique could also be applied to document structural changes to the centrosome throughout the elimination process in oocytes.
With the centrioles eliminated from the cytoplasm, oocytes must use non-centrosomal mechanisms to build a bipolar spindle for the two ensuing meiotic divisions. One important feature of acentrosomal spindle assembly is that chromatin, rather than centrosomes, seems to promote the formation of microtubules. Indeed, early work in centrosome-free Xenopus extracts revealed that even plasmid DNA-coated beads can stabilize and organize microtubules into a bipolar spindle-like structure [127]. At least one factor that is required for the stabilization of microtubules near chromatin in many systems is the small GTPase Ran, which can bind to either GDP or GTP (reviewed in [128]. The chromosomal guanine exchange factor RCC1 is responsible for generating Ran-GTP [129]. Ran-GTP concentration is therefore high near chromatin, but low elsewhere in the cell. Ran-GTP then releases/activates a number of proteins that help to stabilize microtubules near chromatin. In this elegant mechanism, microtubules become stabilized only if they polymerize near chromosomes, but remain dynamic if the plus ends grow away from chromatin. Of note, ran-1 in C. elegans is required for mitotic spindle assembly, but is not required for female meiotic spindle assembly [130, 131]. This is somewhat surprising, given that a microtubule network forms rapidly around the female meiotic chromatin during nuclear envelope breakdown. Instead, C. elegans Ran seems to be specifically required for mitotic spindle assembly (see below).
Katanin and the generation of microtubules by severing
As discussed above, the centrosomes provide a powerful nucleating center for the production of microtubules, but chromatin can also be used to generate microtubules. However, when centrosome function is removed in C. elegans, mitotic chromatin is a poor substitute for the centrosome, suggesting that chromatin-based pathways of microtubule nucleation are under-utilized in the mitotic environment [10, 45, 77]. Therefore, how are microtubule arrays generated to build the acentrosomal meiotic spindle? Surprisingly, microtubule severing by the enzyme katanin has been implicated in the generation of microtubules near female meiotic chromatin during spindle assembly [8, 132].
The existence of microtubule-severing activity was first reported in mitotically active Xenopus egg extracts [133]. This activity was not found in the interphase extract, suggesting that the component(s) responsible might disassemble interphase microtubules in preparation for mitosis. Subsequent work identified a single heterodimeric protein that exhibited ATP-dependent microtubule severing activity. This complex was named katanin, after the Japanese samurai sword, katana [134, 135].
The C. elegans MEI-1/MEI-2 katanin complex is required specifically for the assembly of the acentrosomal female meiotic spindles [136, 137]. After meiosis is complete, katanin is degraded by ubiquitin-mediated proteolysis; if it is not degraded, mitotic spindle function is severely perturbed (reviewed in [138]. In the absence of katanin, microtubules form around meiotic chromatin, but they fail to assemble a bipolar spindle [136, 137, 139]. Polar bodies are often not generated due to the failure to segregate chromosomes, and those that do form are usually extremely large.
Electron tomography and 3D modeling, used to compare female meiotic spindles in wild-type and katanin-depleted embryos, revealed some interesting structural features of the spindle microtubules [132]. The wild-type spindles are composed of many short microtubules, most of which are aligned and oriented along the spindle axis. As such, most of the microtubules do not span the entire chromatin-to-pole distance, and many of the pole–proximal microtubule ends are distributed throughout the volume of the spindle. The majority of the microtubule ends in the spindle display a flared or open morphology, indicating that they are likely either growing or transiting between dynamic states. Consistent with this observation, fluorescence speckle microscopy of spindles created in Xenopus extracts also suggests that most microtubules are shorter than the pole-chromatin distance [140]. In addition, mature Drosophila meiosis I spindles could contain microtubules of mixed polarity, based on anti-parallel trajectories of the microtubule plus-end marker EB1::GFP within the spindle [141]. Therefore, meiotic spindle assembly/function might not depend on individual microtubules making a connection between chromatin and the poles. Such a connection may not be required, if, for instance, chromosomes are pushed rather than pulled apart during anaphase. Alternatively, many microtubules of different lengths could become closely associated or bundled in order to form the bipolar array (Fig. 3e).
Fig. 3.
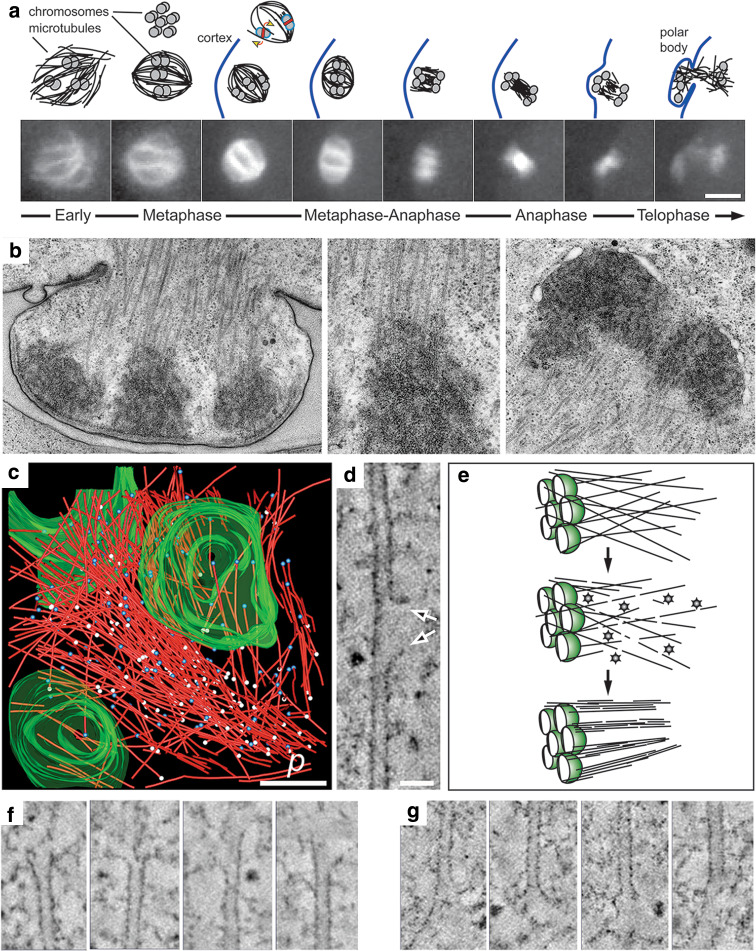
Acentrosomal spindle organization in C. elegans female meiosis. a Schematic representation (above) of the first meiotic division (β-tubulin::GFP; lower panels). Microtubules form around the chromatin and become organized into a bipolar array (only three bivalents are drawn). At this stage, KLP-18 is required for microtubule bundling and pole focusing. Chromosomes align on the metaphase plate in a rosette pattern (shown above), and KLP-19 (red strip) is implicated in providing an anti-poleward force that could exert a torque on the paired chromosomes to ensure their proper alignment (yellow arrows). The redistribution of microtubules in metaphase-anaphase and telophase is represented schematically. By telophase, microtubules begin to accumulate around chromatin to assemble the second meiotic spindle. b Electron micrographs showing the formation of the first polar body with an enlarged view of a kinetochore region (mid panel). c Three-dimensional reconstruction of one half of a meiotic wild-type spindle (microtubules in red, pole–proximal ends as white spheres, pole–distal ends as blue spheres, chromatin in green; p is spindle pole, from [132]). d Tomographic slice showing lateral disruption of the lattice of a spindle microtubule (arrows). e Model explaining the role of katanin (shown as stars) in female meiotic spindle assembly. One half spindle is shown; chromatin is green. Katanin converts a few long microtubules into many short microtubule fragments that may become bundled and arranged into the bipolar array. f Gallery of microtubule pole–distal, and g gallery of microtubule pole–proximal ends showing a variety of open, flared morphologies. Scale bars are 5 μm in a, 500 nm in c, and 50 nm in d
A close look at the microtubule polymers in the meiotic spindle also revealed lateral defects in the microtubule wall, which likely represent the initial severing process by katanin; these defects were not observed in spindles from mei-1(null) mutants [132]. Furthermore, in mei-1(null) mutant embryos only a few long microtubules were found around the chromatin as compared to wild-type spindles. Thus, katanin-induced severing likely involves an initial perforation of the microtubule lattice followed by complete severing of the laterally attacked microtubules [132, 135]. In an acentrosomal environment where chromatin is a relatively inefficient microtubule nucleator [10, 45, 77], katanin-induced severing likely functions to increase the total number of microtubules [132]. Interestingly, double-mutant analysis has indicated that microtubule severing and γ-tubulin-dependent nucleation may provide redundant mechanisms for increasing microtubule number [8]. A useful analogy to this increase in microtubule number is the Scotch® tape dispenser: from a single role of tape, one can pull out a long strip or, with periodic tearing, any number of shorter fragments that can be used for a multitude of tasks. Therefore, the meiotic chromatin does not need to be efficient at generating microtubules if length is not an issue. Severing of a few long polymers generates many short fragments and likely facilitates their incorporation into a bipolar spindle array.
Organizing the microtubules into a bipolar array
After the microtubules form around chromatin, an acentrosomal spindle begins to take shape as microtubules organize into a recognizably bipolar array [7, 116, 142–144]. Depletion/inactivation of specific microtubule-motor proteins from Xenopus extracts and analysis of mutations that affect the assembly of acentrosomal spindles in Drosophila have shed light on the question of how such microtubule arrays can be converted into a bipolar spindle [19, 145, 146]. Microtubule bundling and sorting is most likely achieved by plus-end directed motor proteins, implicated in pushing microtubule minus ends away from the chromatin [145, 147–149] and non-motor MAPs, such as PRC1 and CLASP families [150]. The resulting radially symmetric microtubule array is then focused into two opposing poles, likely by minus-end directed motors with microtubule tethering properties [145, 151]. Interestingly, electron tomography of meiotic spindles in C. elegans showed no characteristic pole structure or electron-dense material [132].
The chromatin is the most likely location for microtubule-organizing factors required during the early stages of acentrosomal spindle assembly. In meiosis I, the six bivalent chromosomes are oriented in an end-to-end manner [7, 152], and the six bivalents usually adopt a characteristic rosette pattern when viewed down the spindle axis in metaphase (Fig. 3a). As noted from electron micrographs of C. elegans meiotic chromatin (Fig. 3b), microtubules appear to embed directly into the surface of the uniformly electron-dense structure. Furthermore, these sites lack any obvious structural features that mark them as kinetochore versus non-kinetochore regions. Despite this, kinetochore proteins clearly decorate the outer pole-facing surface of the condensed meiotic chromatin [153]. The C. elegans chromosomes are holocentric, with the kinetochore spanning the entire length of the chromosome [7, 154]. The holocentric kinetochore presents an important challenge to the fidelity of chromosome segregation because it is a large target for multiple microtubules. This property suggests that merotelic attachment could occur readily [17, 71, 155].
KLP-19 is a kinesin-like protein that might help to limit merotelic attachment in both meiosis and mitosis by providing a constant anti-poleward force on chromosomes [155]. KLP-19, closely related to motors of the kinesin-4 family, locates to a specific region of non-kinetochore chromatin, circumferentially between bivalents in meiosis I and sister chromatids in meiosis II [155, 156]. Therefore, the two kinetochore regions of the paired chromosomes flank an internal KLP-19 zone. Provided lateral contact with microtubules occurs, this internal zone would tend to pull the chromosome pair along microtubules in an anti-poleward direction towards the metaphase plate. The kinetochore region adjacent to the KLP-19 zone would pull the chromosome in the opposite direction. This could generate torque on the chromosomes and result in their alignment on the metaphase plate, with the two paired kinetochore regions facing opposite poles [155]. In vivo imaging of meiotic spindles with GFP::β-tubulin revealed that prominent microtubule bundles run along the sides of the chromatin [156]. The microtubule bundles are proposed to aid in the orientation and alignment of chromosomes. Some association between the lateral surfaces of microtubules and the chromatin has been observed in EM tomograms of the meiotic spindle (E. O’Toole, unpublished observation). Further observations of microtubule ultrastructure should reveal more information about the nature of the microtubule–chromatin interface.
Another kinesin-like protein that is involved in organizing microtubules is KLP-18 (kinesin-12 family member) [142]. During meiotic prometaphase, metaphase and anaphase, KLP-18 becomes concentrated at the poles. Depletion of KLP-18 by RNAi prevents parallel bundling/bipolar organization of the microtubules that accumulate around the female meiotic chromosomes, leading to chromosome mis-segregation [142, 156]. A similar role in promoting the bipolar arrangement of microtubules has also been ascribed to the mesp-1 (meiotic spindle) gene [156]. Although the molecular mechanism is not clear, these proteins could participate in microtubule cross-linking, as has been suggested for the kinesin-5 proteins (formerly BimC) in other organisms [157, 158]. The sole C. elegans kinesin-5 member BMK-1 seems to provide a resistance force against pole separation during anaphase of mitosis [159]; otherwise, obvious defects are not apparent in meiosis in the null mutant [160].
Throughout the assembly process, the meiotic spindle also exhibits a stereotypical translocation to the cortex accompanied by a rotational movement [7, 161]. The fact that these anastral spindles exhibit rotational movement suggests an interaction between the cortex and at least one of the poles. At the start of the process, the spindle translocates to the cortex in a parallel orientation. This movement requires the kinesin-1 microtubule plus-end directed motor protein UNC-116 and the microtubule minus-end directed motor protein dynein [161, 162]. The 90° rotation of the spindle and final juxtaposition of one pole to the cortical surface requires LIN-5 (Mud/NuMA-related), ASPM-1 (abnormal spindle-like, microcephaly-associated) and CMD-1 (calmodulin), which form a complex that is concentrated at the spindle poles [163]. One function for this complex is to localize dynein. Dynein could in turn provide a pulling force to maintain contact between the spindle and the cortex [163].
During the metaphase-to-anaphase transition, the distribution of microtubules in the spindle rapidly transforms from a prominent chromatin-to-pole location to a primarily inter-chromosomal, or midzone location (‘inside-out’ spindle) [7, 8]. In telophase, chromosomes separate to a maximal distance of 4 μm, and microtubules are detected only in the midzone. In this extremely asymmetric cell division, the bivalent chromosome set that is closest to the cortex becomes packaged into a small polar body that is extruded. After a brief interphase, the process reiterates in meiosis II. Interestingly, a similar pattern of microtubule reorganization is observed in the mouse oocyte [164]. The predominance of inter-chromosomal microtubules during anaphase suggests that segregation of the chromosomes in C. elegans is achieved by a pushing mechanism. During the anaphase-telophase transition, further remodeling of the microtubules occurs. The cytokinesis furrow bisects the midzone between the separated chromosomes, and the microtubules rapidly disappear as the polar body is formed. An analysis of a weak katanin mutant indicated that microtubule severing is also required for the redistribution of microtubules at this stage [8]. However, other microtubule regulators could also play a role. As with the weak katanin mutant, when the microtubule-depolymerizing kinesin KLP-7 is reduced, large polar bodies are often created during meiosis (M. Srayko, unpublished observations). Although there could be many different reasons for this phenotype, a simple explanation based on the expected biochemical activity of both KLP-7 and katanin is that their inhibition prevents the normal removal of microtubules from the polar regions during the anaphase-telophase transition, when polar bodies are being extruded.
Centrosome-based spindle assembly in the one-cell embryo of C. elegans
At the end of meiosis, the centrosomes begin to nucleate microtubules in preparation for mitosis. In addition to being a center for microtubule production, the centrosome also provides a platform from which modulators of microtubule dynamics and stability can act (for a review, see [65]. Several proteins have been identified that play key roles in spindle assembly in mitosis, and most of them locate to the centrosome or the spindle microtubules. Near the top of the hierarchy for regulators of mitotic spindle assembly are the Aurora kinases. These kinases are required for many mitotic processes, including centrosome maturation, spindle assembly, chromosome segregation and cytokinesis [165–168]. Metazoans have two distinct Aurora kinases, Aurora A and Aurora B (i.e., AIR-1 and AIR-2 in C. elegans) [169, 170]. Aurora B kinases behave as “chromosomal passengers” in meiosis and mitosis by associating with kinetochores until anaphase, whereupon they translocate to central spindle microtubules (for a review on chromosomal passengers, see [171]). Aurora B in C. elegans is also required for the release of chromosome cohesion in meiosis. Aurora A kinases, however, locate to the centrosome and spindle microtubules, and have at least three major roles in C. elegans: (1) centrosome maturation and microtubule nucleation [45, 95, 103, 169], (2) bipolar spindle formation and stability of spindle microtubules [172], and (3) nuclear envelope breakdown [173]. It is still largely unknown how Aurora kinases facilitate these important steps in mitotic spindle assembly. Part of the difficulty in studying Aurora kinase functions (particularly Aurora A) is that their complete depletion results in pleiotropic defects, thus limiting insight into any single function within a related process (for a review on Aurora A’s many functions at the spindle pole, see [174]).
One protein, TPXL-1, has been implicated in a subset of Aurora A’s functions during C. elegans spindle assembly [172]. TPXL-1 is a worm orthologue of the well-characterized microtubule-stabilizing protein TPX2 [175–177]. Strikingly, in tpxl-1(RNAi) embryos, the assembling mitotic spindle collapses soon after nuclear envelope break-down, because the centrosomes are pulled towards the chromatin [172]. TPXL-1 activates and localizes the Aurora A kinase to spindle microtubules. This targeting likely facilitates Aurora A-dependent phosphorylation of downstream substrates required for microtubule stability [172]. One candidate is the TAC-1 (transforming acidic coiled-coil-like) protein. TAC-1 is a targeting component of the XMAP215 homologue ZYG-9, and both proteins are required for microtubule plus-end growth [178–181]. Furthermore, TAC-1 homologues are Aurora A substrates [182]. Because TPX2 is also a microtubule binding protein [183], it is possible that phosphorylation of TPXL-1 by Aurora A also contributes to stabilization of the spindle microtubules. It is likely that the identification of other Aurora A substrates will provide further insights into the mechanisms of spindle assembly.
The protein phosphatase complex RSA (regulator of spindle assembly) is also important for centrosome-based microtubule functions, but unlike Aurora A, it is not required for centrosome maturation per se [57]. This PP2A phosphatase complex includes RSA-1 (a regulatory B subunit) and RSA-2 (a putative structural component), which are specifically required for microtubule outgrowth from centrosomes and for spindle microtubule stability during mitotic spindle assembly. The RSA complex is localized to the centrosome through an RSA-2-mediated physical interaction with the core PCM component SPD-5 [57, 96]. Furthermore, RSA seems to mediate spindle assembly functions in part by down-regulating the microtubule depolymerase KLP-7 (to allow robust microtubule growth) and up-regulating the microtubule-stabilizing protein TPXL-1 (to stabilize these microtubules during spindle assembly) [57]. A direct interaction between the RSA complex and TPXL-1 suggests that this protein could be a substrate of the phosphatase; RSA is required for the proper targeting of TPXL-1 to the centrosome. Therefore, RSA could coordinate the activities of at least two different microtubule regulators to facilitate spindle assembly.
Early spindle assembly: making contact with the kinetochore
After nuclear envelope breakdown, a subset of microtubules makes contact with kinetochores on chromosomes to begin the process of spindle assembly. In some organisms, a subset of microtubules that grow toward chromatin do not establish connection with kinetochores, but instead interact with microtubules from the opposite pole. These interpolar microtubules could supply pushing forces to help separate chromatids during anaphase, or help to regulate the separation of chromatids by resisting pulling forces on the spindle poles. Depending on the organism or cell type, a varying proportion of non-kinetochore (or astral) microtubules also grows out from the centrosome.
Many organisms seem to utilize chromatin-based pathways to help generate kinetochore microtubules during spindle assembly, even when centrosomes are present. In Drosophila the production of kinetochore microtubules during spindle assembly is facilitated by a protein complex called augmin. Kinetochore microtubules are initially generated via the centrosome or chromatin pathway; these microtubules are then targeted by augmin, which in turn recruits and activates γ-TuRC for further microtubule nucleation [184]. A human augmin-like complex (HAUS) that regulates centrosome and spindle integrity has also been reported [185].
In many systems, the Ran-GTP gradient also contributes to the production and stabilization of microtubules around chromatin, even if centrosomes are present [128]. However, the role of Ran in C. elegans spindle assembly might not involve chromatin-based production of microtubules. In ran-1(RNAi) embryos, robust microtubule growth from centrosomes is observed during spindle assembly, but the centrosomes gradually drift apart, and chromosome separation fails in anaphase despite relatively normal separation of centrosomes [45, 130]. A sensitive assay that measures the numbers of microtubule plus ends over time revealed that more microtubules grow towards chromatin than away from chromatin during the early stages of mitotic spindle assembly [45]. When Ran is depleted, microtubule nucleation and outgrowth rates are no longer anisotropic. The reason for this anisotropy in C. elegans is not clear, but Ran is also required for nuclear envelope formation [130], and it has been suggested that the nuclear envelope might contribute to microtubule growth anisotropy [45]. Unlike many higher eukaryotic systems, spindle assembly in C. elegans begins with the nuclear envelope largely intact, except for two fenestrae, through which microtubules grow towards chromatin [72]. Eventually, the envelope breaks down completely, but the delay could be important for efficient spindle assembly in at least two ways. First, the nuclear membrane could simply force all microtubules within the nuclear environment to grow towards the chromatin, thus increasing the probability of contact with the kinetochores. Second, if microtubules within this region do not contact a kinetochore, they would continue to grow past chromatin and be directed into the opposite pole, where they would be depolymerized, perhaps by KLP-7. In this way, the chromatin would be surrounded by a highly dynamic population of microtubules constantly growing from the centrosomes and quickly shrinking again if contact with the kinetochore is not achieved.
Maintaining stable microtubules during spindle assembly
After microtubules make contact with kinetochores on chromosomes, these microtubules can exhibit some addition and subtraction of tubulin subunits from either the plus ends or the minus ends. In many types of interphase cells, microtubule minus ends in the centrosome tend to be stable; they do not lose or gain tubulin subunits, and most of the tubulin turnover takes place at the plus ends. In mitosis, however, while much of the microtubule turnover is still at the plus ends, the dynamics of minus ends becomes more complex (for a review on microtubule minus ends, see [186]). Photobleaching and fluorescence speckle microscopy used to follow the movement of polymers have revealed that spindle microtubules slowly lose subunits from their minus ends, but gain subunits at the plus ends, a process termed microtubule flux. This could contribute to the regulation of spindle length and chromosome movement towards the centrosome in anaphase A [59, 187, 188]. However, similar photobleaching experiments have so far not detected any significant flux within C. elegans spindle microtubules in anaphase [189]. This suggests that once formed, the kinetochore microtubules are relatively stable during anaphase. This may explain why anaphase A-specific chromosome segregation in the C. elegans one-cell embryo is relatively subtle [154].
EM tomography has also been used to study the minus ends of microtubules near the centrosome. The kinetochore microtubule minus ends near the centrosome often display an open morphology [71], which suggests that these microtubules could be in a dynamic state (Fig. 4d, e). This is in contrast to the photobleaching experiments mentioned above; however, it could be that the amount of flux is below the limit of detection via light microcopy techniques. One hint for a potential source of the open microtubule minus ends comes from the study of tpxl-1(RNAi) embryos. Electron tomography of the tpxl-1(RNAi) collapsed spindle phenotype revealed a drastic shortening of the kinetochore microtubules. Compared to wild-type spindles, these microtubules exhibited more open minus ends near the centrosome [172]. Therefore, the tpxl-1(RNAi) embryos provide support for the idea that microtubule instability manifests as an open, pole-proximal end morphology. It is also possible, however, that the open ends arise as a consequence of excessive pulling forces from the kinetochore microtubules.
Pulling forces on microtubules during anaphase
Astral microtubules are able to make contact with the cell cortex and thus provide a mechanism to orient and position the mitotic spindle relative to cortical polarity cues [30, 190]. The mitotic spindle in the one-cell C. elegans embryo provides a particularly striking example of extensive astral microtubule arrays. Pulling forces, likely located at the cortex, act on astral microtubules to separate the two centrosomes during anaphase [189–193]. Unlike many other systems, most of the mitotic anaphase chromosome movement in the C. elegans embryo is due to the separation of the two centrosomes (anaphase B), rather than by decreasing the distance between the centrosomes and the chromosomes (anaphase A) [154].
With a shift of the first mitotic spindle towards the posterior of the embryo, both daughter cells end up with a copy of the replicated diploid genome, but the anterior cell (AB) is slightly larger than the posterior one (P0). At this point polarization and chromosome segregation during the first mitotic division is achieved, and the early embryo continues with its developmental program.
Conclusions
The nematode worm develops in an amazingly stereotypical manner [194]. This feature has been a critical advantage for the study of cell fate patterns, and this is mirrored in the cell biology of the one-cell embryo. Complicated morphological changes to the microtubule cytoskeleton are required for spindle assembly, and cell division and these changes occur in each individual embryo with almost perfect reproducibility. This aspect of C. elegans biology contributed to the success of the first genome-wide RNAi screen, which allowed a classification of cellular defects from a collection of over 100,000 videos [195]. Moreover, this genome-wide screen has stimulated systematic structure and function analyses by electron tomography [57, 68, 71, 132, 172]. The predictable nature of the cellular events will continue to be a compelling reason to use the early C. elegans embryo to study assembly and 3D architecture of centrosomal and acentrosomal spindles.
Acknowledgments
We thank Drs. Carrie Cowan and Kevin O’Connell for a critical reading of the manuscript. Work in the authors’ laboratories is supported by the Deutsche Forschungsgemeinschaft (DFG MU 1423/2-1 and 1423/3-1) to T.M.-R and by an Alberta Heritage Foundation for Medical Research (AHFMR) Scholar Award, a Canadian Institutes of Health (CIHR) New Investigator Award and operating grants from the CIHR and the Natural Sciences and Engineering Research Council of Canada to M.S. This work was also supported in part by Grant RR-00592 from the National Center for Research Resources of the National Institutes of Health to Andreas Hoenger (Boulder).
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
Contributor Information
Thomas Müller-Reichert, Phone: +49-351-2101763, FAX: +49-351-2102000, Email: mueller-reichert@tu-dresden.de.
Martin Srayko, Phone: +1-780-4929858, FAX: +1-780-4929234, Email: srayko@ualberta.ca.
References
- 1.Kimble JE, White JG. On the control of germ cell development in Caenorhabditis elegans . Dev Biol. 1981;81:208–219. doi: 10.1016/0012-1606(81)90284-0. [DOI] [PubMed] [Google Scholar]
- 2.Miller MA, Nguyen VQ, Lee MH, Kosinski M, Schedl T, Caprioli RM, Greenstein D. A sperm cytoskeletal protein that signals oocyte meiotic maturation and ovulation. Science. 2001;291:2144–2147. doi: 10.1126/science.1057586. [DOI] [PubMed] [Google Scholar]
- 3.Miller MA, Ruest PJ, Kosinski M, Hanks SK, Greenstein D. An Eph receptor sperm-sensing control mechanism for oocyte meiotic maturation in Caenorhabditis elegans . Genes Dev. 2003;17:187–200. doi: 10.1101/gad.1028303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ward S, Carrel JS. Fertilization and sperm competition in the nematode Caenorhabditis elegans . Dev Biol. 1979;73:304–321. doi: 10.1016/0012-1606(79)90069-1. [DOI] [PubMed] [Google Scholar]
- 5.Yang HY, McNally K, McNally FJ. MEI-1/katanin is required for translocation of the meiosis I spindle to the oocyte cortex in C. elegans . Dev Biol. 2003;260:245–259. doi: 10.1016/S0012-1606(03)00216-1. [DOI] [PubMed] [Google Scholar]
- 6.McNally KL, McNally FJ. Fertilization initiates the transition from anaphase I to metaphase II during female meiosis in C. elegans . Dev Biol. 2005;282:218–230. doi: 10.1016/j.ydbio.2005.03.009. [DOI] [PubMed] [Google Scholar]
- 7.Albertson DG, Thomson JN. Segregation of holocentric chromosomes at meiosis in the nematode, Caenorhabditis elegans . Chromosome Res. 1993;1:15–26. doi: 10.1007/BF00710603. [DOI] [PubMed] [Google Scholar]
- 8.McNally K, Audhya A, Oegema K, McNally FJ. Katanin controls mitotic and meiotic spindle length. J Cell Biol. 2006;175:881–891. doi: 10.1083/jcb.200608117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Connell KF, Maxwell KN, White JG. The spd-2 gene is required for polarization of the anteroposterior axis and formation of the sperm asters in the Caenorhabditis elegans zygote. Dev Biol. 2000;222:55–70. doi: 10.1006/dbio.2000.9714. [DOI] [PubMed] [Google Scholar]
- 10.Hamill DR, Severson AF, Carter JC, Bowerman B. Centrosome maturation and mitotic spindle assembly in C. elegans require SPD-5, a protein with multiple coiled-coil domains. Dev Cell. 2002;3:673–684. doi: 10.1016/S1534-5807(02)00327-1. [DOI] [PubMed] [Google Scholar]
- 11.Cuenca AA, Schetter A, Aceto D, Kemphues K, Seydoux G. Polarization of the C. elegans zygote proceeds via distinct establishment and maintenance phases. Development. 2003;130:1255–1265. doi: 10.1242/dev.00284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cowan CR, Hyman AA. Centrosomes direct cell polarity independently of microtubule assembly in C. elegans embryos. Nature. 2004;431:92–96. doi: 10.1038/nature02825. [DOI] [PubMed] [Google Scholar]
- 13.Sadler PL, Shakes DC. Anucleate Caenorhabditis elegans sperm can crawl, fertilize oocytes and direct anterior-posterior polarization of the 1-cell embryo. Development. 2000;127:355–366. doi: 10.1242/dev.127.2.355. [DOI] [PubMed] [Google Scholar]
- 14.Gonczy P, Schnabel H, Kaletta T, Amores AD, Hyman T, Schnabel R. Dissection of cell division processes in the one cell stage Caenorhabditis elegans embryo by mutational analysis. J Cell Biol. 1999;144:927–946. doi: 10.1083/jcb.144.5.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schmidt DJ, Rose DJ, Saxton WM, Strome S. Functional analysis of cytoplasmic dynein heavy chain in Caenorhabditis elegans with fast-acting temperature-sensitive mutations. Mol Biol Cell. 2005;16:1200–1212. doi: 10.1091/mbc.E04-06-0523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Malone CJ, Misner L, Le Bot N, Tsai MC, Campbell JM, Ahringer J, White JG. The C. elegans hook protein, ZYG-12, mediates the essential attachment between the centrosome and nucleus. Cell. 2003;115:825–836. doi: 10.1016/S0092-8674(03)00985-1. [DOI] [PubMed] [Google Scholar]
- 17.Albertson DG. Formation of the first cleavage spindle in nematode embryos. Dev Biol. 1984;101:61–72. doi: 10.1016/0012-1606(84)90117-9. [DOI] [PubMed] [Google Scholar]
- 18.Galli M, van den Heuvel S. Determination of the cleavage plane in early C. elegans embryos. Annu Rev Genet. 2008;42:389–411. doi: 10.1146/annurev.genet.40.110405.090523. [DOI] [PubMed] [Google Scholar]
- 19.Merdes A, Cleveland DW. Pathways of spindle pole formation: different mechanisms; conserved components. J Cell Biol. 1997;138:953–956. doi: 10.1083/jcb.138.5.953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Scholey JM, Brust-Mascher I, Mogilner A. Cell division. Nature. 2003;422:746–752. doi: 10.1038/nature01599. [DOI] [PubMed] [Google Scholar]
- 21.Lloyd C, Chan J. Not so divided: the common basis of plant and animal cell division. Nat Rev Mol Cell Biol. 2006;7:147–152. doi: 10.1038/nrm1831. [DOI] [PubMed] [Google Scholar]
- 22.Mitchison T, Kirschner M. Dynamic instability of microtubule growth. Nature. 1984;312:237–242. doi: 10.1038/312237a0. [DOI] [PubMed] [Google Scholar]
- 23.Kirschner MW, Mitchison T. Microtubule dynamics. Nature. 1986;324:621. doi: 10.1038/324621a0. [DOI] [PubMed] [Google Scholar]
- 24.Howard J, Hyman AA. Growth, fluctuation and switching at microtubule plus ends. Nat Rev Mol Cell Biol. 2009;10:569–574. doi: 10.1038/nrm2713. [DOI] [PubMed] [Google Scholar]
- 25.Chretien D, Fuller SD, Karsenti E. Structure of growing microtubule ends: two-dimensional sheets close into tubes at variable rates. J Cell Biol. 1995;129:1311–1328. doi: 10.1083/jcb.129.5.1311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Arnal I, Karsenti E, Hyman AA. Structural transitions at microtubule ends correlate with their dynamic properties in Xenopus egg extracts. J Cell Biol. 2000;149:767–774. doi: 10.1083/jcb.149.4.767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mandelkow EM, Mandelkow E, Milligan RA. Microtubule dynamics and microtubule caps: a time-resolved cryo-electron microscopy study. J Cell Biol. 1991;114:977–991. doi: 10.1083/jcb.114.5.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Muller-Reichert T, Chretien D, Severin F, Hyman AA. Structural changes at microtubule ends accompanying GTP hydrolysis: information from a slowly hydrolyzable analogue of GTP, guanylyl (α,β)methylenediphosphonate. Proc Natl Acad Sci USA. 1998;95:3661–3666. doi: 10.1073/pnas.95.7.3661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nogales E, Wolf SG, Downing KH. Visualizing the secondary structure of tubulin: three-dimensional map at 4 A. J Struct Biol. 1997;118:119–127. doi: 10.1006/jsbi.1997.3841. [DOI] [PubMed] [Google Scholar]
- 30.Bringmann H, Hyman AA. A cytokinesis furrow is positioned by two consecutive signals. Nature. 2005;436:731–734. doi: 10.1038/nature03823. [DOI] [PubMed] [Google Scholar]
- 31.Glotzer M. The molecular requirements for cytokinesis. Science. 2005;307:1735–1739. doi: 10.1126/science.1096896. [DOI] [PubMed] [Google Scholar]
- 32.Howard J, Hyman AA. Microtubule polymerases and depolymerases. Curr Opin Cell Biol. 2007;19:31–35. doi: 10.1016/j.ceb.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 33.Kellogg DR, Field CM, Alberts BM. Identification of microtubule-associated proteins in the centrosome, spindle, and kinetochore of the early Drosophila embryo. J Cell Biol. 1989;109:2977–2991. doi: 10.1083/jcb.109.6.2977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Shiina N, Moriguchi T, Ohta K, Gotoh Y, Nishida E. Regulation of a major microtubule-associated protein by MPF and MAP kinase. EMBO J. 1992;11:3977–3984. doi: 10.1002/j.1460-2075.1992.tb05491.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gard DL, Kirschner MW. A microtubule-associated protein from Xenopus eggs that specifically promotes assembly at the plus-end. J Cell Biol. 1987;105:2203–2215. doi: 10.1083/jcb.105.5.2203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Faruki S, Karsenti E. Purification of microtubule proteins from Xenopus egg extracts: identification of a 230 K MAP4-like protein. Cell Motil Cytoskeleton. 1994;28:108–118. doi: 10.1002/cm.970280203. [DOI] [PubMed] [Google Scholar]
- 37.Andersen SS, Buendia B, Dominguez JE, Sawyer A, Karsenti E. Effect on microtubule dynamics of XMAP230, a microtubule-associated protein present in Xenopus laevis eggs and dividing cells. J Cell Biol. 1994;127:1289–1299. doi: 10.1083/jcb.127.5.1289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Andersen SS, Karsenti E. XMAP310: a Xenopus rescue-promoting factor localized to the mitotic spindle. J Cell Biol. 1997;139:975–983. doi: 10.1083/jcb.139.4.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hughes JR, Meireles AM, Fisher KH, Garcia A, Antrobus PR, Wainman A, Zitzmann N, Deane C, Ohkura H, Wakefield JG. A microtubule interactome: complexes with roles in cell cycle and mitosis. PLoS Biol. 2008;6:e98. doi: 10.1371/journal.pbio.0060098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amos LA, Schlieper D. Microtubules and maps. Adv Protein Chem. 2005;71:257–298. doi: 10.1016/S0065-3233(04)71007-4. [DOI] [PubMed] [Google Scholar]
- 41.Brouhard GJ, Stear JH, Noetzel TL, Al-Bassam J, Kinoshita K, Harrison SC, Howard J, Hyman AA. XMAP215 is a processive microtubule polymerase. Cell. 2008;132:79–88. doi: 10.1016/j.cell.2007.11.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kerssemakers JW, Munteanu EL, Laan L, Noetzel TL, Janson ME, Dogterom M. Assembly dynamics of microtubules at molecular resolution. Nature. 2006;442:709–712. doi: 10.1038/nature04928. [DOI] [PubMed] [Google Scholar]
- 43.Slep KC, Vale RD. Structural basis of microtubule plus end tracking by XMAP215, CLIP-170, and EB1. Mol Cell. 2007;27:976–991. doi: 10.1016/j.molcel.2007.07.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Matthews LR, Carter P, Thierry-Mieg D, Kemphues K. ZYG-9, a Caenorhabditis elegans protein required for microtubule organization and function, is a component of meiotic and mitotic spindle poles. J Cell Biol. 1998;141:1159–1168. doi: 10.1083/jcb.141.5.1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Srayko M, Kaya A, Stamford J, Hyman AA. Identification and characterization of factors required for microtubule growth and nucleation in the early C elegans embryo. Dev Cell. 2005;9:223–236. doi: 10.1016/j.devcel.2005.07.003. [DOI] [PubMed] [Google Scholar]
- 46.Desai A, Verma S, Mitchison TJ, Walczak CE. Kin I kinesins are microtubule-destabilizing enzymes. Cell. 1999;96:69–78. doi: 10.1016/S0092-8674(00)80960-5. [DOI] [PubMed] [Google Scholar]
- 47.Moores CA, Milligan RA. Lucky 13-microtubule depolymerisation by kinesin-13 motors. J Cell Sci. 2006;119:3905–3913. doi: 10.1242/jcs.03224. [DOI] [PubMed] [Google Scholar]
- 48.Helenius J, Brouhard G, Kalaidzidis Y, Diez S, Howard J. The depolymerizing kinesin MCAK uses lattice diffusion to rapidly target microtubule ends. Nature. 2006;441:115–119. doi: 10.1038/nature04736. [DOI] [PubMed] [Google Scholar]
- 49.Hunter AW, Caplow M, Coy DL, Hancock WO, Diez S, Wordeman L, Howard J. The kinesin-related protein MCAK is a microtubule depolymerase that forms an ATP-hydrolyzing complex at microtubule ends. Mol Cell. 2003;11:445–457. doi: 10.1016/S1097-2765(03)00049-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Mulder AM, Glavis-Bloom A, Moores CA, Wagenbach M, Carragher B, Wordeman L, Milligan RA. A new model for binding of kinesin 13 to curved microtubule protofilaments. J Cell Biol. 2009;185:51–57. doi: 10.1083/jcb.200812052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Kline-Smith SL, Khodjakov A, Hergert P, Walczak CE. Depletion of centromeric MCAK leads to chromosome congression and segregation defects due to improper kinetochore attachments. Mol Biol Cell. 2004;15:1146–1159. doi: 10.1091/mbc.E03-08-0581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Andrews PD, Ovechkina Y, Morrice N, Wagenbach M, Duncan K, Wordeman L, Swedlow JR. Aurora B regulates MCAK at the mitotic centromere. Dev Cell. 2004;6:253–268. doi: 10.1016/S1534-5807(04)00025-5. [DOI] [PubMed] [Google Scholar]
- 53.Lan W, Zhang X, Kline-Smith SL, Rosasco SE, Barrett-Wilt GA, Shabanowitz J, Hunt DF, Walczak CE, Stukenberg PT. Aurora B phosphorylates centromeric MCAK and regulates its localization and microtubule depolymerization activity. Curr Biol. 2004;14:273–286. doi: 10.1016/j.cub.2004.01.055. [DOI] [PubMed] [Google Scholar]
- 54.Knowlton AL, Lan W, Stukenberg PT. Aurora B is enriched at merotelic attachment sites, where it regulates MCAK. Curr Biol. 2006;16:1705–1710. doi: 10.1016/j.cub.2006.07.057. [DOI] [PubMed] [Google Scholar]
- 55.Huang H, Feng J, Famulski J, Rattner JB, Liu ST, Kao GD, Muschel R, Chan GK, Yen TJ. Tripin/hSgo2 recruits MCAK to the inner centromere to correct defective kinetochore attachments. J Cell Biol. 2007;177:413–424. doi: 10.1083/jcb.200701122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Bakhoum SF, Genovese G, Compton DA. Deviant kinetochore microtubule dynamics underlie chromosomal instability. Curr Biol. 2009;19:1937–1942. doi: 10.1016/j.cub.2009.09.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Schlaitz AL, Srayko M, Dammermann A, Quintin S, Wielsch N, MacLeod I, de Robillard Q, Zinke A, Yates JR, III, Muller-Reichert T, Shevchenko A, Oegema K, Hyman AA. The C. elegans RSA complex localizes protein phosphatase 2A to centrosomes and regulates mitotic spindle assembly. Cell. 2007;128:115–127. doi: 10.1016/j.cell.2006.10.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zhang X, Ems-McClung SC, Walczak CE. Aurora A phosphorylates MCAK to control ran-dependent spindle bipolarity. Mol Biol Cell. 2008;19:2752–2765. doi: 10.1091/mbc.E08-02-0198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rogers GC, Rogers SL, Schwimmer TA, Ems-McClung SC, Walczak CE, Vale RD, Scholey JM, Sharp DJ. Two mitotic kinesins cooperate to drive sister chromatid separation during anaphase. Nature. 2004;427:364–370. doi: 10.1038/nature02256. [DOI] [PubMed] [Google Scholar]
- 60.Kinoshita K, Arnal I, Desai A, Drechsel DN, Hyman AA. Reconstitution of physiological microtubule dynamics using purified components. Science. 2001;294:1340–1343. doi: 10.1126/science.1064629. [DOI] [PubMed] [Google Scholar]
- 61.Bre MH, Kreis TE, Karsenti E. Control of microtubule nucleation and stability in Madin-Darby canine kidney cells: the occurrence of noncentrosomal, stable detyrosinated microtubules. J Cell Biol. 1987;105:1283–1296. doi: 10.1083/jcb.105.3.1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Jaspersen SL, Winey M. The budding yeast spindle pole body: structure, duplication, and function. Annu Rev Cell Dev Biol. 2004;20:1–28. doi: 10.1146/annurev.cellbio.20.022003.114106. [DOI] [PubMed] [Google Scholar]
- 63.O’Toole ET, Winey M, McIntosh JR. High-voltage electron tomography of spindle pole bodies and early mitotic spindles in the yeast Saccharomyces cerevisiae . Mol Biol Cell. 1999;10:2017–2031. doi: 10.1091/mbc.10.6.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Marshall WF. What is the function of centrioles? J Cell Biochem. 2007;100:916–922. doi: 10.1002/jcb.21117. [DOI] [PubMed] [Google Scholar]
- 65.Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- 66.Wolf N, Hirsh D, McIntosh JR. Spermatogenesis in males of the free-living nematode, Caenorhabditis elegans . J Ultrastruct Res. 1978;63:155–169. doi: 10.1016/S0022-5320(78)80071-9. [DOI] [PubMed] [Google Scholar]
- 67.O’Connell KF, Caron C, Kopish KR, Hurd DD, Kemphues KJ, Li Y, White JG. The C. elegans zyg-1 gene encodes a regulator of centrosome duplication with distinct maternal and paternal roles in the embryo. Cell. 2001;105:547–558. doi: 10.1016/S0092-8674(01)00338-5. [DOI] [PubMed] [Google Scholar]
- 68.Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans . Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 69.Kochanski RS, Borisy GG. Mode of centriole duplication and distribution. J Cell Biol. 1990;110:1599–1605. doi: 10.1083/jcb.110.5.1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dutcher SK. Long-lost relatives reappear: identification of new members of the tubulin superfamily. Curr Opin Microbiol. 2003;6:634–640. doi: 10.1016/j.mib.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 71.O’Toole ET, McDonald KL, Mantler J, McIntosh JR, Hyman AA, Muller-Reichert T. Morphologically distinct microtubule ends in the mitotic centrosome of Caenorhabditis elegans . J Cell Biol. 2003;163:451–456. doi: 10.1083/jcb.200304035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/S0092-8674(03)00117-X. [DOI] [PubMed] [Google Scholar]
- 73.Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 74.Kemp CA, Kopish KR, Zipperlen P, Ahringer J, O’Connell KF. Centrosome maturation and duplication in C. elegans require the coiled-coil protein SPD-2. Dev Cell. 2004;6:511–523. doi: 10.1016/S1534-5807(04)00066-8. [DOI] [PubMed] [Google Scholar]
- 75.Leidel S, Gonczy P. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/S1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 76.Delattre M, Leidel S, Wani K, Baumer K, Bamat J, Schnabel H, Feichtinger R, Schnabel R, Gonczy P. Centriolar SAS-5 is required for centrosome duplication in C. elegans . Nat Cell Biol. 2004;6:656–664. doi: 10.1038/ncb1146. [DOI] [PubMed] [Google Scholar]
- 77.Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 78.Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 79.Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 81.Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 82.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 83.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 84.Zhu F, Lawo S, Bird A, Pinchev D, Ralph A, Richter C, Muller-Reichert T, Kittler R, Hyman AA, Pelletier L. The mammalian SPD-2 ortholog Cep192 regulates centrosome biogenesis. Curr Biol. 2008;18:136–141. doi: 10.1016/j.cub.2007.12.055. [DOI] [PubMed] [Google Scholar]
- 85.Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 86.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 88.Yabe T, Ge X, Pelegri F. The zebrafish maternal-effect gene cellular atoll encodes the centriolar component sas-6 and defects in its paternal function promote whole genome duplication. Dev Biol. 2007;312:44–60. doi: 10.1016/j.ydbio.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 91.Song MH, Aravind L, Muller-Reichert T, O’Connell KF. The conserved protein SZY-20 opposes the Plk4-related kinase ZYG-1 to limit centrosome size. Dev Cell. 2008;15:901–912. doi: 10.1016/j.devcel.2008.09.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Kemp CA, Song MH, Addepalli MK, Hunter G, O’Connell K. Suppressors of zyg-1 define regulators of centrosome duplication and nuclear association in Caenorhabditis elegans . Genetics. 2007;176:95–113. doi: 10.1534/genetics.107.071803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Hannak E, Kirkham M, Hyman AA, Oegema K. Aurora-A kinase is required for centrosome maturation in Caenorhabditis elegans . J Cell Biol. 2001;155:1109–1116. doi: 10.1083/jcb.200108051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Boxem M, Maliga Z, Klitgord N, Li N, Lemmens I, Mana M, de Lichtervelde L, Mul JD, van de Peut D, Devos M, Simonis N, Yildirim MA, Cokol M, Kao HL, de Smet AS, Wang H, Schlaitz AL, Hao T, Milstein S, Fan C, Tipsword M, Drew K, Galli M, Rhrissorrakrai K, Drechsel D, Koller D, Roth FP, Iakoucheva LM, Dunker AK, Bonneau R, Gunsalus KC, Hill DE, Piano F, Tavernier J, van den Heuvel S, Hyman AA, Vidal M. A protein domain-based interactome network for C. elegans early embryogenesis. Cell. 2008;134:534–545. doi: 10.1016/j.cell.2008.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/S0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 98.Job D, Valiron O, Oakley B. Microtubule nucleation. Curr Opin Cell Biol. 2003;15:111–117. doi: 10.1016/S0955-0674(02)00003-0. [DOI] [PubMed] [Google Scholar]
- 99.Oakley BR. Gamma-tubulin. Curr Top Dev Biol. 2000;49:27–54. doi: 10.1016/S0070-2153(99)49003-9. [DOI] [PubMed] [Google Scholar]
- 100.Oakley CE, Oakley BR. Identification of gamma-tubulin, a new member of the tubulin superfamily encoded by mipA gene of Aspergillus nidulans . Nature. 1989;338:662–664. doi: 10.1038/338662a0. [DOI] [PubMed] [Google Scholar]
- 101.Gunawardane RN, Martin OC, Cao K, Zhang L, Dej K, Iwamatsu A, Zheng Y. Characterization and reconstitution of Drosophila gamma-tubulin ring complex subunits. J Cell Biol. 2000;151:1513–1524. doi: 10.1083/jcb.151.7.1513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Murphy SM, Urbani L, Stearns T. The mammalian gamma-tubulin complex contains homologues of the yeast spindle pole body components spc97p and spc98p. J Cell Biol. 1998;141:663–674. doi: 10.1083/jcb.141.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Hannak E, Oegema K, Kirkham M, Gonczy P, Habermann B, Hyman AA. The kinetically dominant assembly pathway for centrosomal asters in Caenorhabditis elegans is gamma-tubulin dependent. J Cell Biol. 2002;157:591–602. doi: 10.1083/jcb.200202047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Moritz M, Braunfeld MB, Sedat JW, Alberts B, Agard DA. Microtubule nucleation by gamma-tubulin-containing rings in the centrosome. Nature. 1995;378:638–640. doi: 10.1038/378638a0. [DOI] [PubMed] [Google Scholar]
- 105.Oegema K, Wiese C, Martin OC, Milligan RA, Iwamatsu A, Mitchison TJ, Zheng Y. Characterization of two related Drosophila gamma-tubulin complexes that differ in their ability to nucleate microtubules. J Cell Biol. 1999;144:721–733. doi: 10.1083/jcb.144.4.721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Wiese C, Zheng Y. A new function for the gamma-tubulin ring complex as a microtubule minus-end cap. Nat Cell Biol. 2000;2:358–364. doi: 10.1038/35014051. [DOI] [PubMed] [Google Scholar]
- 107.Keating TJ, Borisy GG. Immunostructural evidence for the template mechanism of microtubule nucleation. Nat Cell Biol. 2000;2:352–357. doi: 10.1038/35014045. [DOI] [PubMed] [Google Scholar]
- 108.Moritz M, Braunfeld MB, Guenebaut V, Heuser J, Agard DA. Structure of the gamma-tubulin ring complex: a template for microtubule nucleation. Nat Cell Biol. 2000;2:365–370. doi: 10.1038/35014058. [DOI] [PubMed] [Google Scholar]
- 109.Erickson HP, Stoffler D. Protofilaments and rings, two conformations of the tubulin family conserved from bacterial FtsZ to alpha/beta and gamma tubulin. J Cell Biol. 1996;135:5–8. doi: 10.1083/jcb.135.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 111.Byers B, Shriver K, Goetsch L. The role of spindle pole bodies and modified microtubule ends in the initiation of microtubule assembly in Saccharomyces cerevisiae . J Cell Sci. 1978;30:331–352. doi: 10.1242/jcs.30.1.331. [DOI] [PubMed] [Google Scholar]
- 112.Rout MP, Kilmartin JV. Components of the yeast spindle and spindle pole body. J Cell Biol. 1990;111:1913–1927. doi: 10.1083/jcb.111.5.1913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Bullitt E, Rout MP, Kilmartin JV, Akey CW. The yeast spindle pole body is assembled around a central crystal of Spc42p. Cell. 1997;89:1077–1086. doi: 10.1016/S0092-8674(00)80295-0. [DOI] [PubMed] [Google Scholar]
- 114.Moritz M, Braunfeld MB, Fung JC, Sedat JW, Alberts BM, Agard DA. Three-dimensional structural characterization of centrosomes from early Drosophila embryos. J Cell Biol. 1995;130:1149–1159. doi: 10.1083/jcb.130.5.1149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Strome S, Powers J, Dunn M, Reese K, Malone CJ, White J, Seydoux G, Saxton W. Spindle dynamics and the role of gamma-tubulin in early Caenorhabditis elegans embryos. Mol Biol Cell. 2001;12:1751–1764. doi: 10.1091/mbc.12.6.1751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Theurkauf WE, Hawley RS. Meiotic spindle assembly in Drosophila females: behavior of nonexchange chromosomes and the effects of mutations in the nod kinesin-like protein. J Cell Biol. 1992;116:1167–1180. doi: 10.1083/jcb.116.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Gueth-Hallonet C, Antony C, Aghion J, Santa-Maria A, Lajoie-Mazenc I, Wright M, Maro B. Gamma-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci. 1993;105(Pt 1):157–166. doi: 10.1242/jcs.105.1.157. [DOI] [PubMed] [Google Scholar]
- 118.Sathananthan AH. Mitosis in the human embryo: the vital role of the sperm centrosome (centriole) Histol Histopathol. 1997;12:827–856. [PubMed] [Google Scholar]
- 119.Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/S0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 120.Tulu US, Rusan NM, Wadsworth P. Peripheral, non-centrosome-associated microtubules contribute to spindle formation in centrosome-containing cells. Curr Biol. 2003;13:1894–1899. doi: 10.1016/j.cub.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 121.Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/S0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 122.Rebollo E, Llamazares S, Reina J, Gonzalez C. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2004;2:E8. doi: 10.1371/journal.pbio.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Moutinho-Pereira S, Debec A, Maiato H. Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol Biol Cell. 2009;20:2796–2808. doi: 10.1091/mbc.E09-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Hoyer-Fender S (2009) Centriole maturation and transformation to basal body. Semin Cell Dev Biol doi:10.1016/j.semcdb.2009.07.002 [DOI] [PubMed]
- 125.Ward S, Klass M. The location of the major protein in Caenorhabditis elegans sperm and spermatocytes. Dev Biol. 1982;92:203–208. doi: 10.1016/0012-1606(82)90164-6. [DOI] [PubMed] [Google Scholar]
- 126.Kim DY, Roy R. Cell cycle regulators control centrosome elimination during oogenesis in Caenorhabditis elegans . J Cell Biol. 2006;174:751–757. doi: 10.1083/jcb.200512160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Heald R, Tournebize R, Blank T, Sandaltzopoulos R, Becker P, Hyman A, Karsenti E. Self-organization of microtubules into bipolar spindles around artificial chromosomes in Xenopus egg extracts. Nature. 1996;382:420–425. doi: 10.1038/382420a0. [DOI] [PubMed] [Google Scholar]
- 128.Kalab P, Heald R. The RanGTP gradient—a GPS for the mitotic spindle. J Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Bischoff FR, Ponstingl H. Catalysis of guanine nucleotide exchange on Ran by the mitotic regulator RCC1. Nature. 1991;354:80–82. doi: 10.1038/354080a0. [DOI] [PubMed] [Google Scholar]
- 130.Askjaer P, Galy V, Hannak E, Mattaj IW. Ran GTPase cycle and importins alpha and beta are essential for spindle formation and nuclear envelope assembly in living Caenorhabditis elegans embryos. Mol Biol Cell. 2002;13:4355–4370. doi: 10.1091/mbc.E02-06-0346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Bamba C, Bobinnec Y, Fukuda M, Nishida E. The GTPase Ran regulates chromosome positioning and nuclear envelope assembly in vivo. Curr Biol. 2002;12:503–507. doi: 10.1016/S0960-9822(02)00741-8. [DOI] [PubMed] [Google Scholar]
- 132.Srayko M, O’Toole ET, Hyman AA, Muller-Reichert T. Katanin disrupts the microtubule lattice and increases polymer number in C. elegans meiosis. Curr Biol. 2006;16:1944–1949. doi: 10.1016/j.cub.2006.08.029. [DOI] [PubMed] [Google Scholar]
- 133.Vale RD. Severing of stable microtubules by a mitotically activated protein in Xenopus egg extracts. Cell. 1991;64:827–839. doi: 10.1016/0092-8674(91)90511-V. [DOI] [PubMed] [Google Scholar]
- 134.McNally FJ, Vale RD. Identification of katanin, an ATPase that severs and disassembles stable microtubules. Cell. 1993;75:419–429. doi: 10.1016/0092-8674(93)90377-3. [DOI] [PubMed] [Google Scholar]
- 135.Hartman JJ, Mahr J, McNally K, Okawa K, Iwamatsu A, Thomas S, Cheesman S, Heuser J, Vale RD, McNally FJ. Katanin, a microtubule-severing protein, is a novel AAA ATPase that targets to the centrosome using a WD40-containing subunit. Cell. 1998;93:277–287. doi: 10.1016/S0092-8674(00)81578-0. [DOI] [PubMed] [Google Scholar]
- 136.Clark-Maguire S, Mains PE. mei-1, a gene required for meiotic spindle formation in Caenorhabditis elegans, is a member of a family of ATPases. Genetics. 1994;136:533–546. doi: 10.1093/genetics/136.2.533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Srayko M, Buster DW, Bazirgan OA, McNally FJ, Mains PE. MEI-1/MEI-2 katanin-like microtubule severing activity is required for Caenorhabditis elegans meiosis. Genes Dev. 2000;14:1072–1084. [PMC free article] [PubMed] [Google Scholar]
- 138.Bowerman B, Kurz T. Degrade to create: developmental requirements for ubiquitin-mediated proteolysis during early C. elegans embryogenesis. Development. 2006;133:773–784. doi: 10.1242/dev.02276. [DOI] [PubMed] [Google Scholar]
- 139.Clark-Maguire S, Mains PE. Localization of the mei-1 gene product of Caenorhaditis elegans, a meiotic-specific spindle component. J Cell Biol. 1994;126:199–209. doi: 10.1083/jcb.126.1.199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Yang G, Houghtaling BR, Gaetz J, Liu JZ, Danuser G, Kapoor TM. Architectural dynamics of the meiotic spindle revealed by single-fluorophore imaging. Nat Cell Biol. 2007;9:1233–1242. doi: 10.1038/ncb1643. [DOI] [PubMed] [Google Scholar]
- 141.Liang ZY, Hallen MA, Endow SA. Mature Drosophila meiosis I spindles comprise microtubules of mixed polarity. Curr Biol. 2009;19:163–168. doi: 10.1016/j.cub.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Segbert C, Barkus R, Powers J, Strome S, Saxton WM, Bossinger O. KLP-18, a Klp2 kinesin, is required for assembly of acentrosomal meiotic spindles in Caenorhabditis elegans . Mol Biol Cell. 2003;14:4458–4469. doi: 10.1091/mbc.E03-05-0283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Tournebize R, Andersen SS, Verde F, Doree M, Karsenti E, Hyman AA. Distinct roles of PP1 and PP2A-like phosphatases in control of microtubule dynamics during mitosis. EMBO J. 1997;16:5537–5549. doi: 10.1093/emboj/16.18.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Gard DL. Microtubule organization during maturation of Xenopus oocytes: assembly and rotation of the meiotic spindles. Dev Biol. 1992;151:516–530. doi: 10.1016/0012-1606(92)90190-R. [DOI] [PubMed] [Google Scholar]
- 145.Walczak CE, Vernos I, Mitchison TJ, Karsenti E, Heald R. A model for the proposed roles of different microtubule-based motor proteins in establishing spindle bipolarity. Curr Biol. 1998;8:903–913. doi: 10.1016/S0960-9822(07)00370-3. [DOI] [PubMed] [Google Scholar]
- 146.Walczak CE. Microtubule dynamics and tubulin interacting proteins. Curr Opin Cell Biol. 2000;12:52–56. doi: 10.1016/S0955-0674(99)00056-3. [DOI] [PubMed] [Google Scholar]
- 147.Afshar K, Barton NR, Hawley RS, Goldstein LS. DNA binding and meiotic chromosomal localization of the Drosophila nod kinesin-like protein. Cell. 1995;81:129–138. doi: 10.1016/0092-8674(95)90377-1. [DOI] [PubMed] [Google Scholar]
- 148.Vernos I, Raats J, Hirano T, Heasman J, Karsenti E, Wylie C. Xklp1, a chromosomal Xenopus kinesin-like protein essential for spindle organization and chromosome positioning. Cell. 1995;81:117–127. doi: 10.1016/0092-8674(95)90376-3. [DOI] [PubMed] [Google Scholar]
- 149.Wang SZ, Adler R. Chromokinesin: a DNA-binding, kinesin-like nuclear protein. J Cell Biol. 1995;128:761–768. doi: 10.1083/jcb.128.5.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Akhmanova A, Hoogenraad CC. Microtubule plus-end-tracking proteins: mechanisms and functions. Curr Opin Cell Biol. 2005;17:47–54. doi: 10.1016/j.ceb.2004.11.001. [DOI] [PubMed] [Google Scholar]
- 151.Matthies HJ, McDonald HB, Goldstein LS, Theurkauf WE. Anastral meiotic spindle morphogenesis: role of the non-claret disjunctional kinesin-like protein. J Cell Biol. 1996;134:455–464. doi: 10.1083/jcb.134.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Herman RK, Madl JE, Kari CK. Duplications in Caenorhabditis elegans . Genetics. 1979;92:419–435. doi: 10.1093/genetics/92.2.419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Monen J, Maddox PS, Hyndman F, Oegema K, Desai A. Differential role of CENP-A in the segregation of holocentric C. elegans chromosomes during meiosis and mitosis. Nat Cell Biol. 2005;7:1248–1255. doi: 10.1038/ncb1331. [DOI] [PubMed] [Google Scholar]
- 154.Oegema K, Desai A, Rybina S, Kirkham M, Hyman AA. Functional analysis of kinetochore assembly in Caenorhabditis elegans . J Cell Biol. 2001;153:1209–1226. doi: 10.1083/jcb.153.6.1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Powers J, Rose DJ, Saunders A, Dunkelbarger S, Strome S, Saxton WM. Loss of KLP-19 polar ejection force causes misorientation and missegregation of holocentric chromosomes. J Cell Biol. 2004;166:991–1001. doi: 10.1083/jcb.200403036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Wignall SM, Villeneuve AM. Lateral microtubule bundles promote chromosome alignment during acentrosomal oocyte meiosis. Nat Cell Biol. 2009;11:839–844. doi: 10.1038/ncb1891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.Kashina AS, Baskin RJ, Cole DG, Wedaman KP, Saxton WM, Scholey JM. A bipolar kinesin. Nature. 1996;379:270–272. doi: 10.1038/379270a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Kashina AS, Scholey JM, Leszyk JD, Saxton WM. An essential bipolar mitotic motor. Nature. 1996;384:225. doi: 10.1038/384225a0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Saunders AM, Powers J, Strome S, Saxton WM. Kinesin-5 acts as a brake in anaphase spindle elongation. Curr Biol. 2007;17:R453–R454. doi: 10.1016/j.cub.2007.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Bishop JD, Han Z, Schumacher JM. The Caenorhabditis elegans Aurora B kinase AIR-2 phosphorylates and is required for the localization of a BimC kinesin to meiotic and mitotic spindles. Mol Biol Cell. 2005;16:742–756. doi: 10.1091/mbc.E04-08-0682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Yang HY, Mains PE, McNally FJ. Kinesin-1 mediates translocation of the meiotic spindle to the oocyte cortex through KCA-1, a novel cargo adapter. J Cell Biol. 2005;169:447–457. doi: 10.1083/jcb.200411132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Ellefson ML, McNally FJ. Kinesin-1 and cytoplasmic dynein act sequentially to move the meiotic spindle to the oocyte cortex in Caenorhabditis elegans . Mol Biol Cell. 2009;20:2722–2730. doi: 10.1091/mbc.E08-12-1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.van der Voet M, Berends CW, Perreault A, Nguyen-Ngoc T, Gonczy P, Vidal M, Boxem M, van den Heuvel S. NuMA-related LIN-5, ASPM-1, calmodulin and dynein promote meiotic spindle rotation independently of cortical LIN-5/GPR/Galpha. Nat Cell Biol. 2009;11:269–277. doi: 10.1038/ncb1834. [DOI] [PubMed] [Google Scholar]
- 164.Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 165.Gopalan G, Chan CS, Donovan PJ. A novel mammalian, mitotic spindle-associated kinase is related to yeast and fly chromosome segregation regulators. J Cell Biol. 1997;138:643–656. doi: 10.1083/jcb.138.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.Glover DM, Leibowitz MH, McLean DA, Parry H. Mutations in aurora prevent centrosome separation leading to the formation of monopolar spindles. Cell. 1995;81:95–105. doi: 10.1016/0092-8674(95)90374-7. [DOI] [PubMed] [Google Scholar]
- 167.Chan CS, Botstein D. Isolation and characterization of chromosome-gain and increase-in-ploidy mutants in yeast. Genetics. 1993;135:677–691. doi: 10.1093/genetics/135.3.677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Andrews PD, Knatko E, Moore WJ, Swedlow JR. Mitotic mechanics: the auroras come into view. Curr Opin Cell Biol. 2003;15:672–683. doi: 10.1016/j.ceb.2003.10.013. [DOI] [PubMed] [Google Scholar]
- 169.Schumacher JM, Ashcroft N, Donovan PJ, Golden A. A highly conserved centrosomal kinase, AIR-1, is required for accurate cell cycle progression and segregation of developmental factors in Caenorhabditis elegans embryos. Development. 1998;125:4391–4402. doi: 10.1242/dev.125.22.4391. [DOI] [PubMed] [Google Scholar]
- 170.Schumacher JM, Golden A, Donovan PJ. AIR-2: An Aurora/Ipl1-related protein kinase associated with chromosomes and midbody microtubules is required for polar body extrusion and cytokinesis in Caenorhabditis elegans embryos. J Cell Biol. 1998;143:1635–1646. doi: 10.1083/jcb.143.6.1635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Ruchaud S, Carmena M, Earnshaw WC. Chromosomal passengers: conducting cell division. Nat Rev Mol Cell Biol. 2007;8:798–812. doi: 10.1038/nrm2257. [DOI] [PubMed] [Google Scholar]
- 172.Ozlu N, Srayko M, Kinoshita K, Habermann B, O’Toole ET, Muller-Reichert T, Schmalz N, Desai A, Hyman AA. An essential function of the C. elegans ortholog of TPX2 is to localize activated aurora A kinase to mitotic spindles. Dev Cell. 2005;9:237–248. doi: 10.1016/j.devcel.2005.07.002. [DOI] [PubMed] [Google Scholar]
- 173.Portier N, Audhya A, Maddox PS, Green RA, Dammermann A, Desai A, Oegema K. A microtubule-independent role for centrosomes and aurora a in nuclear envelope breakdown. Dev Cell. 2007;12:515–529. doi: 10.1016/j.devcel.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Barr AR, Gergely F. Aurora-A: the maker and breaker of spindle poles. J Cell Sci. 2007;120:2987–2996. doi: 10.1242/jcs.013136. [DOI] [PubMed] [Google Scholar]
- 175.Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr Biol. 2004;14:1801–1811. doi: 10.1016/j.cub.2004.10.002. [DOI] [PubMed] [Google Scholar]
- 176.Gruss OJ, Carazo-Salas RE, Schatz CA, Guarguaglini G, Kast J, Wilm M, Le Bot N, Vernos I, Karsenti E, Mattaj IW. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/S0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- 177.Schatz CA, Santarella R, Hoenger A, Karsenti E, Mattaj IW, Gruss OJ, Carazo-Salas RE. Importin alpha-regulated nucleation of microtubules by TPX2. EMBO J. 2003;22:2060–2070. doi: 10.1093/emboj/cdg195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Srayko M, Quintin S, Schwager A, Hyman AA. Caenorhabditis elegans TAC-1 and ZYG-9 form a complex that is essential for long astral and spindle microtubules. Curr Biol. 2003;13:1506–1511. doi: 10.1016/S0960-9822(03)00597-9. [DOI] [PubMed] [Google Scholar]
- 179.Lee MJ, Gergely F, Jeffers K, Peak-Chew SY, Raff JW. Msps/XMAP215 interacts with the centrosomal protein D-TACC to regulate microtubule behaviour. Nat Cell Biol. 2001;3:643–649. doi: 10.1038/35083033. [DOI] [PubMed] [Google Scholar]
- 180.Le Bot N, Tsai MC, Andrews RK, Ahringer J. TAC-1, a regulator of microtubule length in the C. elegans embryo. Curr Biol. 2003;13:1499–1505. doi: 10.1016/S0960-9822(03)00577-3. [DOI] [PubMed] [Google Scholar]
- 181.Bellanger JM, Gonczy P. TAC-1 and ZYG-9 form a complex that promotes microtubule assembly in C. elegans embryos. Curr Biol. 2003;13:1488–1498. doi: 10.1016/S0960-9822(03)00582-7. [DOI] [PubMed] [Google Scholar]
- 182.Giet R, McLean D, Descamps S, Lee MJ, Raff JW, Prigent C, Glover DM. Drosophila Aurora A kinase is required to localize D-TACC to centrosomes and to regulate astral microtubules. J Cell Biol. 2002;156:437–451. doi: 10.1083/jcb.200108135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 183.Wittmann T, Wilm M, Karsenti E, Vernos I. TPX2, A novel Xenopus MAP involved in spindle pole organization. J Cell Biol. 2000;149:1405–1418. doi: 10.1083/jcb.149.7.1405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Goshima G, Mayer M, Zhang N, Stuurman N, Vale RD. Augmin: a protein complex required for centrosome-independent microtubule generation within the spindle. J Cell Biol. 2008;181:421–429. doi: 10.1083/jcb.200711053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Lawo S, Bashkurov M, Mullin M, Ferreria MG, Kittler R, Habermann B, Tagliaferro A, Poser I, Hutchins JR, Hegemann B, Pinchev D, Buchholz F, Peters JM, Hyman AA, Gingras AC, Pelletier L. HAUS, the 8-subunit human Augmin complex, regulates centrosome and spindle integrity. Curr Biol. 2009;19:816–826. doi: 10.1016/j.cub.2009.04.033. [DOI] [PubMed] [Google Scholar]
- 186.Dammermann A, Desai A, Oegema K. The minus end in sight. Curr Biol. 2003;13:R614–R624. doi: 10.1016/S0960-9822(03)00530-X. [DOI] [PubMed] [Google Scholar]
- 187.Rath U, Rogers GC, Tan D, Gomez-Ferreria MA, Buster DW, Sosa HJ, Sharp DJ. The Drosophila kinesin-13, KLP59D, impacts pacman and flux-based chromosome movement. Mol Biol Cell. 2009;20:4696–4705. doi: 10.1091/mbc.E09-07-0557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Buster DW, Zhang D, Sharp DJ. Poleward tubulin flux in spindles: regulation and function in mitotic cells. Mol Biol Cell. 2007;18:3094–3104. doi: 10.1091/mbc.E06-11-0994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Labbe JC, McCarthy EK, Goldstein B. The forces that position a mitotic spindle asymmetrically are tethered until after the time of spindle assembly. J Cell Biol. 2004;167:245–256. doi: 10.1083/jcb.200406008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Grill SW, Howard J, Schaffer E, Stelzer EH, Hyman AA. The distribution of active force generators controls mitotic spindle position. Science. 2003;301:518–521. doi: 10.1126/science.1086560. [DOI] [PubMed] [Google Scholar]
- 191.Kozlowski C, Srayko M, Nedelec F. Cortical microtubule contacts position the spindle in C. elegans embryos. Cell. 2007;129:499–510. doi: 10.1016/j.cell.2007.03.027. [DOI] [PubMed] [Google Scholar]
- 192.Grill SW, Gonczy P, Stelzer EH, Hyman AA. Polarity controls forces governing asymmetric spindle positioning in the Caenorhabditis elegans embryo. Nature. 2001;409:630–633. doi: 10.1038/35054572. [DOI] [PubMed] [Google Scholar]
- 193.Pecreaux J, Roper JC, Kruse K, Julicher F, Hyman AA, Grill SW, Howard J. Spindle oscillations during asymmetric cell division require a threshold number of active cortical force generators. Curr Biol. 2006;16:2111–2122. doi: 10.1016/j.cub.2006.09.030. [DOI] [PubMed] [Google Scholar]
- 194.Sulston JE, Horvitz HR. Post-embryonic cell lineages of the nematode, Caenorhabditis elegans . Dev Biol. 1977;56:110–156. doi: 10.1016/0012-1606(77)90158-0. [DOI] [PubMed] [Google Scholar]
- 195.Sonnichsen B, Koski LB, Walsh A, Marschall P, Neumann B, Brehm M, Alleaume AM, Artelt J, Bettencourt P, Cassin E, Hewitson M, Holz C, Khan M, Lazik S, Martin C, Nitzsche B, Ruer M, Stamford J, Winzi M, Heinkel R, Roder M, Finell J, Hantsch H, Jones SJ, Jones M, Piano F, Gunsalus KC, Oegema K, Gonczy P, Coulson A, Hyman AA, Echeverri CJ. Full-genome RNAi profiling of early embryogenesis in Caenorhabditis elegans . Nature. 2005;434:462–469. doi: 10.1038/nature03353. [DOI] [PubMed] [Google Scholar]