Abstract
Centrioles are cylinders made of nine microtubule (MT) triplets present in many eukaryotes. Early studies, where centrosomes were seen at the poles of the mitotic spindle led to their coining as “the organ for cell division”. However, a variety of subsequent observational and functional studies showed that centrosomes might not always be essential for mitosis. Here we review the arguments in this debate. We describe the centriole structure and its distribution in the eukaryotic tree of life and clarify its role in the organization of the centrosome and cilia, with an historical perspective. An important aspect of the debate addressed in this review is how centrioles are inherited and the role of the spindle in this process. In particular, germline inheritance of centrosomes, such as their de novo formation in parthenogenetic species, poses many interesting questions. We finish by discussing the most likely functions of centrioles and laying out new research avenues.
Keywords: Centriole, Centrosome, Cilia, Mitosis, Microtubule, Parthenogenesis
Introduction
Since it was first discovered by Boveri and Van Beneden in the 1880s, the centriole remains a surprisingly mysterious organelle. Although there have been many recent advances in the comprehension of its structure and functions, it remains, as quoted by Paoletti and Bornens [1] a kind of “terra incognita” of the cell.
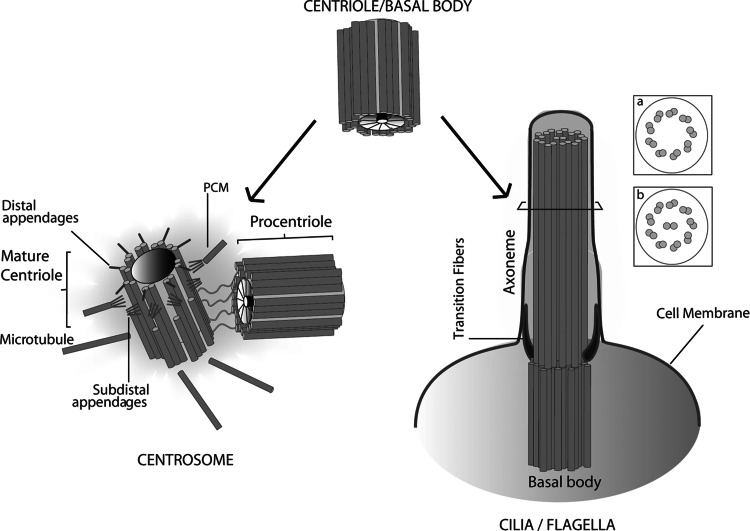
Centrioles are cylinders made of nine microtubule (MT) triplets organized in a nine-fold symmetrical configuration (Fig. 1), which are present in many eukaryotes and essential for the formation of several microtubule-organizing structures including centrosomes and cilia (Fig. 1) [2, 3]. The centrosome is the primary microtubule-organizing center (MTOC) in many animal cells, participating in a variety of processes such as cell polarity, intracellular traffic, and cell division. At the heart of each centrosome there are two centrioles, which define centrosome properties.
Fig. 1.
Centriole/basal body structure and function. The centriole/basal body is a structural constituent of the centrosome and of cilia and/or flagella. The canonical centriole has nine MT triplets and is ~0.5 μm long and 0.2 μm in diameter. Each centrosome is composed of a mother and daughter centriole, in an orthogonal configuration, surrounded by a matrix of proteins, the pericentriolar material (PCM). The older mother shows appendages, where Mts are docked. In certain cells, the centriole migrates and tethers to the membrane via its appendages and seeds the growth of cilia and flagella that enable cell movement and/or chemoreception. The skeleton of cilia and flagella, called the axoneme, results from a continuation of the basal body structure and might be composed of nine doublets with no central pair (a) as it is in the case of most immotile cilia; or nine MT doublets with a central MT pair as it is for most motile cilia (b)
Cilia and flagella are microtubule-based cell projections, which can be motile, and are involved in a plethora of processes, from sensing extracellular signals to moving fluid and cell motility. Centrioles are called basal bodies when anchored at the membrane to nucleate the axoneme, the core microtubule structure of cilia (Fig. 1). Basal body properties define many cilia characteristics, such as its location and symmetry.
Early studies, where centrosomes were seen at the poles of the mitotic spindle led to their coining as “the organ for cell division” coordinating karyokinesis and cytokinesis [4, 5]. However, a variety of subsequent observational and functional studies showed that centrosomes might not always be essential for spindle establishment, and to the hypothesis that “the spindle of Metazoan cells is a basal body distributor that guarantees the accurate segregation of both chromosomes and centrioles (basal bodies)”, being cilia formation centriole’s major function [6, 7].
Here we review the arguments in this debate. We start by describing the centriole structure and its distribution in the eukaryotic tree of life. We clarify its role in the organization of the centrosome and cilia. We then present historical and current arguments used in this debate with particular attention to the role of centrioles in mitosis and to its modes of inheritance and distribution to daughter cells. We finish by discussing the most likely functions of centrioles and what avenues of research are open in this field.
Centriole structure and distribution in the eukaryotic tree of life
Centrioles/basal bodies are ~0.2 μm in diameter and their length may vary according to species and tissue type, around 0.5 μm (Fig. 1) [3, 8]. They are polarized along the proximo-distal axis [3, 8, 9]. Electron microscopy has revealed that at their proximal end, immature centrioles, and in some species even mature ones, have a feature called the cartwheel, a structure made of a central hub linked by spokes to the inner tubule of each triplet [10, 11] (Fig. 1). Older centrioles have sub-distal appendages and distal appendages/transition fibers, which conserve the nine-fold symmetry of the centriole and dock cytoplasmic MTs and anchor centrioles to the cell membrane, respectively [3, 8] (Fig. 1). Basal bodies may have additional appendages including rootlets at their base and a basal foot formed along precise microtubule triplets [12] [9]. These structures may themselves radiate MTs and organize basal body position and orientation in relation to other cellular components, which is important for coordinating the movement of cilia and organization of the intricate cytoskeleton in many protists [9, 12]. Basal bodies have a transition zone at their distal end, contiguous with the axoneme, which is important for the nucleation of ciliary microtubules, as a docking site for protein transport into the cilia, and for organized disassembly/assembly of the cilia. Centrioles/basal bodies are extremely stable structures, and their MTs are resistant to cold, depolymerizing drugs, and detergents. This stability may be provided by post-translational modifications of centriolar tubulin, such as polyglutamylation [13, 14], and by other structural components of the centriole, such as tektins and ribbon proteins [15, 16].
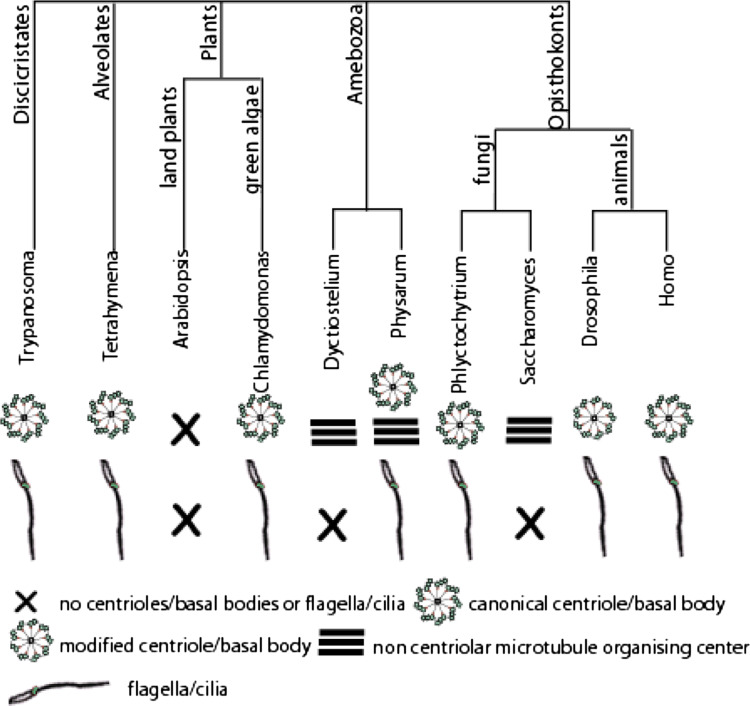
Centrioles and basal bodies (CBBs) are found in all major eukaryotic groups, suggesting their presence in the Last Eukaryotic Common Ancestor (LECA; [17, 18]; Fig. 2). In most studied organisms, centriole microtubules are organized in a nine-fold symmetry and triplet arrangement, although there are some exceptions. For example, depending on the tissue type, Drosophila melanogaster presents doublet or triplet MTs, while Caenorhabditis elegans also presents singleton MTs [2]. Different species and tissues within a single species can also present basal body appendages such as rootlets and basal feet, with different shapes and sizes. The symmetry of the axonemal structure can deviate from the prototypical nine-fold. This is common in invertebrates, such as worms and insects [19].
Fig. 2.
Centrioles, basal bodies, and flagella in the eukaryotic tree of life. Note that in all represented branches there are organisms that have cilia/flagella suggesting they were present in the last common ancestor. These structures are associated with the presence of centrioles/basal bodies. Whenever flagella/cilia are absent, the centriole is lost. In some cases, other MTOCs exist, such as the spindle pole body of yeasts or the nuclear associated body of dictyostelium. Certain organisms have modified centriole structures, such as Drosophila
The presence of CBBs correlates with the occurrence of a flagellated stage in the organism life cycle. CBBs were lost throughout evolution from several eukaryotic Phyla [17] in species that do not have flagella, such as amoebas (e.g., Dictyostelium discoidum), yeasts (e.g., Schizosaccharomyces pombe, Saccharomyces cerevisiae) and higher plants (e.g., Arabidopsis thaliana). Some of those species that have no flagella have modified centrosomes with no centrioles, such as the nuclear associated body (NAB) in amoeba and the spindle pole body (SPB) in the yeasts [2]. These MTOCs help in the organization of the spindle in mitosis and despite their difference in structure, they share several components with canonical centrosomes (reviewed in Ref. [20]). The correlation between the occurrence of CBBs and the presence of flagella/cilia but not centrosome-like structures, suggests CBBs are mostly essential for axoneme formation (Fig. 2).
Centrioles as centrosome organizers and nucleators of cilia and flagella
The centrosome is comprised of two centrioles, the older one, called mother or mature, and a younger one, called the daughter or immature, which are surrounded by an electron-dense matrix, the pericentriolar material (PCM) (Fig. 1). The PCM harbors molecules that anchor cytoplasmic MTs in interphase and mitosis, such as members of the pericentrin and AKAP450 family of proteins. These molecules are enriched in coiled-coil domains and are thought to form a lattice-like structure (reviewed in Ref. [3]), which docks molecules that mediate the nucleation of MTs, such as γ-tubulin [21]. γ-Tubulin is also associated with the walls of the centrioles [142] and its levels increase considerably prior to mitosis [22]. The architecture of the microtubule array generated by the centrosome depends on the balance between nucleation, anchoring, and severing of the MTs. For example, while a fibroblast displays a radial array of microtubules focused at the centrosome, epithelial cells, whose centrosome display less anchoring/nucleation activity, may show non-focused arrays perpendicular to the apical membrane [23]. At the centrosome, it is mostly the mother centriole that is able to anchor MTs on its associated sub-distal appendages [24]. However, microtubule anchoring complexes also exist in the PCM and in pericentrosomal satellites, electron dense granules composed of PCM1 protein. Ninein, centriolin, dynactin, EB1, CAP, FOP and others (reviewed in Refs. [3, 25, 26]) play a role in microtubule anchoring, while microtubule-severing proteins, such as katanin, release MTs into the cytoplasm [25]. In conclusion, the centrioles provide a location where the PCM is assembled and where MTs can be anchored, leading to the formation of an MTOC.
Centriolar characteristics determine most properties of the centrosome, such as stability, capacity to reproduce and polarity. Single centrioles [27], and even small centrioles such as the ones generated upon mutation of proteins involved in their assembly [28–30], can recruit the PCM, but centriole loss leads in general to PCM dispersal [13, 31, 32]. The ability of centrioles to duplicate is therefore central to the centrosome’s reproductive capacity [33].
The other role of centrioles is the nucleation of the axoneme, the skeleton and motor machinery of cilia and flagella. There are different types of cilia and flagella, depending on the structure of the axoneme. Differences in the structure of axonemes may be reflected in their properties, for example, whether they are motile or not (reviewed in Ref. [34]). The structure of motile cilia is generally conserved with nine doublets surrounding a central microtubule pair, while non-motile cilia may show a greater diversity of doublet microtubule numbers and the quite often absence of the central pair ([35], Fig. 1). Cilia exist in different numbers and shapes in different cell types, such as the unique tail of spermatozoa and the 200–300 cilia of epithelial cells in our airways. Non-motile cilia, such as primary cilia, are microtubule-based sensory organelles, present in most of our cells, that play important roles in development and disease [36]. As centrioles define many of the centrosome properties, so do basal bodies in relation to cilia and flagella. Basal bodies provide a nine-fold symmetric template, from which the ciliary axoneme extends. The junction of the basal body to the ciliary axoneme is the transition zone, where the most external microtubule of the centriole does not continue, a central pair of MTs is nucleated in the majority of motile cilia and distal appendages are found (Fig. 1). The distal appendages, also called transition fibers, might provide a filter for molecules that go inside the cilium, since protein synthesis does not occur inside cilia [35]. Transport of cilia/flagella components and signaling proteins inside the cilium depends very often on motor proteins, such as dyneins and kinesins, that move on the axoneme, and is called intraflagellar transport [36, 37]. The position of the basal body and its orientation within the cell ensures that cilia are formed and oriented in the correct fashion to move fluid flow in a certain direction or to sense signals from particular surrounding cells. Structures at the basal body, such as the rootlets and the basal feet orient cilia beating and localization within the cytoplasmic membrane. The orientation of the basal bodies, while initially influenced by tissue polarity cues [38], is then influenced by the direction of fluid flow set by the newly formed cilia in a positive feedback loop [39]. In cells where CBBs play a role in axoneme and centrosome formation, the duality of function is clear (Fig. 1). For example, in cycling ciliated animal cells, such as fibroblasts, upon centrosome maturation at the G2/M transition, the cilium is reabsorbed and the CBBs move to the center to participate in mitotic spindle assembly [40] in the context of the centrosome. At the exit of mitosis, the CBB can migrate to the membrane, participating again in cilia formation. A Golgi-derived vesicle appears at the distal end of the mother CBB, which then fuses with the cytoplasmic membrane. The migration of the centriole to the surface of the cell is dependent on a variety of proteins and the actin cytoskeleton [41]. If nutrients are removed, cells might become quiescent (G0), and the cilium can grow further, up to 10 μm [26]. Upon serum stimulation, cells re-enter the cell cycle and the cilia are normally reabsorbed. Little is known about the molecular mechanisms governing the switch from a centriole to a basal body and vice versa. A variety of molecules and cytoskeleton filaments have been recently shown to play a role. For example, CP110, a centriolar component also involved in centriole biogenesis and length control, might cap the centriolar structure, regulating cilia formation [42]. ODF2, a protein located at the distal appendages, allows the mother centriole to become a basal body, being important for tethering at the membrane [43]. Recent evidence suggests that cilia may be involved in determining the cell division axis and centrosome positioning, at least in kidney cells, linking the two centriolar functions [44]. The duality of the centriole, as a basal body that nucleates axonemes and as a core component of the centrosome, raises important questions regarding its function that have been addressed by researchers for more than half a century.
On the role of centrioles in cell division: discovery and controversy
The earlier work on the centrosome by 19th-century scientists was nicely described in Wilson’s book The Cell in Development and Inheritance (1896) [45]. More recent description of the first years of characterization of the centriole/centrosome can also be found in Wheatley [46] and in the excellent reviews of Fulton [47] and Gall [48]. Boveri was the first to coin the term centrosome in his observations although Van Beneden clearly identified the same structure. The name centriole was proposed by Boveri to define a central granule constantly found inside the centrosome. However, as these structures are at the limit of the resolution of a light microscope, other early cytologists attributed the terms centriole or centrosome (or sometimes centrosphere or central body) to similar structures. Ultrastructural observations then revealed the nature of the centriole and allowed scientists to distinguish clearly between the centriole (the central cylindrical structure with nine-fold symmetry) and the centrosome (the centriole(s) and PCM (Fig. 1)). One of the difficulties in analyzing and defining the centrosome has been the absence of a membrane delimiting this organelle. Ten years after the discovery of centrioles, Henneguy [49] and Lenhossék [50] independently found that centrioles can give rise to basal bodies. Later EM observations proved the identical structure of CBBs cartwheels, and many cases are now known of the interconversion of CBBs as discussed above.
Rise and fall of centrioles as the division center of the cell
As early as 1887, Boveri and Van Beneden independently identified a spherical structure lying at the mitotic spindle poles in the first cleavages of Ascaris embryos. Both of them characterized this structure as “the organ for cell division”. This conclusion was supported not only by the obvious strategic position of the centrosome at the spindle pole, but also by a careful reconstitution of mitotic spindle formation during which the centrosomes move ritually to antipodes on the nucleus and seem to participate in the elaboration of the spindle. This structure-organizing aspect was also supported by the apparent dominant force of the centrosome in abnormal situations such as multipolar spindles in polyspermic sea urchin eggs. Even more surprising, when we realize that they had no tools for following the microtubule cytoskeleton, these authors also concluded that the centrosome can be seen as “the dynamic centre of the cell” a statement which many decades later was found largely to be true for most cell types. Following this logic and still with an intuition incredibly ahead of its time Boveri (as well as other early cytologists like Hansemann and Galeotti) proposed that abnormalities in centriole duplication could be at the origin of the genome instability so characteristic of cancer cells [4, 5, 51]. This prophetic hypothesis is today at the heart of many current studies showing centrosome amplification as a signature of many tumors [52–54].
During the exciting period of pioneering studies on the centrosome, the centriole was first seen as essential for mitosis. In later years however, reports of its absence in many species of fungi and seed plants as well as in many classes of protists for acting at the mitotic spindle poles seriously challenged this point of view [7]. As commented by Fulton [55] many cell biologists of this period actually did encounter at some point in their carrier a question related to this organelle, but the lack of available tools and the minuteness of the object often precluded a clear conclusion worthy of publication: “Many of us have had a transient affair with centrioles and retain a profound affection for them, but few have committed ourselves to a long-term relationship, perhaps because, in spite of our enchantment, we could not find a way to build a meaningful relationship-that is, to break through the enigmas”. Although the general belief had prevailed that centrioles were absolutely essential for division in animal cells, eventually this last bastion was finally attacked. Dietz succeeded in experimentally dislocating the centrosome from the nucleus in the primary spermatocytes of the crane fly (Pales ferruginea) by flattening the cells [56]. He observed that in these conditions, during meiosis the centrosome is lacking at one pole (and even sometimes both) but the spindle forms perfectly and allows the correct migration of the chromosomes and cytokinesis. Dietz concluded that at least in this system, centrioles are “completely dispensable” for spindle formation. Even more provocative, Friedlander and Warhman proposed that “the spindle of Metazoan cells is a basal body distributor that guarantees the accurate segregation of both chromosomes and centrioles (basal bodies)” [6]. They founded their conclusion on observations of meiotic spindles in spermatocytes of the silkworm Bombyx mori where the centrioles are clearly separated from the spindle microtubules (Fig. 3). In his paper “The autonomy of the centriole: fact or fallacy”, Pickett-Heaps reviewed most ideas concerning the mitotic role of the centriole [7]. He approached two different problems concerning the centriole: the question of its autonomy and its function in spindle microtubule assembly. This author strongly questioned the role of the centriole in establishment of a mitotic spindle and he proposed that in most cases centrioles are not involved in the formation of the spindle: “The centrioles instead appear more likely to be inert passengers ensured of being partitioned equally between daughter cells by being attached to the spindle apparatus” [7].
Fig. 3.
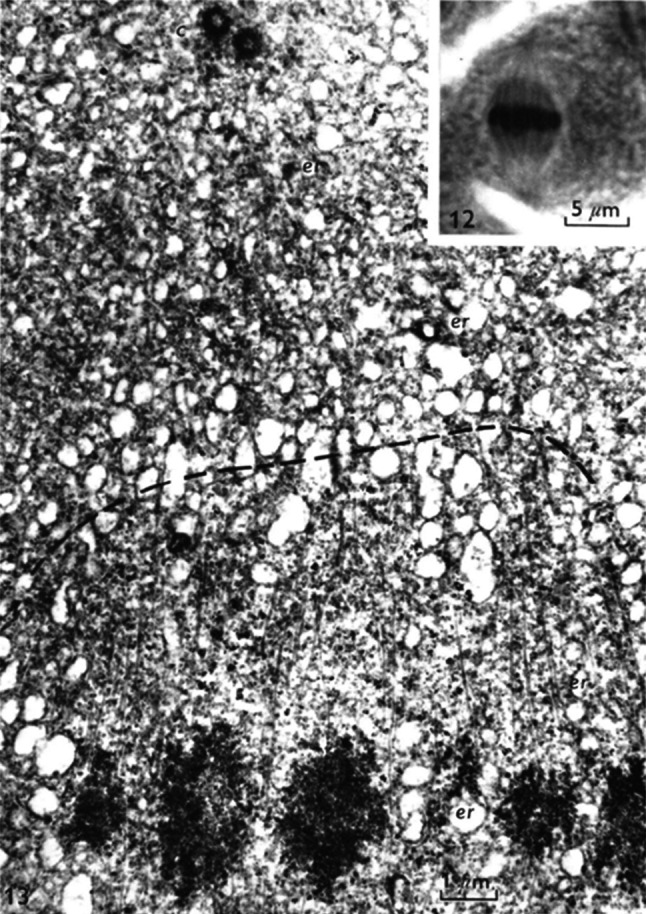
Male meiosis metaphase in Bombyx mori (From Friedlander and Warhman 1970). The spindle terminating is underlined by the broken line. The centrioles (which will become basal bodies) are clearly found at distance from the spindle microtubules. The authors proposed that the spindle is a basal body distributor: We consider the position of the centrioles at the poles of the spindle as a consequence rather than a cause of spindle formation. Main figure, TEM ×13500 Inset: components of the spindle in the light microscope. ×3000
Starting from the mid 1970s the centriole was bound to arouse a revival of interest, mainly because it became evident that one key role of the centrosome was the nucleation and organization of microtubule arrays. An important conceptual advance brought by Pickett-Heaps is the notion of MTOC a structure which can clearly exist with or without centrioles [7]. In our point of view, this concept reconciles the contradictory opinions regarding the role of the centrioles. It is worth to note however that whereas a centrosome is necessarily an MTOC, the reverse is not true ([18], footnote 2).
In a key paper, Gould and Borisy [57] using the remarkable presence of virus like particles (VLPs) in Chinese hamster ovary cells as a marker of the PCM demonstrate that microtubules are actually nucleated mostly from this material and not by centrioles themselves. Importantly, PCM aggregates without centrioles are able to nucleate MTs in vitro. This work explained the presence of both centriolar and acentriolar MTOCs, and showed that in some biological models, an MTOC can be comprised of a focal accumulation of PCM. A question remains however: what does the presence of centrioles contribute to an MTOC? More accurate knowledge of the function of the centriole during mitosis was obtained progressively in parallel with an understanding of the mechanism of microtubule nucleation. Previously it was difficult to address these questions and only recent markers, knowledge of players, and modern technology have allowed their elucidation.
Spindles can form without centrioles in animals
Natural absence of centrioles in dividing Metazoan cells
The most obvious case of an animal spindle lacking centrioles is that of some female meiosis. This has been known for a long time but it was first considered as a very special case adapted to the mechanism of fertilization. If the natural elimination of centrioles has occasionally been documented in somatic cells, it was restricted to non-dividing cells such as myotubes [58–60] and insect wing epidermal cells [61]. During differentiation of some epithelial cell types non centrosomal microtubule arrays are generated but the centrioles remain in the cell [62] reviewed in Ref. [23].
However, as far as we know, there have been no reports of a natural and continuous absence of centrioles in a proliferating Metazoan tissue. A very interesting case is that of the mouse embryo where centrioles are transiently absent from blastomeres during the first cleavages [63]. Centrioles are detected by EM for the first time at the 64-cell stage [64].
Another interesting case occurs in the fungal gnat Sciara in which unfertilized eggs initiate development, forming anastral spindles and successfully segregating sister chromatids [65].
Experimental inactivation of the centrosome in Metazoan cells
Destabilizing centrioles with antibodies
Glutamylation is a posttranslational modification of tubulin which is essentially restricted to the centrioles in non neuronal cells [66]. In HeLa cells, electroporation of an antibody specific for polyglutamylated tubulin led to a transient disappearance of centrioles as well as dispersal of the PCM [13]. During the period when centrioles were absent, monopolar and normal bipolar spindles formed. Centrioles then reformed after about 48 h and the cell population displayed a normal centrosome labeling after 84 h. This elegant study showed that: (1) centrioles are responsible for the organization of PCM; (2) animal cells can proceed to mitosis and form a spindle without centrioles; (3) the biogenesis of a procentriole does not require the structural integrity of the parent one.
Ectopic spindle poles with no centrioles generated by microtubule poisons
Multipolar mitosis can be induced in Chinese hamster ovary cells by release from a colchicine block [67]. In tripolar mitosis, centrioles are distributed in either a 2:1:1 or in a 2:2:0 pattern. In the 2:2:0 situation the pole without a centriole presents a cloud of PCM, recognizable in these cells by the presence of VLPs. These cells accomplish mitosis perfectly up until telophase and reformation of the nucleus. It seems nevertheless that the daughter cells have more difficulty separating than the tripolar cells with a 2:1:1 centriole pattern. This study suggested an unpredicted function for the centriole in the completion of midbody cleavage.
Ablation of the centrioles by laser irradiation and microdissection
Michael Berns and collaborators were among the first to irradiate the centrosome with a laser microbeam [68, 69]. UV and laser microbeam irradiation of the centrioles is now facilitated by in vivo labeling with fusion protein constructs such as centrin-GFP [70] or γ tubulin-GFP [71–74] for a review see Ref. [75]. Laser microsurgery is nowadays so precise that only one centriole of the diplosome can be specifically ablated [76]. Microsurgical removal of the centrioles can also be achieved by dissecting a cell into two separate parts, a karyoplast containing the nucleus and a cytoplast containing the centrioles [77, 78]. In all cases these acentriolar cells can assemble a mitotic spindle in the absence of centrioles, thus confirming the dispensability of centrioles for this process. A high frequency of aberrations is observed however including mono- and multi-polar spindles and also defects in the separation of the daughter cells, which support the idea that centrioles are involved in the bipolarity of the spindle as well as in completion of cytokinesis.
Animal cell lines without centrioles
In Drosophila, a cell line (1182–4) which constitutively lacks centrioles [79], has been obtained from haploid embryos laid by females homozygous for the lethal mh 1182 mutation [80, 81]. This characteristic has been firmly established by two extensive ultrastructural studies, including high-voltage electron microscopy from serial thick sections [82]. In these cells, mitotic spindles generally present broad or scattered poles, and present no or only few astral microtubules. There is no recruitment of PCM proteins like γ-tubulin, centrosomin, D-PLP, or centriolar proteins such as DSas-4 to a centrosomal structure at the spindle poles. Even if the more polarized acentriolar spindles present a focal concentration of γ-tubulin at the poles, this structure disperses after microtubule disassembly [83, 84]. These cells divide quite well in culture. However, there is a high frequency of daughter cells linked by a cytoplasmic bridge suggesting that abscission process is more difficult [70]. The cause of the acentriolar nature of the 1182–4 cell line remains unclear, but their existence supports the total expendability of centrioles for mitosis in Drosophila.
Acentriolar Drosophila mutants
The first report of the complete development of a Metazoan with non functional centrosomes came from the centrosomin mutant in Drosophila. Centrosomin is a key component of the PCM and is recruited to the centrosome during mitosis. A loss of function mutation in cnn prevents the assembly of many components of the centrosome such as gamma tubulin and impairs the ability of the centrosome to nucleate microtubules [85, 86]. A similar role has been recently attributed to the CNN fission yeast and human orthologues—mto1 [87] and CDK5RAP2 [88], respectively. Surprisingly Drosophila individuals mutant for cnn (but born from heterozygous mothers) develop normally without any morphological aberrations, although they are sterile [89]. These observations were so surprising that it raised some doubts about the non functional status of centrosomin-depleted centrosomes, since in this case centrioles were still present. Some years later Basto et al. [32] and Bettencourt-Dias et al. [31] studied Drosophila mutants lacking DSas-4 or with truncation of SAK/PLK4, two proteins essential for centriole duplication. In both cases the adult individuals lack centrioles (almost total absence for DSas-4 and partial for SAK/PLK4) but they successfully go through development without morphological abnormalities. The adults however are devoid of cilia and flagella structures and die very early after emergence, as the absence of mechanosensory organs leads to a severe lack of coordination. Precise examination of asymmetrical divisions in neuroblasts revealed a higher frequency of abnormal mitosis showing that centrioles are helpful in this mechanism [32, 89]. Moreover, some cells failed cytokinesis [32]. Adult males are sterile, showing defects in meiotic divisions and having no sperm axonemes [32, 90]. The case of females is interesting: it has been possible to obtain eggs of DSas-4 and SAK/PLK4 genotype either by germinal clones in heterozygous females or by hand feeding of homozygous females. In both cases eggs appear normal but corresponding embryos arrest very early in development after only a few abnormal mitoses [90, 91], showing that centrioles are necessary for syncytial mitoses. New Drosophila cell lines homozygous for the mutation DSas-4 were recently established (N. Lecland, S. Pereira, N. Malmanche, H. Maiato A. Guichet and A. Debec, unpublished results). As in the case of the 1182–4 line, these cells do not have centrioles and there is no recruitment of PCM proteins to a centrosomal structure at the spindle poles, corroborating the total expendability of centrioles for mitotic division in Drosophila. To conclude, centrioles are not needed in vivo for the assembly of a mitotic/meiotic spindle; however these structures might be needed for the fidelity and asymmetry of cell division in specialized tissue types.
Centrosomes are not the only players in spindle formation: the existence of complementary pathways
The debate on the role of the centriole in mitosis is also long winded due to the fact that animal cells use several cooperative strategies to build a spindle. At least two other pathways have been shown in recent years to be used in spindle assembly: the chromatin and the augmin pathways. The nuclear envelope may also contribute to microtubule nucleations [92]. These topics have been reviewed extensively in recent years.
A role for centrioles in cell division?
In the previous sections we showed evidence that centrosomes are not needed to nucleate and organize the mitotic spindle, because cooperative pathways can assemble this structure. However, the experiments discussed above also revealed the importance of centrioles for certain aspects of cell division, such as cell cycle progression, proper chromosome segregation, asymmetric cell division and cytokinesis, depending on tissue type.
Cell cycle progression
Many signaling proteins localize to centrosomes. Experiments were conducted on vertebrate cells to address the possible control of the cell cycle by centrioles. Often acentriolar cells entered mitosis but then arrested in G1 in the next cell cycle. It was first suspected that this arrest could reflect a control of cell cycle progression by centrioles [77, 78, 93], but this point is still debated. It has recently been proposed that such an effect is the result of cumulative stress resulting from both centriole removal and experimental stress (e.g. excessive light) and is not the consequence of a real centriole checkpoint [74, 94] also discussed in Ref. [95]. The fact that acentriolar cells can normally progress through the cell cycle suggests that there is no conserved checkpoint that depends on the presence of centrosomes [31, 32, 83].
Chromosome segregation and cytokinesis
Studies of cnn mutants suggested that centrosomes are required for correct spindle function in male meiosis [96] and for mitosis in the syncytial embryo [85, 97]. The same is true for Drosophila mutants that lack centrioles as it is the case for spermatocytes of SAK/PLK4 and SAS4, and early embryos devoid of SPD2, SAK/PLK4 and SAS4 [31, 90, 91, 98]. In testes, acentriolar meiotic spindles are largely abnormal, resulting in dramatic chromosome segregation and cytokinetic defects. Formation of bipolar spindles in spermatocytes occurs through both chromosome and centrosome dependent mechanisms [92]. The presence of a weak Spindle Assembly Checkpoint might be at the origin of precocious chromosome segregation and aberrant meiotic products [92, 99]. In the case of syncytial embryos, the extremely rapid mitotic cycles may rely very heavily on centrosome-dependent mechanisms for bipolar spindle assembly and spindle localization. Abnormal acentriolar spindles interact with each other leading to wide-spread abnormalities which may lead to a mitotic arrest [100]. Indeed all SAK/PLK4 mutant embryos die arrested in a mitotic-like stage, suggestive of an active checkpoint arrest [90]. In summary, whereas centrioles may be dispensable for cell division in some tissues of the fly, they are absolutely essential in others, perhaps due to tissue specificity constraints, such as weaker checkpoints or the context of a syncytium. The same is true in other organisms, such as the C. elegans embryo [101] and fission yeast [102], where the centrosome and its equivalent, the spindle pole body, are essential for bipolar spindle assembly and cytokinesis, respectively.
In human cultured cells, centrosomes are also important to ensure mitotic fidelity (reviewed in Ref. [2]), in particular cytokinesis. Centrosomes may interact with the cortex to induce furrow formation. This was first and mostly elegantly demonstrated in a series of experiments by Rappaport in which he experimentally induced ectopic furrows in syncytial sand dollar embryos [103, 104]. These furrows formed between neighboring centrosomes and their astral microtubule arrays in spite of the fact this region did not contain chromatin or a central spindle. These studies lead to the proposal that signals from opposing overlap centrosome-based microtubules induce cleavage furrow formation. Recent studies have demonstrated that centrosome activities in addition to microtubule nucleation induce furrow formation. The centrosome-associated recycling endosome is required for both vesicle mediated membrane delivery and actin organization during cleavage furrow ingression [105]. An increasing number of centrosomal proteins have been reported to participate in cytokinesis, such as centriolin, Cep55, CP110 and BBS6, some of which localize both to centrosomes and the midbody. In addition, a recent study identified the CNN-binding protein centrocortin, which is not required for normal centrosome based microtubule organization or recycling endosome organization, but it is required for normal cleavage furrow formation [106]. Such a signaling role in cytokinesis has been suggested in HeLa and L929 cells [70] where tracking of centrioles by centrin-GFP shows that the mother centriole transiently moves close to the intercellular bridge and then moves back to the cell center. This suggests centrioles may carry a signal that triggers abscission. It is possible that in each cell type, cell-specific characteristics selected for distinct contributions of the different spindle assembly mechanisms, leading to the evolution of tissue-specific, spindle-assembly mechanisms [90].
Developmental mechanisms
More generally, centrioles could be responsible for precise regulation of mitosis, either in space (the plane of division, for example during the asymmetrical divisions in some cell types but not all), in time (gating the decision to divide, deciding the final number of divisions) and even possibly in regulation of cell fate (Drosophila male stem cells and rodent neural progenitors [107, 108]) and could be therefore strongly implicated in development and morphogenesis. In this light, it is important to refer that redundant mechanisms might also be at play, as in Drosophila male germline stem cells are more dependent on centrosomes for asymmetric cell division [107], as compared to neuroblasts and female germline stem cells [32, 91] (for reviews on other mechanisms in asymmetric cell division see Refs. [109–111]).
Recent work suggests that the inherited difference in age of centrioles within a single cell might be important in signaling. By virtue of the centriole cycle, a cell in mitosis has four centrioles that belong to different generations: a grandmother and a mother, each associated with one daughter. The cell inheriting the grandmother centriole after cytokinesis grows the primary cilia first and is able to respond to signaling cues [112]. Those results suggest that the centriole age carries developmental information, playing an important role in breaking the symmetry between two daughter cells, and in keeping stem cells associated with their niche. Finally ciliary function might be important in the positioning of mitotic spindles, as it is the case in kidney development [44].
How are centrioles inherited? The question of continuity
An important aspect of the debate addressed in this review is how centrioles are inherited and the role of the spindle in this process. Is the spindle a necessary machine for centrioles to be equally distributed to daughter cells? Does a cell need to inherit centrioles in order to have them? The notion of centrosome continuity, and that centriole inheritance is needed for cells to have centrioles, has been challenged by peculiar situations where centrioles can appear without a pre-existing centriole (“de novo” formation). That is the case in many organisms upon flagella formation, such as the ameboflagellate Naegleria [113], or the fern Marsilea during spermatogenesis [114], or parthenogenesis in several insects as we will further discuss in this review. Because of the important differences we have divided this section in two parts: inheritance of centrioles in somatic and in germ cells.
Inheritance in somatic cells
Concomitant with the first observations of the centrosome as a division organ of the cell, Boveri and Van Beneden defined the centriole as a permanent cell organelle. This was later found to be true in many animal cell types. One of the best illustrations of this situation were the observations of Huettner [115] in the syncytial divisions of the early Drosophila embryo. In this case Huettner was able to follow with very good resolution the duplication of the centrioles and their continuity during the successive nuclear divisions. In addition Huettner stated a strong argument “that the centrioles are here the causative agents for astral formation” (page 132) an important point as some doubtful biologists raised the concern that centrioles might be artifacts of fixation (discussed also in Ref. [116].
The centrosome and centriole cycle
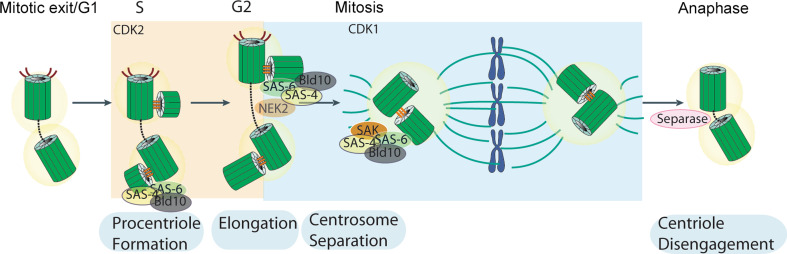
During the 1970s and 1980s, microscopists studied the centriolar cycle at the ultrastructural level [117–122]. They confirmed the former optical observations, i.e. that there is a tight correlation between the replication of the chromosomes and the duplication of the centrioles in S phase. During the G1/S phase of the cell cycle, a nascent centriole (the procentriole) forms at the proximal end of each parent centriole. The formation of a procentriole occurs after the loss of orthogonal configuration of the parent centrioles. The daughter centrioles grow progressively, and have acquired most of their final length by mitosis. Each mother and daughter centriole pair constitutes a new centrosome. In G2, centrioles recruit many proteins in preparation for mitosis, including microtubule regulators; this is called centrosome maturation. Centrosomes then separate physically at the entrance of mitosis, each one migrating to one of the poles of the spindle, to which they remain associated. Cytokinesis then formally separates each centrosome to each daughter cell. The centriolar cycle is also reflected in the organization of the PCM around centrioles. Rieder and Borisy [121] followed the dynamics of the PCM in PtK2 cells and found that the PCM is mostly associated with the parent centriole. The daughter centriole acquires PCM only in the next cell cycle when it becomes itself a parent. These authors determined the parent–daughter centriole relationship first by the configuration of the two centrioles; they also identified the older centriole by its association with the primary cilium.
When thinking about centriole inheritance in somatic cells, it is clear that processes such as centriole biogenesis, control of centriole number and control of centriole segregation play a very important role in determining how many centrioles are inherited by daughter cells.
Daughter centriole formation
The first structures observed include an electron dense material, which forms close to the mother centriole, and a cartwheel, with nine spokes, that forms on top of the electron dense material. Then microtubules are recruited to the cartwheel, the number and order of addition depending on which organism [10, 123, 124]. Elongation of the centriole is dependent on the elongation of those microtubules. Whether there is a cap that regulates centriolar length it is still not known. While the molecular composition and function of the electron dense disc is not known, recent work has shed some light onto the function and composition of the cartwheel, and regulators of microtubule “anchoring” and elongation at the centriole structure. SAS-6 and Bld10/CEP135 are two components of the cartwheel, the first being part of the hub and spokes and the second forming the spoke tips. These molecules were originally identified in C. elegans and Chlamydomonas, respectively, but their role in centriole formation has been validated in many different organisms [30, 125–136] (Fig. 4). Work in C. elegans and more recently in human cells has shown that SAS-4/CPAP is involved in microtubule recruitment to the centriole and elongation of this structure. Its role might be counteracted by yet another centriole component CP110 [137–139]. Other players have been involved in the initial steps of centriole formation, such as asterless [140, 141], gamma tubulin [135, 142–145], and in centriole elongation, such as POC5 [146]. Exactly how they work and interact is not yet clear.
Fig. 4.
The centriole duplication cycle is coupled with the chromosome replication and segregation cycle. Procentriole formation begins in S phase upon recruitment of SAS-6 and BLD10/CEP135, which are needed to form the cartwheel, a structure that helps in defining the centriole nine-fold symmetry. CDK2 activity may be necessary for speeding up procentriole formation and elongation, hence coordinating this event with DNA replication. In G2, the daughter centriole reaches full elongation and maturation with the recruitment of several molecules to the pericentriolar material (PCM). SAS4/CPAP plays a role in centriole elongation. CDK1 activity increases in G2 regulating a variety of molecules and processes needed for entry into mitosis, such as changes in microtubule dynamics. Through the concerted action of molecules such as the kinase Nek2, the two centrosomes separate. When a cell exits mitosis, the centrioles within the centrosome disengage through the action of separase. That process may allow recruitment or activation of molecules necessary for duplication. SAK/PLK4 is essential and can trigger procentriole formation. Upon proper alignment of the sister chromatids, the spindle assembly checkpoint is satisfied, Cyclin B is degraded and separase is activated. Sister chromatid separation and centriole disengagement are triggered. The mitotic spindle segregates the chromosomes equally to the two daughter cells
Control of centriole number
At least two controls operate to restrict centriole duplication in a normal cycling animal cell. “One and only one” new centriole forms orthogonally to each pre-existing one in a conservative fashion. Moreover, the biogenesis of centrioles is coordinated with DNA replication, so that they are synthesized only once per cell cycle (Fig. 4): when one cycle is delayed, the other stops, avoiding mistakes. It is not known how procentriole number is restricted to one and whether there is a preferred location close to the mature centriole for the new structure to be formed. Two situations are known to override this control. SAK/PLK4 is a protein kinase of the polo-like family necessary for centriole biogenesis. Overexpression of SAK/PLK4 in humans and Drosophila and SAS-6 in humans leads to the formation of “flower-like structures” [127, 133, 147–149], a single mother surrounded by several daughters. There is a very tight control of the protein levels of SAS-6 and SAK/PLK4 in cycling cells, via two major ubiquitin ligase complexes involved in protein degradation in the cell cycle. SAS-6 is degraded via APC/Cdh1 [127], while SAK/PLK4 is degraded by SCF/Slimb complex [147, 149]. In the absence of this regulation, the biogenesis of many centrioles leads to the presence of many centrosomes and problems in cell division.
Little is known about the coordination between the centriole and DNA replication cycles, but it is clear that the cell cycle machinery must play an important role. Wong and Stearns found that centrosome duplication is controlled extrinsically, since S-phase cytoplasm advances the duplication of a G1 centrosome upon cell fusion [150]. This activity might be related to CDK2, a kinase involved in promoting DNA replication, which accelerates the centriole assembly process (Fig. 4). Wong and Stearns also reported a centrosome-intrinsic block to re-duplication, as the centrosome of a G2 cell does not duplicate in an S-phase cytoplasm. This is not due to an inhibitory effect from the cytoplasm because, when G2 cells were fused to G1 cells, the G1 centrosomes duplicated. So, once centrioles have duplicated in S phase, they cannot duplicate again until the next S phase [150]. One possible explanation for the intrinsic block to reduplication is the control of centriole duplication by their disengagement, as this constitutes one prerequisite for the growth of daughter centrioles [76, 151, 152]. Vidwans et al. [153] showed that centriole disengagement is dependent on the APC/C-Cdc20. Tsou and Stearns [154] and Tsou et al. [155] suggested that PLK1 and Separase, a protease that is activated by APC/C-Cdc20 at the metaphase-to-anaphase transition to trigger separation of sister chromatids, also trigger centriole disengagement (Fig. 4). This may license the centrioles for a new round of duplication, perhaps by relieving a pre-existing block to duplication or exposing sites where new centrioles can bud. Mitosis is most likely a non-permissive state for procentriole formation and elongation as when mitosis is prolonged in a variety of cell types, centrosome duplication cannot proceed [153]. However, it is not known how many of those inputs are read by the centriole assembly machinery to confine its activity to a particular window of time.
In some cells in our body the regulation of the arithmetic of centriole biogenesis is different, one of the most emblematic cases occurring during gametogenesis as we will discuss below. Another case is seen in mammalian epithelial multiciliated cells, where 200–300 basal bodies are formed in each cell after differentiation [125, 156–158]. This arises in the developing vertebrate respiratory and reproductive tracts and the ventricular system of the brain. Multiple centrioles form around a mother centriole, breaking the control of “only one” centriole forming per mother. Centrioles can also form around less characterized, non-microtubule-based dense structures of heterogeneous size, called deuterosomes [156].
Segregation of centrioles to daughter cells: is the mitotic spindle a distributor?
Central to the question discussed in this review is the separation of the centrosomes, their association with the spindle poles and their inheritance. In certain species, centrioles do not associate with the spindle poles, they remain at the membrane where they nucleate axonemes. That is the case of ciliates. Centrioles, when present in most species that have a small number of flagella or cilia, associate with the spindle poles upon cell division.
Centrosome separation at the entry of mitosis plays an important role in their normal segregation. Several molecules play a role in centrosome separation. First, the two centrosomes are physically associated by a mesh of fibers composed by rootletin, amongst other components, which are tethered to the proximal end of centrioles, through a molecule called c-NAP1. Upon entry into mitosis, phosphorylation of C-NAP1 by a mitotic kinase called NEK2, releases it from the centrioles, leading to centrosome separation [2]. The activity of NEK2 is counteracted by a phosphatase, PP1. Another substrate of NEK2 is β-catenin, which is also localized at the proximal end of centrioles, and expression of a stabilized version of that protein leads to premature centrosome separation [159]. Microtubule pushing forces are also very important in centrosome separation and are driven by Eg5, a plus end directed kinesin that binds antiparallel microtubules and slides them in opposite directions. Dynein, a minus end directed motor, also plays a role in the separation of centrosomes and in mammals dynein, Lis1 and CLIP-170 counteract Eg5 activity [160]. Cortical and kinetochore forces also play a role in the establishment of a bipolar spindle. For example, cortical microtubules and their contact with the actin/myosin or focal adhesions present in the cortex may play an important role in centrosome separation [161]. Finally, upon nuclear envelope breakdown, kinetochore pushing forces might also be important in the establishment of a bipolar spindle [162]. During centrosome separation, centrosomes remain associated with the nuclear envelope until it breaks down. Association between the nucleus and the centrosome is provided by dynein and proteins on the outer nuclear membrane, such as the SUN and KASH proteins.
Centrosomes normally remain associated with the poles of the spindle. This is a highly dynamic process, dependent on spindle pole focusing by motors, such as dynein, HSET, microtubule associated molecules such as ASP and NUMA, and microtubule nucleation capacity of centrioles, which is dependent on molecules such as CNN/CDK5RAP2 [2]. Depletion of dynein, HSET, ASP or NUMA leads to highly unfocused spindle poles and centrosome detachment [53]. Finally, depletion of CNN/CDK5RAP2 leads to detachment of centrioles from PCM, with subsequent centriole segregation errors [86].
However, upon mitotic exit, the two centrioles disengage and in certain cell types, one of the centrioles migrates to the cleavage site during cytokinesis [70]. It is possible that a physical association still exists between mother and daughter centrioles [24, 70], which may help to avoid centriole loss during cell division.
Germline inheritance of centrosomes
Unlike the symmetric form of centrosome inheritance found in the somatic divisions, centrosome inheritance through the male and female germline is highly asymmetric and goes against the argument that a spindle is necessary for normal centriole distribution to occur. Just as the chromosomes undergo reductional divisions during gametogenesis in order to maintain diploidy, centrosomes must also undergo reductional divisions. In contrast to the chromosomes, in which reduction of chromosome number is equivalent in the two sexes, centrosome reduction occurs differently in males and females.
Centrosome reduction in the germline
During spermiogenesis in many sexually reproducing species, centrosome reduction occurs primarily through the loss of the PCM, while during oogenesis centrosome reduction primarily involves centriolar loss with retention of the PCM albeit dispersed throughout the egg cytoplasm [163]. This complementary loss enables restoration of an intact fully functional centrosome following fertilization with the sperm supplying the centrioles and the PCM supplied by the egg cytoplasm.
This pattern of inheritance in which the fertilizing sperm supplies a centriole into an oocyte lacking a centriole constitutes Boveri’s hypothesis of fertilization (1901). In the vast majority of animal species studied, the oocyte looses centrioles at some point during its maturation. This situation comforted many early cell biologists in the idea that there is a necessity to bring one centriole to generate the following ones. For example for Drosophila Sonnenblick [164], p 71) observed “…in the maturation division figures no centrioles or asters are present…The centrioles seen in all mitoses from the first cleavage on are, in all likelihood, derived from the division center or centers originally associated with the spermatozoon which contributed the fusion nucleus” and King [165] concludes “No centrioles or asters are present in the maturation division figures. Therefore, the centrioles seen in all mitoses (from the first cleavage on) presumably are derived from a centriole contributed by the entering sperm”. Support for this view comes from the finding that amphibian eggs require the presence of an injected centrosome to trigger parthenogenesis [166].
This rule of the sperm providing a centrosome is not universal. There are exceptions such as the case of the mouse embryo where the sperm centriole disintegrates after entering the egg and the first zygotic cleavages occur without centrioles [63]. Such de novo formation of centrioles has been also observed in rabbit oocytes parthenogenetically activated [167]. Similarly, artificially activated sea urchin eggs display cytasters with real centrioles [168–170].
Centrosome reduction during spermiogenesis
The extent and specific events of centrosome reduction during spermiogenesis are highly variable among species. The germline stem cells of the male contain centrosomes and degeneration of the centrosomes and formation of the axoneme generally occurs after meiosis and formation of haploid spermatids. As described by Manandhar, the first stage of centrosome reduction is loss of microtubule nucleation, followed by loss of key PCM proteins and finally partial or complete degeneration of centrioles [163]. Immunofluorescent studies in Drosophila provide a good illustration of this degeneration pattern [171, 172]. During the initial stages of spermatogenesis, centrosomes appear intact containing γ-tubulin [171], CNN [173], centrin [174] and DSPD2 [98]. In addition, they are associated with robust astral microtubule arrays. However, in haploid post-meiotic spermatids there are no robust astral microtubule arrays [173, 175] and γ-tubulin is subsequently lost. Immediately following fertilization, the male derived centrosomes again contain gamma-tubulin, CNN, CP190 [174] associated with the centriole, and their activity depends on PCM recruitment proteins, such as DSPD2.
Studies in mammals demonstrate centriole degeneration in addition to loss of key PCM components. Perhaps the most extreme example of centriole degeneration occurs in mice and other rodents in which both members of the centrosome degenerate [176]. In contrast in humans and other mammals only one member of the centriole pair fully degenerates. The difference between mouse and human with respect to centriole degeneration is illustrated in a recent study of the mammalian centriolar protein TSKS, a substrate of the centriolar associated kinase TSSK1 [177]. In both mice and humans, TSKS is highly concentrated at the centrioles during spermiogenesis and in post-meiotic spermatids. While this localization persists in fully mature human spermatozoa, TSKS is no longer present on mature mouse spermatozoa. These patterns likely reflect complete degeneration of centrioles in mice but not humans.
Thus, during spermatogenesis, centrosomes not only experience loss of the PCM but the centrioles are often degraded as well. Therefore, upon fertilization the egg cytoplasm supplies components for restoring the centrioles as well as the PCM. Because of the complete loss of centrioles during spermatogenesis in mice, centrioles must arise de novo from components of the egg cytoplasm [178]. Because de novo centriole formation from maternally supplied components is a prerequisite for development with no fertilization (parthenogenesis), it is surprising that complete parthenogenesis leading to viable organisms has not been found in mice. This is likely because extensive chromosomal imprinting in mice requires one set of chromosomes to be passed through the male germline for successful zygotic development [179].
Centrosome reduction during oogenesis
Centrosome reduction during oogenesis primarily involves centriole loss. In the mouse, centrioles are lost during meiosis I [63]. Although lacking centrioles, the spindle poles contain key centrosomal proteins including gamma tubulin, pericentrin and NuMA [180–182]. Centriole loss during oogenesis occurs in rabbits, cows, sheep and humans and thus is likely a conserved feature of mammalian oogenesis.
PCM components are not eliminated during mouse oogenesis but they are redistributed throughout the oocyte. Prior to oocyte maturation, structures known as multivesicular aggregates (MVA) form and concentrate at the centrosome [180]. These structures are enriched in gamma tubulin. However, they do not nucleate microtubules. During oocyte maturation, the MVA migrates toward the germinal vesicle and breaks down into smaller aggregates some of which nucleate microtubules forming MTOCs. As the nucleus enters metaphase of meiosis I, the poles of the spindle are organized by these MTOCs [183]. Another core centrosomal component, pericentrin, is assembled into small aggregates in the cytoplasm. Later in meiosis, pericentrin becomes focused at the spindle poles [184].
Centrosome reduction has also been well studied in the Drosophila oogenesis. During the initial stages of Drosophila oogenesis, four rounds of mitotic divisions with incomplete cytokinesis produce a cyst of 16 interconnected cells. One of these will form the oocyte, while the other 15 will form nurse cells supplying nutrients and key developmental factors to the oocyte [185]. A striking initial event in oogenesis is the migration of the centrioles from the 15 nurse cells into the cell destined to become the oocyte [186]. Surprisingly centrosome migration into the oocyte is independent of the microtubule cytoskeleton and depends on a properly polarized fusome, a large organelle distributed throughout the interconnected 16 cyst cells [187]. These migrating centrosomes do not act as an MTOC in the oocyte and they disappear later in oogenesis. Rather, MTOC activity is broadly distributed throughout the oocyte cytoplasm. A recent report has specifically studied the fate of the centrosome associated with the oocyte nucleus [188]. It was found that a centriole containing centrosome remains associated with the oocyte nucleus late into oogenesis in stage 9. When this centriole degenerates is unclear but it is likely no longer present by metaphase of meiosis I [186]. Consequently the poles of the meiotic spindle lack centrioles and centrosome components such as gamma tubulin, CNN, CP60, CP190 [172] and D-PLP [30].
We must underscore the point that definitive proof of the absence of centrioles can only be provided by electron microscopy on serial sections. Such an analysis can constitute a kind of nightmare for the biologist with regard to the size of most animal oocytes, and so in many species the real status of the oocyte centriole remains unclear.
Restoration of centrosome function immediately following fertilization
In most mammalian species, the sperm contains a proximal centriole and a degenerate distal centriole. Upon fertilization the proximal centriole forms a large astral microtubule array which is required for pronuclear migration and recruitment of maternally supplied PCM components [189]. During recruitment of PCM components, transformation of the sperm into the male pronucleus occurs. Duplication of the paternally derived centrioles also occurs during this period. These events ultimately result in the production of two functional centrosomes that migrate to the poles of the juxtaposed male and female pronuclei.
Experimental evidence that the paternally derived centrioles rapidly acquire a PCM through recruitment of maternally supplied components comes from studies in Drosophila. Immunofluorescent analysis demonstrates exclusion of gamma tubulin, CP60, CP190 and likely most PCM components from the mature sperm [96, 173, 174]. Immediately following fertilization these components are again detected associated with the sperm derived centrioles. The paternally derived centrioles likely recruit these and other PCM components from the maternal cytoplasm.
Insights into the mechanisms by which PCM components are recruited to the paternally derived centriole following fertilization, come from studies of SDP2, a member of a core group conversed centriole duplication factors [98, 101]. The Drosophila studies demonstrate that DSPD2 is a component of both the centrioles and PCM and plays a key role in recruiting PCM components specifically to the sperm derived centriole immediately following fertilization. In SPD2 Drosophila mutants, fertilization occurs normally, but the paternally derived centriole fails to recruit CNN and generate an MTOC [98].
This asymmetric biparental origin of the zygotic centrosome has significant implications for the outcome of somatic cell nuclear transfer (SCNT) experiments. SCNT refers to the animal cloning technique in which a somatic nucleus is injected into an enucleated oocyte. Significantly, centrosomes are transferred along with the somatic nucleus into the host oocyte [190]. In principle, incompatibility between the host cytoplasm and donor centrosomes could result in aborted embryonic development. For example the injected oocyte contains donor centrosomal material associated with the somatically derived centrosomes as well as large amounts of centrosomal material remaining in the cytoplasm of the host oocyte. How the remodeling of the donor centrosome occurs in this environment is unclear. In addition, centrosome composition and duplication is cell cycle regulated. If the somatic and oocyte cell cycles are mismatched this may disrupt centrosome remodeling and duplication. In fact, in a recent study examining SCNT in bovine oocytes, found that 50% of the embryos had defects in centrosome number or distribution [191]. Many centrosomes failed to associate with the nucleus and some embryos contained up to six centrosomes at the first mitosis. These centrosome defects readily explain the aneuploidy observed in SCNT embryos.
Centrosome formation and inheritance during parthenogenesis
The complementary forms of centrosome reduction during gametogenesis in the two sexes present one of a number of formidable barriers to parthenogenetic development. Thus, it is surprising that parthenogenesis has evolved numerous times throughout the phyla [192]. Perhaps the most extreme example occurs in Hymenoptera (ants, wasps, and bees), where parthenogenesis is universal and tightly linked to sex determination: fertilized embryos develop into diploid females and unfertilized embryos develop into haploid males [193].
A key issue regarding parthenogenesis is whether centrosomes are formed and if so what is their origin. While not required for the formation of a functional mitotic spindle, centrosomes play a critical role in the initial syncytial embryonic divisions following fertilization in insects. These divisions are rapid, synchronous and syncytial; that is nuclei divide without accompanying cytokinesis. After a series of syncytial divisions in the interior of the embryos, nuclei migrate to the cortex where they undergo four more rounds of division and finally cellularize [194]. Cellularization transforms the syncytial embryo into one organism containing thousands of cells. Studies in which parthenogenesis was artificially induced in the fungal gnat Sciara demonstrate that a functional centrosome is critical for these initial divisions following fertilization [65]. In parthenogenetic Sciara embryos, functional bipolar spindles form and development proceeds normally for up to three or four rounds of division. However, because these nuclei are on a syncytial environment, neighboring spindles eventually collide resulting in aneuploid division products. Careful analysis of the spindles reveals that they lack astral microtubules. It appears that the astral microtubules are essential for maintaining separation between neighboring nuclei once crowding of the dividing nuclei occurs. Following cellularization the cleavage furrow produces a membrane barrier between dividing nuclei and astral microtubules are no longer required for maintaining separation.
Elegant genetic analysis of CNN and DSAS4 mutants that specifically disrupt centrosomes and centrioles confirm and extend this analysis. Embryos derived from females homozygous for either of these mutants produce anastral spindles [85, 89, 91], fused spindles and aneuploid nuclei, similar to that found in Sciara syncytial embryos.
De novo centrosome formation in parthenogenetic hymenoptera
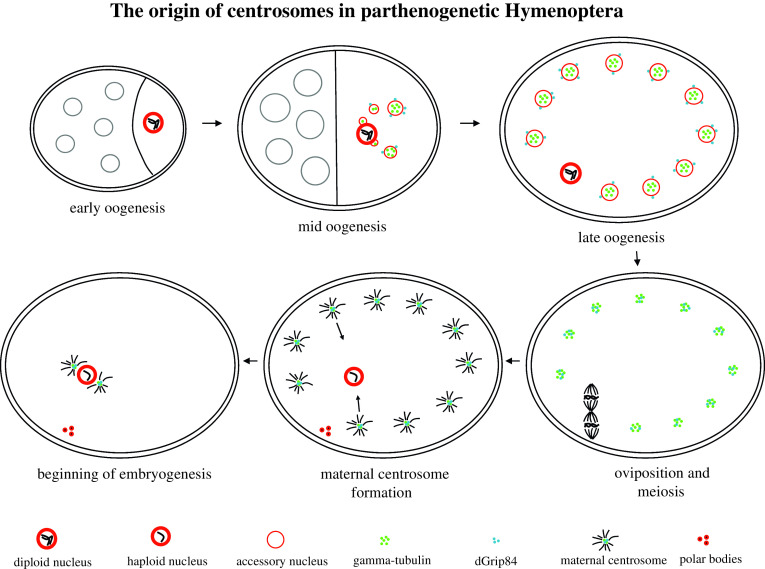
Although many aspects of development occur normally in the absence of a centrosome, adult flies lacking centrosomes do not live long. In addition, centrosomes are necessary for normal syncytial embryonic development and spermiogenesis in insects. Therefore, one would expect that centrosomes are present in parthenogenetically derived insects. The origin of centrosomes during insect parthenogenesis has been explored in a number of species including the jewel wasp, Nasonia vitripennis. In Nasonia, centrosomes originate de novo during the late stages of oogenesis. Centrosome genesis relies on the formation of membrane structures derived from the female oocyte known as accessory nuclei [195]. Through an unknown process, the oocyte nucleus buds off hundreds of nuclear envelope-based vesicles [196]. These vesicles contain nuclear lamin however they do not contain nucleic acids. As oogenesis proceeds, these vesicles form an even distribution along the cortex of the oocyte. γ-Tubulin accumulates to high levels in the accessory nuclei. This accumulation is most striking in the honey bee in which γ-tubulin is localized in discrete internal patches. In addition members of the γ–TuRC complex localize outside of the nuclear envelope. Breakdown of the accessory nuclei envelope occurs in synchrony with activation and breakdown of the oocyte nucleus. Immediately following breakdown of the accessory nuclei, microtubule-based asters form along the cortex. This breakdown is thought to enable the γ–TuRC complex to associate with gamma tubulin initiating aster formation. EM analysis demonstrated that these asters contain canonical centrioles [197]. Thus, it is likely that these accessory nuclei are the site of de novo centriole formation.
In addition to de novo formation of centrioles in parthenogenetic embryos, the de novo centriole formation is observed in other biological contexts, like amebo-flagellates, lower plants or multiciliated cells of vertebrates, as we mentioned before.
Controlling centrosome number in parthenogenetic hymenoptera
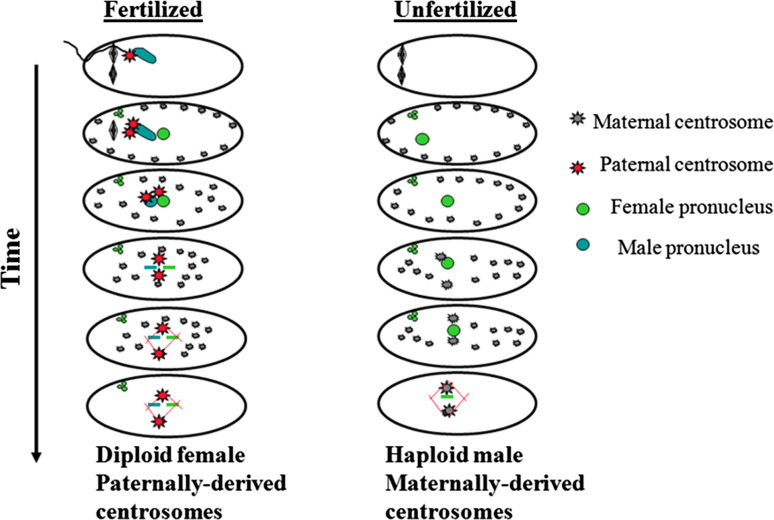
A freshly laid hymenopteran egg with its hundreds of active centrosomes and a single female pronucleus presents a precarious situation for the mitotically active embryo. Additional centrosomes increase the likelihood of forming non functional multipolar spindles. To prevent this, the female pronuclei in Nasonia captures only two centrosomes [198]. Live imaging reveals many cortical asters rapidly migrating toward the female pronucleus. However, once the pronucleus captures two asters, one from each side, additional asters are prevented from associating with it. The mechanism it not known but one possibility is that the asters form a protective array of microtubules encompassing the female pronucleus. Once the pronucleus has two asters, the hundreds of unassociated asters in the cytoplasm degenerate. Thus, an embryo is created containing a haploid nucleus associated with two centrosomes equipped to enter mitosis with a bipolar spindle (Figs. 5, 6).
Fig. 5.
The origin of centrosomes in parthenogenetic Hymenoptera. Centrosome in Hymenoptera, maternal centrosomes originate from accessory nuclei, specialized organelles that bud off from the oocyte nucleus. These organelles contain high concentrations and foci of gamma tubulin with the centrosomal component Dgrip84 localized on the outer surface. During late oogenesis these organelles break down enabling the gamma-tubulin foci to seed formation of hundreds of centrosomes. Modified from Ferree et al. [195]
Fig. 6.
Centrosomes are reciprocally inherited in Hymenoptera. In both fertilized and unfertilized embryos, hundreds of maternally derived centrosomes are evenly distributed along the cortex and upon egg activation, these centrosomes migrate toward the female pronucleus. In unfertilized embryos, once two centrosomes associate with the female pronucleus, additional centrosomes are repelled. In fertilized embryos the male pronucleus is associated with two paternal basal-body derived centrosomes. At pronuclear fusion, these repel all maternally derived centrosomes. Shortly after centrosome association with the nuclear envelope in both fertilized and unfertilized embryos, unassociated centrosomes disappear (modified from Tram and Sullivan [198])
Analysis of the fertilized diploid Nasonia embryos reveals that like the unfertilized embryos, cortical asters still form and migrate toward the female oocyte. However, the male pronucleus, with its associated centrosomes, migrates toward and fuses with the female pronucleus prior to the arrival of the cytoplasmic centrosomes. The centrosomes associated with the male pronucleus prevent maternally derived centrosomes from associating with the fused pronuclei. As in the unfertilized embryos, the unassociated centrosomes rapidly degenerate. Thus, unfertilized male embryos inherit maternally derived centrosomes while fertilized female embryos inherit paternally derived centrosomes. While instances of paternal and maternal centrosome inheritance have been documented, this reciprocal form of centrosome inheritance may be unique to Hymenoptera [198] (Figs. 5, 6).
The importance of eliminating the many ectopic cytoplasmic centrosomes is highlighted in studies of the parthenogenetic Drosophila species, D. mercatorum [199]. In this species parthenogenetic embryos develop although inefficiently and many embryos die. Cellular analysis indicates that like hymenoptera numerous maternally derived cytoplasmic centrosomes form in the unfertilized embryos. Some of these will stably associate with the female pronucleus. However, in contrast to Hymenoptera, degeneration of the unassociated cytoplasm centrosomes does not occur. These ectopic centrosomes often associate with the normally dividing nuclei producing multipolar spindles, aneuploidy and ultimately embryonic lethality. Thus, parthenogenesis not only requires mechanisms of de novo centrosome assembly, but mechanisms to efficiently eliminate unassociated excess centrosomes.
The insect bacterial endosymbiont, Arsenophonus nasoniae, has taken advantage of the reciprocal pattern of inheritance found in Hymenoptera. Because A. nasoniae is maternally inherited, it favors an increased proportion of females in the population. To achieve this, it relies on the fact that hymenoptera males but not females require maternally inherited centrosomes for normal development. Arsenophonus inhibits the formation of maternally derived centrosomes, thus specifically preventing development of unfertilized male embryos [200]. Accessory nuclei form in Arsenophonus infected hymenoptera oocytes. Therefore, the effect of A. nasoniae on centrosomes formation is likely downstream of accessory nuclei formation.
Active players or passengers: can we ever end the debate?
The old question of the role of centrioles in cell division is still outstanding. Are the centrioles dispensable for mitosis? What we have discussed in this review is that the centriole is a polyfunctional organelle and cannot be classified simply as “essential” or “dispensable” for this complex process. When looking at the eukaryotic tree of life, centrioles do not participate in mitotic spindle formation in many species, suggesting that this was not the ancestral function although information is lacking for many organisms (Fig. 2). It is thus possible that in the last common ancestor, which contained centrioles as required in flagella formation, centrioles were passengers in cell division. Perhaps centrioles were more recently co-opted to take part in different mechanisms associated with mitosis such as positioning of the spindle or a differential control of cytokinesis completion in different tissues. The experiments cited in previous chapters are starting to suggest answers to this question: in Metazoans, centrioles are strongly involved in the bipolar nature of the spindle and, at least for some cell types, in cytokinesis. Less obvious roles are also suspected, such as controlling the fidelity of chromosome segregation. More generally, centrioles could be responsible for precise regulation of mitosis, either in space (the plane of division, for example during the asymmetrical divisions in some cell types but not all) or in time (gating the decision to divide, deciding the final number of divisions) and even possibly in regulation of cell fate (Drosophila male stem cells, [107]) and could be therefore strongly implicated in development and morphogenesis. In this light, the recent findings concerning Drosophila acentriolar mutants are surprising and suggest that morphogenesis in Drosophila could rely more on acentrosomal pathways, like for example in wing epidermal cells [61] or tracheal cells (V. Brodu personal communication). In vertebrates, which have a much more developed cytoskeleton, the centrioles are probably essential organelles, required for other cellular functions in addition to cell division.
No case of a permanent acentriolar cell line has been reported in vertebrates, and pathological mitoses of cancerous cells present centrioles. It is symptomatic that cancerous cells retain these organelles, although they are in most cases precisely affected in centrosome number and function, suggesting that the presence of these organelles provides some selective advantage [201–205]. An interesting example of co-option of the centrioles that we discussed in this review is in the case of sexual reproduction. Here, there is a complementarity between the egg and the sperm, the later providing the centriole upon fertilization. Even in Drosophila, where centrioles are dispensable for somatic cell divisions, centrioles are needed for early embryonic development. Perhaps it was not difficult to co-opt centrioles for this role, as the flagellated sperm normally brings a basal body at its base. This role may also have been important in preventing parthenogenetic development in many species.
Is the mitotic spindle a distributor of centrioles?
Clearly that is not always the case. Since centrioles can form de novo, there is no conceptual need for continuity. However, de novo centriole formation in general leads to lack of control in localization, number and timing of newborn structures, so “canonical” duplication may provide more control of the centriole biogenesis in proliferating cells. Moreover, canonical duplication provides an additional property that cells may explore in developmental decisions, which is centriole age [107, 112]. Contrary to abundant organelles like mitochondria which can be easily shared between the two daughter cells, there is a need for a precise partition of the two centrioles duplexes of the mother cell. And so paradoxically even if the centrioles are almost certainly active players of mitosis, their strategic position at the spindle poles also guarantees their correct distribution; this by no means imply they are inert.
What interesting avenues for future?
It will be very important in the future to precise the role of the centriole during mitosis, and to unravel how centriole biogenesis is regulated in development and human disease, such as cancer, ciliopathies and microcephalies. It will be important to better understand other modes of assembling spindles and contextualize that knowledge in the light of evolution, answering how cell division and centriole function co-evolved. Finally, we need to explore the functional diversity of centrioles in cell division and in fertilization. One century after Boveri and pioneering cell biologists, we are still puzzled by the same questions but there has been a clear acceleration of knowledge on centrioles during the last 10 years. So we can reasonably hope that next years will bring exciting findings.
Acknowledgments
The authors thank Ines Bento, Michel Bornens, Janet Chenevert and Marie Delattre for critical reading of the manuscript in draft form. We are also grateful to Patrick Ferree’s for making one of the figures.
Open Access
This article is distributed under the terms of the Creative Commons Attribution Noncommercial License which permits any noncommercial use, distribution, and reproduction in any medium, provided the original author(s) and source are credited.
References
- 1.Paoletti A, Bornens M. Organisation and functional regulation of the centrosome in animal cells. Prog Cell Cycle Res. 1997;3:285–299. doi: 10.1007/978-1-4615-5371-7_23. [DOI] [PubMed] [Google Scholar]
- 2.Bettencourt-Dias M, Glover DM. Centrosome biogenesis and function: centrosomics brings new understanding. Nat Rev Mol Cell Biol. 2007;8:451–463. doi: 10.1038/nrm2180. [DOI] [PubMed] [Google Scholar]
- 3.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr Opin Cell Biol. 2002;14:25–34. doi: 10.1016/S0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 4.Boveri T. Concerning the origin of malignant tumors. Translated and annotated by H. Harris. J Cell Sci. 2008;121:1–84. doi: 10.1242/jcs.025742. [DOI] [PubMed] [Google Scholar]
- 5.Boveri T. The origin of malignant tumors. Baltimore, Maryland: Williams and Wilkins; 1914. [Google Scholar]
- 6.Friedlander M, Wahrman J. The spindle as a basal body distributor. A study in the meiosis of the male silkworm moth, Bombyx mori . J Cell Sci. 1970;7:65–89. doi: 10.1242/jcs.7.1.65. [DOI] [PubMed] [Google Scholar]
- 7.Pickett-Heaps JD. The autonomy of the centriole: fact or fallacy? Cytobios. 1971;3:205–214. [Google Scholar]
- 8.Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-X. [DOI] [PubMed] [Google Scholar]
- 9.Bornens M. Organelle positioning and cell polarity. Nat Rev Mol Cell Biol. 2008;9:874–886. doi: 10.1038/nrm2524. [DOI] [PubMed] [Google Scholar]
- 10.Dippell RV. The development of basal bodies in paramecium. Proc Natl Acad Sci USA. 1968;61:461–468. doi: 10.1073/pnas.61.2.461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vorobjev IA, Nadezhdina ES. The centrosome and its role in the organization of microtubules. Int Rev Cytol. 1987;106:227–293. doi: 10.1016/S0074-7696(08)61714-3. [DOI] [PubMed] [Google Scholar]
- 12.Beisson J, Jerka-Dziadosz M. Polarities of the centriolar structure: morphogenetic consequences. Biol Cell. 1999;91:367–378. doi: 10.1016/S0248-4900(99)80097-5. [DOI] [PubMed] [Google Scholar]
- 13.Bobinnec Y, Khodjakov A, Mir LM, Rieder CL, Edde B, Bornens M. Centriole disassembly in vivo and its effect on centrosome structure and function in vertebrate cells. J Cell Biol. 1998;143:1575–1589. doi: 10.1083/jcb.143.6.1575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Janke C, Rogowski K, van Dijk J. Polyglutamylation: a fine-regulator of protein function? ‘Protein modifications: beyond the usual suspects’ review series. EMBO Rep. 2008;9:636–641. doi: 10.1038/embor.2008.114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Steffen W, Linck RW. Evidence for tektins in centrioles and axonemal microtubules. Proc Natl Acad Sci USA. 1988;85:2643–2647. doi: 10.1073/pnas.85.8.2643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hinchcliffe EH, Linck RW. Two proteins isolated from sea urchin sperm flagella: structural components common to the stable microtubules of axonemes and centrioles. J Cell Sci. 1998;111(Pt 5):585–595. doi: 10.1242/jcs.111.5.585. [DOI] [PubMed] [Google Scholar]
- 17.Cavalier-Smith T. The phagotrophic origin of eukaryotes and phylogenetic classification of Protozoa. Int J Syst Evol Microbiol. 2002;52:297–354. doi: 10.1099/00207713-52-2-297. [DOI] [PubMed] [Google Scholar]
- 18.Azimzadeh J, Bornens M. The centrosome in evolution. In: Nigg EA, editor. Centrosomes in development and disease. Weinheim: Wiley; 2004. pp. 93–122. [Google Scholar]
- 19.Mencarelli C, Lupetti P, Dallai R. New insights into the cell biology of insect axonemes. Int Rev Cell Mol Biol. 2008;268:95–145. doi: 10.1016/S1937-6448(08)00804-6. [DOI] [PubMed] [Google Scholar]
- 20.Adams IR, Kilmartin JV. Spindle pole body duplication: a model for centrosome duplication? Trends Cell Biol. 2000;10:329–335. doi: 10.1016/S0962-8924(00)01798-0. [DOI] [PubMed] [Google Scholar]
- 21.Zheng Y, Wong ML, Alberts B, Mitchison T. Nucleation of microtubule assembly by a gamma-tubulin-containing ring complex. Nature. 1995;378:578–583. doi: 10.1038/378578a0. [DOI] [PubMed] [Google Scholar]
- 22.Khodjakov A, Rieder CL. The sudden recruitment of gamma-tubulin to the centrosome at the onset of mitosis and its dynamic exchange throughout the cell cycle, do not require microtubules. J Cell Biol. 1999;146:585–596. doi: 10.1083/jcb.146.3.585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Bartolini F, Gundersen GG. Generation of noncentrosomal microtubule arrays. J Cell Sci. 2006;119:4155–4163. doi: 10.1242/jcs.03227. [DOI] [PubMed] [Google Scholar]
- 24.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Azimzadeh J, Bornens M. Structure and duplication of the centrosome. J Cell Sci. 2007;120:2139–2142. doi: 10.1242/jcs.005231. [DOI] [PubMed] [Google Scholar]
- 26.Badano JL, Teslovich TM, Katsanis N. The centrosome in human genetic disease. Nat Rev Genet. 2005;6:194–205. doi: 10.1038/nrg1557. [DOI] [PubMed] [Google Scholar]
- 27.Sluder G, Rieder CL. Centriole number and the reproductive capacity of spindle poles. J Cell Biol. 1985;100:887–896. doi: 10.1083/jcb.100.3.887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Leidel S, Gonczy P. SAS-4 is essential for centrosome duplication in C. elegans and is recruited to daughter centrioles once per cell cycle. Dev Cell. 2003;4:431–439. doi: 10.1016/S1534-5807(03)00062-5. [DOI] [PubMed] [Google Scholar]
- 29.Kirkham M, Muller-Reichert T, Oegema K, Grill S, Hyman AA. SAS-4 is a C. elegans centriolar protein that controls centrosome size. Cell. 2003;112:575–587. doi: 10.1016/S0092-8674(03)00117-X. [DOI] [PubMed] [Google Scholar]
- 30.Rodrigues-Martins A, Bettencourt-Dias M, Riparbelli M, Ferreira C, Ferreira I, Callaini G, Glover DM. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr Biol. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 31.Bettencourt-Dias M, Rodrigues-Martins A, Carpenter L, Riparbelli M, Lehmann L, Gatt MK, Carmo N, Balloux F, Callaini G, Glover DM. SAK/PLK4 is required for centriole duplication and flagella development. Curr Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 32.Basto R, Lau J, Vinogradova T, Gardiol A, Woods CG, Khodjakov A, Raff JW. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 33.Sluder G, Miller FJ, Rieder CL. Reproductive capacity of sea urchin centrosomes without centrioles. Cell Motil Cytoskeleton. 1989;13:264–273. doi: 10.1002/cm.970130405. [DOI] [PubMed] [Google Scholar]
- 34.Satir P, Christensen ST. Overview of structure and function of mammalian cilia. Annu Rev Physiol. 2006;69:14.1–14.24. doi: 10.1146/annurev.physiol.69.040705.141236. [DOI] [PubMed] [Google Scholar]
- 35.Hoyer-Fender S (2009) Centriole maturation and transformation to basal body. Semin Cell Dev Biol [DOI] [PubMed]
- 36.Fliegauf M, Benzing T, Omran H. When cilia go bad: cilia defects and ciliopathies. Nat Rev Mol Cell Biol. 2007;8:880–893. doi: 10.1038/nrm2278. [DOI] [PubMed] [Google Scholar]
- 37.Rosenbaum JL, Witman GB. Intraflagellar transport. Nat Rev Mol Cell Biol. 2002;3:813–825. doi: 10.1038/nrm952. [DOI] [PubMed] [Google Scholar]
- 38.Park TJ, Mitchell BJ, Abitua PB, Kintner C, Wallingford JB. Dishevelled controls apical docking and planar polarization of basal bodies in ciliated epithelial cells. Nat Genet. 2008;40:871–879. doi: 10.1038/ng.104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Mitchell B, Jacobs R, Li J, Chien S, Kintner C. A positive feedback mechanism governs the polarity and motion of motile cilia. Nature. 2007;447:97–101. doi: 10.1038/nature05771. [DOI] [PubMed] [Google Scholar]
- 40.Rieder CL, Jensen CG, Jensen LC. The resorption of primary cilia during mitosis in a vertebrate (PtK1) cell line. J Ultrastruct Res. 1979;68:173–185. doi: 10.1016/S0022-5320(79)90152-7. [DOI] [PubMed] [Google Scholar]
- 41.Dawe HR, Farr H, Gull K. Centriole/basal body morphogenesis and migration during ciliogenesis in animal cells. J Cell Sci. 2007;120:7–15. doi: 10.1242/jcs.03305. [DOI] [PubMed] [Google Scholar]
- 42.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 43.Ishikawa H, Kubo A, Tsukita S, Tsukita S. Odf2-deficient mother centrioles lack distal/subdistal appendages and the ability to generate primary cilia. Nat Cell Biol. 2005;7:517–524. doi: 10.1038/ncb1251. [DOI] [PubMed] [Google Scholar]
- 44.Jonassen JA, San Agustin J, Follit JA, Pazour GJ. Deletion of IFT20 in the mouse kidney causes misorientation of the mitotic spindle and cystic kidney disease. J Cell Biol. 2008;183:377–384. doi: 10.1083/jcb.200808137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson EB (1896) The cell in development and inheritance. In: Osborn HF (ed) Macmillan, New York
- 46.Wheatley DN. The centriole: a central enigma of cell biology. Amsterdam: Elsevier Biomedical Press; 1982. [Google Scholar]
- 47.Fulton C. Centrioles. In: Reinert J, Ursprung H, editors. Origin and continuity of cell organelles. Berlin Heidelberg New York: Springer; 1971. [Google Scholar]
- 48.Gall JG. Early studies on centrioles and centrosomes. In: Nigg EA, editor. Centrosome in development and disease. Weinheim: Wiley-VCH Verlag GmbH & Co. KGaA; 2004. [Google Scholar]
- 49.Henneguy LF. Sur les rapports des cils vibratiles avec les centrosomes. Arch Anat Microsc. 1897;1:481–496. [Google Scholar]
- 50.Lenhossék MV. Üeber Flimmerzellen. Verh Anat Ges. 1897;12:106. [Google Scholar]
- 51.Brinkley BR, Goepfert TM. Supernumerary centrosomes and cancer: Boveri’s hypothesis resurrected. Cell Motil Cytoskeleton. 1998;41:281–288. doi: 10.1002/(SICI)1097-0169(1998)41:4<281::AID-CM1>3.0.CO;2-C. [DOI] [PubMed] [Google Scholar]
- 52.Fukasawa K. Centrosome amplification, chromosome instability and cancer development. Cancer Lett. 2005;230:6–19. doi: 10.1016/j.canlet.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 53.Godinho SA, Kwon M, Pellman D. Centrosomes and cancer: how cancer cells divide with too many centrosomes. Cancer Metastasis Rev. 2009;28:85–98. doi: 10.1007/s10555-008-9163-6. [DOI] [PubMed] [Google Scholar]
- 54.Nigg EA. Origins and consequences of centrosome aberrations in human cancers. Int J Cancer. 2006;119:2717–2723. doi: 10.1002/ijc.22245. [DOI] [PubMed] [Google Scholar]
- 55.Fulton C. Pinwheels as cell’s doodlebugs. Cell. 1982;30:341–343. doi: 10.1016/0092-8674(82)90229-X. [DOI] [Google Scholar]
- 56.Dietz R. The dispensability of the centrioles in the spermatocyte division of Pales ferruginea (Nematocera) In: Darlington CD, Lewis KR, editors. Chromosomes today. London: Oliver and Boyd; 1966. pp. 161–166. [Google Scholar]
- 57.Gould RR, Borisy GG. The pericentriolar material in Chinese hamster ovary cells nucleates microtubule formation. J Cell Biol. 1977;73:601–615. doi: 10.1083/jcb.73.3.601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tassin AM, Maro B, Bornens M. Fate of microtubule-organizing centers during myogenesis in vitro. J Cell Biol. 1985;100:35–46. doi: 10.1083/jcb.100.1.35. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Connolly JA, Kiosses BW, Kalnins VI. Centrioles are lost as embryonic myoblasts fuse into myotubes in vitro. Eur J Cell Biol. 1986;39:341–345. [PubMed] [Google Scholar]
- 60.Srsen V, Fant X, Heald R, Rabouille C, Merdes A. Centrosome proteins form an insoluble perinuclear matrix during muscle cell differentiation. BMC Cell Biol. 2009;10:28. doi: 10.1186/1471-2121-10-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Mogensen MM, Tucker JB. Evidence for microtubule nucleation at plasma membrane-associated sites in Drosophila . J Cell Sci. 1987;88(Pt 1):95–107. doi: 10.1242/jcs.88.1.95. [DOI] [PubMed] [Google Scholar]
- 62.Bellett G, Carter JM, Keynton J, Goldspink D, James C, Moss DK, Mogensen MM. Microtubule plus-end and minus-end capture at adherens junctions is involved in the assembly of apico-basal arrays in polarised epithelial cells. Cell Motil Cytoskeleton. 2009;66:893–908. doi: 10.1002/cm.20393. [DOI] [PubMed] [Google Scholar]
- 63.Szöllösi D, Calarco P, Donahue RP. Absence of centrioles in the first and second meiotic spindles of mouse oocytes. J Cell Sci. 1972;11:521–541. doi: 10.1242/jcs.11.2.521. [DOI] [PubMed] [Google Scholar]
- 64.Gueth-Hallonet C, Antony C, Aghion J, Santa-Maria A, Lajoie-Mazenc I, Wright M, Maro B. γ-Tubulin is present in acentriolar MTOCs during early mouse development. J Cell Sci. 1993;105(Pt 1):157–166. doi: 10.1242/jcs.105.1.157. [DOI] [PubMed] [Google Scholar]
- 65.de Saint Phalle B, Sullivan W. Spindle assembly and mitosis without centrosomes in parthenogenetic Sciara embryos. J Cell Biol. 1998;141:1383–1391. doi: 10.1083/jcb.141.6.1383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Bobinnec Y, Moudjou M, Fouquet JP, Desbruyeres E, Edde B, Bornens M. Glutamylation of centriole and cytoplasmic tubulin in proliferating non-neuronal cells. Cell Motil Cytoskeleton. 1998;39:223–232. doi: 10.1002/(SICI)1097-0169(1998)39:3<223::AID-CM5>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 67.Keryer G, Ris H, Borisy GG. Centriole distribution during tripolar mitosis in Chinese hamster ovary cells. J Cell Biol. 1984;98:2222–2229. doi: 10.1083/jcb.98.6.2222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Berns MW, Richardson SM. Continuation of mitosis after selective laser microbeam destruction of the centriolar region. J Cell Biol. 1977;75:977–982. doi: 10.1083/jcb.75.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Peterson SP, Berns MW. Evidence for centriolar region RNA functioning in spindle formation in dividing PTK2 cells. J Cell Sci. 1978;34:289–301. doi: 10.1242/jcs.34.1.289. [DOI] [PubMed] [Google Scholar]
- 70.Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- 71.Khodjakov A, Cole RW, Oakley BR, Rieder CL. Centrosome-independent mitotic spindle formation in vertebrates. Curr Biol. 2000;10:59–67. doi: 10.1016/S0960-9822(99)00276-6. [DOI] [PubMed] [Google Scholar]
- 72.Khodjakov A, Cole RW, Rieder CL. A synergy of technologies: combining laser microsurgery with green fluorescent protein tagging. Cell Motil Cytoskeleton. 1997;38:311–317. doi: 10.1002/(SICI)1097-0169(1997)38:4<311::AID-CM1>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 73.Khodjakov A, Rieder CL, Sluder G, Cassels G, Sibon O, Wang CL. De novo formation of centrosomes in vertebrate cells arrested during S phase. J Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Uetake Y, Loncarek J, Nordberg JJ, English CN, La Terra S, Khodjakov A, Sluder G. Cell cycle progression and de novo centriole assembly after centrosomal removal in untransformed human cells. J Cell Biol. 2007;176:173–182. doi: 10.1083/jcb.200607073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Magidson V, Loncarek J, Hergert P, Rieder CL, Khodjakov A. Laser microsurgery in the GFP era: a cell biologist’s perspective. Methods Cell Biol. 2007;82:239–266. doi: 10.1016/S0091-679X(06)82007-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Maniotis A, Schliwa M. Microsurgical removal of centrosomes blocks cell reproduction and centriole generation in BSC-1 cells. Cell. 1991;67:495–504. doi: 10.1016/0092-8674(91)90524-3. [DOI] [PubMed] [Google Scholar]
- 78.Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G. Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science. 2001;291:1547–1550. doi: 10.1126/science.1056866. [DOI] [PubMed] [Google Scholar]
- 79.Debec A, Szöllösi A, Szöllösi D. A Drosophila melanogaster cell line lacking centriole. Biol Cell. 1982;44:133–138. [Google Scholar]
- 80.Gans M, Audit C, Masson M. Isolation and characterization of sex-linked female-sterile mutants in Drosophila melanogaster . Genetics. 1975;81:683–704. doi: 10.1093/genetics/81.4.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Debec A. Haploid cell cultures of Drosophila melanogaster . Nature. 1978;274:255–256. doi: 10.1038/274255a0. [DOI] [PubMed] [Google Scholar]
- 82.Szöllösi A, Ris H, Szollosi D, Debec A. A centriole-free Drosophila cell line. A high voltage EM study. Eur J Cell Biol. 1986;40:100–104. [PubMed] [Google Scholar]
- 83.Debec A, Detraves C, Montmory C, Geraud G, Wright M. Polar organization of gamma-tubulin in acentriolar mitotic spindles of Drosophila melanogaster cells. J Cell Sci. 1995;108(Pt 7):2645–2653. doi: 10.1242/jcs.108.7.2645. [DOI] [PubMed] [Google Scholar]
- 84.Moutinho-Pereira S, Debec A, Maiato H. Microtubule cytoskeleton remodeling by acentriolar microtubule-organizing centers at the entry and exit from mitosis in Drosophila somatic cells. Mol Biol Cell. 2009;20:2796–2808. doi: 10.1091/mbc.E09-01-0011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Megraw TL, Li K, Kao LR, Kaufman TC. The centrosomin protein is required for centrosome assembly and function during cleavage in Drosophila . Development. 1999;126:2829–2839. doi: 10.1242/dev.126.13.2829. [DOI] [PubMed] [Google Scholar]
- 86.Lucas EP, Raff JW. Maintaining the proper connection between the centrioles and the pericentriolar matrix requires Drosophila centrosomin. J Cell Biol. 2007;178:725–732. doi: 10.1083/jcb.200704081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Samejima I, Lourenco PC, Snaith HA, Sawin KE. Fission yeast mto2p regulates microtubule nucleation by the centrosomin-related protein mto1p. Mol Biol Cell. 2005;16:3040–3051. doi: 10.1091/mbc.E04-11-1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Fong KW, Choi YK, Rattner JB, Qi RZ. CDK5RAP2 is a pericentriolar protein that functions in centrosomal attachment of the gamma-tubulin ring complex. Mol Biol Cell. 2008;19:115–125. doi: 10.1091/mbc.E07-04-0371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Megraw TL, Kao LR, Kaufman TC. Zygotic development without functional mitotic centrosomes. Curr Biol. 2001;11:116–120. doi: 10.1016/S0960-9822(01)00017-3. [DOI] [PubMed] [Google Scholar]
- 90.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. From centriole biogenesis to cellular function: centrioles are essential for cell division at critical developmental stages. Cell Cycle. 2008;7:11–16. doi: 10.4161/cc.7.1.5226. [DOI] [PubMed] [Google Scholar]
- 91.Stevens NR, Raposo AA, Basto R, St Johnston D, Raff JW. From stem cell to embryo without centrioles. Curr Biol. 2007;17:1498–1503. doi: 10.1016/j.cub.2007.07.060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rebollo E, Llamazares S, Reina J, Gonzalez C. Contribution of noncentrosomal microtubules to spindle assembly in Drosophila spermatocytes. PLoS Biol. 2004;2:E8. doi: 10.1371/journal.pbio.0020008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Khodjakov A, Rieder CL. Centrosomes enhance the fidelity of cytokinesis in vertebrates and are required for cell cycle progression. J Cell Biol. 2001;153:237–242. doi: 10.1083/jcb.153.1.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Srsen V, Merdes A. The centrosome and cell proliferation. Cell Div. 2006;1:26. doi: 10.1186/1747-1028-1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Wilson PG. Centriole inheritance. Prion. 2008;2:9–16. doi: 10.4161/pri.2.1.5064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Li K, Kaufman TC. The homeotic target gene centrosomin encodes an essential centrosomal component. Cell. 1996;85:585–596. doi: 10.1016/S0092-8674(00)81258-1. [DOI] [PubMed] [Google Scholar]
- 97.Vaizel-Ohayon D, Schejter ED. Mutations in centrosomin reveal requirements for centrosomal function during early Drosophila embryogenesis. Curr Biol. 1999;9:889–898. doi: 10.1016/S0960-9822(99)80393-5. [DOI] [PubMed] [Google Scholar]
- 98.Dix CI, Raff JW. Drosophila Spd-2 recruits PCM to the sperm centriole, but is dispensable for centriole duplication. Curr Biol. 2007;17:1759–1764. doi: 10.1016/j.cub.2007.08.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Rebollo E, Gonzalez C. Visualizing the spindle checkpoint in Drosophila spermatocytes. EMBO Rep. 2000;1:65–70. doi: 10.1093/embo-reports/kvd011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Perez-Mongiovi D, Malmanche N, Bousbaa H, Sunkel C. Maternal expression of the checkpoint protein BubR1 is required for synchrony of syncytial nuclear divisions and polar body arrest in Drosophila melanogaster . Development. 2005;132:4509–4520. doi: 10.1242/dev.02028. [DOI] [PubMed] [Google Scholar]
- 101.Pelletier L, Ozlu N, Hannak E, Cowan C, Habermann B, Ruer M, Muller-Reichert T, Hyman AA. The Caenorhabditis elegans centrosomal protein SPD-2 is required for both pericentriolar material recruitment and centriole duplication. Curr Biol. 2004;14:863–873. doi: 10.1016/j.cub.2004.04.012. [DOI] [PubMed] [Google Scholar]
- 102.Magidson V, Chang F, Khodjakov A. Regulation of cytokinesis by spindle-pole bodies. Nat Cell Biol. 2006;8:891–893. doi: 10.1038/ncb1449. [DOI] [PubMed] [Google Scholar]
- 103.Rappaport R. Establishment of the mechanism of cytokinesis in animal cells. Int Rev Cytol. 1986;105:245–281. doi: 10.1016/S0074-7696(08)61065-7. [DOI] [PubMed] [Google Scholar]
- 104.Rappaport R. Cytokinesis in animal cells. Int Rev Cytol. 1971;31:169–213. doi: 10.1016/S0074-7696(08)60059-5. [DOI] [PubMed] [Google Scholar]
- 105.Riggs B, Rothwell W, Mische S, Hickson GR, Matheson J, Hays TS, Gould GW, Sullivan W. Actin cytoskeleton remodeling during early Drosophila furrow formation requires recycling endosomal components nuclear-fallout and Rab11. J Cell Biol. 2003;163:143–154. doi: 10.1083/jcb.200305115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Kao LR, Megraw TL. Centrocortin cooperates with centrosomin to organize Drosophila embryonic cleavage furrows. Curr Biol. 2009;19:937–942. doi: 10.1016/j.cub.2009.04.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Yamashita YM, Mahowald AP, Perlin JR, Fuller MT. Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science. 2007;315:518–521. doi: 10.1126/science.1134910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Wang X, Tsai JW, Imai JH, Lian WN, Vallee RB, Shi SH. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Januschke J, Gonzalez C. Drosophila asymmetric division, polarity and cancer. Oncogene. 2008;27:6994–7002. doi: 10.1038/onc.2008.349. [DOI] [PubMed] [Google Scholar]
- 110.Siller KH, Doe CQ. Spindle orientation during asymmetric cell division. Nat Cell Biol. 2009;11:365–374. doi: 10.1038/ncb0409-365. [DOI] [PubMed] [Google Scholar]
- 111.Gonczy P. Mechanisms of asymmetric cell division: flies and worms pave the way. Nat Rev Mol Cell Biol. 2008;9:355–366. doi: 10.1038/nrm2388. [DOI] [PubMed] [Google Scholar]
- 112.Anderson CT, Stearns T. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr Biol. 2009;19:1498–1502. doi: 10.1016/j.cub.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Dingle AD, Fulton C. Development of the flagellar apparatus of Naegleria. J Cell Biol. 1966;31:43–54. doi: 10.1083/jcb.31.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Mizukami I, Gall J. Centriole replication II. Sperm formation in the fern, Marsilea, and the cycad, Zamia. J Cell Biol. 1966;29:97–111. doi: 10.1083/jcb.29.1.97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Huettner AF. Continuity of the centrioles in Drosophila melanogaster . Z Zellforsch.u mikroskop Anat. 1933;19:119–134. doi: 10.1007/BF00587553. [DOI] [Google Scholar]
- 116.Wilson EB, Huettner AF. The central bodies again. Science. 1931;73:447–448. doi: 10.1126/science.73.1895.447. [DOI] [PubMed] [Google Scholar]
- 117.Kuriyama R, Borisy GG. Centriole cycle in Chinese hamster ovary cells as determined by whole-mount electron microscopy. J Cell Biol. 1981;91:814–821. doi: 10.1083/jcb.91.3.814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Robbins E, Jentzsch G, Micali A. The centriole cycle in synchronized HeLa cells. J Cell Biol. 1968;36:329–339. doi: 10.1083/jcb.36.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Alvey PL. An investigation of the centriole cycle using 3T3 and CHO cells. J Cell Sci. 1985;78:147–162. doi: 10.1242/jcs.78.1.147. [DOI] [PubMed] [Google Scholar]
- 120.Rattner JB, Phillips SG. Independence of centriole formation and DNA synthesis. J Cell Biol. 1973;57:359–372. doi: 10.1083/jcb.57.2.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Rieder CL, Borisy GG. The centrosome cycle in PtK2 cells: asymmetric distribution and structural changes in the pericentriolar material. Biol Cell. 1982;44:117–132. [Google Scholar]
- 122.Stubblefield E (1968) Centriole replication in mammalian cells. In: The proliferation and spread of neoplastic cells. Williamson and Wilkins Co., Baltimore, pp 175–193
- 123.Dutcher SK. Elucidation of basal body and centriole functions in Chlamydomonas reinhardtii . Traffic. 2003;4:443–451. doi: 10.1034/j.1600-0854.2003.00104.x. [DOI] [PubMed] [Google Scholar]
- 124.Cavalier-Smith T. Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardtii . J Cell Sci. 1974;16:529–556. doi: 10.1242/jcs.16.3.529. [DOI] [PubMed] [Google Scholar]
- 125.Vladar EK, Stearns T. Molecular characterization of centriole assembly in ciliated epithelial cells. J Cell Biol. 2007;178:31–42. doi: 10.1083/jcb.200703064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Yabe T, Ge X, Pelegri F. The zebrafish maternal-effect gene cellular atoll encodes the centriolar component sas-6 and defects in its paternal function promote whole genome duplication. Dev Biol. 2007;312:44–60. doi: 10.1016/j.ydbio.2007.08.054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Strnad P, Leidel S, Vinogradova T, Euteneuer U, Khodjakov A, Gonczy P. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans . Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 130.Mottier-Pavie V, Megraw TL. Drosophila Bld10 is a centriolar protein that regulates centriole, basal body, and motile cilium assembly. Mol Biol Cell. 2009;20:2605–2614. doi: 10.1091/mbc.E08-11-1115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Matsuura K, Lefebvre PA, Kamiya R, Hirono M. Bld10p, a novel protein essential for basal body assembly in Chlamydomonas: localization to the cartwheel, the first ninefold symmetrical structure appearing during assembly. J Cell Biol. 2004;165:663–671. doi: 10.1083/jcb.200402022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Leidel S, Delattre M, Cerutti L, Baumer K, Gonczy P. SAS-6 defines a protein family required for centrosome duplication in C. elegans and in human cells. Nat Cell Biol. 2005;7:115–125. doi: 10.1038/ncb1220. [DOI] [PubMed] [Google Scholar]
- 133.Kleylein-Sohn J, Westendorf J, Le Clech M, Habedanck R, Stierhof YD, Nigg EA. Plk4-induced centriole biogenesis in human cells. Dev Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 134.Hiraki M, Nakazawa Y, Kamiya R, Hirono M. Bld10p constitutes the cartwheel-spoke tip and stabilizes the 9-fold symmetry of the centriole. Curr Biol. 2007;17:1778–1783. doi: 10.1016/j.cub.2007.09.021. [DOI] [PubMed] [Google Scholar]
- 135.Dammermann A, Muller-Reichert T, Pelletier L, Habermann B, Desai A, Oegema K. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 136.Delattre M, Canard C, Gonczy P. Sequential protein recruitment in C. elegans centriole formation. Curr Biol. 2006;16:1844–1849. doi: 10.1016/j.cub.2006.07.059. [DOI] [PubMed] [Google Scholar]
- 137.Kohlmaier G, Loncarek J, Meng X, McEwen BF, Mogensen MM, Spektor A, Dynlacht BD, Khodjakov A, Gönczy P. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Schmidt TI, Kleylein-Sohn J, Westendorf J, Le Clech M, Lavoie SB, Stierhof YD, Nigg EA. Control of centriole length by CPAP and CP110. Curr Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 139.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated protein that controls centriole length. Nat Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 140.Blachon S, Gopalakrishnan J, Omori Y, Polyanovsky A, Church A, Nicastro D, Malicki J, Avidor-Reiss T. Drosophila asterless and vertebrate Cep152 are orthologs essential for centriole duplication. Genetics. 2008;180:2081–2094. doi: 10.1534/genetics.108.095141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Varmark H, Llamazares S, Rebollo E, Lange B, Reina J, Schwarz H, Gonzalez C. Asterless is a centriolar protein required for centrosome function and embryo development in Drosophila . Curr Biol. 2007;17:1735–1745. doi: 10.1016/j.cub.2007.09.031. [DOI] [PubMed] [Google Scholar]
- 142.Fuller SD, Gowen BE, Reinsch S, Sawyer A, Buendia B, Wepf R, Karsenti E. The core of the mammalian centriole contains gamma-tubulin. Curr Biol. 1995;5:1384–1393. doi: 10.1016/S0960-9822(95)00276-4. [DOI] [PubMed] [Google Scholar]
- 143.Ruiz F, Beisson J, Rossier J, Dupuis-Williams P. Basal body duplication in Paramecium requires gamma-tubulin. Curr Biol. 1999;9:43–46. doi: 10.1016/S0960-9822(99)80045-1. [DOI] [PubMed] [Google Scholar]
- 144.Shang Y, Tsao CC, Gorovsky MA. Mutational analyses reveal a novel function of the nucleotide-binding domain of gamma-tubulin in the regulation of basal body biogenesis. J Cell Biol. 2005;171:1035–1044. doi: 10.1083/jcb.200508184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the gamma-tubulin-mediated addition of centriolar microtubules. J Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Azimzadeh J, Hergert P, Delouvee A, Euteneuer U, Formstecher E, Khodjakov A, Bornens M. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol. 2009;185:101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Cunha-Ferreira I, Rodrigues-Martins A, Bento I, Riparbelli M, Zhang W, Laue E, Callaini G, Glover DM, Bettencourt-Dias M. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 148.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 149.Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Wong C, Stearns T. Centrosome number is controlled by a centrosome-intrinsic block to reduplication. Nat Cell Biol. 2003;5:539–544. doi: 10.1038/ncb993. [DOI] [PubMed] [Google Scholar]
- 151.Kuriyama R, Terada Y, Lee KS, Wang CL. Centrosome replication in hydroxyurea-arrested CHO cells expressing GFP-tagged centrin2. J Cell Sci. 2007;120:2444–2453. doi: 10.1242/jcs.008938. [DOI] [PubMed] [Google Scholar]
- 152.Sluder G. Centrosome duplication and its regulation in the higher animal cell. In: Nigg EA, editor. Centrosomes in development and disease. Weinheim: Wiley; 2004. pp. 167–189. [Google Scholar]
- 153.Vidwans SJ, Wong ML, O’Farrell PH. Mitotic regulators govern progress through steps in the centrosome duplication cycle. J Cell Biol. 1999;147:1371–1378. doi: 10.1083/jcb.147.7.1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 155.Tsou MF, Wang WJ, George KA, Uryu K, Stearns T, Jallepalli PV. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156.Dirksen ER. Centriole and basal body formation during ciliogenesis revisited. Biol Cell. 1991;72:31–38. doi: 10.1016/0248-4900(91)90075-X. [DOI] [PubMed] [Google Scholar]
- 157.Sorokin SP. Reconstructions of centriole formation and ciliogenesis in mammalian lungs. J Cell Sci. 1968;3:207–230. doi: 10.1242/jcs.3.2.207. [DOI] [PubMed] [Google Scholar]
- 158.Dirksen ER. Centriole morphogenesis in developing ciliated epithelium of the mouse oviduct. J Cell Biol. 1971;51:286–302. doi: 10.1083/jcb.51.1.286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Bahmanyar S, Kaplan DD, Deluca JG, Giddings TH, Jr, O’Toole ET, Winey M, Salmon ED, Casey PJ, Nelson WJ, Barth AI. β-Catenin is a Nek2 substrate involved in centrosome separation. Genes Dev. 2008;22:91–105. doi: 10.1101/gad.1596308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Tanenbaum ME, Macurek L, Galjart N, Medema RH. Dynein, Lis1 and CLIP-170 counteract Eg5-dependent centrosome separation during bipolar spindle assembly. EMBO J. 2008;27:3235–3245. doi: 10.1038/emboj.2008.242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Xu FL, Saunders WS. Actin and microtubules: working together to control spindle polarity. Cancer Cell. 2008;14:197–199. doi: 10.1016/j.ccr.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 162.Toso A, Winter JR, Garrod AJ, Amaro AC, Meraldi P, McAinsh AD. Kinetochore-generated pushing forces separate centrosomes during bipolar spindle assembly. J Cell Biol. 2009;184:365–372. doi: 10.1083/jcb.200809055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Manandhar G, Schatten H, Sutovsky P. Centrosome reduction during gametogenesis and its significance. Biol Reprod. 2005;72:2–13. doi: 10.1095/biolreprod.104.031245. [DOI] [PubMed] [Google Scholar]
- 164.Sonnenblick BP. The early embryology of Drosophila melanogaster. In: Demerec M, editor. Biology of Drosophila. New York: Hafner Publishing Company; 1950. pp. 62–167. [Google Scholar]
- 165.King RC. Ovarian development in Drosophila melanogaster. New York: Academic Press; 1970. [Google Scholar]
- 166.Maller J, Poccia D, Nishioka D, Kidd P, Gerhart J, Hartman H. Spindle formation and cleavage in Xenopus eggs injected with centriole-containing fractions from sperm. Exp Cell Res. 1976;99:285–294. doi: 10.1016/0014-4827(76)90585-1. [DOI] [PubMed] [Google Scholar]
- 167.Szöllösi D, Ozil JP. De novo formation of centrioles in parthenogenetically activated, diploidized rabbit embryos. Biol Cell. 1991;72:61–66. doi: 10.1016/0248-4900(91)90079-3. [DOI] [PubMed] [Google Scholar]
- 168.Dirksen ER. The presence of centrioles in artificially activated sea urchin eggs. J Biophys Biochem Cytol. 1961;11:244–247. doi: 10.1083/jcb.11.1.244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Miki-Noumura T. Studies on the de novo formation of centrioles: aster formation in the activated eggs of sea urchin. J Cell Sci. 1977;24:203–216. doi: 10.1242/jcs.24.1.203. [DOI] [PubMed] [Google Scholar]
- 170.Kuriyama R, Borisy GG. Cytasters induced within unfertilized sea-urchin eggs. J Cell Sci. 1983;61:175–189. doi: 10.1242/jcs.61.1.175. [DOI] [PubMed] [Google Scholar]
- 171.Riparbelli MG, Whitfield WG, Dallai R, Callaini G. Assembly of the zygotic centrosome in the fertilized Drosophila egg. Mech Dev. 1997;65:135–144. doi: 10.1016/S0925-4773(97)00066-X. [DOI] [PubMed] [Google Scholar]
- 172.Megraw TL, Kaufman TC. The centrosome in Drosophila oocyte development. Curr Top Dev Biol. 2000;49:385–407. doi: 10.1016/S0070-2153(99)49019-2. [DOI] [PubMed] [Google Scholar]
- 173.Li K, Xu EY, Cecil JK, Turner FR, Megraw TL, Kaufman TC. Drosophila centrosomin protein is required for male meiosis and assembly of the flagellar axoneme. J Cell Biol. 1998;141:455–467. doi: 10.1083/jcb.141.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Callaini G, Riparbelli MG, Dallai R. Centrosome inheritance in insects: fertilization and parthenogenesis. Biol Cell. 1999;91:355–366. doi: 10.1016/S0248-4900(99)80096-3. [DOI] [PubMed] [Google Scholar]
- 175.Giansanti MG, Bucciarelli E, Bonaccorsi S, Gatti M. Drosophila SPD-2 is an essential centriole component required for PCM recruitment and astral-microtubule nucleation. Curr Biol. 2008;18:303–309. doi: 10.1016/j.cub.2008.01.058. [DOI] [PubMed] [Google Scholar]
- 176.Schatten H, Schatten G, Mazia D, Balczon R, Simerly C. Behavior of centrosomes during fertilization and cell division in mouse oocytes and in sea urchin eggs. Proc Natl Acad Sci USA. 1986;83:105–109. doi: 10.1073/pnas.83.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177.Xu B, Hao Z, Jha KN, Zhang Z, Urekar C, Digilio L, Pulido S, Strauss JF, 3rd, Flickinger CJ, Herr JC. TSKS concentrates in spermatid centrioles during flagellogenesis. Dev Biol. 2008;319:201–210. doi: 10.1016/j.ydbio.2008.03.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Schatten G. The centrosome and its mode of inheritance: the reduction of the centrosome during gametogenesis and its restoration during fertilization. Dev Biol. 1994;165:299–335. doi: 10.1006/dbio.1994.1256. [DOI] [PubMed] [Google Scholar]
- 179.Kono T. Genetic modification for bimaternal embryo development. Reprod Fertil Dev. 2009;21:31–36. doi: 10.1071/RD08213. [DOI] [PubMed] [Google Scholar]
- 180.Calarco PG. Centrosome precursors in the acentriolar mouse oocyte. Microsc Res Tech. 2000;49:428–434. doi: 10.1002/(SICI)1097-0029(20000601)49:5<428::AID-JEMT4>3.0.CO;2-K. [DOI] [PubMed] [Google Scholar]
- 181.Meng XQ, Fan HY, Zhong ZS, Zhang G, Li YL, Chen DY, Sun QY. Localization of gamma-tubulin in mouse eggs during meiotic maturation, fertilization, and early embryonic development. J Reprod Dev. 2004;50:97–105. doi: 10.1262/jrd.50.97. [DOI] [PubMed] [Google Scholar]
- 182.Tang CJ, Hu HM, Tang TK. NuMA expression and function in mouse oocytes and early embryos. J Biomed Sci. 2004;11:370–376. doi: 10.1007/BF02254442. [DOI] [PubMed] [Google Scholar]
- 183.Schuh M, Ellenberg J. Self-organization of MTOCs replaces centrosome function during acentrosomal spindle assembly in live mouse oocytes. Cell. 2007;130:484–498. doi: 10.1016/j.cell.2007.06.025. [DOI] [PubMed] [Google Scholar]
- 184.Can A, Semiz O, Cinar O. Centrosome and microtubule dynamics during early stages of meiosis in mouse oocytes. Mol Hum Reprod. 2003;9:749–756. doi: 10.1093/molehr/gag093. [DOI] [PubMed] [Google Scholar]
- 185.Deng W, Lin H. Asymmetric germ cell division and oocyte determination during Drosophila oogenesis. Int Rev Cytol. 2001;203:93–138. doi: 10.1016/S0074-7696(01)03005-4. [DOI] [PubMed] [Google Scholar]
- 186.Mahowald AP, Strassheim JM. Intercellular migration of centrioles in the germarium of Drosophila melanogaster. An electron microscopic study. J Cell Biol. 1970;45:306–320. doi: 10.1083/jcb.45.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Bolivar J, Huynh JR, Lopez-Schier H, Gonzalez C, St Johnston D, Gonzalez-Reyes A. Centrosome migration into the Drosophila oocyte is independent of BicD and egl, and of the organisation of the microtubule cytoskeleton. Development. 2001;128:1889–1897. doi: 10.1242/dev.128.10.1889. [DOI] [PubMed] [Google Scholar]
- 188.Januschke J, Gervais L, Gillet L, Keryer G, Bornens M, Guichet A. The centrosome–nucleus complex and microtubule organization in the Drosophila oocyte. Development. 2006;133:129–139. doi: 10.1242/dev.02179. [DOI] [PubMed] [Google Scholar]
- 189.Schatten H, Sun QY. The functional significance of centrosomes in mammalian meiosis, fertilization, development, nuclear transfer, and stem cell differentiation. Environ Mol Mutagen. 2009;50(8):620–636. doi: 10.1002/em.20493. [DOI] [PubMed] [Google Scholar]
- 190.Sun QY, Schatten H. Centrosome inheritance after fertilization and nuclear transfer in mammals. Adv Exp Med Biol. 2007;591:58–71. doi: 10.1007/978-0-387-37754-4_4. [DOI] [PubMed] [Google Scholar]
- 191.Dai Y, Wang L, Wang H, Liu Y, Li N, Lyu Q, Keefe DL, Albertini DF, Liu L. Fate of centrosomes following somatic cell nuclear transfer (SCNT) in bovine oocytes. Reproduction. 2006;131:1051–1061. doi: 10.1530/rep.1.01058. [DOI] [PubMed] [Google Scholar]
- 192.Engelstadter J. Constraints on the evolution of asexual reproduction. Bioessays. 2008;30:1138–1150. doi: 10.1002/bies.20833. [DOI] [PubMed] [Google Scholar]
- 193.Heimpel GE, de Boer JG. Sex determination in the hymenoptera. Annu Rev Entomol. 2008;53:209–230. doi: 10.1146/annurev.ento.53.103106.093441. [DOI] [PubMed] [Google Scholar]
- 194.Mazumdar A, Mazumdar M. How one becomes many: blastoderm cellularization in Drosophila melanogaster . Bioessays. 2002;24:1012–1022. doi: 10.1002/bies.10184. [DOI] [PubMed] [Google Scholar]
- 195.Ferree PM, McDonald K, Fasulo B, Sullivan W. The origin of centrosomes in parthenogenetic hymenopteran insects. Curr Biol. 2006;16:801–807. doi: 10.1016/j.cub.2006.03.066. [DOI] [PubMed] [Google Scholar]
- 196.Bilinski SM, Kloc M. Accessory nuclei revisited: the translocation of snRNPs from the germinal vesicle to the periphery of the future embryo. Chromosoma. 2002;111:62–68. doi: 10.1007/s00412-002-0186-4. [DOI] [PubMed] [Google Scholar]
- 197.Riparbelli MG, Giordano R, Callaini G. Centrosome inheritance in the parthenogenetic egg of the collembolan Folsomia candida . Cell Tissue Res. 2006;326:861–872. doi: 10.1007/s00441-006-0253-x. [DOI] [PubMed] [Google Scholar]
- 198.Tram U, Sullivan W. Reciprocal inheritance of centrosomes in the parthenogenetic hymenopteran Nasonia vitripennis . Curr Biol. 2000;10:1413–1419. doi: 10.1016/S0960-9822(00)00795-8. [DOI] [PubMed] [Google Scholar]
- 199.Eisman R, Kaufman TC. Cytological investigation of the mechanism of parthenogenesis in Drosophila mercatorum . Fly (Austin) 2007;1:317–329. doi: 10.4161/fly.5408. [DOI] [PubMed] [Google Scholar]
- 200.Ferree PM, Avery A, Azpurua J, Wilkes T, Werren JH. A bacterium targets maternally inherited centrosomes to kill males in Nasonia . Curr Biol. 2008;18:1409–1414. doi: 10.1016/j.cub.2008.07.093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.D’Assoro AB, Lingle WL, Salisbury JL. Centrosome amplification and the development of cancer. Oncogene. 2002;21:6146–6153. doi: 10.1038/sj.onc.1205772. [DOI] [PubMed] [Google Scholar]
- 202.Doxsey S. Duplicating dangerously: linking centrosome duplication and aneuploidy. Mol Cell. 2002;10:439–440. doi: 10.1016/S1097-2765(02)00654-8. [DOI] [PubMed] [Google Scholar]
- 203.Duensing S, Munger K. Centrosomes, genomic instability, and cervical carcinogenesis. Crit Rev Eukaryot Gene Expr. 2003;13:9–23. doi: 10.1615/CritRevEukaryotGeneExpr.v13.i1.20. [DOI] [PubMed] [Google Scholar]
- 204.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat Rev Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 205.Sluder G, Nordberg JJ. The good, the bad and the ugly: the practical consequences of centrosome amplification. Curr Opin Cell Biol. 2004;16:49–54. doi: 10.1016/j.ceb.2003.11.006. [DOI] [PubMed] [Google Scholar]