Abstract
The present study examines the temporal dynamics of macrophage activation marker expression in response to variations in stimulation. We demonstrate that markers can be categorized as ‘early’ (expressed most abundantly at 6 h post-stimulation) or ‘late’ (expressed at 24 h post-stimulation). Thus nos2 and p40 (IL-12/IL-23) are early markers of innate and classical activation, while dectin-1 and mrc-1 are early markers and fizz1 (found in inflammatory zone-1) and ym1 are late markers of alternative activation. Furthermore, argI is a late marker of both innate and alternative activation. The ability of interferon (IFN)-γ to alter these activation markers was studied at both the protein level and gene level. As reported previously, IFN-γ was able to drive macrophages towards the classical phenotype by enhancing nos2 gene expression and enzyme activity and p40 (IL-12/IL-23) gene expression in lipopolysaccharide (LPS)-stimulated macrophages. IFN-γ antagonized alternative macrophage activation, as evident by reduced expression of dectin-1, mrc-1, fizz1 and ym1 mRNA transcripts. In addition, IFN-γ antagonized arginase activity irrespective of whether macrophages were activated innately or alternatively. Our data explain some apparent contradictions in the literature, demonstrate temporal plasticity in macrophage activation states and define for the first time ‘early’ and ‘late’ markers associated with anti-microbial/inflammatory and wound healing responses, respectively.
Keywords: alternative activation, classical, innate, macrophage
Introduction
Recent studies indicate a previously unknown level of complexity in macrophage activation states dependent on the nature of the stimulant or combination of stimulants to which they are exposed. Stimulation of macrophages with lipopolysaccharide (LPS) has been termed ‘innate’ and is associated with the expression of a number of proinflammatory mediators, including nos2 and p40 (IL-12/IL-23). In the presence of interferon (IFN)-γ, the expression of these mediators has been noted to increase; conditions that have been termed classical activation. Both these activation pathways induce the expression of anti-microbial and proinflammatory products. One such product is the enzyme inducible nitric oxide synthase 2 (NOS2), which acts to oxidize the terminal guanidine group of l-arginine to produce nitric oxide (NO). In the murine system, a wide variety of functions for NO have been defined within the immune system [1], including the ability to kill or reduce the replication of a variety of pathogens through a direct effect or by depletion of required substrates [2]. In contrast, stimulation of macrophages with interleukin (IL)-4 or IL-13 is termed ‘alternative’ activation and has been associated with the up-regulation of the mannose receptor (MR, encoded for by mrc-1) [3] and dectin-1 [4] that are associated with anti-microbial activity as well as products associated with tissue repair and a healing response such as fizz1 (found in inflammatory zone-1/RELM-α), ym1 and arginase I [5–7].
Nevertheless, some apparent anomalies have been described which suggest that the dichotomy in marker segregation between the different macrophage activation states may not always be entirely polarized. For example, as the induction of arginase I can be mediated by IL-4/IL-13 in macrophages this enzyme has often been described as a marker for alternative activation [5,8–10]. However, LPS, a potent inducer of innate activation through Toll-like receptor (TLR)-4 ligation, has also been shown in some studies to induce both isoforms of arginase in addition to NOS2 activity [11–14]. At the same time other studies also exist, suggesting that LPS is less capable than IL-4 of arginase induction [6,7]. Furthermore, although the ability of IFN-γ to prime macrophages and augment many of the effects of LPS on macrophage activation has been examined, the effects of IFN-γ on products of alternatively activated macrophages has not been studied in detail.
Herein, these apparent contradictions and shortcomings are addressed through a series of studies that examine the temporal gene expression of nos2, p40 (IL-12/IL-23), argI, dectin-1, mrc-1, fizz1 and ym1 mRNA in murine bone marrow-derived (BMD) macrophages stimulated with LPS or IL-4 in the presence or absence of IFN-γ. We demonstrate for the first time a biphasic response in activation marker expression whereby anti-microbial and inflammatory markers of both innate and alternative macrophages are maximally expressed early (6 h) following activation, while markers associated with tissue repair are expressed maximally late (>24 h) post-activation. Furthermore, while co-stimulation with IFN-γ down-regulates all IL-4-induced activation markers measured, it up-regulates LPS-induced anti-microbial and inflammatory markers selectively while down-regulating arginase activity. The implications for these observations are discussed.
Materials and methods
Culture of BMD macrophages
Bone marrow-derived (BMD) macrophages were prepared as described previously [15]. Briefly, bone marrow cells were flushed from the femurs of 8-week-old male BALB/c mice (maintained at the University of Strathclyde). The BMD precursor cells were grown in Petri dishes for 8 days in Dulbecco's modified Eagle's medium (Gibco-BRL, Paisley, UK) that had been supplemented with 30% (v/v) l-cell conditioned medium, 20% (v/v) heat-inactivated fetal calf serum (FCS) (Sigma-Aldrich, St. Louis, MO, USA), 2 mM l-glutamine (Cambrex BioScience, Veniers, Belgium), 100 U/ml penicillin (Cambrex BioScience) and 100 µg/ml streptomycin (Cambrex BioScience). l-cell conditioned medium was derived from the supernatants of confluent L929 cells and provides a source of macrophage colony-stimulating factor (M-CSF) and granulocyte–macrophage colony-stimulating factor (GM-CSF).
Treatment of cells
Cells were then harvested and plated at 106 cell/well in 24-well plates in RMPI-1640 medium (Gibco-BRL) supplemented with 10% (v/v) heat-inactivated FCS (Sigma-Aldrich), 2 mM l-glutamine, 100 U/ml penicillin and 100 µg/ml streptomycin. Cells were stimulated with mouse recombinant IL-4 (10, 100 or 1000 U/ml; BD Pharmingen™, Franklin Lakes, NJ, USA), LPS from Escherichia coli 055:B5 (20, 200 or 2000 ng/ml; Sigma, Poole, UK) and mouse recombinant IFN-γ (20 U/ml; BD Pharmingen™) for up to 48 h. Throughout this period, cells were harvested for quantitative real-time polymerase chain reaction (PCR) analysis or for use in arginase assays, and supernatants were collected for analysis of nitrite levels.
Preparation of cDNA
Cells were harvested in 1 ml of Trizol™ reagent (Invitrogen, Paisley, UK) and total RNA isolated according to the manufacturer's instructions. Two µg of total RNA was used to make cDNA, first by incubation for 5 min at 65°C with 300 ng random primers (Promega, Madison, WI, USA). The mixture was then cooled for 10 min at room temperature before the addition of 2 µl of AffinityScriptTM RT Buffer (Stratagene, Stockport, UK), 10 mM dithiothreitol (DTT), 4 mM 2′-deoxynucleosides 5′-triphosphate (dNTP) mix (Promega) and 1 µl of AffinityScript™ Multiple Reverse Transcriptase (Stratagene). Samples were then incubated for 10 min at 25°C, 1 h at 50°C and then 15 min at 70°C.
Quantitative PCR (qRT–PCR)
The Stratagene Mx3000p system was used to perform and analyse qRT–PCR experiments. Each reaction contained 10 µl of SYBR® Green JumpStart™Taq ReadyMix™ (Sigma), 25 pmol of forward primer and 25 pmol of reverse primer, 8 µl of molecular grade water (Sigma) and 1 µl of cDNA template (primers used are shown in Table 1). Reactions were performed under the following conditions: 1 cycle at 95°C for 10 min, and 40 cycles of 1 min at 95°C, 1 min at the appropriate annealing temperature and 1 min at 72°C. Samples were then subjected to 1 min at 95°C, 30 s at 55°C and 30 s at 95°C to allow generation of a dissociation curve to ensure amplification of a specific product. Relative gene expression levels were calculated by using an included standard curve for each individual gene and normalized to the housekeeping gene. The maximum gene expression for each experiment was assigned as 100% and all other treatments calculated in comparison to this. Data are presented as the mean [± standard error of the mean (s.e.m.)] expression for n = 3.
Table 1.
Primer sequences.
| Primer name | Sequence (5′ to 3′) | Optimal annealing temperature (°C) |
|---|---|---|
| argI forward | TGACATCAACACTCCCCTGACAAC | 61 |
| argI reverse | GCCTTTTCTTCCTTCCCAGCAG | |
| dectin-1 forward | GGAATCCTGTGCTTTGTGGTAGTAG | 64 |
| dectin-1 reverse | GGAAGGCAAGACTGAGAAAAACCTC | |
| fizz1 forward | ACCTTTCCTGAGATTCTGCCCC | 64 |
| fizz1 reverse | CAGTGGTCCAGTCAACGAGTAAGC | |
| p40(IL-12/IL-23) forward | CCTGGTTTGCCATCGTTTTG | 62 |
| p40(IL-12/IL-23) reverse | TCAGAGTCTCGCCTCCTTTGTG | |
| mrc-1 forward | TCTTTTACGAGAAGTTGGGGTCAG | 64 |
| mrc-1 reverse | ATCATTCCGTTCACCAGAGGG | |
| nos2 forward | GGTCTTTGACGCTCGGAACTGTAG | 64 |
| nos2 reverse | CACAACTGGGTGAACTCCAAGGTG | |
| Tbp forward | AACAGCAGCAGCAACAACAGCAGG | 64 |
| Tbp reverse | TGATAGGGGTCATAGGAGTCATTGG | |
| ym1 forward | GGCTACACTGGAGAAAATAGTCCCC | 64 |
| ym1 reverse | CCAACCCACTCATTACCCTGATAG |
The nucleotide sequences of the primers used for analysis of gene expression by real-time polymerase chain reaction.
Arginase assays
Macrophage arginase activity was determined using a reaction with α-isonitrosopropiophenon, as described previously [16]. Arginase activity was calculated using an internal standard curve generated from known quantities of urea. One unit (U) of arginase activity was defined as the enzyme activity that catalyses the production of 1 µmol urea/min.
Greiss assays for determination of NO production
Quantification of nitrite accumulation, using a method similar to that of Tsai and colleagues [17], was used as a measure of NOS2 activity. To 50 µl of well supernatant, 50 µl of Greiss reagent (equal volumes of 2% sulphanilamide in 5% H3PO4 and 0·2% naphylene diamine HCl in water) was added. After incubation for 10 min at room temperature in darkness, absorbance was read at 540 nm using a SPECTRAmax 190 microtitre spectrophotometer and softmax pro version 3·0 software (Moelcular Devices, Hove, UK). Nitrite production was determined by comparison to a standard curve generated using NaNO2.
Statistical analysis
Data are shown as mean ± s.e.m. of n = 3 repeats. Statistically significant differences were calculated using the Mann–Whitney U-test with a value of P < 0·05 accepted as significant.
Results
Arginase is induced by both innate (LPS) and alternative (IL-4) activation
BMD macrophage arginase enzyme activity, which is associated normally with alternative macrophage activation, was significantly (P < 0·05) raised at 3 h, reached plateau at 18 h and remained constant over a 48 h time-period, irrespective of whether macrophages were stimulated with 200 ng/ml LPS or 100 U/ml IL-4 (Fig. 1a). Treatment with IL-4 in combination with LPS increased arginase activity significantly by approximately two times compared LPS alone and four times compared with IL-4 alone, albeit with similar kinetics. Over the 48-h time-period, no nitrite was detectable in supernatants from unstimulated or IL-4-treated cells. As expected, LPS caused a time-dependent increase in nitrite levels accumulating within the supernatant (Fig. 1b). The addition of IL-4 significantly (P < 0·05) delayed the LPS-induced production of NO as measured by nitrite concentration (Fig. 1b).
Fig. 1.
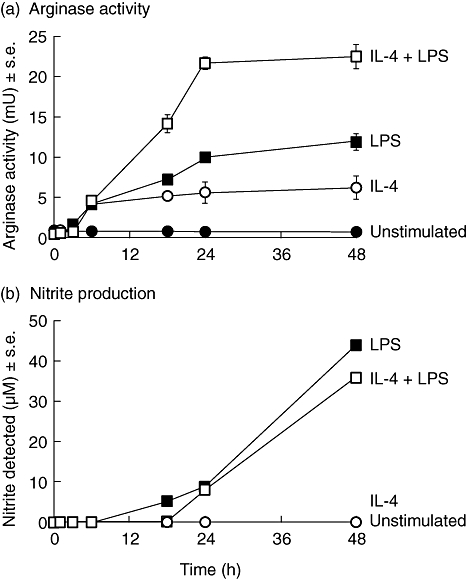
Arginase activity (a) and nitric oxide (NO) production (b) by unstimulated bone marrow-derived (BMD) macrophages (•), or BMD macrophages stimulated with 100 U/ml interleukin (IL)-4 (○), 200 ng/ml lipopolysaccharide (LPS) ( ), or with both 100 U/ml IL-4 and 200 ng/ml LPS (□) over a 48-h time-period. Results show mean ± standard error of n = 3.
), or with both 100 U/ml IL-4 and 200 ng/ml LPS (□) over a 48-h time-period. Results show mean ± standard error of n = 3.
Maximal nos2 expression precedes maximal arg1 expression in innately activated macrophages
The induction of arg1 mRNA transcript expression and arginase activity by IL-4 and LPS follows a similar pattern over a wide range of concentrations (Fig. 2). Thus, arginase activity was induced after 48 h of treatment with 100 U/ml and 1000 U/ml of IL-4, while LPS induced arginase activity at all concentrations assessed (20–2000 ng/ml) (Fig. 2a). Indeed, overall, LPS at the concentrations employed induced considerably higher levels of arginase than those obtained by IL-4 stimulation. As would be anticipated LPS, but not IL-4, induced NO production as measured by nitrite in the supernatant at all concentrations used (Fig. 2b).
Fig. 2.
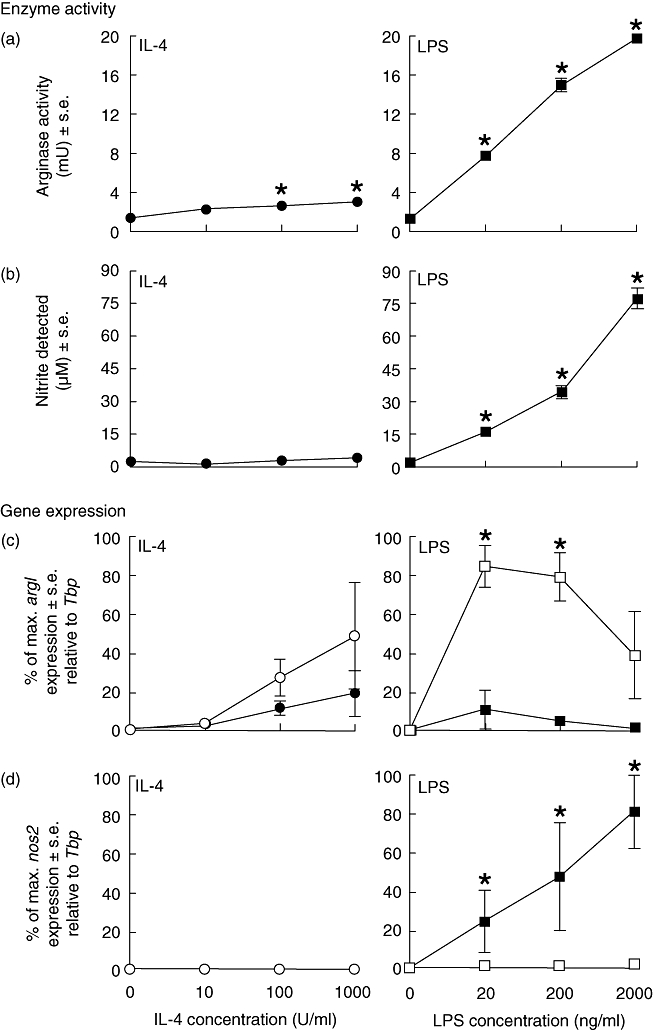
Arginase activity (a) and nitric oxide (NO) production (b) by bone marrow-derived (BMD) macrophages that were stimulated with interleukin (IL)-4 (0–1000 U/ml) (•) or lipopolysaccharide (LPS) (0–2000 ng/ml) ( ) for 48 h. Results show mean ± standard error (s.e.) of n = 3 samples. *P < 0·05 in comparison with unstimulated controls. argI (c) and nos2 (d) mRNA expression in BMD macrophages that have been stimulated for 6 (closed circles/squares) or 24 h (open circles/squares) with 0–1000 U/ml IL-4 (circles) or 0–2000 ng/ml LPS (squares). Expression levels for argI and nos2 mRNA transcripts were normalized to the housekeeping gene Tbp for each experimental run. The maximum gene expression for each run was designated 100% and all treatments calculated in comparison to this. Data is presented as the mean expression ± s.e. of n = 3. *P < 0·05 for values between time-points of the same treatment.
) for 48 h. Results show mean ± standard error (s.e.) of n = 3 samples. *P < 0·05 in comparison with unstimulated controls. argI (c) and nos2 (d) mRNA expression in BMD macrophages that have been stimulated for 6 (closed circles/squares) or 24 h (open circles/squares) with 0–1000 U/ml IL-4 (circles) or 0–2000 ng/ml LPS (squares). Expression levels for argI and nos2 mRNA transcripts were normalized to the housekeeping gene Tbp for each experimental run. The maximum gene expression for each run was designated 100% and all treatments calculated in comparison to this. Data is presented as the mean expression ± s.e. of n = 3. *P < 0·05 for values between time-points of the same treatment.
Of particular note, transcripts for arg1 were induced in a dose- and time-dependent manner by either IL-4 or LPS in BMD macrophages (Fig. 2c). While argI mRNA transcripts were induced by IL-4 treatment (100–1000 U/ml) at ‘early’ (6 h post-treatment) time-points, induction was maximal at ‘late’ (24 h post-treatment) time-points (Fig. 2c). Following LPS stimulation, macrophage argI mRNA transcript expression was induced by 6 h, but increased significantly by 24 h post-treatment with either 20 ng/ml or 200 ng/ml of LPS (Fig. 2c). Stimulation with 2000 ng/ml of LPS did not induce arg1 transcripts ‘early’ at 6 h, although induction was apparent by 24 h. Indeed, arg1 transcript expression continued to increase in LPS-stimulated macrophage up to 48 h post-activation (data not shown). As anticipated, LPS but not IL-4 induced nos2 mRNA transcript expression in BMD macrophages. In stark contrast to arg1 mRNA expression, nos2 expression was maximal ‘early’ at 6 h but not present ‘late’ at 24 h post-activation (Fig. 2d).
nos2, p40 (IL-12/IL-23), mrc-1 and dectin-1 are early markers and arg1, fizz1 and ym1 are late markers of macrophage activation
In order to determine further the time-dependent expression of genes associated with innate and alternative activation, BMD macrophages were stimulated with IL-4 (100 U/ml) or LPS (200 ng/ml) for 6 or 24 h and RT–PCR performed (Fig. 3) to measure expression of nos2, p40 (IL-12/IL-23), mrc-1, dectin-1, argI, fizz1 and ym1. In agreement with the study above, argI mRNA transcripts were induced by either IL-4 or LPS, with expression of transcripts being highest after LPS stimulation for 24 h (Fig. 3a). Both nos2 and p40 (IL-12/IL-23) demonstrated highest levels of transcript expression ‘early’ (6 h) after exposure to LPS although, by 24 h with LPS stimulation, relative transcript levels were reduced significantly (Fig. 3b and c). IL-4 did not induce significant expression of either nos2 or p40 (IL-12/IL-23) at either time-point examined. The alternative macrophage activation-associated markers mrc-1 and dectin-1 were maximally up-regulated ‘early’ within 6 h of stimulation with IL-4 and expression maintained throughout 24 h (Fig. 3d and e). LPS had no effect on the expression of mrc-1 and dectin-1 genes (Fig. 3d and e). Similarly, LPS had no effect on fizz1 and ym1 expression (Fig. 3f and g). Significantly increased expression of fizz1 and ym1 transcripts was observed ‘late’ (24 h) but not ‘early’ (6 h) following IL-4 stimulation (Fig. 3f and g).
Fig. 3.
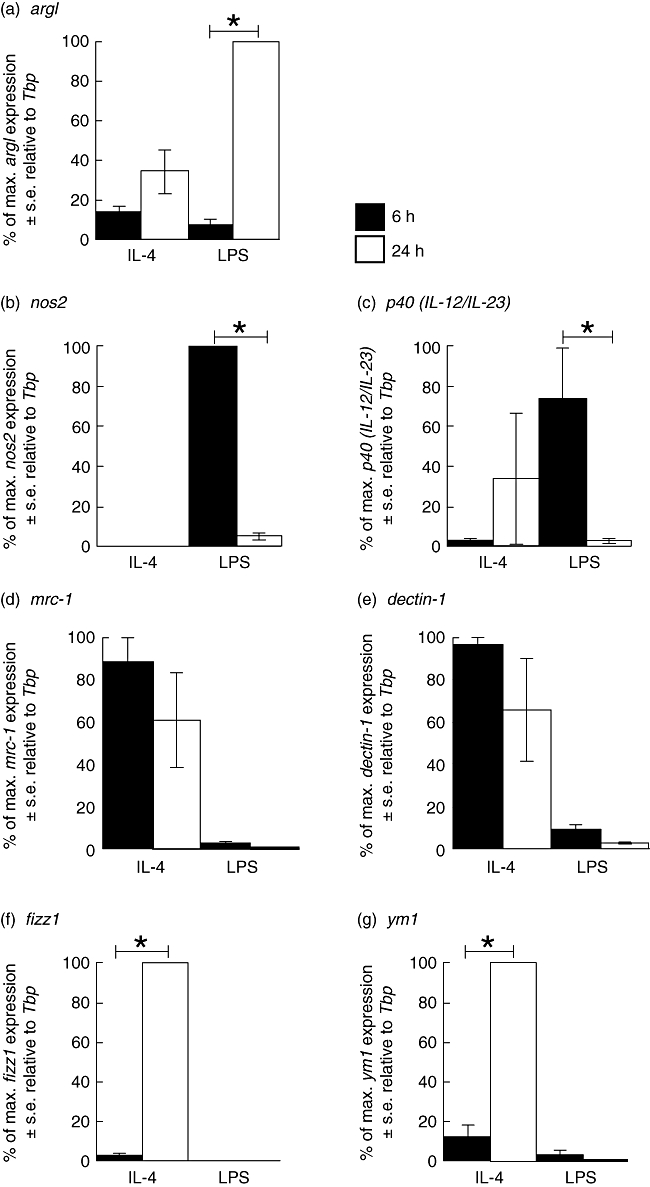
Analysis of argI (a), nos2 (b), p40 (IL-12/IL-23) (c), mrc-1 (d), dectin-1 (e), fizz1 (f) and ym1 (g) mRNA expression by real time polymerase chain reaction (PCR) in bone marrow-derived (BMD) macrophages that have been stimulated for 6 (closed bar) or 24 h (open bar) with 100 U/ml interleukin (IL)-4 or 200 ng/ml lipopolysaccharide (LPS). Expression levels for each gene were normalized to the housekeeping gene Tbp for each experimental run. The maximum gene expression for each run was designated 100% and all treatments calculated in comparison to this. Data are presented as the mean expression ± standard error of n = 3. *P < 0·05 for values between time-points of the same treatment.
Arginase activity is inhibited by IFN-γ and thus not induced by classical activation
The ability of IFN-γ to modulate arginase activity and NO production was measured in BMD macrophages stimulated with IL-4, LPS or both IL-4 and LPS (Fig. 4). The addition of IFN-γ to the cultures reduced significantly detectable arginase enzyme activity induced by LPS or by co-stimulation with both IL-4 and LPS (Fig. 4b). The ability of IL-4 to induce significant macrophage arginase activity (Fig. 4a) was inhibited by IFN-γ (Fig. 4b). The addition of IL-4 to LPS-stimulated cultures resulted in a twofold reduction (P < 0·05) of nitrite production (Fig. 4c), whereas IFN-γ induced a significant increase in the levels of nitrite whether IL-4 was present or absent (Fig. 4d).
Fig. 4.
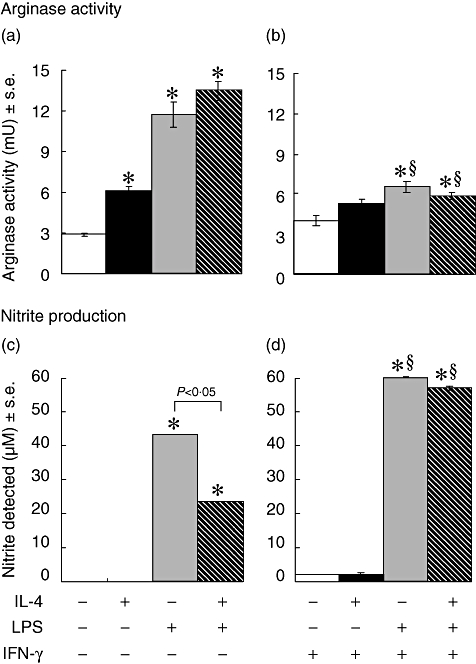
Arginase activity (a, b) and nitric oxide (NO) production (c,d) in bone marrow-derived (BMD) macrophages that were stimulated with 100 U/ml interleukin (IL)-4, 200 ng/ml lipopolysaccharide (LPS) and 20 U/ml interferon (IFN)-γ for 48 h. Results show mean ± standard error of n = 3 samples. *P < 0·05 in comparison with control (open bar) §P < 0·05 in comparison with no IFN-γ in culture.
IFN-γ modulates gene expression to drive macrophage activation towards the classical phenotype
The ability of IFN-γ to modulate LPS- and/or IL-4-induced expression of nos2, p40 (IL-12/IL-23), mrc-1 and dectin-1 mRNA transcripts was studied after 6 h of stimulation (Fig. 5), as we have identified them as ‘early’ markers of activation. IFN-γ enhanced LPS-induced (with or without IL-4 co-stimulation) expression of both nos2 (Fig. 5a) and p40 (IL-12/IL-23) (Fig. 5b). IL-4 inhibited (P < 0·05) LPS-induced nos2 significantly, but not LPS- and IFN-γ-induced nos2 (Fig. 5a). By comparison, IL-4 inhibited LPS- and IFN-γ-induced p40 (IL-12/IL-23) expression (Fig. 5b). mrc-1 and dectin-1 mRNA transcript expressions, induced by IL-4 stimulation, were inhibited significantly by co-stimulation with LPS and IFN-γ (Fig. 5c and d).
Fig. 5.
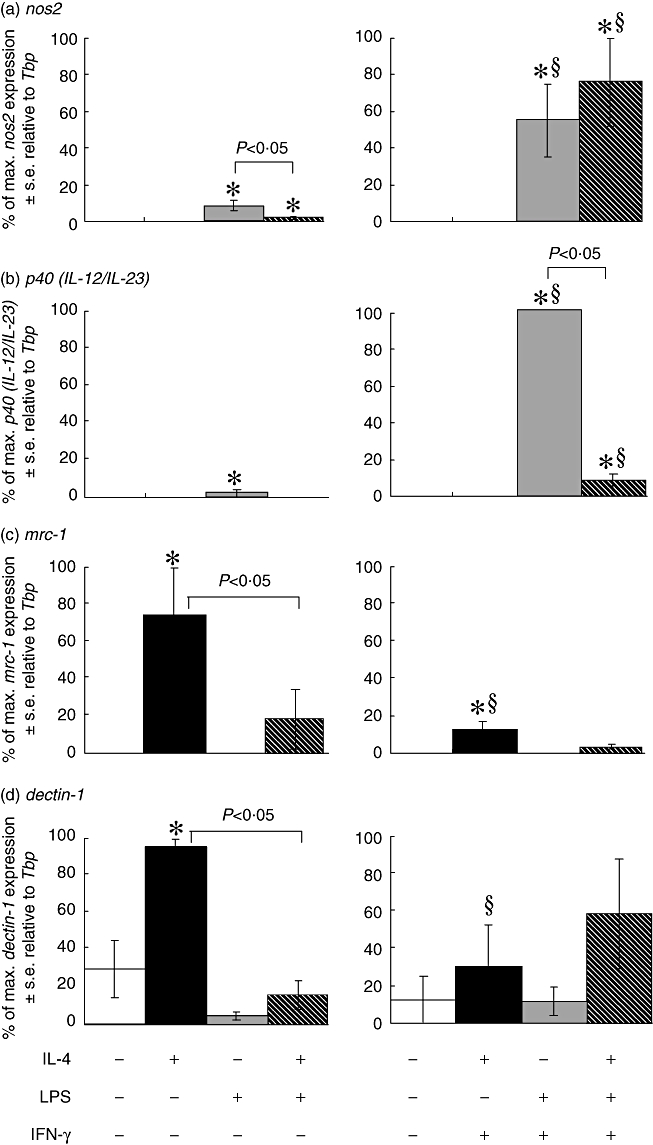
Analysis of early time-point markers nos2 (a), p40 (IL-12/IL-23) (b), mrc-1 (c) and dectin-1 (d) mRNA expression by real-time polymerase chain reaction (PCR) in bone marrow-derived (BMD) macrophages that have been stimulated for 6 h with 100 U/ml interleukin (IL)-4, 200 ng/ml lipopolysaccharide (LPS) and 20 U/ml interferon (IFN)-γ. Expression levels for each gene were normalized to the housekeeping gene Tbp for each experimental run. The maximum gene expression for each run was designated 100% and all treatments calculated in comparison to this. Data are presented as the mean ± standard error of n = 3. *P < 0·05 in comparison with control (open bar), §P < 0·05 in comparison with no IFN-γ in culture.
The ability of IFN-γ to modulate expression of arg1, fizz1 and ym1, which we have identified as ‘late’ activation markers, were analysed after 24 h by RT–PCR (Fig. 6). Twenty-four h of stimulation with IL-4 induced a low level of arg1 expression, which was increased significantly (P < 0·05) with the addition of LPS to these cultures. IFN-γ inhibited the ability of IL-4 or LPS, but not treatment with both LPS and IL-4, to stimulate macrophage arg1 expression significantly over background (Fig. 6a). LPS and IFN-γ both down-regulated IL-4 induced macrophage fizz1 and ym1 mRNA transcript expression significantly (Fig. 6b and c).
Fig. 6.
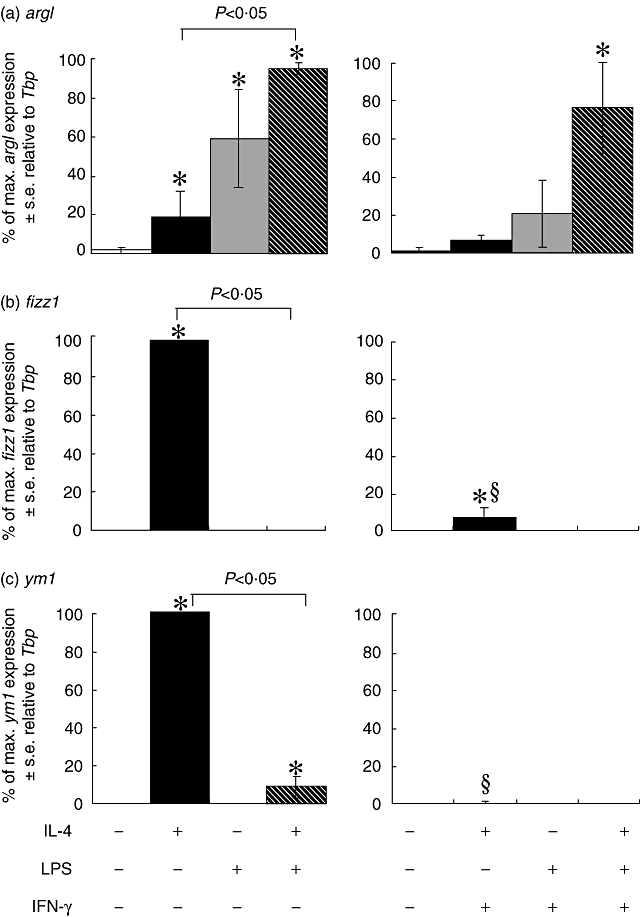
Analysis of late time-point markers argI (a), fizz1 (b) and ym1 (c) mRNA expression by real-time polymerase chain reaction (PCR) in bone marrow-derived (BMD) macrophages that have been stimulated for 24 h with 100 U/ml interleukin (IL)-4, 200 ng/ml lipopolysaccharide (LPS) and 20 U/ml interleukin (IFN)-γ. Expression levels for each gene were normalized to the housekeeping gene Tbp for each experimental run. The maximum gene expression for each run was designated 100% and all treatments calculated in comparison to this. Data are presented as the mean ± standard error of n = 3. *P < 0·05 in comparison with control (open bar), §P < 0·05 in comparison with no IFN-γ in culture.
Discussion
Depending on the stimulus, macrophage activation states have (historically) been designated in a number of ways; innate (stimulated with TLR ligand – usually LPS), classical (stimulated with TLR ligand and IFN-γ) or alternative (stimulated with IL-4 or IL-13) [18–22]. Each activation state has been associated with the expression of different genes, gene products and biochemical properties in studies involving rodents and humans [1,3,4,10,23–27]. Innate activation has been associated with production of anti-pathogen and inflammatory products such as tumour necrosis factor (TNF)-α, IL-12, NOS2 and cyclooxygenase (COX)-2, as well as reactive oxygen and nitrogen intermediates. Classical activation has been demonstrated to augment the production of these mediators [28]. In contrast, alternative activation has been associated with arginase 1, fizz1 and ym1 that are involved in tissue repair, control of some parasitic infections and control of reactive nitrogen intermediates [6,7,10,27,29,30]. However, these macrophages are also associated with the expression of the mannose receptor [3] and dectin-1 [4], which are important in pathogen recognition and have been demonstrated to play a beneficial role in fungal infections [31–35]. More recently it has been suggested that activated macrophages comprise a spectrum based on three major activities; classically activated (as described previously), wound-healing (stimulated with IL-4 or IL-13) and regulatory, which are induced by TLR ligation in combination with other stimuli, including immune complexes [21]. The current study not only supports the idea of a spectrum of activation states with a degree of overlap in relative gene expression, but also demonstrates that these activation states are subject to significant temporal plasticity.
An example of this is the expression of arg1 and downstream arginase activity that we have demonstrated in both innately and alternatively activated macrophages. While much of the literature defines argI expression as a hallmark of murine alternative activation [5,9,10,36,37], other studies describe argI expression or arginase activity in macrophages following LPS stimulation [12–14,38,39]. Further studies speculate that the ability of LPS to influence arginase is species-dependent [40]. The present study demonstrates that arg1 is induced by both innate (LPS) and alternative (IL-4) activation of murine bone marrow-derived macrophages, but is predominantly found ‘late’ (> 24 h) post-stimulation and demonstrates for the first time that the effects of innate and alternative activation on macrophage arginase activity are at least additive. The timing of arg1 expression relative to nos2 expression reported in the current murine studies is consistent with that reported in rat macrophages [13]. It has been suggested that the delay in onset of argI expression after nos2 expression is a homeostatic mechanism to limit the inflammation caused by endotoxin-induced NO production and to allow the healing process to begin [38].
The underlying molecular mechanisms for the apparent switch from nos2 to argI expression after LPS stimulation have been investigated in recent years, and it has been shown that mitogen-activated protein kinase phosphatase-1 (MKP-1) may function as a molecular switch to drive arginine metabolism away from NO production towards urea and ornithine production via the action of arginase [38]. In these studies RAW264·7 macrophages were transfected stably to over-express MKP-1, and it was found that LPS stimulation led to a significant reduction in NOS2 induction, whereas LPS stimulation of macrophages from Mkp-1–/– mice resulted in enhanced NOS2 protein production [41].
Despite both IL-4 and LPS inducing macrophage arginase gene expression and enzyme activity, it is probable that the underlying molecular mechanisms are somewhat different for each stimulant. Kasmi et al. [42] have shown recently that macrophages possess two distinct pathways for arginase I induction. The first pathway is linked to alternative activation and is signal transducer and activator of transcription 6 (STAT6)-dependent, and the second pathway is regulated by myeloid differentiation primary response gene 88 (MyD88)-dependent TLR signalling. In order to induce arginase I, both these pathways lead ultimately to induction of CCAAT-enhancer-binding proteins (C/EBP)-β[43]. These observations would be consistent with our present findings demonstrating enhanced bone marrow-derived macrophage arginase activity induced by both IL-4 and LPS treatment compared with stimulation with either IL-4 or LPS independently.
Promoting the concept of macrophage plasticity in function, our results demonstrate that the presence of multiple stimuli alters gene expression. Thus arginase activity was inhibited by IFN-γ and was not found in classically activated macrophages. Similarly, IFN-γ down-regulated dectin-1, mrc-1, fizz1 and ym1 mRNA transcripts, all of which are specific markers of alternative activation. This observation is similar to others [3,4,6,7]. Interestingly, it has also been found that despite IL-4 and IFN-γ having opposing effects on the surface expression of the mannose receptor, both these cytokines can act to enhance macrophage mannose receptor-mediated phagocytosis [44]. The ability of IFN-γ to modulate IL-4-induced macrophage activities may be due to the ability of this Th1 type cytokine to inhibit STAT6 signal transduction [45].
IL-4 is known to have an array of effects on macrophages, including morphological changes, as well as inducing the expression and activity of a number of receptors and molecules [3]. Up-regulation of the mannose receptor (mrc-1), dectin-1 receptor and the fizz1 and ym1 genes are recognized as indicators that the alternative pathway of activation has been induced [3,4,6,7]. The data presented show that the expression of dectin-1, mrc-1, fizz1 and ym1 are IL-4-dependent in murine BMD macrophages. This finding is supported further by in vivo studies using murine models for parasite infections which induce T helper type 2 (Th2) responses. In these systems, the Th2-driven, chronic stage of infection is characterized by the presence of alternatively activated macrophages indicated by the IL-4-dependent expression of fizz1 and ym1[7,9,27,29,46]. Recently, Mylonas et al. [47] have used these in vivo models of parasite infection to demonstrate further the plasticity of macrophage activation. This study has shown that even after long-term exposure to Th2 cytokines, macrophages could still respond to LPS and IFN-γ stimulation to become anti-microbial in function.
The studies reported herein demonstrate that the major markers of macrophage activation can be categorized as early (maximum at 6 h) or late (evident at 24 h) and their temporal and sequential expression can be rationalized according to their ascribed functions. Thus, both nos2 and p40 (IL-12/IL-23), involved in microbicidal killing and activation of natural killer (NK) cells to produce IFN-γ, respectively, are early markers of innate macrophage activation. Similarly, the mannose and dectin-1 receptors are involved in the recognition of mannose residues on pathogens [20] and β-glucans on fungal pathogens [32–34], respectively, and are early markers of alternative activation. In contrast, the ym1 and fizz1 gene products, chitnase [48] and resistin-like proteins [49], respectively, that are involved in wound healing and tissue repair are late markers of alternative activation. Finally, argI, which has the ability to reduce NO production through competition for arginine [50], is expressed late in alternative and innate activated macrophages. Arginase has been implicated in wound healing and tissue repair mechanisms [51,52] due to its function of converting arginine to proline (required for collagen synthesis and deposition) and polyamines (such as putrescine, spermidine and spermine) [53–55], which are required during cell proliferation.
In the course of the present study the expression of macrophage activation markers has been described for the first time in terms of their temporal plasticity. In all studies, mRNA transcripts for arg1 and nos2 were detectable prior to their enzyme activities, demonstrating that, perhaps not surprisingly, transcriptional changes are evident before translational changes. These results add a further dimension to the suggestions by Mosser and Edwards [22], who indicate that macrophages have a spectrum of activation with overlapping characteristics. Not only is expression of these overlapping characteristics time-dependent, but these activities are totally appropriate for the required functions of the macrophages at specific time-points.
Disclosure
None.
References
- 1.Bogdan C. Nitric oxide and the immune response. Nat Immunol. 2001;2:907–16. doi: 10.1038/ni1001-907. [DOI] [PubMed] [Google Scholar]
- 2.James SL. Role of nitric oxide in parasitic infections. Microbiol Rev. 1995;59:533–47. doi: 10.1128/mr.59.4.533-547.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Stein M, Keshav S, Harris N, Gordon S. Interleukin 4 potently enhances murine macrophage mannose receptor activity: a marker of alternative immunologic macrophage activation. J Exp Med. 1992;176:287–92. doi: 10.1084/jem.176.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Willment JA, Lin HH, Reid DM, et al. Dectin-1 expression and function are enhanced on alternatively activated and GM-CSF-treated macrophages and are negatively regulated by IL-10, dexamethasone, and lipopolysaccharide. J Immunol. 2003;171:4569–73. doi: 10.4049/jimmunol.171.9.4569. [DOI] [PubMed] [Google Scholar]
- 5.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3:23–35. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 6.Raes G, De Baetselier P, Noël W, Beschin A, Brombacher F, Hassanzadeh Ghassabeh G. Differential expression of FIZZ1 and Ym1 in alternatively activated versus classically activated macrophages. J Leukoc Biol. 2002;71:597–602. [PubMed] [Google Scholar]
- 7.Raes G, Noël W, Beschin A, Brys L, De Baetselier P, Hassanzadeh Ghassabeh G. FIZZ1 and Ym as tools to discriminate between differentially activated macrophages. Dev Immunol. 2002;9:151–9. doi: 10.1080/1044667031000137629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Munder M, Eichmann K, Morán JM, Centeno F, Soler G, Modolell M. Th1/Th2-regulated expression of arginase isoforms in murine macrophages and dendritic cells. J Immunol. 1999;163:3771–7. [PubMed] [Google Scholar]
- 9.Loke P, Nair MG, Parkinson J, Guilliano D, Blaxter M, Allen JE. IL-4 dependent alternatively activated macrophages have a distinctive in vivo gene expression phenotype. BMC Immunol. 2002;3:7–17. doi: 10.1186/1471-2172-3-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Noël W, Raes G, Hassanzadeh Ghassabeh G, De Baetselier P, Beschin A. Alternatively activated macrophages during parasitic infections. Trends Parasitol. 2004;20:126–33. doi: 10.1016/j.pt.2004.01.004. [DOI] [PubMed] [Google Scholar]
- 11.Corraliza IM, Soler G, Eichmann K, Modolell M. Arginase induction by suppressors of nitric oxide synthesis (IL-4, IL-10 and PGE2) in murine bone-marrow-derived macrophages. Biochem Biophys Res Commun. 1995;206:667–73. doi: 10.1006/bbrc.1995.1094. [DOI] [PubMed] [Google Scholar]
- 12.Wang WW, Jenkinson CP, Griscavage JM, et al. Co-induction of arginase and nitric oxide synthase in murine macrophages activated by lipopolysaccharide. Biochem Biophys Res Commun. 1995;210:1009–16. doi: 10.1006/bbrc.1995.1757. [DOI] [PubMed] [Google Scholar]
- 13.Sonoki T, Nagasaki A, Gotoh T, et al. Coinduction of nitric oxide synthase and arginase I in cultured rat peritoneal macrophages and rat tissues in vivo by lipolysaccharide. J Biol Chem. 1997;272:3689–93. doi: 10.1074/jbc.272.6.3689. [DOI] [PubMed] [Google Scholar]
- 14.Sosroseno W, Musa M, Ravichandran M, Fikri Ibrahim M, Bird PS, Seymour GJ. Arginase activity in a murine macrophage cell line (RAW264.7) stimulated with lipopolysaccharide from Actinobacillus actinomycetemcomitans. Oral Microbiol Immunol. 2006;21:145–50. doi: 10.1111/j.1399-302X.2006.00262.x. [DOI] [PubMed] [Google Scholar]
- 15.Jones LA, Anthony JP, Henriquez FL, et al. Toll-like receptor-4-mediated macrophage activation is differentially regulated by progesterone via the glucocorticoid and progesterone receptors. Immunology. 2008;125:59–69. doi: 10.1111/j.1365-2567.2008.02820.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Corraliza IM, Campo ML, Soler G, Modolell M. Determination of arginase activity in macrophages: a micromethod. J Immunol Methods. 1994;174:231–5. doi: 10.1016/0022-1759(94)90027-2. [DOI] [PubMed] [Google Scholar]
- 17.Tsai SH, Lin-Shiau SY, Lin JK. Suppression of nitric oxide synthase and the down-regulation of the activation of NFκB in macrophages by resveratol. Br J Pharmacol. 1999;126:673–80. doi: 10.1038/sj.bjp.0702357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stout RD, Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. J Leukoc Biol. 2004;76:509–13. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–64. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 20.Taylor PR, Martinez-Pomares L, Stacey M, Lin HH, Brown GD, Gordon S. Macrophages receptors and immune recognition. Annu Rev Immunol. 2005;23:901–44. doi: 10.1146/annurev.immunol.23.021704.115816. [DOI] [PubMed] [Google Scholar]
- 21.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. J Leukoc Biol. 2006;80:1298–307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mosser DM, Edwards JP. Exploring the full spectrum of macrophage activation. Nat Rev Immunol. 2008;8:958–69. doi: 10.1038/nri2448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fenton MJ, Buras JA, Donnelly RP. IL-4 reciprocally regulates IL-1 and IL-1 receptor antagonist expression in human monocytes. J Immunol. 1992;149:1283–8. [PubMed] [Google Scholar]
- 24.Orino E, Sone S, Nii A, Ogura T. IL-4 upregulates IL-1 receptor antagonist gene expression and its production in human blood monocytes. J Immunol. 1992;149:925–32. [PubMed] [Google Scholar]
- 25.Kodelja V, Müller C, Politz O, Hakij N, Orfanos CE, Goerdt S. Alternative macrophage activation-associated CC-chemokine-1, a novel structural homologue of macrophage inflammatory protein-1α with a Th2-associated expression pattern. J Immunol. 1998;160:1411–18. [PubMed] [Google Scholar]
- 26.Brombacher F, Kastelein RA, Alber G. Novel IL-12 family members shed light on the orchestration of Th1 responses. Trends Immunol. 2003;24:207–12. doi: 10.1016/S1471-4906(03)00067-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nair MG, Cochrane DW, Allen JE. Macrophages in chronic type 2 inflammation have a novel phenotype characterised by the abundant expression of YM1 and FIZZ1 that can be partly replicated in vitro. Immunol Lett. 2003;85:173–80. doi: 10.1016/s0165-2478(02)00225-0. [DOI] [PubMed] [Google Scholar]
- 28.Mosser DM. The many faces of macrophage activation. J Leukoc Biol. 2003;73:209–12. doi: 10.1189/jlb.0602325. [DOI] [PubMed] [Google Scholar]
- 29.Nair MG, Gallagher IJ, Taylor MD, et al. Chitinase and Fizz family members are a generalised feature of nematode infection with selective upregulation of Ym1 and Fizz1 by antigen presenting cells. Infect Immun. 2005;73:385–94. doi: 10.1128/IAI.73.1.385-394.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raes G, Beschin A, Ghassabeh GH, De Baetselier P. Alternatively activated macrophages in protozoan infections. Curr Opin Immunol. 2007;19:1–6. doi: 10.1016/j.coi.2007.05.007. [DOI] [PubMed] [Google Scholar]
- 31.Yamamoto Y, Klein TW, Friedman H. Involvement of mannose receptor in cytokine interleukin-1 beta (IL-1beta), IL-6, and granulocyte–macrophage colony-stimulating factor responses, but not in chemokine macrophage inflammatory protein 1beta (MIP-1beta), MIP-2, and KC responses, caused by attachment of Candida albicans to macrophages. Infect Immun. 1997;65:1077–82. doi: 10.1128/iai.65.3.1077-1082.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown GD, Gordon S. A new receptor for β-glucans. Nature. 2001;413:36–7. doi: 10.1038/35092620. [DOI] [PubMed] [Google Scholar]
- 33.Taylor PR, Tsoni SV, Willment JA, et al. Dectin-1 is required for β-glucan recognition and control of fungal infection. Nature Immunol. 2007;8:31–8. doi: 10.1038/ni1408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kimberg M, Brown GD. Dectin-1 and its role in antifungal immunity. Med Mycol. 2008;46:631–6. doi: 10.1080/13693780802140907. [DOI] [PubMed] [Google Scholar]
- 35.van de Veerdonk FL, Marijnissen RJ, Kullberg BJ, et al. The macrophage mannose receptor induces IL-17 in response to Candida albicans. Cell Host Microbe. 2009;5:329–40. doi: 10.1016/j.chom.2009.02.006. [DOI] [PubMed] [Google Scholar]
- 36.Raes G, Van den Bergh R, De Baetselier P, Ghassabeh GH. Arginase-1 and Ym1 are markers for murine, but not human, alternatively activated myeloid cells. J Immunol. 2005;174:6561–2. doi: 10.4049/jimmunol.174.11.6561. [DOI] [PubMed] [Google Scholar]
- 37.Van Ginderachter JA, Movahedi K, Ghassabeh GH, et al. Classical and alternative activation of mononuclear phagocytes: picking the best of both worlds for tumor promotion. Immunobiology. 2006;211:487–501. doi: 10.1016/j.imbio.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 38.Salimuddin, Nagasaki A, Gotoh T, Isobe H, Mori M. Regulation of the genes for arginase isoforms and related enzymes in mouse macrophages by lipopolysaccharide. Am J Physiol Endocrinol Metab. 1999;277:110–17. doi: 10.1152/ajpendo.1999.277.1.E110. [DOI] [PubMed] [Google Scholar]
- 39.Klasen S, Hammermann R, Fuhrmann M, et al. Glucocorticoids inhibit lipopolysaccharide-induced up-regulation of arginase in rat alveolar macrophages. Br J Pharmacol. 2001;132:1349–57. doi: 10.1038/sj.bjp.0703951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bronte V, Zanovello P. Regulation of immune responses by l-arginine metabolism. Nat Rev Immunol. 2005;5:641–54. doi: 10.1038/nri1668. [DOI] [PubMed] [Google Scholar]
- 41.Nelin LD, Wang X, Zhao Q, et al. MKP-1 switches arginine metabolism from nitric oxide synthase to arginase following endotoxin challenge. Am J Physiol Cell Physiol. 2007;293:C632–C640. doi: 10.1152/ajpcell.00137.2006. [DOI] [PubMed] [Google Scholar]
- 42.Kasmi KCE, Qualls JE, Pesce JT, et al. Toll-like receptor-induced arginase 1 in macrophages thwarts effective immunity against intracellular pathogens. Nat Immunol. 2008;9:1399–406. doi: 10.1038/ni.1671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Albina JE, Mahoney EJ, Daley JM, Wesche DE, Morris SM, Jr, Reichner JS. Macrophage arginase regulation by CCAAT/enhancer-binding protein beta. Shock. 2005;23:168–72. doi: 10.1097/01.shk.0000148054.74268.e2. [DOI] [PubMed] [Google Scholar]
- 44.Raveh D, Kruskal BA, Farland J, Ezekowitz RAB. Th1 and Th2 cytokines cooperate to stimulate mannose-receptor-mediated phagocytosis. J Leukoc Biol. 1998;64:108–13. [PubMed] [Google Scholar]
- 45.Heller NM, Matsukura S, Georas SN, et al. Interferon-γ inhibits STAT6 signal transduction and gene expression in human airway epithelial cells. Am J Respir Cell Mol Biol. 2004;31:573–82. doi: 10.1165/rcmb.2004-0195OC. [DOI] [PubMed] [Google Scholar]
- 46.Reyes JL, Terrazas LI. The divergent roles of alternatively activated macrophages in helminthic infections. Parasite Immunol. 2007;29:609–19. doi: 10.1111/j.1365-3024.2007.00973.x. [DOI] [PubMed] [Google Scholar]
- 47.Mylonas KJ, Nair MG, Preito-Lafuente L, Paape D, Allen JE. Alternatively activated macrophages elicited by helminth infection can be reprogrammed to enable microbial killing. J Immunol. 2009;182:3084–94. doi: 10.4049/jimmunol.0803463. [DOI] [PubMed] [Google Scholar]
- 48.Owhashi M, Kirai N, Horii Y. Eosinophil chemotactic factor release from neutrophils induced by stimulation with Schistosoma janonicum eggs. Parasitol Res. 1997;83:42–6. doi: 10.1007/s004360050205. [DOI] [PubMed] [Google Scholar]
- 49.Holcomb IN, Kabakoff RC, Chan B, et al. FIZZ1, a novel cysteine-rich secreted protein associated with pulmonary inflammation, defines a new gene family. EMBO J. 2000;19:4046–55. doi: 10.1093/emboj/19.15.4046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang CI, Liao JC, Kuo L. Arginase modulates nitric oxide production in activated macrophages. Am J Physiol Heart Circ Physiol. 1998;274:H342–H348. doi: 10.1152/ajpheart.1998.274.1.H342. [DOI] [PubMed] [Google Scholar]
- 51.Witte MB, Barbul A, Schick MA, Vogt N, Becker HD. Upregulation of arginase expression in wound-derived fibroblasts. J Surg Res. 2002;105:35–142. doi: 10.1006/jsre.2002.6443. [DOI] [PubMed] [Google Scholar]
- 52.Witte MB, Barbul A. Arginine physiology and its implication for wound healing. Wound Rep Reg. 2003;11:419–23. doi: 10.1046/j.1524-475x.2003.11605.x. [DOI] [PubMed] [Google Scholar]
- 53.Kepka-Lenhart D, Mistry SK, Wu G, Morris SM., Jr. Arginase I: a limiting factor for nitric oxide and polyamine synthesis by activated macrophages? Am J Physiol Reg Int Comp Physiol. 2000;279:2237–42. doi: 10.1152/ajpregu.2000.279.6.R2237. [DOI] [PubMed] [Google Scholar]
- 54.Li H, Meininger CJ, Hawker JR, et al. Regulatory role of arginase I and II in nitric oxide, polyamine, and proline syntheses in endothelial cells. Am J Physiol Endocrinol Metabol. 2001;280:E75–E82. doi: 10.1152/ajpendo.2001.280.1.E75. [DOI] [PubMed] [Google Scholar]
- 55.Deignan JL, Livesay JC, Yoo PK, et al. Ornithine deficiency in the arginase double knockout mouse. Mol Genet Metabol. 2006;89:87–96. doi: 10.1016/j.ymgme.2006.04.007. [DOI] [PubMed] [Google Scholar]


