Abstract
Background:
LIM and SH3 protein 1 (LASP-1) is a nucleo-cytoplasmatic signalling protein involved in cell proliferation and migration and is upregulated in breast cancer in vitro studies have shown that LASP-1 might be regulated by prostate-derived ETS factor (PDEF), p53 and/or LASP1 gene amplification. This current study analysed the prognostic significance of LASP-1 on overall survival (OS) in 177 breast cancer patients and addressed the suggested mechanisms of LASP-1-regulation.
Methods:
Nucleo-cytoplasmatic LASP-1-positivity of breast carcinoma samples was correlated with long-term survival, clinicopathological parameters, Ki67-positivity and PDEF expression. Rate of LASP1 amplification was determined in micro-dissected primary breast cancer cells using quantitative RT–PCR. Cell-phase dependency of nuclear LASP-1-localisation was studied in synchronised cells. In addition, LASP-1, PDEF and p53 expression was compared in cell lines of different tumour entities to define principles for LASP-1-regulation.
Results:
We showed that LASP-1 overexpression is not due to LASP1 gene amplification. Moreover, no correlation between p53-mutations or PDEF-expression and LASP-1-status was observed. However, nuclear LASP-1-localisation in breast carcinomas is increased during proliferation with peak in G2/M-phase and correlated significantly with Ki67-positivity and poor OS.
Conclusion:
Our results provide evidence that nuclear LASP-1-positivity may serve as a negative prognostic indicator for long-term survival of breast cancer patients.
Keywords: LASP-1, Ki67, PDEF, p53, breast cancer
Breast cancer is expected to account for 26% of all new cancer cases among women in the western world of which 89% will survive 5 years after diagnosis (Jemal et al, 2007). In spite of the significant improvements in diagnostic and therapeutic modalities for the treatment of cancer patients, metastasis still composes the major cause of mortality being responsible for 60% of breast cancer deaths (Hanahan and Weinberg, 2000). Metastatic disease remains generally incurable with a median survival time of only a few years. Regardless of the increase in its incidence, mortality related to breast cancer is decreasing because of raised awareness and screening, as well as multidisciplinary treatment. The introduction of endocrine therapy and the treatment with trastuzumab (Herceptin) in patients with HER-2/neu overexpression reduced the rates of recurrence by 50% and significantly improved survival (Widakowich et al, 2007). Nevertheless, breast cancer remains a multi-step process linked to more than one single molecular alteration. Therefore, elucidating genes that are overexpressed in breast cancer cells may yield promising targets for novel therapeutic agents (Mauriac et al, 2005).
The LIM and SH3 domain protein (LASP-1) was initially identified from a cDNA library of breast cancer metastases. It became the first member of a newly defined LIM-protein subfamily of the nebulin group characterised by the combined presence of LIM and SH3 domains (Grunewald and Butt, 2008). LASP-1 is localised within multiple sites of dynamic F-actin assembly such as focal contacts, lamellipodia and membrane ruffles it binds to the specific shuttle proteins Zyxin and lipoma preferred partner (LPP) and is involved in cell migration and proliferation (Schreiber et al, 1998; Chew et al, 2002; Butt et al, 2003; Nakagawa et al, 2006). Silencing of LASP-1 by RNA-interference in various cancer cell lines resulted in strong inhibition of proliferation and migration with cell cycle arrest in G2/M-phase (Grunewald et al, 2006, 2007b).
LIM and SH3 protein 1 mRNA is expressed ubiquitously at low basal levels in all normal human tissues, but is overexpressed in metastatic human breast cancer (Grunewald and Butt, 2008) and ovarian cancer (Grunewald et al, 2007b; Dimova et al, 2009). In a recent case–control study, LASP-1-expression correlated significantly with tumour size and nodal-positivity (Grunewald et al, 2007a). Albeit the protein is predominantly situated at focal adhesions, nuclear localisation of the protein could be clearly detected by confocal microscopy and western blots of cytosolic and nuclear preparations from various breast cancer cell lines (Grunewald et al, 2007a). These data prompted us to further investigate the long-term survival in relation to nuclear and cytosolic LASP-1-localisation in a large well-characterised cohort of breast cancer patients and to analyse the nuclear LASP-1-localisation in the different phases of the cell cycle.
In invasive breast cancer cells, LASP-1-expression was significantly inversely affected by prostate-derived ETS factor (PDEF), a transcription factor known to repress a variety of genes that are possibly involved in oncogenesis, such as the apoptosis regulator survivin (Ghadersohi et al, 2007) and the pro-invasive protease uPA (Turner et al, 2008).
Real-time PCR analysis confirmed upregulation of LASP1 mRNA in PDEF-deficient invasive and highly metastatic breast cancer cells (MBA-MB-231, BT-549), while in the non-invasive MCF-7 breast cancer cell line, endogenously expressing PDEF, a reduced LASP-1 protein level was detected (Turner et al, 2008).
In a study conduced with hepatocellular carcinoma (HCC), LASP-1 was repressed by wild-type p53 at the transcriptional level (Wang et al, 2009). Functional negative p53 mutations led to increased LASP-1-expression and to a more aggressive HCC phenotype (Wang et al, 2009).
In this study, we aimed to determine whether the PDEF level or mutations of the tumour suppressor p53 represent a general mechanism of LASP-1 deregulation in human cancer. Thus, we analysed several tumour cell lines of different entities as well as breast cancer tissue for LASP-1-expression in correlation to PDEF protein concentration and p53 status.
As LASP1 gene amplification was reported earlier in one breast cancer cell line (Tomasetto et al, 1995b) and in 40% of crude extracts of lymph nodes derived from metastatic breast cancer (Tomasetto et al, 1995a) and was accounted for the principal cause of LASP-1 overexpression, we also re-analysed the rate of LASP1 gene amplification in individual micro-dissected primary breast cancer cells. It is noteworthy that the LASP1 gene maps to a region (17q12) that is altered in 20–30% of human breast cancers (Tomasetto et al, 1995a, 1995b) and tumours bearing amplifications of 17q11-21 are associated with an adverse prognosis because of increased resistance to chemotherapy and endocrine therapy (Ross and Fletcher, 1999).
Materials and methods
Tissue samples
The studies were performed with approval of the ethics committee of the University of Wuerzburg. Paraffin-embedded tissue samples of surgical biopsies from 177 patients with invasive breast cancer were obtained from the Department of Pathology of the University of Wuerzburg. The patients were aged from 32 to 85 (mean 55.3±11.9) years. All carcinomas were mainly collected from patients that underwent wide excisions.
Grading of malignancy of ductal carcinomas was evaluated according to the Scarff, Bloom and Richardson criteria as suggested by Nottingham City Hospital Pathologists (Dalton et al, 2000). Tumour staging was performed according to parameters of the TNM system (Singletary and Connolly, 2006).
Ten paraffin-embedded breast tissue samples from reduction mammoplasties were used as control tissue to obtain reference DNA for the evaluation of the quantitative RT–PCR results.
Immunohistochemistry
For immunohistochemical staining procedures tissue sections were cut from regular paraffin-embedded tissue at 2–3 μm. Sections were placed onto APES (3-amino-propyltriethoxy-silane; Roth, Karlsruhe, Germany) coated slides, dewaxed twice in xylene for 10 min, rehydrated in graded ethanol (two changes in 96%, one change in 70%, one change with distilled water) and in TRIS-buffered saline (25 mM TRIS/HCl, pH 7.4, 137 mM NaCl, 2.7 mM KCl) for 1 min each. For antigen retrieval, sections were subjected to heat pretreatment by boiling in 0.01 M of sodium citrate buffer (pH 6.0) for 5 min in a microwave oven (800 W s–1). Endogenous peroxidase was blocked by incubation in 3.0% hydrogen peroxide in methyl alcohol for 5 min, washed in PBS and incubated in Beriglobin (Aventis-Behring GmbH, Marburg, Germany) 1 : 10 in PBS at room temperature (RT) for 15 min to prevent unspecified attachments. Slides were then incubated with the polyclonal anti-LASP-1 antibody (Butt et al, 2003) diluted 1 : 1000 in antibody diluent (DAKO, Hamburg, Germany) or with Ki67 antibody (DAKO) diluted 1 : 100 in antibody diluent at 4 °C overnight followed by EnVision/rabbit detection system (DAKO) for 30 min at RT. 3,3′-Diaminobenzidine (DAB; DAKO) was used as chromogen and sections were counterstained in haematoxylin (Mayers, Sigma, Deisenhofen, Germany), dehydrated through graded ethanol (in the inverse way as described above) and embedded in Entelan (Merck, Darmstadt, Germany). The specificity of the LASP-1 antibody is shown in Supplementary Figure 1.
For PDEF staining, slices were incubated in Beriglobin, diluted 1 : 50 in PBS at RT for 15 min before incubation with polyclonal anti-PDEF antibody (Invitrogen, Karlsruhe, Germany) diluted 1 : 50 in ‘antibody diluent’ (DAKO) at 4 °C overnight. After washing with PBS, the slides were incubated for 15 min with biotinylated secondary antibody followed by 15 min incubation with streptavidin-HRP (both LSAB2 system DAKO). The specificity of the PDEF antibody is shown in Supplementary Figure 2.
Evaluation of immunohistochemical LASP-1 staining and LASP-1-IRS
After staining procedure, the slides were screened and scored as previously described (Grunewald et al, 2007a). To assess the role of LASP-1 in human breast cancer, we examined its expression in 177 breast carcinoma samples from patients selected randomly from January 1985 to December 2007. Semi-quantitative evaluation of LASP-1 immunostaining was carried out by defining the percentage of positive cells and the staining intensity as described below. For positive controls, breast cancer sections previously described as highly LASP-1-positive (Grunewald et al, 2006, 2007a) were used. No staining was observed in negative controls with omitted primary antibody or with pre-immune serum (data not shown).
Scoring of cytosolic LASP-1-expression was carried out in analogy to the scoring of hormone receptor Immune Reactive Score (IRS) ranging from 0 to 12 according to Remmele et al (Remmele and Stegner, 1987), which is used routinely in surgical pathology for the quantification of hormone receptor expression in mammary carcinoma.
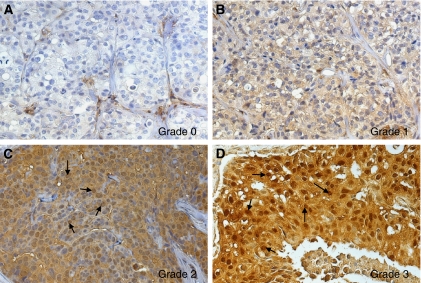
The percentage of LASP-1-postitive stained cells was scored in five grades (grade 0=0–19%, grade 1=20–39%, grade 2=40–59%, grade 3=60–79% and grade 4=80–100% LASP-1-expressing tumour cells) by examining 10 high-power fields ( × 40 magnification) in each tissue sample. In addition, the intensity of LASP-1-expression by the tumour cells was determined (score 0=none, score 1=low, score 2=moderate, score 3=strong). The multiplication of these two grading scores (% LASP-1-positive tumour cells × staining intensity) calculates the immunoreactive score for LASP-1-expression (LASP-1-IRS). Examples for the very heterogeneous LASP-1-expression in invasive breast cancer are given in Figure 1.
Figure 1.
Representative images of heterogeneous LASP-1-expression in human invasive breast cancer. Immunohistochemical staining of LASP-1 (DAB, brown, magnification × 20). (A) No LASP-1-expression (grade 0). (B) Low LASP-1-expression (grade 1). (C) Medium LASP-1-expression (grade 2). (D) High LASP-1-expression (grade 3). Arrows in (C) and (D) point to LASP-1-positive nuclei.
For better statistical discrimination, samples scored with cytosolic LASP-1-IRS <5 were classified as LASP-1-negative, those with LASP-1-IRS >5 as LASP-1-positive.
Nuclear LASP-1-staining was scored by determining percentage of positive nuclei regardless of cytosolic LASP-1-expression and cytosolic staining intensity. Samples were considered as nuclear-positive when 10% or more cells showed nuclear LASP-1 staining. Examples for nuclear LASP-1 staining are observed in Figures 1C and D.
The immunomarkers c-erbB2 (HER-2/neu), oestrogen receptor and progesterone receptor assessed in this study had been previously detected by standard immunohistochemistry and were drawn from the archival database of the Department of Pathology of the University of Wuerzburg.
Scoring of Ki67-positivity
Immunohistochemical scoring was performed by counting 10 randomly selected 40 × high-power fields containing representative sections of tumour and calculated as the percentage of positively stained cells to total cells by counting at least 1000 malignant cells. Ki67 ⩾10% nuclear staining was required for a positive classification (Tan et al, 2005).
Statistical analysis
Associations between nuclear or cytosolic LASP-1-localisation were evaluated by multivariate non-parametric analysis using Fisher's exact (F) and Mann–Whitney (M) test. These tests were conducted using Graph Pad Prism Software for Windows (GraphPad Software, Inc., La Jolla, CA, USA). P<0.05 were regarded as statistically significant.
Tissue preparation, micro-dissection, DNA preparation
In all, 64 formalin-fixed paraffin-embedded breast cancer tissue samples of the patient cohort and 10 control breast tissues were placed onto PEN-membrane coated slides (polyethylene naphthalate; Leica, Wetzlar, Germany) were deparaffinised (two changes of xylene, two changes of 96% ethanol, one change of 70% ethanol, one change with distilled water, 1 min each). After staining with haematoxylin for 90 s and eosin for 60 s, all sections were rinsed 3 × with distilled water. The slides were then air-dried at RT and used for micro-dissection. Incubation and staining times were kept as short as possible to enhance DNA recovery and proteinase K digestion (Godfrey et al, 2000; Ehrig et al, 2001).
Laser capture micro-dissection was performed using the Laser MicroBeam System (Leica LMD 6000; Leica). Tumour tissue (2.0–2.5 mm2) was captured into the lid of a 0.5 ml reaction tube and digested with 30 μl proteinase K digestion buffer (50 mM Tris, pH 8.1; 1 mM ethylenediamine tetraacetic acid; 0.5% Tween 20; 3 mU ml–1 proteinase K). Subsequently, the tubes were closed in this inverted position and incubated for 50–60 h at 37 °C. Undigested debris was removed by centrifugation at 14 000 g for 5 min, and proteinase K was inactivated by incubation at 95 °C for 10 min. The samples can be stored safely for months at −20 °C (Lehmann and Kreipe, 2001).
Quantitative RT–PCR
Quantitative analysis of genomic LASP1 DNA was performed by monitoring the increase in fluorescence of the dye SYBR Green (SYBR Green Supermix, Bio-Rad, Munich, Germany) using the iCycler iQ System (Bio-Rad).
Primers were designed to meet specific criteria by using Primer3 software (http://frodo.wi.mit.edu) (Rozen and Skaletsky, 2000) and were obtained from Operon Biotechnologie GmbH (Cologne, Germany). The sequences of the primers used for LASP1 DNA amplification were 5′-TGTCTCCTGACTGGTTGCGT-3′, and 5′-TGATCTGGTCCTGGGTCTTC-3′. Primers for GAPDH were used as internal reference gene: 5′-ATCAAGAAGGTGGTGAAGCAG-3′ and 5′-TACTCCTTGGAGGCCATGTG-3′.
SYBR Green PCR was performed in optical caps for a 96-well tray (Bio-Rad) using a 25 μl final reaction mixture containing 1 μl of each primer pair (stock 5 μM), 1 μl of the micro-dissected lysed tissue sample, 12.5 μl iQ SYBR Green (Bio-Rad) and sterile water. The reaction mixture was preheated at 95 °C for 5 min, followed by 40 cycles at 95 °C, 57 °C and 72 °C for 30 s each.
All amplification curves generated with SYBR Green from stained tissue showed the typical sigmoid curve. In every run, a negative control was included to exclude false-positive results. Melting curve analysis was implemented to ensure the correct PCR product. Each tissue sample was analysed at least twice for LASP1 and twice for GAPDH.
The relative gene copy number was evaluated on the basis of the threshold cycles (CT values) of the gene of interest CT(LASP1) and of the internal reference gene CT(GAPDH).
The CT(GAPDH)/CT(LASP1) ratio in benign control breast tissues will reflect non-amplified LASP1 conditions. In case of a LASP1 gene amplification in tumour samples, the threshold cycle number will decrease while the values for the CT(GAPDH)/CT(LASP1) ratio will increase. The reference range from 10 micro-dissected normal breast tissues was determined as 0.9699±0.0743. Therefore, the expectation interval can be calculated as: μ±2 s=(0.821278502↔1.118530727) with μ (arithmetic mean) and s (s.d.).
Cell lines and cell culture conditions
Hepatocellular carcinoma cell lines Hep-3B and Hep-G2, breast cancer cell lines BT-20, MCF-7 and MDA-MB-231, urothelial cancer cell lines T24 and RT-4, glioblastoma cell lines U251MG, U138MG and U87MG, medulloblastoma cell lines DAOY and D283 as well as chorioncarcinoma cell lines JEG-3 and JAR were obtained from Cell Line Services (Heidelberg, Germany) and grown in plastic cell culture flasks in a humidified incubator at 37 °C under 5% CO2 atmosphere in RPMI 1640 medium containing 10% heat-inactivated fetal bovine serum (PAA, Linz, Austria) and 1% streptomycin/ampicillin (Invitrogen). Cells were cultured until homogeneous morphology of cells was reached (passage 3–4) because LASP-1 belongs to a group of several differential expressed proteins that are upregulated after later passages (Sun et al, 2006).
p53 Mutations
Cell lines with known p53 mutations are listed in the database: http://p53.free.fr. All mutations result in a non-functional p53 protein: http://p53.iarc.fr.
Cell cycle synchronisation and FACS analysis
BT-20 cells were rendered quiescent (G0) by serum deprivation in RPMI 1640 with 0.1% FCS for 24 h followed by incubation with medium supplemented with 10% FCS to allow cell cycle re-entry in G1. To block S-phase transition, cells were incubated in medium supplemented with 10% FCS and 2 μg ml–1 aphidicolin for 24 h (Sigma). To synchronise the culture at G2/M phase, cells in the S-phase were released in RPMI 1640 medium supplemented with 10% FCS for 12 h.
Cell cycle distribution was monitored by propidium iodide staining and measuring fluorescence in a FACScan 2 (Becton Dickinson, Heidelberg, Germany). BT-20 cells were harvested by trypsination and fixed in 70% ethanol (4 °C) for 1 h followed by incubated in a solution containing 50 μg ml–1 RNase in PBS for 30 min. For staining, 50 μg ml–1 propidium iodide was added for another 30 min. Cells were analysed by FACS, and the proportion in G0/G1, S and G2/M phases was estimated using Modfit cell cycle analysis programme. Measurements were performed on at least three independent synchronisation experiments.
Preparation of nuclear and cytosolic cell fractions
Human breast cancer cell lines were harvested at 80% confluence through trypsination. Isolation of nuclei and cytosol was carried out using NE-PER nuclear and cytoplasmic extraction Reagents (Pierce, Bonn, Germany) following the manufacturer's instructions. Samples were solved in Laemmli sample buffer at a final concentration of 1 × 106 ml–1 and stored at −20 °C before western blot electrophoresis.
Western blot analysis
For western blotting, cells were lysed in Laemmli sample buffer. Equal amounts of protein, according to cell count, were resolved by 12% SDS–PAGE. After blotting on nitrocellulose membrane and blocking with 3% non-fat dry milk in 10 mM Tris, pH 7.5, 100 mM NaCl, 0.1% (w/v) Tween 20, the membrane was incubated with the antibody raised against LASP-1 (1 : 20 000) (Butt et al, 2003) or PDEF antibody (1 : 1000) followed by incubation with horseradish peroxidase-coupled goat anti-rabbit IgG (Bio-Rad), diluted 1 : 5000, and visualised by ECL (Amersham Biosciences, Freiburg, Germany). Quantification of autoradiography signals was carried out by densitometry using the ImageJ software (NIH, Bethesda, MD, USA).
GAPDH (1 : 1000; Santa Cruz, Heidelberg, Germany) was used as a specific cytosolic marker to exclude cytoplasmic contamination of the nuclei preparation. Anti-Lamin A+C antibody (1 : 50; Abcam, Cambridge, UK) served as a specific nuclear marker to exclude nuclear contamination in cytoplasmic cell samples. At least three independent experiments have been carried out and representative results are shown.
Results
Overexpression of LASP-1 in breast cancer is not due to gene amplification
As increasing amounts of contaminating non-malignant cells lead to a significant decrease in detection sensitivity (Kallioniemi et al, 1994), we used individual micro-dissected breast cancer cells to examine the LASP1 copy number in DNA samples from 64 patients with known invasive breast carcinoma selected randomly from January 2000 to December 2007.
We detected only 1 out of 64 tissue samples (1.5%) with a CT(GAPDH)/CT(LASP1) ratio higher than the expectation interval, showing a negligible rate of LASP1 gene amplification. Therefore, the observed overexpression of the LASP-1 protein in more than 55% of human breast cancers (Grunewald et al, 2007a) is likely due to reasons distinct from gene amplification.
LASP-1 overexpression neither correlates with PDEF expression nor with p53 mutations
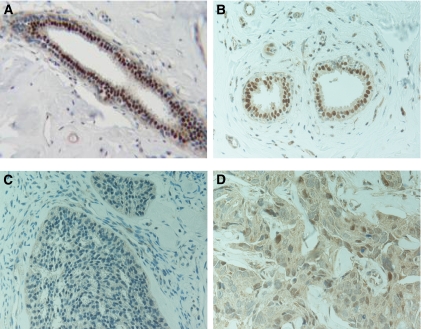
Commonly, the inner layer of benign ductal luminal epithelial cells show a high nuclear PDEF staining while in invasive ductal carcinoma, a weak PDEF staining is detected mainly in the cytosol (Feldman et al, 2003), (Figure 2). To determine whether the reported reciprocal effect of PDEF on LASP-1 in non-invasive and invasive breast cancer cell lines (Turner et al, 2008) is transferable to breast tumour patient samples, we evaluated 35 primary breast cancer tissues for PDEF expression; 17 with known high (⩾8) LASP-1 IRS and 18 specimens with low (⩽3) LASP-1-IRS.
Figure 2.
Immunohistochemical staining of PDEF (DAB, brown, magnification × 400) in benign breast tissue (A and B) and invasive ductal breast cancer samples (C and D) showing a nuclear PDEF staining in normal tissue and a more cytosolic PDEF localisation in tumour cells.
In all 35 tested human breast cancer samples, we observed a comparable cytosolic PDEF localisation without significant differences in staining intensity. Only two out of the tested samples showed in parts additional nuclear PDEF staining. No correlation between the PDEF levels in invasive ductal carcinoma and high or low LASP-1 expression could be detected.
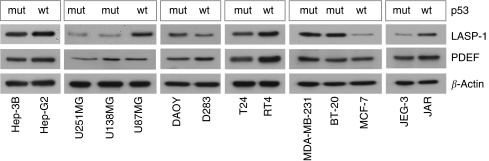
When examining tumour cell lines of different entities, an overall PDEF expression is observed that does not correlate per se with low LASP-1 protein concentration (Figure 3). For example, while the PDEF levels are similar in all three tested breast cancer cell lines (MDA-MB-231, BT-20, MCF-7) LASP-1 expression is only reduced in MCF-7 cells. Analogous differences are also observed with glioblastoma and chorioncarcinoma cell lines (Figure 3).
Figure 3.
PDEF and LASP-1 protein expression in various cancer cell lines with (mut) and without (wt) known p53 mutations that lead to functionally inactive tumour suppressor protein. β-Actin blotting was used as a control for equal protein loading. Cell lines: hepatocellular carcinoma (Hep-G2, Hep-3B), glioblastoma (U251, U13898, U87), medulloblastoma (DAOY, D238), urothelial carcinoma (T24, RT4), breast cancer (MDA-MB-231, BT-20, MCF-7) and chorioncarcinoma (JEG-3, JAR). There is neither a correlation between PDEF and LASP-1 expression nor between LASP-1 protein levels and p53 mutations.
To test whether LASP1 is transcriptionally regulated by p53, we analysed several human cancer cell lines of different tumour entities with and without known p53 mutations for LASP-1-expression by western blot. All mutations result in a functionally inactive tumour suppressor. In summary, the analysis showed no correlation between high LASP-1-expression and p53 mutations (Figure 3). For instance, in spite of the total loss of function because of an additional stop codon in the p53 gene of the urothelial cell line T24, both, p53 wild-type RT-4 and p53 mutant T24 urothelial cancer cell lines express high levels of LASP-1. In contrast, the chorioncarcinoma cell line JEG-3 with a p53 mutation and the breast carcinoma cell line MCF-7-expressing wild-type p53 show low LASP-1-expression.
Nuclear localisation of LASP-1 correlates with poor long-term survival
In an earlier case–control study, a strong cytoplasmic staining for LASP-1 was detected in >55% of the invasive tumours, which correlated significantly with increased tumour size and rate of nodal-positivity. In addition, we observed a distinct nuclear LASP-1-localisation pattern that was absent in benign tissue (Grunewald et al, 2007a).
We therefore performed a retrospective study (January 1985 until December 2007) with samples of 177 archival cases of confirmed histological diagnosis of invasive breast carcinomas to evaluate the long-term survival of breast tumour patients in relation to nuclear and cytoplasmic LASP-1-positivity.
Cytoplasmic LASP-1 protein expression was detected in 95% of the breast carcinomas; thereof 31% showed an additional nuclear LASP-1 staining (Figure 1, Table 1). A low cytoplasmic expression correlated with negative/low nuclear staining and a high cytoplasmic LASP-1-expression with high nuclear localisation (Table 1).
Table 1. Analysis of overall survival in relation to LASP-1 staining intensity.
| LASP-1 staining intensity | No. of patients | Positive nuclear LASP-1 staining | No. of deceased patients | Overall survival (%) |
|---|---|---|---|---|
| Score 0 | 9 | 0 (0%) | 0 | 100 |
| Score 1 | 46 | 6 (13%) | 7 | 84.8 |
| Score 2 | 109 | 46 (42.2%) | 27 | 75.2 |
| Score 3 | 13 | 4 (28.5%) | 4 | 69.2 |
| Total | 177 | 56 (31%) | 38 | 89.6 |
Abbreviation: LASP-1=LIM and SH3 protein 1.
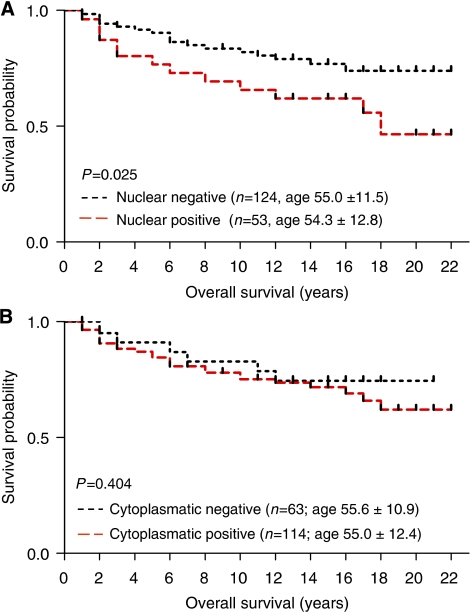
The prognostic effect of nuclear and cytosolic LASP-1 staining was further tested using the Kaplan–Meier survival analysis. There was a significant correlation (P=0.025) between patients with positive nuclear LASP-1 staining and poor overall survival (OS) (Figure 4A, Table 2) while there was no significant association between OS and cytoplasmic LASP-1 staining (Figure 4B, P=0.404). Surprisingly, a relationship between high positive nuclear staining and low grading (P=0.025) was observed, whereas all other clinical parameters analysed (i.e., nodal status, oestrogen and progesterone receptor status, recurrence) did not correlate with nuclear LASP-1-localisation (Table 3).
Figure 4.
Outcome of patients with nuclear and cytosolic LASP-1 localisation. Kaplan–Meier plots measuring patients OS in years against cumulative survival for nuclear (A) and cytosolic (B) LASP-1 staining. The analysis included patients diagnosed from 1985 to 2007 (n=177). Nuclear LASP-1-positivity is associated with poor OS.
Table 2. Analysis of overall survival in relation to nuclear LASP-1-positivity.
| Nuclear LASP-1-positivity | No. of patients | No. of deceased patients | Overall survival (%) |
|---|---|---|---|
| No | 121 | 21 | 82.6 |
| Yes | 56 | 17 | 69.6 |
Abbreviation: LASP-1=LIM and SH3 protein 1.
Table 3. Univariate analysis of positive nuclear LASP-1 staining and clinicopathological parameters.
| Parameters | Positive nuclear LASP-1 staining | P-value |
|---|---|---|
| Nodal status | ||
| N+ (n=83) | 31 (37.3%) | |
| N− (n=94) | 25 (26.6%) | 0.2 (F) |
| Tumour size | ||
| Tis (n=3) | 0 (0.0%) | |
| T1 (n=108) | 37 (34.3%) | |
| T2 (n=51) | 15 (29.4%) | |
| T3 (n=5) | 2 (40.0%) | |
| T4 (n=10) | 2 (20.0%) | 0.69 (M) |
| Metastasis | ||
| M+ (n=6) | 1 (16.7%) | |
| M− (n=171) | 55 (32.2%) | 0.67 (F) |
| Grading | ||
| G1 (n=7) | 4 (57.1%) | |
| G2 (n=69) | 27 (39.1% | |
| G3 (n=45) | 9 (20.0%) | 0.03 (M) |
| Recurrence | ||
| Yes (n=36) | 8 (22.2%) | |
| No (n=87) | 27 (31.0%) | 0.38 (F) |
| ER | ||
| ER+ (n=98) | 28 (28,6%) | |
| ER− (n=62) | 23 (37.1%) | 0.3 (F) |
| Progesteron receptor | ||
| PR+ (n=86) | 22 (25.6%) | |
| PR− (n=71) | 27 (38.0%) | 0.13 (F) |
| HER2/neu | ||
| Her+ (n=27) | 6 (33.3%) | |
| Her− (n=47) | 18 (28.1%) | 0.77 (F) |
Abbreviations: ER=oestrogen receptor; F=Fisher's exact test; LASP-1=LIM and SH3 protein 1; M=Mann–Whitney Test; PR=progesterone receptor.
Statistical significance is assumed when P<0.05.
Rate of nuclear LASP-1-localisation increases during proliferation and augments in G2/M phase
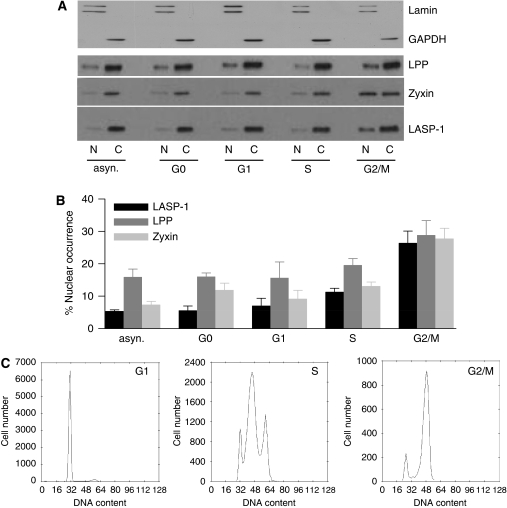
There are two facts regarding LASP-1 that seem to be linked: (a) the prominent nuclear localisation of LASP-1 in primary breast cancer (Figure 1) (Grunewald et al, 2007a) and (b) cell cycle arrest at G2/M accompanied by reduced cell proliferation after knockdown of LASP-1 in breast carcinoma cell lines (Grunewald et al, 2006). Therefore, it was tempting to speculate that the rate of nuclear LASP-1-localisation might be cell cycle dependent. To test this hypothesis, BT-20 cell lysates were subjected to cytoplasmic and nuclear fractioning. Purity of the fractions was confirmed by probing western blot membranes for the nucleus marker Lamin A+B and the cytoplasmic marker GAPDH. Figure 5A shows that there was virtually no cross-contamination.
Figure 5.
Cell cycle-dependent nuclear and cytosolic LASP-1 distribution. (A) Western blotting of nuclear (N) and cytosolic (C) proteins levels of LPP, Zyxin and LASP-1 in BT-20 cells synchronised to G0, G1, S and G2/M phase. Purity of nuclear and cytosolic fractions was controlled by Lamin A+B and GAPDH western blots, respectively. (B) Percentage of nuclear LPP, Zyxin and LASP-1 protein concentration (expressed as percentage of total cellular LPP, Zyxin and LASP-1 protein levels, respectively) plotted against cell cycle phases. (C) Flow-cytometric analysis of cell cycle phases (propidium iodide staining of total DNA).
In cells with an asynchronous cell cycle, in non-proliferating G0 phase cells (resting/senescent) and in G1 phase cells (G1/S-checkpoint), LASP-1 was found primarily within the cytoplasm (95%) (Figures 5A and B). During proliferation (S-phase) the nuclear LASP-1 concentration increased up to 10% and reached a peak at G2/M phase (G2/M) although the overall LASP-1 protein level did not change (data not shown).
As LPP and Zyxin are known binding partners of LASP-1 (Keicher et al, 2004; Li et al, 2004) and are discussed as possible shuttle proteins to transfer LASP-1 into the nucleus we also controlled their distribution during cell cycle phases. Consistently, like LASP-1, both proteins showed a cell phase-dependent nuclear increase in G2/M (Figure 5B) without changes in absolute protein concentration. All cell cycle phases were controlled by flow cytometry (Figure 5C).
To further validate the influence of nuclear LASP-1 occurence on cell proliferation, we quantified the number of positive stained cells for the proliferation marker Ki67 in 30 breast cancer tissue samples with known high LASP-1 IRS⩾8 and either nuclear or cytosolic LASP-1 localisation.
Although only 30.7% of the invasive ductal carcinoma samples with cytosolic LASP-1 expression are positive for Ki67 staining, 68.7% of the breast cancer tissues with nuclear LASP-1 occurrence show positive staining for the proliferation marker Ki67 (χ2 test; P=0.04).
Discussion
The LASP1 gene was initially identified from a cDNA library of metastatic axillary lymph nodes (MLN) from human breast cancer and therefore called MLN50. The gene was mapped to chromosomal region 17q11-q21.3, a region known to contain the c-erbB-2 (HER-2/neu) and the BRCA1 oncogene and to be altered in 20–30% of all breast cancers (Tomasetto et al, 1995a, 1995b). Since its discovery in 1995, several experimental approaches have been carried out to determine the cause of LASP-1 overexpression and its regulatory mechanisms. For instance, LASP-1 overexpression was reported to be due to LASP1 gene amplification detected in 12 out of 98 tested whole breast cancer samples (Bieche et al, 1996) while Tomasetto et al detected an amplification of LASP1 only in one (BT-474) out of eight different breast cancer cell lines (Tomasetto et al, 1995b). Others observed deregulation of normal LASP-1-expression in relation to changes in PDEF and urokinase-type plasminogen activator (uPA) concentration or because of loss of p53 tumour suppressor activity (Turner et al, 2008; Salvi et al, 2009; Wang et al, 2009).
However, in this work, we analysed the expression pattern of LASP-1 in primary invasive breast cancers using micro-dissected tissues. Our data clearly show that the LASP1 gene is not amplified in the vast majority of human breast cancers (only 1 out of 64 cases), suggesting that LASP-1 overexpression is mediated through transcriptional regulation rather than gene amplification. In the context of transcriptional regulation, we revealed that LASP-1 overexpression does not correlate per se with defects in the tumour suppressor protein p53 transcriptionally repressing LASP-1 (Wang et al, 2009). Although the data for the regulation of LASP1 gene expression by p53 are convincing, there are clearly additional mechanisms involved in LASP-1 protein upregulation such as transcriptional cofactors and decay rates than just functional defects in p53.
As for PDEF, we could not confirm an association between low PDEF protein expression and high LASP-1 levels although Turner et al showed that re-expression of PDEF in cells with low PDEF protein expression resulted in reduced LASP-1 levels (Turner et al, 2008).
However, PDEF mRNA concentration and protein expression in breast cancer cell lines is discussed controversially. As reported earlier, PDEF protein detection did not always correspond to PDEF mRNA levels. Although some studies showed increased PDEF mRNA (Turcotte et al, 2007) or protein expression in invasive ductal carcinoma (Sood et al, 2007) others observed reduced protein expression in breast cancer cells (Feldman et al, 2003; Doane et al, 2006; Ghadersohi et al, 2007; Turner et al, 2008). Recently, this discrepancy was explained by the identification of two microRNAs in human breast tumour samples that directly repressed PDEF protein expression in spite of the detection of high PDEF mRNA concentration (Findlay et al, 2008).
In a recent paper by Grunewald et al, LASP-1 was reported to be highly expressed in invasive breast carcinomas compared with fibroadenomas. Strong cytoplasmic staining for LASP-1 was found in 55.4% of the invasive breast tumours (Grunewald et al, 2007a). In addition to the reported localisation at focal contacts and lamellipodia, a perinuclear and nuclear distribution of the protein was observed. These data hint to a potential additional signalling function of LASP-1 as a shuttle protein thereby transducing growth signals from the sites of cellular contacts with the ECM into the nucleus.
In support of this hypothesis, this work shows a cell cycle-dependent increase of nuclear LASP-1 during the mitotic G2/M phase in proliferating tumour cells (Figure 5C) while serum-starved quiescent cells (G0) as well as cells in G1 and S-phase show only minor levels of the protein in the nucleus. Our observations are consistent with earlier data showing a specific cell cycle arrest at G2/M and inhibition of cell proliferation after LASP-1 knockdown in breast and ovarian cancer cell lines (Grunewald et al, 2006, 2007a, 2007b). In reverse, a high LASP-1 concentration in the nucleus would show sustained cell proliferation. In fact, we found that approximately 70% of the patient samples with nuclear LASP-1 staining were positive for the cell proliferation marker Ki67 while only 30% of the patients with cytosolic LASP-1 expression showed positive Ki67 staining.
Consistently, earlier studies revealed a correlation between LASP-1-expression and tumour size as well as nodal-positivity in human breast carcinoma (Grunewald et al, 2007a). The present continuative long-term follow-up strengthens the assumed link between increased nuclear LASP-1-localisation and poor survival of patients with breast cancer suggesting an effect of nuclear LASP-1 on cell proliferation, especially because the absolute amount of cytosolic LASP-1-expression does not correlate with patients' OS.
Unexpectedly, we found a high nuclear localisation of LASP-1 in differentiated G1 tumours while in parallel nuclear LASP-1 abundance was correlated with worse prognosis. It is possible that tumours with a high nuclear LASP-1-expression represent a subgroup with poor survival irrespective of the grading. This could, for example, be due to a decreased response to endocrine or chemotherapeutic treatment. However, the number of available G1 tumours was very low. Therefore, we will not draw definitive conclusions regarding these data.
On the molecular level, the zinc-finger containing LIM-domain of LASP-1 offers a possibility for direct binding to DNA (Hammarstrom et al, 1996). LASP-1 may even form heterodomains to become a nuclear transcription factor (Kadrmas and Beckerle, 2004).
Although LASP-1 sequence analysis revealed no nuclear localisation signal, the classical import pathway for the nucleus (Kutay and Guttinger, 2005), LASP-1 binds to the well-characterised shuttle proteins and transcription factors LPP and Zyxin that are upregulated in a wide variety of human cancers (Beckerle, 1997; Petit et al, 2003; Keicher et al, 2004; Li et al, 2004; Grunewald et al, 2009). For Zyxin, it is known that during mitosis a fraction of the cytoplasmic-dispersed protein becomes phosphorylated (most likely by Cdc2 kinase) and associates with the tumour suppressor h-warts (LATS1), a key governor of G2/M-progression, at the mitotic apparatus (Hirota et al, 2000).
Our data suggest that pathophysiological localisation of LASP-1 in the nucleus of malignant cells may induce mitosis and thereby enhance cell proliferation, possibly in concert with Zyxin and LPP. Further work will be needed to identify the nuclear shuttle partner(s) of LASP-1, the mechanism of nuclear translocation and the regulation of cell cycle progression.
The present continuative long-term follow-up provides evidence for the relation of increased nuclear LASP-1-localisation and poor survival of patients leading to the question whether nuclear LASP-1-positivity defines a subgroup of patients with unfavourable prognosis that is not responding to conventional treatment approaches. Future work is on the way to elucidate the precise molecular and clinical effect of LASP-1 nuclear overexpression.
Acknowledgments
We thank Dr Christian Korn for his help with the LEICA laser micro-dissection microscope and Petra Thalheimer for technical assistance.
Footnotes
Supplementary Information accompanies the paper on British Journal of Cancer website (http://www.nature.com/bjc)
Supplementary Material
References
- Beckerle MC (1997) Zyxin: zinc fingers at sites of cell adhesion. Bioessays 19: 949–957 [DOI] [PubMed] [Google Scholar]
- Bieche I, Tomasetto C, Regnier CH, Moog-Lutz C, Rio MC, Lidereau R (1996) Two distinct amplified regions at 17q11-q21 involved in human primary breast cancer. Cancer Res 56: 3886–3890 [PubMed] [Google Scholar]
- Butt E, Gambaryan S, Gottfert N, Galler A, Marcus K, Meyer HE (2003) Actin binding of human LIM and SH3 protein is regulated by cGMP- and cAMP-dependent protein kinase phosphorylation on serine 146. J Biol Chem 278: 15601–15607 [DOI] [PubMed] [Google Scholar]
- Chew CS, Chen X, Parente Jr JA, Tarrer S, Okamoto C, Qin HY (2002) Lasp-1 binds to non-muscle F-actin in vitro and is localized within multiple sites of dynamic actin assembly in vivo. J Cell Sci 115: 4787–4799 [DOI] [PubMed] [Google Scholar]
- Dalton LW, Pinder SE, Elston CE, Ellis IO, Page DL, Dupont WD, Blamey RW (2000) Histologic grading of breast cancer: linkage of patient outcome with level of pathologist agreement. Mod Pathol 13: 730–735 [DOI] [PubMed] [Google Scholar]
- Dimova I, Orsetti B, Negre V, Rouge C, Ursule L, Lasorsa L, Dimitrov R, Doganov N, Toncheva D, Theillet C (2009) Genomic markers for ovarian cancer at chromosomes 1, 8 and 17 revealed by array CGH analysis. Tumori 95: 357–366 [DOI] [PubMed] [Google Scholar]
- Doane AS, Danso M, Lal P, Donaton M, Zhang L, Hudis C, Gerald WL (2006) An estrogen receptor-negative breast cancer subset characterized by a hormonally regulated transcriptional program and response to androgen. Oncogene 25: 3994–4008 [DOI] [PubMed] [Google Scholar]
- Ehrig T, Abdulkadir SA, Dintzis SM, Milbrandt J, Watson MA (2001) Quantitative amplification of genomic DNA from histological tissue sections after staining with nuclear dyes and laser capture microdissection. J Mol Diagn 3: 22–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feldman RJ, Sementchenko VI, Gayed M, Fraig MM, Watson DK (2003) Pdef expression in human breast cancer is correlated with invasive potential and altered gene expression. Cancer Res 63: 4626–4631 [PubMed] [Google Scholar]
- Findlay VJ, Turner DP, Moussa O, Watson DK (2008) MicroRNA-mediated inhibition of prostate-derived Ets factor messenger RNA translation affects prostate-derived Ets factor regulatory networks in human breast cancer. Cancer Res 68: 8499–8506 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ghadersohi A, Pan D, Fayazi Z, Hicks DG, Winston JS, Li F (2007) Prostate-derived Ets transcription factor (PDEF) downregulates survivin expression and inhibits breast cancer cell growth in vitro and xenograft tumor formation in vivo. Breast Cancer Res Treat 102: 19–30 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Godfrey TE, Kim SH, Chavira M, Ruff DW, Warren RS, Gray JW, Jensen RH (2000) Quantitative mRNA expression analysis from formalin-fixed, paraffin-embedded tissues using 5′ nuclease quantitative reverse transcription-polymerase chain reaction. J Mol Diagn 2: 84–91 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald TG, Butt E (2008) The LIM and SH3 domain protein family: structural proteins or signal transducers or both? Mol Cancer 7: 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald TG, Kammerer U, Kapp M, Eck M, Dietl J, Butt E, Honig A (2007a) Nuclear localization and cytosolic overexpression of LASP-1 correlates with tumor size and nodal-positivity of human breast carcinoma. BMC Cancer 7: 198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald TG, Kammerer U, Schulze E, Schindler D, Honig A, Zimmer M, Butt E (2006) Silencing of LASP-1 influences zyxin localization, inhibits proliferation and reduces migration in breast cancer cells. Exp Cell Res 312: 974–982 [DOI] [PubMed] [Google Scholar]
- Grunewald TG, Kammerer U, Winkler C, Schindler D, Sickmann A, Honig A, Butt E (2007b) Overexpression of LASP-1 mediates migration and proliferation of human ovarian cancer cells and influences zyxin localisation. Br J Cancer 96: 296–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grunewald TG, Pasedag SM, Butt E (2009) Cell adhesion and transcriptional activity—defining the role of the novel protooncogene LPP. Transl Oncol 2: 107–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammarstrom A, Berndt KD, Sillard R, Adermann K, Otting G (1996) Solution structure of a naturally-occurring zinc-peptide complex demonstrates that the N-terminal zinc-binding module of the Lasp-1 LIM domain is an independent folding unit. Biochemistry 35: 12723–12732 [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg RA (2000) The hallmarks of cancer. Cell 100: 57–70 [DOI] [PubMed] [Google Scholar]
- Hirota T, Morisaki T, Nishiyama Y, Marumoto T, Tada K, Hara T, Masuko N, Inagaki M, Hatakeyama K, Saya H (2000) Zyxin, a regulator of actin filament assembly, targets the mitotic apparatus by interacting with h-warts/LATS1 tumor suppressor. J Cell Biol 149: 1073–1086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jemal A, Siegel R, Ward E, Murray T, Xu J, Thun MJ (2007) Cancer statistics, 2007. CA Cancer J Clin 57: 43–66 [DOI] [PubMed] [Google Scholar]
- Kadrmas JL, Beckerle MC (2004) The LIM domain: from the cytoskeleton to the nucleus. Nat Rev Mol Cell Biol 5: 920–931 [DOI] [PubMed] [Google Scholar]
- Kallioniemi OP, Kallioniemi A, Piper J, Isola J, Waldman FM, Gray JW, Pinkel D (1994) Optimizing comparative genomic hybridization for analysis of DNA sequence copy number changes in solid tumors. Genes Chromosomes Cancer 10: 231–243 [DOI] [PubMed] [Google Scholar]
- Keicher C, Gambaryan S, Schulze E, Marcus K, Meyer HE, Butt E (2004) Phosphorylation of mouse LASP-1 on threonine 156 by cAMP- and cGMP-dependent protein kinase. Biochem Biophys Res Commun 324: 308–316 [DOI] [PubMed] [Google Scholar]
- Kutay U, Guttinger S (2005) Leucine-rich nuclear-export signals: born to be weak. Trends Cell Biol 15: 121–124 [DOI] [PubMed] [Google Scholar]
- Lehmann U, Kreipe H (2001) Real-time PCR analysis of DNA and RNA extracted from formalin-fixed and paraffin-embedded biopsies. Methods (San Diego, CA) 25: 409–418 [DOI] [PubMed] [Google Scholar]
- Li B, Zhuang L, Trueb B (2004) Zyxin interacts with the SH3 domains of the cytoskeletal proteins LIM-nebulette and Lasp-1. J Biol Chem 279: 20401–20410 [DOI] [PubMed] [Google Scholar]
- Mauriac L, Debled M, MacGrogan G (2005) When will more useful predictive factors be ready for use? Breast (Edinburgh, Scotland) 14: 617–623 [DOI] [PubMed] [Google Scholar]
- Nakagawa H, Terasaki AG, Suzuki H, Ohashi K, Miyamoto S (2006) Short-term retention of actin filament binding proteins on lamellipodial actin bundles. FEBS Lett 580: 3223–3228 [DOI] [PubMed] [Google Scholar]
- Petit MM, Meulemans SM, Van de Ven WJ (2003) The focal adhesion and nuclear targeting capacity of the LIM-containing lipoma-preferred partner (LPP) protein. J Biol Chem 278: 2157–2168 [DOI] [PubMed] [Google Scholar]
- Remmele W, Stegner HE (1987) Recommendation for uniform definition of an immunoreactive score (IRS) for immunohistochemical estrogen receptor detection (ER-ICA) in breast cancer tissue. Pathologe 8: 138–140 [PubMed] [Google Scholar]
- Ross JS, Fletcher JA (1999) The HER-2/neu oncogene: prognostic factor, predictive factor and target for therapy. Semin Cancer Biol 9: 125–138 [DOI] [PubMed] [Google Scholar]
- Rozen S, Skaletsky H (2000) Primer3 on the WWW for general users and for biologist programmers. Methods Mol Biol (Clifton, NJ) 132: 365–386 [DOI] [PubMed] [Google Scholar]
- Salvi A, Bongarzone I, Micciche F, Arici B, Barlati S, De Petro G (2009) Proteomic identification of LASP-1 down-regulation after RNAi urokinase silencing in human hepatocellular carcinoma cells. Neoplasia (New York, NY) 11: 207–219 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schreiber V, Moog-Lutz C, Regnier CH, Chenard MP, Boeuf H, Vonesch JL, Tomasetto C, Rio MC (1998) Lasp-1, a novel type of actin-binding protein accumulating in cell membrane extensions. Mol Med 4: 675–687 [PMC free article] [PubMed] [Google Scholar]
- Singletary SE, Connolly JL (2006) Breast cancer staging: working with the sixth edition of the AJCC cancer staging manual. CA Cancer J Clin 56: 37–47; quiz 50–1 [DOI] [PubMed] [Google Scholar]
- Sood AK, Saxena R, Groth J, Desouki MM, Cheewakriangkrai C, Rodabaugh KJ, Kasyapa CS, Geradts J (2007) Expression characteristics of prostate-derived Ets factor support a role in breast and prostate cancer progression. Human Pathol 38: 1628–1638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun HJ, Bahk YY, Choi YR, Shim JH, Han SH, Lee JW (2006) A proteomic analysis during serial subculture and osteogenic differentiation of human mesenchymal stem cell. J Orthop Res 24: 2059–2071 [DOI] [PubMed] [Google Scholar]
- Tan PH, Bay BH, Yip G, Selvarajan S, Tan P, Wu J, Lee CH, Li KB (2005) Immunohistochemical detection of Ki67 in breast cancer correlates with transcriptional regulation of genes related to apoptosis and cell death. Mod Pathol 18: 374–381 [DOI] [PubMed] [Google Scholar]
- Tomasetto C, Moog-Lutz C, Regnier CH, Schreiber V, Basset P, Rio MC (1995a) Lasp-1 (MLN 50) defines a new LIM protein subfamily characterized by the association of LIM and SH3 domains. FEBS Lett 373: 245–249 [DOI] [PubMed] [Google Scholar]
- Tomasetto C, Regnier C, Moog-Lutz C, Mattei MG, Chenard MP, Lidereau R, Basset P, Rio MC (1995b) Identification of four novel human genes amplified and overexpressed in breast carcinoma and localized to the q11-q21.3 region of chromosome 17. Genomics 28: 367–376 [DOI] [PubMed] [Google Scholar]
- Turcotte S, Forget MA, Beauseigle D, Nassif E, Lapointe R (2007) Prostate-derived Ets transcription factor overexpression is associated with nodal metastasis and hormone receptor positivity in invasive breast cancer. Neoplasia (New York, NY) 9: 788–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner DP, Findlay VJ, Kirven AD, Moussa O, Watson DK (2008) Global gene expression analysis identifies PDEF transcriptional networks regulating cell migration during cancer progression. Mol Biol Cell 19: 3745–3757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang B, Feng P, Xiao Z, Ren EC (2009) LIM and SH3 protein 1 (Lasp1) is a novel p53 transcriptional target involved in hepatocellular carcinoma. J Hepatol 50: 528–537 [DOI] [PubMed] [Google Scholar]
- Widakowich C, de Azambuja E, Gil T, Cardoso F, Dinh P, Awada A, Piccart-Gebhart M (2007) Molecular targeted therapies in breast cancer: where are we now? Int J Biochem Cell Biol 39: 1375–1387 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.