Abstract
The effect of radiation on the mitochondrial genome in vivo is largely unknown. Though mitochondrial DNA (mtDNA) is vital for cellular survival and proliferation, it has little DNA repair machinery compared with nuclear DNA (nDNA). A better understanding of how radiation affects mtDNA should lead to new approaches for radiation protection. We have developed a new system using real-time PCR that sensitively detects the change in copy number of mtDNA compared with nDNA. In each sample, the DNA sequence coding 18S rRNA served as the nDNA reference in a run simultaneously with a mtDNA sequence. Small bowel collected 24 hours after 2 Gy or 4 Gy total body irradiation (TBI) exhibited increased levels of mtDNA compared with control mice. A 4 Gy dose produced a greater effect than 2 Gy. Similarly, in bone marrow collected 24 hours after 4 Gy or 7 Gy TBI, 7 Gy produced a greater response than 4 Gy. As a function of time, a greater effect was seen at 48 hours compared with 24 hours. In conclusion, we found that radiation increased the ratio of mtDNA:nDNA and that this effect seems to be tissue independent and seems to increase with radiation dose and duration following radiation exposure.
1. INTRODUCTION
Breakage of cellular DNA following radiation is a dose dependent phenomenon and occurs in both the nuclear and extra-nuclear DNA. Thus, while the vast majority of DNA is located in the nucleus (nDNA), on a mass basis, mitochondrial DNA (mtDNA) is equally affected. After radiation damage, the nuclear DNA has many advantages over mtDNA. These have been reviewed previously1 and include 1) rapid and efficient double strand break repair,1, 2 and 2) rapid and near perfect single strand break repair.3 Following irradiation, mtDNA has few repair mechanisms and continued mitochondrial function is preserved primarily due to its high copy number.4, 5
The cellular response to radiation with regard to the protection of mtDNA has rarely been studied. It is predictable that unchecked mtDNA damage will prevent the replication of properly functioning mitochondria over time, leading to mitochondrial depletion, and then late cell dysfunction.6 The latter would be most pronounced for long-lived cell types such as myocardial, renal, or neuronal cells. Consistent with this hypothesis, these tissues begin to demonstrate serious dysfunction 2 to 8 years after radiation exposure.7 Thus, as a means of self-protection, cells will want to respond to radiation to reduce the frequency and delay the onset of lethal radiation damage to the mitochondria. One such mechanism is enhanced replication of mtDNA.
In this study, we examined the early effects of radiation on the mtDNA copy number of different cell types using doses similar to those that might be experienced by bioterror victims or patients undergoing a bone marrow transplant. Two sets of primers were designed: one set coding for the 12S rRNA gene of the murine mitochondrial genome, and the other set coding for the 18S rRNA gene, an abundant housekeeping gene in the nDNA. The ratio of these two gene frequencies was compared at different times following different radiation doses to look for evidence of enhanced replication of mtDNA following irradiation.
2. MATERIALS AND METHODS
2.1. Animals and Treatments
Male Balb/c mice, 6 to 8 weeks old, were divided into groups of 5 animals. Control mice were not irradiated, while other groups received total body irradiation (TBI) via a J. L. Shepherd Irradiator with a 137Cs γ-ray source. The dose rate was 1.84 Gy/min and homogeneity was better than ±6.5%. The mice were then sacrificed at 24 or 48 hours post-exposure. Mice were housed in a pathogen-free barrier facility. All protocols were in accordance with USPHS guidelines and approved by the University of Rochester Committee on Animal Research.
2.2. DNA Extraction from Mouse Tissues
The small bowel and bone marrow tissues were collected and frozen immediately at -70°C until use. Total DNA was extracted from these tissues by standard proteolytic digestion followed by phenol/chloroform/isoamyl alcohol purification. Absorbance at 260 nm was used for quantification of the extracted DNA.
2.3. Real-time Quantitative PCR to Estimate mtDNA
The PCR primers for detecting the 12S rRNA gene of murine mitochondrial genome were designed on the basis of the GenBank nucleotide sequence (NC_006914, Mus musculus domesticus mitochondrion). The sequences for the primers were 5’-ACC GCG GTC ATA CGA TTA AC-3’ (forward) and 5’-CCC AGT TTG GGT CTT AGC TG-3’ (reverse), producing a 177 bp amplicon. The primer sequences amplifying mouse nuclear 18S rRNA gene are 5’-CGC GGT TCT ATT TTG TTG GT-3’ (forward) and 5’-AGT CGG CAT CGT TTA TGG TC-3’ (reverse), which produce a 219 bp product. The real-time quantitative PCR was carried out using separate tubes for mtDNA and nDNA amplification. Each tube contained 30 ng of total DNA from the same extract as well as 20 μl of reaction mixture consisting of 2 × iQ™ SYBR Green Supermix (1 × final), and one pair of the primers directed against either mtDNA or nDNA, with each primer being 0.5 μM. Each reaction was subjected to an initial denaturation of 5 min at 95°C followed by 40 amplification cycles of denaturation at 95°C for 30 s, 51°C to anneal for 20 s, and 1 min of extension at 72°C. A Bio-Rad i-cycler iQ real-time PCR detection system was employed to run the reactions. A conventional PCR was first run to develop the cycling conditions using the same primers. PCR assays were performed in duplicate or triplicate for each DNA sample. The cycle number (Ct) at which the fluorescent signal of a given reaction crossed the threshold value was used as a basis for quantification of mtDNA and nDNA copy numbers. The difference in Ct and the assumption of a doubling of DNA per cycle were used to calculate the mtDNA:nDNA ratio.
3. RESULTS AND DISCUSSION
3.1. Optimization of Real-Time Quantitative PCR
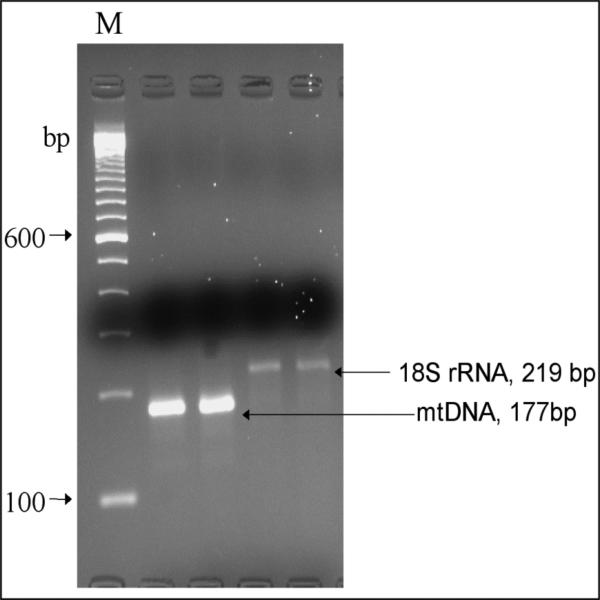
Conventional PCR was first performed to optimize the real-time PCR conditions. Duplicate PCR were run using the mtDNA and nDNA primers added to purified mouse small bowel DNA (Figure 1). As predicted, the size of the amplicon derived from the mtDNA was 177 bp, and from the nDNA is 219 bp. The band size and intensity were visualized with ethidium bromide after electrophoresis in 4% agarose gel; analysis indicated a high mtDNA:nDNA ratio.
Figure 1.
Conventional PCR for mtDNA and nDNA. PCR conditions were optimized to produce amplicons of the expected size and intensity.
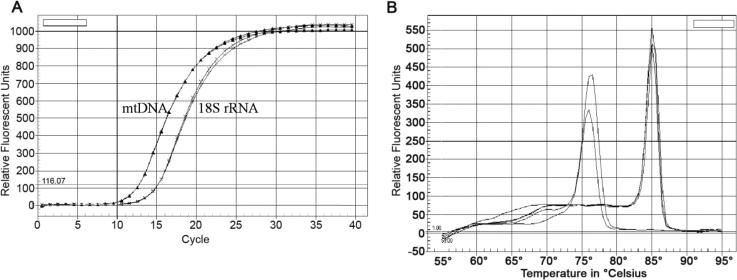
The real-time PCR amplification plot in Figure 2A shows the typical two phases, an exponential phase followed by a plateau phase. A threshold is set during the early exponential phase, as shown. The lower number of cycles needed to reach the threshold level indicates a greater copy number of the mtDNA compared with the nDNA. The 18S rRNA gene is present in multiple copies in the nuclear genome, and therefore the ratios obtained by this assay are not meant to be equivalent to the average copies of mtDNA per cell.8 Melt curves are shown in Figure 2B. The expected single, narrow peaks indicate that the mtDNA and nDNA products are sufficiently pure and specific. Taken together, the data confirm that the real-time PCR conditions are satisfactorily optimized and can be used to determine relative changes in copy numbers of the respective transcripts.
Figure 2.
Optimized real-time quantitative PCR. A. Sample amplification curves obtained using the real-time quantitative PCR system for mtDNA and nDNA genes. B. Sample melt-curve analysis of mtDNA and nDNA products from a SYBR Green I assay.
3.2. Dose-Dependent Increase of mtDNA Copy Number in Small Bowel
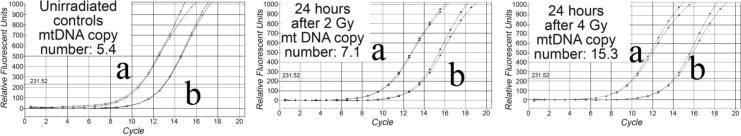
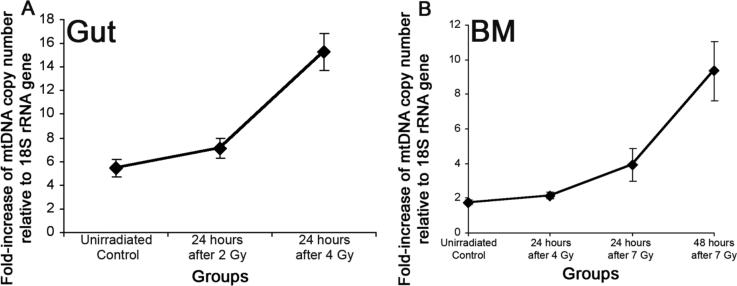
The small bowel mucosa is a rapidly renewing tissue. Within a few days of radiation there is complete replacement of the entire luminal epithelium.9 Thus loss of mtDNA could seriously impact recovery given the potential for hemi-depletion with each cell division. It was therefore interesting to see how mtDNA might respond to low dose radiation. To test this, radiation doses that have no obvious histological effects were used. The small bowel tissues were collected from mice 24 hr after 0 Gy, 2 Gy, or 4 Gy irradiation. The total DNA was extracted from each sample and subjected to real-time PCR. Each cycle leads to almost a doubling in DNA copies, thus the difference between cycle numbers measured at the threshold (designated ß) can be used to calculate the relative ratio of mtDNA:nDNA using the equation: 2ß. At 24 hours after irradiation there was an increase in the mtDNA:nDNA ratio compared with non-irradiated tissues (Figures 3 and 4A). Tissues irradiated to 4 Gy indicated a clear increase in copy ratio compared with 2 Gy, consistent with a dose response. This result is also consistent with a compensatory replication of mtDNA to replace lost or damaged mtDNA. Other explanations include a lowering of the copy number of chromosomes per cell by redistribution of the cell cycle; however, this is very unlikely since the proportion of cells in S-phase at 24 hr after irradiation is stable or moderately increased, which would lead to a decrease rather than the observed increase in mtDNA:nDNA ratio.
Figure 3.
Representative real-time PCR of mtDNA and nDNA extracted from small bowel of mice 24 hours after 2 or 4 Gy TBI. (a) denotes change in relative copy number of mtDNA; (b) denotes change of relative copy number of nDNA.
Figure 4.
A: Increased mtDNA in the irradiated gut is dose and time dependent. Mice were exposed to 2 or 4 Gy TBI. The small bowel was harvested at 24 hours. B: Increased mtDNA in the irradiated BM is dose- and time dependent. Mice were exposed to 4 or 7 Gy TBI. The BM was harvested at either 24 or 48 hours. Values are mean ± 1 SD.
3.3. Dose- and Time-Dependent Increase in mtDNA Copy Number in Bone Marrow
Bone marrow (BM) is probably the most sensitive organ to radiation. Within a few days of TBI, the bone marrow has only a few microscopically detectable hematopoietic cells, yet within two or three weeks of sublethal exposure, the bone marrow repopulates. This extraordinary cellular replication must include a matching mtDNA replacement. Damage to bone marrow mtDNA could otherwise lead to devastating replication-related hemi-depletion. To examine for changes in mtDNA of the bone marrow following irradiation, animals were given sublethal TBI (4 Gy) or near LD50/30 doses (7 Gy). Marrow was harvested at 24 or 48 hours after exposure and analyzed as above. Again the copy ratio of mtDNA in BM of mice increased after 4 Gy.
As in the previous small bowel studies, the amplification of mtDNA seems to be dose dependent, here 7 Gy produced a higher mtDNA:nDNA ratio. The ratio seems to increase further at 48 hr compared to 24 hr (Figure 4B).
4. CONCLUSIONS
We report a set of primers and procedures that can be used to measure relative copy number of mtDNA to nDNA, and which can be used on DNA purified from murine tissues. Sequence modification would make this technology applicable to other animal tissues. Further, we demonstrate that small bowel and bone marrow collected from irradiated mice have an increase in mtDNA copy number after irradiation when measured at 24 or 48 hours and that the copy number may increase with radiation dose. These results are consistent with our previous hypothesis that some radiation effects are due to damage of mitochondrial DNA offset in part by adaptive replication of mtDNA.1, 10
5. ACKNOWLEDGEMENTS
This research was supported in part by the Center for Medical Countermeasures Against Radiation Program, U19-AI067733, National Institute of Allergy and Infectious Diseases. The authors wish to thank Amy K. Huser for editorial assistance.
REFERENCES
- 1.Okunieff P, Swarts S, Keng P, Sun W, Wang W, Kim J, Yang S, Zhang H, Liu C, Williams JP, Huser AK, Zhang L. Antioxidants reduce consequences of radiation exposure. Adv. Exp. Med. Biol. 2007 doi: 10.1007/978-0-387-74911-2_20. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iliakis G, Wang H, Perrault AR, Boecker W, Rosidi B, Windhofer F, Wu W, Guan J, Terzoudi G, Pantelias G. Mechanisms of DNA double strand break repair and chromosome aberration formation. Cytogenet. Genome Res. 2004;104(1-4):14–20. doi: 10.1159/000077461. [DOI] [PubMed] [Google Scholar]
- 3.Purkayastha S, Milligan JR, Bernhard WA. On the chemical yield of base lesions, strand breaks, and clustered damage generated in plasmid DNA by the direct effect of x rays. Radiat. Res. 2007;168(3):357–366. doi: 10.1667/RR0964.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Larsen NB, Rasmussen M, Rasmussen LJ. Nuclear and mitochondrial DNA repair: similar pathways? Mitochondrion. 2005;5(2):89–108. doi: 10.1016/j.mito.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 5.Croteau DL, Stierum RH, Bohr VA. Mitochondrial DNA repair pathways. Mutat. Res. 1999;434(3):137–148. doi: 10.1016/s0921-8777(99)00025-7. [DOI] [PubMed] [Google Scholar]
- 6.Weissman L, de Souza-Pinto NC, Stevnsner T, Bohr VA. DNA repair, mitochondria, and neurodegeneration. Neuroscience. 2007;145(4):1318–1329. doi: 10.1016/j.neuroscience.2006.08.061. [DOI] [PubMed] [Google Scholar]
- 7.Gaya AM, Ashford RF. Cardiac complications of radiation therapy. Clin. Oncol. (R. Coll. Radiol.) 2005;17(3):153–159. doi: 10.1016/j.clon.2004.09.016. [DOI] [PubMed] [Google Scholar]
- 8.Bross K, Krone W. On the number of ribosomal RNA genes in man. Humangenetik. 1972;14(2):137–141. doi: 10.1007/BF00273298. [DOI] [PubMed] [Google Scholar]
- 9.Denham JW, Hauer-Jensen M, Kron T, Langberg CW. Treatment-time-dependence models of early and delayed radiation injury in rat small intestine. Int. J. Radiat. Oncol. Biol. Phys. 2000;48(3):871–887. doi: 10.1016/s0360-3016(00)00708-2. [DOI] [PubMed] [Google Scholar]
- 10.Wolff S. Environ. Health. Perspect. Vol. 106. Suppl 1: 1998. The adaptive response in radiobiology: evolving insights and implications; pp. 277–283. [DOI] [PMC free article] [PubMed] [Google Scholar]