Abstract
Objectives
Our aims were to identify and functionally characterize coding region nonsynonymous single nucleotide polymorphisms in the hepatic efflux transporter, bile salt export pump (BSEP; ABCB11) and to assess interindividual variability in BSEP expression.
Methods
We identified 24 single nucleotide polymorphisms, including nine nonsynonymous variants, in ABCB11 from genomic DNA of approximately 250 ethnically diverse healthy individuals using denaturing high-performance liquid chromatography analysis and DNA sequencing. Wild type and variant BSEP were generated and functionally characterized for taurocholate transport activity in vitro in HeLa cells using a recombinant vaccinia-based method. BSEP expression was assessed by real-time mRNA analysis, western blot analysis, and immunofluorescence confocal microscopy.
Results
For the most part, polymorphisms were rare and ethnicity dependent. In-vitro functional studies revealed several rare variants, including 616A>G, 1674G>C, 1772A>G, and 3556G>A, to be associated with significantly impaired taurocholate transport activity while the 890A>G variant trended towards impaired function but was not statistically significant. The 3556G>A variant was associated with reduced cell surface – total protein expression compared with wild-type BSEP. Expression of BSEP by mRNA and protein analysis was determined from a bank of human liver samples. Wide interindividual variability was noted in both mRNA (19-fold) and protein (31-fold) expression levels. The common variant 1331T>C was associated with significantly reduced hepatic BSEP mRNA levels.
Conclusion
Accordingly, our study indicates that there are functionally relevant polymorphisms in ABCB11, which may be of potential relevance in the predisposition to acquired liver disorders such as drug-induced cholestasis.
Keywords: ABCB11, bile acid, bile salt export pump, cholestasis, pharmacogenetics, polymorphism, transporter
Introduction
Bile formation and secretion are a critical function of mammalian liver and essential for enabling a number of physiologic functions such as intestinal digestion and absorption of lipids, elimination of toxins, carcinogens, drugs and other xenobiotic compounds, and excretion of endobiotics like cholesterol, bilirubin, and hormones [1]. Bile acids, synthesized from the enzymatic catabolism of cholesterol, are the major solutes in bile, essential for the maintenance of bile flow and biliary lipid secretion [2]. In fact, the vectorial excretion of bile salts from blood into bile, also known as bile salt-dependent bile flow, represents the major driving force for bile flow. Bile secretion across the canalicular membrane of hepatocytes represents the rate-limiting step in bile formation and consists primarily of active transport mechanisms driven by ATP hydrolysis as biliary constituents are often translocated against steep concentration gradients in the range of 100–1000-fold [3,4].
The efficient biliary excretion of monovalent bile acids across the canalicular membrane of the hepatocyte is mediated by the bile salt export pump (BSEP; ABCB11), an ATP-binding cassette (ABC) transmembrane transporter. Originally identified from pig liver partial cDNA sequence, BSEP had been initially named sister of P-glycoprotein (Spgp) because of its homology to P-gp (50% sequence identity) [5]. Based on this sequence, a full-length homolog of Spgp was cloned from rat liver. In membrane vesicles from transfected insectderived Sf9 cells, the rat Spgp was shown to exhibit a 5-fold stimulation of ATP-dependent taurocholate transport compared with control vesicles, indicating its true function as a bile salt transporting ABC protein [6]. Subsequent immunogold labeling and in-situ immunofluorescence of rat liver showed selective localization of the 160λkDa protein to the canalicular membrane of the hepatocyte [6]. Accordingly, it became clear that Spgp is the major canalicular BSEP of mammalian liver and it was thus renamed BSEP. Rat BSEP is highly homologous to the human ortholog, with 82.3% sequence identity and 88% amino acid similarity [6,7]. In-vitro studies evaluating human BSEP expressed in membrane vesicles confirmed bile salts to be its major substrates [8,9]. Although bile salts are its primary substrates, BSEP has also been shown to transport a limited number of nonbile acid substrates, including vinblastine, calcein-AM, and pravastatin [10,11].
The critical importance of BSEP to normal human hepatic function is illustrated by the progressive familial intrahepatic cholestases (PFIC), a group of rare inherited disorders characterized by progressive liver disease that typically manifest in childhood as impaired bile flow in the absence of hepatobiliary structural abnormality. Loss of function mutations in ABCB11 has been identified in patients with PFIC type 2 (PFIC2), typically resulting in absence of hepatic BSEP expression [7,12]. These infants typically present with severe jaundice, hepatomegaly, failure to thrive, and pruritus [13,14]. Laboratory studies show direct hyperbilirubinemia, elevated serum aminotransferases, and paradoxically normal serum gamma glutamyl transpeptidase and cholesterol concentrations [15]. Biliary bile acid concentrations are less than 1% of the normal. There is a rapid progressive course to cirrhosis and liver failure, which can only be cured by liver transplantation. Mechanisms for loss of function associated with PFIC2 mutations have been studied in vitro and include impaired trafficking of protein to the membrane, protein misfolding, loss of substrate recognition, or increased proteasomal degradation [16–20].
Although complete loss of function BSEP mutations as seen with PFIC2 is very rare, more recently genetic heterogeneity has been noted in the BSEP gene with the identification of coding region polymorphisms identified from genomic DNA of ethnically diverse healthy individuals (http://www.pharmgkb.org). Several polymorphisms are common (>10% allele frequency) and, in general, allele frequencies tended to be ethnic dependent. Indeed, recent studies would suggest that additional mutations and/or polymorphisms in ABCB11 may be associated with various disease processes such as benign recurrent intrahepatic cholestasis (BRIC), intrahepatic cholestasis of pregnancy (ICP), and risk for drug-induced cholestasis (DC) [21–25]. A recent study suggested aberrant splicing events and mechanisms that promote exon skipping might explain defects associated with some BSEP mutations and/or polymorphic variants [6]. However, to date, there has been no comprehensive study of the functional relevance of naturally occurring polymorphisms in BSEP to protein function. Clearly, functionally relevant BSEP polymorphisms, if found to be present, would significantly add to our knowledge of the genetic basis of altered bile acid physiology, hepatocyte function, and risk for cholestatic disorders including drug-induced liver injury. In this study we report the systematic functional characterization of nonsynonymous polymorphisms in human BSEP.
Methods
Materials
[3H] taurocholate (3.4 Ci/mmol, >97% purity) and unlabeled taurocholate were purchased from PerkinElmer Life Sciences (Boston, Massachusetts, USA). Recombinant vaccinia virus containing the T7-RNA polymerase gene (vtf-7) was a gift provided by Dr Bernard Moss (National Institutes of Health, Bethesda, Maryland, USA). The pEF6/V5-His-TOPO expression vector and monoclonal mouse Anti-V5 antibody were purchased from Invitrogen Corporation (Carlsbad, California, USA). Human liver samples were kindly provided by Dr F.P. Guengerich at Vanderbilt University. All other chemical and reagents, unless stated otherwise, were obtained from Sigma-Aldrich Research (St. Louis, Missouri, USA) and were of the highest grade available.
Identification of single nucleotide polymorphisms in bile salt export pump ABCB11
Genomic DNA samples were provided by the Coriell Cell Repository (Camden, New Jersey, USA). ABCB11 variants were identified by direct sequencing of genomic DNA as described earlier from an ethnically diverse population of 247 healthy individuals: 100 European–Americans, 100 African–Americans, 30 Asian–Americans, 10 Mexican–Americans, and seven Pacific Islanders [26]. The reference cDNA sequence of ABCB11 was obtained from GenBank (NM_003742). Primers were designed manually to span the exons and 50–200λbp of flanking intronic sequence. The primer sequences can be found at http://www.pharmgkb.org. Variant positions are relative to the ATG start site and are based on the reference cDNA sequence of ABCB11.
Wild type and variant bile salt export pump plasmid construction
The full open reading frame of human BSEP cDNA was obtained by PCR using Expand Long Template PCR System (Roche Applied Science, Indianapolis, Indiana, USA) from a cDNA library synthesized from human liver mRNA using oligonucleotide primers 5′-TGTGGGTTGCAATTACCATGTCTGACTCAG-3′ as the forward and 5′-GCATTGGGTCAACTGATGGGGGATCCAGTG-3′ as the reverse (melting temperature 94°C × 30λs, annealing temperature 57°C × 45λs, extension temperature 68°C × 6λmin for 30 cycles). A single PCR product (approximately 4.1λkb) was visualized on an ethidium bromide-stained 0.8% agarose gel. An aliquot of the PCR product was ligated into the pEF6/V5-His-TOPO plasmid expression vector (Invitrogen). After transformation and growth in Escherichia coli, individual colonies containing the pEF6/V5-His-TOPO/BSEP construct in the sense orientation were identified with an appropriate restriction endonuclease digest. The only full-length clone tolerant of propagation in E. coli was found to have seven nonsynonymous mutations when fully sequenced with an ABI 3700 DNA analyzer (Applied Biosystems Inc., Foster City, California, USA). It was subsequently found that BSEP-transformed bacteria showed no or minimal growth due to the presence of a cryptic bacterial promoter between positions 59 and 81 downstream of the start codon of BSEP cDNA [9]. The cryptic promoter was inactivated by introducing synonymous base pair changes at positions 76–81 (TATAAT->TACAAC) and each of the seven nonsynonymous mutations was reverted to the wild-type reference allele using QuikChange site-directed mutagenesis kit (Stratagene, La Jolla, California, USA) and confirmed by direct DNA sequencing, resulting in a fully functional wild-type BSEP clone with 100% amino acid identity to the BSEP reference sequence (GenBank NM_003742).
Site-directed mutagenesis was used to generate missense PFIC2 mutants, nonsynonymous BSEP variants, and a null function BSEP variant for in-vitro functional analysis. The appropriate point mutations were individually introduced into wild-type BSEP packaged into pEF6/V5-His-TOPO using the QuikChange site-directed mutagenesis kit (Stratagene) using the following PCR conditions: melting temperature 95°C × 30λs, annealing temperature 55°C × 60λs, extension temperature 68°C × 22λmin for 17 cycles. The presence of the point mutations was confirmed by direct DNA sequencing. For the null function BSEP (truncated) variant, a single base pair, G, was deleted at position 908 of the coding strand of BSEP cDNA using oligonucleotide primers 5′-GAGAAAAGAGAGGTTGAAAGTATGAGAAAAATCTTGTG-3′ as the forward and 5′-CACAAGATTTTTCTCATACTTTCAACCTCTCTTTTCTC-3′ as the reverse, resulting in the creation of the stop codon TAG at amino acid position 320 before both ATP hydrolysis sites.
Cell culture and virus preparation
HeLa (American Type Culture Collection) cells were cultured in Dulbecco's modified Eagle's medium supplemented with 5% fetal bovine serum (FBS) and 1% penicillin/streptomycin. For preparation of viral stock of vtf-7 virus, HeLa cells grown to near confluency in 25-cm tissue culture plates were infected with 1 plaque-forming unit/10 cells. After an incubation period of 48λh at 37°C, the infected cells were pelleted, homogenized, and recovered through centrifugation, followed by titering of viral stock as described by Blakely et al. [27].
Human hepatoma (American Type Culture Collection) cells (HepG2) were cultured in Dulbecco's modified Eagle's medium supplemented with L-glutamine, 10% FBS, and 1% penicillin/streptomycin. For transient transfection, cells were grown on sterile uncoated 35λmm glass-bottomed microwell dishes (MatTek, Ashland, Massachusetts, USA) and transfected at 70–80% confluency with 1λμg of V5-tagged wild type or variant plasmid DNA (BSEP) using FuGENE HD Transfection Reagent (Roche Applied Science). After 48λh incubation, cells were analyzed by immunofluorescence confocal microscopy.
Transport studies using recombinant vaccinia virus
HeLa cells grown in 12-well plates (approximately 0.8×106 cells/well) were infected with vaccinia (vtf-7) at a multiplicity of infection of 10 plaque-forming units/cell in serum-free Opti-MEM I medium (Life Technologies Inc.) and allowed to adsorb for 30λmin at 37°C. A double-transfection assay was developed as follows: variant BSEP was evaluated in a 3-well row. Well 1 consisted of a double transfection with 0.05λμg of wild-type NTCP cDNA packaged into pEF6/V5-His-TOPO vector (Invitrogen) generated as described earlier [28] and 0.95λμg wild-type BSEP (in pEF6/V5-His-TOPO vector). Well 2 was the negative control sample and consisted of a double transfection with 0.05λμg of wild-type NTCP and 0.95λμg null-function (truncated) BSEP. Well 3 consisted of a double transfection with 0.05λμg of wild-type NTCP and 0.95λμg variant BSEP. Double transfections were performed with Lipofectin (Life Technologies Inc.) and incubated at 37°C for 16λh. Transport was then evaluated using labeled taurocholate as outlined earlier [29]. Therefore, each row consisted of variant BSEP, wild-type BSEP, and its own negative control–null function BSEP. Absolute taurocholate (5λμmol/l) uptake (pmol/mg protein) at 10λmin was assessed. Taurocholate transport efficiency for each variant BSEP was normalized to control null function BSEP and expressed as a percentage of wild-type BSEP transport. All experiments were carried out in duplicate on two experimental days (n=4) to minimize and account for sample-to-sample, plate-to-plate, and day-to-day variability.
Epitope-tagged wild type and variant bile salt export pump construction
We used a 14 amino acid epitope (V5) in the pEF6/V5-His-TOPO vector (Invitrogen) to generate V5-tagged wild type, variant, and null function BSEP. V5-tagged wild-type NTCP was generated as described earlier [28]. The QuikChange site-directed mutagenesis kit (Stratagene) was used to introduce a point mutation which converted the stop codon of BSEP to a cysteine residue (TGA→TGC), using the following oligonucleotide sense and antisense primers: 5′-CTGGATCCCCCATCAGTTGCCCAATGCAAGGGCGAATTC-3′ and 5′-GAATTCGCCCTTGCATTGGGCAACTGATGGGGGATCCAG-3′. The presence of the point mutation was verified by full sequencing. The V5 tag was generated for null function BSEP, wild-type BSEP and each of the nonsynonymous BSEP variants, and these were used for characterization of total protein and cell surface expression as well as for immunofluorescence confocal microscopy.
Bile salt export pump and NTCP expression in HeLa cells
HeLa cells double transfected with null-function BSEP plus wild-type NTCP, wild-type BSEP plus wild-type NTCP, variant BSEP plus wild-type NTCP cDNA tagged with the V5 epitope were scraped off plates, and the resulting suspension was centrifuged at 21λ000g for 3λmin. The cell pellet was reconstituted with HED buffer (25λmmol/l HEPES, 1.5λmmol/l EDTA, 1λmmol/l dithiothreitol, pH 7.4) containing protease inhibitors (Complete, Roche Molecular Biochemicals) and lysed by sonication. Samples were diluted with Laemmli buffer, and 3.75λμg of total cell protein were separated by SDS-polyacrylamide gel electrophoresis on 10% gels. Following transfer onto nitrocellulose membranes, blots were probed with a monoclonal anti-V5 antibody (1λ:λ5000 dilution, Invitrogen) and appropriate secondary antibody. To normalize sample loading, blots were stripped and reprobed with anticalnexin antibody (StressGen, Victoria, British Columbia, Canada). Bands were visualized using enhanced chemiluminescence (Amersham Pharmacia Biotech, Piscataway, New Jersey, USA). Densitometric analysis was performed using ImageJ (http://rsb.info.nih.gov/ij/).
Bile salt export pump and NTCP cell surface expression
HeLa cells were grown on 6-well plates and double transfected with the null function, wild type and variant BSEP along with wild-type NTCP cDNAs tagged with the V5 epitope using a similar protocol for transport experiments. Sixteen hours posttransfection, cells were washed with ice-cold phosphate-buffered saline (PBS), Ca/Mg (138λmmol/l NaCl2, 2.7λmmol/l KCl, 1.5λmmol/l KH2PO4, 9.6λmmol/l Na2HPO4, 1λmmol/l MgCl2, 0.1λmmol/l CaCl2, pH 7.3) and then treated with a membrane-impermeable biotinylating agent (sulfo-NHS-SS-biotin, 1.5λmg/ml, Pierce, Rockford, Illinois, USA) at 4°C for 1λh. Subsequently, the cells were washed three times with ice-cold PBS, Ca/Mg containing 100λmmol/l glycine and then incubated for 20λmin at 4°C with the same buffer to remove the remaining labeling agent. After washing with PBS, Ca/Mg, cells were disrupted with 700λμl of lysis buffer (10λmmol/l Tris–base, 150λmmol/l NaCl, 1λmmol/l EDTA, 0.1% SDS, 1% Triton X-100, pH 7.4) containing protease inhibitors (Complete, Roche Molecular Biochemicals) at 4°C for 1λh with constant agitation. After centrifugation, 140λμl of streptavidin agarose beads (Pierce) were added to 600λμl of cell lysate and incubated for 1λh at room temperature. Beads were washed four times with ice-cold lysis buffer, and the biotinylated proteins were released by incubation of the beads with 2× Laemmli buffer for 30λmin at room temperature. Similar to total cell lysates, samples of the biotinylated fractions (25λμl) were subjected to western blot analysis for the detection of immunodetectable BSEP and NTCP with monoclonal anti-V5 antibody (1:5000 dilution) and the intracellular, endoplasmic reticulum-resident protein calnexin, as described earlier. Densitometric analysis was performed using ImageJ (http://rsb.info.nih.gov/ij/).
Immunofluorescence confocal microscopy
HepG2 cells transiently transfected with V5-tagged wild-type or variant BSEP plasmid DNA were fixed for 15λmin in room temperature 4% paraformaldehyde, then washed 3× for 10λmin each with PBS. Cells were then blocked with 10% BSA in PBS plus 0.01% Triton X-100 for 1λh at room temperature. Anti-V5 antibody (Invitrogen) was diluted 1λ:λ1000 in 5% FBS–PBS, placed on the slides, and incubated overnight at 4°C. Cells were then washed 6× for 10λmin each in PBS, then blocked again for 1λh at room temperature with 10% BSA in PBS+0.01% Triton X-100. Cells were then incubated at 1λ:λ200 dilution with secondary fluorescent dye (Alexa Fluor 488) labeled goat anti-mouse whole antibody (Molecular Probes, Eugene, Oregon, USA) and 1λμl TO-PRO-3 (642/661) iodide nuclear stain (Invitrogen) at a 1λ:λ4000 dilution at room temperature for 1λh. Cells were then washed 5× at 5λmin each and 5× at 10λmin each with PBS. Cells were mounted with antifading mounting medium (Vectashield, Vector Laboratories Inc., Burlingame, California, USA) and viewed by confocal immunofluorescence microscopy. HepG2 cells transfected with plasmid alone and cells transfected with V5-tagged BSEP wild-type plasmid DNA without incubation in primary antibody were used as two separate controls. Samples were imaged using a Zeiss LSM510 confocal microscope with a 63×/1.4 plan apochromat objective (digital zoom 2×). Multitrack scans (488λnm excitation, 505–550λnm emission filter and 633λnm excitation, 650λnm long-pass emission filter) were used to eliminate detection bleed through. For slow frame scanning, confocal images were obtained by scanning laterally (top view; x–y scans) across the cell. Image analysis and processing were performed with Zeiss LSM and Adobe Photoshop software.
Real-time mRNA quantification from human liver samples
Human liver samples (Nashville Regional Organ Procurement Agency, Nashville, Tennessee, USA) from Caucasian donors without liver pathology were used. The study protocol was approved by the Institutional Review Board at Vanderbilt University Medical Center. Total RNA was extracted with a Versagene RNA Tissue Kit (Gentra Systems Inc., Minneapolis, Minnesota, USA). The integrity of RNA was assessed by 260/280λnm absorbance ratio and agarose gel electrophoresis or by an automated microfluidics-based system (Bioanalyzer 2100, Agilent, Palo Alto, California, USA). First-strand liver cDNA was synthesized using iScript cDNA Synthesis Kit (Bio-Rad, Hercules, California, USA). Real-time PCR was performed using iCycler iQ Real-Time Detection System (Bio-Rad). PCR reactions were carried out with iQ SYBR Green Supermix (Bio-Rad) and a specific primer pair in a final concentration of 200λnmol/l. Specific primer pairs are as follows: BSEP, forward 5′-GTCGGACCTGCATTGTCATTG-3′ and reverse 5′-ATGTGTGTCTGAGATTCTTGCATT-3′; NTCP, forward 5′-ACTGGTCCTGGTTCTCATTCC-3′ and reverse 5′-GTGGCAATCAAGAGTGGTGTC-3′; and 18S rRNA, forward 5′-CGGCTACCACATCCAAGGAA-3′ and reverse 5′-GCTGGAATTACCGCGGCT-3′. The cycling conditions were as follows: single cycle at 95°C for 3λmin, 40 cycles at 95°C for 15λs, and at 60°C for 1λmin (BSEP and NTCP); single cycle at 95°C for 3λmin, 34 cycles at 95°C for 15λs and 65°C for 1λmin (18S rRNA).
To confirm the amplification of a specific product, melting curve analysis was performed and PCR products directly visualized on 2% low-melting agarose gels. Quantification was calculated using a standard curve plotting log amount of DNA against Ct value. DNA standards were amplified from liver cDNA using iQ SYBR Green Supermix (Bio-Rad). PCR products were purified and DNA concentration was determined using Picogreen assay (Molecular Probes). Standard samples with known copy numbers were prepared and gene expression was normalized to 18S rRNA content.
Genotyping of human liver samples for bile salt export pump polymorphisms
Total genomic DNA was isolated from human liver samples through the DNeasy tissue kit (Qiagen, Valencia, California, USA). Samples were genotyped for the ABCB11 1331T>C and 2029A>G polymorphisms using a validated 5′ nuclease PCR based assay with allele specific fluorescent probes (TaqMan SNP Genotyping Assays, Applied Biosystems, Foster City, California, USA) as previously described [30,31].
Immunoblots of bile salt export pump from human liver samples
Each liver sample was placed in 500λμl HED buffer (25λmmol/l HEPES, 1.5λmmol/l EDTA, 1λmmol/l dithiothreitol, pH 7.4), homogenized, sonicated for 30λs and placed on ice. Samples were centrifuged at 4000g for 5λmin. Supernatant was saved and total protein quantified using BCA Protein Assay Kit (Pierce). Samples were diluted with NuPAGE reducing agent (Invitrogen) and 50λμg of total protein was loaded onto 10% gels and separated by SDS-polyacrylamide gel electrophoresis. Following transfer onto nitrocellulose membranes, blots were probed with a commercially available rabbit derived polyclonal antibody to the C-terminal peptide of BSEP (1λ:λ100 dilution; Abgent Envision Proteomics, San Diego, California, USA) and appropriate secondary antibody. To normalize sample loading, blots were stripped (Restore, Pierce) and reprobed with a rabbit polyclonal antibody to GAPDH (FL-335) (1λ:λ1000 dilution) (Santa Cruz Biotechnology, Inc., Santa Cruz, California, USA). Bands were visualized with enhanced chemiluminescence (Amersham Pharmacia Biotech). Densitometric analysis was performed using ImageJ (http://rsb.info.nih.gov/ij/).
Statistical analysis
Determination of statistical differences between group parameters was determined using Student's t-test, Mann–Whitney U test, one way analysis of variance (using Tukey–Kramer multiple comparison test), Pearson correlation, or Fisher's exact test, as appropriate using Prism software (GraphPad Software, Inc., San Diego, California, USA). A P value of less than 0.05 was taken to be the minimum level of statistical significance.
Results
Single nucleotide polymorphisms in ABCB11
Ethnically identified DNA samples from the Coriell Cell Repository (Camden) were evaluated for the presence of ABCB11 polymorphisms. From approximately 250 DNA samples, a total of 24 coding region single nucleotide polymorphisms (SNPs) were identified in our screening. Of these, 15 were synonymous and nine were nonsynonymous SNPs (Table 1). For the most part, SNPs in ABCB11 were relatively rare and population specific, with few exceptions. Of the nonsynonymous SNPs, the 1331T>C variant was very common in all ethnic populations tested while the 2029A>G variant was noted to be a common polymorphism in African–Americans.
Table 1.
Single-nucleotide polymorphisms in ABCB11
| Allele frequencies (%) |
|||||||
|---|---|---|---|---|---|---|---|
| SNP | rs number | Amino acid change | African–American | European–American | Asian–American | Mexican-American | Pacific Islanders |
| λ108T>C | rs3815675 | Synonymous | 1.5 | 1.5 | 25 | 5.0 | 21.4 |
| 167C>T | rs11568361 | Ser56Leu | 0.5 | 0 | 0 | 0 | 0 |
| 174C>T | rs11568362 | Synonymous | 0.5 | 0 | 0 | 0 | 0 |
| 270T>C | rs414877 | Synonymous | 3.0 | 3.5 | 5.0 | 0 | 7.1 |
| 402C>T | rs11568377 | Synonymous | 3.5 | 0 | 0 | 0 | 0 |
| 585G>C | rs11568365 | Synonymous | 0.5 | 0 | 0 | 0 | 0 |
| 616A>G | rs11568357 | Ile206Val | 2.5 | 0 | 0 | 0 | 0 |
| 696G>T | rs11568358 | Synonymous | 0 | 0.5 | 0 | 0 | 0 |
| 807T>C | rs2287616 | Synonymous | 2.0 | 0.5 | 23.3 | 5.0 | 21.4 |
| 890A>G | rs11568372 | Glu297Gly | 0 | 0.5 | 0 | 0 | 0 |
| 957A>G | rs7563233 | Synonymous | 31.5 | 0.5 | 0 | 15.0 | 0 |
| 1281C>T | rs11568360 | Synonymous | 0.5 | 0 | 0 | 0 | 0 |
| 1331T>C | rs2287622 | Val444Ala | 53.0 | 57.1 | 66.7 | 50.0 | 92.9 |
| 1671C>T | rs11568368 | Synonymous | 0 | 0.5 | 0 | 0 | 0 |
| 1674G>C | rs11568369 | Gln558His | 0 | 0.5 | 0 | 0 | 0 |
| 1772A>G | rs11568367 | Asn591Ser | 0 | 0.5 | 0 | 0 | 0 |
| 1774G>C | rs11568370 | Glu592Gln | 0 | 0.5 | 0 | 0 | 0 |
| 1791G>T | rs11568371 | Synonymous | 0 | 0.5 | 0 | 0 | 0 |
| 2029A>G | rs11568364 | Met677Val | 15.0 | 5.5 | 1.7 | 5.0 | 0 |
| 2412A>G | rs11568373 | Synonymous | 8.0 | 0 | 0 | 5.0 | 0 |
| 3084A>G | rs97692 | Synonymous | 28.6 | 54.6 | 63.3 | 37.5 | 21.4 |
| 3258A>G | rs11568359 | Synonymous | 7.0 | 0 | 0 | 0 | 0 |
| 3435A>G | rs11568366 | Synonymous | 1.0 | 0 | 0 | 0 | 0 |
| 3556G>A | rs1521808 | Glu1186Lys | 2.5 | 0 | 0 | 0 | 0 |
Allele frequencies for single-nucleotide polymorphisms (SNPs) in ABCB11 were determined from a DNA panel of ethnically defined healthy individuals – African–Americans (n=100), European–Americans (n=100), Asian–Americans (n=30), Mexican–Americans (n=10) and Pacific Islanders (n=7).
Functional analysis of bile salt export pump variants
A panel of expression plasmids comprising wild-type BSEP, nine nonsynonymous BSEP variants, and two known PFIC2 BSEP missense mutants, 2944G>A (Gly982Arg) and 3457C>T (Arg1153Cys), was constructed for functional studies. A recombinant vaccinia-based method was used to transiently express both the bile acid uptake transporter sodium/taurocholate cotransporting polypeptide (NTCP) and bile acid efflux transporter BSEP in HeLa cells for transport studies. NTCP was used to deliver bile acids intracellularly as efflux transporters such as BSEP can only transport intracellular substrates. Therefore, wild type or variant BSEP was transfected concomitant with wild-type NTCP for cellular loading of the conjugated bile acid taurocholate. BSEP activity was assessed by comparing the accumulation of substrate in cells transfected with NTCP and variant BSEP with cells transfected with NTCP and wild-type BSEP.
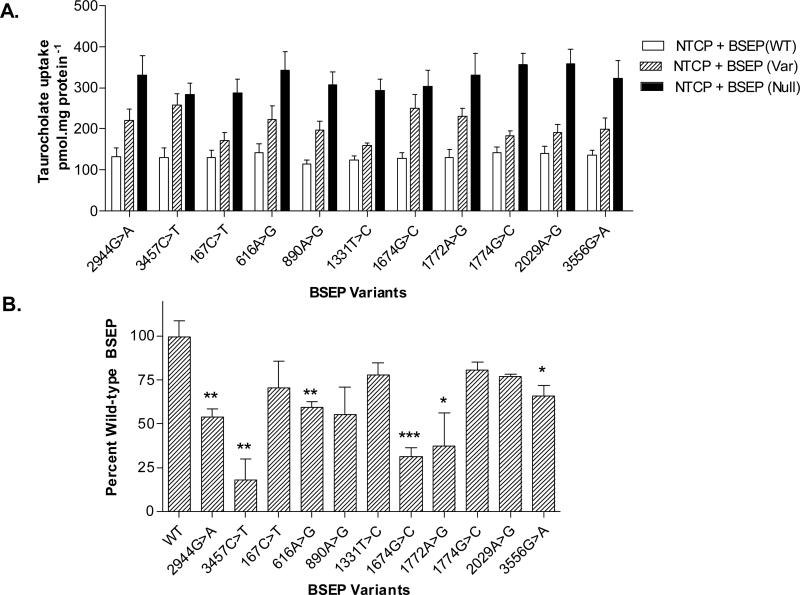
As expected, the PFIC2-associated missense mutations (2944G>A and 3457C>T) showed significantly impaired transport activity for the prototypical bile acid substrate taurocholate (Fig. 1b). In addition, several rare BSEP polymorphisms, including 616A>G (Ile206Val), 1674G>C (Gln558His), 1772A>G (Asn591Ser), and 3556G>A (Glu1186Lys) were also associated with significantly impaired taurocholate transport compared with wild-type BSEP, exhibited by retained increased intracellular taurocholate accumulation (P<0.05). Another rare BSEP variant, 890A>G (Glu297Gly), tended to have impaired taurocholate transport function, but did not reach statistical significance (P=0.051). Two common BSEP polymorphisms, 1331T>C (Val444Ala) and 2029A>G (Met677Val), were not associated with significant changes in taurocholate transport activity.
Fig. 1.
Taurocholate transport by bile salt export pump (BSEP) variants expressed in HeLa cells using a recombinant vaccinia-based method. (a) Absolute taurocholate (5λμmol/l) uptake (pmol/mg protein) in HeLa cells transfected with combination of NTCP/variant BSEP, NTCP/wild-type BSEP and NTCP/null function BSEP. (b) Taurocholate uptake (5λμmol/l) at 10λmin expressed as percentage of wild-type BSEP taurocholate transport activity. Data shown as mean ± SE (n=4). *P<0.05; **P<0.01; **P<0.001 wild-type BSEP.
Total and cell surf ace expression of bile salt export pump variants
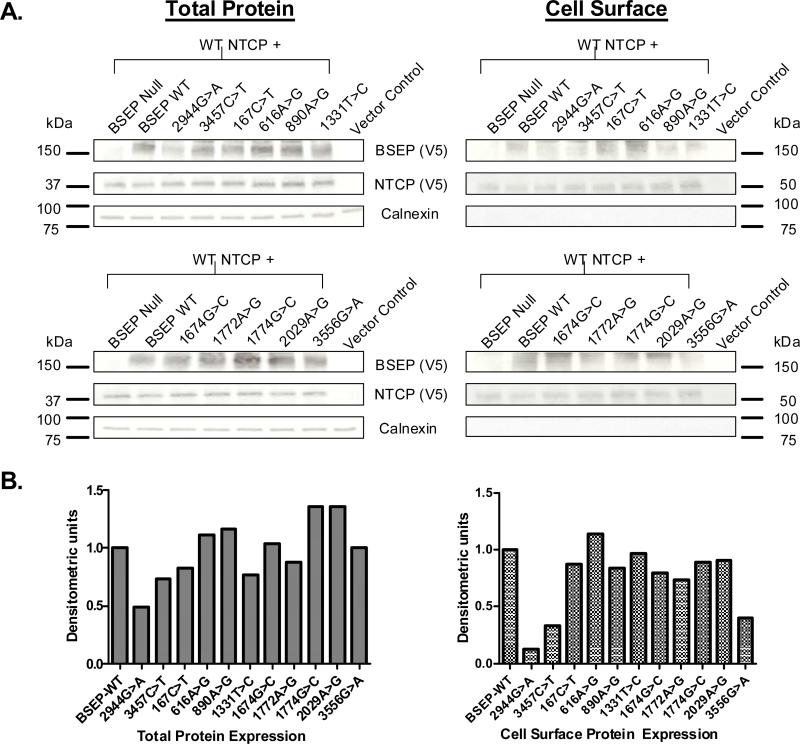
To determine whether the observed differences in transport activity of the BSEP polymorphisms were potentially as a result of altered protein expression or cell surface mistrafficking, western blot analysis of total cell lysates and cell surface biotinylated fractions were performed (Fig. 2a). V5 epitope-tagged wild-type and variant BSEP and wild-type NTCP constructs were generated and the same recombinant vaccinia-based method as used for transport experiments was used to express NTCP and wild-type or variant BSEP in HeLa cells. Taurocholate transport experiments revealed the V5-tagged variants to be functionally equivalent to the nontagged BSEP variants (data not shown).
Fig. 2.
Total and cell surface protein expression of bile salt export pump (BSEP) variants in HeLa cells. (a) Total (left panels) and cell surface (right panels) protein expression from HeLa cells double transfected with V5-tagged wild-type (WT) NTCP (approximately 40λkDa) and wild-type/variant BSEP (approximately 160λkDa) or control (null BSEP or vector alone) was evaluated by western blot analysis (left panels). Blots were probed with anti-V5 antibody, then stripped and probed with anticalnexin antibody (bottom panels). (b) Semiquantitative variant BSEP total and cell surface protein expression (densitometric units) in reference to wild-type BSEP expression.
Densitometric analysis revealed the average total and cell surface expression of NTCP (approximately 5% variability, after normalization to calnexin expression; n=3 blots) was equivalent amongst cells transfected with wild type or variant BSEP. In contrast, variant to wild-type BSEP expression varied widely amongst both total (approximately 80% variability) and cell surface (approximately 90% variability) samples. Significantly reduced total and cell surface protein expression was noted for both of the PFIC-associated mutants 2944G>A and 3457C>T (Fig. 2b) compared with wild-type BSEP, suggesting protein instability and/or cell surface mistrafficking as mechanisms for loss of action. The BSEP variant 3556G>A also showed reduced cell surface expression relative to its total protein expression, suggesting mistrafficking defects may contribute to impaired transport function. Interestingly, the other BSEP variants associated with impaired protein function, 616G>A, 1674G>C, and 1772G>A, all showed comparable total and cell surface protein expression relative to wild-type BSEP suggesting mechanisms other than altered protein expression or mistrafficking contribute to their impaired function. Cell surface-expressed NTCP was enriched in a larger apparent molecular mass (approximately 50λkDa) major band compared with that observed in total cell lysates (approximately 37λkDa) due to glycosylation of NTCP on the cell surface, which we have previously shown [28]. Calnexin, an intracellular resident endoplasmic reticulum-associated protein, was expressed in wild-type and variant total lysate samples, but as expected, was not enriched within the cell surface biotinylated samples.
Plasma membrane localization of bile salt export pump variants in HepG2 cells
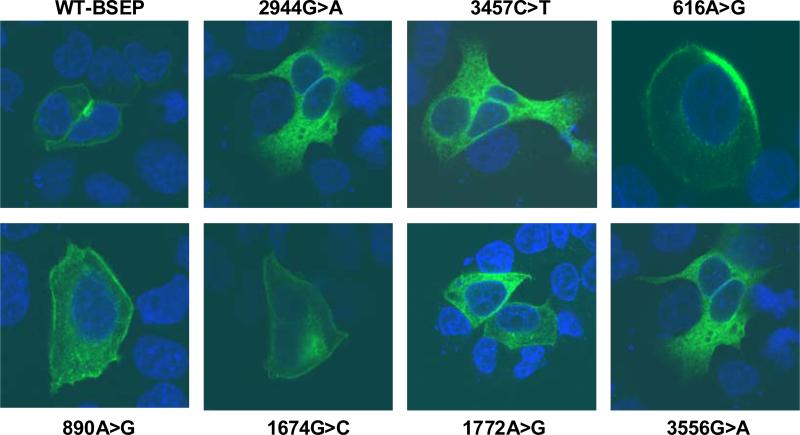
Cell surface biotinylation experiments and western analysis of total cell lysates in transfected HeLa cells suggested that the reduced function associated with the BSEP PFIC2 mutants and the 3556G>A variant may be because of altered trafficking of the protein to the cell surface and increased protein degradation or instability, respectively. To further assess our findings, we performed immunofluorescence confocal microscopy in transiently transfected HepG2 cells. Moreover, unlike HeLa cells, as HepG2 is a liver-derived human cell line, membrane expression seen in this cell line may better recapitulate in-vivo physiology.
HepG2 cells were transiently transfected with V5-tagged wild-type or variant BSEP plasmid DNA. Using a secondary fluorescent-labeled antibody, we demonstrate that wild-type BSEP was targeted to the plasma membrane of HepG2 cells when viewed laterally (x–y scans) across the cells by confocal microscopy (Fig. 3). In contrast, expression of the PFIC2 mutants, 2944G>A, and 3457C>T, in HepG2 cells show marked retention of protein intracellularly and essentially the absence of plasma membrane expression, consistent with western blot analysis described earlier. The 3556G>A variant also showed both increased retention of intracellular protein and markedly reduced plasma membrane localization. Furthermore, other BSEP variants associated with impaired taurocholate transport, 616A>G, 1674G>C, and 1772A>G, all seem to localize appropriately to the plasma membrane. The 890A>G variant, which trended towards impaired function, also seemed to be targeted to the plasma membrane.
Fig. 3.
Immunofluorescence confocal microscopy of wild-type and variant bile salt export pump (BSEP) in HepG2 cells. Immunofluorescent-labeled secondary antibody (Alexa Fluor 488) and TO-PRO-3 iodide were used to detect V5-tagged wild-type/variant
BSEP (green) and nuclei (blue), respectively. Wild-type BSEP (top left panel) was targeted to the cell surface in transiently transfected HepG2 cells as demonstrated by lateral (x–y scan) confocal imaging across the cell. The familial intrahepatic cholestases type 2 mutants, 2944G>A and 3457C>T, show marked intracellular protein accumulation without appreciable membrane staining. The 3556G>A variant shows both intracellular retention of protein and reduced membrane staining. The remaining BSEP variants seem to be appropriately targeted to the plasma membrane.
Real-time quantification of bile salt export pump mRNA in human liver samples
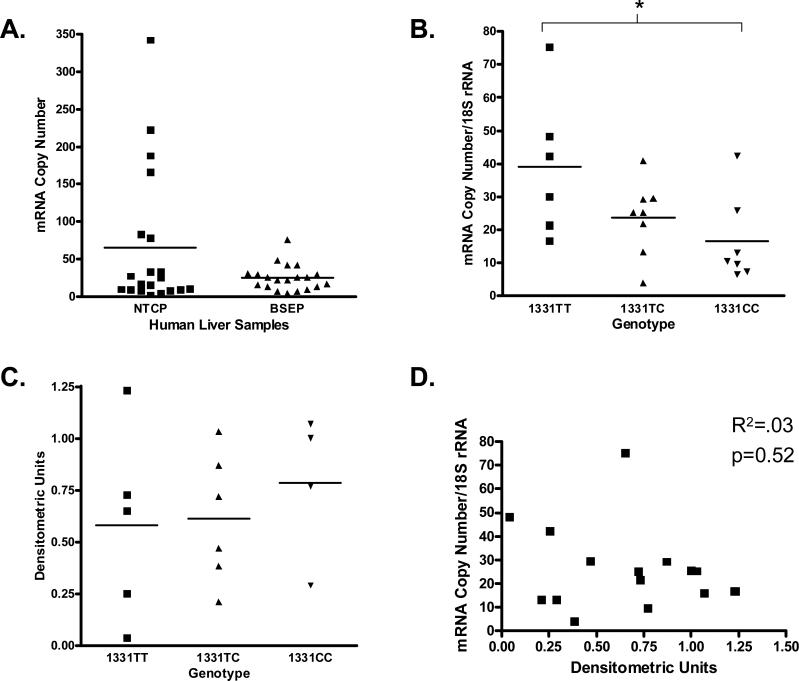
Real-time PCR quantification was performed on cDNA samples from a bank of 21 human livers (13 males, eight females). When normalized to 18S rRNA, the relative mean mRNA expression of BSEP tended to be lower than NTCP (24.9 vs. 64.6 copies/18S rRNA, P=0.07) (Fig. 4a). Interindividual variability was greater for NTCP (180-fold) versus BSEP (19-fold). Sex analysis revealed no significant sex differences in mRNA levels (28.1 male vs. 21.7 female copies/18S rRNA, P=0.42) (data not shown).
Fig. 4.
mRNA analysis of bile salt export pump (BSEP) and NTCP and mRNA: genotype, protein: genotype, mRNA: protein correlation from human liver samples. (a) mRNA copy number for NTCP and BSEP, normalized to 18S rRNA. Data expressed as mean ± SEM. (b) BSEP mRNA copy number grouped by BSEP 1331TT/TC/CC genotype. Data expressed as mean ± SEM, *P<0.05. (c) Semiquantitative BSEP protein expression (densitometric units) grouped by BSEP 1331TT/TC/CC genotype. Data expressed as mean ± SEM. (d) Scatter plot of BSEP mRNA copy number versus densitometric units.
DNA was extracted from the same human liver samples and genotyped for the commonly occurring BSEP polymorphism, 1331T>C. Out of a total of 42 alleles, the 1331T variant had a 48% allele frequency while the 1331C variant had a 52% allele frequency, consistent with previously published report and our own analysis in a larger cohort [32]. Interestingly, there was a significant association of genotype to mRNA expression. Individuals homozygous for the 1331TT genotype had the highest mRNA levels (39.1 copies/18S rRNA), those heterozygous for the 1331TC genotype had intermediate mRNA levels (23.7 copies/18S rRNA), and those homozygous for the variant 1331CC genotype had the lowest mRNA levels (16.5 copies/18S rRNA) (Fig. 4b; P<0.05).
Bile salt export pump protein expression in human liver samples
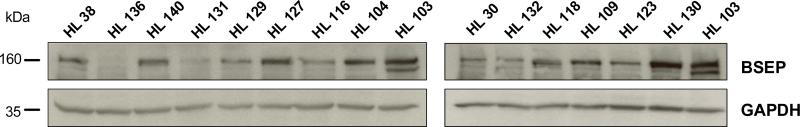
Human liver samples were analyzed by western blot analysis for BSEP protein expression to assess interindividual variability. Of the 21 human liver samples for which BSEP mRNA expression and genotyping analysis were performed, 15 had available tissue for western blot analysis. There was wide intersubject variation in BSEP expression amongst liver samples (Fig. 5) with approximately 31-fold variation in protein expression determined by densitometric analysis after normalization for GAPDH expression. Although there was wide overall intersubject variability in both BSEP mRNA and protein expression, there was no significant correlation between BSEP mRNA levels and protein expression (Fig. 4d, R2=0.03, P=0.52). Furthermore, while BSEP 1331T>C genotype was significantly associated with mRNA levels, genotype was not significantly associated with BSEP protein expression (Fig. 4c, P=0.71).
Fig. 5.
bile salt export pump (BSEP) protein expression from human liver samples. BSEP protein expression (approximately 160λkDa) (top panels) from a panel of normal human liver samples was assessed by western blot analysis. Samples were normalized for loading using anti-GAPDH antibody (bottom panels).
Discussion
To date, little information has been available regarding the functional relevance of genetic polymorphisms in ABCB11, despite increasing evidence supporting a crucial role of this transporter in the hepatic efflux of bile acids and its inhibition as a likely cause for drug-induced liver injury. In this study, we identified 24 SNPs in the coding region of ABCB11, including nine nonsynonymous polymorphisms, from a genomic DNA repository of approximately 250 ethnically defined healthy individuals. Although, for the most part, our data would suggest that nonsynonymous SNPs in ABCB11 are uncommon, certain SNPs were found to have relatively high allele frequencies in an ethnic-dependent manner. Functional characterization of BSEP variants indicated that several polymorphisms were associated with impaired transport function.
Lang et al. [33] previously evaluated approximately 200 DNA samples of European–Caucasian, African–American, Japanese and Korean origin for polymorphisms in ABCB11. Similar to our findings, ABCB11 polymorphisms tended to be rare and ethnic dependent. The two common nonsynonymous polymorphisms found in our study, 1331T>C and 2029A>G, were also found with similar allele frequencies in the Lang study. However, with the exception of the 616A>G variant, rare nonsynonymous polymorphisms identified from our study (167C>T, 890A>G, 1674G>C, 1772A>G, 1774G>C, and 3556G>A) were not found in this earlier study. Vice versa, we did not identify several rare nonsynonymous polymorphisms identified from the earlier study.
The critical importance of BSEP to normal liver physiology is illustrated by individuals who possess loss of function mutations in BSEP resulting in PFIC2, characterized by progressive liver disease with impairment of bile flow in the absence of hepatobiliary structural abnormality [13,14]. However, the phenotypic variability of genetic heterogeneity in ABCB11 is evidenced by the recent association of additional ABCB11 variants to less severe cholestatic syndromes, including BRIC, ICP, and DC [21–23,25]. Byrne et al. [8] recently evaluated a number of BSEP mutations and/or polymorphisms associated with PFIC2, BRIC, ICP or DC for alterations in pre-messenger RNA splicing, abnormal processing of BSEP protein or impaired BSEP protein function. When analyzed using an in-vitro minigene system in HepG2 cells, a number of variants were associated with differential splicing or exon skipping events.
We developed a double-transporter expression model using a recombinant vaccinia expression system to express the bile acid uptake transporter NTCP in combination with wild-type or variant BSEP for the standardized evaluation of transport efficiency. Two loss of function missense BSEP mutants associated with PFIC2, 2944G>A, and 3457C>T, were included in our model system for validation of our functional assay. As expected, the PFIC2 mutants were associated with significantly impaired taurocholate transport activity compared with wild-type BSEP. Moreover, additional rare BSEP polymorphisms were also associated with significantly reduced taurocholate transport activity. The 616A>G, 1674G>C, 1772A>G, and 3556G>A variants showed significantly impaired function for taurocholate transport while the 890A>G variant tended to also have reduced taurocholate transport activity. Two commonly occurring BSEP polymorphisms, 1331T>C and 2029A>G, seemed to have no significant effect on taurocholate transport efficiency when compared with wild-type BSEP.
Protein expression was assessed by western blot analysis of total cell lysates and cell surface-enriched fractions. The PFIC2-associated mutants, 2944G>A and 3457C>T, showed both significantly impaired total and cell surface protein expression compared with wild-type BSEP, suggesting increased protein degradation/instability and/or mistrafficking defects are responsible for the complete loss of function associated with these mutants in vivo and consistent with previously published reports showing similar mechanisms for loss of activity [18]. Similarly, immunofluorescence confocal microscopy in HepG2 cells showed loss of cell membrane expression and increased intracellular accumulation of protein for the BSEP mutants 2944G>A and 3457C>T. Interestingly, the 3556G>A variant showed significantly decreased cell surface expression but comparable total protein expression compared with wild-type BSEP, suggesting mistrafficking of this variant to the cell surface as a mechanism for impaired function. When analyzed by confocal immunofluorescence microscopy in HepG2 cells, portions of the 3556G>A variant seemed to be retained intracellularly as well as at the plasma membrane, suggesting the mistrafficking defects may not be complete for this variant. Byrne et al. [8] found that the 3556G>C variant was associated with mild exon skipping but protein expression was not assessed for this variant. The 616A>G, 1674G>C, and 1772A>G variants seemed to have comparable total and cell surface protein expression compared with wild-type BSEP, suggesting impairment of function associated with these variants may be related to substrate binding or specificity.
The clinical implication of less than complete loss of bile acid transport function in BSEP is unclear. It is biologically plausible that drug-mediated inhibition of BSEP may lead to hepatotoxic adverse effects such as cholestasis or transaminitis. In vitro and in vivo studies using rat BSEP showed that drugs such as the insulin sensitizer troglitazone, the endothelin antagonist bosentan, cyclosporine A, and the antibiotic rifampin mediated DC throigh direct (cis) inhibition of Bsep [34–36]. The fact that only a minority of patients develops cholestasis during therapy with potentially cholestatic drugs may be due to the existence of ABCB11 polymorphisms associated with constitutive impairments of BSEP function in susceptible patients. Indeed, Lang et al. [24] found that a common ABCB11 variant, 1331T>C (Val444Ala), was more frequently observed in DC patients than in drug-induced hepatocellular injury patients or healthy controls. However, when expressed and functionally characterized in Sf9 membrane vesicles, there were no significant differences in protein expression or taurocholate transport activity in the Val444Ala variant versus wild-type BSEP, consistent with our data.
The same group previously reported that the Val444Ala variant was associated with low-protein expression from a bank of 110 liver samples from individuals undergoing liver resection for various hepatic diseases [32], though it was not statistically significant. In contrast, we demonstrated that, when expressed in HeLa cells, protein expression of the 444Ala variant was comparable with wild-type BSEP expression. We also assessed interindividual variability in BSEP expression by real-time mRNA analysis and protein expression by western blot analysis in a smaller human liver bank. There was high interindividual variability with 19-fold and 31-fold differences in BSEP mRNA and protein expression levels, respectively. Although the BSEP mRNA expression level was significantly associated with 1331T>C genotype (TT>TC>CC, P<0.05), there was no significant correlation between mRNA amount and protein expression or 1331T>C genotype and protein expression. One possible reason for the discrepancy in our results is the sample population. Our liver samples were procured from healthy donors secondary to trauma while the majority of liver samples from the study by Meier et al. [32] were acquired from individuals undergoing liver resection secondary to primary liver cancer or liver metastases. Although pathologically normal liver tissue was used in their experiments, the underlying disease state or biochemical derangements due to liver cancer or metastasis could have significant implications for normal liver physiology and homeostasis. In addition, although in-vitro studies show no differences taurocholate transport activity for the 1331T>C variant, it is possible that this SNP is located in linkage disequilibrium with a promoter SNP that may alter basal or inducible expression of this transporter. The possibility that this SNP is associated with allele-specific expression of ABCB11 can also be not ruled out. Clearly, given the remarkably high allele frequency, additional studies in larger numbers of liver samples would aid in clarifying the importance of this variant to in-vivo BSEP function and its potential role as a genetic determinant of predisposition to DC or hepatocellular injury.
Interestingly, our screening identified a variant 890A>G which had been previously documented in individuals with PFIC2 and more recently BRIC2 [7,25]. There are conflicting data pertaining to the functional relevance of this variant. When introduced in rat Bsep, it was associated with mistrafficking to the plasma membrane in MDCK cells and almost abolished taurocholate transport activity when assessed in Sf9 membrane vesicles [18]. Additional studies of this variant expressed in rat Bsep showed reduced, but not absent, plasma membrane BSEP expression and significantly impaired catalytic activity when assessed in membrane vesicles or MDCK II cells [37,38]. In contrast, Hayashi et al. [16] expressed the 890A>G variant in human BSEP and assessed localization in MDCK II cells by cell surface biotinylation studies and taurocholate transport activity in membrane vesicles from HEK 293 cells. They found both intracellular and plasma membrane localization for this variant in MDCK II cells but when normalized for protein expression, this variant had equivalent taurocholate transport activity to wild-type BSEP. Furthermore, Strautnieks et al. [39] conducted immunohistochemical analysis of a large group of PFIC2 patients with various mutations, including 890A>G. Although more than 90% of samples had absent or abnormal BSEP staining, expression varied most for the 890A>G and 1445A>G mutations with some BSEP expression detected in 45% of patients with these mutations. Interestingly, Byrne et al. [8] demonstrated that incubation of the 890A>G variant transfected in CHO-K1 cells at 28°C alone or in combination with 10% glycerol, conditions which promote the correct processing of membrane proteins trapped in the endoplasmic reticulum or golgi, resulted in equivalent protein expression as wild-type BSEP. Based on our protein expression and transport studies in HeLa cells, our data suggests this variant has comparable total protein expression and cell surface expression to wild-type BSEP and tends to be associated impaired taurocholate transport activity, although it did not react statistical significance in our study. Further studies will be needed to confirm this variant's functional capacity and its apparent phenotypic variability that range from severe cholestasis (PFIC2) to more benign cholestatic states (BRIC2).
In conclusion, we report the systematic identification and in-vitro functional characterization of nonsynonymous polymorphisms in ABCB11. To our knowledge, this represents the first detailed functional examination of BSEP polymorphisms present in major ethnic populations and creates the framework for further investigations of the consequences of BSEP polymorphisms in vivo. We demonstrate that several polymorphisms in BSEP have impaired taurocholate transport activity. In human livers, marked intersubject variability in expressed levels of BSEP mRNA and protein was observed. Clearly, additional studies, especially among those individuals with congenital or acquired cholestatic syndromes, are warranted to better determine the role and clinical relevance of genetic heterogeneity in ABCB11.
Acknowledgements
This study was supported by National Institutes of Health grants GM61390 (D.L.K.), GM81363 (R.H.H.), GM54724 (R.B.K.) and GM31304 (R.B.K.). Confocal immunofluorescence microscopy experiments, data analysis, and presentation were performed in part through the use of the VUMC Cell Imaging Core Resource (supported by NIH grants CA68485, DK20593, DK58404, HD15052, DK59367, and EY08126). The authors have no conflicts of interest to disclose.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Hofmann AF. Bile secretion and the enterohepatic circulation of bile acids. In: Feldman M, Scharschmidt BF, Sleisenger MH, editors. Gastrointestinal and liver disease. Saunders; Philadelphia/London: 1998. pp. 937–948. [Google Scholar]
- 2.St Pierre MV, Kullak-Ublick GA, Hagenbuch B, Meier PJ. Transport of bile acids in hepatic and non-hepatic tissues. J Exp Biol. 2001;204:1673–1686. doi: 10.1242/jeb.204.10.1673. [DOI] [PubMed] [Google Scholar]
- 3.Trauner M, Boyer JL. Bile salt transporters: molecular characterization, function, and regulation. Physiol Rev. 2003;83:633–671. doi: 10.1152/physrev.00027.2002. [DOI] [PubMed] [Google Scholar]
- 4.Nishida T, Gatmaitan Z, Che M, Arias IM. Rat liver canalicular membrane vesicles contain an ATP-dependent bile acid transport system. Proc Natl Acad Sci U S A. 1991;88:6590–6594. doi: 10.1073/pnas.88.15.6590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Childs S, Yeh RL, Georges E, Ling V. Identification of a sister gene to P-glycoprotein. Cancer Res. 1995;55:2029–2034. [PubMed] [Google Scholar]
- 6.Gerloff T, Stieger B, Hagenbuch B, Madon J, Landmann L, Roth J, et al. The sister of P-glycoprotein represents the canalicular bile salt export pump of mammalian liver. J Biol Chem. 1998;273:10046–10050. doi: 10.1074/jbc.273.16.10046. [DOI] [PubMed] [Google Scholar]
- 7.Strautnieks SS, Bull LN, Knisely AS, Kocoshis SA, Dahl N, Arnell H, et al. A gene encoding a liver-specific ABC transporter is mutated in progressive familial intrahepatic cholestasis. Nat Genet. 1998;20:233–238. doi: 10.1038/3034. [DOI] [PubMed] [Google Scholar]
- 8.Byrne JA, Strautnieks SS, Mieli-Vergani G, Higgins CF, Linton KJ, Thompson RJ. The human bile salt export pump: characterization of substrate specificity and identification of inhibitors. Gastroenterology. 2002;123:1649–1658. doi: 10.1053/gast.2002.36591. [DOI] [PubMed] [Google Scholar]
- 9.Noe J, Stieger B, Meier PJ. Functional expression of the canalicular bile salt export pump of human liver. Gastroenterology. 2002;123:1659–1666. doi: 10.1053/gast.2002.36587. [DOI] [PubMed] [Google Scholar]
- 10.Lecureur V, Sun D, Hargrove P, Schuetz EG, Kim RB, Lan LB, et al. Cloning and expression of murine sister of P-glycoprotein reveals a more discriminating transporter than MDR1/P-glycoprotein. Mol Pharmacol. 2000;57:24–35. [PubMed] [Google Scholar]
- 11.Hirano M, Maeda K, Hayashi H, Kusuhara H, Sugiyama Y. Bile salt export pump (BSEP/ABCB11) can transport a nonbile acid substrate, pravastatin. J Pharmacol Exp Ther. 2005;314:876–882. doi: 10.1124/jpet.105.084830. [DOI] [PubMed] [Google Scholar]
- 12.Jansen PL, Strautnieks SS, Jacquemin E, Hadchouel M, Sokal EM, Hooiveld GJ, et al. Hepatocanalicular bile salt export pump deficiency in patients with progressive familial intrahepatic cholestasis. Gastroenterology. 1999;117:1370–1379. doi: 10.1016/s0016-5085(99)70287-8. [DOI] [PubMed] [Google Scholar]
- 13.Thompson R, Strautnieks S. BSEP: function and role in progressive familial intrahepatic cholestasis. Semin Liver Dis. 2001;21:545–550. doi: 10.1055/s-2001-19038. [DOI] [PubMed] [Google Scholar]
- 14.Alonso EM, Snover DC, Montag A, Freese DK, Whitington PF. Histologic pathology of the liver in progressive familial intrahepatic cholestasis. J Pediatr Gastroenterol Nutr. 1994;18:128–133. doi: 10.1097/00005176-199402000-00002. [DOI] [PubMed] [Google Scholar]
- 15.Whitington PF, Freese DK, Alonso EM, Schwarzenberg SJ, Sharp HL. Clinical and biochemical findings in progressive familial intrahepatic cholestasis. J Pediatr Gastroenterol Nutr. 1994;18:134–141. doi: 10.1097/00005176-199402000-00003. [DOI] [PubMed] [Google Scholar]
- 16.Hayashi H, Takada T, Suzuki H, Akita H, Sugiyama Y. Two common PFIC2 mutations are associated with the impaired membrane trafficking of BSEP/ABCB 11. Hepatology. 2005;41:916–924. doi: 10.1002/hep.20627. [DOI] [PubMed] [Google Scholar]
- 17.Noe J, Kullak-Ublick GA, Jochum W, Stieger B, Kerb R, Haberl M, et al. Impaired expression and function of the bile salt export pump due to three novel ABCB11 mutations in intrahepatic cholestasis. J Hepatol. 2005;43:536–543. doi: 10.1016/j.jhep.2005.05.020. [DOI] [PubMed] [Google Scholar]
- 18.Wang L, Soroka CJ, Boyer JL. The role of bile salt export pump mutations in progressive familial intrahepatic cholestasis type II. J Clin Invest. 2002;110:965–972. doi: 10.1172/JCI15968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Plass JR, Mol O, Heegsma J, Geuken M, de Bruin J, Elling G, et al. A progressive familial intrahepatic cholestasis type 2 mutation causes an unstable, temperature-sensitive bile salt export pump. J Hepatol. 2004;40:24–30. doi: 10.1016/s0168-8278(03)00483-5. [DOI] [PubMed] [Google Scholar]
- 20.Cai SY, Wang L, Ballatori N, Boyer JL. Bile salt export pump is highly conserved during vertebrate evolution and its expression is inhibited by PFIC type II mutations. Am J Physiol Gastrointest Liver Physiol. 2001;281:G316–G322. doi: 10.1152/ajpgi.2001.281.2.G316. [DOI] [PubMed] [Google Scholar]
- 21.Eloranta ML, Hakli T, Hiltunen M, Helisalmi S, Punnonen K, Heinonen S. Association of single nucleotide polymorphisms of the bile salt export pump gene with intrahepatic cholestasis of pregnancy. Scand J Gastroenterol. 2003;38:648–652. doi: 10.1080/00365520310000807. [DOI] [PubMed] [Google Scholar]
- 22.Keitel V, Vogt C, Haussinger D, Kubitz R. Combined mutations of canalicular transporter proteins cause severe intrahepatic cholestasis of pregnancy. Gastroenterology. 2006;131:624–629. doi: 10.1053/j.gastro.2006.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Kubitz R, Keitel V, Scheuring S, Kohrer K, Haussinger D. Benign recurrent intrahepatic cholestasis associated with mutations of the bile salt export pump. J Clin Gastroenterol. 2006;40:171–175. doi: 10.1097/01.mcg.0000196406.15110.60. [DOI] [PubMed] [Google Scholar]
- 24.Lang C, Meier Y, Stieger B, Beuers U, Lang T, Kerb R, et al. Mutations and polymorphisms in the bile salt export pump and the multidrug resistance protein 3 associated with drug-induced liver injury. Pharmacogenet Genomics. 2007;17:47–60. doi: 10.1097/01.fpc.0000230418.28091.76. [DOI] [PubMed] [Google Scholar]
- 25.Van Mil SW, van der Woerd WL, van der BG, Sturm E, Jansen PL, Bull LN, et al. Benign recurrent intrahepatic cholestasis type 2 is caused by mutations in ABCB11. Gastroenterology. 2004;127:379–384. doi: 10.1053/j.gastro.2004.04.065. [DOI] [PubMed] [Google Scholar]
- 26.Leabman MK, Huang CC, De Young J, Carlson EJ, Taylor TR, de la CM, et al. Natural variation in human membrane transporter genes reveals evolutionary and functional constraints. Proc Natl Acad Sci U S A. 2003;100:5896–5901. doi: 10.1073/pnas.0730857100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Blakely RD, Clark JA, Rudnick G, Amara SG. Vaccinia-T7 RNA polymerase expression system: evaluation for the expression cloning of plasma membrane transporters. Anal Biochem. 1991;194:302–308. doi: 10.1016/0003-2697(91)90233-j. [DOI] [PubMed] [Google Scholar]
- 28.Ho RH, Leake BF, Roberts RL, Lee W, Kim RB. Ethnicity-dependent polymorphism in Na+-taurocholate cotransporting polypeptide (SLC10A1) reveals a domain critical for bile acid substrate recognition. J Biol Chem. 2004;279:7213–7222. doi: 10.1074/jbc.M305782200. [DOI] [PubMed] [Google Scholar]
- 29.Kim RB, Leake B, Cvetkovic M, Roden MM, Nadeau J, Walubo A, et al. Modulation by drugs of human hepatic sodium-dependent bile acid transporter (sodium taurocholate cotransporting polypeptide) activity. J Pharmacol Exp Ther. 1999;291:1204–1209. [PubMed] [Google Scholar]
- 30.Ho RH, Choi L, Lee W, Mayo G, Schwarz UI, Tirona RG, et al. Effect of drug transporter genotypes on pravastatin disposition in European- and African-American participants. Pharmacogenet Genomics. 2007;17:647–656. doi: 10.1097/FPC.0b013e3280ef698f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Livak KJ. SNP genotyping by the 5′-nuclease reaction. Methods Mol Biol. 2003;212:129–147. doi: 10.1385/1-59259-327-5:129. [DOI] [PubMed] [Google Scholar]
- 32.Meier Y, Pauli-Magnus C, Zanger UM, Klein K, Schaeffeler E, Nussler AK, et al. Interindividual variability of canalicular ATP-binding-cassette (ABC)-transporter expression in human liver. Hepatology. 2006;44:62–74. doi: 10.1002/hep.21214. [DOI] [PubMed] [Google Scholar]
- 33.Lang T, Haberl M, Jung D, Drescher A, Schlagenhaufer R, Keil A, et al. Genetic variability, haplotype structures, and ethnic diversity of hepatic transporters MDR3 (ABCB4) and bile salt export pump (ABCB11). Drug Metab Dispos. 2006;34:1582–1599. doi: 10.1124/dmd.105.008854. [DOI] [PubMed] [Google Scholar]
- 34.Funk C, Ponelle C, Scheuermann G, Pantze M. Cholestatic potential of troglitazone as a possible factor contributing to troglitazone-induced hepatotoxicity: in vivo and in vitro interaction at the canalicular bile salt export pump (Bsep) in the rat. Mol Pharmacol. 2001;59:627–635. [PubMed] [Google Scholar]
- 35.Stieger B, Fattinger K, Madon J, Kullak-Ublick GA, Meier PJ. Drug- and estrogen-induced cholestasis through inhibition of the hepatocellular bile salt export pump (Bsep) of rat liver. Gastroenterology. 2000;118:422–430. doi: 10.1016/s0016-5085(00)70224-1. [DOI] [PubMed] [Google Scholar]
- 36.Fattinger K, Funk C, Pantze M, Weber C, Reichen J, Stieger B, et al. The endothelin antagonist bosentan inhibits the canalicular bile salt export pump: a potential mechanism for hepatic adverse reactions. Clin Pharmacol Ther. 2001;69:223–231. doi: 10.1067/mcp.2001.114667. [DOI] [PubMed] [Google Scholar]
- 37.Lam P, Pearson CL, Soroka CJ, Xu S, Mennone A, Boyer JL. Levels of plasma membrane expression in progressive and benign mutations of the bile salt export pump (Bsep/Abcb1 1) correlate with severity of cholestatic diseases. Am J Physiol Cell Physiol. 2007;293:C1709–C1716. doi: 10.1152/ajpcell.00327.2007. [DOI] [PubMed] [Google Scholar]
- 38.Kagawa T, Watanabe N, Mochizuki K, Numari A, Ikeno Y, Itoh J, et al. Phenotypic differences in PFIC2 and BRIC2 correlate with protein stability of mutant Bsep and impaired taurocholate secretion in MDCKII cells. Am J Physiol Gastrointest Liver Physiol. 2008;294:G58–G67. doi: 10.1152/ajpgi.00367.2007. [DOI] [PubMed] [Google Scholar]
- 39.Strautnieks SS, Byrne JA, Pawlikowska L, Cebecauerova D, Rayner A, Dutton L, et al. Severe bile salt export pump deficiency: 82 different ABCB11 mutations in 109 families. Gastroenterology. 2008;134:1203–1214. doi: 10.1053/j.gastro.2008.01.038. [DOI] [PubMed] [Google Scholar]