Abstract
The production of mouse chimeras is a common step in the establishment of genetically modified animal strains. Chimeras also provide a powerful experimental tool for following cell behavior during both prenatal and postnatal development. This protocol outlines a simple and economical technique for the production of large numbers of mouse chimeras using traditional diploid morula ↔ diploid embryonic stem (ES) cell aggregations. Additional steps are included to describe the procedures necessary to produce specialized tetraploid chimeras using tetraploid morula ↔ diploid ES cell aggregations. This increasingly popular form of chimera produces embryos of nearly complete ES cell derivation that can be used to speed transgenic production or ask developmental questions. Using this protocol, mouse chimeras can be generated and transferred to pseudopregnant surrogate mothers in a 5-d period.
INTRODUCTION
The mouse, Mus musculus, is the preeminent mammalian genetic model system. This is due, in part, to the relative ease with which preimplantation embryos can be manipulated in vitro and later returned to pseudopregnant foster females. One of the more powerful manipulations is the generation of chimeric mice by combining a host embryo with genetically dissimilar, or modified, embryo-derived cells. In many cases, the germ line in these chimeras will contain cells derived from both the host and transplanted cell genomes. Thus, these chimeras can pass either genome to their progeny. Although this represents the traditional manner by which genetically modified ES cells are used to establish transgenic mice1,2, chimeric animals may also be used directly for phenotypic analyses to determine the developmental potential or fate of wild-type and mutant embryonic cells3, as well as the developmental potential of other embryo-derived stem cells4,5. Over the past decade, chimeras have undergone a renaissance in their use and now, in combination with improved techniques for manipulating the mouse genome and for deriving nuclear transfer ES cells, provide a powerful way to study mouse development6–8.
Chimeric embryos can be produced by a number of techniques, including simply culturing embryos on a lawn of ES cells9. The two most common methods of producing chimeric mice from ES cells are by the injection of cells into the blastocoel cavity of a blastocyststage host embryo6,10 or by the aggregation of ES cells with a morula-stage host embryo6,11. Although blastocyst injection allows the investigator to scrutinize each cell that will be injected and select only those cell morphologies that are most likely to colonize the chimeric animal efficiently, it requires access to specialized equipment and is considered by some to be technically more demanding. Aggregation, on the other hand, requires no specialized equipment and is inexpensive and relatively easily learned, but it sacrifices the ability to select ES cells individually. This can result in the reduction of chimera quality, particularly for higher-passage or otherwise problematic ES cells12. These differences aside, the choice of technique is largely a matter of individual preference.
In the case of certain hybrid ES cell lines such as R1, a 129/Sv×129/J F1 hybrid line12, and various other hybrid ES cell lines13,14 (predominantly 129×C57BL/6 Fls hybrids), the contribution of the ES cells to embryonic regions15 can be elevated by the use of tetraploid (4n) host embryos and the generation of tetraploid embryo ↔ diploid ES cell chimeras16 (reviewed in ref. 17). In these chimeras, the tetraploid cells become principally restricted to the extraembryonic membranes with minimal contribution to the embryo proper18,19. This results in almost completely ES cell–derived embryos, which give rise to animals that are viable and fertile. This technique, which was developed for mice, has been adapted and used in other species, such as cattle20.
In diploid embryo ↔ diploid ES cell chimeras, the fetus will be comprised of a mixture of ES-derived cells along with a high proportion of host embryo–derived cells. The use of tetraploid ↔ diploid chimeras can allow for the analysis of experiments in which the host diploid embryo derivatives of diploid ↔ diploid chimeras might otherwise mask phenotypes in the ES-derived cell populations. In addition to producing high-percentage chimeras, this technique can also be used to determine embryonic versus extraembryonic phenotypes experimentally or to analyze embryos derived from ES cells produced by other manipulations including lentiviral or RNAi modifications.
Tetraploidy may be induced by a number of methods (reviewed in ref. 17). The most common being by electrofusing the plasma membranes of a two-cell diploid embryo to produce a temporarily binucleate one-cell embryo. An alternative protocol is to inhibit the first zygotic division with cytochalasin21,22. A limitation of this latter approach is that there is no visual confirmation that the procedure has worked, whereas successful electrofusion may be readily confirmed.
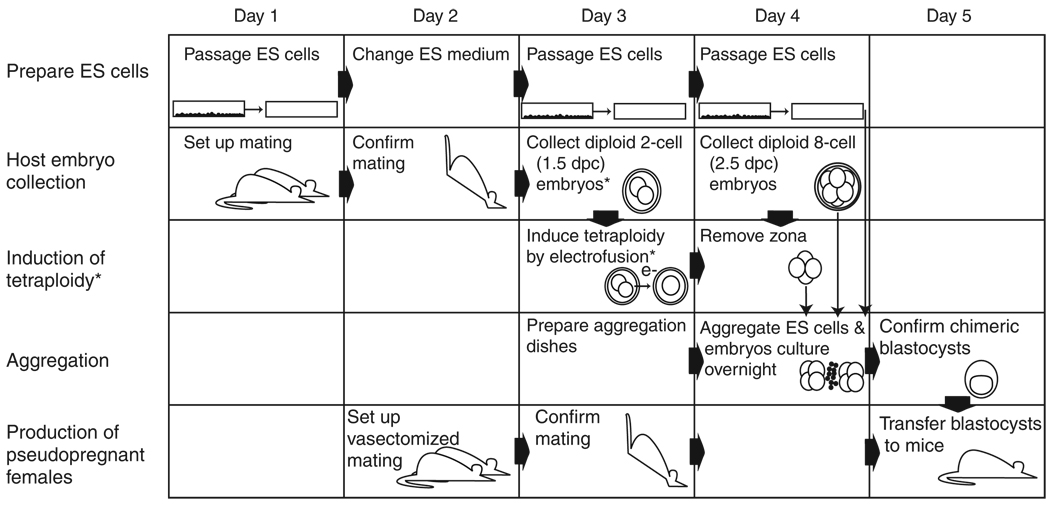
The method described below requires that several individual protocols be coordinated. Figure 1 presents a general timeline for the procedures described in this protocol. The protocol describes procedures that are common to the production of either diploid ↔ diploid or diploid ↔ tetraploid chimeras. Box 1 describes steps that are specific to the production of diploid ↔ tetraploid chimeras. Typically, this protocol will take less than 3 h each day over the course of the experiment.
Figure 1.
Timeline and integration of the protocols. The exact regimen for the preparation of ES cells varies with different cell lines. Usually, ES cell medium is changed daily, and cells are passaged on the second or third day once they reach 70% confluency. For the generation of aggregation chimeras, it is imperative that cells are passaged in such a way that they form clumps of cells that can be recovered from the dish after brief treatment with trypsin. Experiments with mice need to be carefully planned; matings to produce the embryos required for the aggregations and pseudopregnant females for the transfer of chimeric blastocysts need to be synchronized. For the routine production of diploid embryo ↔ diploid ES cell chimeras, embryos are recovered at the eight-cell stage at 2.5 dpc when they appear as uncompacted morulae. The zona is immediately removed, and the embryos are then used to set up the aggregation. Embryos can, however, also be recovered 1 or 2 d earlier at 1.5 or 0.5 dpc and cultured until they form morulae, at which time the zona is removed before aggregation setup. The same applies for embryos that are to be rendered tetraploid, which can be recovered at 0.5 dpc and cultured overnight until they reach the two-cell stage. Asterisks indicate the steps that are unique to the production of tetraploid (4n) embryo ↔ diploid ES cell chimeras.
INDUCTION OF TETRAPLOIDY BY ELECTROFUSION
On day 3 (Fig. 1), rinse a microscope slide bearing the electrode with 70% ethanol and then sterile dH2O. This is absolutely critical, as debris such as microscopic particles from the foam packaging material and any buildup of salts will severely affect the electrical conductance across the electrode, causing the embryos to lyse. Place the electrode in a clean 10-cm tissue culture dish, using tape to prevent the electrode from moving within the dish.
-
On the underside of a 3.5-cm tissue culture dish, draw lines to divide the dish into three sections. Label one side ‘unfused’, another ‘fused’ and the last ‘>2 cell’ or ‘cleaved’ (Fig. 3, part 7). Place one to three drops of KSOM in each section. Cover all the drops with a layer of mineral oil and store the dish at 37 °C and 5% CO2 in a tissue culture incubator. When embryos are flushed, some may be older than the 3- to 4-cell stage and therefore cannot be used for electrofusion. These can be used for diploid aggregation experiments and can be temporarily stored in the >2-cell section of the dish.
 As in all experiments involving embryo culture, mice from different genetic backgrounds develop at different rates. Thus, the time of day at which to harvest two-cell embryos will need to be empirically determined. In general, two-cell embryos are found before noon of 1.5 dpc.
As in all experiments involving embryo culture, mice from different genetic backgrounds develop at different rates. Thus, the time of day at which to harvest two-cell embryos will need to be empirically determined. In general, two-cell embryos are found before noon of 1.5 dpc. -
Hook up the electrofusion device to its electrode. Turn the machine on and make sure that the switch on the back of the device is set to “non-electrolyte.” Alternatively, one-cell embryos can be incubated with cytochalasin B to induce polyploidy21,22. Although this does not require specialized electrofusion equipment, it does require careful optimization of dosage and incubation times. Electrofusion of mouse embryos has been shown to produce only tetraploid embryos18,39, but cytochalasin B may, in some situations, also result in 4n:2n mosaicism within a single embryo (reviewed in ref. 17).
 By switching to “electrolyte,” it becomes possible to carry out fusion directly in growth medium (e.g., KSOM). In doing this, however, one sacrifices the ability to use the weak A/C orientation field. In this case, embryos must be oriented manually.
By switching to “electrolyte,” it becomes possible to carry out fusion directly in growth medium (e.g., KSOM). In doing this, however, one sacrifices the ability to use the weak A/C orientation field. In this case, embryos must be oriented manually. This protocol was written for use with a CF-150B pulse gererator. Alternative devices such as those produced by BTX can also be used. The key parameter for electrofusion is that a voltage differential of roughly 0.09–0.15 V µm–1 be applied across the embryos. This can be calculated based on the gap distance of the electrode but will ultimately require empirical optimization. Electrodes of different gap distances are commercially available, ranging from 250-µm to 1-mm gap distances. For the electrofusion of two-cell-stage mouse embryos for the induction of tetraploidy, we routinely use an electrode with a 250-µm gap. The settings described below are for such an electrode.
This protocol was written for use with a CF-150B pulse gererator. Alternative devices such as those produced by BTX can also be used. The key parameter for electrofusion is that a voltage differential of roughly 0.09–0.15 V µm–1 be applied across the embryos. This can be calculated based on the gap distance of the electrode but will ultimately require empirical optimization. Electrodes of different gap distances are commercially available, ranging from 250-µm to 1-mm gap distances. For the electrofusion of two-cell-stage mouse embryos for the induction of tetraploidy, we routinely use an electrode with a 250-µm gap. The settings described below are for such an electrode.
On the electrofusion device, increase the “repeat” to 2. Pressing the “mode” button toggles the displays needed for the next steps. The square pulse voltage should be at 35 V (this is adjusted by the “amplitude”). The pulse duration should be 35 µs and is adjustable with the “width” dial. The AC voltage should be set to 0.5–1.0 V; this will require that the attenuator be turned on (press the “Apl./10” button). Make sure the “engage” light is off. These settings will deliver two pulses of 35 V (DC current) for 35 µs with an A/C orientation field strength of about 0.5–1 V. All this occurs across a 250-µm channel between the electrodes. Best fusion efficiency is obtained when the machine is left on for 1–2 h before doing the fusion.
Fill the electrode channel with the 0.3 M mannitol solution. Place a few more drops of mannitol on a clean 6-cm tissue culture dish. Place an additional one or two drops of mannitol on the unused area of the microscope slide. These drops will serve as embryo reservoirs. The function of mannitol is to act as an osmolyte to maintain the osmotic balance of the cells without the need for electrically conductive salt ions.
Transfer 20–30 two-cell embryos to the first of three mannitol drops in the 6-cm plate. The embryos will float initially but will gradually sink to the bottom (Fig. 3, part 1).
Transfer the embryos through the next two drops, once again waiting for them to sink. Once equilibrated, transfer them to the reservoir drop on the microscope slide (Fig. 3, part 2).
-
Load 10–15 embryos in a transfer pipette. Carefully place the embryos in a single-file line between the electrodes (Fig. 3, part 3).
 Although many more embryos can be loaded onto most electrodes at any one time, manipulation of 10–15 at any given time has ultimately proven the most convenient in terms of overall fusion efficiency.
Although many more embryos can be loaded onto most electrodes at any one time, manipulation of 10–15 at any given time has ultimately proven the most convenient in terms of overall fusion efficiency. Press the “engage” button on the fusion apparatus. This will cause each embryo to rotate in the electrode channel so that it is aligned in such a fashion that its plane of cleavage is parallel to the electrodes (Fig. 3, part 4). Proper alignment is essential to high fusion efficiency, but leaving the embryos for too long in the mannitol may damage them.
Once aligned, press the trigger to initiate the electrical pulse (Fig. 3, part 5).
-
Transfer these embryos back to a mannitol drop on the slide and repeat the procedure with the remaining embryos. Ideally, change the mannitol solution between each electrofusion. The investigator should pay attention to the evaporation of the mannitol drops. If the embryos that are transferred from one drop to another do not sink immediately, replace the solutions with fresh mannitol. Failure to do this will result in poor fusion efficiency or cell lysis.
 Transferring the embryos to a mannitol droplet allows the user to continue to use the same pipette for the next batch of embryos, but it does keep the embryos in the mannitol for a longer period of time. An alternative is to transfer the embryos to culture medium. In this case, the user will need to prevent contamination of the mannitol droplets by small amounts of growth medium. This is typically done by frequently changing transfer pipettes.
Transferring the embryos to a mannitol droplet allows the user to continue to use the same pipette for the next batch of embryos, but it does keep the embryos in the mannitol for a longer period of time. An alternative is to transfer the embryos to culture medium. In this case, the user will need to prevent contamination of the mannitol droplets by small amounts of growth medium. This is typically done by frequently changing transfer pipettes.
-
Immediately transfer the embryos in a minimal volume of mannitol to a clean tissue culture dish containing three drops of M2 (or FHM). Wash embryos through M2 drops (Fig. 3, part 6). Then wash embryos though two or three drops of KSOM.
 Prolonged exposure to the mannitol solution will harm the embryos. This will become evident by the gradual development of a rough or gray appearance to the cell membranes.
Prolonged exposure to the mannitol solution will harm the embryos. This will become evident by the gradual development of a rough or gray appearance to the cell membranes. After washing, place all embryos in the KSOM drop on the unfused side of the remaining tissue culture dish and return this dish to the 37 °C incubator under 5% CO2.
-
Every 10–15 min, check the embryos for fusion under the stereo dissecting microscope. Once they have fused completely to a single cell (Fig. 4c–e; Supplementary Video 1), transfer them to the fused side of the dish (Fig. 3, part 7). Before you transfer an embryo to the fused side, vary the lighting of the microscope. Occasionally, you will find that embryos that seemed to have fused under one lighting condition are clearly still two-cell embryos when subjected to different lighting conditions. It is important to move embryos soon after fusion, as the next round of cell division will produce a two-cell embryo that is indistinguishable from unfused diploid two-cell embryos (Fig. 4f–h). It will take between 1 and 2 h for all fusion events to occur.
 Some embryos, depending on the time of day and strain used, will divide before fusion. These can be moved to a separate drop and used as diploid embryos in parallel aggregations.
Some embryos, depending on the time of day and strain used, will divide before fusion. These can be moved to a separate drop and used as diploid embryos in parallel aggregations.
Return embryos to the incubator to culture to the four-cell stage (overnight).
Return to Step 18 of the main protocol.
The techniques described here have existed for many years, and variations may be found in several original publications, review papers and book chapters. The protocols described in most publications, including the one here, are derived in large part from methods12,23,24 that are themselves derivates of an array of experimental embryological protocols documented in earlier works (e.g., ref. 25). The reader is referred in particular to two sources6,11 for alternative descriptions of these and other techniques that pertain to mouse experimental embryology. Additional information regarding extensions of these techniques as well as alternative protocols describing zona pellucida removal or ES cell and embryo culture can be found26,27. A methodical description of the generation, troubleshooting and subsequent analyses of mouse mutations is also available7. Although this protocol is focused on making mouse chimeras, many of these techniques can be adapted to other mammalian species. As a final note, although these techniques are commonly used in a variety of laboratories around the world, special permission to use these protocols may be required by institutional animal care and use committees. All experiments with live mice should be carried out in accordance with relevant guidelines and regulations.
MATERIALS
REAGENTS
Aggregation plates (see REAGENT SETUP)
 Should be made 5–24 h before aggregation.
Should be made 5–24 h before aggregation.ES cells (see REAGENT SETUP)
ES cell medium6 (see REAGENT SETUP)
0.1% (w/v) gelatin (made up in sterile dH2O and autoclaved to dissolve)
PBS (Ca2+- and Mg2+-free; GIBCO, cat. no. 13151-014), sterile
0.25% (v/v) trypsin/0.2% (w/v) EDTA in PBS (GIBCO, cat. no. 25200-056)
KSOM+aa (Specialty Media, cat. no. MR-121-D)
 KSOM+aa (KSOM) should be stored frozen (−20 °C) for no more than 3 months and thawed (4 °C) for no more than 2 weeks.
KSOM+aa (KSOM) should be stored frozen (−20 °C) for no more than 3 months and thawed (4 °C) for no more than 2 weeks.Mineral oil (Sigma, cat. no. M8410)
70% (v/v) ethanol
 Ethanol is flammable and may be harmful by inhalation or ingestion. The use of gloves is prescribed.
Ethanol is flammable and may be harmful by inhalation or ingestion. The use of gloves is prescribed.dH2O, sterile
Acid Tyrode’s solution (Sigma, cat. no. T1788)
 Low pH.
Low pH.Polyvinylpyrrolidone (PVP; Sigma, cat. no. P0930) to reduce stickiness of zona pellucida and embryos (Step 21; optional)
M2 or FHM medium (Specialty Media, cat. no. MR-122-D)
Pregnant mice for production of host mouse embryos (see REAGENT SETUP): 2.5 days postcoital (dpc), for the generation of diploid ↔ diploid chimeras only, or 1.5 dpc, for the generation of tetraploid ↔ diploid chimeras only
Recipient pseudopregnant female mouse (set up for vasectomized mating on day 2; mating confirmed on day 3)
0.3 M mannitol (Sigma, cat. no. M4125), for induction of tetraploidy by electrofusion only
EQUIPMENT
3.5-, 6- and 10-cm tissue culture dishes (Falcon, cat. nos. 35-3001, 35-3004 and 35-3003, respectively)
80- to 100-µm internal bore diameter transfer pipettes for manipulating embryos (see ref. 6 for instructions on how to make and use)
Transfer pipettes pulled finely enough to manipulate ES cell clumps (internal bore diameter, ~50 µm)
Stereo dissecting microscope
Inverted microscope
Aggregation needles (BLS; http://www.bls-ltd.com/) for generating depression wells in plastic plates (department store darning needles can also be used, but several variants may need to be tested for the optimal curvature for producing depressions in the plates)
Electrofusion device and electrode (e.g., CF150B pulse generator, BLS or Electro Cell Manipulator ECM 2001, BTX; http://www.btxonline.com/; with microscope slide electrode), for use with induction of tetraploidy by electrofusion only
REAGENT SETUP
Aggregation plates
On day 3 (5–24 h before aggregation), place 14 drops of KSOMon a 3.5-cmtissue culture dish. The drops should be 2–3 mm in diameter and should be covered with a layer of mineral oil. Use the pattern indicated (Fig. 2, part 3). Sterilize the aggregation needle with 70% (v/v) ethanol and rinse in sterile dH2O. While viewing the plate through a stereo dissecting microscope, press the needle firmly into the plastic, making six depressions per drop (Fig. 2, part 3). Place aggregation plates in the 37 °C, 5% CO2 incubator 5–24 h before aggregation to equilibrate.
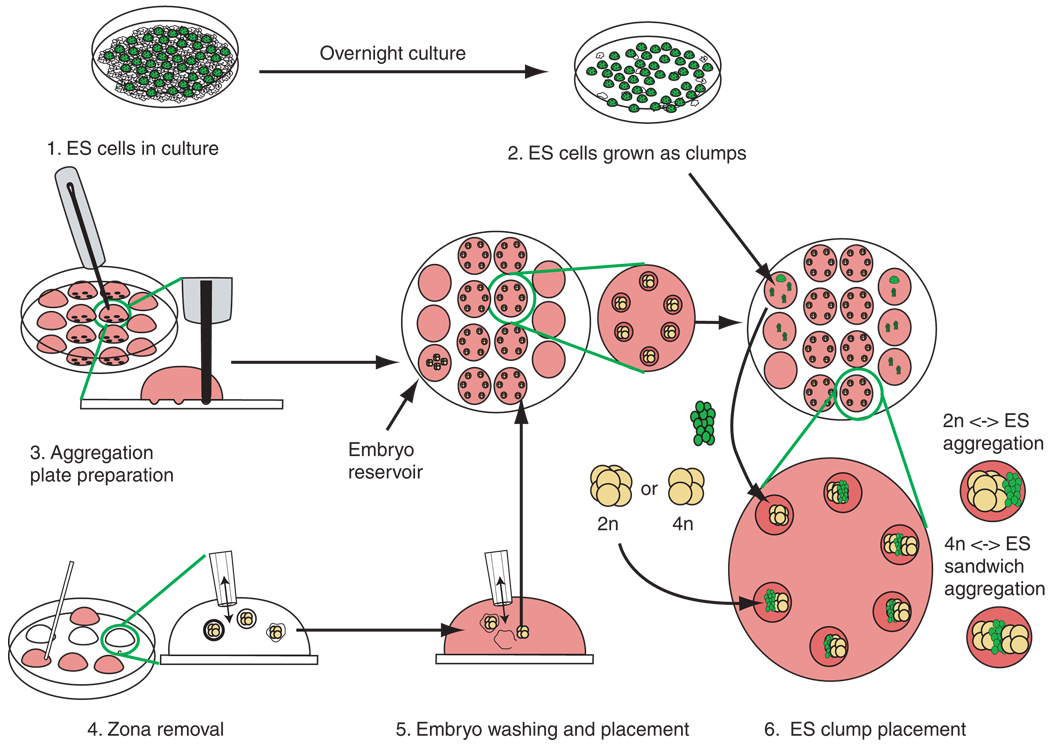
Figure 2.
Generalized scheme for the production of aggregation chimeras. The procedure can be divided into six parts. 1. ES cells are passaged to deplete feeders. 2. ES cells are lightly trypsinized to release small clumps. ES cell clumps are transferred to drops serving as ES reservoirs on the outer rows of the aggregation plate. 3. The aggregation plate routinely contains four rows of KSOM drops: two outer rows of three drops that serve as ES cell and embryo reservoirs and two inner rows of four drops that contain depressions required for the aggregation of embryos with ES cells. Once made, the drops are overlayed with mineral oil. Depression wells are then made in the central two rows of wells of the aggregation plate. We routinely make six depressions per drop. It is advisable that the depressions are made and the plate be equilibrated in an incubator before the addition of cells or embryos. 4. The zona pellucida of the embryos is dissolved in Acid Tyrode’s solution. It is important to keep pipetting the embryos, as they tend to become sticky and should not make prolonged contact with the plastic surface of the dish, otherwise they can be difficult to dislodge. 5. The embryos are then washed through M2 medium. Zona-free embryos are transferred to a reservoir drop on the aggregation plate. Single embryos are immediately transferred into each depression. This must be done quickly, as zona-free embryos will aggregate with each other if left in contact. If tetraploid chimeras are being set up, then half of the embryos should be left in a reservoir until the ES cell clumps have been added to the embryos in the depressions. 6. ES cell clumps are transferred from the reservoirs to the depression wells. Care must be taken that the ES cells are in physical contact with the embryos, otherwise the embryos may form blastocysts that will not have incorporated the ES cells. For tetraploid aggregations, a second embryo is then placed in the depression so that the ES cells are sandwiched between two embryos.
 KSOM typically contains penicillin and streptomycin. Simple ethanol sterilization of the aggregation needle in combination with the use of antibiotics is generally sufficient to maintain an aseptic culture.
KSOM typically contains penicillin and streptomycin. Simple ethanol sterilization of the aggregation needle in combination with the use of antibiotics is generally sufficient to maintain an aseptic culture.
 Larger drops will be more affected by vibration and movement. This will lead to greater fluid movement in the depression wells. Generally, six embryos will be placed per drop, with six drops per plate maintained as ES cell or embryo reservoirs.
Larger drops will be more affected by vibration and movement. This will lead to greater fluid movement in the depression wells. Generally, six embryos will be placed per drop, with six drops per plate maintained as ES cell or embryo reservoirs.
 The malleability of the plastic directly influences the depth of the depression one can make in a dish. This, in turn, affects the probability that cells and embryos will come into contact in the well. Falcon 35-3001 3.5-cm tissue culture dishes have proven to be sufficiently malleable. Pressing too hard will crack the plastic, but gentle pressure will produce shallow depressions that will not result in efficient juxtaposition of the embryos and ES cells. The goal is to form the deepest depression possible, without cracking the plastic. In practice, one will find that placing the depressions away from the center of the drop facilitates manipulation of embryos and ES cells in later steps.
The malleability of the plastic directly influences the depth of the depression one can make in a dish. This, in turn, affects the probability that cells and embryos will come into contact in the well. Falcon 35-3001 3.5-cm tissue culture dishes have proven to be sufficiently malleable. Pressing too hard will crack the plastic, but gentle pressure will produce shallow depressions that will not result in efficient juxtaposition of the embryos and ES cells. The goal is to form the deepest depression possible, without cracking the plastic. In practice, one will find that placing the depressions away from the center of the drop facilitates manipulation of embryos and ES cells in later steps.
 Although usually made on day 3, aggregation plates can be made on the day of aggregation (day 4). Although at least 5 h of equilibration is typical, the minimum time required for maximum efficiency has not been tested. Plates over 24 h old should not be used.
Although usually made on day 3, aggregation plates can be made on the day of aggregation (day 4). Although at least 5 h of equilibration is typical, the minimum time required for maximum efficiency has not been tested. Plates over 24 h old should not be used.
ES cells
ES cells should be thawed approximately 5 d before beginning the protocol and passaged once (Fig. 1) ES cells are often grown on mitotically inactive fibroblast feeder cell layers. The two most commonly used feeder cells are primary mouse embryo fibroblasts (MEFs) and STO cells, a thioguanine- and ouabain-resistant subline of SIM mouse fibroblasts28. In many cases, the addition of leukemia inhibitory factor (LIF) to the ES cell medium and the plating of cells on a gelatin substrate alleviate the requirement of the feeder cell layer6. To retain full developmental potential, especially of hybrid cell lines used for generating completely ES cell–derived embryos through the generation of tetraploid chimeras, we would, however, recommend the use of both feeders and medium supplemented with LIF. If you are generating conditional alleles, it is recommended that you test the loxP or FRT sites using transient transfection of the appropriate site-specific recombinase7.
ES cell medium
Prepare a solution of the following: 15% (v/v) FCS (e.g., Hyclone, cat. no. A-1115-L; we routinely batch test for optimum performance), 2 mM glutamine (GIBCO, cat. no. 35050-61), 0.1 mM MEM non-essential amino acids (GIBCO, cat. no. 11140-050), 0.1 mM β-mercaptoethanol (Sigma, cat. no. M7522), 1 mM sodium pyruvate (GIBCO, cat. no. 11360-070), 50 U ml–1 penicillin, 50 µg ml–1 streptomycin (GIBCO, cat. no. 15070-063), 1000 U ml–1 LIF/ESGRO (Chemicon, cat. no. ESG-1107). Media should be prepared in a tissue culture hood and should require no further sterilization. Store up to 1 month at 4 °C.
 Preparation of ES cell medium may present exposure to ingestion or absorption hazards.
Preparation of ES cell medium may present exposure to ingestion or absorption hazards.
Pregnant mice for production of host mouse embryos
The choice of strain used as a host embryo is important7. Generally, one chooses a strain with a genetic reporter that is different from the ES cell background. This will most typically be a coat color polymorphism or an allele that encodes a ubiquitously expressed fluorescent protein in either the host embryo or donor ES cell7,29,30. Thus, individual cells that arise from either the ES or host embryo compartment of the chimera can be distinguished. In addition, the offspring of the chimera can be rapidly identified as progeny derived from either the host embryo or ES cell genome. It has been shown in chimeras produced by morula ↔ morula aggregation that, in some strain combinations, one strain will be represented in a higher percentage of cells than the other31. Although this has not been rigorously tested, it is commonly believed that ES cell ↔ morula aggregation chimeras will have similar behaviors. In recent years, the use of 129B6 hybrid ES cells has proven particularly efficient for the production of tetraploid ↔ diploid chimeras6,32,33. Pups derived from this technique show essentially wild-type characteristics, although they tend to be slightly larger and have an elevated hematocrit when compared with diploid ↔ diploid chimeras34. For tetraploid hosts, it should be noted that although the embryonic portion of a midgestation tetraploid ↔ diploid chimera will be >99% diploid, a few tetraploid cells do persist throughout the fetus, especially in tissues such as the heart18. If the study requires that the chimeras themselves be analyzed, it is recommended that the embryonic stem cells or host embryo be marked by a ubiquitously expressed, genetically encoded reporter. The most commonly used reporters are fluorescent proteins or chromogenic substrates such as alkaline phosphatase or lacZ (reviewed in ref. 35). This allows the investigator to distinguish between the two populations of cells and therefore to measure the extent of (tetraploid) host embryo contribution to the tissue of interest. F1 CBA/B6 and outbred stocks have commonly been used for induction of tetraploidy by electrofusion (reviewed in ref. 17).
PROCEDURE
Preparation of mouse embryonic stem (ES) cells (days 1–4)
-
Set up an ES cell culture on day 1 by plating about 2 × 106 cells on a 10-cm tissue culture dish. Plate the cells so that they will reach 70% confluency by day 3. Change the ES cell medium on day 2. Using an inverted microscope with DIC or phase contrast optics, confirm that on day 3 (the day before aggregation; Fig. 1) ES cells are 70% confluent (Fig. 2 part 1).
 For the best ES cell competency, be kind to your ES cells. You are asking a small number of ES cells to generate every tissue of a mouse. ES cell cultures, in general, should be tended to meticulously. Change the medium once per day. The phenol red pH indicator in the ES medium should not be allowed to develop a bright yellow color. Although this protocol assumes that the investigators already have genetically modified ES cells, it is important to establish that the ES cell line in which the mutation was introduced is itself competent for germline contribution. This is established by generating chimeras using genetically unmodified ES cells and carrying out test matings.
For the best ES cell competency, be kind to your ES cells. You are asking a small number of ES cells to generate every tissue of a mouse. ES cell cultures, in general, should be tended to meticulously. Change the medium once per day. The phenol red pH indicator in the ES medium should not be allowed to develop a bright yellow color. Although this protocol assumes that the investigators already have genetically modified ES cells, it is important to establish that the ES cell line in which the mutation was introduced is itself competent for germline contribution. This is established by generating chimeras using genetically unmodified ES cells and carrying out test matings. Add 2–5 ml of 0.1% gelatin solution to a 6-cm tissue culture dish. Swirl to coat the bottom of the dish and then aspirate. Some investigators like to incubate dishes coated with gelatin for up to 2 h at room temperature (20–25 °C), or until the gelatin dries.
-
Remove the medium from the ES cells and replace with fresh ES medium and incubate in a humidified incubator at 37 °C, 5% CO2 for 2 h.
 Here and throughout, all incubations at 37 °C should be carried out in a humidified 37 °C, 5% CO2 incubator.
Here and throughout, all incubations at 37 °C should be carried out in a humidified 37 °C, 5% CO2 incubator. Aspirate ES medium and wash cells with 2–3 ml of sterile PBS (without Ca2+ and Mg2+) to remove residual proteins. Remove PBS and add 0.5 ml 0.25% trypsin/0.2% EDTA in PBS.
Incubate culture for 3–4 min at 37 °C.
Neutralize the trypsin with 3 ml of ES cell medium. Pipet the solution up and down a few times to reduce the solution to clumps of three to eight cells.
Wash the gelatinized dish once with PBS.
Add 1 ml of trypsinized ES cells to the gelatinized dish.
Bring the ES cell solution volume to 5 ml with ES medium and incubate at 37 °C overnight. This step reduces the number of feeder cells present in the subsequent steps.
Prepare aggregation plates to be used on day 4.
On day 4 (Fig. 1), remove the medium from the ES cells and replace with fresh ES medium. Incubate at 37 °C for 2 h.
Repeat Steps 4–6.
Using a 50-µm transfer pipette, transfer 20–30 µl of the ES cell suspension to a drop of KSOM on an aggregation plate (Fig. 2, part 2).
Host embryo preparation (day 3 for tetraploid ↔ diploid chimeras; day 4 for diploid ↔ diploid chimeras)
-
14
Prepare KSOM microdrop embryo culture dishes by placing three to four drops of KSOM in a 6-cm tissue culture dish, using a transfer pipette. Cover all the drops with a layer of mineral oil and incubate at 37 °C.
 For tetraploid ↔ diploid chimeras, harvest two-cell embryos (Steps 14–17) and induce tetraploidy by electrofusion (Box 1) on day 3 of the protocol. For diploid ↔ diploid chimeras, harvest eight-cell embryos (Steps 14–17) on day 4.
For tetraploid ↔ diploid chimeras, harvest two-cell embryos (Steps 14–17) and induce tetraploidy by electrofusion (Box 1) on day 3 of the protocol. For diploid ↔ diploid chimeras, harvest eight-cell embryos (Steps 14–17) on day 4. In theory, it is best to prepare all microdrop cultures a few hours before the procedure so as to let them equilibrate in the incubator. In practice, it is usually acceptable to prepare these dishes immediately before the technique.
In theory, it is best to prepare all microdrop cultures a few hours before the procedure so as to let them equilibrate in the incubator. In practice, it is usually acceptable to prepare these dishes immediately before the technique. -
15
Wash dissecting tools with 70% ethanol and sterile dH2O.
 Minute quantities of salts, detergents, fixatives and/or ethanol can affect the viability of preimplantation embryos. It is best to have a dedicated set of surgical instruments available for harvesting live embryos.
Minute quantities of salts, detergents, fixatives and/or ethanol can affect the viability of preimplantation embryos. It is best to have a dedicated set of surgical instruments available for harvesting live embryos. -
16
Flush host embryos from oviducts of 2.5-dpc pregnant mice (eight-cell embryos for the generation of diploid ↔ diploid chimeras) or 1.5-dpc pregnant mice (two-cell embryos for the generation of tetraploid ↔ diploid chimeras ) as described6 (this reference includes a description of this technique and general protocols for handling and transferring preimplantation mouse embryos).
 The timing of the light/dark cycle and the injection of hormones for superovulation (reviewed in ref. 6) have the most significant effects on the timing of fertilization, and subsequently on the developmental stage of the embryos at any particular time during the day. In addition, genetic background also affects the rate of development. The investigator will need to determine empirically the best time of day for harvesting a particular stage of embryo. In general, two- and four-cell-stage embryos can be harvested between 9 a.m. and 9 p.m. 1.5 dpc; eight-cell embryos are present at 2.25–2.5 dpc. Embryos that are too young may be cultured in KSOM microdrops until they develop to the appropriate stage.
The timing of the light/dark cycle and the injection of hormones for superovulation (reviewed in ref. 6) have the most significant effects on the timing of fertilization, and subsequently on the developmental stage of the embryos at any particular time during the day. In addition, genetic background also affects the rate of development. The investigator will need to determine empirically the best time of day for harvesting a particular stage of embryo. In general, two- and four-cell-stage embryos can be harvested between 9 a.m. and 9 p.m. 1.5 dpc; eight-cell embryos are present at 2.25–2.5 dpc. Embryos that are too young may be cultured in KSOM microdrops until they develop to the appropriate stage. -
17
Transfer embryos to microdrop cultures with an 80- to 100-µm transfer pipette and incubate at 37 °C. For tetraploid ↔ diploid chimeras, continue with Box 1 (Fig. 3) before proceeding with Step 18.
 Depending on the age of the embryos, the procedure may need to be interrupted to allow the embryos to develop in vitro to the appropriate cell number for either diploid ↔ diploid or tetraploid ↔ diploid aggregations.
Depending on the age of the embryos, the procedure may need to be interrupted to allow the embryos to develop in vitro to the appropriate cell number for either diploid ↔ diploid or tetraploid ↔ diploid aggregations. -
18
In a 10-cm tissue culture dish, place four large (6–10 mm) drops of M2 medium (FHM can be used instead of M2) and three large drops of Acid Tyrode’s (AT) solution (Fig. 2, part 4). The latter is used to remove the proteinaceous zona pellucida from the embryos. Refracted light at the rounded borders of the drops can obscure or even hide embryos. One can minimize this effect by making ‘square’ drops. This is accomplished by making four tiny drops that define the corners of the square and then flooding the intervening space with the appropriate medium. An evaluation of alternative methods for the removal of the zona pellucida, including use of Pronase (Sigma, cat no. P8811), is described36.
-
19
Place 20–30 embryos in the first drop of M2.
-
20
Transfer five to ten embryos, depending on skill, to the first drop of AT. Immediately transfer these embryos to the second and then third AT drops.
 The buffers present in M2 will neutralize the Acid Tyrode’s solution. It is therefore important to minimize the volume of medium transferred with embryos when pipetting.
The buffers present in M2 will neutralize the Acid Tyrode’s solution. It is therefore important to minimize the volume of medium transferred with embryos when pipetting. -
21
In the third AT drop, observe the zona pellucida. It should gradually erode to a thin flexible shell (Fig. 2, part 5), which will begin to crumple in the few seconds before it completely disappears. Gentle pipetting of the embryos should rupture the zona pellucida and free the embryos. With practice, by carefully controlling the embryos it is possible to remove the zona pellucida without letting the embryos sink and ever allowing them to touch the plastic (Fig. 2, part 4). Once they are free of the zona pellucida, immediately transfer them through the drops of M2. To reduce stickiness, PVP can be added to the medium and used to coat instruments37.
 This is the most difficult step of the entire procedure. It should be attempted beforehand on practice preimplantation embryos of any stage. Once in AT solution, the digestion of the zona pellucida occurs quickly and is coincident with an increased stickiness of the embryos. The zona pellucida, and especially denuded embryos, will stick firmly to the plastic dish and even inside the transfer pipette. This can be minimized by using small batches of embryos, constantly moving the embryos while they are in AT by pipetting and removing the embryos to M2 immediately after zona pellucida rupture.
This is the most difficult step of the entire procedure. It should be attempted beforehand on practice preimplantation embryos of any stage. Once in AT solution, the digestion of the zona pellucida occurs quickly and is coincident with an increased stickiness of the embryos. The zona pellucida, and especially denuded embryos, will stick firmly to the plastic dish and even inside the transfer pipette. This can be minimized by using small batches of embryos, constantly moving the embryos while they are in AT by pipetting and removing the embryos to M2 immediately after zona pellucida rupture. -
22
Repeat Steps 20 and 21 with the remaining embryos.
-
23
Wash the zona pellucida–free embryos through three drops of KSOM.
-
24
Transfer embryos to depression wells of the aggregation dish. For each well, place one diploid embryo inside the well. For tetraploid aggregations (Fig. 4), a second embryo should be placed in each embryo reservoir drop (Fig. 2, part 5).

-
25
Gently transfer the aggregation plates to the incubator for 1 h. This incubation step, while not essential, gives the embryos in the depression well time to adhere weakly to the plastic. This will facilitate movement of the plates and subsequent addition of ES cells.
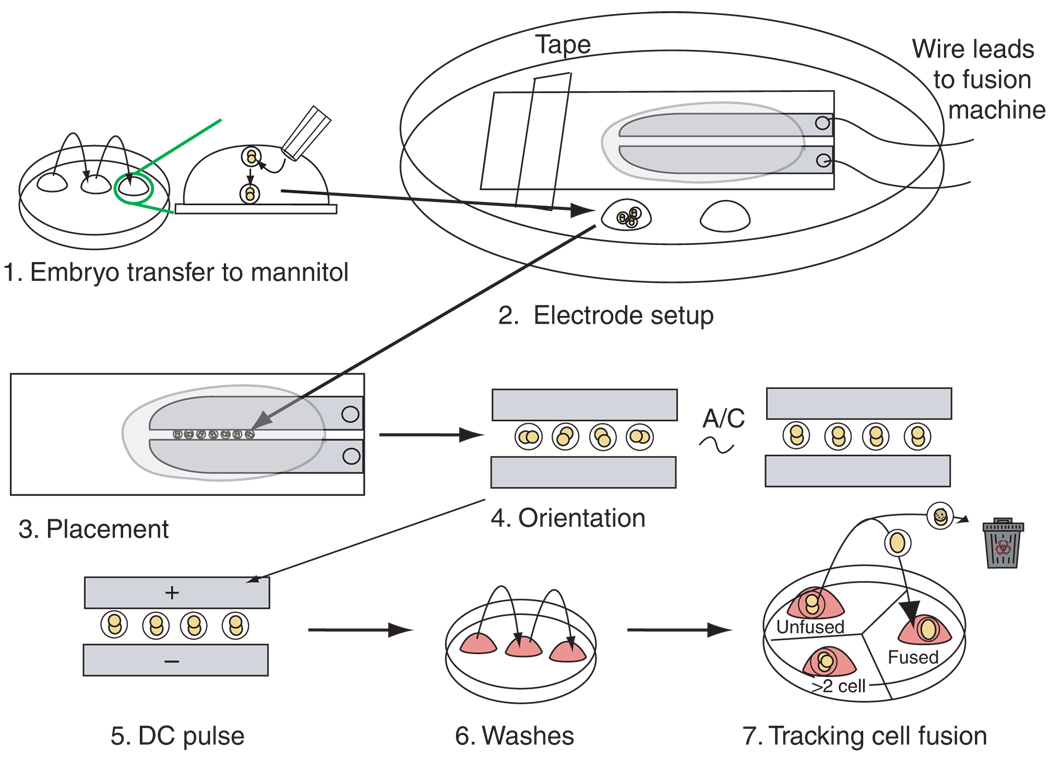
Figure 3.
Protocol for generating tetraploid embryos by electrofusion. The procedures can be divided into seven parts. 1. Embryos are washed through mannitol solution, allowing them to sink to the bottom of the drop. 2. The electrode slide is placed (taped or clipped) to the lid of a 10-cm tissue culture dish. The embryos are transferred to a reservoir drop of mannitol placed on the plastic dish to the side of the electrode slide. 3. The embryos (generally 15–20 at a time) are lined up in single file between the electrodes. 4. Embryos are oriented with the interface between the two blastomeres perpendicular to the electrodes using an A/C orientation field. 5. The aligned embryos are pulsed with DC current to initiate blastomere fusion. 6. Immediately after pulsing, embryos are washed through sequential drops of M2 medium to remove any remaining mannitol. Embryos are then sequentially washed into KSOM medium. 7. They are then placed in a drop of KSOM in a dish containing three drops of KSOM. The drops are labeled ‘unfused’, ‘fused’ and ‘cleaved’ or ‘>2 cell’. The KSOM drops are covered in mineral oil, and the plate is placed in an incubator. The plate is removed from the incubator at 10- to 15-min intervals to monitor the process of fusion. As blastomeres fuse, individual embryos are immediately transferred from the unfused drop to the fused drop. Unhealthy or lysing embryos are discarded. The drop labeled ‘>2 cell’ can be used for placing older, unfused (diploid) embryos. These embryos will begin to cleave soon after pulsing and should immediately be removed from the pool of fusing embryos so as not to be confused with embryos that may have fused and then cleaved.
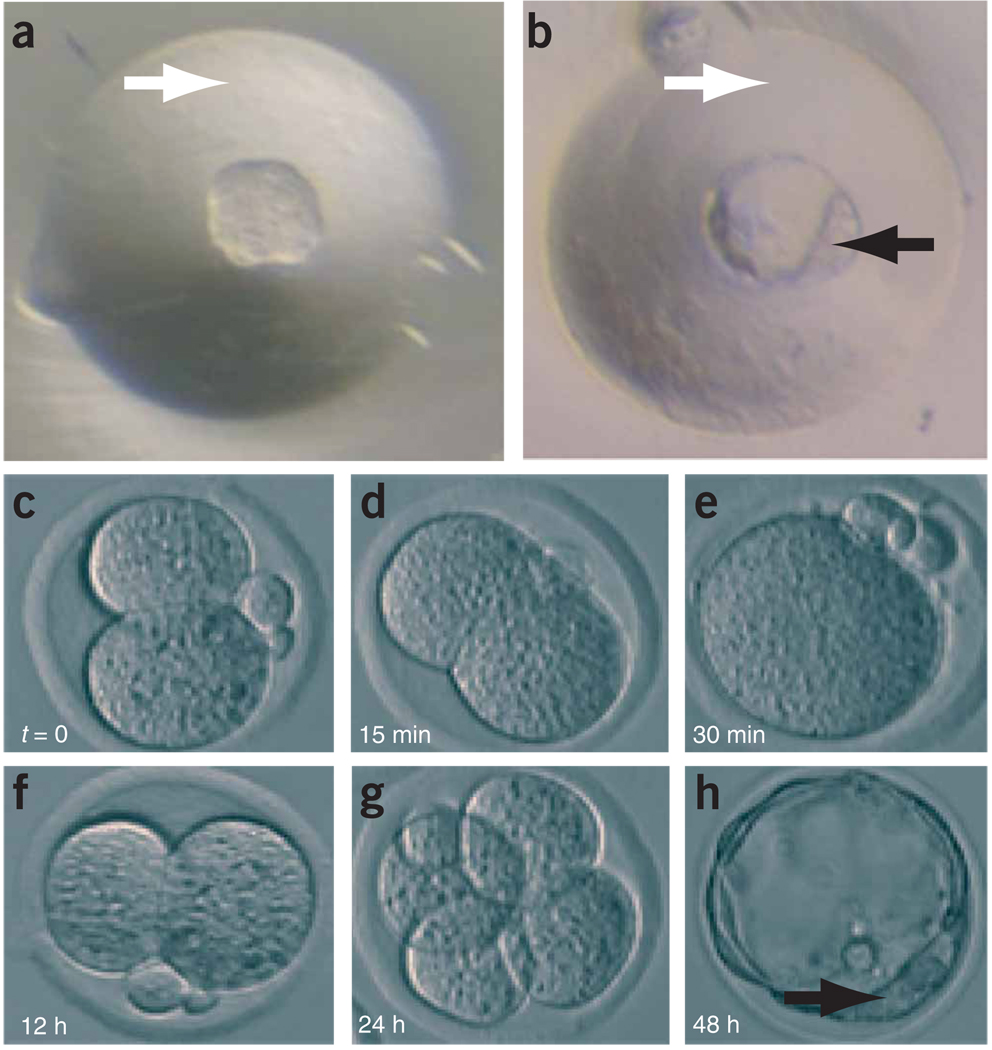
Figure 4.
Morphology of embryos throughout the procedure. (a,b) Development of zona-free embryos in depression wells: zona-free morula (a) and zona-free blastocyst after 24 h in culture (b). White arrow, depression well; black arrow, inner cell mass of blastocyst. (c–e) Induction of tetraploidy by electrofusion. Also see Supplementary Video 1. Two-cell diploid embryo before fusion (c). Initial stages of fusion as plasma membrane breaks down (d). Fusion is completed, which results in a one-cell tetraploid embryo (e). (f–h) In vitro development of tetraploid embryos. Two-cell tetraploid embryo 12 h after fusion (f). Four-cell tetraploid embryo 24 h after fusion (g). Tetraploid blastocyst 48 h after fusion (h). A series of images that shows the progression of both a diploid embryo ↔ diploid ES cell aggregation and a tetraploid embryo ↔ diploid ES cell aggregation, where the ES cells are expressing a GFP transgene, has been reported30.
Aggregation
-
26
Using a 50-µm transfer pipette, select a small (8–15 cells) clump of ES cells (from Step 13; even though in theory only one to three ES cells contribute to the somatic lineages of chimeras38).
-
27
Place the tip of the pipette slightly above the intended depression well, about midway between the surface of the plastic and the top of the microdrop. Gently expel the ES cell clump. The clump should slowly sink toward the embryo into the depression well (Fig. 2, part 6).

-
28
For tetraploid aggregations only, select a second zona pellucida–free embryo from the embryo reservoir and carefully drop it into the depression well such that the two embryos ‘sandwich’ the ES cell clump (Fig. 2, part 6).

-
29
Repeat procedure for the remaining wells, until you use all the embryos.

-
30
Carefully transfer aggregation plates to the incubator and incubate undisturbed overnight.
 Any vibration or rapid movement of the aggregation plates at this point may result in separation of the ES cells from the embryos.
Any vibration or rapid movement of the aggregation plates at this point may result in separation of the ES cells from the embryos.
Transfer embryos to recipient pseudopregnant mouse
-
31
On day 5 (Fig. 1), transfer embryos to a recipient female mouse (a detailed description of this may be found in ref. 6).
 By this time, the investigator will have invested a considerable amount of time and resources into the project. All of this can be wasted by a poor embryo transfer. Thus, it is wise that the procedure be practiced on sacrificed females with AffiGel Blue Gelbeads (Bio-Rad, cat. no. 153-7301), in lieu of actual embryos. Later, the procedure should be practiced several times with living embryos and pseudopregnant females before attempting to transfer experimentally manipulated embryos. The efficiency should be such that over 90% of transferred embryos develop into fetuses or pups.
By this time, the investigator will have invested a considerable amount of time and resources into the project. All of this can be wasted by a poor embryo transfer. Thus, it is wise that the procedure be practiced on sacrificed females with AffiGel Blue Gelbeads (Bio-Rad, cat. no. 153-7301), in lieu of actual embryos. Later, the procedure should be practiced several times with living embryos and pseudopregnant females before attempting to transfer experimentally manipulated embryos. The efficiency should be such that over 90% of transferred embryos develop into fetuses or pups. Although blastocysts are normally present at 3.5 dpc, the greatest blastocyst transfer efficiency is obtained by the surgical transfer of in vitro–cultured blastocysts to the uteri of 2.5-dpc pseudopregnant females. The pseudopregnant female’s gestational age should be used to estimate the progression of the pregnancy. Thus, on the day after a 3.5-dpc blastocyst is transferred to a 2.5-dpc female, the pregnancy would be considered the same stage as a naturally mated 3.5-dpc pregnancy.
Although blastocysts are normally present at 3.5 dpc, the greatest blastocyst transfer efficiency is obtained by the surgical transfer of in vitro–cultured blastocysts to the uteri of 2.5-dpc pseudopregnant females. The pseudopregnant female’s gestational age should be used to estimate the progression of the pregnancy. Thus, on the day after a 3.5-dpc blastocyst is transferred to a 2.5-dpc female, the pregnancy would be considered the same stage as a naturally mated 3.5-dpc pregnancy.
![]()
Although the procedure must be coordinated over several days, the timing of individual steps is as follows. The preparation of aggregation plates on day 3 requires 10 min. Preparation of ES cells on days 1–4 requires the following time: 15 min on days 1 and 3 but 5 min on day 2 for Step 1, 5 min for Steps 2–3, 10 min for Steps 4–9 and 10 min for Steps 11–13. Host embryo preparation (on day 3 or 4, depending on whether tetraploid or diploid host embryos are to be used) requires 20 min to 1 h for Steps 14–17 and 30 min to 1.5 h for Steps 18–25. Aggregation on day 4 requires 30 min to 1 h for Steps 26–30. Transferring embryos to recipient pseudopregnant mice on day 5 requires 40 min to 1.5 h for Step 31. If the induction of tetraploidy by electrofusion is carried out on day 3, this will require 10 min for Steps 1–5, 20 min for Steps 6–13 and 1–2 h for Steps 14–16.
![]()
In Step 24, if the blastomeres of the embryo separate, simply pool the four (diploid) or two (tetraploid) blastomeres into a single depression well; there is a good chance they will re-adhere.
By careful breath control, one can guide the ES clump to rest against the embryo in Steps 27 and 28. This is done by gently expelling and aspirating fluid against the ES cell clump during its descent. If few embryos are available, it is possible to aggregate the ES cells with only one zona pellucida–free embryo each in Steps 28 and 29, but this will lead to a reduced efficiency.
Although the greatest blastocyst transfer efficiencies are obtained with 2.5-dpc pseudopregnant females (Step 31), a 0.5-dpc pseudopregnant female can be used in an emergency40. In this case, blastocysts should be transferred to the infundibulum of the oviduct rather than to the uterus. Transfer of blastocysts to 3.5-dpc uteri is also possible but results in the lowest efficiency.
Chimeras derived from tetraploid embryos (Box 1) can be delivered vaginally, but they are at risk for complications. Many investigators choose to deliver by caesarian section. This requires that a pregnant foster mother of the same gestational age as the pseudopregnant female be available to raise the pups.
During induction of tetraploidy by electrofusion (Box 1), if there is protein buildup on the electrode (Step 3), it can be removed by digestion with trypsin. If the embryos lyse in Step 11, check to make sure the attenuator (Apl./10) function is on. Because of variability in the apparatus, it may be necessary to optimize the voltages on a given piece of equipment. Do not let the embryos remain in mannitol longer than the minimum required time to equilibrate and transfer them. Use fresh mannitol after every one or two batches of embryos. If only a few embryos fuse (Step 14), try decreasing the number of embryos placed between the two electrodes at any one time. Unfused embryos can be subjected to electrofusion additional times.
ANTICIPATED RESULTS
The successful production of blastocysts depends to the greatest degree on the quality of the culture medium. Under optimal conditions, almost all aggregation wells should produce late-stage morula or blastocysts after overnight culture (Fig. 4b). For the tetraploidy protocol (Box 1), the efficiency of electrofusion is also variable. Electrofusion should occur in over 50–95% of embryos. Unfused embryos may be subjected to additional pulses as needed.
Supplementary Material
ACKNOWLEDGMENTS
We thank R.R. Behringer, E.H. Lacy and V.E. Papaioannou for advice, discussions and support.
Footnotes
COMPETING INTERESTS STATEMENT The authors declare that they have no competing financial interests.
Note: Supplementary information is available via the HTML version of this article.
Reprints and permissions information is available online at http://npg.nature.com/reprintsandpermissions
References
- 1.Bradley A, Evans M, Kaufman MH, Robertson E. Formation of germ-line chimeras from embryo-derived teratocarcinoma cell lines. Nature. 1984;309:255–256. doi: 10.1038/309255a0. [DOI] [PubMed] [Google Scholar]
- 2.Thompson S, Clarke AR, Pow AM, Hooper ML, Melton DW. Germ line transmission and expression of a corrected HPRT gene produced by gene targeting in embryonic stem cells. Cell. 1989;56:313–321. doi: 10.1016/0092-8674(89)90905-7. [DOI] [PubMed] [Google Scholar]
- 3.Tam PP, Rossant J. Mouse embryonic chimeras: tools for studying mammalian development. Development. 2003;130:6155–6163. doi: 10.1242/dev.00893. [DOI] [PubMed] [Google Scholar]
- 4.Kunath T, et al. Imprinted X-inactivation in extra-embryonic endoderm cell lines from mouse blastocysts. Development. 2005;132:1649–1661. doi: 10.1242/dev.01715. [DOI] [PubMed] [Google Scholar]
- 5.Tanaka S, Kunath T, Hadjantonakis AK, Nagy A, Rossant J. Promotion of trophoblast stem cell proliferation by FGF4. Science. 1998;282:2072–2075. doi: 10.1126/science.282.5396.2072. [DOI] [PubMed] [Google Scholar]
- 6.Nagy A, Gertsenstein M, Vinterstein K, Behringer RR. Manipulating the mouse embryo: a laboratory manual. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2003. [Google Scholar]
- 7.Papaioannou VE, Behringer RR. Mouse Phenotypes: A handbook of mutation analysis. Cold Spring Harbor, New York: Cold Spring Harbor Laboratory Press; 2005. [Google Scholar]
- 8.Wakayama T, et al. Differentiation of embryonic stem cell lines generated from adult somatic cells by nuclear transfer. Science. 2001;292:740–743. doi: 10.1126/science.1059399. [DOI] [PubMed] [Google Scholar]
- 9.Wood SA, et al. Simple and efficient production of embryonic stem cell-embryo chimeras by coculture. Proc. Natl. Acad. Sci. USA. 1993;90:4582–4585. doi: 10.1073/pnas.90.10.4582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Papaioannou V, Johnson R. Production of chimeras by blastocyst and morula injection of targeted ES cells. In: Joyner AL, editor. Gene Targeting: A Practical Approach. 2nd edn. Oxford: Oxford Univ. Press; 2000. pp. 133–175. [Google Scholar]
- 11.Nagy A, Rossant J. Production and analysis of ES cell aggregation chimeras. In: Joyner AL, editor. Gene Targeting: A Practical Approach. 2nd edn. Oxford: Oxford Univ. Press; 2000. pp. 177–206. [Google Scholar]
- 12.Nagy A, Rossant J, Nagy R, Abramow-Newerly W, Roder JC. Derivation of completely cell culture-derived mice from early-passage embryonic stem cells. Proc. Natl. Acad. Sci. USA. 1993;90:8424–8428. doi: 10.1073/pnas.90.18.8424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li X, et al. The genetic heterozygosity and fitness of tetraploid embryos and embryonic stem cells are crucial parameters influencing survival of mice derived from embryonic stem cells by tetraploid embryo aggregation. Reproduction. 2005;130:53–59. doi: 10.1530/rep.1.00667. [DOI] [PubMed] [Google Scholar]
- 14.Eggan K, et al. Hybrid vigor, fetal overgrowth, and viability of mice derived by nuclear cloning and tetraploid embryo complementation. Proc. Natl. Acad. Sci. USA. 2001;98:6209–6214. doi: 10.1073/pnas.101118898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Beddington RS, Robertson EJ. An assessment of the developmental potential of embryonic stem cells in the midgestation mouse embryo. Development. 1989;105:733–737. doi: 10.1242/dev.105.4.733. [DOI] [PubMed] [Google Scholar]
- 16.Nagy A, et al. Embryonic stemcells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- 17.Eakin GS, Behringer RR. Tetraploid development in the mouse. Dev. Dyn. 2003;228:751–766. doi: 10.1002/dvdy.10363. [DOI] [PubMed] [Google Scholar]
- 18.Eakin GS, Hadjantonakis AK, Papaioannou VE, Behringer RR. Developmental potential and behavior of tetraploid cells in the mouse embryo. Dev. Biol. 2005;288:150–159. doi: 10.1016/j.ydbio.2005.09.028. [DOI] [PubMed] [Google Scholar]
- 19.Mackay GE, West JD. Fate of tetraploid cells in 4n ↔ 2n chimeric mouse blastocysts. Mech. Dev. 2005;122:1266–1281. doi: 10.1016/j.mod.2005.09.001. [DOI] [PubMed] [Google Scholar]
- 20.Iwasaki S, Campbell KH, Galli C, Akiyama K. Production of live calves derived from embryonic stem-like cells aggregated with tetraploid embryos. Biol. Reprod. 2000;62:470–475. doi: 10.1095/biolreprod62.2.470. [DOI] [PubMed] [Google Scholar]
- 21.Tarkowski AK, Witkowska A, Opas J. Development of cytochalasin in B-induced tetraploid and diploid/tetraploid mosaic mouse embryos. J. Embryol. Exp. Morphol. 1977;41:47–64. [PubMed] [Google Scholar]
- 22.Ueda O, Jishage K, Kamada N, Uchida S, Suzuki H. Production of mice entirely derived from embryonic stem (ES) cell with many passages by coculture of ES cells with cytochalasin B induced tetraploid embryos. Exp. Anim. 1995;44:205–210. doi: 10.1538/expanim.44.205. [DOI] [PubMed] [Google Scholar]
- 23.Kubiak JZ, Tarkowski AK. Electrofusion of mouse blastomeres. Exp. Cell Res. 1985;157:561–566. doi: 10.1016/0014-4827(85)90143-0. [DOI] [PubMed] [Google Scholar]
- 24.McLaughlin KJ. Production of tetraploid embryos by electrofusion. Methods Enzymol. 1993;225:919–930. doi: 10.1016/0076-6879(93)25058-a. [DOI] [PubMed] [Google Scholar]
- 25.Mintz B. Allophenic mice of mutiple embryo origin. In: Daniel JC Jr, editor. Methods in Mammalian Embryology. San Francisco: W.H. Freeman and Company; 1971. pp. 186–214. [Google Scholar]
- 26.Kishigami S, et al. Production of cloned mice by somatic cell nuclear transfer. Nat. Protocols. 2006;1:125–138. doi: 10.1038/nprot.2006.21. [DOI] [PubMed] [Google Scholar]
- 27.Singer O, Tiscornia G, Ikawa M, Verma IM. Rapid generation of knockdown transgenic mice by silencing lentiviral vectors. Nat. Protocols. 2006;1:1–7. doi: 10.1038/nprot.2006.44. [DOI] [PubMed] [Google Scholar]
- 28.Ware LM, Axelrad AA. Inherited resistance to N- and B-tropic murine leukemia viruses in vitro: evidence that congenic mouse strains SIM and SIM.R differ at the Fv-1 locus. Virology. 1972;50:339–348. doi: 10.1016/0042-6822(72)90385-6. [DOI] [PubMed] [Google Scholar]
- 29.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 30.Hadjantonakis AK, Gertsenstein M, Ikawa M, Okabe M, Nagy A. Generating green fluorescent mice by germline transmission of green fluorescent ES cells. Mech. Dev. 1998;76:79–90. doi: 10.1016/s0925-4773(98)00093-8. [DOI] [PubMed] [Google Scholar]
- 31.Mullen RJ, Whitten WK. Relationship of genotype and degree of chimerism in coat color to sex ratios and gametogenesis in chimeric mice. J. Exp. Zool. 1971;178:165–176. doi: 10.1002/jez.1401780203. [DOI] [PubMed] [Google Scholar]
- 32.Eggan K, Jaenisch R. Differentiation of F1 embryonic stem cells into viable male and female mice by tetraploid embryo complementation. Methods Enzymol. 2003;365:25–39. doi: 10.1016/s0076-6879(03)65002-0. [DOI] [PubMed] [Google Scholar]
- 33.Eggan K, et al. Male and female mice derived from the same embryonic stem cell clone by tetraploid embryo complementation. Nat. Biotechnol. 2002;20:455–459. doi: 10.1038/nbt0502-455. [DOI] [PubMed] [Google Scholar]
- 34.Schwenk F, et al. Hybrid embryonic stem cell-derived tetraploid mice show apparently normal morphological, physiological, and neurological characteristics. Mol. Cell. Biol. 2003;23:3982–3989. doi: 10.1128/MCB.23.11.3982-3989.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hadjantonakis AK, Dickinson ME, Fraser SE, Papaioannou VE. Technicolour transgenics: imaging tools for functional genomics in the mouse. Nat. Rev. Genet. 2003;4:613–625. doi: 10.1038/nrg1126. [DOI] [PubMed] [Google Scholar]
- 36.Nijs M, Camus M, Van Steirteghem AC. Evaluation of different biopsy methods of blastomeres from 2-cell mouse embryos. Hum. Reprod. 1988;3:999–1003. doi: 10.1093/oxfordjournals.humrep.a136831. [DOI] [PubMed] [Google Scholar]
- 37.Hartshorn C, Rice JE, Wangh LJ. Differential pattern of Xist RNA accumulation in single blastomeres isolated from 8-cell stage mouse embryos following laser zona drilling. Mol. Reprod. Dev. 2003;64:41–51. doi: 10.1002/mrd.10223. [DOI] [PubMed] [Google Scholar]
- 38.Wang Z, Jaenisch R. At most three ES cells contribute to the somatic lineages of chimeric mice and of mice produced by ES-tetraploid complementation. Dev. Biol. 2004;275:192–201. doi: 10.1016/j.ydbio.2004.06.026. [DOI] [PubMed] [Google Scholar]
- 39.James RM, Kaufman MH, Webb S, West JD. Electrofusion of mouse embryos results in uniform tetraploidy and not tetraploid/diploid mosaicism. Genet. Res. 1992;60:185–194. doi: 10.1017/s0016672300030937. [DOI] [PubMed] [Google Scholar]
- 40.Bronson RA, McLaren A. Transfer to the mouse oviduct of eggs with and without the zona pellucida. J. Reprod. Fertil. 1970;22:129–137. doi: 10.1530/jrf.0.0220129. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.