Abstract
The production of complex, yet well defined materials offers many opportunities in regenerative medicine, in which the mechanical and biological properties of the matrix must meet stringent requirements. Here we report the recombinant production of modular polypeptidic materials, based on the highly resilient protein resilin, which are equipped with multiple biologically active domains. The recombinant materials exhibit useful mechanical and cell adhesion behavior.
The significance of mechanical forces1 and biological cues2 in directing tissue remodeling is well established and has accentuated the need to appropriately engineer both the mechanical and biological properties of a matrix. Much attention has focused on mimicking the desirable mechanical behavior and biocompatibility of materials such as elastin3 and collagen,4 to produce materials that mimic the extra-cellular matrix, support dynamic interactions with cells, and stimulate explicit cell and tissue responses. Thus, modular designs incorporating both mechanically and biologically active domains, to sense and aptly respond to bio-mechanical demands or changes in the environment, have emerged as a desirable approach in the engineering of biomaterials,5,6 although such modularity can be difficult to achieve with synthetic approaches.
The engineering of synthetic elastomers for use in complex biological environments has been limited owing to difficulties in the incorporation of cell-adhesive, cell-degradable, and protein-binding domains. In contrast, many hydrophilic polymers have found wide-spread use in biomedical applications, with poly(ethylene glycol) used to a great extent because of its biologically and chemically inert nature and antifouling properties.7 The physicochemical properties of these polymers offer advantages, compared to their hydrophobic counterparts, in the immobilization of hydrophilic drugs or active growth factors, as they allow aqueous loading rather than exposure to hydrophobic solvents or polymers, which protects protein drugs from instability and denaturation.8,9 Consequently, elastomeric proteins, which actively respond to dynamic mechanical and physical environments, present a progressive outlook for the design of mechanically active materials. Interest in emulating the highly elastomeric behavior of the natural elastins has motivated enormous growth in research on elastin-like polypeptides (ELPs) over the last few decades;10,11 peptide- and polypeptide-based ELPs have found a variety of biotechnological applications such as drug delivery,12 protein purification13 and tissue engineering.14,15
The engineering of hydrophilic elastomers with specified biological activity therefore continues to offer enormous opportunities in the production of hydrogels for tissue engineering applications. We turned our attention to the insect protein resilin, a very efficient elastomeric protein, that is characterized by low stiffness and high extensibility. It functions as both an energy store in sound-producing organs in insects,16 and as a damper of vibrations in insect flight,17,18 responding to mechanical deformation at unusually high frequencies over the insect’s lifetime. Although resilin’s outstanding properties were discovered nearly five decades ago,17 only recently have useful quantities (300 mg/L) of recombinant resilin, with a putative 15 amino acid repeat of GGRPSDSYGAPGGGN, been synthesized from E. coli cultures; the resulting polypeptide has a reported solubility of up to 300 mg/mL, and once crosslinked through the tyrosine residues, the hydrated material shows a relatively low modulus (2.5 kPa), excellent extensibility (300%), and unmatched resilience (up to 97%).19,20 In contrast to the elastins, which tolerate many amino acid substitutions21,22 and the addition of biologically active domains,5,23 the flexibility of the resilin repeat sequence toward amino acid substitutions and addition of biologically active domains has not yet been explored. Given the excellent hydrophilicity and mechanical properties of recombinantly derived resilins, such development offers unique opportunities in the design of a new class of mechanically active biomaterials for engineering tissues such as the heart and vocal fold.
Here we describe the design and biosynthesis of novel modular, resilin-like hydrophilic elastomers with modules intended to control mechanical properties, cell adhesion, growth factor delivery, and material degradation. We report that slight modifications to the amino acid sequence of the putative resilin repeat and incorporation of biologically active domains do not measurably alter the poly-peptide conformational properties (as assessed via circular dichroic (CD) and Fourier transform infrared (FTIR) spectroscopies) or mechanical properties (as assessed in both oscillatory rheology and tensile testing). The materials show good cytocompatibility and cell adhesion, suggesting the use of these new polypeptide-based, hydrophilic elastomers in both tissue regeneration and drug delivery.
A schematic of the modular protein and its amino acid sequence is shown in Fig 1. The 15 amino acid-long repeating unit derived from the N-terminal exon of the Drosophila melanogaster resilin gene with the putative repeat motif GGRPSDSYGAPGGGN was used as a structural domain to impart mechanical strength to the scaffold. An RLP containing this domain and biologically active domains (RLP12) was cloned into a pET-28a-based expression vector (ESI, Fig. S1 and S2†) for facile expression from E. coli. We modified the resilin repeat by replacing the tyrosine (Y) with phenylalanine (F), in order to provide future strategies for photochemical crosslinking of the polypeptide without compromising the biological activity of the other domains.24 Lysine residues have also been included outside the putative resilin repeat as additional crosslinking sites in the work here. The capacity to support the attachment of human cells to the scaffold was provided by the introduction of the cell-binding ligand RGDSP, derived from the tenth type-III domain of fibronectin, owing to its relevance in vascularization.25 A matrix metalloproteinase (MMP)-sensitive sequence, GPQG↓IWGQ, derived from the human α1(I) collagen chain and readily cleaved by multiple MMPs, was included to promote proteolytic degradation,26 and the heparin-binding domain, CKAAKRPKAAKDKQTK, was included in the sequence for the noncovalent immobilization of heparin to permit sequestration and controlled release of growth factors.27 We have demonstrated previously that this and other heparin-binding domains are useful for mediating both heparin binding and growth factor (VEGF and bFGF) binding and release from other polymeric materials.28
Fig. 1.
Schematic and amino acid sequence of highly elastic and biologically active resilin-like polypeptide (RLP). This sequence contains 12 repeats of the resilin consensus sequence and is hence referred to as RLP12 in the text.
In the production of these materials, the Studier auto-induction of the expression host BL21Star™(DE3)/pET28a-RLP12 was employed to attain higher cell mass and higher yields as an alternative to IPTG induction;20 these protocols have yielded ca. 70–80 mg of RLP12 per litre of culture. Improvements in yields were attained via cell lysis under denaturing conditions (8 M urea), with subsequent buffer exchange and elution under native conditions during purification via Ni-NTA affinity chromatography. Purified RLP12 was analyzed via SDS-PAGE and MALDI-MS; SDS-PAGE results are shown in Fig. 2a. SDS-PAGE of the purified RLP12 shows a pure (>99% purity) polypeptide and MALDI-MS confirmed the expected molecular mass of the RLP12 (27.5 kDa, ESI, Fig. S3†). Amino acid analysis of the polypeptide also demonstrated that the composition of the expressed polypeptide was, for all amino acids, within 5% of the expected values (ESI, Table S4†).
Fig. 2.
(a) SDS PAGE of RLP12. Lane 1—purified protein via Ni-affinity chromatography; lane 2—molecular weight marker. (b) CD spectrum of RLP12 in 10 mM PBS, pH 7.4. (c) Amide I region of the FTIR spectrum of RLP12. The experimental data (open circles) was fit with three peaks corresponding to β-turn (blue), random coil (red), and β-sheet (green) structures. The resulting fit is shown in black; three peaks provided the best fit (R2 = 0.99985).
The effects of specific amino acid substitutions in the putative resilin repeat sequence on its conformational properties and its elasticity have not been previously investigated. Both CD and FTIR spectroscopies were employed to probe the secondary structure of RLP12 before and after crosslinking; results are presented for uncrosslinked polypeptide in Fig. 2b–c. The CD and FTIR spectra of RLP12 in solution and in the crosslinked gel are similar (ESI, Fig S5 and S6†) and the RLP12 is therefore suggested to possess similar secondary structures and conformational flexibility under both conditions. A CD spectrum of RLP12 (50 µM in PBS, pH 7.4; Fig. 2b) at 25 °C is dominated by a strong negative band near 195 nm, indicating a largely unordered structure, as anticipated. Second derivative deconvolution of the CD spectrum amplifies the minor contributions from coexisting secondary structures; the negative peak near 230 nm and the positive peak near 213 nm in the second derivative of the CD signal indicates the presence of type-II β-turns in addition to the largely unordered structure. The contribution from β-turns was confirmed in a CD spectrum of the uncrosslinked RLP12 in trifluoroethanol at 25 °C (ESI, Fig. S7†). Similar results have been observed by Tamburro and coworkers for short peptides (15–45 amino acids) comprising 1–3 resilin repeats with an almost identical sequence but lacking any biologically active domains,29 and for resilin-like peptides (also lacking any biologically active domains, Figure S8†), suggesting the similarity in the conformational properties of native resilin sequences and the modifiedRLP12 reported here. FTIR spectra were recorded for more concentrated RLP12 solutions (3.6 mM in deuterated PBS, pH 7.4); an expanded amide-I region of the spectra of the uncrosslinked RLP12 is shown in Fig. 2c. The broad nature of the amide-I band, centered at slightly below 1650 cm−1, suggests contributions from a range of multiple conformations, and may include water-bound carbonyls.30 Deconvolution of the amide-I region of the spectrum (Fig. 2c) yields three different peaks at approximately 1670 cm−1, 1645 cm−1 and 1622 cm−1, corresponding to those reported for β-turns, random coil and β-sheet structures respectively.31 Despite the breadth of the peak, the FTIR confirms the mainly random-coil and unordered nature of the RLP12, with secondary structures reminiscent of conformers observed in resilin-like peptides and in other elastomeric proteins such as elastin, abductin and wheat gluten.29,32 The contribution of β-sheet structure observed in the IR spectra ofRLP12, although quite small, with no such contribution detected in the CD spectra, likely relates to the different protein polymer concentrations probed in the two different spectroscopic experiments. Nevertheless, these data indicate that alterations in the repeat sequence of resilin and the addition of cell-binding, heparin-binding, and MMP-sensitive domains do not appreciably alter the expected conformational behaviors of the polypeptide.
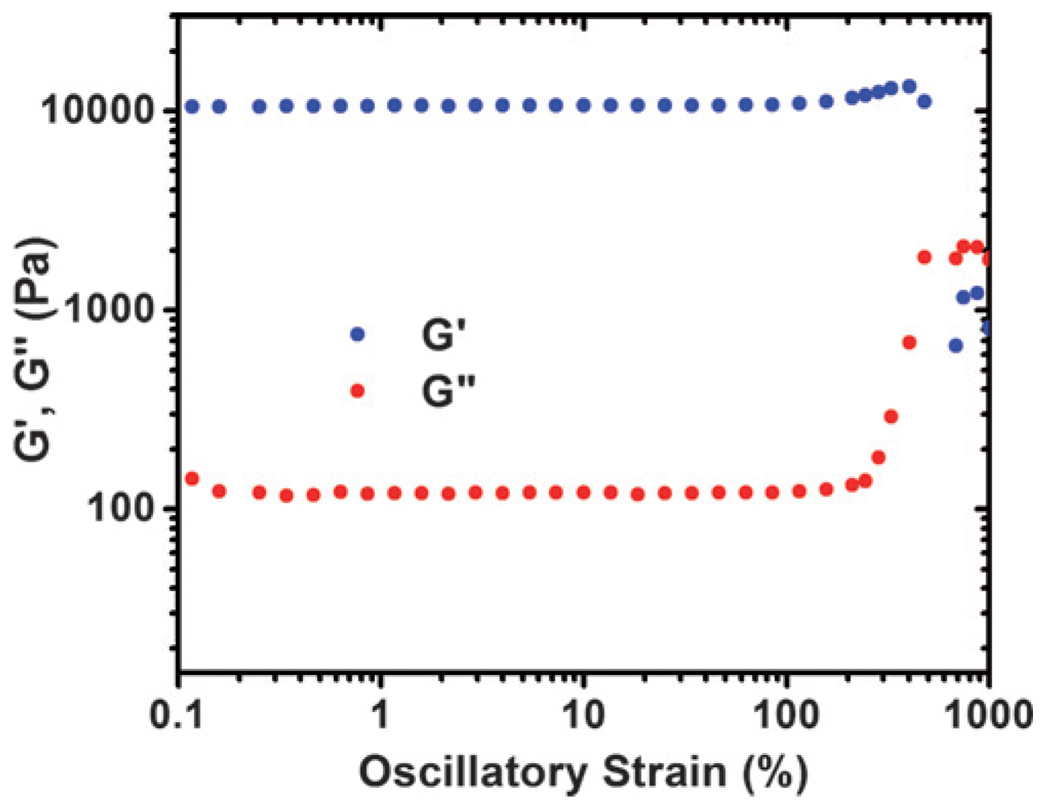
The similarities in the contribution of unordered conformations in the uncrosslinked and crosslinked RLP12 suggest that the crosslinked films of the RLP12 should retain sufficient chain mobility for desired elastomeric behavior, a critical need for the application of these and other biologically active, resilin-based materials in biomaterials applications. The mechanical properties of bulk hydrogels of the RLP12 (rheological and tensile) were thus characterized after crosslinking of the RLP12 under mild aqueous conditions via the Mannich-type reaction of [tris(hydroxymethyl)phosphino]propionic acid (THPP) with primary amines of lysine residues present outside of the resilin repeat in the polypeptide.33 The rheological properties of crosslinked RLP12 gels were obtained in dynamic torsional shear mode at 25 °C using a cone-on-plate configuration (ESI, Fig S9† and Fig. 3). To ensure homogeneous mixing of the crosslinker, the 25wt% solution of RLP12 was pre-mixed with THPP, with an equimolar ratio of reactive hydroxymethylphosphine (HMP) groups to lysines. The solution was observed to gel rapidly after the temperature was increased to 25 °C, as indicated by the considerable increase in viscosity and dynamic storage (G′) modulus. The storage modulus reached a steady value of 10 kPa in approximately 2 h, with a 100-order difference between the storage and loss moduli (G″ = 100 Pa), confirming the formation of a hydrogel; similar results were obtained at 37 °C. The equilibrium shear moduli of these RLP12 gels are comparable to those of elastin-like polypeptide gels comprising chemically crosslinked ELPs of similar molecular weight.34 The loss angle, δ, which provides a relative measure of viscous to elastic effects in a material (δ = 0° indicating an elastic solid and δ = 90° indicating a Newtonian viscous fluid) is <1°, suggesting that the RLP12 hydrogels are highly elastic, energy-storing solids. The insensitivity of G′ to frequency for RLP12 demonstrates the solid-like behavior of these permanently crosslinked gels (ESI, Fig S9†). Strain sweeps conducted on 25 wt% gels maintain linear stress-strain behavior until 200% strain and withstand strain up to 450%, indicating that the RLP12 forms a highly elastomeric crosslinked network (Fig. 3). Moreover, the mechanical properties of the scaffold can easily be modulated by varying the amount of crosslinker used.
Fig. 3.
Strain sweep data for RLP12 gels (25 wt%, ω = 6 rad/s).
Typical stress-strain responses of crosslinked and hydrated bulk RLP12 films, upon tensile testing, are shown in Fig 4. The films exhibit Young’s moduli of ~30–60 kPa with an average extension-to-break ratio of 180%; the heterogeneity of these results likely results from variabilities in the gripping of the swollen samples as well as some variation in sample thickness and crosslinking. The tensile test results indicate that these RLP12 films exhibit an 8-fold lower modulus compared to resilin tendon from dragonfly,35 show a ca. 10-fold higher modulus than previously reported recombinant resilin,19 and compare favorably to elastin-like polypeptides chemically crosslinked within the elastic domains;36 the data thus suggest flexible crosslinking approaches for resilin-like sequences that do not rely on crosslinking at the tyrosine sites of the consensus repeat sequence. The combination of these mechanical properties with the high water content (85%) of the films also suggests opportunities in the engineering of hydrophilic elastomers; optimization of crosslinking and mechanical properties is underway. Estimates of the molecular weight between crosslinks (MWc) in the RLP12 films were obtained from stress-strain data using the statistical theory of rubber elasticity;37 these data suggested a MWc of 7653 g/mol, indicating the reaction of 3.6 lysine residues per chain, which is in excellent agreement with the number of reacted lysine residues (3.5) indicated from amino acid analysis of crosslinked films (ESI, Table S10†). The rheology and tensile tests suggest that the RLP12 hydrogels and films are highly elastomeric despite the alteration in the amino acid sequence of resilin and the introduction of the biological domains.
Fig. 4.
Tensile testing data for hydrated RLP12 films. Stress-strain data were recorded at a strain rate of 10% gauge length per minute at 25 °C. (Data shown for three separate films of identical composition.)
Adhesion and proliferation of mouse NIH 3T3 fibroblasts to the crosslinked scaffolds were investigated by the microscopic observation of cell morphology and by alamarBlue® assay (Invitrogen). An increase in the fluorescence of alamarBlue® reagent correlates to an increase in metabolic intermediates (e.g. NADH, NADPH, and FADH) produced by viable cells; therefore an increase in fluorescence signal is commonly used as a quantitative measure of the number of cells present in the medium. Mouse fibroblast NIH 3T3 cells seeded on RLP12 gels showed increased metabolic activity levels on days 1–3, as shown in Fig. 5a, and the number of cells adherent to the scaffold increased with time. In addition, absorbance values for cells cultured indirectly, with RLP12 suspended above the cells, were not statistically different than values for standard culture indicating that there are no toxic leachable components in the gel. The spreading of NIH 3T3 cells on this substrate was visualized by staining for F-actin; Fig. 5b shows spread cells with well-formed and organized stress fibers on both the RLP12 sample and on the TCPS (tissue culture polystyrene) positive control, indicating the good adhesion of cells to the crosslinked RLP12 substrate, which is consistent with the observed cell survival of mouse fibroblast cells encapsulated within THPP-crosslinked, elastin-like polypeptides.33 Both cell morphology and metabolic activity levels thus suggest that the RLP12 crosslinked by THPP is cytocompatible.
Fig. 5.
Proliferation data (day 1–3) and fluorescence microscopy images (day 1) of NIH-3T3 cells on control TCPS and on films formed from RLP12. The scale bar represents 100 µm.
In summary, we present a new elastomeric and cell-adhesive recombinant polypeptide-based material with the additional potential for growth factor delivery and proteolytic remodeling by cell-associated MMPs. The RLP12 exhibits a mainly unordered conformation with small contribution from type-II β-turns, consistent with previous studies of resilin and other elastomeric proteins. The crosslinked material is highly elastic and has the capacity for cell attachment and proliferation. The mechanical properties of this matrix recommend its use in mechanically demanding tissue engineering applications. Detailed characterization of the rheological properties of these hydrogels, in the presence of heparin and growth factors and at low and high frequencies, as well as the interaction of these materials with multiple cell types, are under investigation.
Supplementary Material
Acknowledgments
This work was supported by grants from the University of Delaware Research Foundation and the National Science Foundation (DMR 0239744), as well as by the National Center for Research Resources (NCRR), a component of the National Institutes of Health (instrument facilities: P20-RR017716 and P20-RR015588). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NCRR or NIH. Amino acid analyses were conducted at UC Davis’ molecular structure facility.
Footnotes
Electronic Supplementary Information (ESI) available: Synthesis procedures and experimental details; MALDI-MS; amino acid analysis; and frequency sweep oscillatory rheology.
Notes and references
- 1.Discher DE, Janmey P, Wang YL. Science. 2005;310:1139–1143. doi: 10.1126/science.1116995. [DOI] [PubMed] [Google Scholar]
- 2.Lutolf MP, Hubbell JA. Nat. Biotechnol. 2005;23:47–55. doi: 10.1038/nbt1055. [DOI] [PubMed] [Google Scholar]
- 3.Welsh ER, Tirrell DA. Biomacromolecules. 2000;1:23–30. doi: 10.1021/bm0002914. [DOI] [PubMed] [Google Scholar]
- 4.Ito H, Steplewski A, Alabyeva T, Fertala A. J. Biomed. Mater. Res., Part A. 2006;76a:551–560. doi: 10.1002/jbm.a.30551. [DOI] [PubMed] [Google Scholar]
- 5.Straley KS, Heilshorn SC. Soft Matter. 2009;5:114–124. [Google Scholar]
- 6.Halstenberg S, Panitch A, Rizzi S, Hall H, Hubbell JA. Biomacromolecules. 2002;3:710–723. doi: 10.1021/bm015629o. [DOI] [PubMed] [Google Scholar]
- 7.Harris JM. In: Poly(Ethylene Glycol) Chemistry: Biotechnical and Biomedical Applications. Harris JM, editor. New York: Plenum Press; 1992. [Google Scholar]
- 8.Huang X, Lowe TL. Biomacromolecules. 2005;6:2131–2139. doi: 10.1021/bm050116t. [DOI] [PubMed] [Google Scholar]
- 9.Zhang YL, Chu CC. J. Biomed. Mater. Res. 2001;54:1–11. doi: 10.1002/1097-4636(200101)54:1<1::aid-jbm1>3.0.co;2-m. [DOI] [PubMed] [Google Scholar]
- 10.Simnick AJ, Lim DW, Chow D, Chilkoti A. Polym. Rev. 2007;47:121–154. [Google Scholar]
- 11.Wright ER, Conticello VP. Adv. Drug Delivery Rev. 2002;54:1057–1073. doi: 10.1016/s0169-409x(02)00059-5. [DOI] [PubMed] [Google Scholar]
- 12.Dreher MR, Raucher D, Balu N, Colvin OM, Ludeman SM, Chilkoti A. J. Controlled Release. 2003;91:31–43. doi: 10.1016/s0168-3659(03)00216-5. [DOI] [PubMed] [Google Scholar]
- 13.Banki MR, Feng LA, Wood DW. Nat. Methods. 2005;2:659–661. doi: 10.1038/nmeth787. [DOI] [PubMed] [Google Scholar]
- 14.McHale MK, Setton LA, Chilkoti A. Tissue Eng. 2005;11:1768–1779. doi: 10.1089/ten.2005.11.1768. [DOI] [PubMed] [Google Scholar]
- 15.Heilshorn SC, Liu JC, Tirrell DA. Biomacromolecules. 2005;6:318–323. doi: 10.1021/bm049627q. [DOI] [PubMed] [Google Scholar]
- 16.Skals N, Surlykke A. J. Exp. Biol. 1999;202:2937–2949. doi: 10.1242/jeb.202.21.2937. [DOI] [PubMed] [Google Scholar]
- 17.Weisfogh T. J. Exp. Biol. 1960;37:889. [Google Scholar]
- 18.Vincent JFV, Wegst UGK. Arthropod. Struct. Dev. 2004;33:187–199. doi: 10.1016/j.asd.2004.05.006. [DOI] [PubMed] [Google Scholar]
- 19.Elvin CM, Carr AG, Huson MG, Maxwell JM, Pearson RD, Vuocolo T, Liyou NE, Wong DCC, Merritt DJ, Dixon NE. Nature. 2005;437:999–1002. doi: 10.1038/nature04085. [DOI] [PubMed] [Google Scholar]
- 20.Kim M, Elvin C, Brownlee A, Lyons R. Protein Expression Purif. 2007;52:230–236. doi: 10.1016/j.pep.2006.11.003. [DOI] [PubMed] [Google Scholar]
- 21.Urry DW, Gowda DC, Parker TM, Luan CH, Reid MC, Harris CM, Pattanaik A, Harris RD. Biopolymers. 1992;32:1243–1250. doi: 10.1002/bip.360320913. [DOI] [PubMed] [Google Scholar]
- 22.Urry DW, Luan CH, Parker TM, Gowda DC, Prasad KU, Reid MC, Safavy A. J. Am. Chem. Soc. 1991;113:4346–4348. [Google Scholar]
- 23.Panitch A, Yamaoka T, Fournier MJ, Mason TL, Tirrell DA. Macromolecules. 1999;32:1701–1703. [Google Scholar]
- 24.Nowatzki PJ, Franck C, Maskarinec SA, Ravichandran G, Tirrell DA. Macromolecules. 2008;41:1839–1845. [Google Scholar]
- 25.Hersel U, Dahmen C, Kessler H. Biomaterials. 2003;24:4385–4415. doi: 10.1016/s0142-9612(03)00343-0. [DOI] [PubMed] [Google Scholar]
- 26.Nagase H, Fields GB. Biopolymers. 1996;40:399–416. doi: 10.1002/(SICI)1097-0282(1996)40:4%3C399::AID-BIP5%3E3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 27.Liu SC, Zhou FY, Hook M, Carson DD. Proc. Natl. Acad. Sci. U. S. A. 1997;94:1739–1744. doi: 10.1073/pnas.94.5.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yamaguchi N, Kiick KL. Biomacromolecules. 2005;6:1921–1930. doi: 10.1021/bm050003+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bochicchio B, Pepe A, Tamburro AM. Chirality. 2008;20:985–994. doi: 10.1002/chir.20541. [DOI] [PubMed] [Google Scholar]
- 30.Blout ER, Doyle BB, Traub W, Lorenzi GP. Biochemistry. 1971;10:3052–3060. doi: 10.1021/bi00792a011. [DOI] [PubMed] [Google Scholar]
- 31.Torii H, Tasumi M. In: Infrared Spectroscopy of Biomolecules. Mantsch HH, Chapman D, editors. New York: John Wiley & Sons; 1996. [Google Scholar]
- 32.VanDijk AA, VanWijk LL, VanVliet A, Haris P, VanSwieten E, Tesser GI, Robillard GT. Protein Sci. 1997;6:637–648. doi: 10.1002/pro.5560060313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lim DW, Nettles DL, Setton LA, Chilkoti A. Biomacromolecules. 2008;9:222–230. doi: 10.1021/bm7007982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lim DW, Nettles DL, Setton LA, Chilkoti A. Biomacromolecules. 2007;8:1463–1470. doi: 10.1021/bm061059m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Weisfogh T. J. Mol. Biol. 1961;3:648–667. [Google Scholar]
- 36.Di Zio K, Tirrell DA. Macromolecules. 2003;36:1553–1558. [Google Scholar]
- 37.Flory PJ. Principles of Polymer Chemistry. Ithaca, NY: Cornell University Press; 1953. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.