Abstract
The use of biomarkers ensures breast cancer patients receive optimal treatment. Established biomarkers such as estrogen receptor (ER) and progesterone receptor (PR) have been playing significant roles in the selection and management of patients for endocrine therapy. HER2 is a strong predictor of response to trastuzumab. Recently, the roles of ER as a negative and HER2 as a positive indicator for chemotherapy have been established. Ki67 has traditionally been recognized as a poor prognostic factor, but recent studies suggest that measurement of Ki67-positive cells during treatment will more effectively predict treatment efficacy for both anti-hormonal and chemotherapy. p53 mutations are found in 20–35% of human breast cancers and are associated with aggressive disease with poor clinical outcome when the DNA-binding domain is mutated. The utility of cyclin D1 as a predictor of breast cancer prognosis is controversial, but cyclin D1b overexpression is associated with poor prognosis. Likewise, overexpression of the low molecular weight form of cyclin E1 protein predicts poor prognosis. Breast cancers from BRCA1/2 carriers often show high nuclear grades, negativity to ER/PR/HER2, and p53 mutations, and thus, are associated with poor prognosis. The prognostic values of other molecular markers, such as p14ARF, TBX2/3, VEGF in breast cancer are also discussed. Careful evaluation of these biomarkers with current treatment modality is required to determine whether their measurement or monitoring offer significant clinical benefits.
Keywords: breast cancer, prognosis, molecular marker, Ki67, ER, PR, HER2, cyclin D1, cyclin E, p53, ARF, TBX2/3, BRCA1/2, VEGF
Introduction
Breast cancer is the major common malignancy in women, and its treatment is possible if diagnosed at an early stage.1–4 Traditional prognostic factors include the axillary lymph node status, the tumor size, and the nuclear grade and histological grade.1,3 The prognosis for breast cancer generally depends on its stage, typically graded as I to IV with sub-stages (Table 1). Malignant progression of breast cancer involves the conversion of “benign proliferative lesions” and carcinomas in situ to early (stages I and II) disease, to locally advanced (stage III) disease, and then to metastasis to bone, brain, lungs, and other sites (stage IV).
Table 1.
Histological stages of human breast cancer.
| Stage | TNM | Description | 5-year survival |
|---|---|---|---|
| 0 | Tis N0 M0 | Carcinoma in situ. No tumor in regional lymph nodes, No distant metastases. | 99% |
| I | T1 N0 M0 | Tumor is less than or equal to 2 centimeters, No tumor in regional lymph nodes, No distant metastases. | 92% |
| IIA | T0 N1 M0 | • No evidence of primary tumor, metastases to movable ipsilateral nodes, No distant metastases. | 82% |
| T1 N1 M0 | • Tumor is less than or equal to 2 centimeters, metastases to movable ipsilateral nodes, No distant metastases. | ||
| T2 N0 M0 | • Tumor is between 2 and 5 centimeters, No tumor is regional lymph nodes, No distant metastases. | ||
| IIB | T2 N1 M0 | • Tumor is between 2 and 5 centimeters, metastases to movable ipsilateral nodes, No distant metastases. | 65% |
| T3 N0 M0 | • Tumor is over 5 centimeters, No tumor in regional lymph nodes, No distant metastases. | ||
| IIIA | T0 N2 M0 | • No evidence of primary tumor, metastases to fixed ipsilateral nodes, No distant metastases. | 47% |
| T1 N2 M0 | • Tumor is less than or equal to 2 centimeters, metastases to fixed ipsilateral nodes, No distant metastases. | ||
| T2 N2 M0 | • Tumor is between 2 and 5 centimeters, metastases to fixed ipsilateral nodes, No distant metastases. | ||
| T3 N1, N2 M0 | • Tumor is over 5 centimeters, metastases to movable or fixed ipsilateral nodes, No distant metastases. | ||
| IIIB | T4 Any N M0 | • Tumor extends to chest wall, any nodal involvement, No distant metastases. | 44% |
| Any T N3 M0 | • Any primary tumor involvement, metastases. to ipsilateral internal mammary nodes, No distant metastases. | ||
| IV | Any T Any N M1 | Any primary tumor involvement, any nodal involvement, distant metastases. | 14% |
Abbreviations: T, status of primary tumor; N, regional lymph nodes; M, distant metastases.
Fundamental to malignant progression are the heterotypic processes regulating epithelial-mesenchymal transition, hypoxia, desmoplasia, and angiogenesis5,6 The development of cancer involves dysregulation of proliferative-signaling and growth-inhibitory factors, activation of oncogenes, and loss of tumor suppressor genes, all of which result in suppression of apoptosis and senescence.6 The increasing understanding of the pathophysiological background of breast cancer is associated with new molecular techniques, improved risk assessment, targeted therapy, and individualized treatment.1,3,7 Gene expression profiling may provide predictive and prognostic gene signatures which could help characterize tumors and enable more tailored therapies.8,9
Interest in novel prognostic markers is based on the fact that a significant number of patients with early-stage breast cancer harbor microscopic metastasis at the time of diagnosis. Many molecular markers that have been studied have both prognostic and predictive values. Prognostic markers are indicators of aggressiveness, invasiveness, extent of spread of tumors, and thus, correlate with survival independent of systemic therapy and can be used to select patients at risk. On the other hand, predictive markers are reports which allow clinicians to expect therapeutic outcomes and decide future treatment plans. This review summarizes mainly the prognostic values of classical (Ki67, ER, PR, HER2) and novel molecular factors (p53, p14ARF, cyclin D1, cyclin E, TBX2/3, BRCA1/2, and VEGF) for breast cancer. These novel molecular markers have been chosen from the viewpoint of involvement in the regulation of the p53 and RB tumor suppressor pathways, DNA damage response, and angiogenesis/metastasis, which play critical roles in human breast cancer development.
Ki67
Proliferative markers have been broadly evaluated as prognostic factor for early stage breast cancer patients. Ki67, a nuclear non-histone protein, was identified by Gerdes et al as after immunization of mice with the Hodgkin’s lymphoma.10–13 Ki67 is expressed only in cells in the proliferative phases of the cell cycle (G1, S, G2, and M phases). Ki67 is vital for cell proliferation, since downregulation of Ki67 using antisense nucleotides prevents cell proliferation.10 Ki67 is tightly controlled and regulated, implying a fundamental role in cell proliferation. However, it has been very difficult to determine its function because of its lack of obvious homology with known proteins. Bridger et al suggested a role of Ki67 in organizing DNA, based on its localization to extranucleolar sites during early G1; these sites contain centromeric and satellite DNA.14 Ki67 is also known to bind to DNA. Mac-Callum and Hall suggested a structural role for Ki67 within the nucleolus, based on its ability to interact with other proteins and bind with RNA and DNA.14,15 They also suggested that Ki67 is an essential factor in the synthesis of ribosomes during cell division.15 Further studies should be conducted to elucidate the roles of Ki67 in cell proliferation and tumorigenesis.
Ki67 expression is usually estimated as the percentage of tumor cells positively stained by the antibody, with nuclear staining being the most common criterion of proliferative index. Numerous studies have shown that Ki67 is of prognostic value in many types of malignant tumors. In breast cancer, most studies show a strong, statistically significant correlation with clinical outcomes, both on univariate and multivariate analyses. A strong correlation has been noted between the percentage of cells positive for Ki67 and the nuclear grade, age, and mitotic rate.16,17 Multiple studies have indicated that breast cancer overexpress Ki67 in more than 20–50% of the cells are at high risk of developing recurrent disease, showing a statistically significant correlation with clinical outcome, such as disease-free survival or overall survival.18–30
The predictive value of Ki67 on the survival of breast cancer patients has also been studied. To determine the clinical significance of the level of tumor cell proliferation during endocrine therapy for breast cancer, Dowsett et al31 measured the expression of Ki67 in tumor biopsy samples taken before and after 2 weeks of pre-surgical treatment with anastrozole or tamoxifen alone, or the combination, in 158 patients with hormone receptor-positive breast cancer. In a multivariable analysis, they found that higher Ki67 expression after 2 weeks of endocrine therapy was associated with lower recurrence-free survival (P = 0.004) whereas higher Ki67 expression at baseline was not.31 To compare the prognostic significance of Ki67 expression in breast cancer before and after neoadjuvant chemotherapy, the expression of Ki67 was assessed using immunohistochemistry (IHC) in pre-therapy core-needle biopsy and post-therapy surgical excision specimens.32 In a matched cohort of 103 patients, post-therapy Ki67 was the only significant independent prognostic factor among Ki67, ER, PR, HER2, clinical stage, histology on multivariate analysis of relapse-free survival. On multivariate analysis for overall survival, both pre- and excision Ki67 expression was significant independent predictors, but the latter showed a stronger prognostic impact. In a cohort of 284 patients with only excision samples, post-therapy Ki67 was a significant independent prognostic factor. They concluded that post-chemotherapy Ki67 is a strong predictor of outcome for patients not achieving a pathological complete response.32,33 In addition, Ki67 correlates with other well-characterized proliferation markers, such as the proliferating cell nuclear antigen, which is a target of E2Fs34 (Fig. 1). Ki67 staining will continued to be used as a useful laboratory test to predict the prognosis of breast cancer patients since it is technically easier and more closely associated with clinical outcomes than DNA ploidy analysis or S phase measurement by flow cytometry.
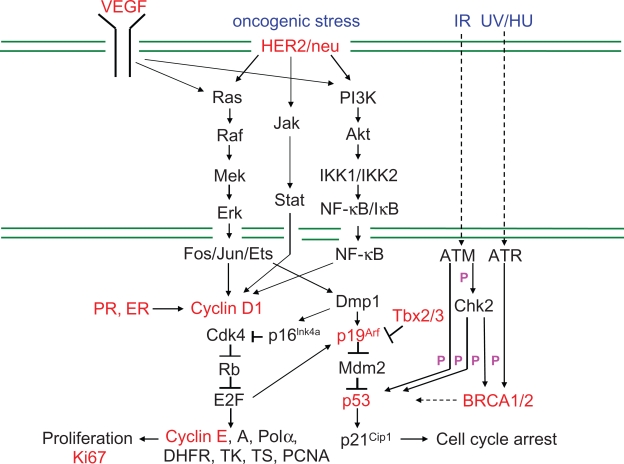
Figure 1.
Signaling pathways involving molecular markers for human breast cancer. The green lines show cytoplasmic and nuclear membranes. HER2/neu is an orphan receptor that can be activated by overexpression or mutation of the transactivating domain (rat neu). Overexpression of HER2/neu results in enhanced cell survival and mitogenicity and its deregulation can lead to breast tumorigenesis. VEGF shows mitogenic activity by stimulating the Ras-Raf-Mapk and PI3K pathways. Erk activation by the Ras-Raf pathway leads to activation of the cyclin D1 promoter by Fos/Jun/Ets transcription factors. Cyclin D1 makes a complex with Cdk4/6 to phosphorylate Rb and release E2F proteins that regulate G1-S transition. Both cyclins E and A2 are direct targets for activating E2Fs. Activation of the PI3K-Akt pathway results in enhanced anti-apoptotic action through inhibition of the pro-apoptosis proteins (e.g. Bad, GSK3 and the transcription factor FKHR-L1, not shown). In addition, activation of the JAK-STAT pathway by HER2 leads to cell proliferation. A major mitogenic player acting downstream of HER2 is cyclin D1.215 As indicated, a number of pathways lead from the receptors to enhanced activation of cyclin D1, thereby promoting cell cycle progression. Of note, cyclin D1 also interacts with the ERα to promote its transcriptional activity in Cdk-independent fashion.57,58 Dmp1 (cyclin D binding myb-like protein 1; also named Dmtf1) is a haplo-insufficient tumor suppressor that regulates the Arf-Mdm2-p53 tumor surveillance pathway.126,138,216 Cyclin D1 is a negative regulator for Dmp1: however, it synergizes with Dmp1 on the Arf promoter.217 BRCA1/2 proteins are directly or indirectly phosphorylated by ATM/ATR kinases in response to DNA damage, which interact with p53 to stop the cell cycle by activating the p21Cip1/WAF1 promoter.
Abbreviations: IKK, IκB kinase; Polα, DNA polymerase α; DHFR, dihydrofolate reductase; TK, thymidine kinase; TS, thymidylate synthase; PCNA, proliferating cell nuclear antigen; ER, estrogen receptor; PR, progesterone receptor; VEGF, vascular endothelial growth factor; IR, ionizing radiation; HU, hydroxyurea; ATM, ataxia-telangiectasia mutated; ATR, Ataxia telangiectasia and Rad3 related; Chk2, checkpoint homolog 2.
Estrogen and Progesterone Receptors (ER and PR)
The ER and PR are dimeric, gene-regulatory proteins. Estrogen and progesterone are well-established endocrine steroid regulators that modulate multiple aspects of mammary gland pathology. These two hormones work together to direct mammary epithelial growth, differentiation, and survival.35 Although both steroids are commonly thought to be of primary importance for tumors arising in the reproductively competent years, between puberty and menopause, local aromatization of adrenal androgens provides additional estrogens in the postmenopausal years. Estrogen and progesterone act through their nuclear receptors to modulate transcription of target genes.35,36 Genes encoding the receptors for each class of steroids are members of a single large superfamily of transcription-modulating factors. ERs may exist either in homodimeric or heterodimeric species, composed of α and β receptors acting as hormone-dependent transcriptional regulators. 37 The ER pathway plays a critical role in the pathophysiology of human breast cancer. Although it is known that ERα is of key importance in the mammary ductal elongation of puberty, PR and ERβ appear to be more involved with lactational differentiation of the lobules.38,39 From knockout studies in mice, it is apparent that PR plays important role in ductal and lobuloalveolar development of normal mammary gland.40
Overexpression of ERα is a well-established prognostic factor in breast cancer patients. Generally, ERα-positive breast cancers are associated with slow tumor growth, lower histology grade, DNA diploidy, and thus a better overall prognosis.35 More than 90% of lobular breast carcinomas are ER-positive; while medullary and inflammatory carcinomas are predominantly ER-negative. ER/PR-negative tumors are often associated with aggressive disease, and these tumors frequently show amplification of HER2, c-Myc, and Int2 oncogenes, and mutations of the p53 tumor suppressor gene.
The value of ERα status as an independent prognostic variable has been diminished by its association with other established indicators of favorable prognosis. These include older age of the patient, low-grade histology, a favorable nuclear grade, a low S-phase fraction, a normal content of DNA, a low proliferative index, and a low thymidine labeling index.41 In addition, because adjuvant or palliative hormone therapy is a common treatment for patients with ER-positive tumors receives, it is difficult to evaluate the prognostic value of their ER status alone. In some studies, the longer duration of disease-free survival (DFS) and overall survival rates of patients with ERα-positive tumors are seen only in the presence of hormone therapy. In addition, the favorable effect of ERα-positive status as a discriminant often is lost after several years, suggesting that treatment benefit is temporary.42,43 When node-positive patients not receiving adjuvant hormone therapy were studied, the 5-year DFS rate was 20% higher for ERpositive patients compared with that for ER-negative patients. However, the 5-year DFS rate of the most favorable subgroup (i.e. patients with one to three positive nodes and ER-positive tumors) was below 60%.42 Among node-negative patients, small but statistically significant differences in DFS and overall survival rates have been found between ER-positive cases and ER-negative cases after various periods of follow-up. The results of a multivariate analysis of prognostic factors in over 3,000 patients showed ER status to be more important for prognosis than tumor size in node-negative cases, but not in node-positive cases.43 In one study, the ER status was less important for predicting duration of DFS or overall survival than the nuclear grade and the number of positive nodes.44 Allred and colleagues showed that tamoxifen decreased the risk of local regional recurrence in patients with ER-positive ductal carcinoma in situ.45
The prognostic significance of ERβ is not well defined.46–48 Honma et al49 studied archival materials from 442 invasive breast cancers treated with adjuvant tamoxifen monotherapy and with a long follow-up period (median: 11.1 years) using three antibodies to detect ERβN, ERβ1, and ERβcx (ERβ2). Positive staining for ERβN or ERβ1 was associated with significantly better survival. By contrast, ERβcx status showed no association with length of survival. ERβ1status was significantly associated with longer survival in postmenopausal, but not premenopausal, women. ERβ1 positivity was associated with significantly better survival in patients with ERα(–)PR(–) or ERα(–)PR(–)HER2(–) (triple-negative) tumors, which are widely believed to have a poor prognosis. Another study also showed that higher expression of ERβ in ERα/PR-positive breast cancer was associated with longer overall survival (70% for high ERβ vs. 30% for low ERβ at 100 months) compared with the same group of patients expressing much lower levels of ERβ. Differential expression of ERβ had no prognostic value in patients with ERα/PR negative breast cancer.50 Further studies will be required to establish the value of ERβ to predict the prognosis of breast cancer patients.
Because the growth of breast cancer is often regulated by the female sex steroids, determinations of the cellular concentrations of ER and PR in the tumor continue to be used as predictors of good prognosis and of potential benefit from anti-hormonal therapy. To improve the value of ER determinations for tumor prognosis, tests for the presence of the estrogen-regulated PR protein are routinely performed. In many breast tumor cell lines—and in normal tissues containing ER, such as the endometrium and brain—PR expression is induced by estrogen.51,52 It is still not known whether ER regulates PR in normal human mammary epithelium in precisely the same subpopulation of ductal and lobular luminal cells, although this assumption is considered to be likely. It is of interest that the ER and PR appear to be strongly up-regulated in ductal carcinoma in situ and in hormone-dependent breast cancer, relative to normal mammary epithelium.
PR is a heterodimeric protein with A and B subunits. Overexpression of the PR indicates that the ER pathway is intact, even if the tumor is reported as ER-negative.51,52 When biochemical ligand-binding assays indicate concentrations of 10 fmol/mg cytosol protein or more, the tumors are generally considered ER-positive and PR-positive for clinical purposes. The ER and PR status can be measured using immunohistochemistry. These results correlate closely with biochemical ligand binding assays and clinical response rates to endocrine therapy.53,54 Furthermore, PR promoter methylation is commonly observed in primary breast cancer cases and may be a mechanism by which this protein is downregulated.55 Importantly, higher PR levels are negatively correlated with tumor size and grade. In a recent study, Liu et al showed that expression of PR in ER-positive tumors improved survival of patients receiving estrogen receptor therapy. Hence, PR may be used as prognostic factor in this group of patients.56 In summary, ER will be used as a marker to predict the response to hormonal therapy and PR will be used as a predictor of response to hormonal therapy as well as a prognostic factor in ER+ breast cancers.
Estrogen and progesterone are well known as direct modulators of expression of growth factor receptor pathways and downstream, cell-cycle regulatory genes known as nuclear protooncogenes. The nuclear proto-oncoproteins and other cell-cycle regulatory proteins, such as AIB-1, c-Myc, and cyclin D1, represent points of regulatory convergence of steroid and growth factor pathways in cells. Of considerable interest is the observation that the cell-cycle regulator cyclin D1 also interacts with the ERα to promote its transcriptional activity57,58 (Fig. 1). Parallel observations have also been made with the AIB-1 (amplified in breast cancer 1; SRC-3) protein, regarding both sensitization of ER transactivation and growth factor signal transduction.59 AIB1 is an ER coregulatory protein that together with other co-activators, like transcription intermediary factor 2 (TIF2) and nuclear receptor co-repressor (NCoR), is implicated in the estrogen signaling pathway and estrogen- regulated tumor progression. An alternatively spliced variant form of AIB-1 is even more potent for these effects.59 It has been reported that ERα activation and nuclear localization, as well as coactivator interactions, are regulated through its phosphorylation.5,60
Harigopal et al studied the prognostic significance of AIB1, TIF2 and NCoR protein expression in 670 breast cancer specimens by breast tissue microarray, and demonstrated the relationship of coregulatory proteins to ER, PR and HER2/neu.61 High AIB1 expression was associated with poor patient outcome (P = 0.002), while no association was noted for TIF2 (P = 0.376) or NCoR (P = 0.12). When subclassified by nodal or ER status, AIB1 was not prognostic in the node-positive and ER-positive subsets. However, in the ER-negative and node-negative subsets, high AIB1 expression was associated with poor patient outcome (P = 0.02 and P = 0.007 respectively).61 There was significant positive correlation between AIB1 and ER/PR status and with other cofactors (TIF2 and NCoR), but not with HER2/neu status. Thus, high AIB1 expression predicted worse overall survival in this study, suggesting that AIB1 may be involved in breast carcinogenesis.
HER2
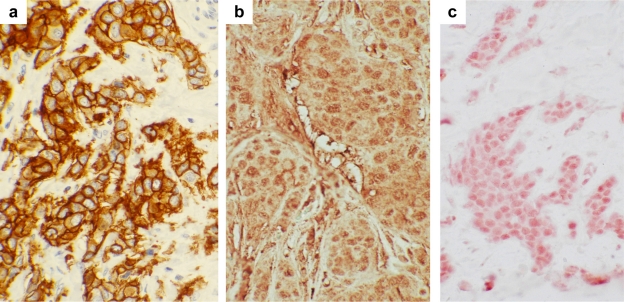
HER2 (also known as c-erbB-2 or neu) is a protooncogene that encodes a 185-kDa tyrosine kinase glycoprotein belonging to the EGFR family.62–65 The extracellular domain in HER2 is 44% homologous to the corresponding region in the epidermal growth factor receptor (EGFR), and the internal domain is 88% homologous to this region in EGFR.66 In addition to EGFR, the type I subfamily includes HER2, HER3, and HER4.62,63 These receptors all possess a large glycosylated extracellular ligand-binding domain, a single hydrophobic transmembrane domain, and a cytoplasmic tyrosine kinase domain. It is overexpressed in ∼30 % of breast cancer cases, primarily due to gene amplification. Within these cases, 60% are ductal carcinomas in situ and 20–30% are infiltrating breast carcinomas.60,67,68 A representative picture of HER2 overexpression in human breast cancer is shown in Figure 2a.
Figure 2.
Immunohistochemical staining of HER2, cyclin D1, and TBX2 proteins in human breast cancer samples. Human breast cancer tissues were stained for HER2 (panel a), cyclin D1 (panel b), and TBX2 proteins (panel c) with specific antibodies. For HER2: stained with A0485 (Dako, rabbit polyclonal); for cyclin D1: sc-20044 (Santa Cruz Biotech, mouse monoclonal IgG2a); and for TBX2: sc-17880 (Santa Cruz Biotech, goat polyclonal). Magnification, x40.
The HER2 status can be determined in human tumor samples using immunohistochemistry or fluorescent in situ hybridization. Amplification and/or overexpression of the HER2 oncogene are/is associated with a poor DFS rate in patients with axillary node-positive breast cancer.64,69 To investigate the prognostic significance of HER2 overexpression, immunohistochemical staining for HER2 was performed on sections from paraffin blocks of 292 primary invasive breast cancers obtained from women enrolled in the National Surgical Adjuvant Breast and Bowel Project protocol B-06.70 A positive reaction indicative of HER2 overexpression was observed in 21% of their breast cancer samples. Patients with HER2 overexpression had a significantly worse overall survival (P = 0.0012) with twice the mortality rate of women without detectable HER2 expression.70 In multivariate analysis, detection of HER2 overexpression was the second most predictive independent variable for survival after nodal status. Overexpression of HER2 was more common among tumors of high nuclear grade (29%) than those of low nuclear grade (12%). The association of HER2 overexpression with decreased survival was evident only among women with tumors of low nuclear grade. In this subgroup, HER2 overexpression was associated with an approximately fivefold increase in mortality rate (P = 0.00001). Thus, the combined predictive value of HER2 overexpression and nuclear grade was evident regardless of lymph node status.71 Allred and colleagues71 evaluated HER2 expression using immunohistochemistry in 613 patients with node-negative breast cancer enrolled in the Intergroup Study 0011. In their study, patients were stratified into low-risk groups (n = 307) and high-risk groups (n = 306). Lowrisk patients were defined as having small (<3 cm), ER-positive tumors and were observed without additional treatment after initial surgery. High-risk patients had either ER-negative tumors or large (≥3 cm), ER-positive tumors; they were randomized to be observed (n = 146) or to receive adjuvant chemotherapy (n = 160) after surgery. In their study, HER2 was overexpressed in 14.3% of all tumors combined, and overexpression was higher in invasive carcinomas associated with an extensive in situ component (21.5%) than in carcinomas without a significant noninvasive or in situ histological component (11.2%; P < 0.0001). When patients with low-risk lesions not containing a significant in situ component (n = 179) were analyzed, HER2 was a strong prognostic factor. Patients in this group with HER2-positive tumors had only a 40% DFS at 5 years, compared with more than 80% in patients with HER2-negative tumors (P < 0.0001).71
Using cDNA microarrays, Perou and colleagues classified invasive breast carcinomas into five subtypes based on their distinct gene expression profile (Norway/Stanford dataset).72 These included a luminal epithelial cell phenotype (subtypes A and B), a basal epithelial cell type phenotype, a HER2(+) phenotype, and a group of cancers expressing a ‘ normal-like’ gene profile. Patients whose tumors exhibited the basal-like and HER2-positive subtypes had the worst survival rates, while those with the luminal epithelial type had improved survival rates.73
HER2 expression is scored as 0, 1+, 2+, or 3+, depending on the number of cells with membrane staining and on the intensity of the staining. If the tumor is 0 or 1+, it is considered HER2-negative. If the tumor is 2+ or 3+, it is considered HER-2-positive. The humanized monoclonal antibody, trastuzumab (Herceptin®, Genentech, CA) is used for treatment of HER2-positive patients.68,74 Since HER2 is relatively uniformly overexpressed in human breast cancers (Fig. 2a), monoclonal antibodies have been developed to the extracellular domain to treat carcinomas overexpressing HER2. Trastuzumab is also approved for treating patients with metastatic breast cancer.75
A randomized phase III trial of 469 women with metastatic breast cancer with HER2 expression demonstrated that chemotherapy plus trastuzumab was associated with a longer time to disease progression (median, 7.4 vs. 4.6 months; P < 0.001), a higher rate of objective response (50 percent vs. 32 percent, P < 0.001), a longer duration of response (median, 9.1 vs. 6.1 months; P < 0.001), a lower rate of death at 1 year (22 percent vs. 33 percent, P = 0.008), longer survival (median survival, 25.1 vs. 20.3 months; P = 0.01), and a 20% reduction in the risk of death.68 In a phase II study, HER2 status was measured weekly in women with HER2-normal and HER2-overexpressing metastatic breast cancer given trastuzumab and paclitaxel. Efficacy was correlated with immunohistochemical and fluorescent in situ hybridization (FISH) assay results.76 HER2 status was evaluated using four different immunohistochemical assays and FISH. Patients received a median of 25 weekly infusions (range, one to 85 infusions). The intent-to-treat response rate for all 95 patients enrolled was 56.8% (95% confidence interval, 47% to 67%). A response rate of 61.4% (4.5% complete response, 56.8% partial response) was observed in 88 fully assessable patients.76 In patients with HER2-overexpressing tumors, overall response rates ranged from 67% to 81% compared with 41% to 46% in patients with HER2-normal expression (ranges reflect the different assay methods used to assess HER2 status). Differences in response rates between patients with HER2-overexpressing tumors and those with normal HER2 expression were statistically significant for all assay methods, with CB11 and TAB250 antibodies and FISH having the strongest significance. Given the established efficacy of trastuzumab for the treatment of advanced breast cancer, the HER2 testing will continue to be used as a standard laboratory test in breast cancer clinics.
p53
The p53 tumor suppressor gene, located on chromosome 17p13, has a regulatory function in the defense against various kinds of cancer, including breast cancer.77–83 The p53 gene transcription is activated by genetic, environmental, and metabolic stimuli, which direct cells to different pathways including cell cycle arrest, DNA repair, and apoptosis. It plays a central role in sensing genotoxic and non-genotoxic stresses and transducing an anti-proliferative effect80,82 (Fig. 1). p53 is activated and regulated by post-translational modifications to both the N- and C-terminal regions (e.g. phosphorylation, ubiquitination, and acetylation) and binds as a tetramer to specific DNA sequences via a central DNA-binding core region. p53 exerts its primary biologic function by modulating the transcription of dozens of genes.81,84 In mice, loss of p53 is associated with multiple spontaneous tumors77 and p53-loss accelerates the appearance of mammary tumors in murine mammary tissue that also overexpresses Myc, HER2/neu, IGF1, and/or Wnt1.85,86 Inheritance of a p53 mutant allele causes the rare familial Li-Fraumeni syndrome, characterized by multiple early-onset cancers, including breast cancer in women who survive childhood cancers. Germline mutations in ATM and CHEK2, which are involved in p53 activation, also increase susceptibility to breast cancer.87
Twenty to 35% of sporadic cases of breast cancers harbor somatic mutations in p53, based primarily on sequencing of exons 5 through 8 (the DNA-binding core), where approximately 90% of mutations are found.78,88,89 A total of 2,274 mutations in breast cancers are listed in the p53 database maintained by the International Agency for Research on Cancer.90
p53, when mutated, accumulates in the nucleus of neoplastic cells. Mutation of p53 is characterized by a common amino acid substitution resulting in a shift from an arginine (Arg) to a proline (Pro) at codon 72.91 Due to increased half-life and an altered conformational structure, the mutant form of p53 protein accumulates within malignant cells. Polymorphic variants have different biological properties; for example, the Arg/Arg genotype has been reported to induce apoptosis more effectively than the Pro/Pro genotype.92–94
The significance of p53 in carcinogenesis is manifested by the frequent genetic alterations in human cancers, found as mutations within the DNA binding domain and as loss of heterozygosity (LOH) in the loci of the gene. Several studies have reported LOH at codon 72 with preferential loss of the Pro allele both in breast cancer and other malignancies.93,95–97 The p53 genotype, in addition to the LOH status of codon 72, influences the prognosis in cancer. Two groups have studied this polymorphism in relation to breast cancer prognosis; they found a reduced survival for homozygous carriers of the Pro allele,98,99 consistent with the idea that the Pro72 variant of p53 is less efficient in inducing apoptosis than the Arg 72 variant.
Initial studies trying to elucidate the role of a mutated p53 gene in breast cancer prognosis were based on detecting p53 accumulation using immunohistochemistry. However, even in a meta-analysis of more than 9,000 breast cancer patients, the prognostic value of the p53 overexpression appeared weak.100 Conversely, sequencing studies have shown strong prognostic significance of p53 mutations in breast cancer. In one study where p53 mutations from 1,794 European patients with at least 10 years of follow-up were analyzed, mutations in p53 exons 5 through 8 were more common in ductal and medullary tumors with an aggressive phenotype (i.e. high grade, large size, node-positive cases, and low hormone receptor content), and in women less than 60 years old. Furthermore, the presence of a mutation conferred an overall 2.27-fold increased relative risk of breast cancer-specific mortality, independently of other known prognostic markers (e.g. tumor size, node status, and ER/PR expression).101 In a comprehensive meta-analysis of 16 studies including over 3,500 patients,102 the relative hazard of dying of breast cancer for unselected patients with a p53 mutation in their tumor was 2.0 (95% confidence interval, 1.7–2.5). For nodenegative patients the relative hazard was 1.7 (95% confidence interval, 1.2–2.3), and for node-positive patients the relative hazard was 2.6 (95% confidence interval, 1.7–3.9).
This and later studies have confirmed that p53 mutations are associated with worse overall and disease-free survival in breast cancer cases, and this effect is independent of other risk factors. In several studies, the presence of a p53 mutation was the single, most powerful prognostic indicator for both recurrence and death. Whether the prognostic significance of all types of mutations is the same, is still controversial. Borresen et al103 reported that patients with mutations affecting or disrupting the zinc-binding domains L2 and L3 (p53 codons 163–195 and 236–251) have a worse prognosis than patients with mutations elsewhere. In a later study, mutations affecting amino acids directly involved in DNA-binding—many in the zinc binding domain—were related with the poorest prognosis.104 These findings were confirmed in a different study, where patients with missense mutations affecting DNA-binding or zinc-binding domains had a very aggressive phenotype with a short survival.105 Another group reported that the prognosis for mutations in the conserved regions II and V was worse than for mutations in the conserved regions III and IV and non-conserved regions.106 The poor prognosis for patients with specific p53 mutations could mean that they have a gain-of-function effect or a particularly strong dominant-negative phenotype.107
Recent reports have described the detection of tumor-specific DNA circulating in plasma from breast cancer patients. Garcia et al108 examined plasma DNA from the breast cancer patients; in 61 of the 142 patients, with an average 58 months of follow-up, similar molecular signatures in tumor and plasma DNA was detected. For patients with tumor DNA in their plasma, the hazard ratio for recurrence was 2.5. Disease-free survival was 37% for positive patients and 75% for negative patients (P = 0.005). Among the 35 recurrences observed, 25 were positive for tumor plasma DNA and 10 were negative, (P < 0.001). These results indicate that tumor plasma DNA at diagnosis can serve as a prognostic marker of the overall survival of breast cancer patients. Further work is required to determine the role of the p53 mutations in plasma DNA in breast cancer prognosis.
Although both HER2 overexpression and p53 mutations are important prognostic factors for breast cancer, only a few reports have described the signaling cascades that link HER2 and p53. It was reported that HER2 overexpression in p53 wild-type human ovarian carcinoma cell line induced apoptosis shortly after transfection, while HER2 expression was associated with proliferation in cells with mutated p53.109 It was also reported that HER2/neu induces p53 ubiquitination via Akt-mediated Mdm2 phosphorylation and inactivates p53 in Arf-deficient cells.110 Thus, both ARF and p53 are important genetic determinants in susceptibility to HER2 overexpression in breast cancer. Further experiments should be conducted to characterize the signaling pathway between HER2 and ARF/p53.
ARF
The INK4a/ARF locus encodes two unrelated tumor suppressor proteins, p16INK4a and p14ARF, which participate in the two main cell-cycle control pathways, p16INK4a-Rb and p14ARF-p53111–115 (Fig. 1). p14ARF (p19Arf in mice), is a 14 kDa (19 kDa) protein predominantly localized in the nucleolus. It blocks the cell cycle in both G1 and G2 phases and inhibits the growth of incipient cancer cells by indirectly activating p53. It also inhibits ribosomal RNA processing and interacts with topoisomerase I.116 Arf triggers sumoylation of many cellular proteins, including Mdm2 and nucleophosmin (NPM/B23), with which p19Arf physically interacts in vivo. This occurs equally well in cells expressing or lacking functional p53.117 Thus, Arf’s p53-independent effects on gene expression and tumor suppression might depend on Arf-induced sumoylation.117
Methylation of CpG promoter islands has been described as a mechanism of gene silencing. Exon 1 of the p16INK4a gene and the p14ARF promoter gene reside within CpG islands. Therefore, both can become methylated de novo and silenced. However, genetic alterations that selectively inactivate p14ARF have been poorly analyzed in breast cancer. One comprehensive analysis of the inactivation mechanisms (mutation, homozygous and hemizygous deletion, and promoter hypermethylation) in 100 primary breast carcinomas118,119 used RT-PCR to document variable expression of the p14ARF transcript, with 17% demonstrating overexpression and 26% demonstrating decreased expression. No detectable alterations were observed in most cases with overexpressed p14ARF mRNA, but 77% of tumors with decreased expression had at least one of these genetic/epigenetic alterations. A different group analyzed the methylation status of p16INK4a and p14ARF by methylation-specific PCR in 100 breast, 95 colon and 27 bladder carcinomas. 120 Clinicopathological parameters were obtained from the medical records of the patients, and p14ARF showed a higher rate of hypermethylation than p16INK4a in all three tumor types.120 Sharma et al also found promoter hypermethylation of p14ARF in serum and tumor DNA from breast cancer patients. Aberrant methylation was significantly correlated with poor prognosis when analyzed with clinicopathological parameters of the breast tumor subtypes.121 Thus, both p16INK4a and p14ARF hypermethylation may be useful markers to predict the prognosis of breast cancer patients.
TBX2/3
T-box proteins contain a T-domain that affects dimerization and DNA binding. TBX2 belongs to the Tbx subfamily of T-box transcription factors.122,123 Other subfamilies of T-box genes are Brachyury, T-brain1, Tbx1 and Tbx6. TBX2, TBX3, TBX4 and TBX5 belong to the TBX2 subfamily. TBX2 and TBX3 are the only mammalian T-box factors with reported transcriptional repressor functions. Tbx2 and Tbx3 are closely related T-box proteins that have been implicated in tissue development in different sites, including the mammary gland. TBX3 is required for normal mammary development in mouse models and in patients with ulnar-mammary syndrome (UMS).123 TBX2 and TBX3 also have been implicated in tumor development through downregulation of the ARF tumor suppressor and an associated bypass of senescence.124,125 Overexpression of Bmi1, Pokemon, and Twist also contribute to tumor formation by repressing the INK4a/ARF locus.126 The TBX2 gene encoding a key developmental transcription factor is amplified and overexpressed in BRCA1 and BRCA2-mutated breast tumors.127 However, it is not known how Tbx2 mediates its repressive effect, nor whether endogenous Tbx2 or Tbx3 have a similar anti-senescence function in transformed cells. This is a particularly important question because the loss of CDKN2 A in many human cancers would, in principle, bypass the requirement for Tbx2/3-mediated repression of ARF in suppressing senescence (Fig. 1).
It has been reported that TBX2 is amplified in 8.6 to 21.6 % of sporadic human breast carcinomas, where the protein is overexpressed.122 A representative picture of TBX2 overexpression in human breast cancer is shown in Figure 2c. Ectopic expression of Tbx2 results in DNA polyploidy and cisplatin resistance. 128 Thus, overexpression of Tbx2 contributes to breast carcinogenesis by accelerating cell proliferation, changing DNA ploidy, and making cells resistant to chemotherapy. More studies are needed to elucidate the mechanism of TBX2/3 overexpression in breast cancer.
Cyclin D1
D-type cyclins (cyclin D1, D2, and D3) are other key regulator proteins of the G1 phase progression.113,115,129,130 There are three cyclins in this family with differential effects on the development of the normal mammary gland.131 The cyclin D1 protein is synthesized in response to growth factors; its levels peak in the mid-G1 phase of the cell cycle. The association of cyclin D1 to Cdk is crucial to drive cells to the restriction point where the cell is committed to divide.129,130 D-type cyclins bind to Cdks 4 and 6 and phosphorylate downstream substrates, mainly pRb, and these complexes can also sequester Cdk inhibitors (p21Cip1 and p27 Kip1) in the G1/S transition115,129,130 (Fig. 1). Furthermore, cyclin D1, the first member identified, can have Cdk-independent functions and can act as a co-factor for ERα independently of the ligand.57,58 D-type cyclins inhibit the activity of the Dmp1 transcription factor, which is a critical regulator of the Arf-p53 pathway.126,131–138
Human cyclin D1 is the product of a gene (CCND1) located on chromosome 11q13. Overexpression of cyclin D1 has been observed in many human tumors and is likely to promote cell proliferation and differentiation by shortening the G1/S transition.129,130,139 Amplification of the cyclin D1 gene has been detected in about 15% of breast cancers, while overexpression of cyclin D1 at mRNA and protein levels is seen in up to 50% of primary breast cancers, mostly ER-positive and well-differentiated tumors.129–131 A representative picture of cyclin D1 overexpression in human breast cancer is shown in Figure 2b.
Apparently the frequency of cyclin D1 gene amplification (∼15%) is lower than the incidence of overexpression (∼50%). Recently, Barbash et al140 demonstrated that attenuation of cyclin D1 SCFFbx4 E3 ubiquitin ligase activity occurs frequently in human cancer and may represent a common mechanism of overexpression and deregulation of cyclin D1. Significantly, inactivation of the Fbx4 ligase is the result of mutations in N-terminal regulatory regions of Fbx4 that disrupt ligase dimerization, thereby revealing the biological significance of SCF ligase oligomerization. They established Fbx4 as a tumor suppressor in human cancer, the function of which is abrogated by a unique category of mutations that target E3 ligase activity.140
The relationship between overexpression of cyclin D1 and breast cancer outcome has been controversial, with studies reporting both positive and negative findings.141–143 Subgroup analyses within relatively few patients have also hampered definitive conclusions. In addition, other molecules in the RB pathway have not been measured simultaneously. In spite of these limitations, there appears to be a relationship between cyclin D1 gene amplification and poor disease outcome in ER-positive patients.144–146 In contrast, cyclin D1 protein expression is associated with a good prognosis in some studies,141,147 potentially as a consequence of its positive relationship with ER expression and negative relationship with RB mutations. However, other studies failed to confirm this relationship.142,148
Interpretation is further complicated by suggestions that under some circumstances, cyclin D1 overexpression may worsen clinical outcome by conferring resistance to endocrine treatments.149,150 Consistent with this possibility, one small clinical study suggested that the duration of response to tamoxifen was significantly longer in ER-positive patients with low cyclin D1 than those with high cyclin D1.150 Resolution of these issues must await more detailed analysis of cyclin D1 expression and patient outcomes in the context of prospective randomized clinical trials.
The CCND1 locus encodes two gene products, cyclin D1a and cyclin D1b, which have discrete mechanisms of regulation and impact on cell behavior.151,152 A polymorphism at nucleotide 870 in the CCND1 gene, rs603965, influences the relative production of the encoded proteins and can reveal increased risk for tumor development. Millar et al153 studied the impact of both the G/A870 polymorphism and cyclin D1b protein production on breast cancer risk, disease phenotype and patient outcome. In a large multiethnic case-control study, the G/A870 polymorphism conferred no significant risk for breast cancer overall, by stage or ER status. However, the cyclin D1b protein was upregulated in breast cancer, independent of cyclin D1a levels, and exhibited heterogeneous levels in breast cancer specimens. High cyclin D1a expression inversely correlated with the Ki67 proliferation marker, but was not associated with clinical outcome. In contrast, elevated cyclin D1b expression was independently associated with adverse outcomes, including recurrence, distant metastasis, and decreased survival.153 Cyclin D1b overexpression was particularly associated with poor outcome in ER-negative breast cancer. Thus, specific cyclin D1 isoforms are associated with discrete forms of breast cancer and high cyclin D1b protein levels hold prognostic potential.
Cyclin D2 is more ubiquitously expressed in tissues than cyclin D1. Cyclin D2 is expressed in normal human mammary epithelial cells, but interestingly, its overexpression is rare in breast cancers.154,155 This is due to promoter hypermethylation in most cases.156 Its functional significance, if any, in breast oncogenesis has yet to be determined, although potential roles of cyclin D2 in terminal differentiation and senescence of human breast epithelium have been proposed. Indeed, MMTV-cyclin D2 mice show increased proliferation of mammary glands in pregnant females, but alveolar differentiation is partially or completely inhibited.157 In one report, cyclin D3 was overexpressed in breast cancers, but there are limited data on its relationship to the phenotype of breast cancer and prognosis of patients.158 Keyomarsi et al showed that the absence of cyclin D1 or cyclin D3 protein expression was associated with improved disease-specific and overall survival, but the correlations were less striking than those for cyclin E.159
Cyclin E
Cyclin E is the limiting factor for G1 phase progression and S phase entry. The cyclin E gene is a target of E2Fs and the protein associates with Cdk2 and activates its kinase activity shortly before entry of cells into the S phase113,115,129 (Fig. 1). The restriction point “R” at the transition between G1- and S-phases of the cell cycle has been recognized as the endpoint of regulatory pathways that are critical for growth control, and thus also for prevention of excessive or unrestricted growth. Deregulation of these pathways—or the deletion or overexpression of particular factors that are substantial for these pathways— have been linked to malignant transformation of cells and the development of cancer. While there is evidence of the importance of cyclin D1 in mammary tumorigenesis,160 the role of cyclin E in this respect has only recently been established. Cyclin E is expressed in supra-physiological levels in many human cancers and its genomic locus (19q12-q13) is frequently amplified.161,162 High levels of cyclin E and low levels of the G1-specific cell cycle inhibitor p27KIP1 exhibit a good correlation.159,163–169 Another clue for the importance of cyclin E in breast carcinoma is the finding of centrosome amplifications in these tumors, which could pave the way for genomic instability.170–172
Two proteins codified by two different genes but with high homology, called cyclin E1 (formerly cyclin E) and cyclin E2 have been identified.173–177 Cyclin E2, like cyclin E1, associates with Cdk2 and activates its kinase activity at the G1/S boundary.176 Cyclin E2 shares 47% overall similarity to cyclin E1; whether its structural features outside the conserved regions reveal unique functions is unknown.176
Increased expression of cyclin E1 has been reported in approximately 40% of breast cancer cases,159,178,179 and of cyclin E2 in 38% of primary ER-negative breast cancers.176 Elevated levels of both cyclins were more frequently found in ER-negative than in ER-positive tumors.176 The prognostic role of cyclin E (E1) has been retrospectively evaluated in a number of studies.180–187 Keyomarsi et al found that the overexpression of the cyclin E protein was accompanied by the appearance of low molecular weight (LMW) isoforms, and both were a reliable prognostic marker in stage I–III breast cancer patients. In fact, the hazard ratio for death due to breast cancer in patients with high levels of cyclin E was higher than associated with any other biological marker examined (seven times higher than the hazard ratio associated with lymph node metastases). Of note, all node-negative patients with high levels of cyclin E (12 out of 114) died of breast cancer. In some studies, a correlation between cyclin E and high histological grade181,182,184,186 or ER negativity180,187 has been reported, while an inverse correlation with p27KIP1 was observed in a study where both factors were prognostic.168
To study the oncogenic potential of the LMW forms of cyclin E in breast carcinogenesis, transgenic mice expressing full-length cyclin E alone, full-length and the EL4 isoforms, or the EL2/3 isoforms of cyclin E were generated under the control of MMTVLTR.188,189 Both primary mammary tumor formation and metastasis were markedly enhanced in LMW cyclin E-transgenic mice. LMW cyclin E overexpression in mammary epithelial cells of mice was sufficient to induce mammary carcinomas in 34 of 124 (27%) animals compared with 7 of 67 (10%) mice expressing only the full-length cyclin E, suggesting higher oncogenic activity of the LMW form.188 In addition, metastasis was more frequently found in LMW cyclin E tumor-bearing animals compared with tumors in mice with the full-length cyclin E background (P < 0.05). They also reported that LMW cyclin E overexpression selected for inactivation of p53 by LOH and frequent inactivation of p19Arf, canceling the protective checkpoint function of the Arf-p53 pathway and accelerating progression to mammary carcinomas.188,189
BRCA1 and BRCA2
Mutations of BRCA1 and BRCA2 located in chromosome 17q21 and 13q13, respectively, are involved in breast carcinogenesis.190 The gene was subsequently cloned and found to be novel, containing an aminoterminal, zinc- and DNA-binding “ring finger” motif; a carboxyl-terminal BCRT domain; and a nuclear localization sequence. Interestingly, mutations are particularly prevalent in breast cancers of Ashkenazi Jewish women and other select populations,191 but some families carrying BRCA mutations have few afflicted members. In other studies comparing different carrier populations, BRCA1 mutations were reported (but rarely) in women with no familial association of the disease.192 BRCA1 mutations were also associated with shorter survival (63% vs. 91%, P = 0.04) in a study of 249 European patients (36 patients with BRCA1 mutations, 8 with BRCA2 mutations, and 205 controls).193 These studies emphasize the variable penetrance of inherited risk conferred by this gene.
Although mutations in the BRCA1 gene are widely prevalent in patients with familial breast and ovarian cancer, mutations are rarely detected in sporadic breast cancers. BRCA2 is associated with familial cancers of the female and male breast and, to a lesser extent, the ovaries. Its gene shares homology with BRCA1, and its encoded protein has similar biochemical functions to BRCA1. However, BRCA2 mutation appears less involved in risk of ovarian cancer compared to BRCA1. Mutations of BRCA2 confer risk of male breast cancer and (to a more limited extent) several other cancers, such as prostate cancer, pancreatic cancer, non-Hodgkin’s lymphoma, basal cell carcinoma, bladder carcinoma, and fallopian tube tumors.
Current studies using CGH and cDNA microarray analysis suggest a distinct signature of chromosome gains and losses and gene expression for the different classes of familial breast cancers (BRCA1, BRCA2, and BRCAX) compared to sporadic breast cancers.194 BRCA1 appears to interact with the p53 protein (leading to induction of the cell-cycle inhibitor p21Cip1/WAF1; Fig. 1)195 directly, with the RNA polymerase holoenzyme, with the transcription factor CREB, with two proteins termed BAP1 and BARD1, with ERα (suppressing its function), with the promoter of c-MYC (suppressing expression of this protooncogene), and with promoters of other genes. BRCA1 and BRCA2 proteins are also found in complexes with Rad51, a protein important for the cellular response to DNA damage.192,194 In addition, BRCA1 is phosphorylated by the ATM/ATR kinases, in response to DNA damage (Fig. 1).
Thus, both BRCA proteins are now emerging as central gatekeepers of genomic stability.190,195 In studies of mice bearing a conditional knockout of the Brca1 gene, it has been implicated in mammary ductal morphogenesis and checkpoint control in the G1/S and G2/M phases of the cell cycle. Perhaps the most interesting among BRCA1 protein-protein interactions in mammary epithelial cells is the one with the ERα.196 This apparently key anti-estrogenic effect may place the BRCA proteins on center stage for control of the sex steroid-regulated pathways, long suspected to induce breast cancer.
Breast cancers of BRCA1/2 carriers more often have a high nuclear grade, poorly differentiated morphology, negativity to ER/PR/HER2, positivity to cytokeratins, overexpression of cyclin E, low expression of p27KIP1, and p53 mutations.194 BRCA1 mutations are associated with a poor prognosis in breast cancer within an Ashkenazi Jewish population in several studies.197 Chappuis et al198 reported that BRCA1/BRCA2 mutations were associated with shorter survival (58% vs. 82%, P = 0.003) in a Canadian Ashkenazi Jewish population. In another Canadian Ashkenazi Jewish population, Foulkes et al199 showed that the survival rate was 50% in those with BRCA1 mutations and 90% in their controls. Later, Robson et al200 studied 496 archived tissue blocks (43 cases of BRCA1 mutations, 14 cases of BRCA2 mutations, and 440 control patients); the overall 10-year survival rate was 62% in the BRCA1 mutation group and 86% in the control (P < 0.0001), but there was no difference in the BRCA2 mutation group versus controls.200
A recent study of BRCA1/2 mutations in young breast cancer patients (age 36 or less)201 showed that carriers of BRCA1/2 mutations had a ∼25% lower 3-year survival rate and were less likely to express ER, PR, and HER2, the so-called “triple-negative” form of the disease associated with poor prognosis. Another study showed that patients who were mutant BRCA1/2 carriers had increased 25-year contralateral breast cancer recurrence compared with patients without those mutations; this recurrence was 1.6-fold higher in patients with BRCA1 mutations compared to BRCA2 mutations.202 Additionally, patients diagnosed prior to age 40 had a significantly better chance of survival compared to those diagnosed later in life.202 However, other studies have failed to demonstrate the prognostic values of BRCA1/BRCA2 mutations in breast cancer, possibly because 1) the sample sizes of those with BRCA1/2 mutations was too small, 2) studies were biased by including only surviving patients, 3) there was no adjustment for therapy or other factors.
Vascular endothelial growth factor (VEGF)
Vascular endothelial growth factor (VEGF) is composed of a family of five isoforms (VEGFA, VEGFB, VEGFC, VEGFD, and PLGF) which act as ligands for tyrosine kinase receptors (VEGF-Rs).203 Upon binding of VEGF to its receptors (primarily VEGFR2), intracellular signaling pathways, including MEK-ERK and PI3K-Akt, are activated that mediate angiogenic switches (Fig. 1). This activation of angiogenesis in both normal and cancerous tissue is dependent on increased endothelial cell proliferation and invasion, increased vessel permeability, and recruitment of other support cells that make up the vessel architecture, such as pericytes.
VEGF and angiogenesis are important to tumor growth and metastasis across a range of solid tumor types.204–206 VEGF has been implicated as a key mediator of angiogenesis in breast cancer.206,207 In one study, Yoshiji et al207 found that VEGF expression was markedly upregulated compared with surrounding normal tissue in each of the 18 human breast tissue samples evaluated. In a separate study assessing the role of VEGF in breast cancer, Brown et al208 observed VEGF to be expressed at high levels in ductal carcinoma (comedo-type, invasive, and metastatic), but not in infiltrating lobular carcinoma. In a series of elegant experiments, Relf and colleagues209 isolated selected angiogenic factors from 64 primary breast tumors and investigated their relationship to tumor growth and progression. These factors included VEGF, transforming growth factor-β1 (TGF-β1), pleiotrophin, acidic and basic fibroblast growth factor (FGF), and platelet-derived endothelial cell growth factor. Overall, VEGF was one of the most important mediators of tumor angiogenesis in human breast cancer tissue, and elevated VEGF levels correlated with poor survival.
Toi and colleagues210 demonstrated that VEGF expression is associated with microvessel density in breast tumor biopsies. Their postoperative tissue analysis of 328 primary breast cancer patients showed a positive correlation between the rate of VEGF expression and microvessel density. While many pro-angiogenic factors have been identified in breast cancer, VEGF appears to be the only factor expressed throughout the entire life cycle of a breast tumor. Hence all the above reports have confirmed that VEGF is associated with increased microvessel density in breast cancer.
The most important factor that determines survival of breast cancer patients is dissemination of cancer cells from primary site into distant organs and establishment of metastatic colonies. Comparison of gene signatures from primary tumor, regional, and distant metastasis indicates that VEGF is only overexpressed in distant metastasis and is associated with poor survival. More importantly and clinically significant, Shivakumar et al211 showed that increased serum VEGF levels were detectable in patients with metastatic compared to benign lesions and were positively correlated with tumor grade. In a retrospective study of 679 patients, Linderholm et al212 showed significantly higher levels of intra-tumoral VEGF in patients with triple-negative breast cancers compared to other types of breast cancers.
Jacobs and colleagues showed that an increased risk of invasive breast cancer was correlated with 2 VEGF gene polymorphisms, VEGF-2578C and VEGF-1154G,213 both hypothesized to increase expression of VEGF. Intense angiogenic activity has been observed in inflammatory breast cancer. In an attempt to characterize the angiogenic phenotype of this disease, van der Auwera et al214 measured mRNA expression of tumor angiogenesis and lymphangiogenesis factors in patients with both inflammatory (n = 16) and noninflammatory (n = 20) breast cancer. Although both forms of the disease exhibited high levels of angiogenic activity, inflammatory breast cancer showed significantly higher mRNA expression of angiogenic and lymphangiogenic genes, including those encoding for VEGF-C and VEGF-D. These observations may help clarify why inflammatory breast cancer has a high metastatic potential via hematogenous and lymphatic routes.
Potential use of molecular prognostic markers for early detection, diagnosis, treatment and prevention of breast cancer
The use of classical markers such as Ki67, ER, PR, and HER2 for the prediction of patients’ survival and treatment response of breast cancer has been well established, and thus, they will be continued to be used as useful laboratory tests. Although numerous genetic and phenotypic alterations have been reported in breast cancer, only a handful have been fully identified and brought to clinical studies. Among molecules regulating the p53 and RB tumor suppressor pathways, mutation of p53, overexpression of cyclin D1b, LMW cyclin E are apparently associated with poor clinical outcome of patients. Experiments using transgenic/knockout mice for these molecules have provided unique perspectives of these molecules to the biology of breast cancer. Although p53 mutations have been extensively studied in human breast cancer, only a few studies have been conducted on the prognostic values of the upstream regulators for p53, such as overexpression of Hdm2, TBX2/3, Pokemon, or inactivation of p14ARF, hDMP1. It is expected that p53 will remain intact when upstream p53 regulators are altered in tumor cells. Examination of human breast cancer samples for the alterations of these molecules and determination of their prognostic values will be useful for future diagnosis, development of novel therapies, and prevention of breast cancer.
Acknowledgments
We are grateful to K. Klein (Research Support Core, Wake Forest University Health Sciences) for editing the manuscript. K. Inoue is supported by ACS RSG-07-207-01-MGO, NIH/NCI 2R01CA106314–06, and P30CA12197. P. Taneja is supported by the Susan G. Komen Foundation postdoctoral fellowship KG080179.
Footnotes
Disclosures
This manuscript has been read and approved by all authors. This paper is unique and is not under consideration by any other publication and has not been published elsewhere. The authors and peer reviewers of this paper report no conflicts of interest. The authors confirm that they have permission to reproduce any copyrighted material.
References
- 1.Andreopoulou E, Hortobagyi GN. Prognostic Factors in Metastatic Breast Cancer: Successes and Challenges Toward Individualized Therapy. J Clin. Oncol. 2008;26:3660–2. doi: 10.1200/JCO.2008.16.1026. [DOI] [PubMed] [Google Scholar]
- 2.Jemal A, Siegel R, Ward E, Hao Y, Thun MJ. Cancer statistics, 2009. Cancer J Clin. 2009;59:225–49. doi: 10.3322/caac.20006. [DOI] [PubMed] [Google Scholar]
- 3.Masood S. Prognostic/predictive factors in breast cancer. Clin Lab Med. 2005;25:809–25. doi: 10.1016/j.cll.2005.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Morrow PK, Hortobagyi GN. Management of breast cancer in the genome era. Annu Rev Med. 2009;60:153–65. doi: 10.1146/annurev.med.60.061107.145152. [DOI] [PubMed] [Google Scholar]
- 5.Dickson RB, Lippman ME. Autocrine and paracrine growth factors in the normal and neoplastic breast. In: Harris JR, Lippman ME, Morrow M, et al., editors. Philadelphia: Lippincott Williams & Wilkins; 2000. p. 303. [Google Scholar]
- 6.Pakkiri P, Lakhani SR, Smart CE.Current and future approach to the pathologist’s assessment for targeted therapy in breast cancer Pathology 20094189–99.Review. [DOI] [PubMed] [Google Scholar]
- 7.Debies MT, Welch DR. Genetic basis of human breast cancer metastasis. J Mammary Gland Biol Neoplasia. 2001;6:441–51. doi: 10.1023/a:1014739131690. [DOI] [PubMed] [Google Scholar]
- 8.de Snoo F, Bender R, Glas A, Rutgers E.Gene expression profiling: decodingbreast cancer Surg Oncol 200918366–78.Review. [DOI] [PubMed] [Google Scholar]
- 9.Kuderer NM, Lyman GH.Gene expression profile assays as predictors of distant recurrence-free survival in early-stage breast cancer Cancer Invest 200927885–90.Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Brown DC, Gatter KC. Ki67 protein: the immaculate deception? Histopathology. 2002;40:2–11. doi: 10.1046/j.1365-2559.2002.01343.x. [DOI] [PubMed] [Google Scholar]
- 11.Gerdes J, Schwab U, Lemke H, Stein H. Production of a mouse monoclonal antibody reactive with a human nuclear antigen associated with cell proliferation. Int J Cancer. 1983;31:13–20. doi: 10.1002/ijc.2910310104. [DOI] [PubMed] [Google Scholar]
- 12.Gerdes J, Li L, Schlueter C, et al. Immunobiochemical and molecular biologic characterization of the cell proliferation-associated nuclear antigen that is defined by monoclonal antibody Ki-67. Am J Pathol. 1991;138:867–73. [PMC free article] [PubMed] [Google Scholar]
- 13.Bridger JM, Kill IR, Lichter P. Association of pKi-67 with satellite DNA of the human genome in early G1 cells. Chromosome Res. 1998;6:13–24. doi: 10.1023/a:1009210206855. [DOI] [PubMed] [Google Scholar]
- 14.MacCallum DE, Hall PA. The location of pKi67 in the outer dense fibrillary compartment of the nucleolus points to a role in ribosome biogenesis during the cell division cycle. J Pathol. 2000a;190:537–44. doi: 10.1002/(SICI)1096-9896(200004)190:5<537::AID-PATH577>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 15.MacCallum DE, Hall PA. The biochemical characterization of the DNA binding activity of pKi67. J Pathol. 2000b;191:286–98. doi: 10.1002/1096-9896(2000)9999:9999<::AID-PATH628>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 16.Sahin AA, Ro J, Ro JY, et al. Ki-67 immunostaining in node-negative stage I/II breast carcinoma. Significant correlation with prognosis. Cancer. 1991;68:549–57. doi: 10.1002/1097-0142(19910801)68:3<549::aid-cncr2820680318>3.0.co;2-j. [DOI] [PubMed] [Google Scholar]
- 17.Keshgegian AA, Cnaan A. Proliferation markers in breast carcinoma. Mitotic figure count, S-phase fraction, proliferating cellnuclear antigen, Ki-67 and MIB-1. Am J Clin Pathol. 1995;104:42–9. doi: 10.1093/ajcp/104.1.42. [DOI] [PubMed] [Google Scholar]
- 18.Assersohn L, Salter J, Powles TJ, et al. Studies of the potential utility of Ki67 as a predictive molecular marker of clinical response in primary breast cancer. Breast Cancer Res Treat. 2003;82:113–23. doi: 10.1023/B:BREA.0000003968.45511.3f. [DOI] [PubMed] [Google Scholar]
- 19.Beck T, Weller EE, Weikel W, Brumm C, Wilkens C, Knapstein PG. Usefulness of immunohistochemical staining for p53 in the prognosis of breast carcinomas: correlations with established prognosis parameters and with the proliferation marker, MIB-1. Gynecol Oncol. 1995;57:96–104. doi: 10.1006/gyno.1995.1104. [DOI] [PubMed] [Google Scholar]
- 20.Bottini A, Berruti A, Bersiga A, et al. Relationship between tumour shrinkage and reduction in Ki67 expression after primary chemotherapy in human breast cancer. Br J Cancer. 2001;85:1106–12. doi: 10.1054/bjoc.2001.2048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bouzubar N, Walker KJ, Griffiths K, et al. Ki67 immunostaining in primary breast cancer: pathological and clinical associations. Br J Cancer. 1989;59:943–7. doi: 10.1038/bjc.1989.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ceccarelli C, Trere D, Santini D, Taffurelli M, Chieco P, Derenzini M. AgNORs in breast tumours. Micron. 2000;31:143–9. doi: 10.1016/s0968-4328(99)00071-2. [DOI] [PubMed] [Google Scholar]
- 23.Chang J, Powles TJ, Allred DC, et al. Biologic markers as predictors of clinical outcome from systemic therapy for primary operable breast cancer. J Clin Oncol. 1999;17:3058–63. doi: 10.1200/JCO.1999.17.10.3058. [DOI] [PubMed] [Google Scholar]
- 24.Colleoni M, Zahrieh D, Gelber RD, et al. Preoperative systemic treatment:prediction of responsiveness. Breast. 2003;12:538–42. doi: 10.1016/s0960-9776(03)00163-2. [DOI] [PubMed] [Google Scholar]
- 25.Dettmar P, Harbeck N, Thomssen C, et al. Prognostic impact of proliferation-associated factors MIB1 (Ki-67) and S-phase in node-negative breast cancer. Br J Cancer. 1997;75:1525–33. doi: 10.1038/bjc.1997.261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Jacquemier JD, Penault-Llorca FM, Bertucci F, et al. Angiogenesis as a prognostic marker in breast carcinoma with conventional adjuvant chemotherapy: a multiparametric and immunohistochemical analysis. J Pathol. 1998;184:130–5. doi: 10.1002/(SICI)1096-9896(199802)184:2<130::AID-PATH19>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- 27.Locker AP, Birrell K, Bell JA, et al. Ki67 immunoreactivity in breast carcinoma: relationships to prognostic variables and short term survival. Eur J Surg Oncol. 1992;18:224–9. [PubMed] [Google Scholar]
- 28.MacGrogan G, Mauriac L, Durand M, et al. Primary chemotherapy in breast invasive carcinoma: predictive value of the immunohistochemicaldetection of hormonal receptors, p53, c-erbB-2, MiB1, pS2 and GST pi. Br J Cancer. 1996;74:1458–65. doi: 10.1038/bjc.1996.565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Midulla C, De Iorio P, Nagar C, et al. Immunohistochemical expression of p53, nm23-HI, Ki67 and DNA ploidy: correlation with lymph node status and other clinical pathologic parameters in breast cancer. Anticancer Res. 1999;19:4033–7. [PubMed] [Google Scholar]
- 30.Veronese SM, Gambacorta M, Gottardi O, Scanzi F, Ferrari M, Lampertico P. Proliferation index as a prognostic marker in breast cancer. Cancer. 1993;71:3926–31. doi: 10.1002/1097-0142(19930615)71:12<3926::aid-cncr2820711221>3.0.co;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Dowsett M, Smith IE, Ebbs SR, et al. IMPACT Trialists Group Prognostic value of Ki67 expression after short-term presurgical endocrine therapy for primary breast cancer. J Natl Cancer Inst. 2007;99:167–70. doi: 10.1093/jnci/djk020. [DOI] [PubMed] [Google Scholar]
- 32.Jones RL, Salter J, A’Hern R, et al. The prognostic significance of Ki67 before and after neoadjuvant chemotherapy in breast cancer Breast Cancer Res Treat 200911653–68.Epub 2008 Jul 1. [DOI] [PubMed] [Google Scholar]
- 33.Dawsett M, Dunbier AK. Emerging biomarkers and new understanding of traditional markers in personalized therapy for breast cancer. Clin. Cancer Res. 2008;14:8019–26. doi: 10.1158/1078-0432.CCR-08-0974. [DOI] [PubMed] [Google Scholar]
- 34.DeGregori J, Kowalik T, Nevins JR. Cellular targets for activation by the E2F1 transcription factor include DNA synthesis- and G1/S-regulatory genes. Mol Cell Biol. 1995;15:4215–24. doi: 10.1128/mcb.15.8.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ross JS, Hortobagyi GN. Molecular Oncology of Breast Cancer. Jones and Bartlett Publishers; Sudsbury, Massachusetts: 2005. [Google Scholar]
- 36.Jensen EV, Jordan VC. The estrogen receptor: a model for molecular medicine. Clin Cancer Res. 2003;9:1980. [PubMed] [Google Scholar]
- 37.Sommer S, Fuqua SA. Estrogen receptor and breast cancer. Semin. Cancer Biol. 2001;11:339–52. doi: 10.1006/scbi.2001.0389. [DOI] [PubMed] [Google Scholar]
- 38.Su JL, McKee DD, Ellis B, et al. Production and characterization of an estrogen receptor beta subtype specific mouse monoclonal antibody. Hybridoma. 2000;19:481–7. doi: 10.1089/027245700750053977. [DOI] [PubMed] [Google Scholar]
- 39.Palmieri C, Cheng GJ, Saji S, et al. Estrogen receptor beta in breast cancer. Endocr. Relat Cancer. 2002;9:1–13. doi: 10.1677/erc.0.0090001. [DOI] [PubMed] [Google Scholar]
- 40.Humphreys RC, Lydon JP, O’Malley BW, Rosen JM. Use of PRKO mice to study the role of progesterone in mammary gland development. J Mammary Gland Biol Neoplasia. 1997;2:343–54. doi: 10.1023/a:1026343212187. [DOI] [PubMed] [Google Scholar]
- 41.Thorpe SM, Rose C, Rasmussen BB, et al. Steroid hormone receptors as prognostic indicators in primary breast cancer. Breast Cancer Res Treat. 1986;7:91–8. [PubMed] [Google Scholar]
- 42.Crowe JP, Jr, Gordon NH, Hubay CA, et al. Estrogen receptor determination and long term survival of patients with carcinoma of the breast. Surg Gynecol Obstet. 1991;173:273–8. [PubMed] [Google Scholar]
- 43.McGuire WL, Tandon AK, Allred DC, Chamness GC, Clark GM. How to use prognostic factors in axillary node-negative breast cancer patients. J Natl Cancer Inst. 1990;82:1006–15. doi: 10.1093/jnci/82.12.1006. [DOI] [PubMed] [Google Scholar]
- 44.Fisher B, Fisher ER, Redmond C, Brown A. Tumor nuclear grade, estrogen receptor, and progesterone receptor: their value alone or in combination as indicators of outcome following adjuvant therapy for breast cancer. Breast Cancer Res Treat. 1986;7:147–60. doi: 10.1007/BF01806245. [DOI] [PubMed] [Google Scholar]
- 45.Allred DC, Bryant J, Land S, et al. Estrogen receptor expression as a predictive marker of the effectiveness of tamoxifen in the treatment of DCIS: findings from NSABP Protocol B-24 [abstract] Breast Cancer Res Treat. 2002;76:A36. [Google Scholar]
- 46.Dotzlaw H, Leygue E, Watson PH, Murphy LC. Estrogen receptor- beta messenger RNA expression in human breast tumor biopsies: relationship to steroid receptor status and regulation by progestins. Cancer Res. 1999;59:529–32. [PubMed] [Google Scholar]
- 47.Fuqua SA, Schiff R, Parra I, et al. Expression of wild-type estrogen receptor beta and variant isoforms in human breast cancer. Cancer Res. 1999;59:5425–8. [PubMed] [Google Scholar]
- 48.Speirs V, Kerin MJ. Prognostic significance of oestrogen receptor beta in breast cancer. Br J Surg. 2008;87:405–09. doi: 10.1046/j.1365-2168.2000.01402.x. [DOI] [PubMed] [Google Scholar]
- 49.Honma N, Horii R, Iwase T, et al. Clinical importance of estrogen receptorbeta evaluation in breast cancer patients treated with adjuvant tamoxifen therapy. J Clin Oncol. 2000;26:3727–34. doi: 10.1200/JCO.2007.14.2968. [DOI] [PubMed] [Google Scholar]
- 50.Maehle BO, Collett K, Tretli S, Akslen LA, Grotmol T. Estrogen receptor beta—an independent prognostic marker in estrogen receptor alpha and progesterone receptor-positive breast cancer? APMIS. 2009;117:644–50. doi: 10.1111/j.1600-0463.2009.02510.x. [DOI] [PubMed] [Google Scholar]
- 51.Donegan WL. Tumor-related prognostic factors for breast cancer. CA Cancer J Clin. 1997;47:28–51. doi: 10.3322/canjclin.47.1.28. [DOI] [PubMed] [Google Scholar]
- 52.Holmes FA, Fritsche HA, Loewy JW, et al. Measurement of estrogen and progesterone receptors in human breast tumors: enzyme immunoassay versus binding assay. J Clin Oncol. 1990;8:1025–35. doi: 10.1200/JCO.1990.8.6.1025. [DOI] [PubMed] [Google Scholar]
- 53.Hahnel R, Woodings T, Vivian AB. Prognostic value of estrogen receptors in primary breast cancer. Cancer. 1979;44:671–5. doi: 10.1002/1097-0142(197908)44:2<671::aid-cncr2820440238>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- 54.Shek LL, Godolphin W. Survival with breast cancer: the importance of estrogen receptor quantity. Eur J Cancer Clin Oncol. 1989;25:243–50. doi: 10.1016/0277-5379(89)90015-1. [DOI] [PubMed] [Google Scholar]
- 55.McCormack O, Chung WY, Fitzpatrick P, et al. Progesterone receptor B (PRB) promoter hypermethylation in sporadic breast cancer: progesterone receptor B hypermethylation in breast cancer. Breast Cancer Res Treat. 2008;111:45–53. doi: 10.1007/s10549-007-9757-7. [DOI] [PubMed] [Google Scholar]
- 56.Liu S, Chia SK, Mehl E, et al. Progesterone receptor is a significant factor associated with clinical outcomes and effect of adjuvant tamoxifen therapy in breast cancer patients. Breast Cancer Res Threat. 2010;119:53–61. doi: 10.1007/s10549-009-0318-0. [DOI] [PubMed] [Google Scholar]
- 57.Neuman E, Ladha MH, Lin N, et al. Cyclin D1 stimulation of estrogen receptor transcriptional activity independent of cdk4. Mol Cell Biol. 1997;17:5338–47. doi: 10.1128/mcb.17.9.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Zwijsen RM, Wientjens E, Klompmaker R, van der Sman J, Bernards R, Michalides RJ. CDK-independent activation of estrogen receptor by cyclin D1. Cell. 1997;88:405–15. doi: 10.1016/s0092-8674(00)81879-6. [DOI] [PubMed] [Google Scholar]
- 59.Font de Mora J, Brown M. AIB1 is a conduit for kinase-mediated growth factor signaling to the estrogen receptor. Mol Cell Biol. 2000;20:5041–7. doi: 10.1128/mcb.20.14.5041-5047.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lee H, Bai W. Regulation of estrogen receptor nuclear export by ligandinduced and p38-mediated receptor phosphorylation. Mol Cell Biol. 2002;22:5835–45. doi: 10.1128/MCB.22.16.5835-5845.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Harigopal M, Heymann J, Ghosh S, Anagnostou V, Camp RL, Rimm DL. Estrogen receptor co-activator (AIB1) protein expression by automated quantitative analysis (AQUA) in a breast cancer tissue microarray and association with patient outcome. Breast Cancer Res Treat. 2009;115:77–85. doi: 10.1007/s10549-008-0063-9. [DOI] [PubMed] [Google Scholar]
- 62.Holbro T, Civenni G, Hanes NE. The ErbB receptors and their role in cancer progression. Exp Cell Res. 2003;284:99–110. doi: 10.1016/s0014-4827(02)00099-x. [DOI] [PubMed] [Google Scholar]
- 63.Hynes NE, Lane HA. ERBB receptors and cancer: the complexity of target inhibitors. Nat Rev Cancer. 2005;5:341–54. doi: 10.1038/nrc1609. [DOI] [PubMed] [Google Scholar]
- 64.Slamon DJ, Clark GM, Wong SG, Levin WJ, Ullrich A, McGuire WL. Human breast cancer: correlation of relapse and survivalwith amplification of the HER-2/neu oncogene. Science. 1987;235:177–82. doi: 10.1126/science.3798106. [DOI] [PubMed] [Google Scholar]
- 65.Zhou BP, Hung MC. Dysregulation of cellular signaling by HER2/neu in breast cancer. Sem Oncol. 2003;30:38–48. doi: 10.1053/j.seminoncol.2003.08.006. [DOI] [PubMed] [Google Scholar]
- 66.Yamamoto T, Ikawa S, Akiyama T, et al. Similarity of protein encoded by the human c-erb-B-2 gene to epidermal growth factor receptor. Nature. 1986;319:230–4. doi: 10.1038/319230a0. [DOI] [PubMed] [Google Scholar]
- 67.Bacus SS, Gudkov AV, Esteva FJ, Yarden Y. Expression of erb-B receptors and their ligands in breast cancer: implications to biological behavior and therapeutic response. Breast Dis. 2000;11:63–75. doi: 10.3233/bd-1999-11106. [DOI] [PubMed] [Google Scholar]
- 68.Slamon DJ, Leyland-Jones B, Shak S, et al. Use of chemotherapy plus a monoclonal antibody against HER2 for metastatic breast cancer that overexpresses HER2. N Engl J Med. 2001;344:783–92. doi: 10.1056/NEJM200103153441101. [DOI] [PubMed] [Google Scholar]
- 69.Borg A, Tandon AK, Sigurdsson H, et al. HER-2/neu amplification predictspoor survival in node-positive breast cancer. Cancer Res. 1990;50:4332–7. [PubMed] [Google Scholar]
- 70.Paik S, Hazan R, Fisher ER, et al. Pathologic findings from the National Surgical Adjuvant Breast and Bowel Project: prognostic significance of erbB-2 protein overexpression in primary breast cancer. J Clin Oncol. 1990;8:103–12. doi: 10.1200/JCO.1990.8.1.103. [DOI] [PubMed] [Google Scholar]
- 71.Allred DC, Clark GM, Tandon AK, et al. Her-2/neu in node-negative breast cancer: prognostic significance of overexpression influenced by the presence of in situ carcinoma. J Clin Oncol. 1992;10:599–605. doi: 10.1200/JCO.1992.10.4.599. [DOI] [PubMed] [Google Scholar]
- 72.Perou CM, Sorlie T, Eisen MB, et al. Molecular portraits of human breast tumours. Nature. 2000;406:747–52. doi: 10.1038/35021093. [DOI] [PubMed] [Google Scholar]
- 73.Sorlie T, Tibshirani R, Parker J, et al. Repeated observation of breast tumor subtypes in independent gene expression data sets. Proc Natl Acad Sci USA. 2003;100:8418–23. doi: 10.1073/pnas.0932692100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Press M, Anderson S, Dybdal N, Lieberman G, Mass R. Fluorescencein situ hybridization (FISH) is superior to immunohistochemistry (IHC) for determining HER2 status: the Herceptin (R) experience [abstract] Mod Pathol. 2002;15:A47. [Google Scholar]
- 75.Kruser TJ, Wheeler DL.Mechanisms of resistance to HER family targeting antibodies Exp Cell Res 2010January11[Epub ahead of print]. [DOI] [PubMed] [Google Scholar]
- 76.Seidman AD, Fornier M, Esteva FJ, et al. Weekly trastuzumab and paclitaxel therapy for metastatic breast cancer with analysis of efficacy by HER2 immunophenotype and gene amplification. J Clin Oncol. 2001;19:2587–95. doi: 10.1200/JCO.2001.19.10.2587. [DOI] [PubMed] [Google Scholar]
- 77.Attardi LD, Donehower LA. Probing p53 biological functions through the use of genetically engineered mouse models. Mutat Res. 2005;576:4–21. doi: 10.1016/j.mrfmmm.2004.08.022. [DOI] [PubMed] [Google Scholar]
- 78.Børresen-Dale AL.TP53 and breast cancer Hum Mutat 200321292–300.Review. [DOI] [PubMed] [Google Scholar]
- 79.Gasco M, Yulug IG, Crook T.TP53 mutations in familial breast cancer: functional aspects Hum Mutat 200321301–6.Review. [DOI] [PubMed] [Google Scholar]
- 80.Giaccia AJ, Kastan MB. The complexity of p53 modulation: emerging patterns from divergent signals. Genes Dev. 1998;12:2973–83. doi: 10.1101/gad.12.19.2973. [DOI] [PubMed] [Google Scholar]
- 81.Lacroix M, Toillon RA, Leclercq G. p53 and breast cancer, an update. Endocr Relat Cancer. 2006;13:293–325. doi: 10.1677/erc.1.01172. [DOI] [PubMed] [Google Scholar]
- 82.Meek DW.Tumour suppression by p53: a role for the DNA damage response? Nat Rev Cancer 20099714–23.Review. [DOI] [PubMed] [Google Scholar]
- 83.Toledo F, Wahl GM. Regulating the p53 pathway: in vitro hypotheses, in vivo veritas. Nat Rev Cancer. 2006;6:909–23. doi: 10.1038/nrc2012. [DOI] [PubMed] [Google Scholar]
- 84.Menendez D, Igna A, Resnick MA.The expanding universe of p53 targets Nat Rev Cancer 20099724–37.Review. [DOI] [PubMed] [Google Scholar]
- 85.Gunther EJ, Moody SE, Belka GK, et al. Impact of p53 loss on reversal and recurrence of conditional Wnt-induced tumorigenesis. Genes Dev. 2003;17:488–501. doi: 10.1101/gad.1051603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Ventura A, Kirsch DG, McLaughlin ME, et al. Restoration of p53 function leads to tumour regression in vivo. Nature. 2007;445:661–5. doi: 10.1038/nature05541. [DOI] [PubMed] [Google Scholar]
- 87.Lacroix M, Leclercq G. The “portrait” of hereditary breast cancer. Breast Cancer Res Treat. 2005;89:297–304. doi: 10.1007/s10549-004-2172-4. [DOI] [PubMed] [Google Scholar]
- 88.Bertheau P, Espié M, Turpin E, et al. TP53 status and response to chemotherapy in breast cancer Pathobiology 200875132–9.Epub 2008 Jun 10. Review. [DOI] [PubMed] [Google Scholar]
- 89.Vousden KH, Lu X. Live or let die: the cell’s response to p53. Nat Rev Cancer. 2002;2:594–604. doi: 10.1038/nrc864. [DOI] [PubMed] [Google Scholar]
- 90.Olivier M, Eeles R, Hollstein M, Khan MA, Harris CC, Hainaut P. The IARC TP53 database: new online mutation analysis and recommendations to users. Hum Mutat. 2002;19:607–14. doi: 10.1002/humu.10081. [DOI] [PubMed] [Google Scholar]
- 91.Thomas M, Kalita A, Labrecque S, Pim D, Banks L, Matlashewski G. Two polymorphic variants of wild-type p53 differ biochemical and biologically. Mol Cell Biol. 1999;19:1092–100. doi: 10.1128/mcb.19.2.1092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Dumont P, Leu JI, Della Pietra AC, 3rd, George DL, Murphy M. The codon72 polymorphic variants of p53 have markedly different apoptotic potential. Nat Genet. 2003;33:357–65. doi: 10.1038/ng1093. [DOI] [PubMed] [Google Scholar]
- 93.Pim D, Banks L. p53 polymorphic variants at codon72 exerts different effects on cell cycle progression. Int J Cancer. 2004;108:196–9. doi: 10.1002/ijc.11548. [DOI] [PubMed] [Google Scholar]
- 94.Sullivan A, Syed N, Gasco M, et al. Polymorphism in wild-type p53 modulates response to chemotherapy in vitro and in vivo. Oncogene. 2004;23:3328–37. doi: 10.1038/sj.onc.1207428. [DOI] [PubMed] [Google Scholar]
- 95.Bonafe M, Ceccarelli C, Farabegoli F, et al. Retention of the codon72 Arginine allele is associatedwith a reduction of disease-free and overall survival in Arginine/Proline heterozygous breast cancer patients. Clin Cancer Res. 2003;9:4860–4. [PubMed] [Google Scholar]
- 96.Mitra S, Banerjee S, Misra C, et al. Interplay between human papilloma virus infection and p53 gene alterations in head and neck squamous cell carcinoma of an Indian population. J Clin Pathol. 2007;60:1040–7. doi: 10.1136/jcp.2005.034835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Schneider-Stock R, Boltz C, Peters B, et al. Selective loss of codon72 Proline p53 and frequent mutational inactivation of retained Arginine allele in colorectal cancer. Neoplasia. 2004;6:529–35. doi: 10.1593/neo.04178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Tommiska J, Eerola H, Heinonen M, et al. Breast cancer patients with p53 Pro 72 homozygous genotype have a poorer survival. Clin Cancer Res. 2005;11:5098–103. doi: 10.1158/1078-0432.CCR-05-0173. [DOI] [PubMed] [Google Scholar]
- 99.Toyama T, Zhang Z, Nishio M, et al. Association of TP53 codon 72 polymorphism and the outcome of adjuvant therapy in breast cancer patients. Breast Cancer Res. 2007;9:R34. doi: 10.1186/bcr1682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Barbareschi M. Prognostic value of the immunohistochemical expression of p53 inbreast carcinomas—a review of the literature involving over 9,000 patients. Appl Immunohistochem. 1996;4:106–16. [Google Scholar]
- 101.Olivier M, Langerød A, Carrieri P, et al. The clinical value of somatic TP53 gene mutations in 1,794 patients with breast cancer. Clin Cancer Res. 2006;12:1157–1167. doi: 10.1158/1078-0432.CCR-05-1029. [DOI] [PubMed] [Google Scholar]
- 102.Pharoah PD, Day NE, Caldas C. Somatic mutations in the p53 gene and prognosis in breast cancer: a meta-analysis. Br J Cancer. 1999;80:1968–73. doi: 10.1038/sj.bjc.6690628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Børresen AL, Andersen TI, Eyfjörd JE, et al. TP53 mutations and breast cancer prognosis: particularly poor survival rates for cases with mutations in the zinc-binding domains. Genes Chromosomes Cancer. 1995;14:71–5. doi: 10.1002/gcc.2870140113. [DOI] [PubMed] [Google Scholar]
- 104.Berns EM, van Staveren IL, Look MP, Smid M, Klijn JG, Foekens JA. Mutations in residues of TP53 that directly contact DNA predict poor outcome in human primary breast cancer. Br J Cancer. 1998;77:1130–6. doi: 10.1038/bjc.1998.187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Alsner J, Yilmaz M, Guldberg P, Hansen LL, Overgaard J. Heterogeneity in the clinical phenotype of TP53 mutations in breast cancer patients. Clin Cancer Res. 2000;6:3923–31. doi: 10.1186/bcr109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Bergh J, Norberg T, Sjögren S, Lindgren A, Holmberg L. Complete sequencing of the p53 gene provides prognostic information in breast cancer patients, particularly in relation to adjuvant systemic therapy and radiotherapy. Nat Med. 1995;1:1029–34. doi: 10.1038/nm1095-1029. [DOI] [PubMed] [Google Scholar]
- 107.Brosh R, Rotter V. When mutants gain new powers: news from the p53 field. Nat Rev Cancer. 2009;9:701–23. doi: 10.1038/nrc2693. [DOI] [PubMed] [Google Scholar]
- 108.Garcia JM, Garcia V, Silva J, et al. Extracellular tumor DNA in plasma and overall survival in breast cancer patients. Genes Chromosomes Cancer. 2006;45:692–701. doi: 10.1002/gcc.20334. [DOI] [PubMed] [Google Scholar]
- 109.Casalini P, Botta L, Menard S. Role of p53 in HER2-induced proliferation or apoptosis. J Biol Chem. 2001;276:12449–53. doi: 10.1074/jbc.M009732200. [DOI] [PubMed] [Google Scholar]
- 110.Zhou BP, Liao Y, Xia W, Zou Y, Spohn B, Huang MC. Her2/Neu induces p53 ubiquitination via Akt-mediated MDM2 phosphorylation. Nat Cell Biol. 2001;3:973–82. doi: 10.1038/ncb1101-973. [DOI] [PubMed] [Google Scholar]
- 111.Agrawal A, Yang J, Murphy RF, Agrawal DK. Regulation of the p14 ARF-Mdm2-p53pathway: an overview in breast cancer. Exp Mol Pathol. 2006;81:115–22. doi: 10.1016/j.yexmp.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 112.Sharpless NE. INK4a/ARF: A multifunctional tumor suppressor locus. Mutat Res. 2005;576:22–38. doi: 10.1016/j.mrfmmm.2004.08.021. [DOI] [PubMed] [Google Scholar]
- 113.Sherr CJ.Principles of tumor suppression Cell 2004116235–46.Review. [DOI] [PubMed] [Google Scholar]
- 114.Sherr CJ. Divorcing ARF and p53: an unsettled case. Nat Rev Cancer. 2006;6:663–73. doi: 10.1038/nrc1954. [DOI] [PubMed] [Google Scholar]
- 115.Sherr CJ, McCormick F. The RB and p53 pathways in cancer. Cancer Cell. 2002;2:103–12. doi: 10.1016/s1535-6108(02)00102-2. [DOI] [PubMed] [Google Scholar]
- 116.Sugimoto M, Kuo ML, Roussel MF, Sherr CJ. Nucleolar Arf tumor suppressor inhibits ribosomal RNA processing. Mol Cell. 2003;11:415–24. doi: 10.1016/s1097-2765(03)00057-1. [DOI] [PubMed] [Google Scholar]
- 117.Tago K, Chiocca S, Sherr CJ. Sumoylation induced by the ARF tumor suppressor: A p53-independent function. Proc Natl Acad Sci USA. 2005;21:7689–94. doi: 10.1073/pnas.0502978102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Silva J, Domínguez G, Silva JM, et al. Analysis of genetic and epigenetic processes that influence p14 ARF expression in breast cancer. Oncogene. 2001;20:4586–90. doi: 10.1038/sj.onc.1204617. [DOI] [PubMed] [Google Scholar]
- 119.Silva J, Silva JM, Domínguez G, et al. Concomitant expression of p16INK4a and p14 ARF in primary breast cancer and analysis of inactivation mechanisms. J Pathol. 2003;199:289–97. doi: 10.1002/path.1297. [DOI] [PubMed] [Google Scholar]
- 120.Dominguez G, Silva J, Garcia JM, et al. Prevalence of aberrantmethylation of p14 ARF over p16INK4a in some human primary tumors. Mutat Res. 2003;530:9–17. doi: 10.1016/s0027-5107(03)00133-7. [DOI] [PubMed] [Google Scholar]
- 121.Sharma G, Mirza S, Prasad CP, Srivastava A, Gupta SD, Ralhan R. Promoter hypermethylation of p16INK4A, p14ARF, Cyclin D2 and Slit2 in serum and tumor DNA from breast cancer patients. Life Sci. 2007;80:1873–81. doi: 10.1016/j.lfs.2007.02.026. [DOI] [PubMed] [Google Scholar]
- 122.Rowley M, Grothey E, Couch FJ.The role of Tbx2 and Tbx3 in mammary development and tumorigenesis J Mammary Gland Biol Neoplasia 20049109–18.Review. [DOI] [PubMed] [Google Scholar]
- 123.Vance KW, Carreira S, Brosch G, Goding CR. Tbx2 is overexpressed and plays an important role in maintaining proliferationand suppression of senescence in melanomas. Cancer Res. 2005;65:2260–8. doi: 10.1158/0008-5472.CAN-04-3045. [DOI] [PubMed] [Google Scholar]
- 124.Jacobs JJ, Keblusek P, Robanus-Maandag E, et al. Senescence bypass screen identifies TBX2, which represses Cdkn2a (p19(ARF)) and is amplifiedin a subset of human breast cancers. Nat Genet. 2000;26:291–9. doi: 10.1038/81583. [DOI] [PubMed] [Google Scholar]
- 125.Lingbeek ME, Jacobs JJ, van Lohuizen M. The T-box repressors TBX2 and TBX3 specifically regulate the tumor suppressor gene p14 ARF via a variant T-site in the initiator. J Biol Chem. 2002;277:26120–7. doi: 10.1074/jbc.M200403200. [DOI] [PubMed] [Google Scholar]
- 126.Sugiyama T, Frazier DP, Taneja P, et al. Signal transduction involving the Dmp1 transcription factor and its alteration in human cancer. Clinical Medicine Oncology. 2008;2:209–19. doi: 10.4137/cmo.s548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Sinclair CS, Adem C, Naderi A, et al. TBX2 is preferentially amplified in BRCA1- and BRCA2-related breast tumors. Cancer Res. 2002;62:3587–91. [PubMed] [Google Scholar]
- 128.Davis E, Teng H, Bilican B, et al. Ectopic Tbx2 expression results in polyploidy and cisplatin resistance. Oncogene. 2008;27:976–84. doi: 10.1038/sj.onc.1210701. [DOI] [PubMed] [Google Scholar]
- 129.Sherr CJ. Cancer cell cycles. Science. 1996;274:1672–7. doi: 10.1126/science.274.5293.1672. [DOI] [PubMed] [Google Scholar]
- 130.Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–809. doi: 10.1038/sj.onc.1208619. [DOI] [PubMed] [Google Scholar]
- 131.Sutherland RL, Musgrove EA. Cyclins and breast cancer. J Mammary Gland Biol Neoplasia. 2004;9:95–104. doi: 10.1023/B:JOMG.0000023591.45568.77. [DOI] [PubMed] [Google Scholar]
- 132.Inoue K, Sherr CJ. Gene expression and cell cycle arrest mediated by transcription factor DMP1 is antagonized by D-type cyclins through a cyclin-dependent-kinase-independent mechanism. Mol Cell Biol. 1998;18:1590–600. doi: 10.1128/mcb.18.3.1590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Inoue K, Sherr CJ, Shapiro LH. Regulation of the CD13/aminopeptidase N gene by DMP1, a transcription factor antagonized by D-type cyclins. J Biol Chem. 1998;273:29188–94. doi: 10.1074/jbc.273.44.29188. [DOI] [PubMed] [Google Scholar]
- 134.Inoue K, Roussel MF, Sherr CJ. Induction of ARF tumor suppressor gene expression and cell cycle arrest by transcription factor DMP1. Proc Natl Acad Sci USA. 1999;96:3993–8. doi: 10.1073/pnas.96.7.3993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Inoue K, Wen R, Rehg JE, et al. Disruption of the ARF transcriptional activator DMP1 facilitates cell immortalization, Ras transformation, and tumorigenesis. Genes Dev. 2000;14:1797–809. [PMC free article] [PubMed] [Google Scholar]
- 136.Inoue K, Zindy F, Randle DH, Rehg JE, Sherr CJ. Dmp1 is haplo-insufficient for tumor suppression and modifies the frequencies of Arf and p53 mutations in Myc-induced lymphomas. Genes Dev. 2001;15:2934–9. doi: 10.1101/gad.929901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Mallakin A, Sugiyama T, Taneja P, et al. Mutually exclusive inactivation of DMP1 and ARF/p53 in lung cancer. Cancer Cell. 2007;12:381–94. doi: 10.1016/j.ccr.2007.08.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Inoue K, Mallakin A, Frazier DP.Dmp1 and tumor suppression Oncogene 2007264329–35.Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Arnold A, Papanikolau A. Cyclin D1 in breast pathogenesis. J Clin. Oncol. 2006;23:4215–24. doi: 10.1200/JCO.2005.05.064. [DOI] [PubMed] [Google Scholar]
- 140.Barbash O, Zamfirova P, Lin DI, et al. Mutations in Fbx4 inhibit dimerization of the SCF (Fbx4) ligase and contribute to cyclin D1 overexpression in human cancer. Cancer Cell. 2008;14:68–78. doi: 10.1016/j.ccr.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Gillett C, Smith P, Gregory W, et al. Cyclin D1 and prognosis in human breast cancer. Int J Cancer. 1996;69:92–9. doi: 10.1002/(SICI)1097-0215(19960422)69:2<92::AID-IJC4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- 142.McIntosh GG, Anderson JJ, Milton I, et al. Determination of the prognostic value of cyclin D1 overexpression in breast cancer. Oncogene. 1995;11:885–91. [PubMed] [Google Scholar]
- 143.Michalides R, Hageman P, van Tinteren H, et al. A clinicopathological study on overexpression of cyclin D1and of p53 in a series of 248 patients with operable breast cancer. Br J Cancer. 1996;73:728–34. doi: 10.1038/bjc.1996.128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Berns EM, Foekens JA, van Staveren IL, et al. Oncogene amplification and prognosis in breast cancer: Relationship with systemic treatment. Gene. 1995;159:11–8. doi: 10.1016/0378-1119(94)00534-y. [DOI] [PubMed] [Google Scholar]
- 145.Bieche I, Olivi M, Nogues C, Vidaud M, Lidereau R. Prognostic value of CCND1 gene status in sporadic breast tumours, as determined by real-time quantitative PCR assays. Br J Cancer. 2002;86:580–6. doi: 10.1038/sj.bjc.6600109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Seshadri R, Lee CS, Hui R, McCaul K, Horsfall DJ, Sutherland RL. Cyclin D1 application is not associated with reduced overall survival in primary breast cancer but may predict early relapse in patients with features of good prognosis. Clin Cancer Res. 1996;2:1177–84. [PubMed] [Google Scholar]
- 147.Hwang TS, Han HS, Hong YC, Lee HJ, Paik NS. Prognostic value of combined analysis of cyclin D1 and estrogen receptor status in breast cancer patients. Pathol Int. 2003;53:74–80. doi: 10.1046/j.1440-1827.2003.01441.x. [DOI] [PubMed] [Google Scholar]
- 148.Barnes DM, Gillett CE. Cyclin D1 in breast cancer. Breast Cancer Res. Treat. 1998;52:1–15. doi: 10.1023/a:1006103831990. [DOI] [PubMed] [Google Scholar]
- 149.Han S, Park K, Bae BN, et al. Cyclin D1 expression and patient outcome after tamoxifen therapy in estrogen receptor positive metastatic breast cancer. Oncol Rep. 2003a;10:141–4. [PubMed] [Google Scholar]
- 150.Kenny FS, Hui R, Musgrove EA, et al. Overexpression of cyclin D1 messenger RNA predicts for poor prognosis in estrogen receptor-positive breast cancer. Clin Cancer Res. 1999;5:2069–76. [PubMed] [Google Scholar]
- 151.Knudsen KE. The cyclin D1b splice variant: an old oncogene learns new tricks. Cell Div. 2006;1:15. doi: 10.1186/1747-1028-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Knudsen KE, Diehl JA, Haiman CA, Knudsen ES.Cyclin D1: polymorphism, aberrant splicing and cancer risk Oncogene 2006251620–8.Review. [DOI] [PubMed] [Google Scholar]
- 153.Millar EK, Dean JL, McNeil CM, et al. Cyclin D1b protein expression in breast cancer is independent of cyclin D1a and associated with poor disease outcome Oncogene 2009281812–20.Epub 2009 Mar 16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Buckley MF, Sweeney KJ, Hamilton JA, et al. Expression and amplification of cyclin genes in human breast cancer. Oncogene. 1993;8:2127–213. [PubMed] [Google Scholar]
- 155.Lukas J, Bartkova J, Welcker M, et al. Cyclin D2 is a moderately oscillating nucleoprotein required for G1 phase progression in specific cell types. Oncogene. 1995;10:2125–34. [PubMed] [Google Scholar]
- 156.Evron E, Umbricht CB, Korz D, et al. Loss of cyclin D2 expression in the majority of breast cancers is associated with promoter hypermethylation. Cancer Res. 2001;61:2782–7. [PubMed] [Google Scholar]
- 157.Kong G, Chua SS, Yijun Y, et al. Functional analysis of cyclin D2 and p27 Kip1 in cyclin D2 transgenic mouse mammary gland during development. Oncogene. 2002;21:7214–25. doi: 10.1038/sj.onc.1205895. [DOI] [PubMed] [Google Scholar]
- 158.Russell A, Thompson MA, Hendley J, Trute L, Armes J, Germain D. Cyclin D1 and D3 associate with the SCF complex and are coordinately elevated in breast cancer. Oncogene. 1999;18:1983–91. doi: 10.1038/sj.onc.1202511. [DOI] [PubMed] [Google Scholar]
- 159.Keyomarsi K, Tucker SL, Buchholz TA, et al. Cyclin E and survival in patients with breast cancer. N Engl J Med. 2002;347:1566–75. doi: 10.1056/NEJMoa021153. [DOI] [PubMed] [Google Scholar]
- 160.Donnellan R, Chetty R.Cyclin D1 and human neoplasia Mol Pathol 1998511–7.Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161.Akama Y, Yasui W, Yokozaki H, et al. Frequent amplification of the cyclin E gene in human gastric carcinomas. Jpn J Cancer Res. 1995;86:617–21. doi: 10.1111/j.1349-7006.1995.tb02442.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Demetrick DJ, Matsumoto S, Hannon GJ, et al. Chromosomal mapping of the genes for the human cell cycle proteins cyclin C (CCNC), cyclin E (CCNE), p21 (CDKN1) and KAP (CDKN3) Cytogenet Cell Genet. 1995;69:190–2. doi: 10.1159/000133960. [DOI] [PubMed] [Google Scholar]
- 163.Catzavelos C, Bhattacharya N, Ung YC, et al. Decreased levels of the cellcycle inhibitor p27 Kip1 protein: prognostic implications in primary breast cancer. Nat Med. 1997;3:227–30. doi: 10.1038/nm0297-227. [DOI] [PubMed] [Google Scholar]
- 164.Fredersdorf S, Burns J, Milne AM, et al. High level expression of p27(kip1) and cyclin D1 in some human breast cancer cells: inverse correlation between the expression of p27(kip1) and degree of malignancy in human breast and colorectal cancers. Proc Natl Acad Sci USA. 1997;94:6380–5. doi: 10.1073/pnas.94.12.6380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Keyomarsi K, Conte D, Jr, Toyofuku W, Fox MP. Deregulation of cyclin E in breast cancer. Oncogene. 1995;11:941–50. [PubMed] [Google Scholar]
- 166.Nielsen NH, Arnerlov C, Emdin SO, Landberg G. Cyclin E overexpression, a negative prognostic factor in breast cancer with strong correlation to oestrogen receptor status. Brit J Cancer. 1996;74:874–80. doi: 10.1038/bjc.1996.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Nielsen NH, Emdin SO, Cajander J, Landberg G. Deregulation of cyclin E and D1 in breast cancer is associated with inactivation of the retinoblastoma protein. Oncogene. 1997;14:295–304. doi: 10.1038/sj.onc.1200833. [DOI] [PubMed] [Google Scholar]
- 168.Porter PL, Malone KE, Heagerty PJ, et al. Expression of cell-cycle regulators p27 Kip1 and cyclin E, alone and in combination, correlate with survival in young breast cancer patients. Nat Med. 1997;3:222–5. doi: 10.1038/nm0297-222. [DOI] [PubMed] [Google Scholar]
- 169.Tan P, Cady B, Wanner M, et al. The cell cycle inhibitor p27 is an independent prognostic marker in small (T1 a, b) invasive breast carcinomas. Cancer Res. 1997;57:1259–63. [PubMed] [Google Scholar]
- 170.Carroll PE, Okuda M, Horn HF, et al. Centrosome hyperamplification in human cancer: Chromosome instability induced by p53 mutation and/or Mdm2 overexpression. Oncogene. 1999;18:1935–44. doi: 10.1038/sj.onc.1202515. [DOI] [PubMed] [Google Scholar]
- 171.Lingle WL, Lutz WH, Ingle JN, Maihle NJ, Salisbury JL. Centrosome hypertrophy in human breast tumors: Implications for genomic stability and cell polarity. Proc Natl Acad Sci USA. 1998;95:2950–5. doi: 10.1073/pnas.95.6.2950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Spruck CH, Won KA, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 173.Gudas JM, Payton M, Thukral S, et al. Cyclin E2, a novel G1 cyclin that binds Cdk2 and is aberrantly expressed in human cancers. Mol Cell Biol. 1999;19:612–22. doi: 10.1128/mcb.19.1.612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Lauper N, Beck AR, Cariou S, et al. Cyclin E2: a novel CDK2 partner in the late G1 and S phases of the mammalian cell cycle. Oncogene. 1998;17:2637–43. doi: 10.1038/sj.onc.1202477. [DOI] [PubMed] [Google Scholar]
- 175.Payton M, Coats S. Cyclin E2, the cycle continues. Int. J. Biochem. Cell Biol. 2002;34:315–20. doi: 10.1016/s1357-2725(01)00137-6. [DOI] [PubMed] [Google Scholar]
- 176.Payton M, Scully S, Chung G, Coats S. Deregulation of cyclin E2 expression and associated kinase activity in primary breast tumors. Oncogene. 2002;21:8529–34. doi: 10.1038/sj.onc.1206035. [DOI] [PubMed] [Google Scholar]
- 177.Zariwala M, Liu J, Xiong Y. Cyclin E2, a novel human G1 cyclin and activating partner of CDK2 and CDK3, is induced by viral oncoproteins. Oncogene. 1998;17:2787–98. doi: 10.1038/sj.onc.1202505. [DOI] [PubMed] [Google Scholar]
- 178.Lodén M, Stighall M, Nielsen NH, et al. The cyclin D1 high and cyclin E high subgroups of breast cancer: separate pathways in tumorogenesis based on pattern of genetic aberrations and inactivation of the pRb node. Oncogene. 2002;21:4680–90. doi: 10.1038/sj.onc.1205578. [DOI] [PubMed] [Google Scholar]
- 179.Wingate H, Bedrosian I, Akli S, Keyomarsi K. The low molecular weight (LMW) isoforms of cyclin E deregulate the cell cycle of mammary epithelial cells. Cell Cycle. 2003;2:461–6. [PubMed] [Google Scholar]
- 180.Donnellan R, Kleinschmidt I, Chetty R. Cyclin E immunoexpression in breast ductal carcinoma: pathologic correlations and prognostic implications. Hum Pathol. 2001;32:89–94. doi: 10.1053/hupa.2001.21141. [DOI] [PubMed] [Google Scholar]
- 181.Foulkes WD, Brunet JS, Stefansson IM, et al. The prognostic implication of the basal-like (cyclin E high/p27 low/p53+/glomeruloidmicrovascularproliferation+) phenotype of BRCA1-related breast cancer. Cancer Res. 2004;64:830–5. doi: 10.1158/0008-5472.can-03-2970. [DOI] [PubMed] [Google Scholar]
- 182.Han S, Park K, Bae BN, et al. Prognostic implication of cyclin E expression and its relationship with cyclin D1 and p27 Kip1 expression on tissue microarrays of node negative breast cancer. J Surg Oncol. 2003b;83:241–7. doi: 10.1002/jso.10268. [DOI] [PubMed] [Google Scholar]
- 183.Kim HK, Park IA, Heo DS, et al. Cyclin E overexpression as an independent risk factor of visceral relapse in breast cancer. Eur J Surg Oncol. 2001;27:464–71. doi: 10.1053/ejso.2001.1137. [DOI] [PubMed] [Google Scholar]
- 184.Kühling H, Alm P, Olsson H, et al. Expression of cyclins E, A, and B, and prognosis in lymph node-negative breast cancer. J Pathol. 2003;199:424–31. doi: 10.1002/path.1322. [DOI] [PubMed] [Google Scholar]
- 185.Lindahl T, Landberg G, Ahlgren J, et al. Overexpression of cyclin E protein is associated with specific mutation types in the p53 gene and poor survival in human breast cancer. Carcinogenesis. 2004;25:375–80. doi: 10.1093/carcin/bgh019. [DOI] [PubMed] [Google Scholar]
- 186.Rudolph P, Kühling H, Alm P, et al. Differential prognostic impact of the cyclins E and B in premenopausal and postmenopausal women with lymph node-negative breast cancer. Int J Cancer. 2003;105:674–80. doi: 10.1002/ijc.11132. [DOI] [PubMed] [Google Scholar]
- 187.Span PN, Tjan-Heijnen VC, Manders P, Beex LV, Sweep CG. Cyclin-E is a strong predictor of endocrine therapy failure in human breast cancer. Oncogene. 2003;22:4898–04. doi: 10.1038/sj.onc.1206818. [DOI] [PubMed] [Google Scholar]
- 188.Akli S, Van Pelt CS, Bui T, et al. Overexpression of the low molecular weight cyclin E in transgenic mice induces metastatic mammary carcinomas through the disruption of the ARF-p53 pathway. Cancer Res. 2007;67:7212–22. doi: 10.1158/0008-5472.CAN-07-0599. [DOI] [PubMed] [Google Scholar]
- 189.Taneja P, Frazier DP, Kendig RD, et al. MMTV mouse models and the diagnostic values of MMTV-like sequences in human breast cancer. Expert Rev Mol Diagn. 2009;9:423–40. doi: 10.1586/ERM.09.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 190.Allain DC, Sweet K, Agnese DM. Management options after prophylactic surgeries in women with BRCA mutations: a review. Cancer Control. 2007;14:330–7. doi: 10.1177/107327480701400403. [DOI] [PubMed] [Google Scholar]
- 191.Mor P, Oberle K.Ethical issues related to BRCA gene testing in orthodox Jewish women Nurs Ethics 200815512–22.Review. [DOI] [PubMed] [Google Scholar]
- 192.García MJ, Benítez J. The Fanconi anaemia/BRCA pathway and cancer susceptibility. Searching for new therapeutic targets. Clin Transl Oncol. 2008;10:78–84. doi: 10.1007/s12094-008-0160-6. [DOI] [PubMed] [Google Scholar]
- 193.Moller P, Borg A, Evans DG, et al. Survival in prospectively ascertained familial breast cancer: analysis of a series stratified by tumour characteristics, BRCA mutations and oophorectomy. Int J Cancer. 2002;101:555–9. doi: 10.1002/ijc.10641. [DOI] [PubMed] [Google Scholar]
- 194.Hedenfalk I, Ringner M, Ben-Dor A, et al. Molecular classification of familial non-BRCA1/BRCA2 breast cancer. Proc Natl Acad Sci USA. 2003;100:2532–7. doi: 10.1073/pnas.0533805100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Somasundaram K, Zhang H, Zeng YX, et al. Arrest of the cell cycle by the tumour-suppressor BRCA1 requires the CDK-inhibitor p21 WAF1/CiP1. Nature. 1997;389:187–90. doi: 10.1038/38291. [DOI] [PubMed] [Google Scholar]
- 196.Fan S, Wang J, Yuan R, et al. BRCA1 inhibition of estrogen receptor signaling in transfected cells. Science. 1999;284:1354–6. doi: 10.1126/science.284.5418.1354. [DOI] [PubMed] [Google Scholar]
- 197.Bordeleau L, Panchal S, Goodwin P. Prognosis of BRCA-associated breast cancer: a summary of evidence. Breast Cancer Res Treat. 2010;119:13–24. doi: 10.1007/s10549-009-0566-z. [DOI] [PubMed] [Google Scholar]
- 198.Chappuis PO, Kapusta L, Begin LR, et al. Germline BRCA1/2 mutations and p27(Kip1) protein levels independently predict outcome after breast cancer. J Clin Oncol. 2000;18:4045–52. doi: 10.1200/JCO.2000.18.24.4045. [DOI] [PubMed] [Google Scholar]
- 199.Foulkes WD, Chappuis PO, Wong N, et al. Primary node negative breast cancer in BRCA1 mutation carriers has a poor outcome. Ann Oncol. 2000;11:307–13. doi: 10.1023/a:1008340723974. [DOI] [PubMed] [Google Scholar]
- 200.Robson ME, Chappuis PO, Satagopan J, et al. A combined analysis of outcome following breast cancer: differences in survival based on BRCA1/BRCA2 mutation status and administration of adjuvant treatment. Breast Cancer Res. 2004;6:R8–R17. doi: 10.1186/bcr658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 201.Zakhartseva LM, Gorovenko NG, Podolskaya SV, et al. Breast cancer immunohistochemical features in young women with BRCA 1/2 mutations. Exp Oncol. 2009;31:174–8. [PubMed] [Google Scholar]
- 202.Graeser MK, Engel C, Rhiem K, et al. Contralateral breast cancer risk in BRCA1 and BRCA2 mutation carriers. J Clin Oncol. 2009;27:5887–92. doi: 10.1200/JCO.2008.19.9430. [DOI] [PubMed] [Google Scholar]
- 203.Ellis LM, Hicklin DJ. VEGF-targeted therapy: mechanism of anti-tumor activity. Nature Rev Cancer. 2008;8:579–91. doi: 10.1038/nrc2403. [DOI] [PubMed] [Google Scholar]
- 204.Hsu JY, Wakelee HA. Monoclonal antibodies targeting vascular endothelial growth factor: current status and future challenges in cancer therapy. Bio Drugs. 2009;23:289–304. doi: 10.2165/11317600-000000000-00000. [DOI] [PubMed] [Google Scholar]
- 205.Sakakibara S, Tosato G. Regulation of angiogenesis in malignancies associated with Epstein-Barr virus and Kaposi’s sarcoma-associated herpes virus. Future Microbiol. 2009;4:903–17. doi: 10.2217/fmb.09.49. [DOI] [PubMed] [Google Scholar]
- 206.Toomey DP, Murphy JF, Conlon KC. COX-2, VEGF and tumour angiogenesis. Surgeon. 2009;7:174–80. doi: 10.1016/s1479-666x(09)80042-5. [DOI] [PubMed] [Google Scholar]
- 207.Yoshiji H, Gomez DE, Shibuya M, Thorgeirsson UP. Expression of vascular endothelial growth factor, its receptor, and other angiogenic factors in human breast cancer. Cancer Res. 1996;56:2013–6. [PubMed] [Google Scholar]
- 208.Brown LF, Berse B, Jackman RW, et al. Expression of vascular permeability factor (vascular endothelial growth factor) and its receptors in breast cancer. Hum Pathol. 1995;26:86–91. doi: 10.1016/0046-8177(95)90119-1. [DOI] [PubMed] [Google Scholar]
- 209.Relf M, LeJeune S, Scott PA, et al. Expression of the angiogenic factors vascular endothelial cell growth factor, acidic and basic fibroblast growth factor, tumor growth factor beta-1, platelet-derived endothelial cell growth factor, placenta growth factor, and pleiotrophin in human primary breast cancer and its relation to angiogenesis. Cancer Res. 1997;57:963–9. [PubMed] [Google Scholar]
- 210.Toi M, Inada K, Suzuki H, Tominaga T. Tumor angiogenesis in breast cancer: itsimportance as a prognostic indicator and the association with vascular endothelial growth factor expression. Breast Cancer Res Treat. 1995;36:193–204. doi: 10.1007/BF00666040. [DOI] [PubMed] [Google Scholar]
- 211.Shivakumar S, Prabhakar BT, Jayashree K, Rajan MG, Salimath BP. Evaluation of serum vascular endothelial growth factor (VEGF) and microvessel density (MVD) as prognostic indicators in carcinoma breast. J Cancer Res Clin Oncol. 2009;135:627–36. doi: 10.1007/s00432-008-0497-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Linderholm BK, Hellborg H, Johansson U, et al. Significantly higher levels of vascular endothelial growth factor (VEGF) and shorter survival times for patients with primary operable triple-negative breast cancer. Ann Oncol. 2009;10:1639–46. doi: 10.1093/annonc/mdp062. [DOI] [PubMed] [Google Scholar]
- 213.Jacobs EJ, Feigelson HS, Bain EB, et al. Polymorphisms in the vascular endothelial growth factor gene and breast cancer in the Cancer Prevention Study II cohort. Breast Cancer Res. 2006;8:R22. doi: 10.1186/bcr1400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.van der Auwera I, van Laere SJ, van den Eynden GG, et al. Increased angiogenesis and lymphangiogenesis in inflammatory versus noninflammatory breast cancer by real-time reverse transcriptase-PCR gene expression quantification. Clin Cancer Res. 2004;10:7965–71. doi: 10.1158/1078-0432.CCR-04-0063. [DOI] [PubMed] [Google Scholar]
- 215.Yu Q, Geng Y, Sicinski P. Specific protection against breast cancers by cyclin D1 ablation. Nature. 2001;411:1017–21. doi: 10.1038/35082500. [DOI] [PubMed] [Google Scholar]
- 216.Inoue K, Sugiyama T, Taneja P, Morgan RL, Frazier DP.Emerging roles of DMP1 in lung cancer Cancer Res 2008684487–90.Review. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 217.Sreeramaneni R, Chaudhry A, McMahon M, Sherr CJ, Inoue K. Ras-Raf-Arf signaling critically depends on the Dmp1 transcription factor. Mol Cell Biol. 2005;25:220–32. doi: 10.1128/MCB.25.1.220-232.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]