Abstract
The ubiquitin ligase anaphase-promoting complex (APC/C) is essential for cell division in all eukaryotes. Loss of APC/C-activity arrests cells at metaphase and results in severe aberrations of the mitotic spindle, but how the APC/C regulates spindle formation is not understood. Here, we report that the APC/C promotes the ubiquitination and degradation of four proteins required for Ran-dependent spindle assembly: Bard1, Hmmr, HURP, and NuSAP. Among these substrates, HURP and NuSAP can be degraded during spindle formation, when the spindle checkpoint is active. Their degradation requires additional layers of regulation, and both SAFs are only degraded after being released from their inhibitor importin-β by RanGTP. Our findings reveal a tightly regulated mechanism, by which the APC/C and the GTPase Ran control the abundance of active spindle assembly factors to achieve the accurate formation of the mitotic spindle.
Introduction
Faithful chromosome segregation depends on the robust assembly of the mitotic spindle. In dividing cells of higher eukaryotes, the microtubules comprising the spindle originate from centrosomes and chromosomes. While centrosomes act as microtubule-organizing centers, from where microtubules grow until being captured by kinetochores (Kirschner and Mitchison, 1986), the chromosomes promote growth of microtubules by regulating a conserved GTPase, Ran (Clarke and Zhang, 2008; Kalab and Heald, 2008).
Similar to most GTPases, Ran is active when bound to GTP (RanGTP), but inactive when loaded with GDP (RanGDP). The charging of Ran with GTP requires the guanine-nucleotide exchange factor Rcc1, which accumulates on mitotic chromosomes (reviewed in Kalab and Heald, 2008). Conversely, cytoplasmic RanGAP and RanBP1 increase the GTPase-activity of Ran to produce RanGDP. The spatial separation of Ran GTP-binding and hydrolysis results in a gradient of RanGTP with its highest concentration around chromatin (Kalab et al., 2002, 2006; Li and Zheng, 2004; Caudron et al., 2005).
RanGTP exerts its function in spindle formation by triggering the dissociation of spindle assembly factors (SAFs) from nuclear transport receptors of the importin-β family (Gruss et al., 2001; Nachury et al., 2001; Wiese et al., 2001). The importins are inhibitors of Ran-dependent SAFs: for example, importin-α/β interferes with the capacity of Tpx2 to promote microtubule polymerization and Aurora A-activation (Gruss et al., 2001), and importin-β inhibits the activity of the SAFs HURP and NuSAP to nucleate and crosslink spindle microtubules (Ribbeck et al., 2007; Koffa et al., 2006; Sillje et al., 2006). The local, Ran-dependent release of SAFs from importins provides a molecular explanation for microtubule nucleation in the vicinity of mitotic chromosomes.
Although importins are highly abundant in human cells (Ribbeck et al., 1998), their regulatory capacity can be overwhelmed, and the concentration of SAFs during mitosis has to be tightly controlled. Increasing the levels of Ran-dependent SAFs leads to defective spindle formation and chromosome missegregation (Stewart and Fang, 2005; Li et al., 2007; Wong et al., 2008). As a consequence, the aberrant expression of Ran-pathway components, such as the tumor suppressor Brca1-Bard1 or the oncogene Hmmr, has been linked to the development of tumors with abnormal spindle structures (Joukov et al., 2006; Pujana et al., 2007). Surprisingly, except for Tpx2, mechanisms regulating the abundance of Ran-dependent SAFs during mitosis are not known.
Most eukaryotic cell cycle regulators are controlled by ubiquitin-dependent proteolysis, which depends on the recognition of substrates by E3 enzymes (Deshaies and Joazeiro, 2009; Wickliffe et al., 2009). Among the ~600 human E3s, the anaphase-promoting complex (APC/C) is an attractive candidate for regulating the turnover of SAFs. The APC/C and its physiological E2s UbcH10 and Ube2S are required for progression of cells through mitosis, when spindle formation takes place (Peters, 2006; Williamson et al., 2009). In addition, the APC/C localizes to the poles of a growing spindle (Tugendreich et al., 1995; Kraft et al., 2003), and depletion of APC/C-subunits, co-depletion of UbcH10 and Ube2S, or expression of APC/C-inhibitors results in spindle defects (Ban et al., 2007; Goshima et al., 2007; Somma et al., 2008; Williamson et al., 2009). Moreover, the APC/C triggers the degradation of Tpx2 after completion of spindle assembly (Stewart and Fang, 2005). However, whether the APC/C controls SAFs during spindle formation has not been determined, and its role in regulating spindle assembly is not well understood.
Here, we report that the APC/C is responsible for the degradation of four Ran-dependent SAFs: Bard1, Hmmr, HURP, and NuSAP. The proteolysis of HURP and NuSAP can occur during spindle formation and is regulated by a mechanism centered on their inhibitor importin-β and RanGTP. Our results suggest that spindle formation relies on the proper activation and correctly timed degradation of SAFs, which is brought about by a unique interplay between the GTPase Ran and the APC/C.
Results
Bard1, Hmmr, HURP, and NuSAP are substrates of the APC/C
To identify substrates of the APC/C required for spindle assembly, we performed an in vitro-expression cloning screen. We synthesized ~40 spindle-binding proteins by in vitro transcription/translation (IVT/T) and monitored their ubiquitination by the APC/C and its E2s UbcH10 and Ube2S (Williamson et al., 2009). To test for specificity, we ensured that the ubiquitination of potential substrates was blocked by the APC/C-inhibitor Emi1 or by a ubiquitin mutant lacking Lys11, as expected for human APC/C (Jin et al., 2008). Using this approach, we identified Bard1, Hmmr, HURP, and NuSAP as candidate substrates, which were modified with K11-linked ubiquitin chains in an APC/C-dependent manner (Fig. 1A-D). Bard1, a subunit of the Brca1-Bard1 tumor suppressor, acts in spindle pole formation (Joukov et al., 2006); Hmmr regulates the localization of Tpx2 at the spindle pole (Groen et al., 2004); and HURP and NuSAP nucleate and crosslink microtubules in the vicinity of chromatin (Raemaekers et al., 2003; Koffa et al., 2006; Sillje et al., 2006; Wong and Fang, 2006).
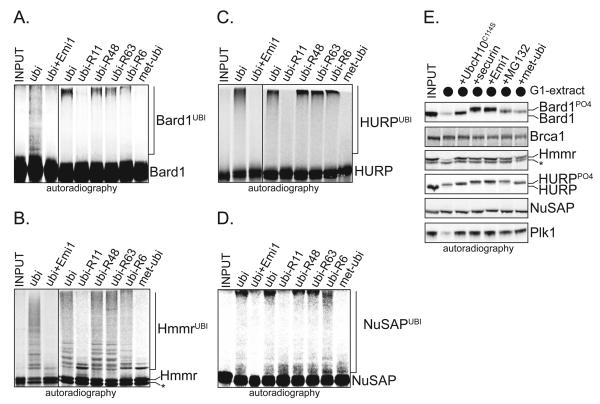
Figure 1. Bard1, Hmmr, HURP, and NuSAP are APC/C-substrates in vitro.
A. Bard1 is ubiquitinated by APC/C. 35S-Bard was synthesized by IVT/T and added to APC/CCdc20, UbcH10, Ube2S, and p31comet. As indicated, reactions were supplemented with Emi1 or ubiquitin mutants (ubi-R11: Lys11 of ubiquitin is changed to Arg). Reaction products were analyzed by autoradiography. B. Hmmr is ubiquitinated by APC/C. The ubiquitination of 35S-Hmmr by APC/CCdh1, UbcH10, and Ube2S, was analyzed as described above. The asterisk marks a truncation product of the Hmmr-IVT. C. HURP is ubiquitinated by APC/C. The ubiquitination of 35S-HURP by APC/CCdh1, UbcH10, and Ube2S, was analyzed as described above. D. NuSAP is ubiquitinated by APC/C. The ubiquitination of 35S-NuSAP by APC/CCdh1, UbcH10, and Ube2S, was analyzed as described above. E. Bard1 and Hmmr are degraded in an APC/C-dependent manner. The turnover of 35S-labeled proteins was analyzed in G1-extracts supplemented with UbcH10. UbcH10C114S (dominant-negative E2), securin (competitive inhibitor), Emi1 (APC/C-inhibitor), MG132 (proteasome inhibitor), and methylubiquitin (chain-formation inhibitor) were added as indicated. Reactions were incubated for 2h, and analyzed by autoradiography. In extracts, Bard1 and HURP were phosphorylated by cyclin-CDKs, when endogenous cyclin B1 was stabilized due to strong APC/C-inhibition. We ensured that CDK-dependent phosphorylation is not responsible for stabilization of Bard1 in the absence of APC/C (Supp. Fig. 1C). The asterisk marks a truncation product of the Hmmr-IVT/T.
To test whether these substrates are degraded in an APC/C-dependent manner, we monitored their stability in human extracts. Bard1 and Hmmr were turned over in G1-extracts with active APC/CCdh1, but not if APC/C was inhibited by a dominant-negative mutant of the APC/C-E2 UbcH10 (UbcH10C114S), the APC/C-inhibitor Emi1, an excess of a competing APC/C-substrate, or the proteasome inhibitor MG132 (Fig. 1E). Bard1 and Hmmr were also degraded in mitotic extracts with active APC/CCdc20, but both proteins were stable in S phase extracts or in extracts of asynchronous cells with inactive APC/C (Supp. Fig. 1A, B). Surprisingly, despite being strongly ubiquitinated by purified APC/C, HURP and NuSAP were only incompletely degraded in G1- or mitotic extracts with active APC/C, suggesting that their turnover is subject to additional layers of regulation, as discussed below (Fig. 1E; Supp. Fig. 1A).
To determine whether the APC/C is able to promote the degradation of these substrates in vivo, we overexpressed the APC/C-activators Cdc20 or Cdh1 in 293T cells. As previously observed for other APC/C-substrates, this treatment triggered the degradation of Bard1 (Fig. 2A), and further experiments implied that Bard1 was recognized by the APC/C when bound to its partner Brca1 (Supp. Fig. 2A). Brca1 was also degraded following APC/C-activation in cells. However, we did not observe Brca1- degradation in extracts (Fig. 1E) nor its ubiquitination by purified APC/C (data not shown), suggesting that it might not be a direct APC/C-substrate. In addition, increased levels of Cdh1 also resulted in the degradation of Hmmr, HURP, and NuSAP, which could be blocked by parallel expression of the APC/C-inhibitor Emi1 (Fig. 2B-D). Thus, the APC/C is able to trigger the proteolysis of Bard1, Hmmr, HURP, and NuSAP in cells.
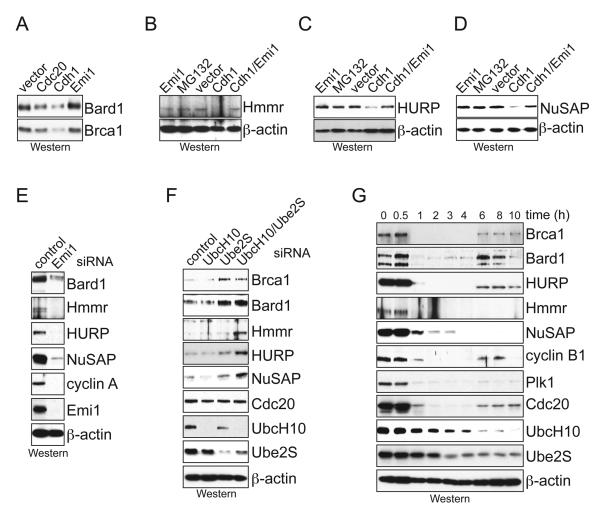
Figure 2. Bard1, Hmmr, HURP, and NuSAP are APC/C-substrates in vivo.
A. Bard1 is degraded by APC/C in vivo. 293T cells were transfected with Bard1, HABrca1 (amino acids 1-400), and either the APC/C-activators Cdc20/Cdh1 or the APC/C-inhibitor Emi1. The levels of Bard1 and Brca1 were determined by Western blot using αBard1- and αHA-antibodies. B. Hmmr is degraded by APC/C in vivo. Hmmr was transfected with Cdh1 and Emi1, as indicated, and levels of Hmmr were analyzed by Western blot using αHmmr-antibodies. C. HURP is degraded by APC/C in vivo. 293T cells were transfected with HURP, Cdh1, and Emi1, as indicated, and the levels of HURP were analyzed by Western blot using αHURP-antibodies. D. HANuSAP is degraded by the APC/C in vivo. NuSAP was transfected with Cdh1 and Emi1 as described above, and levels of NuSAP were analyzed by Western blot using αHA-antibodies. E. APC/C-activation depletes Bard1, Hmmr, HURP, and NuSAP from cells. Emi1 was depleted from HeLa cells using a characterized siRNA, and the abundance of indicated proteins was analyzed by Western blot. F. Bard1, Hmmr, HURP, and NuSAP-levels increase in cells upon APC/C-inhibition caused by depletion of UbcH10 and Ube2S. The levels of indicated proteins were analyzed by Western blot. G. Bard1, Hmmr, HURP, and NuSAP are degraded upon exit from mitosis. HeLa cells arrested in mitosis by thymidine/nocodazole were released into fresh medium. Samples were taken at different time points, and the levels of indicated proteins were determined by Western blot.
For the analysis of endogenous proteins, we altered the APC/C-activity in HeLa cells by depleting APC/C-regulators. To activate the APC/C, we decreased the levels of its inhibitor Emi1 using a characterized siRNA (Williamson et al., 2009). This treatment markedly reduced the abundance of Bard1, Hmmr, HURP, and NuSAP, as well as that of known APC/C-substrates (Fig. 2E). If the APC/C-E2s UbcH10 and Ube2S were depleted in conjunction with Emi1, no degradation was observed (Supp. Fig. 2B). Conversely, the inhibition of the APC/C by depletion of UbcH10 and Ube2S increased the levels of Bard1, Hmmr, HURP, and NuSAP (Fig. 2F), strongly suggesting that the endogenous SAFs are degraded by the APC/C.
We next tested whether the candidate substrates are regulated during cell cycle progression in parallel with known APC/C-substrates. Similar to most substrates of the APC/C, Bard1, Hmmr, HURP, and NuSAP accumulated in HeLa cells arrested in mitosis with nocodazole, and they were degraded upon exit from mitosis (Fig. 2G). All candidate substrates were also absent in quiescent cells with active APC/C, but they were co-expressed with APC/C-substrates upon cell cycle entry (Supp. Fig. 2C). Thus, Bard1, Hmmr, HURP, and NuSAP are co-regulated with known APC/C-substrates during cell cycle progression. Based on these experiments, we conclude that Bard1, Hmmr, HURP, and NuSAP, are substrates of the APC/C.
Identification of APC/C-recognition motifs
As a first step towards dissecting the mechanism underlying the degradation of these SAFs, we determined their APC/C-recognition motifs, such as D-, KEN-, or TEK-boxes (Peters, 2006; Jin et al., 2008). We first identified truncation mutants of the candidate substrates, which showed resistance against APC/C-dependent degradation in extracts (Supp. Fig. 3). Subsequently, we introduced point mutations into the SAF-domains required for degradation to disrupt D-, KEN-, and TEK-boxes, and then analyzed the turnover of the mutant SAFs in extracts and cells.
This analysis identified specific APC/C-recognition motifs, whose mutation led to stabilization of each substrate. We found two D-boxes in the N-terminus of Bard1 (Fig. 3A-C); D-, KEN-, and TEK-boxes in the C-terminus of Hmmr (Fig. 3D-F); D-, KEN-, and TEK-boxes in the N-terminus of HURP (Fig. 3G-I); and a D- and KEN-box in the C-terminus of NuSAP (Fig. 3J-M). For all SAFs, multiple APC/C-recognition motifs had to be mutated in combination to stabilize the protein in extracts and cells. The importance of degrons recognized by the APC/C further supports our conclusion that Bard1, Hmmr, HURP, and NuSAP are substrates of this ubiquitin ligase.
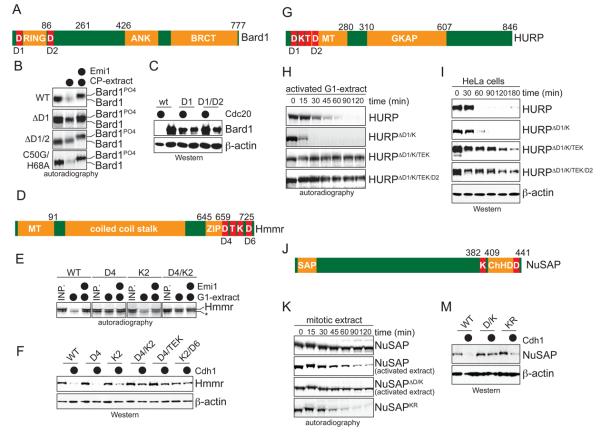
Figure 3. The SAF-degrons are adjacent to domains required for spindle assembly.
A. Schematic overview of Bard1 (RING: RING-domain; ANK: ankyrin repeats; BRCT: Brct-domain; D: D-box). B. Mutation of D-boxes stabilizes Bard1 in mitotic extracts. 35S-labeled mutants were tested for APC/C-dependent degradation in mitotic extracts with active APC/CCdc20. The C50G/H68A-mutant ablates the function of the RING-domain, but has no effect on APC/C-dependent degradation. C. Mutation of D-boxes stabilizes Bard1 in vivo. 293T cells were co-transfected with Bard1-mutants and Cdc20, and Bard1-levels were determined by Western blot. D. Schematic overview of Hmmr. (MT: microtubule-binding domain; ZIP: leucine zipper; T: TEK-box; K: KEN-box) E. Mutation of a D- and KEN-box stabilizes Hmmr in G1-extracts. The indicated 35S-labeled point mutants were tested for APC/C-dependent degradation in G1-extracts as described above. Reaction products were analyzed by autoradiography. F. Mutation of D-, KEN-, and TEK-boxes stabilize Hmmr in vivo. mycHmmr or the indicated mutants were co-expressed in 293T cells with Cdh1, and protein levels were analyzed by Western blot using αmyc-antibodies. G. Schematic overview of HURP. (MT: microtubule binding domain; GKAP: GKAP-domain). H. The degradation of HURP in G1-extracts depends on D-, KEN-, and TEK-boxes. To ensure degradation of wt-HURP, G1-extracts were treated with RanQ69L (see later). The indicated 35S-labeled mutants were incubated in G1-extracts before being analyzed by autoradiography. I. Degradation of HAHURP and its mutants in HeLa cells after release from nocodazole arrest. Samples were taken at the indicated times and analyzed for HURP-levels by Western blot. J. Schematic overview of NuSAP. The SAP- and ChHD-domains were defined by Rijmaekers et al., 2003. K. Degradation of NuSAP in mitotic extracts depends on D- and KEN-boxes. If indicated, the mitotic extract was supplemented with RanGTP. 35S-labeled mutants were incubated in the extracts for different times, before being analyzed by autoradiography. In NuSAPKR, a stretch of Lys residues has been changed to alanines, leading to its degradation in the absence of RanGTP. M. The degradation of NuSAP in cells depends on D- and KEN-boxes. The respective HANuSAP mutants (wt; DK: mutation of D- and KEN-box; KR: mutation of a Lys-rich cluster) were co-expressed with Cdh1 in 293T cells, as indicated. The levels of NuSAP and its mutants were detected by Western blot.
Interestingly, all APC/C-recognition motifs are in close proximity to SAF-domains required for spindle assembly. The D-boxes in Bard1 flank the RING-domain required for it to function at the spindle pole (Joukov et al., 2006); the degrons of Hmmr are at its C-terminus, which plays key roles during spindle pole maturation (Joukov et al., 2006); and the D-, KEN- and TEK-boxes of HURP and NuSAP are in their respective microtubule-binding domains (Raemaekers et al., 2003; Wong et al., 2008).
Importin-β and RanGTP regulate the ubiquitination and degradation of SAFs
The proximity of degrons to functionally important domains suggested that activity and stability of SAFs might be co-regulated. HURP and NuSAP, two Ran-dependent SAFs, are inhibited by importins and activated by RanGTP (Sillje et al., 2006; Koffa et al., 2006; Ribbeck et al., 2006). Strikingly, despite being efficiently ubiquitinated by purified APC/C, these two SAFs are stable in extracts, which contain high levels of importins. By contrast, Bard1 and Hmmr, which are not known to bind importins, are effectively degraded in these extracts. These observations suggested that importins and RanGTP might not only control the activation, but also the degradation of HURP and NuSAP.
To test this hypothesis, we added recombinant importins to the APC/C-dependent ubiquitination of HURP and NuSAP. Strikingly, importin-β, but not -α, blocked the ubiquitination of both SAFs in a dose dependent manner (Fig. 4A-C). The mutants importin-β3W and importin-β462, which bind substrates with low affinity (Moore et al., 1999; Cingolani et al., 2002), did not significantly impair HURP-ubiquitination (Fig. 4D). In contrast to the SAFs, importins did not affect the ubiquitination of Plk1 or cyclin B1 (Supp. Fig. 4A, B). When synthesized by IVT/T, HURP is bound to importin-β, which is abundant in reticulocyte lysate (Supp. Fig. 4C). RanGTP dissociated HURP from importin-β and promoted its APC/C-dependent ubiquitination (Fig. 4C; Supp. Fig. 4C). Moreover, RanGTP allowed the APC/C to ubiquitinate HURP and NuSAP in the presence of excess importin-β (Fig. 4E, G). RanGTP promoted the ubiquitination of HURP slightly more efficiently than its dissociation from importin-β, suggesting that ubiquitination might interfere with the rebinding of HURP to importin-β. Thus, importin-β and RanGTP are ableto regulate the APC/C-dependent ubiquitination of HURP and NuSAP.
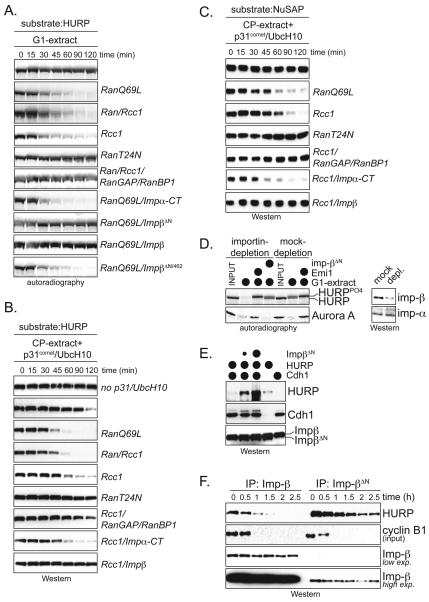
Figure 4. Importin-β and RanGTP regulate the ubiquitination of HURP and NuSAP.
A. Importin-β blocks the APC/C-dependent ubiquitination of HURP. 35S-HURP was incubated with APC/CCdh1, Ube2S, and UbcH10. As indicated, Emi1, importin-β, importin-α-CT, or importin-α/β were added. Reactions were analyzed by autoradiography. B. Importin-β inhibits HURP-ubiquitination in a dose-dependent manner. 35S-HURP was incubated with APC/CCdh1, increasing concentrations of RanQ69L, and importin-β, as indicated. C. Quantification of effects of RanGTP on HURP-degradation in mitotic extracts (red); HURP-ubiquitination by APC/CCdh1 (green) and HURP-dissociation from importin-β (blue). D. An intact cargo-binding domain is required for importin-β to act as APC/C-inhibitor. Importin-β and two mutants with reduced cargo affinity (importin-β3WA; importin-β462) were analyzed for their effect on ubiquitination of HURP by APC/C as described above. E. RanGTP allows HURP-ubiquitination in the presence of importin-β. 35S-HURP was incubated with APC/CCdh1, Ube2S, and UbcH10. Importin-β/α and GTP-bound RanQ69L were added as indicated. Reactions were analyzed by autoradiography. F. Importin-β blocks the APC/C-dependent ubiquitination of NuSAP. 35S-NuSAP was analyzed for APC/C-dependent ubiquitination as described above. G. RanGTP allows NuSAP-ubiquitination in the presence of importin-β. 35S-NuSAP was incubated with APC/CCdh1, Ube2S, and UbcH10. Importin-β/α and RanQ69L were added as indicated, and reactions were analyzed by autoradiography.
We next tested whether higher concentrations of RanGTP trigger the degradation of HURP and NuSAP in extracts, as suggested by the in vitro ubiquitination studies. We added constitutively GTP-bound RanQ69L, Ran and its GEF Rcc1, or Rcc1 (to activate endogenous Ran), to G1-extracts with APC/CCdh1 or to mitotic extracts with APC/CCdc20 and monitored the stability of HURP and NuSAP. Indeed, RanGTP strongly accelerated the degradation of both SAFs, whereas it had no effects on the degradation of other APC/C-substrates, such as cyclin B (Fig. 5A-C; Supp. Fig. 4D-G). RanGTP induced the degradation of HURP at concentrations that also promoted its ubiquitination by the APC/C and its dissociation from importin-β (Fig. 4C). RanT24N, which cannot be charged with GTP, was unable to promote SAF-degradation (Fig. 5A-C), and increasing the rate of GTP-hydrolysis by adding RanGAP/RanBP1 stabilized HURP and NuSAP even in the presence of Ran/Rcc1. Thus, RanGTP strongly promotes the degradation of HURP and NuSAP in extracts, consistent with importin-β stabilizing these SAFs.
Figure 5. Importins and RanGTP control the degradation of HURP and NuSAP.
A. Importin-β and RanGTP control the stability of HURP in G1-extracts. The degradation of 35S-HURP in G1-extracts was analyzed by autoradiography. When indicated, extracts were supplemented with GTP-bound RanQ69L; Ran and its GEF Rcc1; Rcc1 to activate endogenous Ran; nucleotide-free RanT24N; RanGAP/RanBP1 to generate RanGDP; importin-α-CT; importin-βΔN; importin-β; and the cargo-binding deficient importin-βΔ N/462. B. Importin-β and RanGTP control the stability of HURP in mitotic extracts. APC/CCdc20 was activated in mitotic extracts by UbcH10 and p31comet, and degradation of endogenous HURP was monitored by Western blot using αHURP-antibodies. Ran- or importin-proteins were added as described above. C. Importin-β and RanGTP control the stability of NuSAP in mitotic extracts. The degradation of endogenous NuSAP was monitored in mitotic extracts with active APC/C by Western blot using αNuSAP-antibodies. As indicated, Ran- or importin-proteins were added. D. Depletion of importin-β-like proteins promotes degradation of HURP in G1-extracts. Importin-β-like proteins were depleted from G1-extracts by a Ran-affinity column, as described (Ribbeck et al., 1998). The extent of depletion is shown by Western blot (right panel). 35S-HURP or 35S-Aurora A were added to importin- and mock-depleted extracts, and analyzed for APC/C-dependent degradation after 2h by autoradiography. Extracts were supplemented with importin-βΔN or Emi1, as indicated. E. Importin-β stabilizes HURP in vivo. The APC/C-dependent degradation of HURP was induced in 293T cells by Cdh1-expression. As indicated, increasing amounts of an importin-βΔN plasmid were co-transfected, which stabilized HURP in a dose-dependent manner. F. Importin-β stabilizes HURP upon exit from mitosis. U2OS cells stably expressing FLAGimportin-β (left) or FLAGimportin-βΔN (right) were released from a nocodazole-induced mitotic arrest into G1. As FLAG-tagged importins are expressed well below the level of the endogenous importin-β, we monitored the stability of HURP bound to affinity-purified FLAGimportins. HURP was detected by Western blot.
We addressed the role of importin-β in stabilizing SAFs by altering its abundance in extracts and cells. Consistent with the assays described above, HURP and NuSAP, but not cyclin B1, were stabilized in RanGTP-treated extracts by the Ran-insensitive importin-βΔN or by an excess of importin-β over RanGTP (Fig. 5A-C; Supp. Fig. 4F, G). By contrast, the SAFs were not stabilized by importin-α or by the importin-β462 mutant defective in cargo-binding (Fig. 5A-C). Moreover, if importin-β was depleted from G1-extracts, HURP was turned over in an APC/C-dependent manner even without additional RanGTP (Fig. 5D). The results from extracts were reproduced in 293T cells, where importin-βΔN protected HURP against APC/C-dependent degradation caused by Cdh1-overexpression (Fig. 5E). Binding to importin-βΔN also delayed the APC/C-dependent degradation of endogenous HURP upon exit of U2OS cells from mitosis (Fig. 5F). We conclude that RanGTP and importin-β control the ubiquitination and degradation of HURP and NuSAP.
Importin-β is a substrate-specific APC/C-inhibitor
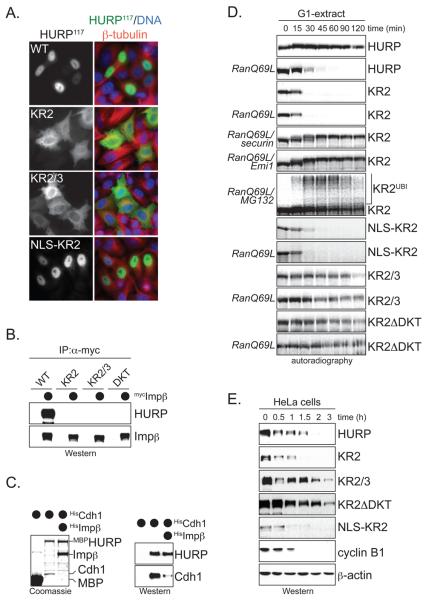
Importin-β controls the degradation of HURP and NuSAP, but not cyclin B1, and thus, likely regulates the SAFs rather than the APC/C. If this were the case, mutating the importin-binding sites of HURP should result in its APC/C-dependent degradation in the absence of RanGTP. Deletion analysis showed that the N-terminal 117 residues of HURP (HURP117) were sufficient to mediate importin-β-binding in vitro (Supp. Fig. 5A). The mutation of two stretches of basic amino acids (KR2, KR3) to alanine residues impaired the binding of HURP117 to importin-β, and the simultaneous mutation of both (KR2/3) abrogated this interaction (Supp. Fig. 5A). As expected for loss of importin-binding, mutation of KR2 or KR2/3 ablated the nuclear accumulation of HURP117 in HeLa cells (Fig. 6A). In 293T or HeLa cells, the mutation of KR2 or KR2/3 in full-length HURP dramatically reduced its binding to importin-β, but not to Tpx2 or Aurora A (Fig. 6B; Supp. Fig. 5B, C). Thus, KR2 and KR3 are crucial importin-β binding sites in HURP.
Figure 6. Importin-β stabilizes HURP by direct binding.
A. Mutation of KR2 or KR2/3 ablates the nuclear accumulation of HURP117. HeLa cells transfected with HAHURP117 or indicated mutants were analyzed by fluorescence microscopy using αHA-antibodies. In NLS-KR2, the SV40-NLS was fused to the N-terminus of HURP117-KR2. The right panel shows staining of HURP (green); tubulin (red); and DNA (DAPI; blue). B. Mutation of KR2 or KR2/3 abrogates binding of HURP to importin-β. HeLa cells were transfected with HAHURP, HAHURPKR2, HAHURPKR2/3, or HAHURPΔDKT (lacking D-, KEN-, and TEK-boxes), and mycimportin-β, and synchronized in mitosis with nocodazole. Importin-β was precipitated by αmyc-beads, and co-purifying HURP was detected by Western blot using αHA-antibodies. C. Importin-β and APC/C compete for access to HURP. MBP or MBPHURP were bound to amylose resin. As indicated, MBPHURP was pre-incubated with Hisimportin-β. HisCdh1 was added to all reactions, and the beads were analyzed for bound HisCdh1, using Coomassie (left) and Western blot (right). D. Mutation of its importin-β-binding site triggers APC/C-dependent degradation of HURP in the absence of RanGTP. The degradation of 35S-labeled HURP-mutants in G1-extracts was monitored by autoradiography. As indicated, RanQ69L, securin as competitive APC/C-inhibitor, Emi1, or MG132 were added. E. Degradation of importin-β-binding mutants of HURP in cells. HeLa cells expressing HAHURP or indicated mutants were released from a nocodazole-dependent mitotic arrest, and degradation of HURP was analyzed by Western blot.
KR2 and KR3 overlap with the APC/C-binding motifs in HURP (Supp Fig. 6A). Accordingly, the mutation of both KR2 and KR3 not only abrogated the association of HURP with importin-β, but also its APC/C-dependent ubiquitination and degradation (Supp. Fig. 6B-D). Similarly, loss of APC/C-recognition motifs (HURPΔDKT) interfered with HURP-binding to importin-β (Fig. 6B). These findings imply that importin-β and APC/C compete for access to HURP, and indeed, addition of importin-β blocked the interaction between HURP and the APC/C-substrate targeting subunit Cdh1 (Fig. 6C). This strongly suggests that importin-β stabilizes HURP by shielding its APC/C-binding sites.
Thus, loss of importin-binding should trigger HURP-degradation in the absence RanGTP. We were able to test this hypothesis, because mutation of KR2 strongly reduced the affinity of HURP to importin-β, but still allowed its ubiquitination by APC/C. Indeed, HURPKR2 was quickly degraded in G1-extracts without additional RanGTP (Fig. 6D), and importin-βΔN failed to stabilize HURPKR2 under these conditions (Supp. Fig. 6C). The degradation of HURPKR2 required APC/C and the proteasome, as HURPKR2 was stabilized by addition of APC/C- and proteasome-inhibitors, or by mutation of its APC/C-recognition sites (HURPKR2/3, HURPKR2ΔDKT; Fig. 6D). Similar degradation kinetics as in extracts were observed, when the stability of HURP-mutants was analyzed in cells (Fig. 6E). NuSAP appears to be regulated in a similar manner, as deletion of a Lys-rich region adjacent to its KEN-box led to NuSAP-degradation in the absence of RanGTP (Fig. 3K). Thus, importin-β stabilizes HURP, and most likely NuSAP, by direct binding to the SAFs, rather than by regulating the APC/C or the proteasome.
The above findings also implied that importin-β needs to bind to sites overlapping with the APC/C-recognition motifs of HURP to stabilize this SAF. In agreement with this observation, HURPKR2 was not stabilized in extracts or cells, if an importin-binding motif was fused to the N-terminus of HURPKR2 (NLS-KR2; Fig. 6D), even though this fusion rescued nuclear import of HURP117/KR2 in cells (Fig. 6A). Thus, importin-β acts as a substrate-specific APC/C-inhibitor, which masks the APC/C-recognition motifs of SAFs.
Importin-β stabilizes SAFs during mitosis
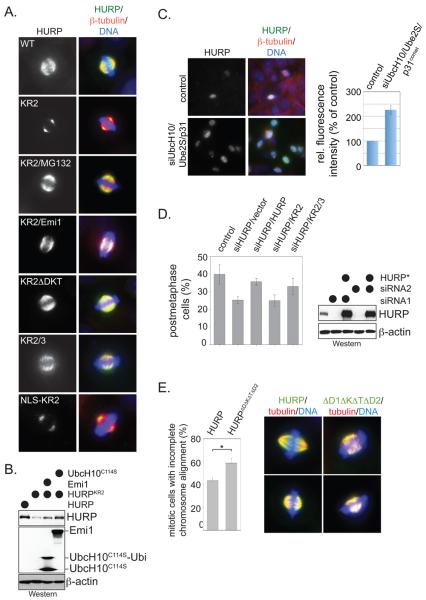
When in the cell cycle does importin-β regulate the stability of SAFs? To address this question, we compared the abundance of HURP to that of HURPKR2, which has lost regulation by importin-β, yet is still recognized by the APC/C. While HURP and HURPKR2 were expressed at similar levels in interphase HeLa cells (Supp. Fig. 7A, B), we found strong differences in their abundance during mitosis. Consistent with previous reports (Wong and Fang, 2006; Sillje et al., 2006), HURP accumulated in prometaphase cells on spindle microtubules (Fig. 7A). By contrast, only low amounts of the importin-binding deficient HURPKR2 were detected under these conditions, and in the majority of cells, HURPKR2 was absent from spindle microtubules. In addition, only low concentrations of HURPKR2 were observed in HeLa cells synchronized in prometaphase with nocodazole, as detected by Western blotting (Fig. 7B; Supp. Fig. 7B). HURPKR2 re-accumulated under these conditions, when cells were treated with the proteasome inhibitor MG132 (Fig. 7A; Supp. Fig. 7B). These findings suggest that importin-β is able to protect HURP from proteasomal degradation during early stages of mitosis.
Figure 7. Importin-β stabilizes SAFs during mitosis.
A. Stabilization by importin-β is required for accumulation of HURP on spindle microtubules. HAHURP and mutants were expressed in HeLa cells, and their intracellular location was determined in pre-anaphase cells by immunofluorescence using αHA-antibodies (green). The spindle was stained with antibodies against β-tubulin (red), DNA was detected by DAPI (blue). MG132 was added as indicated. B. APC/C-inhibition stabilizes HURPKR2 in prometaphase. HeLa cells expressing HAHURP or HAHURPKR2 were synchronized in prometaphase by nocodazole treatment. As indicated, UbcH10C114S or mycEmi1, which inhibit the APC/C, were co-expressed. The levels of HURP or HURPKR2 were determined by Western blot. C. Some endogenous HURP is degraded in an APC/C-dependent manner before metaphase. HeLa cells were treated with siRNA against UbcH10, Ube2S, and p31comet, which inhibits the APC/C and arrests cells in prometaphase. Endogenous HURP was detected by fluorescence microscopy using αHURP-antibodies (left panel). The fluorescence intensity in pre-anaphase cells was measured using ImageJ. The quantification of three independent experiments is shown on the right. D. Stabilization by importin-β is required for HURP-function in early mitosis. HURP was depleted from HeLa cells by siRNA, and cells were arrested in mitosis by nocodazole. As indicated, depleted HeLa cells were transfected with siRNA-resistant HURP mutants. 1h after release into new medium, the number of cells that had initiated anaphase was determined. The bottom panel shows HURP depletion by siRNA and rescue by siRNA-resistant HURP*. E. Degradation of HURP is important for spindle formation. HAHURP or HAHURPΔD1ΔKΔTΔD2 were expressed in HeLa cells and detected by immunofluorescence against HA (green). The percentage of HURP-positive pre-anaphase cells with incomplete chromosome alignment was determined 48h post transfection in three independent experiments (p-value < 0.05; left). Most spindles in cells expressing the stable HAHURPΔD1ΔKΔTΔD2 were defective, examples for which are shown on the right.
To test whether the APC/C is responsible for the reduced levels of HURPKR2, we mutated the APC/C-recognition motifs of HURP in addition to its KR2-site (HURPKR2ΔDKT; HURPKR2/3). Both mutants were stabilized in comparison to HURPKR2 and detected at high levels on spindle microtubules (Fig. 7A). Accordingly, HURPKR2/3 was abundant in lysates of synchronized HeLa cells, as measured by Western blotting (Supp. Fig. 7B). In addition, HURPKR2 was stabilized during prometaphase by co-expression of Emi1 or a dominant-negative version of the APC/C-specific E2 UbcH10 (UbcH10C114S; Fig. 7B). Confirming our mechanistic analysis in extracts, the fusion of an importin-binding site to the N-terminus of HURPKR2 did not inhibit its degradation (Fig. 7A); this also shows that the premature degradation of HURPKR2 in cells did not result from nuclear exclusion prior to mitosis. Our findings therefore provide evidence that importin-β protects HURP from APC/C-dependent degradation during early mitosis.
Consistent with these results, endogenous HURP and NuSAP bind importin-β in U2-OS cells synchronized in prometaphase (Supp. Fig. 7C, D). Moreover, immobilized importin-βΔN efficiently captured HURP and NuSAP from lysates of prometaphase cells, only if these lysates were pretreated with RanGTP to dissociate endogenous SAF-importin complexes, suggesting that most of HURP and NuSAP is bound by importins at this time of mitosis (Supp. Fig. 7E). However, as RanGTP dissociates HURP and NuSAP from importin-β in the vicinity of chromatin, a fraction of the endogenous SAFs should be degraded in an APC/C-dependent manner. To test this assumption, we inhibited the APC/C by depleting UbcH10, Ube2S, and the APC/C-activator p31comet, as reported previously (Williamson et al., 2009), and then analyzed the abundance of endogenous, spindle-bound HURP in pre-anaphase cells by fluorescence microscopy. Importantly, the levels of HURP in pre-anaphase cells strongly increased upon APC/C-inhibition, which was accompanied by spindle defects (Fig. 7C). To corroborate these findings, we synchronized HeLa cells in prometaphase using nocodazole or taxol and inhibited the proteasome with MG132 (Supp. Fig. 7F). In taxol-treated cells, HURP and NuSAP accumulated upon addition of MG132. HURP and NuSAP were more stable in nocodazole-treated cells, as seen before, suggesting that microtubules might regulate the degradation of SAFs. Together, these findings show that importin-β is able to protect HURP and NuSAP from APC/C-dependent degradation during prometaphase, at the same time when it regulates the activation of both SAFs.
The regulated degradation of SAFs is important for mitosis
We finally determined whether the regulated degradation of SAFs is important for cell cycle progression. As loss of importin-β binding leads to the degradation of HURP before anaphase, the mutation of the KR2 sites in HURP should interfere with its role in spindle formation. We depleted HURP in prometaphase cells with spindles disassembled with nocodazole, released the cells into fresh medium to allow spindle formation, and then counted the number of cells entering anaphase as a measure of successful spindle formation. Confirming earlier reports (Wong and Fang, 2006), the loss of HURP by siRNAs delayed anaphase entry, which could be rescued by expression of siRNA-resistant HURP (Fig. 7D). By contrast, HURPKR2, which does not bind importin-β yet is recognized by the APC/C, did not rescue the phenotypes caused by depletion of endogenous HURP, suggesting that the stabilization of HURP by importin-β is required for efficient spindle formation.
To determine the importance of the SAF-degradation, we analyzed spindle structures in cells expressing HURP or NuSAP at higher levels. The expression of the stable HURPΔD1ΔKΔTEKΔD2 increased the number of prometaphase cells with incomplete chromosome alignment, suggesting that stabilization of HURP delayed the establishment of a metaphase plate (Fig. 7E). In addition, almost all spindles in cells expressing HURPΔD1ΔKΔTEKΔD2 were highly aberrant. We often observed short spindles with broadened spindle poles and highly unstructured spindle microtubules, which is reminiscent of spindle aberrations observed upon overexpression of the APC/C-inhibitor Emi1 (Ban et al., 2007). These effects were more pronounced with NuSAP, where overexpression of the wild-type protein was sufficient to induce strong aberrations in the structure of the mitotic spindle (Supp. Fig. 7G). We conclude that the regulated degradation of HURP and NuSAP is important for cells to achieve accurate spindle assembly.
Discussion
Our search for novel APC/C-substrates was motivated by the aberrant spindle assembly in the absence of APC/C and the sparse information on SAFs known to be regulated by this machine. Here, we report the identification of four APC/C-substrates with roles in regulating spindle formation: Bard1, Hmmr, HURP, and NuSAP. Among these substrates, HURP and NuSAP can be degraded during spindle assembly, although the spindle checkpoint is active. Consequently, the degradation of these SAFs requires additional layers of regulation, and they are only degraded after being released from importin-β by RanGTP. If this unique mechanism of regulation is lost, spindle formation is impaired. Our findings reveal a unique mode of regulation for APC/C-dependent ubiquitination and provide a molecular basis for the role of APC/C in spindle formation.
The APC/C controls Ran-dependent spindle assembly
All APC/C-substrates identified in this study have functions in Ran-dependent spindle assembly. Bard1 and Hmmr localize to spindle poles, and loss of their activity results in unfocused poles or multipolar spindles (Groen et al., 2004; Maxwell et al., 2005; Joukov et al., 2006). HURP and NuSAP, which are enriched on kinetochore fibers, promote the nucleation and crosslinking of spindle microtubules (Sillje et al., 2006; Wang et al., 2006; Ribbeck et al., 2006). Aberrant levels of any of these SAFs cause spindle defects, which demonstrates that regulating their abundance is important for proper spindle formation (Fig. 7E; Supp. Fig. 7G; Raemaekers et al., 2003; Tsou et al., 2003; Maxwell et al., 2005; Sillje et al., 2006; Wong and Fang, 2006; Joukov et al., 2006; Li et al., 2007). As similar spindle abnormalities are observed upon depletion of APC/C-subunits (Goshima et al., 2007; Somma et al., 2008), co-depletion of UbcH10 and Ube2S (Williamson et al., 2009), or overexpression of APC/C-inhibitors (Ban et al., 2007), the stabilization of these components of the Ran-pathway likely contributes to the spindle defects caused by APC/C-inhibition.
What is the reason for the APC/C regulating so many substrates within the same pathway? One explanation might be found in the observation that many components of Ran-dependent spindle assembly function in dynamic protein interaction networks. For example, Bard1 binds Hmmr and Tpx2 to keep the activity of Hmmr in spindle assembly in check, and HURP, Tpx2, and Aurora A were suggested to be part of a complex in X. laevis extracts (Joukov et al., 2006; Pujana et al., 2007; Koffa et al., 2006). It is possible that degradation of a single complex subunit could disturb the balance between distinct activities. Consistent with this hypothesis, depleting either Brca1/Bard1 or Hmmr results in centrosome amplification, while co-depleting these proteins has less dramatic effects (Pujana et al., 2007). Targeting multiple SAFs could also allow the APC/C to regulate spindle assembly more robustly than it would by degrading a single protein, and spindle formation might be less sensitive to the aberrant expression of a single SAF.
The APC/C thus targets active HURP and NuSAP for degradation during spindle assembly, and it removes Tpx2, Bard1, Hmmr, and the remaining HURP and NuSAP, from cells once spindle formation has been completed, thereby resetting this pathway for a new round of cell division. We conclude that the APC/C is an important regulator of Ran-dependent spindle formation.
Regulation of APC/C-dependent degradation by importin-β and RanGTP
Our findings also reveal a reciprocal function of Ran in regulating APC/C-dependent degradation events. Similar to its role in spindle formation, RanGTP controls the turnover of SAFs by counteracting importin-β. As we have shown in detail for HURP, importin-β competitively inhibits the APC/C-recognition of HURP by directly binding to its degrons, and our experiments with NuSAP suggest that it is regulated in a similar manner. By contrast, importin-β interacts with, but does not stabilize cyclin B1, and accordingly, the binding sites for importin-β and APC/C localize to different regions of cyclin B1 (Hagting et al., 1999). We have tested multiple APC/C-substrates without functions in spindle assembly, but none was stabilized by importin-β (data not shown). Our results, therefore, identify importin-β as a substrate-specific inhibitor and RanGTP as a substrate-specific activator of the APC/C.
Importin-β and RanGTP likely regulate SAF-degradation prior to anaphase, when spindle formation takes place. HURPKR2, which does not associate with importin-β yet is recognized by the APC/C, is degraded despite incomplete spindle assembly. As a consequence, the expression of HURPKR2 in prometaphase cells failed to rescue the delay in anaphase entry caused by lack of endogenous HURP. Consistent with these observations, most of HURP and NuSAP appear to be bound by importins during prometaphase. To act in spindle formation, HURP and NuSAP have to be dissociated from importin-β by RanGTP (Sillje et al., 2006; Ribbeck et al., 2006), which should expose their degrons. Accordingly, we observed that some endogenous HURP was degraded in an APC/C-dependent manner during prometaphase. Thus, activation and degradation of HURP can occur during prometaphase, and both events are under control of importin-β and RanGTP.
The tight relationship between the RanGTP-dependent activation and degradation of SAFs is reminiscent of transcription factors, which are often simultaneously activated and marked for degradation by ubiquitin (Lipford and Deshaies, 2003). Similar to the temporal delay between the ubiquitin-dependent activation of transcription factors and their degradation, the proteolysis of HURP and NuSAP has to wait until the SAFs have fulfilled their role in spindle formation, or otherwise the cycle of activation and degradation would be futile. We speculate that several factors might contribute to the proper timing of SAF-degradation during mitosis. It is possible that in the presence of an active spindle checkpoint, the APC/C ubiquitinates HURP and NuSAP with slow kinetics. Spindle-bound deubiquitinating enzymes might also delay the degradation of SAFs, as suggested for APC/C-regulation after DNA damage (Bassermann et al., 2008). However, we favor the hypothesis that the degradation of HURP and NuSAP during prometaphase requires microtubules or an unknown microtubule-dependent activity. As both HURP and NuSAP promote microtubule nucleation (Koffa et al., 2006; Ribbeck et al., 2006), their microtubule-dependent degradation would ensure that the SAFs had been sufficiently active before being turned over. Experiments are underway to test this hypothesis.
Supplementary Material
Acknowledgements
We are very grateful to Rebecca Heald, Karsten Weis, and Petr Kalab for reagents, invaluable advice, and critically reading the manuscript. We thank Julia Schaletzky for her insight throughout the project and for carefully reading the manuscript; Michelle Yasukawa and Ann Fischer for tissue culture support; and members of the Drubin and Barnes laboratories for help with fluorescence microscopy. We thank all members of the Rape lab for many discussions, suggestions, and encouragement. This work was funded by a grant from the NIH MR (RO1 GM083064) and an NIH Director New Innovator Award to MR. MR is a Pew fellow.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Methods
A detailed description of the used constructs, siRNAs, antibodies, and procedures can be found in the Supplementary Information.
References
- Ban KH, et al. The END network couples spindle pole assembly to inhibition of the anaphase-promoting complex/cyclosome in early mitosis. Dev. Cell. 2007;13:29–42. doi: 10.1016/j.devcel.2007.04.017. [DOI] [PubMed] [Google Scholar]
- Bassermann F, Frescas D, Guardavaccaro D, Busino L, Peschiaroli A, Pagano M. The Cdc14B-Cdh1-Plk1 axis controls the G2 DNA-damage-response checkpoint. Cell. 2008;134:256–67. doi: 10.1016/j.cell.2008.05.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caudron M, Bunt G, Bastiaens P, Karsenti E. Spatial coordination of spindle assembly by chromosome-mediated signaling gradients. Science. 2005;309:1373–1376. doi: 10.1126/science.1115964. [DOI] [PubMed] [Google Scholar]
- Clarke PR, Zhang C. Spatial and temporal coordination of mitosis by Ran GTPase. Nat. Rev. Mol. Cell Biol. 2008;9:464–477. doi: 10.1038/nrm2410. [DOI] [PubMed] [Google Scholar]
- Cingolani G, Bednenko J, Gillespie MT, Gerace L. Molecular basis for the recognition of a nonclassical nuclear localization signal by importin beta. Mol Cell. 2002;10:1345–53. doi: 10.1016/s1097-2765(02)00727-x. [DOI] [PubMed] [Google Scholar]
- Deshaies RJ, Joazeiro CA. RING-domain E3 ubiquitin ligases. Annu Rev. Biochem. 2009:399–434. doi: 10.1146/annurev.biochem.78.101807.093809. [DOI] [PubMed] [Google Scholar]
- Goshima G, Wollman R, Goodwin SS, Zhang N, Scholey JM, Vale RD, Stuurman N. Genes required for mitotic spindle assembly in Drosophila S2 cells. Science. 2007;316:417–421. doi: 10.1126/science.1141314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Groen AC, Cameron LA, Coughlin M, Miyamoto DT, Mitchison TJ, Ohi R. XRHAMM functions in ran-dependent microtubule nucleation and pole formation during anastral spindle assembly. Curr. Biol. 2004;14:1801–1811. doi: 10.1016/j.cub.2004.10.002. [DOI] [PubMed] [Google Scholar]
- Gruss OJ, et al. Ran induces spindle assembly by reversing the inhibitory effect of importin alpha on TPX2 activity. Cell. 2001;104:83–93. doi: 10.1016/s0092-8674(01)00193-3. [DOI] [PubMed] [Google Scholar]
- Hagting A, Jackman M, Simpson K, Pines J. Translocation of cyclin B1 to the nucleus at prophase requires a phosphorylation-dependent nuclear import signal. Curr. Biol. 1999;9:680–9. doi: 10.1016/s0960-9822(99)80308-x. [DOI] [PubMed] [Google Scholar]
- Jin L, Williamson A, Banerjee S, Phillip I, Rape M. Mechanism of ubiquitin chain formation by the human Anaphase-Promoting Complex. Cell. 2008;133:653–665. doi: 10.1016/j.cell.2008.04.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joukov V, et al. The BRCA1/BARD1 heterodimer modulates ran-dependent mitotic spindle assembly. Cell. 2006;127:539–552. doi: 10.1016/j.cell.2006.08.053. [DOI] [PubMed] [Google Scholar]
- Kalab P, Weis K, Heald R. Visualization of a Ran-GTP gradient in interphase and mitotic Xenopus egg extracts. Science. 2002;295:2452–2456. doi: 10.1126/science.1068798. [DOI] [PubMed] [Google Scholar]
- Kaláb P, Pralle A, Isacoff EY, Heald R, Weis K. Analysis of a RanGTP-regulated gradient in mitotic somatic cells. Nature. 2006;440:697–701. doi: 10.1038/nature04589. [DOI] [PubMed] [Google Scholar]
- Kalab P, Heald R. The RanGTP gradient - a GPS for the mitotic spindle. J. Cell Sci. 2008;121:1577–1586. doi: 10.1242/jcs.005959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirschner MW, Mitchison T. Microtubule dynamics. Nature. 1986;324:621. doi: 10.1038/324621a0. [DOI] [PubMed] [Google Scholar]
- Koffa MD, Casanova CM, Santarella R, Kocher T, Wilm M, Mattaj IW. HURP is part of a Ran-dependent complex involved in spindle formation. Curr. Biol. 2006;16:743–754. doi: 10.1016/j.cub.2006.03.056. [DOI] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. EMBO J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Zhou Y, Sun L, Xing G, Tian C, Sun J, Zhang L, He F. NuSAP is degraded by APC/C-Cdh1 and its overexpression results in mitotic arrest dependent of its microtubules' affinity. Cell Signal. 2007;19:2046–55. doi: 10.1016/j.cellsig.2007.05.017. [DOI] [PubMed] [Google Scholar]
- Lipford JR, Deshaies RJ. Diverse roles for ubiquitin-dependent proteolysis in transcriptional activation. Nat. Cell Biol. 2003;5:845–50. doi: 10.1038/ncb1003-845. [DOI] [PubMed] [Google Scholar]
- Maxwell CA, Keats JJ, Belch AR, Pilarski LM, Reiman T. Receptor for hyaluronan-mediated motility correlates with centrosome abnormalities in multiple myeloma and maintains mitotic integrity. Cancer Res. 2005;65:850–860. [PubMed] [Google Scholar]
- Moore JD, Yang J, Truant R, Kornbluth S. Nuclear import of Cdk/cyclin complexes: identification of distinct mechanisms for import of Cdk2/cyclin E and Cdc2/cyclin B1. J Cell Biol. 1999;144:213–24. doi: 10.1083/jcb.144.2.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nachury MV, Maresca TJ, Salmon WC, Waterman-Storer CM, Heald R, Weis K. Importin beta is a mitotic target of the small GTPase Ran in spindle assembly. Cell. 2001;104:95–106. doi: 10.1016/s0092-8674(01)00194-5. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat. Rev. Mol. Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pujana MA, et al. Network modeling links breast cancer susceptibility and centrosome dysfunction. Nat. Genet. 2007;39:1338–1349. doi: 10.1038/ng.2007.2. [DOI] [PubMed] [Google Scholar]
- Raemaekers T, et al. NuSAP, a novel microtubule-associated protein involved in mitotic spindle organization. J. Cell Biol. 2003;162:1017–1029. doi: 10.1083/jcb.200302129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reddy SK, Rape M, Margansky WA, Kirschner MW. Ubiquitination by the anaphase-promoting complex drives spindle checkpoint inactivation. Nature. 2007;446:921–925. doi: 10.1038/nature05734. [DOI] [PubMed] [Google Scholar]
- Ribbeck K, Lipowsky G, Kent HM, Stewart M, Görlich D. NTF2 mediates nuclear import of Ran. EMBO J. 1998;17:6587–6598. doi: 10.1093/emboj/17.22.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, et al. NuSAP, a mitotic RanGTP target that stabilizes and cross-links microtubules. Mol. Biol. Cell. 2006;17:2646–2660. doi: 10.1091/mbc.E05-12-1178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribbeck K, Raemaekers T, Carmeliet G, Mattaj IW. A role for NuSAP in linking microtubules to mitotic chromosomes. Curr. Biol. 2007;17:230–236. doi: 10.1016/j.cub.2006.11.050. [DOI] [PubMed] [Google Scholar]
- Sillje HHW, Nagel S, Korner R, Nigg EA. HURP is a Ran-importin β-regulated protein that stabilizes kinetochore microtubules in the vicinity of chromosomes. Curr. Biol. 2006;16:731–742. doi: 10.1016/j.cub.2006.02.070. [DOI] [PubMed] [Google Scholar]
- Somma MP, et al. Identification of Drosophila mitotic genes by combining co-expression analysis and RNA interference. PLoS Genet. 2008;4:e1000126. doi: 10.1371/journal.pgen.1000126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stewart S, Fang G. Anaphase-promoting complex/cyclosome controls the stability of TPX2 during mitotic exit. Mol. Cell Biol. 2005;25:10516–10527. doi: 10.1128/MCB.25.23.10516-10527.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsou AP, et al. Identification of a novel cell cycle regulated gene, HURP, overexpressed in human hepatocellular carcinoma. Oncogene. 2003;22:298–307. doi: 10.1038/sj.onc.1206129. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Wickliffe K, Williamson A, Jin L, Rape M. The multiple layers of ubiquitin-dependent cell cycle control. Chem. Rev. 2009;109:1537–48. doi: 10.1021/cr800414e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiese C, Wilde A, Moore MS, Adam SA, Merdes A, Zheng Y. Role of importin-beta in coupling Ran to downstream targets in microtubule assembly. Science. 2001;291:653–656. doi: 10.1126/science.1057661. [DOI] [PubMed] [Google Scholar]
- Williamson A, Wickliffe KE, Mellone BG, Song L, Karpen GH, Rape M. Identification of a physiological E2 module for the human anaphase-promoting complex. Proc. Natl. Acad. Sci. USA. 2009;106:18213–8. doi: 10.1073/pnas.0907887106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Fang G. HURP controls spindle dynamics to promote proper interkinetochore tension and efficient kinetochore capture. J. Cell Biol. 2006;173:879–891. doi: 10.1083/jcb.200511132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J, Lerrigo R, Jang CY, Fang G. Aurora A regulates the activity of HURP by controlling the accessibility of its microtubule-binding domain. Mol. Biol. Cell. 2008;19:2083–91. doi: 10.1091/mbc.E07-10-1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.