Abstract
Lipases are acyl hydrolases that represent a diverse group of enzymes present in organisms ranging from prokaryotes to humans. This article focuses on an evolutionarily related family of extracellular lipases that include lipoprotein lipase, hepatic lipase and endothelial lipase. As newly synthesized proteins, these lipases undergo a series of co- and post-translational maturation steps occurring in the endoplasmic reticulum, including glycosylation and glycan processing, and protein folding and subunit assembly. This article identifies and discusses mechanisms that direct early and late events in lipase folding and assembly. Lipase maturation employs the two general chaperone systems operating in the endoplasmic reticulum, as well as a recently identified lipase-specific chaperone termed lipase maturation factor 1. We propose that the two general chaperone systems act in a coordinated manner early in lipase maturation in order to help create partially folded monomers; lipase maturation factor 1 then facilitates final monomer folding and subunit assembly into fully functional homodimers. Once maturation is complete, the lipases exit the endoplasmic reticulum and are secreted to extracellular sites, where they carry out a number of functions related to lipoprotein and lipid metabolism.
Keywords: chaperone, endoplasmic reticulum, endothelial lipase, glycosylation, hepatic lipase, lipase maturation factor 1, lipoprotein lipase, protein folding
Lipase function & molecular structure
This article focuses on the maturation (i.e., folding and assembly) of a family of extracellular lipases that play essential roles in circulating lipid metabolism. The family is comprised of evolutionarily conserved members that include pancreatic lipase (PL; gene name PNLIP), lipoprotein lipase (LPL; gene name LPL), hepatic lipase (HL; gene name LIPC) and endothelial lipase (EL; gene name LIPG). All are secreted N-linked glycoproteins that hydrolyze two principal lipid substrates, triglycerides (TGs) and phospholipids, but with widely varying efficiencies [1]. PL and LPL are largely TG lipases, while HL can hydrolyze both substrates and EL is principally a phospholipase. Unlike many other lipases, these lipases cleave their substrates at the sn-1 position, releasing fatty acids and monoglycerides or lysophospholipids [1,2]. While this article mainly focuses on mechanisms of lipase maturation, lipase folding and assembly require an appreciation of lipase function and molecular structure, which will be briefly summarized, along with the role of these lipases in disease. For a more comprehensive discussion of these topics, a number of excellent reviews are available [2–18].
Pancreatic lipase functions in the absorption of dietary fat; it is secreted by pancreatic acinar cells, acting in the intestinal lumen to hydrolyze bile-emulsified TGs. The released fatty acids and monoglycerides are taken up by intestinal enterocytes, and then re-esterified into TGs and packaged, along with vitamin and cholesteryl esters, into the hydrophobic core of chylomicrons. During fasting, the liver synthesizes a similar TG-rich lipoprotein termed VLDL [19]. In the circulation, TGs sequestered in the core of chylomicrons and VLDL are inaccessible to tissues except through the action of LPL. This lipase is synthesized by a variety of cell types including adipocytes and myocytes from skeletal muscle and heart, respectively. Once secreted, LPL is transported to the lumenal face of capillaries, where it is bound to the surface of the endothelium by heparin sulfate proteoglycans and the recently identified GPIHBP1 [20] – a glycosylphosphatidylinositol-anchored protein that acts as a platform for LPL lipolysis of TG-rich lipoproteins. At this site, TGs are hydrolyzed by LPL into its more polar products (i.e., free fatty acids and monoglycerides), which are then accessible to subjacent tissues through receptors such as CD36 [6]. Once lipolyzed by LPL, chylomicrons and VLDL are released from the capillary beds of peripheral tissues and circulate to liver sinusoids where they become further hydrolyzed by HL [11]. The result is the creation of lipoprotein remnants with much of their TGs removed – a form that is then taken up by the liver via receptor-mediated endocytosis. This process is also assisted by LPL and HL; in a noncatalytic fashion, these lipases act as ligands that facilitate uptake of remnant particles by the LDL-receptor related protein [3,9,11].
Along with their roles in the metabolism of TG-rich lipoproteins, the lipases also have important functions in the formation and metabolism of HDL. For example, the hydrolysis of TG-rich lipoproteins by LPL causes the core to shrink, leaving redundant surface lipids which, along with ApoA1, break off and form nascent HDL [3,4]. In addition, HL and EL have important roles in HDL remodeling and uptake that are essential to the process of reverse cholesterol transport [11,14]. Indeed, all the lipases emerge as important genetic determinants of circulating lipid levels in human populations and are thought to contribute to a number of pathogenic states including atherosclerosis and inflammation, obesity and insulin secretion and sensitivity (discussed later) [11,21–27]. However, regardless of the role, lipases can function only when secreted, a process that first requires their maturation in the endoplasmic reticulum (ER).
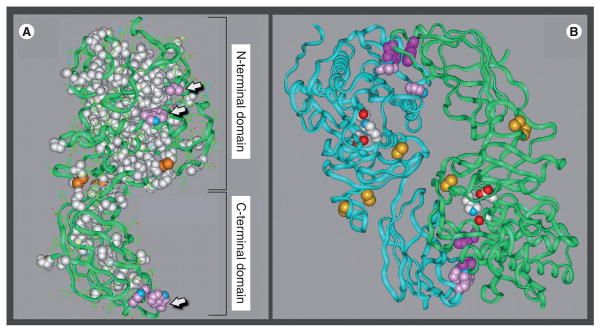
Lipase maturation involves the folding of newly synthesized polypeptides into fully functional enzymes, a process requiring attainment of a full 3D conformation. While only PL has a solved crystal structure [28], domain-exchange experiments indicate that all family members share folding domains with structures very similar to PL [17]. Indeed, the crystal structure of PL has been used to model LPL [8], EL [29] and HL (Figure 1A). All members of the lipase family share a basic tertiary structure comprised of two folding domains, dividing the lipase protein into a large N-terminal domain and smaller C-terminal domain separated by a short ‘hinge’ region (Figure 1A). The primary function of the N-terminal domain is catalysis, while the C-terminal domain provides ancillary functions aiding catalysis by facilitating the binding of lipid substrates and enzyme cofactors, while also providing binding sites for GPIHBP1 and heparan sulfate proteoglycans [2].
Figure 1. Structural model of hepatic lipase.
(A) The hepatic lipase (HL) monomer is modeled after the crystal structure of pancreatic lipase (PL). Model building and short molecular dynamics runs were performed on a Silicon Graphics Indigo 2 computer with Insight II and Discover 2005 software (Accelrys Inc., CA, USA). The side chains of HL were mapped on to the backbone according to a multiple sequence alignment for HL and PL generated by using the MULTALIN multiple alignment algorithm. The location of the two major folding domains of the HL model is indicated by brackets. Amino acids that are conserved between HL and PL and are depicted in a space-filling form as white; the green ribbons are comprised of residues unique to HL. Also shown are conserved disulfides (orange); arrows point to side chains of surface-located hydrophobic residues (pink). (B) The noncovalent HL homodimer, with the two individual monomers is depicted as ribbons colored blue and green. The side chains of active-site residues are colored red, white and blue; conserved disulfides are highlighted orange; and the side chains of hydrophobic residues comprising putative dimer interaction sites are shown in pink and purple.
Models are courtesy of Keith Munson and Howard Wong.
While the C-terminal domain is comprised of a series of β-sheets that form a sandwich-like fold [8], the N-terminal domain features the active site cleft covered by a lid domain. Upon association with the lipid substrate, the lid opens to provide access to the Ser/Asp/His catalytic triad (Figure 1B, colored red, white and blue). The active site cleft is comprised of secondary structures termed the α/β hydrolase fold, a key feature of esterases, lipases and thioesterases [30,31]. It is comprised of a fan of internal β-sheets surrounded by α-helices, and comprised of amino acids that are conserved between all members of the lipase family (Figure 1A, white). Notably, the residues unique to HL are found to be located on the surface (Figure 1A, green ribbons), including hydrophobic residues at both N- and C-terminal domains that are seemingly in contact with the aqueous phase (Figure 1A, arrows). While such an arrangement would be thermodynamically unstable, these surface hydrophobic residues are shielded when two monomers assemble into a homodimer, and appear to form intermolecular contact points that may hold the monomers together in a head-to-tail arrangement (Figure 1B).
Notably, among the lipase family members, only PL functions as a monomer [2,32]; LPL, HL and EL are active only as noncovalent homodimers with monomers arranged in a head-to-tail orientation [29,33,34]. As discussed later, we believe that this difference in subunit structure marks a fundamental divergence in the maturation pathway between PL and the rest of the lipase family. Specifically, LPL, HL and EL probably share a common pathway that is tailored to the unique characteristics of monomer structure (e.g., surface hydrophobic residues) required for successful homodimer assembly. In fact, unlike PL, the folding and assembly of all three lipase homodimers require a specialized maturation factor: lipase maturation factor 1 (LMF1; see later). Moreover, the evolutionary relationships between the lipase family members suggest that PL diverged from an ancestral lipase prior to HL, LPL and EL, which share a more recent root [35]; like PL, the ancient bacterial and fungal lipases also function as monomers [2]. Indeed, it is tempting to speculate that a specialized maturation pathway co-evolved with this more recent branch of the lipase gene family to facilitate attainment of its unique structural requirements.
Lipases in disease
Lipase maturation, along with other transcriptional, post-transcriptional and post-translational mechanisms [3,4,9], regulates the expression and secretion of lipase activity, and thus contributes to pathophysiological conditions that have been attributed to lipase deficiency, overexpression and altered function. For example, naturally occurring mutations in mice and humans that affect LPL [36,37], LMF1 [38] or the extracellular lipolytic platform GPIHBP1 [39], result in LPL deficiency, which in turn causes massive hypertriglyceridemia (chylomicronemia) and low HDL levels. Although such mutations are rare, it is now clear from genome-wide association studies that variation in LPL is among the strongest genetic determinants of plasma TG and HDL-C levels in normal human populations [40–43]. Thus, genetic variation of both cis- and trans-acting factors affecting LPL activity levels, including lipase maturation, can elicit profound changes in plasma lipid levels that are known to contribute to atherosclerosis [44], pancreatitis [45] and metabolic syndrome [46,47].
In addition to LPL deficiency, genetically engineered mouse models have also shed light on the pathophysiological consequences of LPL overexpression in various tissues [48]. Transgenic mice expressing elevated LPL activity in skeletal muscle [49] and liver [50] demonstrate that TG oversupply to these tissues – with concomitant decreased delivery of circulating TGs to adipose tissue – causes insulin resistance [27,51,52] and resistance to diet-induced obesity [53]. Consistent with these mouse models, genetic variation in the human LPL gene has been associated with insulin resistance [54,55], as well as obesity [56,57], in some human populations. LPL overexpression in the heart causes cardiomyopathy [58] owing, at least in part, to the accumulation of toxic lipid intermediates [59], which raises the possibility of a potential role for LPL in cardiac lipid accumulation and dilated cardiomyopathy associated with obesity and diabetes [60–62]. In mouse models, LPL deficiency and overexpression, specifically in macrophages, is associated with respective protection against, or acceleration of, atherosclerosis [63,64], raising the possibility of an involvement of LPL in cardiovascular disease beyond its effects on plasma lipid levels. Indeed, a recent genetic analysis demonstrated association between a polymorphism of the LPL gene and myocardial infarction [65].
Similar to LPL, rare forms of HL deficiency [66,67] as well as commonly occurring variation in HL [40–43], have been linked to altered plasma TG and cholesterol levels. As HDL appears to be the primary lipoprotein affected by HL, the role of this enzyme in atherosclerosis and coronary artery disease has been intensively investigated (for a review, see [11]). These studies suggest both pro- and antiatherogenic properties of HL, and indicate complex, context-dependent involvement of HL in cardiovascular disease [13]. EL is also emerging as an important factor in the control of lipoprotein metabolism and inflammation in humans [23,24,68–70]. EL has recently been identified as an important determinant of plasma HDL-C levels [40–43], reflecting its primary role in HDL catabolism. Taken together, it is clear that LPL, HL and EL contribute significantly to circulating lipid levels and related metabolic traits; moreover, factors such as lipase maturation are now emerging as important determinants in their expression.
Lipase glycosylation & processing in the endoplasmic reticulum
The majority of newly synthesized proteins in the ER transverse the secretory pathway with at least one glycan chain covalently attached to an asparagine residue in the consensus sequence Asn-X-Ser/Thr (NXS/T), where X is any amino acid except proline [71–73]. The lipases are no exception; members of the lipase gene family contain at least one N-linked glycan chain in each folding domain. Glycosylation has multiple roles: site-specific glycosylation decreases regional hydrophobicity, increasing solubility and influencing local folding behavior; glycan chains also act as ligands, guiding proteins to appropriate chaperones in the ER. Such ligands are uncovered when glycan chains are processed through the specific removal of glucose and mannose residues by ER-specific glycosidases. For example, glucose cleavage controls binding and release from lectin chaperones (see later), while mannose trimming limits the time allotted for protein folding before initiating ER-associated degradation (ERAD) [71,72,74].
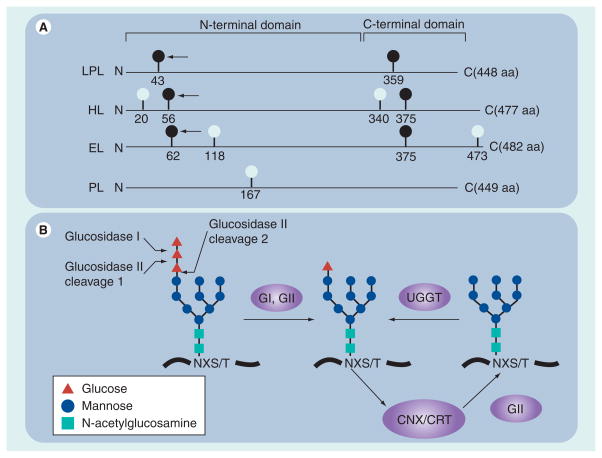
The importance of site-specific glycosylation in lipase maturation can be appreciated by the conservation of NXS/T sites residing in both N- and C-terminal folding domains of LPL, HL and EL (Figure 2A; dark circles). By contrast, PL is the only lipase family member lacking such conserved sites (Figure 2A), suggesting that its maturation may diverge in some aspects from other members of the lipase family. In fact, PL folding is much more efficient than HL and LPL [75], and its maturation proceeds without the apparent need for robust interaction with ER-folding factors [76]. Unlike PL, the maturation of LPL, HL and EL requires folding and assembly into homodimers, which increases their structural complexity. Maturation of these lipases proceed efficiently only if nascent chains engage with specific folding factors whose interactions are controlled in part by site-specific glycosylation of the N-terminal domain. In particular, N-linked glycosylation at positions 43, 56 and 62 of LPL, HL and EL (Figure 2A, arrows) is critical for efficient maturation. Studies have demonstrated that removal of these conserved sites by site-directed mutagenesis results in severe maturation defects, as evidenced by ER retention of lipase proteins and, at least for LPL and HL, the abolishment of enzyme activity [77–79]. These critical glycosylation sites are adjacent to a pair of cysteine residues forming the first disulfide bridge, and near the lid domain that shields hydrophobic residues within the active site cleft [8,18]. Moreover, in vitro LPL refolding experiments indicate that establishing native tertiary structure to the N-terminal domain is the rate-limiting step in the formation of the homodimer [80]. Thus, folding of the N-terminal domain requires the intramolecular assembly of key peptide surfaces whose efficiency can be greatly increased if chaperones are present in order to limit nonproductive interactions (see later).
Figure 2. Glycosylation sites and processing events that are important in lipase maturation.
(A) Glycan attachment sites (consensus sequence NXS/T) among members of the human lipase gene family are depicted by light and dark balls. Dark balls represent evolutionarily conserved sites. The arrow points to sites of glycosylation essential in lipase maturation. Also shown are the relative positions of both N- and C-terminal folding domains. (B) Cleavage sites for GI and GII on the unprocessed high mannose chain; this chain is added to NXS/T sites shortly after they emerge from the ribosome during translation elongation. GI and GII cleavages occur rapidly and result in a processed monoglucosylated chain that can bind to CNX or CRT cotranslationally; release occurs after the second cleavage by GII. If the nascent protein becomes misfolded, a single glucose can be added to the unglucosylated chain by the ER lumenal protein, UGGT 1; thus, the lipase can reattach to CNX/CRT in a process termed chaperone cycling.
aa: Amino acid; CNX: Calnexin; CRT: Calreticulin; EL: Endothelial lipase; ER: Endoplasmic reticulum; G: Glucosidase; HL: Hepatic lipase; LPL: Lipoprotein lipase; PL: Pancreatic lipase; UGGT: UDP-glucose: glycoprotein glucosyltransferase.
Along with the addition of glycan chains to conserved sites in the N-terminal domain, their initial processing by ER glucosidases is critical for efficient lipase maturation. Glycosylation occurs by transferring a preformed ‘high-mannose’ glycan chain from dolichol phosphate to the NXS/T sequence of a growing polypeptide chain by the oligosaccharyl transferase complex in the ER [72]. As shown in Figure 2B, the carbohydrate composition of this preformed glycan chain is predominated by nine mannose residues (circles) topped by three glucose residues (triangles). After transfer to the growing polypeptide chain, glucosidases I (GI) and II (GII) sequentially remove the glucose residues (Figure 2B). Removal of the two outermost glucose residues results in a monoglucosylated high-mannose chain that binds with high affinity to the ER chaperones calnexin or calreticulin (CNX/CRT). Cleavage of the last glucose residue by GII releases the glycan chain from CNX/CRT; however, it can rebind after addition of a glucose by the ER lumenal protein UDP-glucose: glycoprotein glucosyltransferase 1 (UGGT; Figure 2B). Thus, the innermost glucose residue acts as a ligand on N-linked glycoproteins that promotes its engagement with CNX/CRT; its release and reattachment is controlled by GII and UGGT, respectively. Using such a mechanism, nascent N-linked glycoproteins can undergo repeated rounds of chaperone binding (also known as cycling) in order to increase folding efficiency. As expected, inhibition of CNX/CRT binding by inhibiting ER glucosidases results in deficient lipase maturation, as evidenced by loss of lipolytic activity and lipase aggregation [75,81–83]. These results underscore the importance of lectin chaperones in lipase folding (see later), and explain why specific glycan attachment sites have remained conserved during the evolution of the lipase gene family.
Lipase maturation factors
Chaperone and folding factors aid in the maturation of most proteins, from the time they first emerge from the Sec61 translocon (i.e., cotranslocationally) to the end of their folding and assembly cycle in the ER lumen (i.e., post-translationally). These factors also provide surveillance of maturation, known as ER quality control [72,84–87], ensuring that immature proteins remain in the ER until their folding is complete. Moreover, they direct terminally misfolded proteins to degradative pathways, and provide safeguards against massive protein misfolding by regulating the ER stress response [88]. As most proteins utilize the functions provided by these ER factors during the course of their maturation, they are considered ‘general’ maturation factors. However, some proteins also engage with specialized proteins when folding or assembly requires the attainment of unique structural characteristics; these are often known as ‘client-specific’ factors. Indeed, lipase maturation requires both.
A recent proteomics study found that HL maturation uses general maturation factors originating from both major chaperone systems operating in the ER: the CNX/CRT and binding protein (BiP)/Grp94 systems [76]. Both systems are comprised of component factors that form large, multiprotein complexes in the ER [89]. Table 1 lists some of the component factors that were found to associate with HL during its sojourn in the ER. In particular, HL associates with the lectin chaperone CNX, as would be expected from the maturation defects encountered when glucosidase inhibitors prevent binding of HL and LPL to CNX (discussed earlier); other studies using co-immunoprecipitation have also detected HL–CNX association [75,90]. Moreover, the maturation efficiency of transfected human LPL in insect cells is greatly increased by cotransfection with mammalian CNX/CRT [91]. Along with CNX, components of the CNX/CRT system were also identified as being associated with HL during its maturation [76], including UGGT, GII and ERp57 (Table 1). UGGT and GII provide repeated access of HL to CNX (discussed earlier), while ERp57 functions in the formation (oxidation), elimination (reduction) and transfer (isomerization) of disulfide bonds among cysteine residues of the CNX-bound nascent polypeptide.
Table 1.
General factors in lipase maturation.
| Chaperone system | Component | Location | Functions in lipase maturation |
|---|---|---|---|
| CNX/CRT | CNX | ER membrane | Stabilize unfolded and misfolded forms; provide platform for folding factors. |
| UGGT, GII | ER lumen | Chaperone cycling, providing repeated access of HL to CNX. | |
| ERp57 | ER lumen | Oxidation, reduction and isomerization of disulfide bonds. | |
| BiP/Grp94 | BiP, Grp94 | ER lumen | Stabilize unfolded and misfolded forms; entry into the ERAD pathway. |
| PDI | ER lumen | Oxidation, reduction and isomerization of disulfide bonds. | |
| PPIase | ER lumen | Isomerization of peptidyl–prolyl bonds. |
BiP: 78-kDa binding protein; CNX: Calnexin; CRT: Calreticulin; ER: Endoplasmic reticulum; ERAD: Endoplasmic reticulum-associated degradation; ERp57: 57-kDa endoplasmic reticulum protein; GII: Glucosidase II; Grp94: 94-kDa glucose-regulated protein; HL: Hepatic lipase; PDI: Protein disulfide isomerase; PPIase: Peptidyl-prolyl cis–trans isomerase; UGGT: UDP-glucose: glycoprotein glucosyltransferase 1.
Along with the CNX/CRT system, HL also associates with BiP/Grp94 (Table 1). Unlike CNX, BiP (also termed the 78-kDa glucose-regulated protein; Grp78) binds directly to the polypeptide backbone of nascent and misfolded proteins, a process independent of N-linked glycosylation [92–94]. BiP has an affinity for extended hydrophobic domains encountered in unfolded or misfolded proteins, with a preference for alternating aromatic and hydrophobic amino acids [95], estimated to occur on average every 36 residues [72]. Thus, it is not surprising that most proteins bind transiently with BiP during their sojourn in the ER. Indeed, BiP was found to co-isolate with every member of the lipase gene family (including PL) during a proteomics screen [Doolittle M, Unpublished Data, 76]. Along with BiP, components of the BiP/Grp94 system were also found to associate with HL (Table 1), including the 94 kDa glucose-regulated protein Grp94, protein disulfide isomerase and peptidyl-prolyl cis-trans isomerase (PPIase) [76]. PPIase converts cis peptidyl–prolyl bonds into the favored trans configuration comprising the majority of peptide linkages in native proteins; notably, such isomerization is a rate-limiting step in the in vitro refolding of chemically denatured LPL [80]. UGGT is also found to be associated with BiP [89], as with HL [76]; thus, while serving a function within the CNX/CRT chaperone cycle (Figure 2B), UGGT is also a component of the BiP/Grp94 chaperone system. Besides its role in protein maturation, BiP has additional functions, including ER quality control, regulation of the ER-stress response, and targeting terminally misfolded proteins to ERAD [86,92–94].
Besides general factors, lipase maturation also requires a client-specific chaperone, which has recently been identified as the protein affected by the combined lipase deficiency (cld) mutation. Mice homozygous for cld exhibit massive hypertriglyceridemia immediately after birth [38,96], and die shortly after owing to complications arising from the absence of LPL activity [97]; HL activity is also diminished [38,96,98]. The lack of lipase activity is not due to decreased levels of lipase mRNA or protein; rather, newly synthesized LPL remains inactive and is retained in the ER [83,99–101]. In fact, the inactive LPL protein expressed in cld/cld cells is highly aggregated, resembling misfolded LPL resulting from lec23 [83] – a mutation affecting glucosidase I activity and thus preventing proteins from entering into the CNX cycle (Figure 2B). Nevertheless, the cld mutation is not genetically localized to lipase structural genes, lectin chaperones or any other general factors discussed above [102]. Moreover, the effects of cld appear to be limited to lipase maturation, as no other affected nonlipase proteins have been identified. We have recently shown that, along with LPL and HL, EL maturation is also defective in cld/cld cells [Peterfy M, Unpublished Data], increasing the ‘combined lipase deficiency’ phenotype to include all members of the lipase gene family that are known to form homodimers. Conspicuously, the maturation of PL, which is functional as a monomer, is unaffected by the mutation [38,99]. By screening genes within the critical chromosomal region of cld, only one candidate gene, a hypothetical transmembrane protein (Tmem112), was found to rescue the lipase maturation defect occurring in cld/cld cells. The gene was renamed ‘lipase maturation factor’ to reflect its function in mice [38]. Loss-of-function mutations (Y439X and W464X) in the human LMF1 ortholog also causes combined lipase deficiency [38,103], verifying its function in humans as well.
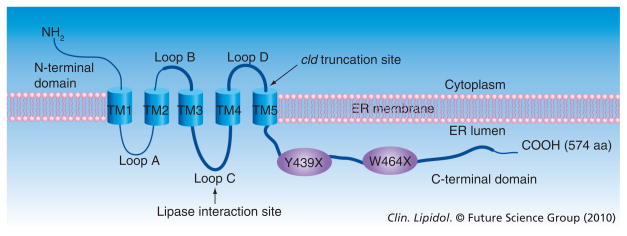
As demonstrated by immunofluorescence microscopy [38], LMF1 colocalizes with CNX in the ER membrane. Figure 3 illustrates the topology of this client-specific LMF, which is a polytopic, multipass membrane protein localized exclusively to the ER [104]. LMF1 contains a large evolutionarily conserved domain of unknown function, referred to as DUF1222 in the Pfam database (Figure 3, thick line) [105]. This domain is found in a number of hypothetical proteins from bacteria to humans, with LMF1 being the first member of the DUF family with a known function. LMF1 has five α-helical transmembrane domains dividing the protein into three domains facing the cytoplasm, and three others oriented toward the ER lumen (Figure 3). The cld mutation causes truncation of the large C-terminal domain (Figure 3, arrow), as do Y439X and W464X nonsense mutations (Figure 3, ovals), suggesting that this domain is essential in carrying out the function of lipase maturation. Moreover, we have recently identified loop C as the site where both LPL and HL physically bind to LMF1 (Figure 3, arrow); notably, PL associates very poorly to this site. Thus, two domains important in carrying out the function of lipase maturation, loop C and the C-terminal domain, reside within DUF1222; both face the ER lumen where they are in proximity to nascent lipase polypeptides.
Figure 3. Structure of the recently identified lipase maturation factor 1.
The five α-helical transmembrane segments divide the protein into six separate domains. The approximate size of each domain is 49 aa, N-terminal domain; 56 aa, loops A, B; 71 aa, loop C; 46 aa, loop D; and 188 aa, C-terminal domain. The evolutionarily conserved domain of unknown function (DUF1222) is depicted as a thick line. Arrows point to the cld-induced truncation site and the site of lipase interaction. Y439X and W464X are nonsense mutations causing combined lipase deficiency in human patients. aa: Amino acid; ER: Endoplasmic reticulum; TM: Transmembrane.
Mechanisms of lipase maturation
While the key factors in lipase maturation are now fairly well established (see earlier), the mechanisms involved in the process have yet to be fully elucidated. Nevertheless, as shown in Figure 4, a hypothetical model can be envisioned based on evidence gleaned from in vivo studies detailing the maturation of well-studied protein substrates, such as viral and some host proteins [72,106,107], and from in vitro studies monitoring the structure of folding intermediates occurring during the refolding of chemically denatured LPL [80,108,109]. In particular, LPL refolding experiments have demonstrated that folding of the smaller C-terminal domain happens quickly and completely, whereas folding of the N-terminal domain is much less efficient and occurs through folding intermediates exhibiting a degree of disordered tertiary structure [80]. Thus, the N-terminal domain would be expected to be subject to much more misfolding in vivo, with its proper folding constituting the rate-limiting step in lipase maturation. It is not surprising, therefore, that glycosylation at the conserved site in the N-terminal domain, as opposed to the C-terminal domain, is required for lipase maturation (Figure 2; see arrows).
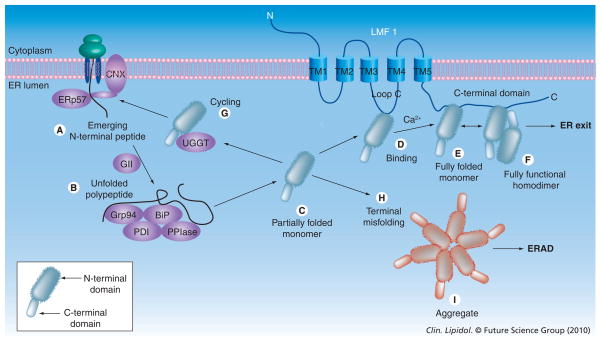
Figure 4. Model of lipase maturation.
The N- and C-terminal folding domains comprising the lipase monomer; wavy lines indicate an N-terminal domain that is partially folded (C, D) or misfolded (G, I). Only the homodimer exhibits lipolytic activity and exits the ER; all other lipase forms are inactive and are retained in the ER. Terminally misfolded forms, such as the aggregate, are destined for ERAD. BiP: 79 kDa binding protein; CNX: Calnexin; ER: Endoplasmic reticulum; ERAD: Endoplasmic reticulum-associated degradation; ERp56: 57 kDa endoplasmic reticulum protein; GrP94; Grp94: 94 kDa glucose-regulated protein; HL: Hepatic lipase; PDI: Protein disulfide isomerase; PPIase: Peptidyl-prolyl cis–trans isomerase; TM: Transmembrane; UGGT: UDP-glucose: glycoprotein glucosyltransferase 1.
The mechanism of lipase maturation can be considered to occur in two fundamental stages: the first stage would involve folding of the monomer (Figures 4A–E), which begins soon after the N-terminus of the lipase emerges through the Sec61 translocon (i.e., cotranslocationally); the second stage would entail assembly of fully folded monomers into homodimers (Figure 4F). In vitro evidence indicates that the first stage is rate limiting [80]; however, lipase maturation takes hours in vitro, as opposed to minutes in vivo [82]. Such disparate kinetics strongly indicates the involvement of chaperones and folding factors that facilitate folding of the monomer in vivo. While the second stage occurs rapidly in vitro (and likely in vivo as well), the product of the second step, lipase homodimers, are considered to be in a high-free-energy state that is inherently unstable and subject to dissociation [80,108]. Thus, both homodimers and folding intermediates are envisioned as being associated with chaperones and folding factors in the ER lumen, acting to stabilize and catalyze these two main steps in lipase maturation (Figure 4). Indeed, disruption of Ca2+ levels in the ER, an ion required for proper chaperone function [72], causes cessation of LPL maturation until appropriate levels are restored [110].
Folding of the monomer begins with chaperone stabilization of the lipase N-terminal domain as it emerges into the ER lumen through the Sec61 translocon (Figure 4A). Such stabilization would eliminate intermolecular associations leading to nonproductive aggregation [111], and would delay folding until lipase translation is finished. While either BiP or CNX/CRT could carry out this function, we believe CNX is the likely chaperone, based on the proximal position of the first conserved glycosylation site within the lipase N-terminal domain (Figure 2). Proteins with glycan attachment sites falling within approximately the first 50 amino acids (such as LPL, HL and EL) have been found to engage with CNX prior to BiP; proteins without such N-glycans at their N-terminus tend to associate with BiP first [72,107]. While both CNX and CRT carry out very similar functions, CNX is the lectin chaperone of choice during HL maturation [76]. CNX is better situated for cotranslational association with target proteins, since CNX (unlike CRT) is a membrane-bound protein, as is the Sec61 translocon, and only CNX is found to be associated with the ribosome during protein synthesis [112]. As the protein disulfide isomerase ERp57 physically associates with CNX, it seems likely that formation of intramolecular disulfide linkages also begins at this time (Figure 4A).
Glucosidase II cleavage would release the full-length lipase polypeptide into the ER lumen, which would likely be captured quickly as an unfolded intermediate by the BiP/Grp94 multi-protein complex (Figure 4B). Among its component proteins, the PPIase would catalyze the rate-limiting isomerization of cis peptidyl–prolyl bonds to the trans configuration. Upon release from the BiP–Grp94 complex, folding of the lipase polypeptide would happen quickly, with establishment of the β-sandwich fold comprising the C-terminal domain occurring efficiently and completely [80]. By contrast, the N-terminal domain would remain in a ‘molten globule’ state (Figure 4C). In LPL-refolding experiments, such a state was found to exhibit a native-like secondary structure, as determined by far-UV circular dichroism, but a less-ordered tertiary structure, as assessed by increased intrinsic tryptophan fluorescence and elevated binding of the hydrophobic molecule 4,4′-dianilino-1,1′-binaphthyl-5,5′-disulfonic acid [80]. In Figure 4C, we consider such an intermediate as a ‘partially folded’ monomer with the potential to form either a fully folded monomer (Figure 4E) or to undergo misfolding (Figure 4H), which would prevent further maturation. We propose that LMF1 would capture a partially folded monomer exhibiting native structure (Figure 4D) but not misfolded forms; rather, misfolding would cause the monomer to recycle back to CNX through UGGT (Figure 4G) or to enter into the ERAD pathway (Figure 4H). The slow kinetics of HL maturation suggests that CNX cycling indeed occurs [75], and this is supported by the finding that HL associates with UGGT [76]. UGGT can sense localized regions of polypeptide disorder in misfolded proteins [113,114], adding a glucose residue to the high-mannose chain in order to re-establish CNX binding (Figure 2B). Unlike HL, however, repeated CNX cycling of LPL seems unlikely, owing to the much faster kinetics of LPL maturation [81]. In fact, LPL aggregates appear in pulse experiments nearly simultaneously with the formation of homodimers; these aggregates exhibit properties of terminally misfolded forms (Figure 4I), including intermolecular disulfide bonding, ER retention and eventual ERAD [81].
Lipoprotein lipase refolding in vitro is capable of forming homodimers with an efficiency of approximately 40%, a process taking hours at 25°C and requiring the presence of Ca2+ ions and other stabilizing factors [80]; notably, the fully folded monomer was not detected in vitro, indicating that it is a very short-lived intermediate. In the case of in vivo lipase maturation, there are several reasons to suspect that chaperone association would stabilize the partially folded intermediate (Figure 4D), the short-lived fully folded monomer (Figure 4E) and the fully functional homodimer (Figure 4F). First, in vivo lipase maturation occurs with an efficiency of approximately 70% at 37°C [75,81] – a temperature that greatly reduces the efficiency of LPL refolding in vitro by increasing LPL aggregation [80]. Aggregation occurs by inappropriate intermolecular interactions that are prevented by chaperone association [111]. Second, the short-lived nature of the fully folded monomer may result from surface hydrophobic regions required for noncovalent homodimer association (Figure 1A, arrows); such surface hydrophobicity would be thermodynamically unstable unless shielded from the aqueous environment by chaperone association (Figure 4E). Finally, the high-free-energy state of the homodimer favors its rapid dissociation into misfolded monomers unless stabilized in vitro by binding to factors such as heparin [80]. However, studies have indicated that the LPL dimer is extraordinarily stable in the ER lumen [81,82], suggesting that an ER-specific stabilizing factor prevents such dissociation in vivo (Figure 4F).
What is the stabilizing factor that facilitates these later steps in lipase maturation, as shown in Figure 4D–F? Considering that such steps involve the attainment of structural features unique to lipase proteins, we propose that they are chaperoned by the client-specific factor LMF1. LMF1 has the ability to bind lipases in the ER, and in its absence, LPL homodimers decline severely while LPL aggregates increase dramatically [83]. Thus, without functional LMF1, maturation steps Figure 4D–F occur very inefficiently indeed. By contrast, early lipase maturation steps occurring through CNX and BiP seem to be independent of LMF1. For example, LPL in cld/cld cells is properly glycosylated and processed [83], indicating that these early maturation steps remain unaffected (Figure 2). Thus, LMF1 most likely functions in later stages of lipase maturation, when lipase monomers and homodimers may need to be shielded from an ER environment conducive to their misfolding, dissociation and aggregation.
Conclusion
Lipase maturation is a process occurring in the ER that involves the folding and assembly of newly synthesized (nascent) lipase polypeptides into fully functional enzymes. The structural similarities of LPL, HL and EL, particularly in their requirement for a homodimer configuration, suggest that these lipase family members share a similar maturation pathway. This pathway utilizes the two major chaperone systems operating in the ER, the CNX and BiP/Grp94 multiprotein complexes, which probably stabilize and catalyze folding of nascent lipase polypeptides into partially folded monomers. Final maturation of the partially folded monomers to fully assembled homodimers may require the client-specific chaperone, LMF1. This lipase-specific chaperone may also stabilize the homodimer from its dissociation into a lower free energy state of misfolded monomers. The absence or functional loss of any of these ER chaperones, such as LMF1, results in combined lipase deficiency that has profound impacts on pathophysiological phenotypes such as hypertriglyceridemia.
Future perspective
The most challenging aspect of lipase maturation is the elucidation of LMF1 function, both as a maturation factor and as a candidate in lipase regulation. As a maturation factor, a number of questions remain unanswered. What is the precise role of LMF1 in the mechanism of lipase folding; does it stabilize the partially folded monomer and homodimer as proposed in Figure 4? What are the functions of the various LMF1 domains in this and related processes, such as ER localization? The polytopic nature of LMF1 suggests a complexity of functions involving associations with both cytoplasmic and ER lumenal proteins; what is the identity of these binding partners and how do they assist in lipase maturation? Is LMF1 also involved in the effective exit of homodimers from the ER, possibly by associating with the cytosolic coat protein II vesicles budding from the ER?
As a result of its essential role in lipase maturation, LMF1 may regulate lipase activity levels in vivo. It is the only known factor affecting the activity and secretion of every member of the lipase gene family except PL. The consequence of LMF1 deficiency on systemic and cell-associated phenotypes is a challenge for the future. The cld/cld mouse has limited utility in this regard, owing to the lethal nature of LPL deficiency occurring shortly after birth. Thus, inducible and tissue-specific models of LMF1 deficiency are needed to overcome the lethality and to ascertain cell-associated phenotypes in isolation from overwhelming systemic effects, such as massive hypertriglyceridemia. Furthermore, LMF1 may have roles other than lipase maturation; unlike lipase proteins, it is expressed in nearly all tissues and early during development [Peterfy M, Unpublished Data]. Moreover, naturally occurring LMF1 splice forms exist that lack the DUF1222 domain, and thus cannot function in lipase maturation. What are the functions of these splice variants, and what are the roles of LMF1 in tissues or during developmental times when lipases are not expressed? Could LMF1 be involved in some aspect of ER homeostasis, such as contributing to the ER-stress response? While questions abound concerning LMF1 function, expression and regulation, it is abundantly clear that it plays a pivotal role in lipase maturation.
Executive summary
Lipase function & molecular structure
Lipase maturation is defined as the folding and assembly of a family of proteins consisting of pancreatic lipase (PL), lipoprotein lipase (LPL), hepatic lipase (HL) and endothelial lipase (EL).
While all lipases hydrolyze triglycerides and/or phospholipids, each has specific functions in vivo, including absorption of dietary fats (PL), remodeling and uptake of lipoproteins (LPL, HL and EL) and regulation of fatty acid influx in adipose tissue, muscle and the heart. The lipase monomer is divided into an N- and C-terminal folding domain. Except for PL, monomers must assemble into homodimers in order to form fully functional enzymes.
Lipases in disease
Lipoprotein lipase, HL and EL have emerged as some of the strongest genetic determinants of circulating triglyceride and HDL-C levels in human populations.
Mutations affecting trans-acting factors that affect lipase expression, such as lipase maturation, can also have profound affects on triglyceride and HDL-C levels.
Mouse models of LPL overexpression indicate that lipase dysregulation can contribute to metabolic traits related to obesity, cardiomyopathy and insulin resistance.
Lipase glycosylation & processing in the endoplasmic reticulum
Lipoprotein lipase, HL and EL exhibit conserved glycosylation sites in both folding domains. Glycosylation of conserved N-terminal sites is necessary for efficient lipase maturation.
Glycan processing by glucose trimming is an early but essential step in lipase maturation, facilitating cycles of lipase binding and release of chaperone calnexin (CNX) from the endoplasmic reticulum (ER).
Lipase maturation factors
The folding and assembly of lipases in the ER requires general and specific maturation factors.
General factors include the CNX/calreticulin and BiP/Grp94 chaperone systems, each comprised of multiple folding factors.
The ER membrane protein lipase maturation factor (LMF) 1 is a lipase-specific factor required in LPL, HL and EL folding and assembly.
Mechanisms of lipase maturation
Maturation begins cotranslationally by association of growing lipase chains with CNX; after its translation and release, the unfolded lipase is likely captured by the BiP/Grp94 chaperone system.
Folding of the large N-terminal domain is a rate-limiting step, and proceeds through partially folded intermediates that can misfold prior to final folding and assembly.
Final lipase folding and assembly produce fully functional homodimers, a process likely aided by LMF1. LMF1 may also stabilize lipase homodimers until their exit from the ER.
Footnotes
For reprint orders, please contact: reprints@futuremedicine.com
Financial & competing interests disclosure
We acknowledge the support of the David Geffen School of Medicine, University of California, Los Angeles, the Cedars-Sinai Medical Center, the Department of Veterans Affairs and grant HL24841 from the National Institutes of Health. We would also like to acknowledge Drs Howard Wong and Keith Munson for their generous contribution of the structural model of hepatic lipase presented in Figure 1. The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
No writing assistance was utilized in the production of this manuscript.
Bibliography
Papers of special note have been highlighted as:
▪ of interest
▪▪ of considerable interest
- 1.McCoy MG, Sun GS, Marchadier D, Maugeais C, Glick JM, Rader DJ. Characterization of the lipolytic activity of endothelial lipase. J Lipid Res. 2002;43:921–929. [PubMed] [Google Scholar]
- 2.Wong H, Schotz MC. The lipase gene family. J Lipid Res. 2002;43:993–999. doi: 10.1194/jlr.r200007-jlr200. [DOI] [PubMed] [Google Scholar]
- 3.Mead JR, Irvine SA, Ramji DP. Lipoprotein lipase: structure, function, regulation, and role in disease. J Mol Med. 2002;80:753–769. doi: 10.1007/s00109-002-0384-9. [DOI] [PubMed] [Google Scholar]
- 4.Merkel M, Eckel RH, Goldberg IJ. Lipoprotein lipase: genetics, lipid uptake, and regulation. J Lipid Res. 2002;43:1997–2006. doi: 10.1194/jlr.r200015-jlr200. [DOI] [PubMed] [Google Scholar]
- 5.Stein Y, Stein O. Lipoprotein lipase and atherosclerosis. Atherosclerosis. 2003;170:1–9. doi: 10.1016/s0021-9150(03)00014-5. [DOI] [PubMed] [Google Scholar]
- 6.Goldberg IJ, Eckel RH, Abumrad NA. Regulation of fatty acid uptake into tissues: lipoprotein lipase- and CD36-mediated pathways. J Lipid Res. 2009;50(Suppl):S86–S90. doi: 10.1194/jlr.R800085-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Otarod JK, Goldberg IJ. Lipoprotein lipase and its role in regulation of plasma lipoproteins and cardiac risk. Curr Atheroscler Rep. 2004;6:335–342. doi: 10.1007/s11883-004-0043-4. [DOI] [PubMed] [Google Scholar]
- 8.van Tilbeurgh H, Roussel A, Lalouel JM, Cambillau C. Lipoprotein lipase. Molecular model based on the pancreatic lipase x-ray structure: consequences for heparin binding and catalysis. J Biol Chem. 1994;269:4626–4633. [PubMed] [Google Scholar]
- 9.Wang H, Eckel RH. Lipoprotein lipase: from gene to obesity. Am J Physiol Endocrinol Metab. 2009;297:E271–E288. doi: 10.1152/ajpendo.90920.2008. [DOI] [PubMed] [Google Scholar]
- 10.Perret B, Mabile L, Martinez L, Terce F, Barbaras R, Collet X. Hepatic lipase: structure/function relationship, synthesis, and regulation. J Lipid Res. 2002;43:1163–1169. [PubMed] [Google Scholar]
- 11.Santamarina-Fojo S, Gonzalez-Navarro H, Freeman L, Wagner E, Nong Z. Hepatic lipase, lipoprotein metabolism, and atherogenesis. Arterioscler Thromb Vasc Biol. 2004;24:1750–1754. doi: 10.1161/01.ATV.0000140818.00570.2d. [DOI] [PubMed] [Google Scholar]
- 12.Cohen JC, Vega GL, Grundy SM. Hepatic lipase: new insights from genetic and metabolic studies. Curr Opin Lipidol. 1999;10:259–267. doi: 10.1097/00041433-199906000-00008. [DOI] [PubMed] [Google Scholar]
- 13.Jansen H, Verhoeven AJ, Sijbrands EJ. Hepatic lipase: a pro- or anti-atherogenic protein? J Lipid Res. 2002;43:1352–1362. doi: 10.1194/jlr.r200008-jlr200. [DOI] [PubMed] [Google Scholar]
- 14.Rader DJ, Jaye M. Endothelial lipase: a new member of the triglyceride lipase gene family. Curr Opin Lipidol. 2000;11:141–147. doi: 10.1097/00041433-200004000-00006. [DOI] [PubMed] [Google Scholar]
- 15.Broedl UC, Jin W, Rader DJ. Endothelial lipase: a modulator of lipoprotein metabolism upregulated by inflammation. Trends Cardiovasc Med. 2004;14:202–206. doi: 10.1016/j.tcm.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 16.Badellino KO, Rader DJ. The role of endothelial lipase in high-density lipoprotein metabolism. Curr Opin Cardiol. 2004;19:392–395. doi: 10.1097/01.hco.0000130161.89169.02. [DOI] [PubMed] [Google Scholar]
- 17.Wong H, Davis RC, Hill JS, Yang D, Schotz MC. Lipase engineering: a window into structure–function relationships. Methods Enzymol. 1997;284:171–184. doi: 10.1016/s0076-6879(97)84011-6. [DOI] [PubMed] [Google Scholar]
- 18.Derewenda ZS, Cambillau C. Effects of gene mutations in lipoprotein and hepatic lipases as interpreted by a molecular model of the pancreatic triglyceride lipase. J Biol Chem. 1991;266:23112–23119. [PubMed] [Google Scholar]
- 19.Williams KJ. Molecular processes that handle – and mishandle – dietary lipids. J Clin Invest. 2008;118:3247–3259. doi: 10.1172/JCI35206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Young SG, Davies BS, Fong LG, et al. GPIHBP1: an endothelial cell molecule important for the lipolytic processing of chylomicrons. Curr Opin Lipidol. 2007;18:389–396. doi: 10.1097/MOL.0b013e3281527914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hasham SN, Pillarisetti S. Vascular lipases, inflammation and atherosclerosis. Clin Chim Acta. 2006;372:179–183. doi: 10.1016/j.cca.2006.04.020. [DOI] [PubMed] [Google Scholar]
- 22.Pappan KL, Pan Z, Kwon G, et al. Pancreatic β-cell lipoprotein lipase independently regulates islet glucose metabolism and normal insulin secretion. J Biol Chem. 2005;280:9023–9029. doi: 10.1074/jbc.M409706200. [DOI] [PubMed] [Google Scholar]
- 23.Paradis ME, Badellino KO, Rader DJ, et al. Endothelial lipase is associated with inflammation in humans. J Lipid Res. 2006;47:2808–2813. doi: 10.1194/jlr.P600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 24.Paradis ME, Badellino KO, Rader DJ, et al. Visceral adiposity and endothelial lipase. J Clin Endocrinol Metab. 2006;91:3538–3543. doi: 10.1210/jc.2006-0766. [DOI] [PubMed] [Google Scholar]
- 25.Pentikainen MO, Oksjoki R, Oorni K, Kovanen PT. Lipoprotein lipase in the arterial wall: linking LDL to the arterial extracellular matrix and much more. Arterioscler Thromb Vasc Biol. 2002;22:211–217. doi: 10.1161/hq0102.101551. [DOI] [PubMed] [Google Scholar]
- 26.Preiss-Landl K, Zimmermann R, Hammerle G, Zechner R. Lipoprotein lipase: the regulation of tissue specific expression and its role in lipid and energy metabolism. Curr Opin Lipidol. 2002;13:471–481. doi: 10.1097/00041433-200210000-00002. [DOI] [PubMed] [Google Scholar]
- 27.Pulawa LK, Eckel RH. Overexpression of muscle lipoprotein lipase and insulin sensitivity. Curr Opin Clin Nutr Metab Care. 2002;5:569–574. doi: 10.1097/00075197-200209000-00017. [DOI] [PubMed] [Google Scholar]
- 28.Winkler FK, D’Arcy A, Hunziker W. Structure of human pancreatic lipase. Nature. 1990;343:771–774. doi: 10.1038/343771a0. [DOI] [PubMed] [Google Scholar]
- 29.Griffon N, Jin W, Petty TJ, et al. Identification of the active form of endothelial lipase, a homodimer in a head-to-tail conformation. J Biol Chem. 2009;284:23322–23330. doi: 10.1074/jbc.M109.037002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schrag JD, Cygler M. Lipases and α/β hydrolase fold. Methods Enzymol. 1997;284:85–107. doi: 10.1016/s0076-6879(97)84006-2. [DOI] [PubMed] [Google Scholar]
- 31.Reis P, Holmberg K, Watzke H, Leser ME, Miller R. Lipases at interfaces: a review. Adv Colloid Interface Sci. 2009;147–148:237–250. doi: 10.1016/j.cis.2008.06.001. [DOI] [PubMed] [Google Scholar]
- 32.Lowe ME. The triglyceride lipases of the pancreas. J Lipid Res. 2002;43:2007–2016. doi: 10.1194/jlr.r200012-jlr200. [DOI] [PubMed] [Google Scholar]
- 33.Hill JS, Davis RC, Yang D, Schotz MC, Wong H. Hepatic lipase: high-level expression and subunit structure determination. Methods Enzymol. 1997;284:232–246. doi: 10.1016/s0076-6879(97)84015-3. [DOI] [PubMed] [Google Scholar]
- 34.Wong H, Yang D, Hill JS, Davis RC, Nikazy J, Schotz MC. A molecular biology-based approach to resolve the subunit orientation of lipoprotein lipase. Proc Natl Acad Sci USA. 1997;94:5594–5598. doi: 10.1073/pnas.94.11.5594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hide WA, Chan L, Li WH. Structure and evolution of the lipase superfamily. J Lipid Res. 1992;33:167–178. [PubMed] [Google Scholar]
- 36.Brunzell JD. Familial lipoprotein lipase deficiency and other causes of chylomicronemia syndromes. In: Scriver RS, Beaudet AL, Sly WS, editors. The Metabolic Basis of Inherited Disease. McGraw Hill; NY, USA: 1995. pp. 1913–1932. [Google Scholar]
- 37.Weinstock PH, Bisgaier CL, Aalto-Setala K, et al. Severe hypertriglyceridemia, reduced high density lipoprotein, and neonatal death in lipoprotein lipase knockout mice. Mild hypertriglyceridemia with impaired very low density lipoprotein clearance in heterozygotes. J Clin Invest. 1995;96:2555–2568. doi: 10.1172/JCI118319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38▪▪.Peterfy M, Ben-Zeev O, Mao HZ, et al. Mutations in LMF1 cause combined lipase deficiency and severe hypertriglyceridemia. Nat Genet. 2007;39:1483–1487. doi: 10.1038/ng.2007.24. Identifies the only known lipase-specific chaperone, lipase maturation factor 1 (LMF1), as the protein affected by the cld mutation. It also identifies the first human patient with combined lipase deficiency resulting from a mutation in LMF1. [DOI] [PubMed] [Google Scholar]
- 39.Beigneux AP, Davies BS, Gin P, et al. Glycosylphosphatidylinositol-anchored high-density lipoprotein-binding protein 1 plays a critical role in the lipolytic processing of chylomicrons. Cell Metab. 2007;5:279–291. doi: 10.1016/j.cmet.2007.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Aulchenko YS, Ripatti S, Lindqvist I, et al. Loci influencing lipid levels and coronary heart disease risk in 16 European population cohorts. Nat Genet. 2009;41:47–55. doi: 10.1038/ng.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Kathiresan S, Willer CJ, Peloso GM, et al. Common variants at 30 loci contribute to polygenic dyslipidemia. Nat Genet. 2009;41:56–65. doi: 10.1038/ng.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sabatti C, Service SK, Hartikainen AL, et al. Genome-wide association analysis of metabolic traits in a birth cohort from a founder population. Nat Genet. 2009;41:35–46. doi: 10.1038/ng.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Willer CJ, Sanna S, Jackson AU, et al. Newly identified loci that influence lipid concentrations and risk of coronary artery disease. Nat Genet. 2008;40:161–169. doi: 10.1038/ng.76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Austin MA, Hokanson JE, Edwards KL. Hypertriglyceridemia as a cardiovascular risk factor. Am J Cardiol. 1998;81:B7–B12. doi: 10.1016/s0002-9149(98)00031-9. [DOI] [PubMed] [Google Scholar]
- 45.Pejic RN, Lee DT. Hypertriglyceridemia. J Am Board Fam Med. 2006;19:310–316. doi: 10.3122/jabfm.19.3.310. [DOI] [PubMed] [Google Scholar]
- 46.Groop L. Genetics of the metabolic syndrome. Br J Nutr. 2000;83(Suppl 1):S39–S48. doi: 10.1017/s0007114500000945. [DOI] [PubMed] [Google Scholar]
- 47.Ginsberg HN, Stalenhoef AF. The metabolic syndrome: targeting dyslipidaemia to reduce coronary risk. J Cardiovasc Risk. 2003;10:121–128. doi: 10.1097/01.hjr.0000060840.46106.ea. [DOI] [PubMed] [Google Scholar]
- 48.Zechner R, Strauss J, Frank S, et al. The role of lipoprotein lipase in adipose tissue development and metabolism. Int J Obes Relat Metab Disord. 2000;24(Suppl 4):S53–S56. doi: 10.1038/sj.ijo.0801506. [DOI] [PubMed] [Google Scholar]
- 49.Levak-Frank S, Radner H, Walsh A, et al. Muscle-specific overexpression of lipoprotein lipase causes a severe myopathy characterized by proliferation of mitochondria and peroxisomes in transgenic mice. J Clin Invest. 1995;96:976–986. doi: 10.1172/JCI118145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Merkel M, Weinstock PH, Chajek-Shaul T, et al. Lipoprotein lipase expression exclusively in liver. A mouse model for metabolism in the neonatal period and during cachexia. J Clin Invest. 1998;102:893–901. doi: 10.1172/JCI2912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Ferreira LD, Pulawa LK, Jensen DR, Eckel RH. Overexpressing human lipoprotein lipase in mouse skeletal muscle is associated with insulin resistance. Diabetes. 2001;50:1064–1068. doi: 10.2337/diabetes.50.5.1064. [DOI] [PubMed] [Google Scholar]
- 52.Kim JK, Fillmore JJ, Chen Y, et al. Tissue-specific overexpression of lipoprotein lipase causes tissue-specific insulin resistance. Proc Natl Acad Sci USA. 2001;98:7522–7527. doi: 10.1073/pnas.121164498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jensen DR, Schlaepfer IR, Morin CL, et al. Prevention of diet-induced obesity in transgenic mice overexpressing skeletal muscle lipoprotein lipase. Am J Physiol. 1997;273:R683–R689. doi: 10.1152/ajpregu.1997.273.2.R683. [DOI] [PubMed] [Google Scholar]
- 54.Goodarzi MO, Guo X, Taylor KD, et al. Lipoprotein lipase is a gene for insulin resistance in Mexican Americans. Diabetes. 2004;53:214–220. doi: 10.2337/diabetes.53.1.214. [DOI] [PubMed] [Google Scholar]
- 55.Holzl B, Iglseder B, Sandhofer A, et al. Insulin sensitivity is impaired in heterozygous carriers of lipoprotein lipase deficiency. Diabetologia. 2002;45:378–384. doi: 10.1007/s00125-001-0771-8. [DOI] [PubMed] [Google Scholar]
- 56.Jemaa R, Tuzet S, Portos C, Betoulle D, Apfelbaum M, Fumeron F. Lipoprotein lipase gene polymorphisms: associations with hypertriglyceridemia and body mass index in obese people. Int J Obes Relat Metab Disord. 1995;19:270–274. [PubMed] [Google Scholar]
- 57.Radha V, Vimaleswaran KS, Ayyappa KA, Mohan V. Association of lipoprotein lipase gene polymorphisms with obesity and Type 2 diabetes in an Asian Indian population. Int J Obes (Lond) 2007;31:913–918. doi: 10.1038/sj.ijo.0803547. [DOI] [PubMed] [Google Scholar]
- 58.Yagyu H, Chen G, Yokoyama M, et al. Lipoprotein lipase (LpL) on the surface of cardiomyocytes increases lipid uptake and produces a cardiomyopathy. J Clin Invest. 2003;111:419–426. doi: 10.1172/JCI16751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Park TS, Hu Y, Noh HL, et al. Ceramide is a cardiotoxin in lipotoxic cardiomyopathy. J Lipid Res. 2008;49:2101–2112. doi: 10.1194/jlr.M800147-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.McGavock JM, Lingvay I, Zib I, et al. Cardiac steatosis in diabetes mellitus: a 1H-magnetic resonance spectroscopy study. Circulation. 2007;116:1170–1175. doi: 10.1161/CIRCULATIONAHA.106.645614. [DOI] [PubMed] [Google Scholar]
- 61.Peterson LR, Herrero P, Schechtman KB, et al. Effect of obesity and insulin resistance on myocardial substrate metabolism and efficiency in young women. Circulation. 2004;109:2191–2196. doi: 10.1161/01.CIR.0000127959.28627.F8. [DOI] [PubMed] [Google Scholar]
- 62.Sharma S, Adrogue JV, Golfman L, et al. Intramyocardial lipid accumulation in the failing human heart resembles the lipotoxic rat heart. FASEB J. 2004;18:1692–1700. doi: 10.1096/fj.04-2263com. [DOI] [PubMed] [Google Scholar]
- 63.Babaev VR, Fazio S, Gleaves LA, Carter KJ, Semenkovich CF, Linton MF. Macrophage lipoprotein lipase promotes foam cell formation and atherosclerosis in vivo. J Clin Invest. 1999;103:1697–1705. doi: 10.1172/JCI6117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Wilson K, Fry GL, Chappell DA, Sigmund CD, Medh JD. Macrophage-specific expression of human lipoprotein lipase accelerates atherosclerosis in transgenic apolipoprotein E knockout mice but not in C57BL/6 mice. Arterioscler Thromb Vasc Biol. 2001;21:1809–1815. doi: 10.1161/hq1101.097805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Izar MC, Helfenstein T, Ihara SS, et al. Association of lipoprotein lipase D9N polymorphism with myocardial infarction in Type 2 diabetes: the genetics, outcomes, and lipids in Type 2 diabetes (GOLD) study. Atherosclerosis. 2009;204:165–170. doi: 10.1016/j.atherosclerosis.2008.08.006. [DOI] [PubMed] [Google Scholar]
- 66.Hegele RA, Little JA, Vezina C, et al. Hepatic lipase deficiency. Clinical, biochemical, and molecular genetic characteristics. Arterioscler Thromb. 1993;13:720–728. doi: 10.1161/01.atv.13.5.720. [DOI] [PubMed] [Google Scholar]
- 67.Knudsen P, Antikainen M, Uusi-Oukari M, et al. Heterozygous hepatic lipase deficiency, due to two missense mutations R186H and L334F, in the HL gene. Atherosclerosis. 1997;128:165–174. doi: 10.1016/s0021-9150(96)05999-0. [DOI] [PubMed] [Google Scholar]
- 68.Jin W, Millar JS, Broedl U, Glick JM, Rader DJ. Inhibition of endothelial lipase causes increased HDL cholesterol levels in vivo. J Clin Invest. 2003;111:357–362. doi: 10.1172/JCI16146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Broedl UC, Maugeais C, Millar JS, et al. Endothelial lipase promotes the catabolism of ApoB-containing lipoproteins. Circ Res. 2004;94:1554–1561. doi: 10.1161/01.RES.0000130657.00222.39. [DOI] [PubMed] [Google Scholar]
- 70.Edmondson AC, Brown RJ, Kathiresan S, et al. Loss-of-function variants in endothelial lipase are a cause of elevated HDL cholesterol in humans. J Clin Invest. 2009;119:1042–1050. doi: 10.1172/JCI37176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Hebert DN, Garman SC, Molinari M. The glycan code of the endoplasmic reticulum: asparagine-linked carbohydrates as protein maturation and quality-control tags. Trends Cell Biol. 2005;15:364–370. doi: 10.1016/j.tcb.2005.05.007. [DOI] [PubMed] [Google Scholar]
- 72▪.Hebert DN, Molinari M. In and out of the ER: protein folding, quality control, degradation, and related human diseases. Physiol Rev. 2007;87:1377–1408. doi: 10.1152/physrev.00050.2006. Comprehensive review of protein folding in the endoplasmic reticulum (ER), with excellent explanations of co- and post-translational folding events, including the general folding factors and chaperones involved. [DOI] [PubMed] [Google Scholar]
- 73.Trombetta ES, Helenius A. Conformational requirements for glycoprotein reglucosylation in the endoplasmic reticulum. J Cell Biol. 2000;148:1123–1129. doi: 10.1083/jcb.148.6.1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Caramelo JJ, Parodi AJ. Getting in and out from calnexin/calreticulin cycles. J Biol Chem. 2008;283:10221–10225. doi: 10.1074/jbc.R700048200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75▪.Ben-Zeev O, Doolittle MH. Maturation of hepatic lipase. Formation of functional enzyme in the endoplasmic reticulum is the rate-limiting step in its secretion. J Biol Chem. 2004;279:6171–6181. doi: 10.1074/jbc.M310051200. Analysis of the relative maturation rates of the lipase gene family, and the identification of the physical characteristics of hepatic lipase intermediates in the ER and their association with calnexin. [DOI] [PubMed] [Google Scholar]
- 76▪▪.Doolittle MH, Ben-Zeev O, Bassilian S, Whitelegge JP, Peterfy M, Wong H. Hepatic lipase maturation: a partial proteome of interacting factors. J Lipid Res. 2009;50:1173–1184. doi: 10.1194/jlr.M800603-JLR200. Proteomic-based study that identifies proteins associated with hepatic lipase during its folding and assembly. This is the first study to demonstrate that lipase maturation uses components of both major chaperone systems operating in the ER. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Ben-Zeev O, Stahnke G, Liu G, Davis RC, Doolittle MH. Lipoprotein lipase and hepatic lipase: the role of asparagine-linked glycosylation in the expression of a functional enzyme. J Lipid Res. 1994;35:1511–1523. [PubMed] [Google Scholar]
- 78.Miller GC, Long CJ, Bojilova ED, et al. Role of N-linked glycosylation in the secretion and activity of endothelial lipase. J Lipid Res. 2004;45:2080–2087. doi: 10.1194/jlr.M400162-JLR200. [DOI] [PubMed] [Google Scholar]
- 79.Wolle J, Jansen H, Smith LC, Chan L. Functional role of N-linked glycosylation in human hepatic lipase: asparagine-56 is important for both enzyme activity and secretion. J Lipid Res. 1993;34:2169–2176. [PubMed] [Google Scholar]
- 80▪▪.Zhang L, Lookene A, Wu G, Olivecrona G. Calcium triggers folding of lipoprotein lipase into active dimers. J Biol Chem. 2005;280:42580–42591. doi: 10.1074/jbc.M507252200. Elegant and important study examining lipase refolding from chemically denatured lipoprotein lipase (LPL). It describes the physical characteristics of LPL-folding intermediates and identifies factors essential in the folding and assembly of homodimers in vitro. [DOI] [PubMed] [Google Scholar]
- 81.Ben-Zeev O, Doolittle MH, Davis RC, Elovson J, Schotz MC. Maturation of lipoprotein lipase. Expression of full catalytic activity requires glucose trimming but not translocation to the cis-Golgi compartment. J Biol Chem. 1992;267:6219–6227. [PubMed] [Google Scholar]
- 82▪.Ben-Zeev O, Mao HZ, Doolittle MH. Maturation of lipoprotein lipase in the endoplasmic reticulum. Concurrent formation of functional dimers and inactive aggregates. J Biol Chem. 2002;277:10727–10738. doi: 10.1074/jbc.M108128200. Analysis of the physical forms of LPL occurring in the ER, as well as measurement of their maturation kinetics. This is the first description of aggregation as a misfolded end point of LPL maturation in vivo; this study also demonstrates the stability of the LPL homodimer in the ER. [DOI] [PubMed] [Google Scholar]
- 83.Briquet-Laugier V, Ben-Zeev O, White A, Doolittle MH. cld and lec23 are disparate mutations that affect maturation of lipoprotein lipase in the endoplasmic reticulum. J Lipid Res. 1999;40:2044–2058. [PubMed] [Google Scholar]
- 84.Dejgaard S, Nicolay J, Taheri M, Thomas DY, Bergeron JJ. The ER glycoprotein quality control system. Curr Issues Mol Biol. 2004;6:29–42. [PubMed] [Google Scholar]
- 85.Sitia R, Braakman I. Quality control in the endoplasmic reticulum protein factory. Nature. 2003;426:891–894. doi: 10.1038/nature02262. [DOI] [PubMed] [Google Scholar]
- 86.Trombetta ES, Parodi AJ. Quality control and protein folding in the secretory pathway. Annu Rev Cell Dev Biol. 2003;19:649–676. doi: 10.1146/annurev.cellbio.19.110701.153949. [DOI] [PubMed] [Google Scholar]
- 87.Ruddock LW, Molinari M. N-glycan processing in ER quality control. J Cell Sci. 2006;119:4373–4380. doi: 10.1242/jcs.03225. [DOI] [PubMed] [Google Scholar]
- 88.Schroder M, Kaufman RJ. ER stress and the unfolded protein response. Mutat Res. 2005;569:29–63. doi: 10.1016/j.mrfmmm.2004.06.056. [DOI] [PubMed] [Google Scholar]
- 89.Meunier L, Usherwood YK, Chung KT, Hendershot LM. A subset of chaperones and folding enzymes form multiprotein complexes in endoplasmic reticulum to bind nascent proteins. Mol Biol Cell. 2002;13:4456–4469. doi: 10.1091/mbc.E02-05-0311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Boedeker JC, Doolittle MH, White AL. Differential effect of combined lipase deficiency (cld/cld) on human hepatic lipase and lipoprotein lipase secretion. J Lipid Res. 2001;42:1858–1864. [PubMed] [Google Scholar]
- 91.Zhang L, Wu G, Tate CG, Lookene A, Olivecrona G. Calreticulin promotes folding/dimerization of human lipoprotein lipase expressed in insect cells (sf21) J Biol Chem. 2003;278:29344–29351. doi: 10.1074/jbc.M300455200. [DOI] [PubMed] [Google Scholar]
- 92.Buck TM, Wright CM, Brodsky JL. The activities and function of molecular chaperones in the endoplasmic reticulum. Semin Cell Dev Biol. 2007;18:751–761. doi: 10.1016/j.semcdb.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Ni M, Lee AS. ER chaperones in mammalian development and human diseases. FEBS Lett. 2007;581:3641–3651. doi: 10.1016/j.febslet.2007.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Gething MJ. Role and regulation of the ER chaperone BiP. Semin Cell Dev Biol. 1999;10:465–472. doi: 10.1006/scdb.1999.0318. [DOI] [PubMed] [Google Scholar]
- 95.Blond-Elguindi S, Cwirla SE, Dower WJ, et al. Affinity panning of a library of peptides displayed on bacteriophages reveals the binding specificity of BiP. Cell. 1993;75:717–728. doi: 10.1016/0092-8674(93)90492-9. [DOI] [PubMed] [Google Scholar]
- 96.Paterniti JR, Jr, Brown WV, Ginsberg HN, Artzt K. Combined lipase deficiency (cld): a lethal mutation on chromosome 17 of the mouse. Science. 1983;221:167–169. doi: 10.1126/science.6857276. [DOI] [PubMed] [Google Scholar]
- 97.Reue K, Doolittle MH. Naturally occurring mutations in mice affecting lipid transport and metabolism. J Lipid Res. 1996;37:1387–1405. [PubMed] [Google Scholar]
- 98.Davis RC, Ben-Zeev O, Martin D, Doolittle MH. Combined lipase deficiency in the mouse. Evidence of impaired lipase processing and secretion. J Biol Chem. 1990;265:17960–17966. [PubMed] [Google Scholar]
- 99.Scow RO, Schultz CJ, Park JW, Blanchette-Mackie EJ. Combined lipase deficiency (cld/cld) in mice affects differently post-translational processing of lipoprotein lipase, hepatic lipase and pancreatic lipase. Chem Phys Lipids. 1998;93:149–155. doi: 10.1016/s0009-3084(98)00039-5. [DOI] [PubMed] [Google Scholar]
- 100.Olivecrona T, Chernick SS, Bengtsson-Olivecrona G, Paterniti JR, Jr, Brown WV, Scow RO. Combined lipase deficiency (cld/cld) in mice. Demonstration that an inactive form of lipoprotein lipase is synthesized. J Biol Chem. 1985;260:2552–2557. [PubMed] [Google Scholar]
- 101.Blanchette-Mackie EJ, Wetzel MG, Chernick SS, Paterniti JR, Jr, Brown WV, Scow RO. Effect of the combined lipase deficiency mutation (cld/cld) on ultrastructure of tissues in mice. Diaphragm, heart, brown adipose tissue, lung, and liver. Lab Invest. 1986;55:347–362. [PubMed] [Google Scholar]
- 102.Peterfy M, Mao HZ, Doolittle MH. The cld mutation: narrowing the critical chromosomal region and selecting candidate genes. Mamm Genome. 2006;17:1013–1024. doi: 10.1007/s00335-006-0045-3. [DOI] [PubMed] [Google Scholar]
- 103.Cefalu AB, Noto D, Arpi ML, et al. Novel LMF1 nonsense mutation in a patient with severe hypertriglyceridemia. J Clin Endocrin Metab. 2009;94:4584–4590. doi: 10.1210/jc.2009-0594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104▪▪.Doolittle MH, Neher SB, Ben-Zeev O, et al. Lipase maturation factor 1 (LMF1): Membrane topology and interaction with lipase proteins in the endoplasmic reticulum. J Biol Chem. 2009;284:33623–33633. doi: 10.1074/jbc.M109.049395. Recent study characterizing the membrane topology of LMF1, demonstrating the domain structure of this lipase-specific chaperone. It also demonstrates, for the first time, that lipases in the ER physically associate with LMF1 and locates the binding site to a specific LMF1 domain. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 105.Sonnhammer EL, Eddy SR, Durbin R. Pfam: a comprehensive database of protein domain families based on seed alignments. Proteins. 1997;28:405–420. doi: 10.1002/(sici)1097-0134(199707)28:3<405::aid-prot10>3.0.co;2-l. [DOI] [PubMed] [Google Scholar]
- 106.Molinari M, Eriksson KK, Calanca V, et al. Contrasting functions of calreticulin and calnexin in glycoprotein folding and ER quality control. Mol Cell. 2004;13:125–135. doi: 10.1016/s1097-2765(03)00494-5. [DOI] [PubMed] [Google Scholar]
- 107.Molinari M, Helenius A. Chaperone selection during glycoprotein translocation into the endoplasmic reticulum. Science. 2000;288:331–333. doi: 10.1126/science.288.5464.331. [DOI] [PubMed] [Google Scholar]
- 108.Lookene A, Zhang L, Hultin M, Olivecrona G. Rapid subunit exchange in dimeric lipoprotein lipase and properties of the inactive monomer. J Biol Chem. 2004;279:49964–49972. doi: 10.1074/jbc.M407419200. [DOI] [PubMed] [Google Scholar]
- 109.Lookene A, Zhang L, Tougu V, Olivecrona G. 1,1′-bis(anilino)-4-,4′-bis(naphtalene)-8,8′-disulfonate acts as an inhibitor of lipoprotein lipase and competes for binding with apolipoprotein CII. J Biol Chem. 2003;278:37183–37194. doi: 10.1074/jbc.M303894200. [DOI] [PubMed] [Google Scholar]
- 110.Osibow K, Frank S, Malli R, Zechner R, Graier WF. Mitochondria maintain maturation and secretion of lipoprotein lipase in the endoplasmic reticulum. Biochem J. 2006;396:173–182. doi: 10.1042/BJ20060099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Rozema D, Gellman SH. Artificial chaperone-assisted refolding of denatured-reduced lysozyme: modulation of the competition between renaturation and aggregation. Biochemistry. 1996;35:15760–15771. doi: 10.1021/bi961638j. [DOI] [PubMed] [Google Scholar]
- 112.Chevet E, Wong HN, Gerber D, et al. Phosphorylation by CK2 and MAPK enhances calnexin association with ribosomes. Embo J. 1999;18:3655–3666. doi: 10.1093/emboj/18.13.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ritter C, Quirin K, Kowarik M, Helenius A. Minor folding defects trigger local modification of glycoproteins by the ER folding sensor GT. Embo J. 2005;24:1730–1738. doi: 10.1038/sj.emboj.7600645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Taylor SC, Ferguson AD, Bergeron JJ, Thomas DY. The ER protein folding sensor UDP-glucose glycoprotein glucosyltransferase modifies substrates distant to local changes in glycoprotein conformation. Nat Struct Mol Biol. 2004;11:128–134. doi: 10.1038/nsmb715. [DOI] [PubMed] [Google Scholar]