Summary
Cik1, in association with the kinesin Kar3, controls both the mitotic spindle and nuclear fusion during mating. Here, we show that there are two Cik1 isoforms and that the mitotic form includes an N-terminal domain required for ubiquitination by the Anaphase Promoting Complex/Cyclosome (APC/C). During vegetative growth, Cik1 is expressed during mitosis and regulates the mitotic spindle, allowing for accurate chromosome segregation. Following mitosis, APC/CCdh1 targets Cik1 for ubiquitin-mediated proteolysis. Upon exposure to the mating pheromone α factor, a smaller APC/C-resistant Cik1 isoform is expressed from an alternate transcriptional start site. This shorter Cik1 isoform is stable and cannot be ubiquitinated by APC/CCdh1. Moreover, the two Cik1 isoforms are functionally distinct. Cells that express only the long isoform have defects in nuclear fusion, while cells expressing only the short isoform have an increased rate of chromosome loss. These results demonstrate a coupling of transcriptional regulation and APC/C-mediated proteolysis.
Introduction
Microtubules regulate many cellular processes such as organelle transport, chromosome segregation, and nuclear fusion during mating. In the budding yeast Saccharomyces cerevisiae, one dynein and six kinesin-related motor proteins mediate these microtubule-based processes (Hildebrandt and Hoyt, 2000). Kar3 is a C-terminal kinesin with minus end-directed motility (Endow et al., 1994). During vegetative growth, Kar3 localizes to the nuclear face of the spindle pole body (SPB) and the mitotic spindle and has roles in forming, positioning, and maintaining the spindle (Manning et al., 1999; Page et al., 1994; Saunders et al., 1997). Kar3 also functions in the cytoplasm during mating, where it localizes to the cytoplasmic face of the SPB and the plus ends of cytoplasmic microtubules that extend into the mating projection or shmoo tip (Meluh and Rose, 1990; Page et al., 1994). Here, Kar3 catalyzes the depolymerization of microtubules, bringing the two nuclei together (Maddox et al., 2000; Sproul et al., 2005).
Kar3 function depends on its binding to one of two accessory proteins, Cik1 or Vik1 (Manning et al., 1999; Page et al., 1994; Page and Snyder, 1992). Each of these proteins is structurally similar to Kar3, with a central coiled-coil domain that mediates heterodimerization (Barrett et al., 2000). However, Cik1 and Vik1 differ in their N-terminal domains. In particular, Cik1 has a putative nuclear localization signal (NLS) at its N-terminus, which helps target Kar3 within the cell (Manning et al., 1999). Although both are important for Kar3 function, cik1Δ and vik1Δ strains do not exactly phenocopy kar3Δ strains, suggesting that each protein acts with Kar3 for a subset of its functions. Moreover, Cik1 expression patterns and cik1Δ phenotypes suggest that it acts during both vegetative growth and mating, whereas Vik1 is not expressed during mating. During mitotic growth, both kar3Δ and cik1Δ cells are temperature sensitive and have an increased rate of chromosome loss (Page and Snyder, 1992; Saunders et al., 1997). In addition, strains with either deletion have longer and more prominent microtubule arrays, suggesting that the Kar3-Cik1 complex promotes the depolymerization of microtubules.
The Anaphase Promoting Complex/Cyclosome (APC/C) is a multi-subunit E3 ubiquitin ligase that targets a number of cell-cycle regulators for proteasomal degradation (Peters, 2006; Thornton and Toczyski, 2006). Two activator subunits, Cdc20 and Cdh1, mediate substrate recognition by the APC/C and act sequentially during the cell cycle (Schwab et al., 1997; Visintin et al., 1997). Cdc20 expression peaks during mitosis, when it is required for the turnover of securin and B-type cyclins to allow progression through the metaphase-anaphase transition (Cohen-Fix et al., 1996; Shirayama et al., 1999). Cdh1 is expressed throughout the cell cycle. However its activity is negatively regulated by Cdk phosphorylation so that it only becomes active when cyclin levels drop at the end of mitosis (Jaspersen et al., 1999; Zachariae et al., 1998), remaining active through the next G1 phase.
As part of a larger screen to identify ubiquitin ligases that target cyclical proteins for degradation, we identified Cik1 as a candidate Cdh1 target. Here we demonstrate that Cik1 is an APC/CCdh1 target that has an interesting form of regulation. During vegetative growth, Cik1 is cyclically expressed and is targeted for degradation by Cdh1 in each cell cycle. However, when cells arrest in a specialized G1 phase during mating, CIK1 is transcribed and translated from alternate start sites, producing a smaller protein that lacks the APC/C-targeting sequence required for its destruction. Several ubiquitin ligase substrates are protected from turnover by phosphorylation under special circumstances, such as DNA damage or osmotic stress (Agarwal et al., 2003; Escote et al., 2004). Similarly, altering transcription under different environmental conditions, in order to alter the stability of the translated protein, may prove to be a useful mechanism by which cells can alter their physiology in response to various stimuli.
Results
APC/CCdh1 regulates Cik1 stability
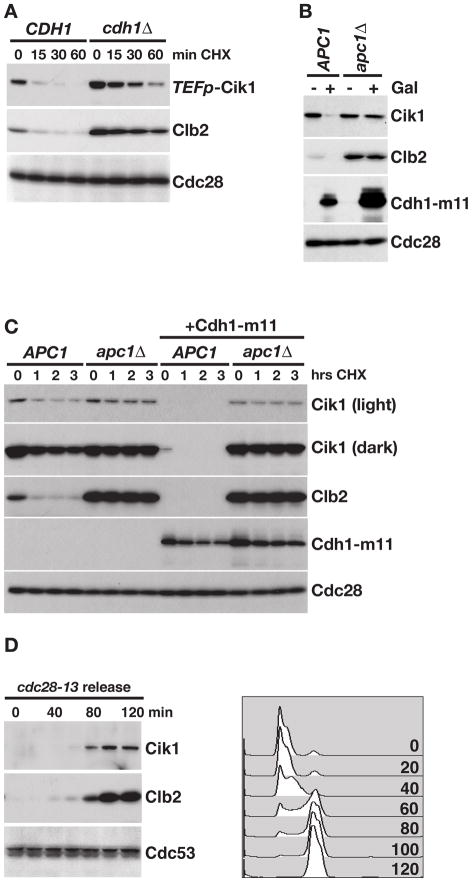
In order to gain a greater understanding of how ubiquitin-mediated proteolysis regulates the cell cycle, we designed a high-throughput microscopy screening approach to identify ubiquitin ligases that target a panel of proteins whose levels fluctuate throughout the cell cycle (to be described elsewhere). In a pilot experiment, we mated strains carrying GFP-tagged alleles of 73 unstable proteins to a strain in which CDH1 was replaced with RFP, and generated haploids that expressed each GFP-tagged protein and were cdh1Δ::RFP. We then used high-throughput microscopy to compare expression of each GFP fusion protein between wild type and cdh1Δ::RFP strains (as in Benanti et al., 2007). Using this approach, we identified the kinesin-associated protein Cik1 as a candidate target of APC/CCdh1 (Figure S1A). To determine if Cdh1 regulates Cik1 proteolysis, the half-life of Cik1 was compared between CDH1 and cdh1Δ cells. Cik1 was stabilized in cdh1Δ cells, similar to the APC/CCdh1 target Clb2 (Figure 1A).
Figure 1. APC/CCdh1 regulates Cik1 levels and stability.
(A) Cycloheximide-chase assay showing the levels of Cik1-GFP and Clb2 after the addition of cycloheximide to asynchronous CDH1 and cdh1Δ cells for the indicated number of minutes (min CHX). Since Cik1 is expressed at very low levels, the strong, constitutive TEF1 promoter (TEFp) was introduced upstream of the gene. Cdc28 is shown as a loading control. Cell cycle profiles are shown in Figure S1B. (B) APC1 and apc1Δ strains were treated with galactose (Gal) to induce expression of Cdh1-m11 and levels of Cik1-3FLAG, Clb2, HA-Cdh1-m11 and Cdc28 were analyzed by Western blotting. Cell cycle profiles are shown in Figure S1C. (C) Cycloheximide-chase assay showing the levels of Cik1-3FLAG, Clb2, and HA-Cdh1-m11 in APC1 and apc1Δ cells grown in glucose (Cdh1-m11 off) or galactose (Cdh1-m11 on). Samples were collected after the indicated number of hours in cycloheximide (hrs CHX). (D) cdc28-13 cells expressing Cik1-GFP were arrested at 37°C for 3 hours and then released at 23°C. Levels of Cik1-GFP and Clb2 were followed by Western blot and cell cycle position was monitored by flow cytometry. Cdc53 levels are shown as a loading control.
Cdh1 is expressed throughout the cell cycle, but its association with the core APC/C complex and its nuclear localization are blocked by cyclin B/Cdk phosphorylation (Jaquenoud et al., 2002; Jaspersen et al., 1999; Zachariae et al., 1998). For this reason, APC/CCdh1 is activated as cells exit mitosis and B-type cyclin levels decrease. We tested whether expression of a constitutively active Cdh1 that lacks Cdk phosphorylation sites (Cdh1-m11)(Zachariae et al., 1998) affected levels of Cik1, and found that expression of Cdh1-m11 protein led to decreased levels of Cik1 and Clb2 (Figure 1B). To ensure that this effect was due to increased APC/C activity, we utilized strains in which the APC/C is rendered non-essential through deletion of CLB5 and PDS1 and overexpression of SIC1 (Thornton and Toczyski, 2003). In strains lacking the core APC/C subunit Apc1, Cdh1-m11 expression had no effect on Cik1 levels, confirming that Cdh1 regulates Cik1 in an APC/C-dependent manner. When we measured half-life (Figure 1C), we saw that there was an increase in stability of Cik1 in apc1Δ cells, similar to Clb2, and this difference was greatly enhanced upon expression of Cdh1-m11. The fact that Cik1 was more strongly stabilized in apc1Δ strains (Figure 1C) compared to cdh1Δ strains (Figure 1A) suggests that Cdc20 may contribute to Cik1 turnover in the absence of Cdh1.
During vegetative growth, Cik1 transcription is cell-cycle regulated, peaking in mitosis like other Cdh1 targets such as Clb2 (Pramila et al., 2006; Spellman et al., 1998). To confirm that Cik1 protein levels also increase in mitosis, cells were arrested in G1 phase with a temperature-sensitive allele of Cdc28 (cdc28-13) and released at the permissive temperature. Like Clb2, Cik1 levels were low in G1-arrested cells and rose as cells entered mitosis (Figure 1D), confirming that its levels are low at the time in the cell cycle when Cdh1 is active.
An alternate form of Cik1 is expressed during mating
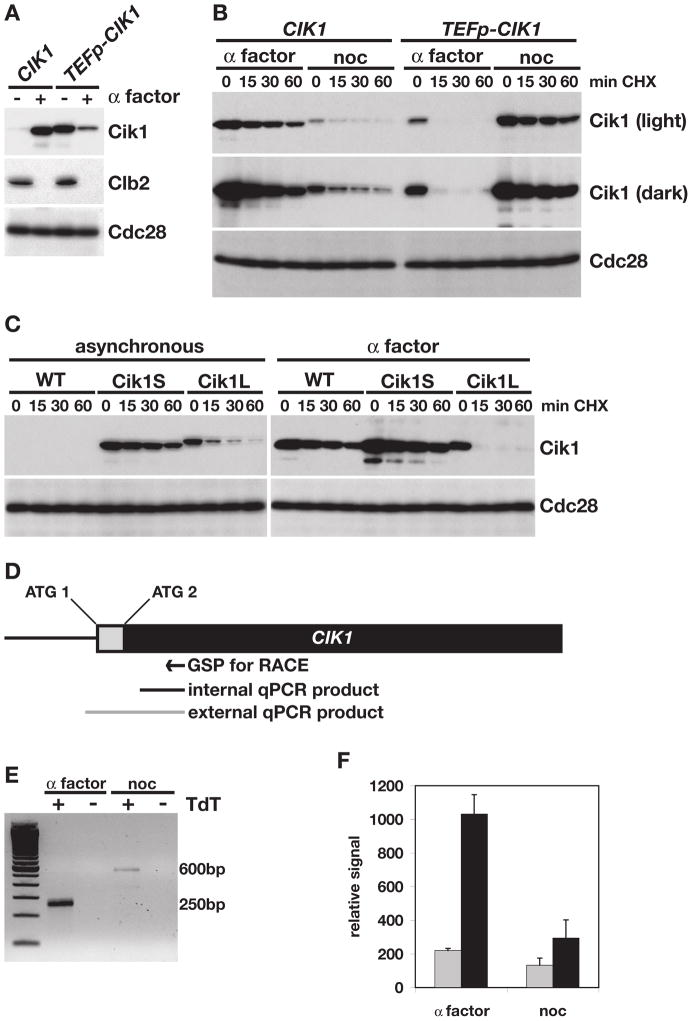
Cik1 and Kar3 are both required for cells to undergo karyogamy during mating, and both genes are transcriptionally induced when cells are exposed to the mating pheromone α factor (Kurihara et al., 1996). However, exposure to α factor also arrests cells in G1 phase, the time of the cell cycle when APC/CCdh1 is most active. This led us to test whether Cik1 turnover was blocked when cells were arrested with α factor. Consistent with published results (Manning et al., 1999; Page and Snyder, 1992), α factor treatment led to a strong increase in Cik1 protein in wild type cells, while levels of another Cdh1 target, Clb2, decreased (Figure 2A). In contrast to what was observed in asynchronous cells (Figure 1A), Cik1 was stable in α factor-treated cells (Figure 2B, left side), suggesting that Cik1 was protected from APC/CCdh1-mediated turnover. Cik1 was also stable when cells were treated with nocodazole and the APC/C is rendered inactive by the spindle checkpoint (Figure 2B).
Figure 2. A smaller, stable form of Cik1 is expressed during mating.
(A) Western blot comparing Cik1-GFP and Clb2 levels in asynchronous (−) or α factor-treated (+) cells expressing Cik1 from its endogenous promoter (CIK1), or the TEF1 promoter (TEFp-CIK1). Cdc28 is shown as a loading control. (B) Cycloheximide-chase assay showing levels of Cik1-GFP in wild type (CIK1) or TEFp-CIK1 cells that were arrested with α factor or nocodazole and then treated with cycloheximide for the indicated time (min CHX). Light and dark exposures of the GFP blot are shown. Cell cycle profiles are shown in Figure S2B. (C) Cycloheximide-chase assay of Cik1 in wild type (WT), TEFp-CIK1 (Cik1L), or TEFp-ΔATG-CIK1 (Cik1S) cells. Cells were either asynchronous or α factor arrested before cycloheximide addition. Cell cycle profiles are shown in Figure S2C. (D) Diagram of the CIK1 open reading frame. The grey box indicates the sequence that encodes the N-terminal 34 amino acids not included in the Cik1S protein. The relative position of the first (ATG 1) and second (ATG 2) ATG codons are indicated. GSP indicates the gene specific primer used for 5′ RACE in (E). Black and grey bars represent the qPCR products generated in (F). (E) Ethidium-bromide stained gel comparing 5′RACE products from α factor and nocodazole (noc) arrested cells. As a control, reactions were carried out with (+) and without (−) the TdT enzyme in the last step of the RACE protocol. The two products are approximately 250 and 600 base pairs as indicated. (F) Comparison of internal and external RT-qPCR products (shown in (D)) from α factor- and nocodazole- (noc) arrested cells. qPCR reactions were done in triplicate in two separate experiments and all 6 values were averaged. Error bars represent standard deviations.
These observations led us to ask how Cdh1-mediated turnover of Cik1 was blocked in α factor-treated cells. Similar to previous reports (Barrett et al., 2000; Manning et al., 1999; Page and Snyder, 1992), we noted that Cik1 displayed a slight increase in mobility upon α factor treatment (Figure 2A,B), which could have been the result of a post-translational modification. However, an alternate explanation was suggested when we examined the levels and stability of Cik1 in G1 cells that expressed Cik1 from the TEF1 promoter (TEFp), which is not regulated by α factor. In contrast to what we observed in wild type cells, levels of Cik1 in TEFp-CIK1 cells were low in α factor-treated cells (Figure 2A, right side) and Cik1 expressed from the TEFp did not undergo a mobility shift following α factor treatment (Figure 2A,B), arguing against an α factor specific post-translational modification. Moreover, Cik1 expressed from the TEFp was turned over rapidly in α factor-treated cells (Figure 2B, right side). Unlike half-life experiments carried out in asynchronous cells, some of which are in a phase of the cell cycle with low APC/CCdh1 activity (Figure 1A, S1B), examination of TEFp-Cik1 in G1-arrested cells revealed very rapid Cik1 turnover.
Since the only difference between the wild type and TEFp-CIK1 strains was the promoter driving CIK1 expression, we hypothesized that α factor induced CIK1 transcription produces an alternate Cik1 protein. Upon examination of the Cik1 amino acid sequence, we noted a second methionine located 35 amino acids downstream of the first, which might serve as an alternative translational initiation site from an α factor induced transcript. To test whether translation could initiate from this internal methionine, we reconstructed the TEFp-CIK1 strain and deleted the first ATG codon of the CIK1 open reading frame upon knocking in the TEFp sequence. Confirming our hypothesis, this ATG knock out strain (hereafter Cik1S, for short isoform) expressed a stable Cik1 protein that appeared to be the same molecular weight as the protein expressed in wild type cells treated with α factor (Figure 2C). This protein was smaller than the protein produced from the TEFp-CIK1 strain that retained the first ATG codon (hereafter Cik1L, for long isoform). Importantly, in α factor-treated cells, which have active APC/CCdh1, wild type Cik1 and Cik1S were stable, but Cik1L was highly unstable (Figure 2C).
One possibility suggested by these data is that CIK1 transcription initiates downstream of the first ATG codon following α factor treatment, so that translation must initiate at the second ATG. To address this possibility, RNA was isolated from cells arrested with α factor or nocodazole (which arrests cells in mitosis, the cell cycle phase when CIK1 is transcribed) (Figure S2D) and transcription start sites were mapped using 5′RACE (Figure 2D,E). As expected, there was a greater amount of CIK1 RNA in α factor-treated compared to nocodazole-treated cells (Figure 2E). In addition, the 5′ end of the CIK1 transcript in α factor-treated cells was approximately 350 base pairs shorter than the transcript found in nocodazole-treated cells. Cloning and sequencing of the 5′ RACE products revealed that in nocodazole-treated cells, CIK1 transcription initiated between 280 and 295 base pairs upstream of the first ATG codon (at base pairs −280, −285, −292, and −295 relative to the first ATG). In contrast, 5 of 6 clones from the α factor sample began downstream of the first ATG codon (at base pairs +75, +72, +41, +40, +37 and −21), confirming the hypothesis that most CIK1 transcription in α factor excludes the first ATG codon.
To quantify the relative amount of product from each transcriptional initiation site, RT-qPCR was also performed on the RNA samples from α factor- and nocodazole-treated cells. A common reverse primer, downstream of the second CIK1 ATG, was paired with each of two upstream primers: one primer that spanned the first ATG and would not be included in the α factor transcript (producing the external product, grey, Figure 2D,F), and a second primer that was downstream of the second ATG (producing the internal product, black, Figure 2D,F). In nocodazole-treated cells, a similar amount of CIK1 transcript was detected with both primer sets. However, in α factor-treated cells approximately 5 times more mRNA was detected using the internal primer set than the external primer set (Figure 2F), confirming the 5′RACE results. These data argue that mating-induced transcription produces a CIK1 mRNA that excludes the first ATG codon and that translation initiates downstream, producing a Cik1 protein that lacks the first 34 amino acids.
Kar3 enhances Cik1 turnover in vivo
During vegetative growth, Cik1 localizes to the nuclear face of the SPB and regulates the mitotic spindle (Page et al., 1994; Page and Snyder, 1992). However, when cells are exposed to α factor, Cik1 relocalizes to the cytoplasmic face of the SPB and the microtubules that extend into the shmoo tip. Our identification of two Cik1 isoforms suggests a mechanism by which this change in localization might be mediated. The N-terminal sequence that is specific to the mitotic Cik1L protein contains a putative NLS (Manning et al., 1999) that is absent from the Cik1S protein, which might explain the difference in localization. To examine this, we used our strains expressing individual GFP-tagged Cik1 isoforms to analyze their localization during vegetative growth.
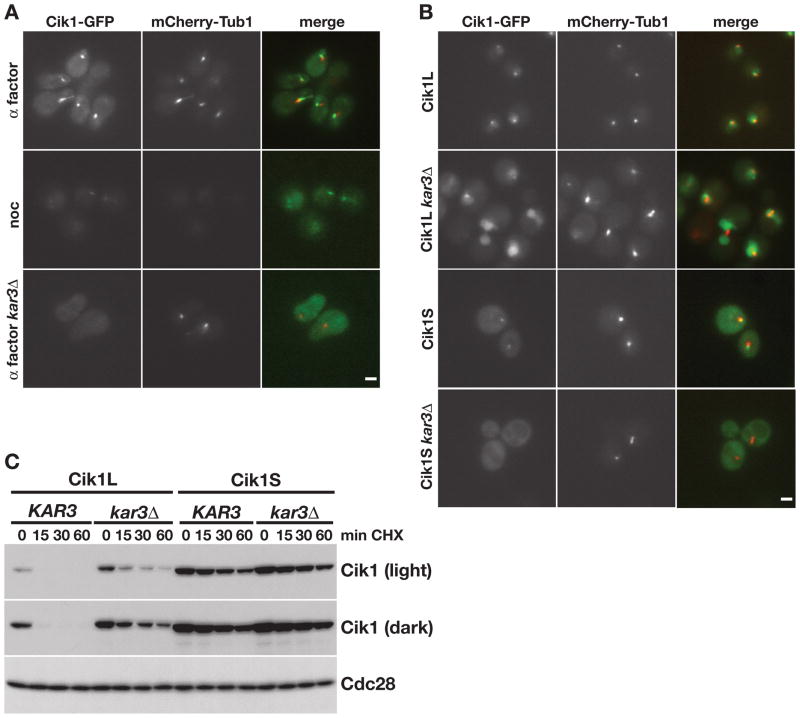
First, to confirm the localization of Cik1-GFP expressed from its endogenous promoter, we examined its localization in cells co-expressing mCherry-tagged tubulin. Consistent with previous reports (Page et al., 1994; Page and Snyder, 1992), in α factor-treated cells Cik1 was observed at the SPB, on microtubules extending into the shmoo tip, and in the cytoplasm (Figure 3A, upper panels). Levels of Cik1 were very low during vegetative growth, as previously noted (Page and Snyder, 1992), although faint SPB staining could also be observed when cells were arrested with nocodazole (Figure 3A, middle panels). In addition, the SPB and microtubule association in α factor-treated cells was lost in cells lacking Kar3 (Figure 3A, lower panels), which is required for Cik1 association with microtubules (Barrett et al., 2000; Page et al., 1994).
Figure 3. Kar3 regulates the localization and stability of Cik1.
(A) Cik1-GFP and mCherry-Tub1 expression in live cells arrested for 3 hours with α factor or nocodazole (noc), as indicated. GFP, mCherry, and merged images are shown. Scale bar represents 2μm. (B) Localization of GFP-tagged Cik1 isoforms and mCherry-Tub1 in wild type and kar3Δ strains. GFP, mCherry, and merged images are show. (C) Cycloheximide-chase assay of Cik1L (TEFp-CIK1) and Cik1S (TEFp-ΔATG-CIK1) in KAR3 and kar3Δ strains. Strains were arrested in α factor for 3 hours and cycloheximide was added for the indicated number of minutes. Two exposures of the GFP blot are shown. Cell cycle profiles are shown in Figure S3.
We next analyzed the localization of the two Cik1 isoforms, Cik1L and Cik1S. As predicted, the Cik1L isoform that includes the putative NLS was observed in the nucleus and at the SPB (Figure 3B, Cik1L). Upon deletion of KAR3, the SPB association was lost and Cik1L was diffuse in the nucleus (Cik1L kar3Δ). The Cik1S isoform was also observed at the SPB (Cik1S). However, in cells lacking KAR3, Cik1S protein was cytoplasmic (Cik1S kar3Δ), confirming that this isoform normally localizes to the cytoplasmic face of the SPB. These data suggest that Cik1 localizes to the nucleus during mitotic growth, but not during mating, because only the mitotic isoform contains the NLS required for nuclear localization.
The APC/C targets several proteins that regulate and localize to the mitotic spindle (Gordon and Roof, 2001; Hildebrandt and Hoyt, 2001; Juang et al., 1997; Woodbury and Morgan, 2007). In addition, the APC/C itself localizes to the mitotic spindle in yeast and mammals, as well as to centrosomes in mammalian cells (Kraft et al., 2003; Melloy and Holloway, 2004; Tugendreich et al., 1995). Since Kar3 is required for Cik1 microtubule and SPB localization, we tested whether deletion of KAR3 affected Cik1 turnover in vivo. Cik1L was partially stabilized in kar3Δ cells (Figure 3C), suggesting that localization to the SPB and/or microtubules makes Cik1L a better target for the APC/C. Consistent with the fact that Cik1S is stable, its turnover was unchanged in kar3Δ cells.
Cik1L is a direct target of APC/CCdh1
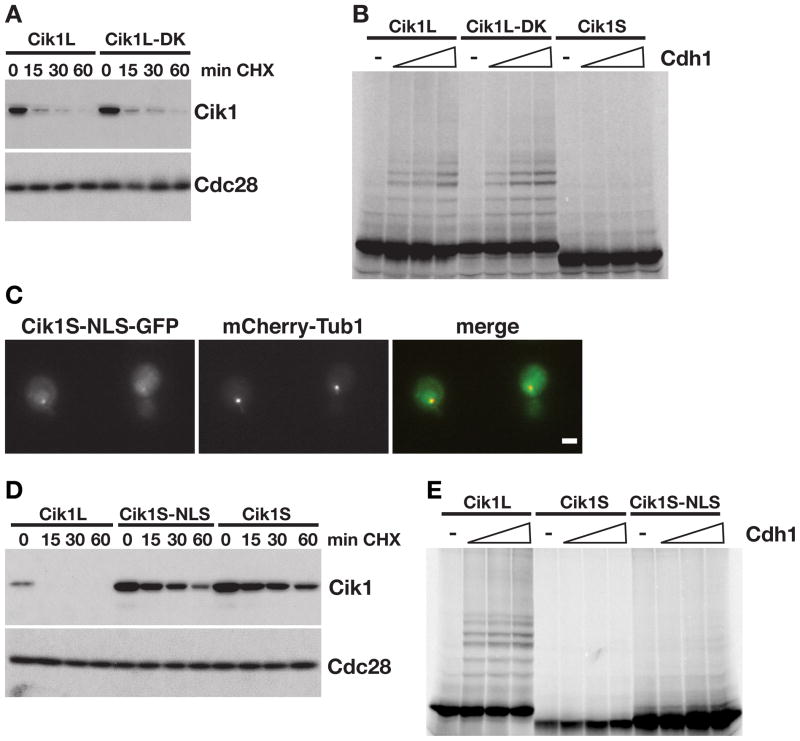
Many APC/C substrates contain recognizable destruction sequences termed D-boxes (RxxLxxxxN, where x is any amino acid) (Glotzer et al., 1991) and KEN boxes (KENxxxN/D) (Pfleger and Kirschner, 2000). No discernable D-box or KEN box motifs could be identified in the N-terminal 34 amino acids of Cik1L, which are required for APC/C-mediated turnover in vivo. However, 3 partial D-boxes (RxxL, starting at amino acids 259, 364, and 416) and one partial KEN-box (KEN at amino acid 284) were identified in the coiled-coil and motor homology domains of the protein. Consistent with our observation that the N-terminus of Cik1L mediates turnover, mutation of all 4 of these sequences in the Cik1L protein (Cik1L-DK) did not alter Cik1L stability (Figure 4A).
Figure 4. Cik1L is a direct target of APC/CCdh1.
(A) Cycloheximide-chase assay showing levels of Cik1 in Cik1L (TEFp-CIK1) and Cik1L-DK (TEFp-CIK1-DK) cells after addition of cycloheximide for the indicated times (min CHX). Cell cycle profiles are shown in Figure S4A. (B) Cik1L, Cik1L-DK and Cik1S were translated in vitro and labeled with 35S-methionine. Equivalent amounts were incubated with purified E1, E2, ATP, ubiquitin, and APC/C in the absence (−) or presence of Cdh1 (100, 200, or 400nM). Reactions were analyzed by SDS-PAGE and visualized with a phosphorimager, quantitation is shown in Figure S4B. (C) Cik1S-NLS-GFP and mCherry-Tub1 expression. GFP, mCherry and merged images are shown. Scale bar represents 2μm. (D) Cycloheximide-chase assay showing levels of Cik1 in Cik1L (TEFp-CIK1), Cik1S-NLS (TEFp-ΔATG-CIK1-NLS), and Cik1S (TEFp-ΔATG-CIK1) cells after α factor arrest followed by the addition of cycloheximide (min CHX). Cell cycle profiles are shown in Figure S4C. (E) In vitro ubiquitination of Cik1L, Cik1S and Cik1S-NLS by the APC/C as described in (B), except that 50, 150, or 300nM Cdh1 was added. Quantitation is shown in Figure S4D.
Since mutation of the discernable APC/C-recognition motifs did not affect Cik1L turnover in vivo, we next confirmed that Cik1L is a direct target of APC/CCdh1. Cik1L, Cik1L-DK, and Cik1S were translated in vitro and tested in APC/CCdh1 ubiquitination reactions. Consistent with the in vivo data, both Cik1L and the Cik1L-DK mutant were ubiquitinated by APC/CCdh1 in vitro, however Cik1S ubiquitination by APC/CCdh1 was not detectable (Figure 4B). This result suggests that a unique destruction sequence in the N-terminus of Cik1L mediates APC/C-dependent turnover. Moreover, these data argue that the alternate localization of the Cik1S and Cik1L isoforms in the cell cannot explain their stability differences. To confirm that the N-terminus of Cik1L provides a sequence required for destruction, we fused the exogenous SV40 NLS to the C-terminus of the Cik1S protein to restore its nuclear localization (Figure 4C). Targeting Cik1S to the nucleus did not restore its turnover in vivo (Figure 4D). Moreover, addition of the SV40 NLS did not restore ubiquitination of Cik1S by the APC/C in vitro (Figure 4E). Therefore, we conclude that the N-terminus of Cik1L both confers nuclear localization and includes a sequence that is necessary for APC/C-mediated ubiquitination.
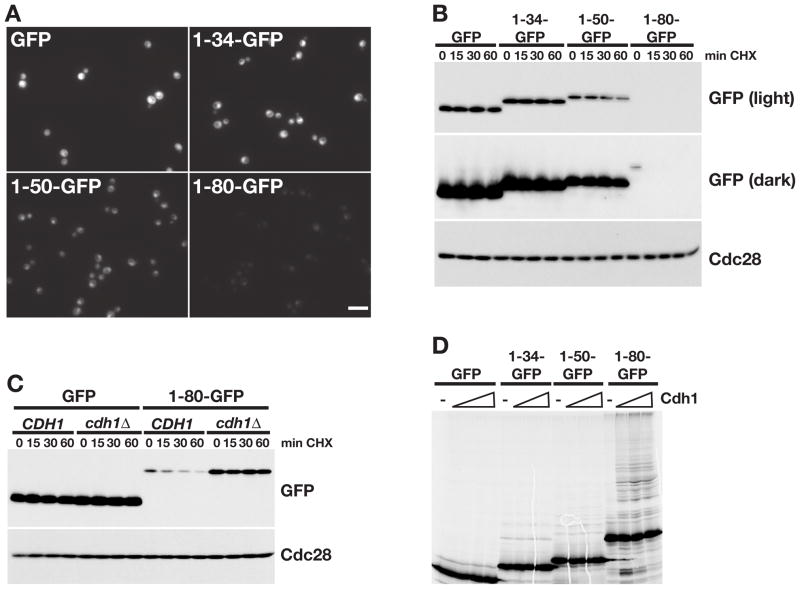
We next tested whether the N-terminus of Cik1L is sufficient to target an unrelated protein for APC/C-mediated destruction. Three segments of the Cik1L N-terminus (34, 50, or 80 amino acids) were fused to GFP and levels examined in vivo (Figure 5A). Although all fusion proteins targeted GFP to the nucleus, only the 80 amino acid sequence (1-80-GFP) destabilized GFP (Figure 5A,B). The 1-80-GFP protein was stable in cdh1Δ cells, confirming that its turnover was Cdh1-dependent. Moreover, only 1-80-GFP was ubiquitinated by APC/CCdh1 in vitro (Figure 5D), demonstrating that the N terminus of Cik1L acts as a transferrable degron.
Figure 5. The N-terminus of Cik1L is an APC/C degron.
(A) GFP expression in cells expressing GFP or Cik1-GFP fusions (1-34-GFP, 1-50-GFP, 1-80-GFP) from the TEF promoter. Scale bar represents 10μm. (B) Cycloheximide chase assay of the indicated GFP proteins. Cells from (A) were arrested with α factor and treated with cycloheximide for the indicated number of minutes (min CHX). Proteins were analyzed by GFP Western blot (light and dark exposures are shown). Cdc28 is shown as a loading control. Cell cycle profiles are shown in Figure S5A. (C) Cycloheximide-chase assay of GFP and 1-80-GFP in asynchronous CDH1 and cdh1Δ cells. Cell cycle profiles are shown in Figure S5B. (D) GFP and Cik1-GFP fusion proteins were translated in vitro and equivalent amounts were incubated with purified E1, E2, ATP, ubiquitin, and APC/C in the absence (−) or presence of Cdh1. Reactions were analyzed by SDS-PAGE and visualized with a phosphorimager, quantitation is shown in Figure S5C.
Cik1L and Cik1S have separable functions
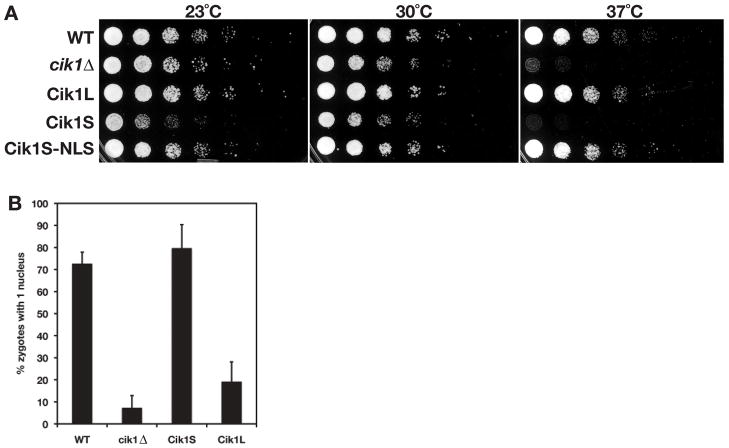
During vegetative growth, Cik1 is required for accurate chromosome segregation. Cells lacking Cik1 have mitotic defects that render cells sensitive to growth at high temperature and cause cells to lose chromosomes (Page and Snyder, 1992). Since Cik1S localizes to the cytoplasm and not the mitotic spindle, we sought to determine if cells that express only the Cik1S isoform have mitotic defects, similar to those of cik1Δ cells. First, we compared growth of wild type (WT), cik1Δ Cik1L, Cik1S, and Cik1S-NLS cells at different temperatures. Similar to cik1Δ cells, Cik1S cells grew poorly at 37°C (Figure 6A), however this sensitivity was reversed upon addition of the exogenous NLS to the C-terminus of Cik1S. Cells expressing only the mitotic Cik1L isoform grew as well as wild type cells at all temperatures. Next, the frequency of chromosome loss was calculated in strains carrying an extra copy of chromosome VII (Sandell and Zakian, 1993). Like cik1Δ cells, Cik1S cells lost chromosomes at an increased frequency compared to wild type and Cik1L cells (Table 1). These data confirm that the mating specific Cik1S isoform cannot compensate for loss of the Cik1L isoform during vegetative growth.
Figure 6. Cik1L and Cik1S have separable functions.
(A) Cik1S cells are temperature sensitive, similar to cik1Δ cells. 5-fold dilutions of strains expressing the indicated Cik1 proteins were plated and incubated at 23°C, 30°C, or 37°C for 2 days (23°C) or 1 day (30 and 37°C). (B) MATa and MATα strains of the same genotype were mated and the number of zygotes with 1 or >1 nuclei were counted. Shown is the average of 3 or 4 experiments. Error bars represent standard deviations.
Table 1.
Cik1L is required for accurate chromosome segregation.
| frequency chromosome loss/cella | standard deviation | |
|---|---|---|
| WT | 4.9E-04 (1) | 3.8E-04 |
| cik1Δ | 4.0E-02 (82) | 1.8E-02 |
| Cik1S | 5.8E-02 (118) | 4.5E-02 |
| Cik1L | 1.2E-03 (2.4) | 1.7E-03 |
Average of 3 experiments is shown, fold change compared to wild type (WT) is indicated in parentheses.
Following formation of a zygote, cik1Δ cells are unable to undergo karyogamy (nuclear fusion). To determine if cells expressing the mitotic Cik1L isoform could carry out this function in the absence of the Cik1S protein, we measured karyogamy in bilateral matings (i.e. where both mating partners have the same genotypes outside of the mating locus). Karyogamy was disrupted in Cik1L cells, similar to cik1Δ cells, whereas karyogamy in Cik1S cells was similar to wild type cells (Figure 6B). These data confirm that Cik1L and Cik1S have distinct functions, in mitosis and mating respectively, and cannot compensate for loss of one another.
Discussion
Here, we present evidence that there are two developmentally regulated and functionally distinct isoforms of the kinesin-associated protein Cik1. During vegetative growth, CIK1 is transcribed in a cell cycle-regulated manner, similar to the B-type cyclin Clb2 (Pramila et al., 2006; Spellman et al., 1998), leading to the expression of Cik1L. Cik1L binds to and targets the kinesin Kar3 to SPBs, regulates mitotic spindle dynamics, and is required for accurate chromosome segregation. At the end of mitosis, Cik1L is ubiquitinated by APC/CCdh1, which targets it for proteasomal degradation. The synthesis and destruction of Cik1L are similar to those of other APC/CCdh1 targets, such as Clb2, and are likely important for proper assembly and disassembly of the mitotic spindle during the cell cycle.
Unlike other APC/CCdh1 targets, Cik1 also has an essential function when cells mate and undergo karyogamy. This presents a problem for the cell because in order to carry out this karyogamy function, Cik1 must be expressed at high levels when cells arrest in G1 in response to mating pheromone, a time when APC/CCdh1 remains active. Our data suggests that Cik1 regulation has evolved such that Cik1 is resistant to APC/C-mediated degradation during mating. In response to α factor treatment, the transcription factors Kar4 and Ste12 bind to the CIK1 promoter and induce high levels of transcription (Kurihara et al., 1996). We have shown that α factor induced transcription initiates downstream of the first ATG codon of the CIK1 open reading frame, forcing translation of CIK1 to initiate at a second ATG codon and generating a short isoform (Cik1S) that lacks the N-terminal 34 amino acids of Cik1L. The N-terminal sequence that is missing in the Cik1S protein contains an NLS, and this explains how Cik1S localizes to the cytoplasmic face of the SPB and to cytoplasmic microtubules (instead of the nucleus) during mating. We also show that this N-terminal sequence in Cik1L is required for APC/C-mediated ubiquitination. Cik1S protein lacks this sequence and is therefore resistant to APC/C-mediated turnover during α factor arrest. This coupling of transcription to translation and proteolysis represents an uncharacterized mode of regulating ubiquitination by the APC/C.
Importantly, we find that the two Cik1 isoforms have distinct functions. The mitotic Cik1L protein is required for growth at high temperatures, and is important for accurate chromosome segregation. While cells expressing exclusively the Cik1L protein are similar to wild type cells, cells that express only the Cik1S isoform are temperature sensitive and lose chromosomes more frequently (Figure 6A, Table 1). Conversely, the Cik1L isoform cannot carry out the essential karyogamy function of Cik1S. Cik1L cells have defects in nuclear fusion during mating, similar to cik1Δ cells (Figure 6B).
Previous reports have demonstrated that Cik1 binds to Kar3 both during vegetative growth and in α factor (Barrett et al., 2000; Page et al., 1994), and that the N-terminus of Cik1 is dispensable for the Cik1-Kar3 interaction (Barrett et al., 2000). In addition, our data shows that the SPB and microtubule association of both Cik1L and Cik1S are lost in kar3Δ cells (Figure 4B). Together, these data suggest that both Cik1 isoforms form functional complexes with Kar3. Therefore, it is likely that the separation of function of Cik1L and Cik1S are due to the differential localization of the two forms. This is supported by the fact that restoring nuclear localization of Cik1S reverses the temperature sensitivity of Cik1S cells (Figure 6A). Therefore, Cik1S cannot compensate for Cik1L function because it does not localize to the nucleus, and cannot associate with the mitotic spindle. Conversely, Cik1L cannot function in karyogamy because it is not in the cytoplasm (and is destroyed in α factor-treated cells).
Interestingly, analogous regulation of transcriptional and translational initiation in response to α factor has also been described for the KAR4 gene (Gammie et al., 1999). Kar4 is a transcription factor that is required for transcriptional induction of CIK1 and KAR3 in response to α factor (Kurihara et al., 1996). Like CIK1, in α factor KAR4 transcription initiates downstream of the first ATG and translation begins at an internal methionine. This suggests that this type of regulation may be common within the mating pathway. However, there are some notable differences between the regulation of Cik1 and Kar4 isoforms. In the case of Kar4, the extra N-terminal sequence in the longer, mitotic isoform does not include a regulatable destruction motif. Also unlike Cik1, both Kar4 isoforms are thought to function in the same way when expressed at similar levels. Internal translational start sites resulting from alternate transcriptional initiation have also been observed for other yeast genes. In several cases, such as HTS1, VAS1, TRM1, MOD5 and SUC2 (Danpure, 1995), longer isoforms contain targeting sequences that are missing from their shorter isoforms. Therefore, alternative translational initiation appears to be a common mechanism for producing differentially localized isoforms.
The N-terminal domain of Cik1L is required for both APC/CCdh1- mediated ubiquitination and nuclear localization. Since the Cik1S protein that lacks the 34 N-terminal amino acids is not a substrate for APC/CCdh1 in vitro (Figure 4B), this demonstrates that these residues are necessary for the targeting of Cik1 by APC/CCdh1. However, these 34 amino acids are not sufficient to confer APC/C-mediated ubiquitination of an unrelated protein. Instead, 80 amino acids of Cik1L are required to confer APC/C-targeting. This domain does not contain any canonical APC/C recognition motifs, such as D-boxes or KEN boxes, and therefore represents a new type of recognition motif. Although localization is not a factor in ubiquitination reactions in vitro, nuclear localization may contribute to Cik1L turnover in vivo. Cdh1 is thought to associate with the core APC/C and be active in the nucleus (Jaquenoud et al., 2002; Zhou et al., 2003). However, Cdh1 does target the non-nuclear protein Hsl1 (Burton and Solomon, 2000), suggesting that it may act in the cytoplasm as well. It is possible that removal of a destruction motif, coupled with cytoplasmic localization, may both play a part in protecting Cik1S from APC/CCdh1-mediated turnover.
Our findings also suggest that localization of Cik1L to the SPB may help facilitate its APC/C-mediated turnover. In kar3Δ cells, Cik1L is nuclear but it is not observed at SPBs (Figure 3B). Kar3 cannot be essential for ubiquitination of Cik1L by the APC/C, since it is not present in in vitro reactions. Yet, Cik1L is partially stabilized in the absence of KAR3 in vivo (Figure 3C). One possibility is that the APC/C coordinates turnover of mitotic spindle regulators, such as Ase1, Fin1, Kip1 and Cin8 (Gordon and Roof, 2001; Hildebrandt and Hoyt, 2001; Juang et al., 1997; Woodbury and Morgan, 2007), by interacting directly with the mitotic spindle and/or SPBs. This model is supported by evidence that several APC/C subunits localize to the spindle in yeast (Melloy and Holloway, 2004), flies (Huang and Raff, 2002), and mammalian cells (Kraft et al., 2003; Tugendreich et al., 1995; Zhou et al., 2003). The APC/C may target several proteins at the mitotic spindle and thereby help drive spindle breakdown after mitosis. It has been difficult to test this model since only minor defects in spindle breakdown have been observed in budding yeast strains that carry non-destructable alleles of some Cdh1 targets, such as Cin8 and Fin1 (Hildebrandt and Hoyt, 2001; Woodbury and Morgan, 2007). One possibility is that stabilization of many components simultaneously may be required to observe a strong phenotype. Here, we show that expression of the stable Cik1S protein negatively affects cell growth and delays cells in mitosis (Figure 5A, Figure S2C). However, this phenotype is likely the result of the localization of Cik1S to the cytoplasm, since restoring nuclear localization reverses the temperature sensitivity of Cik1S strains. Similar to what has been observed in cells expressing stable Cin8 or Fin1, we have not detected any significant mitotic defects in cells expressing the stable Cik1S-NLS protein.
Substrate targeting by the APC/C was initially thought to be regulated through the control of Cdc20 and Cdh1. These adaptors are regulated both by their association with the APC/C, and also by protein inhibitors (Bharadwaj and Yu, 2004; Martinez et al., 2006; Reimann et al., 2001). However, recent data suggests that the interaction between substrates and the APC/C is also regulated. Phosphorylation blocks the association of yeast securin (Agarwal et al., 2003; Holt et al., 2008), vertebrate Aurora A (Littlepage and Ruderman, 2002), and Cdc6 (Mailand and Diffley, 2005) with the APC/C. We have now identified a new form of regulation of APC/C-mediated proteolysis in response to environmental cues: the coupling of transcriptional initiation to the synthesis of an APC/C destruction motif. While the scope of this mechanism of substrate regulation is currently unknown, the transcriptional induction of many yeast genes following exposure to mating phermone or nitrogen starvation results in alternate transcriptional and translational start site usage, suggesting it may be widespread (Gammie et al., 1999; Law et al., 2005). Moreover, a similar mechanism of APC/C substrate regulation may occur in multicellular eukaryotes, where alternative splicing may produce similar results. For example, alternative splicing produces an isoform of the Nek2 protein, Nek2B, which lacks the APC/C-targeting domains found in Nek2A (Hames et al., 2001). The inclusion or exclusion of a degron motif may be a common form of regulating APC/C-mediated proteolysis.
Experimental Procedures
Strains and plasmids
A complete strains list is provided in Supplemental Table S1. Details of strain and plasmid construction are described in the Supplemental Data.
Microscopy
Images of GFP fusion proteins and mCherry-tubulin were obtained on a Leica DMRXA fluorescence microscope using a 100X 1.4NA PlanApo oil immersion objective. Images were taken with a Hamamatsu C4742-95 CCD camera. Data was analyzed using OpenLab software from Improvision. Black and white images were all captured for the same exposure times and contrast enhancement was performed equivalently on all panels. For merged color panels, contrast was optimized for each panel.
Half-life analysis and Western blotting
For cycloheximide-chase assays cells were grown to mid-log phase and treated with 50μg/ml cycloheximide. Samples were collected at increasing time points for Western blot and fixed for flow cytometry. In the indicated experiments, mid-log phase cells were first arrested with 10μg/ml α factor or 10μg/ml nocodazole prior for 2–3 hours before addition of cycloheximide. Flow cytometry to examine cell cycle profile was carried out as described (Benanti et al., 2007). In all experiments multiple exposures of the Western blots were compared to confirm half-life differences. Samples for Western blots were prepared as described (Benanti et al., 2007) and performed with antibodies against GFP (clone JL8, BD Biosciences), FLAG (clone M2, Sigma), Clb2 (provided by D. Kellogg), HA (clone 16B12, Covance), Cdc53 (yC-17, Santa Cruz Biotechnology) and Cdc28 (yC-20, Santa Cruz Biotechnology).
For the experiment shown in Figure 1C, Cdh1-m11 was induced by shifting cells from rich media containing 2% dextrose to rich media containing 2% galactose for 3 hours in the APC1 strain, and for 5 hours in apc1Δ strain. Since these strains have a significant difference in doubling time longer inductions are necessary in apc1Δ for equivalent induction of the GAL promoter. For the half-life analysis shown in Figure 1D, cells were grown for 20 hours in 2% dextrose (Cdh1-m11 off) or 2% galactose (Cdh1-m11 on) before the addition of cycloheximide.
RNA analysis
RNA was purified from CIK1-GFP cells that were arrested in α factor or nocodazole for 3 hours. Expression of the two isoforms was confirmed by GFP Western blot and cell cycle position was confirmed by flow cytometry (Figure S2D). 5′ RACE was carried out with the 5′ RACE system from Invitrogen, per kit instructions, except that a Qiagen Qiaquick column was used for purification of cDNA. A CIK1 primer located just downstream of the second ATG codon (CCCTTTCAATAATCTTGGGGT) was used for the reverse transcriptase reaction (Figure 2D). For final PCR analysis the Abridged Anchor Primer provided with the kit was used in combination with a nested CIK1 primer (GTCCATGGGTTTCTTAGTATACGTAGCGGAG). 5′ RACE products were cloned into the pGEMT-Easy vector (Promega) and sequenced. RT-qPCR analysis was carried out as described (Benanti et al., 2007). A common CIK1 reverse primer (CCCTTTCAATAATCTTGGGGT) was used with either an external forward primer (CATAAGCTTTATTATTAGGATGAATAACTCC), or an internal forward primer (CTAACTCCTAATAACAACAAG) to generate the products diagramed in Figure 2D.
In vitro ubiquitination
Ubiquitination assays contain E1, E2, ubiquitin, ATP, Cdh1, APC/C and substrate. An equal mix of purified Ubc4-6xHis and Ubc1-6xHis was used as E2, 6xHis-Cdh1 was purified from Sf9 cells, and APC/C was purified from yeast via a TAP-tag on Cdc16. Cik1 and GFP substrates were produced by coupled in vitro transcription and translation in rabbit reticulocyte extracts, as described by the manufacturer (Promega). All purifications and reactions were performed as previously described (Carroll and Morgan, 2005). Reactions were subjected to SDS-PAGE and visualized using a phosphorimager. To quantitation ubiquitinated products, all bands larger than the unmodified substrate, in each lane, were quantitated using Imagequant software. Quantitation of the lanes from reacations lacking Cdh1 were subtracted from those lanes from reactions containing Cdh1, to account for background signal.
Chromosome loss assay
Chromosome loss assays were performed as in Galgoczy and Toczyski (2001), Table 1 (Galgoczy and Toczyski, 2001). YDPT180-3 (cik1Δ), YDPT182-1 (Cik1S), and YDPT183-1 (Cik1L) were derived from 18–20 and maintained at 23°C on 2% glucose SD lacking tyrosine and lysine, to maintain selection for both copies of chromosome VII. Cells were sonicated and plated onto rich plates or rich plates containing cycloheximide. Chromosome loss events were scored at white Ura− Lys− colonies. Frequencies of chromosome loss per cell were calculated for each strain in three independent experiments and averaged together.
Karyogamy assay
Matings were carried out by mixing equal numbers of mid-log phase MATa and MATα strains of the same genotype, pelleting the cells, and incubating at 30°C. After 4 hours, cells were fixed in 70% ethanol and resuspended in mounting medium containing DAPI. Cells were imaged and zygotes containing 1 or >1 nuclei were counted. At least 100 zygotes/experiment were counted for wild type and Cik1L matings. At least 50 zygotes/experiment were counted for cik1Δ and Cik1S matings. Cik1S and cik1Δ strains have a reduced mating efficiency so fewer zygotes are formed in each experiment. An average of 3 experiments is shown. Error bars represent standard deviations.
Supplementary Material
Acknowledgments
We thank Fred Schaufele and the UCSF Diabetes Center Microscopy Core for assistance with high-throughput microscopy; Steve Reed for providing the pRS406-mCherry-Tub1 plasmid; members of the Toczyski and Morgan labs for their continued support and suggestions; and Thomas Fazzio, Michael Downey, and Ian Foe for comments on the manuscript. M.E.M. was supported by an NSF predoctoral fellowship. High-throughput microscopy was supported in part by NIH grant P30DK06720. This work was funded by NIH grants T32CA108462 (J.A.B.), K99GM085013 (J.A.B.), GM053270 (D.O.M.), and GM70539 (D.P.T).
References
- Agarwal R, Tang Z, Yu H, Cohen-Fix O. Two distinct pathways for inhibiting pds1 ubiquitination in response to DNA damage. J Biol Chem. 2003;278:45027–45033. doi: 10.1074/jbc.M306783200. [DOI] [PubMed] [Google Scholar]
- Barrett JG, Manning BD, Snyder M. The Kar3p kinesin-related protein forms a novel heterodimeric structure with its associated protein Cik1p. Mol Biol Cell. 2000;11:2373–2385. doi: 10.1091/mbc.11.7.2373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benanti JA, Cheung SK, Brady MC, Toczyski DP. A proteomic screen reveals SCFGrr1 targets that regulate the glycolytic-gluconeogenic switch. Nat Cell Biol. 2007;9:1184–1191. doi: 10.1038/ncb1639. [DOI] [PubMed] [Google Scholar]
- Bharadwaj R, Yu H. The spindle checkpoint, aneuploidy, and cancer. Oncogene. 2004;23:2016–2027. doi: 10.1038/sj.onc.1207374. [DOI] [PubMed] [Google Scholar]
- Burton JL, Solomon MJ. Hsl1p, a Swe1p inhibitor, is degraded via the anaphase-promoting complex. Mol Cell Biol. 2000;20:4614–4625. doi: 10.1128/mcb.20.13.4614-4625.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll CW, Morgan DO. Enzymology of the anaphase-promoting complex. Methods Enzymol. 2005;398:219–230. doi: 10.1016/S0076-6879(05)98018-X. [DOI] [PubMed] [Google Scholar]
- Cohen-Fix O, Peters JM, Kirschner MW, Koshland D. Anaphase initiation in Saccharomyces cerevisiae is controlled by the APC-dependent degradation of the anaphase inhibitor Pds1p. Genes Dev. 1996;10:3081–3093. doi: 10.1101/gad.10.24.3081. [DOI] [PubMed] [Google Scholar]
- Danpure CJ. How can the products of a single gene be localized to more than one intracellular compartment? Trends Cell Biol. 1995;5:230–238. doi: 10.1016/s0962-8924(00)89016-9. [DOI] [PubMed] [Google Scholar]
- Endow SA, Kang SJ, Satterwhite LL, Rose MD, Skeen VP, Salmon ED. Yeast Kar3 is a minus-end microtubule motor protein that destabilizes microtubules preferentially at the minus ends. EMBO J. 1994;13:2708–2713. doi: 10.1002/j.1460-2075.1994.tb06561.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escote X, Zapater M, Clotet J, Posas F. Hog1 mediates cell-cycle arrest in G1 phase by the dual targeting of Sic1. Nat Cell Biol. 2004;6:997–1002. doi: 10.1038/ncb1174. [DOI] [PubMed] [Google Scholar]
- Galgoczy DJ, Toczyski DP. Checkpoint adaptation precedes spontaneous and damage-induced genomic instability in yeast. Mol Cell Biol. 2001;21:1710–1718. doi: 10.1128/MCB.21.5.1710-1718.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gammie AE, Stewart BG, Scott CF, Rose MD. The two forms of karyogamy transcription factor Kar4p are regulated by differential initiation of transcription, translation, and protein turnover. Mol Cell Biol. 1999;19:817–825. doi: 10.1128/mcb.19.1.817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glotzer M, Murray AW, Kirschner MW. Cyclin is degraded by the ubiquitin pathway. Nature. 1991;349:132–138. doi: 10.1038/349132a0. [DOI] [PubMed] [Google Scholar]
- Gordon DM, Roof DM. Degradation of the kinesin Kip1p at anaphase onset is mediated by the anaphase-promoting complex and Cdc20p. Proc Natl Acad Sci U S A. 2001;98:12515–12520. doi: 10.1073/pnas.231212498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hames RS, Wattam SL, Yamano H, Bacchieri R, Fry AM. APC/C-mediated destruction of the centrosomal kinase Nek2A occurs in early mitosis and depends upon a cyclin A-type D-box. EMBO J. 2001;20:7117–7127. doi: 10.1093/emboj/20.24.7117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA. Mitotic motors in Saccharomyces cerevisiae. Biochim Biophys Acta. 2000;1496:99–116. doi: 10.1016/s0167-4889(00)00012-4. [DOI] [PubMed] [Google Scholar]
- Hildebrandt ER, Hoyt MA. Cell cycle-dependent degradation of the Saccharomyces cerevisiae spindle motor Cin8p requires APC(Cdh1) and a bipartite destruction sequence. Mol Biol Cell. 2001;12:3402–3416. doi: 10.1091/mbc.12.11.3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt LJ, Krutchinsky AN, Morgan DO. Positive feedback sharpens the anaphase switch. Nature. 2008;454:353–357. doi: 10.1038/nature07050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang JY, Raff JW. The dynamic localisation of the Drosophila APC/C: evidence for the existence of multiple complexes that perform distinct functions and are differentially localised. J Cell Sci. 2002;115:2847–2856. doi: 10.1242/jcs.115.14.2847. [DOI] [PubMed] [Google Scholar]
- Jaquenoud M, van Drogen F, Peter M. Cell cycle-dependent nuclear export of Cdh1p may contribute to the inactivation of APC/C(Cdh1) EMBO J. 2002;21:6515–6526. doi: 10.1093/emboj/cdf634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jaspersen SL, Charles JF, Morgan DO. Inhibitory phosphorylation of the APC regulator Hct1 is controlled by the kinase Cdc28 and the phosphatase Cdc14. Curr Biol. 1999;9:227–236. doi: 10.1016/s0960-9822(99)80111-0. [DOI] [PubMed] [Google Scholar]
- Juang YL, Huang J, Peters JM, McLaughlin ME, Tai CY, Pellman D. APC-mediated proteolysis of Ase1 and the morphogenesis of the mitotic spindle. Science. 1997;275:1311–1314. doi: 10.1126/science.275.5304.1311. [DOI] [PubMed] [Google Scholar]
- Kraft C, Herzog F, Gieffers C, Mechtler K, Hagting A, Pines J, Peters JM. Mitotic regulation of the human anaphase-promoting complex by phosphorylation. Embo J. 2003;22:6598–6609. doi: 10.1093/emboj/cdg627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurihara LJ, Stewart BG, Gammie AE, Rose MD. Kar4p, a karyogamy-specific component of the yeast pheromone response pathway. Mol Cell Biol. 1996;16:3990–4002. doi: 10.1128/mcb.16.8.3990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Law GL, Bickel KS, MacKay VL, Morris DR. The undertranslated transcriptome reveals widespread translational silencing by alternative 5′ transcript leaders. Genome Biol. 2005;6:R111. doi: 10.1186/gb-2005-6-13-r111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Littlepage LE, Ruderman JV. Identification of a new APC/C recognition domain, the A box, which is required for the Cdh1-dependent destruction of the kinase Aurora-A during mitotic exit. Genes Dev. 2002;16:2274–2285. doi: 10.1101/gad.1007302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox PS, Bloom KS, Salmon ED. The polarity and dynamics of microtubule assembly in the budding yeast Saccharomyces cerevisiae. Nat Cell Biol. 2000;2:36–41. doi: 10.1038/71357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–926. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- Manning BD, Barrett JG, Wallace JA, Granok H, Snyder M. Differential regulation of the Kar3p kinesin-related protein by two associated proteins, Cik1p and Vik1p. J Cell Biol. 1999;144:1219–1233. doi: 10.1083/jcb.144.6.1219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez JS, Jeong DE, Choi E, Billings BM, Hall MC. Acm1 is a negative regulator of the CDH1-dependent anaphase-promoting complex/cyclosome in budding yeast. Mol Cell Biol. 2006;26:9162–9176. doi: 10.1128/MCB.00603-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melloy PG, Holloway SL. Changes in the localization of the Saccharomyces cerevisiae anaphase-promoting complex upon microtubule depolymerization and spindle checkpoint activation. Genetics. 2004;167:1079–1094. doi: 10.1534/genetics.103.025478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meluh PB, Rose MD. KAR3, a kinesin-related gene required for yeast nuclear fusion. Cell. 1990;60:1029–1041. doi: 10.1016/0092-8674(90)90351-e. [DOI] [PubMed] [Google Scholar]
- Page BD, Satterwhite LL, Rose MD, Snyder M. Localization of the Kar3 kinesin heavy chain-related protein requires the Cik1 interacting protein. J Cell Biol. 1994;124:507–519. doi: 10.1083/jcb.124.4.507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page BD, Snyder M. CIK1: a developmentally regulated spindle pole body-associated protein important for microtubule functions in Saccharomyces cerevisiae. Genes Dev. 1992;6:1414–1429. doi: 10.1101/gad.6.8.1414. [DOI] [PubMed] [Google Scholar]
- Peters JM. The anaphase promoting complex/cyclosome: a machine designed to destroy. Nat Rev Mol Cell Biol. 2006;7:644–656. doi: 10.1038/nrm1988. [DOI] [PubMed] [Google Scholar]
- Pfleger CM, Kirschner MW. The KEN box: an APC recognition signal distinct from the D box targeted by Cdh1. Genes Dev. 2000;14:655–665. [PMC free article] [PubMed] [Google Scholar]
- Pramila T, Wu W, Miles S, Noble WS, Breeden LL. The Forkhead transcription factor Hcm1 regulates chromosome segregation genes and fills the S-phase gap in the transcriptional circuitry of the cell cycle. Genes Dev. 2006;20:2266–2278. doi: 10.1101/gad.1450606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reimann JD, Gardner BE, Margottin-Goguet F, Jackson PK. Emi1 regulates the anaphase-promoting complex by a different mechanism than Mad2 proteins. Genes Dev. 2001;15:3278–3285. doi: 10.1101/gad.945701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandell LL, Zakian VA. Loss of a yeast telomere: arrest, recovery, and chromosome loss. Cell. 1993;75:729–739. doi: 10.1016/0092-8674(93)90493-a. [DOI] [PubMed] [Google Scholar]
- Saunders W, Hornack D, Lengyel V, Deng C. The Saccharomyces cerevisiae kinesin-related motor Kar3p acts at preanaphase spindle poles to limit the number and length of cytoplasmic microtubules. J Cell Biol. 1997;137:417–431. doi: 10.1083/jcb.137.2.417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwab M, Lutum AS, Seufert W. Yeast Hct1 is a regulator of Clb2 cyclin proteolysis. Cell. 1997;90:683–693. doi: 10.1016/s0092-8674(00)80529-2. [DOI] [PubMed] [Google Scholar]
- Shirayama M, Toth A, Galova M, Nasmyth K. APC(Cdc20) promotes exit from mitosis by destroying the anaphase inhibitor Pds1 and cyclin Clb5. Nature. 1999;402:203–207. doi: 10.1038/46080. [DOI] [PubMed] [Google Scholar]
- Spellman PT, Sherlock G, Zhang MQ, Iyer VR, Anders K, Eisen MB, Brown PO, Botstein D, Futcher B. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9:3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sproul LR, Anderson DJ, Mackey AT, Saunders WS, Gilbert SP. Cik1 targets the minus-end kinesin depolymerase kar3 to microtubule plus ends. Curr Biol. 2005;15:1420–1427. doi: 10.1016/j.cub.2005.06.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat Cell Biol. 2003;5:1090–1094. doi: 10.1038/ncb1066. [DOI] [PubMed] [Google Scholar]
- Thornton BR, Toczyski DP. Precise destruction: an emerging picture of the APC. Genes Dev. 2006;20:3069–3078. doi: 10.1101/gad.1478306. [DOI] [PubMed] [Google Scholar]
- Tugendreich S, Tomkiel J, Earnshaw W, Hieter P. CDC27Hs colocalizes with CDC16Hs to the centrosome and mitotic spindle and is essential for the metaphase to anaphase transition. Cell. 1995;81:261–268. doi: 10.1016/0092-8674(95)90336-4. [DOI] [PubMed] [Google Scholar]
- Visintin R, Prinz S, Amon A. CDC20 and CDH1: a family of substrate-specific activators of APC-dependent proteolysis. Science. 1997;278:460–463. doi: 10.1126/science.278.5337.460. [DOI] [PubMed] [Google Scholar]
- Woodbury EL, Morgan DO. Cdk and APC activities limit the spindle-stabilizing function of Fin1 to anaphase. Nat Cell Biol. 2007;9:106–112. doi: 10.1038/ncb1523. [DOI] [PubMed] [Google Scholar]
- Zachariae W, Schwab M, Nasmyth K, Seufert W. Control of cyclin ubiquitination by CDK-regulated binding of Hct1 to the anaphase promoting complex. Science. 1998;282:1721–1724. doi: 10.1126/science.282.5394.1721. [DOI] [PubMed] [Google Scholar]
- Zhou Y, Ching YP, Chun AC, Jin DY. Nuclear localization of the cell cycle regulator CDH1 and its regulation by phosphorylation. J Biol Chem. 2003;278:12530–12536. doi: 10.1074/jbc.M212853200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.