Abstract
Purpose of review
The proteome is the pool of proteins expressed at a given time and circumstance. The word “proteomics” summarizes several technologies for visualization, quantitation and identification of these proteins. Recent advances in these techniques are helping to elucidate platelet processes which are relevant to bleeding and clotting disorders, transfusion medicine and regulation of angiogenesis.
Recent findings
Over 1100 platelet proteins have been identified using proteomic techniques. Various subproteomes have been characterized, including platelet releasates (the “secretome”), alpha and dense granules, membrane and cytoskeletal proteins, platelet-derived microparticles, and the platelet “phosphoproteome”. Proteomic data about platelets have become increasingly available in integrated databases.
Summary
Proteomic experiments in resting and activated platelets have identified novel signaling pathways and secreted proteins which may represent therapeutic targets, as well as potential cancer biomarkers.
Keywords: proteomics, platelets, mass spectrometry, releasate
Introduction
The primary function of platelets is to stop hemorrhage after tissue trauma and vascular injury. Platelets act not only through the immediate release of a variety of lipid and protein mediators but also through signal-dependent pre-mRNA splicing and the translation of constitutively expressed mRNA[1**]. These post-transcriptional pathways are potential targets for molecular intervention in atherothrombosis[2]. Proteomic experiments can provide data about localization, interactions, posttranslational modifications, and activation states of gene products.
Proteomic Techniques: A Brief Introduction
Proteomic experiments begin with a protein mixture which is digested to a peptide mixture, either in a gel or in solution. Two-dimensional electrophoresis (2-DE) has been available since the 1970s, and was used in the 1990s to construct early maps of platelet proteins. Newer gel-based methods include differential in-gel electrophoresis (DIGE), which allows for quantitation and comparison of samples from different proteomic states. In DIGE, samples are differentially labeled with fluorescent dyes and simultaneously analyzed in a single gel. Alternatively, protein spots may be excised from a gel, digested, and analyzed by mass spectrometry (MS).
Gel-free methods, which use in-solution proteolytic digestion, can detect proteins not well represented by 2-DE, such as transmembrane and basic proteins. Multidimensional protein identification technology (MudPIT) employs ion-exchange and reverse-phase liquid chromatography (LC) for peptide separation[3]. Mass spectrometry (MS) has the capacity to identify proteins in a high throughput manner, using bioinformatics approaches linked to protein sequence databases. The two main mass-spectrometric options are electrospray ionization (ESI) and matrix-assisted laser desorption/ionization (MALDI). Both MALDI and a related method, surface-enhanced laser desorption/ionization (SELDI), employ a matrix to ionize proteins; SELDI uses protein chips with chromatographic surfaces. LC-MS/MS is the method now chosen by most proteomics researchers[4]. Isotope affinity tagging techniques help to increase the sensitivity of detection of smaller peptide fragments and provide quantitative information about protein concentrations. Isotope coded affinity tags (ICAT) selectively label cysteine residues of peptide fragments following tryptic digest of the protein sample; this technique increases the depth of protein coverage but is limited by selective labeling. Isotope tags for relative and absolute quantitation (iTRAQ) label every tryptic fragment, thus allowing for measurement of absolute changes in protein composition[5]. Figure 1 summarizes the various proteomic approaches which have been used to study platelets.
Figure 1. Overview of proteomic techniques.
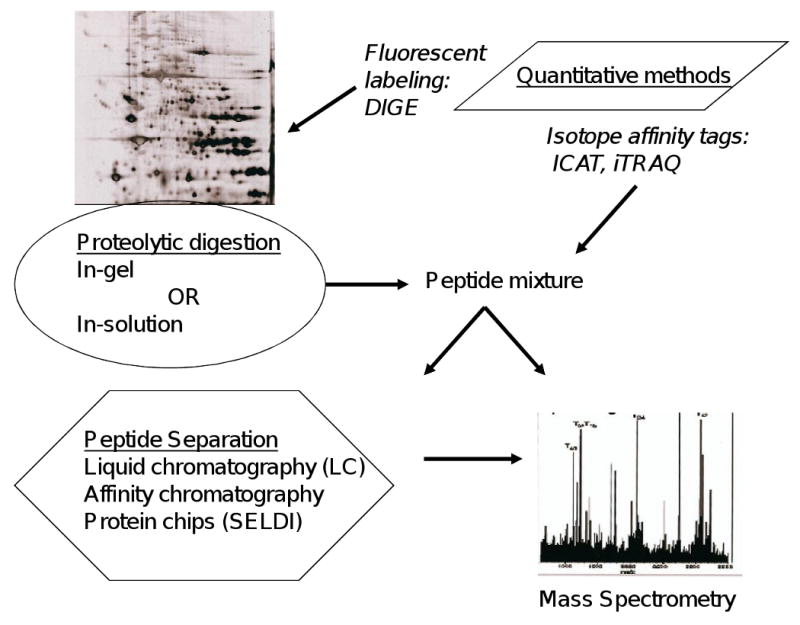
Proteolytic digestion is performed either in a gel or in solution to produce peptides for mass spectrometry (MS) analysis. Differential in-gel electrophoresis (DIGE) employs fluorescent labeling of peptides. Isotope affinity tags can be used in gel-free proteomic experiments to improve peptide quantitation. Various chromatographic methods are available for peptide separation and selective enrichment of proteins. SELDI uses ProteinChip arrays to segregate protein populations. Proteins are identified by database searches following MS analysis.
ICAT - isotope-coded affinity tag, iTRAQ - isotope tagging for relative and absolute quantitation, SELDI - Surface-enhanced laser desorption/ionization
As discussed in the next sections, proteomic data have been published for resting and activated platelets, platelet alpha granules, platelet dense granules, platelet membranes, and platelet-derived microparticles.
The Platelet Releasate: Activation-Dependent Changes
Quiescent platelets display minimal translational activity. Platelet activation leads to the rapid translation of pre-existing mRNA, with the release or derivation of platelet-secreted proteins, cytokines, exosomes, and microparticles. Using thrombin-stimulated platelets, a combination of MALDI-TOF and MudPIT identified more than 300 released proteins[6]. Several of the secreted proteins have been identified in atherosclerotic lesions but are absent in normal vasculature; these include secretogranin III, cyclophilin A, and calumenin. In another study, the use of 2-DE, MALDI-TOF and LC-ESI MS/MS to study platelet dense granules led to the identification of an extracellular role for 14-3-3 zeta protein in atherosclerosis[7*]. Such secreted proteins are potential targets for future drug development, given their extracellular localization, without the risk of bleeding that complicates direct inhibition of platelet activation.
Piersma et al.[8] obtained an activated platelet releasate proteome using high resolution, high mass accuracy hybrid ion-trap Fourier transform mass spectrometry. This group combined 1-D gel electrophoresis and LC-MS/MS in a strategy known as GeLC-MS/MS. They identified nearly 400 proteins previously reported in platelet alpha granules and microparticles, along with over 300 novel proteins. Only 5% of the proteins overlapped with previously reported platelet releasate datasets[6;9*]. Proteins smaller than 8 kDa were not detected, due to the limitations of the 10% acrylamide gel and use of a cut-off filter to concentrate the samples.
DellaCorte et al.[10] combined 2D-DIGE and MALDI-TOF and identified 36 proteins differentially regulated in the platelet releasate following thrombin activation. They detected proteins ranging between 220 kDa and 15 kDa. Coppinger et al.[9] compared platelet releasates induced by ADP, collagen and TRAP stimulation in the presence and absence of aspirin using GeLC-MS/MS. An overall decrease in protein expression was observed in the presence of aspirin, and the secretion profiles were agonist-dependent. This group also employed cytokine antibody arrays to quantitate platelet-derived cytokines in the presence and absence of aspirin.
Greening et al.[11] co-identified 41 proteins in both non-activated platelets and plasma, and concluded that analyses of the human plasma proteome must take into account the contribution of platelet-derived proteins. This is because some ex vivo platelet activation occurs during blood collection, and platelet-derived microparticles circulate in normal plasma.
Proteome of Platelet Microparticles
Microparticles (MP) are small membrane vesicles that are released from cells upon activation or during apoptosis. The platelet microparticle proteome has been characterized[12], and appears to have a composition distinct from the plasma proteome. This could be significant because while plasma-derived MPs imply cellular activation and possible damage, platelet-derived MPs appear to be present in healthy individuals. Smalley et al.[13] compared the proteomic profiles of plasma-derived and platelet-derived MPs using two methods, spectral count analysis and isotope-coded affinity tag (ICAT) labeling of proteins. Proteins present only in the plasma MPs included several associated with apoptosis, iron transport, complement components, and the coagulation process.
Signaling Pathways: Platelet Membrane Proteome and Phosphoproteome
Novel membrane proteins that signal during platelet aggregation have been identified using LC-MS/MS and other chromatographic techniques[14-17]. Some of these proteins become phosphorylated on tyrosine or serine residues on platelet aggregation. Maguire et al.[18] profiled platelet proteins associated with detergent-resistant membrane lipid rafts. Lipid rafts may act as concentrating platforms that co-cluster receptors and signaling molecules, thus coordinating platelet activation and secretion. Proteins recruited upon von Willebrand factor activation included platelet GPIbα, glucose transporter 14 and C-terminal LIM protein 36.
Garcia et al. identified 41 proteins which were differentially phosphorylated following stimulation by thrombin receptor activating peptide (TRAP)[19]. These included a novel protein, the adapter downstream of tyrosine kinase 2 (Dok-2), which may play a role in thrombus formation[20]. The G6b-B protein, which contains an immunoreceptor tyrosine-based inhibitory motif, may play a role in limiting platelet activation[17]. Zahedi et al.[21*] noted the limitations of gel-based proteomic techniques for the detection of hydrophobic, alkaline, very small or large proteins, or proteins with less abundant phosphorylation sites. The investigators adopted a two-pronged chromatographic strategy coupled with nano-LC-MS/MS, and identified 278 phosphorylated proteins in resting platelets. Their finding that GP1bα is phosphorylated on a protein kinase A/protein kinaseG consensus site may be important for the regulation of the GPIb-IX-V signaling pathway.
Platelet Storage Lesion
The platelet storage lesion refers to the in vitro changes that occur when donated platelets are stored for several days before transfusion to a recipient. These include loss of disk shape, increased release of alpha-granules and cytosolic proteins, increased procoagulant activity, and altered glycoprotein expression[22*]. To characterize the platelet storage lesion in more detail, investigators used 2-DE[23], differential in-gel electrophoresis (DIGE)[22;24], isotope-coded affinity tagging (ICAT)[22], and isotope tagging for relative and absolute quantitation (iTRAQ)[22]. As expected, the gel-free methods were better than in-gel methods at detecting low-abundance molecules and hydrophobic proteins. Both beta-actin and septin2 were found to be altered by storage in separate studies, highlighting the importance of cytoskeletal reorganization and apoptosis in the platelet storage lesion. These approaches hold promise for finding biomarkers that might be useful in judging platelet quality[25*].
Platelet Proteome and Disease
Recently, proteomic techniques identified a diagnosis of Quebec Platelet Disorder in a family with severe bleeding problems of unknown origin[26**]. While clinical features were not clearly suggestive of the disorder, LC-MS/MS revealed reduced amounts of alpha granule proteins which led to the correct diagnosis. This work relied on the earlier proteomic characterization of platelet alpha granules by Maynard et al.[27*].
Investigators have examined the effects of drugs, illnesses and biological variation on platelet protein profiles. An ELISA-based study of carbonyl groups and 3-nitrotyrosine residues in platelet proteins suggested that in patients with schizophrenia, reactive oxygen species and reactive nitrogen species may stimulate oxidative and nitrative modifications of platelet proteins[28]. Another group, using 2-DE, found that treatment with an antihypertensive drug modified the platelet protein profile of hypertensive patients [29]. Winkler et al.[30] cautioned that clinical studies of platelet proteomes should employ an analytical method that can detect small quantitative differences, such as DIGE.
Cervi et al.[31**] compared the platelet proteome of mice injected with tumor cells compared with saline-injected controls using SELDI-TOF. They identified platelet factor 4 (PF4) as a biomarker for a variety of tumor types, even though plasma levels of PF4 did not change. Next, they demonstrated that increases in platelet PF4 can be measured using the more accessible method of ELISA. Thus, PF4 in platelets (but not plasma) represents both a potentially useful cancer biomarker and a potential target for early cancer therapy. In a different SELDI-TOF study[32**], the platelet (but not plasma) concentrations of angiogenesis regulatory proteins were modified by and reflected the presence of tumors in mice. These differences were attributed to the ability of platelets to selectively take up angiogenesis regulators in cancer bearing hosts. The authors suggested that the “platelet angiogenesis proteome” could be used for early detection of tumor establishment or recurrence.
Integrating Transcriptomic and Proteomic Studies
The degree of correlation between platelet proteomic and transcriptomic data in the end points of detection and identification varies by study[11;33**;34]. Some investigators have found that about one third of platelet proteins identified by proteomic methods are not reflected in the transcriptome[11;35]. The discordance may be due to i) the limited mRNA stability of these genes, ii) failure of microarray analysis to detect very low levels of RNA, iii) the occurrence of proteins which may be synthesized in megakaryocytes, after which mRNA is degraded, and iv) the fact that some proteins may be taken up from plasma or from other cells rather than synthesized in megakaryocytes or platelets[36].
Conversely, mRNA transcripts identified by transcriptomic methods sometimes are not detected by proteomic techniques. When proteins are not identified, this may be due to the failure of current proteomic methods to identify proteins with certain structural or biochemical characteristics, and/or to lack of translation of mRNA. To capture and concentrate the “low-abundance” proteome, Guerrier et al.[37**] used a combinatorial ligand library composed of millions of diverse hexapeptide baits. This approach led to the addition of 147 proteins to the list of over 1100 proteins previously identified in the platelet proteome. In contrast, platelets contain approximately 1,600-3,000 individual transcripts[38].
Activity Based Proteomic Profiling of Platelets
Activity based proteomics is a functional proteomics technology where molecular probes are used to target a select group of functionally related proteins. Wong et al.[39*] applied such a strategy to differentially profile the nucleotide-binding proteome of active and resting platelets. The investigators performed affinity chromatography using immobilized nucleotides and employed LC-MS/MS to identify the recovered proteins. Nearly all of the differentially expressed nucleotide binding proteins were associated with the cytoskeleton. The dynamic range of the study was negatively affected by abundant nucleotide-binding cytoskeletal proteins such as actin and myosin. Future work should include better removal of cytoskeletal proteins from platelet lysates or alternative strategies to enhance the detection of low abundance proteins.
Data Resources for the Platelet Proteome
Proteomic data about platelets have become increasingly available in various databases. The Human Proteome Organization (HUPO) Proteomics Standards Initiative has produced a document, known as the minimum information about a proteomics experiment (MIAPE), which enumerates several integrated databases[40]. For example, the SWISS-PROT Web page has published the platelet proteome on a 2D gel (http://ca.expasy.org). The Reactome web site (http://www.reactome.org) includes information about the complex pathways which operate in platelets[41]. The Human Protein Reference Database (http://www.hprd.org) includes features such as PhosphoMotif Finder and Human Proteinpedia[42]. Dittrich et al.[33**] created a central resource for the platelet proteome, interactome and phosphorylation state known as PlateletWeb (http://plateletweb.bioapps.biozentrum.uni-wuerzburg.de). This “virtual platelet” knowledge base also includes a characterization of the platelet protein kinase repertoire (kinome).
Conclusions and Future Perspectives
Proteomic approaches have uncovered novel signaling pathways and secreted proteins involved in platelet activation and inhibition. Proteomic discoveries have identified potential therapeutic targets in atherothrombosis, quality markers in platelets stored for transfusion, diagnostic approaches to bleeding dyscrasias, and biomarkers for early detection of cancer.
Acknowledgments
This work was supported by NIH grants HL091939 and HL086378.
Abbreviations
- MS
Mass spectrometry
- LC
Liquid chromatography
- MP
Microparticles
- MudPIT
Multidimensional protein identification technology
- ICAT
isotope-coded affinity tag
- iTRAQ
isotope tagging for relative and absolute quantitation
- 2-DE
Two-dimensional electrophoresis
- DIGE
Differential in-gel electrophoresis
- ELISA
Enzyme-linked immunosorbent assay
- MALDI-TOF
Matrix-assisted laser desorption/ionization – Time of flight
- SELDI
Surface-enhanced laser desorption/ionization
Contributor Information
Lisa Senzel, Email: lsenzel@notes.cc.sunysb.edu, Department of Pathology, State University of New York, Stony Brook, University Hospital, Level 3, Rm 532, Stony Brook, NY 11794-7300, Phone: 631-444-2601, Fax: 631-444-2653.
Dmitri V. Gnatenko, Email: dgnatenko@notes.cc.sunysb.edu, Department of Medicine, State University of New York, Stony Brook, Division of Hematology, HSC T15/030, Stony Brook, N.Y. 11794-8151, Phone: 631-444-1260, FAX: 631-444-7530.
Wadie F. Bahou, Email: wbahou@notes.cc.sunysb.edu, Department of Medicine, State University of New York, Stony Brook, Division of Hematology, HSC T15/030, Stony Brook, N.Y. 11794-8151, Phone: 631-444-2059, FAX: 631-444-7530.
Reference List
- **1.Weyrich AS, Schwertz H, Kraiss LW, et al. Protein synthesis by platelets: historical and new perspectives. J Thromb Haemost. 2009;7:241–246. doi: 10.1111/j.1538-7836.2008.03211.x. [DOI] [PMC free article] [PubMed] [Google Scholar]; This paper reviews the mechanisms by which platelet synthesize proteins and describes how protein synthesis alters the phenotype and functions of platelets. Signal-dependent pre-mRNA splicing allows platelets to alter their transcriptome and proteome profiles in response to cellular activation.
- 2.Davi G, Patrono C. Platelet activation and atherothrombosis. N Engl J Med. 2007;357:2482–2494. doi: 10.1056/NEJMra071014. [DOI] [PubMed] [Google Scholar]
- 3.Delahunty CM, Yates JR., III MudPIT: multidimensional protein identification technology. Biotechniques. 2007;43:563, 565, 567. [PubMed] [Google Scholar]
- 4.Kocher T, Superti-Furga G. Mass spectrometry-based functional proteomics: from molecular machines to protein networks. Nat Methods. 2007;4:807–815. doi: 10.1038/nmeth1093. [DOI] [PubMed] [Google Scholar]
- 5.Arab S, Gramolini AO, Ping P, et al. Cardiovascular proteomics: tools to develop novel biomarkers and potential applications. J Am Coll Cardiol. 2006;48:1733–1741. doi: 10.1016/j.jacc.2006.06.063. [DOI] [PubMed] [Google Scholar]
- 6.Coppinger JA, Cagney G, Toomey S, et al. Characterization of the proteins released from activated platelets leads to localization of novel platelet proteins in human atherosclerotic lesions. Blood. 2004;103:2096–2104. doi: 10.1182/blood-2003-08-2804. [DOI] [PubMed] [Google Scholar]
- *7.Hernandez-Ruiz L, Valverde F, Jimenez-Nunez MD, et al. Organellar proteomics of human platelet dense granules reveals that 14-3-3zeta is a granule protein related to atherosclerosis. J Proteome Res. 2007;6:4449–4457. doi: 10.1021/pr070380o. [DOI] [PubMed] [Google Scholar]; This is the first study to describe the soluble protein composition of platelet dense granules. The 14-3-3zeta protein may represent a therapeutic target in atherosclerosis.
- 8.Piersma SR, Broxterman HJ, Kapci M, et al. Proteomics of the TRAP-induced platelet releasate. J Proteomics. 2009;72:91–109. doi: 10.1016/j.jprot.2008.10.009. [DOI] [PubMed] [Google Scholar]
- *9.Coppinger JA, O'Connor R, Wynne K, et al. Moderation of the platelet releasate response by aspirin. Blood. 2007;109:4786–4792. doi: 10.1182/blood-2006-07-038539. [DOI] [PubMed] [Google Scholar]; The authors profiled the platelet releasate after stimulation with 3 different agonists before and after aspirin. They used GelC-tandem MS as well as cytokine antibody arrays to complement the detection of small molecules.
- 10.Della CA, Maugeri N, Pampuch A, et al. Application of 2-dimensional difference gel electrophoresis (2D-DIGE) to the study of thrombin-activated human platelet secretome. Platelets. 2008;19:43–50. doi: 10.1080/09537100701609035. [DOI] [PubMed] [Google Scholar]
- 11.Greening DW, Glenister KM, Kapp EA, et al. Comparison of human platelet membrane-cytoskeletal proteins with the plasma proteome: Towards understanding the platelet-plasma nexus. Proteomics Clin Appl. 2008;2:63–77. doi: 10.1002/prca.200780067. [DOI] [PubMed] [Google Scholar]
- 12.Garcia BA, Smalley DM, Cho H, et al. The platelet microparticle proteome. J Proteome Res. 2005;4:1516–1521. doi: 10.1021/pr0500760. [DOI] [PubMed] [Google Scholar]
- 13.Smalley DM, Root KE, Cho H, et al. Proteomic discovery of 21 proteins expressed in human plasma-derived but not platelet-derived microparticles. Thromb Haemost. 2007;97:67–80. [PubMed] [Google Scholar]
- 14.Martens L, Van DP, Van DJ, et al. The human platelet proteome mapped by peptide-centric proteomics: a functional protein profile. Proteomics. 2005;5:3193–3204. doi: 10.1002/pmic.200401142. [DOI] [PubMed] [Google Scholar]
- 15.Moebius J, Zahedi RP, Lewandrowski U, et al. The human platelet membrane proteome reveals several new potential membrane proteins. Mol Cell Proteomics. 2005;4:1754–1761. doi: 10.1074/mcp.M500209-MCP200. [DOI] [PubMed] [Google Scholar]
- 16.Nanda N, Bao M, Lin H, et al. Platelet endothelial aggregation receptor 1 (PEAR1), a novel epidermal growth factor repeat-containing transmembrane receptor, participates in platelet contact-induced activation. J Biol Chem. 2005;280:24680–24689. doi: 10.1074/jbc.M413411200. [DOI] [PubMed] [Google Scholar]
- 17.Senis YA, Tomlinson MG, Garcia A, et al. A comprehensive proteomics and genomics analysis reveals novel transmembrane proteins in human platelets and mouse megakaryocytes including G6b-B, a novel immunoreceptor tyrosine-based inhibitory motif protein. Mol Cell Proteomics. 2007;6:548–564. doi: 10.1074/mcp.D600007-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maguire PB, Foy M, Fitzgerald DJ. Using proteomics to identify potential therapeutic targets in platelets. Biochem Soc Trans. 2005;33:409–412. doi: 10.1042/BST0330409. [DOI] [PubMed] [Google Scholar]
- 19.Garcia A, Prabhakar S, Hughan S, et al. Differential proteome analysis of TRAP-activated platelets: involvement of DOK-2 and phosphorylation of RGS proteins. Blood. 2004;103:2088–2095. doi: 10.1182/blood-2003-07-2392. [DOI] [PubMed] [Google Scholar]
- 20.Garcia A. Proteome analysis of signaling cascades in human platelets. Blood Cells Mol Dis. 2006;36:152–156. doi: 10.1016/j.bcmd.2005.12.013. [DOI] [PubMed] [Google Scholar]
- *21.Zahedi RP, Lewandrowski U, Wiesner J, et al. Phosphoproteome of resting human platelets. J Proteome Res. 2008;7:526–534. doi: 10.1021/pr0704130. [DOI] [PubMed] [Google Scholar]; This study used clever chromatographic techniques to expand upon previous studies of the platelet phosphoproteome, which had been done with 2DE and tandem MS. They improved the size range of protein detection, the resolution of hydrophobic and alklaline proteins and of those with less abundant phosphorylation sites.
- *22.Thon JN, Schubert P, Duguay M, et al. Comprehensive proteomic analysis of protein changes during platelet storage requires complementary proteomic approaches. Transfusion. 2008;48:425–435. doi: 10.1111/j.1537-2995.2007.01546.x. [DOI] [PubMed] [Google Scholar]; Investigators found 5 platelet proteins which changed concentration over a 7-day storage period using DIGE, ICAT and iTRAQ. While iTRAQ was able to detect a wide variety of proteins altered by storage, ICAT appeared to select for certain lower-abundance proteins not easily captured by iTRAQ.
- 23.Glenister KM, Payne KA, Sparrow RL. Proteomic analysis of supernatant from pooled buffy-coat platelet concentrates throughout 7-day storage. Transfusion. 2008;48:99–107. doi: 10.1111/j.1537-2995.2007.01487.x. [DOI] [PubMed] [Google Scholar]
- 24.Thiele T, Steil L, Gebhard S, et al. Profiling of alterations in platelet proteins during storage of platelet concentrates. Transfusion. 2007;47:1221–1233. doi: 10.1111/j.1537-2995.2007.01255.x. [DOI] [PubMed] [Google Scholar]
- *25.Perrotta PL. Proteomics: moving from a discovery to a quality assurance tool. Transfusion. 2008;48:408–410. doi: 10.1111/j.1537-2995.2007.01636.x. [DOI] [PubMed] [Google Scholar]; Discusses strengths and weaknesses of 2-DE, DIGE, ICAT and iTRAQ and how these techniques can complement each other in exploring the platelet proteome.
- **26.Maurer-Spurej E, Kahr WH, Carter CJ, et al. The value of proteomics for the diagnosis of a platelet-related bleeding disorder. Platelets. 2008;19:342–351. doi: 10.1080/09537100802010547. [DOI] [PubMed] [Google Scholar]; This is the first report using proteomics to identify a familial platelet defect. The alteration in the alpha granule protein profile led to the diagnosis of Quebec Platelet Disorder.
- *27.Maynard DM, Heijnen HF, Horne MK, et al. Proteomic analysis of platelet alpha-granules using mass spectrometry. J Thromb Haemost. 2007;5:1945–1955. doi: 10.1111/j.1538-7836.2007.02690.x. [DOI] [PubMed] [Google Scholar]; The authors identified 284 non-redundant alpha-granule proteins, 44 of which were not previously known. This work provided a foundation for the medical diagnosis of the familial bleeding disorder described above.
- 28.Dietrich-Muszalska A, Olas B. Modifications of blood platelet proteins of patients with schizophrenia. Platelets. 2009;20:90–96. doi: 10.1080/09537100802641499. [DOI] [PubMed] [Google Scholar]
- 29.Sacristan D, Marques M, Zamorano J, et al. Modifications by Olmesartan medoxomil treatment of the platelet protein profile of moderate hypertensive patients. Proteomics Clin Appl. 2008:1300–1312. doi: 10.1002/prca.200700021. [DOI] [PubMed] [Google Scholar]
- 30.Winkler W, Zellner M, Diestinger M, et al. Biological variation of the platelet proteome in the elderly population and its implication for biomarker research. Mol Cell Proteomics. 2008;7:193–203. doi: 10.1074/mcp.M700137-MCP200. [DOI] [PubMed] [Google Scholar]
- **31.Cervi D, Yip TT, Bhattacharya N, et al. Platelet-associated PF-4 as a biomarker of early tumor growth. Blood. 2008;111:1201–1207. doi: 10.1182/blood-2007-04-084798. [DOI] [PubMed] [Google Scholar]; This paper shows that platelet, but not plasma, levels of PF-4 are selectively elevated in mice bearing human tumors. The authors used SELFI-TOF as a discovery tool, followed by ELISA as a more accessible method which could be clinically useful for early cancer detection.
- **32.Klement GL, Yip TT, Cassiola F, et al. Platelets actively sequester angiogenesis regulators. Blood. 2009;133:2835–2842. doi: 10.1182/blood-2008-06-159541. [DOI] [PMC free article] [PubMed] [Google Scholar]; While plasma levels of angiogenesis regulators have not been shown to be reliable markers of cancer presence or therapeutic response, this paper shows that platelets sequester these proteins selectively in cancer bearing hosts. The authors used SELDI-TOF as a discovery tool.
- **33.Dittrich M, Birschmann I, Mietner S, et al. Platelet protein interactions: map, signaling components, and phosphorylation groundstate. Arterioscler Thromb Vasc Biol. 2008;28:1326–1331. doi: 10.1161/ATVBAHA.107.161000. [DOI] [PubMed] [Google Scholar]; The authors describe a database, PlateletWeb, that houses a “virtual platelet.” They inventory platelet-specific proteins with annotations about protein domains and functions. They establish connections between platelet proteins via reported interactions from other cells.
- 34.Hillmann AG, Harmon S, Park SD, et al. Comparative RNA expression analyses from small-scale, single-donor platelet samples. J Thromb Haemost. 2006;4:349–356. doi: 10.1111/j.1538-7836.2006.01684.x. [DOI] [PubMed] [Google Scholar]
- 35.Gnatenko DV, Perrotta PL, Bahou WF. Proteomic approaches to dissect platelet function: Half the story. Blood. 2006;108:3983–3991. doi: 10.1182/blood-2006-06-026518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bugert P, Ficht M, Kluter H. Towards the identification of novel platelet receptors: Comparing RNA and proteome approaches. Transfus Med Hemother. 2006;33:236–243. [Google Scholar]
- **37.Guerrier L, Claverol S, Fortis F, et al. Exploring the platelet proteome via combinatorial, hexapeptide ligand libraries. J Proteome Res. 2007;6:4290–4303. doi: 10.1021/pr0703371. [DOI] [PubMed] [Google Scholar]; The authors use millions of diverse hexapeptide baits to capture and concentrate the “low-abundance” proteome. Using this bead-based approach, abundant species quickly saturate their ligand, while rare species keep being adsorbed onto their respective ligand. This results in a “normalization” of the relative abundance ratios.
- 38.Gnatenko DV, Dunn JJ, Schwedes J, et al. Transcript profiling of human platelets using microarray and serial analysis of gene expression (SAGE) Methods Mol Biol. 2009;496:245–272. doi: 10.1007/978-1-59745-553-4_16. [DOI] [PubMed] [Google Scholar]
- *39.Wong JW, McRedmond JP, Cagney G. Activity profiling of platelets by chemical proteomics. Proteomics. 2009;9:40–50. doi: 10.1002/pmic.200800185. [DOI] [PubMed] [Google Scholar]; This paper is the first to apply activity based (chemical) proteomics to platelets. Activity profiling uses molecular probes to target a select group of functionally related proteins. The authors performed a preliminary investigation of the platelet nucleotide-binding proteome.
- 40.Taylor CF, Paton NW, Lilley KS, et al. The minimum information about a proteomics experiment (MIAPE) Nat Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- 41.Vastrik I, D'Eustachio P, Schmidt E, et al. Reactome: a knowledge base of biologic pathways and processes. Genome Biol. 2007;8:R39. doi: 10.1186/gb-2007-8-3-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Keshava Prasad TS, Goel R, Kandasamy K, et al. Human Protein Reference Database--2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]


