Abstract
Understanding catecholamine metabolism is crucial for elucidating the pathogenesis of hereditary hypertension. Here we integrated transcriptional and biochemical profiling with physiologic quantitative trait locus (eQTL and pQTL) mapping in adrenal glands of the HXB/BXH recombinant inbred (RI) strains, derived from the spontaneously hypertensive rat (SHR) and normotensive Brown Norway (BN.Lx). We found simultaneous down-regulation of five heritable transcripts in the catecholaminergic pathway in young (6 weeks) SHRs. We identified cis-acting eQTLs for Dbh, Pnmt (catecholamine biosynthesis) and Vamp1 (catecholamine secretion); enzymatic activities of Dbh and Pnmt paralleled transcripts, with pQTLs for activities mirroring eQTLs. We also detected trans-regulated expression of Vmat1 and Chga (both involved in catecholamine storage), with co-localization of these trans-eQTLs to the Pnmt locus. Pnmt re-sequencing revealed promoter polymorphisms that result in decreased response of the transfected SHR promoter to glucocorticoid, compared with BN.Lx. Of physiological pertinence, Dbh activity negatively correlated with systolic blood pressure in RI strains, whereas Pnmt activity was negatively correlated with heart rate. The finding of such cis- and trans-QTLs at an age before the onset of frank hypertension suggests that these heritable changes in biosynthetic enzyme expression represent primary genetic mechanisms for regulation of catecholamine action and blood pressure control in this widely studied model of hypertension.
INTRODUCTION
Excessive sympathoadrenal activity is implicated in the pathogenesis of both essential (1,2) and acquired (3) hypertension, in humans (4,5) and experimental animals (6), and adrenergic receptor antagonists are a mainstay of antihypertensive therapy (7,8). Increased sympatho-neuronal activity was observed in normotensive humans with family history of hypertension (2), as well as young spontaneously hypertensive rats [SHRs (9–11)]. The present study focuses primarily on genes involved in adrenal medullary physiology and their role in the pathogenesis of essential hypertension. Implicating evidence comes from studies showing that adrenal medullectomy in young SHR results in a decrease in blood pressure in the adult (12–14), but the exact mechanism by which this occurs is not well established. Understanding mechanisms that influence catecholamine biosynthesis, storage, secretion and degradation in the adrenal medulla of young SHRs may give clues as to which processes are dysregulated early in the development of hypertension, before full manifestation of the disease, and may shed light on factors that mediate disease susceptibility.
Previous studies focusing on catecholamine biosynthetic enzyme activities in the adrenal medulla of the SHR showed inconsistent results, possibly owing to the different normotensive controls used in the experiments. Tyrosine hydroxylase (Th) was in turn reported to be decreased (10,15,16), unchanged (17) and increased (18,19) in young SHRs. However, studies involving adult SHRs report an increased Th activity when compared with age- and sex-matched controls (15,20). Dopamine beta-hydroxylase (Dbh), phenylethanolamine N-methyltransferase (Pnmt) and dopa decarboxylase (Ddc) activities were decreased (15,21) or increased (9,22) in the young SHRs, but unchanged in the adult SHRs (9,15). Because of the apparent correlation of enzyme activities of Th, Dbh and Pnmt, some authors suggested that these genes may be co-regulated by a single locus (23).
In order to characterize the effect of genetic variation on catecholamine physiology, we integrated gene expression profiling and biochemical phenotyping with linkage mapping. We used the HXB/BXH panel of recombinant inbred (RI) strains, derived from SHRs and the normotensive BN.Lx inbred progenitor strains (24), and used widely for genetic dissection of phenotypes segregating in this panel. Young (6-week-old) males were used for all experiments to avoid confounding the results by the consequences of fully developed hypertension or by the effects of female sex hormones. We first quantified the variability across 29 RI strains and the two progenitor strains of transcript levels of chromaffin cell-expressed genes that are responsible for crucial aspects of catecholamine biology. We then collected biochemical phenotypes reflecting the protein products (i.e. enzymes) of these genes, as well as the intermediates synthesized by them. Subsequently, linkage mapping was performed for both sets of phenotypes (i.e. transcript levels and biochemical traits). Quantitative trait loci (QTLs) were detected, which suggested a regulatory network of cis- and trans-acting loci. Positional candidates from QTL regions were re-sequenced in the progenitor strains to discover specific DNA sequence variants that were subsequently tested for their functional significance by transfection. Correlation of key catecholamine biosynthetic enzymes with systolic blood pressure and heart rate was established.
RESULTS
mRNA expression differences in hypertension pathway genes
A total of 211 adrenal gene expression profiles of hypertension candidate genes from Affymetrix RAE230A were examined for heritable differences in transcript levels in the RI strains (see Supplementary Material, Fig. S1). Candidate genes were grouped according to their function and/or pathway to which they belong. For each expressed gene, heritability (H2), normalized ratio (NR) and t-test P-values were calculated. One hundred and twenty-five hypertension candidate genes were found expressed in the adrenal data set, with 64 of those exhibiting heritable gene expression at H2 ≥ 20%.
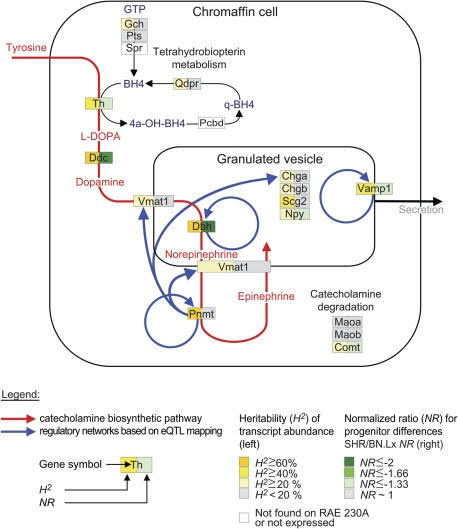
Catecholaminergic genes stood out as the most represented group with 13 out of the 64 heritable transcripts (see Fig. 1). Some of them exhibited significant progenitor differences (six genes), and those were uniformly in the direction of underexpression in the SHR. Gene expression levels for all four catecholamine biosynthetic enzymes—Th, Ddc, Dbh and Pnmt—were highly heritable (H2 ≥ 60% for Ddc, Dbh and Pnmt, H2 ∼ 50% for Th) and, with the exception of Pnmt, also significantly underexpressed in the SHR. Other significantly SHR-underexpressed genes with heritable transcripts included vesicle-associated membrane protein 1 (Vamp1), neuropeptide Y (Npy) and catechol-O-methyltransferase (Comt). Gene transcript levels for catecholamine biosynthetic enzymes (Th, Ddc, Dbh, Pnmt) and granins (Chga, Chgb, Scg2) from the microarray data were validated by quantitative real-time reverse transcription-polymerase chain reaction (qRT-PCR) in tissues from a separate harvesting of age- and sex-matched progenitors (see Supplementary Material, Fig. S2). Significant differences were confirmed for Th, Ddc and Dbh gene expression, with all three genes showing underexpression in SHRs compared with BN.Lx.
Figure 1.
Chromaffin cell genes involved in catecholamine biosynthesis, storage, secretion and degradation. Depicted are important chromaffin cell-expressed genes and functional relationships between them. Two different parameters are color coded: the heritability (H2) of transcript abundance as computed from the RI panel and the normalized ratio (NR) for differences between the progenitors (on the left and right side of each box, respectively). The red curved arrow represents the catecholamine biosynthetic pathway, with the intermediates also in red. The blue curved arrows represent the detected gene regulatory networks from eQTL analysis, pointing from the regulator to the regulated gene.
Enzymatic activity of Dbh and Pnmt in adrenal medulla
To explore whether differences in mRNA levels across the RI panel translate into differences in enzyme activities, Dbh and Pnmt were assayed in adrenal gland homogenate. Compared with the BN.Lx, the activity of both enzymes was significantly decreased in the SHR. Dbh exhibited nearly a 2-fold decrease (NR = −1.8, P = 0.001), with H2 = 46% in the RI strains. Pnmt enzyme activity showed a smaller (NR = −1.3), but significant (P = 0.001), decrease in the SHR, with H2 = 34%.
Adrenal catecholamine content
Dbh and Pnmt are involved in the two final steps of catecholamine biosynthesis in the chromaffin cell. In order to investigate the effect of Dbh and Pnmt activity changes on catecholamine concentrations, their respective substrates and products, i.e. dopamine (DA), norepinephrine (NE) and epinephrine (EPI), were measured in adrenal tissue of the RI strains and their progenitors. SHR DA content was significantly increased (NR = 1.4, P = 0.048), whereas NE was significantly decreased (NR = −1.4, P = 0.002), when compared with the BN.Lx. EPI content did not show differences between the parental strains. Adrenal DA demonstrates much higher heritability (H2 = 63%) then NE, or EPI (H2 = 35% and H2 = 36%, respectively). Corticosterone is known to stimulate the gene expression of Pnmt, and to a lesser degree Dbh as well. Adrenal tissue corticosterone content was significantly different between the progenitors, with NR = 1.9 and P = 0.024 (Table 1). The H2 of adrenal corticosterone content in the RI strains was 15%. The results are summarized in Table 1. In sympathetic nerve (vas deferens), norepinephrine and dopamine displayed parental strain patterns similar to the adrenal results: with norepinephrine lower in SHR than BN.Lx (6.28 ± 0.41 versus 8.50 ± 1.84 ng/mg, NR = −1.4, P = 0.033), but dopamine higher in SHR than BN.Lx (0.21 ± 0.02 versus 0.09 ± 0.01 ng/mg, NR = 2.3, P = 0.001).
Table 1.
Biochemical phenotypes measured in adrenal tissue homogenate
| Phenotype | SHR (mean ± SD) | BN.Lx (mean ± SD) | SHR versus BN.Lx t-test P-value | NRSHR/BN.Lx | H2 in RI strains |
|---|---|---|---|---|---|
| DA (ng/mg) | 1.92 ± 0.46 | 1.41 ± 0.23 | 0.048 | 1.4 | 63% |
| Dbh (nmol/h/mg) | 1.24 ± 0.24 | 2.23 ± 0.35 | 0.001 | −1.8 | 46% |
| NE (ng/mg) | 67.6 ± 10.9 | 97.6 ± 8.07 | 0.002 | −1.4 | 35% |
| Pnmt (pmol/h/mg) | 33.0 ± 3.09 | 43.3 ± 3.29 | 0.001 | −1.3 | 34% |
| EPI (ng/mg) | 191 ± 27.8 | 191 ± 20.6 | 0.994 | 1.0 | 36% |
| Cort (IU/mg) | 466 ± 146 | 244 ± 73.1 | 0.024 | 1.9 | 15% |
HXB/BXH RI strains and the progenitor strains, SHR and BN.Lx, were typed for catecholaminergic biochemical parameters in the adrenal gland. The table shows progenitor strain means ± SD, t-test P-value for progenitor strain differences, normalized ratio (NR) to indicate the direction of the progenitor strain difference, and heritability (H2) calculated from RI strain data. Biochemical parameters were normalized to milligram adrenal weight. No significant differences in the mean adrenal gland weight (maw) were observed between the progenitor strains, with mawSHR = 16.0 ± 3.0 mg, mawBN.Lx = 13.5 ± 1.9 mg and t-test P = 0.18.
DA, dopamine; NE, norepinephrine; EPI, epinephrine; Cort, corticosterone.
Correlations among biochemical, physiological and gene expression phenotypes
To examine the extent to which biochemical phenotypes, transcript levels and cardiovascular physiological phenotypes are codetermined in the RI panel, Spearman's rank correlations were performed. The results can be found in Supplementary Material, Table S1. Adrenal DA, NE and EPI were significantly correlated across the RI panel. Adrenal DA content was negatively correlated (ρ = −0.408, P = 0.031) with adrenal Dbh activity. Adrenal EPI correlated positively with adrenal Pnmt (ρ = 0.429, P = 0.023). Dbh transcript abundance (probe set ID 1370564_at) was significantly positively (ρ = 0.570, P = 0.002) correlated with the adrenal Dbh enzyme activity. There was also a significant negative (ρ = −0.551, P = 0.002) correlation between Dbh transcript abundance and adrenal tissue DA concentration. Pnmt transcript abundance (probe set ID, 1371054_at) correlated positively (ρ = 0.436, P = 0.02) with adrenal Pnmt enzyme activity. Cardiovascular phenotypes were measured in the RI strains at age 12 weeks using telemetry. Dbh enzymatic activity was negatively correlated with systolic blood pressure (ρ = −0.476, P = 0.01). Pnmt enzyme activity was negatively correlated with heart rate (ρ = −0.383, P = 0.044), as was adrenal EPI (ρ = −0.550, P = 0.002) and DA (ρ = −0.382, P = 0.045).
Mapping QTLs for chromaffin cell-expressed genes
Gene transcripts (obtained from microarrays) of chromaffin cell-expressed genes with heritable transcript level variation were treated as quantitative traits and subjected to expression quantitative trait locus (eQTL) mapping. The results are summarized in Table 2. There were a total of six significant (including marginally significant) eQTLs (bolded P-values in Table 2). Uniformly, for each of these QTLs, the SHR allele at the eQTL peak locus was associated with a decrease in transcript level. Cis- and trans-eQTLs were determined by examining the relative position of a gene and its associated eQTL. Cis-eQTLs were mapped for Dbh, Pnmt and Vamp1, whereas trans-eQTLs were detected for Ddc, Vmat1 Chga transcripts.
Table 2.
Expression QTL mapping results for chromaffin cell-expressed genes with heritable transcript levels in the HXB/BXH RI strains
| Gene transcript |
Mapped expression QTLs |
|||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Transcript (probe ID) | gene symbol | gene position (Chr:Mbp) | progenitor t-test P-val | H2 in RI (%) | eQTL position (Chr:Mbp) | eQTL type | LOD | P-value | % variance attributable to eQTL | genes of interest in the 95% CI |
| 1387075_at | Th | 1:203 | 0.0365 | 48 | 8:80 | Trans | 2.51 | 0.176 | ||
| 1368064_a_at | Ddc | 14:93 | 0.0002 | 70 | 14:45 | Trans | 3.43 | 0.040 | 42 | |
| 1370564_at | Dbh | 3:6 | 0.0216 | 64 | 3:6 | Cis | 5.58 | 0.006 | 60 | Dbh |
| 1371054_at | Pnmt | 10:87 | 0.4492 | 60 | 10:90 | Cis | 2.95 | 0.049 | 36 | Pnmt |
| 1387999_at | Vmat1 | 16:22 | 0.4263 | 23 | 10:90 | Trans | 3.33 | 0.041 | 43 | Pnmt |
| 1387235_at | Chga | 6:127 | 0.1854 | 36 | 10:87 | Trans | 2.68 | 0.093 | 35 | Pnmt |
| 1373510_at | Vamp1 | 4:161 | 0.0213 | 53 | 4:161 | Cis | 3.50 | 0.072 | 43 | Vamp1 |
| 1368826_at | Comt | 11:85 | 0.0130 | 24 | 4:149 | Trans | 2.71 | 0.318 | ||
| 1368034_at | Chgb | 3:121 | 0.7601 | 25 | 8:80 | Trans | 3.11 | 0.124 | ||
| 1368044_at | Scg2 | 9:79 | 0.1274 | 49 | 5:16 | Trans | 2.11 | 0.674 | ||
| 1387154_at | Npy | 4:78 | 0.0031 | 26 | 17:78 | Trans | 2.14 | 0.426 | ||
| 1387221_at | Gch | 15:23 | 0.1691 | 39 | 19:48 | Trans | 2.12 | 0.320 | ||
| 1367695_at | Qdpr | 14:71 | 0.2837 | 28 | 10:99 | Trans | 3.12 | 0.149 | ||
Genes with heritable transcripts (see Fig. 1, coded yellow/orange) were included. Results for each transcript consist of LOD-peak position, eQTL type, LOD score and P-value. For the significant—including marginally significant—eQTLs (bolded P-values), the proportion of the variance in transcript level attributable to the eQTL is also given. The last column indicates chromaffin cell-expressed genes within the 95% CI of the mapped eQTLs.
Probe ID, affymetrix RAE 230A probe set ID; eQTL, expression QTL; H2 = heritability calculated in the HXB/BXH RI strains; LOD, logarithm of odds; 95% CI, 95% confidence interval for eQTL position.
The Vamp1 cis-eQTL mapped to chromosome 4 at 152–173 Mbp (95% CI), with a peak LOD = 3.50 and P = 0.072, explaining ∼43% of the transcript abundance variability. Ddc trans-eQTL mapped to chromosome 14 at 30–68 Mbp (95% CI), with a peak LOD = 3.43 and P = 0.040, explaining 42% of the transcript abundance variability.
Results of integrative analysis of transcript abundance and biochemical parameters for Dbh and Pnmt are described in the following two subheadings. Vmat1 and Chga trans-eQTLs co-localized with Pnmt cis-eQTL and are described together with Pnmt.
QTLs for adrenal Dbh transcript, Dbh activity and DA concentration cluster on chromosome 3p12
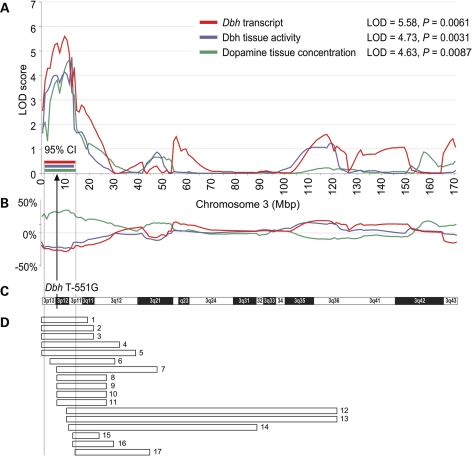
A Dbh cis-eQTL mapped to chromosome 3 at 1–14 Mbp (95% CI), with a peak LOD = 5.58 and P = 0.0061, explaining 60% of the Dbh transcript variability (see Fig. 2A, red curve). Dbh enzyme activity in adrenal tissue mapped to chromosome 3 at 1–12 Mbp (95% CI), LOD = 4.73 and P = 0.0031 (see Fig. 2A, blue curve). The QTL explains 54% of the variability in Dbh enzyme activity (see Fig. 2B, blue curve). The SHR allele at the peak locus was associated with a decrease in phenotypic value for Dbh transcript as well as Dbh enzyme activity. Adrenal DA concentration mapped to chromosome 3 at 2–14 Mbp (95% CI), with a peak LOD = 4.26 and P = 0.0354 (see Fig. 2A, green curve). The QTL explains 50% of adrenal DA concentration variability and the SHR allele at the peak locus was associated with an increase in adrenal DA (see Fig. 2B, green curve). It is important to note that the QTL confidence intervals for adrenal DA, Dbh activity and Dbh transcript are nearly identical, and contain the Dbh gene (see Fig. 2A and C).
Figure 2.
Co-localization of physiological and expression QTLs on the short arm of chromosome 3. (A) LOD plots for dopamine tissue concentration, Dbh tissue enzymatic activity and Dbh gene expression are shown. Peak LOD values are given in the inset. 95% confidence intervals (95% CIs), determined by 2-LOD drop, are shown beneath the peaks. (B) Additive/directional effect of the SHR allele on each trait at different RNO 3 loci is expressed as percent deviation from the overall trait mean. The arrow marks the location of the Dbh promoter SNP T − 551G. (C) Chromosome 3 idiogram is provided for reference. (D) Analysis of previously mapped cardiovascular QTLs overlapping with the conflated 95% CIs for the three QTLs reported here. Each bar represents a previously localized physiological QTL (data from http://rgd.mcw.edu). Bar numbers refer to Supplementary Material, Table S2 online, where details for each QTL can be found.
Regulation of Pnmt, Chga and Vmat1 from the Pnmt locus on chromosome 10q31
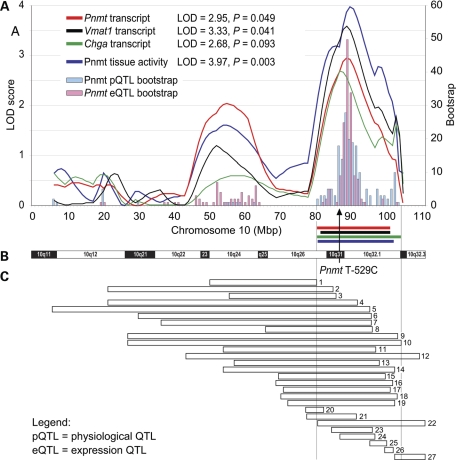
The Pnmt cis-eQTL is located on chromosome 10 at 81–101 Mbp (95% CI), with a peak LOD = 2.95, P = 0.049, and explains 36% of the Pnmt transcript variability (see Fig. 3A, red curve). Pnmt enzyme activity also demonstrated linkage to chromosome 10 at 81–102 Mbp (95% CI), with a peak LOD = 3.97 and P = 0.003 (see Fig. 3A, blue curve). The QTL explains 48% of Pnmt enzyme activity variability. For both the Pnmt transcript abundance and the Pnmt enzyme activity, the SHR allele at the peak locus was associated with a decrease in trait value. Because of the bimodal distribution of the LOD scores (most likely due to linkage disequilibrium within RNO 10), bootstrap tests were also performed. The results suggested that the evidence for linkage was most pronounced in the more telomeric peak, which we therefore regarded as the most likely position for the QTL. The 95% CI for these LOD peaks contain the Pnmt promoter SNP T-529C (see Fig. 3A and B). Both Vmat1 and Chga expression profiles mapped to the Pnmt region (see Fig. 3A). Vmat1 trans-eQTL mapped to chromosome 10 at 82–101 Mbp (95% CI), with a peak LOD = 3.33, P = 0.041, and with 43% of transcript variability attributable to the eQTL. Chga trans-eQTL mapped to chromosome 10 at 80–104 Mbp (95% CI), with a peak LOD = 3.50 and P = 0.093, explaining 35% of the transcript abundance variability. Importantly, this confidence interval contains the Pnmt gene, marked by the promoter SNP T-529C, and is nearly identical with the confidence intervals for Pnmt, Vmat1 and Chga eQTLs reported above. The SHR allele at the peak locus was associated with a decrease in transcript levels for both Vmat1 and Chga.
Figure 3.
Co-localization of physiological and expression QTLs on chromosome 10. (A) LOD plots for adrenal Pnmt tissue enzymatic activity and Pnmt, Vmat1 and Chga transcript abundance. Peak LOD values are given in the inset. Bootstrap test was used to estimate the 95% confidence intervals (95% CIs) for Pnmt tissue activity and Pnmt transcript abundance—see Materials and Methods for details. Horizontal bars represent 2-LOD drop CIs; different traits are colored same as the LOD plots. SHR allele at the peak locus was associated with a trait value decrease for all four traits. The arrow points to the location of the Pnmt promoter SNP T − 529G. (B) Idiogram of rat chromosome 10 is provided for reference. (C) Analysis of previously mapped cardiovascular QTLs overlapping with the conflated 95% CI for the QTLs reported here. Each bar represents a physiological QTL (data from http://rgd.mcw.edu). Bar numbers refer to Supplementary Material, Table S3 online, where details can be found.
Polymorphism discovery in the Dbh and the Pnmt genes
Systematic discovery of sequence polymorphism in Dbh and Pnmt between SHR and BN.Lx strains by re-sequencing yielded several single-nucleotide polymorphisms (SNPs) and one insertion/deletion (in/del). The results are described below. Numbers refer to the basepair distance from the CAP site; nucleotide change is given as BN.Lx → SHR. Re-sequencing in the Dbh gene (see Supplementary Material, Fig. S3) revealed eight variants, all of them SNPs: three in the promoter region (T-892G, T-885C and T-551G), four in the intronic regions (G2354A, T2513G, G4258T and A15184G), and one located downstream of the 3′-UTR (G18530A). Re-sequencing in the Pnmt gene (see Supplementary Material, Fig. S4) showed seven variants: four SNPs (T-529C, T-404C, C-396T and C-351T) and one in/del (-457 A/-) in the promoter region, one SNP in exon 1 (A209G, synonymous at codon 63, Ala63Ala) and one SNP in exon 3 (T1475C, coding for a non-synonymous amino acid change Val285Ala). Non-coding SNPs were located outside the highly conserved regions across species http://genome.ucsc.edu. The non-synonymous coding SNP T1475C identified in Pnmt, exon 3, alters the enzyme's last amino acid from Val to Ala; however, this substitution is outside the catalytic domain and is thus likely to be neutral (25).
In vitro studies on Dbh and Pnmt promoter polymorphism function
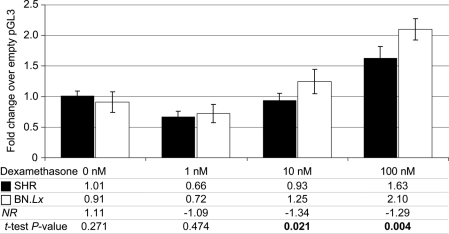
Bioluminescent activity of luciferase was measured in rat PC12 pheochromocytoma cells transfected with Dbh or Pnmt promoter/luciferase reporter plasmids, with or without activation by the glucocorticoid dexamethasone, or the secretory stimuli PACAP or nicotine. For each gene, two variants, which differed in the SNPs identified between SHR and BN.Lx, were tested and compared. Dbh promoter SNPs did not cause significantly different expression of the luciferase reporter under several conditions (see Supplementary Material, Fig. S5). Pnmt promoter SNPs, however, demonstrated a significant functional effect evident after stimulation with dexamethasone, resulting in a lesser increase of luciferase expression in the cells carrying the SHR promoter variant (see Supplementary Material, Fig. S6), congruent with the directional changes of the Pnmt biochemical phenotype. This was replicated in a separate experiment, proving a dose-dependent effect (see Fig. 4).
Figure 4.
Functional studies on the single-nucleotide polymorphisms identified in the Pnmt promoter. Bioluminescent activity of luciferase was measured in rat PC12 pheochromocytoma cells transfected with ∼1 kb segment of Pnmt promoter/luciferase reporter in pGL3-Basic vector (Promega) after 16 h incubation with incremental doses of the glucocorticoid dexamethasone. Each experiment was conducted in four replicates, with luciferase results normalized to cell protein in each plate. Results are presented as -fold augmentation by dexamethasone stimulus over the signal from cells transfected with a promoterless (empty) pGL3-Basic vector. Dexamethasone stimulation elicited significant differences in promoter activity. Differences between SHR and BN.Lx promoters were dose dependent and were significant starting at concentrations of 10 nm of dexamethasone.
Analysis of QTLs previously mapped to genomic regions supporting the Dbh and Pnmt QTLs
To explore the extent to which cardiovascular phenotypes studied by others are linked to chromosomal regions delimited by the 95% confidence intervals for the Dbh and Pnmt QTLs reported here, the Rat Genome Database http://rgd.mcw.edu was searched. The results of this analysis are summarized in Supplementary Material, Table S2 for Dbh, and in Supplementary Material, Table S3 for Pnmt. In the Dbh region, a total of 17 QTLs overlap wholly, or in part, with the interval spanning 1–14 Mbp, which corresponds to the conflated 95% CIs for the three QTLs on chromosome 3 (see Fig. 2D). There are 14 cardiovascular QTLs (eight blood pressure, four cardiac mass, one heart rate, one for aerobic running capacity), 2 for alcohol consumption and 1 for body weight. QTL symbols, progenitor strains of the populations in which QTLs were mapped, genomic positions, LOD scores and/or P-values are detailed in Supplementary Material, Table S2. In the Pnmt region, a total of 27 QTLs overlap wholly, or in part, with the interval 81–104 Mbp corresponding to the conflated 95% CIs for the four QTLs on chromosome 10 (see Fig. 3C). There are 20 blood pressure QTLs, 4 cardiac mass QTLs, 2 stress response QTLs and 1 heart rate QTL. QTL symbols, progenitor strains of the mapping populations, genomic positions, LOD scores and/or P-values are detailed in Supplementary Material, Table S3.
DISCUSSION
We have taken an integrative approach to the hereditary basis of complex traits, utilizing both gene expression and protein-level phenotypes to identify sequence variations that influence catecholamine biosynthesis and storage. Our study focused on heritable gene expression and biochemical traits of the sympathoadrenal system that may shed light on the genetic basis of cardiovascular pathology observed in the SHR. We utilized the HXB/BXH RI population to follow the segregation of these traits among strains of varying genetic makeup. The genetic determinants of regulatory networks in the chromaffin cell of the adrenal medulla were examined by linkage analysis. Through integration of gene expression profiling, biochemical phenotyping and quantitative trait locus mapping, cis-acting regulatory mechanisms were identified for the Dbh, Pnmt and Vamp1 genes (see Fig. 1). Analysis of tissue catecholamine concentrations and enzyme activities corroborated these findings for Dbh and Pnmt and provided a more complete picture of relevant regulatory networks and the interplay between various genes involved in catecholamine biosynthesis. Genetic variants were discovered in Dbh and Pnmt, which were subsequently tested in vitro for functional effects. The Pnmt locus also appeared to trans-regulate two additional genes, Chga and Vmat1, both being functionally linked with Pnmt.
Vamp1 is a small integral membrane protein of the SNARE class within secretory granules that plays a key role in membrane fusion and exocytosis (26). The cis-mediated decrease in Vamp1 expression described here is likely to result in changes in catecholamine secretion rate. To establish such effect, precise measurement of blood catecholamines would be required. In this study, we did not pursue this line of research. However, it is important to note that a decrease in Vamp1 transcript was associated with the SHR allele—a common observation in other findings reported here.
Dbh regulation and dopamine concentration in the adrenal gland
Dbh transcript levels positively correlated with Dbh tissue activity, suggesting that the source of the observed biochemical trait variation lies with differences in gene expression/mRNA stability, rather than enzyme structure. QTL analysis of both gene expression and biochemical data yielded highly significant, overlapping QTLs centered on the Dbh gene on chr 3 at 6 Mbp, implying that the differential gene expression is regulated in cis (see Fig. 2). The SHR allele was associated with a lower value for both of these traits, accompanied by an increase in DA, which is a substrate for Dbh. Viewed from a pathway perspective, increased DA can result either from an increase in quantity or turnover of the upstream enzymes (Th, Ddc), or from a decrease in quantity or turnover of the downstream enzymes (Dbh, Pnmt). Because DA concentration was negatively correlated with Dbh tissue activity, as well as with Dbh mRNA levels, the most parsimonious explanation of these findings is that decreased Dbh gene expression leads to decreased enzyme level, which in turn results in DA accumulation in the adrenal tissue of the SHR. This is further supported by a QTL for DA tissue concentration mapping to precisely the same region as the Dbh QTLs, but with an opposite allelic association. Furthermore, in the adrenals of the SHR, the Dbh transcript abundance shows the lowest levels (by RT–PCR) of all catecholamine biosynthetic enzymes—about half the value for Th that is traditionally considered the rate-limiting step in catecholamine biosynthesis. In contrast, the Dbh and Th levels are comparable in BN.Lx (see Supplementary Material, Fig. S2).
We therefore propose that lower Dbh in the young SHR presents a ‘bottleneck’ in catecholamine biosynthesis, leading to DA accumulation (and catecholamine depletion) in adrenergic cells, which then contributes to the pathogenesis of hypertension. Consistent with this notion, adrenal Dbh activity and SBP correlate negatively in the RI strains. Even though re-sequencing (see Supplementary Material, Fig. S3) and subsequent functional in vitro testing of discovered Dbh SNPs did not lead to identification of a causative sequence variant (see Supplementary Material, Fig. S5), the evidence for Dbh region involvement in the adrenal tissue Dbh enzyme activity, and the resulting DA concentration changes, is strong. In addition, the Dbh region is enriched in cardiovascular QTLs mapped in various crosses by multiple groups (see Fig. 2D, Supplementary Material, Table S2), suggesting the presence of an as-yet-to-be-identified important cardiovascular regulatory variant in this immediate region.
A strong cis-eQTL is likely to operate in all tissues where the gene is expressed (27). This viewpoint is supported by the findings by others of low Dbh activity in young SHR not only in adrenal glands, but also in heart ventricle and spleen (28), as well as in the brain (29). Low Dbh activity seems to lead to increased DA, accompanied by decreased NE. Increased brain DA was indeed described in young SHR (30). Dopamine systems in the brain are known to be involved in central blood pressure control and in integrating limbic information with cardiovascular homeostasis (31). Stimulation of the region of origin of the mesolimbic dopamine system in the brain, the ventral tegmental area, causes a long-lasting increase in blood pressure (32). Therefore, an increase in central DA may contribute to the development of hypertension. However, the other consequence of low central Dbh activity, i.e. the decrease in NE, can by itself contribute to a rise in blood pressure. NE has an inhibitory effect in the nucleus tractus solitarius on blood pressure elevation (33), and diminished central norepinephrine concentrations have been reported in the SHR (34); thus a decrease in NE may be involved in the development and progression of hypertension in the SHR. Decreased noradrenergic activity of sympathoinhibitory neurons in the anterior hypothalamus may also contribute to exacerbation of hypertension in the SHR (35).
Borderline hypertensive humans, in concordance with our rat data, appear to be characterized by increased DA coupled with Dbh suppression (36,37). Thus, genetically determined variation in Dbh activity, ultimately influenced by a promoter (enhancer) variant, may affect the DA/NE ratio in various tissues, including the basal ganglia, where such changes may have effects on central blood pressure regulation. Indeed, naturally occurring genetic variation at the human DBH locus can have profound effects upon blood pressure (38).
Regulation of Pnmt activity and gene expression: Chga and Vmat1 co-regulation
The neurotransmitter and hormone EPI is a physiologically active neuroregulator that assumes an important role in the stress response and is a major factor in the pathogenesis of cardiovascular and neuropsychiatric illnesses (39). The mechanisms by which Pnmt, the biosynthetic enzyme of EPI, is genetically regulated have been of interest in an effort to better understand the pathogenesis of these disorders. The general Pnmt region on rat chromosome 10 harbors many cardiovascular QTLs, implying the involvement of the region in blood pressure control (see Fig. 3C). In addition, significant difference in allelic frequencies of PNMT promoter SNPs were described between hypertensive and normotensive individuals, suggesting that genetic variation at the PNMT locus may play a role in the development of human essential hypertension (40,41).
Pnmt was previously examined as a hypertension candidate gene in the rat (42), but no polymorphisms were discovered between SHRSP and WKY in the Pnmt coding region. In the present study, we searched for polymorphisms between SHR and BN.Lx strains on the grounds of the observed differences in gene expression levels and enzyme activity, which mapped in cis (see Fig. 3), thus implying a causative variant in (or close to) the Pnmt genic region. Positive correlation of Pnmt gene expression levels with Pnmt enzyme activity, the discovery of SNP polymorphisms in the promoter and the lack of functionally significant polymorphism in the coding regions is consistent with the changes in quantity, rather than quality of the enzyme and point in toward differences in transcriptional regulation. This is further supported by the discovery that promoter SNPs caused differential gene expression in response to glucocorticoid (dexamethasone) stimulation in vitro (see Fig. 4). Glucocorticoids are the main regulators of Pnmt expression in vivo (43). The intra-adrenal portal vascular system provides the medulla with uniquely high concentrations of glucocorticoids, which are needed to induce Pnmt expression resulting in EPI synthesis (44). Despite the significant differences in Pnmt activity between the parental strains, we did not detect difference in the Pnmt transcript levels between the two parental strains in this study. However, there are clear cis-acting Pnmt transcript level-influencing allelic effects that segregate in the RI strains, as demonstrated by the significant cis-eQTL. Variation at the Pnmt locus is therefore likely to be functionally significant for determination of both Pnmt transcription and Pnmt enzymatic activity in the SHR.
The mechanism by which a decrease in Pnmt contributes to hypertension is illuminated by EPI-deficient, Pnmt knock-out mice (45), which become hypertensive under stress, possibly due to decreased central activity of vasodepressor adrenergic neurons of the brainstem, coupled with diminished peripheral (EPI-mediated) vasodilatation. In the present study, we did not find a significant correlation between Pnmt activity and blood pressure; however, adrenal Pnmt and its product EPI correlated negatively with heart rate.
An integrative finding of our study is mapping of trans-eQTLs for Chga and Vmat1 to the Pnmt region. Chga is crucial for the formation of secretory granules, within which it stabilizes catecholamines in a storage complex (46). Vmat1 function (47) involves transporting DA into the secretory granule for hydroxylation by Dbh to form NE, which is then transported by Vmat1 out of the granule for methylation by Pnmt to form EPI, which is finally transported—again by Vmat1—into the granule for storage, where it is stabilized by Chga (see Fig. 1). Chga also participates in autonomic control of blood pressure though its fragment catestatin, which causes inhibition of cholinergic-stimulated catecholamine release. Chga knock-out results in hypertension in the mouse (48), which is consistent with our results showing low adrenal Chga expression associated with the SHR allele. Vmat1 has not so far been implicated in hypertension but was found associated with neuropsychiatric diseases (49,50).
Given the functional coupling of these three genes, it is plausible to put forward a hypothesis that Pnmt, Chga and Vmat1 expression are regulated jointly by a variant within the Pnmt locus which thereby controls the formation of EPI. Our eQTL mapping results implicate the Pnmt genomic region as being central to this proposed co-regulation. Mapping the gene expression levels of Pnmt, Chga and Vmat1 to the same locus (see Fig. 3), as well as the uniform association of low gene expression with the SHR allele for all three genes seems to corroborate such co-regulation. However, despite mapping in cis to the Pnmt gene, trans-control of Pnmt expression itself from such a joint regulatory locus should also be considered when searching for the molecular mechanisms underlying the QTL co-localization at hand (see Fig. 3).
Conclusions and perspectives
We present evidence for simultaneous down-regulation of the transcription of five genes in the catecholamine biosynthetic pathway in the SHR. Decreased levels of catecholamine biosynthetic genes in the SHR have been described before (15). The novelty of our study lies in the examination of the genetic basis of gene expression differences of these genes in the HXB/BXH RI strains and finding simultaneous down-regulation of several key genes in the young SHR. Furthermore, we established that such down-regulations are heritable traits and that decreases in transcript levels are associated with the SHR genotype in each case. We also measured activities of Dbh and Pnmt and found that enzyme activity mirrored gene expression in that they were regulated in cis, implying primary, genetically regulated changes in expression of these genes between SHR and BN.Lx. Because the tissues for microarray analysis came from different animals from the tissues for biochemistries and RT–PCR, the finding of co-localized expression and biochemical QTLs for Dbh and Pnmt can be regarded as a confirmation of the genetic underpinnings of these traits. Furthermore, the finding of differential gene expression for Dbh, Pnmt and Vamp1 in young SHR supports control by inherited variants in the genic regions, rather then gene suppression in response to long-standing blood pressure increase. The down-regulation of these genes is therefore temporally independent of the development of hypertension, and may contribute to the pathogenesis of blood pressure elevation, possibly in part via central mechanisms, since catecholamines exert central vasodepressor actions in the brain stem (33,35). These results suggest new approaches to characterizing the role of the sympathochromaffin system in essential hypertension.
MATERIALS AND METHODS
Animals
HXB/BXH RI rat strains were produced by inbreeding between F2 generation males and females resulting from a sex-reciprocal cross of two highly inbred strains: BN.Lx (BN.Lx/Cub) and SHR (SHR/Ola) (24,51). In this study, we used 29 RI strains (HXB and BXH) at F60. Animals were housed in an air-conditioned animal facility and allowed free access to standard laboratory chow and water. All experiments were done in agreement with the Animal Protection Law of the Czech Republic (311/1997) and were approved by the Ethics Committee of the Institute of Physiology, Czech Academy of Sciences, Prague. Males were killed by cervical dislocation at 6 weeks of age. At this age the blood pressure is still normal, thus increasing the likelihood of separating the causative pathogenetic mechanisms from the consequential effects of fully developed hypertension.
Tissues for gene expression profiling, biochemical profiling and qRT-PCR validation
The animal materials originated from two separate tissue harvests: (i) adrenal glands harvested for global gene expression profiling and (ii) adrenal glands and post-ganglionic sympathetic nerve termini (from vas deferens) harvested for biochemical phenotyping and RT–PCR verification of gene expression profiling data. Both sets consisted of tissue harvested from the same HXB/BXH RI strains, but on two separate occasions, and thus from different individual members of the respective RI strains. The sets were similar in size: 29 RI strains, 2 progenitor strains, and 4–6 individuals per strain.
Gene expression profiling
RI strains were profiled for gene expression in adrenal tissue using the Affymetrix Gene Chip array RAE 230A (Affymetrix, Santa Clara, CA, USA). The original experimental design is discussed by Hubner et al. (52). The adrenal dataset has been submitted to ArrayExpress.
Biochemical profiling
Adrenal glands and sympathetic nerve termini (male vas deferens) were harvested, immediately frozen and stored at −80°C. From each pair of adrenal glands per harvested animal, one adrenal was used for biochemical phenotyping and the other was used to extract mRNA for RT–PCR validation of microarray data. Tissues for biochemical phenotyping were homogenized in 1.9 ml of 10 mm MES buffer (pH = 6.0), using a Tissuemizer (Tekmar, Cincinnati, OH, USA). Frozen tissues were placed into pre-cooled (4°C) buffer, homogenized, spun at 13 000 G for 1 min to clear debris, the supernatants divided into aliquots and placed immediately on dry ice to freeze. Aliquots were stored at −80°C. The following analyses were performed in aliquots of tissue homogenates: Dbh spectrophotometric enzymatic activity assay (53), Pnmt enzymatic activity assay (54), radioenzymatic catecholamine assay based on O-methylation (55,56), corticosterone assay (competitive immunoassay, Assay Designs, Inc., Ann Arbor, MI, USA); in plasma, catecholamines (dopamine, norepinephrine, epinephrine) were measured by radioenzymatic assay based on O-methylation (55,56). The results were normalized to milligram tissue wet weight. To explore whether differences in mRNA levels across the RI panel translate into differences in enzyme activities, Dbh and Pnmt were assayed in adrenal gland homogenate and normalized to milligram adrenal weight; no significant differences in the mean adrenal gland weight (maw) were observed between the progenitor strains, with mawSHR = 16.0 ± 3.0 mg, and mawBN.Lx = 13.5 ± 1.9 mg, and t-test P = 0.18.
Validation of microarray data by qRT–PCR
Total RNA was prepared from the freshly frozen adrenal glands of the progenitor strains (median yield: 55 µg/one adrenal gland). RNA was extracted by the RNAzol (guanidinium thiocyanate) method (TelTest, Friendswood, TX, USA), followed by RNAse-free DNAseI (Qiagen, Valencia, CA, USA) treatment (to eliminate residual genomic DNA). Integrity of the RNA was confirmed by the appearance of 28S and 18S rRNA bands on ethidium bromide-stained gels. Total RNA was quantitated using a Ribogreen Quantitation kit (Molecular Probes, Invitrogen). First-strand cDNA was prepared from 1 µg of total RNA template by reverse transcription with the ‘SuperScript™ first-strand synthesis system for RT-PCR’, using SuperScript II reverse transcriptase (Invitrogen, Carlsbad, CA, USA), and random hexamer primers. Samples were randomized. RT–PCR was performed using real-time TaqMan technology with a Sequence Detection System, model 7700 (Perkin Elmer) and fluorescent plate reader, using the Amplifluor™ universal detection system (Serologicals Corporation; Norcross, GA). Quantitative RT–PCR primers for Th, Ddc, Dbh and Pnmt were designed using Primer Express V.2.0 (PE Applied Biosystems, Foster City, CA, USA). For primer detail, see Supplementary Material, Table S4. Normalization was performed by quantitating the endogenous 18S rRNA and transcript abundance expressed as fold modulation over 18S rRNA.
Pathway annotation
Pathways, in which the differentially expressed genes are involved, were identified using Kyoto Encyclopedia of Genes and Genomes (KEGG, www.genome.jp/kegg) and annotated with Gene Map Annotator and Pathway Profiler (GenMAPP 2.1, www.genmapp.org) (57).
Blood pressure measurement
Arterial blood pressure and heart rate were measured using radiotelemetry in 12-week-old unanesthetized, unrestrained males from the progenitor SHR and BN.Lx strains as well as from RI strains (N = 6–8 males per strain). All rats were allowed to recover for at least 7 days after surgical implantation of radiotelemetry transducers (Data Sciences International, Inc.) before the start of blood pressure recordings. Pulsatile pressures were recorded in 5 s bursts every 10 min throughout the day and night and 12 and 24 h averages for systolic and diastolic arterial blood pressures were calculated for each rat for a 1 week period. The results from each rat in the same group were then averaged to obtain the group means.
Statistical analysis and heritability (H2) calculation
Phenotype data were checked for outliers, using a method described by Grubbs (58), and optimized for three to seven observations, corresponding to the number of individual rats within each strain. Manifest outliers were removed. Data from all ∼150 samples were grouped according to strain resulting in 31 groups (29 RI strains + 2 progenitors). T-tests were performed to detect significant differences between the progenitor strains. H2 for gene expression profiles and biochemical phenotypes was calculated using a technique that has been designed for use in RI strains that corrects for the inbreeding incurred during the RI strain production (59). Pearson's product–moment correlation and Spearman's rank correlation coefficients were computed to assess the degree of correlation among phenotypes.
Normalized ratio
NR is used here to compare biochemical phenotype means and gene expression profile means for the two progenitor strains, the SHR and the BN.Lx. It is a variant of the simple fold-change value, e.g. [SHR]/[BN.Lx], but it transforms the values between 0 and 1 into values between −INF(infinity) and −1 by inverting them and multiplying by negative one. It is calculated as follows: if [SHR] ≥ [BN.Lx], then NR = [SHR]/[BN.Lx]; if [SHR] < [BN.Lx], then NR = −[BN.Lx]/[SHR]. The main advantage of NR is its symmetry about zero.
QTL analysis
A new SNP genotype-based linkage map (60) of the HXB/BXH RI set was employed to perform genome-wide scans to detect QTLs for measured biochemical phenotypes using QTL Cartographer (61) and Map Manager QTX (62), and to detect expression QTLs using QTL Reaper (63) software packages. Permutation analysis (64) was carried out to assess the probabilistic significance of the linkages and to correct for multiple testing across genetic markers to obtain a genome-wide corrected P-value. To estimate the size of the chromosomal segment that, with a 95% confidence, contains detected QTLs (95% CI), two complementary strategies were implemented: the 2-LOD support interval method (65) and the bootstrap test (66). The bootstrap test estimates a confidence interval by creating multiple bootstrap data sets by randomly choosing strains with replacement from the original RI set; each bootstrap data set is then used for QTL mapping, and the location of the strongest QTL for each set is recorded. Here, 200 bootstrap data sets per phenotype were used. These locations are summarized in a histogram that shows the size of the region in which the QTL would be expected to be found.
Definition of cis- and trans-acting eQTLs
Cis-eQTLs were defined as eQTLs that map within 10 Mbp upstream or downstream of the physical location of the probe set on the genomic sequence (20 Mbp total window size). All eQTLs that fell outside of this window were by default classified as trans. Physical locations of probe sets were downloaded from the UCSC Genome Browser website http://genome.ucsc.edu.
Polymorphism discovery at the Pnmt and Dbh genes in the SHR and Bn.Lx strains
The progenitors of the HXB/BXH RI strains, the Brown Norway rat (BN.Lx/Cub) and the Spontaneously Hypertensive Rat (SHR/Ola) were used. Liver was collected from one male per each strain at 6 weeks of age. Progenitor strain DNA was extracted from liver tissue, using the DNeasy Blood & Tissue Kit (Qiagen) and adhering to the manufacturer's protocol. To eliminate residual RNA, samples were treated with RNase A (Qiagen). Primer3 (67) web-based application was used to design PCR primers for amplification of ∼800 bp-long overlapping segments, spanning all exons, exon/intron borders, ∼1.75 kb of proximal promoter and ∼1.75 kb of the 3′ [downstream] sequence of the Dbh gene. However, in the case of Pnmt, the extent of re-sequencing was limited by the extent of known genomic sequence (gb: X75333.1; gi: 414186) in this locus, resulting in only ∼1000 bp upstream and ∼100 bp downstream segments were re-sequenced. Initial amplification to enhance the target sequences was carried out in 25 µl PCR containing 25 ng genomic DNA, 2 mm MgCl2, 10 mm Tris–HCl, 200 µm dNTP, 0.5 U Amplitaq Gold DNA Polymerase (PE Applied Biosystems) and 50 pmol of each primer. PCR was performed by Peltier Tetrad Thermal Cycler (MJ Research, Watertown, MA, USA). The Touchdown Profile program (MJ Research) was used, which begins at annealing temperatures of 66°C and runs down to 50°C at 1°C/cycle for the first 16 PCR cycles, followed by a uniform three-step amplification profile (94°C denaturing step for 30 s, 50°C annealing step for 30 s, 72°C extension step for 30 s) for another 24 cycles, finally holding at 10°C. Enzymatic purification (Exo-SAP) was then pursued using Exonuclease I (Fermentas, Inc., ON, Canada) and Shrimp Alkaline Phosphatase (Fermentas, Inc.). Fifteen microliters of each PCR product were mixed with 0.225 µl of Exonuclease I (20 U/µl), 1.2 µl of SAP (1 U/µl) and 4.35 µl of water. The Exo-SAP program (MJ Research) was used, which runs at 37°C for 30 min, followed by 15 min at 85°C. The cycle-sequencing reaction was performed following the Big Dye Terminator Version 3.1 (PE Applied Biosystems) protocol on 2.5 µl of the template (Exo-SAP product), using either the forward or the reverse primer for the Big Dye amplification origin. The cycle-sequencing reaction was subsequently performed with both primers to achieve better coverage. The cycle-sequencing reaction program begun at 96°C for 1 min., followed by a uniform three-step amplification profile (96°C denaturing step for 10 s, 50°C annealing step for 5 s, 60°C extension step for 4 min) for another 29 cycles, holding at 4°C. Finally, the product of the cycle-sequencing reaction was purified on soaked Sephadex G50-50 beads (Sigma-Aldrich, St. Louis, MO, USA), and placed in multiscreen filtration plates (Fisher Scientific, Pittsburgh, PA, USA). After spinning into clean plates, loading dye was added and samples were then loaded into an ABI PRISM 3100 capillary DNA Analyzer (PE Applied Biosystems) according to the manufacturer's instructions.
Pyrosequencing
Single-nucleotide polymorphisms at Dbh T-550G and Pnmt T-529C were scored using Pyrosequencing (Biotage, Charlottesville, VA, USA), performed according to the described protocol (68). Primers were designed using the PSQ Assay Design software (Biotage) and ordered from Sigma-Proligo (http://www.proligo.com). A PCR product from each sample was generated in a 10 µl reaction containing 1× AmpliTaq Gold Master Mix, 2 U of AmpliTaq Gold (Applied Biosystems), 250 µm dNTPs, 0.25 µm forward primer, 0.25 µm reverse primer and 5 µl of bisulphite-modified sample DNA. The amplifications were carried out at 95°C for 10 min, followed by a six-cycle touchdown PCR protocol of 95°C for 1 min, 63°C for 1 min with −1°C for each cycle to 58°C, and 72°C for 1 min. This was followed by 44 cycles at 95°C for 1 min, 60°C for 1 min, 72°C for 1 min and a 10 min extension at 72°C. Single-stranded DNA of 10 µl of each PCR sample was generated following the PSQ 96 sample preparation guide using a vacuum filtration sample device following the manufacturer's instructions (Biotage). The single-stranded product was annealed to 0.4 µm of the sequencing primer placed at 85°C for 2 min and cooled to room temperature for 5 min. Pyrosequencing was performed on a PSQ96 HS 96A system (Biotage) with the Pyro Gold Reagent kit (Biotage) according to the manufacturer's instructions.
Sequence analysis
Output data from the ABI PRISM 3100 DNA Analyzer were read by the BioEdit sequence alignment editor (Ibis Therapeutics, Carlsbad, CA, USA). SHR and BN.Lx sequences were aligned and between-strain polymorphisms identified. MacVector (MacVector, Inc., Cary, NC, USA) was used to analyze the sequence for differences in transcription factor binding sites resulting from such polymorphisms.
Candidate gene promoter/reporter transfection assay
Construction of the promoter/reporter plasmids
Primer3 (67) http://frodo.wi.mit.edu/cgi-bin/primer3/primer3_www.cgi was used to design PCR primers to amplify a ∼1.1 kb fragment of Dbh proximal promoter, or a ∼1 kb fragment of the Pnmt proximal promoter from genomic DNA of the SHR and BN.Lx. Computational sequence analysis of the Dbh and Pnmt promoters revealed the absence of SacI and XhoI restriction enzyme sites. Therefore, an SacI (5′-GAGCTC-3′) or XhoI (5′-CTCGAG-3′) restriction enzyme site was inserted in the 5′ end of the forward and reverse PCR primers to allow cloning. The Dbh or Pnmt promoters were PCR amplified from SHR and BN.Lx genomic DNA and subsequently digested with SacI and XhoI. T4 DNA ligase (Invitrogen) was used to insert the promoter fragments between the SacI and XhoI sites in the polylinker region of the firefly luciferase reporter vector, pGL3-Basic (Promega, Madison, WI, USA), which lacks eukaryotic promoter and enhancer sequences, and contains the cDNA for firefly luciferase. Creation of SNP variants of the Pnmt promoter/luciferase reporter constructs was accomplished using the QuickChange Site-Directed Mutagenesis Kit (Stratagene, La Jolla, CA, USA). Correct insertion of the promoters was confirmed by DNA sequencing. Plasmid DNA for transfection was prepared and purified using the QIAfilter Plasmid Midi Kit (Qiagen).
Transfection of the promoter/reporter constructs: Rat PC12 pheochromocytoma cells [grown in DMEM high glucose (Invitrogen) with 5% heat-inactivated fetal bovine serum (Gemini Bioproducts, Woodland, CA, USA), 10% heat-inactivated horse serum (Gemini Bioproducts), penicillin (100 U/ml), streptomycin (100 µg/ml), and l-glutamine (0.292 mg/ml)] were transfected (at 50–60% confluence, 1 day after splitting 1:4) with Dbh or Pnmt promoter reporter plasmid DNA [1 µg supercoiled DNA per well; 12-well polystyrene plates (coated with poly-l-lysine; Sigma), 2.2 cm diameter wells, Corning Inc., Corning, NY, USA] using the liposome method (Superfect; Qiagen). Cells were incubated with or without dexamethasone (different concentrations: 1 nm, 10 nm or 100 nm; Calbiochem), 100 nm pituitary adenylate cyclase-activating peptide [PACAP (ovine; Calbiochem)] or 1 mm nicotine (Sigma). Cells were lysed 16 h after transfection with lysis buffer (300 µl per well) [0.1 m phosphate buffer (K2HPO4 + KH2PO4), pH 7.8, 1 mm DTT and 0.1% Triton-X 100].
Luciferase reporter activity assay
The bioluminescent activity of luciferase in 80 µl of transfected cell lysates was determined using the AutoLumat LB 953 luminometer (EG&G Berthold, Nashua, NH, USA) to measure light emission (incubation time = 0 s, measure time = 10 s, temperature = 25°C) after addition of assay buffer [100 µl per sample; 100 mm Tris-acetate (pH 7.8), 10 mm Mg-acetate, 1 mm EDTA (pH 8.0), 3 mm ATP and 100 µm luciferin (Sigma-Aldrich)]. As a control for varying cell number within individual wells, the total protein content was measured in the cell lysate using the Bio-Rad Protein Assay [coomassie blue dye absorbance shift; based on the Bradford method (Bio-Rad, Hercules, CA, USA)]. Luciferase activity in the cell lysate is expressed as the normalized ratio of (luciferase activity)/(total protein content) or (RLU/µg protein).
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health and the Veterans Administration (D.T.O.C., N.J.S., S.K.M.) and by EU funding via the EURATools consortium (T.J.A., N.H., M.P.). M.L.J. was supported by an American Heart Association pre-doctoral fellowship award. M.P. is supported by grant IAA500110604 from the Grant Agency of the Czech Academy of Sciences. M.P. is an international research scholar of the Howard Hughes Medical Institute.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Dr Kelly A. Frazer for reading this manuscript carefully and providing insightful comments.
Conflict of Interest statement. None declared.
REFERENCES
- 1.DeQuattro V., Feng M. The sympathetic nervous system: the muse of primary hypertension. J. Hum. Hypertens. 2002;16(Suppl. 1):S64–S69. doi: 10.1038/sj.jhh.1001346. [DOI] [PubMed] [Google Scholar]
- 2.Esler M. The sympathetic system and hypertension. Am. J. Hypertens. 2000;13:99S–105S. doi: 10.1016/s0895-7061(00)00225-9. doi:10.1016/S0895-7061(00)00225-9. [DOI] [PubMed] [Google Scholar]
- 3.Greifenkamp J.D., DiPette D.J. Adrenal medulla. Curr. Hypertens. Rep. 1999;1:241–245. doi: 10.1007/s11906-999-0028-3. doi:10.1007/s11906-999-0028-3. [DOI] [PubMed] [Google Scholar]
- 4.Anderson E.A., Sinkey C.A., Lawton W.J., Mark A.L. Elevated sympathetic nerve activity in borderline hypertensive humans. Evidence from direct intraneural recordings. Hypertension. 1989;14:177–183. doi: 10.1161/01.hyp.14.2.177. [DOI] [PubMed] [Google Scholar]
- 5.Mancia G., Grassi G., Giannattasio C., Seravalle G. Sympathetic activation in the pathogenesis of hypertension and progression of organ damage. Hypertension. 1999;34:724–728. doi: 10.1161/01.hyp.34.4.724. [DOI] [PubMed] [Google Scholar]
- 6.Cabassi A., Vinci S., Calzolari M., Bruschi G., Borghetti A. Regional sympathetic activity in pre-hypertensive phase of spontaneously hypertensive rats. Life Sci. 1998;62:1111–1118. doi: 10.1016/s0024-3205(98)00034-4. doi:10.1016/S0024-3205(98)00034-4. [DOI] [PubMed] [Google Scholar]
- 7.Wenzel R.R., Bruck H., Noll G., Schafers R.F., Daul A.E., Philipp T. Antihypertensive drugs and the sympathetic nervous system. J. Cardiovasc. Pharmacol. 2000;35:S43–S52. doi: 10.1097/00005344-200000004-00006. doi:10.1097/00005344-200000004-00006. [DOI] [PubMed] [Google Scholar]
- 8.Rabbia F., Martini G., Cat Genova G., Milan A., Chiandussi L., Veglio F. Antihypertensive drugs and sympathetic nervous system. Clin. Exp. Hypertens. 2001;23:101–111. doi: 10.1081/ceh-100001201. doi:10.1081/CEH-100001201. [DOI] [PubMed] [Google Scholar]
- 9.Nagatsu T., Kato T., Numata Y., Keiko I., Umezawa H. Serum dopamine beta-hydroxylase activity in developing hypertensive rats. Nature. 1974;251:630–631. doi: 10.1038/251630a0. doi:10.1038/251630a0. [DOI] [PubMed] [Google Scholar]
- 10.Grobecker G., Roizen M.F., Weise V., Saavedra J.M., Kopin I.J. Sympathoadrenal medullary activity in young, spontaneously hypertensive rats. Nature. 1975;258:267–268. doi: 10.1038/258267a0. doi:10.1038/258267a0. [DOI] [PubMed] [Google Scholar]
- 11.Nagaoka A., Lovenberg W. Plasma norepinephrine and dopamine-beta-hydroxylase in genetic hypertensive rats. Life Sci. 1976;19:29–34. doi: 10.1016/0024-3205(76)90370-2. doi:10.1016/0024-3205(76)90370-2. [DOI] [PubMed] [Google Scholar]
- 12.Borkowski K.R., Quinn P. Adrenaline and the development of spontaneous hypertension in rats. J. Auton. Pharmacol. 1985;5:89–100. doi: 10.1111/j.1474-8673.1985.tb00109.x. doi:10.1111/j.1474-8673.1985.tb00109.x. [DOI] [PubMed] [Google Scholar]
- 13.Borkowski K.R. Effect of adrenal demedullation and adrenaline on hypertension development and vascular reactivity in young spontaneously hypertensive rats. J. Auton. Pharmacol. 1991;11:1–14. doi: 10.1111/j.1474-8673.1991.tb00239.x. doi:10.1111/j.1474-8673.1991.tb00239.x. [DOI] [PubMed] [Google Scholar]
- 14.Lee R.M., Borkowski K.R., Leenen F.H., Tsoporis J., Coughlin M. Combined effect of neonatal sympathectomy and adrenal demedullation on blood pressure and vascular changes in spontaneously hypertensive rats. Circ. Res. 1991;69:714–721. doi: 10.1161/01.res.69.3.714. [DOI] [PubMed] [Google Scholar]
- 15.Grobecker H., Saavedra J.M., Weise V.K. Biosynthetic enzyme activities and catecholamines in adrenal glands of genetic and experimental hypertensive rats. Circ. Res. 1982;50:742–746. doi: 10.1161/01.res.50.5.742. [DOI] [PubMed] [Google Scholar]
- 16.Moura E., Pinho Costa P.M., Moura D., Guimaraes S., Vieira-Coelho M.A. Decreased tyrosine hydroxylase activity in the adrenals of spontaneously hypertensive rats. Life Sci. 2005;76:2953–2964. doi: 10.1016/j.lfs.2004.11.017. doi:10.1016/j.lfs.2004.11.017. [DOI] [PubMed] [Google Scholar]
- 17.Nakamura K. Enhanced sympathetic activity in young spontaneously hypertensive rats is not the trigger mechanism for genetic hypertension. Naunyn Schmiedebergs Arch. Pharmacol. 1977;299:143–148. doi: 10.1007/BF00498555. doi:10.1007/BF00498555. [DOI] [PubMed] [Google Scholar]
- 18.Teitelman G., Ross R.A., Joh T.H., Reis D.J. Differences in utero in activities of catecholamine biosynthetic enzymes in adrenals of spontaneously hypertensive rats. Clin. Sci. (Lond.) 1981;61(Suppl. 7):227s–230s. doi: 10.1042/cs061227s. [DOI] [PubMed] [Google Scholar]
- 19.Kumai T., Tanaka M., Watanabe M., Kobayashi S. Elevated tyrosine hydroxylase mRNA levels in the adrenal medulla of spontaneously hypertensive rats. Jpn. J. Pharmacol. 1994;65:367–369. doi: 10.1254/jjp.65.367. doi:10.1254/jjp.65.367. [DOI] [PubMed] [Google Scholar]
- 20.Kumai T., Tanaka M., Tateishi T., Watanabe M., Nakura H., Asoh M., Kobayashi S. Effects of anti-androgen treatment on the catecholamine synthetic pathway in the adrenal medulla of spontaneously hypertensive rats. Naunyn Schmiedebergs Arch. Pharmacol. 1998;357:620–624. doi: 10.1007/pl00005216. doi:10.1007/PL00005216. [DOI] [PubMed] [Google Scholar]
- 21.Yamabe H., De Jong W., Lovenberg W. Further studies on catecholamine synthesis in the spontaneously hypertensive rat: catecholamine synthesis in the central nervous system. Eur. J. Pharmacol. 1973;22:91–98. doi: 10.1016/0014-2999(73)90188-x. [DOI] [PubMed] [Google Scholar]
- 22.Nagatsu T., Ikuta K., Numata Y., Kato T., Sano M. Vascular and brain dopamine beta-hydroxylase activity in young spontaneously hypertensive rats. Science. 1976;191:290–291. doi: 10.1126/science.1858. [DOI] [PubMed] [Google Scholar]
- 23.Ciaranello R.D., Hoffman H.J., Shire J.G., Axelrod J. Genetic regulation of the catecholamine biosynthetic enzymes. II. Inheritance of tyrosine hydroxylase, dopamine-beta-hydroxylase, and phenylethanolamine N-methyltransferase. J. Biol. Chem. 1974;249:4528–4536. [PubMed] [Google Scholar]
- 24.Pravenec M., Klir P., Kren V., Zicha J., Kunes J. An analysis of spontaneous hypertension in spontaneously hypertensive rats by means of new recombinant inbred strains. J. Hypertens. 1989;7:217–221. [PubMed] [Google Scholar]
- 25.Betts M.J., Russell R.B. Bioinformatics for Geneticists. UK: Wiley, West Sussex; 2003. Amino Acid Properties and Consequences of Subsitutions. In Barnes, M.R. and Gray, I.C. [Google Scholar]
- 26.Ungermann C., Langosch D. Functions of SNAREs in intracellular membrane fusion and lipid bilayer mixing. J. Cell Sci. 2005;118:3819–3828. doi: 10.1242/jcs.02561. doi:10.1242/jcs.02561. [DOI] [PubMed] [Google Scholar]
- 27.Petretto E., Mangion J., Dickens N.J., Cook S.A., Kumaran M.K., Lu H., Fischer J., Maatz H., Kren V., Pravenec M., et al. Heritability and tissue specificity of expression quantitative trait loci. PLoS Genet. 2006;2:e172, 1625–1633. doi: 10.1371/journal.pgen.0020172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuchel O., Racz K., Debinski W., Buu N.T. A defective beta-hydroxylation of dopamine may precede the full development of hypertension in spontaneously hypertensive rats. Can. J. Cardiol. 1989;5:327–331. [PubMed] [Google Scholar]
- 29.Saavedra J.M., Grobecker H., Axelrod J. Changes in central catecholaminergic neurons in the spontaneously (genetic) hypertensive rat. Circ. Res. 1978;42:529–534. doi: 10.1161/01.res.42.4.529. [DOI] [PubMed] [Google Scholar]
- 30.Howes L.G., Rowe P.R., Summers R.J., Louis W.J. Age related changes of catecholamines and their metabolites in central nervous system regions of spontaneously hypertensive (SHR) and normotensive Wistar–Kyoto (WKY) rats. Clin. Exp. Hypertens. A. 1984;6:2263–2277. doi: 10.3109/10641968409052207. doi:10.3109/10641968409052207. [DOI] [PubMed] [Google Scholar]
- 31.Cornish J.L., Wilks D.P., Van den Buuse M. A functional interaction between the mesolimbic dopamine system and vasopressin release in the regulation of blood pressure in conscious rats. Neuroscience. 1997;81:69–78. doi: 10.1016/s0306-4522(97)00157-7. doi:10.1016/S0306-4522(97)00157-7. [DOI] [PubMed] [Google Scholar]
- 32.van den Buuse M. Pressor responses to brain dopaminergic stimulation. Clin. Exp. Pharmacol. Physiol. 1997;24:764–769. doi: 10.1111/j.1440-1681.1997.tb02129.x. [DOI] [PubMed] [Google Scholar]
- 33.Takami T., Ito H., Suzuki T. Decreased norepinephrine content in the medulla oblongata in severely hypertensive rats. Clin. Exp. Pharmacol. Physiol. 1993;20:161–167. doi: 10.1111/j.1440-1681.1993.tb01664.x. doi:10.1111/j.1440-1681.1993.tb01664.x. [DOI] [PubMed] [Google Scholar]
- 34.Yamori Y., Lovenberg W., Sjoerdama A. Norepinephrine metabolism in brainstem of spontaneously hypertensive rats. Science. 1970;170:544–546. doi: 10.1126/science.170.3957.544. [DOI] [PubMed] [Google Scholar]
- 35.Oparil S., Yang R.H., Jin H.K., Wyss J.M., Chen Y.F. Central mechanisms of hypertension. Am. J. Hypertens. 1989;2:477–485. doi: 10.1093/ajh/2.6.477. [DOI] [PubMed] [Google Scholar]
- 36.Shigetomi S., Buu N.T., Kuchel O. Dopaminergic abnormalities in borderline essential hypertensive patients. Hypertension. 1991;17:997–1002. doi: 10.1161/01.hyp.17.6.997. [DOI] [PubMed] [Google Scholar]
- 37.DeQuattro V., Campese V., Lurvey A., Yen G., Kypridakis G. Low response of serum dopamine beta-hydroxylase to stimuli in primary hypertension. Decreased DbetaH response in hypertension. Biochem. Med. 1976;15:1–9. doi: 10.1016/0006-2944(76)90067-3. doi:10.1016/0006-2944(76)90067-3. [DOI] [PubMed] [Google Scholar]
- 38.Zabetian C.P., Romero R., Robertson D., Sharma S., Padbury J.F., Kuivaniemi H., Kim K.S., Kim C.H., Kohnke M.D., Kranzler H.R., et al. A revised allele frequency estimate and haplotype analysis of the DBH deficiency mutation IVS1 + 2T→C in African- and European-Americans. Am. J. Med. Genet. A. 2003;123:190–192. doi: 10.1002/ajmg.a.20300. [DOI] [PubMed] [Google Scholar]
- 39.Her S., Bell R.A., Bloom A.K., Siddall B.J., Wong D.L. Phenylethanolamine N-methyltransferase gene expression. Sp1 and MAZ potential for tissue-specific expression. J. Biol. Chem. 1999;274:8698–8707. doi: 10.1074/jbc.274.13.8698. doi:10.1074/jbc.274.13.8698. [DOI] [PubMed] [Google Scholar]
- 40.Cui J., Zhou X., Chazaro I., DeStefano A.L., Manolis A.J., Baldwin C.T., Gavras H. Association of polymorphisms in the promoter region of the PNMT gene with essential hypertension in African Americans but not in whites. Am. J. Hypertens. 2003;16:859–863. doi: 10.1016/s0895-7061(03)01026-4. doi:10.1016/S0895-7061(03)01026-4. [DOI] [PubMed] [Google Scholar]
- 41.Rana B.K., Insel P.A., Payne S.H., Abel K., Beutler E., Ziegler M.G., Schork N.J., O'Connor D.T. Population-based sample reveals gene–gender interactions in blood pressure in White Americans. Hypertension. 2007;49:96–106. doi: 10.1161/01.HYP.0000252029.35106.67. doi:10.1161/01.HYP.0000252029.35106.67. [DOI] [PubMed] [Google Scholar]
- 42.Koike G., Jacob H.J., Krieger J.E., Szpirer C., Hoehe M.R., Horiuchi M., Dzau V.J. Investigation of the phenylethanolamine N-methyltransferase gene as a candidate gene for hypertension. Hypertension. 1995;26:595–601. doi: 10.1161/01.hyp.26.4.595. [DOI] [PubMed] [Google Scholar]
- 43.Wong D.L. Epinephrine biosynthesis: hormonal and neural control during stress. Cell Mol. Neurobiol. 2006;26:891–900. doi: 10.1007/s10571-006-9056-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Wurtman R.J. Stress and the adrenocortical control of epinephrine synthesis. Metabolism. 2002;51:11–14. doi: 10.1053/meta.2002.33185. doi:10.1053/meta.2002.33185. [DOI] [PubMed] [Google Scholar]
- 45.Bao X., Lu C.M., Liu F., Gu Y., Dalton N.D., Zhu B.Q., Foster E., Chen J., Karliner J.S., Ross J., Jr, et al. Epinephrine is required for normal cardiovascular responses to stress in the phenylethanolamine N-methyltransferase knockout mouse. Circulation. 2007;116:1024–1031. doi: 10.1161/CIRCULATIONAHA.107.696005. doi:10.1161/CIRCULATIONAHA.107.696005. [DOI] [PubMed] [Google Scholar]
- 46.Taupenot L., Harper K.L., O'Connor D.T. The chromogranin–secretogranin family. N. Engl. J. Med. 2003;348:1134–1149. doi: 10.1056/NEJMra021405. doi:10.1056/NEJMra021405. [DOI] [PubMed] [Google Scholar]
- 47.Henry J.P., Botton D., Sagne C., Isambert M.F., Desnos C., Blanchard V., Raisman-Vozari R., Krejci E., Massoulie J., Gasnier B. Biochemistry and molecular biology of the vesicular monoamine transporter from chromaffin granules. J. Exp. Biol. 1994;196:251–262. doi: 10.1242/jeb.196.1.251. [DOI] [PubMed] [Google Scholar]
- 48.Mahapatra N.R., O'Connor D.T., Vaingankar S.M., Hikim A.P., Mahata M., Ray S., Staite E., Wu H., Gu Y., Dalton N., et al. Hypertension from targeted ablation of chromogranin A can be rescued by the human ortholog. J. Clin. Invest. 2005;115:1942–1952. doi: 10.1172/JCI24354. doi:10.1172/JCI24354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Richards M., Iijima Y., Kondo H., Shizuno T., Hori H., Arima K., Saitoh O., Kunugi H. Association study of the vesicular monoamine transporter 1 (VMAT1) gene with schizophrenia in a Japanese population. Behav. Brain. Funct. 2006;2:39. doi: 10.1186/1744-9081-2-39. doi:10.1186/1744-9081-2-39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lohoff F.W., Dahl J.P., Ferraro T.N., Arnold S.E., Gallinat J., Sander T., Berrettini W.H. Variations in the vesicular monoamine transporter 1 gene (VMAT1/SLC18A1) are associated with bipolar i disorder. Neuropsychopharmacology. 2006;31:2739–2747. doi: 10.1038/sj.npp.1301196. doi:10.1038/sj.npp.1301196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pravenec M., Kren V., Krenova D., Bila V., Zidek V., Simakova M., Musilova A., van Lith H.A., van Zutphen L.F. HXB/Ipcv and BXH/Cub recombinant inbred strains of the rat: strain distribution patterns of 632 alleles. Folia Biol. (Praha) 1999;45:203–215. [PubMed] [Google Scholar]
- 52.Hubner N., Wallace C.A., Zimdahl H., Petretto E., Schulz H., Maciver F., Mueller M., Hummel O., Monti J., Zidek V., et al. Integrated transcriptional profiling and linkage analysis for identification of genes underlying disease. Nat. Genet. 2005;37:243–253. doi: 10.1038/ng1522. doi:10.1038/ng1522. [DOI] [PubMed] [Google Scholar]
- 53.O'Connor D.T., Frigon R.P., Stone R.A. Human pheochromocytoma dopamine-beta-hydroxylase: purification and molecular parameters of the tetramer. Mol. Pharmacol. 1979;16:529–538. [PubMed] [Google Scholar]
- 54.Kennedy B., Bigby T.D., Ziegler M.G. Nonadrenal epinephrine-forming enzymes in humans. Characteristics, distribution, regulation, and relationship to epinephrine levels. J. Clin. Invest. 1995;95:2896–2902. doi: 10.1172/JCI117996. doi:10.1172/JCI117996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ziegler M.G., Kennedy B., Elayan H. A sensitive radioenzymatic assay for epinephrine forming enzymes. Life Sci. 1988;43:2117–2122. doi: 10.1016/0024-3205(88)90361-x. doi:10.1016/0024-3205(88)90361-X. [DOI] [PubMed] [Google Scholar]
- 56.Kennedy B., Ziegler M.G. A more sensitive and specific radioenzymatic assay for catecholamines. Life Sci. 1990;47:2143–2153. doi: 10.1016/0024-3205(90)90314-h. doi:10.1016/0024-3205(90)90314-H. [DOI] [PubMed] [Google Scholar]
- 57.Doniger S.W., Salomonis N., Dahlquist K.D., Vranizan K., Lawlor S.C., Conklin B.R. MAPPFinder: using Gene Ontology and GenMAPP to create a global gene-expression profile from microarray data. Genome Biol. 2003;4:R7. doi: 10.1186/gb-2003-4-1-r7. doi:10.1186/gb-2003-4-1-r7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Grubbs F.E. Procedures for detecting outlying observations in samples. Technometrics. 1969;11:1–21. doi:10.2307/1266761. [Google Scholar]
- 59.Belknap J.K. Effect of within-strain sample size on QTL detection and mapping using recombinant inbred mouse strains. Behav. Genet. 1998;28:29–38. doi: 10.1023/a:1021404714631. doi:10.1023/A:1021404714631. [DOI] [PubMed] [Google Scholar]
- 60.Saar K., Beck A., Bihoreau M.T., Birney E., Brocklebank D., Chen Y., Cuppen E., Demonchy S., Dopazo J., Flicek P., et al. SNP and haplotype mapping for genetic analysis in the rat. Nat. Genet. 2008;40:560–566. doi: 10.1038/ng.124. doi:10.1038/ng.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Basten C.J., Weir B.S., Zeng Z.B. QTL Cartographer: A Reference Manual and Tutorial for QTL Mapping. Raleigh, NC: Department of Statistics, North Carolina State University; 2002. [Google Scholar]
- 62.Manly K.F., Cudmore R.H., Jr, Meer J.M. Map manager QTX, cross-platform software for genetic mapping. Mamm. Genome. 2001;12:930–932. doi: 10.1007/s00335-001-1016-3. doi:10.1007/s00335-001-1016-3. [DOI] [PubMed] [Google Scholar]
- 63.Manly K.F., Wang J., Williams R.W. QTL Reaper. Memphis, TN: University of Tennessee Health Science Center; 2004. [Google Scholar]
- 64.Doerge R.W., Churchill G.A. Permutation tests for multiple loci affecting a quantitative character. Genetics. 1996;142:285–294. doi: 10.1093/genetics/142.1.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Van Ooijen J.W. Accuracy of mapping quantitative trait loci in autogamous species. Theor. Appl. Genet. 1992;84:803–811. doi: 10.1007/BF00227388. [DOI] [PubMed] [Google Scholar]
- 66.Visscher P.M., Thompson R., Haley C.S. Confidence intervals in QTL mapping by bootstrapping. Genetics. 1996;143:1013–1020. doi: 10.1093/genetics/143.2.1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Rozen S., Skaletsky H. Primer3 on the WWW for general users and for biologist programmers. Methods Mol. Biol. 2000;132:365–386. doi: 10.1385/1-59259-192-2:365. [DOI] [PubMed] [Google Scholar]
- 68.Colella S., Shen L., Baggerly K.A., Issa J.P., Krahe R. Sensitive and quantitative universal pyrosequencing methylation analysis of CpG sites. Biotechniques. 2003;35:146–150. doi: 10.2144/03351md01. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.