Abstract
The constitutional t(11;22) is the most frequent recurrent non-Robertsonian translocation in humans, the breakpoints of which are located within palindromic AT-rich repeats on 11q23 and 22q11 (PATRR11 and PATRR22). Genetic variation of the PATRR11 was found to affect de novo t(11;22) translocation frequency in sperm derived from normal healthy males, suggesting the hypothesis that polymorphisms of the PATRR22 might also influence the translocation frequency. Although the complicated structure of the PATRR22 locus prevented determining the genotype of the PATRR22 in each individual, genotyping of flanking markers as well as identification of rare variants allowed us to demonstrate an association between the PATRR22 allele type and the translocation frequency. We found that size and symmetry of the PATRR22 affect the de novo translocation frequency, which is lower for the shorter or more asymmetric versions. These data lend support to our hypothesis that the PATRRs form secondary structures in the nucleus that induce genomic instability leading to the recurrent translocation.
INTRODUCTION
The constitutional t(11;22) is the most frequent recurrent non-Robertsonian translocation in humans. Carriers of this balanced translocation usually have no clinical symptoms and are often identified after the birth of offspring with an unbalanced form of the translocation, the supernumerary-der(22)t(11;22) syndrome (Emanuel syndrome). Patients with the supernumerary-der(22) syndrome have a distinctive phenotype, which consists of severe mental retardation, preauricular tag or sinus, ear anomaly, cleft or high-arched palate, micrognathia, heart defects and genital abnormalities in the male (1–3).
The t(11;22) translocation represents a good model for studying the molecular mechanisms that contribute to genomic rearrangements. There are several reports describing multiple cases of t(11;22) balanced carriers including de novo cases (1,2,4). The recurrent nature of this translocation prompted the examination of the t(11;22) breakpoints for a specific genomic structure, culminating in the identification of palindromic AT-rich repeats at both breakpoint regions on chromosomes 11q23 and 22q11 (PATRR11 and PATRR22) (5–8). All the breakpoints were located within the PATRR11 and PATRR22, near the center of the PATRR but different at a nucleotide level among individual families, confirming that the rearrangement is recurrent (9). Indeed, translocation-specific PCR using the sequences flanking the translocation junction fragments from both derivative translocation chromosomes detected multiple de novo t(11;22)s in sperm derived from normal healthy males (10).
In our previous study, we demonstrated that the nucleotide sequence of the PATRR11 was hypervariable and varied among individual alleles. We calculated the de novo translocation frequency based on the occurrence of positive PCRs and reported that the polymorphisms of the PATRR11 affected the de novo translocation frequency (11). It is reasonable to hypothesize that PATRR22 polymorphisms might be another important factor in the generation of the t(11;22) translocation, although the data also showed that the PATRR22 has little influence on the frequency. Our recent studies have indicated that the PATRR22 also manifests variation at the nucleotide sequence level, but we did not find any large-scale size polymorphism similar to the insertion/deletion polymorphism seen for the PATRR11 (6). Indeed, the translocation frequency varies moderately among individuals with the same PATRR11 genotype, promoting the speculation that subtle polymorphisms in the PATRR22 affect this variation.
In this study, we analyzed the de novo t(11;22) translocation frequency as a function of the PATRR22 allele type. It is well documented that the PATRR22 is located at one of the chromosome 22-specific low-copy repeats (LCR22-B), which have been identified at multiple loci on 22q11 (12–14). The duplicated sequences share 97–98% sequence homology with each other, preventing genotyping of the PATRR22. This is due to competing sequence that produces excessive background during PCR amplification (6). In the current study, we utilized the genotypes of PATRR22-flanking marker to determine whether the de novo t(11;22) translocation frequency could be associated with variation of the PATRR22. Further, we optimized PCR conditions for amplification of the PATRR22 using new LCR-specific primers. This approach identified rare PATRR22 variants facilitating the ability to analyze the association between PATRR22 variation and t(11;22) formation.
RESULTS
PATRR22 genotype did not affect the total frequency of de novo t(11;22)s
To investigate the effect of PATRR22 polymorphisms on the frequency of de novo t(11;22) formation in sperm from normal healthy males, the translocation frequency in individuals with various PATRR22 genotypes was determined. To avoid the potential effects introduced by PATRR11 polymorphisms, individuals homozygous for the L-PATRR11, the allele most commonly seen in the Japanese population, were selected (11). Among these subjects, the frequency of translocation ranged from 6.01 × 10−6 to 1.65 × 10−4, a relatively narrow range when compared with variation of greater than three orders of magnitude induced by polymorphisms of the PATRR11. The AT-rich region flanking the PATRR22 manifests size polymorphisms (A allele, 355 bp; B allele, 179 bp; C allele, 121 bp) (6). The initial analyses of eight PATRR22 alleles from four individuals suggested that these polymorphisms appeared to be linked to the PATRR22 polymorphisms. This linkage disequilibrium was confirmed by further analyses of additional 16 alleles (3 A alleles, 6 B alleles and 7 C alleles; data not shown). Thus, we utilized these polymorphisms as a surrogate for the genotype of the PATRR22, which is difficult to determine. We identified three major genotypes, A/C, B/C and C/C. Although the total translocation frequency varies among individuals (Table 1), we found no statistical difference among the A/C, B/C and C/C groups (P = 0.14).
Table 1.
Association between the type of PATRR22 polymorphism and total frequency of de novo translocations
| Case | Genotype of AT-rich region | PATRR11 genotype | Template DNA (ng/µl) | Positive PCR | Translocation frequencya |
|---|---|---|---|---|---|
| 1 | A/C | L/L | 100 | 23/44 | 2.24 × 10−5 |
| 2b | A/C | L/L | 50 | 30/50 | 5.55 × 10−5 |
| 3 | A/C | L/L | 100 | 10/50 | 6.76 × 10−6 |
| 4b | B/C | L/L | 50 | 39/50 | 9.18 × 10−5 |
| 5 | B/C | L/L | 100 | 35/51 | 3.51 × 10−5 |
| 6b | B/C | L/L | 10 | 21/50 | 1.65 × 10−4 |
| 7 | C/C | L/L | 100 | 33/71 | 1.89 × 10−5 |
| 8 | C/C | L/L | 100 | 17/26 | 3.21 × 10−5 |
| 9 | C/C | L/L | 100 | 18/39 | 1.88 × 10−5 |
| 14b | A/A | L/AS | 10 | 7/50 | 4.57 × 10−5 |
aThe total frequency was calculated based on the total number of positive PCR.
bThe first two examinations of these samples showed positive results in almost all of the aliquots, precluding the estimation of the exact frequency of the translocation. We therefore used 50 ng (Cases 2 and 4) and 10 ng (Cases 6 and 14) of DNA as a template in these cases.
PATRR22 allele type affects the frequency of de novo t(11;22)s
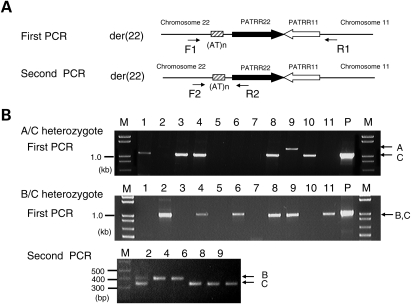
To circumvent the effect of various factors that potentially affect translocation generation, we determined the allelic origin of each translocation product and determined the allele-specific translocation frequencies in each individual (Fig. 1). In the case of A/C heterozygotes, the translocation was more frequently generated from the C allele than from the A allele (P = 0.0033; Table 2). The PATRR22, regardless of the genotype of the AT-rich region, manifests an almost perfect palindromic structure, showing >98% homology between the proximal and distal arms (Table 3). However, the size of the A allele PATRR22 is 583 bp, which is 14 bp shorter than that of C allele PATRR22 (597 bp). The A allele PATRR22 carries a short asymmetric region at its center, whereas the C allele of the PATRR22 does not. Thus, short size or central asymmetry might influence the relatively low translocation frequency of the A allele.
Figure 1.
Analysis of the allelic origin of de novo t(11;22) translocations. (A) Nested PCR strategy for determining allelic origin of the der(22) of the t(11;22). PATRRs are shown by thick arrows, and the AT-rich region flanking the PATRR22 is indicated by a hatched box. PCR primers are shown by thin arrows to indicate the location and orientation. (B) Representative results of the nested PCR. For an A/C heterozygote, the first PCR can distinguish the allelic origin of the PCR products based on product size (upper panel). For a B/C heterozygote, the size of the first PCR product is similar to one another (middle panel). The second PCR can clearly distinguish between the allelic origins (lower panel). Two bands were observed in lane 2, suggesting that the template DNA includes two or more translocations. M, size markers; P, DNA from t(11;22) translocation carrier served as positive controls.
Table 2.
Association between allele type of the PATRR22 polymorphism and frequency of de novo translocation
| Case | Genotype of the AT-rich region | PATRR11 genotype | Frequency | Positive PCR | Allelic origin |
||
|---|---|---|---|---|---|---|---|
| A | B | C | |||||
| 1 | A/C | L/L | 2.24 × 10−5 | 23/44 | 1 (4%) | — | 22 (96%) |
| 2 | A/C | L/L | 5.55 × 10−5 | 30/50 | 5 (17%) | — | 25 (83%) |
| 3 | A/C | L/L | 6.01 × 10−6 | 9/50 | 2 (22%) | — | 7 (78%) |
| 10 | A/C | L/AS | 1.45 × 10−5 | 19/50 | 1 (5%) | — | 18 (95%) |
| 4 | B/C | L/L | 9.18 × 10−5 | 39/50 | — | 20 (51%) | 19 (49%) |
| 5 | B/C | L/L | 2.55 × 10−5 | 35/51 | — | 17 (49%) | 18 (51%) |
| 6 | B/C | L/L | 1.65 × 10−4 | 21/50 | — | 9 (43%) | 12 (57%) |
| 11 | B/C | L/S | 1.35 × 10−5 | 18/50 | — | 7 (39%) | 11 (61%) |
| 12 | B/C | L/S | 1.60 × 10−5 | 30/73 | — | 14 (47%) | 16 (53%) |
| 13 | B/C | L/AS | 8.72 × 10−6 | 22/88 | — | 10 (45%) | 12 (55%) |
L, long PATRR; S, symmetric short PATRR; AS, asymmetric short PATRR.
Table 3.
Characterization of the polymorphic PATRR22 alleles
| PATRR22 | Size (bp) | Accession number | Homology between proximal and distal arms (%) | ΔG (kcal/mol) | ΔG/nt (kcal/mol) |
|---|---|---|---|---|---|
| Standard A allele | 583 | AB261997a | 98.6 | 12.57 | 0.022 |
| Standard B allele | 595 | AB538236 | 98.7 | 9.66 | 0.016 |
| Variant B allele (Case 5) | 595 | AB538238 | 98.0 | 14.03 | 0.024 |
| Standard C allele | 597 | AB538237 | 98.3 | 13.24 | 0.022 |
| Variant B1 allele (Case 15) | 553 | AB533274 | 100.0 | 2.71 | 0.005 |
| Variant C1 allele (Case 15) | 509 | AB533275 | 98.8 | 5.47 | 0.011 |
| Variant C2 allele (Case 16) | 539 | AB533276 | 99.6 | 5.67 | 0.011 |
| Variant C3 allele (Case 16) | 457 | AB533277 | 96.9 | 19.90 | 0.044 |
| Flanking AT-rich region (A allele) | 355 | AB261997a | 89.3 | 49.35 | 0.139 |
aAB261997 includes sequence information of both PATRR22 and flanking AT-rich region of A allele.
Although the percentage of translocations from the C allele was marginally greater than that from the B allele in B/C heterozygotes, statistical analysis indicated no significant difference (P = 0.06). The size difference between the B and the C alleles is only 2 bp (595 bp versus 597 bp) and both are similar in their symmetry (Table 3). Since allelic preference varied among individuals, it is hypothesized that subtle nucleotide alteration among the same allele type might influence the variation of the translocation frequency. We sequenced both the B and the C allele PATRR22s in all six B/C heterozygotes. All the sequences from the B and C allele PATRR22s were identical to one another except one case that manifested a two-nucleotide substitution in the B allele (Case 5). Although the substitution slightly reduces the symmetry of the B allele, the difference did not appear to alter the allelic preference for the C allele.
To investigate the potential role of secondary structure on translocation frequency, we calculated ΔG and the ΔG value normalized with size. It is assumed that PATRRs with a lower ΔG value would show a greater propensity for secondary structure leading to a higher translocation frequency. However, these values did not appear to correlate with the translocation frequency (Table 3).
Identification of rare variants that produced t(11;22) less frequently
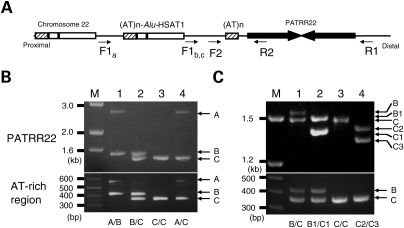
To elucidate the effect of size and symmetry more completely, we attempted to identify individuals carrying shorter or more asymmetric PATRR22 variants, which are rare. The LCR22s share 97–98% sequence homology with each other. Therefore, polymorphisms of the PATRR22 are difficult to analyze because PCR amplification is always accompanied by multiple PCR products emanating from other LCR22s (6). PCR primers located at sites with subtle nucleotide difference among the LCR22s allowed us to reduce the background amplification (Fig. 2A and B). Occasionally, the PCR amplified non-specific products from other LCR22s, such that we could not use this system to accurately determine the PATRR22 genotype. However, we could utilize this PCR for the identification of rare size variants of the PATRR22 by screening individuals who did not carry the standard ∼597 bp PATRR22 on either allele. Over 200 male volunteers were screened and we eventually identified two individuals (Fig. 2C).
Figure 2.
Screening and genotyping of rare short PATRR22 variants. (A) Genomic structure of the PATRR22 region. LCR-specific primers are designed to amplify the PATRR22 (arrows). Proximal region represents the ‘AT-rich region-HSAT1-Alu’ cassette repeated three or four times (6,29). Sites for the forward primer (F) carry sequence variations among the AT-rich region polymorphic alleles. In the B and C alleles, the PCR product originates from the closest F site, whereas it can anneal with the second closest site in the A allele. (B) LCR-specific PCR for amplifying the PATRR22. Upper panel represents the LCR-specific PCR for the PATRR22, whereas the lower panel indicates the genotyping of the AT-rich region flanking the PATRR22. Genotypes of the AT-rich region are given at the bottom. The PATRR22-specific PCR product from the A allele is much longer than the others for the reason described above. M, size markers. (C) Rare variants. Lane 1, standard B/C heterozygote; lane 2, compound heterozygote for B/C rare variants; lane 3, C/C homozygote; lane 4, compound heterozygote for C rare variants.
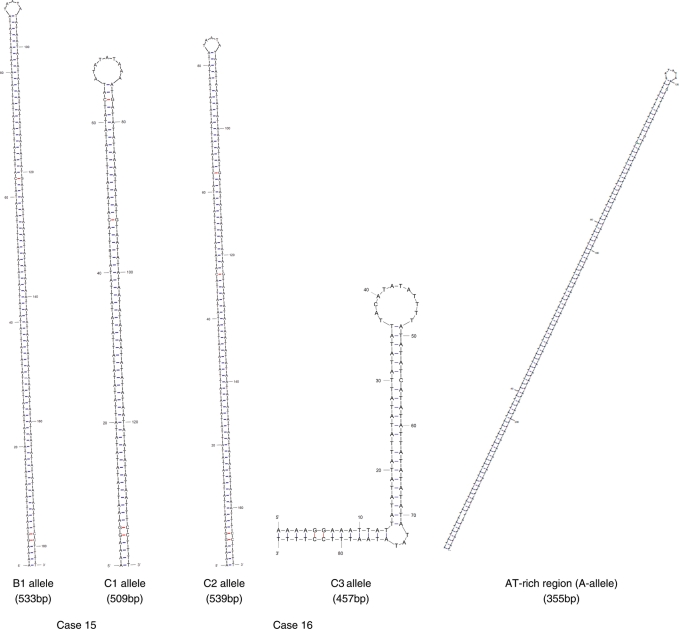
One B/C heterozygote (Case 15) was found to carry two short PATRR22s. Sequence analysis revealed that both the B and C alleles manifest a symmetric structure and the sizes were 553 and 509 bp, respectively (Table 4). The total translocation frequency for this male was similar to that of others, but the shorter 509 bp variant produced fewer translocations than the 553 bp variant (18 versus 82%). This result clearly indicates that the size of the PATRR22 affects the translocation frequency. Another case, a C/C homozygote (Case 16), was also found to carry two short PATRR22s, the sizes of which are 539 and 457 bp (Table 4). The 539 bp allele was found to be symmetric, whereas the 457 bp allele manifests an asymmetric structure. The total translocation frequency for this male was also similar to that of others, but the translocations were exclusively derived from symmetric 539 bp PATRR22. Predicted secondary structures for these alleles are shown in Figure 3. The 457 bp PATRR22 manifests an asymmetric structure with a large single-stranded loop at the center. However, we did not determine whether size or symmetry affected the allelic bias.
Table 4.
De novo translocation frequency in sperm from two individuals with rare short PATRR22 on both alleles
| Case | Genotype of AT-rich region | PATRR11 genotype | Genotype of PATRR22 |
De novo translocation |
|||
|---|---|---|---|---|---|---|---|
| Template DNA (ng/µl) | Positive PCR | Allelic origin | Frequency | ||||
| 15 | B/C | L/L | B1 (553 bp, symmetry) | 5 | 28/96 | 23 (82%) | 1.66 × 10−4 |
| C1 (509 bp, symmetry) | 5 (18%) | 3.24 × 10−5 | |||||
| 16 | C/C | L/L | C2 (539 bp, symmetry) | 10 | 14/58 | 14 (100%) | 2.62 × 10−5 |
| C3 (457 bp, asymmetry) | 0 (0%) | 0 | |||||
Figure 3.
M-fold analysis of the secondary structure of rare short variants of the PATRR22 and proximal AT-rich region flanking the PATRR22 (A allele). For the AT-rich region flanking the PATRR22, only the central 229 bp region is shown.
The AT-rich region flanking the PATRR22 also produces translocations
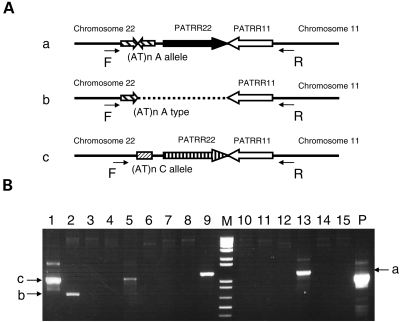
During the course of analysis of the de novo t(11;22)s derived from A/C heterozygotes, we unexpectedly identified a variant der(22) product, which originated from the A allele (Fig. 4). The breakpoint on chromosome 11 was located at the center of the PATRR11, which is consistent with the standard t(11;22) breakpoints. However, the breakpoint on chromosome 22 was located at the center of the 355 bp flanking AT-rich region of the A allele (AB538239). The 355 bp AT-rich region forms a quasi-palindromic structure similar to the PATRR. When the entire 355 bp was analyzed, the homology between the arms was 89.3% (Table 3). However, when the central 229 bp region was analyzed separately, the homology was 93% forming an almost perfect palindrome (Fig. 3).
Figure 4.
Rare der(22) junction derived from an A allele. (A) Schematic representation of the organization of junction from the der(22) from an A/C heterozygote. The A/C heterozygote produces three types of der(22); a standard der(22) with the breakpoint at the center of the PATRR22 of the A allele (a), a rare der(22) with the breakpoint at the center of the AT-rich region of the A allele (b), and a standard der(22) with the breakpoint at the center of PATRR22 of the C allele (c). (B) The der(22)-specific PCR. The allelic origin of the der(22) can be determined by its size. Lanes 1 and 5, a der(22) derived from a C allele; lanes 9 and 13, a der(22) derived from an A allele; lane 2, a rare der(22) with the breakpoint at the center of AT-rich region of the A allele. M, size markers; P, DNA from a t(11;22) translocation carrier served as a positive control.
We increased the number of PCRs to estimate the frequency of this rare translocation. This rare translocation was observed twice in a total of 400 PCRs (1.52 × 10−7). On the other hand, we did not identify any translocations from the 457 bp asymmetric PATRR22 in 400 PCRs (<7.59 × 10−8; data not shown). The fact that the 355 bp symmetric AT-rich region produced translocations, but the 457 bp asymmetric PATRR22 did not indicates that not only size but also symmetry is an important factor in determining the translocation frequency.
DISCUSSION
The data obtained in this study demonstrate that variation of the PATRR22 does not affect the total frequency of de novo t(11;22) generation in sperm. This was clearly demonstrated by the observation of a similar total translocation frequency even in compound heterozygotes of short variants. This is in contrast to the finding obtained for the PATRR11 (11). Translocation frequency appears to be stable in sperm samples from the same individual obtained at different times, suggesting that the frequency is not influenced by environmental factors (15). In the present study, when the translocation frequency was compared among individuals with the same PATRR11 genotype, which would be likely to affect the frequency, the overall frequency was similar among the group with different PATRR22 genotypes. One likely reason is that PATRR22 variation is less pronounced relative to the insertion/deletion polymorphisms of the PATRR11 resulting in less change in the translocation frequency.
Another reason might be that variation of the translocation frequency based on the genotype of the PATRR22 was so subtle that the effects of variation in other genetic factors obscured any association. Such factors might include inter-individual differences in genetic predisposition of fidelity in DNA repair (16). For example, although the A allele produced a significantly smaller number of translocations than C alleles, one rare A/A homozygote, which was predicted to produce translocations less frequently, was actually found to manifest a frequency comparable to others such as the C/C homozygotes (Table 1). In this subject, a certain genetic factor that promotes generation of translocations possibly raised his translocation frequency in spite of the adverse effect of the A/A genotype.
In contrast, comparison of the allelic origin of translocations within an individual allowed us to compare the translocation frequency within a similar genetic background, which clearly demonstrated allelic preference. (i) The A allele (583 bp) produced less translocations than either the B or the C allele (595 and 597 bp, respectively). (ii) A short rare variant (509 bp) produced less translocations than the opposing allele (553 bp). (iii) An asymmetric short variant (457 bp) did not produce any translocations. (iv) An AT-rich flanking region (A allele) that manifested a symmetric palindrome produced translocations despite its short length (355 bp). Taken together, our data indicate that size and symmetry of the PATRR22 affect the translocation frequency, which is consistent with the findings obtained for the PATRR11.
The mechanism of PATRR-mediated chromosomal translocation in humans is still unknown. Analyses of several other recurrent and non-recurrent translocations demonstrated the presence of PATRR-like genomic structures at the breakpoint region, suggesting that at least a certain subset of translocations are mediated by the PATRRs (17,18). Palindromic DNA can generate intra-strand base pairing, which produces a single-stranded hairpin structure or a double-stranded cruciform structure (19,20). Such secondary structures are likely to be generated during the DNA replication and likely form hotspot for genomic rearrangement (21–23). It is possible that the PATRRs also adopt a cruciform structure that induces genomic instability leading to the recurrent translocations (24). In vitro analysis has demonstrated that longer and more symmetric PATRR variants show a greater potential to adopt a cruciform conformation (25). The data that the size and symmetry of the PATRR11 and PATRR22 affect the translocation frequency indirectly provide a strong suggestion that the PATRRs form cruciforms in living cells and that the cruciform secondary structure mediates the recurrent chromosomal translocations. We recently established a plasmid-based model system for PATRR-mediated translocations in human cell lines and demonstrated that the DNA secondary structure was necessary for initiation of rearrangement between the PATRRs (26). A more thorough analysis of PATRR-mediated translocations would reinforce the hypothesis that DNA secondary structure governs the gross chromosomal rearrangements.
MATERIALS AND METHODS
Genotyping of the PATRR22
All human samples were provided from individual volunteers in a Japanese population after obtaining the appropriate informed consent. The study was approved by the Ethical Review Board for Human Genome Studies at Fujita Health University. For the analysis of polymorphisms of the PATRR, genomic DNA was extracted from blood samples or cheek swabs using PureGene (Gentra). The PATRR11 was genotyped by PCR using primers described previously (11). For the PATRR22, the size polymorphisms of the AT-rich region flanking the PATRR22 (A allele, 355 bp; B allele, 179 bp; C allele, 121 bp) were first determined. This polymorphism is known to link to polymorphisms of the PATRR22 (6).
Next, the PATRR22s were amplified using LCR-specific primers designed for this study. Primer −469F: 5′-CCATATGCAGTTATAAATATGTTTCATGGTTAG-3′, +440R: 5′-ACAAGTAAACAGGTTTTCAAAGCT-3′. The purified PCR products were cloned into pT7Blue vector (Novagen). We used the SURE strain (Stratagene) to maintain the unstable PATRR insert. The plasmid inserts were sequenced as described previously (27). Sequences were determined from at least three plasmid clones to avoid misinterpretation as a result of PCR artifacts.
Assessment of de novo translocation frequency
Sperm samples were provided from individual volunteers with various genotypes of the PATRR22. DNA was extracted using PureGene according to the manufacturer's protocol. Translocation-specific PCR was performed with the appropriate primers as described previously (4). PCR was performed using a protocol of 40 cycles of 10 s at 98°C and 5 min at 60°C. The frequency of de novo translocation events was calculated based on the presence of positive PCRs (10). Briefly, we amplified multiple batches of 100 ng sperm DNA each containing 33 000 haploids (n). We counted the number of positive PCRs per total reactions (p). The translocation frequency (q) was calculated on the basis that the probability of a positive PCR corresponds to a total sum of a binomial series of the translocation frequency calculated using the equation p = 1 − (1 − q)n as described previously.
The frequency of de novo t(11;22) was determined using the der(22)-specific PCR so that the allelic origin of the translocation could be easily assessed (Fig. 1). The PCR was performed using one primer located flanking the PATRR11 (11 + 290R2: 5′-CTTTCTTAACATAGCTTCTAC-AC-3′) and the other flanking the PATRR22 (22 − 394F: 5′-TCAGTTTATTCCCAAACTCCCAAAT-3′). In the case of an A/C heterozygote, the allelic origin of the translocation is easily identified by the size of the PCR products (Fig. 1B). In the case of a B/C heterozygote, a second PCR was performed using primer pair, 22 − 394F and 22b (5′-CTGCATCCTTCAACGTTCCATC-3′; Fig. 1B).
In silico cruciform extrusion assay
The potential secondary structure of the PATRR22 as well as of the rare variants identified in the current study (Table 3) was analyzed using the m-fold server (http://www.bioinfo.rpi.edu/applications/mfold). A free energy value (GCRUCIFORM) was obtained (28). Similarly, a free energy value for the same sequence annealed to its complementary strand (GDS) was obtained and then halved. Free energy for the formation of secondary structure (ΔG) is the GDS − GCRUCIFORM difference, and this value was normalized by the nucleotide number (ΔG/nucleotide). The most negative free energy value was used for subsequent analyses when numerous values were obtained.
Statistical analysis
Statistical comparisons between groups were performed using the Student's t-test and one-way analysis of variance (ANOVA). Differences were considered to be significant at P < 0.05.
Conflict of Interest statement. None declared.
FUNDING
These studies were supported by a grant-in-aid for Scientific Research, Genome, and for 21st Century COE Program of Fujita Health University from the Ministry of Education, Culture, Sports, Science, and Technology of Japan, by a grant from the Ministry of Health, Labour and Welfare of Japan (to H.K.) and by a grant (CA39926) from the National Institutes of Health, USA (to B.S.E.).
REFERENCES
- 1.Fraccaro M., Lindsten J., Ford C.E., Iselius L. The 11q;22q translocation: a European collaborative analysis of 43 cases. Hum. Genet. 1980;56:21–51. doi: 10.1007/BF00281567. [DOI] [PubMed] [Google Scholar]
- 2.Zackai E.H., Emanuel B.S. Site-specific reciprocal translocation, t(11;22)(q23;q11), in several unrelated families with 3:1 meiotic disjunction. Am. J. Med. Genet. 1980;7:507–521. doi: 10.1002/ajmg.1320070412. doi:10.1002/ajmg.1320070412. [DOI] [PubMed] [Google Scholar]
- 3.Carter M.T., St Pierre S.A., Zackai E.H., Emanuel B.S., Boycott K.M. Phenotypic delineation of Emanuel syndrome (supernumerary derivative 22 syndrome): clinical features of 63 individuals. Am. J. Med. Genet. A. 2009;149A:1712–1721. doi: 10.1002/ajmg.a.32957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kurahashi H., Shaikh T.H., Zackai E.H., Celle L., Driscoll D.A., Budarf M.L., Emanuel B.S. Tightly clustered 11q23 and 22q11 breakpoints permit PCR-based detection of the recurrent constitutional t(11;22) Am. J. Hum. Genet. 2000a;67:763–768. doi: 10.1086/303054. doi:10.1086/303054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kurahashi H., Shaikh T.H., Hu P., Roe B.A., Emanuel B.S., Budarf M.L. Regions of genomic instability on 22q11 and 11q23 as the etiology for the recurrent constitutional t(11;22) Hum. Mol. Genet. 2000b;9:1665–1670. doi: 10.1093/hmg/9.11.1665. doi:10.1093/hmg/9.11.1665. [DOI] [PubMed] [Google Scholar]
- 6.Kurahashi H., Inagaki H., Hosoba E., Kato T., Ohye T., Kogo H., Emanuel B.S. Molecular cloning of a translocation breakpoint hotspot in 22q11. Genome Res. 2007;17:461–469. doi: 10.1101/gr.5769507. doi:10.1101/gr.5769507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edelmann L., Spiteri E., Koren K., Pulijaal V., Bialer M.G., Shanske A., Goldberg R., Morrow B.E. AT-rich palindromes mediate the constitutional t(11;22) translocation. Am. J. Hum. Genet. 2001;68:1–13. doi: 10.1086/316952. doi:10.1086/316952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tapia-Páez I., Kost-Alimova M., Hu P., Roe B.A., Blennow E., Fedorova L., Imreh S., Dumanski J.P. The position of t(11;22)(q23;q11) constitutional translocation breakpoint is conserved among its carriers. Hum. Genet. 2001;109:167–177. doi: 10.1007/s004390100560. [DOI] [PubMed] [Google Scholar]
- 9.Kurahashi H., Emanuel B.S. Long AT-rich palindromes and the constitutional t(11;22) breakpoint. Hum. Mol. Genet. 2001a;10:2605–2617. doi: 10.1093/hmg/10.23.2605. doi:10.1093/hmg/10.23.2605. [DOI] [PubMed] [Google Scholar]
- 10.Kurahashi H., Emanuel B.S. Unexpectedly high rate of de novo constitutional t(11;22) translocations in sperm from normal males. Nat. Genet. 2001b;29:139–140. doi: 10.1038/ng1001-139. doi:10.1038/ng1001-139. [DOI] [PubMed] [Google Scholar]
- 11.Kato T., Inagaki H., Yamada K., Kogo H., Ohye T., Kowa H., Nagaoka K., Taniguchi M., Emanuel B.S., Kurahashi H. Genetic variation affects de novo translocation frequency. Science. 2006;311:971. doi: 10.1126/science.1121452. doi:10.1126/science.1121452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edelmann L., Pandita R.K., Morrow B.E. Low-copy repeats mediate the common 3-Mb deletion in patients with velo-cardio-facial syndrome. Am. J. Hum. Genet. 1999;64:1076–1086. doi: 10.1086/302343. doi:10.1086/302343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dunham I., Shimizu N., Roe B.A., Chissoe S., Hunt A.R., Collins J.E., Bruskiewich R., Beare D.M., Clamp M., Smink L.J., et al. The DNA sequence of human chromosome 22. Nature. 1999;402:489–495. doi: 10.1038/990031. doi:10.1038/990031. [DOI] [PubMed] [Google Scholar]
- 14.Shaikh T.H., Kurahashi H., Saitta S.C., O'Hare A.M., Hu P., Roe B.A., Driscoll D.A., McDonald-McGinn D.M., Zackai E.H., Budarf M.L., et al. Chromosome 22-specific low copy repeats and the 22q11.2 deletion syndrome: genomic organization and deletion endpoint analysis. Hum. Mol. Genet. 2000;9:489–501. doi: 10.1093/hmg/9.4.489. doi:10.1093/hmg/9.4.489. [DOI] [PubMed] [Google Scholar]
- 15.Kato T., Yamada K., Inagaki H., Kogo H., Ohye T., Emanuel B.S., Kurahashi H. Age has no effect on de novo constitutional t(11;22) translocation frequency in sperm. Fertil. Steril. 2007;88:1446–1448. doi: 10.1016/j.fertnstert.2007.01.019. doi:10.1016/j.fertnstert.2007.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Baer C.F., Miyamoto M.M., Denver D.R. Mutation rate variation in multicellular eukaryotes: causes and consequences. Nat. Rev. Genet. 2007;8:619–631. doi: 10.1038/nrg2158. doi:10.1038/nrg2158. [DOI] [PubMed] [Google Scholar]
- 17.Kurahashi H., Inagaki H., Ohye T., Kogo H., Kato T., Emanuel B.S. Palindrome-mediated chromosomal translocations in humans. DNA Repair (Amst.) 2006a;5:1136–1145. doi: 10.1016/j.dnarep.2006.05.035. doi:10.1016/j.dnarep.2006.05.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kurahashi H., Inagaki H., Ohye T., Kogo H., Kato T., Emanuel B.S. Chromosomal translocations mediated by palindromic DNA. Cell Cycle. 2006b;5:1297–1303. doi: 10.4161/cc.5.12.2809. [DOI] [PubMed] [Google Scholar]
- 19.Leach D.R. Long DNA palindromes, cruciform structures, genetic instability and secondary structure repair. Bioessays. 1994;16:893–900. doi: 10.1002/bies.950161207. doi:10.1002/bies.950161207. [DOI] [PubMed] [Google Scholar]
- 20.Bzymek M., Lovett S.T. Evidence for two mechanisms of palindrome-stimulated deletion in Escherichia coli: single-strand annealing and replication slipped mispairing. Genetics. 2001;158:527–540. doi: 10.1093/genetics/158.2.527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lobachev K.S., Gordenin D.A., Resnick M.A. The Mre11 complex is required for repair of hairpin-capped double-strand breaks and prevention of chromosome rearrangements. Cell. 2002;108:183–193. doi: 10.1016/s0092-8674(02)00614-1. doi:10.1016/S0092-8674(02)00614-1. [DOI] [PubMed] [Google Scholar]
- 22.Bacolla A., Jaworski A., Larson J.E., Jakupciak J.P., Chuzhanova N., Abeysinghe S.S., O'Connell C.D., Cooper D.N., Wells R.D. Breakpoints of gross deletions coincide with non-B DNA conformations. Proc. Natl Acad. Sci. USA. 2004;101:14162–14167. doi: 10.1073/pnas.0405974101. doi:10.1073/pnas.0405974101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lemoine F.J., Degtyareva N.P., Lobachev K., Petes T.D. Chromosomal translocations in yeast induced by low levels of DNA polymerase a model for chromosome fragile sites. Cell. 2005;120:587–598. doi: 10.1016/j.cell.2004.12.039. doi:10.1016/j.cell.2004.12.039. [DOI] [PubMed] [Google Scholar]
- 24.Kurahashi H., Inagaki H., Yamada K., Ohye T., Taniguchi M., Emanuel B.S., Toda T. Cruciform DNA structure underlies the etiology for palindrome-mediated human chromosomal translocations. J. Biol. Chem. 2004;279:35377–35383. doi: 10.1074/jbc.M400354200. doi:10.1074/jbc.M400354200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kogo H., Inagaki H., Ohye T., Kato T., Emanuel B.S., Kurahashi H. Cruciform extrusion propensity of human translocation-mediating palindromic AT-rich repeats. Nucleic Acids Res. 2007;35:1198–1208. doi: 10.1093/nar/gkm036. doi:10.1093/nar/gkm036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Inagaki H., Ohye T., Kogo H., Kato T., Bolor H., Taniguchi M., Shaikh T.H., Emanuel B.S., Kurahashi H. Chromosomal instability mediated by non-B DNA: cruciform conformation and not DNA sequence is responsible for recurrent translocation in humans. Genome Res. 2009;19:191–198. doi: 10.1101/gr.079244.108. doi:10.1101/gr.079244.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Inagaki H., Ohye T., Kogo H., Yamada K., Kowa H., Shaikh T.H., Emanuel B.S., Kurahashi H. A palindromic AT-rich repeat in the NF1 gene is hypervariable in humans and evolutionarily conserved among primates. Hum. Mutat. 2005;26:332–342. doi: 10.1002/humu.20228. doi:10.1002/humu.20228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gotter A.L., Shaikh T.H., Budarf M.L., Rhodes C.H., Emanuel B.S. A palindrome-mediated mechanism distinguishes translocations involving LCR-B of chromosome 22q11.2. Hum. Mol. Genet. 2004;13:103–115. doi: 10.1093/hmg/ddh004. doi:10.1093/hmg/ddh004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Babcock M., Yatsenko S., Stankiewicz P., Lupski J.R., Morrow B.E. AT-rich repeats associated with chromosome 22q11.2 rearrangement disorders shape human genome architecture on Yq12. Genome Res. 2007;17:451–460. doi: 10.1101/gr.5651507. doi:10.1101/gr.5651507. [DOI] [PMC free article] [PubMed] [Google Scholar]