Abstract
Myotubularins are a family of dual-specificity phosphatases that act to modify phosphoinositides and regulate membrane traffic. Mutations in several myotubularins are associated with human disease. Sequence changes in MTM1 and MTMR14 (also known as Jumpy) have been detected in patients with a severe skeletal myopathy called centronuclear myopathy. MTM1 has been characterized in vitro and in several model systems, while the function of MTMR14 and its specific role in muscle development and disease is much less well understood. We have previously reported that knockdown of zebrafish MTM1 results in significantly impaired motor function and severe histopathologic changes in skeletal muscle that are characteristic of human centronuclear myopathy. In the current study, we examine zebrafish MTMR14 using gene dosage manipulation. As with MTM1 knockdown, morpholino-mediated knockdown of MTMR14 results in morphologic abnormalities, a developmental motor phenotype characterized by diminished spontaneous contractions and abnormal escape response, and impaired excitation–contraction coupling. In contrast to MTM1 knockdown, however, muscle ultrastructure is unaffected. Double knockdown of both MTM1 and MTMR14 significantly impairs motor function and alters skeletal muscle ultrastructure. The combined effect of reducing levels of both MTMR14 and MTM1 is significantly more severe than either knockdown alone, an effect which is likely mediated, at least in part, by increased autophagy. In all, our results suggest that MTMR14 is required for motor function and, in combination with MTM1, is required for myocyte homeostasis and normal embryonic development.
INTRODUCTION
Membrane traffic is the term used to describe the regulated movement of vesicles and organelles between subcellular compartments (1). It is a highly regulated process, with multiple layers of positive and negative effectors. Phosphoinositides, or PIs, are key regulatory molecules of membrane traffic (2). PIs are low abundance phospholipids that serve as a molecular road map for various organelles and vesicles, providing specific recognition motifs for enzymes and regulatory proteins that control the fission or fusion of membrane bound structures. The importance of membrane traffic and PIs is underscored by the growing list of human diseases caused by abnormalities in their regulation (3).
PIs are dynamically regulated by a large group of lipid kinases and lipid phosphatases (3). Myotubularins are a family of 15 related dual-specificity phosphatases that act primarily to dephosphorylate PIs (4). They act specifically on PI3P and PI3,5P2, dephosphorylating them to form PIP and PI5P, respectively. PI3P and PI3,5P2 are concentrated on endosomes (5); myotubularins as a group are thus assumed to participate in the regulation of membrane traffic within and between endosomes and other organelles (4,6).
Mutations in several myotubularins are associated with human neurologic disease (6). MTMR2 and MTMR13 mutations are associated with Charcot-Marie-Tooth disease, the most common form of inherited peripheral neuropathy (7). Mutations in MTM1 are the only known genetic cause of myotubular myopathy (8). Myotubular myopathy is a severe disorder of skeletal muscle characterized by profound weakness in infancy. It is one of a group of muscle diseases called centronuclear myopathies that share common histopathologic findings (9). Two other gene products, dynamin-2 and amphiphysin-2, are associated with centronuclear myopathy (10,11); both genes are known regulators of membrane traffic (12). In addition, two patients with centronuclear myopathy have been described with sequence changes in MTMR14, a newly characterized myotubularin family member (13). Each patient has a severe clinical phenotype and a unique heterozygous sequence variant in MTMR14 that is predicted to disrupt its phosphatase activity. Of note, one of the patients with MTMR14 mutation also has a mutation in dynamin-2. The dynamin-2 change has previously been reported in cases of clinically mild centronuclear myopathy. The other patient with MTMR14 mutation has an unaffected parent with the same sequence change. Therefore, the exact relationship between these sequence changes and disease in these patients is somewhat unclear.
Until very recently, little was known about MTMR14. It is highly expressed in skeletal muscle, and exogenous GFP-MTMR14 localizes to the Golgi apparatus in vitro (13). It is therefore speculated that MTMR14 functions primarily at the Golgi apparatus instead of at endosomes. In 2009, Shen et al. (14) reported on mice with a targeted deletion in Mtmr14 that resulted in a mild phenotype of early-onset exercise intolerance and late-onset muscle wasting. They also observed a reduction in force production in muscle from the knockout animals, and demonstrated that this change was associated with dysregulation of calcium homeostasis at the sarcoplasmic reticulum. They provided evidence that the changes in calcium homeostasis are due to the effects of excessive PI3,5P2 on the activity of the skeletal muscle ryanodine receptor (RYR1).
Also in 2009, Vergne et al. (15) reported on a novel role for MTMR14/Jumpy in the regulation of autophagy. They discovered that MTMR14 associated with early autophagosomes and served to negatively regulate progression through the steps of autophagy. Knockdown of MTMR14 resulted in increased autophagy, whereas overexpression inhibited the process. MTMR14 phosphatase activity was required for its role as a regulator of autophagy, a finding consistent with the known importance of PI3P in the promotion of early steps of the pathway (16).
The extremely mild phenotype of the knockout mice reported by Shen et al. (14) stands at odds with the severe clinical phenotype reported in patients with centronuclear myopathy and MTMR14 sequence changes. While this may be due to intrinsic differences between mice and humans, other possibilities exist. The primary goal of the present study is to better understand the potential relationship between MTMR14 and human muscle disease. In addition, this study seeks to examine the relative roles in muscle homeostasis for MTMR14 and MTM1, the lipid phosphatase with the best-defined role in the regulation of muscle structure and function (6,17). We recently examined the role of MTM1 in zebrafish muscle development (18). Morpholino knockdown of mtm1 resulted in severe structural and functional abnormalities in skeletal muscle, mirroring the changes seen in patients with myotubular myopathy due to MTM1 mutations. In particular, we identified abnormalities in the organization of the T-tubule/sarcoplasmic reticulum triad and disturbance in triad-mediated excitation–contraction coupling.
To characterize the requirement of MTMR14 in muscle structure and function, we employed morpholino-mediated gene knockdown in the zebrafish. Similar to mtm1 knockdown, zebrafish injected with mtmr14 morpholino (MTMR14 morphants) had an early-onset myopathy, as determined by mild morphologic changes, abnormal embryonic motor behaviors and impairment of excitation–contraction coupling. Unlike MTM1 morphants, however, histopathologic analysis of skeletal muscle did not reveal abnormal nuclei or disturbances in triad structure. We then utilized morpholino-mediated double gene knockdown to examine the potential interrelationship between MTM1 and MTMR14. Double morphants had severe motor abnormalities and widespread ultrastructural changes in the muscle, changes that were significantly more severe than those observed in either morphant alone. Double morphants had increased autophagy, which may explain the severity of the combined phenotype. In summary, we demonstrate that mtmr14 knockdown results in abnormal excitation–contraction coupling and mild myopathy and combined knockdown of two lipid phosphatases causes severe and widespread developmental muscle abnormalities. Based on our data, we hypothesize a model whereby MTMR14 serves not as a primary cause of human centronuclear myopathy but rather as a potent genetic modifier of this childhood muscle disorder.
RESULTS
MTMR14 knockdown in the zebrafish
To study the contribution of MTMR14 to skeletal muscle development and function, we employed morpholino-mediated gene knockdown in the zebrafish. A single MTMR14 ortholog was identified using BLAST analysis with human MTMR14. This ortholog, on zebrafish chromosome 6, has 75% identity with human MTMR14 on the RNA transcript level and 67% on the protein level. We designed a morpholino to the splice donor site of exon 11 of zebrafish mtmr14. This morpholino is predicted to remove exon 11 and produce a truncated mRNA with frameshift and a premature stop codon. To test the efficacy of the morpholino to induce missplicing, three concentrations (4, 8 and 16 ng) of morpholino were injected into the yolk of one to two cell-stage embryos, and RNA was extracted at 48 h post fertilization (hpf). RT–PCR was performed using primers to exons 10–12 (Supplementary Material, Fig. S1). We found that at concentrations of 8 ng and higher, no normal MTMR14 RNA product was detected, while only a faint amount of the truncated product was present (data not shown). This indicated that our morpholino accomplished gene knockdown at low concentration and was suitable for subsequent functional analysis.
MTMR14 knockdown results in developmental motor dysfunction
As a first level of characterization, the effect of mtmr14 knockdown on skeletal muscle function was examined. In zebrafish, the first observable indication of skeletal muscle activity is spontaneous coiling, the alternating contraction of trunk and tail that begins at 17 hpf, peaks at 19 hpf and then decreases over the next 8 h (19). We assessed spontaneous coiling at 24 hpf; embryos injected with mtmr14 morpholino (i.e. MTMR14 morphants) had a small but significant decrease in spontaneous coiling when compared with control-injected embryos (mtmr14 morpholino versus control morpholino, 5.93 ± 0.32 versus 8.58 ± 0.33, n = 150, P < 0.0001) (Table 1 and Supplementary Material, Fig. S2). For comparison, MTM1 morphants were also generated and analyzed (Supplementary Material, Fig. S1) (18). MTM1 morphants were found to exhibit a greater decrease in spontaneous coiling frequency when compared with MTMR14 morphants (4.56 ± 0.37, n = 100 for MTM1 versus 5.93 ± 0.32 for MTMR14, P = 0.0031) (Table 1 and Supplementary Material, Fig. S2).
Table 1.
Comparative motor function as determined by spontaneous coiling in various MTMR14 and MTM1 morphants
| Morpholino | n (no. of trials) | spontaneous coiling (% of control MO) | P-value (versus CTL MO) |
|---|---|---|---|
| Control MO | 273 (12) | 100 | n.s. |
| MTMR14 exon 11 MO | 270 (12) | 69.2 | <0.0001 |
| MTMR14 exon 2 MO | 107 (6) | 69.3 | <0.0001 |
| MTMR14 ATG MO | 94 (6) | 69.2 | <0.0001 |
| MTM1 exon 2 MO | 100 (6) | 54.3 | <0.0001 |
| MTM1/MTMR14 MOs | 142 (9) | 24.1 | <0.0001 |
| Scrambled MTMR14 exon 11 MO | 100 (6) | 97.0 | n.s. |
| MTMR14 exon 11 MO +, WT hMTMR14 RNA | 60 (6) | 95.0 | n.s. |
| MTMR14 exon 11 MO +, R336Q hMTMR14 RNA | 60 (6) | 84.5 | 0.11 |
The next observable skeletal muscle-dependent phenotype in zebrafish is the touch-evoked escape response (19). Touch-evoked escape behaviors begin as rapid alternating contractions of the trunk and tail in response to touch and later (at 27 hpf) incorporate swimming to these rapid alternating contractions, which propel the embryos forward. Touch-evoked escape behaviors grow in strength such that at 48 hpf, embryos are capable of generating bouts of swimming lasting several seconds. We examined the touch-evoked escape response at 48 hpf in MTMR14 morphants when compared with controls and MTM1 morphants. We evaluated greater than 100 embryos per condition, and performed detailed time-lapse video microscopy on 30 from each. As expected, control embryos rapidly swam away from a tactile stimulus, exiting the field of view in approximately 200 ms (Fig. 1 and Supplementary Material, Video S1). MTMR14 morphants, on the other hand, had an obviously abnormal behavioral response that was stereotyped and reproducible (Supplementary Material, Video S2). Specifically, upon touch-evoked stimulation, morphants exhibited an initial burst of apparently normal swimming, but then only moved a short distance from the stimulus before abruptly stopping, unable to swim out of the field of view (Fig. 1). In addition, their escape response was fatigable, as multiple touches resulted in the loss of even the small burst of normal activity. As previously reported, a different pattern of abnormal escape response was observed with MTM1 morphants (Fig. 1). With tactile stimulus, MTM1 morphants either were unable to initiate any significant escape response or else moved away from the stimulus only very slowly (Supplementary Material, Video S3).
Figure 1.
MTMR14 morphants exhibit diminished touch-evoked escape responses. (Top) Tactile stimuli applied to the tail of a wild-type larvae (48–52 hpf) evokes a rapid escape contraction followed by swimming which propels the larvae out of the field of view (n = 27/30). Scale bar 1 mm. (Middle) A similar stimulus applied to the tail of an MTMR14 morphant evokes an escape contraction; however, morphants often failed to generate bouts of swimming sufficient to move out of the field of view (n = 3/30, P = 0.0006). (Bottom) For comparison, an MTM1 morphant performs a reduced escape contraction and low amplitude bending which was insufficient to propel the larvae forward out of the field of view (n = 0/30).
MTMR14 knockdown results in abnormal excitation–contraction coupling
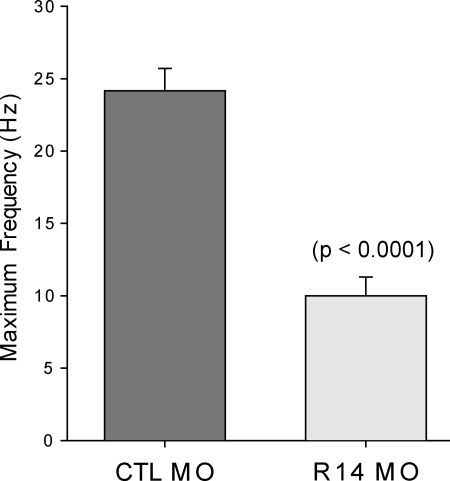
The identification of motor abnormalities in the mtmr14 morpholino-injected embryos prompted us to study muscle electrical properties. Because of the reported association between murine Mtmr14 and RYR1-dependent calcium release (14), we chose to focus on excitation–contraction coupling. We assayed excitation–contraction coupling by using an electrical stimulus to induce tetany at increasingly higher frequencies. We have previously used this technique to reveal excitation–contraction coupling abnormalities in MTM1 morphant fish (18). Control-injected morphant embryos, as we reported previously, sustain tetanic contractions at the highest frequency tested (25 Hz). MTMR14 morphants, conversely, were unable to generate repeated contractions at 15 Hz or higher (Fig. 2). This result is similar to that observed with MTM1 morphants (18), and is consistent with a defect in excitation–contraction coupling.
Figure 2.
MTMR14 morphants fail to respond to high-frequency electrical stimulation. In response to 5 ms depolarizing current injections to +60 mV, skeletal muscle from control embryos routinely contracted above 20 Hz for 10 s (n = 6/6). In contrast, skeletal muscle from MTMR14 morphants failed to contract at 20 Hz (n = 0/6). Values represent the average ± SEM at which muscle could sustain contractions in response to electrical stimuli.
MTMR14 knockdown results in mild morphologic abnormalities
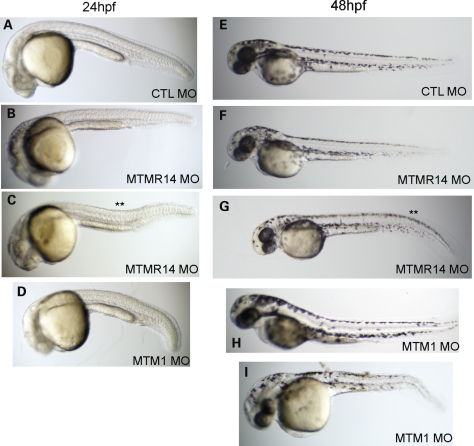
We next analyzed the morphologic appearance of MTMR14 morphant embryos at 24 and 48 hpf. Approximately 50% of all MTMR14 morpholino-injected embryos exhibited some degree of morphologic abnormality (n = 150). The most frequently observed change was a thinned and bent midbody (**, Fig. 3C and G). Midbody shape is largely dependent on structurally and functionally intact muscle, and this morphologic appearance has been reported with knockdowns of other muscle disease-related genes, including MTM1 (18,20–22). For comparison, typical MTM1 morphants at 24 and 48 hpf are shown (Fig. 3D, H and I). The MTM1 morphant morphologic phenotype, as reported previously (18), is more penetrant (∼80% of embryos affected) and in general more severe, with more substantial reduction in body length (C versus D in Fig. 3, for example) and a more severe range of presentation (Fig. 3I).
Figure 3.
MTMR14 morphants exhibit mild morphologic abnormalities. Morphologic appearance at 24 hpf (A–D) and 48 hpf (E–I) of control morphants (CTL; A and E), MTMR14 morphants (MTMR14 MO; B, C, F, G), and MTM1 morphants (MTM1 MO; D, H, I). MTMR14 morphant appearance ranged from essentially normal (B and F) to exhibiting minor defects in the shape of their midbodies (** in C) or tails (** in G). MTM1 morphants, in comparison, exhibit qualitatively more severe changes in overall appearance, body length and midbody changes.
MTMR14 knockdown does not alter skeletal muscle structure
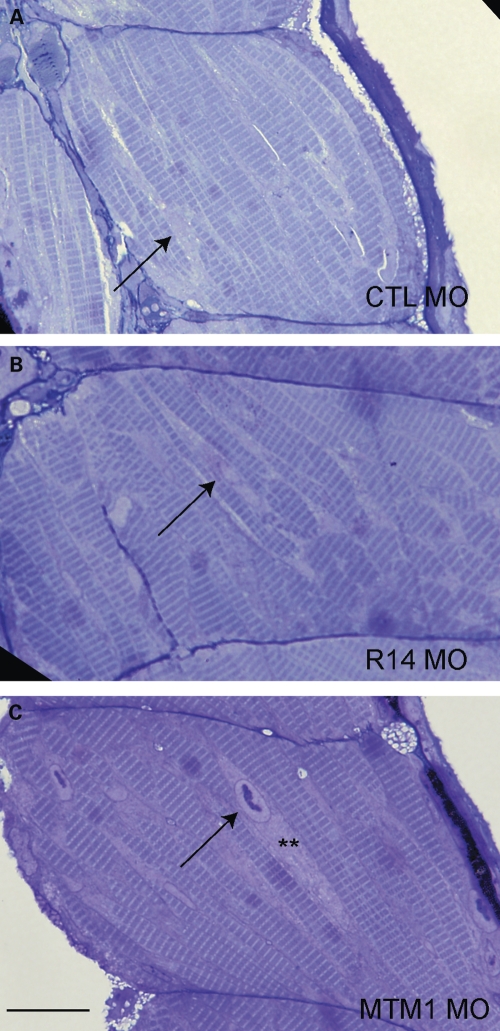
Embryos were further analyzed by histopathologic analysis at 72 hpf. Surprisingly, no abnormalities were detected in MTMR14 morphants by light microscopic examination of muscle structure (Fig. 4). In particular, the perinuclear region was normal without evidence of nuclear dysmorphology or perinuclear disorganization (Fig. 4B). This is in clear contrast to what is observed with mtm1 knockdown, where there are obvious nuclear changes and severe perinuclear abnormalities (Fig. 4C) (18). Nuclear abnormalities and perinuclear disorganization are the structural hallmarks of centronuclear myopathy (9), and the lack of these findings with mtmr14 knockdown supports a conclusion that MTMR14 morphant zebrafish do not exhibit the typical features of centronuclear myopathy.
Figure 4.
MTMR14 knockdown does not result in obvious histopathologic abnormalities. Skeletal muscle was examined in 72 hpf morphants. Muscle from MTMR14 morphants (R14 MO) was histologically normal and similar to muscle from controls (CTL MO). Arrows identify myonuclei. MTM1 morphants (MTM1 MO), on the other hand, exhibit obvious nuclear (arrow) and perinuclear changes (**). n = 4 per condition. Scale bar = 200 µm.
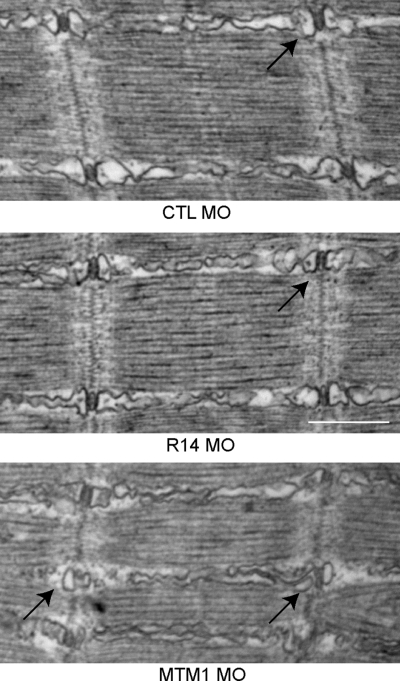
Embryos were lastly examined at 72 hpf for ultrastructural abnormalities using electron microscopy (Fig. 5). MTMR14 morphants had no obvious changes in their muscle ultrastructure. In addition to a normal perinuclear compartment (data not shown), MTMR14 morphants had normal-appearing triads (T-tubules and junctional sarcoplasmic reticulum) and longitudinal sarcoplasmic reticulum (arrow, Fig. 5B). In comparison, as we reported previously (18), MTM1 morphants had obvious ultrastructural abnormalities, including perinuclear disorganization (data not shown) and dilated and dysmorphic T-tubules and sarcoplasmic reticulum (arrows, Fig. 5C). Of note, the Golgi apparatus [the location of MTMR14 expression in vitro (13)] was also structurally unaltered by mtmr14 knockdown, as demonstrated by electron microscopy (data not shown) and by immunofluorescent analysis of isolated myofibers (Supplementary Material, Fig. S3).
Figure 5.
MTMR14 morphant muscle has normal ultrastructure. Ultrastructural analysis of muscle from 48 hpf control (CTL MO), MTMR14 (R14 MO) and MTM1 (MTM1 MO) morphants. Muscle ultrastructure was normal in MTMR14 morphants. In particular, triads were unchanged in appearance (arrow). Conversely, MTM1 morphant muscle has abnormal triads and longitudinal sarcoplasmic reticulum (arrows) (18). n = 4 per condition. Scale bar = 500 nm (34).
The MTMR14 morphant phenotype is specific for MTMR14 knockdown
We performed three control experiments to validate the specificity of the mtmr14 morpholino phenotype. The first was to determine the result of injecting other mtmr14 morpholinos. We tested a translation-blocking morpholino and an exon 2 splice-blocking morpholino. Both gave phenotypes similar to the mtmr14 exon 11 morpholino described earlier. Their injection resulted in a statistically significant decrease in spontaneous coiling (Table 1) and in mild abnormalities in morphologic appearance (Supplementary Material, Fig. S4).
The second control was to examine a morpholino generated with a scrambled version of the exon 11 sequence. This morpholino did not cause a significant reduction in spontaneous coiling when compared with control morpholino-injected embryos (Table 1), and did not cause appreciable morphologic changes (<10% of embryos appeared even mildly abnormal, which is identical to what is observed in control-injected embryos) (Supplementary Material, Fig. S4).
Finally, we attempted to rescue the motor phenotype of the exon 11 morpholino using in vitro transcribed human MTMR14 RNA (Supplementary Material, Fig. S5). Co-injection of human MTMR14 RNA with exon 11 morpholino resulted in significant improvement of spontaneous coiling at 24 hpf, restoring spontaneous coiling to 95% of normal from 75.7% of normal (Table 1; 95 ± 0.26 versus 75.7 ± 0.17%, n = 6 trials, P = 0.05). This result demonstrated that human MTMR14 RNA is biologically active in zebrafish and verified, along with the above two controls, that our morpholino phenotype was specifically due to reduction of MTMR14 levels. We performed an additional experiment to test the requirement of MTMR14 phosphatase activity for rescuing the motor phenotype. We used human MTMR14 RNA with a mutation in the phosphatase domain (R336Q) that has previously been shown to abrogate phosphatase activity (13), co-injecting it with exon 11 morpholino. This resulted in only a small and non-significant improvement of spontaneous coiling (Table 1; 84.5 ± 0.27 versus 75.7 ± 0.17%, n = 6 trials, P = 0.25). Thus, intact phosphatase activity is required for complete rescue.
Overexpression of mutant human MTMR14 RNA does not alter motor function
The data from our MTMR14 morphants suggest that mtmr14 knockdown, while associated with motor dysfunction, is not required for the establishment of normal muscle structure. This is not what would have been predicted from the previous report of patients with MTMR14 sequence variants and severe centronuclear myopathy. A plausible alternative explanation for the mutations is that they exert a dominant negative effect. To test this, we injected RNA derived from human MTMR14 cDNAs containing the published disease-associated sequence variants (R336Q and Y462C) (13). MTMR14 RNA was tagged with GFP and embryos expressing the mutant RNA were selected by the presence GFP expression (data not shown). We examined the touch-evoked escape response at 48 hpf, and did not observe obvious changes in the behavior of embryos injected with the mutated human MTMR14 RNAs or with wild-type MTMR14 RNA. We attempted to determine whether any subtle quantitative difference was present by scoring the touch-evoked behavior of each embryo. Using a 0–3 scale (see Materials and Methods), we failed to uncover any difference between uninjected, wild-type injected and mutant RNA injected embryos (Supplementary Material, Fig. S5) [2.81 ± 0.04 (CTL) versus 2.81 ± 0.06 (WT RNA) versus 2.79 ± 0.13 (R336Q RNA) versus 2.82 ± 0.09 (Y462C RNA)].
Human MTMR14 RNA is unable to rescue the MTM1 morphant phenotype
We also tested the ability of human MTMR14 RNA to rescue the mtm1 knockdown phenotype. We have previously demonstrated that MTMR1 and MTMR2, the closest homologs to MTM1 by sequence identity, are able to rescue the motor abnormalities observed in MTM1 morphants (18). Upon co-injection of wild-type MTMR14 RNA with mtm1 morpholino, however, we observed no alteration in the touch-evoked escape response at 48 hpf (2.28 ± 0.19 versus 2.20 ± 0.22, n = 30, P = 0.78). Therefore, human MTMR14 cannot compensate for the loss of zebrafish MTM1.
Combined MTMR14 and MTM1 knockdown causes a severe motor phenotype
As described earlier, mtmr14 knockdown causes mild but detectable changes in motor function, corroborating the description of a mild myopathy in Mtmr14 knockout mice (14). Mtm1 knockdown, on the other hand, causes a more severe myopathy, particularly in regard to morphologic and ultrastructural abnormalities [above and (18)]. Given that both MTMR14 and MTM1 are highly expressed in muscle and have identical substrates for their enzymatic activity (PI3P and PI3,5P2) (13,23,24), we were interested in understanding the effect on skeletal muscle development of reducing the levels of both genes. We generated ‘double knockdown’ embryos by co-injecting morpholinos to mtm1 and mtmr14 (Supplementary Material, Fig. S1). Simultaneous knockdown of both MTM1 and MTMR14 resulted in severe functional, morphologic and histologic abnormalities not solely restricted to skeletal muscle. As detailed below, we focused the majority of our analysis of double morphants on skeletal muscle structure and function.
We first examined spontaneous coiling as our functional readout for developmental motor activity, and found that double knockdown resulted in a substantial decrease in this behavior (Table 1 and Supplementary Material, Fig. S2). On average, double morphants had only 2.1 coiling events in 15 s, or 24.4% of the number observed in controls (2.1 ± 0.25, n = 142, versus 8.6 ± 0.33, n = 150, P < 0.0001). Combination morphants also had only 46.7 and 35.6% of the number of events observed for MTM1 and MTMR14 morphant embryos, respectively [2.1 versus 4.6 (MTM1) versus 5.9 (MTMR14); P < 0.0001 for both]. Furthermore, 50% of the double morphants exhibited no coiling behavior, indicating a complete arrest in the development of this motor function (71/142 embryos). In contrast, only 15% of MTM1 morphants, 4.7% of MTMR14 morphants and 0% of controls were paralyzed at 24 hpf. To determine if this severe alteration in spontaneous coiling caused by double knockdown was simply an additive effect of the individual morpholinos, or was instead due to a combined effect, we performed a negative binomial regression analysis on our data. Double knockdown produced a reduction in coiling that was 31% more than would be expected for an additive effect (P = 0.04), indicating that knockdown of both MTM1 and MTMR14 resulted in an independent and significantly more severe phenotype than either alone.
Double MTMR14/MTM1 knockdown results in severe morphologic abnormalities
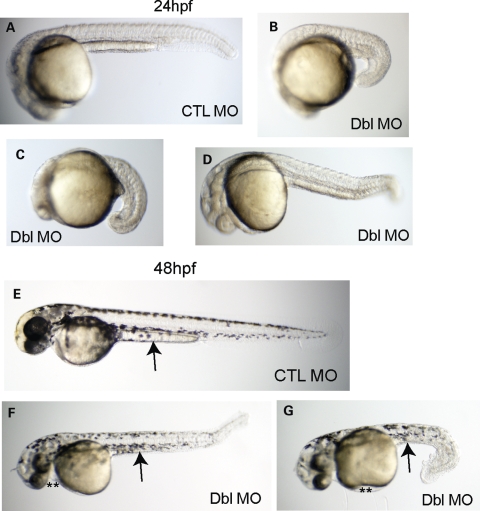
MTMR14/MTM1 double morphants also had profound morphologic abnormalities. These changes were not confined to the muscular compartments of the embryo, but rather reflected global alterations to embryonic development. Depicted in Figure 6 are representative embryos from control morphants (Fig. 6A and E) and MTMR14/MTM1 double morphants at 24 and 48 hpf (Fig. 6B–D, F and G). The majority of MTMR14/MTM1 double morphants at 24 hpf had substantially decreased overall body size (Fig. 6B–D). The heads were hypoplastic, and the bodies and tails were foreshortened and bent. While most embryos exhibited severe morphologic changes, the phenotype was variable and some more mildly affected double morphants were produced. Mild double morphants were less dysmorphic but still shorter, with abnormally appearing somatic compartments (Fig. 6D). Similarly severe changes were observed at 48 hpf (Fig. 6F and G), with additional features including the development of edema (**) and the lack of extension of the yolk sac (arrows).
Figure 6.
MTMR14/MTM1 double morphants have severe disturbances in embryonic development. Morphologic analysis of double morphants at 24 hpf (A–D) and 48 hpf (E–G). When compared with control embryos (CTL MO, A, E), MTMR14/MTM1 double morphants (Dbl MO) exhibited severe alterations in gross morphology. Embryos at 24 hpf were significantly foreshortened with global underdevelopment (B–D). At 48 hpf (F and G), double morphants were small and dysmorphic, with small and bent tails and hypoplastic midbodies. Edema (**) was also present.
Double MTMR14/MTM1 knockdown results in severe alterations in skeletal muscle structure
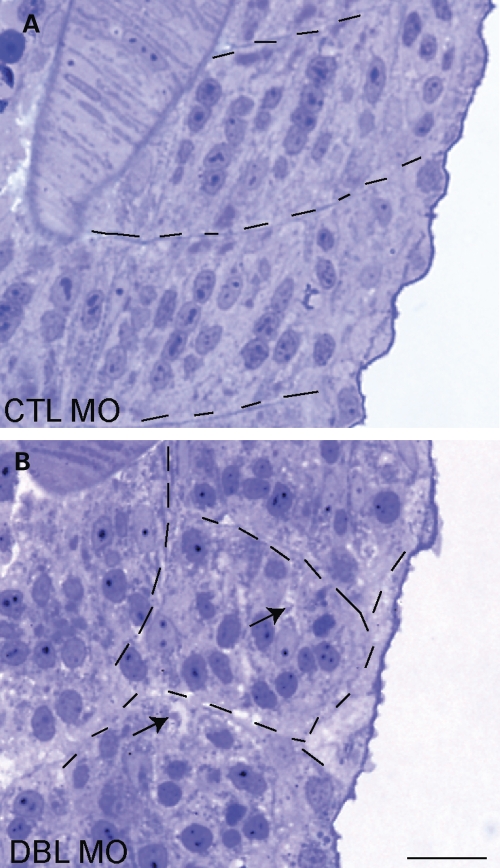
We next analyzed skeletal muscle structure by examining developing muscle compartments using light and electron microscopy. Obvious histopathologic abnormalities were present in 48 hpf double-morphant embryos (Fig. 7). Changes observed included smaller myofiber compartments (delineated by hatched lines), lack of subcellular organization of nuclei (randomly arrayed throughout the muscle as opposed to in chains) and evidence of areas without staining that resemble vacuoles (arrows, also see below).
Figure 7.
MTMR14/MTM1 double morphants have abnormal skeletal muscle structure. Light microscopic analysis of semi thin sections from 48 hpf embryos. (A) Control morphants (CTL) had normal developing skeletal muscle. (B) MTMR14/MTM1 double morphants (Dbl MO) had altered skeletal muscle structure, including abnormal appearance and localization of myonuclei and the presence of many small areas that lacked staining (arrows). Myofiber compartments are delineated by hatched lines. Scale bar = 250 µm.
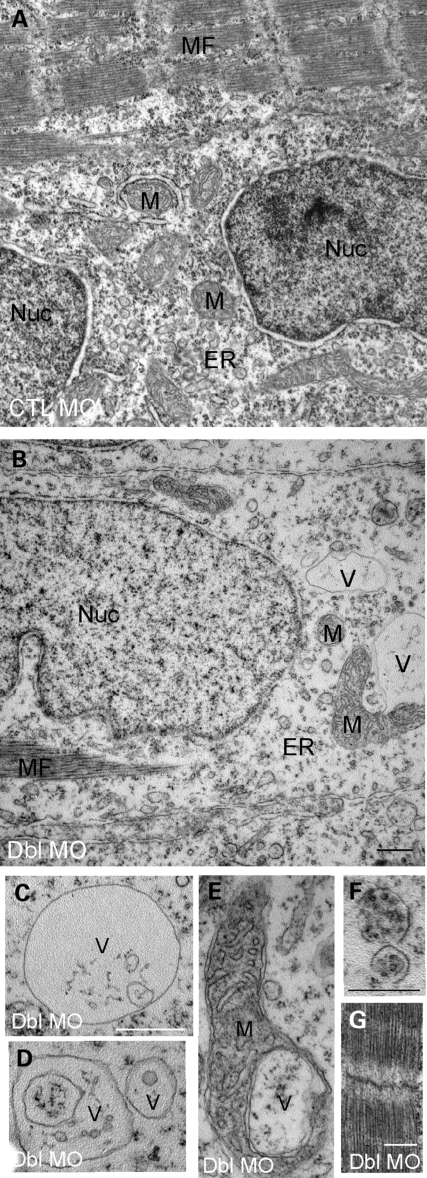
The structural changes were more precisely defined by electron microscopy. In particular, sarcoplasm from the double morphants was disorganized and contained a paucity of electron dense material (Fig. 8A and B). In addition, there were a number of unusual vacuolar structures (Fig. 8, labeled V). These vacuolar structures were large and numerous and were not found in muscle from controls or from either single morphant (Fig. 8C and D). Some of the vacuoles appeared within mitochondria (Fig. 8E). Others contained subcellular material and resembled autophagic vacuoles (see below and Fig. 8F). Despite these widespread cytoplasmic changes, double-morphant muscle did show evidence of myofibril formation (Fig. 8F), indicating that myogenesis was able to proceed at least to the point of the generation of the contractile apparatus.
Figure 8.
Ultrastructural analysis of MTMR14/MTM1 double-morphant skeletal muscle. Electron micrographs of skeletal muscle from 48 hpf embryos. (A and B) Low magnification of MTM1/MTMR14 double morphants (Dbl MO) revealed a paucity of sarcoplasm, decreased endoplasmic reticulum density (ER) and the presence of abnormal vacuolar structures (V) and abnormally appearing mitochondria (M). (C–E) Higher magnification examination of the abnormal vacuolar structures, including a vacuole within a mitochondrion (E) and vacuoles with subcellular contents (F). (G) Despite these changes, myofibrils appeared normal. Scale bars = 500 nm.
Autophagy is increased in MTMR14/MTM1 double morphants
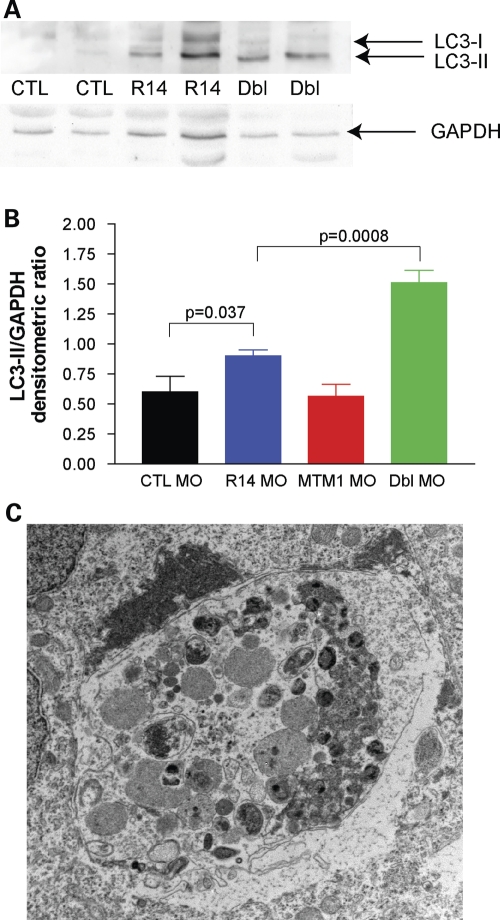
We lastly sought an explanation for the fact that double knockdown of mtmr14 and mtm1 was significantly more severe than the additive combination of the two phenotypes. Given the recent report associating MTMR14 and autophagy (15), we hypothesized that dysregulation of autophagy may provide an explanation. To evaluate the autophagic pathway, we studied LC3 protein levels in control, mtm1, mtmr14 and mtm1/mtmr14 morpholino knockdown embryos at 24 hpf. LC3 is a validated marker of autophagy, with a well-documented shift from a higher molecular species (LC3-I) to a lower molecular weight species (LC3-II) with the induction of autophagy (25). We observed a small but significant increase in LC3-II levels (measured as the ratio of LC3-II to GAPDH) in single MTMR14 morphants when compared with controls (0.90 ± 0.10 versus 0.60 ± 0.26, P = 0.037). We detected a much more robust increase in double-morphant LC3-II levels (1.51 ± 0.21 versus 0.60 ± 0.26, P = 0.0008) (Fig. 9A and B). This result indicated that autophagy was slightly induced with the reduction of mtmr14 and substantially induced when both mtmr14 and mtm1 levels were reduced. LC3-II levels, on the other hand, were not increased in single MTM1 morphants, indicating that autophagy was not induced by knockdown of mtm1 (0.57 ± 0.17 versus 0.60 ± 0.26, P = 0.47).
Figure 9.
Increased autophagy in MTMR14/MTM1 double morphants. (A) Western blot analysis of protein extracted from 48 hpf embryos. Blots were probed with anti-LC3, stripped and then reprobed with anti-GAPDH. Lanes: 1 and 2, control morphants (CTL); lanes 3 and 4, MTMR14 morphants (R14); and lanes 5 and 6 MTMR14/MTM1 double morphants (Dbl). (B) Quantitation of the ratio of LC3-II levels to GAPDH levels. Average values were as follows: CTL MO = 0.60, R14 MO = 0.90, MTM1 MO = 0.57, Dbl MO = 1.51. P-value of one-way ANOVA = 0.0001. (C) Example of an abnormal autophagic compartment revealed by electron microscopic analysis of a 48 hpf double-morphant embryo. Such structures were observed in three independently analyzed double morphants.
We then examined the subcellular ultrastructure of double-morphant embryos at 48 hpf. We observed multiple large vacuolar structures with an appearance consistent with that reported for degradative autophagic compartments (example in Fig. 9C). Such structures were not seen in any age-matched controls or single morphants (n = 3 for each condition). In combination with the LC3 western blot data, the presence of these autophagic vacuoles confirms that autophagy is aberrant in double knockdown embryos.
DISCUSSION
MTMR14 is the most recently identified member of the myotubularin superfamily, a group of dual-specificity phosphatases that dephosphorylate phosphoinositides (6,13). Two patients have been reported with centronuclear myopathy and heterozygous inactivating mutations in MTMR14 (13). In the present study, we address the role of MTMR14 in muscle development and homeostasis with an overarching goal of determining the potential relationship between MTMR14 and human muscle disease. Using the zebrafish as a model system, we found that MTMR14 is required for normal motor function but is dispensable for the proper formation and maintenance of muscle structure. These data demonstrate that knockdown of mtmr14 results in a functional myopathy, likely due to impaired excitation–contraction coupling, but not in the zebrafish equivalent of centronuclear myopathy. Additionally, we discovered that combined knockdown of mtmr14 and mtm1 results in profound developmental motor abnormalities that are more severe than the additive effect of either knockdown alone. This combined effect is likely due, at least in part, to the induction of autophagy. Based on these results, we hypothesize that MTMR14 mutation in humans, while not independently associated with a structural myopathy, may serve as a potent modifier of disease severity in conditions associated with impaired PI signaling, such as human centronuclear myopathy and Charcot-Marie-Tooth disease.
MTMR14 and centronuclear myopathy
It is clear from the results of our zebrafish studies and the previously reported mouse knockout that the loss of Mtmr14 alone is not sufficient to cause vertebrate centronuclear myopathy (14). This is in contrast to Mtm1 knockout mice and knockdown zebrafish, both of which have severe myopathy and histopathologic changes identical to those observed in patients with MTM1 mutations (17,18,26). It is thus unlikely that null mutations in MTMR14 are independently causative in childhood-onset or severe centronuclear myopathy.
Several possibilities exist to reconcile these findings with those of Laporte and colleagues (13), who reported MTMR14 mutations in two cases of severe centronuclear myopathy. One possibility is that these patients have dominant negative mutations in MTMR14 that are significantly more deleterious than the null mutations mimicked by Mtmr14 knockout mice and by our zebrafish morphants (14). The fact that injection of RNA containing either the R336Q or the Y462C mutation in control zebrafish did not have an overt phenotype would argue that this is unlikely to be the case. Another explanation could be related to intrinsic differences between man, mice and zebrafish. The fact that human MTMR14 can rescue the motor phenotype caused by zebrafish mtmr14 argues that MTMR14 function is conserved across species. In addition, the fact that studies of other MTMRs (including MTM1) have faithfully recapitulated disease in vertebrate model systems would also suggest that this is unlikely to be the case.
A third possibility is that MTMR14 is instead a potent modifier of disease severity. This hypothesis is supported by our data, which revealed that the reduction of mtmr14 levels, associated by itself with a mild phenotype, significantly worsened the developmental motor phenotype associated with mtm1 knockdown in the zebrafish. This conclusion would also be consistent with the information available about one of the patients with MTMR14 mutation. One patient has an inactivating variant in MTMR14 (R336Q mutation reduces phosphatase activity) and also a mutation in dynamin-2 (DNM2). This DNM2 mutation has been found to cause a mild, adult onset form of centronuclear myopathy in other patients (11). A logical interpretation of these co-occurring mutations is that the MTMR14 variant potentiates the normally mild phenotype associated with this DNM2 mutation.
An important question then is how MTMR14 may function as a genetic modifier. We believe the answer lies with the fact that MTMR14 is a negative regulator of autophagy (15), and that the loss of this regulation augments the pathology in centronuclear myopathy. Both MTM1 and dynamin-2-related centronuclear myopathies are hypothesized to have disturbed membrane traffic that can result in the aberrant formation or accumulation of organelles or membranes (6,8,12). An attractive model, as discussed below, is that the loss of MTMR14 results in dysregulation of autophagy that, in the setting of increased abnormal membranes, overwhelms the degradative pathway (Fig. 9). Future experimentation is required to establish a definitive link between DNM2-related disease severity and MTMR14. For example, it would be of interest to examine the muscle from the patient with DNM2 mutation and MTMR14 sequence variant for evidence of excessive autophagy.
MTMR14, MTM1 and autophagy
Combined knockdown of mtmr14 and mtm1 results in severe structural and functional abnormalities in co-injected zebrafish. These changes are significantly more severe than would be expected from the additive combination of the independent knockdowns, and additionally are not restricted to skeletal muscle. The double-morphant phenotype is associated with increased induction of autophagy (as determined by measuring LC3-II levels) and the presence of numerous vacuolated membranes. We thus hypothesize that the major explanation for this combined phenotype is dysregulation of membrane and organelle breakdown via aberrant autophagy. Our hypothetical model is that the autophagic system gets overloaded and is unable to perform its normal clearance function, which in turn results in an accumulation of autophagolysosomes, in widespread disturbances in cellular function and in global developmental abnormalities (Fig. 10). This could occur through (at least) two potential pathways. In one, mtmr14 knockdown results in excessive induction of autophagy by increasing PI3P levels, while mtm1 knockdown concurrently causes the accumulation of abnormal membranous structures. This mechanism is favored by our data. We observed a small increase in autophagy with mtmr14 knockdown alone (Fig. 8), consistent with the previously published report demonstrating that MTMR14 is a negative regulator of autophagy (15). We have previously described the accumulation of aberrant membranous structures in mtm1 knockdown zebrafish (18). This model would also be consistent with our speculation that the loss of MTMR14 function adversely modifies the phenotype in human centronuclear myopathy due to DNM2 mutation. DNM2 mutation in this setting would cause the aberrant accumulation of abnormal membranes.
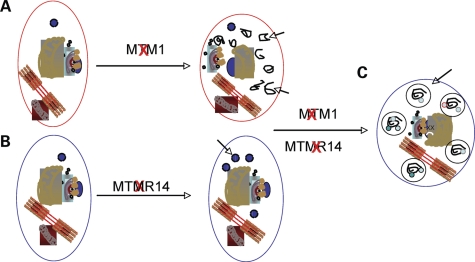
Figure 10.
Model of subcellular dysfunction with MTM1 and MTMR14 knockdown. (A) Knockdown of MTM1 results in the aberrant accumulation of membranes (arrows) as well as the abnormal appearance of organelles [Fig. 4B and (18)]. (B) Knockdown of MTMR14 causes an induction of autophagy [Fig. 8 and (15)]. (C) We hypothesize that the combination of accumulated autophagic substrates with excessive autophagic induction overwhelms the degradative system. This results in the accumulation of abnormal autophagic vacuoles (arrows) (Fig. 8C) and global cellular dysfunction, as reflected by the unexpectedly severe phenotype of double-morphant embryos.
The other model is that knockdown of both genes results in a massive accumulation of PI3P on autophagic isolation membranes, and that increased PI3P in turn causes excessive and inappropriate autophagy. Loss of either MTM1 (18) or MTMR14 (14) has been associated with increased PI3P in vivo. However, only MTMR14 has been directly associated with regulating PI3P on autophagic membranes (15). In fact, Vergne et al. (15) found that, unlike MTMR14, MTM1 knockdown did not result in an induction of autophagy. This is also consistent with our observation that LC3-II levels were not increased with mtm1 knockdown in zebrafish.
MTMR14, MTM1, excitation–contraction coupling and functional redundancy
An additional plausible explanation for the severity of the double-morphant phenotype is that MTM1 and MTMR14 share some redundant or overlapping functions (27). Based on in vitro data, this would not necessarily be predicted. MTMR14 localizes to the Golgi apparatus and the autophagic vacuole, while MTM1 is found at endosomes and plasma membrane ruffles, and each are thought to regulate membrane traffic at these specific locations (13,15,28–30). However, the recent observations from Shen et al. (14) that MTMR14 influences RYR1 function presents a plausible mechanism for how MTMR14 and MTM1 function may intersect. We have previously demonstrated in the zebrafish that MTM1 is expressed at tubulo-reticular junctions and that the loss of mtm1 causes abnormal development of the tubulo-reticular structure and alterations in excitation–contraction coupling (18). A complementary study in Mtm1 knockout mice demonstrated that, in addition to excitation–contraction coupling abnormalities, RYR1 levels are reduced by the loss of Mtm1 (26). We demonstrate here that knockdown of mtmr14 also impairs excitation–contraction coupling to a similar extent to that observed with mtm1 knockdown. This finding corroborates the findings of Shen et al. (14) from Mtmr14 knockouts, and supports the conclusion that both MTM1 and MTMR14 act at the tubulo-reticular junction to influence RYR1 and T-tubule/triad function. Further studies will be required to determine the specific role(s) for each gene product, though the available data would imply that MTM1 acts to maintain the overall triad structure while MTMR14 functions as a direct signal modifier of RYR1-dependent calcium release.
MTMR14 and MTM1 functions, however, are not completely redundant in skeletal muscle. To this point, Mtmr14 knockout mice and knockdown morphants have essentially no ultrastructural changes in their skeletal muscle (14), while Mtm1 knockout mice and morphant zebrafish have marked changes in nuclear appearance, perinuclear organization and tubulo-reticular network organization (17,18,26). Also, it is clear that MTMR14 by itself cannot compensate for the loss of MTM1. This is evident from the fact that Mtm1 knockout mice and patients with MTM1 mutations have a severe myopathy (17,31–33). Also, injection of human MTMR14 RNA is unable to rescue the motor phenotype associated with zebrafish mtm1 knockdown.
Conclusion
In summary, we have demonstrated that MTMR14 is required for motor function, likely by regulating excitation–contraction coupling, and for general morphology in the developing zebrafish embryo, but not required for the formation/maintenance of muscle structure. Double knockdown of mtm1 and mtmr14 results in severe functional, morphologic and ultrastructural abnormalities during development. The severity of the double-morphant phenotype is due at least in part to increases in the autophagic pathway. Based on our results, we speculate that MTMR14 mutations, while not a primary cause of centronuclear myopathy, are instead variants that significantly modify the phenotype caused by mutations in other centronuclear myopathy genes. We would thus advocate screening for MTMR14 sequence variants in cases of centronuclear myopathy with disparate genotype/phenotype correlation.
MATERIALS AND METHODS
Morpholinos
Morpholinos were designed to exon 3 of MTM1 and exon 11 of MTMR14 (GeneTools). Control morpholino was obtained from GeneTools and described previously (18).
MTM1 ex3-CCTGTCAACACACGCAGGAACATTG
MTMR14 ex11-TACTGAACAAATACCTACCGCCCAC
Morpholino injections
4.6 nl of morpholinos were injected into the yolks of one to four cell-stage zebrafish embryos using a Nanoinject (Drummond). MTMR14 and control morpholinos were used at 0.25 mm. MTM1 morpholino was used at 0.70 mm. To control for increased amount of morpholino in double-morphant experiments, control morpholino was co-injected with either MTM1 or MTMR14 morpholino for the single-morphant conditions.
RT–PCR
RNA was isolated from 48 hpf embryos using the RNeasy kit from Qiagen. One microgram of RNA was subsequently used to generate cDNA using the iScript kit (BioRad). PCR was performed on an iCycler (BioRad) using Go-Taq (Promega) and primers to exons 2–4 of zebrafish MTM1, exons 10–12 of zebrafish MTMR14 and control primers to zebrafish dynamin-2.
Spontaneous coiling
Coiling was measured at 24 hpf using a Scienscope dissecting microscope. Coiling was recorded as number of events observed in 15 s for each embryo.
Touch-evoked escape response
Escape response was measured at 48 hpf. It was elicited by touching embryo tails with an eyelash filament. Response was graded from 0 to 3; 0, no response to repeated stimulation; 1, flickers of movement but no swimming; 2, abnormal swimming in response to touch; 3, normal escape response.
Statistical analysis
Coiling and escape response were quantitated with a one-way ANOVA and a post-test Tukey's multiple comparison test using Prism. To determine the significance of effect of double knockdown versus single knockdowns, a negative binomial regression analysis was performed.
Excitation–contraction coupling
Electrophysiological recording from zebrafish was obtained from axial skeletal muscle at room temperature (22°C) using previously described methods (18,34). In brief, larvae were anesthetized in 1X Evans recording solution (in mM): 134 NaCl, 2.9 KCl, 2.1 CaCl2, 1.2 MgCl2, 10 glucose, 10 HEPES, pH 7.8 with NaOH supplemented with 0.02% (w/v) tricaine. The skin of a larvae pinned to a 35 mm Sylgard® coated dish was removed with a pair of No. 5 forceps. The solution was exchanged with 1X Evans containing 15 µM curare throughout the recording session at ∼1 ml/min. The internal recoding solution contained (in mM): 116 K-gluconate, 16 KCl, 2 MgCl2, 10 HEPES, 10 EGTA, at pH 7.2 with KOH. Borosilicate glass electrodes had resistances of 5–8 MΩ when filled with internal recording solution. Recordings were made with an Axoclamp-1D amplifier (Axon Instruments, Union City, CA, USA). Data acquisition was controlled by pClamp 10 software using a Digidata 1440A interface. The initial data analysis was done with Clampfit 10, and figures were prepared using Sigma Plot 11.0.
Morphologic analysis
Embryos were photographed using a Leica MXIII Stereoscope at 2× with a 4× zoom.
RNA-mediated rescue
Wild-type, R336Q and Y462C human MTMR14 cDNAs fused to GFP were a kind gift of Jocelyn Laporte. cDNAs were subcloned from pENTR into pCS2 (kind gift of Chi Bin Chien). Constructs were linearized with NotI and then transcribed using the mMessage mMachine kit (Ambion). Fifty nanogram per microliter of each RNA was injected into embryos and embryos were screened for positive GFP expression at 24 hpf. For experiments, RNA was co-injected with control, MTMR14 or MTM1 morpholinos into one to four cell-stage embryos. Expression of the RNA was again verified by screening embryos at 24 hpf for GFP fluorescence. Rescue was measured in GFP expressing embryos by touch-evoked escape response at 48 hpf.
Ultrastructural analysis
Forty-eight h post fertilization zebrafish embryos were fixed in Karnovsky's fixative and then processed for electron microscopy by the Microscopy and Imaging Laboratory Core facility at the University of Michigan. Images were obtained using a Phillips CM-100 transmission electron microscope.
Western blot analysis
Protein was extracted from batches of 25 embryos at 48 hpf in 250 µl RIPA buffer with complete protease inhibitors (Roche). Protein amount was quantitated using a modified Bradford assay (Pierce). One hundred twenty milligram of sample per batch was resolved by SDS–PAGE and then transferred to Hybond nitrocellulose. Primary antibodies used were anti-LC3 (1:500, Novus) and GAPDH (1:2000, Santa Cruz). Secondary antibodies were diluted 1:2000 (Santa Cruz). Blots were developed using Lumiglo (Cell Signalling) and visualized using a BioRad ChemiDoc XRS. Band densities were quantitated using the Quantity One software suite (BioRad). Significance was determined using an unpaired Student's t-test.
SUPPLEMENTARY MATERIAL
FUNDING
J.J.D. is supported by a career development award from the Muscular Dystrophy Association (MDA4333) and a K08 career development award from National Institute of Health (National Institute of Arthritis and Musculoskeletal and Skin Diseases) (1K08AR054835). We thank the A. Alfred Taubman Medical Research Institute, the Program for Neurology Research and Discovery, and the Janette Ferrantino Award for supporting our research.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jack Iwashyna for help with statistical analysis, Trent Waugh for technical assistance and help with figure design and Dan Klionsky for helpful discussion. We thank Stacey Sakowski for advice and editorial assistance with the manuscript.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Miaczynska M., Stenmark H. Mechanisms and functions of endocytosis. J. Cell Biol. 2008;180:7–11. doi: 10.1083/jcb.200711073. doi:10.1083/jcb.200711073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Di Paolo G., De Camilli P. Phosphoinositides in cell regulation and membrane dynamics. Nature. 2006;443:651–657. doi: 10.1038/nature05185. doi:10.1038/nature05185. [DOI] [PubMed] [Google Scholar]
- 3.McCrea H.J., De Camilli P. Mutations in phosphoinositide metabolizing enzymes and human disease. Physiology (Bethesda) 2009;24:8–16. doi: 10.1152/physiol.00035.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Robinson F.L., Dixon J.E. Myotubularin phosphatases: policing 3-phosphoinositides. Trends Cell Biol. 2006;16:403–412. doi: 10.1016/j.tcb.2006.06.001. doi:10.1016/j.tcb.2006.06.001. [DOI] [PubMed] [Google Scholar]
- 5.De Matteis M.A., Godi A. PI-loting membrane traffic. Nat. Cell Biol. 2004;6:487–492. doi: 10.1038/ncb0604-487. [DOI] [PubMed] [Google Scholar]
- 6.Nicot A.S., Laporte J. Endosomal phosphoinositides and human diseases. Traffic. 2008;9:1240–1249. doi: 10.1111/j.1600-0854.2008.00754.x. doi:10.1111/j.1600-0854.2008.00754.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Previtali S.C., Quattrini A., Bolino A. Charcot-Marie-Tooth type 4B demyelinating neuropathy: deciphering the role of MTMR phosphatases. Expert Rev. Mol. Med. 2007;9:1–16. doi: 10.1017/S1462399407000439. [DOI] [PubMed] [Google Scholar]
- 8.Jungbluth H., Wallgren-Pettersson C., Laporte J. Centronuclear (myotubular) myopathy. Orphanet J. Rare Dis. 2008;3:26. doi: 10.1186/1750-1172-3-26. doi:10.1186/1750-1172-3-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pierson C.R., Tomczak K., Agrawal P., Moghadaszadeh B., Beggs A.H. X-linked myotubular and centronuclear myopathies. J. Neuropathol. Exp. Neurol. 2005;64:555–564. doi: 10.1097/01.jnen.0000171653.17213.2e. [DOI] [PubMed] [Google Scholar]
- 10.Nicot A.S., Toussaint A., Tosch V., Kretz C., Wallgren-Pettersson C., Iwarsson E., Kingston H., Garnier J.M., Biancalana V., Oldfors A., et al. Mutations in amphiphysin 2 (BIN1) disrupt interaction with dynamin 2 and cause autosomal recessive centronuclear myopathy. Nat. Genet. 2007;39:1134–1139. doi: 10.1038/ng2086. doi:10.1038/ng2086. [DOI] [PubMed] [Google Scholar]
- 11.Bitoun M., Maugenre S., Jeannet P.Y., Lacene E., Ferrer X., Laforet P., Martin J.J., Laporte J., Lochmuller H., Beggs A.H., et al. Mutations in dynamin 2 cause dominant centronuclear myopathy. Nat. Genet. 2005;37:1207–1209. doi: 10.1038/ng1657. doi:10.1038/ng1657. [DOI] [PubMed] [Google Scholar]
- 12.Dowling J.J., Gibbs E.M., Feldman E.L. Membrane traffic and muscle: lessons from human disease. Traffic. 2008;9:1035–1043. doi: 10.1111/j.1600-0854.2008.00716.x. doi:10.1111/j.1600-0854.2008.00716.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tosch V., Rohde H.M., Tronchere H., Zanoteli E., Monroy N., Kretz C., Dondaine N., Payrastre B., Mandel J.L., Laporte J. A novel PtdIns3P and PtdIns(3,5)P2 phosphatase with an inactivating variant in centronuclear myopathy. Hum. Mol. Genet. 2006;15:3098–3106. doi: 10.1093/hmg/ddl250. doi:10.1093/hmg/ddl250. [DOI] [PubMed] [Google Scholar]
- 14.Shen J., Yu W.M., Brotto M., Scherman J.A., Guo C., Stoddard C., Nosek T.M., Valdivia H.H., Qu C.K. Deficiency of MIP/MTMR14 phosphatase induces a muscle disorder by disrupting Ca(2+) homeostasis. Nat. Cell Biol. 2009;11:769–776. doi: 10.1038/ncb1884. doi:10.1038/ncb1884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vergne I., Roberts E., Elmaoued R.A., Tosch V., Delgado M.A., Proikas-Cezanne T., Laporte J., Deretic V. Control of autophagy initiation by phosphoinositide 3-phosphatase jumpy. EMBO J. 2009;28:2244–2258. doi: 10.1038/emboj.2009.159. doi:10.1038/emboj.2009.159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Walker S., Chandra P., Manifava M., Axe E., Ktistakis N.T. Making autophagosomes: localized synthesis of phosphatidylinositol 3-phosphate holds the clue. Autophagy. 2008;4:1093–1096. doi: 10.4161/auto.7141. [DOI] [PubMed] [Google Scholar]
- 17.Buj-Bello A., Laugel V., Messaddeq N., Zahreddine H., Laporte J., Pellissier J.F., Mandel J.L. The lipid phosphatase myotubularin is essential for skeletal muscle maintenance but not for myogenesis in mice. Proc. Natl Acad. Sci. USA. 2002;99:15060–15065. doi: 10.1073/pnas.212498399. doi:10.1073/pnas.212498399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dowling J.J., Vreede A.P., Low S.E., Gibbs E.M., Kuwada J.Y., Bonnemann C.G., Feldman E.L. Loss of myotubularin function results in T-tubule disorganization in zebrafish and human myotubular myopathy. PLoS Genet. 2009;5:e1000372. doi: 10.1371/journal.pgen.1000372. doi:10.1371/journal.pgen.1000372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Drapeau P., Saint-Amant L., Buss R.R., Chong M., McDearmid J.R., Brustein E. Development of the locomotor network in zebrafish. Prog. Neurobiol. 2002;68:85–111. doi: 10.1016/s0301-0082(02)00075-8. doi:10.1016/S0301-0082(02)00075-8. [DOI] [PubMed] [Google Scholar]
- 20.Jurynec M.J., Xia R., Mackrill J.J., Gunther D., Crawford T., Flanigan K.M., Abramson J.J., Howard M.T., Grunwald D.J. Selenoprotein N is required for ryanodine receptor calcium release channel activity in human and zebrafish muscle. Proc. Natl Acad. Sci. USA. 2008;105:12485–12490. doi: 10.1073/pnas.0806015105. doi:10.1073/pnas.0806015105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deniziak M., Thisse C., Rederstorff M., Hindelang C., Thisse B., Lescure A. Loss of selenoprotein N function causes disruption of muscle architecture in the zebrafish embryo. Exp. Cell Res. 2007;313:156–167. doi: 10.1016/j.yexcr.2006.10.005. doi:10.1016/j.yexcr.2006.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Nixon S.J., Wegner J., Ferguson C., Mery P.F., Hancock J.F., Currie P.D., Key B., Westerfield M., Parton R.G. Zebrafish as a model for caveolin-associated muscle disease; caveolin-3 is required for myofibril organization and muscle cell patterning. Hum. Mol. Genet. 2005;14:1727–1743. doi: 10.1093/hmg/ddi179. doi:10.1093/hmg/ddi179. [DOI] [PubMed] [Google Scholar]
- 23.Taylor G.S., Maehama T., Dixon J.E. Inaugural article: myotubularin, a protein tyrosine phosphatase mutated in myotubular myopathy, dephosphorylates the lipid second messenger, phosphatidylinositol 3-phosphate. Proc. Natl Acad. Sci. USA. 2000;97:8910–8915. doi: 10.1073/pnas.160255697. doi:10.1073/pnas.160255697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Blondeau F., Laporte J., Bodin S., Superti-Furga G., Payrastre B., Mandel J.L. Myotubularin, a phosphatase deficient in myotubular myopathy, acts on phosphatidylinositol 3-kinase and phosphatidylinositol 3-phosphate pathway. Hum. Mol. Genet. 2000;9:2223–2229. doi: 10.1093/oxfordjournals.hmg.a018913. [DOI] [PubMed] [Google Scholar]
- 25.Klionsky D.J., Abeliovich H., Agostinis P., Agrawal D.K., Aliev G., Askew D.S., Baba M., Baehrecke E.H., Bahr B.A., Ballabio A., et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy. 2008;4:151–175. doi: 10.4161/auto.5338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Qusairi L., Weiss N., Toussaint A., Berbey C., Messaddeq N., Kretz C., Sanoudou D., Beggs A.H., Allard B., Mandel J.L., et al. T-tubule disorganization and defective excitation–contraction coupling in muscle fibers lacking myotubularin lipid phosphatase. Proc. Natl Acad. Sci. USA. 2009;106:18763–18768. doi: 10.1073/pnas.0900705106. doi:10.1073/pnas.0900705106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laporte J., Liaubet L., Blondeau F., Tronchere H., Mandel J.L., Payrastre B. Functional redundancy in the myotubularin family. Biochem. Biophys. Res. Commun. 2002;291:305–312. doi: 10.1006/bbrc.2002.6445. doi:10.1006/bbrc.2002.6445. [DOI] [PubMed] [Google Scholar]
- 28.Cao C., Backer J.M., Laporte J., Bedrick E.J., Wandinger-Ness A. Sequential actions of myotubularin lipid phosphatases regulate endosomal PI(3)P and growth factor receptor trafficking. Mol. Biol. Cell. 2008;19:3334–3346. doi: 10.1091/mbc.E08-04-0367. doi:10.1091/mbc.E08-04-0367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tsujita K., Itoh T., Ijuin T., Yamamoto A., Shisheva A., Laporte J., Takenawa T. Myotubularin regulates the function of the late endosome through the gram domain-phosphatidylinositol 3,5-bisphosphate interaction. J. Biol. Chem. 2004;279:13817–13824. doi: 10.1074/jbc.M312294200. doi:10.1074/jbc.M312294200. [DOI] [PubMed] [Google Scholar]
- 30.Laporte J., Blondeau F., Gansmuller A., Lutz Y., Vonesch J.L., Mandel J.L. The PtdIns3P phosphatase myotubularin is a cytoplasmic protein that also localizes to Rac1-inducible plasma membrane ruffles. J. Cell Sci. 2002;115:3105–3117. doi: 10.1242/jcs.115.15.3105. [DOI] [PubMed] [Google Scholar]
- 31.McEntagart M., Parsons G., Buj-Bello A., Biancalana V., Fenton I., Little M., Krawczak M., Thomas N., Herman G., Clarke A., et al. Genotype–phenotype correlations in X-linked myotubular myopathy. Neuromuscul. Disord. 2002;12:939–946. doi: 10.1016/s0960-8966(02)00153-0. doi:10.1016/S0960-8966(02)00153-0. [DOI] [PubMed] [Google Scholar]
- 32.Herman G.E., Kopacz K., Zhao W., Mills P.L., Metzenberg A., Das S. Characterization of mutations in fifty North American patients with X-linked myotubular myopathy. Hum. Mutat. 2002;19:114–121. doi: 10.1002/humu.10033. doi:10.1002/humu.10033. [DOI] [PubMed] [Google Scholar]
- 33.Laporte J., Hu L.J., Kretz C., Mandel J.L., Kioschis P., Coy J.F., Klauck S.M., Poustka A., Dahl N. A gene mutated in X-linked myotubular myopathy defines a new putative tyrosine phosphatase family conserved in yeast. Nat. Genet. 1996;13:175–182. doi: 10.1038/ng0696-175. doi:10.1038/ng0696-175. [DOI] [PubMed] [Google Scholar]
- 34.Buss R.R., Drapeau P. Physiological properties of zebrafish embryonic red and white muscle fibers during early development. J. Neurophysiol. 2000;84:1545–1557. doi: 10.1152/jn.2000.84.3.1545. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.