Abstract
Genome-wide association (GWA) studies have identified common variants that are associated with a variety of traits and diseases, but most studies have been performed in European-derived populations. Here, we describe the first genome-wide analyses of imputed genotype and copy number variants (CNVs) for anthropometric measures in African-derived populations: 1188 Nigerians from Igbo-Ora and Ibadan, Nigeria, and 743 African-Americans from Maywood, IL. To improve the reach of our study, we used imputation to estimate genotypes at ∼2.1 million single-nucleotide polymorphisms (SNPs) and also tested CNVs for association. No SNPs or common CNVs reached a genome-wide significance level for association with height or body mass index (BMI), and the best signals from a meta-analysis of the two cohorts did not replicate in ∼3700 African-Americans and Jamaicans. However, several loci previously confirmed in European populations showed evidence of replication in our GWA panel of African-derived populations, including variants near IHH and DLEU7 for height and MC4R for BMI. Analysis of global burden of rare CNVs suggested that lean individuals possess greater total burden of CNVs, but this finding was not supported in an independent European population. Our results suggest that there are not multiple loci with strong effects on anthropometric traits in African-derived populations and that sample sizes comparable to those needed in European GWA studies will be required to identify replicable associations. Meta-analysis of this data set with additional studies in African-ancestry populations will be helpful to improve power to detect novel associations.
INTRODUCTION
Genome-wide association (GWA) studies have identified common genetic variants associated with many diseases and related traits (1), including nearly 100 variants associated with anthropometric traits (2–18). These associations have almost all been discovered in European-derived populations. Studies in other ethnicities, particularly African-derived populations, are valuable, because they may help localize the signals of association and because additional variants present at high frequency in African-derived populations may be absent or rare in European samples (19). Furthermore, it is not clear whether associations found in the European samples can be consistently replicated in the samples of predominantly recent African ancestry: genetic, environmental or phenotypic heterogeneity, gene by environment interactions or different recombination histories between populations could all contribute to a lack of replication in African-derived populations.
Moreover, it remains unclear whether GWA studies will achieve the same success in African-derived populations, in which the linkage disequilibrium (LD) among single-nucleotide polymorphisms (SNPs) is weaker. The current available genotyping platforms capture a greater fraction of the common variants in populations of recent European origin (20) than in African-derived populations. Therefore, it is important to examine whether loci associated with phenotypes of interest in European-derived populations remain associated with the same phenotype in an African-derived population.
Another challenge facing GWA studies in African-Americans is that the use of the full range of genomic tools is not well established. Imputation not only expands the set of variants examined (21,22), but also enables the harmonization of data for meta-analysis in large consortia. However, GWA studies of imputed data in African-Americans have not yet been widely reported. Additionally, copy number variants (CNVs) have recently been recognized as a major source of genetic variation among individuals, and their association with polygenic traits has not been widely attempted in African-ancestry populations.
Here, we report a comprehensive GWA study of two anthropometric traits, height and body mass index (BMI), in two population samples of predominantly West African ancestry: African-Americans from Maywood, IL, and Nigerians from Igbo-Ora and Ibadan, Nigeria. On the basis of a meta-analysis of these two studies, we selected the best signals for replication in additional cohorts with predominantly African ancestry. We also focused additional replication efforts on loci near validated SNPs from studies in European-derived populations and on a region in 3q27 that had previously been shown to be linked to BMI in the Maywood cohort (23). Although we did not replicate the top signals identified in the GWA study, nominal replication evidence was observed for variants at several loci identified in European-origin populations. Our results suggest that GWA studies in African-ancestry populations will face similar limitations of power as those in European populations, but that replication and fine-mapping of European signals in African-derived populations may be an important outcome of such studies.
RESULTS
The characteristics of the Maywood and Nigeria samples with available Affymetrix 6.0 genotype data are presented in Table 1. In order to be able to examine the association with anthropometric traits across a denser set of SNPs, we imputed the African-American and Nigerian samples using HapMap reference panels. In particular, we used a combined reference panel of West African [Yoruban from Ibadan, Nigeria (YRI)] and northern European [Utah residents with Northern and Western European ancestry from the CEPH collection (CEU)] samples to impute genotypes for the African-American panel in order to account for the known admixed African and European ancestry of this population [Supplementary Material, Fig. S1; (24–26)] and used the YRI sample as the reference panel for the Nigerian samples. In total, we obtained ∼1.6 and ∼2.1 M imputed genotypes to supplement the ∼860 and ∼800 K directly assayed SNPs in our African-American and Nigerian cohorts, respectively, to test for association with height and BMI (see Materials and Methods).
Table 1.
Descriptive characteristics of phenotypes among study participants from the African-American and the Nigerian cohorts
| No. (%) | Age (years) | Weight (kg) | Height (m) | BMI (kg/m2) | |
|---|---|---|---|---|---|
| African American | |||||
| Female low BMI | 151 (20.32) | 41.14 ± 7.18 | 57.94 ± 7.66 | 1.65 ± 0.07 | 21.36 ± 2.48 |
| Female high BMI | 125 (16.82) | 40.47 ± 8.56 | 100.63 ± 18.78 | 1.63 ± 0.06 | 37.70 ± 6.82 |
| Female all | 276 (37.15) | 40.84 ± 7.83 | 77.27 ± 25.38 | 1.64 ± 0.07 | 28.76 ± 9.52 |
| Male low BMI | 334 (44.95) | 43.26 ± 7.05 | 68.70 ± 7.74 | 1.76 ± 0.07 | 22.14 ± 1.91 |
| Male high BMI | 133 (17.90) | 42.87 ± 8.82 | 104.24 ± 16.70 | 1.76 ± 0.07 | 33.49 ± 4.78 |
| Male all | 467 (62.85) | 43.15 ± 7.59 | 78.82 ± 19.48 | 1.76 ± 0.07 | 25.37 ± 5.95 |
| All | 743 | 42.29 ± 7.75 | 78.24 ± 21.86 | 1.72 ± 0.09 | 26.63 ± 7.65 |
| Nigerian | |||||
| Females | 678 (57.1) | 48.53 ± 14.75 | 62.46 ± 14.61 | 1.60 ± 0.07 | 24.52 ± 5.57 |
| Males | 510 (42.9) | 46.78 ± 16.32 | 63.11 ± 11.98 | 1.70 ± 0.08 | 21.74 ± 3.74 |
| All | 1188 | 47.78 ± 15.46 | 62.74 ± 13.54 | 1.64 ± 0.09 | 23.32 ± 5.06 |
Analysis of SNP association with height and BMI
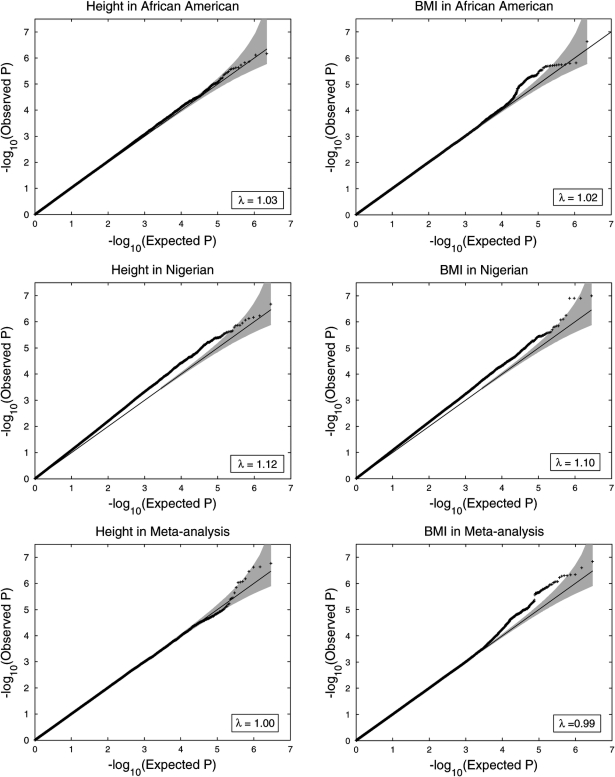
In total, ∼2.9 M SNPs were analyzed in the meta-analysis of the two panels after correcting for the genomic control inflation (GCI) factor, λ (for height and BMI, respectively, λ = 1.03 and 1.02 in the African-American cohort and 1.12 and 1.10 in the Nigerian cohorts, before correction; 1.00 and 0.99 after meta-analysis). No SNP reached a genome-wide significance level for either height or BMI in cohort-specific analysis or in meta-analysis (Fig. 1; Supplementary Material, Figs S2 and S3). We grouped SNPs in moderate LD (r2 ≥ 0.5 using YRI) and considered these as corresponding to the same signal.
Figure 1.
QQ plots for height and BMI in GWA results in the African-American cohort, the Nigerian cohort and the meta-analysis. For height and BMI, respectively, the GC inflation factors = 1.03 and 1.02 in the African-American cohort and 1.12 and 1.10 in the Nigerian cohorts. Studies were individually corrected for GCI (λ) prior to meta-analysis, but not again after meta-analysis. After meta-analysis, the λ's are 1.00 and 0.99, respectively.
For height, a list of 14 independent signals with P < 1 × 10−5 after meta-analysis is shown in Table 2. The SNP with the lowest combined P-value is rs1498759 located in a gene desert on chromosome 12 (P = 1.7 × 10−7). The nearest genes are TBX3 and THRAP2, 0.68 Mb and 0.59 Mb away, respectively. Moreover, a non-synonymous SNP, rs4619, in the IGFBP1 gene reached a P-value of 2.3 × 10−6. For BMI, there were 21 independent signals with P-values <1 × 10−5 (Table 2). The most significant association was with rs2401027, an intronic SNP in C12orf26 gene (P = 1.48 × 10−7).
Table 2.
GWA results for SNPs with P < 1 × 10−5 after meta-analysis and were chosen for stage 1 replication
| Chr | SNP | bp | Genes | A1 | African American |
Nigerian |
Meta-analyzed | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect | P-value | Effect | P-value | Effect | P-value | |||||
| Height | ||||||||||
| 2 | rs6746641 | 219 652 090 | C | −0.120 | 0.0270 | −0.188 | 6.85 × 10−5 | −0.159 | 7.52 × 10−6 | |
| 6 | rs734015 | 25 002 398 | C6orf32 | G | 0.145 | 0.0091 | 0.194 | 9.09 × 10−5 | 0.172 | 3.72 × 10−6 |
| 6 | rs2817723 | 25 007 547 | C6orf32 | C | 0.152 | 0.0048 | 0.185 | 2.53 × 10−4 | 0.170 | 3.53 × 10−6 |
| 7 | rs6942726 | 36 731 148 | ELMO1 | C | −0.170 | 0.0098 | −0.241 | 2.12 × 10−5 | −0.211 | 8.37 × 10−7 |
| 7 | rs4619 | 45 705 909 | IGFBP1, IGFBP3 | A | −0.186 | 7.08 × 10−4 | −0.141 | 0.0036 | −0.167 | 2.30 × 10−6 |
| 12 | rs6582380 | 40 781 393 | GLT8D3 | G | N/A | N/A | 0.592 | 1.05 × 10−5 | 0.592 | 9.99 × 10−6 |
| 12 | rs11169806 | 50 017 258 | BIN2, ELA1, GALNT6 | C | −0.152 | 0.0077 | −0.183 | 2.96 × 10−4 | −0.169 | 8.33 × 10−6 |
| 12 | rs1463833 | 114 279 162 | G | 0.126 | 0.0715 | 0.293 | 1.05 × 10−6 | 0.223 | 8.95 × 10−7 | |
| 12 | rs1498759 | 114 286 802 | T | 0.153 | 0.0120 | 0.234 | 1.76 × 10−5 | 0.206 | 1.70 × 10−7 | |
| 13 | rs1359634 | 30 879 675 | C | 0.341 | 8.07 × 10−6 | 0.144 | 0.0493 | −0.239 | 5.92 × 10−6 | |
| 13 | rs323425 | 104 258 663 | C | 0.272 | 2.26 × 10−4 | 0.212 | 2.28 × 10−4 | −0.235 | 2.33 × 10−7 | |
| 15 | rs10444801 | 53 189 330 | A | 0.234 | 3.60 × 10−5 | N/A | N/A | 0.251 | 6.35 × 10−6 | |
| 15 | rs2994 | 69 359 103 | THSD4 | G | −0.163 | 0.0057 | −0.193 | 1.98 × 10−4 | −0.180 | 4.00 × 10−6 |
| 17 | rs12603456 | 78 532 449 | TBCD, B3GNTL1 | T | 0.190 | 0.0071 | 0.205 | 3.87 × 10−4 | 0.199 | 7.35 × 10−6 |
| BMI | ||||||||||
| 1 | rs1395246 | 63 859 183 | PGM1 | G | N/A | N/A | −0.463 | 7.71 × 10−6 | −0.463 | 8.65 × 10−6 |
| 1 | rs12063584 | 212 441 965 | USH2A | C | −0.227 | 0.0083 | −0.306 | 5.69 × 10−4 | −0.276 | 5.24 × 10−6 |
| 5 | rs16903085 | 36 526 831 | T | −0.588 | 1.67 × 10−4 | −0.492 | 0.0040 | −0.521 | 4.35 × 10−6 | |
| 5 | rs6892149 | 49 961 304 | PARP8 | C | −0.224 | 6.24 × 10−4 | −0.194 | 0.0046 | −0.210 | 9.21 × 10−6 |
| 5 | rs17136138 | 113 326 936 | A | −0.387 | 4.90 × 10−4 | −0.332 | 0.0025 | −0.345 | 6.67 × 10−6 | |
| 6 | rs973089 | 56 543 316 | DST | T | −0.164 | 0.0029 | −0.161 | 0.0039 | −0.170 | 9.52 × 10−6 |
| 7 | rs739750 | 25 035 604 | C7orf31, NPVF | G | −0.273 | 1.41 × 10−5 | −0.127 | 0.0300 | −0.196 | 4.90 × 10−6 |
| 7 | rs11772003 | 100 637 032 | EMID2 | G | 0.155 | 0.0146 | 0.216 | 2.69 × 10−4 | 0.192 | 6.26 × 10−6 |
| 8 | rs7823476 | 33 981 611 | G | −0.184 | 0.0010 | −0.201 | 0.0018 | −0.191 | 5.05 × 10−6 | |
| 9 | rs10981043 | 111 582 297 | C9orf84 | A | −0.327 | 1.70 × 10−6 | −0.121 | 0.1010 | −0.219 | 7.89 × 10−6 |
| 11 | rs4323851 | 1 461 596 | BRSK2 | G | 0.392 | 2.78 × 10−5 | 0.250 | 0.0101 | 0.324 | 1.66 × 10−6 |
| 12 | rs2401027 | 81 276 367 | CCDC59, C12orf26 | C | 0.233 | 0.0014 | 0.277 | 2.80 × 10−5 | 0.257 | 1.48 × 10−7 |
| 15 | rs6495634 | 80 020 621 | T | −0.196 | 0.0420 | −0.472 | 1.27 × 10−6 | −0.323 | 1.56 × 10−6 | |
| 16 | rs6564329 | 74 996 147 | CNTNAP4 | G | 0.236 | 0.0040 | 0.318 | 6.12 × 10−4 | 0.272 | 8.86 × 10−6 |
| 17 | rs8077681 | 68 194 207 | SLC39A11 | G | 0.190 | 0.0011 | 0.162 | 0.0043 | 0.188 | 2.23 × 10−6 |
| 17 | rs492256 | 70 115 771 | C17orf77, CD300E | T | −0.261 | 3.55 × 10−4 | −0.319 | 0.0047 | −0.269 | 9.20 × 10−6 |
| 18 | rs7233501 | 74 334 699 | G | 0.395 | 3.35 × 10−4 | 0.272 | 0.0061 | 0.327 | 8.46 × 10−6 | |
| 20 | rs3003172 | 62 257 372 | OPRL1, NPBWR2, MYT1 | G | −0.199 | 0.0069 | −0.266 | 3.01 × 10−5 | −0.237 | 9.34 × 10−7 |
| 21 | rs415277 | 17 157 265 | G | N/A | N/A | −0.318 | 4.01 × 10−7 | −0.318 | 4.62 × 10−7 | |
| 21 | rs451387 | 17 162 730 | C | N/A | N/A | 0.416 | 1.91 × 10−6 | 0.416 | 1.87 × 10−6 | |
| 22 | rs9608641 | 26 418 266 | G | −0.258 | 2.19 × 10−4 | −0.316 | 0.0010 | −0.278 | 8.58 × 10−7 | |
Genes are within 50 kb upstream and downstream of the index SNP. A1 denotes reference allele with respect to which the effect is given. All SNPs listed under each trait are independent (r2 < 0.5) of each other according to the LD pattern in HapMap YRI individuals. P-values given for African-American and Nigerian cohorts were not GC-corrected, but were GC-corrected before meta-analysis. Meta-analysis was done using Metal weighted by the inverse variance method.
For both anthropometric traits analyzed here, previous GWA studies have identified numerous validated loci in Europeans, but few have been replicated in African-derived populations. Therefore, we examined the association evidence for confirmed loci for both height and BMI (6,7,11,17,18) in our African-derived populations. Because of the different LD patterns in European- and African-derived populations, a particular SNP that shows association in Europeans may be correlated with a nearby causal SNP in European-ancestry but not in African-ancestry samples. Therefore, we examined the association of all SNPs in high LD (r2 ≥ 0.8 in the CEU panel) with confirmed SNPs for height and BMI, but independent in the African genome (r2 < 0.5 in the YRI panel) in our largely African-ancestry samples. For height, a total of 33 independent signals distributed among 13 loci (ACAN, ANAPC13, BMP6, CDK6, DLEU7, DNM3, DYM, HIST1H1D, IHH, LCORL, RBBP8, ZBTB38 and the GDF5 gene cluster) achieved a one-tailed P-value of <0.05; for BMI, a total of six independent signals distributed among four loci (BDNF, ETV5, MC4R and TMEM18) achieved a one-tailed P-value of <0.05. The most significantly associated SNP for eight of the previously known loci (six for height and two for BMI) were selected for stage 1 replication (below, Table 3).
Table 3.
Association results for validated loci in Europeans with P < 0.05 and chosen for stage 1 replication
| Chr | SNP | bp | Genes | A1 | African American |
Nigerian |
Meta-analyzed | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Effect | P-value | Effect | P-value | Effect | P-value | |||||
| Height | ||||||||||
| 2 | rs10445823 | 219 735 669 | CCDC108, IHH, NHEJ1 | C | −0.080 | 0.1401 | −0.097 | 0.0292 | −0.090 | 0.0091 |
| 3 | rs7619451 | 135 715 936 | ANAPC13, CEP63 | T | −0.080 | 0.1450 | −0.108 | 0.0264 | −0.105 | 0.0032 |
| 6 | rs7769495 | 7 657 701 | BMP6 | G | 0.095 | 0.1060 | 0.111 | 0.0353 | 0.103 | 0.0069 |
| 13 | rs201762 | 49 963 837 | DLEU7 | G | −0.398 | 6.75 × 10−4 | −0.009 | 0.9497 | −0.246 | 0.0068 |
| 18 | rs4800455 | 19 002 731 | CABLES1 | G | 0.119 | 0.0338 | 0.091 | 0.0539 | 0.103 | 0.0042 |
| 18 | rs7506861 | 45 139 441 | DYM | A | −0.113 | 0.0782 | −0.133 | 0.0233 | −0.120 | 0.0044 |
| BMI | ||||||||||
| 11 | rs12806736 | 8 460 750 | STK33 | C | −0.090 | 0.1170 | −0.122 | 0.0368 | −0.108 | 0.0075 |
| 18 | rs6567160 | 55 980 115 | MC4R | C | 0.162 | 0.0158 | 0.115 | 0.0814 | 0.138 | 0.0035 |
Genes are within 50 kb upstream and downstream of the index SNP. A1 denotes reference allele with respect to which the effect is given. All SNPs listed under each trait are independent (r2 < 0.5) of each other according to the LD pattern in HapMap YRI individuals. P-values given for African-American and Nigerian cohorts were not GC-corrected, but were GC-corrected before meta-analysis. Meta-analysis was done using Metal weighted by the inverse variance method.
In stage 1 of the follow-up replication analysis, we selected SNPs based on two criteria specified in the Materials and Methods: (i) we selected 14 and 21 uncorrelated (r2 < 0.5) SNPs from the meta-analysis with meta-analyzed P-values <1 × 10−5 for height and BMI, respectively; (ii) we chose six and two uncorrelated SNPs representing the best association evidence from a subset of previously reported loci harboring nominal associations with height and BMI, respectively. We genotyped these SNPs in two independent cohorts of African-Americans: a panel of related individuals also from Maywood, IL (Maywood family, n = 756) and a sample of unrelated individuals from the tails of the height or BMI distributions (GCI; n = 488 for height and 494 for BMI). Among the SNPs we genotyped in stage 1, 15 SNPs showed the same direction of effect on height and BMI as seen in the two GWA discovery panels: 11 novel putative associations (four SNPs for height and seven SNPs for BMI) and 4 SNPs from previously reported loci (three SNPs for height and one for BMI, Supplementary Material, Tables S1 and S2). For stage 2 replication, we genotyped these 15 SNPs in two Jamaican cohorts with a combined total of 2437 individuals (GxE and SPT from Kingston and Spanish Town, Jamaica, respectively; see Materials and Methods).
For the 11 novel putative associations genotyped in stage 2, we observed no compelling evidence of replication (Table 4). One SNP for height (rs12603456) and two SNPs for BMI (rs739750 and rs8077681) reached a nominal significance in one cohort (one-tailed P = 0.044, 0.041, and 0.048, respectively); given 11 SNPs and 2 cohorts, we would expect about one SNP to reach this level of significance by chance. When results from replication genotyping were meta-analyzed by the study design (i.e. quantitative trait and dichotomous trait analyses were separately meta-analyzed), no compelling signals were obtained (data not shown).
Table 4.
Stage 1 and 2 replication results in four independent cohorts for SNPs showing P < 1 × 10−5 in genome-wide analysis for novel loci and that had passed stage 1 replication
| Chr | SNP | Genes | A1 | GCI AA |
Maywood Family |
Jamaica GxE |
Jamaica SPT |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value* | Effect | P-value* | Effect | P-value* | Effect | P-value* | ||||
| Height | |||||||||||
| 6 | rs2817723 | C6orf32 | C | 1.08 (0.83–1.41) | 0.278 | 0.089 | 0.407 | 0.017 | 0.363 | −0.052 | 0.918 |
| 7 | rs4619 | IGFBP1, IGFBP3 | G | 1.06 (0.82–1.37) | 0.333 | N/A | N/A | 0.011 | 0.408 | 0.012 | 0.375 |
| 12 | rs1498759 | T | 0.93 (0.69–1.25) | 0.688 | N/A | N/A | 0.014 | 0.392 | 0.046 | 0.127 | |
| 17 | rs12603456 | TBCD, B3GNTL1 | T | 1.09 (0.83–1.42) | 0.272 | N/A | N/A | 0.010 | 0.421 | 0.065 | 0.044 |
| BMI | |||||||||||
| 7 | rs739750 | C7orf31, NPVF | A | 1.13 (0.81–1.57) | 0.245 | 0.062 | 0.170 | 1.02 (0.80–1.31) | 0.431 | 0.006 | 0.041 |
| 15 | rs6495634 | T | 1.01 (0.60–1.70) | 0.515 | 0.033 | 0.629 | 0.97 (0.67–1.39) | 0.427 | 0.006 | 0.822 | |
| 16 | rs6564329 | CNTNAP4 | G | 0.91 (0.55–1.51) | 0.639 | 0.113 | 0.097 | 0.72 (0.52–1.02) | 0.969 | 0.001 | 0.387 |
| 17 | rs8077681 | SLC39A11 | G | 1.01 (0.60–1.70) | 0.240 | 0.015 | 0.397 | 1.21 (0.97–1.52) | 0.048 | −0.001 | 0.572 |
| 18 | rs7233501 | A | 0.78 (0.55–1.10) | 0.077 | −0.014 | 0.418 | 1.18 (0.90–1.53) | 0.887 | 0.008 | 0.980 | |
| 20 | rs3003172 | OPRL1, NPBWR2, MYT1 | G | 1.02 (0.75–1.37) | 0.540 | −0.124 | 0.018 | 0.89 (0.71–1.11) | 0.150 | 0.004 | 0.871 |
| 21 | rs415277 | C | 1.19 (0.84–1.67) | 0.162 | 0.060 | 0.177 | 0.88 (0.69–1.13) | 0.839 | 0.002 | 0.289 | |
Genes are within 50 kb upstream and downstream of the index SNP. A1 denotes reference allele with respect to which the effect or odds ratio (OR) is given. N/A, the results not available due to failed genotyping. All SNPs listed under each trait are independent (r2 < 0.5) of each other according to the LD pattern in HapMap YRI individuals.
*The one-tailed P-value (Bold indicates P-value < 0.05).
Among the three reported European height associations that passed our stage 1 screening, we observed a strong association with height for one SNP (rs10445823) near IHH (one-tailed P = 0.0036) and nominal associations for one SNP (rs201762) near DLEU7 in the Jamaican SPT (the Jamaican panel from Spanish Town, Jamaica) cohort (one-tailed P = 0.031, Table 5). For the one reported European BMI association near MC4R, we observed nominal association in the Jamaican SPT cohort (rs6567160, one-tailed P = 0.021, Table 5).
Table 5.
Stage 1 and 2 replication results in four independent cohorts for the reported European associations that showed P < 0.05 in our GWA discovery panel and that had passed stage 1 replication
| Chr | SNP | Genes | A1 | GCI AA |
Maywood family |
Jamaica GxE |
Jamaica SPT |
||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| OR (95% CI) | P-value* | Effect | P-value* | Effect | P-value* | Effect | P-value* | ||||
| Height | |||||||||||
| 2 | rs10445823 | CCDC108, IHH, NHEJ1 | C | 0.97 (0.75–1.25) | 0.402 | −0.712 | 0.025 | −0.060 | 0.100 | −0.099 | 0.004 |
| 13 | rs201762 | DLEU7 | G | 0.96 (0.58–1.60) | 0.438 | −1.302 | 0.044 | −0.062 | 0.300 | −0.186 | 0.031 |
| 18 | rs7506861 | DYM | G | 1.17 (0.86–1.59) | 0.153 | 0.642 | 0.050 | 0.010 | 0.431 | 0.033 | 0.222 |
| BMI | |||||||||||
| 18 | rs6567160 | MC4R | C | 1.14 (0.81–1.63) | 0.227 | 0.134 | 0.024 | 1.00 (0.77–1.31) | 0.497 | 0.009 | 0.021 |
Genes are within 50 kb upstream and downstream of the index SNP. A1 denotes reference allele with respect to which the effect or odds ratio (OR) is given. All SNPs listed under each trait are independent (r2 < 0.5) of each other according to the LD pattern in HapMap YRI individuals.
*The one-tailed P-value (Bold indicates P-value < 0.05).
Initial characterization of the genetic architecture of height and BMI in African-derived populations
Although our study is underpowered to detect association with modest effect sizes, we can still use our data to place likely upper bounds on the effect sizes of common variants in African-derived populations. We calculated the power of detecting a true association over a range of effect sizes (Supplementary Material, Fig. S4). Using a significance level of 1 × 10−5 (our threshold for choosing SNPs for further analysis), we would have low (∼3%) power to detect a SNP explaining ∼0.35% of the phenotypic variation, which is approximately the estimated effect size for variation near ZBTB38 for height (6,7,11) and FTO for BMI (17,18). However, at the same significance level, our GWA discovery panel would have ∼50% power to detect variants explaining ∼1% of the phenotypic variation, or ∼80% power to detect variants explaining ∼1.5% of the phenotypic variation (Supplementary Material, Fig. S4). We would have >90% power to replicate findings for variants with such strong effects in our follow-up panel. Thus, we conclude that for height and BMI, there are likely few or no variants of large effect (>1% of variance explained) in African-derived populations.
Moreover, our data also allow us to make an initial estimate of effect sizes at known loci. Specifically, we asked whether the loci found in European-derived populations have similar effect sizes in African-derived populations. Using the published effect size estimates, we calculated the expected number of validated European loci we would detect with one-tailed P < 0.05 in our GWA panel (see Materials and Methods). At a one-tailed significance level of 0.05 in the GWA data, we would expect to bring 22.0 (SD = 3.4) out of the 53 confirmed loci to stage 1 replication. In our study, we observed 17 loci with one-tailed P < 0.05 (13 for height and 4 for BMI). Because of differences between the European and African LD structures, multiple independent signals may underlie each of the 53 confirmed loci in African-derived genomes. Thus we also simulated the number of independent signals expected to attain one-tailed P < 0.05 under the null distribution, matched by allele frequency in the CEU panels and the number of independent signals (with r2 < 0.5 between them using YRI LD estimates) in the YRI panel (see Materials and Methods). Within the 17 loci reported here, a total of 39 independent SNPs were found to have one-tailed P < 0.05. By simulation, a total of 7.4 (5.4, SD = 2.8 for height; 2.0, SD = 1.6 for BMI) were expected. Therefore, the significant enrichment of number of independent signals with one-tailed P < 0.05 observed implies that majority of the loci found to be nominally associated in the GWA phase of our study likely represent real replication.
To further support the notion that variants associated with anthropometric traits have similar effect sizes between African- and European-derived populations, we examined the number of loci expected to show nominal association in either of the Jamaican cohorts among the variants we decided to follow-up. Given the power of our replication panels, we expected to see variants from 2.4 loci (SD = 1.2) to be nominally replicated with one-tailed P < 0.05 in either of the Jamaican cohorts. Considering that we followed up the best signal from eight known loci and genotyped them in four replication panels, the false discovery rate would be 0.024 or 0.20 variants expected to be nominally associated under the null hypothesis. We observed variants from three loci to be nominally associated. Together, our findings again suggest that the effect sizes of variants associated with anthropometric traits are, at least as a class, similar between European- and African-derived populations. As such, one would expect that at least comparable sample sizes will be needed in GWA studies using African-derived populations to identify variants associated with height and BMI.
Analysis of association of common and rare CNVs with BMI
In addition to single-nucleotide variations, structural variations such as CNVs have recently been recognized as a major contributor of genetic variation between individuals. Therefore, to screen more comprehensively for the association of genetic variants with anthropometric traits, we examined the association of both common and rare CNVs with height and BMI. On the basis of the published map of 1319 common CNV (27), we assessed the association of 405 and 431 commonly segregating CNV regions (frequency > 5%) found in the African-American and the Nigerian GWA samples, respectively, with height and BMI. After meta-analysis, none of the common CNVs surpassed our pre-specified Bonferroni threshold of P < 0.00010 (for a total of 482 unique common CNVs tested for association after meta-analysis). The CNV most strongly associated with height had a P = 0.00159, on chromosome 3 at ∼102.2 Mb; the CNV most strongly associated with BMI had a P = 0.0031, on chromosome 3 at ∼167.5 Mb. Neither phenotype showed an appreciable deviation from the null on their respective QQ plots (Supplementary Material, Fig. S5).
One common deletion residing upstream of NEGR1 (chromosome 1 at ∼72.5 Mb), and in perfect LD with SNP rs2815752, had been previously reported to be associated with increasing BMI in European populations (18). The same CNV and SNP were also tested in our study for associations with BMI in our African-derived populations. Both the CNV and the SNP trended in the expected direction, but neither were significantly associated with BMI (meta-analysis one-tailed P = 0.118 for the CNP and one-tailed P = 0.075 for the SNP).
For rare CNVs (frequency < 5%), we focused on the extremes of the BMI distribution (the phenotype for which the African-American cohort was originally ascertained) and evaluated the genome-wide differences in CNV burden between cases and controls using three indicators: the genome-wide CNV rate per individual, the total extent of rare CNVs per individual and average length of rare CNVs (see Materials and Methods). There are no differences in the rate of CNVs (P = 0.342 and 0.311 for African-Americans and Nigerians, respectively; Table 6), but the African-American controls (i.e. lean individuals) carried significantly heavier burden of total CNV span (P = 0.0032) and average CNV length (P = 0.0051). A similar trend was observed in the Nigerian cohort, though the differences were not significant. As such, the collective evidence of difference was enhanced when the two cohorts were meta-analyzed (P = 0.0009 for total CNV span and P = 0.0039 for average CNV length; Table 6). The results are not confounded by genetic ancestry in the African-Americans as the genetic ancestry was not correlated with case–control status (data not shown). We generally observed the same pattern if we restricted our analysis to only the CNVs lying within 20 kb of a gene (Supplementary Material, Table S3) or if we stratified the analysis by gender (Supplementary Material, Table S4). We attempted to replicate this finding by examining a European-derived population similarly genotyped on Affymetrix 6.0 and for which CNV data exist (28). However, we did not observe an increase in burden among the lean individuals in this population (Table 6).
Table 6.
Global rare CNV (frequency <5%) burden analysis
| Cohort | Group | Sample (n) | NSEG (n) | RATE | Total Span (kb) | Avg Span (kb) |
|---|---|---|---|---|---|---|
| African-Americans | Case | 190 | 2321 | 12.22 | 730.1 | 60.58 |
| Control | 435 | 5478 | 12.59 | 897.7 | 73.2 | |
| Empirical P-value | 0.342 | 0.00316 | 0.00512 | |||
| Nigerians | Case | 291 | 3233 | 11.11 | 700.0 | 65.33 |
| Control | 585 | 6830 | 11.68 | 793.8 | 70.74 | |
| Empirical P-value | 0.311 | 0.0665 | 0.158 | |||
| Meta-analyzed P-value | 0.165 | 0.00094 | 0.0039 | |||
| Europeans/European-Americans | Case | 604 | 6142 | 10.17 | 874.8 | 85.76 |
| Control | 1204 | 12 134 | 10.08 | 821.4 | 82.25 | |
| Empirical P-value | 0.692 | 0.144 | 0.274 | |||
Case and control definitions are described in the Materials and Methods. Empirical P is computed from a two-sided test after 50 000 permutations within genotyping plates to control for batch effects across different genotyping plates. The number of individuals (Sample) and the number of rare CNV segments (NSEG) are listed. RATE denotes the number of rare CNV segments per individual in each group. Total Span denotes the average kb of rare CNV segments per individual in each group. Avg Span denotes the average length of rare CNV segments per individual in each group. Meta-analysis was done using Metal weighted by sample size.
A large, rare chromosomal deletion at 16p11.2, spanning SH2B1, was recently reported to be associated with severe early-onset obesity in Caucasians (29). When we examined the BMI-associated region (from ∼28.4 to ∼28.8 Mb on chromosome 16) in our cohorts, we did not observe any deletions or duplications. The nearest CNV we observed was a duplication from ∼29.2 to ∼29.7 Mb in a control individual from Nigeria. Given the size and frequency of the large chromosomal deletion reported (29), we would expect to observe the variant, if present, in our study. Our failing to do so may reflect differences in either the population or in cohort ascertainment. We also examined known BMI loci previously identified by GWA studies in Europeans and did not find any obvious rare deletions or duplications near the loci in individuals at the extremes of the BMI distribution within our population (data not shown).
DISCUSSION
To date, few GWA studies have been attempted in African-derived populations, despite the success in GWA studies based primarily on European-derived participants (19,30). In the present study, we describe the first GWA study assessing the association of imputed genotypes and CNVs with height and BMI in two African-derived populations. We applied imputation methods to test variation at up to 2.9 million HapMap SNPs for association. We note that the optimal approach to impute African-derived genomes is under development; possible approaches include equal weighting of the YRI and CEU panels, using all HapMap reference panels, or sequential imputation using separate reference panels (31–33). Although a rigorous comparison of these approaches is beyond the scope of this paper, our assessment of imputation quality by concordance scores (see Materials and Methods) nonetheless indicated to us that, at least for relatively common SNPs, our imputation results were of high quality and appropriate for use in association testing.
Using information from both imputed and chip-genotyped SNPs, we discovered no common SNPs showing consistent strong evidence of association with either height or BMI in our study. We have previously reported a region near 3q27 showing evidence of linkage to BMI from a cohort sampled from Maywood, IL (23), but no SNP in this region showed elevated association with BMI in our current study (Supplementary Material, Fig. S6). We also examined a total of 53 loci for height (n = 41) (6,7,11) and BMI (n = 12) (17,18) previously reported to be associated in European-derived populations. Among them, we were able to provide some evidence of replication in African-derived populations for IHH and DLEU7 for height and MC4R for BMI. Finally, no strong association of common CNVs with height or BMI was detected. Although lean individuals appeared to have a heavier burden of rare CNVs compared with the heavier individuals, we were not able to replicate this finding in a European data set.
Like many other individual GWA studies, our data set is substantially underpowered to detect novel associations with modest effect sizes, having ∼3% power to detect an SNP with effect size similar to that of ZBTB38 and FTO. Nonetheless, at this significance level, our GWA discovery panel would have ∼50% power to detect SNPs explaining ∼1% of the phenotypic variation or ∼80% power to detect SNPs explaining ∼1.5% of the phenotypic variation, and our replication panels would have >90% power to replicate variants with such strong effects (Supplementary Material, Fig. S4). As such, it appears that there are no common variants with extremely large effects (>1.5%) or multiple common variants with large effects (>1%) in the African-derived genome for the anthropometric traits analyzed here. These represent the initial estimates of the upper bound of effect sizes for variants associated with anthropometric traits in African-derived populations. Increasing sample size and meta-analysis with other data sets of African-derived populations will be necessary to improve power to identify novel associations as well as to refine the estimates of effect sizes present in African-derived populations.
Examples exist where the variants associated with phenotype were shown to have large allele frequency differences between populations [e.g. MYBPC3 for cardiomyopathies (34), KCNQ1 for type 2 diabetes (34) and MYH9 for focal segmental glomerulosclerosis (36)]. Such variants could be missed by GWA studies in a single population, but may be highlighted by admixture mapping, because admixture association signals arise from alleles with large differences in ancestral population frequencies (37,38). Thus, an approach that combines admixture mapping with the GWA data in larger samples may be a fruitful route to identifying variants that were not highlighted by studies in European-derived populations because of low allele frequency (and hence low power). Further, such an approach allows for testing to determine whether an associated variant is able to account for the admixture mapping signal, thus potentially testing for the causality of the variant.
Despite our insufficient power to detect novel associations, we have >60% power to detect true associations explaining ∼0.2% of the variance in our GWA panel at a one-tailed significance level of 0.05. As such, among the 53 confirmed loci for height and BMI previously reported in Europeans, we expected to see nominal associations at ∼22 loci if effect sizes are similar in European- and African-derived populations. In total, we observed 17 loci with at least one independent SNP associated with one-tailed P < 0.05. Our observation of 17 loci nominally associated in the GWA panel is slightly below a priori expectations. However, the estimated percent variance explained is likely inflated due to ‘winner's curse’ for some of the reported loci as effect sizes from independent European-derived replication samples have not been published. Moreover, less than optimal coverage of variants as well as lower correlation of the HapMap SNPs and any actual causal variant in the African-derived genomes would also cause us to overestimate our expectation. Thus, it appears reasonable to anticipate that the effect sizes of variants associated with anthropometric traits in African-derived population will be similar to those observed in European-derived populations thus far, and therefore, assuming equal coverage of variation, at least comparable sample sizes would need to be enrolled in studies conducted in African-ancestry populations to achieve the success of European GWA studies.
Lastly, it was a curious finding that lean individuals in our data set appeared to have heavier burden of rare CNVs compared with the obese individuals (Table 6). We first observed the finding in the African-American panel, which was supported by data from the Nigerian cohort, but was not supported by data from a European-derived cohort that also had Affymetrix 6.0 genotypes and CNV calls (28). Therefore, our finding of heavier CNV burden among lean individuals could represent an effect specific to cohorts of African ancestry, or a statistical fluctuation. If true, and assuming that rare CNVs are generally deleterious from an evolutionary standpoint, this finding could support the thrifty gene hypothesis that heavier individuals are evolutionarily better suited to store energy (39). However, this result would need to be confirmed in additional African-derived cohorts with comparable or larger sample size and genotyped on a similar platform.
In summary, we have described the first GWA study of anthropometric traits in African-derived populations. Although we were not able to identify replicable novel single nucleotide or common CNVs associated with height or BMI, we were able to replicate three of the loci identified in European-derived populations at nominal levels of significance. On the basis of the analysis of power, we have ruled out the existence of multiple variants with large effects (∼1% variance explained) in African-derived samples and presented an initial estimate for the upper bound of effect sizes for anthropometric traits, which will improve with a more comprehensive assessment of signal fine-mapping in African-derived genomes. Given the need for larger sample sizes, the data set described here would represent a useful component of a meta-analysis of African-derived populations, for both SNPs and CNVs, to facilitate mapping of genes for anthropometric traits in African-derived populations.
MATERIALS AND METHODS
Discovery panels
African-American cohort
DNA samples were obtained from a larger cohort of families enrolled in the studies of blood pressure at Loyola University in Maywood, IL. The survey enrolled a representative random sample of the population between the ages of 18 and 74, regardless of obesity phenotype. A subcohort of 775 unrelated participants was selected and oversampled at the upper and lower ends of the BMI distribution to increase the power. The project was reviewed and approved by the IRBs at Loyola University Chicago and Case Western Reserve University. All participants gave the written informed consent.
Nigerian cohort
The sampling frame for the Nigeria cohort was provided by the International Collaborative Study on Hypertension in Blacks (ICSHIB) as described in detail elsewhere (40). Study participants were recruited from Igbo-Ora and Ibadan in southwest Nigeria as part of a long-term study on the environmental and genetic factors underlying hypertension. The sample comprised 1098 unrelated adults with normal or elevated blood pressure and 155 unrelated participants from Ibadan with elevated blood pressure recruited as controls in the Africa-America Diabetes Mellitus (AADM) study (41). Both projects were reviewed and approved by the IRBs of the sponsoring US institutions (Loyola University Chicago and Howard University) and the University of Ibadan. All participants gave written informed consent administered in either English or Yoruba. Phenotype measurements were taken during a screening exam completed by trained research staff using a standardized protocol (42). Body weight was measured to the nearest 0.2 kg on calibrated electronic scales, whereas height was obtained using a stadiometer consisting of a steel tape attached to a straight wall and a wooden headboard. The headboard was positioned with the participant shoeless, feet and back against the wall and head held in the Frankfort horizontal plane and measurement taken to the nearest 0.1 cm. BMI was calculated as the ratio of weight (in kg) to the square of height (in m).
Replication panels
Maywood family cohort
These African-American individuals were recruited from the same neighborhood in Maywood, IL, as the Maywood GWAS discovery panel. The two Maywood panels were a part of larger cohort enrolled to study blood pressure, with no overlap between them. Although the Maywood GWAS discovery panel represented the unrelated individuals, this panel represented a family-based cohort as well as unrelated individuals who were not genotyped in the discovery panel from the original study. In total, 756 individuals (including 704 related individuals from 306 African-American nuclear families) were available. The protocol was approved by the IRB at Loyola University Chicago.
GCI cohorts
Two self-described African-American cohorts representing the extremes of BMI and height were collected by the Genomics Collaborative. The GCI-weight panel consists of 494 individuals representing the 5–12th and 90–97th percentiles of the BMI distribution. The GCI-height panel consists of 488 individuals representing the 5–10th and 90–95th percentiles of the height distribution, as described previously (43,44).
Jamaican cohorts
DNA samples for the Jamaican cohorts were obtained from two sources: Kingston and Spanish Town, Jamaica. The Kingston cohort [Jamaican Gene x Environment panel from Kingston, Jamaica (GxE)] was obtained from a survey conducted in the capital city, Kingston, as part of a larger project to examine gene by environment interactions in the determination of blood pressure among adults 25–74 years. The principal criterion for eligibility was a BMI in either the top or the bottom third of BMI for the Jamaican population (40). Participants were identified principally from the records of the Heart Foundation of Jamaica, a non-governmental organization based in Kingston, which provides low-cost screening services (height and weight, blood pressure, glucose, cholesterol) to the general public. Other participants were identified from among participants in family studies of blood pressure at the Tropical Metabolism Research Unit (TMRU) and from among staff members at the University of the West Indies, Mona. All participants were unrelated. A total of 1039 persons were enrolled, and 959 passed quality-control filters and were used for replication in this study.
The Spanish Town cohort (SPT) was obtained from a survey conducted as part of the International Collaborative Study of Hypertension in Blacks and previously described in detail (40). In this study, a stratified random sample of the Jamaican population aged 25–74 years was recruited from in and around SPT, a stable, residential urban area neighboring the capital city of Kingston. Among 2096 participants enrolled between 1993 and 1998, 1478 were available and passed quality-control filters to comprise the replication panel used here.
Genotyping
GWA genotype data were obtained by applying samples to an Affymetrix 6.0 SNP array. All Affymetrix genotyping was done at the Broad Institute by the Genetic Analysis Platform, and the data exported in cel files. Replication genotyping was done at Children's Hospital Boston using the Sequenom MassArray system.
African-American cohort GWA data cleaning
Seven hundred and seventy-five African-Americans from Maywood, IL, were genotyped with Affymetrix Genome-wide human SNP array 6.0, which provided genotype data at 909 622 SNPs in the human genome. Quality-control efforts were conducted at two levels: exclusion of individuals and exclusion of SNPs. Exclusion of individuals was aimed at removing cryptically related samples, as well as contaminated or poorly genotyped samples. For cryptic relatedness, 17 duplicate individuals with proportion of identity by descent (IBD) sharing >99% (either duplicates or identical twins) were removed. The 17 individuals included eight individuals from four duplicate pairs with discordant reported gender and nine duplicate pairs with concordant reported gender, in which case we kept the individual closer to the extremes of the BMI distribution. For contaminated or poorly genotyped samples, one individual was removed due to excessive heterozygosity (>4 SD above the population mean); three individuals were removed due to apparent wide-spread low-level relatedness (IBD sharing >5%) with 319, 713 and 768 other samples in the population, respectively. Finally, an additional 11 samples with gender discordance between genotypic sex and reported phenotypic sex were excluded. No samples were removed due to genotyping success rate <95%. No population outliers beyond familial clustering on multidimensional scaling analysis were apparent. In total, we removed 32 individuals for a final n = 743.
The following SNPs were removed from analysis: 1176 SNPs known to map to multiple genomic loci, 27 951 SNPs with a proportion of missing genotype >0.05, and 20 687 SNPs with minor allele frequency (MAF) <0.01, 1048 and 83 SNPs showing non-random missingness with respect to neighboring genotype and phenotype (arbitrarily defining cases as those with BMI > 25 and controls as those with BMI ≤ 25; note that BMI was analyzed as a continuous trait in the GWA phase), respectively, and 150 SNPs showing association with plate membership. No SNPs were removed due to significant deviation from the Hardy–Weinberg equilibrium (HWE) because the African-American population is an admixed population, which may result in departure from HWE. However, as a class the association statistics of the 1094 SNPs that would have failed HWE test at P < 1.0 × 10−6 did not exhibit significant departure from expectation under the null (data not shown). In total, 859 332 SNPs genome-wide were analyzed for association. All QC filtering was done using PLINK v1.02 (45).
Nigerian cohort GWA data cleaning
Genomic DNA was available on the 1253 samples. The chip analysis provided data on 909 622 SNPs. Four samples were excluded because of discordance between reported gender and observed gender from the genotype data. Samples with inbreeding coefficients outside of 4 SD of the mean coefficient were removed (n = 11) as well as samples sharing high IBD proportion either due to sample contamination (n = 14), duplicates (n = 18) or cryptic relatedness (n = 1). Additional samples identified as outliers based on multidimensional scaling analysis of the genome-wide IBD pairwise distances (n = 12) or clustering of missing genotypes (n = 5) were dropped.
In total, 1176 SNPs that have been shown to map to several locations in the genome, 34 137 SNPs with a proportion of missing genotypes >0.05, 64 955 SNPs with MAF <1% and 16 616 SNPs with non-random genotype failure, with significant differential missing rates between hypertensives and normotensive, or failing HWE test at P < 1.0 × 10−6 in normotensives were all excluded as part of quality control. In addition, genotypes for 1035 SNPs found to exhibit substantial deviations associated with the assay plates were zeroed out for the samples typed on such affected assay plates. Curated genotype data on 792 857 SNPs were available on 1188 unrelated adults (510 males).
Statistical methods
Imputation
Imputation was performed using the MACH 1.0.16 haplotyper and imputation program (http://www.sph.umich.edu/csg/abecasis/MaCH/), which uses a hidden Markov model to estimate an underlying set of unphased genotypes for each individual in a cohort.
We used reference-phased haplotypes released along with HapMap phase 2 (build 36 release 22) as input for imputation by MACH. Imputation of the Nigeria cohort data set was performed using the YRI (Yoruba) HapMap-phased haplotype panel. The Maywood cohort is an admixed population, for which no adequate single HapMap phase 2 reference panel exists, with proportion of European admixture estimated to be ∼17–19% (24–26). To account for this admixture, an artificial reference panel consisting of equal proportions of the YRI and CEU HapMap-phased haplotypes (using only SNPs found in both YRI and CEU panels, i.e. ∼2.2 M SNPs) was constructed. MACH was run in two rounds to reduce computational load introduced by the large haplotype panel. In the first round, error and recombination rates were estimated from the reference haplotypes and input cohort using 30 iterations of the program. The second round used these rates to give a greedy estimate of the final genotypes in one iteration.
At this time, imputation software does not allow prior probabilities to be assigned to individual reference haplotypes, so some error is expected to be introduced as the program estimates the next best haplotype. We evaluated the performance of imputation on admixed panels with an equal-weight reference panel by calculating concordance rate of imputed genotype with SNPs genotyped previously in the laboratory for a subset of the Maywood individuals. Specifically, among the Maywood individuals genotyped on the Affymetrix 6.0 array, 141 individuals were previously genotyped at 599 SNP loci on chromosome 3q27 not found on the Affymetrix 6.0 array. For each SNP, concordance is calculated as follows: we assigned a score of 1 − 1/2|aG − aD| to each individual, where aG represents the number of the reference allele according to genotyping (aG = 0, 1 or 2) and aD represents the dosage of the reference allele output by MACH (0 < aD < 2); the score averaged over 141 individuals indicates the concordance between actual genotype and the imputed genotype for each SNP. Over all 599 SNPs examined, the average concordance was 95.8%, which is comparable with the accuracy reported when imputing a population of African-Americans using similar approach as well as a population of Nigerians using YRI as the reference panel (31,46). Similar concordance rates were observed when using the ‘best-guess genotype’ output by MACH, rather than dosage (data not shown).
Using the equal-weight reference panel for the Maywood cohort and the YRI reference panel for the Nigerian cohort, we were able to estimate genotypes at ∼1.6 M loci and ∼2.1 M loci with confidence scores ≥0.3 and allele frequencies ≥0.01 in the Maywood and the Nigerian cohorts, respectively, in addition to the directly genotyped SNPs on the Affymetrix 6.0 array. This approach is similar to an approaches proposed previously (31,46). A more extensive validation and comparison of this and related approaches using a weighted reference panel (47) are ongoing. For SNPs that were directly genotyped, we used the direct genotypes rather than the imputed data in analysis.
Association testing
All association testing was done assuming an additive model, in unrelated individuals, as implemented in PLINK (for directly genotyped SNPs; http://pngu.mgh.harvard.edu/~purcell/plink/) and in mach2qtl (for imputed SNPs; http://www.sph.umich.edu/csg/yli/mach/download/). Multivariate linear regression models were constructed for height (cm) controlling for age and stratified by sex, and the residual values were fit to a standard normal distribution to create Z-scores. Multivariate linear regression models for logBMI were constructed controlling for age in each sex separately, and the residual values were fit to a standard normal distribution to create Z-scores.
Principal component adjustment
To examine for possible population stratification, we performed principal component (PC) analysis in both African-American and Nigerian cohorts using all SNPs after QC. Population stratification was characterized using the first 10 PCs calculated based on all SNPs using FamCC (48). As already suggested in the literature, we observed that African-Americans have varying degrees of admixture of African and European ancestries, whereas Nigerians are clustered as a single group, suggesting minimal population stratification in the Nigerian cohort (Supplementary Material, Fig. S1). The phenotype–genotype association was performed using linear regression under an additive model adjusting for the first 10 PCs in the African-American cohort, whereas no PCs were included in the analysis of Nigerian cohort; it has been suggested that the first 10 PCs can appropriately capture the population structure effect in African-American populations (48,49).
Meta-analysis method
Association results from different panels were combined during the discovery phase of the study using Metal (February 2009 release, http://www.sph.umich.edu/csg/abecasis/Metal/index.html), using the inverse variance weighted (fixed effect) meta-analysis mode. Each genome-wide scan was individually corrected for genomic inflation (50) prior to meta-analysis, but not corrected again after meta-analysis. Directly genotyped SNPs and imputed SNPs were meta-analyzed separately; imputed SNPs that were also directly genotyped on the SNP array were not analyzed.
Replication
Replication was completed in two stages, a screening stage (stage 1) in which we looked for consistency of direction of effect in two panels, and a replication stage (stage 2) in which we sought at least nominal (P < 0.05) evidence of replication. Prior to stage 1, two classes of SNPs were selected for replication and genotyped in the family-based replication panel and one of the two African-American replication panels described above. The two classes of SNPs were as follows. (i) Best signals from GWA: the top SNPs from the meta-analyzed results of African-Americans and Nigerians were sequentially checked for other SNPs in LD with r2 ≥ 0.5 in YRI. SNPs in LD exceeding the threshold were clumped together such that each clump was considered to represent an independent signal. Independent signals with P < 1 × 10−5 were selected for stage 1 replication. (ii) Loci associated in European samples: we examined the 53 known loci associated with height or BMI in previous European GWA studies (6,7,11,17,18), for which we have phased HapMap genotype. For each of 53 index SNPs from the known loci, we examined the association evidence of individual clumps (SNPs with r2 ≥ 0.5 in YRI) among all SNPs in high LD (r2 ≥ 0.8) with the index SNP in the HapMap CEU sample. We then selected eight SNPs (one from each locus) with best evidence of association in our GWA panel, along with one or two other proxies within the clump (for protection against failed genotyping), for stage 1 replication. Note that not all clumps with one-tailed P < 0.05 were selected; we preferentially selected SNPs from loci where multiple SNPs appear to be in LD with SNP with P < 0.05. In total, 27 and 25 SNPs for height and BMI, respectively, were selected for stage 1 replication. Because of the relatively loose threshold for passing stage 1 replication, we were able to use stage 1 as an effective screen of our initial GWA results without loss of significant power (see below), particularly for variants with effect sizes >0.3% variance explained.
For stage 2 replication, we selected SNPs that showed the same direction of effect across the four cohorts studied in the GWA and stage 1 replication (Maywood GWA panel, Nigerian GWA panel, Maywood Family panel and GCI panel) and genotyped these in the two additional cohorts (GxE and SPT) from Jamaica. In total, 15 SNPs (7 for height and 8 for BMI) were genotyped in the Jamaican cohorts during stage 2 replication.
The family-based cohort from Maywood, IL, was analyzed using the software Merlin (51). The remaining panels represented unrelated individuals and were analyzed using the assoc command (GCI panels), logistic regression function for dichotomous traits (BMI in the GxE cohort) or linear regression functions for continuous traits (BMI in SPT, height in GxE and SPT) in PLINK (45).
Power calculations
For the GWA panels, power was calculated for continuous trait over a range of effect sizes (percent variance explained, r2) for a two-tailed α level of 1 × 10−5 (for novel associations) or a one-tailed α level of 0.05 (for previously reported European loci) as described previously (52). For replication panels where height or BMI was analyzed as a continuous trait, power at one-tailed α level of 0.5 (stage 1 replication) or 0.05 (stage 2 replication) was calculated. For replication panels where height or BMI was analyzed as a dichotomous trait, the case–control for threshold-selected quantitative trait module of the Genetic Power Calculator (http://pngu.mgh.harvard.edu/~purcell/gpc/) was used to calculate expected allele frequencies in cases and controls, which were in turn used to calculate the one-tailed binomial power at the appropriate α levels using the power.prop.test and bpower functions in R (version 2.6.2, Hmisc package).
To estimate the number of confirmed European loci associated with height and BMI expected to be replicated in our study, the 41 height loci and 12 BMI loci were first categorized according to the percent variance explained (r2) as reported previously (6,7,11,17,18), although these estimates are likely to be inflated by the ‘winner's curse’ in studies where estimates were not generated from independent replication samples. Reported loci were divided into 10 groups by their r2. For each locus, the group median r2 was used as the basis to calculate power and the probability to achieve one-tailed P < 0.05 in the GWA phase or in either of the Jamaican cohorts. Then, the mean number and standard deviation of known European loci expected to achieve one-tailed P < 0.05 in the GWA phase of the study and the number of loci expected to replicate in either of the Jamaican cohorts in our study was assessed over 10 000 rounds of simulation.
Because of differences in the LD pattern between Europeans and Africans, the 53 index SNPs and their correlates in the European genomes may represent more than 53 independent signals in African genomes, possibly due to statistical fluctuations. Therefore, to assess whether the number of nominally associated clumps observed in our GWA panel exceeded the null expectation, we simulate the number of independent clumps we would detect with P < 0.05 by selecting 53 random SNPs in the genome, retrieving all HapMap SNPs with r2 > 0.8 in CEU and counting the number of clumps (grouped with r2 > 0.5 in YRI) passing one-tailed P-value of 0.05. The simulation was matched by the allele frequency of the index SNP (±0.02) and the number of clumps in African genomes (±3 clumps for height and ±1 clump for BMI) and iterated over 1000 simulations.
CNV analysis
Common and rare CNV calling and quality control
Common CNV [sometimes in the literature referred to as copy number polymorphisms (CNPs)] genotypes were called using the CANARY algorithm as part of the Birdsuite package (53), which utilizes a previously defined common CNV map based on HapMap individuals (27). First-degree relatives based on the pairwise whole-genome estimation of allele sharing were removed before running the Birdsuite algorithms. Allele frequency prior files were generated based on YRI HapMap individuals for the Nigerian samples and an 80–20% mix of the YRI and CEU HapMap individuals for the African-American samples.
Quality-control filtering was applied first at the sample level to remove excessively noisy samples. A measure of variability of SNP and copy number (CN) probe intensities, with each standardized per chromosome, is generated by the Birdseye Hidden Markov Model (53). We removed any sample where more than one chromosome failed the metric (>1.5 SD in average estimated SNP or CN probe variability). In total, 693 African-American samples and 1171 Nigerian samples were CN genotyped with the CANARY software (53). We genotyped the cleaned samples for the previously defined set of 1319 common CNVs characterized on the HapMap sample (27). Genotype-level quality controls were applied to remove any variant with <90% call rate by the CANARY algorithm, any variant in which none of the minor allele frequencies is greater than 5%, and any variant on the Y chromosome. This restricted our analyses to 405 and 431 common CNV regions for African-American samples and Nigerian samples, respectively.
To detect rare CNVs (frequency of 5% or less), we used the Birdseye software within the Birdsuite package, which utilizes a hidden Markov model to model the different quantitative response to different CNs of DNA as well as the intrinsic measurement variances across samples for each probe on the array. By considering evidence across neighboring probes, Birdseye assigns, for each sample, a discrete CN at each segment of the genome, along with an LOD score that reflects the confidence of the assignment (53). We removed CNV segments with the Birdseye LOD score <10, with segment length >5 Mb or with a size (kb) to an LOD ratio of <100. We also excluded common CNVs by removing any CNV with >50% of its length spanning a genomic interval with 5% or more of the CNVs in the sample. We then merged nearby type-consistent CNV segments if the combined length of CNVs was at least 80% of the length of those CNVs merged. Finally, we removed additional outliers with >10 Mb of CNVs or >30 CNV segments. In total, 673 African-American samples with 8399 CNVs and 1175 Nigerian samples with 12 341 CNVs passed quality control.
Common and rare CNV analysis
For common CNPs, association testing was performed using a logistic regression model implemented in the PLINK software (version 1.05) (45). In the logistic regression model, CN was used as the predictor variable, and sex- and age-controlled standardized residuals for BMI and height were used as the response variable. Because of the admixed nature of African-Americans, the first 10 PCs that estimated the degree of admixture were used as covariates in the model. Additionally, plate membership was included as a covariate in the analysis for both African-Americans and Nigerians to adjust for possible confounding due to differential calling of CNPs per plate. Samples with sex- and age-adjusted phenotype residual Z-scores >4 SD away from the mean were removed in the respective analysis. Analysis was done in African-Americans and Nigerians separately, each stratified by sex, and then meta-analyzed. Meta-analysis weighted by the number of individuals per variant was performed using Metal (February 2009 release, http://www.sph.umich.edu/csg/abecasis/Metal/index.html).
For rare CNVs, we evaluated genome-wide differences in CNV burden between cases and controls with respect to BMI only. Three indicators were assessed using PLINK, as previously performed for analyses of schizophrenia (54): genome-wide CNV rate per individual, the total extent of rare CNVs per individual and the average length of rare CNVs. For each individual sample, genome-wide CNV rate was calculated by counting the number of CNV segments, as produced by Birdseye, and the total extent of rare CNVs was obtained by summing over the length of each CNV segment. Thus, the average length of rare CNVs is the ratio of the total extent of CNVs to the number of CNVs. Each of these three indicators was then averaged across all case individuals and all control individuals and compared. Statistical significance of case–control differences in each of the parameters tested was determined by 50 000 permutations of case–control status within each genotyping plate as a two-sided test. We also examined specifically the rare CNVs that intersected or fell within 20 kb of genes (RefSeq gene list, hg18 coordinates, ±20 kb around the largest transcript per gene).
For African-Americans, cases were defined as individuals with age- and sex-controlled standardized residual values of 0.6 or greater (n = 190); controls were those with residual values of zero or lower (n = 435). For Nigerians, cases were defined as the top quartile by standardized residuals of BMI (n = 291), and controls were defined as the bottom half (n = 585), in order to approximate the proportion of cases to controls in the African-American samples. Meta-analysis weighted by number of individuals per variant was performed using Metal (February 2009 release, http://www.sph.umich.edu/csg/abecasis/Metal/index.html). For replication, European individuals from the Myocardial Infarction Genetics Consortium (28) were analyzed with the same QC procedure as described above. Cases and controls were also defined as the top quartile and the bottom half, respectively, of the distribution of standardized residuals of BMI.
SUPPLEMENTARY MATERIAL
FUNDING
This work was supported by the National Institutes of Health (R01DK075787 to J.N.H., R01HL074166, R01HG003054 to X.Z., and R37HL45508, R01HL53353 to R.S.C.); the National Center for Research Resources (U54 RR020278 to the Broad Institute Center for Genotyping and Analysis, which provided a grant for subsidized genotyping to R.S.C.); the Intramural Research Program of the Center for Research on Genomics and Global Health, National Human Genome Research Institute (Z01HG200362); National Science Foundation (graduate research fellowship to C.W.K.C.); and an NIH training grant to Case Western Reserve University.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank S. Kathiresan, J.M. Korn, S.A. McCarroll, J. Nemesh, S. Purcell and the Myocardial Infarction Genetics Consortium (MIGen) for help with the CNV analysis and sharing of CNV data, Z.K.Z. Gajdos for critical comments on the manuscript, as well as past and present members of the Hirschhorn laboratory for comments, ideas and discussions.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Hindorff L.A., Junkins H.A., Mehta J.P., Manolio T.A. A Catalog of Published Genome-Wide Association Studies. 2009. www.genome.gov/gwastudies .
- 2.Frayling T.M., Timpson N.J., Weedon M.N., Zeggini E., Freathy R.M., Lindgren C.M., Perry J.R., Elliott K.S., Lango H., Rayner N.W., et al. A common variant in the FTO gene is associated with body mass index and predisposes to childhood and adult obesity. Science. 2007;316:889–894. doi: 10.1126/science.1141634. doi:10.1126/science.1141634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Scuteri A., Sanna S., Chen W.M., Uda M., Albai G., Strait J., Najjar S., Nagaraja R., Orru M., Usala G., et al. Genome-wide association scan shows genetic variants in the FTO gene are associated with obesity-related traits. PLoS Genet. 2007;3:e115. doi: 10.1371/journal.pgen.0030115. doi:10.1371/journal.pgen.0030115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Weedon M.N., Lettre G., Freathy R.M., Lindgren C.M., Voight B.F., Perry J.R., Elliott K.S., Hackett R., Guiducci C., Shields B., et al. A common variant of HMGA2 is associated with adult and childhood height in the general population. Nat. Genet. 2007;39:1245–1250. doi: 10.1038/ng2121. doi:10.1038/ng2121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chambers J.C., Elliott P., Zabaneh D., Zhang W., Li Y., Froguel P., Balding D., Scott J., Kooner J.S. Common genetic variation near MC4R is associated with waist circumference and insulin resistance. Nat. Genet. 2008;40:716–718. doi: 10.1038/ng.156. doi:10.1038/ng.156. [DOI] [PubMed] [Google Scholar]
- 6.Gudbjartsson D.F., Walters G.B., Thorleifsson G., Stefansson H., Halldorsson B.V., Zusmanovich P., Sulem P., Thorlacius S., Gylfason A., Steinberg S., et al. Many sequence variants affecting diversity of adult human height. Nat. Genet. 2008;40:609–615. doi: 10.1038/ng.122. doi:10.1038/ng.122. [DOI] [PubMed] [Google Scholar]
- 7.Lettre G., Jackson A.U., Gieger C., Schumacher F.R., Berndt S.I., Sanna S., Eyheramendy S., Voight B.F., Butler J.L., Guiducci C., et al. Identification of ten loci associated with height highlights new biological pathways in human growth. Nat. Genet. 2008;40:584–591. doi: 10.1038/ng.125. doi:10.1038/ng.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loos R.J., Lindgren C.M., Li S., Wheeler E., Zhao J.H., Prokopenko I., Inouye M., Freathy R.M., Attwood A.P., Beckmann J.S., et al. Common variants near MC4R are associated with fat mass, weight and risk of obesity. Nat. Genet. 2008;40:768–775. doi: 10.1038/ng.140. doi:10.1038/ng.140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.McCarthy M.I., Abecasis G.R., Cardon L.R., Goldstein D.B., Little J., Ioannidis J.P., Hirschhorn J.N. Genome-wide association studies for complex traits: consensus, uncertainty and challenges. Nat. Rev. Genet. 2008;9:356–369. doi: 10.1038/nrg2344. doi:10.1038/nrg2344. [DOI] [PubMed] [Google Scholar]
- 10.Sanna S., Jackson A.U., Nagaraja R., Willer C.J., Chen W.M., Bonnycastle L.L., Shen H., Timpson N., Lettre G., Usala G., et al. Common variants in the GDF5-UQCC region are associated with variation in human height. Nat. Genet. 2008;40:198–203. doi: 10.1038/ng.74. doi:10.1038/ng.74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Weedon M.N., Lango H., Lindgren C.M., Wallace C., Evans D.M., Mangino M., Freathy R.M., Perry J.R., Stevens S., Hall A.S., et al. Genome-wide association analysis identifies 20 loci that influence adult height. Nat. Genet. 2008;40:575–583. doi: 10.1038/ng.121. doi:10.1038/ng.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho Y.S., Go M.J., Kim Y.J., Heo J.Y., Oh J.H., Ban H.J., Yoon D., Lee M.H., Kim D.J., Park M., et al. A large-scale genome-wide association study of Asian populations uncovers genetic factors influencing eight quantitative traits. Nat. Genet. 2009;41:527–534. doi: 10.1038/ng.357. doi:10.1038/ng.357. [DOI] [PubMed] [Google Scholar]
- 13.Estrada K., Krawczak M., Schreiber S., van Duijn K., Stolk L., van Meurs J.B., Liu F., Penninx B.W., Smit J.H., Vogelzangs N., et al. A genome-wide association study of northwestern Europeans involves the C-type natriuretic peptide signaling pathway in the etiology of human height variation. Hum. Mol. Genet. 2009;18:3516–3524. doi: 10.1093/hmg/ddp296. doi:10.1093/hmg/ddp296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Heard-Costa N.L., Zillikens M.C., Monda K.L., Johansson A., Harris T.B., Fu M., Haritunians T., Feitosa M.F., Aspelund T., Eiriksdottir G., et al. NRXN3 is a novel locus for waist circumference: a genome-wide association study from the CHARGE Consortium. PLoS Genet. 2009;5:e1000539. doi: 10.1371/journal.pgen.1000539. doi:10.1371/journal.pgen.1000539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lindgren C.M., Heid I.M., Randall J.C., Lamina C., Steinthorsdottir V., Qi L., Speliotes E.K., Thorleifsson G., Willer C.J., Herrera B.M., et al. Genome-wide association scan meta-analysis identifies three Loci influencing adiposity and fat distribution. PLoS Genet. 2009;5:e1000508. doi: 10.1371/journal.pgen.1000508. doi:10.1371/journal.pgen.1000508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soranzo N., Rivadeneira F., Chinappen-Horsley U., Malkina I., Richards J.B., Hammond N., Stolk L., Nica A., Inouye M., Hofman A., et al. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. doi:10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thorleifsson G., Walters G.B., Gudbjartsson D.F., Steinthorsdottir V., Sulem P., Helgadottir A., Styrkarsdottir U., Gretarsdottir S., Thorlacius S., Jonsdottir I., et al. Genome-wide association yields new sequence variants at seven loci that associate with measures of obesity. Nat. Genet. 2009;41:18–24. doi: 10.1038/ng.274. doi:10.1038/ng.274. [DOI] [PubMed] [Google Scholar]
- 18.Willer C.J., Speliotes E.K., Loos R.J., Li S., Lindgren C.M., Heid I.M., Berndt S.I., Elliott A.L., Jackson A.U., Lamina C., et al. Six new loci associated with body mass index highlight a neuronal influence on body weight regulation. Nat. Genet. 2009;41:25–34. doi: 10.1038/ng.287. doi:10.1038/ng.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cooper R.S., Tayo B., Zhu X. Genome-wide association studies: implications for multiethnic samples. Hum. Mol. Genet. 2008;17:R151–R155. doi: 10.1093/hmg/ddn263. doi:10.1093/hmg/ddn263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bhangale T.R., Rieder M.J., Nickerson D.A. Estimating coverage and power for genetic association studies using near-complete variation data. Nat. Genet. 2008;40:841–843. doi: 10.1038/ng.180. doi:10.1038/ng.180. [DOI] [PubMed] [Google Scholar]
- 21.Li Y., Willer C., Sanna S., Abecasis G. Genotype imputation. Annu. Rev. Genomics Hum. Genet. 2009;10:387–406. doi: 10.1146/annurev.genom.9.081307.164242. doi:10.1146/annurev.genom.9.081307.164242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li Y., Abecasis G.R. Mach 1.0: Rapid Haplotype Reconstruction and Missing Genotype Inference. Am. J. Hum. Genet. 2006;S79:2290. doi:10.2337/diabetes.52.5.1284. [Google Scholar]
- 23.Luke A., Wu X., Zhu X., Kan D., Su Y., Cooper R. Linkage for BMI at 3q27 region confirmed in an African-American population. Diabetes. 2003;52:1284–1287. doi: 10.2337/diabetes.52.5.1284. doi:10.2337/diabetes.52.5.1284. [DOI] [PubMed] [Google Scholar]
- 24.Parra E.J., Marcini A., Akey J., Martinson J., Batzer M.A., Cooper R., Forrester T., Allison D.B., Deka R., Ferrell R.E., et al. Estimating African American admixture proportions by use of population-specific alleles. Am. J. Hum. Genet. 1998;63:1839–1851. doi: 10.1086/302148. doi:10.1086/302148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zhu X., Luke A., Cooper R.S., Quertermous T., Hanis C., Mosley T., Gu C.C., Tang H., Rao D.C., Risch N., et al. Admixture mapping for hypertension loci with genome-scan markers. Nat. Genet. 2005;37:177–181. doi: 10.1038/ng1510. doi:10.1038/ng1510. [DOI] [PubMed] [Google Scholar]
- 26.Chiang C.W.K., Gajdos Z.K.Z., Korn J.M., Kuruvilla F.G., Butler J.L., Hackett R., Guiducci C., Nguyen T.T., Wilks R., Forrester T., et al. Rapid assessment of genetic ancestry in populations of unknown origin by genome-wide genotyping of pooled samples. PLoS Genet. 2010;6:e1000866. doi: 10.1371/journal.pgen.1000866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McCarroll S.A., Kuruvilla F.G., Korn J.M., Cawley S., Nemesh J., Wysoker A., Shapero M.H., de Bakker P.I., Maller J.B., Kirby A., et al. Integrated detection and population-genetic analysis of SNPs and copy number variation. Nat. Genet. 2008;40:1166–1174. doi: 10.1038/ng.238. doi:10.1038/ng.238. [DOI] [PubMed] [Google Scholar]
- 28.Kathiresan S., Voight B.F., Purcell S., Musunuru K., Ardissino D., Mannucci P.M., Anand S., Engert J.C., Samani N.J., et al. Myocardial Infarction Genetics Consortium. Genome-wide association of early-onset myocardial infarction with single nucleotide polymorphisms and copy number variants. Nat. Genet. 2009;41:334–341. doi: 10.1038/ng.327. doi:10.1038/ng.327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bochukova E.G., Huang N., Keogh J., Henning E., Purmann C., Blaszczyk K., Saeed S., Hamilton-Shield J., Clayton-Smith J., O'Rahilly S., et al. Large, rare chromosomal deletions associated with severe early-onset obesity. Nature. 2009;463:666–670. doi: 10.1038/nature08689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Need A.C., Goldstein D.B. Next generation disparities in human genomics: concerns and remedies. Trends Genet. 2009;25:489–494. doi: 10.1016/j.tig.2009.09.012. doi:10.1016/j.tig.2009.09.012. [DOI] [PubMed] [Google Scholar]
- 31.Huang L., Li Y., Singleton A.B., Hardy J.A., Abecasis G., Rosenberg N.A., Scheet P. Genotype-imputation accuracy across worldwide human populations. Am. J. Hum. Genet. 2009;84:235–250. doi: 10.1016/j.ajhg.2009.01.013. doi:10.1016/j.ajhg.2009.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Mathias R.A., Grant A.V., Rafaels N., Hand T., Gao L., Vergara C., Tsai Y.J., Yang M., Campbell M., Foster C., et al. A genome-wide association study on African-ancestry populations for asthma. J. Allergy Clin. Immunol. 2009;125:336–346 e334. doi: 10.1016/j.jaci.2009.08.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shriner D., Adeyemo A., Chen G., Rotimi C.N. Practical considerations for imputation of untyped markers in admixed populations. Genet. Epidemiol. 2009;34:258–265. doi: 10.1002/gepi.20457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dhandapany P.S., Sadayappan S., Xue Y., Powell G.T., Rani D.S., Nallari P., Rai T.S., Khullar M., Soares P., Bahl A., et al. A common MYBPC3 (cardiac myosin binding protein C) variant associated with cardiomyopathies in South Asia. Nat. Genet. 2009;41:187–191. doi: 10.1038/ng.309. doi:10.1038/ng.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Unoki H., Takahashi A., Kawaguchi T., Hara K., Horikoshi M., Andersen G., Ng D.P., Holmkvist J., Borch-Johnsen K., Jorgensen T., et al. SNPs in KCNQ1 are associated with susceptibility to type 2 diabetes in East Asian and European populations. Nat. Genet. 2008;40:1098–1102. doi: 10.1038/ng.208. doi:10.1038/ng.208. [DOI] [PubMed] [Google Scholar]
- 36.Kopp J.B., Smith M.W., Nelson G.W., Johnson R.C., Freedman B.I., Bowden D.W., Oleksyk T., McKenzie L.M., Kajiyama H., Ahuja T.S., et al. MYH9 is a major-effect risk gene for focal segmental glomerulosclerosis. Nat. Genet. 2008;40:1175–1184. doi: 10.1038/ng.226. doi:10.1038/ng.226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Smith M.W., O'Brien S.J. Mapping by admixture linkage disequilibrium: advances, limitations and guidelines. Nat. Rev. Genet. 2005;6:623–632. doi: 10.1038/nrg1657. doi:10.1038/nrg1657. [DOI] [PubMed] [Google Scholar]
- 38.Zhu X., Tang H., Risch N. Admixture mapping and the role of population structure for localizing disease genes. Adv. Genet. 2008;60:547–569. doi: 10.1016/S0065-2660(07)00419-1. doi:10.1016/S0065-2660(07)00419-1. [DOI] [PubMed] [Google Scholar]
- 39.Chakravarthy M.V., Booth F.W. Eating, exercise, and ‘thrifty’ genotypes: connecting the dots toward an evolutionary understanding of modern chronic diseases. J. Appl. Physiol. 2004;96:3–10. doi: 10.1152/japplphysiol.00757.2003. doi:10.1152/japplphysiol.00757.2003. [DOI] [PubMed] [Google Scholar]
- 40.Cooper R., Rotimi C., Ataman S., McGee D., Osotimehin B., Kadiri S., Muna W., Kingue S., Fraser H., Forrester T., et al. The prevalence of hypertension in seven populations of west African origin. Am. J. Public Health. 1997;87:160–168. doi: 10.2105/ajph.87.2.160. doi:10.2105/AJPH.87.2.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rotimi C.N., Dunston G.M., Berg K., Akinsete O., Amoah A., Owusu S., Acheampong J., Boateng K., Oli J., Okafor G., et al. In search of susceptibility genes for type 2 diabetes in West Africa: the design and results of the first phase of the AADM study. Ann. Epidemiol. 2001;11:51–58. doi: 10.1016/s1047-2797(00)00180-0. doi:10.1016/S1047-2797(00)00180-0. [DOI] [PubMed] [Google Scholar]
- 42.Cooper R., Puras A., Tracy J., Kaufman J., Asuzu M., Ordunez P., Mufunda J., Sparks H. Evaluation of an electronic blood pressure device for epidemiological studies. Blood Press. Monit. 1997;2:35–40. [PubMed] [Google Scholar]
- 43.Campbell C.D., Lyon H.N., Nemesh J., Drake J.A., Tuomi T., Gaudet D., Zhu X., Cooper R.S., Ardlie K.G., Groop L.C., et al. Association studies of BMI and type 2 diabetes in the neuropeptide Y pathway: a possible role for NPY2R as a candidate gene for type 2 diabetes in men. Diabetes. 2007;56:1460–1467. doi: 10.2337/db06-1051. doi:10.2337/db06-1051. [DOI] [PubMed] [Google Scholar]
- 44.Lettre G., Butler J.L., Ardlie K.G., Hirschhorn J.N. Common genetic variation in eight genes of the GH/IGF1 axis does not contribute to adult height variation. Hum. Genet. 2007;122:129–139. doi: 10.1007/s00439-007-0385-4. doi:10.1007/s00439-007-0385-4. [DOI] [PubMed] [Google Scholar]
- 45.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., de Bakker P.I., Daly M.J., et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. doi:10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hao K., Chudin E., McElwee J., Schadt E.E. Accuracy of genome-wide imputation of untyped markers and impacts on statistical power for association studies. BMC Genet. 2009;10:27. doi: 10.1186/1471-2156-10-27. doi:10.1186/1471-2156-10-27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Egyud M.R., Gajdos Z.K., Butler J.L., Tischfield S., Le Marchand L., Kolonel L.N., Haiman C.A., Henderson B.E., Hirschhorn J.N. Use of weighted reference panels based on empirical estimates of ancestry for capturing untyped variation. Hum. Genet. 2009;125:295–303. doi: 10.1007/s00439-009-0627-8. doi:10.1007/s00439-009-0627-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Zhu X., Li S., Cooper R.S., Elston R.C. A unified association analysis approach for family and unrelated samples correcting for stratification. Am. J. Hum. Genet. 2008;82:352–365. doi: 10.1016/j.ajhg.2007.10.009. doi:10.1016/j.ajhg.2007.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Price A.L., Patterson N.J., Plenge R.M., Weinblatt M.E., Shadick N.A., Reich D. Principal components analysis corrects for stratification in genome-wide association studies. Nat. Genet. 2006;38:904–909. doi: 10.1038/ng1847. doi:10.1038/ng1847. [DOI] [PubMed] [Google Scholar]
- 50.Devlin B., Roeder K. Genomic control for association studies. Biometrics. 1999;55:997–1004. doi: 10.1111/j.0006-341x.1999.00997.x. doi:10.1111/j.0006-341X.1999.00997.x. [DOI] [PubMed] [Google Scholar]
- 51.Abecasis G.R., Cherny S.S., Cookson W.O., Cardon L.R. Merlin—rapid analysis of dense genetic maps using sparse gene flow trees. Nat. Genet. 2002;30:97–101. doi: 10.1038/ng786. doi:10.1038/ng786. [DOI] [PubMed] [Google Scholar]
- 52.Zar J.H. Biostatistical Analysis. 4th edn. Upper Saddle River, NJ: Prentice Hall, Inc.; 1998. [Google Scholar]
- 53.Korn J.M., Kuruvilla F.G., McCarroll S.A., Wysoker A., Nemesh J., Cawley S., Hubbell E., Veitch J., Collins P.J., Darvishi K., et al. Integrated genotype calling and association analysis of SNPs, common copy number polymorphisms and rare CNVs. Nat. Genet. 2008;40:1253–1260. doi: 10.1038/ng.237. doi:10.1038/ng.237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.International Schizophrenia Consortium. Rare chromosomal deletions and duplications increase risk of schizophrenia. Nature. 2008;455:237–241. doi: 10.1038/nature07239. doi:10.1038/nature07239. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.